- 1College of Veterinary Medicine, Shanxi Agricultural University, Jinzhong, China
- 2College of Animal Science and Technology, Hebei Normal University of Science and Technology, Qinhuangdao, China
- 3Xianyang Regional Wen's Animal Husbandry Co., Ltd., Xianyang, China
- 4School of Management Shanxi Medical University, Taiyuan, China
A Correction on
Isolation, phylogenetics, and characterization of a new PDCoV strain that affects cellular gene expression in human cells
by Yang, X., Yin, H., Liu, M., Wang, X., Song, T., Song, A., Xi, Y., Zhang, T., Sun, Z., Li, W., Niu, S., Zainab, F., Wang, C., Zhang, D., Wang, H., and Yang, B. (2025). Front. Microbiol. 16:1534907. doi: 10.3389/fmicb.2025.1534907
There was a mistake in Figure 9 as published. The bands representing the PDCoV N protein in interferon-related pathway and autophagy-related pathway were the same. This error occurred while we rearranged Figure 9D in the final version by selecting the same band of PDCoV N twice.
The corrected figure and its caption appear below.
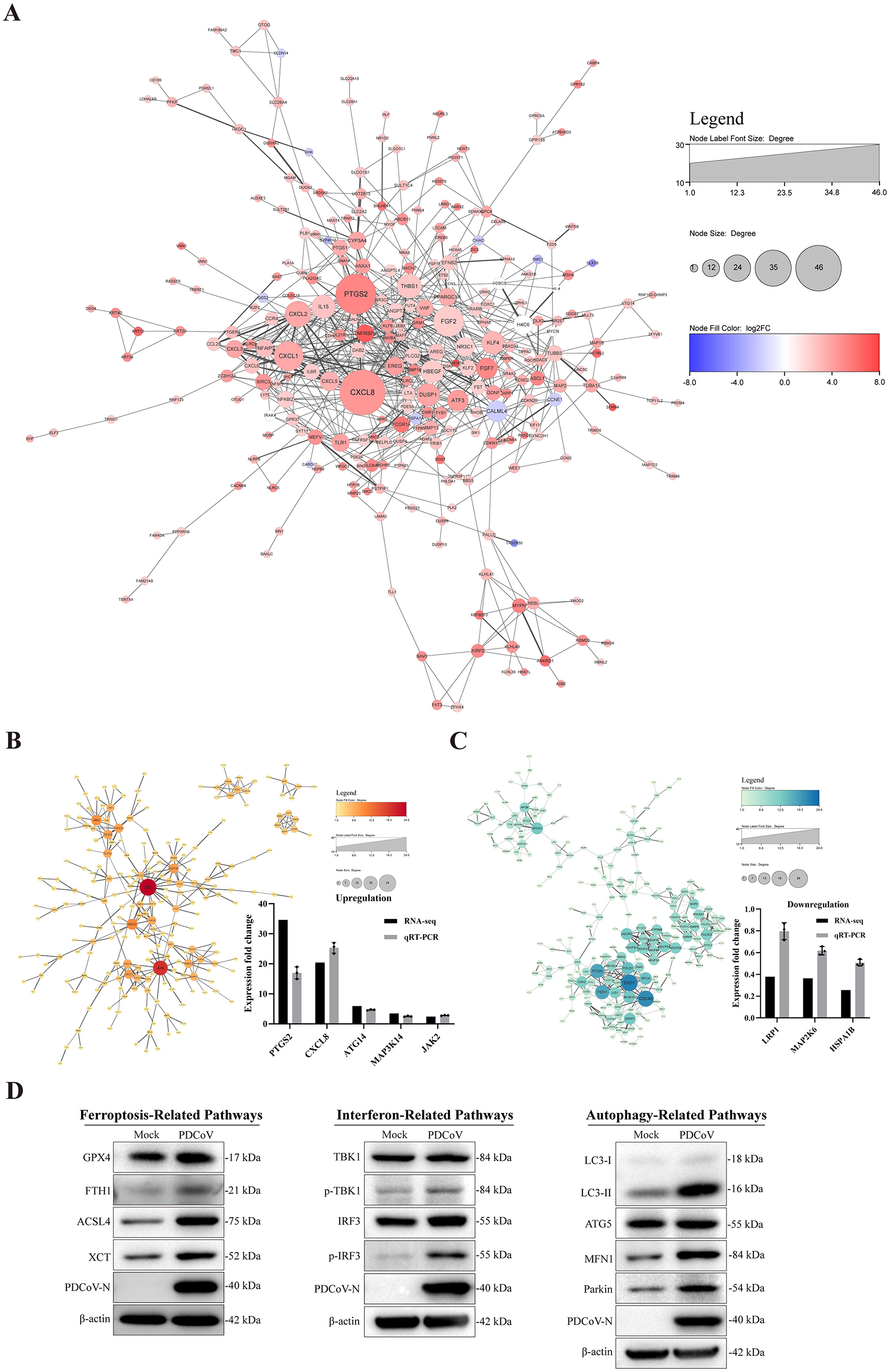
Figure 9. Interacting network of differentially expressed proteins. (A) PPI network of the DEGs. (B, C) PPI network of up-regulated and down-regulated DEGs and RT-qPCR verified results. (D) The related protein expression level of ferroptosis, autophagy and immune response in the context of PDCoV infection using western blotting. β-actin was used as an internal reference. All PPI networks were based on STRING analysis. Each node represented a protein, and each edge represented the interaction between proteins. The upregulated proteins are shown in red shadow, and the downregulated proteins are shown in blue.
The original version of this article has been updated.
Generative AI statement
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: PDCoV, Huh7 cells, phylogenetic tree, transcriptome analysis, immune response
Citation: Yang X, Yin H, Liu M, Wang X, Song T, Song A, Xi Y, Zhang T, Sun Z, Li W, Niu S, Zainab F, Wang C, Zhang D, Wang H and Yang B (2025) Correction: Isolation, phylogenetics, and characterization of a new PDCoV strain that affects cellular gene expression in human cells. Front. Microbiol. 16:1646820. doi: 10.3389/fmicb.2025.1646820
Received: 14 June 2025; Accepted: 25 August 2025;
Published: 08 September 2025.
Edited and reviewed by: Alma Lilian Guerrero-Barrera, Autonomous University of Aguascalientes, Mexico
Copyright © 2025 Yang, Yin, Liu, Wang, Song, Song, Xi, Zhang, Sun, Li, Niu, Zainab, Wang, Zhang, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ding Zhang, emRsdWNreTIwMTRAc2luYS5jb20=; Haidong Wang, d2FuZ2hhaWRvbmdAc3hhdS5lZHUuY24=; Bo Yang, Ym9feWFuZ0BzeGF1LmVkdS5jbg==
†These authors have contributed equally to this work
‡ORCID: Xiaozhu Yang orcid.org/0009-0002-6855-1776
Hanwei Yin orcid.org/0009-0007-6134-4690
 Xiaozhu Yang1†‡
Xiaozhu Yang1†‡ Tao Song
Tao Song Yibo Xi
Yibo Xi Wei Li
Wei Li Farwa Zainab
Farwa Zainab Ding Zhang
Ding Zhang Haidong Wang
Haidong Wang Bo Yang
Bo Yang