- 1Department of Obstetrics and Gynecology, Hunan Provincial Maternal and Child Health Care Hospital, Changsha, China
- 2Tianjin Key Laboratory of Agricultural Animal Breeding and Healthy Husbandry, College of Animal Science and Veterinary Medicine, Tianjin Agricultural University, Tianjin, China
- 3Department of Dermatovenereology, Tianjin Medical University General Hospital/Tianjin Institute of Sexually Transmitted Diseases, Tianjin, China
- 4Department of Obstetrics and Gynecology, 3rd Xiangya Hospital, Central South University, Changsha, Hunan, China
- 5Shanghai Institute of Virology, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 6Key Lab of Molecular Virology and Immunology, Shanghai Institute of Immunity and Infection, Chinese Academy of Sciences, Shanghai, China
Chlamydia trachomatis is the most important infectious cause of tubal infertility and is frequently detected in the human gastrointestinal tract. Chlamydia muridarum, a murine pathogen, closely resembles the human pathogen C. trachomatis. Our previous studies showed that the pGP3-deficient C. muridarum mutant was restricted to the large intestine following intracolonic inoculation, suggesting that the pGP3-deficient mutant was killed by the tissue beyond the large intestine. Here, we report that the intra-ilenum, but not the intra-jejunum, to bypass the gastric barrier rescued the colonization of pGP3-deficient C. muridarum, suggesting that pGP3 is required to overcome host factors of the jejunum to help C. muridarum reach the colon. Moreover, mice genetically deficient in IL-22 not only rescued the colonization of pGP3-deficient C. muridarum following intrajejunal inoculation but also rescued the colonization of pGP3-deficient C. muridarum in the whole gastrointestinal tract tissues following intracolonic inoculation on day 14, suggesting a critical role of IL-22 in regulating chlamydial spread. Importantly, IL-22RA1 flox/flox and Villin-cre mice rescued the colonization of pGP3-deficient C. muridarum following intrajejunal inoculation, suggesting that intestinal epithelial-specific IL-22RA1 signaling is important for the spread of pGP3-deficient C. muridarum from the small intestine to the large intestine. These observations provide a platform for further research on intestinal IL-22RA1 signaling in regulating bacterial spread in the intestine. Therefore, host factors identified in the gastrointestinal tract may also contribute to the female lower genital tract barrier during sexually transmitted diseases.
Highlights
• pGP3 is required for C. muridarum to spread large intestine following intrajejunal inoculation.
• IL-22 is an important factor for blocking the spread of pGP3-deficient C. muridarum from the small intestine to the large intestine.
• Intestinal epithelial IL-22RA1 signaling regulates Chlamydia organisms spreading from small intestine to large intestine.
1 Introduction
Chlamydia trachomatis (C. trachomatis) is a prevalent bacterial pathogen responsible for sexually transmitted infections (STIs) in humans (Budrys et al., 2012). Recent studies have demonstrated that C. trachomatis and Chlamydia muridarum (C. muridarum) colonize the gastrointestinal (GI) tract of their respective hosts (humans and mice) (Yang et al., 2014; Pospischil et al., 2009; Zhang et al., 2015; Craig et al., 2015; Peters et al., 2014; Gratrix et al., 2015; Musil et al., 2016; Gratrix et al., 2014). Notably, murine GI tract colonization by C. muridarum has been shown to influence both infection dynamics and pathogenicity in the genital tract, with these effects dependent on the sequence of exposure to the pathogen. However, the precise mechanisms underlying C. trachomatis colonization of the human gut remain poorly understood.
The murine model of C. muridarum infection has been used to study chlamydial pathogenesis and has revealed numerous chlamydial and host factors required for chlamydial induction or protection of hydrosalpinx (Tan et al., 2018; Wang et al., 2020; Wang et al., 2024; Wang et al., 2023). Chlamydial plasmid-encoded virulence factors essential for infection in the mouse genital tract have been shown to play equally critical roles in GI tract colonization (Zhong, 2018; Shao et al., 2017; Shao et al., 2017; Shao et al., 2017; Koprivsek et al., 2019). The absence of pGP3 in Chlamydia leads to two key deficiencies: attenuated genital tract infection and impaired gastrointestinal colonization (Liu et al., 2014; Lei et al., 2014; O'Connell et al., 2007; Chen et al., 2015). Our previous research revealed that pGP3, an outer membrane-associated protein, is crucial for C. muridarum to overcome the gastric barrier, thereby enabling persistent colonization in the colon (Zhang et al., 2019). Interestingly, when introduced via intracolonic inoculation, pGP3-deficient Chlamydia can still colonize the colon, suggesting that pGP3 triggers enhanced intestinal barrier defense, restricting bacterial dissemination.
Thus, investigating the mechanisms of C. muridarum interaction with the GI tract may enhance our understanding of Chlamydia pathogenesis. In the current study, we used a pGP3-deficient Chlamydia muridarum-spreading mouse model to determine the immunological basis of the intestinal barrier. We reported that mice genetically deficient in IL-22 rescued the colonization of pGP3-deficient C. muridarum following intrajejunal inoculation. Moreover, these mice also rescued the colonization of pGP3-deficient C. muridarum in the whole gastrointestinal tract following intracolonic inoculation on day 14. This suggests that IL-22 is an important factor for pGP3-deficient C. muridarum spread from the small intestine to the large intestine. Interestingly, IL-22RA1 flox/flox and Villin-cre mice rescued the colonization of pGP3-deficient C. muridarum following intrajejunal inoculation, suggesting that pGP3 is important for chlamydial evasion of intestinal epithelial-specific IL-22 expression. Thus, we revealed a novel method for selecting host factors from the GI tract, which may also contribute to the mouse genital tract.
2 Materials and methods
2.1 Chlamydial organisms
All Chlamydia muridarum clones used in this study were derived from the strain Nigg3 (Genbank accession number CP009760.1). The plasmid-free clone CMUT3G5 (GenBank accession# CP006974.1) was initially derived from Nigg3 (Lei et al., 2014), which was used for transformation with the plasmid pCM: GFP to create CM-pGFP (designated as wild type in the current study) or pCM: GFP with a premature stop codon in the pgp3 gene to create CM-pGP3S (designated as mutant in the current study), as described previously (Liu et al., 2014; Liu et al., 2014). The genome and plasmid sequences of CM-pGFP and CM-pGP3S were nearly identical, except for a premature stop codon in pgp3 in CM-pGP3S. As both were transformants, the plasmid copy numbers were similar. Both organisms were propagated in HeLa cells and purified as elementary bodies (EBs), as described previously (Zhang et al., 2015; Fan et al., 1998). Aliquots of the purified EBs were stored in SPG buffer (220 mM sucrose, 12.5 mM phosphate, and 4 mM L-glutamic acid, pH 7.5) at −80 °C until further use.
2.2 Mouse inoculation
Purified C. muridarum EBs were used to infect six-week-old C57BL/6 J mice (Shanghai Lingchang Biotechnology Co., Ltd) intra-jejunum, intra-ileum or intracolon with different inclusion-forming units (IFUs) as indicated in individual experiments.
IL-22 knockout mice were purchased from Cyagen: IL-22 knockout (KO; C57BL/6JCya-Il22em1/Cya, S-KO-10256) and Il22ra1-flox (Il22ra1-flox, S-CKO-07390). Villin-Cre mice were also obtained from Cyagen. The mice were inoculated with CM-pGFP (WT) or CM-pGP3S, as described below in the text.
Intracolonic inoculation was used to deliver 2 × 105 live IFU organisms in 10 μL of SPG buffer to the mouse colon using an inoculation tube (NSET, catalog number 60010; ParaTechs Corp., Lexington, KY, United States).
Intrajejunal or intraileal inoculation: Mice were anesthetized using a mixture of isoflurane and oxygen. Once the mice were unconscious, their abdomens were shaved and sterilized with 70% ethanol. A small incision (approximately 0.25–0.5 in.) was made in the was created in the abdomen using a pair of scissors. The jejunum or ileum was partially pulled out of the body cavity using curved forceps. To the jejunum or ileum, 2 × 105 IFUs of CM-pGP3S in 50 μL of SPG were inoculated using a 1-mL syringe (KDL, Shanghai, China). Care was taken not to remove excess intestines from the animal, completely pierce the intestine, or inject air bubbles. After the injection, the jejunum was placed back into the cavity, and the wound was closed using three to four surgical staples (#ACS- KIT, Braintree Scientific, Inc., Braintree, MA). The mice were resuscitated by placing them on a warm heating pad and supplying them with fresh air.
Note: For example, if the stock titer was 1 × 107 IFU/μL, the stock was diluted 1:10 (10 μL + 90 μL SPG buffer) for a stock titer of 1 × 106 IFU/μl. The stock was diluted 1:50 (20 μL + 980 μL SPG buffer) to a final stock titer of 2 × 104 IFU/μL (2 × 105 IFUs per 10 μL).
2.3 Titrating live chlamydial organisms recovered from swabs and tissue homogenates
To quantify live chlamydial organisms in rectal swabs, each swab was soaked in 0.5 mL of SPG, vortexed with glass beads, and the chlamydial organisms released into the supernatants were titrated on HeLa cell monolayers in duplicates. Infected cultures were processed for immunofluorescence assays as described previously (Tang et al., 2013). Inclusions were counted in five random fields per coverslip under a fluorescence microscope. Coverslips with fewer than one infectious unit (IFU) per field were counted. Coverslips exhibiting cytotoxicity in HeLa cells were excluded. The total number of IFUs per swab was calculated based on the mean number of IFUs per view, the ratio of the view area to that of the well, the dilution factor, and the inoculation volume. Where possible, the mean IFU/swab was derived from serially diluted duplicate samples for each swab. The total number of IFUs/swabs was converted to log10, which was used to calculate the mean and standard deviation across the mice in the same group at each time point.
To quantify live organisms in mouse organs and tissue segments, each organ or tissue segment was transferred to a tube containing 0.5–5 mL SPG, depending on the size of the organ. Each GI tract was cut into seven segments: stomach, duodenum, jejunum, ileum, cecum, colon, and anorectum (rectum). The organs and tissue segments were homogenized in cold SPG using a 2 mL tissue grinder (cat# K885300-0002, Fisher Scientific, Pittsburgh, PA, United States) or an automatic homogenizer (Omni Tissue Homogenizer, TH115; Kennesaw, GA, United States). The homogenates were briefly sonicated and centrifuged at 3,000 rpm for 5 min to pellet the remaining debris. The supernatants were titrated for live C. muridarum on HeLa cells, as described above. The results were expressed as log10 IFUs per organ or tissue segment weight.
2.4 Immunofluorescence assay
For immunofluorescence labeling of C. muridarum in HeLa cells, a rabbit antibody (R1604, raised with purified C. muridarum elementary bodies) was used as the primary antibody, which was visualized using goat anti-rabbit IgG conjugated to Cy2 (green, cat#111-225-144, Jackson ImmunoResearch Laboratories). The DNA dye Hoechst 3328 (blue, Sigma-Aldrich) was used to visualize the nuclei. Doubly labeled samples were used to count C. muridarum using a fluorescence microscope (MIX60, MshOt) with a CCD camera.
2.5 Counting inclusions and calculating IFUs
For each well, IFUs from five random views were counted under an objective lens using the appropriate magnification and were averaged. If one or fewer IFUs per view were found using a 10× objective lens, the entire well was considered. To determine the number of IFUs contained within the sample, the average number of IFUs per view derived from the five views was multiplied by the number of views possible in the total well per magnification, dilution, and factor reflecting the portion of the sample used for titration (Wang et al., 2025). After completing this procedure for each dilution in which IFUs were visible, the average number of IFUs was calculated and expressed as log10 transformed IFUs for statistical analyses.
2.6 Statistical analysis
The experimental data were analyzed using the Wilcoxon rank-sum test to compare the individual tissue types and overall large intestinal chlamydial burden. Fisher’s exact test was used to compare the frequencies of infection between the groups of mice.
3 Results
3.1 pGP3-deficient Chlamydia muridarum failed to spread to large intestine after direct delivery into jejunum
We previously showed that the plasmid-encoded genital tract virulence factor pGP3 is essential for C. muridarum survival in the stomach of the GI tract (Zhang et al., 2019). To further define the organisms from which tissue sites are located beyond the large intestine to prevent C. muridarum from colonizing the large intestine, the different inoculation sites in the mice are shown in Figure 1, and we compared live organisms recovered from rectal swabs following intrajejunal versus intraileal inoculations (Figure 2). Following intrajejunal inoculation, no live organisms were recovered from mice inoculated with pGP3-deficient C. muridarum, although wild-type C. muridarum colonized the GI tract for the two-month period. However, when chlamydial organisms were directly inoculated into the ileum, the pGP3-deficient mutant was rescued and colonized the GI tract. Mice inoculated with the mutant continued to shed live organisms in rectal swabs throughout the two-month period. These results suggest that pGP3 is required for C. muridarum to spread in the large intestine following intrajejunal inoculation.
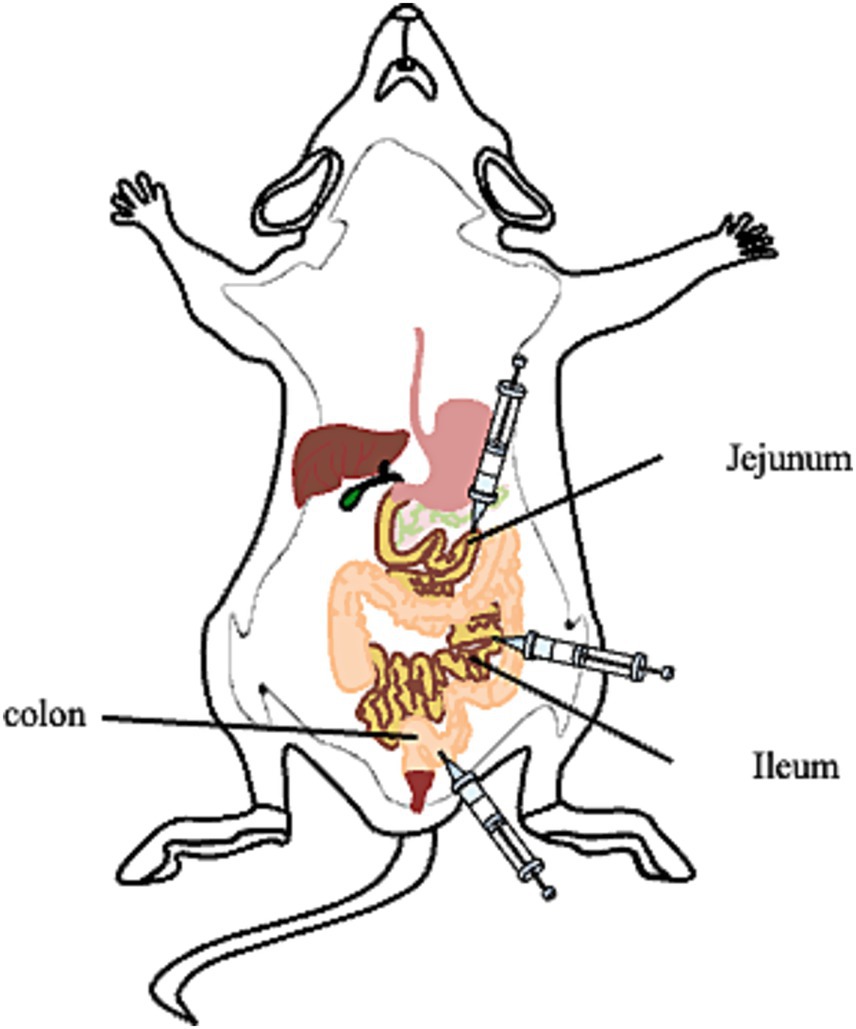
Figure 1. Photograph of the inoculation sites of the mice. Group of mice were inoculated with wild type C. muridarum (CM-pGFP) or pGP3-deficient C. muridarum (CM- pGP3S) following different inoculation sites.
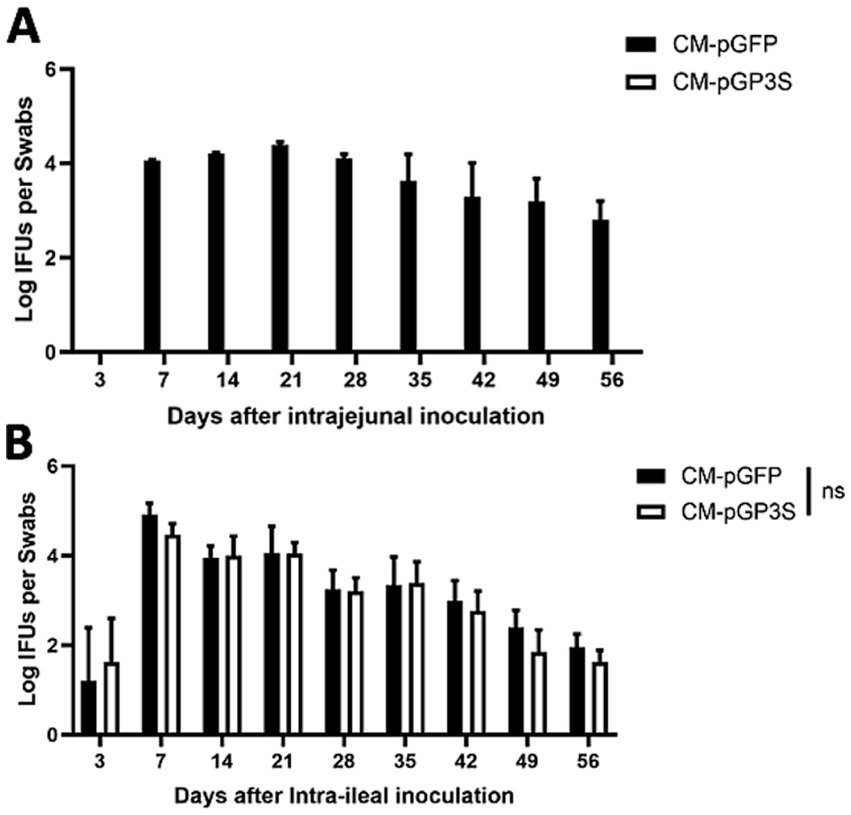
Figure 2. The pGP3-deficient C. muridarum mutant is able to colonize the gastrointestinal tract following an intraileal but not intrajejunal inoculation. Groups of C57BL/6J mice were inoculated with 2 × 105 inclusion forming units (IFUs) of wild-type C. muridarum (CM-pGFP) or pGP3-deficient C. muridarum (CM-pGP3S) either intra-jejunum (A) or intra-ileum (B). At different time points, as shown along the x-axis, mice were monitored for live chlamydial organism recovery from rectal swabs, and the results were expressed as Log10 IFUs per swab, as shown along the Y-axis. Each group consisted of 3–5 mice and data were obtained from 2 to 3 independent experiments.
3.2 IL-22 is an important factor for blocking the spread of pGP3-deficient Chlamydia muridarum from the small intestine to the large intestine
Interleukin-22 (IL-22) is an important cytokine in the intestinal environment that is required to maintain intestinal homeostasis (Zenewicz et al., 2008; Gunasekera et al., 2020; Pavlidis et al., 2022; Huber et al., 2012; Ahlfors et al., 2014; Castleman et al., 2019; Mar et al., 2023). Intestinal tuft cells can also induce anti-Salmonella responses via NKp46 + ILC3 IL22 (Churchill et al., 2025). More importantly, it activates IL-22 production in ILCs to enhance host tissue defense following C. difficile infection (Mears et al., 2025). It is important to determine whether the host factor IL-22 is a key factor in regulating the spread of pGP3-deficient mutants to the large intestine after their direct delivery into the small intestine. Therefore, we compared chlamydial colonization in the GI tract of mice deficient in IL-22.
Following intrajejunal inoculation, the C. muridarum mutant colonized the GI tract of mice deficient in IL-22 (Figure 3A). By day 14 after inoculation, a significant number of live mutants were recovered from the rectal swabs of all IL-22 deficient mice. Overcoming host factor killing is necessary for the detection of intra-jejunum-inoculated chlamydial organisms in rectal swabs, and mice deficient in IL-22 can no longer produce the key host factor IL-22 in their small intestine. These observations suggest that IL-22 regulates the spread of Chlamydia from the small intestine to the large intestine. This conclusion is consistent with the observation that chlamydial mutant organisms were detected in the large intestines of IL-22-deficient mice for at least 56 days post-infection (Figure 3B).
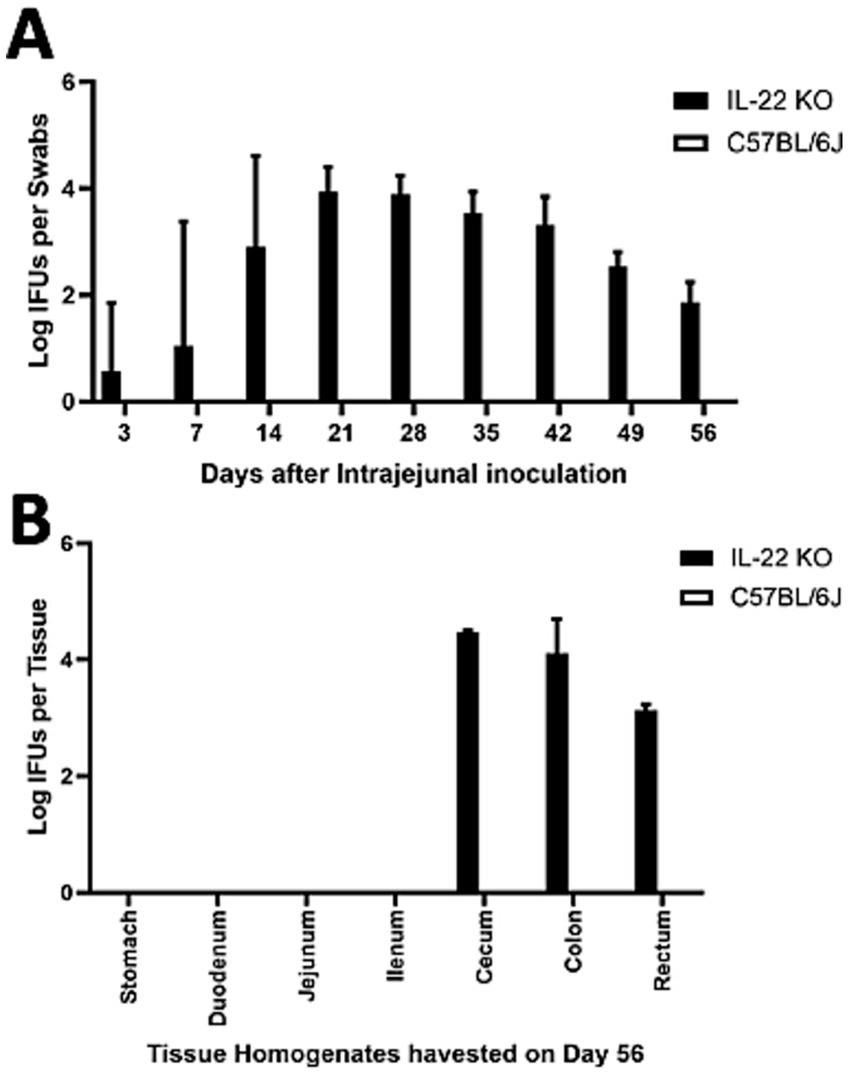
Figure 3. Deficiency in IL-22 rescued the pGP3-deficient C. muridarum mutant to colonize the gastrointestinal tract following an intra-jejunum inoculation. (A) Groups of mice deficient in Interleukin-22 (a key cytokine for regulating gut function, IL-22 KO) or without deficiency (wild-type C57BL/6J) were intra-jejunum inoculated with 2 × 105 IFUs of pGP3-deficient C. muridarum. At different time points, as shown along the X-axis, mice were monitored for live chlamydial organisms in rectal swabs, and the results are expressed as Log10 IFUs per swab, as shown along the Y-axis. N = 5 mice in each group. Data were obtained from two independent experiments. (B) Parallel groups of mice with the same inoculation with pGP3-deficient mutant C. muridarum were sacrificed on day 56 after inoculation, as shown on top of each panel, for monitoring live organism recoveries from different gastrointestinal tissues from the stomach, duodenum to rectum, as indicated along the X-axis. Live organisms are expressed as Log10 IFUs per tissue, as shown along the Y-axis. Each group consisted of 3–5 mice, and the data were obtained from two independent experiments.
We previously demonstrated that pGP3-deficient C. muridarum failed to spread to extra-large intestinal tissues in wild-type mice after intracolonic inoculation (Zhang et al., 2019). By day 14 after intracolonic inoculation in IL-22 deficient mice, the C. muridarum mutant was able to spread to the whole GI tract of mice deficient in IL-22 (Figure 4). This is consistent with the observation that mice deficient in IL-22 rescued pGP3-deficient C. muridarum colonization in the large intestine after direct delivery into the small intestine (Figure 3). Thus, we demonstrated that IL-22 is essential for preventing the spread of Chlamydia from the small intestine to the large intestine.
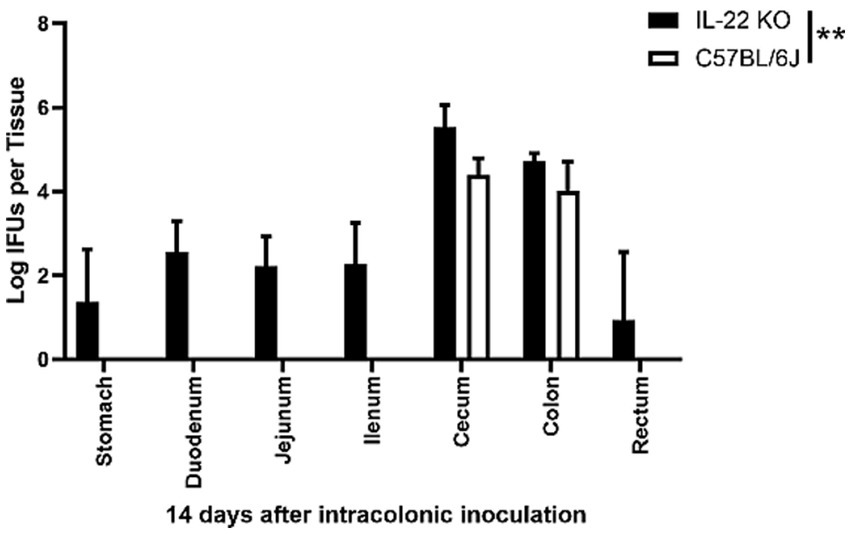
Figure 4. Intracolonically inoculated the pGP3-deficient C. muridarum is able to spread to extra-large intestine tissues in mice deficient in IL-22. Groups of mice deficient in Interleukin-22 or without deficiency (wild-type C57BL/6J) were inoculated with 2 × 105 IFUs of pGP3-deficient C. muridarum intracolonically. Mice were sacrificed on day 14 after inoculation to monitor live organism recoveries from different gastrointestinal tissues from stomach, duodenum to rectum, as indicated along the X-axis. The live organisms were expressed as Log10 IFUs per tissue shown along the Y-axis. Each group has 3 to 5 mice and the data was obtained from 2 independent experiments.
3.3 Intestinal epithelial specific IL-22RA1 deficient mice is sufficient for rescuing the spread of pGP3-deficient C. muridarum from small intestine to large intestine
To further determine whether intestinal IL-22RA1 signaling is necessary for blocking chlamydial spread, we detected chlamydial colonization in the GI tracts of intestinal epithelial-specific IL-22 knockout mice (IL-22ra1 flox/flox mice were bred with Villin-cre mice) (Figure 5). Mice inoculated with the mutant continued to shed live organisms in rectal swabs throughout the 1 month period (Figure 5A), and the pGP3-deficient mutant could spread to the large intestine (cecum and colon) (Figure 5B). These observations suggest that intestinal epithelial-specific IL-22RA1 signaling regulates the spread of Chlamydia from the small intestine to the large intestine.
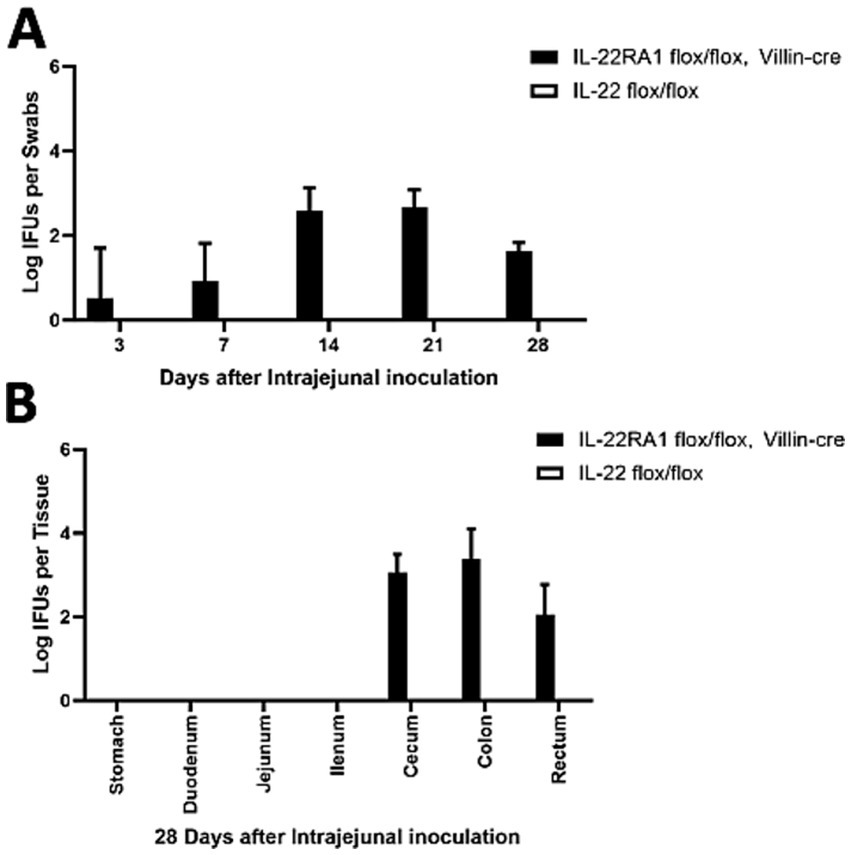
Figure 5. Intestinal epithelial-specific IL-22RA1 knockout mouse rescued the pGP3-deficient C. muridarum colonization following intra-jejunum inoculation. (A) Groups of Intestinal epithelial-specific IL-22RA1 knockout mice (IL-22RA1 flox/flox mice were bred with Villin-cre mice) or control (IL-22 flox/flox mice) were intra-jejunum inoculated with 2 × 105 IFUs of pGP3-deficient C. muridarum. At different time points as shown along the X-axis, mice were monitored for live chlamydial organisms in rectal swabs and the results were expressed as Log10 IFUs per swab as shown along the Y-axis. N = 5 mice for each group. The data was obtained from 2 independent experiments. (B) Parallel groups of the same mice with the same inoculation with pGP3-deficient mutant C. muridarum were sacrificed on day 28 after inoculation, as shown on top of each panel, to monitor live organism recoveries from different gastrointestinal tissues from the stomach, duodenum to the rectum, as indicated along the X-axis. The live organisms were expressed as Log10 IFUs per tissue shown along the Y-axis. Each group has 3 to 5 mice and the data was obtained from 2 independent experiments.
4 Discussion
Although the obligate intracellular bacterium C. trachomatis is a human pathogen of the genital tract, it is frequently detected in the GI tract of mice. However, the significance and mechanisms of C. trachomatis colonization in the gut remain unknown. Since mouse-adapted C. muridarum colonizes the mouse GI tract (Rank and Yeruva, 2014; Dai et al., 2016; Perry and Hughes, 1999), when a naïve mouse is first exposed to C. muridarum in the GI tract, the mouse is orally immunized against subsequent chlamydial infections in the extra-gut tissues. Thus, investigating the mechanisms of C. muridarum interactions with the GI tract may promote our understanding of chlamydial pathogenic mechanisms and facilitate the development of oral vaccines against chlamydial infections in the genital tract. Therefore, murine models have been used to investigate the significance of Chlamydia in the GI tract of these animals.
These interesting models have motivated investigations of the mechanisms of C. muridarum-mouse gut interactions using various methods, such as C. muridarum mutants, knockout mice, or blockade. Using the failure of the CM-pGP3S mutant to spread from the small intestine to the large intestine, we determined that IL-22 and intestinal epithelial-specific IL-22RA1 signaling inhibited CM-pGP3S spread. First, IL-22 knockout mice showed significant spread of CMpGP3S from the small intestine to the large intestine following intrajejunal inoculation. Second, CM-pGP3S did not block the spread from the large intestine to the small intestine in IL-22 knockout mice following intracolonic inoculation. Thus, IL-22 may play a critical role in regulating the spread of bacteria into the large intestine. Finally, the intestinal epithelial-specific IL-22RA1 signaling regulation of chlamydial spreading correlated with intestinal epithelial-specific IL-22RA1 knockout mice rescued from CM-pGP3S spread following intrajejunal inoculation.
These observations led us to hypothesize that CM-pGP3S activates an immune response to block its spread to the large intestine. The current study revealed that intestinal epithelial IL-22RA1 signaling is a critical component of the CMpGP3S-activated barrier. However, the precise relationship between intestinal epithelial IL-22RA1 signaling and CM-pGP3S remains unclear. However, some questions remain unanswered. Which antimicrobial peptide production (CRAMP) by IL-22 or IL-22RA1 signal-mediated immunity is induced by CM-pGP3S? Which microbiome-mediated immunity blocks CMpGP3S spread via IL-22 or IL-22RA1 signals?
Although, knowledge gained from mouse models may not be applicable to C. trachomatis infections in humans. Nevertheless, the mechanistic information obtained from C. muridarum interactions with mouse mucosal tissues may still be informative for understanding how C. trachomatis interacts with human mucosal tissues in the genital tract (Zhao et al., 2015). It is worth noting that the focus of the current study was on C. muridarum spread along the mouse gut. Clearly, more mechanisms are required to investigate the spread of chlamydial organisms from the small intestine to the large intestine.
5 Statistics analyses
The Wilcoxon rank-sum test was used to compare the number of live organisms in the IFUs between different samples.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Animal Welfare and Ethics Committee of Laboratory Animals, Shanghai Institute of Immunity and Infection (Former Institut Pasteur of Shanghai) (A202019). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QT: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Writing – original draft. GF: Formal analysis, Investigation, Writing – review & editing. JM: Data curation, Methodology, Writing – review & editing. LW: Writing – review & editing. ZZ: Funding acquisition, Project administration, Writing – review & editing. TZ: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China for the Youth (Grant no. 32202813) to ZZ (Grant no. 32000138) to TZ and the Changsha Natural Science Foundation (kq2502203).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahlfors, H., Morrison, P. J., Duarte, J. H., Li, Y., Biro, J., Tolaini, M., et al. (2014). IL-22 fate reporter reveals origin and control of IL-22 production in homeostasis and infection. J. Immunol. 193, 4602–4613. doi: 10.4049/jimmunol.1401244
Budrys, N. M., Gong, S., Rodgers, A. K., et al. (2012). Chlamydia trachomatis antigens recognized in women with tubal factor infertility, normal fertility, and acute infection. Obstet. Gynecol. 119, 1009–1016. doi: 10.1097/AOG.0b013e3182519326
Castleman, M. J., Dillon, S. M., Purba, C. M., Cogswell, A. C., Kibbie, J. J., McCarter, M. D., et al. (2019). Commensal and pathogenic bacteria indirectly induce IL-22 but not IFNγ production from human colonic ILC3s via multiple mechanisms. Front. Immunol. 10:649. doi: 10.3389/fimmu.2019.00649
Chen, J., Yang, Z., Sun, X., Tang, L., Ding, Y., Xue, M., et al. (2015). Intrauterine infection with plasmid-free Chlamydia muridarum reveals a critical role of the plasmid in chlamydial Ascension and establishes a model for evaluating plasmid-independent pathogenicity. Infect. Immun. 83, 2583–2592. doi: 10.1128/IAI.00353-15
Churchill, M. J., Pandeya, A., Bauer, R., Christopher, T., Krug, S., Honodel, R., et al. (2025). Enteric tuft cell inflammasome activation drives NKp46+ILC3 IL22 via PGD2 and inhibits Salmonella. J. Exp. Med. 222:e20230803. doi: 10.1084/jem.20230803
Craig, A. P., Kong, F. Y. S., Yeruva, L., Hocking, J. S., Rank, R. G., Wilson, D. P., et al. (2015). Is it time to switch to doxycycline from azithromycin for treating genital chlamydial infections in women? Modelling the impact of autoinoculation from the gastrointestinal tract to the genital tract. BMC Infect. Dis. 15:200. doi: 10.1186/s12879-015-0939-3
Dai, J., Zhang, T., Wang, L., Shao, L., Zhu, C., Zhang, Y., et al. (2016). Intravenous inoculation with Chlamydia muridarum leads to a long-lasting infection restricted to the gastrointestinal tract. Infect. Immun. 84, 2382–2388. doi: 10.1128/IAI.00432-16
Fan, T., Lu, H., Hu, H., Shi, L., McClarty, G. A., Nance, D. M., et al. (1998). Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187, 487–496. doi: 10.1084/jem.187.4.487
Gratrix, J., Singh, A. E., Bergman, J., Egan, C., McGinnis, J., Drews, S. J., et al. (2014). Prevalence and characteristics of rectal chlamydia and gonorrhea cases among men who have sex with men after the introduction of nucleic acid amplification test screening at 2 Canadian sexually transmitted infection clinics. Sex. Transm. Dis. 41, 589–591. doi: 10.1097/OLQ.0000000000000176
Gratrix, J., Singh, A. E., Bergman, J., Egan, C., Plitt, S. S., McGinnis, J., et al. (2015). Evidence for increased chlamydia case finding after the introduction of rectal screening among women attending 2 Canadian sexually transmitted infection clinics. Clin. Infect. Dis. 60, 398–404. doi: 10.1093/cid/ciu831
Gunasekera, D. C., Ma, J., Vacharathit, V., et al. (2020). The development of colitis in Il10(−/−) mice is dependent on IL-22. Mucosal Immunol. 13, 493–506. doi: 10.1038/s41385-019-0252-3
Huber, S., Gagliani, N., Zenewicz, L. A., et al. (2012). IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491, 259–263. doi: 10.1038/nature11535
Koprivsek, J. J., Zhang, T., Tian, Q., et al. (2019). Distinct roles of chromosome- versus plasmid-encoded genital tract virulence factors in promoting Chlamydia muridarum colonization in the gastrointestinal tract. Infect. Immun. 87, e00265–e00219. doi: 10.1128/IAI.00265-19
Lei, L., Chen, J., Hou, S., Ding, Y., Yang, Z., Zeng, H., et al. (2014). Reduced live organism recovery and lack of hydrosalpinx in mice infected with plasmid-free Chlamydia muridarum. Infect. Immun. 82, 983–992. doi: 10.1128/IAI.01543-13
Liu, Y., Chen, C., Gong, S., Hou, S., Qi, M., Liu, Q., et al. (2014). Transformation of Chlamydia muridarum reveals a role for Pgp5 in the suppression of plasmid-dependent gene expression. J. Bacteriol. 2014196, 989–998. doi: 10.1128/JB.01161-13
Liu, Y., Huang, Y., Yang, Z., Sun, Y., Gong, S., Hou, S., et al. (2014). Plasmid-encoded Pgp3 is a major virulence factor for Chlamydia muridarum to induce hydrosalpinx in mice. Infect. Immun. 82, 5327–5335. doi: 10.1128/IAI.02576-14
Mar, J. S., Ota, N., Pokorzynski, N. D., et al. (2023). IL-22 alters gut microbiota composition and function to increase aryl hydrocarbon receptor activity in mice and humans. Microbiome 11:47. doi: 10.1186/s40168-023-01486-1
Mears, K. S., Denny, J. E., Maslanka, J. R., Mdluli, N. V., Hulit, E. N., Matsuda, R., et al. (2025). Therapeutic activation of IL-22 producing innate lymphoid cells enhances host defenses to Clostridioides difficile infection. Cell Rep. 44:115438. doi: 10.1016/j.celrep.2025.115438
Musil, K., Currie, M., Sherley, M., and Martin, S. (2016). Rectal chlamydia infection in women at high risk of chlamydia attending Canberra sexual health Centre. Int. J. STD AIDS 27, 526–530. doi: 10.1177/0956462415586317
O'Connell, C. M., Ingalls, R. R., Andrews, C. W. Jr., Scurlock, A. M., and Darville, T. (2007). Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J. Immunol. 179, 4027–4034. doi: 10.4049/jimmunol.179.6.4027
Pavlidis, P., Tsakmaki, A., Pantazi, E., Li, K., Cozzetto, D., Digby- Bell, J., et al. (2022). Interleukin-22 regulates neutrophil recruitment in ulcerative colitis and is associated with resistance to ustekinumab therapy. Nat. Commun. 13:5820. doi: 10.1038/s41467-022-33331-8
Perry, L. L., and Hughes, S. (1999). Chlamydial colonization of multiple mucosae following infection by any mucosal route. Infect. Immun. 67, 3686–3689. doi: 10.1128/IAI.67.7.3686-3689.1999
Peters, R. P. H., Dubbink, J. H., van der Eem, L., Verweij, S. P., Bos, M. L. A., Ouburg, S., et al. (2014). Cross-sectional study of genital, rectal, and pharyngeal chlamydia and gonorrhea in women in rural South Africa. Sex. Transm. Dis. 41, 564–569. doi: 10.1097/OLQ.0000000000000175
Pospischil, A., Borel, N., Chowdhury, E. H., and Guscetti, F. (2009). Aberrant chlamydial developmental forms in the gastrointestinal tract of pigs spontaneously and experimentally infected with Chlamydia suis. Vet. Microbiol. 135, 147–156. doi: 10.1016/j.vetmic.2008.09.035
Rank, R. G., and Yeruva, L. (2014). Hidden in plain sight: chlamydial gastrointestinal infection and its relevance to persistence in human genital infection. Infect. Immun. 82, 1362–1371. doi: 10.1128/IAI.01244-13
Shao, L., Melero, J., Zhang, N., et al. (2017). The cryptic plasmid is more important for Chlamydia muridarum to colonize the mouse gastrointestinal tract than to infect the genital tract. PLoS One 12:e0177691. doi: 10.1371/journal.pone.0177691
Shao, L., Zhang, T., Liu, Q., Wang, J., and Zhong, G. (2017). Chlamydia muridarum with mutations in chromosomal genes tc0237 and/or tc0668 is deficient in colonizing the mouse gastrointestinal tract. Infect. Immun. 85:e00321–17. doi: 10.1128/IAI.00321-17
Shao, L., Zhang, T., Melero, J., Huang, Y., Liu, Y., Liu, Q., et al. (2017). The genital tract virulence factor pGP3 is essential for Chlamydia muridarum colonization in the gastrointestinal tract. Infect. Immun. 86:e00429–17. doi: 10.1128/IAI.00429-17
Tan, Y., Li, Y., Zhang, Y., et al. (2018). Immunization with Chlamydia psittaci plasmid-encoded protein CPSIT_p7 induces partial protective immunity against chlamydia lung infection in mice. Immunol. Res. 66, 471–479. doi: 10.1007/s12026-018-9018-3
Tang, L., Zhang, H., Lei, L., Gong, S., Zhou, Z., Baseman, J., et al. (2013). Oviduct infection and hydrosalpinx in DBA1/j mice is induced by intra-cervical but not intravaginal inoculation with Chlamydia muridarum. PLoS One 8:e71649. doi: 10.1371/journal.pone.0071649
Wang, Y., Han, Z., Wang, L., Sun, X., Tian, Q., and Zhang, T. (2025). Development and validation of Chlamydia muridarum mouse models for studying genital tract infection pathogenesis. Bio Protoc. 15:e5181. doi: 10.21769/BioProtoc.5181
Wang, C., Jin, Y., Wang, J., et al. (2023). Protective immunity against Chlamydia psittaci lung infection induced by a DNA plasmid vaccine carrying CPSIT_p7 gene inhibits dissemination in BALB/c mice. Int. J. Mol. Sci. 24:7013. doi: 10.3390/ijms24087013
Wang, C., Li, Y., Wang, S., Yan, X., Xiao, J., Chen, Y., et al. (2020). Evaluation of a tandem Chlamydia psittaci Pgp3 multiepitope peptide vaccine against pulmonary chlamydial challenge in mice. Microb. Pathog. 147:104256. doi: 10.1016/j.micpath.2020.104256
Wang, J., Wang, B., Xiao, J., Chen, Y., and Wang, C. (2024). Chlamydia psittaci: a zoonotic pathogen causing avian chlamydiosis and psittacosis. Virulence 15:2428411. doi: 10.1080/21505594.2024.2428411
Yang, R., Jacobson, C., Gardner, G., Carmichael, I., Campbell, A. J. D., and Ryan, U. (2014). Longitudinal prevalence and faecal shedding of Chlamydia pecorum in sheep. Vet. J. 201, 322–326. doi: 10.1016/j.tvjl.2014.05.037
Zenewicz, L. A., Yancopoulos, G. D., Valenzuela, D. M., Murphy, A. J., Stevens, S., and Flavell, R. A. (2008). Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 29, 947–957. doi: 10.1016/j.immuni.2008.11.003
Zhang, Q., Huang, Y., Gong, S., Yang, Z., Sun, X., Schenken, R., et al. (2015). In vivo and ex vivo imaging reveals a long-lasting chlamydial infection in the mouse gastrointestinal tract following genital tract inoculation. Infect. Immun. 83, 3568–3577. doi: 10.1128/IAI.00673-15
Zhang, T., Huo, Z., Ma, J., He, C., and Zhong, G. (2019). The plasmid-encoded pGP3 promotes Chlamydia evasion of acidic barriers in both the stomach and vagina. Infect. Immun. 87, e00844–e00818. doi: 10.1128/IAI.00844-18
Zhao, X., Zhu, D., Ye, J., Li, X., Wang, Z., Zhang, L., et al. (2015). The potential protective role of the combination of IL-22 and TNF-a against genital tract Chlamydia trachomatis infection. Cytokine 73, 66–73. doi: 10.1016/j.cyto.2015.01.027
Keywords: Chlamydia muridarum , pGP3, IL-22, gastrointestinal tract, colonization
Citation: Tian Q, Fang G, Ma J, Wang L, Zuo Z and Zhang T (2025) Intestinal IL-22RA1 signaling regulates Chlamydia deficient in plasmid-encoded pGP3 spreading to large intestine. Front. Microbiol. 16:1647731. doi: 10.3389/fmicb.2025.1647731
Edited by:
Rajesh Mani, The University of Texas Health Science Center at Tyler, United StatesReviewed by:
George W. Liechti, Uniformed Services University of the Health Sciences, United StatesChuan Wang, University of South China, China
Copyright © 2025 Tian, Fang, Ma, Wang, Zuo and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zonghui Zuo, em9uZ2h1aV96dW9AdGphdS5lZHUuY24=; Tianyuan Zhang, enR5MTkxMUAxMjYuY29t
 Qi Tian
Qi Tian Guangchi Fang2
Guangchi Fang2 Zonghui Zuo
Zonghui Zuo