- 1School of Life Sciences, Huzhou University, Huzhou, China
- 2Shandong Center for Disease Control and Prevention, Jinan, China
- 3Deqing County Ecological Forestry Integrated Service Center, Huzhou, China
- 4Laboratory of Invertebrate Pathology, College of Animal Sciences, Zhejiang University, Hangzhou, China
- 5Hynobius Anji National Nature Reserve, The Administration Bureau of Longwangshan Natural Reserve, Anji, China
Introduction: The lactic acid bacteria (LAB) has shown great potential as a sustainable solution to support agriculture through its plant-growth-promoting and biocontrol activities. However, their efficacy as bioinoculants is limited by unpredictable colonization in natural conditions.
Methods: The bacterial strain LP0308, identified as Lactobacillus plantarum (LP0308) based on 16S rRNA sequence analysis, was obtained from rhizosphere soil. The features and colonization strategies of LP0308 were characterized through genome sequencing, tomato seed germination assays, pot experiments, and measurements of soil physicochemical properties and enzyme activities.
Results: LP0308 was introduced into the soil of tomato, and it could stably persist and proliferate for a long-term (0–20 days), as confirmed by colony-forming unit (CFU), quantitative real-time PCR (RT-qPCR), and fluorescence in situ hybridization (FISH) analyses. Further characterization revealed that LP0308 altered the microbial composition of the rhizosphere soil and significantly increased the abundance of Bacillus and potentially pathogenic microorganism. Further analyses revealed that LP0308 altered the rhizosphere soil microbial community, significantly increasing the abundance of Bacillus spp. while decreasing the potential pathogenic microorganisms, such as Ralstonia solanacearum and Fusarium oxysporum. In addition, the successful colonization of LP0308 led to drastically increased expression of encoding biofilm (vpsI1, vpsI2, vpsC, and vpsI3), immune modulation (pbpG, kdtB, and wbpL), and antimicrobial activity gene (farB). L. plantarum strain LP0308 was confirmed as a possible plant growth-promoting rhizobacteria (PGPR), which significantly promoted bud length, plant height, primary root length, root fresh weight, and whole-seedling fresh weight. Additionally, application of LP0308 markedly improved soil nutrient availability and stimulated key enzymatic activities.
Discussion: Together, our findings suggest the LP0308 as a potential target for developing more effective bioinoculants for sustainable agriculture.
Introduction
Global agricultural production and food security have long been fundamental to human survival and development (Singh et al., 2023; Feng et al., 2025). Traditional agriculture often relies on chemical fertilizers and pesticides to boost yields (Carvajal-Yepes et al., 2019). However, by 2030, the global population is expected to reach 8.9 billion, and by 2050, it will grow to 9.86 billion, leading to an estimated 70% increase in the demand for food production (Kah et al., 2019). This increase will put immense pressure on agricultural output and land resources, which are already facing mounting challenges (van Vliet et al., 2017). Among these, accelerated urbanization, soil degradation, and the environmental pollution caused by the excessive use of pesticides and fertilizers are becoming more prominent (Borrelli et al., 2020; Tang et al., 2021). To address these challenges and achieve the United Nations Sustainable Development Goals (SDGs), there is an urgent need to develop alternative fertilizers that can replace traditional chemical fertilizers (Hofmann et al., 2020; McElwee et al., 2020).
Plant growth-promoting rhizobacteria (PGPR) are considered potential beneficial microorganisms capable of forming symbiotic relationships with plants (Wang et al., 2024). PGPR, such as Bacillus, Klebsiella, and Pseudomonas species, play a crucial role in enhancing plant health, improving soil fertility, boosting plant immunity, and facilitating the assimilation of essential nutrients (Wang, 2009; Jansson et al., 2023). However, the inconsistent or suboptimal effectiveness of microbial inoculants in agricultural soils over time may be attributed to the complex soil environment, microbial niche competition, and limited root colonization ability, which hinder the expression of biological activity factors by target strains (Britt, 2021; Khoso et al., 2024). In addition, microbial inoculants are subject to various environmental pressures, such as climate change and abiotic stressors, which can make them seem like “invaders” (Sammauria et al., 2020; Liu et al., 2022). They must also compete for ecological niches with indigenous microbial communities, further reducing their adaptability and limiting largescale use (Moore et al., 2022). Root zone colonization is a critical mechanism through which probiotics exert their beneficial effects, significantly influencing the efficacy of microbial agents utilized as biofertilizers, biopesticides, plant growth stimulants, and bioremediation agents (Knights et al., 2021; Liu et al., 2024). Rhizosphere colonization is one of the most important features of rhizobacteria that determines their survival and propagation, which are prerequisites for versatile bacteria to exert their beneficial functions on host plants (Santoyo et al., 2021). The process of colonization can be delineated into several key stages, including chemotaxis and motility, attachment to the root surface, overcoming plant immunity, proliferation and biofilm development on the rhizoplane, followed by endophytic penetration (Knights et al., 2021). For example, NCT-2 significantly enhanced nitrate removal from the soil and improved plant growth by colonizing the meristematic and elongation zones of the root tip as well as the roots (Chu et al., 2018).
Lactic acid bacteria (LAB), a group of Gram-positive microorganisms, exhibit symbiotic, biological defense, and environmental remediation properties, offering considerable potential for advancing sustainable agriculture and environmental protection (Ayivi et al., 2020; Rama et al., 2024). LAB strains, recognized for their probiotic properties, high safety, and broad applicability, have been granted Generally Recognized as Safe (GRAS) status by the U.S. Food and Drug Administration, making them an ideal candidate for commercial development in food, sustainable agriculture, and pharmaceutical industries (Hati et al., 2013; Sadiq et al., 2019). Studies have shown that LAB are widespread in the phyllosphere, endosphere, and rhizosphere of various plant species, such as maize, pepper, and barley. Furthermore, studies have found that LAB have garnered significant attention in sustainable agriculture due to their presence as components of the epiphytic and/or endophytic microbial communities in feed crops and roots. For example, when inoculated into Lolium perenne, Lactobacilli can colonize the roots at an endophytic level (Lamont et al., 2017). As a fertilizer, LAB can promote biodegradation, accelerate soil organic content, and produce organic acids and metabolic products such as plant growth-promoting (PGP) substances and nitrogen sources, helping plants improve nutrient absorption and stress resistance (Ekundayo, 2014; Minervini et al., 2015). For example, compared to untreated soil, the application of Lactobacillus plantarum H64 biomass combined with wheat bread significantly enhanced the soil organic carbon content (by up to 37%), total nitrogen content (by up to 40%), and the growth of Cichorium endivia in pots (1.7-fold increase) (Cacace et al., 2022). Additionally, LAB has shown antagonistic effects against plant pathogens, even inhibiting pathogenic fungal and bacterial populations in the rhizosphere and phyllosphere (Daranas et al., 2019; Chen et al., 2023). Moreover, recent research has found that LAB exhibits bioremediation efficiency and the ability to detoxify heavy metals and mycotoxins (Kirillova et al., 2017; Kinoshita, 2019). The application of LAB for heavy metal bioremediation in leafy vegetables demonstrated maximum adsorption efficiencies of 79.75% for lead (Pb), 75.28% for cadmium (Cd), and 83.99% for nickel (Ni) (Mostafidi et al., 2023).
Rhizospheric microorganism represent the first line of defense in maintaining soil health and protecting against soil-borne diseases. For example, Burkholderia cenocepacia strain XXVI produces hydroxamate siderophore with biocontrol activity against the pathogen Colletotrichum lindemuthianum ATCC MYA 456, promoting plant growth (Santos-Villalobos et al., 2012; Santoyo et al., 2021). However, limited research has been conducted on the colonization mechanisms of LAB in the rhizosphere and their PGP properties (O’Banion et al., 2023; Sanchez-Gil et al., 2023). In the current study, a LP0308 strain was isolated and purified from the rhizosphere soil of healthy tomatoes, and its characteristics were analyzed through whole genome sequencing. We further investigated the colonization of LP0308 in rhizosphere soil and its effect on rhizosphere microbial community structure of tomato root. In addition, we quantified colonization-related genes following the growth of the bacterium in the soil using quantitative real-time PCR (RT-qPCR). Finally, we analyzed indicators of plant growth promotion, soil physical and chemical properties, and enzyme activity following inoculation with LP0308. The present study enhances our understanding of rhizosphere soil colonization by an indigenous bacterium and may promote the application of probiotic targeting regulation as biofertilizers in agricultural.
Materials and methods
Isolation and identification of LP0308
Rhizosphere soils were collected as described previously (McPherson et al., 2018). Briefly, plants were gently handled to remove loosely adherent soil from their roots, while deliberately retaining an approximately 1-mm-thick layer of soil on the root surfaces (n = 15). Tomato rhizosphere soil was harvested in a tomato field in Zhejiang province, China (120.13°N, 30.87°W). LP0308 strain were isolated from the rhizosphere soil and purified by plate culture with de Man–Rogosa–Sharpe medium (MRS) medium (Hopebio, Qingdao, China). Bacterial colonies were randomly selected from agar plates and subcultured a minimum of three times prior to identification. The growth curve of LP0308 was determined using 96-well honeycomb plates (Corning, United States), with optical density measured at 600 nm at 5 h intervals over a 50 h period.
For scanning electron microscopy (SEM) analysis, LP0308 was initially fixed in 2.5% glutaraldehyde, washed three times with PBS, and subsequently post-fixed in 1% osmium tetroxide. The samples were then dehydrated through a graded ethanol series (50%, 70%, 80%, 95%, and 100%) for 15 min and embedded in epoxy resin (Dr. Spurr’s kit, Electron Microscopy Sciences, Hatfield, PA, United States). Ultrathin sections were stained with 1% uranyl acetate and examined using SEM transmission electron microscope (Hitachi SU8000, Tokyo, Japan) at an accelerating voltage of 80 kV.
L. plantarum stability and proliferation in the soil
The tomato varieties Sweetheart No. 8 were selected for their pomological traits which are highly appreciated by consumers of china. Seeds (n = 100 per variety) were provided by Yinong Agriculture Co., Ltd. (Zhejiang, China). For the seed germination assay, surface-sterile tomato seeds prepared as follows. They were surface-sterilized by immersion in 70% ethanol (Sangon, Shanghai, China) for 1 min, followed by treatment with 10% hypochlorite (Sangon, Shanghai, China) for 5 min, and then subjected to five consecutive rinses with sterile distilled water. For the root colonization assay, surface-sterile tomato seeds prepared as described above. Tomato seedlings at the two- to three-leaf stage, grown under sterile conditions, were immersed in LP0308 bacterial suspension (106 CFU/L) for 4 h before being transplanted into sterile soil and incubated in a climate-controlled sterile growth chamber (Percival growth chambers; CLF Plant Climatics) of 22 °C/18 °C under a 16 h/8 h (day/night), with blank medium (CK) as the control. At 5-day intervals, rhizosphere soil tightly adhering to the root surface was carefully collected and serially diluted with sterile water. All compartments were placed on ice. The number of L. plantarum in rhizosphere soil was quantified by standard colony-forming unit (CFU) enumeration and absolute quantification via RT-qPCR after inoculation with LP0308 5, 10, 15, and 20 days later. CFU counts were determined by serial dilution of each sample (1 × 10–1 to 1 × 10−6), plating, and incubation at 30 °C for 48 h. Colonies were then manually counted (n = 5). The experiment was repeated five times. The colony-forming units per gram of soil (CFU/g) were then enumerated to assess bacterial colonization dynamics. For qPCR, the rhizosphere soil was collected and transferred to PowerBead Tubes (QIAGEN DNeasy PowerSoil Kit, Hilden, Germany) for LP0308 counts analysis. PowerBead Tubes containing soil samples were homogenized in a Precellys-24 tissue Homogenizer (Bertin Technologies, Aixen, France) at 5,000 rpm for 45 s. Total DNA was extracted using QIAGEN Dneasy PowerSoil Kit for soil according to the manufacturer’s instructions. The final DNA concentration and purity were determined by NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, NC, United States), and DNA quality was checked by 1% agarose gel electrophoresis.
Quantitative PCR was performed by Roche LightCycler 480 system (Roche, Basel, Switzerland) using the SYBR qPCR master mix (Vazyme Biotech, Nanjing, China). The primer sequences for all amplified genes are shown in Supplementary Table 1: LP for LP0308 and BA1 for Bacillus (Chen et al., 2010; Bodenhausen et al., 2014; Roosa et al., 2014). RT-qPCR was conducted using the following program: 5 min of denaturation at 95 °C, followed by 40 cycles of 10 s at 95 °C, 10 s of annealing at an appropriate temperature, and 10 s of elongation at 72 °C, followed by a final melting curve step (from 65 °C to 92°C, 0.5 °C/s). The copies were calculated by comparing Cp values to a standard curve amplified from a series of 10-fold dilutions of containing purified genomic DNA for individual gene quantification. The same approach to confirm the accuracy of Bacillus copies number. Each group included at least three technical replicates and five biological replicates.
Fluorescence in situ hybridization analysis of LP0308 in soil
Fluorescence in situ hybridization (FISH) analysis of LP0308 in rhizosphere soil. LP0308 was fixed with FISH fixative for 30 min at room temperature, permeabilized in PBS with 0.2% Triton X-100 for 5 min, and then incubated in pre-hybridization solution at 37 °C for 1 h. The fixed cells were then hybridized with the fluorescently probe LbpV3 (5′-CCGTCAATACCTGAACAG-3′) lactobacillus–specific probe overnight at 4 °C (Gosiewski et al., 2012). Then, the cells were washed five times with PBS and stained with 1:1,000 DAPI solution for 5 min. Fluorescence images were captured using a fluorescence microscope (CryoStar, NX70, France).
The rhizosphere soil microbiota analysis by 16S rRNA gene sequencing
Two rhizosphere soil samples (CK and LP0308 group, 5 g each) per plant were collected and immediately processed for DNA extraction. For each plant, DNA was extracted from 0.3 g of rhizosphere soil using the DNeasy PowerSoil kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. To construct an Illumina sequencing library, the V4–V5 hypervariable regions of the 16S rRNA gene were amplified from 1 μl of purified DNA using the universal primers 515F and 806R, as listed in Supplementary Table 1. Purified PCR productions were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, CS, United States) following the standard protocols provided by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Twenty bacterial samples were successfully sequenced.
To investigate the rhizosphere soil bacterial community structure, alpha-diversity indices (Shannon and Simpson indices; the number of observed species) were calculated using Mothur. Bray–Curtis distances between samples were used for principal coordinate analysis (PCoA, cmdscale function in R). To test the effect of location and compartment on the estimated explained variance, a permutational multivariate analysis of variance (PERMANOVA) analysis was performed (Adonis function from the vegan package, in R). Network analyses were used to explore the co-occurrence patterns of rhizosphere soil bacterial microorganisms in each niche. Moreover, linear discriminant analysis (LDA) effect size (LEfSe) was conducted using the normalized ASV table.
To assess the difference in rhizosphere microorganisms between the CK and LP0308 colonization groups (LP), the molecular ecological networks among the top 100 genera with the highest abundance of bacterial taxa were described through network analysis using the Molecular Ecological Network Analyses Pipeline (MENAP) (Deng et al., 2012). The correlation matrix was calculated by the Pearson correlation coefficient. Networks were constructed using random matrix theory-based methods with a correlation threshold of 0.65 (Zhou et al., 2010; Zhou et al., 2011). The resulting correlations were imported into the Gephi platform and then visualized by the Fruchterman Reingold algorithms (Bastian et al., 2009). The topological features of every node, including clustering, average degree, degree, and closeness centrality, were calculated for each graph (Shannon et al., 2003).
Finally, phylogenetic investigation of communities by Functional Annotation of Prokaryotic Taxa (FAPROTAX) was employed for stringent predictions of microbial functional metabolic pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The phenotypes of rhizosphere soil bacteria in each group (CK and LP0308) were predicted using the BugBase database. Sequencing data were deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) under accession number PRJNA1270370.
Antagonistic effect of LP0308 against soil borne pathogens
To evaluate the antagonistic activity of LP0308 against common soil-borne pathogens of tomato, five phytopathogenic strains were selected from laboratory collection: Ralstonia solanacearum, Fusarium oxysporum, Cladosporium fulvum, Alternaria solani, and Botrytis cinerea. Uninoculated liquid medium and sterile water were used as a negative control. The LP0308 strain was cultured in MRS medium at 37 °C for 24 h. The supernatant was collected after centrifugation at 10,000 × g for 3 min and filtered through a 0.22 mm PVDF membrane (Millipore Corp., Billerica, MA, United States) to obtain the cell free fermentation liquid. Antimicrobial activity of LP0308 was evaluated using the agar well diffusion method (Zhang et al., 2022).
Germination of tomato seeds and pot experiment
The seeds were treated with sterile LP0308 fermentation supernatant. Bud length was measured with a ruler for plants in the CK (sterile MRS liquid medium) and LP0308 (LP) groups. Germinated seedlings were transferred into 10 cm × 8.5 cm pot with each pot containing one seedling. Each seedling pot was filled with approximately 150 g of sterilized soil. After 20 days, plant height and root length were measured with ruler. The root fresh weight and seedling fresh weight were measured with a precision balance. Then, the 0.2 g fresh root sample was extracted with 2 ml of phosphate buffer (pH 7.4) and then centrifuged at 1,000 g for 10 min. The contents of indoleacetic acid (IAA) of tomato root were measured via enzyme-linked immunosorbent assay (ELISA) according to the protocols (ELISA kits BH100, Zoonbio Biotechnology, Nanjing, China). The experiment was repeated 5 times, each time with 15 tomato plants.
Determination of soil physicochemical properties and enzyme activities
Soil samples were collected using a sterile dry brush to gently scrape the surface soil from the roots of control and LP0308-treated tomato plants. Soil pH was measured using a pH meter on an appropriate amount of air-dried soil (sieved through a 20-mesh sieve), which was suspended in a 1:2.5 soil-to-water ratio. The soil organic matter (OM) was measured using the wet oxidation redox titration. Ammonium and nitrate were analyzed by a continuous-flow analyzer. The available phosphorus (AP) was measured using the molybdenum blue method. In addition, we measured the alkaline nitrogen (AN), and available potassium (AK), following the method (Bao, 2000; Nichols and Wright, 2006). The activities of several soil enzymes were assessed, including polyphenol oxidase (POX), urease, β-glutaminase (GLS), peroxidase (POD), alkaline phosphatase (ALP), cellulase (CE), N-acetyl-β-D-glucosaminidase (NAG), and β-1,4-glucosidase (BG). All soil enzyme activities were measured with a commercial activity assay kit (Solebao, Beijing, China) according to the manufacturer’s instructions.
Genome sequencing, assembly, and annotation
The whole genome of LP0308 was sequenced using the PacBio RSII platform at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China), generating 2,266 Mbp of high-quality reads. To ensure data integrity, sequences with non-standard bases at the 5′ ends, lengths under 25 bp, more than 10% ambiguous bases (Ns), or a quality score ≤ 20 were filtered out. Genome assembly was conducted using SOAPdenovo v1.05 with various k-mer sizes, and remaining gaps were closed through PCR amplification (Liang et al., 2018; Xie et al., 2019). The raw sequencing data, including both single-end and paired-end reads, are available in GenBank under BioProject ID PRJNA1269284. Gene prediction for strain LP0308 was carried out using Glimmer v3.02.
Genes of strain LP0308 prediction was performed using Glimmer, while tRNA and rRNA genes were identified with tRNA-scan-SE and Barrnap, respectively. The predicted CDSs were then annotated by aligning sequences against multiple databases, including NR, Swiss-Prot, Pfam, GO, COG, and KEGG, utilizing tools such as BLAST, Diamond, and HMMER. In brief, query proteins were compared to these databases, and gene annotations were assigned based on the best matches with an e-value threshold of less than 105 (Ashburner et al., 2000). The raw dataset including both single and paired-end reads, is deposited at GenBank under BioProject and accession number ID: PRJNA763083.
Quantitative real-time PCR validation of genes
After inoculating the soil with LP0308 (107 CFU/ml), the expression levels of colonization-related genes [exopolysaccharide biosynthesis glycosyltransferase VpsI (gene1112, vpsI1), D-alanyl-D-alanine endopeptidase PBP7/8 (gene1112, pbpG), exopolysaccharide biosynthesis glycosyltransferase VpsI (vpsI2), exopolysaccharide biosynthesis acetyltransferase VpsC (vpsC), exopolysaccharide biosynthesis glycosyltransferase VpsI (vpsI3), lipopolysaccharide core biosynthesis protein (kdtB), glycosyltransferase WbpL (wbpL) and fatty acid efflux system protein FarB (farB)] were determined at 0, 5, 10, 15, and 20 days using RT-qPCR (n = 5).
Total RNA from the rhizosphere soil was extracted using the RNeasy® PowerSoil® Total RNA Kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol. The RNA quality and concentration were measured with a Qubit Fluorometer (Thermo Fisher, United States). RNA of the desired quality (1 mg) was reverse transcribed into cDNA using QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). The specific primers used for RT-qPCR of colonization-related genes are listed in Supplementary Table 1. RT-qPCR was performed according to a previously described of this study. Gene expression was calculated relative to the housekeeping gene 23S rRNA using the 2–ΔΔCt method.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism (version 9.0). Following assessment of data normality and homogeneity of variance, appropriate statistical tests (parametric or non-parametric) were selected. One-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test or Student’s t-test was performed to compare the quantification of soil bacteria, bugbase predictions, topological features of network analysis, IAA contents, plant growth indicators, soil physical properties, and enzyme activities and gene expression between the experimental and control groups were analyzed. A p-value of <0.05 was considered statistically significant. All experiments were conducted with a minimum of three independent replicates.
Results
General features of the LP0308
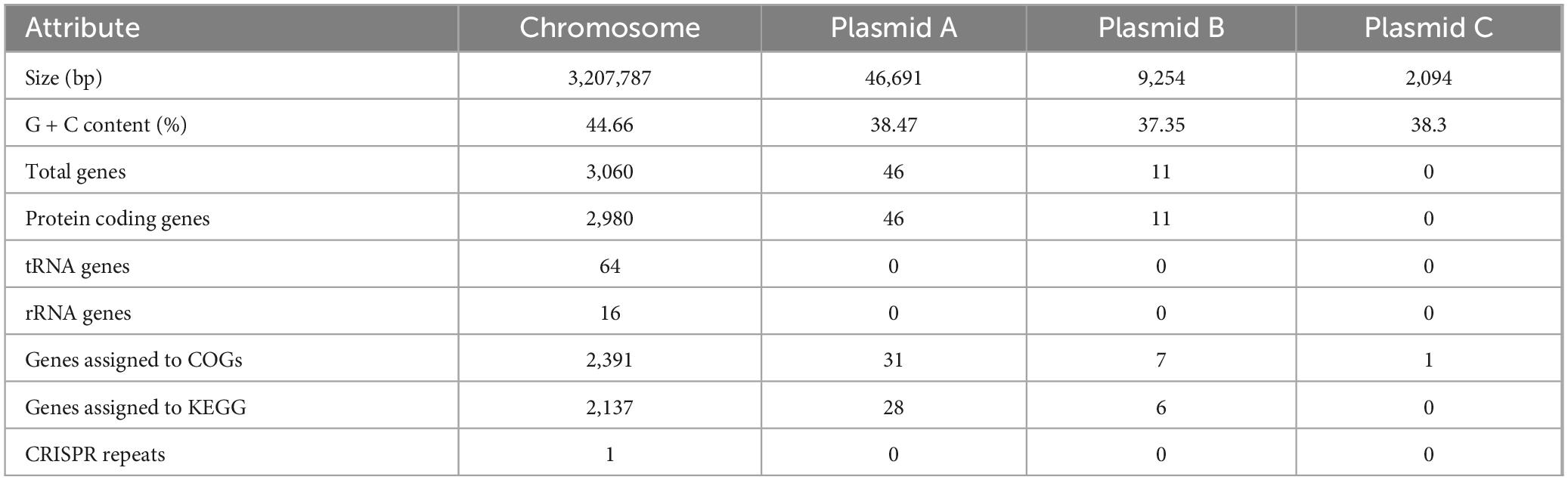
A phylogenetic tree was constructed using whole-genome sequences to elucidate relationships among LAB strains. It showed that strain LP0308 was closely related to L. plantarum and was therefore designated LP0308 (Supplementary Figure 1). Electron microscopy revealed the bacterium was rod shaped and 1.5–2 μm in length, with a diameter of 0.6–1 μm (Figure 1A). FISH with a Cy3-labeled LP0308-specific probe (red) showed large amounts of L. plantarum colonizing to the rhizosphere soil (blue), compared to the rhizosphere soil of normal group (Figures 1B, C). The growth curve exhibits a typical S-shaped pattern. The strain remains in the lag phase for the first 10 h post-inoculation, enters the exponential phase from 10 to 20 h, and reaches the late logarithmic phase at 25 h (Figure 1D).
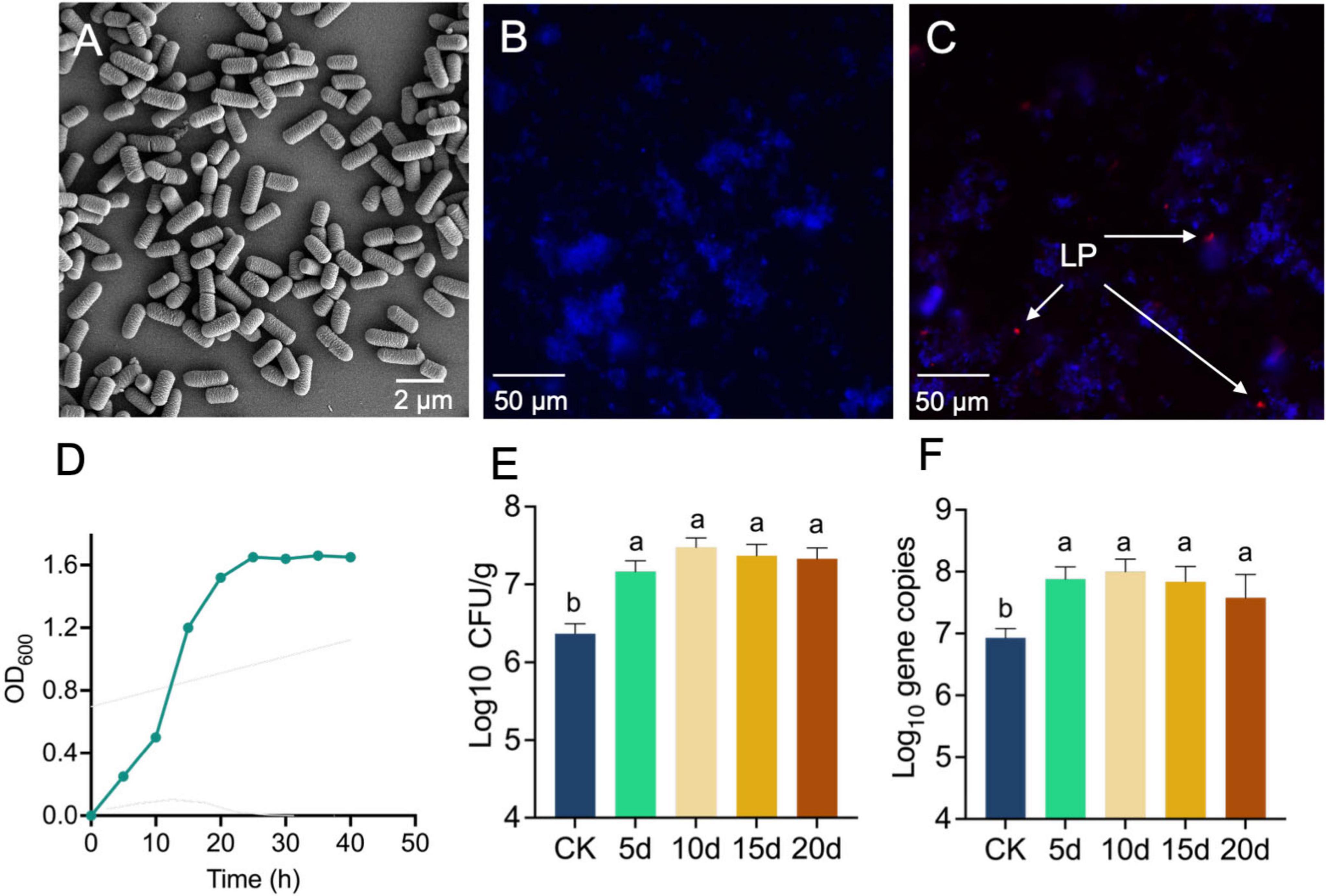
Figure 1. Characteristics and colonization of LP0308 in the tomato rhizosphere soil. (A) Scanning electron microscopy (SEM) images of LP0308. Scale bars are 2 μm. (B) Compared to the rhizosphere soil of normal group, FISH with a Cy3–labeled Lactobacillus plantarum-specific probe (red) showed large amounts of L. plantarum colonizing to the rhizosphere soil (blue) (C). (D) The growth curve of LP0308 exhibits a typical S-shaped pattern. LP0308 colonizing the rhizosphere was quantified by CFU (E) and RT-qPCR (F). Statistically significant changes were determined by ANOVA and the post hoc Tukey honestly significant difference (HSD) test. Different letters represent statistically significant differences (P < 0.05, n = 5).
The LP0308 strain colonizing the rhizosphere was quantified by CFU and RT-qPCR (Figures 1E, F). Over time, there was a statistically significant increase in bacterial levels in the rhizosphere soil. The percentage of LP0308 bacterial increased from 2.32 × 106 CFU/g to 3.0 × 107 CFU/g during this period (0–10 days) (t = 6.302, df = 8, P = 0.0002). The density of strain LP0308 colonizing rhizosphere soil also reached 2.147 × 107 CFU/g soil in 20 days, showing the long-term stability of LP0308 (Figure 1F). Independent experimental replicates, performed using RT-qPCR with universal 16S rRNA gene primers, yielded consistent results across different time points (Figure 1F).
Effect of LP0308 on rhizosphere soil microorganisms
We observed 885 unique bacterial ASV (CK) and 1,678 unique ASV (LP) on collected samples, respectively. Alpha diversity (Shannon index, Faith’s PD) indices indicated a gradual increase of microbial diversity from the CK to the LP group (Figures 2A, B). We found that the bacterial communities were divided into two main clusters along with the first PCoA axis according to the compartment, whereas the major factor explaining communities was the inoculation of LP0308. The two groups were significantly different (P = 0.001, PERMANOVA), and were treated differently as the particular spatial scales (Figures 2B, C and Supplementary Figures 2A, B). Lefse analysis was performed to explain the differences between rhizosphere soil microorganism composition between the two groups. Based on LDA effect size, Lactobacillus, Bacillales, Bacillus, and Sporosarcina were significantly enriched in LP0308 group, while Aliifodinibius, Halobacteriaceae, Halococcus, Balneolaceae, and Haladaptatus were mostly related to the uninoculated plants individuals (Figure 2D). The total number of Lactobacillus and Bacillus were estimated using DNA copy numbers measured by RT-qPCR (Figure 2E). Moreover, there was a statistically significant positive correlation between the total Lactobacillus number and the Bacillus number in the rhizosphere soil (R2 = 0.91) (Figure 2E).
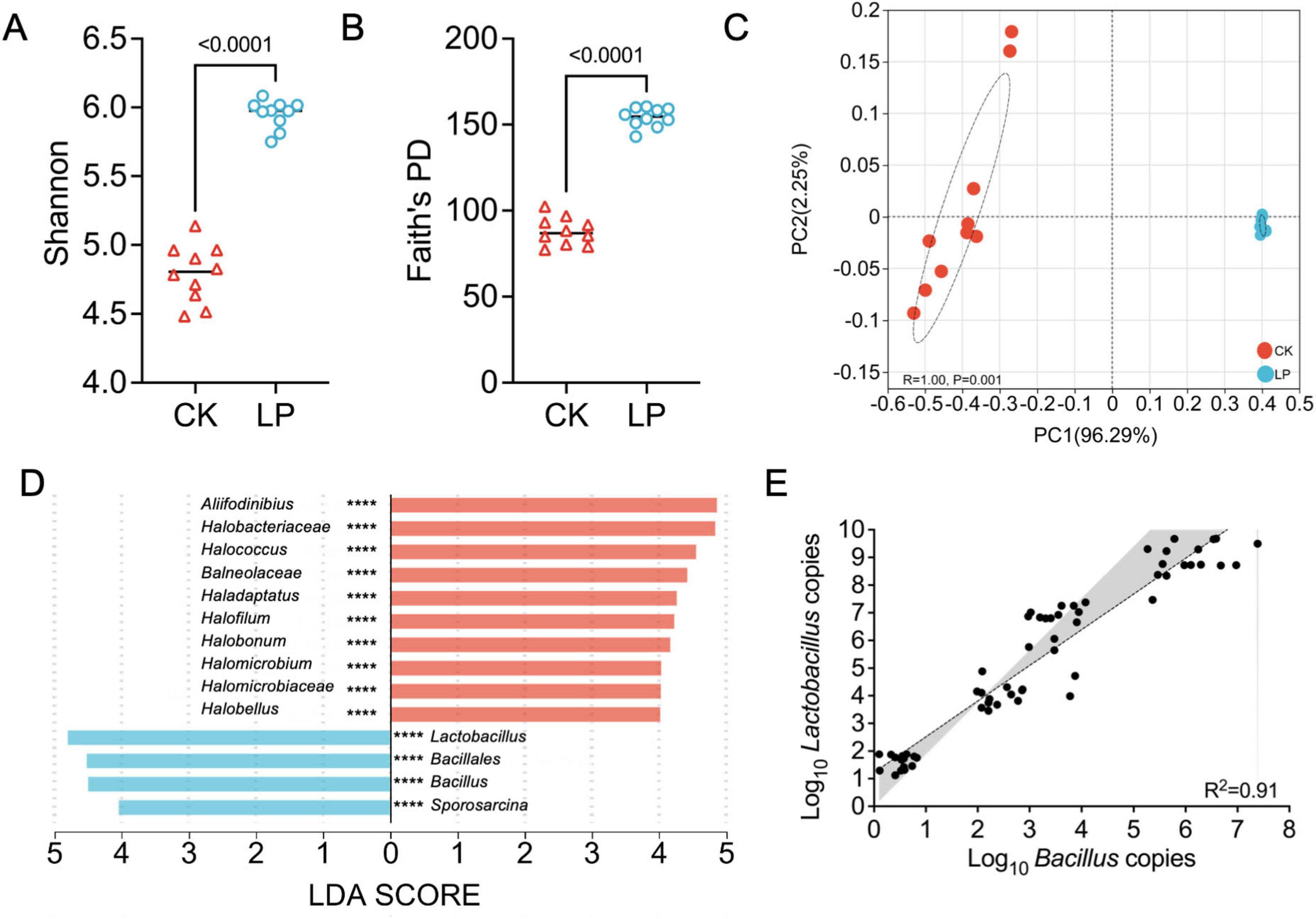
Figure 2. Colonization of LP0308 changed the composition of rhizosphere soil bacteria. (A,B) Alpha diversity and (C) beta diversity were investigated. Alpha diversity (Shannon and Faith’s PD) of bacterial communities (based on ASVs) at CK and LP0308 group (10 days). PCoA plot showing variation in community structure among CK and LP0308 group [permutational multivariate analysis of variance (PERMANOVA) test with 999 permutations, P > 0.05] based on Bray–Curtis distance. Each point represents an individual sample. (D) LDA effect size (Lefse) analysis was conducted to investigate the variations in rhizosphere soil microbial composition between the two groups (CK and LP0308). (E) Correlation between Lactobacillus plantarum and Bacillus at rhizosphere soil after inoculation with LP0308. Dotted lines denote the linear regressions. Statistically significant differences were calculated by using unpaired Student’s t-test (P < 0.05). **** indicates P < 0.0001.
Assembly processes and coexistence in the rhizosphere soil microorganisms
We then conducted neutral and null model analyses to quantitatively decipher the relative contribution of ecological processes including homogeneous selection (HoS), heterogeneous selection (HeS), dispersal limitation (DL), homogenizing dispersal (HD), and drift in driving the assembly of rhizosphere soil-associated bacterial communities (Figure 3A). Null model analysis showed that the assembly of normal group (CK) was dominantly shaped by HoS (38%), followed by HD (52%), drift and non-dominant processes (10%). However, the rare subcommunity assembly was shaped almost equally by deterministic (10% of HoS and 2% of HeS) and stochastic (68% of HD, 20% of drift and non-dominant) processes (Figure 3A). The two sample treatments (CK vs. LP) exhibit different βNTI patterns. βNTI values were significantly higher for bacterial communities of LP than LP0308 treatments (P < 0.05) (Supplementary Figure 2C). Finally, the neutral community model (NCM) successfully estimated a large fraction of the relationship between the occurrence frequency of ASV and their relative abundance variations, with 88.15% and 90.65% of explained community variance for CK and LP group, respectively (Figure 3B). The explained variation of NCM remained fairly consistent across taxonomic ranks from phylum to genus, suggesting that taxa within the same phylogenetic lineage generally respond similarly to stochastic processes. The m value was estimated to be 0.18 in LP group and 0.13 in CK, respectively. These results indicated that species dispersal of rhizosphere soil microorganism was higher after colonization of LP0308 (Figure 3B).
Figure 3. Community assembly and microbial network in rhizosphere soil bacteria mediated by Lactobacillus plantarum 0308. (A) Community-level drivers of rhizosphere soil-associated microbial communities assembly over study regions. The four components were described with to classify community pairs into underlying drivers “Selection” implying environmental filtering; “Homogenizing Dispersal” representing strong dispersal; “Dispersal limitation” suggesting weak dispersal; and “Drift” indicating stochasticity (“undominated”). (B) Fit of the neutral community model (NCM) of community assembly. The predicted occurrence frequencies for CK and LP0308 group representing bacteria communities, respectively. The dark purple dotted lines indicate the best fit to the NCM and the light purple dotted lines represent 95% confidence intervals around the model prediction. ASVs that occur more or less frequently than predicted by the NCM are shown in different colors. Nm indicates the metacommunity size times immigration, R2 indicates the fit to this model. (C) The correlation-based network of rhizosphere soil-associated microbial genus was detected in two different group (CK and LP0308). Each node corresponds to a genus, and edges between nodes correspond to either positive (green) or negative (red) correlations inferred from OUT abundance profiles using the random matrix theory-based methods with a correlation threshold of 0.65 in MENA. The size of each node is proportional to the abundance of the genus. Lactobacillus and Bacillus had the highest abundance in the LP0308 group, respectively. (D) Unique node-level topological features of different taxa categories, including clustering, average degree, degree, and the closeness centrality.
Subsequently, the constructed phylogenetic molecular ecological networks (pMENs) consisted of 414 edges and 358 edges from genera in the top 50 abundance in 60 samples. Interestingly, we observed that, in addition to the successful colonization of Lactobacillus, the abundance of Bacillus increased significantly compared to the control group (Figure 3C). The number of clustering (0.57 vs. 0.50), average degree (8.63 vs. 7.16), degree (414 vs. 358), and the closeness centrality (0.38 vs. 0.36) of the rhizosphere soil microorganism network (CK) were also considerably higher than for the treatment group network (Figure 3D). These finding, together with the network global properties, indicated that the CK group exhibited much closer interconnections than the treatment group at the phylum level.
In addition, functional profiles of rhizosphere soil bacterial community were predicted using FAPROTAX analyses, and significant differences among all groups were determined (Figure 4A). The heatmap result showed that the dominant functional groups were chemoheterotrophy (18.96% ± 2.82% vs. 40.90% ± 1.45%, P < 0.001), fermentation (1.09% ± 0.69% vs. 29.31% ± 2.54%, P < 0.001), aerobic chemoheterotrophy (15.42% ± 1.96% vs. 10.75% ± 1.42%, P < 0.001), phototrophy (11.52% ± 1.52% vs. 0.53% ± 0.09%, P < 0.001), and photoautotrophy (11.43% ± 1.52% vs. 0.53% ± 0.08%, P < 0.001) between the CK and LP0308 treatment groups (Figure 4A).

Figure 4. The functional content of the rhizosphere soil-associated microbial communities. (A) The heatmap result showed that the dominant functional groups by using FAPROTAX based on the baseline and endpoint 16S amplicon sequencing data between the CK and LP0308 treatment groups. (B) The phenotypes of rhizosphere soil bacteria in each group (CK and LP0308) were predicted using the BugBase database. Statistically significant differences were calculated by using unpaired Student’s t-test (P < 0.05).
Phenotypes of the rhizosphere soil bacterial in each group were predicted based on the BugBase database. Interestingly, the sum of anaerobic (6.12% ± 0.83% vs. 47.49% ± 5.54%), mobile elements (87.81% ± 1.51% vs. 99.30% ± 0.20%), potentially pathogenic (69.50% ± 2.80% vs. 93.81% ± 0.87%), and stress tolerant (2.94% ± 0.57% vs. 15.05% ± 2.07%) were particularly lower in the LP0308-statins group than the control group (P < 0.001) (Figure 4B). However, a higher proportion of aerobic (48.30% ± 3.12% vs. 10.88% ± 0.98%) and a stronger potential to synthesize biofilms (57.73% ± 3.05% vs. 23.24% ± 2.02%) in the LP were observed when compared with the CK (P < 0.001) (Figure 4B).
Then, five representative soil-borne tomato pathogens were used to assess antagonistic effect of LP0308. R. solanacearum and F. oxysporum was inhibited by the LP0308 cell-free supernatant in the agar well diffusion assay; no growth suppression was detected for C. fulvum, B. cinerea, or A. solani (Supplementary Figure 3).
LP0308 genomic features and rhizosphere colonization genes
LP0308 possesses a chromosome and three plasmids (Figure 5). The chromosome genome spans 3,207,787 base pairs and harbors 3,060 total genes. Of these, 2,391 and 2,137 genes are annotated in the COG and KEGG databases, respectively. The complete genome contains 2,980 protein-encoding sequences, 64 tRNA genes, 16 rRNA genes, and a CRISPR region. In addition, three circular plasmids, respectively, with the size of 245,534 and 89,508 base pairs were found (Table 1).
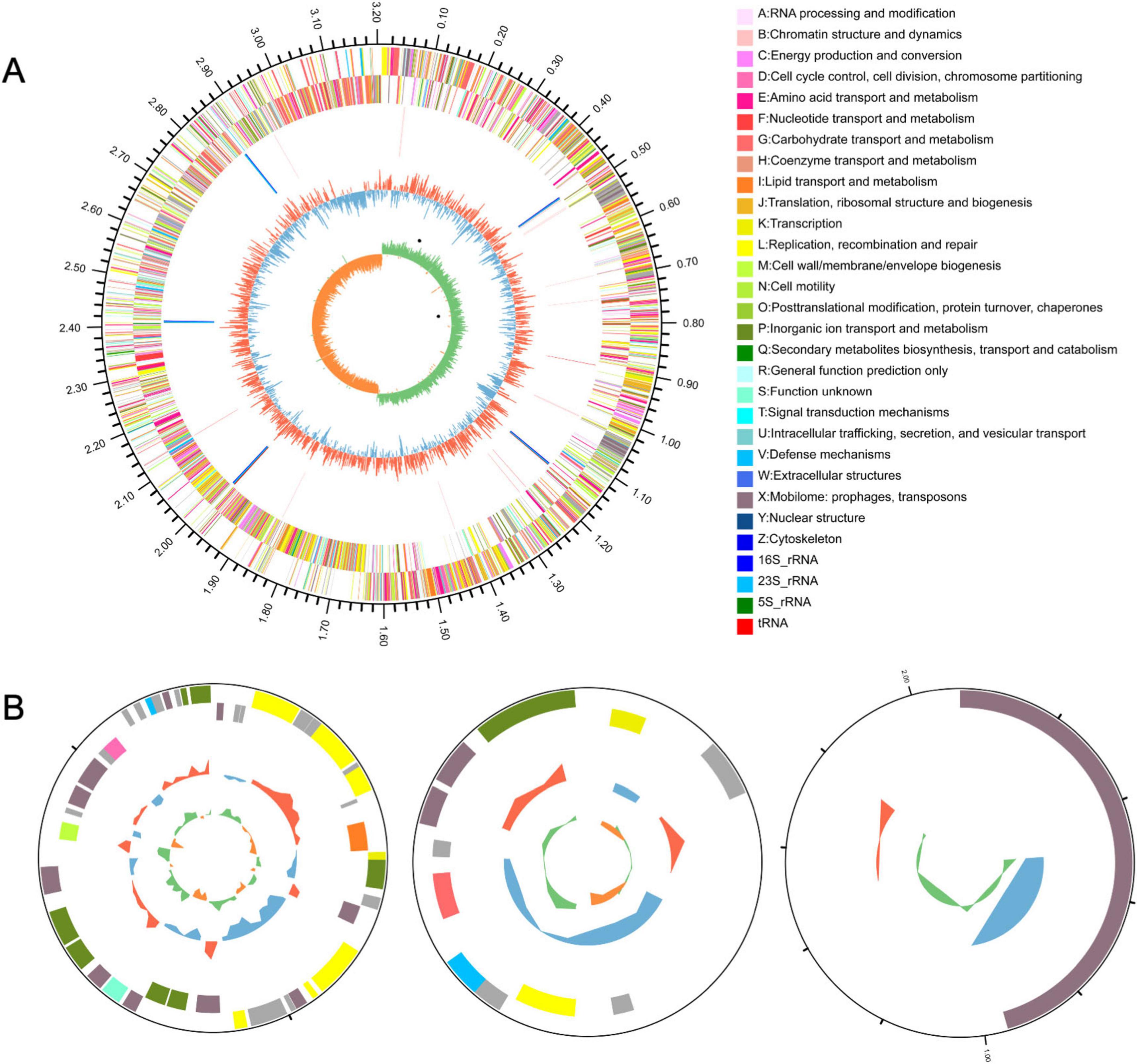
Figure 5. LP0308 strain genome resources and characteristics. Circular representation of the chromosome (A) and plasmid (B) of Lactobacillus plantarum strain LP0308. The outermost circle represents the total genome size. The second and third circles display sequences encoding protein amino acids on the + and– strands, with different colors indicating distinct COG functional classifications. The fourth circle denotes rRNA and tRNA. The fifth circle illustrates GC content, where outward red segments indicate regions with GC content higher than the genome-wide average, while inward blue segments indicate regions with lower GC content; the peak height reflects the magnitude of deviation from the average GC content. The sixth circle represents the GC skew value (G – C/G + C), where a positive value suggests a higher likelihood of CDS transcription from the positive strand, whereas a negative value indicates transcription from the negative strand.
Quantitative real-time PCR was utilized to determine how the response of LP0308 gene expression after colonization in soil on tomato plant. The expression of genes encoding biofilm (vpsI1, vpsI2, vpsC, and vpsI3), immune modulation (pbpG, kdtB, and wbpL), and antimicrobial activity (farB) proteins was measured based on their colonization-related functions (Figure 6). After LP0308 successfully colonized the rhizosphere soil, three biofilm-related, three immune modulation-related, and one antimicrobial activity-related showed substantially increased expression (Figure 6). In particular, compared with the level observed at 0 day, the expression of pbpG (fivefold), vpsI3 (eightfold), and farB (sevenfold) reached extremely significant levels at 15 days (t = 14.08, df = 8, P < 0.0001), 10 days (t = 29.66, df = 8, P < 0.0001), and 10 days (t = 19.41, df = 8, P < 0.0001), respectively (Figure 6).
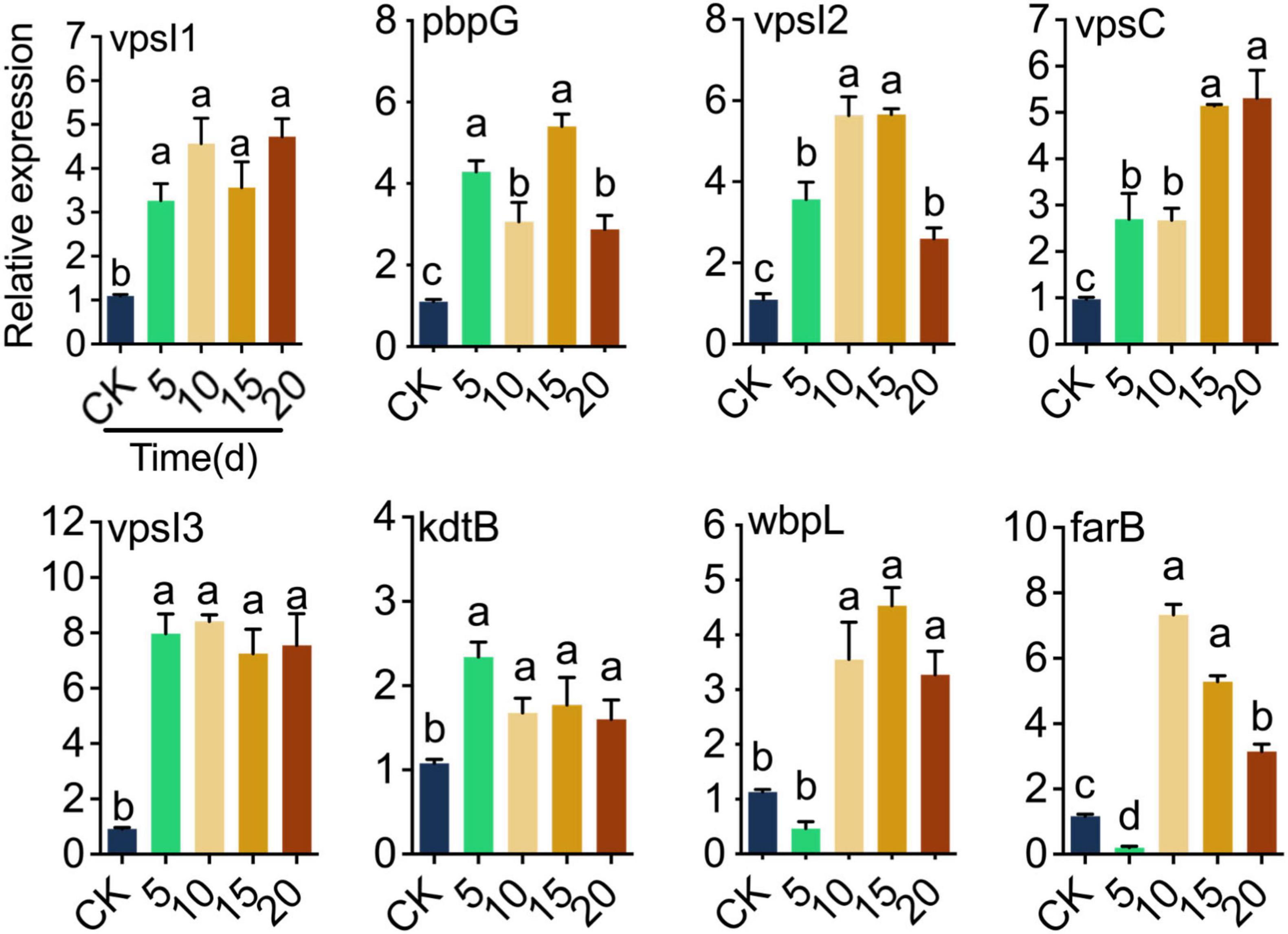
Figure 6. Genes specifically contributing to tomato rhizosphere soil microorganism colonization. Colonization-related gene expression at different times (0, 5, 10, 15, and 20 days) after inoculation with LP0308 (107 CFU/ml) by RT-qPCR. The mean ± standard error of the mean (SEM) of experiments performed in quintuplicate, with five tomato plants/experiment, are shown. Variation analysis was performed by one-way ANOVA followed by Tukey’s post-hoc test. Different letters represent significant differences at P < 0.05. Encoding biofilm (vpsI1, vpsI2, vpsC, and vpsI3), immune modulation (pbpG, kdtB, and wbpL) and antimicrobial activity (farB) proteins.
Growth-promoting effects of LP0308 on tomato buds and seedlings
Tomato seed treatment with LP0308 increased the seed germination and seedling growth compared to the control treatments (Figures 7A, B). Moreover, LP0308 significantly increased bud length (Figure 7C), plant height (Figure 7D), primary root length (Figure 7E), root fresh weight (Figure 7F), and whole-seedling fresh weight (Figure 7G) (P < 0.05; n = 15). For example, primary root length was 4.58 ± 0.41 cm in LP0308-treated seedlings vs. 3.07 ± 0.45 cm in the control (CK) group (t = 6.866, df = 28, P < 0.0001). In addition, root indole-3-acetic acid (IAA) concentration rose from 14.35 ± 1.90 to 22.67 ± 2.44 ng/g, representing a 1.58-fold increase (t = 6.029, df = 28, P = 0.0003) (Figure 7H).

Figure 7. Effects of LP0308 on the growth and development of tomato. The tomato seedlings were treated with either the control or different concentrations of LP0308 (1 × 107 CFU/ml) and sampled after 20 days. The promoting effect of LP0308 on the germination of tomato seeds (A) and seedlings (B). The following parameters were measured: (C) bud length, (D) plant height and (E) root length, (F) root fresh weight, and (G) seedling fresh weight. (H) The contents of indoleacetic acid (IAA) of tomato root. Data were analyzed from 15 seedlings for each treatment. The error bars indicate the ±SDs of the means. Statistical significance was determined by two-sided Student’s t-test.
Effect of LP0308 on soil properties and soil enzyme activities
LP0308 treatment led to a pronounced enrichment of soil OM (37.52 ± 3.91 vs. 25.72 ± 3.82) and AK (407.33 ± 14.47 vs. 332.33 ± 8.02), AP (343.66 ± 40.41 vs. 259.67 ± 14.15), and AN (380.22 ± 4.55 vs. 275.30 ± 4.55) compared with the control group (P < 0.05). However, soil pH (6.80 ± 0.25 vs. 6.77 ± 0.20) and soil temperature (19.69 ± 0.58 vs. 18.33 ± 1.53) were statistically indistinguishable from the control (P > 0.05) (Figures 8A–F). Concurrently, enzymatic profiling revealed significant upregulation of POX (1.48 ± 0.13 vs. 0.95 ± 0.06), urease (8.48 ± 1.38 vs. 5.20 ± 0.42), GLS (0.25 ± 0.04 vs. 0.16 ± 0.01), POD (0.13 ± 0.01 vs. 0.09 ± 0.01), CE (1.20 ± 0.17 vs. 0.97 ± 0.19), and NAD (2.01 ± 0.29 vs. 1.39 ± 0.07) activities under LP0308 inoculation (P < 0.05) compared with the control group (P < 0.05), whereas ALP (0.31 ± 0.08 vs. 0.33 ± 0.08) and BG (0.25 ± 0.04 vs. 0.23 ± 0.04) did not differ statistically from the control (P = 0.76 and 0.25, respectively) (Figures 9A–H).
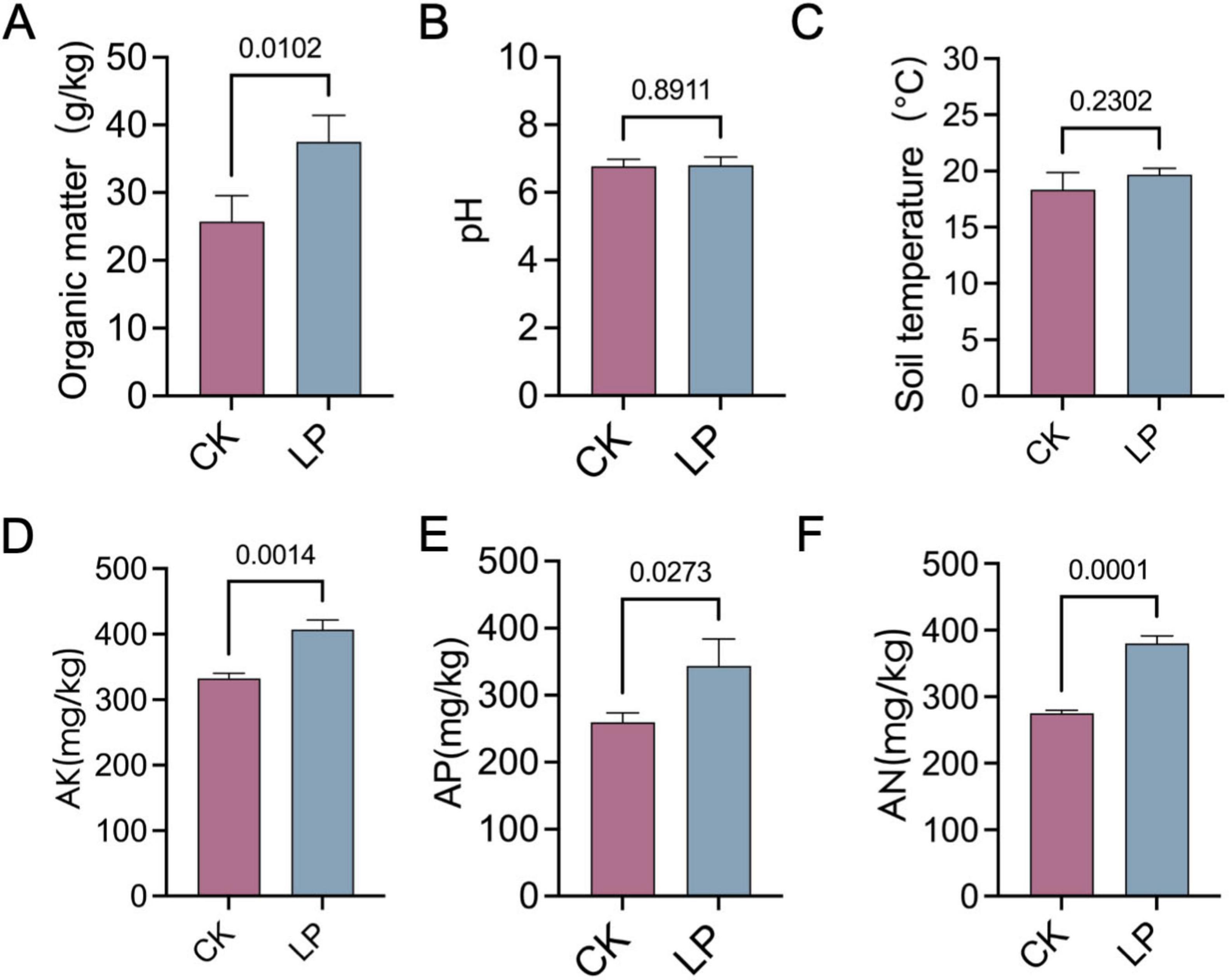
Figure 8. Effects of LP0308 on rhizosphere soil physicochemical properties. Soil from tomato rhizospheres was analyzed after treatment with LP0308 (LP) or sterile control (CK). (A) Organic matter, (B) soil pH, (C) soil temperature, (D) available potassium (AK), (E) available phosphorus (AP), and (F) available nitrogen (AN). Bars represent means ± SD (n = 15). P values derive from two-tailed Student’s t-tests comparing LP with CK for each parameter.

Figure 9. Effects of LP0308 inoculation on rhizosphere soil enzyme activities. Activities of eight functionally diverse enzymes were quantified in tomato rhizosphere soils following LP0308 (LP) or control (CK) treatment: polyphenol oxidase (POX, A), urease (B), β-glucosidase (GLS, C), peroxidase (POD, D), alkaline phosphatase (ALP, E), cellulase (CEU, F), N-acetyl-β-D-glucosaminidase (NAG, G), and β-glucosidase (BG, H). Points represent individual soil samples; bars (or horizontal lines) denote means ± SD (n = 10). P values are from two-tailed Student’s t-tests comparing LP with CK for each enzyme.
Discussion
The successful colonization of the in soil on tomato plant by probiotics is an essential step in controlling beneficial microbe ecosystems and plant health or soil fertility. In this study, LP0308 was introduced into the soil of tomato, and it could stably persist and proliferate. LP0308 levels in the rhizosphere soil significantly increased and demonstrated long-term stability during this period (0–25 days). This colonization efficiency was considerably stronger than the effect reported by Qian et al. (2022), who found that the density of strain Hansschlegelia zhihuaiae S113 colonizing rhizosphere soil reached maximum level (4.7 × 108 cells/g) on the 5th day, but the colonization effect of strain S113 gradually weakened after the 10th day. The high survival rate of LP0308 in soil means the L. plantarum might be used as probiotics. L. plantarum strain LP0308 was confirmed as a possible plant PGPR, which significantly promoted bud length, plant height, primary root length, root fresh weight, and whole-seedling fresh weight. According to Amprayna et al. (2016), LAB exhibit PGP characteristics, as they can produce the coenzyme IAA and solubilize minerals. LAB colonizing the plant rhizosphere can enhance the plant’s biological traits and boost yield. For instance, rice seeds coated with Lactococcus lactis significantly increased root and shoot lengths (Amprayna et al., 2016). Moreover, L. lactis notably promoted both growth and yield in cabbage (Somers et al., 2007). However, soil sterilization inherently alters physicochemical gradients and excludes biotic interactions, which may overestimate strain persistence; therefore, future work will assess LP0308’s colonization and resilience in natural, non-sterile soils to confirm its agronomic relevance.
In addition, we found that LP0308 altered the microbial composition of the rhizosphere soil and significantly increased the abundance of Bacillus. LAB are widespread microorganisms that hold promising benefits for enhancing crop and livestock production (Ikeda et al., 2013). Together with LAB, other Firmicutes (e.g., Bacillus species) are ubiquitous, often found as soil inhabitants and easily transferred from soil to roots (Arkhipova and Anokhina, 2009). Studies have found that some strains of Bacillus can inhibit the pathogenic soil-borne fungus Fusarium graminearum and produce cytokinins, acting as PGP probiotics (Arkhipova and Anokhina, 2009; Zhao et al., 2014). Wang et al. (2019) found that Bacillus stearothermophilus increases the relative abundance of LAB strains in the soil and exhibits antagonistic effects against plant pathogens in the soil. Similar studies have found that Pediococcus pentosaceus positively correlated with L. plantarum (Jiang et al., 2021). Recently, LAB and other Bacillus-based biofertilizers have been successfully integrated with established microorganisms in environmental and agriculture applications. These microbial biofertilizers are known to boost crop yields, facilitate the replenishment of essential minerals in plant roots, and improve the metabolic processes involved in OM decomposition (Khoso et al., 2024).
Interestingly, LP0308 significantly reduced potentially pathogenic and promoted biofilm formation of rhizosphere soil microorganism. We found that the cell-free supernatant of LP0308 significantly inhibited R. solanacearum and F. oxysporum. PGPR are known to produce secondary metabolites antagonistic to various soil-borne pathogens (Uwaremwe et al., 2021). These results support previous research that shows that L. plantarum significantly reduces the virulence factors, biosurfactants formation of pathogenic bacteria such as Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli (Anupama et al., 2009a; Ahn et al., 2018). Research by Anupama et al. (2009b) revealed that L. plantarum exhibits antagonistic activity against plant pathogenic bacteria Xanthomonas campestris and R. solanacearum (Visser et al., 1984). Further studies have shown that LAB eradicate the formation of pathogenic microorganism surface adhesion by producing glycolipid biosurfactants (Manjarres Melo et al., 2021; Guan et al., 2023). L. plantarum isolated from traditional sourdoughs has shown significant antagonism activity against Erwinia amylovora, Penicillium digitatum, and B. cinerea in vitro (Tremonte et al., 2017; Sadanov et al., 2023). In addition, Lactobacillus buchneri 6M1 and Lactobacillus brevis 5M2 isolated from corn silages showed antifungal activity against F. graminearum and carboxylesterase activities (Paradhipta et al., 2021). The anti-pathogenic microorganism activity of LAB is typically expressed through competition for essential nutrients and the production of bioactive substances such as organic acids, carbon dioxide, ethanol, amines, hydrogen peroxide, fatty acids, acetaldehyde, diacetyl, cyclic dipeptides, vitamin, exopolysaccharides bacteriocins, or bacteriocin-like inhibitory substances (Rouse et al., 2008; Arena et al., 2019). A key mechanism for probiotic colonization is the formation of biofilms, enabling survival under stress conditions in the host, while facilitating nutrient exchange and coordinating host activities (Rasooly et al., 2022). In this study, LP0308 may enhance its competitive advantage in the rhizosphere microbiome through biofilm formation. However, reliance on in vitro assays and functional predictions means that the antagonistic effects and biofilm-mediated competitive advantage remain unvalidated under the rhizosphere’s complex biotic and abiotic conditions.
The successful colonization of LP0308 led to drastically increased expression of encoding biofilm-associated gene (vpsI1, vpsI2, vpsC, and vpsI3), immune modulation (pbpG, kdtB, and wbpL) and antimicrobial activity (farB) in the rhizosphere soil on a genome-wide scale. For example, Vibrio cholerae enhances the production of exopolysaccharide (EPS) by regulating the transcription of VPS genes on its chromosome, thereby enabling the formation of well-developed, mature biofilms that improve its resistance to environmental stress (Casper-Lindley and Yildiz, 2004; Butz et al., 2021). Penicillin-binding proteins (PBPs), which are serine-based enzymes, act as redox sensors to regulate cell wall synthesis, as well as nutrient-driven cell growth, division, and proliferation, and are essential for plant growth (Sacco et al., 2022). Another study showed that the farB gene is involved in maintaining cell membrane integrity, regulating intracellular fatty acid levels, and contributing to bacterial resistance, particularly against antimicrobial long-chain fatty acids (Yin et al., 2019; Khoder et al., 2022). Genes involved in core enterococcal and genome plasticity are key drivers of rhizosphere soil colonization.
Additionally, application of LP0308 markedly improved soil nutrient availability and stimulated key enzymatic activities, indicating its potential to enhance rhizosphere health and fertility. In this study, OM content increased from 25.72 g/kg in the control to 37.52 g/kg following LP0308 treatment. Higher OM could facilitate the growth and physiological activity of plant roots, as well as the metabolism of microorganisms, thus promoting enzymatic secretion by plants and microbes (Fanin et al., 2014). Lee et al. (2022) found that soil inoculation with Rhodopseudomonas palustris PS3 enhanced both soil nutrient availability and tomato endogenous nitrogen content, raising soil nutrient availability from 35% to 56% in an organic field. In addition to nutrients, we also found that the activities of some soil enzymes, such as POX, urease and ALP, were significantly increased. Since soil enzyme activity is regarded as an indicator of microbial activity (Xiao et al., 2020). We hypothesize that LP0308 inoculation can stimulate microbial activity in the soil. Research found that Sinorhizobium meliloti CCNWSX0020 application improved soil microbial parameters, such as soil enzyme activity (i.e., β-glucosidase activity and ALP) and microbial biomass, thereby promoting alfalfa growth (Duan et al., 2021). For example, phosphatases play a critical role in catalyzing the hydrolysis of organic phosphorus compounds and are essential for enhancing overall soil quality (Yang et al., 2017; Wang et al., 2020). Therefore, we presume that LP0308 not only proliferates within the soil microbiome but also activates native microbial functional guilds, thereby accelerating nutrient turnover and availability.
Conclusion
The conducted study allowed a holistic assessment of the LP0308 colonization strategy and function in the tomato rhizosphere soil. Through a deeper understanding and utilization of L. plantarum, we can better address challenges in agriculture and environmental sectors, paving the way for a more sustainable future. These microorganisms offer a means of working synergistically with nature, contributing to soil improvement, plant protection, and the enhancement of environmental quality. The findings in our study provide evidence of the fundamental basis of rhizosphere colonization and signify an important step toward the development of robust bioinoculants in sustainable agriculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
XZ: Writing – original draft, Conceptualization, Resources, Writing – review & editing, Funding acquisition, Data curation. HL: Software, Writing – review & editing, Investigation. TC: Conceptualization, Writing – review & editing, Supervision. PC: Formal analysis, Methodology, Writing – review & editing. XWu: Writing – review & editing, Validation. ZW: Writing – review & editing, Validation. HM: Validation, Writing – review & editing. GQ: Writing – review & editing, Project administration. MZ: Writing – review & editing, Formal analysis. XL: Writing – review & editing, Writing – original draft, Project administration. XWa: Visualization, Writing – review & editing. CW: Writing – review & editing, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Zhejiang Provincial Natural Science Foundation of China under Grant No. LQN25C170001, Huzhou Science and Technology Bureau (2024YZ07), Scientific Research Fund of Zhejiang Provincial Education Department (Y20235377), Environmental Risk Factor Distribution Characteristics and Safety Assessment of the Crested Ibis Habitat in Deqing, Zhejiang (ZJXRDQ-2024-JC61), and Microbial Resource Survey and Application Research Project of Longwangshan Natural Reserve Project (TCCS2023-014).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1652881/full#supplementary-material
References
Ahn, K. B., Baik, J. E., Park, O. J., Yun, C. H., and Han, S. H. (2018). Lactobacillus plantarum lipoteichoic acid inhibits biofilm formation of Streptococcus mutans. PLoS One 13:e0192694. doi: 10.1371/journal.pone.0192694
Amprayna, K., Supawonga, V., Kengkwasingha, P., and Getmalab, A. (2016). “Plant growth promoting traits of lactic acid bacterium isolated from rice rhizosphere and its effect on rice growth,” in Proceedings of the 5th Burapha University International Conference STP-029-10, (Pattaya), 181–186.
Anupama, S. K., Chun Keun Lim, S. C., Hyun Hur, J., and Cho, S. (2009b). Antagonistic Effect of Lactobacillus sp. Strain KLF01 Against Plant Pathogenic Bacteria Ralstonia solanacearum. Korean J. Pesticide Sci. 13, 45–33.
Anupama, S. K., Eun-Chang, K., Chuen Keun L, Sae-Youil, C., Hyun Hur, J., and Park, D. H. (2009a). Biological control of soft rot on chinese cabbage using beneficial bacterial agents in greenhouse and field. Korean J. Pesticide Sci. 13, 325–331.
Arena, M. P., Russo, P., Spano, G., and Capozzi, V. (2019). Exploration of the microbial biodiversity associated with north apulian sourdoughs and the effect of the increasing number of inoculated lactic acid bacteria strains on the biocontrol against fungal spoilage. Fermentation 5:97. doi: 10.3390/fermentation5040097
Arkhipova, T. N., and Anokhina, N. L. (2009). Effects of wheat plant inoculation with cytokinin-producing microorganisms on plant growth at increasing level of mineral nutrition. Russian J. Plant Physiol. 56, 814–819. doi: 10.1134/s1021443709060119
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Ayivi, R. D., Gyawali, R., Krastanov, A., Aljaloud, S. O., Worku, M., Tahergorabi, R., et al. (2020). Lactic acid bacteria: Food safety and human health applications. Dairy 1, 202–232. doi: 10.3390/dairy1030015
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: An open source software for exploring and manipulating networks,” in Proceedings of the 3rd international AAAI conference on weblogs and social media, ICWSM 2009. Washington, DC: AAAI Press, 361–362. doi: 10.1609/icwsm.v3i1.13937
Bodenhausen, N., Bortfeld-Miller, M., Ackermann, M., and Vorholt, J. A. (2014). A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 10:e1004283. doi: 10.1371/journal.pgen.1004283
Borrelli, P., Robinson, D. A., Panagos, P., Lugato, E., Yang, J. E., Alewell, C., et al. (2020). Land use and climate change impacts on global soil erosion by water (2015-2070). Proc. Natl. Acad. Sci. U S A. 117, 21994–22001. doi: 10.1073/pnas.2001403117
Britt, D. W. (2021). Plug-and-play bioinspired seed coatings. Nat. Food 2, 456–457. doi: 10.1038/s43016-021-00325-6
Butz, H. A., Mey, A. R., Ciosek, A. L., Crofts, A. A., Davies, B. W., and Payne, S. M. (2021). Regulatory effects of CsrA in Vibrio cholerae. mBio 12:e03380-20. doi: 10.1128/mBio.03380-20
Cacace, C., Rizzello, C. G., Brunetti, G., Verni, M., and Cocozza, C. (2022). Reuse of wasted bread as soil amendment: Bioprocessing, effects on alkaline soil and escarole (Cichorium endivia) production. Foods 11:189. doi: 10.3390/foods11020189
Carvajal-Yepes, M., Cardwell, K., Nelson, A., Garrett, K. A., Giovani, B., Saunders, D. G. O., et al. (2019). A global surveillance system for crop diseases. Science 364, 1237–1239. doi: 10.1126/science.aaw1572
Casper-Lindley, C., and Yildiz, F. H. (2004). VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186, 1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004
Chen, H., Yan, X., Du, G., Guo, Q., Shi, Y., Chang, J., et al. (2023). Recent developments in antifungal lactic acid bacteria: Application, screening methods, separation, purification of antifungal compounds and antifungal mechanisms. Crit. Rev. Food Sci. Nutr. 63, 2544–2558. doi: 10.1080/10408398.2021.1977610
Chen, Y., Zhang, W. Z., Liu, X., Ma, Z. H., Li, B., Allen, C., et al. (2010). A Real-time PCR assay for the quantitative detection of Ralstonia solanacearum in the horticultural soil and plant tissues. J. Microbiol. Biotechnol. 20, 193–201. doi: 10.4014/jmb.0906.06019
Chu, S., Zhang, D., Zhi, Y., Wang, B., Chi, C. P., Zhang, D., et al. (2018). Enhanced removal of nitrate in the maize rhizosphere by plant growth-promoting Bacillus megaterium NCT-2, and its colonization pattern in response to nitrate. Chemosphere 208, 316–324. doi: 10.1016/j.chemosphere.2018.05.189
Daranas, N., Rosello, G., Cabrefiga, J., Donati, I., Frances, J., Badosa, E., et al. (2019). Biological control of bacterial plant diseases with Lactobacillus plantarum strains selected for their broad-spectrum activity. Ann. Appl. Biol. 174, 92–105. doi: 10.1111/aab.12476
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113
Duan, C., Mei, Y., Wang, Q., Wang, Y., Li, Q., Hong, M., et al. (2021). Rhizobium inoculation enhances the resistance of alfalfa and microbial characteristics in copper-contaminated soil. Front. Microbiol. 12:781831. doi: 10.3389/fmicb.2021.781831
Ekundayo, F. O. (2014). Isolation and identification of lactic acid bacteria from rhizosphere soils of three fruit trees, fish and ogi. Int. J. Curr. Microbiol. Appl. Sci. 3, 991–998. doi: 10.1016/B978-0-12-384730-0.18001-2
Fanin, N., Hättenschwiler, S., Schimann, H., Fromin, N., and Bailey, J. K. (2014). Interactive effects of C, N and P fertilization on soil microbial community structure and function in an Amazonian rain forest. Funct. Ecol. 29, 140–150. doi: 10.1111/1365-2435.12329
Feng, Q., Luo, Y., Liang, M., Cao, Y., Wang, L., Liu, C., et al. (2025). Rhizobacteria protective hydrogel to promote plant growth and adaption to acidic soil. Nat. Commun. 16:1684. doi: 10.1038/s41467-025-56988-3
Gosiewski, T., Chmielarczyk, A., Strus, M., Brzychczy-Wloch, M., and Heczko, P. B. (2012). The application of genetics methods to differentiation of three Lactobacillus species of human origin. Ann. Microbiol. 62, 1437–1445. doi: 10.1007/s13213-011-0395-2
Guan, C., Zhang, W., Su, J., Li, F., Chen, D., Chen, X., et al. (2023). Antibacterial and antibiofilm potential of Lacticaseibacillus rhamnosus YT and its cell-surface extract. BMC Microbiol. 23:12. doi: 10.1186/s12866-022-02751-3
Hati, S., Mandal, S., and Prajapati, J. B. (2013). Novel starters for value added fermented dairy products. Curr. Res. Nutr. Food Sci. 1, 83–91. doi: 10.12944/CRNFSJ.1.1.09
Hofmann, T., Lowry, G. V., Ghoshal, S., Tufenkji, N., Brambilla, D., Dutcher, J. R., et al. (2020). Technology readiness and overcoming barriers to sustainably implement nanotechnology-enabled plant agriculture. Nat. Food. 1, 416–425. doi: 10.1038/s43016-020-0110-1
Ikeda, D. W. E., Chang, K., McGinn, J., Miller, S., Keliihoomalu, C., and DuPonte, M. (2013). Natural farming: Lactic acid bacteria. Sustain Agric. doi: 10.1159/000106089
Jansson, J. K., McClure, R., and Egbert, R. G. (2023). Soil microbiome engineering for sustainability in a changing environment. Nat. Biotechnol. 41, 1716–1728. doi: 10.1038/s41587-023-01932-3
Jiang, S., Cai, L., Lv, L., and Li, L. (2021). Pediococcus pentosaceus, a future additive or probiotic candidate. Microb. Cell. Fact. 20:45. doi: 10.1186/s12934-021-01537-y
Kah, M., Tufenkji, N., and White, J. C. (2019). Nano-enabled strategies to enhance crop nutrition and protection. Nat. Nanotechnol. 14, 532–540. doi: 10.1038/s41565-019-0439-5
Khoder, M., Osman, M., Kassem, I., Rafei, R., Shahin, A., Fournier, P. E., et al. (2022). Whole genome analyses accurately identify neisseria spp. and limit taxonomic ambiguity. Int. J. Mol. Sci. 23:13456. doi: 10.3390/ijms232113456
Khoso, M. A., Wagan, S., Alam, I., Hussain, A., Ali, Q., Saha, S., et al. (2024). Impact of plant growth-promoting rhizobacteria (PGPR) on plant nutrition and root characteristics: Current perspective. Plant Stress 11:100341. doi: 10.1016/j.stress.2023.100341
Kinoshita, H. (2019). Biosorption of heavy metals by lactic acid bacteria for detoxification. Methods Mol. Biol. 1887, 145–157. doi: 10.1007/978-1-4939-8907-2_13
Kirillova, A. V., Danilushkina, A. A., Irisov, D. S., Bruslik, N. L., Fakhrullin, R. F., Zakharov, Y. A., et al. (2017). Assessment of resistance and bioremediation ability of Lactobacillus strains to lead and cadmium. Int. J. Microbiol. 2017:9869145. doi: 10.1155/2017/9869145
Knights, H. E., Jorrin, B., Haskett, T. L., and Poole, P. S. (2021). Deciphering bacterial mechanisms of root colonization. Environ. Microbiol. Rep. 13, 428–444. doi: 10.1111/1758-2229.12934
Lamont, J. R., Wilkins, O., Bywater-Ekegärd, M., and Smith, D. L. (2017). From yogurt to yield: Potential applications of lactic acid bacteria in plant production. Soil Biol. Biochem. 111, 1–9. doi: 10.1016/j.soilbio.2017.03.015
Lee, S. K., Chiang, M. S., Hseu, Z. Y., Kuo, C. H., and Liu, C. T. (2022). A photosynthetic bacterial inoculant exerts beneficial effects on the yield and quality of tomato and affects bacterial community structure in an organic field. Front. Microbiol. 13:959080. doi: 10.3389/fmicb.2022.959080
Liang, X., Sun, C., Chen, B., Du, K., Yu, T., Luang-In, V., et al. (2018). Insect symbionts as valuable grist for the biotechnological mill: An alkaliphilic silkworm gut bacterium for efficient lactic acid production. Appl. Microbiol. Biotechnol. 102, 4951–4962. doi: 10.1007/s00253-018-8953-1
Liu, X., Le Roux, X., and Salles, J. F. (2022). The legacy of microbial inoculants in agroecosystems and potential for tackling climate change challenges. iScience 25:103821. doi: 10.1016/j.isci.2022.103821
Liu, Y., Xu, Z., Chen, L., Xun, W., Shu, X., Chen, Y., et al. (2024). Root colonization by beneficial rhizobacteria. FEMS Microbiol. Rev. 48:fuad066. doi: 10.1093/femsre/fuad066
Manjarres Melo, J. J., Alvarez, A., Ramirez, C., and Bolivar, G. (2021). Antagonistic activity of lactic acid bacteria against phytopathogenic fungi isolated from cherry tomato (Solanum lycopersicum var. cerasiforme). Curr. Microbiol. 78, 1399–1408. doi: 10.1007/s00284-021-02416-w
McElwee, P., Calvin, K., Campbell, D., Cherubini, F., Grassi, G., Korotkov, V., et al. (2020). The impact of interventions in the global land and agri-food sectors on Nature’s contributions to people and the UN sustainable development goals. Glob. Chang Biol. 26, 4691–4721. doi: 10.1111/gcb.15219
McPherson, M. R., Wang, P., Marsh, E. L., Mitchell, R. B., and Schachtman, D. P. (2018). Isolation and analysis of microbial communities in soil, rhizosphere, and roots in perennial grass experiments. J. Vis. Exp. 137:57932. doi: 10.3791/57932
Minervini, F., Celano, G., Lattanzi, A., Tedone, L., De Mastro, G., Gobbetti, M., et al. (2015). Lactic acid bacteria in durum wheat flour are endophytic components of the plant during its entire life cycle. Appl. Environ. Microbiol. 81, 6736–6748. doi: 10.1128/AEM.01852-15
Moore, J. A. M., Abraham, P. E., Michener, J. K., Muchero, W., and Cregger, M. A. (2022). Ecosystem consequences of introducing plant growth promoting rhizobacteria to managed systems and potential legacy effects. New Phytol. 234, 1914–1918. doi: 10.1111/nph.18010
Mostafidi, M., Sanjabi, M. R., Mojgani, N., Eskandari, S., Arbabi Bidgoli, S., and Faisal Manzoor, M. (2023). Heavy metal bioremediation potential of autochthonous lactic acid bacteria for use in edible leafy vegetables. J. Food Quality 2023, 1–9. doi: 10.1155/2023/8730676
Nichols, K. A., and Wright, S. F. (2006). Carbon and nitrogen in operationally defined soil organic matter pools. Biol. Fertility Soils 43, 215–220. doi: 10.1007/s00374-006-0097-2
O’Banion, B. S., Jones, P., Demetros, A. A., Kelley, B. R., Knoor, L. H., Wagner, A. S., et al. (2023). Plant myo-inositol transport influences bacterial colonization phenotypes. Curr. Biol. 33, 3111–3124.e3115. doi: 10.1016/j.cub.2023.06.057
Paradhipta, D. H. V., Joo, Y. H., Lee, H. J., Lee, S. S., Noh, H. T., Choi, J. S., et al. (2021). Effects of inoculants producing antifungal and carboxylesterase activities on corn silage and its shelf life against mold contamination at feed-out phase. Microorganism. 9:558. doi: 10.3390/microorganisms9030558
Qian, Y., Zhao, G., Zhou, J., Zhao, H., Mutter, T. Y., and Huang, X. (2022). Combined bioremediation of bensulfuron-methyl contaminated soils with arbuscular mycorrhizal fungus and Hansschlegelia zhihuaiae S113. Front. Microbiol. 13:843525. doi: 10.3389/fmicb.2022.843525
Rama, G. R., Bucker, F., Salazar, M. M., Ray, S., and Granada, C. E. (2024). “Lactic acid bacteria: Taxonomy, characteristic features, physiology, and diversity,” in Antimicrobial Peptides from Lactic Acid Bacteria, eds S. Ray, P. Kumar, and M. Mandai (Berlin: Springer Nature), 1–32.
Rasooly, R., Howard, A. C., Balaban, N., Hernlem, B., and Apostolidis, E. (2022). The effect of tannin-rich witch hazel on growth of probiotic lactobacillus plantarum. Antibiotics 11:395. doi: 10.3390/antibiotics11030395
Roosa, S., Wauven, C. V., Billon, G., Matthijs, S., Wattiez, R., and Gillan, D. C. (2014). The Pseudomonas community in metal-contaminated sediments as revealed by quantitative PCR: A link with metal bioavailability. Res. Microbiol. 165, 647–656. doi: 10.1016/j.resmic.2014.07.011
Rouse, S., Harnett, D., Vaughan, A., and van Sinderen, D. (2008). Lactic acid bacteria with potential to eliminate fungal spoilage in foods. J. Appl. Microbiol. 104, 915–923. doi: 10.1111/j.1365-2672.2007.03619.x
Sacco, M. D., Wang, S., Adapa, S. R., Zhang, X., Lewandowski, E. M., Gongora, M. V., et al. (2022). A unique class of Zn(2+)-binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile. Nat. Commun. 13:4370. doi: 10.1038/s41467-022-32086-6
Sadanov, A., Alimzhanova, M., Ismailova, E., Shemshura, O., Ashimuly, K., Molzhigitova, A., et al. (2023). Antagonistic and protective activity of Lactobacillus plantarum strain 17 M against E. amylovora. World J. Microbiol. Biotechnol. 39:314. doi: 10.1007/s11274-023-03765-3
Sadiq, F. A., Yan, B., Tian, F., Zhao, J., Zhang, H., and Chen, W. (2019). Lactic acid bacteria as antifungal and anti-mycotoxigenic agents: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 18, 1403–1436. doi: 10.1111/1541-4337.12481
Sammauria, R., Kumawat, S., Kumawat, P., Singh, J., and Jatwa, T. K. (2020). Microbial inoculants: Potential tool for sustainability of agricultural production systems. Arch. Microbiol. 202, 677–693. doi: 10.1007/s00203-019-01795-w
Sanchez-Gil, J. J., Poppeliers, S. W. M., Vacheron, J., Zhang, H., Odijk, B., Keel, C., et al. (2023). The conserved iol gene cluster in Pseudomonas is involved in rhizosphere competence. Curr. Biol. 33, 3097–3110.e3096. doi: 10.1016/j.cub.2023.05.057
Santos-Villalobos, S., Barrera-Galicia, G. C., Miranda-Salcedo, M. A., and Pena-Cabriales, J. J. (2012). Burkholderia cepacia XXVI siderophore with biocontrol capacity against Colletotrichum gloeosporioides. World J. Microbiol. Biotechnol. 28, 2615–2623. doi: 10.1007/s11274-012-1071-9
Santoyo, G., Urtis-Flores, C. A., Loeza-Lara, P. D., Orozco-Mosqueda, M. D. C., and Glick, B. R. (2021). Rhizosphere colonization determinants by plant growth-promoting rhizobacteria (PGPR). Biology 10:475. doi: 10.3390/biology10060475
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Singh, B. K., Delgado-Baquerizo, M., Egidi, E., Guirado, E., Leach, J. E., Liu, H., et al. (2023). Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 21, 640–656. doi: 10.1038/s41579-023-00900-7
Somers, E., Amke, A., Croonenborghs, A., van Overbeek, L. S., and Vanderleyden, J. (2007). “Lactic acid bacteria in organic agricultural soils,” in Poster session presented at Rhizosphere 2, Montpellier, (France), 26–31. doi: 10.1038/s41396-018-0174-1
Tang, F. H. M., Lenzen, M., McBratney, A., and Maggi, F. (2021). Risk of pesticide pollution at the global scale. Nat. Geosci. 14, 206–210. doi: 10.1038/s41561-021-00712-5
Tremonte, P., Pannella, G., Succi, M., Tipaldi, L., Sturchio, M., Coppola, R., et al. (2017). Antimicrobial activity of Lactobacillus plantarum strains isolated from different environments: A preliminary study. Int. Food Res. J. 24, 852–859. doi: 10.3389/fmicb.2016.00464
Uwaremwe, C., Yue, L., Wang, Y., Tian, Y., Zhao, X., Liu, Y., et al. (2021). An Endophytic strain of Bacillus amyloliquefaciens suppresses Fusarium oxysporum infection of chinese wolfberry by altering its rhizosphere bacterial community. Front. Microbiol. 12:782523. doi: 10.3389/fmicb.2021.782523
van Vliet, J., Eitelberg, D. A., and Verburg, P. H. (2017). A global analysis of land take in cropland areas and production displacement from urbanization. Global Environ. Change 43, 107–115. doi: 10.1016/j.gloenvcha.2017.02.001
Visser, R., Holzapfel, W. H., and Kotze, J. M. (1984). Antagonism of lactic acid bacteria against phytopathogenic bacteria. Appl. Environ. Microbiol. 52, 552–555. doi: 10.1128/aem.52.3.552-555
Wang, J., Wang, X., Liu, G., Wang, G., Wu, Y., and Zhang, C. (2020). Fencing as an effective approach for restoration of alpine meadows: Evidence from nutrient limitation of soil microbes. Geoderma 363:114148. doi: 10.1016/j.geoderma.2019.114148
Wang, N., Wang, T., Chen, Y., Wang, M., Lu, Q., Wang, K., et al. (2024). Microbiome convergence enables siderophore-secreting-rhizobacteria to improve iron nutrition and yield of peanut intercropped with maize. Nat. Commun. 15:839. doi: 10.1038/s41467-024-45207-0
Wang, S. (2009). Molecular mechanism of plant growth promotion and induced systemic resistance to tobacco mosaic virus by Bacillus spp. J. Microbiol. Biotechnol. 19, 1250–1258. doi: 10.4014/jmb.0901.008
Wang, Y., Bi, L., Liao, Y., Lu, D., Zhang, H., Liao, X., et al. (2019). Influence and characteristics of Bacillus stearothermophilus in ammonia reduction during layer manure composting. Ecotoxicol. Environ. Saf. 180, 80–87. doi: 10.1016/j.ecoenv.2019.04.066
Xiao, L., Liu, G., Li, P., Li, Q., and Xue, S. (2020). Ecoenzymatic stoichiometry and microbial nutrient limitation during secondary succession of natural grassland on the Loess Plateau, China. Soil Tillage Res. 200:104605. doi: 10.1016/j.still.2020.104605
Xie, S., Vallet, M., Sun, C., Kunert, M., David, A., Zhang, X., et al. (2019). Biocontrol potential of a novel endophytic bacterium from mulberry (Morus) tree. Front. Bioeng. Biotechnol. 7:488. doi: 10.3389/fbioe.2019.00488
Yang, W., Li, P., Rensing, C., Ni, W., and Xing, S. (2017). Biomass, activity and structure of rhizosphere soil microbial community under different metallophytes in a mining site. Plant Soil 434, 245–262. doi: 10.1007/s11104-017-3546-9
Yin, Z., Yuan, C., Du, Y., Yang, P., Qian, C., Wei, Y., et al. (2019). Comparative genomic analysis of the Hafnia genus reveals an explicit evolutionary relationship between the species alvei and paralvei and provides insights into pathogenicity. BMC Genomics 20:768. doi: 10.1186/s12864-019-6123-1
Zhang, X., Feng, H., He, J., Muhammad, A., Zhang, F., and Lu, X. (2022). Features and colonization strategies of Enterococcus faecalis in the gut of Bombyx mori. Front. Microbiol. 13:921330. doi: 10.3389/fmicb.2022.921330
Zhao, Y., Selvaraj, J. N., Xing, F., Zhou, L., Wang, Y., Song, H., et al. (2014). Antagonistic action of Bacillus subtilis strain SG6 on Fusarium graminearum. PLoS One 9:e92486. doi: 10.1371/journal.pone.0092486
Zhou, J., Deng, Y., Luo, F., He, Z., and Yang, Y. (2011). Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio 2:e00122-11. doi: 10.1128/mBio.00122-11
Keywords: colonization, Lactobacillus plantarum, microbial composition, rhizosphere soil, tomato
Citation: Zhang X, Liao H, Cai T, Cai P, Wu X, Wang Z, Ma H, Qiu G, Zhao M, Lu X, Wang X and Wu C (2025) Features and rhizosphere colonization strategies of Lactobacillus plantarum 0308 in soil-tomato systems. Front. Microbiol. 16:1652881. doi: 10.3389/fmicb.2025.1652881
Received: 24 June 2025; Accepted: 12 August 2025;
Published: 29 August 2025.
Edited by:
Grace Anna Hoysted, Maynooth University, IrelandReviewed by:
Shuangxi Zhu, Northwest A&F University, ChinaHuiming Wu, Zhejiang Agriculture and Forestry University, China
Copyright © 2025 Zhang, Liao, Cai, Cai, Wu, Wang, Ma, Qiu, Zhao, Lu, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Choufei Wu, MDI0MDdAempodS5lZHUuY24=; Xianting Wang, NzU4ODQ4MEBxcS5jb20=
 Xiancui Zhang
Xiancui Zhang Haoran Liao1
Haoran Liao1 Tong Cai
Tong Cai