- 1Department of Biotechnology, Vel Tech Rangarajan Dr. Sagunthala R&D Institute of Science and Technology, Chennai, Tamil Nadu, India
- 2Park’s Biolabs LLP, Chennai, Tamil Nadu, India
- 3Arqgene, Vellore, Tamil Nadu, India
Introduction: Vitamin B12 (B12) is an essential cofactor for key metabolic processes in most living organisms, yet only certain bacteria can synthesize it de novo. Common forms of B12 include adenosylcobalamin (AdoCbl), methylcobalamin (MeCbl) and cyanocobalamin (CNCbl). This study presents the B12 production capability of an extremophile—Ectopseudomonas alcaliphila MSJ19, and a multilevel evaluation of bioactivity of various B12 forms.
Methods: B12 extracted from Ectopseudomonas alcaliphila MSJ19 was initially analyzed by bioassay and LC–MS to confirm the presence of natural B12 forms, followed by in vitro enzyme activity assays with glycerol dehydratase (GD) and diol dehydratase (DD). The functionality of various B12 forms on these enzymes was further evaluated using in-silico molecular docking studies. The bioactivity at the in vivo level was assessed by introducing a coenzyme B12-dependent 3-hydroxypropionic acid (3-HP) biosynthetic pathway in E. coli W and Ectopseudomonas alcaliphila MSJ19 for their ability to transform glycerol into 3-HP.
Results: Bioassay and LC–MS analysis confirmed the presence of ~7 μg/g cdw B12 in the processed extract and specific precursor-product ion transitions, indicated the production of natural B12 forms. To functionally validate the bioactivity of the crude B12 extract, the coenzyme B12-dependent 3-HP biosynthesis pathway was employed in recombinant E. coli W. Supplementation with different B12 forms revealed a hierarchical GD and DD activity (AdoCbl > MeCbl > CNCbl) and a dose-dependent increase in 3-HP production, with an optimal threshold around 500 nM. The conformational specificity of AdoCbl and competitive inhibition of CNCbl and MeCbl were supported by molecular docking of all 3 B12 forms with GD and DD. Notably, crude B12 extract at 0.35 nM yielded 5.9 mM 3-HP titer, closely matching the 7.8 mM obtained with AdoCbl, confirming its bioactive equivalence. Furthermore, recombinant Ectopseudomonas alcaliphila MSJ19 (EaMr) harboring the 3-HP pathway produced up to 3.3 mM 3-HP without external B12 supplementation, highlighting innate capability of the host to produce and utilize bioactive B12in vivo.
Discussion: Collectively—in vitro, in silico and in vivo approaches establish a functional framework for certifying B12 bioactivity and demonstrating EaM as a potent chassis for production of value-added chemicals.
1 Introduction
Vitamin B12 (B12) is a unique cobalt-containing tetrapyrrole cofactor essential for diverse metabolic processes in prokaryotes and eukaryotes (Spataru, 2024). Clinically, B12 holds significant importance, as its deficiency is prevalent among all age groups and linked to pernicious anemia and several neurological diseases (Niklewicz et al., 2023). Despite its critical role in most living organisms, only bacteria are capable of synthesizing it de novo in two biologically active forms, such as adenosylcobalamin (AdoCbl) and methylcobalamin (MeCbl). Due to very low thermostability and high photosensitivity, these natural forms are often chemically modified into a stable cyanocobalamin (CNCbl) form. Hydroxocobalamin is another commonly found B12 form; however, its applications aren’t widespread compared to others. Among these, MeCbl acts as a cofactor only for methionine synthase (MS) in mammals and bacteria. AdoCbl, on the other hand, supports a broad range of coenzyme B12-dependent enzymes known as isomerases, which include mutases, eliminases, and amino mutases. Methyl malonyl-CoA mutase (MMUC) is a well-recognized coenzyme B12-dependent enzyme in humans, while other enzymes have been identified in bacteria, including β-lysine-5,6-aminomutase (LAM), 2-methylene glutarate mutase (MGM), diol dehydratase (DD), D-ornithine-4,5-aminomutase (OAM), ethanolamine ammonia lyase (EAL), glutamate mutase (GM), glycerol dehydratase (GD), isobutyryl-CoA mutase (IM), and ribonucleoside triphosphate reductase (RTPR) (Montoya and Escobar-Briones, 2025). Several of these have been characterized well, in which two isofunctional enzymes—glycerol dehydratase and diol dehydratase are of particular interest in this study, due to their role beyond bacterial metabolism, as catalysts for platform chemical production such as 1,3-propanediol, 3-hydroxypropionic acid, 1-propanol and butanone (Madavi et al., 2024; Brown, 2005). Generally, coenzyme B12-dependent enzymes catalyze intramolecular 1,2—rearrangements mediated through the 5′-deoxyadenosyl radical of coenzyme. Substrate binding to the enzymes generates the active radical by homolytic cleavage of the Co-C bond. This highlights the role of active B12 forms in mediating such radical-based catalysis (Brunold, 2005).
Over the years, a wide range of B12 quantification and characterization methods have been developed including: Microbiological assay, High-performance liquid chromatography (HPLC)—Diode Array Detector (DAD), Liquid chromatography—Mass Spectrometer (LC–MS), UV–vis spectrometry, Raman scattering, atomic absorption spectrometry, Immunoassay, Fluorescence detection, chemiluminescence, capillary electrophoresis, surface plasmon resonance and induced coupled plasma-MS (ICP-MS) (Yang et al., 2024; Trad et al., 2025; Guo et al., 2024; Kansay et al., 2024; Fan et al., 2025). These techniques have been instrumental in analyzing B12 from various samples such as pharmaceutical, nutraceutical, and food products, bacterial cultures, serum, seaweeds, algae and mushrooms. Common challenges encountered in B12 quantification are low B12 concentration in samples often below the limit of detection (LOD) of many methods, stability and sensitivity factors, sample matrix interference, complexity of extraction, sample pretreatment and analytical procedures, and co-detection of B12 analogs like cobinamide, cobamide, cobyric acid and pseudo-B12 (Santos et al., 2024; Lu et al., 2025; Konings et al., 2024; Deptula et al., 2017). B12 analogs are majorly found in bacterial fermentation extracts, hence sample pretreatment steps like solid phase extraction (SPE) and immunoaffinity purification, along with LC–MS, were beneficial in distinguishing bioactive B12 forms from B12 analogs. Though chromatographic methods can distinguish and quantify active B12 forms, they offer little insight into the biological functionality of the B12 present (Xie et al., 2019; Chamlagain et al., 2024; Koseki et al., 2023). In contrast, bioactivity assay of B12 extracts can be obtained only through the measurement of biological output such as cell growth, protein expression, enzyme activity and biochemical production. Conventional microbiological assay using Lactobacillus leichmannii and auxotrophic mutants of Salmonella typhimurium, and Escherichia coli serve as perfect examples for both quantification and bioactivity evaluation of B12 (Raux et al., 1996; Bhushan et al., 2016). In addition, recent developments on PCR-based strategies provide confirmation for B12 production on a genotypic level (Venkatesan et al., 2024). Yet they fail to distinguish various forms of B12 and are prone to false positives by B12 analogs and sample matrix, thus requiring extensive sample pretreatment (Kong et al., 2017; Li et al., 2017). Each B12 quantification method has its pros and cons; most importantly, this study does not aim to replace or challenge well-established B12 analytical methods. Rather, it focuses on the lacuna in evaluating the bioactivity of various forms of B12 from a natural producer in terms of functional biological output.
This work aims to analyze the bioactivity of crude B12 extracted from a novel extremophilic B12 producer. Through confirming the production of natural B12 forms by Ectopseudomonas alcaliphila MSJ19 (EaM), the study navigates toward in vitro, in silico and in vivo approaches to evaluate B12 bioactivity and shed light on the effect of various B12 forms on bioactivity. The outcomes provide valuable insights into the functionalities of B12 from natural producers and the significance of B12 dose and forms in clinical and industrial applications. The developed framework to functionally characterize B12 is intended to trigger more research focus toward the development of high-throughput biological output-based B12 quantification. Finally, the host’s capability to produce biologically active B12 has been channeled toward 3-hydroxypropionic acid production in E. coli W and EaM by metabolic engineering approaches. Ectopseudomonas alcaliphila MSJ19 is an extremophile with psychrophilic (growth at 4–40 °C) and alkaliphilic (optimal pH 9–10) properties. To our knowledge, this represents the first report evaluating B12 bioactivity from an extremophilic strain (Venkatesan et al., 2024; Yumoto et al., 2001). The alkaliphilic nature provides revolutionary bioprocess advantages such as pH-based bio-containment that prevents mesophilic contamination, elimination of complex buffering systems and potential compatibility with non-sterile fermentation infrastructure (Zeng et al., 2023; Wernick et al., 2016). Thus, providing a scope for Ectopseudomonas alcaliphila MSJ19 as a potent microbial chassis for the sustainable production of value-added biochemicals.
2 Materials and methods
2.1 Chemicals, strains, and plasmids
All chemicals, reagents and media were correspondingly purchased from SRL-India, Sigma Aldrich, TCI chemicals and Himedia. Yeast alcohol dehydrogenase (yADH) was purchased from Sigma-Aldrich. Ectopseudomonas alcaliphila MSJ19 was isolated in our previous study, and its 16S rRNA sequence has been deposited in GenBank (ID: PX397011). Plasmid pDK7 (p15a)/pddCDE, gdrAB was developed by amplification of pddCDE genes from genomic DNA isolated from Klebsiella pneumoniae 109 and subsequently cloned into KpnI and HindIII restriction sites of pDK7 (p15a)/dhaB123, gdrAB plasmid. The plasmids were transformed into appropriate hosts following the protocol adopted from (Zhou et al., 2013). All strains, plasmids and primers used in this study are listed in Table 1.
2.2 Shake flask production of vitamin B12 by Ectopseudomonas alcaliphila MSJ19
Overnight lysogeny broth (LB) EaM culture was pre-cultured in LB medium until mid-late log phase of growth. Subsequently, 0.1 OD600 of exponentially grown cells was reinoculated appropriately in LB production medium containing precursors: CoCl2 (5 mg/L), DMBI (75 mg/L), and Betaine (1 g/L) and incubated under aerobic conditions at 37 °C, 200 rpm. Wild-type E. coli W were cultivated under similar conditions to serve as a negative control wherever appropriate in this study. Cell growth was measured at regular intervals by a UV–Vis spectrophotometer, and after 18 h, cells were harvested for B12 extraction (4,500 rpm, 15 min). One OD600 corresponds to 0.33 g (±0.05 g) of dried cell mass per liter (Arasu et al., 2013).
2.3 Extraction and quantification of B12
Cells were washed twice with 100 mM potassium phosphate buffer (pH 7.0) and resuspended in the same buffer for B12 extraction under ice with minimal light exposure. Cell concentration was measured before and after lysis. Cells were lysed by ultrasonication at 30% amplitude for 6 min with a 10-s ON/OFF cycle (VCX 130, Sonics; 20 kHz), centrifuged (4,500 rpm, 10 min), supernatant filtered through a 0.22 μm syringe filter and used as crude B12 extract for further analysis.
For B12 quantification by bioassay, the protocol mentioned in our previously published study was followed exactly (Venkatesan et al., 2024). To convert natural B12 forms into the more stable CNCbl form, 0.1% w/v NaCN was added to the crude B12 extract, and after 5 min incubation (37°C), the mixture was autoclaved (121 °C, 15 min) and cooled on ice. The samples were centrifuged (4,500 rpm, 20 min) and the supernatant was passed through a 0.22 μm syringe filter prior to LC–MS analysis. The LC–MS analysis was performed for both crude B12 extract and cyano-converted extract, using a Waters TQD LC–MS/MS system equipped with a Kinetex (2.6 μm, XB C18 Column, 2.1 × 100 mm). 20 mM ammonium formate in water (Mobile Phase A) and methanol (Mobile Phase B) were used for sample elution under the following linear gradient: 90% mobile phase A for 0–2 min, 90% mobile phase A for 2–4 min, 10% mobile phase A for 4–5 min, 90% mobile phase A for 5–7 min. The flow rate was maintained at 0.3 mL/min, the column temperature was set to 35 °C, and the injection volume was 10 μL. Mass spectrometry was conducted in positive electrospray ionization (ESI) mode with a source temperature of 140°C, desolvation temperature of 300 °C, cone gas flow of 10 L/h and desolvation gas flow of 1,000 L/h. The capillary voltage was set at 26 V, and the cone voltage was 35 V. The quantification of CNCbl was performed using multiple reaction monitoring (MRM) transitions, monitoring the precursor ion at m/z 678.5 and the product ions at m/z 147.0 and 358.9, with collision energies of 34 eV and 24 eV, respectively (Stumpf et al., 2024; Kahoun et al., 2022). This targeted MRM setting was chosen to achieve high specificity and sensitivity for CNCbl, while avoiding cross-detection of other B12 forms in crude extracts (Reddy KotamReddy et al., 2023). The same LC–MS method was also adopted for crude B12 extracts.
2.4 Enzyme activity assay
The modified M9 medium used for shake flask studies in EcW GD and EcW DD contained: MgSO4·7H2O, 0.5 g/L; NaCl, 1.0 g/L; NH4Cl, 1.0 g/L; yeast extract, 1 g/L; glycerol, 100 mM; potassium phosphate buffer (pH 7.0), 100 mM; kanamycin 50 mg/L; and chloramphenicol 25 mg/L. Unless stated otherwise, LB medium and the same modified M9 medium with appropriate antibiotics were used for primary inoculum and secondary inoculum, respectively. Shake flask cultivation was carried out with a working volume of 50 mL culture with an inoculum of 0.1 OD600 in a 250 mL Erlenmeyer flask at 37°C, 250 rpm under aerobic conditions. For enzyme production, the cultures were induced at 0.6 ± 0.05 OD600 with 0.5 mM IPTG. After 6 h incubation, cells were harvested (5,000 rpm, 15 min) and washed once with 20 mM potassium phosphate buffer. Subsequently, cells were resuspended in the same buffer and subjected to ultrasonication under ice at 30% amplitude for 4 min with a 10-s ON/OFF cycle. The obtained lysate was centrifuged (13,000 rpm, 30 min), and the supernatant was collected to measure total protein concentration (by the Bradford method), glycerol dehydratase (GD) and diol dehydratase (DD) activity, respectively.
GD activity was measured by following the protocol developed by Sankaranarayanan et al. (2017), and the same method was employed to measure DD activity. Briefly, the substrate mixture (~1.8 mL), containing 20 mM potassium phosphate buffer (pH 8.0), 3 mM MgCl2 and 40 mM 1,2-PDO, was placed in a 1-cm path length spectrophotometer cuvette. B12 solution (100 μL) was added to this assay mixture, containing 0.15 mM NADH and 1.5 mM ATP. B12 concentration and type were varied individually to study their effects. Then, the coupling enzyme (40 μL) yADH (12 U/mL) was added using an air-tight gas chromatography syringe, and the cuvette was incubated for 3 min in a water bath at 37 °C. The enzymatic reaction was initiated by injecting 50 μL of crude GD or crude DD enzyme solution, appropriately (typically <0.03 U/mL). The NADH concentration was determined at 340 nm with the extinction coefficient (ε340) of 6.22 mM−1 cm−1 on a UV spectrophotometer. One unit of GD or DD activity was defined as the amount of enzyme required to convert 1 μmol of 1,2-PDO to propionaldehyde per minute under given assay conditions.
2.5 Molecular docking of various B12 forms with GD and DD
Molecular docking was performed between each B12 form—AdoCbl, CNCbl, MeCbl and the active sites of GD and DD. High-resolution crystal structures of GD (PDB ID: 1IWP) and DD (PDB ID: 1DIO) were retrieved from the protein data bank (PDB) (Yamanishi et al., 2002; Shibata et al., 1999). The crystal structures were refined by eliminating water molecules and ligands using PyMOL software (version 3.1.3). Refined proteins were subsequently processed using Autodock tools (v 1.5.7) by setting grid parameters for both GD and DD based on reference active site coordinates reported already (Yamanishi et al., 2002; Masuda et al., 2000). 3D structure of all the ligand molecules AdoCbl, CNCbl, and MeCbl were procured from protein structures (PDB ID: 5C8A, 5NP4, 3SC0) co-crystalized with respective ligands. Each B12 ligand was assigned its respective charges and docked into the aforementioned active site grid. Docked conformations exhibiting higher binding and similar interactions with key active site residues were considered for further evaluation.
2.6 Shake flask 3-HP production in EcW GD and EcW DD
Shake flask 3-HP production with the respective host was carried out aerobically using the same modified M9 medium (50 mL) with a starting inoculum of 0.1 OD600 in a 250-ml Erlenmeyer flask incubated at 37°C, 250 rpm. The cultures were induced at 0.6 ± 0.05 OD600 with 0.1 mM IPTG and supplemented with various forms and concentrations of B12, respectively, at 3, 6, 9, and 12 h of cultivation. The details on B12 form and concentration supplemented for each shake flask experiment were furnished in Supplementary Table 1. Samples were collected periodically to determine the cell mass, residual substrate and metabolites. Briefly, the collected culture samples were centrifuged (10,000 rpm, 10 min), then the supernatant was diluted appropriately and filtered using a 0.22 μm PVDF membrane filter (Millipore). Then the samples were passed through an HPLC system equipped with an Aminex HPX-87H column (300 mm × 7.8 mm, Bio-Rad, United States) maintained at 65°C. The mobile phase consisted of 2.5 mM H2SO4 with a flow rate of 0.5 mL/min, and metabolite concentrations were analyzed using a Refractive Index Detector (RID) and a Photo-diode array detector (PDA) (Ravi and Sankaranarayanan, 2024).
2.7 Shake flask 3-HP production in recombinant Ectopseudomonas alcaliphila MSJ19 (EaMr)
Shake flask cultivation of the EaMr for 3-HP production was carried out in the same B12 production medium with the addition of Kanamycin (30 mg/L). The cultivation was carried out aerobically at 37°C, 200 rpm, with an initial cell concentration of 0.1 OD600. 100 mM of glycerol (carbon source for 3-HP production) was added when the cell concentration reached 0.7 ~ 1 OD600. The samples were withdrawn periodically to determine the cell mass, glycerol, 3-HP and other metabolites.
3 Results
3.1 Production of natural forms of B12 by Ectopseudomonas alcaliphila MSJ19
Consistent with our previous study, the B12 levels of Ectopseudomonas alcaliphila MSJ19 (EaM) quantified by bioassay were 7.18 μg/g cdw (Venkatesan et al., 2024). To validate B12 forms, LC–MS analysis was performed for: (a) crude B12 extract, (b) cyano-converted B12 extract, and (c) crude B12 extract spiked with 0.5 μM each of standard AdoCbl and MeCbl (Figure 1). A distinct peak at RT 4.32 min was observed for cyano-converted B12 extract corresponding to MRM transitions m/z 147.0 and 358.9, matching precisely with the standard CNCbl profile. These transitions were selected based on their high sensitivity and specificity for CNCbl quantification. The concentration of B12 was quantified as 6.93 μg/g cdw. In contrast, no corresponding peaks were observed at RT 4.3 min for either the crude extract or the spiked crude extract, indicating that CNCbl was not natively present in the bacterial extract. These results collectively support that EaM produces only the natural (coenzyme) forms of B12—namely, MeCbl and AdoCbl—which are not detected in this LC–MS method due to their distinct transition requirements (such as m/z 685.6 and 665.6, respectively) (Heal et al., 2014). By providing a clear distinction between the presence of natural and non-natural forms of B12, the present strategy offers a streamlined and scalable framework for both qualitative and quantitative assessment of B12 production in industrially relevant microbial strains.
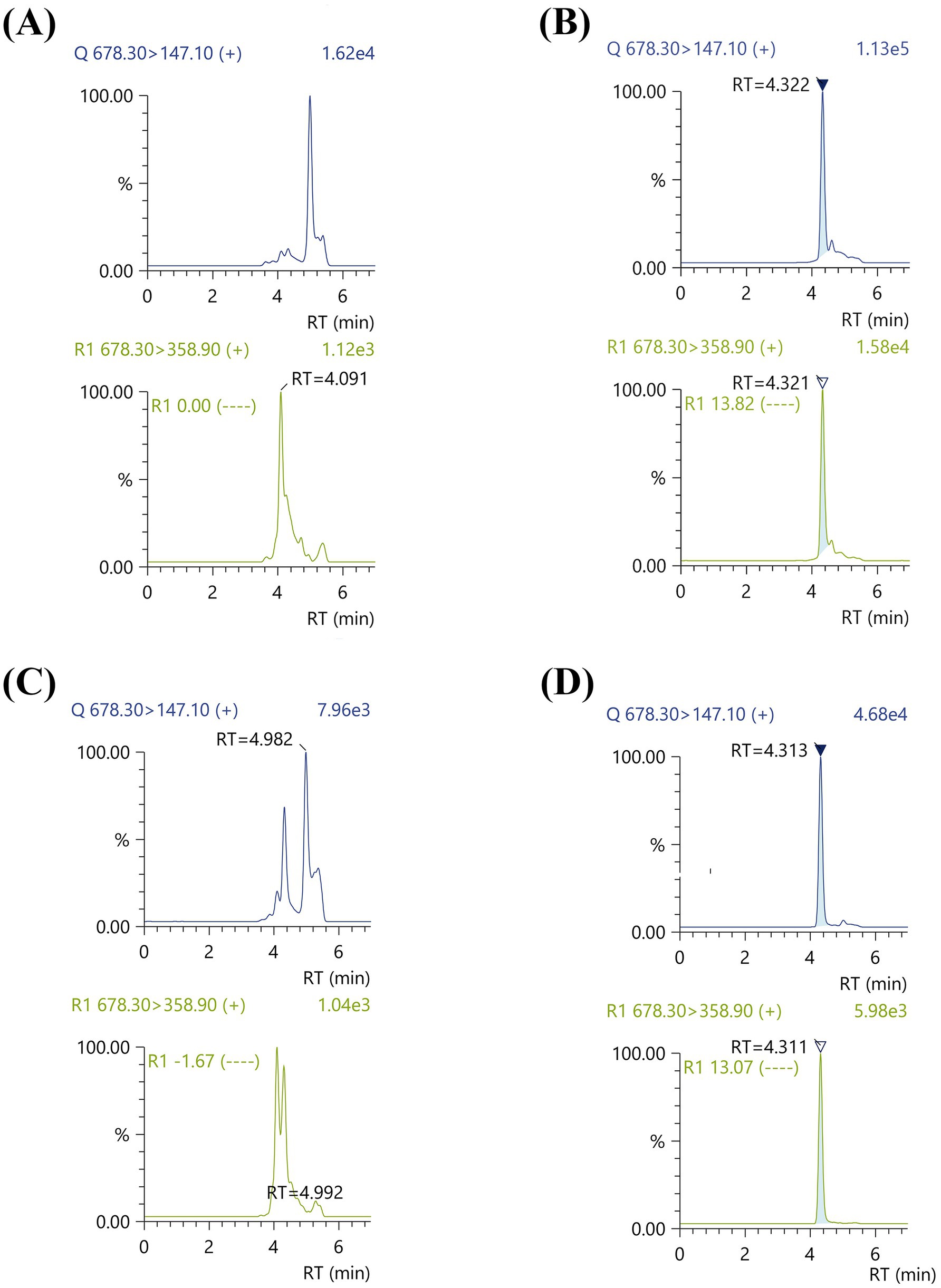
Figure 1. LC–MS/MS chromatograms for confirmation of natural B12 forms in Ectopseudomonas alcaliphila MSJ19 extract under specific MRM transitions: (A) Crude B12 extract (no peak observed at RT ~ 4.32 min), (B) Cyano-converted B12 extract (distinct peak observed at RT 4.32 min, matching CNCbl standard), (C) Crude B12 extract spiked with 0.5 μM MeCbl and AdoCbl (no peak observed at RT ~ 4.32 min) and (D) CNCbl (concentration = 5 ppb; clear peak at RT 4.31 min).
3.2 In-vitro bioactivity evaluation of crude B12 extract
Glycerol dehydratase (GD) and diol dehydratase (DD) are isofunctional, coenzyme B12-dependent enzymes, whose characteristics and in vitro assays have been well studied (Toraya et al., 2022; Nasir et al., 2020). These enzymes are known to be catalytically active only in the presence of AdoCbl with varying degrees of sensitivity (Marsh and Meléndez, 2012; Toraya et al., 1979). While other B12 forms, such as MeCbl and CNCbl, are often reported as competitive inhibitors (Poppe and Rétey, 1997; Toraya and Ishida, 1991). These features make GD and DD valuable in vitro tools for evaluating the functional bioactivity of B12 from bacterial extracts.
Generally, activity assays for these isofunctional enzymes are performed at a saturated coenzyme B12 concentration of around 10–20 μM (Kumar et al., 2016; Wei et al., 2014). However, due to the low concentration of crude B12 used for this study (0.35 nM), a preliminary investigation was carried out to study the effect of AdoCbl concentration on GD and DD activity. Maximal activities were observed at 15 μM AdoCbl, yielding 14.32 U/mg for GD and 6.79 U/mg for DD. At 0.35 nM, the enzyme activity dropped to 0.32 U/mg for GD, while no significant activity was observed for DD (Figure 2A). The difference in activities between GD and DD correlates with their known kinetic parameters, specifically the reported Km values of GD (~ 8 nM to 20 nM) and DD (~ 0.7 μM) from Klebsiella sp. (Yamanishi et al., 2002; Wang et al., 2007). According to previous reports, GD attained 95% of its maximum activity and DD only 4% at 120 nM AdoCbl (Yamada et al., 2004). Relatively, the current study shows that GD and DD attained 81% and 7% of their respective maximum activities at 100 nM AdoCbl, confirming the accuracy of the assay and reinforcing AdoCbl sensitivity among the enzymes. As anticipated, no significant enzyme activity was observed when the assay was performed with CNCbl and MeCbl, even at 15 μM, the saturated concentration used for AdoCbl (Figure 2B). The missing 5′-deoxyadenosyl radical upon binding of CNCbl and MeCbl to the enzyme is expected to be the sole reason for their inability to support GD and DD activity (Toraya, 2000; Bucher et al., 2012). Previous reports support this by showing that MeCbl and CNCbl act as competitive inhibitors for DD (Ki of 0.73 μM and 1.8 μM, respectively) (Toraya et al., 1977) and CNCbl for GD (Ki = 21.6 nM) (Poppe and Rétey, 1997). Notably, with 0.35 nM of crude B12 extract, the GD activity measured was 0.21 U/mg, which was slightly lower than 0.32 U/mg obtained with standard AdoCbl at the same concentration. While this suggests that the crude B12 extract predominantly contains AdoCbl, it confirms that EaM has dominantly produced AdoCbl; the lower activity could likely be due to the presence of some MeCbl, which may exert competitive inhibition. However, further studies are required to confirm this hypothesis. These results collectively demonstrate that the B12 produced by Ectopseudomonas alcaliphila MSJ19 is functionally bioactive, with in vitro-based assays providing indirect but reliable confirmation of AdoCbl as the dominant form in the crude extract.
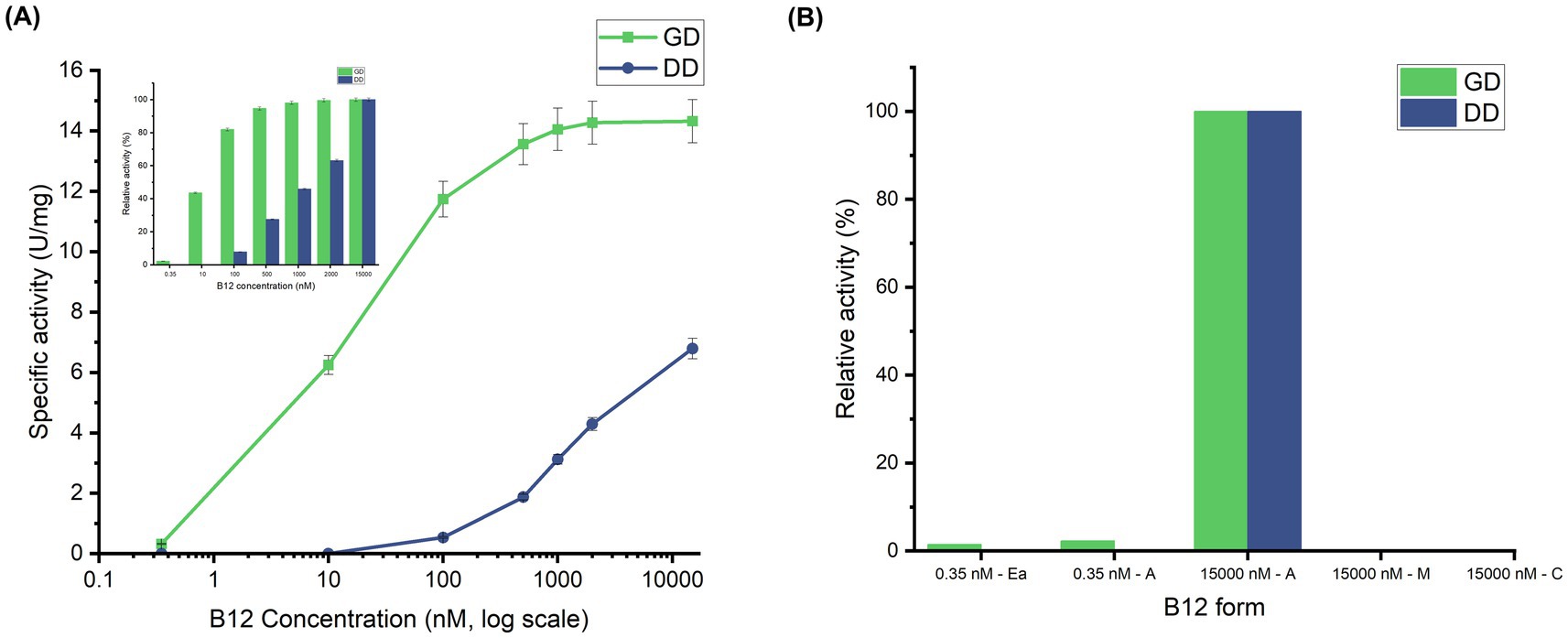
Figure 2. Harnessing in vitro enzyme activity assays of GD and DD to evaluate B12 bioactivity: (A) AdoCbl concentration-dependent variation of GD and DD activity (U/mg)—Concentration (nM) is plotted on a logarithmic scale. The inset shows relative activities (%) of GD and DD normalized to their respective maximum activities. (B) Effect of B12 forms on GD and DD activity; Experimental groups: Ea—Ectopseudomonas alcaliphila MSJ19 crude B12 extract; A, Adeonsylcobalamin; M, MeCbl; C, CNCbl. Error bars represent standard deviation from three independent biological replicates (n = 3).
3.3 In silico prediction of various B12 forms reactive specificity with GD and DD
To complement the differential catalytic activity of B12 forms on a structural basis, molecular docking was performed between three B12 ligands—AdoCbl, CNCbl, and MeCbl—and the known crystal structures of GD and DD. A total of six docking combinations were generated, and a complete summary of interactions and H-bond distances for each docking conformation is provided in Supplementary Table 2. Based on previous crystallographic studies, 12 key active site residues were defined for GD (Yamanishi et al., 2002) and 7 for DD (Masuda et al., 2000) to assess the binding capability of ligands within the functionally active site.
In GD, AdoCbl exhibited the most favorable binding conformation for catalytic function, forming hydrogen bonds with five active site residues (SER122, THR104, SER225, THR173, LYS102) and a binding energy of −3.93 kcal/mol (Figures 3A1,D1). These interactions span both the corrin ring and adenosyl moiety, positioning the ligand in a favorable conformation for Co-C homolysis and radical exchange. CNCbl exhibited a significantly lower binding energy (−14.5 kcal/mol) and formed four interactions with active site residues (SER122, ASP235, ALA124, THR173) (Figures 3B1,E1). While such tight binding reflects higher affinity, the absence of the adenine moiety blocks its catalytic ability and complements its role as a competitive inhibitor. MeCbl also interacted only with one active residue (SER122) and an intermediate binding energy (−5.81 kcal/mol) (Figures 3C1,F1), further reflecting its non-catalytic but potentially competitive inhibitory role.
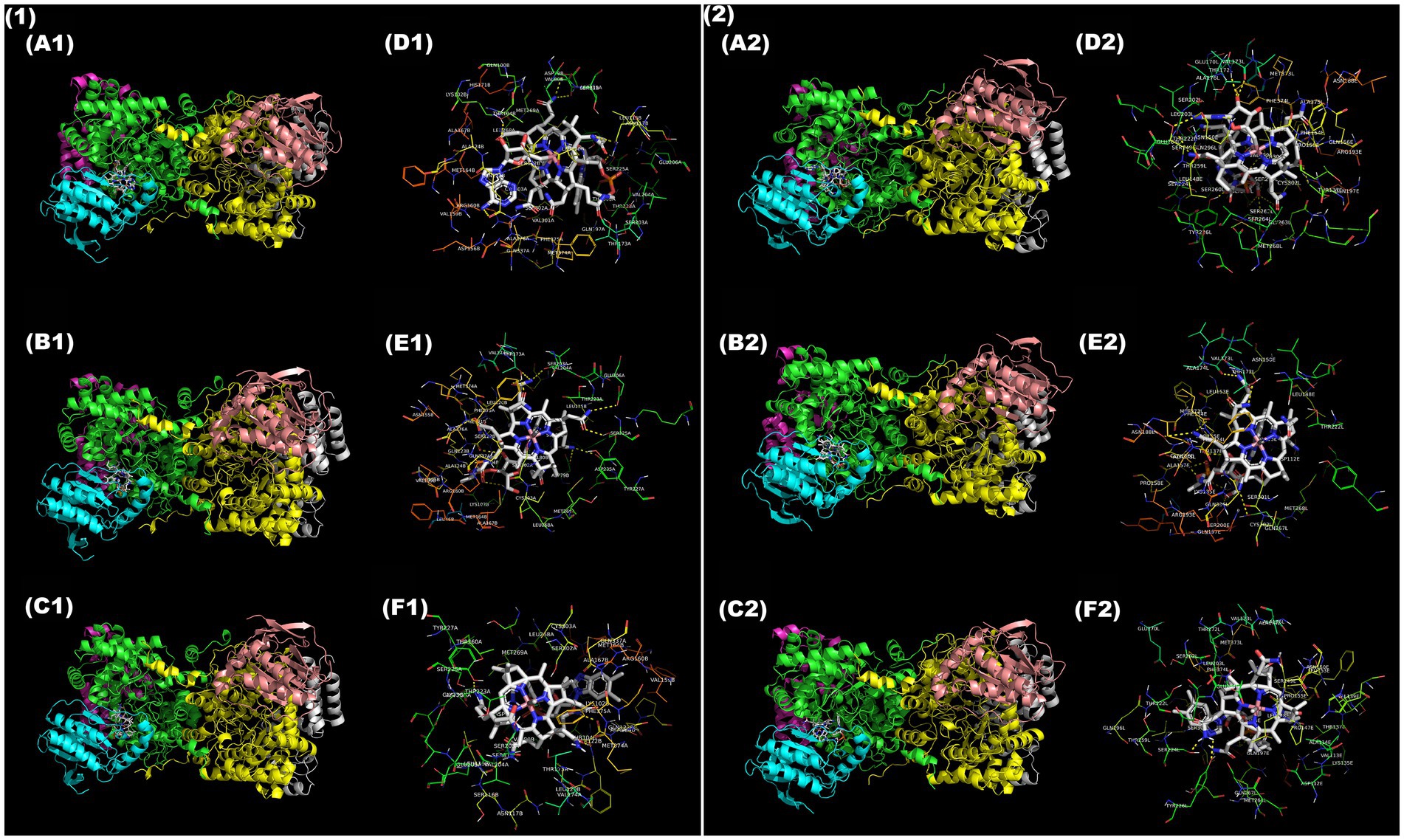
Figure 3. Molecular docking of B12 ligands with GD (1) and DD (2). Stereo views showing overall GD (A1–C1) and DD (A2–C2) structure in complex with (A1,A2) AdoCbl, (B1,B2) CNCbl, and (C1,C2) MeCbl. Chains of the GD heterotrimer are colored as follows: A—green, B—cyan, C—magenta, D—yellow, E—salmon, F—grey; Chains of the DD heterotrimer are colored as follows: A—yellow, B—salmon, E—cyan, G—grey, L—green, M—magenta. Zoomed-in interaction maps of ligands with active site residues of GD (D1—F1) and DD (D2—F2): (D1, D2) AdoCbl, (E1, E2) CNCbl, and (F1, F2) MeCbl. Hydrogen bonds and polar interactions are visualized between ligand atoms and neighboring amino acid residues. Ligand atom color scheme: C—grey, N—Navy blue, O—red, S—Orange, Co—pink.
In DD, AdoCbl again exhibited the highest number of active site interactions, yet fewer than in GD. Only two out of six interactions matched with key active side residues (THR172, SER301) following an intermediate binding energy (−7.23 kcal/mol) (Figures 3A2,D2). This observation aligns with the in vitro enzyme activity assay, where DD activity expressed a higher Km than GD, thus justifying the lower specificity of AdoCbl with DD. CNCbl had only one matching residue (THR172) among five interactions with a lower binding energy (−12.9 kcal/mol) (Figures 3B2,E2). MeCbl had only one matching residue (SER224) among its two interactions with a higher binding energy (−6.99 kcal/mol) among all 3 B12 forms with DD (Figures 3C2,F2). These binding predictions reflect the inferiority of B12-driven catalysis with DD as compared to GD.
Importantly, these findings elucidate the conserved structural and functional preference of GD and DD for AdoCbl. Evidently, comparison of available crystal structures and docking combinations of this study has shown that CNCbl and MeCbl are also capable of binding within the active site, but they lack the adenine moiety necessary to trigger Co-C bond homolysis and substrate rearrangements (Shibata et al., 2018). Hence, the adenine moiety not only acts as a radical initiator, but also participates in key interactions to position the cofactor in an appropriate spatial conformation for catalytic activity. Therefore, the in-silico findings support the in vitro enzymatic assay, confirming that only AdoCbl positions itself in a catalytically active conformation in both GD and DD in a conserved manner. Meanwhile, CNCbl and MeCbl are capable of competitive inhibition due to their catalytically inactive binding conformation.
3.4 In-vivo bioactivity evaluation of crude B12 extract
In the two-step catalytic pathway for 3-HP production, Coenzyme B12 (AdoCbl) serves as an essential cofactor for glycerol dehydratase (Kumar et al., 2012). Therefore, 3-HP production can act as a reliable qualitative metric for assessment of B12 bioactivity, offering a more meaningful output than conventional microbiological assay. To evaluate this, EcW GD and EcW DD were supplemented individually with AdoCbl, MeCbl, and CNCbl for 3-HP production. Among these, AdoCbl yielded the highest 3-HP production in EcW GD, confirming it as the most effective cofactor for GD activity. Whereas MeCbl resulted in only 63% of this maximum, and CNCbl only 36%. A similar trend was observed for EcW DD, yet its maximum 3-HP titre was only 52% of that achieved with EcW GD. Such a low 3-HP titre of DD in this expression system is obvious due to the following well-documented reasons: (i) 1,2-PDO is the preferred substrate for DD over glycerol (Sauvageot et al., 2002), (ii) absence of diol dehydratase reactivase in this expression system, making DD prone to suicide inactivation like GD in the presence of glycerol (Bilić et al., 2019), (iii) gdrAB is known to be ineffective in reactivating DD (Kajiura et al., 2007), and (iv) less B12 specificity of DD as observed in enzyme activity analysis (Poppe and Rétey, 1997). These results further demonstrate a substantial decline in GD and DD activity with synthetic B12 forms and justify the functional superiority of the natural B12 forms.
Notably, the modest 3-HP production with CNCbl suggests that E. coli may possess intrinsic metabolic mechanisms to convert CNCbl into biologically active forms, analogous to human metabolic pathways (Kelly, 1997). However, the relatively low 3-HP titer (70% lower than AdoCbl) indicates that this intracellular conversion is likely rate-limiting. The difference in 3-HP production between MeCbl and CNCbl also reflects the metabolic complexity of their respective conversion process, as CNCbl conversion is mediated by a four-step enzymatic process, while MeCbl requires only a single step (Rizzo et al., 2016).
While earlier studies typically employed 2000 nM AdoCbl for optimal 3-HP production in E. coli (Nguyen-Vo et al., 2019), the concentration of crude B12 extract used is comparatively less (0.35 nM). Therefore, this study also evaluated the effect of B12 concentration on 3-HP production across a wide range (0.35–2,000 nM) for each B12 form (Figures 4A,B). Interestingly, B12 concentration had a significant effect on 3-HP production, similar to the enzyme activity. Remarkably, the highest 3-HP titer of 50.2 mM was observed at 500 nM AdoCbl for EcW GD, beyond which no significant increase in titre could be observed. The optimal 3-HP production at 500 nM reflects seamlessly with the Vmax of GD, embarking on the critical impact of B12 on the rate-limiting step of the 3-HP catalytic pathway. The 3-HP production titer (41.5 mM) with 2 μM AdoCbl was also consistent with previous reports (Sankaranarayanan et al., 2017). Similar trends were observed for other B12 forms. Owing to the higher Km of DD, 3-HP production was maximal (26.2 mM) only at 2,000 nM and suggesting that further increase in B12 may still enhance activity (typically close to the Vmax of DD (>7 μM)).
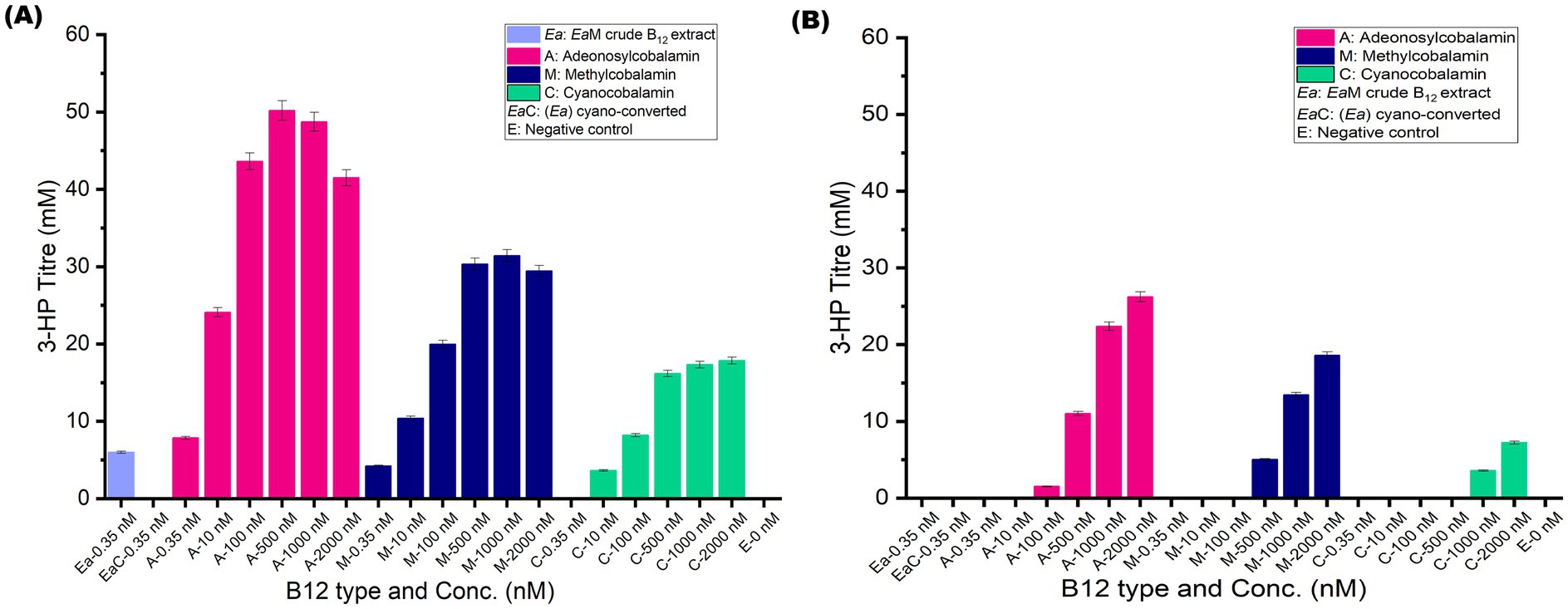
Figure 4. Functional evaluation of B12 forms and concentration on 3-HP production by EcW GD and EcW DD, respectively. (A) Summary of 3-HP production titre (mM) of EcW GD under supplementation with different B12 forms at varying concentrations (nM). (B) Summary of 3-HP production titre (mM) of EcW DD under supplementation with different B12 forms at varying concentrations (nM). Error bars represent standard deviation from three independent biological replicates.
Of particular interest, crude B12 extract at 0.35 nM supported a 3-HP titre of 5.9 mM in EcW GD—closely matching the 7.8 mM titer at 0.35 nM AdoCbl. This confirms the presence of active B12 forms in the extract. The marginal difference could be attributed to the presence of some MeCbl in the extract, as only the total B12 concentration was quantified. Consistently, MeCbl at 0.35 nM attained a lower 3-HP titer of 4.2 mM. In contrast, no measurable 3-HP production was observed at 0.35 nM cyano-converted extract, despite a very low 3-HP titer of 1.1 mM at 0.35 nM CNCbl. This suggests potential interference from matrix effects during conversion (Nakos et al., 2017) or simply the titre falling to the limit of detection (LOD = 0.8–1.0 mM). As expected, no 3-HP production was observed in the negative control with E. coli W extract, thus justifying that any potential impurities in crude bacterial extracts do not affect 3-HP production. These findings reinforce the presence of a biologically active form of B12 in crude extract, and the chemical conversion process has led to a non-natural/synthetic B12 form, which obviously has led to a decrease in or no 3-HP production. Collectively, these results strongly establish the utility of 3-HP production as an in vivo functional assay for B12 bioactivity. Building on these findings, the next section validates the in vivo B12 bioactivity using recombinant Ectopseudomonas alcaliphila MSJ19 itself.
3.5 Assessment of the 3-HP production capability of EaMr
To evaluate the in vivo bioactivity of endogenously produced B12, Ectopseudomonas alcaliphila MSJ19 was engineered to express the 3-HP biosynthetic pathway via plasmid pUCPK harboring dhaB123, gdrAB, and KGSADH. Shake flask cultivation was performed with and without an exogenous supply of 2 μM AdoCbl, thereby ensuring that any observed 3-HP production is solely dependent on the host’s innate B12 biosynthesis capability. Correspondingly, EaMr produced a maximum 3-HP titre of 3.2 mM without external B12, indicating the endogenous production of coenzyme B12 was sufficient to activate GD and enable 3-HP biosynthesis (Figure 5A). As expected, no 3-HP production was observed in control flasks without glycerol supplementation (data not shown), confirming that 3-HP originated exclusively from glycerol metabolism and not from medium components or endogenous carbon sources. 3-HP production was improved (9.5 mM) under B12 supplementation, indicating that 3-HP flux can be further enhanced through B12 supplementation (Figure 5B). In addition, glycerol consumption and 3-HP production were relatively low in either case compared to EcW GD. This could be presumed due to intrinsic regulatory barriers, such as the presence of transcriptional repressors in the host’s native glycerol catabolic pathway and/or limited compatibility between the heterologous plasmid system and the host transcriptional or translational machinery (Thi Nguyen et al., 2021; Prieto-de Lima et al., 2024). Although elucidating these factors was beyond the scope of this study, the results clearly establish the functional bioavailability of naturally synthesized B12 in EaMr. These findings not only validate Ectopseudomonas alcaliphila MSJ19 as a biologically competent B12 producer but also highlight its potential as a versatile microbial chassis for value-added chemical production beyond vitamin B12.
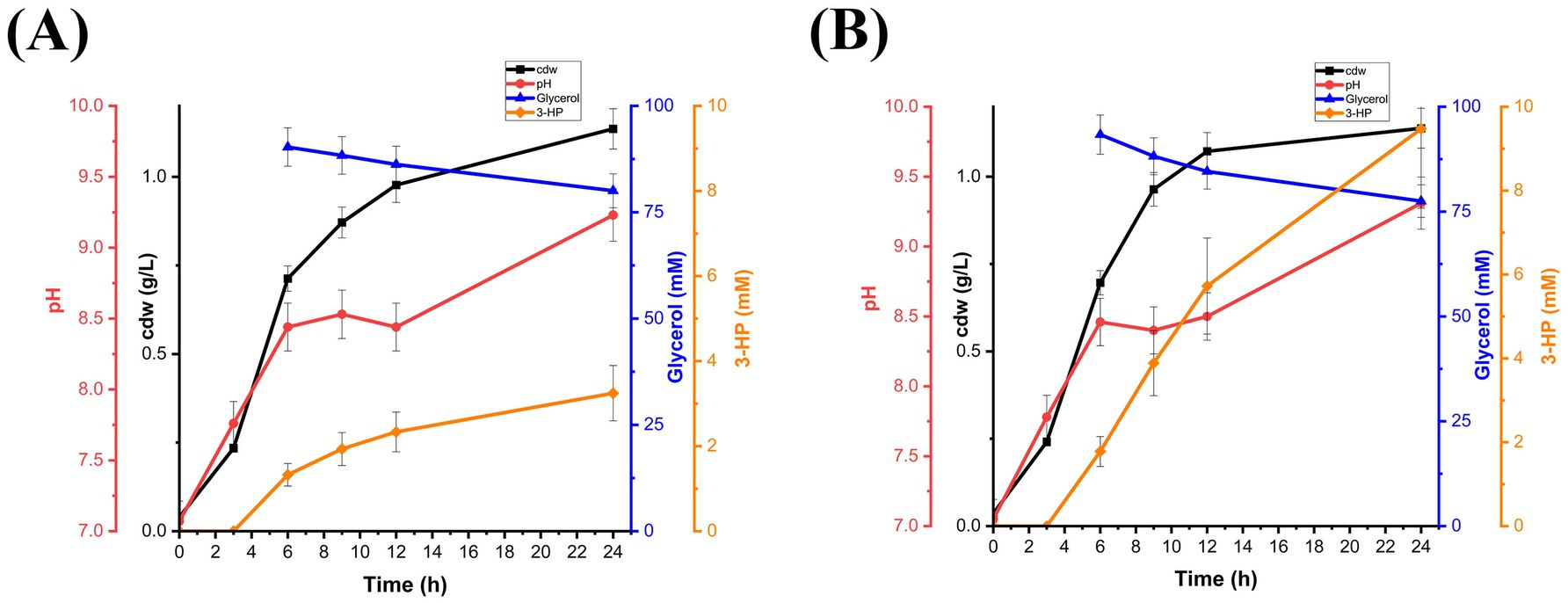
Figure 5. Time-course profile of EaMr showing cell growth (cdw g/L), pH variation, glycerol consumption (mM), and 3-HP production (mM): (A) without exogenous AdoCbl supplementation and (B) with supplementation of 2 μM AdoCbl. Error bars represent standard deviation from three independent biological replicates.
4 Discussion
Vitamin B12 is structurally complex and exists in several natural and synthetic forms. Among them, only AdoCbl and MeCbl are biologically active, serving as cofactors in radical-based and methyl-transfer enzymatic reactions, respectively. Bacteria are the sole workhorses for industrial scale production of this essential vitamin; however, they can produce inactive B12 analogs (Thirupathaiah et al., 2012). Therefore, assessing the bioactivity of B12 rather than relying only on total B12 quantification is essential to grade its functional bioavailability. Recent advancements in chromatographic and immunoassay methods have played a significant role in classifying the forms of B12 (Möller et al., 2022; Balabanova et al., 2022). However, studies are limited in evaluating the activity of crude extracts of natural B12 producers using a valid biological output (Chamlagain et al., 2021). This study details a biologically integrated workflow combining in vitro, in silico and in vivo approaches to uncover the potential of active forms of B12 produced by a novel extremophilic strain (EaM).
Initially, conventional bioassay using Salmonella typhimurium ΔmetE ΔcbiB and LC–MS were valuable in confirming the production of natural form (AdoCbl & MeCbl) of B12 (~7 μg/g cdw) by Ectopseudomonas alcaliphila MSJ19. The establishment of enzymatic assay methods for coenzyme B12-dependent enzymes such as GD and DD paved a plausible approach to further study the bioactivity of crude B12 extract. The coupled enzymatic method to measure GD activity also stood reliable for DD activity measurement, particularly due to its increased substrate preference to 1,2-PDO (Toraya et al., 2022). Substrate binding to the holoenzyme triggers Co-C bond homolysis, leading to the formation of cob(II)alamin and 5′-deoxyadenosyl radical. Theoretically, this radical is essential to mediate 1,2-rearrangements in the substrate during enzyme catalysis (Giedyk et al., 2015). Justifiable to this, both GD and DD were capable of product formation only in the presence of AdoCbl, while no notable enzyme activity was observed for CNCbl and MeCbl even at very high concentrations (15 μM) due to their inability to form an adenosyl radical.
Interestingly, GD activity with 0.35 nM crude extract was nearly equivalent to that of standard AdoCbl, confirming the dominant presence of AdoCbl in the crude extract. The lack of DD activity with crude extract is attributed to its higher Km of ~0.8 μM for AdoCbl, further validating the reliability of such enzyme activity assays to confirm B12 bioactivity. Despite the non-catalytic activity of other B12 forms, they play a larger role as competitive inhibitors, and it is to be realized that their presence in sample extracts tends to underestimate the bioactivity of actual AdoCbl present. The potential inhibitory effects of other B12 forms were supported by molecular docking, which revealed comparable binding energies across all B12 forms, suggesting competitive inhibition. Thus, in vitro assays combined with in silico insights reinforce the fact that B12 bioactivity is not defined by binding affinity alone, but also by the ability to support 5′-deoxyadenosyl radical generation and substrate rearrangements.
Transitioning toward the applicability of the coenzyme B12-dependent 3-HP production pathway in recombinant E. coli as an in vivo model system for B12 bioactivity enlightened the fate of other B12 forms beyond competitive inhibition. Contrarily, 3-HP production in recombinant E. coli was observed under supplementation of all 3 B12 forms individually with varying degrees (AdoCbl > MeCbl > CNCbl). Although in vitro enzyme assay and in silico models have strongly backed the competitive nature of other B12 forms on GD and DD, this discrepancy likely arises from the host’s intracellular B12 salvage and conversion mechanisms, enabling conversion of other B12 forms into AdoCbl. Haptocorrin-based B12 binding, absorption by intrinsic factors, innate mechanisms to convert various B12 forms into a metabolically active form and bioavailability were well documented in humans (Vincenti et al., 2021). While such B12 conversion mechanisms were very scarcely reported in bacterial systems (Reynolds et al., 1980), this study is the first of its kind to report their impact on coenzyme B12-dependent platform chemical synthesis. Future work should investigate the regulation of these conversion mechanisms and their fine-tuning to improve the flux of AdoCbl for platform chemical production.
A concentration-dependent variation in 3-HP titre across B12 forms, paralleled GD and DD enzymatic activity trends. These outcomes not only validate the presence of natural B12 form in Ectopseudomonas alcaliphila MSJ19 extract but also demonstrate that AdoCbl is indispensable for GD/DD—mediated bioconversion. Furthermore, the distinct functional differences between natural and synthetic B12 forms, along with the concentration thresholds observed, provide the groundwork for future studies to develop a quantitative enzyme-based assay for B12. These insights also offer a valuable framework for optimizing 3-HP production.
Finally, recombinant expression of the 3-HP pathway in Ectopseudomonas alcaliphila MSJ19 confirms that the host’s endogenously synthesized B12 is not only biologically active but also sufficient to support product formation without external B12 supply. This approach effectively bypasses the tedious processes for B12 extraction, purification and quantification, which often suffer from factors like sample instability, interference from analogs, and the need for advanced instrumentation (Avrămia et al., 2024; Pakeeza et al., 2024). To our knowledge, this is the first instance to report 3-HP production in an extremophilic Ectopseudomonas strain without an exogenous supply of B12. This acts as a beacon for future avenues to improve B12 production in this host and explore its possibilities as a reliable and efficient non-model microbial chassis for other value-added chemicals production.
5 Conclusion
This study presented a comprehensive multifaceted approach to evaluate the bioactivity of B12 from a natural producer. In vitro and in silico investigations have shed light on the specificity of GD and DD toward AdoCbl and revealed the competitive inhibitory effects of other B12 forms, likely due to the absence of the essential 5′-deoxyadenosyl radical. An in vivo approach to evaluate B12 bioactivity has further uncovered the effect of bacterial innate metabolic capability to convert various B12 forms into the catalytically active form. Crude B12 extract from Ectopseudomonas alcaliphila MSJ19 demonstrated 66% of the enzyme activity and 76% of 3-HP production compared to standard AdoCbl, reinforcing its high bioactive potential. Finally, the extremophilic host was able to produce 3.2 mM 3-HP without external B12 supplementation, validating its endogenous B12 biosynthetic capability and eliminating the need for complex B12 extraction procedures. This positions Ectopseudomonas alcaliphila MSJ19 as a promising microbial chassis not only for sustainable B12 production but also for broader application in the production of value-added chemicals. Overall, the study offers a scalable, biologically relevant pipeline to assess B12 bioactivity across microbial systems and diverse sample sources. Thus, it provides both methodological innovation and foundational insights for metabolic engineering of coenzyme B12-dependent pathways.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SV: Writing – original draft, Data curation, Methodology, Visualization, Conceptualization, Investigation, Software, Validation, Project administration, Writing – review & editing, Formal analysis. MS: Supervision, Investigation, Conceptualization, Writing – review & editing, Funding acquisition, Project administration, Writing – original draft, Formal analysis, Resources, Data curation, Validation, Methodology. KL: Validation, Visualization, Formal analysis, Investigation, Writing – review & editing, Data curation, Software.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors gratefully acknowledged the financial support received from Vel Tech Rangarajan Dr. Sagunthala R&D Institute of Science and Technology, under the Seed Fund, Grant No. VTU/Seed Fund/FY 2023-24/019. This support was instrumental in facilitating the successful execution of this research work.
Acknowledgments
The authors would like to thank Sunghoon Park, Biochemical Engineering Laboratory, School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology, Republic of Korea, for providing bacterial strains. The authors are grateful to Guhan Jayaraman, IIT Madras, India, for his generous support in providing access to sophisticated analytical equipment.
Conflict of interest
MS was employed by Park’s Biolabs LLP. KL was employed by Arqgene.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1654548/full#supplementary-material
References
Arasu, M. V., Sarkar, R., Sekar, B. S., Kumar, V., Rathnasingh, C., Choi, J. D. R., et al. (2013). Isolation of a novel Pseudomonas species SP2 producing vitamin B 12 under aerobic condition. Biotechnol. Bioprocess Eng. 18, 43–51. doi: 10.1007/s12257-012-0518-z
Ashok, S., Sankaranarayanan, M., Ko, Y., Jae, K., Ainala, S. K., Kumar, V., et al. (2013). Production of 3-Hydroxypropionic Acid From Glycerol by Recombinant Klebsiella pneumoniae D dhaT D yqhD Which Can Produce Vitamin B 12 Naturally. Biotechnol. Bioeng. 110, 511–524. doi: 10.1002/bit.24726
Avrămia, I., Oroian, M.-A., and Oiţă, R.-C. (2024). A review of current trends of vitamin identification and quantification by chromatography from food samples. J. Food Compos. Anal. 131:106244. doi: 10.1016/j.jfca.2024.106244
Balabanova, L., Pentekhina, I., Nedashkovskaya, O., Degtyarenko, A., Grigorchuk, V., Yugay, Y., et al. (2022). Shift of choline/betaine pathway in recombinant Pseudomonas for cobalamin biosynthesis and abiotic stress protection. Int. J. Mol. Sci. 23:13934. doi: 10.3390/ijms232213934
Bhushan, B., Tomar, S. K., and Mandal, S. (2016). Phenotypic and genotypic screening of human-originated lactobacilli for vitamin B12production potential: process validation by micro-assay and UFLC. Appl. Microbiol. Biotechnol. 100, 6791–6803. doi: 10.1007/s00253-016-7639-9
Bilić, L., Barić, D., Banhatti, R. D., Smith, D. M., and Kovačević, B. (2019). Computational study of glycerol binding within the active site of coenzyme B12-dependent diol dehydratase. J. Phys. Chem. B 123, 6178–6187. doi: 10.1021/acs.jpcb.9b04071
Brown, K. L. (2005). Chemistry and enzymology of vitamin B12. Chem. Rev. 105, 2075–2149. doi: 10.1021/cr030720z
Brunold, T. C. (2005). “Computational studies: B 12 cofactors and their interaction with enzyme active sites” in Encyclopedia of inorganic chemistry. Chichester: John Wiley & Sons.
Bucher, D., Sandala, G. M., Durbeej, B., Radom, L., and Smith, D. M. (2012). The elusive 5′-deoxyadenosyl radical in coenzyme-B 12-mediated reactions. J. Am. Chem. Soc. 134, 1591–1599. doi: 10.1021/ja207809b
Chamlagain, B., Edelmann, M., Katina, K., Varmanen, P., and Piironen, V. (2024). Vitamin B12 production in solubilized protein extract of bioprocessed wheat bran with Propionibacterium freudenreichii. Lwt 192:115731. doi: 10.1016/j.lwt.2024.115731
Chamlagain, B., Peltonen, L., Edelmann, M., Ramos-Diaz, J. M., Kemppinen, A., Jouppila, K., et al. (2021). Bioaccessibility of vitamin B12 synthesized by Propionibacterium freudenreichii and from products made with fermented wheat bran extract. Curr. Res. Food Sci. 4, 499–502. doi: 10.1016/j.crfs.2021.07.009
Deptula, P., Chamlagain, B., Edelmann, M., Sangsuwan, P., Nyman, T. A., Savijoki, K., et al. (2017). Food-like growth conditions support production of active vitamin B12 by Propionibacterium freudenreichii 2067 without DMBI, the lower ligand base, or cobalt supplementation. Front. Microbiol. 8:368. doi: 10.3389/fmicb.2017.00368
Fan, Z., Li, Y., Fan, X., Wang, P., Yang, R., and Xie, C. (2025). Simultaneous determination of three active forms of vitamin B12 in situ produced during fermentation by LC-MS/MS. Foods 14:309. doi: 10.3390/foods14020309
Giedyk, M., Goliszewska, K., and Gryko, D. (2015). Vitamin B12 catalysed reactions. Chem. Soc. Rev. 44, 3391–3404. doi: 10.1039/C5CS00165J
Guo, Y., Li, Y., and Xiang, Y. (2024). Advances in fluorescent nanosensors for detection of vitamin B 12. Crit. Rev. Anal. Chem. 55, 1–11. doi: 10.1080/10408347.2024.2328104
Heal, K. R., Carlson, L. T., Ruxa, D. A. H., Armbrust, E. V., Moffett, J. W., Stahl, D. A., et al. (2014). Determination of four forms of vitamin B12 and other B vitamins in seawater by liquid chromatography/tandem mass spectrometry. Rapid. Commun. Mass Spectrom. 28, 2398–2404. doi: 10.1002/rcm.7040
Kahoun, D., Fojtíková, P., Vácha, F., Čížková, M., Vodička, R., Nováková, E., et al. (2022). Development and validation of an LC-MS/MS method for determination of B vitamins and some its derivatives in whole blood. PLoS One 17, 1–15. doi: 10.1371/journal.pone.0271444
Kajiura, H., Mori, K., Shibata, N., and Toraya, T. (2007). Molecular basis for specificities of reactivating factors for adenosylcobalamin-dependent diol and glycerol dehydratases. FEBS J. 274, 5556–5566. doi: 10.1111/j.1742-4658.2007.06074.x
Kansay, V., Sharma, V. D., Chandan, G., Srivastava, V., Batra, N., Mittal, A., et al. (2024). Smartphone platform for low-cost point-of-care quantitative detection of vitamin B 12 utilizing a biodegradable bioplastic nanocomposite-based fluorescence Nanosensor. ACS Appl. Electron. Mater. 6, 1971–1981. doi: 10.1021/acsaelm.3c01843
Kelly, G. (1997). The coenzyme forms of vitamin b12: toward an understanding of their therapeutic potential. Altern. Med. Rev. 2, 459–471.
Kong, D., Liu, L., Song, S., Kuang, H., and Xu, C. (2017). Development of sensitive, rapid, and effective immunoassays for the detection of vitamin B12 in fortified food and nutritional supplements. Food Anal. Methods 10, 10–18. doi: 10.1007/s12161-016-0543-1
Konings, E., Gill, B. D., Jakobsen, J., Joseph, G., Campos-Giménez, E., Deborde, J. L., et al. (2024). Limitations of current analytical reference methods to determine vitamins in foods: challenges to support regulatory compliance and nutritional composition data. Food Chem. 451:139383. doi: 10.1016/j.foodchem.2024.139383
Koseki, K., Yoshimura, R., Ido, K., Katsuura, K., Bito, T., and Watanabe, F. (2023). Determination of vitamin B12 and folate compounds in commercially available edible seaweed products. Front. Biosci. 15:10. doi: 10.31083/j.fbe1502010
Kumar, V., Durgapal, M., Sankaranarayanan, M., Somasundar, A., Rathnasingh, C., Song, H. H., et al. (2016). Effects of mutation of 2,3-butanediol formation pathway on glycerol metabolism and 1,3-propanediol production by Klebsiella pneumoniae J2B. Bioresour. Technol. 214, 432–440. doi: 10.1016/j.biortech.2016.04.032
Kumar, V., Sankaranarayanan, M., Jae, K. E., Durgapal, M., Ashok, S., Ko, Y., et al. (2012). Co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol using resting cells of recombinant Klebsiella pneumoniae J2B strain overexpressing aldehyde dehydrogenase. Appl. Microbiol. Biotechnol. 96, 373–383. doi: 10.1007/s00253-012-4187-9
Li, P., Gu, Q., Wang, Y., Yu, Y., Yang, L., and Chen, J. V. (2017). Novel vitamin B12-producing Enterococcus spp. and preliminary in vitro evaluation of probiotic potentials. Appl. Microbiol. Biotechnol. 101, 6155–6164. doi: 10.1007/s00253-017-8373-7
Lu, Q., Feng, Y., Zhou, Q., Yang, T., Kuang, H., Xu, C., et al. (2025). A time-resolved fluorescent microsphere Immunochromatographic assay for determination of vitamin B12 in infant formula Milk powder. Biosensors 15:65. doi: 10.3390/bios15020065
Madavi, T. B., Chauhan, S., Ravi, S. N., Venkatesan, S. N., Kulothungan, V. K., Bharathiraja, B., et al. (2024). “Whole-cell catalysts: sustainable green-chemical producing entities” in Whole-Cell Biocatal Next-Generation Technol Green Synth Pharm Chem Biofuels. New York: Apple Academic Press.
Marsh, E. N. G., and Meléndez, G. D. R. (2012). Adenosylcobalamin enzymes: theory and experiment begin to converge. Biochim Biophys Acta. 1824, 1154–1164. doi: 10.1016/j.bbapap.2012.03.012
Masuda, J., Shibata, N., Morimoto, Y., Toraya, T., and Yasuoka, N. (2000). How a protein generates a catalytic radical from coenzyme B12: X-ray structure of a diol-dehydratese- adeninylpentylcobalamin complex. Structure 8, 775–788. doi: 10.1016/S0969-2126(00)00164-7
Möller, K., Krock, B., and Koch, F. (2022). Method optimization of the simultaneous detection of B12 congeners leading to the detection of a novel isomer of hydroxycobalamin in seawater. Rapid Commun. Mass Spectrom. 36, 1–10. doi: 10.1002/rcm.9401
Montoya, L., and Escobar-Briones, E. (2025). Unveiling the significance of prokaryotic composition from ferromanganese crusts regarding the interlink between cobalt and vitamin B12 in deep-sea ecosystems. Front. Microbiol. 16, 1–10. doi: 10.3389/fmicb.2025.1524057
Nakos, M., Pepelanova, I., Beutel, S., Krings, U., Berger, R. G., and Scheper, T. (2017). Isolation and analysis of vitamin B12 from plant samples. Food Chem. 216, 301–308. doi: 10.1016/j.foodchem.2016.08.037
Nasir, A., Ashok, S., Shim, J. Y., Park, S., and Yoo, T. H. (2020). Recent Progress in the understanding and engineering of coenzyme B12-dependent glycerol dehydratase. Front. Bioeng. Biotechnol. 8, 1–12. doi: 10.3389/fbioe.2020.500867
Nguyen-Vo, T. P., Liang, Y., Sankaranarayanan, M., Seol, E., Chun, A. Y., Ashok, S., et al. (2019). Development of 3-hydroxypropionic-acid-tolerant strain of Escherichia coli W and role of minor global regulator yieP. Metab. Eng. 53, 48–58. doi: 10.1016/j.ymben.2019.02.001
Niklewicz, A., Smith, A. D., Smith, A., Holzer, A., Klein, A., McCaddon, A., et al. (2023). The importance of vitamin B12 for individuals choosing plant-based diets. Eur. J. Nutr. 62, 1551–1559. doi: 10.1007/s00394-022-03025-4
Pakeeza, N., Draz, M. U., Yaqub, A., Jafry, A. T., Khan, M., and Ajab, H. (2024). Electrochemical sensing of B-complex vitamins: current challenges and future prospects with microfluidic integration. RSC Adv. 14, 10331–10347. doi: 10.1039/D4RA00555D
Poppe, L., and Rétey, J. (1997). Kinetic investigations with inhibitors that mimic the posthomolysis intermediate in the reactions of coenzyme-B12-dependent glycerol dehydratase and diol dehydratase. Eur. J. Biochem. 245, 398–401. doi: 10.1111/j.1432-1033.1997.00398.x
Prieto-de Lima, T. S., Rojas-Jimenez, K., and Vaglio, C. (2024). Strategy for optimizing vitamin B12 production in Pseudomonas putida KT2440 using metabolic modeling. Meta 14:636. doi: 10.3390/metabo14110636
Raux, E., Lanois, A., Levillayer, F., Warren, M. J., Brody, E., Rambach, A., et al. (1996). Salmonella typhimurium cobalamin (vitamin B12) biosynthetic genes: functional studies in S. Typhimurium and Escherichia coli. J. Bacteriol. 178, 753–767. doi: 10.1128/jb.178.3.753-767.1996
Ravi, S. N., and Sankaranarayanan, M. (2023). Redox balanced co-production of Propanediol and 3-Hydroxypropionic acid from glycerol using novel recombinant Klebsiella quasipneumonia MSN12. J. Inorg. Organomet. Polym. Mater. 33, 3833–3844. doi: 10.1007/s10904-023-02676-y
Ravi, S. N., and Sankaranarayanan, M. (2024). Enhanced synthesis of 3-hydroxypropionic acid by eliminating by-products using recombinant Escherichia coli as a whole cell biocatalyst. Top. Catal. 67, 169–180. doi: 10.1007/s11244-023-01796-6
Reddy KotamReddy, R., Pillai, M., Chaitanya Routhu, K., Reddy Gundala, T., Kumar Bathineni, V., Kotikalapudi, S., et al. Simultaneous quantitation of vitamins B9 and B12 in Milk powder extracted with strata™-X PRO and using a Kinetex™ 2.6 μm F5 column by LC-MS/MS. Phenomenex (2023) 5–7.
Reynolds, P. R., Mottur, G. P., and Bradbeer, C. (1980). Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of BtuC and TonB. J. Biol. Chem. 255, 4313–4319. doi: 10.1016/S0021-9258(19)85667-3
Rizzo, G., Laganà, A. S., Rapisarda, A. M. C., La Ferrera, G. M. G., Buscema, M., Rossetti, P., et al. (2016). Vitamin B12 among vegetarians: status, assessment and supplementation. Nutrients 8, 1–23. doi: 10.3390/nu8120767
Sankaranarayanan, M., Ashok, S., and Park, S. (2014). Production of 3-hydroxypropionic acid from glycerol by acid tolerant Escherichia coli. J. Ind. Microbiol. Biotechnol. 41, 1039–1050. doi: 10.1007/s10295-014-1451-2
Sankaranarayanan, M., Seol, E., Kim, Y., Chauhan, A. S., and Park, S. (2017). Measurement of crude-cell-extract glycerol dehydratase activity in recombinant Escherichia coli using coupled-enzyme reactions. J. Ind. Microbiol. Biotechnol. 44, 477–488. doi: 10.1007/s10295-017-1902-7
Sankaranarayanan, M., Somasundar, A., Seol, E., Chauhan, A. S., Kwon, S., Jung, G. Y., et al. (2017). Production of 3-hydroxypropionic acid by balancing the pathway enzymes using synthetic cassette architecture. J. Biotechnol. 259, 140–147. doi: 10.1016/j.jbiotec.2017.07.027
Santos, A. J. M., Khemiri, S., Simões, S., Prista, C., Sousa, I., and Raymundo, A. (2024). Determination of cobalamin (vitamin B12) in selected microalgae and cyanobacteria products by HPLC-DAD. J. Appl. Phycol. 36, 2625–2633. doi: 10.1007/s10811-024-03273-3
Sauvageot, N., Pichereau, V., Louarme, L., Hartke, A., Auffray, Y., and Laplace, J. M. (2002). Purification, characterization and subunits identification of the diol dehydratase of Lactobacillus collinoides. Eur. J. Biochem. 269, 5731–5737. doi: 10.1046/j.1432-1033.2002.03288.x
Shibata, N., Masuda, J., Tobimatsu, T., Toraya, T., Suto, K., Morimoto, Y., et al. (1999). A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase. Structure 7, 997–1008. doi: 10.1016/S0969-2126(99)80126-9
Shibata, N., Sueyoshi, Y., Higuchi, Y., and Toraya, T. (2018). Direct participation of a peripheral side chain of a Corrin ring in coenzyme B12 catalysis. Angew. Chemie 57, 7830–7835. doi: 10.1002/anie.201803591
Spataru, T. (2024). The miracle of vitamin B12 biochemistry. Reactions 5, 20–76. doi: 10.3390/reactions5010002
Stumpf, L., Schildbach, S., and Coffey, A. (2024). Obtaining novel vitamin B 12 production strains Acetobacter malorum HFD 3141 and Acetobacter orientalis HFD 3031 from home-fermented sourdough. Appl. Microbiol. 4, 986–999. doi: 10.1007/s00223-025-01405-6
Thi Nguyen, T., Lama, S., Kumar Ainala, S., Sankaranarayanan, M., Singh Chauhan, A., Rae Kim, J., et al. (2021). Development of Pseudomonas asiatica as a host for the production of 3-hydroxypropionic acid from glycerol. Bioresour. Technol. 329:124867. doi: 10.1016/j.biortech.2021.124867
Thirupathaiah, Y., Rani, C. S., Reddy, M. S., and Venkateswar, R. L. (2012). Effect of chemical and microbial vitamin B12 analogues on production of vitamin B12. World J. Microbiol. Biotechnol. 28, 2267–2271. doi: 10.1007/s11274-012-1011-8
Toraya, T. (2000). Radical catalysis of B12 enzymes: structure, mechanism, inactivation, and reactivation of diol and glycerol dehydratases. Cell. Mol. Life Sci. 57, 106–127. doi: 10.1007/s000180050502
Toraya, T., and Ishida, A. (1991). Roles of the D-ribose and 5,6-dimethylbenzimidazole moieties of the nucleotide loop of adenosylcobalamin in manifestation of coenzymic function in the diol dehydrase reaction. J. Biol. Chem. 266, 5430–5437. doi: 10.1016/S0021-9258(19)67613-1
Toraya, T., Krodel, E., Abeles, R. H., Mildvan, A. S., Toraya, T., and Krodel, E. (1979). Role of peripheral side chains of vitamin B12 coenzymes in the reaction catalyzed by Dioldehydrase. Biochemistry 18, 417–426. doi: 10.1021/bi00570a005
Toraya, T., Tobimatsu, T., Mori, K., Yamanishi, M., and Shibata, N. (2022). Coenzyme B12-dependent eliminases: Diol and glycerol dehydratases and ethanolamine ammonia-lyase. Methods Enzymol. 668, 181–242. doi: 10.1016/bs.mie.2021.11.027
Toraya, T., Ushio, K., Fukui, S., and Hogenkamp, H. P. C. (1977). Studies on the mechanism of the adenosylcobalamin dependent diol dehydrase reaction by the use of analogs of the coenzyme. J. Biol. Chem. 252, 963–970. doi: 10.1016/S0021-9258(19)75192-8
Trad, F. M., AlHamad, T., Younes, N., Abunasser, S., Younes, S., Nizamuddin, P. B., et al. (2025). Accre 8 emerging point of care CLIA system for vitamin B12 assessment compared with three established assays. Sci. Rep. 15, 1–13. doi: 10.1038/s41598-025-97503-4
Venkatesan, S. N., Sankaranarayanan, M., and Bharathiraja, B. (2024). Development of novel method for the precise isolation of vitamin B 12 producing microorganisms from natural sources. Biocatal. Agric. Biotechnol. 2025, 1–13. doi: 10.1016/j.bcab.2025.103512
Vincenti, A., Bertuzzo, L., Limitone, A., D’antona, G., and Cena, H. (2021). Perspective: practical approach to preventing subclinical b12 deficiency in elderly population. Nutrients 13, 1–16. doi: 10.3390/nu13061913
Wang, F., Qu, H., Tian, P., and Tan, T. (2007). Heterologous expression and characterization of recombinant glycerol dehydratase from Klebsiella pneumoniae in Escherichia coli. Biotechnol. J. 2, 736–742. doi: 10.1002/biot.200600101
Wei, X., Meng, X., Chen, Y., Wei, Y., Du, L., and Huang, R. (2014). Cloning, expression, and characterization of coenzyme-B12-dependent diol dehydratase from Lactobacillus diolivorans. Biotechnol. Lett. 36, 159–165. doi: 10.1007/s10529-013-1346-8
Wernick, D. G., Pontrelli, S. P., Pollock, A. W., and Liao, J. C. (2016). Sustainable biorefining in wastewater by engineered extreme alkaliphile Bacillus marmarensis. Sci. Rep. 6, 1–10. doi: 10.1038/srep20224
Xie, C., Coda, R., Chamlagain, B., Varmanen, P., Piironen, V., and Katina, K. (2019). Co-fermentation of propionibacterium freudenreichiiand lactobacillus brevisin wheat bran for in situproduction of vitamin B12. Front. Microbiol. 10, 1–10. doi: 10.3389/fmicb.2019.01541
Yamada, S., Yamada, K., Nishikawa, N., Hioki, R., Nirasawa, M., Kii, K., et al. (2004). Determination of vitamin B12 using the enzyme glycerol dehydrase. Scand. J. Clin. Lab. Invest. 64, 185–194. doi: 10.1080/00365510410001158
Yamanishi, M., Yunoki, M., Tobimatsu, T., Sato, H., Matsui, J., Dokiya, A., et al. (2002). The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol. Eur. J. Biochem. 269, 4484–4494. doi: 10.1046/j.1432-1033.2002.03151.x
Yang, Y., Zhou, B., and Zheng, C. (2024). The fast quantification of vitamin B12 in Milk powder by high-performance liquid chromatography-inductively coupled plasma mass spectrometry. Molecules 29:1795. doi: 10.3390/molecules29081795
Yumoto, I., Yamazaki, K., Hishinuma, M., Nodasaka, Y., Suemori, A., Nakajima, K., et al. (2001). Pseudomonas alcaliphila sp. nov., a novel facultatively psychrophilic alkaliphile isolated from seawater. Int. J. Syst. Evol. Microbiol. 51, 349–355. doi: 10.1099/00207713-51-2-349
Zeng, Q., Man, X., Huang, Z., Zhuang, L., Yang, H., and Sha, Y. (2023). Effects of rice blast biocontrol strain Pseudomonas alcaliphila Ej2 on the endophytic microbiome and proteome of rice under salt stress. Front. Microbiol. :14(March). doi: 10.3389/fmicb.2023.1129614
Keywords: cobalamin, Pseudomonas alcaliphila , bioactivity, 3-hydroxypropionic acid, in vitro , in silico , in vivo
Citation: Venkatesan SN, Sankaranarayanan M and Loganathan K (2025) Functional characterization of vitamin B12 from an extremophile—Pseudomonas alcaliphila and assessment of its microbial chassis potential. Front. Microbiol. 16:1654548. doi: 10.3389/fmicb.2025.1654548
Edited by:
Muthusamy Govarthanan, Kyungpook National University, Republic of KoreaReviewed by:
Jiangxin Wang, Shenzhen University, ChinaSi-Yu Li, National Chung Hsing University, Taiwan
Copyright © 2025 Venkatesan, Sankaranarayanan and Loganathan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mugesh Sankaranarayanan, ZHJtdWdlc2hzQHZlbHRlY2guZWR1Lmlu
 Sathya Narayanan Venkatesan
Sathya Narayanan Venkatesan Mugesh Sankaranarayanan
Mugesh Sankaranarayanan Karthik Loganathan
Karthik Loganathan