- 1Animal Health Laboratory, EU/WOAH and National Reference Laboratory for Brucellosis, Anses/Paris-Est University, Maisons-Alfort, France
- 2ANSES, Ploufragan-Plouzané Niort Laboratory, Zoopôle, Ploufragan, France
- 3Departmental Laboratory of Haute-Garonne (LD31 EVA), Launaguet, France
- 4DDETSPP de l’Aveyron, Bouran, France
- 5DDPP des Pyrénées-Atlantiques, Pau, France
Many species from the genus Brucella are causative agents of the bacterial zoonosis brucellosis. Until recently, it was generally believed that these bacteria exhibit strict host specificity; however, recent findings suggest otherwise. Brucella microti is an atypical Brucella species, no threat to humans, with a broad host spectrum, primarily found in wildlife and rodents, and is the only Brucella species isolated from soil, aquatic environments, and frogs, suggesting its environmental persistence and adaptability to diverse ecological niches. Despite its environmental resilience and wide host range, B. microti has not been detected in domestic animals. This study, for the first time, shows the ability of B. microti to infect domestic small ruminants. During the 2024 prophylaxis campaigns across three farms in two French departments, two sheep and one goat tested positive on classical serological tests for brucellosis. Following bacteriological isolation, HRM-PCR and classical biotyping methods classified the strains as B. microti, rather than the expected zoonotic Brucella spp. (B. abortus, B. suis, and B. melitensis). Hybrid whole-genome sequencing, whole genome single nucleotide polymorphism (wgSNP), and multiple Loci variable-number tandem repeat analysis (MLVA) revealed that the three isolates were genetically closer to the reference B. microti CCM4915 strains, isolated in Central Europe, than previously detected French strains from farmed frogs. The infection of small ruminants by B. microti is even more unusual, as no strain-specific antimicrobial resistance or virulence genes were identified. These findings underscore the need for new diagnostic tools that can identify Brucellae on the species level for proper management and monitoring, particularly in regions with epizootic risks. Further research is essential to clarify the role of B. microti in animal health and risks for public health.
Introduction
Brucellosis is a bacterial zoonotic disease caused by gram-negative bacteria of the Brucella genus, usually transmitted from livestock to humans via the consumption of raw and unpasteurized animal food products. The disease remains a major global public health concern, with an estimated 1.6 to 2.1 million new human cases annually (Laine et al., 2023).
The genus Brucella includes numerous species (LPSN: https://lpsn.dsmz.de/genus/brucella). For clarity, in this study, the term Brucella only includes a unique monophyletic clade (Leclercq et al., 2020), which maintains both IS711 and bcsp31 genes (Bounaadja et al., 2009; Aljanazreh et al., 2023; Sanjuan-Jimenez et al., 2017), that have been used for molecular diagnostics. Previously, the Brucellae carrying both IS711 and bcsp31 comprised 14 species of facultative intracellular bacteria. Six of these species were grouped as classical and recognized as members of the core clade basis on their pathogenicity, host preferences, and adherence to phenotypic characteristics, including B. abortus (cattle), B. melitensis (goats or sheep), B. suis (swine), B. ovis (sheep and goats), B. canis (dogs), and B. neotomae (desert rats) (Occhialini et al., 2022; Olsen and Palmer, 2014; Suárez-Esquivel et al., 2020; Whatmore and Foster, 2021). This clade additionally includes the species B. ceti and B. pinnipedialis, which have been described more recently and isolated from marine mammals (Cloeckaert et al., 2001; Foster et al., 2007; Orsini et al., 2022). In the last two decades, advances in field research, pathogen detection, and molecular typing have made it possible to identify new species in addition to the strains not yet classified (Occhialini et al., 2022; Suárez-Esquivel et al., 2020). Two new species, B. amazoniensis and B. nosferati, isolated from Brazilian gold miners working in the Amazon rainforest (About et al., 2023) and Costa Rican bats, respectively (Hernández-Mora et al., 2023), can be considered part of the core clade Brucella based on phylogenetic analyses. However, we still do not know how these species are transmitted, what the animal reservoirs are, or their evolution in relation to other Brucella species. Additionally, four Brucella species, including B. microti (prevalent in common voles, red foxes, and wild boars), B. inopinata (in humans), B. papionis (in baboons), and B. vulpis (in red foxes), were identified in a wide range of hosts. These species can be distinguished from classical Brucellae by atypical phenotype, which includes altered metabolism, higher metabolic activity, faster growth, different composition of lipopolysaccharide, and presence of flagella (Occhialini et al., 2022). Due to these differences, new strains can be classified as a typical Brucella species, thereby contributing to the increased diversity within this genus (Occhialini et al., 2022). At the same time, applying a second tier of classification based on genetic homology shows that B. microti and B. papionis group more closely with the classical core clade species owing to their higher genomic similarity, whereas other atypical species remain genetically distant and thus appropriately classified as non-core clade Brucella (Occhialini et al., 2022).
B. microti was the first atypical Brucella species to demonstrate a broad host spectrum, with notable occurrences in the common vole (Microtus arvalis) in the Czech Republic (Scholz et al., 2008), mandibular lymph nodes of red foxes (Vulpes vulpes) from Austria (Scholz et al., 2009), and wild boars (Sus scrofa) from Hungary (Rónai et al., 2015). In experimental infections, B. microti exhibited high pathogenic potential, causing death in murine models, similar to classical Brucella spp. known to affect humans, thereby highlighting zoonotic risks (Jiménez et al., 2010). To date, no confirmed human infections have been reported. However, there was a suspected case in which, based on the clinical course, a clear epidemiological link (a bite from an infected rodent), isolation of the pathogen from a sick vole, and a specific serological response in the human patient, led to the conclusion that the disease was likely caused by B. microti (Hubálek et al., 2023).
Furthermore, B. microti was the only Brucella species to be isolated from soil (Scholz et al., 2008), indicating its environmental persistence, which diverges from the facultative intracellular evolution of classical core clade Brucella spp. Additionally, B. microti was also isolated from aquatic environments (Jaÿ et al., 2020), and the first strain was recovered from frogs (Pelophylax ridibundus) raised for human consumption (Jaÿ et al., 2018), further emphasizing its ability to persist in diverse ecological niches. The innate ability of B. microti to withstand acidic environments, ranging from moderate to extreme pH levels (Occhialini et al., 2022; Damiano et al., 2015; Freddi et al., 2017; de la Garza-García et al., 2021; Occhialini et al., 2012), along with its adaptation to anoxic conditions (Freddi et al., 2023), as well as accelerated metabolism, is associated with enhanced nutrient utilization and enzymatic activities (Al Dahouk et al., 2010; Al Dahouk et al., 2012). Furthermore, heterogeneity in LPS genes may explain its adaptability to environmental conditions. This environmental resilience, alongside its presence in a wide range of mammalian hosts, underscores its open lifestyle, making it distinct from more host-restricted Brucella species. However, to date, to the best of our knowledge, no identification of B. microti has been reported in cattle, small ruminants, or pigs, highlighting a gap in understanding its potential to colonize livestock and raising questions about its true zoonotic potential. With this study, the ability of B. microti to infect small ruminants is confirmed, shedding light on the potential risks and the need for proper management and identification of this bacterium in animals, especially from epizootic regions.
Materials and methods
Bacterial cultivation and strain isolation
All collected samples were analyzed at the Department laboratory of Haute-Garonne (LDA31EVA, Launaguet, France) using routine bacteriological procedures in the local veterinary diagnostic laboratories, in accordance with the U47-105 normative and French safety regulations in force. Thus, during slaughter of seropositive animals, three pairs of lymph nodes were collected aseptically, out of which 10 g were homogenized and diluted in 1/2 to 1/5 ratios in phosphate-buffered saline solution (0.9% NaCl PBS). The homogenate was then plated onto four Brucella selective Farrell media. Two plates were incubated at 37°C with 5% CO2, and two plates without CO2 were incubated for up to 10 days. The isolates suspected to be Brucella were transferred to the French National Reference Laboratory for Animal Brucellosis (ANSES, Maisons-Alfort, France) to confirm the Brucella genus and determine the species, by safety regulations.
Phenotypic identification and characterization
Isolates were characterized using standard procedures, following the World Organization for Animal Health (WOAH) guidelines, in a BSL-3 facility. The strains were biotype based on colonial morphology, Gram staining, CO2 requirement, H2S production, oxidase and urease activity, growth on dyes (basic fuchsin and thionin), lysis by phages (Tb, Wb, Iz, R/C), and agglutination with monospecific sera (anti-A, anti-M, and anti-R).
Serological analyses
The Rose Bengal Test (RBT) and Complement Fixation Test (CFT) were performed on sera following the WOAH guidelines. Both diagnostic tests detect antibodies against smooth Brucella spp. CFT results were expressed as a titer (ICFTU/mL) with a positivity threshold of 20 ICFTU/mL.
Molecular analyses
Genomic DNA was extracted using the commercial QIAGEN QIAamp DNA minikit (QIAGEN, Germany) following the manufacturer’s instructions. The real-time PCR (qPCR) was performed using the commercial qualitative Brucella spp. detection kit, ID Gene™ Brucella spp. triplex (Innovative Diagnostics, Montpellier, France), which targets the specific IS711 insertion sequence, according to the manufacturer’s instructions. The PCR mix already contains primers and probes in the kit, and the following PCR program was used: denaturation at 95°C for 2 min, followed by 40 cycles of amplification at 95°C for 10 s and hybridization and elongation at 60°C for 30 s. Multiple Locus Variable-number Tandem Repeat Analysis (MLVA)-16 (Le Flèche et al., 2006) and High-Resolution Melting (HRM)-PCR (Girault et al., 2022) analyses were performed according to previously published protocols (Al Dahouk et al., 2010; Al Dahouk et al., 2012). The individual DNA samples of three isolates were typed with the MLVA-16 panel single-plex PCR, and the agarose gel method was used for amplicon identification. Clustering and congruence analyses were conducted with BioNumerics 7.6.3 (BioMérieux), using data as a character dataset via the categorical distance coefficient and MST (Minimum Spanning Tree) method. A total of 127 MLVA-16 profiles, including core (n = 81) and non-core clade (n = 46) Brucella available in the MLVA database1 or relative publications, were used in the analyses (Supplementary Table S1).
The short and long reads whole genome sequencing (WGS) of isolated strains was performed using Illumina DNA Prep kit (Illumina) and Rapid Barcoding Kit 24 V14 (Oxford Nanopore), respectively. The sequencing runs were performed on NextSeq 2000 equipment (Illumina) at the ANSES sequencing platform facility (ANSES, Ploufragan-Plouzané-Niort laboratory, France) and on MinION using Flow Cell R10.4.1 at the French National Reference Laboratory for Animal Brucellosis (Laboratory for Animal Health, ANSES, Maisons-Alfort, France). Raw reads were trimmed using Trimmomatic 0.36 for Illumina data and Nanofilt 1.10 (De Coster et al., 2018) for MinION data to remove low-quality bases. For Illumina data, trimming was performed with the following parameters: leading 3, trailing 3, sliding window 4:15, and minlen 50. For MinION data, trimming was performed using the -q 10 parameter to remove reads with a quality score below 10. A hybrid de novo assembly, combining Illumina and MinION raw reads, was performed using Unicycler 0.5.0 (Wick et al., 2017) to produce a more accurate and complete assembly. Finally, QUAST 5.2.0 was used to assess assembly robustness by gathering extensive assembly statistics. The three assemblies generated during this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB89093.2
The whole genome single nucleotide polymorphism (wgSNP) analysis was performed using BioNumerics version 7.6.3 (BioMérieux) to trace back the source of infection. The genome of B. melitensis 16 M was used as a reference for comparative analyses across the entire Brucella genus, while the genome of B. microti CCM4915 served as the reference for comparisons within the only B. microti species. A total of 49 available Brucella genome sequences, representing all known Brucella species of the core clade (n = 38) and non-core clade strains (n = 11), were used in this study for comparative analysis (Supplementary Table S1). Chimeric genomes of chromosomes 1 and 2 were generated to compare complete and draft genomes (Huang et al., 2012). A minimum set of position filters was applied on the SNP matrix: (i) contiguous SNPs were removed (if found in a 10 bp-window), (ii) with non-informative SNPs, (iii) a required minimum of 15-fold coverage for each SNP, and (iv) ambiguous (i.e., non-ACGT bases) and unreliable bases (i.e., Ns) were discarded. The refined SNP matrix was used to generate a maximum likelihood tree based on the General Time Reversible model with 200 repetitions for bootstrap using MEGA version 11.0.13 (Tamura et al., 2021).
To target genes and/or regions potentially involved in the AMR, plasmid identification, and virulence (VG), all available B. microti genomes were screened using Abricate version 1.0.13 as described in previous research (Girault et al., 2024). In summary, Abricate was run with entries from seven databases: for antimicrobial resistance genes AMRFinderPlus (NCBI) (Feldgarden et al., 2019), Comprehensive Antibiotic Resistance Database (CARD) (Jia et al., 2017), ResFinder (Zankari et al., 2012), and MEGARes 2.00 (Doster et al., 2020), for the virulence genes virulence factor database (VFDB) (Chen et al., 2016), and in-house database (BRUgenes), while for plasmid presence, PlasmidFinder (Carattoli et al., 2014) was used.
Results
Farms description
The first farm, located in the Aveyron department of France, operates a dual livestock system, combining both cattle and sheep husbandry. The beef herd consists of around forty Aubrac or Aubrac-cross dairy cows, with a grazing system for calves. The dairy sheep flock regroups 750 to 800 Lacaune ewes, depending on the year. The milk from these ewes is collected for the production of local fresh cheese.
The second farm, a dairy sheep farm, located in the Pyrénées-Atlantiques department, has a mixed livestock population, including six breeding rams, 250 ewes (over 18 months), 15 lambs (6–18 months), 80 bovines, and 50 horses during summer pastures. The epidemiological investigation highlighted the presence of domestic pets (three cats and dogs) at the farm, along with possible contact of breeding animals with wildlife species such as wild boar, roe deer, hares, rabbits, foxes, vultures, and bats. Stray animals were not observed on the farm. The type of feed provided included hay, regrowth forage, and complementary feed (corn, alfalfa).
The third farm, a dairy goat farm located in the same Pyrénées-Atlantiques department, which practices hand milking, is exclusively dedicated to dairy production and comprises 42 adult goats (over 12 months) and 14 younger animals (including approximately two bucks). The epidemiological investigation noted regular exposure to wildlife, with sightings of roe deer, foxes, martens, wild boar, and various birds within a fenced wooded area. In addition, about twenty feral goats were observed. These animals had been gathered and restrained by local breeders and were destined for export to Spain at the end of December 2023. Furthermore, the farm operates a communal management system on a shared parcel together with three other local breeders. The feeding regimen consisted primarily of natural forages, including brambles and woodland vegetation, supplemented by corn.
Indirect diagnostics
All indirect diagnostic tests were carried out during the routine national prophylaxis campaign conducted by laboratories in two different French departments in 2024. As part of the farm’s brucellosis control measures, a sample of 50 ewes was collected from the first farm in May. Only one animal resulted serologically positive on RBT and showed an inconclusive result on CFT due to an anti-complementary reaction, without any clinical symptoms evocative of brucellosis. Although it is necessary to repeat the diagnosis after six to 8 weeks, the sanitary authority decided to proceed with the diagnostic slaughter (investigative culling) of seropositive sheep to expedite the process.
On March 18th, blood samples were collected from 57 sheep at the second farm. Serological testing identified one positive sheep in RBT and CFT (with an antibody titer of 853 ICFTU/mL). A follow-up test on April 30th confirmed the seropositive status of the same animal, with a decreased CFT titer of 213 ICFTU/mL. In addition, the animal tested positive again on June 20th, with a significantly increased CFT titer of 1,707 ICFTU/mL, which reduces the likelihood of a false-positive serological result. The Brucella seropositive sheep showed no clinical signs related to Brucellosis infection.
As part of the same prophylaxis campaign in the Pyrénées-Atlantiques department, blood samples were collected from 47 goats on the third farm on April 29th. Of the 47 sera tested, only one was found to be positive on RBT and CFT, with an antibody titer of 26.6 ICFTU/mL. A confirmatory test conducted on June 14th reaffirmed the positive result, although the CFT titer had decreased to 20 ICFTU/mL. As with the sheep, the goat showed no signs of brucellosis infection.
Bacterial isolation and molecular diagnostics
Following the national brucellosis control program, bacteriological investigations were initiated for three animals following the seropositive results. These investigations took place on May 28th at the first farm and on July 27th at two other farms. Three pairs of lymph nodes (retropharyngeal, genital, and retromammary) from each animal were collected and sent to an accredited Department laboratory of Haute-Garonne (LDA31EVA, Launaguet, France) for Brucella culture diagnosis. Following cultivation, four Brucella spp. suspect colonies were isolated from three animals. From genital lymph nodes, one colony was isolated from a sheep from the first farm and a goat from the third farm. From the retromammary lymph nodes, one colony originated from a sheep from the second farm and the goat from the third farm. To confirm the Brucella genus identification and determine the species, all four suspected strains were transferred to the French National Reference Laboratory for brucellosis. After total DNA extraction, the qPCR analysis amplified the IS711 Brucella gene. The targeted gene was amplified in all four strains, resulting in a positive signal indicating that the strains belong to the genus Brucella. When these DNAs were subsequently tested for rapid identification and differentiation of the Brucella genus in HRM-PCR, the melting curve profiles matched with Brucella microti, instead of the expected classical smooth Brucella species like B. abortus, B. suis, and B. melitensis, which were the first expected diagnosis, regarding the host species.
Genomic and bacteriological identification and characterization
The standard bacteriological phenotypic identification of the four isolates (code numbers 24–6,286-7554 for the first farm, 24–6,283-7553 for the second farm, and 24–6,281-7551 and 24–6,281-7552 for the third farm) confirmed the presence of the Brucella genus, with biotyping traits consistent with B. microti (Table 1). In particular, the newly identified strains showed agglutination only with anti-M, but not with anti-A and anti-R monospecific sera, consistent with the reference strain B. microti CCM 4915. In contrast, the 2017 French frog isolate (17–2,122-4144) agglutinated with only anti-A monospecific sera (Table 1).
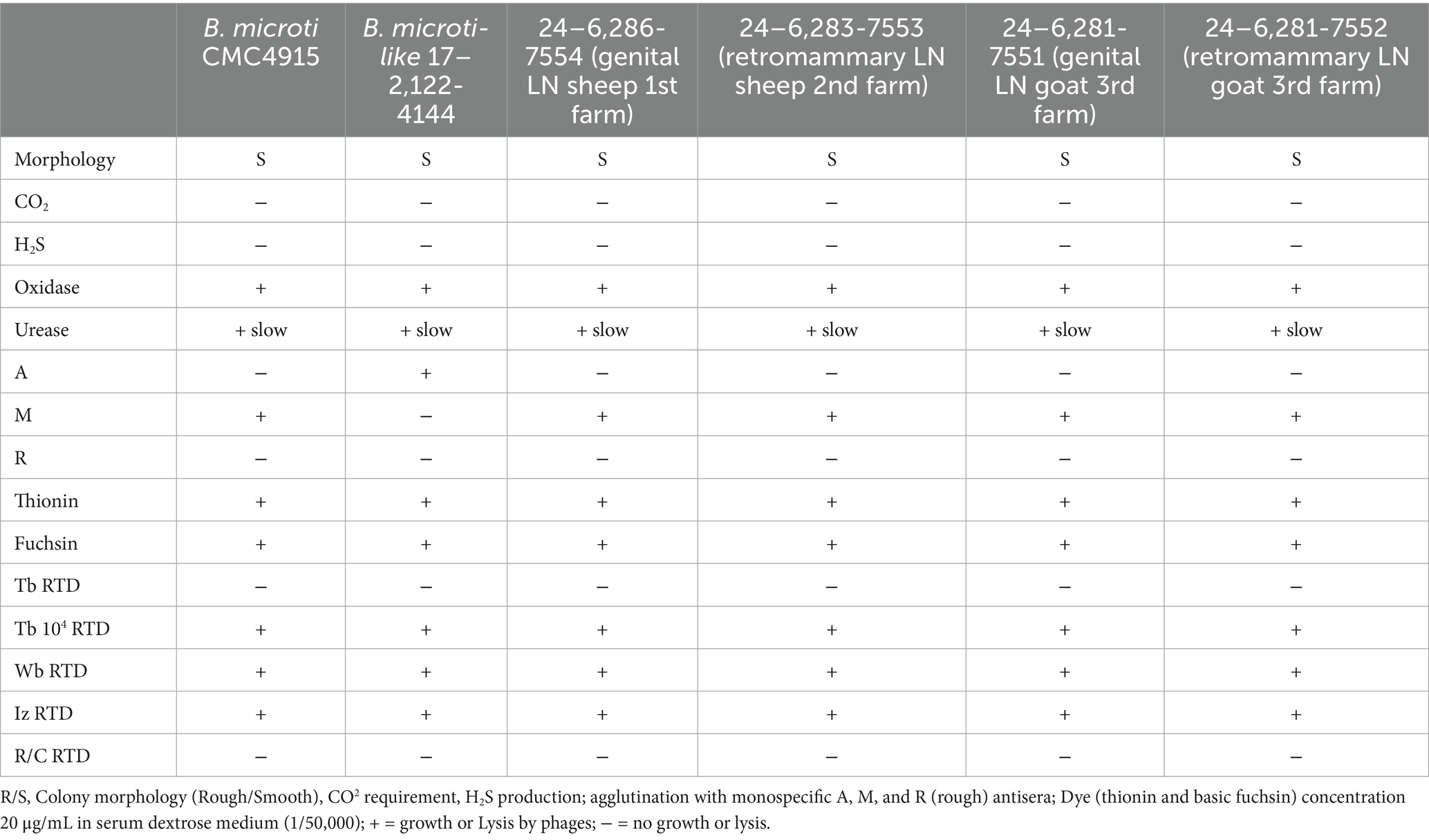
Table 1. Classical phenotypic characterization of the four isolated strains (code numbers 24–6,286-7554 for 1st farm, 24–6,281-7551 and 24–6,281-7552 for 2nd farm, and 24–6,283-7553 for 3rd farm) from the three farms, compared to the reference B. microti CMC4915 strain and B. microti-like 17–2,122-4144 isolated from the marsh frog.
To genotype the strains and potentially determine the source of the infection, phylogenetic investigations were performed on one strain per animal and farm. The MLVA analysis confirmed the B. microti identity of three tested isolates from three farms (Figure 1). All three isolates (colored in blue) clustered together with the known B. microti strains (colored in turquoise) and, in particular, perfectly matched with B. microti CCM 4915, published by Audic et al. (2009) (Supplementary Figure 1).
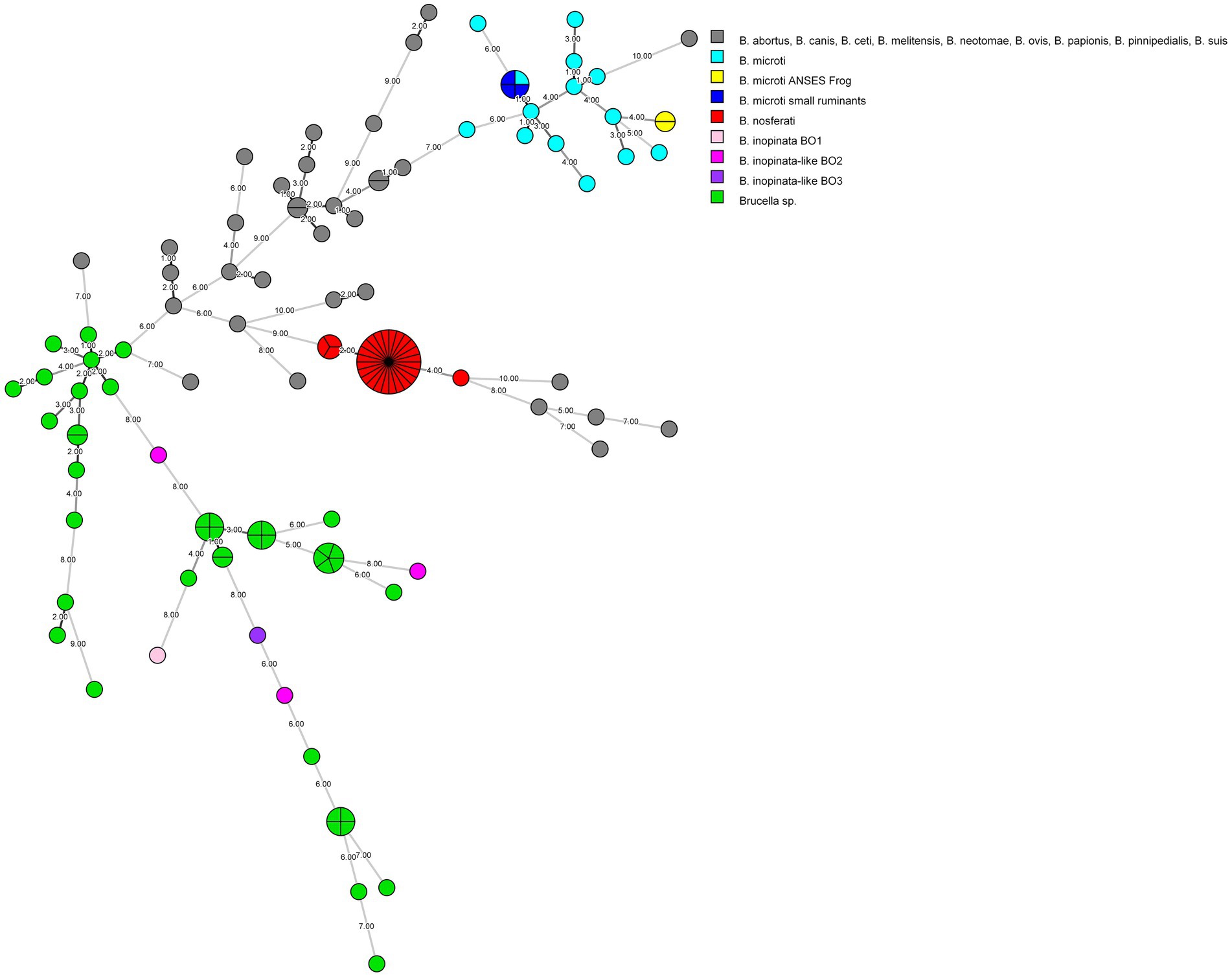
Figure 1. MLVA-16 minimum spanning tree describing relationships of strains investigated in this study, as well as all core (n = 81) and non-core clade (n = 46) Brucella species and strains. Clustering and partitioning were generated with BioNumerics, using data as a character dataset with a categorical distance coefficient and the minimum spanning tree method. Grey circles represent MLVA-16 genotypes of B. abortus, B. canis, B. ceti, B. melitensis, B. neotomae, B. ovis, B. papionis, B. pinnipedialis, B. suis, while others are colored with respect to available strains of B. inopinata, B. inopinata-like, B. microti, B. nosferati, and Brucella sp., including this study. The size of the circle indicates the number of strains corresponding to that genotype. The branch labels and numbers account correspond to the number of differing loci between nodes.
To further characterize the strains, hybrid whole-genome sequencing using both long- and short-read methods was performed, aiming to obtain a high-precision assembly. In total, two contigs corresponding to the two chromosomes were identified for all three sequenced isolates. The total genome sizes were 3,338,075 bp, 3,338,083 bp, and 3,338,107 bp for isolates 24–6,281-7551, 24–6,283-7553, and 24–6,286-7554, respectively. The wgSNP analysis was conducted on available Brucella species sequences representing both core (n = 38) and non-core (n = 11) clades (Supplementary Table 1) to determine the genomic relationship of isolated strains. Phylogenetic comparative whole-genome SNP analysis showed that three strains clustered with other known B. microti strains (Figure 2). Notably, when using B. melitensis 16 M as the reference, out of 31,481 filtered SNPs, the two sheep isolates (24–6,286-7554 and 24–6,283-7553 from farms one and two, respectively) showed no differences compared to the reference B. microti CCM 4915 strain. In contrast, the goat isolate (24–6,281-7551 from the farm three) exhibited only one SNP difference, while the frog isolate (17–2,122-4144) showed 11 SNPs difference, compared to the CCM 4915 strain. When comparing only B. microti isolates aligned to the B. microti CCM 4915 reference genome, out of 146 filtered SNPs, the two sheep isolates (24–6,286-7554 and 24–6,283-7553 from farms one and two, respectively) displayed a difference of seven SNPs relative to the reference strain. The goat isolate (24–6,281-7551 from the farm three) exhibited eight SNPs, while the frog isolate (17–2,122-4144) showed differences in 142 SNPs compared to the CCM 4915 strain.
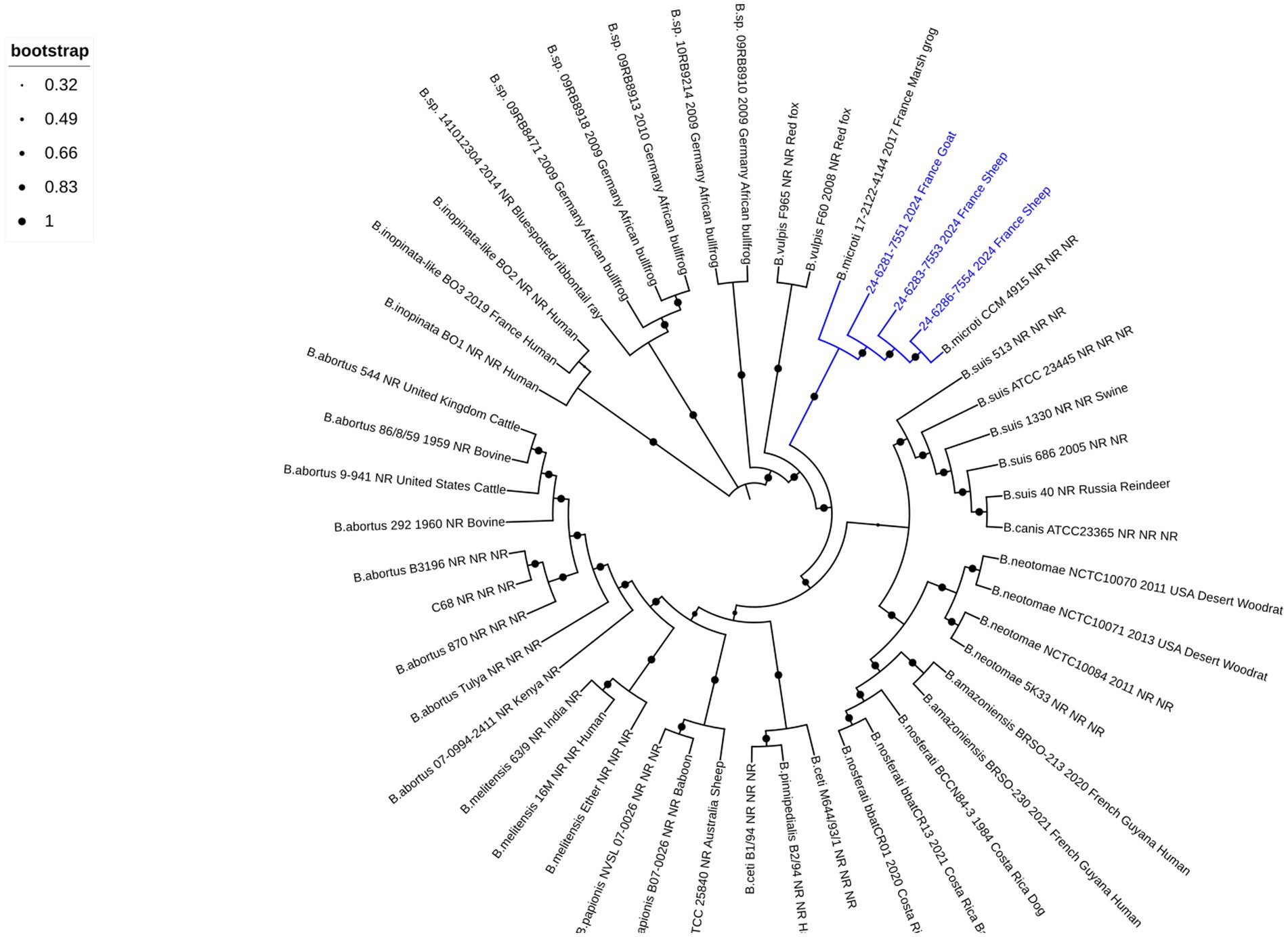
Figure 2. Phylogenetic comparative whole-genome SNP analysis of the small ruminant strains investigated in this study and all Brucella reference strains. The dendrogram was constructed using the maximum likelihood method with 200 bootstrap repetitions based on SNPs matrix (31′481 filtered SNPs). The phylogenetic tree was visualized with the EMBL online tool “Interactive Tree of Life” (iTOL v6) and annotated with four concatenated datasets separated based on underscores (Brucella species, strain name, year of isolation, isolation country, and host), colored in black and blue for all species and three isolates from small ruminants, respectively. The blue color was used to identify the B. microti branches. Not reported data are marked with “NR.” A log scale is used in the tree, allowing a better distinction between isolates.
Finally, to better characterize the B. microti isolates from small ruminants and identify potential differences with other known B. microti species, an in-silico analysis was performed targeting the presence of plasmids, antimicrobial resistance (AMR), and virulence genes. No plasmids were detected in any of the B. microti strains when screening the assemblies against the PlasmidFinder database. Similarly, no antibiotic resistance genes were found when screening the NCBI and ResFinder databases. However, when searching through the CARD and MEGARes databases, six genes (mprF, bep C, D, E, F, and G) involved in AMR mechanisms were identified in all examined genomes, with a minimum identity percentage of 99.64%. Furthermore, all 53 Brucella spp. virulence genes were identified in the four B. microti genomes from this study as well as the previous frog isolate, with a minimal identity percentage of 97.08%, primarily associated with host immune evasion, intracellular survival, and the regulation and expression of the type IV secretion system in Brucella spp., based on the VFDB and BRUgenes databases.
Discussion
Brucellosis is a highly contagious bacterial zoonosis, primarily transmitted to humans through the consumption of contaminated, unpasteurized dairy products, undercooked meat, or direct contact with infected livestock and their reproductive materials. The major species responsible for brucellosis in humans, B. melitensis and B. abortus, are typically associated with livestock, making comprehension of transmission paramount for control measures and public health protection. No B. microti human cases have been reported worldwide, even if a strong exposure to this bacterial species occurred in French farmed frogs destined for human consumption (Jaÿ et al., 2020; Jaÿ et al., 2018). In the case of B. microti, the transmission pathways are unknown. Given the growing recognition of B. microti as a pathogenic species and the presence of all 53 Brucella spp. known virulence genes, the absence of confirmed human infection remains surprising. Indeed, B. microti appears highly pathogenic in common voles and other rodents (Hubálek et al., 2007), as confirmed by experimental infections in murine models (Jiménez et al., 2010; Hubálek et al., 2007; Hanna et al., 2011; Ouahrani-Bettache et al., 2019). The absence of human cases might be partly explained by a lower pathogenicity in non-rodent species (Rudolf et al., 2024). In frogs, only a low percentage of animals showed clinical symptoms (Jaÿ et al., 2020; Jaÿ et al., 2018), as well as in foxes (Scholz et al., 2009) and wild boars (Rónai et al., 2015). According to these data, B. microti is likely to behave as an opportunistic soil bacterium that occasionally infects mammals (other than rodents) through the ingestion of contaminated products, with limited potential for sustained transmission in these hosts (Rudolf et al., 2024).
Our study reports for the first time that B. microti is capable of infecting livestock, particularly small ruminants, thereby expanding the known host range of this emerging pathogen. B. microti exhibits several physiological characteristics, in particular, expanded metabolic activity, compared to classical zoonotic Brucella spp. This enhanced metabolic flexibility suggests a greater ability to survive and proliferate in environmental reservoirs. This may also contribute to the capacity of B. microti to infect a broader range of hosts. Therefore, host hopping could represent a key mechanism driving the spread of these epizootic species (Occhialini et al., 2022). While B. microti has been primarily considered as a pathogenic bacterium for rodents and wildlife, the potential for transmission to livestock has not been thoroughly investigated (Rudolf et al., 2024). The detection of B. microti in livestock may represent a previously overlooked link in the potential transmission pathways, bridging the gap between environmental reservoirs, wildlife, and domestic animals. Cross-species spill-over transmission is strongly influenced by the frequency, duration, and nature of contacts. Opportunities for human intervention strongly impact this parameter (Lloyd-Smith et al., 2009). In Europe, due to the increased size of some wildlife populations in agricultural areas, such as ungulates or suidae, together with a progressive reduction of the usable agricultural area (Perpiña et al., n.d.), the probability of contact between domestic and wild species has increased during the past decades (Ledger et al., 2022). This may lead to potential emergence of wildlife pathogens, as previously observed in domestic dogs infected with B. suis biovar 2, strains presenting high genomic similarities with those circulating in hares and wild boars (Girault et al., 2023).
At the same time, the fact that only one animal per farm was infected may indicate that either these infections were accidental, suggesting the opportunistic nature of B. microti, or that the current surveillance system can identify early onsets of farm infection and therefore protect against greater spread and economic losses. It is important to note that no total culling of the herds was carried out as recommended for control of zoonotic Brucella (B. abortus, B. melitensis, and B. suis), but only the positive animal was removed, and at present, no other positive serological results have been observed. These data suggest that the B. microti infections observed in small ruminants are accidental, indicating the opportunistic nature of infection, similar to Ochrobactrum spp. (Ryan and Pembroke, 2020; Thoma et al., 2009), rather than indicative of widespread transmission within herds, like those of classical Brucella species (Corbel et al., 2006; Godfroid et al., 2013). This raises the need to evolve current regulations by considering all Brucella species, beyond just the controlled zoonotic ones, and defining the appropriate measures based on the bacterial species-host combination. Nevertheless, the performance characteristics of the CFT and RBT tests in ruminants infected with B. microti have not been established. Therefore, the absence of detectable antibodies or intermittent and short-term seropositivity in seronegative animals cannot be excluded. Experimental infection studies in ruminants with B. microti would be valuable to generate validation data for these serological assays, as well as to explore pathogenicity in these hosts, since no clinical symptoms have been observed in these three cases.
The detection of B. microti in small ruminants underscores a significant challenge in current brucellosis monitoring strategies. Traditional serological tests, commonly used to detect classical Brucella infections in livestock, rely on the detection of antibodies against the lipopolysaccharides (LPS), specifically the O-polysaccharide (OPS) component, which links to the outer core and extends into the extracellular environment (Erridge et al., 2002; Mancilla, 2015). These OPS components are found in all smooth Brucella spp., where the relative abundance and distribution of a homopolymeric linear chain of N-formyl-perosamine residues, linked via α1,2 and/or α-1,3 glycosidic bonds, are responsible for the structure and antigenicity (Bundle et al., 1989; Moriyón and López-Goñi, 1998; Zygmunt et al., 2015). Although there are slight structural variabilities in the OPS, they are not crucial in the indirect diagnosis of Brucella infections, since anti-OPS antibodies recognize stable terminal sugar residues. However, this homogeneity in the OPS structure between B. microti and the surveyed highly zoonotic Brucella (B. abortus, B. melitensis, and B. suis) poses a significant challenge for diagnostics, as they share similar antigenic profiles, leading to cross-reactivity in serological tests. As a result, control plans based solely on immuno-serology may fail to accurately identify B. microti-infected animals. Currently, in the EU, ruminants infected with highly zoonotic Brucella sp. have to be slaughtered (Regulation EU 2020/689; ELI: http://data.europa.eu/eli/reg_del/2020/689/oj). In some member states like France, the whole cattle and small ruminant farm has to be slaughtered, which leads to significant economic losses for farmers and low acceptability for citizens. Therefore, the misdiagnosed infection with B. microti may lead to unnecessary culling of healthy livestock. Moreover, spontaneous mutations in B. microti can lead to the conversion of smooth LPS to a truncated rough form as previously identified (Ouahrani-Bettache et al., 2019). The absence of OPS in these rough strains makes the use of classical smooth Brucella sp. antigens ineffective for diagnostics, as is the case with rough strains of B. canis and B. ovis (Djokic et al., 2023). As envisaged by Ouahrani-Bettache et al. (Moriyón et al., 2004) the rough B. microti strain could serve as an interesting candidate for brucellosis vaccination in addition to existing vaccines, since it does elicit an antibody response that is distinguishable from that induced during active infection when complete OPS is present.
France has been officially recognized as brucellosis-free since 2005, following the European regulation (EFSA, 2023). In both departments affected by B. microti infection in small ruminants, no brucellosis outbreaks in cattle, sheep, or goats have been identified since 2003. Only sporadic cases of B. suis biovar two infection have been reported in suidae (Wendling et al., 2020). However, no data are available concerning B. microti prevalence and distribution in France, nor have accidental isolations been reported to the national reference laboratory aside from the previously described frog cases (Jaÿ et al., 2020; Jaÿ et al., 2018). The detection of B. microti in sheep and goats from three geographically distinct French farms highlights the potential for localized outbreaks. Moreover, the fact that these outbreaks occurred in different regions within the same timeframe suggests that B. microti may be more widespread than initially thought. Building on the previous study (Jaÿ et al., 2020), which reported a significant presence of B. microti-like strains in both domestic frogs (Pelophylax ridibundus) and surrounding environments, including water and soil, this study suggests that infection of small ruminants may be linked to environmental reservoirs of this pathogen, even in geographically distant areas, as the affected farms were located in different regions of France. The widespread environmental presence of B. microti in amphibian habitats raises the possibility that similar reservoirs may exist in agricultural environments, potentially facilitating transmission of the pathogen to livestock. The broad-spectrum wgSNP analysis linked the isolated strains from small ruminants in the same subclade with all known B. microti strains (Figure 2). However, there is a greater genetic similarity of these strains with the reference B. microti CCM 4915 isolated in Central Europe (Audic et al., 2009), compared to the frog isolates found in France. Interestingly, compared to the B. microti CCM 4915 reference strain, the two B. microti isolates from sheep in two different French departments exhibited no difference and variations between seven SNPs when aligned with B. melitensis 16 M and B. microti CCM 4915, respectively. In contrast, one and eight SNPs were detected in the goat isolate. These minimal genetic variations among the livestock isolates suggest a high degree of genetic similarity within this population. However, when compared to the frog isolate 17–2,122-4144, a more pronounced divergence was observed, with at least 10 and 134 SNPs distinguishing it from new isolates and B. microti CCM 4915. This genetic difference is also evident in MLVA-16 analysis, where the frog isolate (colored in yellow) clusters separately from the small ruminant strains (colored in blue), which group with B. microti CCM4915 (colored in turquoise) (Figure 1 and Supplementary Figure 1). This indicates that the frog isolate represents a possible distinct lineage, potentially reflecting ecological adaptation or host-specific evolution. The considerable variation of SNPs between the frog and livestock isolates underscores the genetic diversity within B. microti. It highlights the need for further investigation into the ecological and host-associated factors contributing to this diversity.
B. microti was isolated from reproductive tissues, including genital and retromammary lymph nodes, which raises the possibility of bacterial excretion in milk. Although milk samples were not available for direct testing, the presence of the bacterium in these tissues suggests a potential downstream risk associated with the consumption of unpasteurized dairy products from the implicated farms. This underscores the need for strict food safety practices and enhanced public health surveillance in such settings. A further limitation of this study is the inability to conduct environmental sampling in the surrounding areas of the farms or in wildlife habitats to trace the potential source of infection. Given that B. microti has been isolated from wild animals and soil, the possibility of environmental reservoirs contributing to the transmission cycle is high. Interestingly, the first two farms have 40 and 80 cattle, respectively, and no positive serological results were found during the prophylaxis campaigns before and after slaughter of infected ruminants. Additional annual monitoring of these herds has not shown any positive serological results. This suggests that, potentially, B. microti, at least for now, is more adapted to small ruminants. Identification of B. microti in domestic animals emphasizes the importance of monitoring Brucella infections across diverse populations, applying “One Health” approaches at the wildlife-farm-human interface (Lloyd-Smith et al., 2009). Future studies incorporating environmental sampling and broader surveillance of wildlife populations are essential to elucidate the transmission dynamics of this emerging pathogen and to devise effective control measures. Furthermore, the ability of B. microti to persist in wildlife and spill over into domestic animals suggests that targeted strategies are needed to prevent its transmission between these populations. Biosecurity is essential in livestock farming to prevent the spread of diseases, ensure animal welfare, and maintain farm sustainability (Bellini, 2018). In practice, to prevent various infectious diseases, it is recommended to maintain the surroundings of the farm double-fenced, which greatly limits the contact of wild animals with livestock. At the same time, it is recommended to avoid watering in ponds or rivers accessible to wildlife or downstream from other farms. Take precautions when distributing feed in pastures and restrict access to manure in the fields by using a tarpaulin or electric fencing. Domestic carnivores, suidae, and poultry can be sources of many infectious agents for cattle. In extensive and small-scale ruminant farms, biosecurity implementation may be impaired by inadequate premises infrastructure and uncontrolled contacts among different species (Alavedra et al., 2025). Compliance, in these cases, was influenced by farmers’ age, education level, herd size, and production.
In particular, dairy farms showed better biosecurity practices, probably because of improved management and infrastructure. This study highlights the challenges of implementing biosecurity measures on small-scale, extensive farms and shows the ineffectiveness of standardized plans. Biosecurity management in cattle farms consists of so-called protective measures to avoid the introduction of pathogens into the farm and limit the spread and the clinical expression of conditions already present in the farm. It includes a forward flow, management of introductions (animals, feed, workers, and instruments). Their presence should be prohibited in breeding and professional areas where feed is stored. This is true of dogs about neosporosis and poultry about botulism and salmonellosis. Finally, the installation of nets must prevent bird access to open-air feed storage to reduce the risk of contamination of milk by pathogens [Salmonella, HP STEC (highly pathogenic Shiga toxin-producing Escherichia coli), etc.], particularly in the case of raw milk production.
In conclusion, current results in conjunction with previous findings showed that B. microti is a potential pathogen at the interface of livestock, wildlife, and the environment. This highlights the need to improve screening tools for ruminants and to establish surveillance in wildlife and environmental reservoirs to better detect atypical Brucella species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because the Statement of Ethics does not apply to this set of samples taken for diagnostic purposes.
Author contributions
LF: Project administration, Formal analysis, Data curation, Visualization, Methodology, Validation, Investigation, Software, Writing – review & editing, Resources, Supervision, Funding acquisition, Writing – original draft, Conceptualization. VD: Methodology, Validation, Formal analysis, Project administration, Writing – original draft, Funding acquisition, Writing – review & editing, Conceptualization. AD: Data curation, Methodology, Formal analysis, Investigation, Writing – review & editing. MR: Methodology, Investigation, Writing – review & editing, Formal analysis, Data curation. MB: Methodology, Investigation, Writing – review & editing, Formal analysis. FB: Supervision, Writing – review & editing, Resources, Validation, Project administration, Investigation. CPa: Validation, Resources, Investigation, Project administration, Writing – review & editing, Supervision. AL: Supervision, Project administration, Validation, Investigation, Resources, Writing – review & editing. AF: Project administration, Funding acquisition, Formal analysis, Validation, Methodology, Writing – review & editing, Writing – original draft, Conceptualization. CPo: Supervision, Methodology, Software, Validation, Investigation, Conceptualization, Data curation, Resources, Funding acquisition, Writing – review & editing, Writing – original draft, Project administration, Visualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Work Program 2023–2024 of the EU Reference Laboratory for Brucellosis (Project ID 101144103, Topic SMP-FOOD-2023-EURL-EURC-AG-IBA).
Acknowledgments
We would like to thank the technical support team for their help with medium preparation and laboratory management, and the scientists working at the Bacterial Zoonoses Unit for their valuable feedback. We would also like to thank the technical staff at the Departmental Veterinary Authorities for the epidemiological investigations and sampling follow-up, and the Laboratory of Pyrénées and Landes (Lagor, France), as well as the laboratory of Aveyron (Rodez, France), for the serological analysis during prophylaxis campaigns.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1656803/full#supplementary-material
Footnotes
1. ^https://microbesgenotyping.i2bc.paris-saclay.fr/
References
About, F., Pastre, T., Boutrou, M., Martinez, A. Y., Melzani, A., Peugny, S., et al. (2023). Novel species of Brucella causing human brucellosis, French Guiana. Emerg. Infect. Dis. 29, 333–340. doi: 10.3201/eid2902.220725
Al Dahouk, S., Hofer, E., Tomaso, H., Vergnaud, G., Le Flèche, P., Cloeckaert, A., et al. (2012). Intraspecies biodiversity of the genetically homologous species Brucella microti. Appl. Environ. Microbiol. 78, 1534–1543. doi: 10.1128/AEM.06351-11
Al Dahouk, S., Scholz, H. C., Tomaso, H., Bahn, P., Göllner, C., Karges, W., et al. (2010). Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system. BMC Microbiol. 10:269. doi: 10.1186/1471-2180-10-269
Alavedra, M., Moura, D., Cenci-Goga, B., Saraiva, S., Silva, F., Pires, I., et al. (2025). Biosecurity practices in Portuguese small ruminant farms: current status and future directions. Vet. Sci. 12:334. doi: 10.3390/vetsci12040334
Aljanazreh, B., Shamseye, A. A., Abuawad, A., and Ashhab, Y. (2023). Genomic distribution of the insertion sequence IS711 reveal a potential role in Brucella genome plasticity and host preference. Infect. Genet. Evol. 112:105457. doi: 10.1016/j.meegid.2023.105457
Audic, S., Lescot, M., Claverie, J. M., and Scholz, H. C. (2009). Brucella microti: the genome sequence of an emerging pathogen. BMC Genomics 10:352. doi: 10.1186/1471-2164-10-352
Bellini, S. (2018). Application of biosecurity in different production systems at individual, country and regional levels : OIE Commission Régionale Europe.
Bounaadja, L., Albert, D., Chénais, B., Hénault, S., Zygmunt, M. S., Poliak, S., et al. (2009). Real-time PCR for identification of Brucella spp.: a comparative study of IS711, bcsp31 and per target genes. Vet. Microbiol. 137, 156–164. doi: 10.1016/j.vetmic.2008.12.023
Bundle, D. R., Cherwonogrodzky, J. W., Gidney, M. A., Meikle, P. J., Perry, M. B., and Peters, T. (1989). Definition of Brucella a and M epitopes by monoclonal typing reagents and synthetic oligosaccharides. Infect. Immun. 57, 2829–2836. doi: 10.1128/iai.57.9.2829-2836.1989
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Cloeckaert, A., Verger, J. M., Grayon, M., Paquet, J. Y., Garin-Bastuji, B., Foster, G., et al. (2001). Classification of Brucella spp. isolated from marine mammals by DNA polymorphism at the omp2 locus. Microbes Infect. 3, 729–738. doi: 10.1016/S1286-4579(01)01427-7
Corbel, M. J., and Nations, F.World Health Organization (2006). Brucellosis in humans and animals. Geneva: World Health Organization.
Damiano, M. A., Bastianelli, D., Al Dahouk, S., Köhler, S., Cloeckaert, A., De Biase, D., et al. (2015). Glutamate decarboxylase-dependent acid resistance in Brucella spp.: distribution and contribution to fitness under extremely acidic conditions. Appl. Environ. Microbiol. 81, 578–586. doi: 10.1128/AEM.02928-14
De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M., and Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. doi: 10.1093/bioinformatics/bty149
de la Garza-García, J. A., Ouahrani-Bettache, S., Lyonnais, S., Ornelas-Eusebio, E., Freddi, L., Al Dahouk, S., et al. (2021). Comparative genome-wide transcriptome analysis of Brucella suis and Brucella microti under acid stress at pH 4.5: cold shock protein CspA and Dps are associated with acid resistance of B. microti. Front. Microbiol. 12:3770. doi: 10.3389/fmicb.2021.794535
Djokic, V., Freddi, L., de Massis, F., Lahti, E., Van Den, E. M., Whatmore, A., et al. (2023). The emergence of Brucella canis as a public health threat in Europe: what we know, and what we need to learn. Emerg. Microb Infect 12:2249126. doi: 10.1080/22221751.2023.2249126
Doster, E., Lakin, S. M., Dean, C. J., Wolfe, C., Young, J. G., Boucher, C., et al. (2020). MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 48, D561–D569. doi: 10.1093/nar/gkz1010
Erridge, C., Bennett-Guerrero, E., and Poxton, I. R. (2002). Structure and function of lipopolysaccharides. Microbes Infect. 4, 837–851. doi: 10.1016/S1286-4579(02)01604-0
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63, e00483–e00419. doi: 10.1128/AAC.00483-19
Foster, G., Osterman, B. S., Godfroid, J., Jacques, I., and Cloeckaert, A. (2007). Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int. J. Syst. Evol. Microbiol. 57, 2688–2693. doi: 10.1099/ijs.0.65269-0
Freddi, L., Damiano, M. A., Chaloin, L., Pennacchietti, E., Al Dahouk, S., Köhler, S., et al. (2017). The glutaminase-dependent system confers extreme acid resistance to new species and atypical strains of Brucella. Front. Microbiol. 8:2236. doi: 10.3389/fmicb.2017.02236/full
Freddi, L., Garza-García, J. A., Dahouk, S. A., Occhialini, A., and Köhler, S. (2023). Brucella spp. are facultative anaerobic bacteria under denitrifying conditions. Microbiol. Spectr. 11:e0276723. doi: 10.1128/spectrum.02767-23
Girault, G., Djokic, V., Petot-Bottin, F., Perrot, L., Thibaut, B., Sébastien, H., et al. (2023). Molecular investigations of two first Brucella suis biovar 2 infections cases in French dogs. Pathogens 12:792. doi: 10.3390/pathogens12060792
Girault, G., Freddi, L., Jay, M., Perrot, L., Dremeau, A., Drapeau, A., et al. (2024). Combination of in silico and molecular techniques for discrimination and virulence characterization of marine Brucella ceti and Brucella pinnipedialis. Front. Microbiol. 15:1437408. doi: 10.3389/fmicb.2024.1437408
Girault, G., Perrot, L., Mick, V., and Ponsart, C. (2022). High-resolution melting PCR as rapid genotyping tool for Brucella species. Microorganisms 10:336. doi: 10.3390/microorganisms10020336
Godfroid, J., Garin-Bastuji, B., Saegerman, C., and Blasco, J. M. (2013). Brucellosis in terrestrial wildlife. Rev. Sci. Tech. 32, 27–42. doi: 10.20506/rst.32.1.2180
Hanna, N., De, B. J., Pilar, M., Ouahrani-Bettache, S., El Yakhlifi, Z., Köhler, S., et al. (2011). The virB operon is essential for lethality of Brucella microti in the Balb/c murine model of infection. J. Infect. Dis. 203, 1129–1135.
Hernández-Mora, G., Chacón-Díaz, C., Moreira-Soto, A., Barrantes-Granados, O., Suárez-Esquivel, M., Viquez-Ruiz, E., et al. (2023). Virulent Brucella nosferati infecting Desmodus rotundus has emerging potential due to the broad foraging range of its bat host for humans and wild and domestic animals. mSphere 1, e00061–e00023.
Huang, W., Li, L., Myers, J. R., and Marth, G. T. (2012). ART: a next-generation sequencing read simulator. Bioinformatics 28, 593–594. doi: 10.1093/bioinformatics/btr708
Hubálek, Z., Křivanová, A., Nesvadbová, J., and Rudolf, I. (2023). Zoonotic potential of Brucella microti. Vector Borne Zoonotic Dis. 23, 437–439. doi: 10.1089/vbz.2022.0085
Hubálek, Z., Scholz, H. c., Sedláček, I., Melzer, F., Sanogo, Y. o., and Nesvadbová, J. (2007). Brucellosis of the common vole (Microtus arvalis). Vector Borne Zoonot. Dis. 7, 679–688.
Jaÿ, M., Freddi, L., Mick, V., Durand, B., Girault, G., Perrot, L., et al. (2020). Brucella microti-like prevalence in French farms producing frogs. Transbound. Emerg. Dis. 67, 617–625. doi: 10.1111/tbed.13377
Jaÿ, M., Girault, G., Perrot, L., Taunay, B., Vuilmet, T., Rossignol, F., et al. (2018). Phenotypic and molecular characterization of Brucella microti-like bacteria from a domestic marsh frog (Pelophylax ridibundus). Front. Vet. Sci 5:283. doi: 10.3389/fvets.2018.00283
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiménez, M., Ouahrani-Bettache, S., Quintana, J., Mitjana, O., Hanna, N., Bessoles, S., et al. (2010). The new species Brucella microti replicates in macrophages and causes death in murine models of infection. J. Infect. Dis. 202, 3–10. doi: 10.1086/653084
Laine, C. G., Johnson, V. E., Scott, H. M., and Arenas-Gamboa, A. M. (2023). Global estimate of human brucellosis incidence. Emerg. Infect. Dis. 29, 1789–1797. doi: 10.3201/eid2909.230052
Le Flèche, P., Jacques, I., Grayon, M., Al Dahouk, S., Bouchon, P., Denoeud, F., et al. (2006). Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6:9. doi: 10.1186/1471-2180-6-9
Leclercq, S. O., Cloeckaert, A., and Zygmunt, M. S. (2020). Taxonomic organization of the family Brucellaceae based on a phylogenomic approach. Front. Microbiol. 10:83. doi: 10.3389/fmicb.2019.03083
Ledger, S., Rutherford, C., Benham, C., Burfield, I., Deinet, S., and Eaton, M. Wildlife Comeback in Europe: Opportunities and challenges for species recovery. Final report to Rewilding Europe by the Zoological Society of London, BirdLife International and the European Bird Census Council. (2022).
Lloyd-Smith, J. O., George, D., Pepin, K. M., Pitzer, V. E., Pulliam, J. R. C., Dobson, A. P., et al. (2009). Epidemic dynamics at the human-animal interface. Science 326, 1362–1367. doi: 10.1126/science.1177345
Mancilla, M. (2015). Smooth to rough dissociation in Brucella: the missing link to virulence. Front. Cell. Infect. Microbiol. 5:98. doi: 10.3389/fcimb.2015.00098
Moriyón, I., Grilló, M. J., Monreal, D., González, D., Marín, C., López-Goñi, I., et al. (2004). Rough vaccines in animal brucellosis: structural and genetic basis and present status. Vet. Res. 35, 1–38. doi: 10.1051/vetres:2003037
Moriyón, I., and López-Goñi, I. (1998). Structure and properties of the outer membranes of Brucella abortus and Brucella melitensis. Int. Microbiol. 1, 19–26.
Occhialini, A., Bagüés, M. P. J., Saadeh, B., Bastianelli, D., Hanna, N., Biase, D. D., et al. (2012). The glutamic acid decarboxylase system of the new species Brucella microti contributes to its acid resistance and to oral infection of mice. J. Infect. Dis. 206, 1424–1432.
Occhialini, A., Hofreuter, D., Ufermann, C. M., Al Dahouk, S., and Köhler, S. (2022). The retrospective on atypical Brucella species leads to novel definitions. Microorganisms 10:813. doi: 10.3390/microorganisms10040813
Olsen, S. C., and Palmer, M. V. (2014). Advancement of knowledge of Brucella over the past 50 years. Vet. Pathol. 51, 1076–1089. doi: 10.1177/0300985814540545
Orsini, M., Ianni, A., and Zinzula, L. (2022). Brucella ceti and Brucella pinnipedialis genome characterization unveils genetic features that highlight their zoonotic potential. Microbiol Open 11:e1329. doi: 10.1002/mbo3.1329
Ouahrani-Bettache, S., Bagüés, M. P. J. D., Garza, J. D. L., Freddi, L., Bueso, J. P., Lyonnais, S., et al. (2019). Lethality of Brucella microti in a murine model of infection depends on the wbkE gene involved in O-polysaccharide synthesis. Virulence 10, 868–878.
Perpiña, C. C., Kavalov, B., Diogo, V., Jacobs, C., Batista, E. S. F., Baranzelli, C., et al. Trends in the EU agricultural land within 2015–2030. Available online at: https://publications.jrc.ec.europa.eu/repository/handle/JRC113717
Rónai, Z., Kreizinger, Z., Dán, Á., Drees, K., Foster, J. T., Bányai, K., et al. (2015). First isolation and characterization of Brucella microti from wild boar. BMC Vet. Res. 11:147. doi: 10.1186/s12917-015-0456-z
Rudolf, I., Kejíková, R., Kosoy, M., Hubálek, Z., Mravcová, K., Šikutová, S., et al. Brucella microti and rodent-borne brucellosis: a neglected public health threat. Zoonoses Public Health. (2024) Available online at: https://onlinelibrary.wiley.com/doi/abs/10.1111/zph.13188 (accessed Accepted: 4 October 2024).
Ryan, M. P., and Pembroke, J. T. (2020). The genus Ochrobactrum as major opportunistic pathogens. Microorganisms 8:1797. doi: 10.3390/microorganisms8111797
Sanjuan-Jimenez, R., Colmenero, J. D., and Morata, P. (2017). Lessons learned with molecular methods targeting the BCSP-31 membrane protein for diagnosis of human brucellosis. Clin. Chim. Acta 469, 1–9. doi: 10.1016/j.cca.2017.03.014
Scholz, H. C., Hofer, E., Vergnaud, G., Le Fleche, P., Whatmore, A. M., Al Dahouk, S., et al. (2009). Isolation of Brucella microti from mandibular lymph nodes of red foxes, Vulpes vulpes, in lower Austria. Vector Borne Zoonotic Dis. 9, 153–156. doi: 10.1089/vbz.2008.0036
Scholz, H. C., Hubalek, Z., Nesvadbova, J., Tomaso, H., Vergnaud, G., Le Flèche, P., et al. (2008). Isolation of Brucella microti from soil. Emerg. Infect. Dis. 14, 1316–1317. doi: 10.3201/eid1408.080286
Scholz, H. C., Hubalek, Z., Sedláček, I., Vergnaud, G., Tomaso, H., Al Dahouk, S., et al. (2008). Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int. J. Syst. Evol. Microbiol. 58, 375–382. doi: 10.1099/ijs.0.65356-0
Suárez-Esquivel, M., Chaves-Olarte, E., Moreno, E., and Guzmán-Verri, C. (2020). Brucella genomics: macro and micro evolution. IJMS 21:7749. doi: 10.3390/ijms21207749
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thoma, B., Straube, E., Scholz, H. C., Al Dahouk, S., Zöller, L., Pfeffer, M., et al. (2009). Identification and antimicrobial susceptibilities of Ochrobactrum spp. Int. J. Med. Microbiol. 299, 209–220. doi: 10.1016/j.ijmm.2008.06.009
Wendling, S., Jaÿ, M., Pozzi, N., Garin-Bastuji, B., and Ponsart, C.. Bilan de la surveillance de la brucellose porcine en France en 2016. Bulletin épidémiologique, santé animale et alimentation [Internet]. (2020). Available online at: https://www.google.com/url?sa=t&source=web&rct=j&opi=89978449&url=https://be.anses.fr/sites/default/files/N-026_2021-08-04_Brucellose-porc_MaqF.pdf&ved=2ahUKEwjw5p2E39COAxVVTqQEHT2qHwoQFnoECBAQAQ&usg=AOvVaw1mgQt3TqHWbB5ZvtDjAsyS (accessed December 2020).
Whatmore, A. M., and Foster, J. T. (2021). Emerging diversity and ongoing expansion of the genus Brucella. Infect. Genet. Evol. 92:104865. doi: 10.1016/j.meegid.2021.104865
World Organisation for Animal Health (2022). “Brucellosis (infection with B. abortus, B. Melitenis and B. suis)” in WOAH Terrestrial Manual.
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Brucella microti, small ruminants, brucellosis, diagnostics, surveillance strategy
Citation: Freddi L, Djokic V, Dremeau A, Ribeiro M, Berthaud M, Bennasar F, Pailhous C, Lanterne A, Ferreira Vicente A and Ponsart C (2025) First isolation and identification of Brucella microti in sheep and goats: new insights and implications for veterinary medicine. Front. Microbiol. 16:1656803. doi: 10.3389/fmicb.2025.1656803
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Rebekah Tiller, Centers for Disease Control and Prevention (CDC), United StatesHolger C. Scholz, Robert Koch Institute (RKI), Germany
Ana Cristina Ferreira, National Institute for Agricultural and Veterinary Research (INIAV), Portugal
Copyright © 2025 Freddi, Djokic, Dremeau, Ribeiro, Berthaud, Bennasar, Pailhous, Lanterne, Ferreira Vicente and Ponsart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: L. Freddi, bHVjYS5mcmVkZGlAYW5zZXMuZnI=
 L. Freddi
L. Freddi V. Djokic
V. Djokic A. Dremeau
A. Dremeau M. Ribeiro1
M. Ribeiro1 A. Ferreira Vicente
A. Ferreira Vicente C. Ponsart
C. Ponsart