- 1Key Laboratory of Western China’s Environmental Systems (Ministry of Education), Key Scientific Research Base of Bioarchaeology in Cold and Arid Regions (National Cultural Heritage Administration), College of Earth and Environmental Sciences, Lanzhou University, Lanzhou, Gansu, China
- 2MOE Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou, Gansu, China
- 3National Research Center for Conservation of Ancient Wall Paintings and Earthen Sites, Conservation Institute, Dunhuang Academy, Dunhuang, Gansu, China
- 4Key Laboratory of Extreme Environmental Microbial Resources and Engineering, Gansu Province, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, China
- 5Institute of Maijishan Grottoes Art, Dunhuang Academy, Tianshui, Gansu, China
- 6Environmental Science and Engineering Group, Guangdong Technion-Israel Institute of Technology, Shantou, Guangdong, China
- 7Guangdong Provincial Key Laboratory of Materials and Technologies for Energy Conversion, Guangdong Technion-Israel Institute of Technology, Shantou, Guangdong, China
The Maijishan and Mogao Grottoes, both UNESCO World Heritage Sites along the Silk Road, are increasingly threatened by microbial biodeterioration. To characterize bacterial communities of different microbial damages on wall paintings and identify environmental drivers, we combined high-throughput DNA/RNA sequencing with microenvironmental monitoring and conducted a cross-site comparison. At Maijishan, bacterial communities associated with black and white mycelia showed no significant compositional differences within the same cave but varied markedly between caves, indicating site-specific community assembly. Actinobacteria (>50%), particularly Pseudonocardia and Actinomycetospora, predominated, while RNA-based analysis revealed active populations of Escherichia and Stenotrophomonas, likely introduced via exogenous contamination from animal activities. In contrast, black spots from the Mogao Grottoes were dominated by Actinobacteria, Firmicutes, and Proteobacteria, with Rhodococcus as a core genus. No core bacterial OTUs were shared between the sites, suggesting strong microenvironmental filtering. Multivariate analysis identified substrate properties (total organic carbon, total nitrogen, pH) and microclimatic fluctuations (diurnal temperature/humidity ranges) as critical drivers. Maijishan’s persistently humid conditions (RH > 70% for over 180 days/ year) favored Actinobacteria proliferation, whereas Mogao’s arid climate (RH < 70% for over 240 days/year) selected for xerotolerant Firmicutes. These results reveal distinct site-specific microbial colonization patterns and provide a scientific basis for targeted conservation strategies to mitigate microbial damage and preserve these invaluable wall paintings.
1 Introduction
Wall paintings represent one of the most ancient and enduring expressive forms of expression human creativity, bearing exceptional artistic, scientific, and historical significance on a global scale. As prominent components of grotto architecture, they provide rich insights into the evolution of societies and civilizations (Garg et al., 1995; Sansupa et al., 2023; Wu et al., 2022). However, these cultural assets face increasing threats by biodeterioration, a process exacerbated by global climate change characterized by rising temperature variability and intensified hydrological extremes (Leissner et al., 2015; Liu et al., 2020; Ding et al., 2022; Hu and Hewitt, 2024). Among the various degradation agents, microbial activity plays a central role, driving damage through both physicochemical and biochemical mechanisms. Bacteria, in particular, are often the pioneering colonizers on wall-painting surfaces (Cuezva et al., 2012; Ma et al., 2015; Meng et al., 2017; Mihajlovski et al., 2017; Sugiyama et al., 2017; Ma C. et al., 2023). Their metabolites, such as organic acids, pigments and extracellular polymeric substances (EPS), which accelerate the aesthetic value and structural integrity reducing (Abdel-Haliem et al., 2013; Cojoc et al., 2019; Dominguez-Moñino et al., 2017; Elhagrassy, 2018). Through photoautotrophic or chemoautotrophic metabolism, bacterial communities fix carbon and generate organic substrates, facilitating the succession of heterotrophic fungi or archaea (Lan et al., 2010; Li et al., 2021; Liu et al., 2020; Zhang et al., 2019). This trophic cascade fundamentally reshapes microhabitat conditions, promoting secondary colonization by other microorganisms (Xu et al., 2018).
Biodeterioration poses major challenges to wall painting preservation, understanding these microbial communities is thus essential for heritage conservation (Ciferri, 1999; Martin-Sanchez et al., 2012; Ma et al., 2020; Pei et al., 2023). While previous research has relied heavily on culture-dependent methods or DNA-based high-throughput sequencing to characterize microbial diversity, these approaches have limitations. DNA sequencing captures the total community composition, including dormant or dead cells, but does not directly reveal which members are metabolically active. RNA enables the identification of taxa actively participating in biodeterioration at the sampling moment, the integration of DNA and RNA sequencing provides a more complete and dynamic view of microbial communities by resolving both the taxonomic structure (DNA) and the metabolically active fraction (RNA). Applying such an integrated method significantly enhances our ability to investigate the viability of these microbial communities and allows more accurate assessment of microbial threats, thereby improving the scientific basis for preventive conservation (Sanmartín et al., 2018; Meng et al., 2020; Liu W. et al., 2022).
Microbial communities colonizing cultural heritage materials are shaped by a complex interplay of environmental conditions, biological and physicochemical factors (Dornieden et al., 2000; Wu et al., 2021; Zhang et al., 2021). Previous studies have demonstrated that regional climate type and microclimatic fluctuations are key determinants of microbial diversity in heritage sites (Ding et al., 2022), with temperature and precipitation patterns identified as key drivers of microbial diversity (Biagioli et al., 2024; Chen and Gu, 2022; Liu et al., 2018). Bacterial communities, in particular, tend to respond more rapidly and sensitively to environmental variability than fungi, which often display greater adaptability and resilience (Barnard et al., 2013; Chen et al., 2023). This sensitivity means that climate-induced environmental shifts can trigger rapid changes in bacterial assemblages, potentially leading to irreversible deterioration trajectories (Viles and Cutler, 2012; Li et al., 2021; Ding et al., 2022).
This study investigates microbial biodeterioration at two UNESCO World Heritage Sites along the Ancient Silk Road: the Maijishan Grottoes and the Mogao Grottoes. In recent years, microbial proliferations have been observed in Caves No. 28 and No. 30 at Maijishan, appearing as black and white mycelial growths (Ma et al., 2025). While black spot formations have previously been documented in Cave No. 256 at Mogao (Ma W. et al., 2023). By comparing the bacterial communities and microenvironmental conditions across these geographically distinct sites (separated by over 1,400 km), this study aims to: (1) elucidate taxonomic divergences between black and white mycelia in Maijishan Grottoes; (2) identify site-specific bacterial community signatures distinguishing the two sites; and (3) evaluate the relative influence of geographic isolation versus environmental filtering on bacterial community assembly. These findings aim to advance our understanding of microbial biogeography in wall painting environments and provide a scientific foundation for developing site-specific conservation strategies.
2 Materials and methods
2.1 Description of study sites
The Maijishan Grottoes (34°05′–35°10′N, 104°35′–106°44′E), a UNESCO World Cultural Heritage Site, are located in the Tianshui Maiji Mountain, a part of Qinling Mountain range. This cultural site comprises 221 caves, featuring 10,632 clay sculptures and approximately 1,000 m2 of wall paintings. Situated in a transitional zone between subtropical and warm temperate climates, the region is characterized by dense forest coverage, high humidity and stable temperatures year-round. In contrast, the Mogao Grottoes (39°53′–41°35′N, 92°13′–95°30′E), also designated as a UNESCO World Cultural Heritage Site, are positioned along the eastern cliff of Mingsha Mountain, approximately 25 km southeast of Dunhuang. This site includes 735 caves, with over 45,000 m2 of wall paintings and 2,400 sculptures, and is located in a cold desert climate zone marked by low humidity, minimal precipitation, and high potential evaporation (Figures 1A–C).
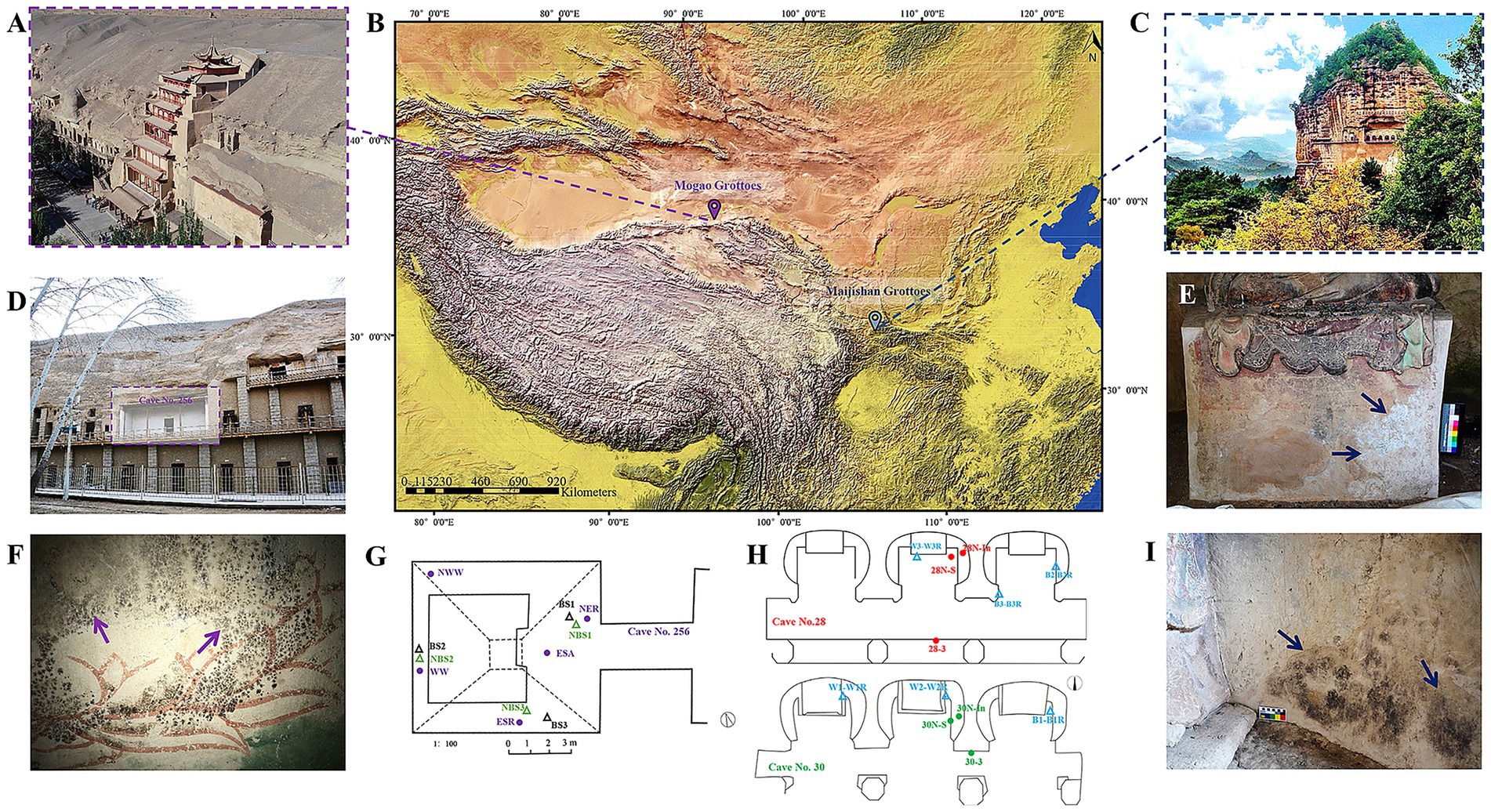
Figure 1. Locations of the study site and typical microbial damages of the Maijishan Grottoes and the Mogao Grottoes in China. (A–C) The geographical locations and exterior illustration of the Maijishan and Mogao Grottoes. (D,F) The appearance of Cave No. 256 and black spots on wall paintings at the Mogao Grottoes. (G) Schematic diagram of sampling and environmental monitoring sites in Cave No. 256, the black and green triangles represent the sampling sites for black-spots samples (BS) and no-spots samples (NBS), respectively, while the purple dots represent the temperature and relative humidity monitoring sites. (E,I) White and black mycelium on the wall paintings of Caves No. 28 and No. 30 at the Maijishan Grottoes. (H) Schematic diagram of sampling and environmental monitoring sites, the blue triangles represent the sampling sites, while the red and green dots represent the temperature and relative humidity monitoring sites.
Both grotto complexes were constructed during the 4th century CE (Maijishan: 384 CE; Mogao: 366 CE), with similar wall painting techniques and materials. These include mineral pigments, natural sediments, original binders (e.g., casein, animal glue, gelatin), and plant fibers (e.g., hemp, wheat straw, cotton) used for structural reinforcement. Microbial colonization and biodeterioration have been observed to varying degrees in both the Maijishan Grottoes and the Mogao Grottoes. This study focused on Cave No. 256 at the Mogao Grottoes, where black spot deterioration is present (Figures 1D,F,G), and Caves No. 28 and No. 30 at the Maijishan Grottoes, which exhibit black and white mycelial growth on wall paintings (Figures 1E,H,I).
2.2 Sampling
Caves No. 28 and No. 30, situated within the middle-lower sections of the Maijishan Grottoes (Supplementary Figures S1B,C,F), with black (Supplementary Figures S1A,D,I) and white (Supplementary Figures S1E,G,H) mycelia colonizing wall paintings. In August 2021, a total of 12 microbial samples (each pair consisting of two samples) were collected from colonized wall painting surfaces. Each sample was collected using sterile forceps within a targeting 10 × 10 cm area (Supplementary Figures S1A,D,E,G–I). Each collected mycelia sample was thoroughly mixed and equally divided: one portion for DNA extraction (designated W1–W3 for white and B1–B3 for black mycelia), and the other preserved in RNAlater (Qiagen, Germany) for RNA analysis (designated W1R-W3R and B1R-B3R) to assess viable metabolism. Sampling involved carefully removing only mycelia without damage to paintings or restored materials, with each weighed approximately 50 mg. Additionally, fragments of non-colonized wall paintings (totaling 10–15 g) were collected from both caves for physicochemical characterization. All samples were stored in sterile Eppendorf tubes and transported to the laboratory at Lanzhou University for further analysis. The sampling was conducted under the authorization and supervision of professionals from the Maijishan Grottoes Art Research Institute.
For comparative analysis, previously collected and sequenced samples from Cave No. 256 at the Mogao Grottoes (Ma W. et al., 2023) were re-analyzed, including black-spot samples (BS) and non-spot (NBS) samples. These samples collected from the wall painting surfaces with the same sampling method, with specific sample information and detailed data provided in the previous publication (Ma W. et al., 2023).
2.3 DNA extraction, RNA extraction, and MiSeq high-throughput sequencing analysis
Total genomic DNA and rRNA from each sample were extracted using the PowerSoil® DNA Isolation Kit and PowerSoil® RNA Kit (MO BIO Laboratories, United States), following the manufacturer’s protocols. RNA was subsequently reverse-transcribed into complementary DNA (cDNA) with the cDNA Synthesis Kit (Sigma-Aldrich, United States). Nucleic acid concentrations (DNA/cDNA) were quantified using a NanoDrop® ND-2000 spectrophotometer (Thermo Scientific, United States), and integrity was verified by 1% agarose gel electrophoresis. The bacterial V3–V4 regions of the 16S rRNA gene were amplified using primers 338F/806R (Peiffer et al., 2013). PCR conditions consisted of initial denaturation at 95°C for 3 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 10 min (detailed reaction mixtures in Supplementary Table S1). Triplicate amplifications per sample were pooled, purified with a Gel Extraction Kit (AXYGEN, China), and quantified via QuantiFluor™-ST Fluorimeter (Promega, United States). Libraries were prepared using the NEXTflex® Rapid DNA-Seq Kit (Bioo Scientific, United States) and sequenced on an Illumina MiSeq PE300 platform (Majorbio Bio-Pharm Technology Co., China).
Raw FASTQ data were processed with Trimmomatic (Bolger et al., 2014) and FLASH (Magoč and Salzberg, 2011), with reads <50 bp discarded and no assembly performed. High-quality sequences were clustered into operational taxonomic units (OTUs) at 97% similarity via UPARSE (Edgar, 2013). Taxonomic classification of bacterial OTUs was performed using the RDP Classifier against the SILVA and UNITE v7.0 databases with a 70% confidence threshold.
2.4 Environmental data collection
Approximately 3 g of homogenized wall painting substrate from the Maijishan Grottoes was oven-dried at 105°C until constant weight to determine gravimetric moisture content (MC). pH and electrical conductivity (EC) were measured using a Sartorius PB-10 pH meter (1:5 w/v in deionized water) and Leici DDSJ-318 conductivity meter (1:5 w/v in 1 M KCl), respectively. Total organic carbon (TOC) and total nitrogen (N) were quantified by high-temperature combustion (450°C and 1,250°C) using a Vario EL cube CHNS analyzer.
Continuous environmental monitoring was conducted from 2021 to 2022 in Caves No. 28 and No. 30 (Figure 1H). HOBO® U23-001 loggers recorded air (28-3, 30-3) and wall surface (28 N-S, 30 N-S) temperature (T) and relative humidity (RH) at hourly intervals. iButton® DS1923 sensors embedded in wall paintings measured internal T and RH every 2 h (28 N-IN, 30 N-IN). The environmental monitoring protocol in Cave No. 256 at the Mogao Grottoes followed the same design as detailed in our previous study (Ma W. et al., 2023), with monitoring locations shown in Figure 1G.
2.5 Statistical analysis
All statistical analyses were performed using SPSS 13.0. The α-diversity indices (Shannon, Simpson, Ace, and Chao) were calculated with MOTHUR (version 1.30.1) (Schloss et al., 2009). Redundancy analysis (RDA) was conducted in R (v4.0.3) using the vegan package to explore relationships between microbial communities and environmental variables. Sankey diagrams were generated with the ggalluvial package within the R environment. Differences between sample groups were tested with the Wilcoxon rank-sum test (p < 0.05). Environmental parameter visualizations were created using Origin 8.0 software.
3 Results and analysis
3.1 Bacterial community diversity and taxonomic composition in Maijishan Grottoes
High-throughput sequencing of the bacterial V3–V4 regions from both DNA and cDNA libraries yielded 782,304 raw sequences, clustered into 526 OTUs, with the specific sequence counts, OTU numbers, and diversity indices for each sample presented in the Supplementary Table S2. Alpha-diversity metrics revealed minimal differences among the four sampling types (Black-DNA, Black-RNA, White-DNA, White-RNA, p < 0.05, Wilcoxon rank-sum test), with the Black-RNA group exhibiting the highest Shannon index, followed by Black-DNA, White-DNA, and White-RNA (Supplementary Table S2). Taxonomic classification identified 21 bacterial phyla, 182 families, and 302 genera. The dominant phyla were Actinobacteria, Proteobacteria, Cyanobacteria, and Firmicutes, with Actinobacteria exceeding 50% relative abundance in all but one RNA sample (B1R). RNA samples showed higher proportions of Proteobacteria, suggesting elevated microbial viability (Figure 2A).
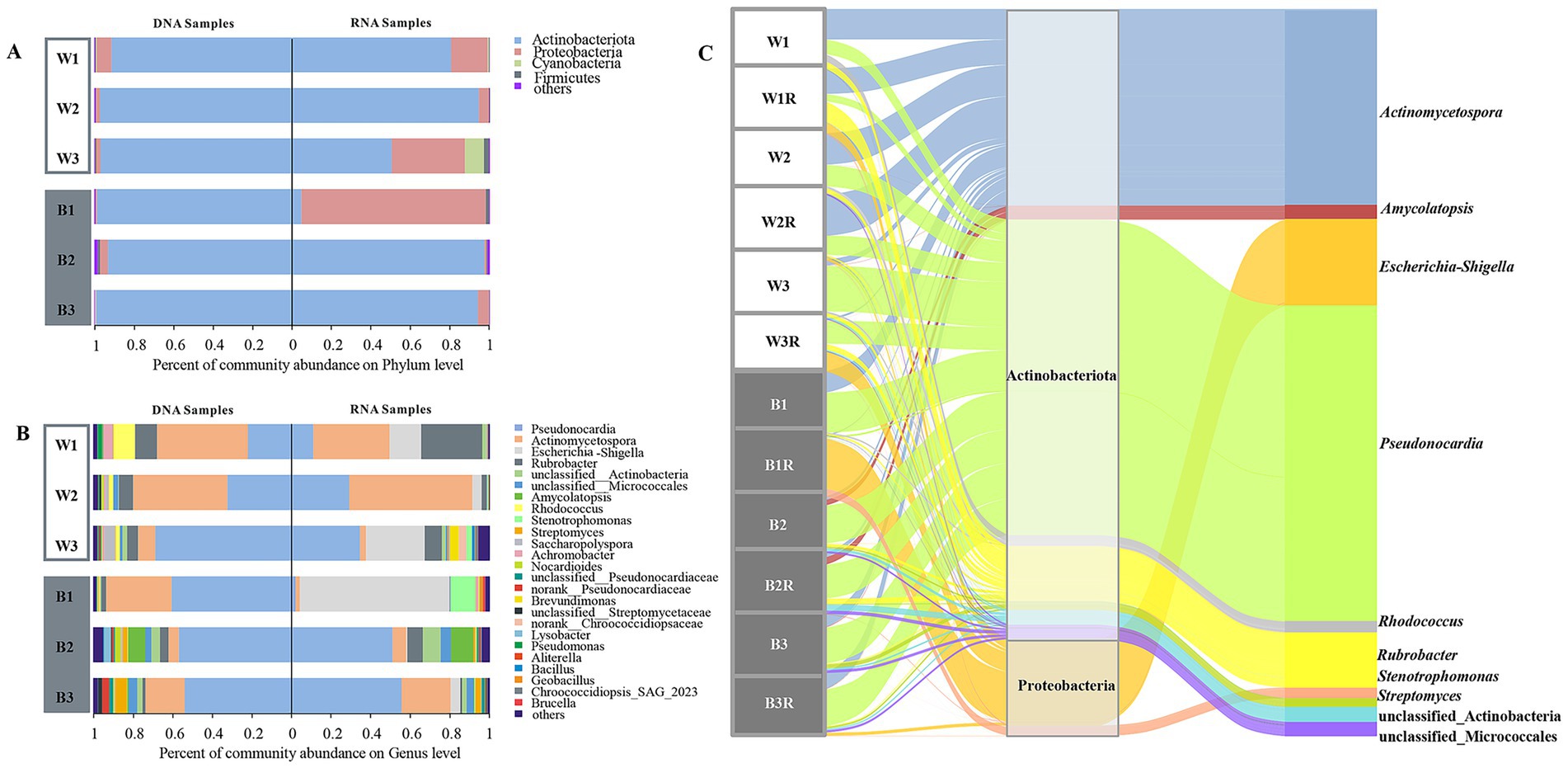
Figure 2. Community structures of bacteria detected at DNA and RNA levels. (A,B) Bacterial phyla and genera. (C) The Sankey diagram of bacterial microbial community structures based on the 16S rRNA gene at DNA and RNA levels.
At the genus level, Pseudonocardia and Actinomycetospora dominated, accounting for >60% relative abundance in most samples (excluding W3R and B1R, Figure 2B). Pseudonocardia was predominant in black mycelial and some white mycelial samples (B1, B2, B2R, B3, B3R, W3, W3R), while Actinomycetospora was more abundant in other white mycelial groups (W1, W1R, W2, W2R) (Figure 2C). Kruskal-Wallis H test revealed no significant taxonomic differences among the four groups. Among the top ten most abundant bacterial genera, Pseudonocardia and Actinomycetospora exhibited similar content in four groups. Notably, RNA samples consistently showed higher levels of Escherichia and Stenotrophomonas in bacterial taxa, indicating higher microbial viability (Supplementary Figure S2).
3.2 Influence of environmental factors and painting materials on bacterial communities in Maijishan Grottoes
The wall painting substrates in Caves No. 28 and No. 30 exhibited weak alkalinity (pH 8–9). Cave No. 28 showed lower moisture content and electrical conductivity than Cave No. 30 but had higher total organic carbon (TOC) and total nitrogen (N) levels (Table 1).
To identify key environmental drivers, variance inflation factor (VIF) analysis employed to eliminate inappropriate factors (e.g., moisture content, MC) with a threshold of 10. Redundancy Analysis (RDA) was then utilized to ascertain the relationships among microbial samples, basic physicochemical characteristics, and environmental factors (Figure 3A). While black and white mycelia showed no significant compositional differences, samples clustered strongly by cave origin regardless of mycelium color. In Cave No. 28, bacterial community composition was primarily shaped by temperature and pH, whereas in Cave No. 30, total nitrogen and electrical conductivity were the dominant factors. To rigorously control the false discovery rate (FDR) in high-dimensional correlation testing, we applied Benjamini-Hochberg correction to all pairwise associations. Heatmap visualization (Figure 3B) revealed that temperature (T) and relative humidity (RH) exerted the most contrasting effects on community structure. Six Actinobacterial genera, including unclassified_c_Actinobacteria, Streptomyces, and norank_f__Pseudonocardiaceae taxa displayed significant positive correlations with T and negative correlations with RH (q < 0.0001, FDR-corrected), while Streptomonospora, unclassified_f__Nocardioidaceae, Amycolatopsis, and unclassified_o__Micrococcales taxa showed positive correlations with T (q < 0.05, FDR-corrected). Furthermore, total nitrogen (N) exhibited significant negative correlations with unclassified_o__Micrococcales, Kribbella and Actinophytocola (q < 0.05, FDR-corrected), whereas total organic carbon (TOC) demonstrated pronounced positive association with Actinophytocola (q < 0.05, FDR-corrected).
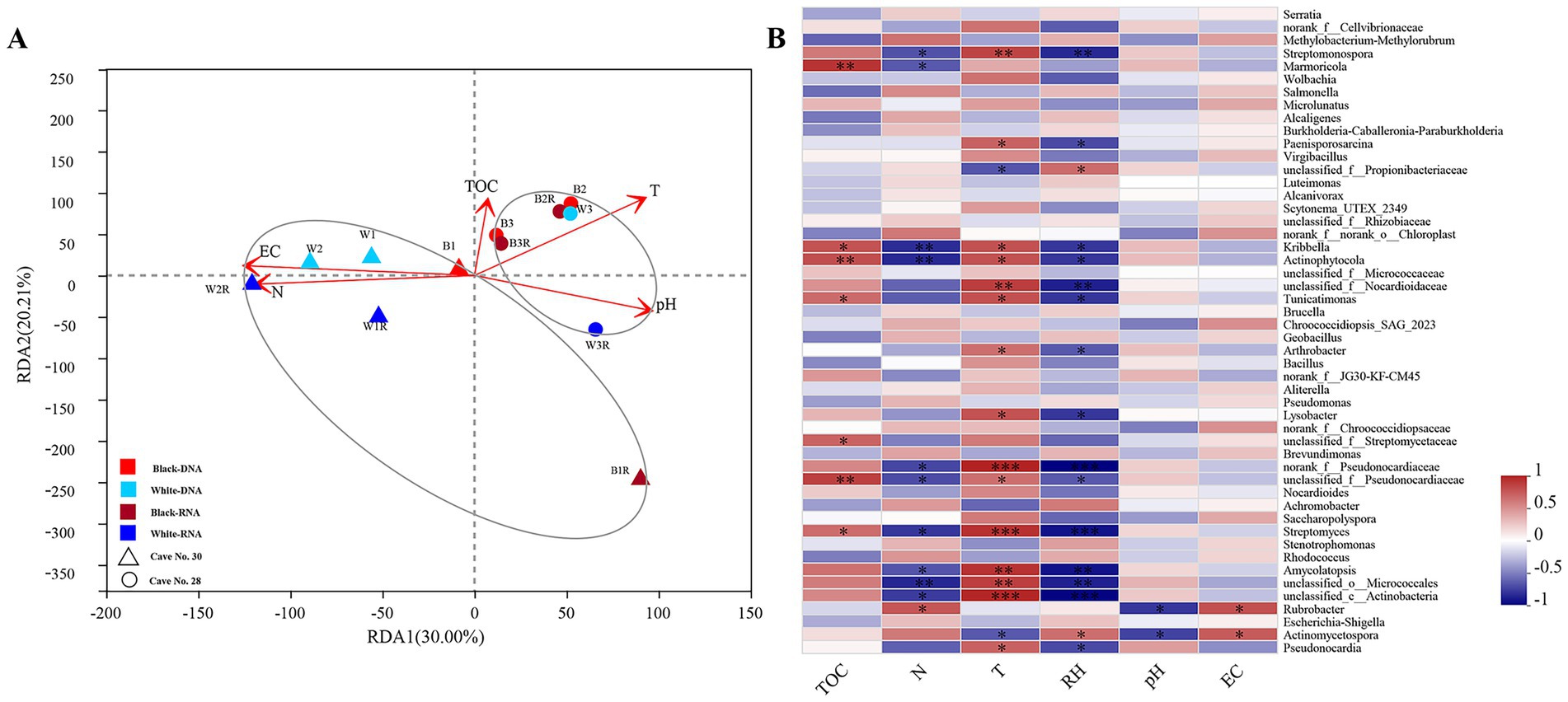
Figure 3. Correlation analysis between microbial community structure and environmental factors. RDA analysis on the genera level of bacteria. (A) Showing the correlations between the sample types and environmental factors. Spearman correlation heatmap analysis showed the associations between the top 50 dominant bacterial (B) genera and the environmental factors. Note: TOC-total organic carbon, N- total nitrogen, T- temperature, RH- relative humidity, EC- electrical conductivity. *: q < 0.05 (FDR-corrected); **: q < 0.01 (FDR-corrected); ***: q < 0.001(FDR-corrected). Red and dark red indicate DNA and RNA samples from black mycelia; sky blue and dark blue indicate DNA and RNA samples from white mycelia. Circles are from Cave No. 28, triangles from Cave No. 30.
3.3 Comparison of bacterial composition in samples from the Maijishan and Mogao Grottoes
Comparative analysis of alpha-diversity revealed statistical differences in bacterial diversity within black microbial samples between the Maijishan Grottoes and the Mogao Grottoes (p < 0.05, Wilcoxon rank-sum test, Figures 4A–D). Both sites were dominated by Actinobacteria, Proteobacteria and Firmicutes, but their relative abundances exhibited site-specific patterns: Actinobacteria were more abundant in Maijishan samples, whereas Proteobacteria and Firmicutes showed higher abundance in Mogao samples (p < 0.05, Wilcoxon rank-sum test, Figure 4E). Upset analysis revealed that Maijishan groups (B-DNA, B-RNA, W-RNA, W-DNA) obtained 163, 175, 187, and 303 OTUs respectively, significantly fewer than the OTU numbers in Mogao samples (467 and 508 OTUs). Among these, 575 OTUs were detected only in the Mogao Grottoes samples, including 289 OTUs detected in both black spots and non-spots samples. Additionally, 209 OTUs were detected only in the Maijishan Grottoes samples, including 65 OTUs detected in both black mycelia DNA/RNA and white mycelia DNA/RNA samples (Figure 5). Only 13 OTUs were shared among the six types of samples from the two sites, none of which could be resolved at the species level, and all had low read counts.
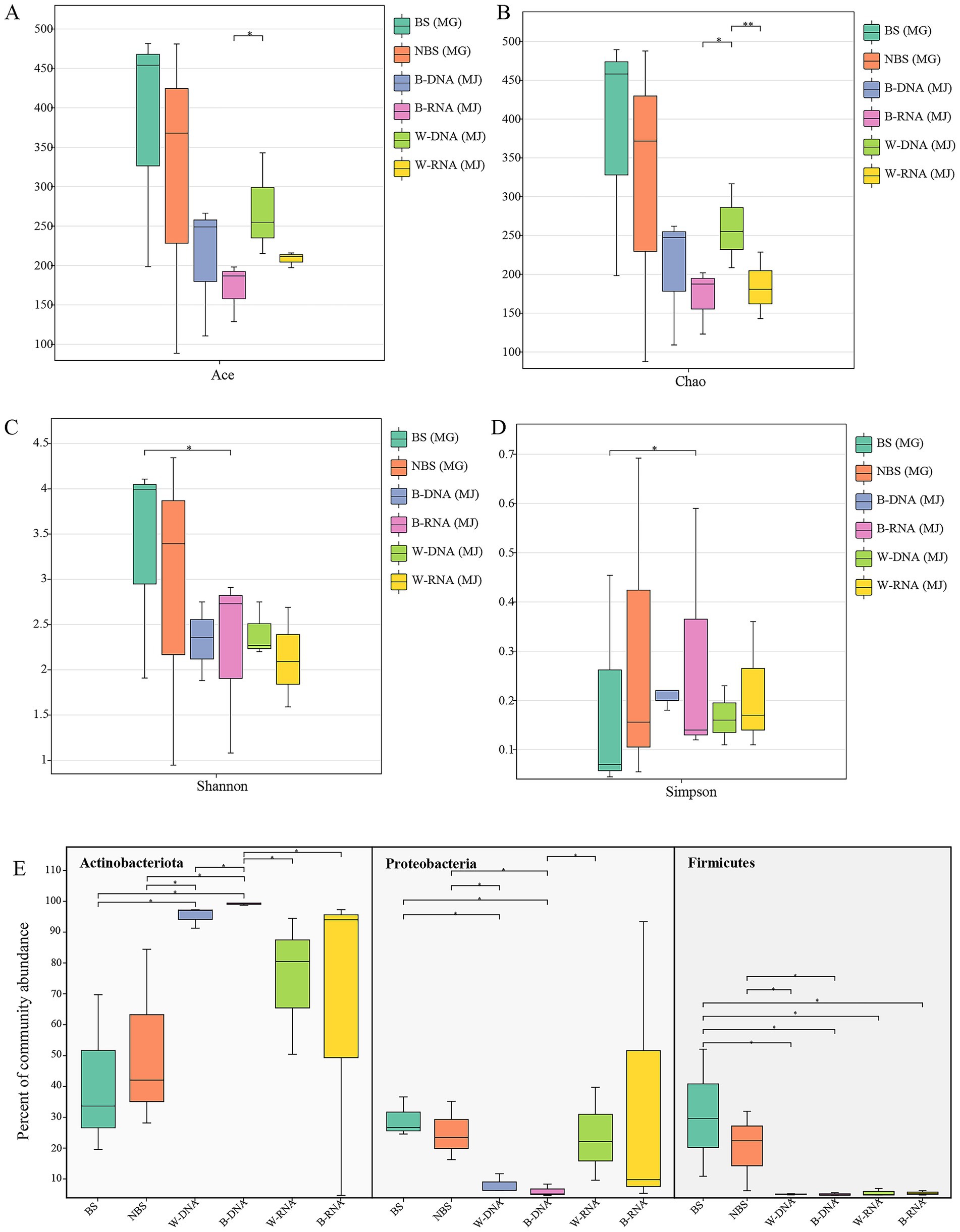
Figure 4. Comparison of bacterial alpha diversity and dominant bacterial phyla between the Maijishan Grottoes and Mogao Grottoes by Wilcoxon rank-sum test. (A,B) Species richness estimators (ACE and Chao indices), (C,D) alpha diversity indices (Shannon and Simpson), and dominant bacterial phyla (E).
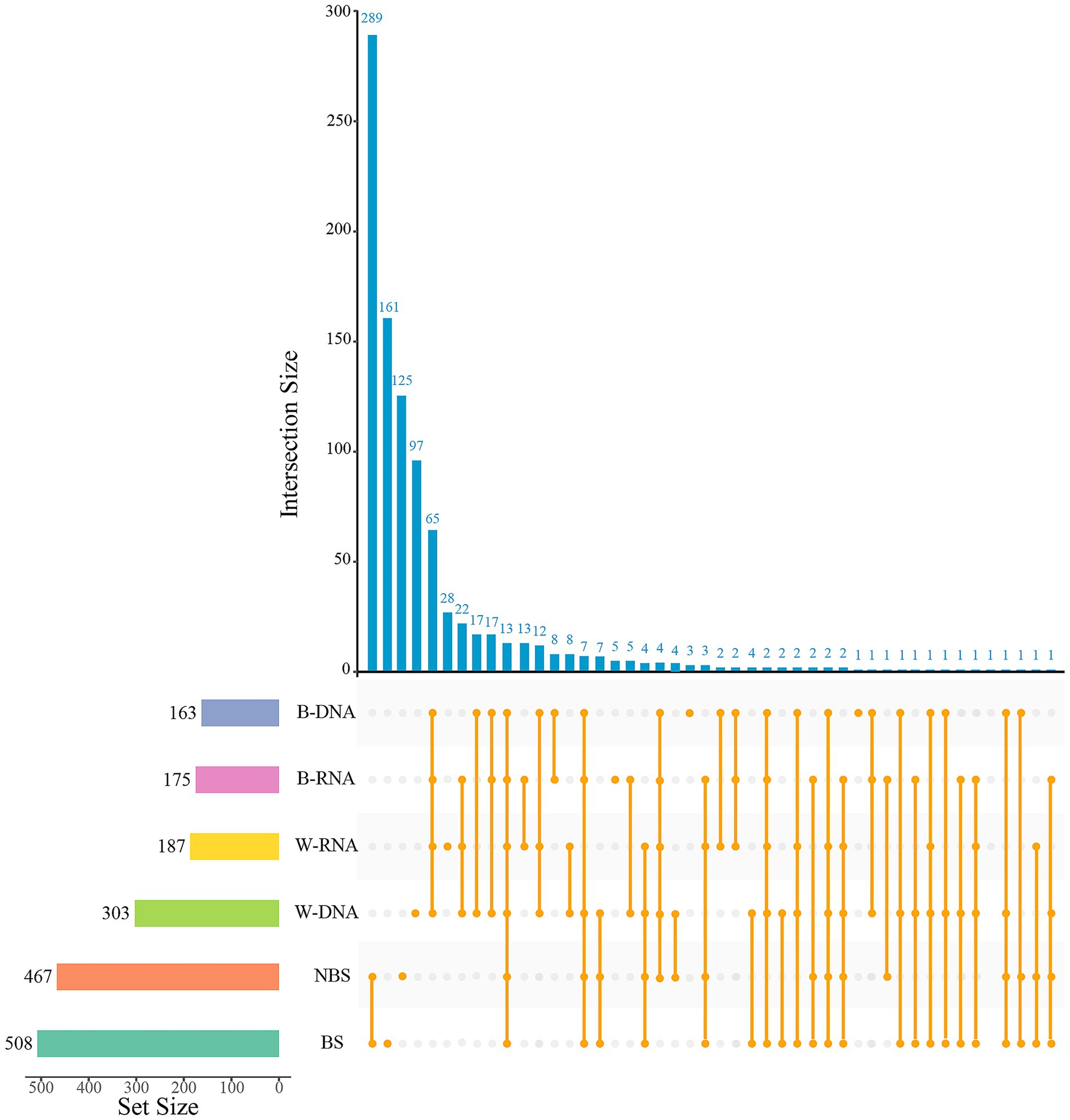
Figure 5. The Upset diagram showing the intersection of bacterial OTUs in different type samples from Mogao Grottoes and Maijishan Grottoes. The dark-blue histogram shows the quantity of intersection OTUs; the histogram of the left shows the total of OTUs in each sample, and the orange dots and lines represent the intersection of samples.
Hierarchical clustering of the top 50 bacterial genera revealed site-specific community composition (Figure 6). Maijishan samples clustered by cave origin, one was for 5 samples collected from Cave No. 30, the other group included 6 samples collected from Cave No. 28 (Figure 6A). Genera were organized into three main clusters, Cluster 1, which included Rubrobacter, Pseudonocardia, Actinomycetospora, Escherichia-Shigella, and Stenotrophomonas, was predominantly present across nearly all samples. Cluster 2, comprising Nocardioides, Lysobacter, Amycolatopsis, etc., was primarily concentrated in samples derived from Cave No. 28. In contrast, Cluster 3, which included Saccharopolyspora, Rhodococcus, Achromobacter, and Pseudomonas, exhibited no discernible distribution pattern. In contrast, Mogao Cave No. 256 samples were divided into two main groups based on sampling locations, the samples located proximal to the tunnel passageway formed one group (BS1, BS3, and NBS1), whereas interior-positioned samples formed another (BS2, NBS2 and NBS3). Dominant genera formed two key clusters: Cluster 1 included Escherichia-Shigella and Achromobacter, Cluster 2 included Rhodococcus, Ralstonia, and Bacillus, distributed across various samples.
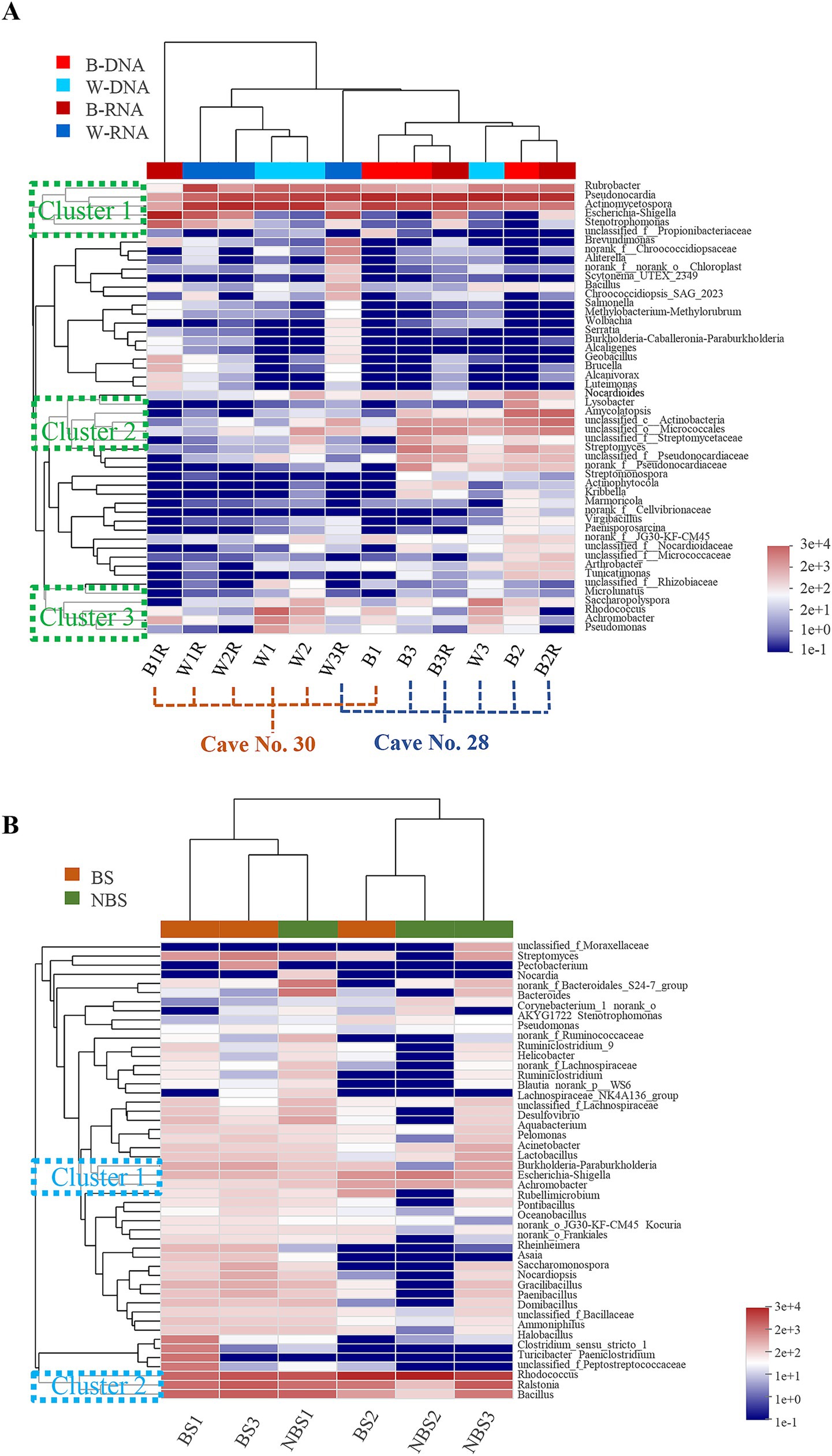
Figure 6. Heatmap and hierarchical cluster analysis of bacterial communities with the top 50 abundant genera in the Maijishan Grottoes (A) and Mogao Grottoes (B), respectively.
3.4 Comparison of diurnal variation of temperature in the Maijishan and Mogao Grottoes
In Maijishan Grottoes, mean diurnal range of air temperature (28-3, 30-3) were significantly higher than those of wall painting surfaces (28 N-S, 30 N-S) and plaster layers (28 N-IN, 30 N-IN). Within each cave system, the mean diurnal range of temperature of wall painting surfaces exceeded plaster layers, indicating that air temperature was more susceptible to external environmental disturbances. Cave No. 30 experienced the most pronounced thermal variability, with the mean diurnal range of air temperature exceeding 5°C annually and peaking at 12°C during November–December, markedly higher than equivalent periods in Cave No. 28. Both caves exhibited minimal diurnal temperature fluctuations (mean range < 2°C) across wall painting surfaces and plaster substrates. However, Cave No. 30 displayed consistently elevated temperature ranges at corresponding monitoring points relative to Cave No. 28 (Supplementary Figure S3A). Contrastingly, the Cave No. 256 of Mogao Grottoes demonstrated smaller mean diurnal range of temperature across five monitoring points. Excluding cave air (ESA), all measurement positions maintained sub-0.5°C fluctuation thresholds, the west wall (WW) exhibited the highest mean diurnal range of temperature in plaster layers for nearly half the year (January, February, August, November, and December, Supplementary Figure S3B).
3.5 Comparison of diurnal variation of relative humidity in Maijishan and Mogao Grottoes
RH data showed that the mean diurnal RH range at all plaster layers monitoring points in both caves of Maijishan Grottoes (2% ~ 10%) exceeded those in Mogao Grottoes Cave No. 256 of (1% ~ 5%). In Maijishan, Cave No. 30 exhibited the highest RH fluctuation in cave air (30-3), peaking at 23.32% in December, while its plaster layer (30 N-IN) remained relatively stable humidity (Figure 7A). Both caves showed higher daily RH variations on wall painting surfaces than in plaster support layers, suggesting that surface RH is more sensitive to ambient changes. At Cave No. 256 of Mogao Grottoes, RH in cave air showed strong seasonal variation associated with precipitation (Figure 7B), with diurnal fluctuations ranging from 2.11% (February) to 9.80% (July). From February to October, RH fluctuations in cave air exceeded those in plaster layers. The west wall (WW) plaster layer displayed exceptional RH stability, with fluctuations remaining below 2%, likely due to its location far from the cave entrance and limited air exchange (Figure 7B).
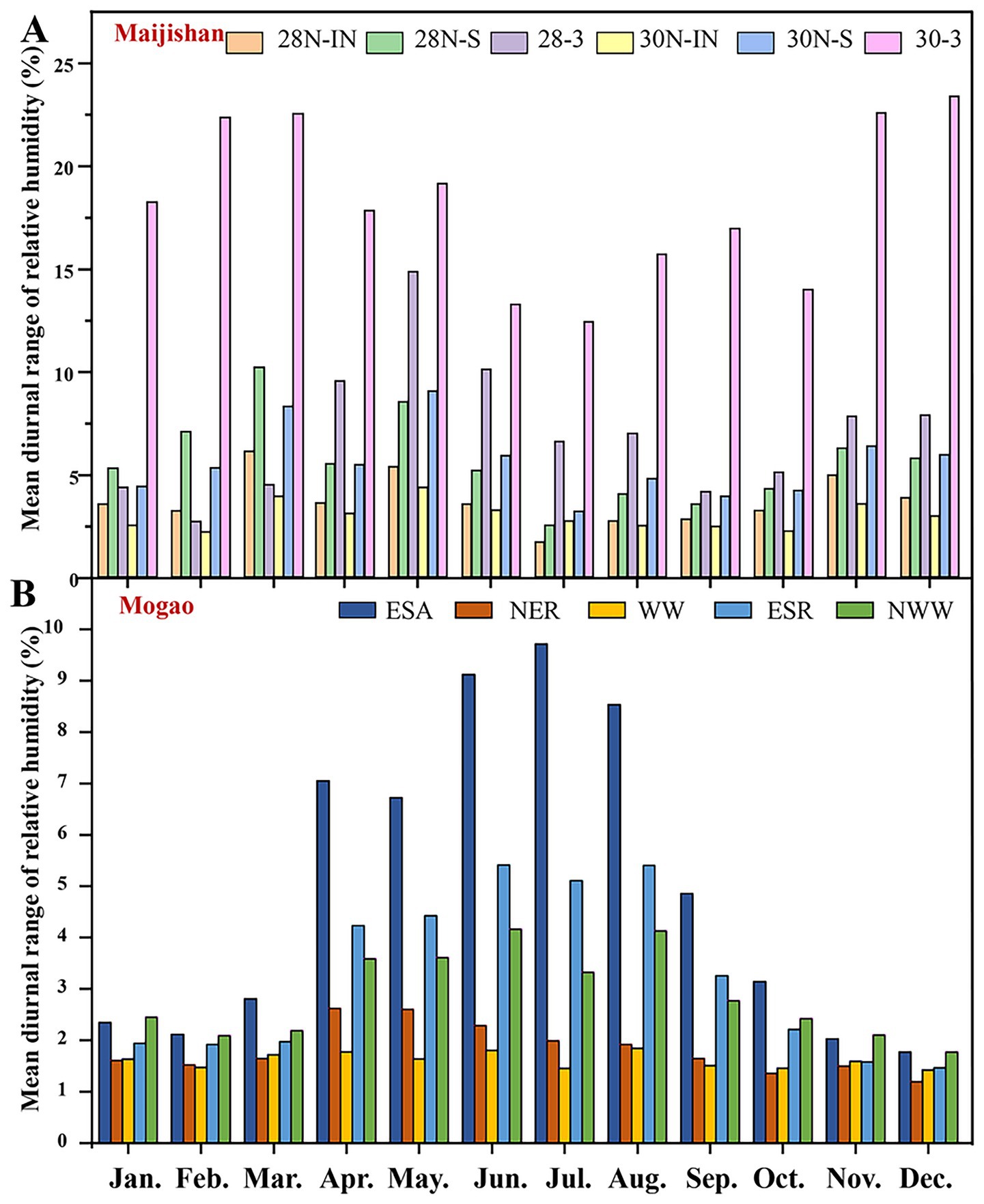
Figure 7. Mean diurnal range of relative humidity within Caves No. 28 and No. 30 of the Maijishan Grottoes (A) and Cave No. 256 of the Mogao Grottoes (B) for entire year. Six monitoring sites of the Maijishan Grottoes comprising wall painting plaster layer of Cave No. 28 and Cave No. 30 (28N-IN, 30N-IN), wall painting surface of Cave No. 28 and Cave No. 30 (28N-S, 30N-S), air of Cave No. 28 and Cave No. 30 (28-3, 30N-3); Five monitoring sites of the Mogao Grottoes comprising one for air at the eastern part of the southern slope (ESA) and four for murals at the north part of the northeast roof (NER), the west wall (WW), the eastern part of southern roof (ESR), and the northwest wall (NWW).
Daily maximum RH frequency analysis showed that in Maijishan Grottoes (Caves No. 28 and 30), RH most often fell within 70–80% at all monitoring points (Table 2). In Cave No. 28, RH > 70% occurred on 214 days at the plaster layer (28 N-IN), 207 days at the wall painting surface (28 N-S), and 185 days in cave air (28-3). In Cave No. 30, corresponding values were 209 days (30 N-IN), 202 days (30 N-S), and 146 days (30-3), respectively. Extreme RH events (>90%) were more common in Cave 28 and more frequent in plaster layers than on wall painting surfaces. In contrast, at Mogao Cave No. 256, daily maximum RH at most points fell within 20–30%, with RH > 70% occurring only on a few days, and never exceeding 80% (Table 2). Notably, the west wall (WW), showing visible water damage, had 73 days with RH > 60%, much higher than other plaster sites. The northeast roof (NER) near the entrance remained RH < 60% year-round.
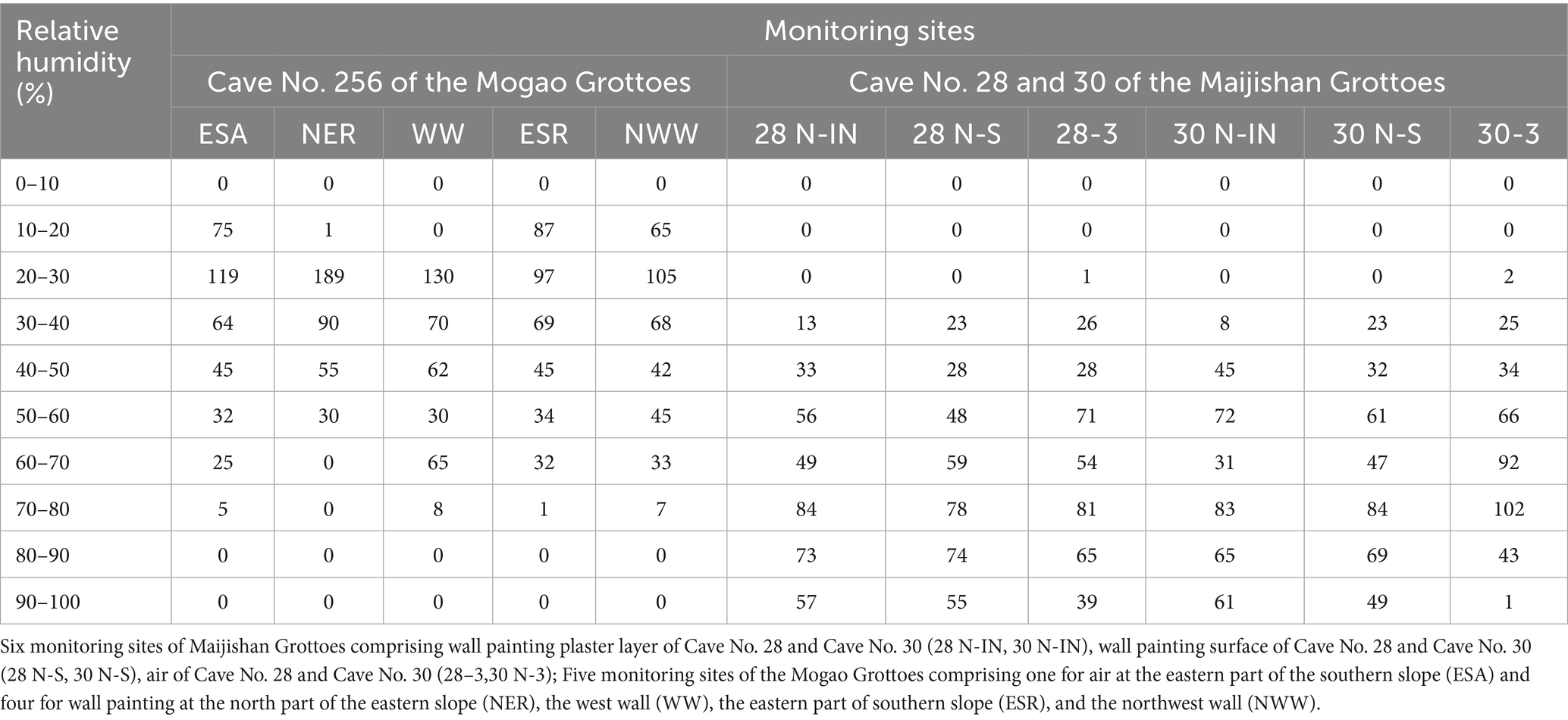
Table 2. Daily maximum relative humidity distribution frequency of monitoring sites in investigative caves of two heritage sites.
4 Discussion
4.1 The core taxa and their biodeterioration potential in Maijishan Grottoes
In this study, combined DNA-based and RNA-based Illumina MiSeq sequencing was employed to comprehensively characterize both total and metabolically active bacterial communities associated with black and white mycelial biofilms in the Maijishan Grottoes. RNA-level sequencing enabled insight into microbial activity, complementing DNA-based community assessments and offering a more dynamic perspective on active biodegradation risks (Meng et al., 2020; Li et al., 2021). Bacterial α-diversity and overall taxonomic profiles showed no significant differences between black and white mycelia within Maijishan Grottoes, suggesting that community composition alone cannot account for the observed phenotypic color variation (Langille et al., 2013; Quince et al., 2017). The distinct coloration of black and white mycelia might arise from a combination of microbial communities, pigment production (e.g., melanins, carotenoids), and the physicochemical properties of extracellular polymeric substances (EPS, Hathaway et al., 2014; Qin and Xia, 2024; Zomer et al., 2024). Moreover, mycelia sampled from the same cave shared highly similar bacterial communities regardless of their color, indicating the dominant influence of cave-specific microenvironmental conditions, and supporting that microenvironmental parameters are the primary determinants of bacterial community assembly (Lavoie et al., 2017).
Microbial biofilms across diverse cultural heritage sites often shared microbial phyla (Hathaway et al., 2014; Duan et al., 2021). Regarding bacterial community structures, Actinobacteria dominated most samples in both DNA and RNA samples (except for sample B1R). Actinobacteria and Proteobacteria are typical phyla have been widely documented in heritage environments such as caves or subterranean sites (Jurado et al., 2005, 2008; Li et al., 2024). As prolific producers of secondary metabolite producers, Actinobacteria contribute to wall painting deterioration by releasing pigments, acids, and antimicrobial compounds, thereby promoting chromatic biofilm formation (Lepinay et al., 2018; Oliveira et al., 2017; Wang et al., 2012). Proteobacteria serve as keystone chemoorganotrophic degraders, functioning as sensitive biological indicators of active biodeterioration (Ai et al., 2022; Khadka et al., 2017).
At the genus level, Pseudonocardia was particularly dominant in samples from Cave No. 30, corroborating its established role as an important taxon in wall paintings at the Maijishan Grottoes (Duan et al., 2017). This genus exhibited acidogenic metabolism capable of modulating local pH to favor oligotrophic proliferation, a critical adaptation enabling dominance in xeric wall painting substrates (Duan et al., 2017; Stomeo et al., 2008). Pseudonocardia thrived in subterranean heritage sites under harsh conditions, such as low light, scarce organic matter, high salinity, elevated humidity and temperature (Laiz et al., 1999), their rapid growth could lead to the formation of white spots on wall paintings (Duan et al., 2021). Another dominant genus, Actinomycetospora, was implicated in microbial outbreaks across multiple caves of the Maijishan Grottoes (He et al., 2021). Importantly, RNA sequencing revealed that Escherichia-Shigella and Stenotrophomonas were substantially more abundant at the RNA-level than in DNA profiles, indicating high metabolic activity despite their relatively lower overall abundance. Thereinto, Escherichia-Shigella (Proteobacteria) often associated with intestinal microbiota (Stoddard et al., 1998; Wang et al., 2013), likely originate from animal vectors such as rodents, birds, or arthropods inhabiting the forested surroundings of the Maijishan Grottoes. These animals are likely to act as vectors for microbial dispersal, introducing exogenous microorganisms (e.g., intestinal microbiota via excreta) into the cave ecosystem. Their activity may exacerbate biodeterioration risks by altering microhabitat conditions or facilitating organic matter accumulation, thereby fostering microbial proliferation on wall paintings (Liu W. et al., 2022; Bastian et al., 2010). Additionally, Rubrobacter was identified in samples, aligning with findings from the Angkor Thom Bayon Temple (Zhang et al., 2018) and other caves of the Maijishan Grottoes (Duan et al., 2017; He et al., 2021), which was recognized as a core genus associated with rosy discoloration on wall paintings, sandstone, and masonry (Schabereiter-Gurtner et al., 2001; Imperi et al., 2007; Nugari et al., 2009). Together, the RNA results demonstrate that a subset of the bacterial community is actively engaged in processes with direct biodeterioration potential, highlighting the importance of integrating RNA data into conservation-oriented microbial risk assessments.
Under favorable growth conditions, microorganisms can colonize on wall paintings by utilizing carbon sources, nitrogen sources, and mineral nutrients derived from original painting materials and restoration plasters (Meng et al., 2020). In this study, several differences were observed in total organic carbon, total nitrogen, and electrical conductivity between Cave No. 28 and Cave No. 30. These differences might alter microenvironmental conditions (Li and Gu, 2022) and provide distinct nutrient substrates for microbial proliferation (Imperi et al., 2007; Sterflinger and Pinzari, 2012). Combining with cave-specific temperature and humidity, microenvironmental conditions might lead the differences in bacterial community composition between the two caves.
4.2 Difference of bacterial community and their adaptions
Across both sites, Actinobacteria, Proteobacteria, and Firmicutes were the core bacterial phyla associated with biodeterioration, consistent with their recognized roles as pioneer colonizers across diverse lithic heritage substrates (Li et al., 2017; Liu et al., 2019; Xing et al., 2023). Actinobacteria was the dominant bacterial phylum at two sites of this study. However, the relative abundance of Actinobacteria in Mogao samples was markedly lower in the Maijishan Grottoes samples. The Maijishan Grottoes exhibited sustained high humidity levels year-round, with extreme humidity events exceeding 90%, which are mainly attributed to their unique geographical setting characterized by dense forest coverage, frequent precipitation, and limited cave ventilation (Yao et al., 2022). The observed variation in Actinobacteria abundance may originate from their hydrophilic properties, the persistently humid microenvironment promoted actinobacterial spore germination and growth (Zvyagintsev et al., 2007). Conversely, samples from the Mogao Grottoes exhibited a higher relative abundance of Firmicutes. Certain members of the phylum Firmicutes, particularly within the classes Bacilli (e.g., Bacillus) and Clostridia (e.g., Clostridium), are capable of forming endospores, which serve as a key mechanism for surviving extreme environments such as drought. These endospores possess a multilayered protective structure that resists desiccation, high temperatures, and UV radiation, significantly enhancing their survival rates under arid conditions (Setlow, 2007; Mckenney et al., 2013). The arid climate of the Dunhuang region selected for the desiccation-resistant metabolic strategies of Firmicutes, enabling their ecological dominance in the Mogao Grottoes’ ecosystems (Ma et al., 2015). Proteobacteria emerged as the dominant taxa driving the biodeterioration processes on cultural heritage substrates, particularly Alpha-proteobacteria, played a pivotal role in heritage biodeterioration through their metabolic versatility and adaptive strategies. Part of Proteobacteria exhibited oligotrophic capabilities, enabling colonization of nutrient-poor stone surfaces via the utilization of diverse organic compounds and UV-resistant pigment production (Pinhassi and Berman, 2003; Yu et al., 2024). At the genus level, Rhodococcus was dominant in samples of the Mogao Grottoes, they were also frequently documented in global heritage biodeterioration cases. Conversely, Pseudonocardia dominated the samples of Maijishan Grottoes, a genus commonly associated with wall paintings in caves and tombs (Stomeo et al., 2008; Portillo et al., 2009; Liu W. et al., 2022). Several studies have confirmed that Actinobacteria (e.g., Pseudonocardia) exhibit strong adaptability to oligotrophic conditions with limited organic nutrients. As pioneering colonizers in natural caves, they initiate early growth and establish material foundations for other microorganisms (Ciferri, 1999). Moreover, Pseudonocardia actively modifies environmental pH through metabolic processes, creating localized slightly alkaline microenvironments that promote rapid proliferation, then contributed to biodeterioration (Portillo and Gonzalez, 2011).
Only 13 OTUs belonged to few overlapping bacterial genera were identified between the two sites, revealing the possible role of site-specific environmental conditions (e.g., humidity, temperature, and substrate composition) in shaping bacterial community composition on wall paintings, the microenvironment exerting selective pressure on bacterial communities (Allen et al., 2020). Microbial damages to wall paintings were typically shaped by the metabolic activities and successional dynamics of diverse microbial communities, comprising bacteria, fungi, and extracellular polymeric substances (EPS). Studies indicated that bacteria play critical roles in microbial biofilms as ecological modulators, dynamically reshaping the composition of other biofilm-associated microorganisms (e.g., fungi, algae) in response to microenvironmental fluctuations (Liu et al., 2020). In our study, bacteria likely serve analogous functions: though not dominant in white/black mycelia, they might respond to microenvironmental variations and influence these mycelia on wall paintings.
4.3 Interplay between environments and bacterial communities
The Maijishan Grottoes, situated in a forested mountainous region with persistent high humidity, the temperature and RH fluctuations might be predominantly influenced by seasonal changes and heat exchange of cliff rock. The Cave No. 28 and Cave No. 30 maintained RH > 70% for over half the year, creating ideal conditions for microbial outbreaks and biodeterioration (He et al., 2022). Rainfall infiltration through cliff fractures and monsoon-driven precipitation (July–September) elevate RH to near 90% for >50 days annually, triggering efflorescence, cracking, pigment layer exfoliation and plaster layers destabilization via thermal expansion-contraction cycles (Bertolin et al., 2020; Li and Gu, 2022; Wang et al., 2025; Rivera et al., 2006). Despite high microbial risk in the Maijishan Grottoes, the bacterial and fungal concentrations (mean: 754 CFU/m3 and 645 CFU/m3, respectively) maintained at a lower level (Duan et al., 2018). Air microbial concentrations between 500 and 1,000 CFU/m3 indicated moderate visitor impact, belonging to balanced level, necessitating moderate crowd control (Porca et al., 2011).
The Mogao Grottoes, located in a desert region with distinct seasons, experienced extreme aridity and significant diurnal temperature fluctuations. Cave No. 256 maintained a relatively lower humidity, with RH consistently below 70%. This stable microenvironment can inhibit microbial colonization, favoring heritage preservation (Loli and Bertolin, 2018). In addition, this cave exhibited minimal daily temperature variation (diurnal range <1°C), which typically supports microbial proliferation, but the arid conditions (RH < 70% for over two-thirds of the year) inhibited numerous microbial growth (Spagnuolo et al., 2019; Sterflinger, 2010). Low precipitation, intense solar radiation, and high evaporation accelerated salt efflorescence on wall paintings via the migration of capillary water, leading efflorescence and then induced painting layers detachment. Microbial communities undergo niche selection on the wall paintings with efflorescence progression, the drought-adapted taxa potentially accelerating growth and deterioration (Ma et al., 2020).
In the Mogao Grottoes samples, more than half of the OTUs were exclusively detected in the two types (BS, NBS) of samples from this location, while at the Maijishan Grottoes, over one-third of the OTUs were shared across all four sample types (W-DNA, B-DNA, W-RNA, B-RNA). The bacterial communities demonstrated significant divergence in both diversity and structural composition at two sites, with community characteristics showing strong spatial dependency on sampling locations. The composition of microbial communities on cultural heritage surfaces are also shaped by dynamic interactions between substrate physicochemical properties (e.g., organic carbon content, nitrogen content, electric conductivity) and site-specific environmental stressors within the microenvironment such as pH, temperature and humidity (Chen and Gu, 2022; Dias et al., 2023; Ding et al., 2021; Duan et al., 2018, 2021; Zhang et al., 2018). Among these variables, environmental conditions emerged as an important determinant, critically modulating both microbial taxonomic dominance and their metabolic functionality across heritage ecosystems (Biagioli et al., 2024). Concurrently, increasing global tourism compounds risks through organic contamination and microclimatic destabilization (Ai et al., 2022; Li et al., 2022). In these semi-enclosed ecosystems, niche filtering overrides dispersal limitations (Saarela et al., 2004; Vellend, 2010). Climatic parameters critically modulate the ecological dominance of microbial taxa in heritage sites (Biagioli et al., 2024). Bacterial communities exhibited remarkable adaptability and resilience under different environmental stress, with their composition dynamically adjusting to maintain stability in response to fluctuant microenvironmental conditions (Chen et al., 2023).
5 Conclusion
This study revealed significant differences in bacterial communities and drivers of microbes on wall paintings in the Maijishan and Mogao Grottoes. Persistent high humidity and cliff seepage in the Maijishan Grottoes might promote Actinobacteria dominance (e.g., Pseudonocardia), their metabolic byproducts (organic acids, pigments) might exacerbate painting layer degradation. In contrast, the arid conditions of the Mogao Grottoes selected for desiccation-tolerant Firmicutes (e.g., Bacillus) and halophilic Proteobacteria (e.g., Halomonas), with salt efflorescence posing a primary threat to integrity of wall paintings. Therefore, site-specific microbial risks call for tailored conservation interventions. For the Maijishan Grottoes, strategies should prioritize humidity regulation (e.g., cyclic dehumidification) and control of Actinobacterial spore. In addition, RNA-level analysis identified metabolically active enteric bacteria (e.g., Escherichia) in the Maijishan Grottoes, potentially introduced by wildlife, thus more attention is necessary for the future preventative conservation. The lack of shared dominant genera between two heritage sites emphasizes the importance of localized microbial surveillance and environment-specific preservation approaches. These findings may advance our understanding of microbial ecology in cultural heritage environments and potentially support the development of evidence-based, sustainable conservation strategies for ancient wall paintings.
Data availability statement
The datasets presented in this study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, accession number PRJNA950462.
Author contributions
WM: Funding acquisition, Data curation, Visualization, Writing – review & editing, Methodology, Writing – original draft, Conceptualization. QC: Validation, Writing – review & editing, Investigation. FW: Writing – review & editing, Methodology, Funding acquisition, Supervision, Resources, Conceptualization. DH: Project administration, Funding acquisition, Supervision, Writing – review & editing, Formal analysis. YD: Software, Funding acquisition, Writing – review & editing. YY: Investigation, Writing – review & editing, Formal analysis. J-DG: Writing – review & editing. XY: Writing – review & editing, Formal analysis. HF: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National Natural Science Foundation of China (No. 32400082; 32460278); National Youth Top Talent Program of China; The Science and Technology Plan of Gansu Province, China (No. 23JRRA644); The Open Project of Gansu Provincial Research Center for Conservation of Dunhuang Cultural Heritage (No. GDW2021ZD08); and The Project of Gansu Provincial Bureau of Cultural Heritage (No. GSWW202229).
Acknowledgments
We would like to give our sincere gratitude to Junjian Hu, Ruibo Pei and Peng Xu from the Institute of Maijishan Grottoes Art for their help during the fieldwork.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1657118/full#supplementary-material
References
Abdel-Haliem, M. E. F., Sakr, A. A., Ali, M. F., Ghaly, M. F., and Sohlenkamp, C. (2013). Characterization of Streptomyces isolates causing colour changes of mural paintings in ancient Egyptian tombs. Microbiol. Res. 168, 428–437. doi: 10.1016/j.micres.2013.02.004
Ai, J., Guo, J., Li, Y., Zhong, X., Lv, Y., Li, J., et al. (2022). The diversity of microbes and prediction of their functions in karst caves under the influence of human tourism activities—a case study of Zhijin cave in Southwest China. Environ. Sci. Pollut. Res. 29, 25858–25868. doi: 10.1007/s11356-021-17783-x
Allen, R., Hoffmann, L. J., Larcombe, M. J., Louisson, Z., and Summerfield, T. C. (2020). Homogeneous environmental selection dominates microbial community assembly in the oligotrophic South Pacific gyre. Mol. Ecol. 29, 4680–4691. doi: 10.1111/mec.15651
Barnard, R. L., Osborne, C. A., and Firestone, M. K. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241. doi: 10.1038/ismej.2013.104
Bastian, F., Jurado, V., Nováková, A., Alabouvette, C., and Saiz-Jimenez, C. (2010). The microbiology of Lascaux cave. Microbiology 156, 644–652. doi: 10.1099/mic.0.036160-0
Bertolin, C., De Ferri, L., Grottesi, G., and Strojecki, M. (2020). Study on the conservation state of wooden historical structures by means of acoustic attenuation and vacuum microbalance. Wood Sci. Technol. 54, 203–226.
Biagioli, F., Coleine, C., Delgado-Baquerizo, M., Feng, Y., Saiz-Jimenez, C., and Selbmann, L. (2024). Outdoor climate drives diversity patterns of dominant microbial taxa in caves worldwide. Sci. Total Environ. 906:167674. doi: 10.1016/j.scitotenv.2023.167674
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen, J., and Gu, J. (2022). The environmental factors used in correlation analysis with microbial community of environmental and cultural heritage samples. Int. Biodeterior. Biodegrad. 173:105460. doi: 10.1016/j.ibiod.2022.105460
Chen, J., Zhao, Q., Li, F., Zhao, X., Wang, Y., Zhang, L., et al. (2023). Nutrient availability and acid erosion determine the early colonization of limestone by lithobiontic microorganisms. Front. Microbiol. 14:1194871. doi: 10.3389/fmicb.2023.1194871
Ciferri, O. (1999). Microbial degradation of paintings. Appl. Environ. Microbiol. 65, 879–885. doi: 10.1128/AEM.65.3.879-885.1999
Cojoc, L. R., Enache, M. I., Neagu, S. E., Lungulescu, M., Setnescu, R., Ruginescu, R., et al. (2019). Carotenoids produced by halophilic bacterial strains on mural paintings and laboratory conditions. FEMS Microbiol. Lett. 366:fnz243. doi: 10.1093/femsle/fnz243
Cuezva, S., Fernandez-Cortes, A., Porca, E., Pašić, L., Jurado, V., Hernandez-Marine, M., et al. (2012). The biogeochemical role of Actinobacteria in Altamira cave, Spain. FEMS Microbiol. Ecol. 81, 281–290. doi: 10.1111/j.1574-6941.2012.01391.x
Dias, L., Pires, V., Sitzia, F., Lisci, C., Candeias, A., Caldeira, A. T., et al. (2023). Evaluating the biosusceptibility of natural stone as an supporting tool to prevent cultural heritage biodeterioration. Eur. Phys. J. Plus 138:570. doi: 10.1140/epjp/s13360-023-04185-w
Ding, X., Lan, W., Li, Y., Yan, A., Katayama, Y., Koba, K., et al. (2021). An internal recycling mechanism between ammonia/ammonium and nitrate driven by ammonia-oxidizing archaea and bacteria (AOA, AOB, and Comammox) and DNRA on Angkor sandstone monuments. Int. Biodeterior. Biodegrad. 165:105328. doi: 10.1016/j.ibiod.2021.105328
Ding, X., Lan, W., Yan, A., Li, Y., Katayama, Y., and Gu, J.-D. (2022). Microbiome characteristics and the key biochemical reactions identified on stone world cultural heritage under different climate conditions. J. Environ. Manag. 302:114041. doi: 10.1016/j.jenvman.2021.114041
Dominguez-Moñino, I., Diaz-Herraiz, M., Jurado, V., Laiz, L., Miller, A. Z., Santos, J. L., et al. (2017). Nature and origin of the violet stains on the walls of a Roman tomb. Sci. Total Environ. 598, 889–899. doi: 10.1016/j.scitotenv.2017.04.017
Dornieden, T., Gorbushina, A. A., and Krumbein, W. E. (2000). Biodecay of cultural heritage as a space/time-related ecological situation—an evaluation of a series of studies. Int. Biodeterior. Biodegrad. 46, 261–270. doi: 10.1016/S0964-8305(00)00107-4
Duan, Y., Wu, F., He, D., Gu, J.-D., Feng, H., Chen, T., et al. (2021). Bacterial and fungal communities in the sandstone biofilms of two famous Buddhist grottoes in China. Int. Biodeterior. Biodegrad. 163:105267. doi: 10.1016/j.ibiod.2021.105267
Duan, Y., Wu, F., Wang, W., Gu, J.-D., Li, Y., Feng, H., et al. (2018). Differences of microbial community on the wall paintings preserved in situ and ex situ of the Tiantishan grottoes, China. Int. Biodeterior. Biodegrad. 132, 102–113. doi: 10.1016/j.ibiod.2018.02.013
Duan, Y., Wu, F., Wang, W., He, D., Gu, J.-D., Feng, H., et al. (2017). The microbial community characteristics of ancient painted sculptures in Maijishan grottoes, China. PLoS One 12:e0179718. doi: 10.1371/journal.pone.0179718
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Elhagrassy, A. F. (2018). Isolation and characterization of actinomycetes from mural paintings of Snu- Sert-ankh tomb, their antimicrobial activity, and their biodeterioration. Microbiol. Res. 216, 47–55. doi: 10.1016/j.micres.2018.08.005
Garg, K. L., Jain, K. K., and Mishra, A. K. (1995). Role of fungi in the deterioration of wall paintings. Sci. Total Environ. 167, 255–271. doi: 10.1016/0048-9697(95)04587-Q
Hathaway, J. J. M., Garcia, M. G., Balasch, M. M., Spilde, M. N., Stone, F. D., Dapkevicius, M. D. L. N. E., et al. (2014). Comparison of bacterial diversity in Azorean and Hawai’ian lava cave microbial mats. Geomicrobiol J. 31, 205–220. doi: 10.1080/01490451.2013.777491
He, D., Wu, F., Ma, W., Gu, J.-D., Xu, R., Hu, J., et al. (2022). Assessment of cleaning techniques and its effectiveness for controlling biodeterioration fungi on wall paintings of Maijishan grottoes. Int. Biodeterior. Biodegrad. 171:105406. doi: 10.1016/j.ibiod.2022.105406
He, D., Wu, F., Ma, W., Zhang, Y., Gu, J.-D., Duan, Y., et al. (2021). Insights into the bacterial and fungal communities and microbiome that causes a microbe outbreak on ancient wall paintings in the Maijishan grottoes. Int. Biodeterior. Biodegrad. 163:105250. doi: 10.1016/j.ibiod.2021.105250
Hu, H., and Hewitt, R. J. (2024). Understanding climate risks to world cultural heritage: a systematic analysis and assessment framework for the case of Spain. Herit. Sci. 12:194. doi: 10.1186/s40494-024-01299-x
Imperi, F., Caneva, G., Cancellieri, L., Ricci, M. A., Sodo, A., and Visca, P. (2007). The bacterial aetiology of rosy discoloration of ancient wall paintings. Environ. Microbiol. 9, 2894–2902. doi: 10.1111/j.1462-2920.2007.01393.x
Jurado, V., Boiron, P., Kroppenstedt, R. M., Laurent, F., Couble, A., Laiz, L., et al. (2008). Nocardia altamirensis sp. nov., isolated from Altamira cave, Cantabria, Spain. Int. J. Syst. Evol. Microbiol. 58, 2210–2214. doi: 10.1099/ijs.0.65482-0
Jurado, V., Groth, I., Gonzalez, J. M., Laiz, L., and Saiz-Jimenez, C. (2005). Agromyces subbeticus sp. Nov., isolated from a cave in southern Spain. Int. J. Syst. Evol. Microbiol. 55, 1897–1901. doi: 10.1099/ijs.0.63637-0
Khadka, B., Adeolu, M., Blankenship, R. E., and Gupta, R. S. (2017). Novel insights into the origin and diversification of photosynthesis based on analyses of conserved indels in the core reaction center proteins. Photosynth. Res. 131, 159–171. doi: 10.1007/s11120-016-0307-1
Lan, W., Li, H., Wang, W.-D., Katayama, Y., and Gu, J.-D. (2010). Microbial community analysis of fresh and old microbial biofilms on Bayon Temple sandstone of Angkor Thom, Cambodia. Microb. Ecol. 60, 105–115. doi: 10.1007/s00248-010-9707-5
Laiz, L., Groth, I., Gonzalez, I., and Saiz-Jimenez, C. (1999). Microbiological study of the dripping waters in Altamira cave (Santillana del Mar, Spain). J. Microbiol. Meth. 36:129.
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lavoie, K. H., Winter, A. S., Read, K. J. H., Hughes, E. M., Spilde, M. N., and Northup, D. E. (2017). Comparison of bacterial communities from lava cave microbial mats to overlying surface soils from lava beds National Monument, USA. PLoS One 12:e0169339. doi: 10.1371/journal.pone.0169339
Leissner, J., Kilian, R., Kotova, L., Jacob, D., Mikolajewicz, U., Broström, T., et al. (2015). Climate for culture: assessing the impact of climate change on the future indoor climate in historic buildings using simulations. Herit. Sci. 3:38. doi: 10.1186/s40494-015-0067-9
Lepinay, C., Mihajlovski, A., Touron, S., Seyer, D., Bousta, F., and Di Martino, P. (2018). Bacterial diversity associated with saline efflorescences damaging the walls of a French decorated prehistoric cave registered as a world cultural heritage site. Int. Biodeterior. Biodegrad. 130, 55–64. doi: 10.1016/j.ibiod.2018.03.016
Li, J., Deng, M., Gao, L., Yen, S., Katayama, Y., and Gu, J.-D. (2021). The active microbes and biochemical processes contributing to deterioration of Angkor sandstone monuments under the tropical climate in Cambodia – a review. J. Cult. Herit. 47, 218–226. doi: 10.1016/j.culher.2020.10.010
Li, J., He, Y., He, C., Xiao, L., Wang, N., Jiang, L., et al. (2024). Diversity and composition of microbial communities in Jinsha earthen site under different degree of deterioration. Environ. Res. 242:117675. doi: 10.1016/j.envres.2023.117675
Li, Q., Zhang, B., Wang, L., and Ge, Q. (2017). Distribution and diversity of bacteria and fungi colonizing ancient Buddhist statues analyzed by high-throughput sequencing. Int. Biodeterior. Biodegrad. 117, 245–254. doi: 10.1016/j.ibiod.2017.01.018
Liu, S.-J., Zhu, H.-Z., Zhang, Z.-F., Zhou, N., Jiang, C.-Y., Wang, B.-J., et al. (2019). Diversity, distribution and co-occurrence patterns of bacterial communities in a karst cave system. Front. Microbiol. 10:1726. doi: 10.3389/fmicb.2019.01726
Liu, W., Zhou, X., Jin, T., Li, Y., Wu, B., Yu, D., et al. (2022). Multikingdom interactions govern the microbiome in subterranean cultural heritage sites. Proc. Natl. Acad. Sci. USA 119:e2121141119. doi: 10.1073/pnas.2121141119
Liu, X., Koestler, R. J., Warscheid, T., Katayama, Y., and Gu, J.-D. (2020). Microbial deterioration and sustainable conservation of stone monuments and buildings. Nat. Sustain. 3, 991–1004. doi: 10.1038/s41893-020-00602-5
Liu, X., Meng, H., Wang, Y., Katayama, Y., and Gu, J.-D. (2018). Water is a critical factor in evaluating and assessing microbial colonization and destruction of Angkor sandstone monuments. Int. Biodeterior. Biodegrad. 133, 9–16. doi: 10.1016/j.ibiod.2018.05.011
Liu, X., Qian, Y., Wang, Y., Wu, F., Wang, W., and Gu, J.-D. (2022). Innovative approaches for the processes involved in microbial biodeterioration of cultural heritage materials. Curr. Opin. Biotechnol. 75:102716. doi: 10.1016/j.copbio.2022.102716
Li, X., Zhou, X., Wu, C., Petropoulos, E., Yu, Y., and Feng, Y. (2022). Temperature and moisture gradients drive the shifts of the bacterial microbiomes in 1000-year-old mausoleums. Atmos. 14:14. doi: 10.3390/atmos14010014
Li, Y.-H., and Gu, J.-D. (2022). A more accurate definition of water characteristics in stone materials for an improved understanding and effective protection of cultural heritage from biodeterioration. Int. Biodeterior. Biodegrad. 166:105338. doi: 10.1016/j.ibiod.2021.105338
Loli, A., and Bertolin, C. (2018). Indoor multi-risk scenarios of climate change effects on building materials in Scandinavian countries. Geosciences 8:347. doi: 10.3390/geosciences8090347
Ma, C., Fang, Z., Li, X., and Liu, X. (2023). Identification of bacterial communities involved in bioweathering crusts on limestone sculptures of the Longmen grottoes. Coatings 13:1506. doi: 10.3390/coatings13091506
Ma, W., Wu, F., He, D., Gu, J.-D., Chen, Y., Yue, Y., et al. (2025). Fungal community structure and viability in biofilms on wall paintings of the Maijishan grottoes. NPJ Herit. Sci. 13:386. doi: 10.1038/s40494-025-01962-x
Ma, W., Wu, F., He, D., Li, J., Zhang, Q., Yang, X., et al. (2023). The biodeterioration outbreak in Dunhuang Mogao grottoes analyzed for the microbial communities and the occurrence time by C-14 dating. Int. Biodeterior. Biodegrad. 178:105533. doi: 10.1016/j.ibiod.2022.105533
Ma, W., Wu, F., Tian, T., He, D., Zhang, Q., Gu, J.-D., et al. (2020). Fungal diversity and its contribution to the biodeterioration of mural paintings in two 1700-year-old tombs of China. Int. Biodeterior. Biodegrad. 152:104972. doi: 10.1016/j.ibiod.2020.104972
Ma, Y., Zhang, H., Du, Y., Tian, T., Xiang, T., Liu, X., et al. (2015). The community distribution of bacteria and fungi on ancient wall paintings of the Mogao grottoes. Sci. Rep. 5:7752. doi: 10.1038/srep07752
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martin-Sanchez, P. M., Nováková, A., Bastian, F., Alabouvette, C., and Saiz-Jimenez, C. (2012). Two new species of the genus Ochroconis, O. Lascauxensis and O. anomala isolated from black stains in Lascaux cave, France. Fungal Biol. 116, 574–589. doi: 10.1016/j.funbio.2012.02.006
Mckenney, P. T., Driks, A., and Eichenberger, P. (2013). The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 11, 33–44. doi: 10.1038/nrmicro2921
Meng, H., Katayama, Y., and Gu, J.-D. (2017). More wide occurrence and dominance of ammonia-oxidizing archaea than bacteria at three Angkor sandstone temples of Bayon, Phnom Krom and wat Athvea in Cambodia. Int. Biodeterior. Biodegrad. 117, 78–88. doi: 10.1016/j.ibiod.2016.11.012
Meng, H., Zhang, X., Katayama, Y., Ge, Q., and Gu, J.-D. (2020). Microbial diversity and composition of the Preah Vihear temple in Cambodia by high-throughput sequencing based on genomic DNA and RNA. Int. Biodeterior. Biodegrad. 149:104936. doi: 10.1016/j.ibiod.2020.104936
Mihajlovski, A., Gabarre, A., Seyer, D., Bousta, F., and Di Martino, P. (2017). Bacterial diversity on rock surface of the ruined part of a French historic monument: the Chaalis abbey. Int. Biodeterior. Biodegrad. 120, 161–169. doi: 10.1016/j.ibiod.2017.02.019
Nugari, M. P., Pietrini, A. M., Caneva, G., Imperi, F., and Visca, P. (2009). Biodeterioration of mural paintings in a rocky habitat: the crypt of the original sin (Matera, Italy). Int. Biodeterior. Biodegrad. 63, 705–711. doi: 10.1016/j.ibiod.2009.03.013
Oliveira, C., Gunderman, L., Coles, C., Lochmann, J., Parks, M., Ballard, E., et al. (2017). 16S rRNA gene-based metagenomic analysis of Ozark cave bacteria. Diversity 9:31. doi: 10.3390/d9030031
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 110, 6548–6553. doi: 10.1073/pnas.1302837110
Pei, S., Wu, F., Chen, Y., Ma, W., He, D., Zhang, Q., et al. (2023). Mechanisms of lead-containing pigment discoloration caused by Naumannella cuiyingiana AFT2T isolated from 1500 years tomb wall painting of China. Int. Biodeterior. Biodegrad. 185:105689. doi: 10.1016/j.ibiod.2023.105689
Pinhassi, J., and Berman, T. (2003). Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient dddition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69, 199–211. doi: 10.1128/AEM.69.1.199-211.2003
Porca, E., Jurado, V., Martin-Sanchez, P. M., Hermosin, B., Bastian, F., Alabouvette, C., et al. (2011). Aerobiology: an ecological indicator for early detection and control of fungal outbreaks in caves. Ecol. Indic. 11, 1594–1598. doi: 10.1016/j.ecolind.2011.04.003
Portillo, M. C., and Gonzalez, J. M. (2011). Moonmilk deposits originate from specific bacterial communities in Altamira cave (Spain). Microb. Ecol. 61, 182–189. doi: 10.1007/s00248-010-9731-5
Portillo, M. C., Saiz-Jimenez, C., and Gonzalez, J. M. (2009). Molecular characterization of total and metabolically active bacterial communities of “white colonizations” in the Altamira cave, Spain. Res. Microbiol. 160, 41–47. doi: 10.1016/j.resmic.2008.10.002
Qin, Y., and Xia, Y. (2024). Melanin in fungi: advances in structure, biosynthesis, regulation, and metabolic engineering. Microb. Cell Factories 23:334. doi: 10.1186/s12934-024-02614-8
Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J., and Segata, N. (2017). Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35, 833–844. doi: 10.1038/nbt.3935
Rivera, A. B., Rainer, L., and Vagts, L. (2006). “Out of their native earth: the history of excavation and cnversation of ancient hopi murals from Awatovi and Kawaika-a” in The conservation of decorated surfaces on earthen architecture. eds. L. Rainer and A. B. Rivera (Los Angeles, CA: Getty Conservation Institute), 53–65.
Saarela, M., Alakomi, H.-L., Suihko, M.-L., Maunuksela, L., Raaska, L., and Mattila-Sandholm, T. (2004). Heterotrophic microorganisms in air and biofilm samples from Roman catacombs, with special emphasis on actinobacteria and fungi. Int. Biodeterior. Biodegrad. 54, 27–37. doi: 10.1016/j.ibiod.2003.12.003
Sanmartín, P., DeAraujo, A., and Vasanthakumar, A. (2018). Melding the old with the new: trends in methods used to identify, monitor, and control microorganisms on cultural heritage materials. Microb. Ecol. 76, 64–80. doi: 10.1007/s00248-016-0770-4
Sansupa, C., Suphaphimol, N., Nonthijun, P., Ronsuek, T., Yimklan, S., Semakul, N., et al. (2023). Life on the wall: the diversity and activity of microbes on 13th – century AD. Lan Na mural painting. Front. Microbiol. 14:1220901. doi: 10.3389/fmicb.2023.1220901
Schabereiter-Gurtner, C., Piñar, G., Vybiral, D., Lubitz, W., and Rölleke, S. (2001). Rubrobacter -related bacteria associated with rosy discolouration of masonry and lime wall paintings. Arch. Microbiol. 176, 347–354. doi: 10.1007/s002030100333
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Setlow, P. (2007). I will survive: DNA protection in bacterial spores. Trends Microbiol. 15, 172–180. doi: 10.1016/j.tim.2007.02.004
Spagnuolo, A., Vetromile, C., Masiello, A., Alberghina, M. F., Schiavone, S., and Lubritto, C. (2019). Climate and cultural heritage: the case study of “real Sito di Carditello”. Heritage 2, 2053–2066. doi: 10.3390/heritage2030124
Sterflinger, K. (2010). Fungi: their role in deterioration of cultural heritage. Fungal Biol. Rev. 24, 47–55. doi: 10.1016/j.fbr.2010.03.003
Sterflinger, K., and Pinzari, F. (2012). The revenge of time: fungal deterioration of cultural heritage with particular reference to books, paper and parchment. Environ. Microbiol. 14, 559–566. doi: 10.1111/j.1462-2920.2011.02584.x
Stoddard, C. S., Coyne, M. S., and Grove, J. H. (1998). Fecal bacteria survival and infiltration through a shallow agricultural soil: timing and tillage effects. J. Environ. Qual. 27, 1516–1523. doi: 10.2134/jeq1998.00472425002700060031x
Stomeo, F., Portillo, M. C., Gonzalez, J. M., Laiz, L., and Saiz-Jimenez, C. (2008). Pseudonocardia in white colonizations in two caves with Paleolithic paintings. Int. Biodeterior. Biodegrad. 62, 483–486. doi: 10.1016/j.ibiod.2007.12.011
Sugiyama, J., Kiyuna, T., Nishijima, M., An, K.-D., Nagatsuka, Y., Tazato, N., et al. (2017). Polyphasic insights into the microbiomes of the Takamatsuzuka tumulus and Kitora tumulus. J. Gen. Appl. Microbiol. 63, 63–113. doi: 10.2323/jgam.2017.01.007
Vellend, M. (2010). Conceptual synthesis in community ecology. Q. Rev. Biol. 85, 183–206. doi: 10.1086/652373
Viles, H. A., and Cutler, N. A. (2012). Global environmental change and the biology of heritage structures. Glob. Change Biol. 18, 2406–2418. doi: 10.1111/j.1365-2486.2012.02713.x
Wang, L., Huang, J., Sanmartín, P., Martino, P. D., Wu, F., Urzì, C. E., et al. (2025). Water determines geomicrobiological impact on stone heritage. Nat. Geosci. 18, 108–111. doi: 10.1038/s41561-024-01631-x
Wang, W. F., Wu, F. S., Ji, A. H., and Feng, H. Y. (2013). Advancement and prospect of bionic techniques in the conservation of the cultural heritage. Appl. Mech. Mater. 461, 469–475. doi: 10.4028/www.scientific.net/AMM.461.469
Wang, W., Ma, Y., Ma, X., Wu, F., Ma, X., An, L., et al. (2012). Diversity and seasonal dynamics of airborne bacteria in the Mogao grottoes, Dunhuang, China. Aerobiologia 28, 27–38. doi: 10.1007/s10453-011-9208-0
Wu, F., Gu, J.-D., Li, J., Feng, H., and Wang, W. (2022). “Microbial colonization and protective management of wall paintings” in Cultural heritage microbiology: Recent developments. eds. R. Mitchell, J. Clifford, and A. Vasanthakumar (London: Archetype Publications), 57–84.
Wu, F., Zhang, Y., He, D., Gu, J.-D., Guo, Q., Liu, X., et al. (2021). Community structures of bacteria and archaea associated with the biodeterioration of sandstone sculptures at the Beishiku Temple. Int. Biodeterior. Biodegrad. 164:105290. doi: 10.1016/j.ibiod.2021.105290
Xing, W., Qi, B., Chen, R., Ding, W., and Zhang, F. (2023). Metagenomic analysis reveals taxonomic and functional diversity of microbial communities on the deteriorated wall paintings of Qinling tomb in the southern tang dynasty, China. BMC Microbiol. 23:140. doi: 10.1186/s12866-023-02887-w
Xu, H.-B., Tsukuda, M., Takahara, Y., Sato, T., Gu, J.-D., and Katayama, Y. (2018). Lithoautotrophical oxidation of elemental sulfur by fungi including Fusarium solani isolated from sandstone Angkor temples. Int. Biodeterior. Biodegrad. 126, 95–102. doi: 10.1016/j.ibiod.2017.10.005
Yao, S., Yan, Z., Ma, Q., Xu, B., Zhang, Z., Bi, W., et al. (2022). Analysis of the annual hygrothermal environment in the Maijishan grottoes by field measurements and numerical simulations. Build. Sci. 221:109229. doi: 10.1016/j.buildenv.2022.109229
Yu, Y., Zhang, J., Chen, R., Coleine, C., Liu, W., Delgado-Baquerizo, M., et al. (2024). Unearthing the global patterns of cultural heritage microbiome for conservation. Int. Biodeterior. Biodegrad. 190:105784. doi: 10.1016/j.ibiod.2024.105784
Zhang, G., Gong, C., Gu, J., Katayama, Y., Someya, T., and Gu, J.-D. (2019). Biochemical reactions and mechanisms involved in the biodeterioration of stone world cultural heritage under the tropical climate conditions. Int. Biodeterior. Biodegrad. 143:104723. doi: 10.1016/j.ibiod.2019.104723
Zhang, X., Ge, Q., Zhu, Z., Deng, Y., and Gu, J.-D. (2018). Microbiological community of the Royal Palace in Angkor Thom and Beng Mealea of Cambodia by Illumina sequencing based on 16S rRNA gene. Int. Biodeterior. Biodegrad. 134, 127–135. doi: 10.1016/j.ibiod.2018.06.018
Zhang, Y., Wu, F., Su, M., He, D., Gu, J.-D., Guo, Q., et al. (2021). Spatial and temporal distributions of microbial diversity under natural conditions on the sandstone stelae of the Beishiku Temple in China. Int. Biodeterior. Biodegrad. 163:105279. doi: 10.1016/j.ibiod.2021.105279
Zomer, A., Ingham, C. J., Von Meijenfeldt, F. A. B., Escobar Doncel, Á., Van De Kerkhof, G. T., Hamidjaja, R., et al. (2024). Structural color in the bacterial domain: the ecogenomics of a 2-dimensional optical phenotype. Proc. Natl. Acad. Sci. USA 121:e2309757121. doi: 10.1073/pnas.2309757121
Keywords: microbial damages, DNA-RNA high-throughput sequencing, relative humidity, environmental monitoring, sustainable preservation
Citation: Ma W, Chen Q, Wu F, He D, Duan Y, Yue Y, Gu J-D, Yang X and Feng H (2025) Difference and environmental drivers of bacterial communities on wall paintings of the Maijishan and Mogao Grottoes, China. Front. Microbiol. 16:1657118. doi: 10.3389/fmicb.2025.1657118
Edited by:
Mai Bingjie, Shanghai University, ChinaCopyright © 2025 Ma, Chen, Wu, He, Duan, Yue, Gu, Yang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fasi Wu, d3Vmc0BkaGEuYWMuY24=; Huyuan Feng, ZmVuZ2h5QGx6dS5lZHUuY24=
 Wenxia Ma
Wenxia Ma Qiqi Chen
Qiqi Chen Fasi Wu
Fasi Wu Dongpeng He2,3
Dongpeng He2,3 Yulong Duan
Yulong Duan Ji-Dong Gu
Ji-Dong Gu Xiaoyan Yang
Xiaoyan Yang Huyuan Feng
Huyuan Feng