- 1Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Beijing, China
Aims: Polyporus umbellatus sclerotium, known for its diuretic properties, relies on a symbiotic association with Armillaria for its growth and quality development. However, the impact of soil microorganisms on this symbiosis remains uncertain and warrants investigation. The primary objective of this research is to characterize the microorganisms capable of enhancing the symbiotic interaction between Armillaria gallica and Polyporus umbellatus sclerotia in the rhizosphere soil.
Methods: Symbiotic cultivation experiments were conducted in woodland habitats with four groups: symbiotic group (Z0), control group (Z1), A. gallica-only group (Z2), and P. umbellatus-only group (Z3). Rhizosphere soil community profiling analysis was conducted using high-throughput sequencing of the bacterial 16S rRNA gene. Subsequently, bacterial strains were isolated, purified, and back-inoculated with A. gallica to assess their effects on this symbiotic relationship.
Results: A total of 10,009 operational taxonomic units (OTUs) were identified, with the symbiotic group (Z0) showing higher bacterial richness and diversity (ACE, Chao1, Shannon indices) compared to Z2 and Z3. Dominant phyla such as Proteobacteria, Acidobacteriota, and Bacteroidota were notably more abundant in Z0. Notably, Rhodococcus sp. Z2-1 significantly promoted A. gallica rhizomorph growth (diameter increased by 112.2%, branches by 160.9%) and symbiosis establishment (100% contact rate in inoculated pots vs. 0–22.2% in controls).
Conclusion: The symbiotic relationship between P. umbellatus and A. gallica shapes rhizosphere bacterial communities, with specific bacteria like Rhodococcus sp. enhancing fungal growth and symbiotic efficiency. This study presents the potential for developing a bio-bacterial fertilizer for cultivation of medicinal material.
Introduction
Polyporus umbellatus (Pers.) Fries, a valuable medicinal fungus utilized in traditional medicine for its therapeutic attributes (Gao et al., 2025), depends significantly on its symbiotic relationship with Armillaria spp., which play a crucial role in influencing the growth rate, yield, and bioactive compound accumulation in P. umbellatus (Liu et al., 2025). Despite its medicinal significance, the extended natural growth cycle and inconsistent yield of P. umbellatus limit its widespread application. Therefore, a comprehensive understanding of the symbiotic mechanisms between P. umbellatus and A. gallica is essential for improving the efficiency and quality of P. umbellatus cultivation.
The rhizosphere microbiome, comprising the diverse microbial communities in the soil surrounding plant roots and fungal mycelia, plays a crucial role in the growth and development of both plants and fungi (Xun et al., 2024). These microbial communities engage in interactions with their hosts, influencing processes such as nutrient uptake, growth promotion, and disease resistance (Elnahal et al., 2022). Recent advancements in microbiomics have provided new methodologies for investigating the structure and functionality of rhizosphere microbial communities. Research indicates that the diversity and functions of rhizosphere microbiota are crucial for shaping symbiotic relationships (Wang et al., 2024). Nevertheless, research on the rhizosphere microbiome associated with P. umbellatus remains limited, especially regarding the dynamic changes and functional roles of microbial communities in the symbiotic system between P. umbellatus and A. gallica.
In the symbiotic relationship between P. umbellatus and A. gallica, the rhizosphere microbiome potentially influences the initiation and maintenance of this symbiosis through various mechanisms. A diverse microbial community can provide a plentiful nutrient source, thereby promoting the growth and development of both P. umbellatus and A. gallica. Moreover, specific microbial taxa may impact the growth of P. umbellatus and A. gallica directly or indirectly by generating plant growth-promoting compounds, facilitating phosphate solubilization, nitrogen fixation, and other functions. Additionally, the interactions within the microbial community can impact the disease resistance and adaptability of P. umbellatus and A. gallica. Nonetheless, the specific composition, functions, and mechanisms of the rhizosphere microbiome in this symbiotic system remain unclear.
This study aims to investigate the structure and diversity of bacterial communities in the rhizosphere soil of P. umbellatus and explore their roles in the symbiotic association of P. umbellatus and A. gallica. Utilizing high-throughput sequencing, we analyzed the bacterial communities across various experimental groups, including the symbiotic group, P. umbellatus-only group, A. gallica-only group, and control group. Furthermore, we isolated and identified specific bacterial strains and conducted co-culture experiments to assess their effects on the growth of P. umbellatus and A. gallica. The results of this study are anticipated to provide valuable theoretical insights into the microbial mechanisms underpinning the symbiotic relationship between P. umbellatus and A. gallica, offering scientific guidance for improving P. umbellatus cultivation and quality.
Materials and methods
The co-cultivation of Polyporus umbellatus sclerotia and Armillaria gallica
P. umbellatus sclerotia were cultivated in woodland habitats in Liuba County, Shaanxi Province, China (33 °30′ N, 106 °59′E). The experimental design consisted of four groups: the experimental group (Z0), the control group (devoid of both P. umbellatus and A. gallica in comparison to the experimental group) (Z1), the A. gallica-only group (containing A. gallica but lacked P. umbellatus compared to the experimental group) (Z2), and the P. umbellatus-only group (including P. umbellatus but lacking A. gallica compared to the experimental group) (Z3). A total of 30 holes were excavated in each group, measuring 50 cm in length, 30 cm in width, and 15 cm in depth. In each hole of the experimental group, one wooden stick (approximately 15 cm in diameter and 30 cm in length) was planted, along with 150 g of immature P. umbellatus sclerotia and 1,500 g of A. gallica-infected smaller wooden sticks (approximately 3 cm in diameter and 8 cm in length). Following a six-month cultivation period, rhizosphere soil samples of P. umbellatus were collected monthly, with five pits sampled during each collection. This approach ensured the acquisition of rhizosphere soil samples throughout the symbiotic phase between P. umbellatus and A. gallica.
Collection of soil samples
The experimental design and sample collection were adapted from the methods described by Zhao et al. (2024), with specific modifications to suit the current study. All soil subsamples collected during the same sampling period and treatment were homogenized and amalgamated into composite samples, which were subsequently transported to the laboratory in a foam box with dry ice. Upon arrival, the samples were stored at −80 °C for DNA extraction and subsequent high-throughput sequencing analysis. In the study design, Z0 represents soil from the experimental group, Z1 denotes soil from the control group, Z2 refers to soil from the A. gallica-only group, and Z3 indicates soil from the P. umbellatus-only group.
DNA extraction and 16S rRNA gene sequencing
Genomic DNA from soil samples was extracted using the cetyltrimethylammonium bromide (CTAB) method. Four soil samples were collected from each habitat. High-throughput sequencing of bacteria was performed using the 16S rRNA gene V3-V4 hypervariable region primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR system and conditions were consistent with prior work (Bo et al., 2024). Following PCR, product quantification, purification, library preparation, and 16S rRNA amplicon sequencing were performed by Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). Sequencing was performed on the Illumina NextSeq 2000 Sequencing Platform, generating 300 bp paired-end reads for 16S rRNA amplicon sequencing. A total of 1,072,758 raw reads were obtained through sequencing, and after quality filtering, 1,040,974 valid reads were obtained. The average sequencing depth of each sample was 52,049 reads/sample (range: 44,154–57,634 reads), ensuring that the sequencing depth of each sample was sufficient to cover community diversity.
Diversity of bacterial communities
The obtained data were processed using the QIIME software platform. The operational taxonomic units (OTUs) representing bacterial species in the 16S OTU table were identified. OTU clustering was performed with a 97% sequence similarity threshold. Species annotation analysis of the OTU sequences was conducted using the QIIME software and the SSU rRNA database SILVA138. Additional analyses of bacterial community diversity were carried out using I-Sanger Bioinformatics Cloud Platform (version 3.0) at https://cloud.majorbio.com. The bacterial co-occurrence interaction network was explored using Networkx (version1.11) to identify significant correlations between bacterial taxa (Maurice et al., 2024).
Isolation and identification of microbial communities associated with Polyporus umbellatus
A total of 10 g soil on the surface of P. umbellatus sclerotia was added in 50 mL of ddH2O with orbital shaker (120 rpm and 30 min). The resulting solutions were then diluted 104-fold. Subsequently, 100 μL of the diluted solution was spread onto various media, including nutrient agar, Luria–Bertani medium (LB), potato dextrose agar (PDA) medium and tryptone soya agar. Incubation of plates occurred at 25 °C for 2–7 days (Gao et al., 2024). Sequencing of the bacterial 16S rRNA gene and fungal ITS gene followed established protocols (Ying et al., 2012; Handique et al., 2024). PCR products were sequenced by Taihe Biotechnology Co., Ltd. (Beijing, China). Sequence identification was performed using EzBioCloud and NCBI BLAST databases (Chalita et al., 2024).
Co-culture experiments of isolated bacteria and Armillaria gallica
The impact of bacteria on growth rate and branching number of the rhizomorph of A. gallica was assessed (Liu et al., 2022). The bacteria were cultured in LB medium overnight at 28 °C with shaking at 180 rpm until reaching an optical density (OD600) of 0.5. Rhizomorph tips of A. gallica (approximately 2 mm in length) were placed at the center of PDA plates, with bacteria spread 2 cm away from both sides of the rhizomorphs. Control plates lacking bacteria were included. All plates were incubated in darkness at 25 °C for 14 days. The experiment was conducted with three replicates (n = 3), and measurements were taken for the diameter and branching number (branch length exceeding 1 mm) of the A. gallica rhizomorphs.
Pot experiment to assess the effect of Rhodococcus sp. strain Z2-1 on symbiosis establishment between Polyporus umbellatus and Armillaria gallica
Rhodococcus sp. strain Z2-1, previously isolated from the rhizosphere soil of P. umbellatus, was used to assess its impact on the symbiotic relationship establishment between P. umbellatus and A. gallica. A pot experiment was conducted with two treatments: a control group (normal cultivation) and an experimental group (inoculated with Z2-1). Each treatment consisted of five replicates planted in glass containers (351 × 211 × 250 mm) filled with sandy soil sterilized at 121 °C for 40 min. Each container was planted with a wooden stick (8 cm in diameter, 25 cm in length), 100 g of immature P. umbellatus sclerotia, and 1,000 g of smaller wooden sticks (5 cm in length, 2 cm in diameter) colonized by A. gallica. Rhodococcus sp. strain Z2-1 was cultured in 200 mL of Luria-Bertani (LB) medium at 28 °C with shaking at 180 rpm for 24 h. Bacterial cells were harvested by centrifugation at 4,000 rpm for 10 min, washed twice with saline, and resuspended in sterile distilled water to a final concentration of 1 × 108 CFU/mL. The experimental group received inoculation with this bacterial suspension, while the control group was treated with an equivalent volume of sterile distilled water. P. umbellatus was cultivated in darkness at room temperature for 6 months. Subsequently, the symbiotic rate between P. umbellatus and A. gallica was assessed to determine the symbiotic-promoting ability of Rhodococcus sp. strain Z2-1.
Statistical analysis
Microbial community analyses were conducted using the R statistical program, version 3.3.0. Student’s t-test was employed to compare the differences between the control and treatment groups. Results are presented as means ± SEM; *p < 0.05; **p < 0.01; ***p < 0.001.
Result
Microbial communities in the soil of the Polyporus umbellatus habitat
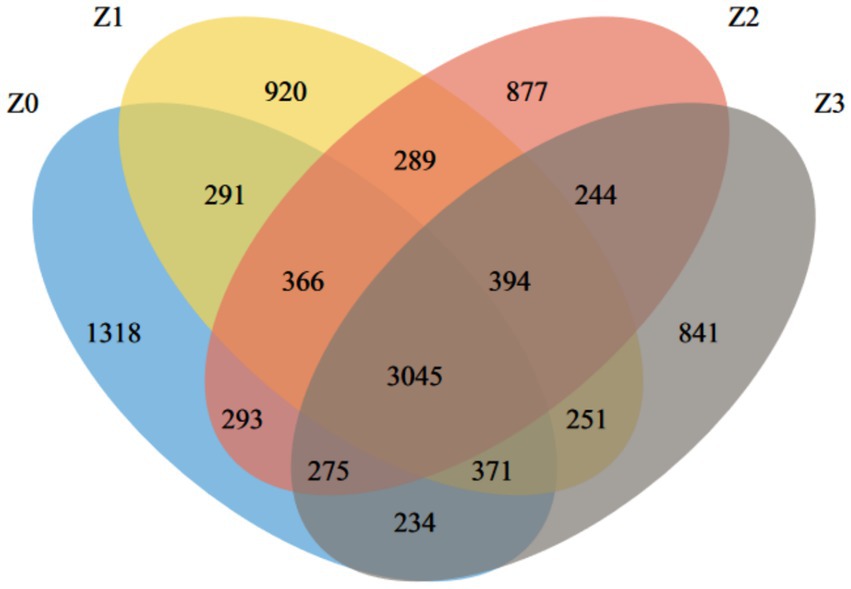
Rarefaction curve analysis showed that the curves for all samples leveled off (Supplementary Figure S1), indicating that the sequencing depth was sufficient to cover the community diversity and further supporting the reliability of the α-diversity and β-diversity analysis results in this study. A total of 10,009 OTUs were identified from the soil samples, with 3,045 core OTUs common to all four samples, representing 30.42% of the total OTUs. The distribution of bacterial OTUs in the samples was as follows: 1,318 in Z0, 920 in Z1, 877 in Z2, and 841 in Z3. The results revealed distinct microbial compositions among the habitats of normally cultivated P. umbellatus (Z0), the blank control (Z1), the A. gallica-only group (Z2) and the P. umbellatus-only group (Z3). Specifically, the number of OTUs in Z0 was significantly higher than that in Z1, Z2, and Z3 (Figure 1), suggesting increased bacterial community richness in the habitat of P. umbellatus and A. gallica during their symbiotic interaction.
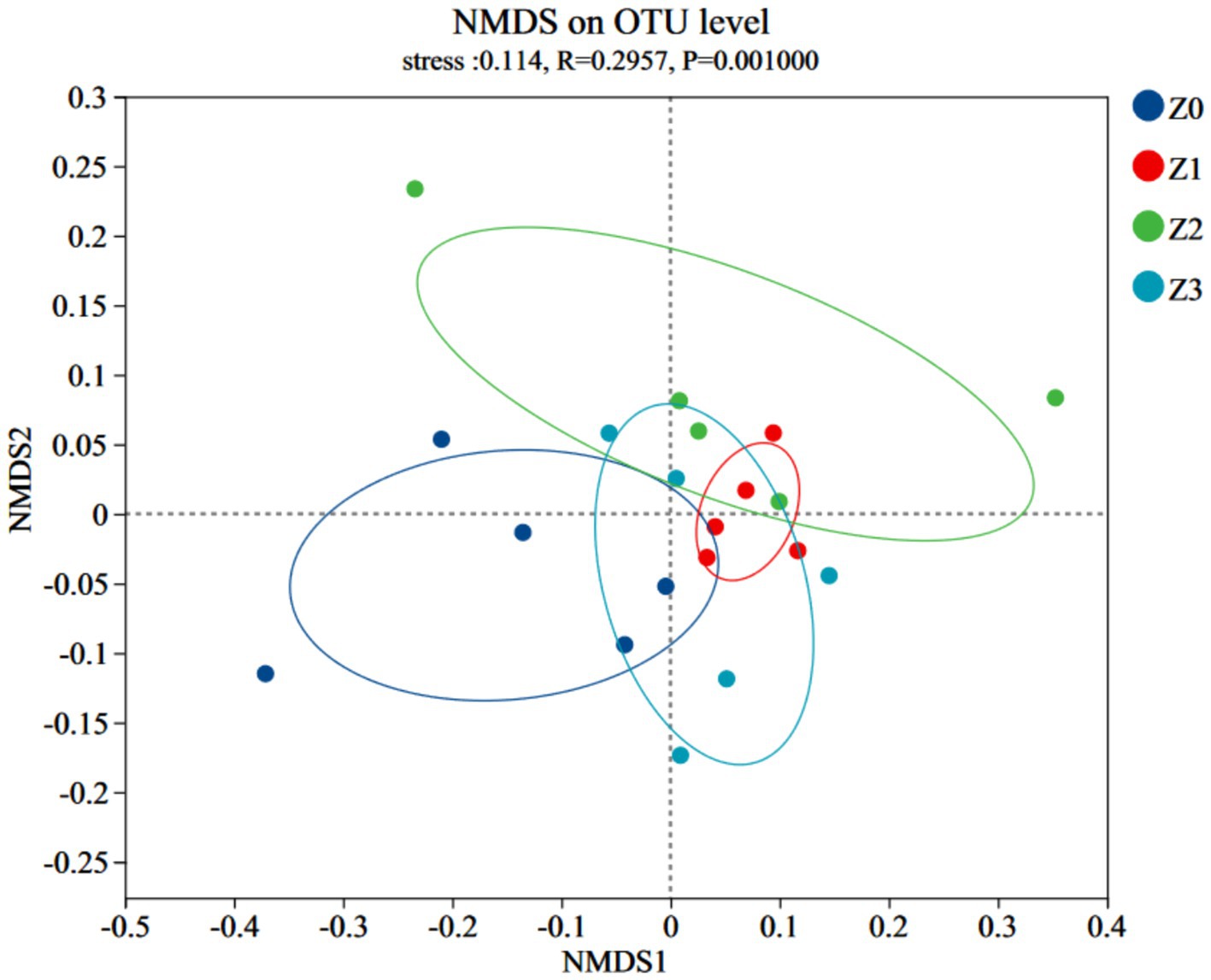
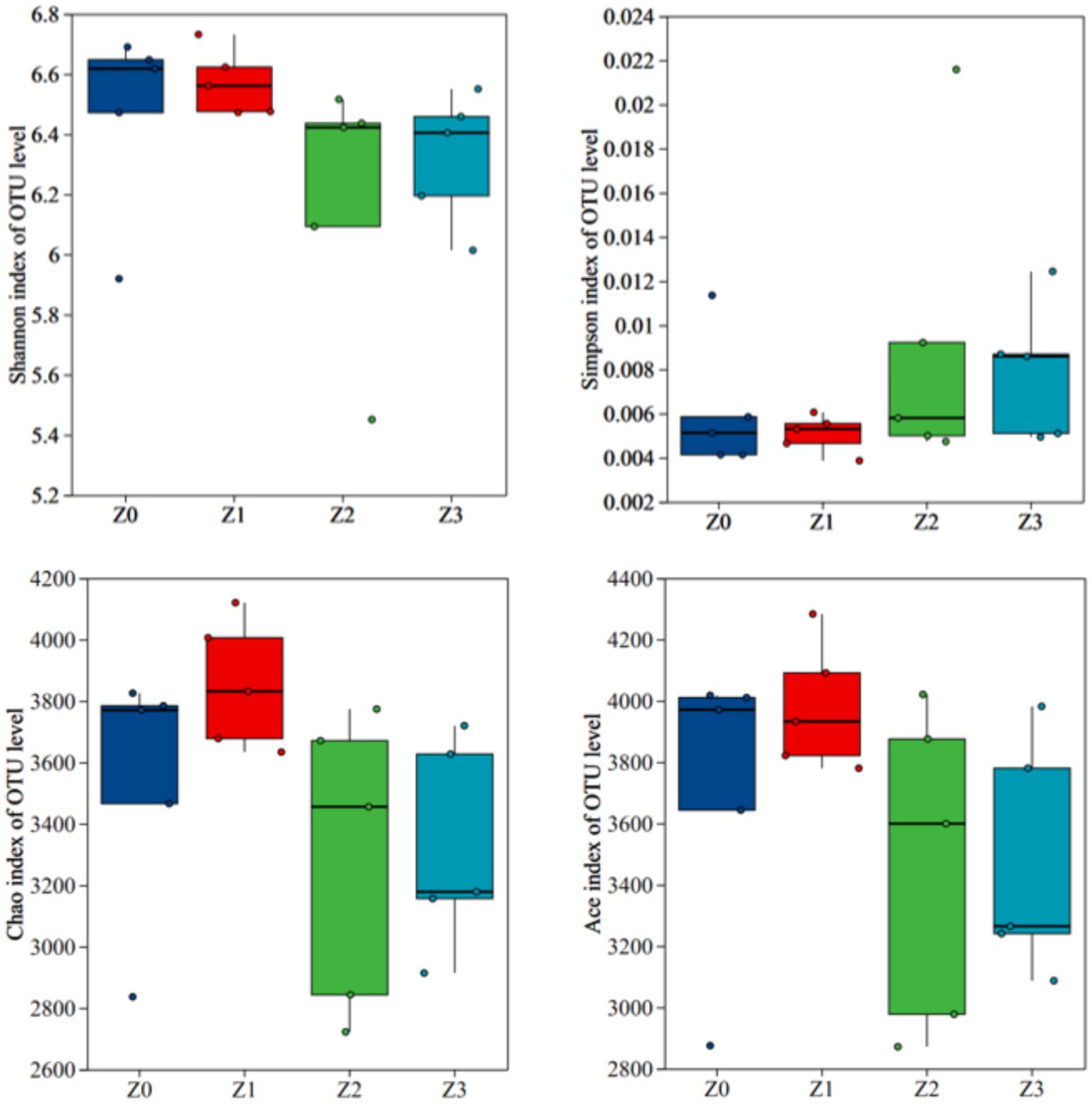
Furthermore, a non-metric multidimensional scaling (NMDS) analysis using the Bray-Curtis distance was conducted to illustrate consistent differences in β-diversity. The stress value for bacterial NMDS was 0.114, which is below the recommended threshold of 0.2 (Figure 2). The distinct clustering of bacterial communities observed in the four soil samples confirmed the impact of symbiosis between P. umbellatusand Armillaria on the bacterial communities during P. umbellatus growth. Figure 3 displays the alpha indices of the four soil samples, encompassing the richness estimator (Chao1 and ACE) and the diversity indices (Shannon and Simpson). The ACE index and Chao1 index of group Z0 exceeded those of groups Z2 and Z3, approximating the values of group Z1. The Shannon index of group Z0 surpasses that of groups Z2 and Z3, akin to group Z1. Conversely, the Simpson index of group Z0 is lower than those of groups Z2 and Z3, but comparable to that of group Z1. Despite these trends, the differences did not reach statistical significance (p > 0.05). These results corroborate the higher bacterial richness in group Z0 compared to groups Z2 and Z3, while being akin to group Z1.
The co-occurring bacterial interaction network indicates the influence of the symbiosis between P. umbellatus and A. gallica on the bacterial interactions in the vicinity of P. umbellatus sclerotia (Figure 4; Supplementary Table S1). The phyla Actinobacteria, Chloroflexi, Proteobacteria, Acidobacteriota, Firmicutes and Bacteroidota frequently interacted in Z0. Moreover, the relative abundance of Proteobacteria, Acidobacteriota, and Bacteroidota was higher in Z0 compared to Z1, Z2, and Z3 (Supplementary Table S2), indicating that the abundance and diversity of Proteobacteria, Acidobacteriota and Bacteroidota may be crucial for establishing a symbiotic relationship between P. umbellatus and A. gallica. The genus-level analysis revealed significant differences in bacterial community composition among the four groups. Specifically, 34, 37, 11, and 13 genera exhibited in groups Z0, Z1, Z2, and Z3, respectively (Supplementary Figure S2). Moreover, certain bacterial genera, such as Mycobacterium, unclassified_f__Xanthobacteraceae, Bradyrhizobium, Flavobacterium, Acidothermus, Clostridium_sensu_stricto_1, Burkholderia-Caballeronia-Paraburkholderia, Devosia, and unclassified_f__Blastocatellaceae, displayed significantly elevated relative abundances in the Z0 group compared to the other control groups, suggesting that these genera may contribute to the promotion of the symbiotic relationship establishment between P. umbellatus and A. gallica (Supplementary Table S3).
Isolation and identification of rhizosphere soil bacteria and their effects on the growth and branching of Armillaria gallica rhizomorphs
A total of 21 bacterial strains were isolated from the rhizosphere soil of P. umbellatus, followed by a co-culture experiment with A. gallica rhizomorphs and the bacterial isolates. The results are summarized in Supplementary Table S3. Among these bacterial isolates, 5 inhibited rhizomorph growth in A. gallica, while 11 reduced the number of rhizomorph branches. Notably, only one bacterial isolate (Z2-1) both promoted rhizomorph growth and increased the number of rhizomorph branches in A. gallica (Figure 5). The rhizomorph growth diameter in the culture dish was 48.18 mm, with 22.7 rhizomorph branches for isolate Z2-1, in contrast to the control group (CK) values of 22.84 mm and 8.7, respectively (Supplementary Table S4). Through EzBioCloud analysis, Z2-1 was identified as Rhodococcus sp. (Supplementary Table S5). Given the negligible recovery of fungal isolates in the preliminary isolation efforts, we propose that the growth of A. gallica is directly facilitated by bacterial associates.
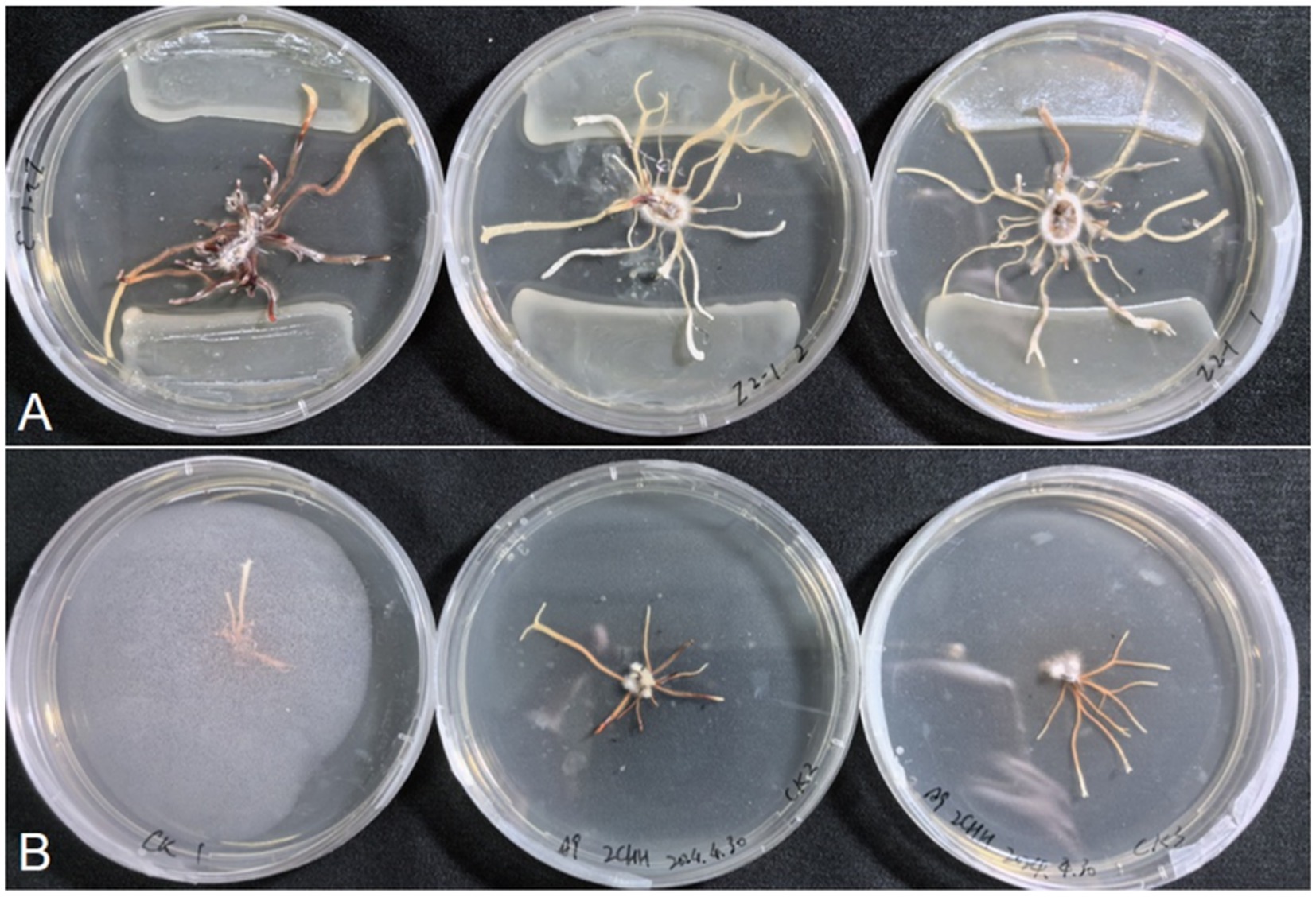
Figure 5. Growth-promoting effects of the strain Z2-1. (A) Co-culture of the strain Z2-1 and A. gallica. (B) CK: A. gallica rhizomorph cultured alone.
Z2-1 promotes the establishment of symbiotic relationship
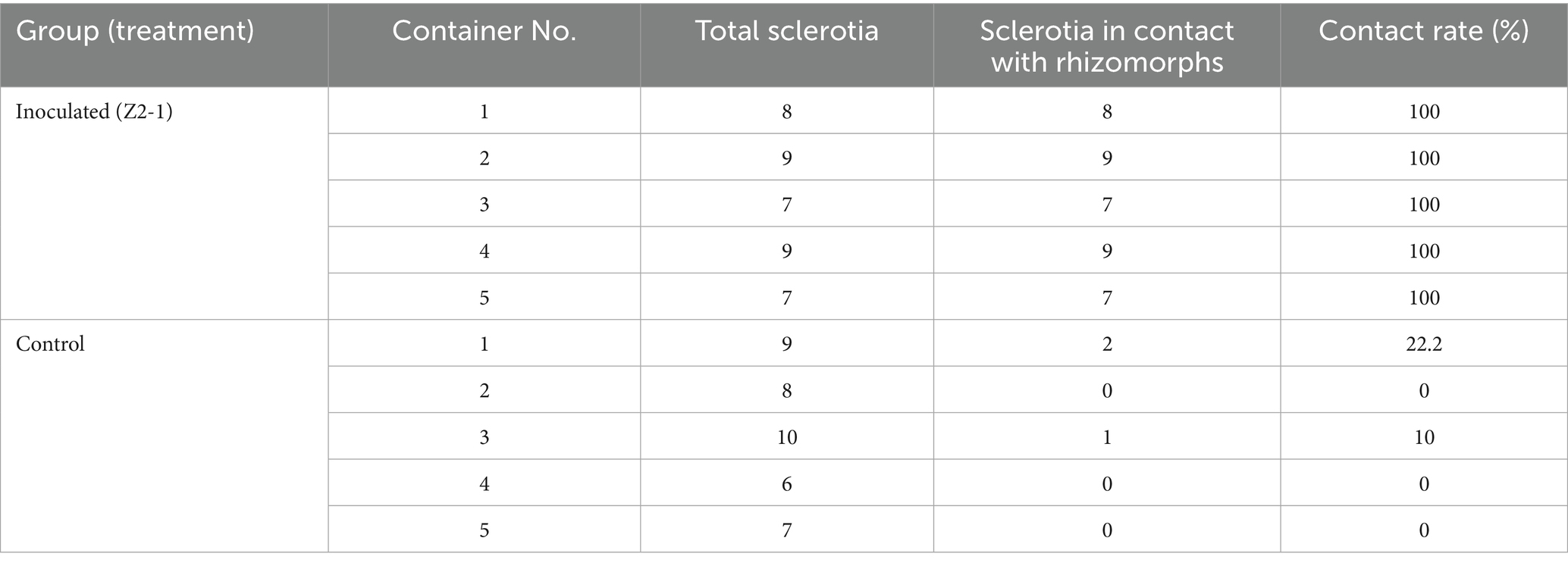
The impact of Z2-1 inoculation on the establishment of symbiosis between P. umbellatus and A. gallica was assessed by examining the contact rate between P. umbellatus sclerotia and A. gallica rhizomorphs in pot experiments. The results demonstrated a significantly higher contact rate in the inoculated group compared to the control group (Supplementary Figure S3). Specifically, in the inoculated group, each container contained 7–9 sclerotia of P. umbellatus, all of which were in contact with A. gallica rhizomorphs, resulting in a contact rate of 100%. In contrast, the control group exhibited much lower contact rates, ranging from 0 to 22.2% (Table 1). These findings indicate that Z2-1 inoculation significantly improved the establishment of symbiosis between P. umbellatus and A. gallica. A chi-square test was performed to assess the significance of the differences in contact rates between the inoculated and control groups. The analysis indicated a highly significant disparity between the two groups (χ2 = 45.0, df = 1, p < 0.001), confirming that Z2-1 inoculation markedly promoted the establishment of symbiosis between P. umbellatus and A. gallica.
Discussion
Our study offers significant insights into the intricate interplay between the medicinal fungus P. umbellatus and the fungus A. gallica, shedding light on the underlying dynamics of the soil microbial community. Specifically, our findings underscore the pivotal role of symbiosis in shaping the bacterial communities within the soil environment of P. umbellatus. The higher number of OTUs in the Z0 group, representing normally cultivated P. umbellatus, compared to the control groups (Z1, Z2, and Z3), suggests that the presence of both P. umbellatus and A. gallica establishes a distinct ecological niche that fosters a more diverse and intricate bacterial community. This observation is further supported by the NMDS analysis, which revealed distinct clustering of bacterial communities in the different soil samples, underscoring the substantial influence of symbiosis between P. umbellatus and A. gallica on the overall structure and composition of the soil microbiome. These findings align with our initial hypothesis positing that symbiosis plays a pivotal role in shaping the soil microbiome, as outlined in the introduction.
The α-diversity indices offer valuable insights into the abundance and diversity of bacterial communities across the experimental groups. Although the differences were not statistically significant (p > 0.05), potentially influenced by sample size or specific statistical methodologies employed, the elevated ACE, Chao1, and Shannon indices in the Z0 group compared to Z2 and Z3 suggest that the presence of P. umbellatus and A. gallica together promotes a higher level of bacterial richness and diversity. This finding aligns with previous studies demonstrating that symbiotic relationships can enhance microbial diversity by providing supplementary resources and habitats for microbial establishment and proliferation (Liu et al., 2021; Albornoz et al., 2022; Bender et al., 2019). The relatively higher abundance of certain bacterial phyla, such as Proteobacteria, Acidobacteriota, and Bacteroidota, in the Z0 group emphasizes their pivotal role in supporting the symbiotic relationship. These phyla are known for their functional capabilities, including nitrogen fixation, organic matter decomposition, and nutrient cycling, which could potentially contribute to the establishment and maintenance of the symbiosis between P. umbellatus and A. gallica (Li et al., 2025; Mukhia et al., 2024; Zhu et al., 2025). Subsequent investigations should encompass larger sample sizes and employ advanced statistical methodologies to validate and expand upon these results.
The co-occurring bacterial interaction network analysis revealed that certain bacterial genera, including Mycobacterium, Bradyrhizobium, and Flavobacterium, exhibited higher abundance in the Z0 group and likely play crucial roles in promoting the symbiotic relationship between P. umbellatus and A. gallica. These genera are known for their beneficial traits, such as the production of plant growth-promoting substances, enhancement of nutrient availability, and suppression of plant pathogens. Notably, Bradyrhizobium is well-known for its nitrogen-fixing capabilities, as they can serve as a vital nutrient source for the plant-fungus symbiotic system (Cheng et al., 2023). Mycobacterium and Flavobacterium are frequently associated with the degradation of complex organic compounds, potentially aiding in nutrient recycling within the soil ecosystem and bolstering the growth of both P. umbellatus and A. gallica (Terzaghi et al., 2021; González et al., 2011). Subsequent investigations should prioritize unraveling the precise mechanisms through which these bacterial genera enhance the symbiotic relationship and investigate their prospective utility in sustainable agricultural practices.
The isolation and identification of bacterial strains from the rhizosphere soil of P. umbellatus provided an opportunity to directly assess the impact of specific bacteria on the growth of A. gallica rhizomorphs. Notably, only one bacterial isolate, identified as Rhodococcus sp., significantly promoted both rhizomorph growth and branching (Ujvári et al., 2025). This finding underscores the potential for specific bacterial taxa to directly and positively influence the fungal symbiont. Given the generally low recovery rate of fungal isolates in the preliminary isolation efforts, this result is particularly intriguing and suggests that bacterial associates may play a predominant role in facilitating the growth and development of A. gallica. Rhodococcus is renowned for its diverse metabolic capabilities, which encompass the production of bioactive compounds and proficiency in degrading a broad spectrum of organic substrates (Cappelletti et al., 2020). These attributes have the potential to enhance nutrient availability and other growth-promoting factors for A. gallica, thereby stimulating its mycelial growth and branching. Future research should focus on elucidating the specific mechanisms by which Rhodococcus sp. promotes the growth of A. gallica, along with exploring the potential applications of this beneficial bacterium in sustainable agriculture. This may involve exploring the impact of bioactive compounds produced by Rhodococcus in nutrient cycling and fungal growth promotion, as well as assessing the broader ecological implications of introducing such beneficial bacteria into agricultural systems.
The significantly higher contact rate in the inoculated group suggests a pivotal role for Z2-1 in facilitating the interaction between P. umbellatus and A. gallica. This influence may stem from bioactive substances generated by Z2-1 or other mechanisms that enhance the growth and branching of A. gallica rhizomorphs, thereby increasing their contact with P. umbellatus sclerotia. These findings highlight the potential of leveraging beneficial bacteria such as Z2-1 to improve the efficiency of symbiotic establishment in agricultural and ecological systems. Subsequent investigations should concentrate on elucidating the specific mechanisms through which Z2-1 promotes symbiosis and exploring its practical applications in sustainable agriculture.
The results of our study carry significant ecological and agricultural implications. From an ecological perspective, the findings emphasize the interconnectedness of macrofungal, symbiotic fungal and bacterial communities within soil ecosystems. This interconnectedness underscores the necessity for a comprehensive comprehension of these relationships to grasp the dynamics of both natural and cultivated ecosystems fully. The symbiotic relationship between P. umbellatus and A. gallica, along with the associated bacterial communities, represents a complex network of interactions that contribute to the overall stability and resilience of the soil ecosystem. In an agricultural context, the identification of specific bacterial strains capable of stimulating the growth of A. gallica and, consequently, the vitality of P. umbellatus, holds substantial promise for sustainable agriculture. For example, these beneficial bacteria could be harnessed as biofertilizers or soil enhancements to boost plant growth and yield, diminish dependence on synthetic fertilizers, and foster more sustainable agricultural methodologies (Asif et al., 2024; Carril et al., 2025; Wang et al., 2022). Future research should focus on exploring the potential applications of these beneficial bacteria in agricultural systems, encompassing their utilization as biofertilizers and their contributions to soil health and plant development.
While our study offers valuable insights into the microbial communities associated with the P. umbellatus-A. gallica symbiosis, it is important to acknowledge several limitations. A notable limitation is the lack of statistical significance in the α-diversity indices (p > 0.05), potentially influenced by the relatively modest sample size or the specific statistical methodologies utilized (Wang et al., 2024). This suggests the importance of future research focusing on augmenting the sample size and employing more sophisticated statistical approaches to validate our findings. Additionally, the challenge of a generally low recovery rate of fungal isolates in the initial isolation endeavors is a common issue encountered in microbial ecology studies (Liu et al., 2024). This underscores the necessity for enhanced isolation techniques and experimental designs to enhance the recovery of fungal isolates. Furthermore, a more in-depth exploration of the functional mechanisms that underlie the observed impacts of specific bacterial genera and isolates on the symbiotic interactions between P. umbellatus and A. gallica is essential. This could involve detailed genomic and metabolic analyses to understand how these bacteria influence fungal growth and symbiotic establishment. The exploration of potential applications of these beneficial bacteria in agricultural systems could pave the way for novel approaches to sustainable crop production and soil management (Sun et al., 2023; Mahmud et al., 2021). Future research should incorporate field trials and long-term assessments to evaluate the practical feasibility and enduring effects of utilizing beneficial bacteria as biofertilizers or soil amendments.
Conclusion
This study reveals the crucial role of specific bacteria in the symbiosis between P. umbellatus and A. gallica. The presence of both fungi significantly enriches rhizosphere bacterial communities, as evidenced by elevated diversity indices (ACE, Chao1, Shannon) in the symbiotic group (Z0). Co-occurrence analysis highlights the abundance of Proteobacteria, Acidobacteriota, and Bacteroidota in Z0. Co-culture experiments demonstrate the growth-promoting effects of Rhodococcus sp. Z2-1, an isolated strain, on A. gallica rhizomorph, leading to a substantial increase in diameter by 112.2% and branches number by 160.9%. Pot experiments validate the symbiotic enhancement by Z2-1, resulting in a 100% contact rate between P. umbellatus and A. gallica. These findings suggest practical applications in sustainable agriculture, including employing beneficial bacteria such as Z2-1 as biofertilizers to improve fungal cultivation. Future research should focus on elucidating the mechanisms of these effects and examining the broader agricultural applications.
Data availability statement
The data presented in the study are deposited in the SRA repository, accession number PRJNA1313114.
Author contributions
LZ: Writing – original draft, Methodology, Data curation, Software, Investigation, Visualization, Formal analysis, Resources, Writing – review & editing. LL: Resources, Writing – review & editing. WG: Conceptualization, Supervision, Writing – review & editing. BL: Writing – review & editing, Funding acquisition, Resources, Methodology, Conceptualization, Supervision. SG: Writing – review & editing, Validation, Project administration, Supervision, Conceptualization, Methodology.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was financially supported by the CAMS Innovation Fund for medical Sciences (2021-I2M-1-031), Key project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302) and The Construction project of the Liuba County Polyporus umbellatus Science and Technology Expert Workstation (422502).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1658060/full#supplementary-material
References
Albornoz, F. E., Prober, S. M., Ryan, M. H., and Standish, R. J. (2022). Ecological interactions among microbial functional guilds in the plant-soil system and implications for ecosystem function. Plant Soil 476, 301–313. doi: 10.1007/s11104-022-05479-1
Asif, K., Shabaan, M., Mahmood, W., Asghar, H. N., Zahir, Z. A., Zulfiqar, U., et al. (2024). Synergistic application of bacterial consortium and organic amendments improves the growth and seed quality of mash bean (Vigna mungo L.). J. Soil Sci. Plant Nutr. 24, 6893–6905. doi: 10.1007/s42729-024-02012-4
Bender, S. F., Schlaeppi, K., Held, A., and Van der Heijden, M. G. A. (2019). Establishment success and crop growth effects of an arbuscular mycorrhizal fungus inoculated into Swiss corn fields. Agric. Ecosyst. Environ. 273, 13–24. doi: 10.1016/j.agee.2018.12.003
Bo, H. J., Li, Z. J., Wang, W., Zhang, R. Z., Wang, H. B., Jin, D. S., et al. (2024). Combining organic and inorganic fertilization enhances soil enzyme activity, the bacterial community, and molecular ecological network complexity in coal mine reclamation areas. Agronomy 14:1427. doi: 10.3390/agronomy14071427
Cappelletti, M., Presentato, A., Piacenza, E., Firrincieli, A., Turner, R. J., and Zannoni, D. (2020). Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 104, 8567–8594. doi: 10.1007/s00253-020-10861-z
Carril, P., Ghorbani, M., Azarnejad, N., Anselmi, S., Renzi, M., and Loppi, S. (2025). Promoting early growth in tomato (Solanum lycopersicum L.) by co-application of biochar and beneficial bacteria. J. Soil Sci. Plant Nutr. 25, 1493–1503. doi: 10.1007/s42729-025-02217-1
Chalita, M., Kim, Y. O., Park, S., Oh, H. S., Cho, J. H., Moon, J., et al. (2024). EzBioCloud: a genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 74:006421. doi: 10.1099/ijsem.0.006421
Cheng, Z. Y., Meng, L. B., Yin, T. J., Li, Y., Zhang, Y. H., and Li, S. M. (2023). Changes in soil rhizobia diversity and their effects on the symbiotic efficiency of soybean intercropped with maize. Agronomy 13:997. doi: 10.3390/agronomy13040997
Elnahal, A. S. M., El-Saadony, M. T., Saad, A. M., Desoky, E. S. M., El-Tahan, A. M., Rady, M. M., et al. (2022). The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: a review. Eur. J. Plant Pathol. 162, 759–792. doi: 10.1007/s10658-021-02393-7
Gao, Z. Z., Li, P. W., Li, C. Z., Tang, R. C., Wang, M. H., Chen, J. Z., et al. (2024). Identification, functional annotation, and isolation of phosphorus-solubilizing bacteria in the rhizosphere soil of Swida wilsoniana (Wanger) Sojak. Appl. Soil Ecol. 194:105207. doi: 10.1016/j.apsoil.2023.105207
Gao, W., Xu, Y. B., Chen, W. H., Wu, J. J., and He, Y. (2025). A systematic review of advances in preparation, structures, bioactivities, structural-property relationships, and applications of P. umbellatus polysaccharides. Food Chem. X 25:102161. doi: 10.1016/j.fochx.2025.102161
González, J. M., Pinhassi, J., Fernández-Gómez, B., Coll-Lladó, M., González-Velázquez, M., Puigbò, P., et al. (2011). Genomics of the proteorhodopsin-containing marine Flavobacterium Dokdonia sp. strain MED134. Appl. Environ. Microbiol. 77, 8676–8686. doi: 10.1128/AEM.06152-11
Handique, M., Bora, P., Ziogas, V., Srivastava, A. K., Jagannadham, P. T. K., and Das, A. K. (2024). Phytophthora infection reorients the composition of rhizospheric microbial assembly in Khasi mandarin (Citrus reticulata Blanco). Agronomy-Basel 14:661. doi: 10.3390/agronomy14040661
Li, J., Li, X. Y., Ma, L., Liu, G. Y., Han, Y. Y., Li, J. Q., et al. (2025). Increased carbon sequestration of different straw return depths varies temporally. Appl. Soil Ecol. 206:105904. doi: 10.1016/j.apsoil.2025.105904
Liu, T. R., Cheng, R., Hua, Z. Y., Gao, H. Y., Wang, C., Li, H., et al. (2024). Identification of growth-promoting bacterial resources by investigating the microbial community composition of P. umbellatussclerotia. J. Fungi 10:386. doi: 10.3390/jof10060386
Liu, T. R., Hua, Z. Y., Han, P. J., Zhao, Y. Y., Zhou, J. H., Jin, Y., et al. (2022). Mycorrhizosphere bacteria, Rahnella sp. HPDA25, promotes the growth of a. gallica and its parasitic host Gastrodia elata. Front. Microbiol. 13:842893. doi: 10.3389/fmicb.2022.842893
Liu, D., Pérez-Moreno, J., He, X. H., Garibay-Orijel, R., and Yu, F. Q. (2021). Truffle microbiome is driven by fruit body compartmentalization rather than soils conditioned by different host trees. mSphere 6, e00039–e00021. doi: 10.1128/mSphere.00039-21
Liu, L., Xing, Y. M., Li, S. J., Zhou, L. S., Li, B., and Guo, S. X. (2025). Different symbiotic species of Armillaria affect the yield and active compound contents of P. umbellatus. Microorganisms 13:228. doi: 10.3390/microorganisms13020228
Mahmud, K., Lee, K., Hill, N. S., Mergoum, A., and Missaoui, A. (2021). Influence of tall fescue Epichloe endophytes on rhizosphere soil microbiome. Microorganisms 9:1843. doi: 10.3390/microorganisms9091843
Maurice, K., Laurent-Webb, L., Bourceret, A., Boivin, S., Boukcim, H., Selosse, M. A., et al. (2024). Networking the desert plant microbiome, bacterial and fungal symbionts structure and assortativity in co-occurrence networks. Environ. Microbiome 19:65. doi: 10.1186/s40793-024-00610-4
Mukhia, S., Kumar, A., and Kumar, R. (2024). Bacterial community distribution and functional potentials provide key insights into their role in the ecosystem functioning of a retreating eastern Himalayan glacier. FEMS Microbiol. Ecol. 100:fiae012. doi: 10.1093/femsec/fiae012
Sun, K., Jiang, H. J., Pan, Y. T., Lu, F., Zhu, Q., Ma, C. Y., et al. (2023). Hyphosphere microorganisms facilitate hyphal spreading and root colonization of plant symbiotic fungus in ammonium-enriched soil. ISME J. 17, 1626–1638. doi: 10.1038/s41396-023-01476-z
Terzaghi, E., Posada-Baquero, R., Di Guardo, A., and Ortega-Calvo, J. J. (2021). Microbial degradation of pyrene in holm oak (Quercus ilex) phyllosphere: role of particulate matter in regulating bioaccessibility. Sci. Total Environ. 786:147431. doi: 10.1016/j.scitotenv.2021.147431
Ujvári, G., Grassi, A., Avio, L., Pagliarani, I., Cristani, C., Giovannetti, M., et al. (2025). Root endophytic bacterial communities are shaped by the specific microbiota associated to mycorrhizal symbionts. Plant Soil 508, 275–292. doi: 10.1007/s11104-024-06801-9
Wang, H. L., Mu, C. C., Yan, G. Y., Xing, Y. J., and Wang, Q. G. (2024). Environmental filtration and host selection effect influence bacterial community assembly in broad-leaved Korean pine forests: a regional scale study. Appl. Soil Ecol. 203:105664. doi: 10.1016/j.apsoil.2024.105664
Wang, Q. Q., Pan, H., Chen, X., Shang, X. T., Yang, Z. S., Yang, X. Y., et al. (2024). Species identification and spatial diversity patterns of the Giant panda National Park (GPNP) in ya'an, Sichuan, China. Glob. Ecol. Conserv. 51:e02938. doi: 10.1016/j.gecco.2024.e02938
Wang, Z. H., Yang, T. J., Mei, X. L., Wang, N. Q., Li, X. G., Yang, Q. S., et al. (2022). Bio-organic fertilizer promotes pear yield by shaping the rhizosphere microbiome composition and functions. Microbiol. Spectr. 10:e0357222. doi: 10.1128/spectrum.03572-22
Xun, W., Liu, Y., Ma, A. Y., Yan, H., Miao, Y. Z., Shao, J. H., et al. (2024). Dissection of rhizosphere microbiome and exploiting strategies for sustainable agriculture. New Phytol. 242, 2401–2410. doi: 10.1111/nph.19697
Ying, Y. X., Ding, W. L., and Li, Y. (2012). Characterization of soil bacterial communities in rhizospheric and nonrhizospheric soil of Panax ginseng. Biochem. Genet. 50, 848–859. doi: 10.1007/s10528-012-9525-1
Zhao, J., Cheng, Y. L., Jiang, N., Qiao, G. H., and Qin, W. T. (2024). Rhizosphere-associated soil microbiome variability in Verticillium wilt-affected Cotinus coggygria. Front. Microbiol. 14:1279096. doi: 10.3389/fmicb.2023.1279096
Keywords: Polyporus umbellatus, Armillaria gallica, symbiosis, rhizosphere bacterial communities, Rhodococcus sp.
Citation: Zhou L, Liu L, Gao W, Li B and Guo S (2025) Symbiotic relationship between Polyporus umbellatus and Armillaria gallica shapes rhizosphere bacterial community structure and promotes fungal growth. Front. Microbiol. 16:1658060. doi: 10.3389/fmicb.2025.1658060
Edited by:
Amit Sinha, New England Biolabs, United StatesReviewed by:
Tianrui Liu, China Academy of Chinese Medical Sciences, ChinaYog Raj, Central Drug Research Institute (CSIR), India
Copyright © 2025 Zhou, Liu, Gao, Li and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Li, enVkZW5ndGlhbnhpYUAxMjYuY29t; Shunxing Guo, c3hndW8xOTg2QDE2My5jb20=
 Lingfeng Zhou
Lingfeng Zhou Liu Liu
Liu Liu Weiwei Gao
Weiwei Gao Bing Li
Bing Li Shunxing Guo1*
Shunxing Guo1*