- 1College of Veterinary Medicine, Qingdao Agricultural University, Qingdao, China
- 2Qingdao Center for Animal Disease Control & Prevention, Qingdao, China
A Commentary on
Development and immunogenicity evaluation of a quadruple-gene-deleted pseudorabies virus strain
by Li, H., Zhang, R., Qu, J., Kang, Y., Zhang, J., Guo, R., et al. (2024) Front. Microbiol. 15:1479794. doi: 10.3389/fmicb.2024.1479794
Introduction
An article, named Development and immunogenicity evaluation of a quadruple-gene-deleted pseudorabies virus strain (doi: 10.3389/fmicb.2024.1479794), was recently published in Frontiers in Microbiology (Li et al., 2024). In this study, Li et al. used the CRISPR/Cas9 technique for modifying a pseudorabies virus (PRV) strain to generate a mutant, albeit deficient in four genes, showing similar growth kinetics to that of its progenitor. After vaccination with the quadruple gene-deficient mutant, neither mice nor piglets displayed obvious clinical signs and pathological alterations. The mutant induced significantly higher levels of gB-specific antibodies, neutralizing antibodies and cytokines than both the Bartha-K61 strain and a triple gene-deficient mutant did. Moreover, compared with other vaccine strains, the quadruple gene-deficient mutant conferred robust protection against challenges with a virulent PRV in mice and piglets. Herein, we would like to express our scientific opinions on this study.
Characteristics of PRV
PRV causes Aujeszky's disease that is particularly devastating to breeding sows and piglets. This virus belongs to the genus Varicellovirus in the family Orthoherpesviridae. An intact PRV virion is composed of four morphologically distinct components (Figure 1): a linear double-stranded DNA genome, an icosahedral capsid, an amorphous tegument layer, and a lipid envelope studded with various glycoproteins, out of which gE, gL, gG, gC, gM, and gN are the main virulence determinants (Peng et al., 2023). The genome includes a unique long region, a unique short region, an internal repeat sequence, and a terminal repeat sequence (Li et al., 2021). Its full-length sequence is approximately 143 kb, comprising at least 72 open reading frames that encode 70–100 proteins (Mettenleiter, 2000; Klupp et al., 2004).
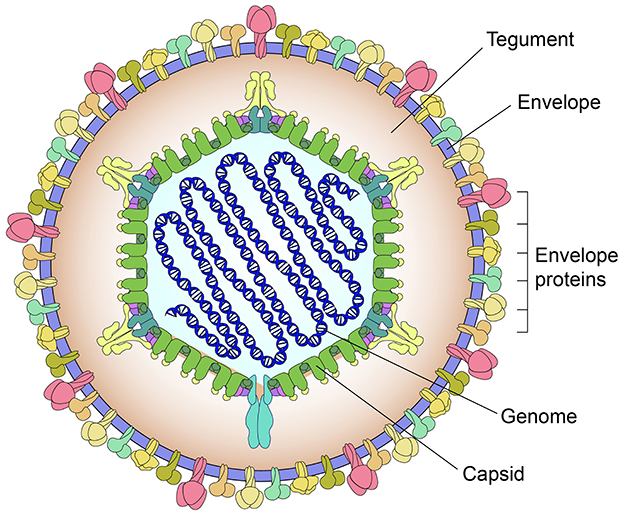
Figure 1. Schematic representation of pseudorabies virion. The virion is composed of four morphologically distinct components: a linear double-stranded DNA genome, an icosahedral capsid, an amorphous tegument layer, and a lipid envelope studded with various glycoproteins.
Development of gene-deleted vaccines against PRV
Conventional live-attenuated vaccines often fail to provide satisfactory effect on controlling PRV variants in recent years (Li et al., 2025). Gene-deleted strains have been extensively explored for the development of anti-PRV vaccine candidates (Zheng et al., 2022). Gene-deleted targets primarily focus on TK, gE, and gI genes, which play essential roles in PRV's neurovirulence, transmission or replication (Moormann et al., 1990; Wu et al., 2016; Sun Y. et al., 2022). Gene deletion alone or combinedly would attenuate the virulence of wild-type strains, facilitating the development of live-attenuated, gene-modified vaccines (Wang et al., 2014). A representative gene-modified strain is the SA215 (ΔgE/ΔgI/ΔTK), licensed in 2003 for producing commercially available vaccines. Other genes, such as gD, gG, US9, and US2, can be also deleted from the PRV genome for constructing vaccine candidates (Mettenleiter et al., 1994; Sun L. et al., 2022).
Potential of quadruple gene-deficient mutant in immunization
PRV UL24 is a nuclear-localized protein. Compared with TK, UL24 functions as a minor virulence-associated factor (Ye et al., 2019), not only antagonizing the antiviral effect mediated by oligoadenylate synthetases-like protein (Chen et al., 2021), but also abrogating the NF-κB activation induced by tumor necrosis factor-α (Wang et al., 2020). Different from the classical vaccine strain SA215, the quadruple gene-deficient mutant, derived from another progenitor SX-10, was constructed by (Li et al. 2024) through deleting one extra UL24 gene.
The authors compared growth curves among different strains, showing that the quadruple gene-deficient mutant was insignificantly affected concerning its growth kinetics. Deletion of UL24 enhanced the level of interferon-β expression, and moreover reduced the PRV virulence in mice. Additionally, the ΔgI/ΔgE/ΔTK/ΔUL24 mutant-vaccinated group exhibited the antibody response significantly stronger than that of the ΔgI/ΔgE/ΔTK mutant-vaccinated group. Both groups revealed the similar titer of neutralizing antibodies at 28 days post-immunization, significantly higher than that of the Bartha-K61-vaccinated group. Further, (Li et al. 2024) evaluated the quadruple gene-deficient mutant regarding its safety and immunogenicity in piglets. The results showed that this mutant caused neither clinical signs nor histopathologic changes, and conferred the optimal protection from the challenge with virulent PRV (Li et al., 2024).
Discussion
Pseudorabies has still been endemic in most parts of the world (Chen et al., 2025; Zhuang et al., 2025). Both gene-modified and -unmodified vaccines are concurrently used in the veterinary field for preventing pseudorabies, whereas the former possesses a better safety profile than the latter. It was reported as early as 1985 that TK-negative PRV was used to develop the gene-deleted vaccine for clinical immunization (Kit et al., 1985). A series of dual gene-deleted strains, such as ΔTK/ΔgI, ΔTK/ΔgE, ΔgD/ΔgI, and ΔgE/ΔgI, were subsequently constructed and also demonstrated to be highly immunogenic and safe in pigs against pseudorabies (Moormann et al., 1990; Peeters et al., 1994; Gu et al., 2015; Wu et al., 2016). Meanwhiles, triple gene-deleted PRVs have been continuously reported to be successfully developed, and also induced strong immune responses in vivo (Ferrari et al., 2000; Zhu et al., 2011; Hu R. M. et al., 2015; Lin et al., 2020).
TK, gE and gI were the three most frequently modified targets. In the study conducted by Li et al., besides the three genes, one extra virulence-related gene, UL24, was deleted from the PRV genome. Because the PRV UL24 functions as an innate immune antagonist in the host (Ye et al., 2019; Wang et al., 2020; Chen et al., 2021), its deletion would theoretically enhance the innate immunity. Indeed, regardless of the PRV UL24 deleted alone or in combination with gI, gE, and TK, the interferon-β transcription always remained at a relatively high level in vivo after the vaccination with UL24-deficient strains. The quadruple gene-deficient mutant could replicate as efficiently as wild-type strains in vitro, and more importantly, was able to elicit more robust immune responses than the triple gene-deficient (ΔTK/ΔgE/ΔgI) strain to some extent (Li et al., 2024). The quadruple gene-deficient mutant was derived from an emerging PRV variant, SX-10. Choosing a variant, rather than a classical strain, is necessary for developing the next-generation vaccine, because classical strains, albeit safe to use, may fail to confer the desirable immune protection (An et al., 2013; Hu D. et al., 2015).
In conclusion, this study is novel, to which similar findings were previously unreported, therefore paving the way for developing new-generation vaccines against pseudorabies in future. Further experiments should be designed for evaluating its immunization efficacy in field conditions.
Author contributions
YL: Writing – original draft. CM: Writing – original draft. FY: Software, Writing – original draft. FL: Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Qingdao Demonstration Project for People-benefit from Science and Techniques (Grant No. 25-1-5-xdny-30-nsh).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, T. Q., Peng, J. M., Tian, Z. J., Zhao, H. Y., Li, N., Liu, Y. M., et al. (2013). Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg. Infect. Dis. 19, 1749–1755. doi: 10.3201/eid1911.130177
Chen, X., Kong, N., Xu, J., Wang, J., Zhang, M., Ruan, K., et al. (2021). Pseudorabies virus UL24 antagonizes OASL-mediated antiviral effect. Virus Res. 295:198276. doi: 10.1016/j.virusres.2020.198276
Chen, Y., Gao, J., Hua, R., and Zhang, G. (2025). Pseudorabies virus as a zoonosis: scientific and public health implications. Virus Genes 61, 9–25. doi: 10.1007/s11262-024-02122-2
Ferrari, M., Brack, A., Romanelli, M. G., Mettenleiter, T. C., Corradi, A., Dal Mas, N., et al. (2000). A study of the ability of a TK-negative and gI/gE-negative pseudorabies virus (PRV) mutant inoculated by different routes to protect pigs against PRV infection. J. Vet. Med. B Infect. Dis. Vet. Public Health 47, 753–762. doi: 10.1046/j.1439-0450.2000.00407.x
Gu, Z., Dong, J., Wang, J., Hou, C., Sun, H., Yang, W., et al. (2015). A novel inactivated gE/gI deleted pseudorabies virus (PRV) vaccine completely protects pigs from an emerged variant PRV challenge. Virus Res. 195, 57–63. doi: 10.1016/j.virusres.2014.09.003
Hu, D., Zhang, Z., Lv, L., Xiao, Y., Qu, Y., Ma, H., et al. (2015). Outbreak of variant pseudorabies virus in Bartha-K61-vaccinated piglets in central Shandong Province, China. J. Vet. Diagn. Invest. 27, 600–605. doi: 10.1177/1040638715593599
Hu, R. M., Zhou, Q., Song, W. B., Sun, E. C., Zhang, M. M., He, Q. G., et al. (2015). Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge. Vaccine 33, 5733–5740. doi: 10.1016/j.vaccine.2015.09.066
Kit, S., Kit, M., and Pirtle, E. C. (1985). Attenuated properties of thymidine kinase-negative deletion mutant of pseudorabies virus. Am. J. Vet. Res. 46, 1359–1367. doi: 10.2460/ajvr.1985.46.06.1359
Klupp, B. G., Hengartner, C. J., Mettenleiter, T. C., and Enquist, L. W. (2004). Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78, 424–440. doi: 10.1128/JVI.78.1.424-440.2004
Li, H., Zhang, J., Guo, R., Li, J., Zhang, X., Han, L., et al. (2025). Immunogenicity evaluation of a recombinant pseudorabies virus co-expressing PCV2 and PCV3 capsid proteins in mice and piglets. Vaccine 60:127307. doi: 10.1016/j.vaccine.2025.127307
Li, H., Zhang, R., Qu, J., Kang, Y., Zhang, J., Guo, R., et al. (2024). Development and immunogenicity evaluation of a quadruple-gene-deleted pseudorabies virus strain. Front. Microbiol. 15:1479794. doi: 10.3389/fmicb.2024.1479794
Li, W., Zhuang, D., Li, H., Zhao, M., Zhu, E., Xie, B., et al. (2021). Recombinant pseudorabies virus with gI/gE deletion generated by overlapping polymerase chain reaction and homologous recombination technology induces protection against the PRV variant PRV-GD2013. BMC Vet. Res. 17:164. doi: 10.1186/s12917-021-02861-6
Lin, J., Li, Z., Feng, Z., Fang, Z., Chen, J., Chen, W., et al. (2020). Pseudorabies virus (PRV) strain with defects in gE, gC, and TK genes protects piglets against an emerging PRV variant. J. Vet. Med. Sci. 82, 846–855. doi: 10.1292/jvms.20-0176
Mettenleiter, T. C. (2000). Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis–state of the art, June 1999. Vet. Res. 31, 99–115. doi: 10.1051/vetres:2000110
Mettenleiter, T. C., Klupp, B. G., Weiland, F., and Visser, N. (1994). Characterization of a quadruple glycoprotein-deleted pseudorabies virus mutant for use as a biologically safe live virus vaccine. J. Gen. Virol. 75 (Pt 7), 1723–1733. doi: 10.1099/0022-1317-75-7-1723
Moormann, R. J., de Rover, T., Briaire, J., Peeters, B. P., Gielkens, A. L., and van Oirschot, J. T. (1990). Inactivation of the thymidine kinase gene of a gI deletion mutant of pseudorabies virus generates a safe but still highly immunogenic vaccine strain. J. Gen. Virol. 71 (Pt 7), 1591–1595. doi: 10.1099/0022-1317-71-7-1591
Peeters, B., Bouma, A., de Bruin, T., Moormann, R., Gielkens, A., and Kimman, T. (1994). Non-transmissible pseudorabies virus gp50 mutants: a new generation of safe live vaccines. Vaccine 12, 375–380. doi: 10.1016/0264-410X(94)90104-X
Peng, Z., Liu, Q., Zhang, Y., Wu, B., Chen, H., and Wang, X. (2023). Cytopathic and genomic characteristics of a human-originated pseudorabies virus. Viruses 15:170. doi: 10.3390/v15010170
Sun, L., Tang, Y., Yan, K., and Zhang, H. (2022). Construction of a quadruple gene-deleted vaccine confers complete protective immunity against emerging PRV variant challenge in piglets. Virol. J. 19:19. doi: 10.1186/s12985-022-01748-8
Sun, Y., Zhao, L., and Fu, Z. F. (2022). Effective cross-protection of a lyophilized live gE/gI/TK-deleted pseudorabies virus (PRV) vaccine against classical and variant PRV challenges. Vet. Microbiol. 267:109387. doi: 10.1016/j.vetmic.2022.109387
Wang, C. H., Yuan, J., Qin, H. Y., Luo, Y., Cong, X., Li, Y., et al. (2014). A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine 32, 3379–3385. doi: 10.1016/j.vaccine.2014.04.035
Wang, T. Y., Yang, Y. L., Feng, C., Sun, M. X., Peng, J. M., Tian, Z. J., et al. (2020). Pseudorabies virus UL24 abrogates tumor necrosis factor alpha-induced NF-κB activation by degrading P65. Viruses 12:51. doi: 10.3390/v12010051
Wu, C. Y., Liao, C. M., Chi, J. N., Chien, M. S., and Huang, C. (2016). Growth properties and vaccine efficacy of recombinant pseudorabies virus defective in glycoprotein E and thymidine kinase genes. J. Biotechnol. 229, 58–64. doi: 10.1016/j.jbiotec.2016.05.009
Ye, C., Chen, J., Cheng, X., Zhou, S., Jiang, S., Xu, J., et al. (2019). Functional analysis of the UL24 protein of suid herpesvirus 1. Virus Genes 55, 76–86. doi: 10.1007/s11262-018-1619-3
Zheng, H. H., Fu, P. F., Chen, H. Y., and Wang, Z. Y. (2022). Pseudorabies virus: from pathogenesis to prevention strategies. Viruses 14:1638. doi: 10.3390/v14081638
Zhu, L., Yi, Y., Xu, Z., Cheng, L., Tang, S., and Guo, W. (2011). Growth, physicochemical properties, and morphogenesis of Chinese wild-type PRV Fa and its gene-deleted mutant strain PRV SA215. Virol. J. 8:272. doi: 10.1186/1743-422X-8-272
Keywords: pseudorabies virus, quadruple-gene-deleted mutant, UL24, vaccine, immunization
Citation: Li Y, Mu C, Yin F and Liu F (2025) Commentary: Development and immunogenicity evaluation of a quadruple-gene-deleted pseudorabies virus strain. Front. Microbiol. 16:1665381. doi: 10.3389/fmicb.2025.1665381
Received: 08 August 2025; Accepted: 26 September 2025;
Published: 13 October 2025.
Edited by:
Junki Maruyama, University of Texas Medical Branch at Galveston, United StatesReviewed by:
Harsh Jogi, Indian Veterinary Research Institute, IndiaCopyright © 2025 Li, Mu, Yin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuxiao Liu, bGF1ZGF3bkAxMjYuY29t
†These authors have contributed equally to this work
 Yan Li1,2†
Yan Li1,2† Fuxiao Liu
Fuxiao Liu