- Department of Proctology, The First Affiliated Hospital of Huzhou University, Huzhou, Zhejiang, China
Intestinal organoids are three-dimensional in vitro models derived from patient-specific tissues, which can recapitulate the structural and functional characteristics of the native intestinal epithelium, including interactions with the gut microbiota. In the study of host-microbiota crosstalk within the context of the Tumor Microenvironment (TME), they have become highly effective tools, providing an opportunity to explore the role of microorganisms in carcinogenic processes, immune regulation, and therapeutic responses. Although organoids can successfully simulate key aspects of the TME, certain features—such as systemic immune interactions, neuroendocrine axes, and dynamic microbial communities—remain difficult to fully replicate. This review primarily covers the advances in organoids applied to the research of the microbiota-TME axis, examines their current limitations, and further advocates for their integration with multi-omics and organ-on-a-chip technologies to enhance physiological relevance and the value of translational applications.
1 Introduction
1.1 The significance of host-microbiome interactions in the TME
Cancer remains a major global health burden, and a central challenge in therapy development is the significant disparity between existing models and the actual TME in human patients. The interaction between the host and the gut microbiota within the TME, a pivotal driver of cancer progression and therapy, has garnered widespread attention in recent years. The tumor microbiome not only significantly influences tumor initiation, progression, and the development of drug resistance (Pich et al., 2025; Qiao et al., 2022) but also plays a crucial role within the TME by modulating immune responses and treatment efficacy (Zhang S. et al., 2025; Grzywa et al., 2017). Advancements in this field have been hampered by the inadequate physiological relevance of conventional models (Williamson et al., 2018), which fail to accurately recapitulate the complex host-microbiota interactions and the dynamics of the TME (Qu et al., 2021; Chen et al., 2024). Within this context, organoids have emerged as a transformative innovation for cancer gut microbiome research. As a novel three-dimensional (3D) in vitro model derived from primary tissues, organoids can highly recapitulate the heterogeneity, architecture, and functional characteristics of tumors (Tardito et al., 2024) while also incorporating relevant elements of the gut microbiota, thereby uniquely facilitating cancer-specific investigations. A defining attribute for cancer applications is the ability of organoids to be coupled with patient-derived samples to establish co-culture systems encompassing epithelial cells, stromal fibroblasts, and microbes. This allows for precise dissection of host-microbiota interactions in the cancer immune microenvironment, for instance, through microinjection or microfluidic techniques (Penarete-Acosta et al., 2024) that enable exposure of tumor cells to gut microbiota. This integration not only revolutionizes the experimental platform for studying the tumor microbiome, enabling investigations into inflammation, carcinogenic mechanisms, and personalized treatment responses, but also provides unprecedented insights into precision intervention strategies for cancer-associated microecology. Thereby, it bridges critical knowledge gaps regarding the gut microbiota in tumor biology and propels cancer research towards new heights of clinical translation. We propose that current research has established host–microbe interactions as the core regulatory axis of the TME, with particularly substantial evidence in the contexts of immunometabolic reprogramming and therapeutic response prediction. The core limitations include insufficient depth in mechanistic understanding, delayed advancement in intratumoral microbiota research, and bottlenecks in clinical translation. This field urgently requires the integration of high-resolution technologies (single-cell sequencing + spatial transcriptomics) with biomimetic models (organoids/organ-on-a-chip) to translate phenotypic correlations into mechanisms of causality, while driving intratumoral microbiota research from the stage of descriptive analysis toward that of functional validation (see Table 1).
2 Introduction to and classification of intestinal organoids
Intestinal organoids are three-dimensional (3D) (Qu et al., 2021), self-organizing micro-organ structures that are generated in vitro by harnessing the regenerative capacity of stem cells. They can highly recapitulate the complex composition and physiological architecture of the native intestinal tissue. Their core structure is formed by a polarized epithelial layer (Chen et al., 2024) that encompasses a variety of functionally differentiated cell subtypes, such as enterocytes (Moll et al., 2022; Pérez-González et al., 2021). These cells organize into crypt-like folds and cellular compartments in three dimensions, mimicking the crypt-villus unit patterning of the intestine (Pérez-González et al., 2021). The organoid structure also includes a supportive mesenchymal cell layer (Chen et al., 2024), and specific models may incorporate additional elements, such as microvascular fragments, to simulate vascular components. The culture conditions profoundly influence the architecture. When cultured within a laminin-rich Matrigel matrix, the organoids exhibit the correct basal-out polarity. Removal of Matrigel can induce a reversal of polarity to an apical-out configuration (Yavitt et al., 2022; Hirota et al., 2021; Yavitt et al., 2023). These 3D structures form through a self-organizing process and can display intestinal cellular diversity, mechanical properties, and functional behaviors, thereby providing a physiologically relevant model for studying intestinal development and disease.
The colorectal cancer (CRC) cell lines refer to populations of cancer cells isolated from human colorectal cancer tissues or metastatic sites (e.g., lymph nodes) that can be cultured long-term in vitro (Luk et al., 2025; Codrich et al., 2021). As essential models in cancer research, their primary principle lies in retaining the molecular characteristics of the primary tumor, thereby laying the groundwork for simulating tumor properties under controlled conditions. Their core advantages include ease of standardized culture, facilitating genetic manipulation, and high-throughput drug screening, which significantly accelerate research into the mechanisms of colorectal carcinogenesis, drug resistance, and the identification of novel therapeutic targets. For instance Somarelli et al. (2020) identified dual CDK2/9 inhibition as a novel combination therapy for colorectal cancer, and Tran et al. (2022) demonstrated the efficacy of BRAF mutation or VEGFR inhibitors in specific cell lines. These models are particularly applied in cancer research to: (1) dissect tumor signaling pathways (Sugito et al., 2022); (2) investigate cancer stem cell properties (Ying et al., 2023; Grigoreva et al., 2025); (3) assess interactions within the immune microenvironment (Teng et al., 2024); (4) develop therapeutic strategies (Zhang S. et al., 2024; Ferreira et al., 2024; Ji et al., 2021); including the validation of tumor-suppressive effects by non-coding RNAs or natural compounds. Notable limitations include: (1) the absence of the complex TME (Castro et al., 2021), limiting their ability to fully recapitulate in vivo conditions; (2) potential genetic drift during long-term passaging (Luk et al., 2025; Codrich et al., 2021; Przybylla et al., 2023), leading to a loss of the original tumor heterogeneity; (3) the loss of critical molecular characteristics in some cell lines (Shi et al., 2024) due to mechanisms such as methylation; Substantial biological differences exist among various cell lines, necessitating careful selection of the most appropriate model for specific research questions.
Adult stem cell-derived colorectal cancer organoids (Nakano et al., 2025; Driehuis et al., 2020) are three-dimensional miniature tumor models generated in vitro from adult stem cells isolated from patient intestinal tissues (e.g., Lgr5-positive stem cells residing at the crypt base). Their generation leverages the self-renewal and multilineage differentiation capabilities of adult stem cells. By reconstituting a physiological microenvironment in vitro (e.g., providing Wnt-signaling niche factors) (Yan et al., 2023), these cells self-organize into organoids that retain the genetic features, cellular heterogeneity, and pathological architecture of the primary tumor (Xu H. et al., 2022). Major advantages include: (1) high preservation of patient tumor’s molecular characteristics and physiological relevance, addressing the limitation of conventional 2D cell lines in modeling tumor heterogeneity (Jin et al., 2025; Wang R. et al., 2022); (2) capability for long-term expansion, enabling the establishment of living tumor biobanks (Mao et al., 2024) (3) facilitation of high-throughput drug screening and personalized therapy evaluation, such as testing chemotherapeutic drug sensitivity (Mao et al., 2024) and developing treatment strategies targeting cancer stem cells (Yan et al., 2023) (e.g., inhibiting inflammatory factors that maintain tumor stemness). Core applications focus on colorectal cancer research: modeling tumorigenesis, investigating specific characteristics of the TME, screening anticancer drugs, and developing effective regenerative medicine strategies. Main limitations include the current absence of non-epithelial components in these models, which impedes research on microenvironmental interactions, and the need for further optimization of standardized culture systems (Figure 1).
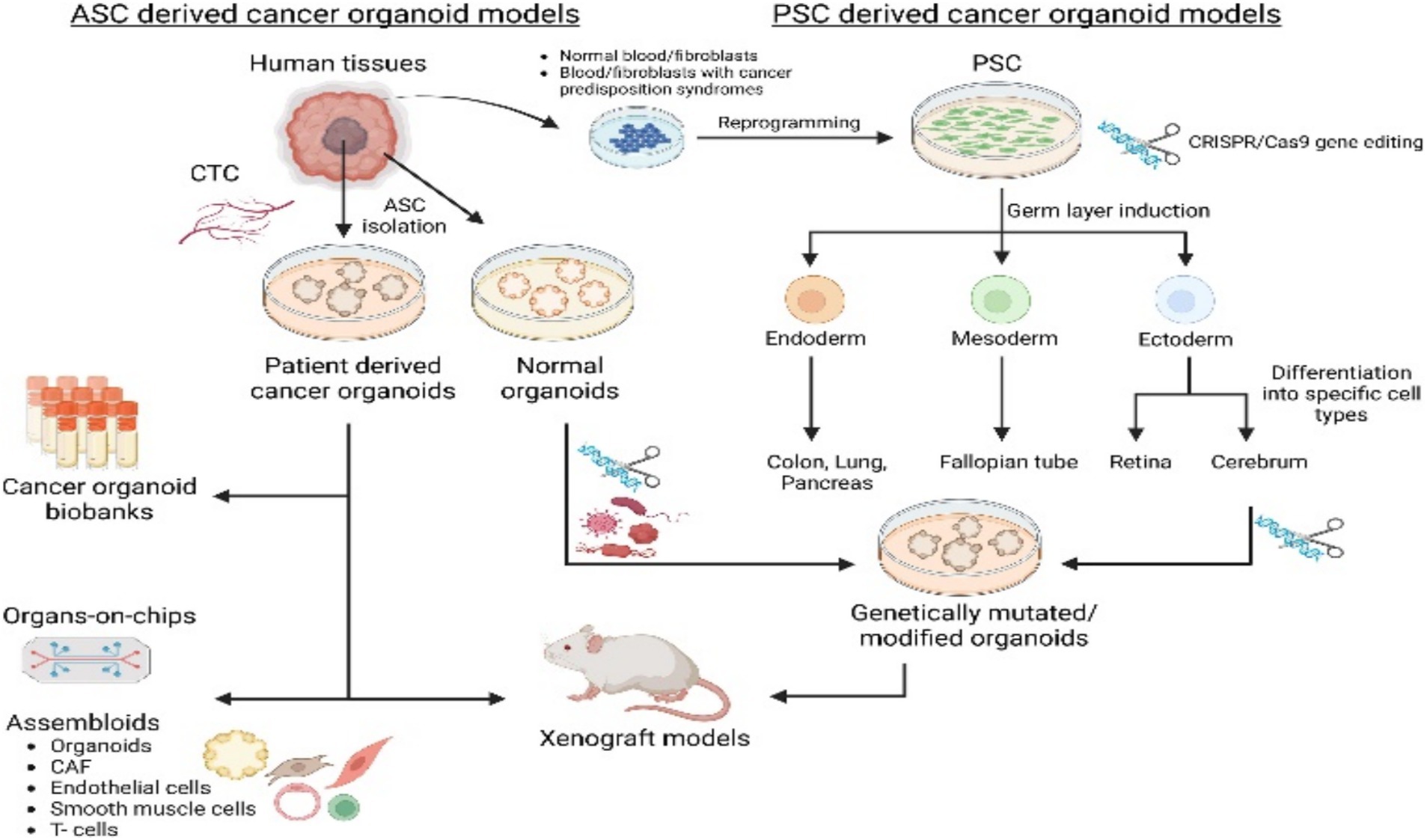
Figure 1. Cancer organoids, derived from either adult stem cells (ASCs) or pluripotent stem cells (PSCs) and incorporating components like circulating tumor cells (CTCs) or cancer-associated fibroblasts (CAFs), are established to model various cancer types and for research applications. Adapted with permission from Yan et al. (2023). Copyright 2023 Cell Stem Cell.
Pluripotent stem cell (PSC)-derived intestinal cancer organoids are generated through the differentiation of pluripotent stem cells (such as induced pluripotent stem cells [iPSCs] or embryonic stem cells [ESCs]) to form three-dimensional micro-organ structures (Xu H. et al., 2022), These structures mimic the cellular composition, tissue architecture, and functional characteristics of human intestinal carcinogenesis, thereby providing a highly physiologically relevant in vitro tumor model; The principle relies on the plasticity of pluripotent stem cells (Wuputra et al., 2021), utilizing directed differentiation (e.g., first inducing definitive endoderm, then further developing into intestinal tissue to reconstruct the intestinal epithelium) (Matkovic Leko et al., 2023), and can be combined with oncogene introduction (e.g., KRAS) or carcinogen treatment (e.g., Azoxymethane) to induce a cancer-like state, thereby elucidating molecular mechanisms of tumor growth (Huang et al., 2021). Its advantages include the precise modeling of tumor heterogeneity and genetic characteristics, providing an efficient platform for drug screening and personalized therapy evaluation with relatively low cost and short time requirements; Limitations include difficulties in modeling non-epithelial components (e.g., immune cells or TME) (Farin et al., 2023), the technical complexity of 3D culture systems that restricts clinical applications (e.g., regenerative medicine) (Zhu et al., 2023), and significant challenges remain in their development as clinical diagnostic or therapeutic tools; In cancer research, such organoids are widely used in colorectal cancer studies to investigate molecular mechanisms of tumor initiation and progression, conduct large-scale drug testing (predicting patient response), stablish biobanks for basic research, and advance the development of precision medicine strategies (Figure 1).
Gut-on-a-chip systems simulate the structure and function of the human intestine by engineering controllable microenvironments within microchips (Özkan et al., 2024), with a particular focus on replicating the three-dimensional morphology of the intestinal epithelium, fluid flow, and mechanical stresses to investigate physiological and pathological mechanisms, The operating principle involves employing microfluidic channels and cell culture techniques with human cells, coupled with precise control of biochemical factors (Xian et al., 2023; Xiang et al., 2020), to recapitulate processes such as intestinal barrier function, nutrient absorption, and pathogen invasion. Furthermore, through interconnection with other organ chips (e.g., liver-chip) (Milani et al., 2022), they model cross-organ signaling. Advantages include a more accurate mimicry of human intestinal physiology, reduced interspecies variation, and higher physiological relevance compared to conventional two-dimensional cultures; they demonstrate improved cost-effectiveness, encounter fewer ethical concerns, and can be applied to drug screening and disease modeling, Limitations of these systems include high manufacturing costs, challenges in model standardization, and difficulties in replicating complex microenvironments such as microbiome interactions, which hinder their universal applicability in broad contexts and scalability for widespread adoption. The core utility of this system lies in modeling intestinal diseases (e.g., inflammatory bowel disease) and drug metabolism processes through dynamic microenvironments (Kaden et al., 2025; Nguyen et al., 2024), with applications in both basic research and clinical practice; particularly in oncology, gut-on-a-chip platforms can establish complex colorectal cancer microenvironment models (Lee et al., 2025), encompassing the replication of the intestinal TME, for analyzing mechanisms of cancer cell infiltration and metastasis, thereby assessing the efficacy and toxicity of therapeutic compounds, ultimately providing a platform for high-throughput prescreening in cancer therapy and an alternative to animal models for metastasis research.
Organotypic explant cultures are defined as three-dimensional (3D) cultures of intact tissue or organ fragments (He and Deng, 2022) directly obtained from an organism, Their principle relies on optimized culture conditions (e.g., air-liquid interface culture, specialized scaffolds, or matrices) (Zhao Y. et al., 2022) to preserve the spatial architecture of the tissue and intercellular signaling networks (Steindl and Valiente, 2025), thereby simulating physiological or pathological conditions in vivo. In cancer research, a key advantage of this model is its faithful preservation of tumor heterogeneity (LeSavage et al., 2022), making it valuable for exploring tumor biology. Limitations include a relatively short culture period that hinders modeling of long-term dynamic processes, technically demanding procedures, and significant challenges in standardization. This model helps reduce the scale of animal experimentation, particularly in cancer pharmacology, by enabling simultaneous assessment of drug effects on both tumor tissue and matched normal tissue to evaluate selective toxicity. It provides a platform for developing targeted therapies and combination immunotherapy strategies.
3 Technical advantages and applications of organoids
3.1 Unique advantages of organoids in microbiome research
As a three-dimensional (3D) in vitro culture system (Kim et al., 2022), organoids can highly simulate the structure, function, and genetic characteristics of human organs, providing an unprecedented platform for studying host–microbe interactions. Compared to traditional models, patient-derived organoids (PDOs) preserve intra- and inter-tumor heterogeneity (Jin et al., 2025) and accurately recapitulate the cellular interaction networks within the original TME. Particularly in gastrointestinal cancer research, organoid technology has demonstrated the ability to recreate key pathological features of tumorigenesis and progression (Idris et al., 2021; Kan et al., 2025), including dynamic interactions between microbes and epithelial cells. Recent studies have found that by integrating artificial intelligence technology with low-biomass microbiome analysis (Han et al., 2021), organoids can reveal intricate interaction mechanisms between microbes and cancer cells. We consider that the evidence for organoids in the simulation of TME and short-term drug response prediction is credible; however, they remain in the proof-of-concept stage regarding dynamic host–microbe interactions and the reconstruction of complex microsystems. Particular caution is warranted: while AI technology can enhance the depth of analysis, the conclusions it infers require dual validation through organoid experiments and clinical data, so as to avoid overinterpreting algorithmic correlations as causal mechanisms.
3.2 Comparison with 2D culture systems: microenvironment modeling capability
Two-dimensional (2D) culture systems exhibit significant limitations in modeling the TME, whereas organoids leverage three-dimensional architecture to better recapitulate the composition and organization of the extracellular matrix (ECM) (Jung et al., 2022; Li K. et al., 2025). The development of air-liquid interface (ALI) organoid culture systems (Atanasova et al., 2023) has enabled the establishment of more physiologically relevant models from colorectal cancer patient tissues. Compared to 2D cultures, organoids can maintain the microenvironmental conditions required for long-term co-culture of tumor cells and microorganisms, encompassing critical parameters such as oxygen gradients, nutrient distribution, and mechanical stresses (Kumarasamy et al., 2024). Notably, intestinal organoids have successfully simulated molecular and microenvironmental characteristics of metastatic tumors (Atanasova et al., 2023), a feat unattainable with conventional monolayer cultures. We recognize that organoids are superior to 2D systems, yet they are not without limitations: the full recapitulation of the in vivo TME remains a persistent challenge, particularly in terms of standardization and cost, which pose bottlenecks.
3.3 Comparison with organ-on-a-Chip Systems: balancing throughput and complexity
Although organ-on-a-chip technology can simulate dynamic microenvironments and tissue-tissue interactions in tumor pathophysiology (Saha et al., 2021; Maity et al., 2025), its limited throughput and operational complexity (Azimian Zavareh et al., 2022) constrain large-scale applications. Organoids, while maintaining sufficient complexity, are better suited for high-throughput drug screening and personalized medicine research (Zhu et al., 2023; Si et al., 2025). Microfluidic chip-integrated organoid systems (Organoid-chip) (Fang et al., 2021) represent a convergence of both technologies’ advantages, enabling the recreation of tissue-specific microenvironments while enhancing experimental throughput and operational feasibility. Particularly in non-small cell lung cancer research (Choi et al., 2023), organoid-immune cell co-culture systems have enabled the observation of dynamic changes in the TME in vitro. We argue that balancing experimental complexity and practicality is a key driving factor for model selection. Organ-on-a-chip systems are highly reliable in simulating physiological dynamics such as hypoxia and fluid shear stress, making them well-suited for in-depth mechanistic exploration. Organoids, on the other hand, possess greater advantages in patient-specific modeling and screening efficiency, rendering them particularly suitable for optimizing personalized treatment of non-small cell lung cancer (NSCLC). Integrated systems (organoid-chip) represent an emerging trend; however, their value remains in the preliminary validation stage.
4 Analysis of host–microbe interactions in the TME using organoids
4.1 Organoid evidence of microbial dysbiosis driving carcinogenesis
4.1.1 Mechanistic evidence for intestinal microbial dysbiosis promoting carcinogenesis by driving chronic inflammation
Intestinal organoid models provide a crucial experimental platform for deciphering chronic inflammation induced by microbial dysbiosis and subsequent carcinogenesis. By remodeling the tumor microenvironment, intestinal microbiota dysbiosis greatly accelerates the infiltration of neutrophils into CRC tissues, thereby leading to mucosal immune dysfunction, stromal cell transformation, and EMT (Li Q. et al., 2025; Tang et al., 2025). This reprogramming of the immune microenvironment enables cancer cells to evade immune surveillance. Disordered microbial communities (e.g., enriched specific pathogenic bacteria) can continuously activate pro-inflammatory signaling pathways (Eiman et al., 2025), resulting in the development of local intestinal and systemic chronic inflammatory states. For example, in mice with TNF receptor deficiency, intestinal organoid experiments have confirmed that TNFR1/2 is targeted to Toll-like receptor 5 (TLR5)-positive Paneth cells and dendritic cells, synergistically contributing to the development of spontaneous ileitis and microbial dysbiosis (Wahida et al., 2021). Notably, the co-culture model of intestinal organoids with dorsal root ganglion (DRG) neurons further demonstrates that inflammation-induced microbial metabolites impair the growth and metabolic functions of colonic organoids while enhancing neuronal excitability; through the release of mediators such as c-Fos and calcitonin gene-related peptide (CGRP), this forms a pro-carcinogenic “microbiota-epithelium-nerve” crosstalk network (Margiotta et al., 2025). Current studies mostly focus on single bacterial species or a limited number of metabolites (Smet et al., 2022; Rivas-Domínguez et al., 2021), and the analysis of the interaction between the overall functional redundancy of microbial communities and the host genetic background remains insufficient. We contend that existing evidence has sufficiently demonstrated the mechanism by which microbial dysbiosis promotes carcinogenesis via inflammatory signaling, whereas evidence related to neural modulation and holistic community interactions remains in the preliminary exploration stage.
4.1.2 Direct evidence of microbial dysbiosis mediating genotoxic effects and bottlenecks in clinical translation
Intestinal organoids exhibit unique advantages in elucidating the mechanisms by which microorganisms exert direct genotoxic carcinogenic effects. Using 3D bioprinted human liver organoids, researchers have verified that bacterial metabolites such as lansoprazole chloride (Lanchlor) can induce significant formation of DNA adducts in organoids, and confirmed their genotoxic damage effects via the comet assay, thereby providing a standardized platform for environmental carcinogen screening (Li et al., 2024). More importantly, the specific mutation signatures (e.g., characteristic base substitution patterns) induced by pks + Escherichia coli in intestinal organoids are highly consistent with the genomic mutation profiles of colorectal cancer patients (Rosendahl Huber et al., 2021; Thomas, 2025). This directly demonstrates the ability of specific bacterial strains to induce carcinogenic mutations. This genotoxicity can be achieved through multiple pathways, including microbial metabolites (e.g., secondary bile acids) inducing DNA oxidative damage and chromosomal aberrations (Fu et al., 2021; Yoon et al., 2025). It also involves the continuous accumulation of reactive oxygen species (ROS) in the chronic inflammatory microenvironment, which leads to genomic instability (Xia et al., 2024). Current studies have obvious limitations: most organoid models still rely on the inoculation of single bacterial strains or simplified microbial communities, making it difficult to simulate the complex in vivo microbial ecology (Li et al., 2024; Hou et al., 2022). The difficulty in translating organoid-based genotoxicity detection results into clinical practice stems from the complexity of metabolic pathways in the human body and the heterogeneity of microbiota-host interactions among individuals. In organoids, KRAS-mutated cancer cells exhibit chemoresistance (van de Haar et al., 2023). This suggests that microbial dysbiosis may indirectly affect the process of carcinogenesis by altering the threshold of cellular response to genotoxicity. This mechanism requires further verification in organoid models integrated with multi-omics approaches. We argue that intestinal organoids provide an irreplaceable targeted verification platform for the direct genotoxicity of microorganisms; however, they have limitations in recapitulating the ecological complexity of microbial communities and host–microbe interactions. This results in evidence for indirect carcinogenic mechanisms (e.g., metabolic remodeling, immune evasion) still remaining in the preliminary stage. In the future, it will be necessary to enhance translational value by means of multicellular co-culture and organoid-animal model coupling.
4.2 Organoid-based validation of tumor-specific microbial features
Organoid platforms provide a key technological foundation for deciphering the spatial and functional heterogeneity of the tumor microbiome (Kan et al., 2025; Poletti et al., 2021), Microfluidic organoid-chips simulate microbial colonization within hypoxic niches, demonstrating that hypoxic microenvironments enrich drug-resistant microbiota through selection pressure, thereby contributing to chemotherapy resistance. In PDO models, low alpha-diversity microbial communities show significant correlation with loss of MHC-I expression (Yáñez-Bartolomé et al., 2023; Zhou et al., 2023), leading to reduced CD8 + T cell infiltration and immunotherapy resistance (Wang et al., 2023). Notably, while organoids can recapitulate tumor-type specificity reflected in microbial composition, they remain limited in simulating multi-species synergistic effects, particularly in maintaining the viability of obligate anaerobes over extended periods. We contend that the value of organoids in simulating tumor heterogeneity and single microbe-host interactions (e.g., hypoxia-induced drug resistance) has been fully validated. However, these mechanisms have mostly been explored through correlational studies, rather than direct causal validation. Its ability to simulate multi-species synergistic effects (e.g., maintenance of anaerobic bacteria) remains in the preliminary exploration stage. And there are significant technical controversies associated with this capability. In the future, interdisciplinary innovation will be required to enhance the integrity of such models.
5 Cancer applications: from mechanistic insights to clinical translation
5.1 Key technological breakthroughs of organoids in cancer microbiome research
5.1.1 Precision co-culture techniques break the confines of traditional models
As a three-dimensional model, organoids address the limitations of traditional two-dimensional cell cultures in simulating tissue architecture and spatial host–microbe interactions. Through direct microinjection of gut microbiota (or specific pathogens) into the lumen of hollow organoids and the establishment of co-culture systems (Verhulsel et al., 2021; Wang et al., 2024), this technology enables the simulation of physical contact and biomolecular exchange between host cells and microbes. More advanced organoid-on-a-chip models incorporate microfluidic technology (Valiei et al., 2023), allowing better recapitulation of physiological dynamics such as intestinal fluid flow and shear stress, thereby making the interaction process more representative of the in vivo environment. In the technological breakthrough of “immune-gut microbiota co-culture systems,” organoids utilize a three-dimensional co-culture environment to integrate intestinal organoids, gut microbiota, and associated immune cells (Shek et al., 2021) (e.g., T cells or NK cells). By employing microfluidic platforms and microinjection techniques (Liu et al., 2024), this system achieves the goal of simulating host–microbe interactions within the TME in vitro. It overcomes the limitations of traditional two-dimensional models and provides physiologically relevant insights into how gut microbiota modulate immunity in colorectal cancer (Golpour et al., 2023), such as microbiota-mediated mechanisms of T cell activation and NK cell cytotoxicity influencing tumor progression, immune evasion, and treatment response. Furthermore, it serves as an experimental platform for developing personalized immunotherapies (Wu et al., 2025) (e.g., microbiota-targeted modulation). However, the system still faces core limitations, including challenges in reconstituting the full spectrum of complex immune cells, standardization issues, and high costs (Chen et al., 2022; Kriaa et al., 2024). We think that 3D organoid models can highly simulate the physical interactions, biomolecular exchange, and basic immune activation between hosts and microorganisms. And their physiological relevance is significantly better than that of 2D models. The bidirectional regulatory role of microorganisms in the tumor immune microenvironment still requires more evidence to confirm. Furthermore, the dynamic reconstruction of complex immune cell networks remains a technical bottleneck.
5.1.2 Multi-omics integration platforms
Research on host-gut microbiota interactions within the TME has achieved significant advancements through the integration of organoids and multi-omics platforms (Kim et al., 2022; Coxon et al., 2025), enabling researchers to accurately simulate the complex characteristics of the colorectal cancer microenvironment in vitro and analyze dynamic interaction mechanisms between gut microbiota and host cells. By integrating data from genomics, transcriptomics, metabolomics, and microbiome studies (Liu B. et al., 2025; Zhang et al., 2023), this approach reveals potential molecular drivers and therapeutic targets. The combination of organoids with single-cell sequencing and metabolomics (Smirnov et al., 2023; Zhao H. et al., 2022) elucidates how microbiota modulate the β-catenin signaling pathway and cellular self-renewal processes under tumor heterogeneity. High-throughput platforms such as bioreactors, combined with machine learning algorithms (Wang Q. et al., 2025), facilitate large-scale drug sensitivity testing. However, multi-omics integration still faces significant challenges, including reduced reproducibility (Su et al., 2025) due to high data heterogeneity and noise, and difficulties in causal inference (Metwaly et al., 2022) (e.g., challenges in determining whether microbiome changes are drivers or consequences of disease) and insufficient standardization (Babu and Snyder, 2023). These limitations impact its broad applicability in clinical translation, but predictive models integrating artificial intelligence are expected to enhance data interpretation and advance the potential for personalized therapy applications. We affirm that organoid-like models can highly accurately simulate tumor-microbe interactions during multi-omics integration and have also become effective tools for drug screening. Their clinical translational value is hampered by data noise, ambiguous causal logic, and a lack of standardization. The “driver or bystander” role of microorganisms still needs to be further verified through spatial multi-omics and longitudinal studies (see Table 2).
5.1.3 Bidirectional regulatory mechanisms of microbial metabolites
5.1.3.1 Carcinogenic effects
The carcinogenic effects of intestinal microbial metabolites are mainly due to the disruption of immune surveillance caused by their abnormal production, with specific mechanisms involving the induction of genetic mutations and alterations to the tumor microenvironment. When the production of metabolites is abnormal, the host immune system fails to recognize and eliminate tumor cells in the early stages of carcinogenesis, leading to tumor progression, and this phenomenon is particularly prominent in colon cancer (Liu et al., 2023). Key metabolites such as short-chain fatty acids (SCFAs), polyamines, and secondary bile acids directly participate in tumorigenesis by regulating immune responses; they promote the occurrence of gene mutations in epithelial cells or impair the ability of immune cells to clear abnormal cells (Liu et al., 2023; Hanus et al., 2021). This pro-carcinogenic effect does not exist in isolation: bacteria such as Fusobacterium alter inflammatory signals in the microenvironment, leading to the emergence of immunosuppressive states, yet the specific molecular pathways remain not fully elucidated. This indicates that there are still inadequacies in mechanistic research—for example, the causal link between intestinal microbiota dysbiosis and colorectal cancer (CRC) risk is currently unclear (Duan et al., 2024; Fan et al., 2025). The application of intestinal organoid models in Lynch syndrome has shown potential, enabling the evaluation of how epigenetic modulation (e.g., EZH2-mediated immune gene modification) drives carcinogenesis (Bowen et al., 2025). However, it is still limited by this constraint: organoids cannot fully simulate the complexity of in vivo host–microbe interactions. Additionally, the dual effects of different metabolites (e.g., certain SCFAs possess both anti-tumor capacity and pro-tumor effects) constitute a major research barrier, and rigorous experimental design is urgently needed to resolve this contradiction.
5.1.3.2 Anti-carcinogenic effects
In the context of cancer resistance, intestinal microbial metabolites remodel the tumor microenvironment through immune regulation, enhancing host defense capabilities and anti-tumor efficacy; however, this process is limited by individual differences and therapeutic variability. Specific metabolites, such as microbial tryptophan catabolites, can activate immune control mechanisms (Jaye et al., 2022), inhibit the progression of obesity-related cancers, and remodel the microenvironment to facilitate the clearance of abnormal cells. These metabolites enhance the anti-tumor efficacy of chemotherapy, radiotherapy, and immunotherapy by regulating immune cell functions (e.g., enhancing T cell activity) (Song et al., 2025; Wang Y. et al., 2025; Yang et al., 2023). In model systems, they exhibit effects of reducing treatment-related side effects and improving prognosis. From a critical perspective, clinical application faces significant challenges: variations in the individual microbiome lead to inconsistent metabolite production, which affects the prediction of therapeutic efficacy (Nazir et al., 2025). Even though engineered bacteria strategies (e.g., designing degrading enzymes to target specific metabolites) have shown potential in mouse models (Wang S. et al., 2022), their translation to humans often fails due to the complex nature of host-immune interactions. By screening probiotic or prebiotic combinations, intestinal organoids can assist in testing the anti-carcinogenic mechanisms of microbe-targeted strategies. However, existing models struggle to capture the full-spectrum effects of metabolites in systemic immune regulation, highlighting the need to integrate high-throughput metabolomics to achieve precision medicine-guided optimization.
5.1.3.3 Immune regulation
The immune regulatory effects of intestinal microbial metabolites manifest as dual regulation of the tumor immune environment; their mechanisms involve the dynamic balance between receptor activation and cytokines, yet the underlying regulatory networks remain highly controversial. Metabolites such as indole-3-acetic acid (IAA) can activate the aryl hydrocarbon receptor (AhR), leading to differential expression of cytokines and changes in immune cell composition, thereby providing support for the regulation of anti-cancer immune checkpoints (Yang et al., 2023; Vaaben et al., 2025). Within the tumor microenvironment, these metabolites can remodel innate and adaptive immune responses (Lu et al., 2022; Kovtonyuk and McCoy, 2023) and influence the efficacy of immunotherapy. At the same time, the consumption of key metabolites by microorganisms (e.g., immune suppression mediated by regulatory T cells (Treg)) (Meza-Perez et al., 2024) may counteract their positive effects, resulting in impairments in immune function. Intestinal organoids in ex vivo models can help simulate the regulation of immune cell dynamics by metabolites; however, critical analysis reveals that the molecular pathways of epigenetic regulation have not yet been fully elucidated (Duan et al., 2024). The complexity of host–microbe interactions often leads to biases in data interpretation (Duan et al., 2024; Li et al., 2022). In the future, it will be necessary to use organoids to integrate multi-omics data, clarify the dual balance of immune regulation, and thereby overcome the bottleneck in precision intervention. We contend that gut microbial metabolites regulate the tumor immune microenvironment and therapeutic responses via receptor-mediated pathways, and their role has received direct experimental support in organ-specific organoid models (e.g., for colon cancer). This evidence is reliable and holds clinical translational potential. However, current organoid technology still has limitations in simulating stromal-immune cell interaction networks. For instance, the dynamic integration of fibroblasts and myeloid cells has not yet been standardized. This may lead to an underestimation of the complexity of microenvironmental pathways.
5.1.4 Analysis of key signaling pathways using organoids
Organoids, as three-dimensional (3D) culture systems, recapitulate the structural, functional, and genetic characteristics of human organs—including epithelial, immune, and microbial components within tumor tissues—thereby providing a highly controlled environment for studying key signaling pathways regulated by the microbiota. For example, studies using organoids have demonstrated that microbiota-derived short-chain fatty acids (SCFAs) activate host signaling pathways (e.g., G protein-coupled receptor pathways) (Xu J. et al., 2022), modulating the tumor immune microenvironment and influencing cancer progression. These metabolites directly act on epithelial cells in organoids, revealing their role in promoting inflammation and carcinogenesis (Boesch et al., 2022). Furthermore, organoids enable in-depth dissection of inflammation-related pathways—such as the TLR4-NF-κB-NLRP3 inflammasome axis (Sheng et al., 2021; Xie and Liu, 2024)—and their roles in driving chronic inflammation and immunosuppression within the TME during microbial dysbiosis. By simulating bacterial interactions, organoids demonstrate how microbiota imbalance amplifies the activation of these pathways, leading to uncontrolled cell proliferation and therapy resistance. Additionally, organoids help elucidate how the microbiota regulates other signaling pathways (Zhang X. Y. et al., 2024; Wu et al., 2023) (e.g., PI3K-AKT and TGFβ signaling) via the intratumoral microbiome, which are critically involved in tumor cell metabolism, metastasis, and response to immunotherapy. For instance, studies combining patient-derived organoids with microbiota transplantation have revealed that microbial metabolites (e.g., lipopolysaccharide or bacterial toxins) indirectly affect immune checkpoint pathways (e.g., PD-1/PD-L1) by remodeling the stromal components of the TME (Si et al., 2025; de Castilhos et al., 2024), providing an experimental foundation for designing microbiota-targeted therapies. Collectively, organoids have revolutionized cancer research by elucidating key signaling pathways, bridging the limitations of traditional cell lines and animal models, offering human-relevant simulations, and advancing our understanding of dynamic host-microbiota collaborations within the TME. We contend that gut microbial metabolites regulate the tumor immune microenvironment and therapeutic responses through receptor-mediated pathways, and their role has received direct experimental support in organ-specific organoid models (e.g., models for colon cancer). This evidence is reliable and holds clinical translational potential. However, current organoid technology still has shortcomings in simulating stromal-immune cell interaction networks. For example, the dynamic integration of fibroblasts and myeloid cells has not yet been standardized. This may lead to an underestimation of the complexity of microenvironmental pathways (Figure 2).
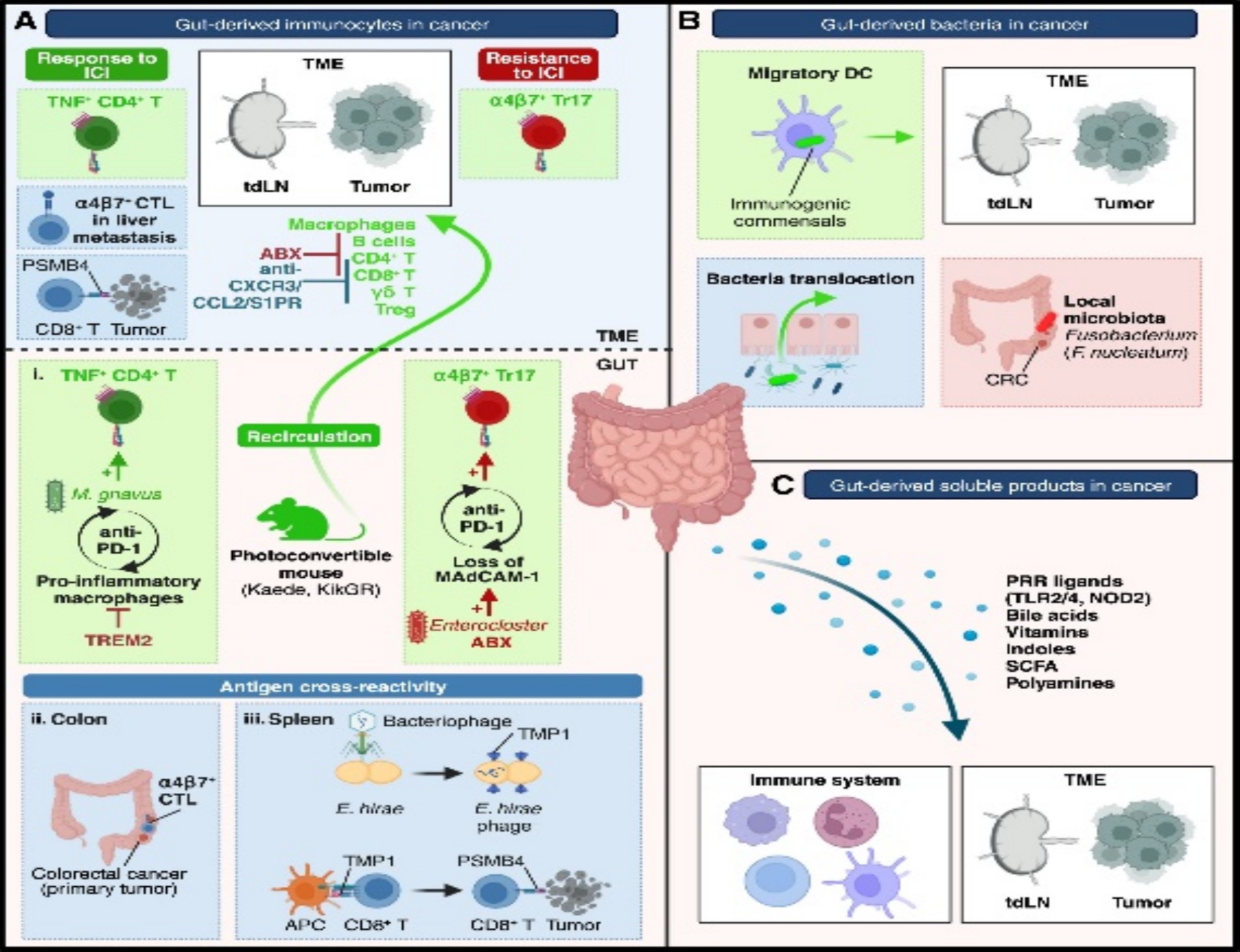
Figure 2. Mechanisms of gut influence on the TME. (A) Gut-derived immunocytes migrate to the TME under microbiota influence. (B) Gut bacteria can translocate to the TME via dendritic cells or barrier leakage. (C) Soluble gut-derived factors modulate cancer–immunity crosstalk. ABX, antibiotics; CRC, colorectal cancer; CTL, cytotoxic T cells; DCs, dendritic cells; TME, TME; Treg, regulatory T cells. Adapted with permission from Pich et al. (2025). Copyright 2024 Immunity.
5.2 Clinical translation scenarios and challenges
5.2.1 Personalized drug screening
Organoids serve as “patient-specific TME-mimicking models” that retain the genetic and phenotypic attributes of the original tumors (Liu et al., 2021; Zitvogel et al., 2024), providing a platform for evaluating individual patient responses to chemotherapy and targeted therapies (Zhou et al., 2025). By exposing patient-derived organoids to various drug conditions (Yan et al., 2024), treatment efficacy can be predicted, thereby reducing the risk of ineffective therapies. A major challenge is that current organoids often lack a complete tumor immune microenvironment (Song et al., 2024), limiting their ability to accurately simulate the impact of microbial metabolites (e.g., short-chain fatty acids, SCFAs) on drug metabolism or immunotherapy responses. Proposed solutions include developing immune-organoid co-culture models by incorporating autologous immune cells or engineering organoids to simulate the tumor immune microenvironment (TIME), alongside advanced microbiota-host co-culture systems (Magré et al., 2023), aiming to reconstitute microbiota-mediated modulation of drug sensitivity in vitro. We contend that the mechanisms underlying the recapitulation of tumor biology and drug sensitivity have received relatively sufficient verification. However, the issue of the lack of immune microenvironment and its remedial strategies are in the preliminary experimental stage. There are controversial issues regarding reproducibility and clinical translation (Figure 3).
Figure 3. Organoids serve as a transformative platform, bridging the gap between basic biological research and clinical practice by facilitating bidirectional translation. Adapted with permission from Wang Y. et al. (2025). Copyright 2023 Cancer Lett.
5.2.2 Optimization of immunotherapy
In this context, organoids serve as a platform (Weng et al., 2023) to investigate how specific microbiota or their metabolites reshape the local tumor immune response (Pich et al., 2025), and to conduct screening of immunotherapy drugs (Grönholm et al., 2021). However, a key challenge is that current organoids struggle to fully recapitulate the complex in vivo immune regulatory network and the systemic microbiota-immune axis. Immune cells cultured in vitro may become functionally exhausted, and organoids often fail to maintain the physical and biochemical integrity of the host mucosal barrier (Hanus et al., 2021), which is a critical interface for microbiota-immune interactions. Proposed solutions include developing microfluidic organ-on-a-chip technology (Grönholm et al., 2021) to integrate immune and microbial components, and employing genetic engineering to modify organoids, thereby enabling more precise simulation of specific immune pathways (Figure 3).
5.2.3 Early disease diagnosis and mechanism studies
5.2.3.1 Microbial biomarkers
Intestinal organoids provide an innovative platform for the discovery of microbial biomarkers in early tumor diagnosis, and elucidate the interaction mechanisms between the intestinal microbiota and tumors by establishing highly physiologically relevant models. A dual-channel system constructed using intestinal organoids differentiated from human induced pluripotent stem cells (hiPSCs) can incorporate patients’ fecal samples for multi-omics analysis, thereby enabling the identification of epithelium-specific biomarkers and key microbial factors associated with the clinical outcomes of melanoma (Ballerini et al., 2025). The advantages of this approach lie in the ability of organoids to more accurately simulate the intestinal microenvironment, which overcomes the limitations of traditional 2D models. It supports the study of host–microbe interactions under controlled conditions and provides an efficient avenue for the high-throughput screening of tumor-associated microbial biomarkers. Organoid models can recapitulate patients’ genetic characteristics (Xiang et al., 2024), enabling more personalized screening for specific tumor markers and thus demonstrating potential in the identification of diagnostic biomarkers and therapeutic targets. Notable limitations of the organoid system remain: it lacks immune components and the in vivo dynamic environment (Özkan et al., 2024; Bouffi et al., 2023), making it difficult to fully recapitulate gut-immune crosstalk or long-term microbial effects. This is likely to affect the clinical translatability of microbial biomarkers. Although organoids can identify associations between changes in microbial communities and disease progression, their predictive capacity is limited in the absence of immune regulation and mechanical stress (e.g., peristalsis) (Özkan et al., 2024), which may lead to biases in the in vivo validation of biomarkers.
We contend that (organoids) possess irreplaceable tool value in the process of revealing microbe-host interaction mechanisms and conducting high-throughput preliminary screening of biomarkers, and their experimental controllability far exceeds that of clinical sample analysis. All organoid-related findings must undergo secondary validation using immune co-culture models or in vivo animal validation; otherwise, there may be an overestimation of the clinical translatability of biomarkers, as these findings have not yet passed the “stress tests” of immune regulation and dynamic physiological environments.
5.2.3.2 Metabolite biomarkers
Regarding metabolite biomarkers, intestinal organoids have provided key clues for early tumor diagnosis by integrating metabolomics techniques, particularly when elucidating tumor-related metabolic reprogramming mechanisms. Relevant studies have confirmed that organoid models can be used to detect intestinal microbial disorders induced by cold stress and their association with liver injury; by analyzing changes in glycerophospholipid metabolism, ABC transporters, and purine metabolism pathways, the correlation between specific microbial taxa and metabolite biomarkers can be identified (Liu A. et al., 2025). The advantage of this approach lies in the high-throughput culture capability of organoids (Xiang et al., 2024), which supports real-time monitoring of metabolic changes. When combined with untargeted metabolomics approaches, it can sensitively detect subtle metabolic abnormalities (e.g., metabolic perturbations caused by toxic exposure) (Xuan et al., 2024), providing a high-sensitivity tool for the discovery of tumor markers. Organoid models are also suitable for studying the effects of dietary or pharmaceutical interventions on metabolite markers. For example, in simulating nutrient metabolism and toxicity analysis, they facilitate the identification of relevant targets for metabolic diseases such as diabetes, thereby providing support for the development of anti-tumor drugs (Nie et al., 2023). Organoids have limitations in simulating metabolite dynamics: they cannot effectively recapitulate in vivo mechanical forces (e.g., digestive fluids) or long-term microbial ecological balance (Özkan et al., 2024), and may overestimate the stability of metabolic pathways (Lu et al., 2024). We affirm that the core advantages of organoid metabolic models lie in their high sensitivity and pathological specificity; however, their simplified physiological simulation and inadequate recapitulation of tumor heterogeneity may diminish their predictive value. In terms of the current evidence chain, the mechanisms underlying the interaction between environmental exposure and microorganisms are relatively sufficient, while the response data generated by pharmaceutical interventions require more targeted validation. The modeling of inter-organ metabolic networks remains in the exploratory stage.
6 Concluding remarks and outlook
Organoids have undoubtedly revolutionized the research paradigm for investigating the complex relationships between the gut microbiota and the TME (TME). They have established a physiologically relevant research platform that breaks through the barriers of traditional models. Organoids are capable of preserving the patient-specific genetic and phenotypic characteristics while supporting co-cultivation with microbial communities. This attribute provides unprecedented perspectives for in-depth understanding of host–microbe interactions, metabolic regulation, and immune modulation mechanisms within the TME. However, this field is still in its initial stages, and multiple challenges must be addressed to fully tap into the potential of organoid technology.
The integration of multi-omics technologies (such as spatially resolved transcriptomics, metabolomics, and proteomics) with advanced organoid systems is crucial for unraveling the spatiotemporal dynamics of interactions between the gut microbiota and the TME. The development of more complex co-culture systems (incorporating immune cells, stromal components, and neuroendocrine elements) is also essential; only in this way can the systemic interaction processes observed in vivo be recapitulated. The rise of organoid-on-a-chip technology, combined with machine learning-driven analytical approaches, holds the potential to construct predictive models that can accelerate personalized drug screening and the development of therapeutic strategies.
To translate organoid-based research findings into clinically applicable outcomes, it is still necessary to establish standardized experimental protocols, promote the construction of scalable biobanks, and conduct rigorous validation in human cohorts. By bridging the gap between experimental models and human pathophysiological processes, organoid technology is likely to lead the next wave of innovations in the field of precision oncology, providing new avenues for cancer therapy targeting the TME-gut microbiota axis.
Author contributions
S-yZ: Writing – original draft, Writing – review & editing. Y-yS: Investigation, Writing – review & editing. F-lC: Investigation, Writing – review & editing. D-fX: Methodology, Writing – review & editing. Y-qX: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Public Welfare Application Research Project (Population Health Category, Key Medical and Health Project) of Huzhou Science and Technology Bureau (Contract No. 2023GZ80) entitled "Clinical Application Research of Colorectal Cancer Organoids in Patients with Colorectal Cancer”.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Atanasova, V. S. , de Jesus Cardona, C. , Hejret, V. , Tiefenbacher, A. , Mair, T. , Tran, L., et al. (2023). Mimicking tumor cell heterogeneity of colorectal cancer in a patient-derived organoid-fibroblast model. Cell. Mol. Gastroenterol. Hepatol. 15, 1391–1419. doi: 10.1016/j.jcmgh.2023.02.014
Azimian Zavareh, V. , Rafiee, L. , Sheikholeslam, M. , Shariati, L. , Vaseghi, G. , Savoji, H., et al. (2022). Three-dimensional in vitro models: a promising tool to scale-up breast Cancer research. ACS Biomater Sci. Eng. 8, 4648–4672. doi: 10.1021/acsbiomaterials.2c00277
Babu, M. , and Snyder, M. (2023). Multi-omics profiling for health. Mol. Cell. Proteomics 22:100561. doi: 10.1016/j.mcpro.2023.100561
Ballerini, M. , Galiè, S. , Tyagi, P. , Catozzi, C. , Raji, H. , Nabinejad, A., et al. (2025). A gut-on-a-chip incorporating human faecal samples and peristalsis predicts responses to immune checkpoint inhibitors for melanoma. Nat. Biomed. Eng. 9, 967–984. doi: 10.1038/s41551-024-01318-z
Boesch, M. , Horvath, L. , Baty, F. , Pircher, A. , Wolf, D. , Spahn, S., et al. (2022). Compartmentalization of the host microbiome: how tumor microbiota shapes checkpoint immunotherapy outcome and offers therapeutic prospects. J. Immunother. Cancer 10:e005401. doi: 10.1136/jitc-2022-005401
Bouffi, C. , Wikenheiser-Brokamp, K. A. , Chaturvedi, P. , Sundaram, N. , Goddard, G. R. , Wunderlich, M., et al. (2023). In vivo development of immune tissue in human intestinal organoids transplanted into humanized mice. Nat. Biotechnol. 41, 824–831. doi: 10.1038/s41587-022-01558-x
Bowen, C. M. , Duzagac, F. , Martel-Martel, A. , Reyes-Uribe, L. , Zaheer, M. , Thompson, J., et al. (2025). Inhibition of histone methyltransferase EZH2 for immune interception of colorectal cancer in lynch syndrome. JCI Insight 10:e177545. doi: 10.1172/jci.insight.177545
Castro, F. , Leite Pereira, C. , Helena Macedo, M. , Almeida, A. , José Silveira, M. , Dias, S., et al. (2021). Advances on colorectal cancer 3D models: the needed translational technology for nanomedicine screening. Adv. Drug Deliv. Rev. 175:113824. doi: 10.1016/j.addr.2021.06.001
Chen, J. , Horiuchi, S. , Kuramochi, S. , Kawasaki, T. , Kawasumi, H. , Akiyama, S., et al. (2024). Human intestinal organoid-derived PDGFRα + mesenchymal stroma enables proliferation and maintenance of LGR4 + epithelial stem cells. Stem Cell Res. Ther. 15:16. doi: 10.1186/s13287-023-03629-5
Chen, H. , Zhang, W. , Maskey, N. , Yang, F. , Zheng, Z. , Li, C., et al. (2022). Urological cancer organoids, patients' avatars for precision medicine: past, present and future. Cell Biosci. 12:132. doi: 10.1186/s13578-022-00866-8
Choi, Y. M. , Lee, H. , Ann, M. , Song, M. , Rheey, J. , and Jang, J. (2023). 3D bioprinted vascularized lung cancer organoid models with underlying disease capable of more precise drug evaluation. Biofabrication 15:034104. doi: 10.1088/1758-5090/acd95f
Codrich, M. , Dalla, E. , Mio, C. , Antoniali, G. , Malfatti, M. C. , Marzinotto, S., et al. (2021). Integrated multi-omics analyses on patient-derived CRC organoids highlight altered molecular pathways in colorectal cancer progression involving PTEN. J. Exp. Clin. Cancer Res. 40:198. doi: 10.1186/s13046-021-01986-8
Coxon, J. , Linder, E. , Sweet, C. , Magness, S. , and Green, L. (2025). Replicating host-microbiome interactions: harnessing organ-on-a-Chip and Organoid technologies to model vaginal and lung physiology. Annu. Rev. Biomed. Eng. 27, 403–423. doi: 10.1146/annurev-bioeng-110122-122343
de Castilhos, J. , Tillmanns, K. , Blessing, J. , Laraño, A. , Borisov, V. , and Stein-Thoeringer, C. K. (2024). Microbiome and pancreatic cancer: time to think about chemotherapy. Gut Microbes 16:2374596. doi: 10.1080/19490976.2024.2374596
Driehuis, E. , Kretzschmar, K. , and Clevers, H. (2020). Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 15, 3380–3409. doi: 10.1038/s41596-020-0379-4
Duan, Y. F. , Dai, J. H. , Lu, Y. Q. , Qiao, H. , and Liu, N. (2024). Disentangling the molecular mystery of tumour-microbiota interactions: microbial metabolites. Clin. Transl. Med. 14:e70093. doi: 10.1002/ctm2.70093
Eiman, L. , Moazzam, K. , Anjum, S. , Kausar, H. , Sharif, E. , and Ibrahim, W. N. (2025). Gut dysbiosis in cancer immunotherapy: microbiota-mediated resistance and emerging treatments. Front. Immunol. 16:1575452. doi: 10.3389/fimmu.2025.1575452
Fan, Y. , Li, Y. , Gu, X. , Chen, N. , Chen, Y. , Fang, C., et al. (2025). Intestinal metabolites in colitis-associated carcinogenesis: building a bridge between host and microbiome. Chin. Med. J. 138, 1961–1972. doi: 10.1097/CM9.0000000000003430
Fang, G. , Lu, H. , Al-Nakashli, R. , Chapman, R. , Zhang, Y. , Ju, L. A., et al. (2021). Enabling peristalsis of human colon tumor organoids on microfluidic chips. Biofabrication 14:a2cef9. doi: 10.1088/1758-5090/ac2ef9
Farin, H. F. , Mosa, M. H. , Ndreshkjana, B. , Grebbin, B. M. , Ritter, B. , Menche, C., et al. (2023). Colorectal Cancer organoid-stroma biobank allows subtype-specific assessment of individualized therapy responses. Cancer Discov. 13, 2192–2211. doi: 10.1158/2159-8290.CD-23-0050
Ferreira, J. , Gonçalves, M. , Preto, A. , and Sousa, M. J. (2024). Anticancer activity of benzo[a]phenoxazine compounds promoting lysosomal dysfunction. Cells 13:385. doi: 10.3390/cells13161385
Fu, C. , Yang, Z. , Yu, J. , and Wei, M. (2021). The interaction between gut microbiome and anti-tumor drug therapy. Am. J. Cancer Res. 11, 5812–5832.
Golpour, F. , Abbasi-Alaei, M. , Babaei, F. , Mirzababaei, M. , Parvardeh, S. , Mohammadi, G., et al. (2023). Short chain fatty acids, a possible treatment option for autoimmune diseases. Biomed. Pharmacother. 163:114763. doi: 10.1016/j.biopha.2023.114763
Grigoreva, T. A. , Kindt, D. N. , Sagaidak, A. V. , Novikova, D. S. , and Tribulovich, V. G. (2025). Cellular Systems for Colorectal Stem Cancer Cell Research. Cells 14:170. doi: 10.3390/cells14030170
Grönholm, M. , Feodoroff, M. , Antignani, G. , Martins, B. , Hamdan, F. , and Cerullo, V. (2021). Patient-derived organoids for precision Cancer immunotherapy. Cancer Res. 81, 3149–3155. doi: 10.1158/0008-5472.CAN-20-4026
Grzywa, T. M. , Paskal, W. , and Włodarski, P. K. (2017). Intratumor and Intertumor heterogeneity in melanoma. Transl. Oncol. 10, 956–975. doi: 10.1016/j.tranon.2017.09.007
Han, X. , Mslati, M. A. , Davies, E. , Chen, Y. , Allaire, J. M. , and Vallance, B. A. (2021). Creating a more perfect union: modeling intestinal Bacteria-epithelial interactions using organoids. Cell. Mol. Gastroenterol. Hepatol. 12, 769–782. doi: 10.1016/j.jcmgh.2021.04.010
Hanus, M. , Parada-Venegas, D. , Landskron, G. , Wielandt, A. M. , Hurtado, C. , Alvarez, K., et al. (2021). Immune system, microbiota, and microbial metabolites: the unresolved triad in colorectal Cancer microenvironment. Front. Immunol. 12:612826. doi: 10.3389/fimmu.2021.612826
He, L. , and Deng, C. (2022). Recent advances in organotypic tissue slice cultures for anticancer drug development. Int. J. Biol. Sci. 18, 5885–5896. doi: 10.7150/ijbs.78997
Hirota, A. , AlMusawi, S. , Nateri, A. S. , Ordóñez-Morán, P. , and Imajo, M. (2021). Biomaterials for intestinal organoid technology and personalized disease modeling. Acta Biomater. 132, 272–287. doi: 10.1016/j.actbio.2021.05.010
Hou, X. , Zheng, Z. , Wei, J. , and Zhao, L. (2022). Effects of gut microbiota on immune responses and immunotherapy in colorectal cancer. Front. Immunol. 13:1030745. doi: 10.3389/fimmu.2022.1030745
Huang, L. , Desai, R. , Conrad, D. N. , Leite, N. C. , Akshinthala, D. , Lim, C. M., et al. (2021). Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell 28, 1090–1104.e6. doi: 10.1016/j.stem.2021.03.022
Idris, M. , Alves, M. M. , Hofstra, R. , Mahe, M. M. , and Melotte, V. (2021). Intestinal multicellular organoids to study colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 1876:188586. doi: 10.1016/j.bbcan.2021.188586
Jaye, K. , Li, C. G. , Chang, D. , and Bhuyan, D. J. (2022). The role of key gut microbial metabolites in the development and treatment of cancer. Gut Microbes 14:2038865. doi: 10.1080/19490976.2022.2038865
Ji, L. , Shen, W. , Zhang, F. , Qian, J. , Jiang, J. , Weng, L., et al. (2021). Worenine reverses the Warburg effect and inhibits colon cancer cell growth by negatively regulating HIF-1α. Cell. Mol. Biol. Lett. 26:19. doi: 10.1186/s11658-021-00263-y
Jin, J. , Chen, W. , Li, J. , Yang, J. , Dai, R. , Tang, J., et al. (2025). Engineered tumor microspheres via microfluidics and decellularized extracellular matrix for high-throughput organoid-based drug screening. Biofabrication 17:045003. doi: 10.1088/1758-5090/adf099
Jung, M. , Ghamrawi, S. , Du, E. Y. , Gooding, J. J. , and Kavallaris, M. (2022). Advances in 3D bioprinting for Cancer biology and precision medicine: from matrix design to application. Adv. Healthc. Mater. 11:e2200690. doi: 10.1002/adhm.202200690
Kaden, T. , Alonso-Román, R. , Stallhofer, J. , Gresnigt, M. S. , Hube, B. , and Mosig, A. S. (2025). Leveraging organ-on-Chip models to investigate host-microbiota dynamics and targeted therapies for inflammatory bowel disease. Adv. Healthc. Mater. 14:e2402756. doi: 10.1002/adhm.202402756
Kan, L. , Yu, Y. , Wang, Y. , Shi, L. , Fan, T. , Chen, H., et al. (2025). The application of organoids in investigating immune evasion in the microenvironment of gastric cancer and screening novel drug candidates. Mol. Cancer 24:125. doi: 10.1186/s12943-025-02328-4
Kim, M. B. , Hwangbo, S. , Jang, S. , and Jo, Y. K. (2022). Bioengineered co-culture of organoids to recapitulate host-microbe interactions. Mater Today Bio. 16:100345. doi: 10.1016/j.mtbio.2022.100345
Kovtonyuk, L. V. , and McCoy, K. D. (2023). Microbial metabolites and immunotherapy: basic rationale and clinical indications. Semin. Immunol. 67:101755. doi: 10.1016/j.smim.2023.101755
Kriaa, A. , Mariaule, V. , De Rudder, C. , Jablaoui, A. , Sokol, H. , Wilmes, P., et al. (2024). From animal models to gut-on-chip: the challenging journey to capture inter-individual variability in chronic digestive disorders. Gut Microbes 16:2333434. doi: 10.1080/19490976.2024.2333434
Kumarasamy, V. , Wang, J. , Frangou, C. , Wan, Y. , Dynka, A. , Rosenheck, H., et al. (2024). The extracellular niche and tumor microenvironment enhance KRAS inhibitor efficacy in pancreatic Cancer. Cancer Res. 84, 1115–1132. doi: 10.1158/0008-5472.CAN-23-2504
Lee, J. , Kim, Y. , Jung, H. I. , Lim, J. , and Kwak, B. S. (2025). Channel-assembling tumor microenvironment on-chip for evaluating anticancer drug efficacy. J. Control. Release 377, 376–384. doi: 10.1016/j.jconrel.2024.11.030
LeSavage, B. L. , Suhar, R. A. , Broguiere, N. , Lutolf, M. P. , and Heilshorn, S. C. (2022). Next-generation cancer organoids. Nat. Mater. 21, 143–159. doi: 10.1038/s41563-021-01057-5
Li, K. , He, Y. , Jin, X. , Jin, K. , and Qian, J. (2025). Reproducible extracellular matrices for tumor organoid culture: challenges and opportunities. J. Transl. Med. 23:497. doi: 10.1186/s12967-025-06349-x
Li, D. , Li, Y. , Yang, S. , Lu, J. , Jin, X. , and Wu, M. (2022). Diet-gut microbiota-epigenetics in metabolic diseases: from mechanisms to therapeutics. Biomed. Pharmacother. 153:113290. doi: 10.1016/j.biopha.2022.113290
Li, Q. , Xiao, Y. , Han, L. , Luo, W. , Dai, W. , Fang, H., et al. (2025). Microbiome dysbiosis, neutrophil recruitment and mesenchymal transition of mesothelial cells promotes peritoneal metastasis of colorectal cancer. Nat Cancer. 6, 493–510. doi: 10.1038/s43018-025-00910-9
Li, Y. , Xu, C. , Zhou, X. , Li, J. , Xu, S. , Tu, Y., et al. (2024). DNA adductomics aided rapid screening of genotoxic impurities using nucleosides and 3D bioprinted human liver organoids. Talanta 273:125902. doi: 10.1016/j.talanta.2024.125902
Liu, A. , Duan, G. , Yang, L. , Hu, Y. , Zhou, H. , and Wang, H. (2025). Low-temperature stress-induced hepatic injury in Darkbarbel catfish (Pelteobagrus vachelli): mediated by gut-liver Axis dysregulation. Antioxidants 14:762. doi: 10.3390/antiox14070762
Liu, B. , Liu, Y. , Xu, S. , Wu, Q. , Wu, D. , Zhan, L., et al. (2025). EasyMultiProfiler: an efficient multi-omics data integration and analysis workflow for microbiome research. Sci. China Life Sci. 8:35. doi: 10.1007/s11427-025-3035-0
Liu, J. , Tian, R. , Sun, C. , Guo, Y. , Dong, L. , Li, Y., et al. (2023). Microbial metabolites are involved in tumorigenesis and development by regulating immune responses. Front. Immunol. 14:1290414. doi: 10.3389/fimmu.2023.1290414
Liu, R. , Wang, J. , Liu, Y. , Gao, Y. , and Yang, R. (2024). Regulation of gut microbiota on immune cell ferroptosis: a novel insight for immunotherapy against tumor. Cancer Lett. 598:217115. doi: 10.1016/j.canlet.2024.217115
Liu, L. , Yu, L. , Li, Z. , Li, W. , and Huang, W. (2021). Patient-derived organoid (PDO) platforms to facilitate clinical decision making. J. Transl. Med. 19:40. doi: 10.1186/s12967-020-02677-2
Lu, J. , Yao, T. , Fu, S. , and Ye, L. (2024). Metabolomic and microbiomic resilience of Hong Kong oysters to dual stressors: zinc oxide nanoparticles and low salinity. Chemosphere 368:143722. doi: 10.1016/j.chemosphere.2024.143722
Lu, Y. , Yuan, X. , Wang, M. , He, Z. , Li, H. , Wang, J., et al. (2022). Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J. Hematol. Oncol. 15:47. doi: 10.1186/s13045-022-01273-9
Luk, I. Y. , Mooi, J. K. , Mouradov, D. , Tan, T. , Scott, C. M. , Chionh, F., et al. (2025). Model systems and unique biological features of high and low-grade colorectal cancer (CRC) revealed by xenografting 84 human CRC cell lines. Commun. Biol. 8:875. doi: 10.1038/s42003-025-08251-0
Magré, L. , Verstegen, M. , Buschow, S. , van der Laan, L. , Peppelenbosch, M. , and Desai, J. (2023). Emerging organoid-immune co-culture models for cancer research: from oncoimmunology to personalized immunotherapies. J. Immunother. Cancer 11:e006290. doi: 10.1136/jitc-2022-006290
Maity, S. , Bhuyan, T. , Jewell, C. , Kawakita, S. , Sharma, S. , Nguyen, H. T., et al. (2025). Recent developments in glioblastoma-on-A-Chip for advanced drug screening applications. Small 21:e2405511. doi: 10.1002/smll.202405511
Mao, Y. , Wang, W. , Yang, J. , Zhou, X. , Lu, Y. , Gao, J., et al. (2024). Drug repurposing screening and mechanism analysis based on human colorectal cancer organoids. Protein Cell 15, 285–304. doi: 10.1093/procel/pwad038
Margiotta, F. , Lucarini, E. , Toti, A. , Curti, L. , Masi, A. , Mello, T., et al. (2025). Gut microbiota dysbiosis affects intestinal sensitivity through epithelium-to-neuron signaling: novel insights from a colon organoid-based model to improve visceral pain therapy. Gut Microbes 17:2547029. doi: 10.1080/19490976.2025.2547029
Matkovic Leko, I. , Schneider, R. T. , Thimraj, T. A. , Schrode, N. , Beitler, D. , Liu, H. Y., et al. (2023). A distal lung organoid model to study interstitial lung disease, viral infection and human lung development. Nat. Protoc. 18, 2283–2312. doi: 10.1038/s41596-023-00827-6
Metwaly, A. , Reitmeier, S. , and Haller, D. (2022). Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 19, 383–397. doi: 10.1038/s41575-022-00581-2
Meza-Perez, S. , Liu, M. , Silva-Sanchez, A. , Morrow, C. D. , Eipers, P. G. , Lefkowitz, E. J., et al. (2024). Proteobacteria impair anti-tumor immunity in the omentum by consuming arginine. Cell Host Microbe 32, 1177–1191.e7. doi: 10.1016/j.chom.2024.06.003
Milani, N. , Parrott, N. , Ortiz Franyuti, D. , Godoy, P. , Galetin, A. , Gertz, M., et al. (2022). Application of a gut-liver-on-a-chip device and mechanistic modelling to the quantitative in vitro pharmacokinetic study of mycophenolate mofetil. Lab Chip 22, 2853–2868. doi: 10.1039/D2LC00276K
Moll, F. , Spaeth, M. , and Schröder, K. (2022). Cre-recombinase induces apoptosis and cell death in enterocyte organoids. Antioxidants 11:1452. doi: 10.3390/antiox11081452
Nakano, K. , Oki, E. , Yamazaki, M. , Suzuki, M. , Kawai, S. , Fujita, T., et al. (2025). Colorectal cancer cell line-derived organoid model with stem cell properties captures the regrowing state of residual cancer cells after neoadjuvant chemotherapy. Cell Death Discov. 11:282. doi: 10.1038/s41420-025-02567-w
Nazir, A. , Hussain, F. , Nadeem Hussain, T. H. , Al Dweik, R. , and Raza, A. (2025). Therapeutic targeting of the host-microbiota-immune axis: implications for precision health. Front. Immunol. 16:1570233. doi: 10.3389/fimmu.2025.1570233
Nguyen, O. , Misun, P. M. , Hierlemann, A. , and Lohasz, C. (2024). A versatile intestine-on-Chip system for deciphering the Immunopathogenesis of inflammatory bowel disease. Adv. Healthc. Mater. 13:e2302454. doi: 10.1002/adhm.202302454
Nie, J. , Liao, W. , Zhang, Z. , Zhang, M. , Wen, Y. , Capanoglu, E., et al. (2023). A 3D co-culture intestinal organoid system for exploring glucose metabolism. Curr Res Food Sci. 6:100402. doi: 10.1016/j.crfs.2022.11.021
Özkan, A. , LoGrande, N. T. , Feitor, J. F. , Goyal, G. , and Ingber, D. E. (2024). Intestinal organ chips for disease modelling and personalized medicine. Nat. Rev. Gastroenterol. Hepatol. 21, 751–773. doi: 10.1038/s41575-024-00968-3
Penarete-Acosta, D. , Stading, R. , Emerson, L. , Horn, M. , Chakraborty, S. , Han, A., et al. (2024). A microfluidic co-culture model for investigating colonocytes-microbiota interactions in colorectal cancer. Lab Chip 24, 3690–3703. doi: 10.1039/D4LC00013G
Pérez-González, C. , Ceada, G. , Greco, F. , Matejčić, M. , Gómez-González, M. , Castro, N., et al. (2021). Mechanical compartmentalization of the intestinal organoid enables crypt folding and collective cell migration. Nat. Cell Biol. 23, 745–757. doi: 10.1038/s41556-021-00699-6
Pich, O. , Bernard, E. , Zagorulya, M. , Rowan, A. , Pospori, C. , Slama, R., et al. (2025). Tumor-infiltrating clonal hematopoiesis. N. Engl. J. Med. 392, 1594–1608. doi: 10.1056/NEJMoa2413361
Poletti, M. , Arnauts, K. , Ferrante, M. , and Korcsmaros, T. (2021). Organoid-based models to study the role of host-microbiota interactions in IBD. J. Crohns Colitis 15, 1222–1235. doi: 10.1093/ecco-jcc/jjaa257
Przybylla, R. , Krohn, M. , Sellin, M. L. , Frank, M. , Oswald, S. , and Linnebacher, M. (2023). Novel in vitro models for cell differentiation and drug transport studies of the human intestine. Cells 12:371. doi: 10.3390/cells12192371
Qiao, H. , Li, H. , Wen, X. , Tan, X. , Yang, C. , and Liu, N. (2022). Multi-omics integration reveals the crucial role of Fusobacterium in the inflammatory immune microenvironment in head and neck squamous cell carcinoma. Microbiol. Spectr. 10:e0106822. doi: 10.1128/spectrum.01068-22
Qu, M. , Xiong, L. , Lyu, Y. , Zhang, X. , Shen, J. , Guan, J., et al. (2021). Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res. 31, 259–271. doi: 10.1038/s41422-020-00453-x
Rivas-Domínguez, A. , Pastor, N. , Martínez-López, L. , Colón-Pérez, J. , Bermúdez, B. , and Orta, M. L. (2021). The role of DNA damage response in Dysbiosis-induced colorectal Cancer. Cells 10:934. doi: 10.3390/cells10081934
Rosendahl Huber, A. , Pleguezuelos-Manzano, C. , and Puschhof, J. (2021). A bacterial mutational footprint in colorectal cancer genomes. Br. J. Cancer 124, 1751–1753. doi: 10.1038/s41416-021-01273-5
Saha, B. , Mathur, T. , Tronolone, J. J. , Chokshi, M. , Lokhande, G. K. , Selahi, A., et al. (2021). Human tumor microenvironment chip evaluates the consequences of platelet extravasation and combinatorial antitumor-antiplatelet therapy in ovarian cancer. Sci. Adv. 7:283. doi: 10.1126/sciadv.abg5283
Shek, D. , Chen, D. , Read, S. A. , and Ahlenstiel, G. (2021). Examining the gut-liver axis in liver cancer using organoid models. Cancer Lett. 510, 48–58. doi: 10.1016/j.canlet.2021.04.008
Sheng, K. , Xu, Y. , Kong, X. , Wang, J. , Zha, X. , and Wang, Y. (2021). Probiotic Bacillus cereus alleviates dextran sulfate sodium-induced colitis in mice through improvement of the intestinal barrier function, anti-inflammation, and gut microbiota modulation. J. Agric. Food Chem. 69, 14810–14823. doi: 10.1021/acs.jafc.1c03375
Shi, Q. , Huang, Z. , Kuang, Y. , Wang, C. , Fang, X. , and Hu, X. (2024). Forkhead box E1, frequently downregulted by promoter methylation, inhibits colorectal cancer cell growth and migration. Cancer Cell Int. 24:169. doi: 10.1186/s12935-024-03352-y
Si, Q. , Tao, S. , Wu, J. , Ma, J. , Li, Z. , Feng, X., et al. (2025). Tumor organoids in immunotherapy: from disease modeling to translational research. J. Immunother. Cancer 13:e011733. doi: 10.1136/jitc-2025-011733
Smet, A. , Kupcinskas, J. , Link, A. , Hold, G. L. , and Bornschein, J. (2022). The role of microbiota in gastrointestinal Cancer and Cancer treatment: chance or curse. Cell. Mol. Gastroenterol. Hepatol. 13, 857–874. doi: 10.1016/j.jcmgh.2021.08.013
Smirnov, A. , Melino, G. , and Candi, E. (2023). Gene expression in organoids: an expanding horizon. Biol. Direct 18:11. doi: 10.1186/s13062-023-00360-2
Somarelli, J. A. , Roghani, R. S. , Moghaddam, A. S. , Thomas, B. C. , Rupprecht, G. , Ware, K. E., et al. (2020). A precision medicine drug discovery pipeline identifies combined CDK2 and 9 inhibition as a novel therapeutic strategy in colorectal Cancer. Mol. Cancer Ther. 19, 2516–2527. doi: 10.1158/1535-7163.MCT-20-0454
Song, T. , Kong, B. , Liu, R. , Luo, Y. , Wang, Y. , and Zhao, Y. (2024). Bioengineering approaches for the pancreatic tumor organoids research and application. Adv. Healthc. Mater. 13:e2300984. doi: 10.1002/adhm.202300984
Song, P. , Peng, Z. , and Guo, X. (2025). Gut microbial metabolites in cancer therapy. Trends Endocrinol. Metab. 36, 55–69. doi: 10.1016/j.tem.2024.06.016
Steindl, A. , and Valiente, M. (2025). Potential of ex vivo organotypic slice cultures in neuro-oncology. Neuro-Oncology 27, 338–351. doi: 10.1093/neuonc/noae195
Su, F. , Su, M. , Wei, W. , Wu, J. , Chen, L. , Sun, X., et al. (2025). Integrating multi-omics data to reveal the host-microbiota interactome in inflammatory bowel disease. Gut Microbes 17:2476570. doi: 10.1080/19490976.2025.2476570
Sugito, N. , Heishima, K. , and Akao, Y. (2022). Chemically modified MIR143-3p exhibited anti-cancer effects by impairing the KRAS network in colorectal cancer cells. Mol. Ther. Nucleic Acids 30, 49–61. doi: 10.1016/j.omtn.2022.09.001
Tang, Y. , Cai, Q. , Tian, Z. , Chen, W. , and Tang, H. (2025). Crosstalk between gut microbiota and Cancer immunotherapy: present investigations and future perspective. Research 8:0600. doi: 10.34133/research.0600
Tardito, S. , Matis, S. , Zocchi, M. R. , Benelli, R. , and Poggi, A. (2024). Epidermal growth factor receptor targeting in colorectal carcinoma: antibodies and patient-derived organoids as a smart model to study therapy resistance. Int. J. Mol. Sci. 25:131. doi: 10.3390/ijms25137131
Teng, H. W. , Wang, T. Y. , Lin, C. C. , Tong, Z. J. , Cheng, H. W. , and Wang, H. T. (2024). Interferon gamma induces higher neutrophil extracellular traps leading to tumor-killing activity in microsatellite stable colorectal Cancer. Mol. Cancer Ther. 23, 1043–1056. doi: 10.1158/1535-7163.MCT-23-0744
Thomas, R. M. (2025). Microbial molecules, metabolites, and malignancy. Neoplasia 60:101128. doi: 10.1016/j.neo.2025.101128
Tran, K. B. , Kolekar, S. , Wang, Q. , Shih, J. H. , Buchanan, C. M. , Deva, S., et al. (2022). Response to BRAF-targeted therapy is enhanced by Cotargeting VEGFRs or WNT/β-catenin signaling in BRAF-mutant colorectal Cancer models. Mol. Cancer Ther. 21, 1777–1787. doi: 10.1158/1535-7163.MCT-21-0941
Vaaben, T. H. , Lützhøft, D. O. , Koulouktsis, A. , Dawoodi, I. M. , Stavnsbjerg, C. , Kvich, L., et al. (2025). Modulating tumor immunity using advanced microbiome therapeutics producing an indole metabolite. EMBO Rep. 26, 1688–1708. doi: 10.1038/s44319-025-00386-9
Valiei, A. , Aminian-Dehkordi, J. , and Mofrad, M. (2023). Gut-on-a-chip models for dissecting the gut microbiology and physiology. APL Bioeng. 7:011502. doi: 10.1063/5.0126541
van de Haar, J. , Ma, X. , Ooft, S. N. , van der Helm, P. W. , Hoes, L. R. , Mainardi, S., et al. (2023). Codon-specific KRAS mutations predict survival benefit of trifluridine/tipiracil in metastatic colorectal cancer. Nat. Med. 29, 605–614. doi: 10.1038/s41591-023-02240-8
Verhulsel, M. , Simon, A. , Bernheim-Dennery, M. , Gannavarapu, V. R. , Gérémie, L. , Ferraro, D., et al. (2021). Developing an advanced gut on chip model enabling the study of epithelial cell/fibroblast interactions. Lab Chip 21, 365–377. doi: 10.1039/D0LC00672F
Wahida, A. , Müller, M. , Hiergeist, A. , Popper, B. , Steiger, K. , Branca, C., et al. (2021). XIAP restrains TNF-driven intestinal inflammation and dysbiosis by promoting innate immune responses of Paneth and dendritic cells. Sci. Immunol. 6:eabf7235. doi: 10.1126/sciimmunol.abf7235
Wang, S. , Fu, W. , Zhao, X. , Chang, X. , Liu, H. , Zhou, L., et al. (2022). Zearalenone disturbs the reproductive-immune axis in pigs: the role of gut microbial metabolites. Microbiome 10:234. doi: 10.1186/s40168-022-01397-7
Wang, Y. , Huang, J. , Tong, H. , Jiang, Y. , Jiang, Y. , and Ma, X. (2025). Nutrient acquisition of gut microbiota: implications for tumor immunity. Semin. Cancer Biol. 114, 88–103. doi: 10.1016/j.semcancer.2025.06.003
Wang, Q. , Liu, Z. , Ma, A. , Li, Z. , Liu, B. , and Ma, Q. (2023). Computational methods and challenges in analyzing intratumoral microbiome data. Trends Microbiol. 31, 707–722. doi: 10.1016/j.tim.2023.01.011
Wang, W. , Liu, Y. , Yao, Z. , Chen, D. , Tang, Y. , Cui, J., et al. (2024). A microfluidic-based gut-on-a-chip model containing the gut microbiota of patients with depression reveals physiological characteristics similar to depression. Lab Chip 24, 2537–2550. doi: 10.1039/D3LC01052J
Wang, R. , Mao, Y. , Wang, W. , Zhou, X. , Wang, W. , Gao, S., et al. (2022). Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis. Genome Biol. 23:106. doi: 10.1186/s13059-022-02673-3
Wang, Q. , Yuan, F. , Zuo, X. , and Li, M. (2025). Breakthroughs and challenges of organoid models for assessing cancer immunotherapy: a cutting-edge tool for advancing personalised treatments. Cell Death Discov. 11:222. doi: 10.1038/s41420-025-02505-w
Weng, G. , Tao, J. , Liu, Y. , Qiu, J. , Su, D. , Wang, R., et al. (2023). Organoid: bridging the gap between basic research and clinical practice. Cancer Lett. 572:216353. doi: 10.1016/j.canlet.2023.216353
Williamson, I. A. , Arnold, J. W. , Samsa, L. A. , Gaynor, L. , DiSalvo, M. , Cocchiaro, J. L., et al. (2018). A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell. Mol. Gastroenterol. Hepatol. 6, 301–319. doi: 10.1016/j.jcmgh.2018.05.004
Wu, Q. , Ge, Z. , Lv, C. , and He, Q. (2025). Interacting roles of gut microbiota and T cells in the development of autoimmune hepatitis. Front. Immunol. 16:1584001. doi: 10.3389/fimmu.2025.1584001
Wu, J. , Zhang, P. , Mei, W. , and Zeng, C. (2023). Intratumoral microbiota: implications for cancer onset, progression, and therapy. Front. Immunol. 14:1301506. doi: 10.3389/fimmu.2023.1301506
Wuputra, K. , Ku, C. C. , Kato, K. , Wu, D. C. , Saito, S. , and Yokoyama, K. K. (2021). Translational models of 3-D organoids and cancer stem cells in gastric cancer research. Stem Cell Res Ther 12:492. doi: 10.1186/s13287-021-02521-4
Xia, K. , Gao, R. , Li, L. , Wu, X. , Wu, T. , Ruan, Y., et al. (2024). Transformation of colitis and colorectal cancer: a tale of gut microbiota. Crit. Rev. Microbiol. 50, 653–662. doi: 10.1080/1040841X.2023.2254388
Xian, C. , Zhang, J. , Zhao, S. , and Li, X. G. (2023). Gut-on-a-chip for disease models. J Tissue Eng. 14:20417314221149882. doi: 10.1177/20417314221149882
Xiang, T. , Wang, J. , and Li, H. (2024). Current applications of intestinal organoids: a review. Stem Cell Res Ther 15:155. doi: 10.1186/s13287-024-03768-3
Xiang, Y. , Wen, H. , Yu, Y. , Li, M. , Fu, X. , and Huang, S. (2020). Gut-on-chip: recreating human intestine in vitro. J Tissue Eng. 11:2041731420965318. doi: 10.1177/2041731420965318
Xie, Y. , and Liu, F. (2024). The role of the gut microbiota in tumor, immunity, and immunotherapy. Front. Immunol. 15:1410928. doi: 10.3389/fimmu.2024.1410928
Xu, H. , Jiao, D. , Liu, A. , and Wu, K. (2022). Tumor organoids: applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 15:58. doi: 10.1186/s13045-022-01278-4
Xu, J. , Moore, B. N. , and Pluznick, J. L. (2022). Short-chain fatty acid receptors and blood pressure regulation: council on hypertension mid-career award for research excellence 2021. Hypertension 79, 2127–2137. doi: 10.1161/HYPERTENSIONAHA.122.18558
Xuan, L. , Luo, J. , Qu, C. , Guo, P. , Yi, W. , Yang, J., et al. (2024). Predictive metabolomic signatures for safety assessment of three plastic nanoparticles using intestinal organoids. Sci. Total Environ. 913:169606. doi: 10.1016/j.scitotenv.2023.169606
Yan, H. , Chan, A. S. , Lai, F. P. , and Leung, S. Y. (2023). Organoid cultures for cancer modeling. Cell Stem Cell 30, 917–937. doi: 10.1016/j.stem.2023.05.012
Yan, X. , Tan, D. , Yu, L. , Li, D. Y. , Huang, W. , Huang, W., et al. (2024). A high-throughput and logarithm-serial-dilution microfluidic Chip for combinational drug screening on tumor organoids. ACS Pharmacol. Transl. Sci. 7, 4135–4143. doi: 10.1021/acsptsci.4c00565
Yáñez-Bartolomé, M. , Macarulla, T. , and Tian, T. V. (2023). The potential of patient-derived organoids in precision medicine of biliary tract cancer. Cell Rep. Med. 4:101294. doi: 10.1016/j.xcrm.2023.101294
Yang, Q. , Wang, B. , Zheng, Q. , Li, H. , Meng, X. , Zhou, F., et al. (2023). A review of gut microbiota-derived metabolites in tumor progression and Cancer therapy. Adv Sci (Weinh). 10:e2207366. doi: 10.1002/advs.202207366
Yavitt, F. M. , Kirkpatrick, B. E. , Blatchley, M. R. , and Anseth, K. S. (2022). 4D materials with Photoadaptable properties instruct and enhance intestinal organoid development. ACS Biomater Sci. Eng. 8, 4634–4638. doi: 10.1021/acsbiomaterials.1c01450
Yavitt, F. M. , Kirkpatrick, B. E. , Blatchley, M. R. , Speckl, K. F. , Mohagheghian, E. , Moldovan, R., et al. (2023). In situ modulation of intestinal organoid epithelial curvature through photoinduced viscoelasticity directs crypt morphogenesis. Sci. Adv. 9:eadd5668. doi: 10.1126/sciadv.add5668
Ying, J. , Zhou, H. , Wang, Z. , You, Q. , Chen, J. , Lu, H., et al. (2023). Aspirin increases chemosensitivity of colorectal cancer cells and inhibits the expression of toll-like receptor 4. Cancer Cell Int. 23:6. doi: 10.1186/s12935-023-02847-4
Yoon, S. J. , Han, S. K. , Kim, T. S. , Suk, K. T. , Choi, D. H. , Kim, Y. D., et al. (2025). The crosstalk between gut microbiota and microbiota-derived metabolites in hepatocellular carcinoma. Crit. Rev. Microbiol. 1:15. doi: 10.1080/1040841X.2025.2501590
Zhang, S. L. , Cheng, L. S. , Zhang, Z. Y. , Sun, H. T. , and Li, J. J. (2023). Untangling determinants of gut microbiota and tumor immunologic status through a multi-omics approach in colorectal cancer. Pharmacol. Res. 188:106633. doi: 10.1016/j.phrs.2022.106633
Zhang, X. Y. , Khakisahneh, S. , Han, S. Y. , Song, E. J. , Nam, Y. D. , and Kim, H. (2024). Ginseng extracts improve circadian clock gene expression and reduce inflammation directly and indirectly through gut microbiota and PI3K signaling pathway. NPJ Biofilms Microbiomes 10:24. doi: 10.1038/s41522-024-00498-5
Zhang, S. , Wen, H. , Chen, Y. , Ning, J. , Hu, D. , Dong, Y., et al. (2025). Crosstalk between gut microbiota and tumor: tumors could cause gut dysbiosis and metabolic imbalance. Mol. Oncol. 19, 1707–1724. doi: 10.1002/1878-0261.13763
Zhang, S. , Xu, S. , Li, D. , Wu, S. , Han, M. , Han, Y., et al. (2024). The small protein LINC01547-ORF inhibits colorectal cancer progression by regulating the CLDN18-FAK-AKT axis. Am. J. Cancer Res. 14, 5504–5520. doi: 10.62347/PNKH7683
Zhao, H. , Jin, K. , Jiang, C. , Pan, F. , Wu, J. , Luan, H., et al. (2022). A pilot exploration of multi-omics research of gut microbiome in major depressive disorders. Transl. Psychiatry 12:8. doi: 10.1038/s41398-021-01769-x
Zhao, Y. , Zhang, B. , Ma, Y. , Zhao, F. , Chen, J. , Wang, B., et al. (2022). Colorectal Cancer patient-derived 2D and 3D models efficiently recapitulate inter- and Intratumoral heterogeneity. Adv. Sci. 9:e2201539. doi: 10.1002/advs.202201539
Zhou, Y. , Dai, Q. , Xu, Y. , Wu, S. , Cheng, M. , and Zhao, B. (2025). PharmaFormer predicts clinical drug responses through transfer learning guided by patient derived organoid. NPJ Precis Oncol. 9:282. doi: 10.1038/s41698-025-01082-6
Zhou, T. , Xie, Y. , Hou, X. , Bai, W. , Li, X. , Liu, Z., et al. (2023). Irbesartan overcomes gemcitabine resistance in pancreatic cancer by suppressing stemness and iron metabolism via inhibition of the hippo/YAP1/c-Jun axis. J. Exp. Clin. Cancer Res. 42:111. doi: 10.1186/s13046-023-02671-8
Zhu, J. , Ji, L. , Chen, Y. , Li, H. , Huang, M. , Dai, Z., et al. (2023). Organoids and organs-on-chips: insights into predicting the efficacy of systemic treatment in colorectal cancer. Cell Death Discov. 9:72. doi: 10.1038/s41420-023-01354-9
Keywords: organoids, gut microbiota, host–microbe interactions, tumor microenvironment, microbial metabolites
Citation: Zheng S-y, Su Y-y, Cai F-l, Xu D-f and Xu Y-q (2025) From mechanisms to precision medicine: the role of organoids in studying the gut microbiota-tumor microenvironment axis. Front. Microbiol. 16:1669482. doi: 10.3389/fmicb.2025.1669482
Edited by:
Abdul Bari Shah, Al-Farabi Kazakh National University, KazakhstanReviewed by:
Yuan Tang, University of Toledo, United StatesKim Fung, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia
Copyright © 2025 Zheng, Su, Cai, Xu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-qiang Xu, MTU4ODgzMDQyMzBAMTYzLmNvbQ==
 Si-yang Zheng
Si-yang Zheng Yong-qiang Xu
Yong-qiang Xu