- 1Nanling Research Institute for Modern Seed Industry, Xiangnan University, Chenzhou, China
- 2School of Chemistry and Environmental Science, Xiangnan University, Chenzhou, China
- 3R&D Centre of Rucheng Small Yellow Ginger Breeding Technology, Xiangnan University, Chenzhou, China
- 4School of Basic Medical Sciences, Xiangnan University, Chenzhou, China
- 5Institute of Botany, Chinese Academy of Sciences, Beijing, China
Introduction: Continuous cropping obstacles, particularly the prevalent ginger wilt disease (bacterial wilt), severely constrain the sustainable development of the small yellow ginger (Zingiber officinale Roscoe) industry in Rucheng County, China. However, the primary pathogen responsible for this disease in the local cultivar and the associated microbiome shifts within the rhizosphere remain unidentified. This study aimed to elucidate the distinctive rhizosphere microbial community changes induced by ginger wilt disease, identify the potential key pathogen responsible for this disease in Rucheng, and provide a scientific basis for overcoming continuous cropping obstacles in small yellow ginger cultivation.
Methods: Soil samples were collected from an uncultivated plot and from the rhizosphere of healthy and wilted small yellow ginger plants. Microbial community structure and composition were analyzed using 16S rRNA gene high-throughput sequencing. Linear Discriminant Analysis (LDA) effect size (LEfSe) was employed to identify differential biomarkers, and functional prediction was performed using BugBase.
Results: Ginger wilt disease significantly altered the rhizosphere soil bacterial community structure and composition: the relative abundance of Proteobacteria increased significantly, primarily due to the enrichment of the genus Ralstonia; conversely, the relative abundance of Acidobacteriota, Firmicutes, and Chloroflexi significantly decreased. Ginger wilt disease also significantly reduced the diversity of the rhizosphere soil bacterial community. LEfSe further confirmed Ralstonia as a diagnostic biomarker for ginger wilt disease. BugBase phenotypic prediction indicated that the microbiota enriched in the diseased ginger rhizosphere exhibited higher capabilities for oxidative stress resistance, pathogenic potential, and mobile element content, attributed to a functional consortium of multiple genera, such as Ralstonia as the dominant contributor alongside major contributors such as Rhodanobacter and Dokdonella.
Discussion: Our findings highlight that the enrichment of Ralstonia is strongly associated with ginger wilt in Rucheng County and concomitant with profound changes in the rhizospheric microbiota of wilted ginger, involving alterations in both community structure and functional potential.
1 Introduction
Ginger (Zingiber officinale Roscoe) is one of the most important economic crops in the world, which is widely used as food, medicine, and cosmetic additives (Benzie and Wachtel-Galor, 2011; Zhang et al., 2021). In China, ginger cultivation spans multiple regions, with Shandong, Yunnan, and Hunan provinces being particularly known for their diverse and distinctive local varieties. Nevertheless, ginger production is severely affected by a wilt disease, which is usually caused by the pathogen Ralstonia solanacearum (Paret et al., 2008; Prameela and Suseela Bhai, 2020; Dang et al., 2023). This devastating pathogen directly reduces yields by 40–50% in endemic areas, with complete crop failure occurring in severe epidemics (Guji et al., 2019; Aysanew and Alemayehu, 2022). Additional pathogens implicated in wilt disease include Achromobacter xylosoxidans (Hua et al., 2024), Enterobacter cloacae (Nishijima et al., 2004; Wang et al., 2008; Liu et al., 2021; Zhao et al., 2022), and Ceratocystis fimbriata (Zhang et al., 2020). The persistence of these pathogens in soil often forces farmers to abandon infected fields in search of new lands for cultivation, creating a critical situation. In Rucheng County, a leading production area for high-quality small yellow ginger in southern Hunan Province, wilt disease poses a serious threat to the local ginger industry, yet the primary pathogen remains unidentified.
Soil microorganisms are the linchpin of soil fertility, affecting not only soil health but also crop disease resistance and agricultural product quality. Rhizosphere microbes play a pivotal role in nutrient cycling and disease suppression, highlighting the significance of microbial diversity and abundance as critical factors for successful cultivation (Bai et al., 2022; Liu et al., 2023; Saud et al., 2023). Continuous monoculture can disrupt microbial community balance, often reducing microbial diversity and promoting disease outbreaks (Li et al., 2021; Haq et al., 2023). While crop rotation is a traditional strategy to mitigate the continuous cropping obstacles (Haq et al., 2023; Liu et al., 2024), the ginger–rice rotation practiced in Rucheng has proven ineffective against wilt disease.
Despite its agricultural significance, ginger’s continuous cropping obstacles and associated shifts in soil microbiota remain poorly understood. A preliminary study in Chongqing, China, reported microbial differences between soils of healthy and disease-affected ginger plants (Liu et al., 2017). Similarly, a study in Taiwan observed microbial community shifts after pathogen infection; however, that study focused primarily on the influence of biocontrol agents and fungicides on the microbiota rather than a direct comparison between healthy and diseased soils (Wang et al., 2022a). Furthermore, an empirical study demonstrated that Ralstonia solanacearum infection can enhance ginger resistance by inducing antibacterial root exudates and enriching beneficial bacteria (Dang et al., 2023). Nevertheless, a detailed comparative analysis of the microbiota in wilt-affected and non-wilt-affected soils is still lacking, and the influence of ginger cultivation and disease progression on the soil microbial composition is not well defined. Consequently, a thorough assessment of soil microbial communities during ginger cultivation and disease incidence is imperative.
This study employs high-throughput 16S rRNA gene sequencing to analyze the diversity of soil bacterial communities in Rucheng County. The goal is to elucidate the influence of ginger cultivation and wilt disease on soil microbial communities, identify the potential pathogen responsible for ginger wilt, and offer insights into the mechanisms that could alleviate the continuous cropping constraints faced by ginger farmers.
2 Materials and methods
2.1 Site description
The study was conducted at a site in Rucheng County, Chenzhou City, Hunan Province (25°30′56″N, 113°36′38″E). The study area falls within a subtropical monsoon humid climate zone, characterized by an average temperature of 16.6 °C, an average annual sunshine duration of 1,731 h, and an annual precipitation of 1545.7 mm. The ginger variety under investigation is the renowned “Rucheng small yellow ginger,” a local cultivar celebrated for its distinctive qualities and adaptability to the region’s unique environmental conditions.
2.2 Soil sample collection
To minimize confounding effects from soil heterogeneity, all ginger rhizosphere soil samples were collected from a single, continuous “small yellow ginger” cultivation field. Soil samples were divided into three groups: soils from three different locations within a continuous uncultivated area adjacent to the field (50–100 cm from the field edge), serving as the Control group (Group C); rhizosphere soil samples from healthy ginger plants, representing the Non-wilt-affected group (Group N); and rhizosphere soil samples from wilted ginger plants, defined as the Sick group (Group S). Each group consisted of three biological replicates. The three replicates for Groups N and S were collected from three independent healthy or wilted plants spaced more than 5 m apart within the same field. This design ensures that observed microbiome differences between N and S groups are primarily driven by plant health status rather than field-scale environmental variation. Sampling was conducted on 28 July 2022.
The rhizosphere soil was collected by carefully excavating around small yellow ginger plants to a depth of 0–20 cm, preserving the complete root systems. Non-adhering soil was gently removed, while soil within 0–0.5 cm of the roots was taken as the rhizosphere sample. After collection, the soil was homogenized, placed in sterile bags, and stored at −80 °C for future analysis (Zhang, 2024).
2.3 DNA extraction and sequencing
Total DNA was extracted from 0.5 g of soil using the TGuide S96 Magnetic Soil/Stool DNA Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s instructions. The DNA concentration was quantified with the Qubit dsDNA HS Assay Kit (Life Technologies, Gaithersburg, MD, United States) and Qubit 4.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Oregon, United States). The V3–V4 region of the bacterial 16S rRNA gene was amplified using universal primer pairs 338F/806R (338F: 5′-ACTCCTACGGGAGGCAGCA-3′; 806R: 5′-GGACTACHVGGGTWTCTAAT-3′) to characterize the composition and structure of the bacterial community (Wang et al., 2022b; Yang et al., 2024). The primers were tailed with sample-specific Illumina index sequences to allow for deep sequencing. The amplification reaction system (total volume, 10 μL) contained 5 μL of KOD FX Neo Buffer, 2 μL of 2 mM dNTPs, 0.3 μL of 10 μM forward primer, 0.3 μL of 10 μM reverse primer, 0.2 μL of KOD FX Neo, 5–50 ng of DNA template, and ddH2O to a final volume of 10 μL. The PCR cycle comprised initial denaturation at 95 °C for 5 min, followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 40 s, with a final extension at 72 °C for 7 min. After amplification, the PCR products were purified using Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN, United States) and quantified by the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer. After quantification, all amplicons were pooled in equal amounts. For library construction, sequencing was performed on an Illumina NovaSeq 6,000 platform (Illumina, San Diego, CA, United States).
2.4 Sequence data analyses
The raw paired-end (PE) sequencing data were in FASTQ format. First, raw data were filtered based on nucleotide quality using Trimmomatic (version 0.33) (Bolger et al., 2014) with the following parameters: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:50. Subsequently, primer sequences were identified and removed using Cutadapt (version 1.9.1) (Martin, 2011), with parameters allowing up to 20% mismatches and a minimum overlap of 15 bp (-e 0.2 -O 15). The clean reads were then processed using the DADA2 pipeline (version 1.26.0) (Callahan et al., 2016) in R for sample inference, which included quality filtering, denoising, merging of paired-end reads, and chimera removal. Specifically, the filterAndTrim function was used to filter reads with the maxEE parameter set to 2, and other parameters were set to their default values. The error model was built from the data using the learnErrors function. Denoising was performed with the dada function, followed by merging of paired-end reads using the mergePairs function (with parameters: minOverlap = 18, maxMismatch = 0.2 × minOverlap). Chimeric sequences were identified and removed with the removeBimeraDenovo function using the “consensus” method. Subsequently, the Amplicon Sequence Variants (ASVs) table was filtered prior to taxonomic assignment to remove any ASVs with a total count less than two across all samples. Taxonomy annotation of the ASVs was performed using the “q2-feature-classifier” plugin in QIIME2 (Bolyen et al., 2019). A pre-trained Naive Bayes classifier based on the SILVA 138.1 database (Quast et al., 2013), specific to the 338F/806R primer pair targeting the V3-V4 region, was used with the “classify-sklearn” method under a confidence threshold of 70%.
2.5 Bioinformatic analysis
The bioinformatics analysis of this study was performed with the aid of the BMK Cloud (Biomarker Technologies Co., Ltd., Beijing, China). Prior to analysis, all samples were rarefied to an even sequencing depth (16S:32,087 sequences per sample) for alpha and beta diversity calculations and a clustered heatmap generation. All other analyses, such as abundance analysis, ternary plot analysis, LEfSe, and BugBase phenotypic prediction, were performed using the non-rarefied data.
Alpha diversity indices (Chao1, ACE, Shannon, Simpson) were calculated using QIIME2 to estimate microbial richness and evenness within each sample (Hagerty et al., 2020). Beta diversity was assessed using Bray–Curtis dissimilarity to measure the similarity among microbial communities across different samples, as implemented in QIIME (Caporaso et al., 2010). Further exploration of beta diversity was achieved through a suite of methods, such as principal coordinate analysis (PCoA), visualization via heatmaps, and non-metric multidimensional scaling (NMDS).
To identify microbial biomarkers significantly associated with wilt-diseased ginger plants, the LEfSe approach was employed (Segata et al., 2011). LEfSe integrates linear discriminant analysis (LDA) with non-parametric Kruskal–Wallis and Wilcoxon rank-sum tests to discern differentially abundant features between groups (Segata et al., 2011; Li et al., 2023). This method was chosen for its ability to account for microbial taxonomic hierarchy and identify consistently enriched taxa across phylogenetic levels. Features with an LDA score > 4.0 were considered discriminative.
To gain functional insights into the potential ecological impacts of the observed microbial community shifts, the organism-level microbiota phenotypes were predicted using BugBase software (Ward et al., 2017). The predicted phenotypes included Gram-positive, Gram-negative, biofilm-forming, pathogenic potential, mobile element-containing, oxygen-utilizing (aerobic, anaerobic, and facultatively anaerobic), and oxidative stress-tolerant. BugBase was run using its default parameters, which employ a data-driven approach to automatically determine the minimum trait coverage threshold for each phenotype by selecting the threshold that maximizes variance across all samples in the dataset.
2.6 Statistical analysis
Taxon abundances (at the phylum and genus levels), bacterial community alpha-diversity, and BugBase phenotypic predictions were analyzed using one-way analysis of variance (ANOVA) with the Bonferroni test (Mukhtar et al., 2021). Given the marked variance heterogeneity in the Ralstonia abundance and Simpson’s index, the non-parametric Kruskal–Wallis (KW) test and Dunn’s post hoc test were used to evaluate intergroup differences. The statistical significance of group clustering in the beta diversity analysis was tested using permutational multivariate analysis of variance (PERMANOVA; Adonis test) with 999 permutations in QIIME2. A significance threshold of a p value of <0.05 was applied for all statistical tests.
3 Results
3.1 Amplicon sequencing and species (ASV) composition
The soil samples (n = 9) were subjected to 16S rRNA gene sequencing to characterize the bacterial communities. Specifically, the V3–V4 regions of the bacterial 16S rRNA genes were amplified and assessed via high-throughput sequencing technology. A total of 1,091,200 raw reads were generated from these nine samples. Following quality control and primer removal, 1,087,336 clean reads were obtained. Further refinement through denoising, merging, and chimera removal culminated in a dataset comprising 663,121 high-quality reads (Table 1). These sequences were further denoised and then clustered into ASVs, which were annotated with taxonomic designations, resulting in the identification of 15,496 distinct ASVs.
The rarefaction analysis illustrates a leveling off of the rarefaction curves upon reaching a certain sequencing depth for all samples (Supplementary Figure S1). This suggests that the sequencing depth was sufficient to capture the majority of the species present within the samples, indicating a high level of coverage and validating the quality of the sequencing data (Xu et al., 2019). Consequently, the sampling depth and sequencing data volume were adequate to provide a reliable representation of the bacterial populations within the soil samples collected from small yellow ginger cultivation sites.
3.2 Abundance analysis of soil bacterial species
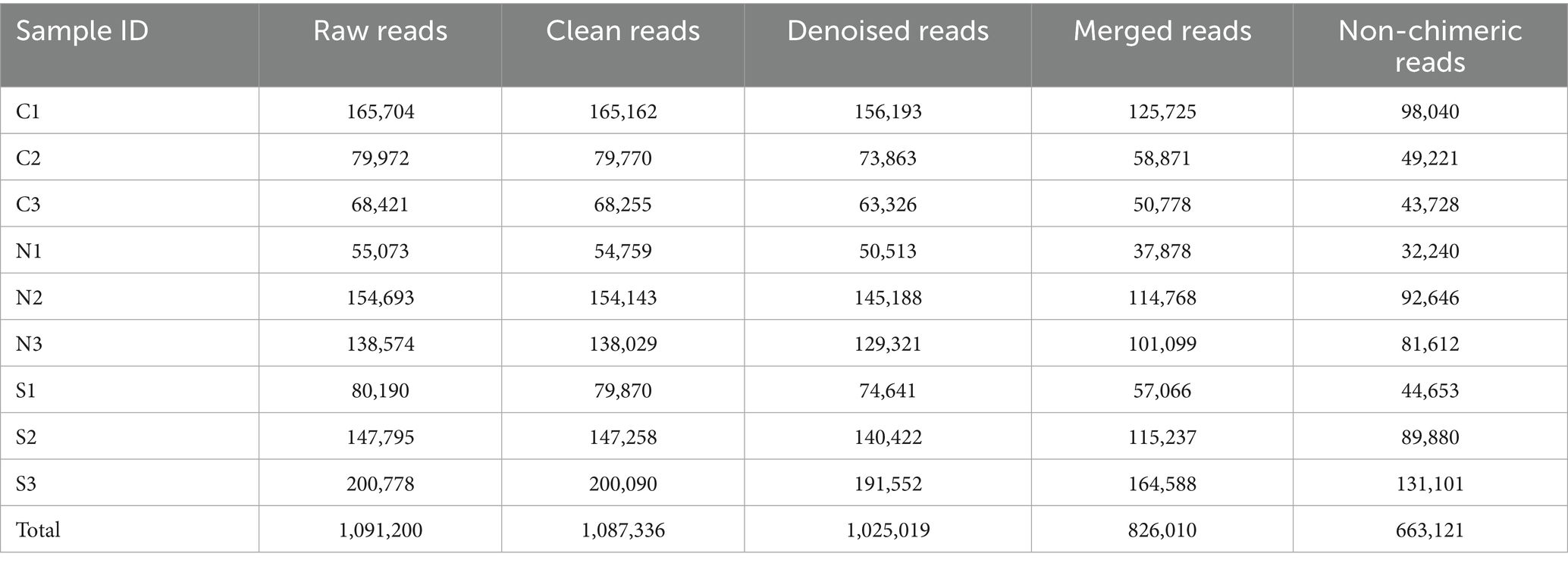
The ASVs were classified into 39 bacterial phyla and 1,019 genera. At the phylum level (Figure 1a), the relatively abundant groups included Proteobacteria, Acidobacteriota, Actinobacteriota, Gemmatimonadota, Bacteroidota, Firmicutes, Chloroflexi, and Myxococcota. Proteobacteria and Acidobacteriota were the dominant phyla, constituting 35.3 to 65.7% of the total bacteria, with a high abundance. Notably, Proteobacteria alone encompassed a significant proportion, from 19.4 to 52.8% of the total bacteria.
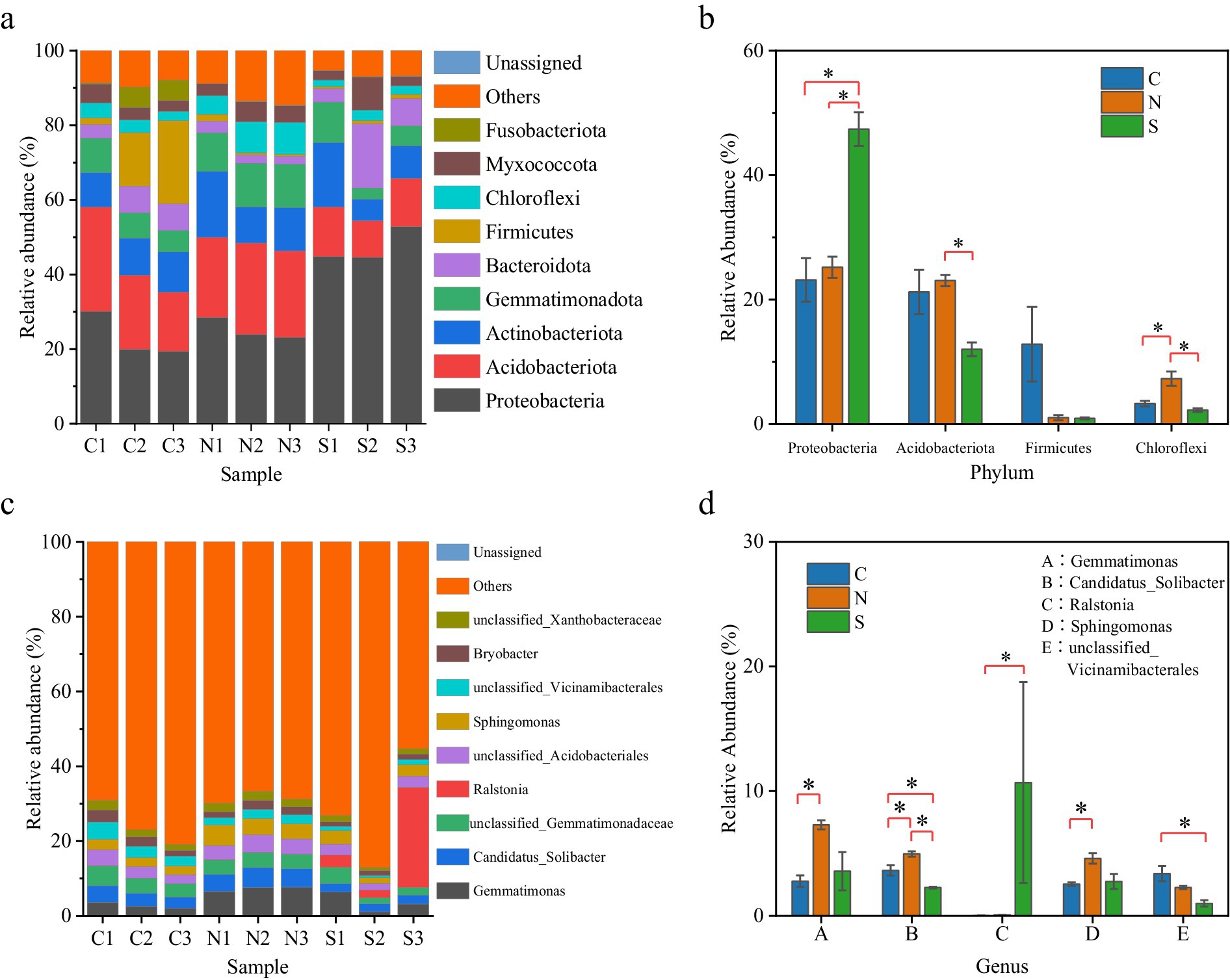
Figure 1. (a) The bacterial community composition at the phylum level. The horizontal coordinate refers to each group of ginger soil bacteria. The vertical coordinate refers to the relative abundance of each group. (b) The relative abundance comparison of the phyla Proteobacteria, Acidobacteriota, Firmicutes, and Chloroflexi among different samples. (c) Main community composition of bacteria at genus level. (d) The relative abundance comparison of the six dominant bacterial genera with significant differences among the top 10. * represents significant difference (p < 0.05, ANOVA).
The analysis of phylum-level abundance variations among the three groups demonstrated significant differences (Figure 1b). Proteobacteria exhibited a significantly elevated abundance in wilt-affected soils (47.41%) compared to the unplanted and the non-wilt-affected soils (23.2 and 25.2%, respectively, p < 0.05, ANOVA). Acidobacteriota were more predominant in the control and non-wilt-affected soils (21.2 and 23.0%, respectively), with a substantial decrease in their abundance in the soils of wilted ginger plants (12.0%, p < 0.05, ANOVA). The abundance of Firmicutes did not show statistically significant differences across the three groups, potentially due to high variability within the data. Nonetheless, elevated abundance levels were noted in soils devoid of the small yellow ginger cultivation (12.8%), contrasting with a downward trend observed in the rhizosphere of both healthy and diseased ginger plants (1.0 and 0.9%, respectively). Additionally, the Chloroflexi phylum displayed a higher abundance specifically in the rhizosphere of healthy small yellow ginger plants (7.3%), significantly contrasting with the reduced abundance in both the unplanted and the wilt-affected soils (3.3 and 2.2%, respectively, p < 0.05, ANOVA).
At the genus level, the analysis highlighted several genera with considerable relative abundance, such as Gemmatimonas, Candidatus_Solibacter, Ralstonia, Sphingomonas, and Unclassified_Vicinamibacteriales (Figures 1c,d). In the rhizosphere of healthy small yellow ginger, Gemmatimonas and Sphingomonas displayed elevated relative abundances compared to the uncultivated soils (p < 0.05, ANOVA). Despite non-significant differences in their abundance between non-wilt-affected and wilt-affected soils, a quantitative reduction is apparent. Candidatus_Solibacter attained its maximum relative abundance in the soil surrounding healthy ginger (5.0%), with a subsequent abundance in unplanted soils (3.6%), and underwent a pronounced decline in diseased conditions (2.3%) relative to both unplanted and non-wilt-affected soils (p < 0.05, ANOVA). Unclassified_Vicinamibacteriales maintained relative abundances in both the uncultivated and the non-wilt-affected soils (3.4 and 2.3%, respectively); yet a significant diminution was observed in their soil prevalence after disease onset (1.0%). Ralstonia, a genus belonging to the Proteobacteria phylum, was detected in all three soil samples from diseased small yellow ginger, with relative abundances ranging from 2.0 to 26.7%. This is in stark contrast to its negligible relative abundance of less than 0.1% in the unplanted and the non-wilt-affected soils. Despite high variability in Ralstonia abundance within wilt-affected soils, which precluded statistical significance when compared to the non-wilt-affected soils, a pronounced quantitative increase in Ralstonia was observed in the wilt-affected soils, which was markedly higher than in both the uncultivated and the non-wilt-affected soils (Figure 1d).
3.3 Alpha-diversity analysis of soil microbial communities
The diversity indices of the soil microbial communities revealed no significant disparities in the ACE values and Chao1 indices when comparing the wilt-affected ginger soil with the unplanted and non-wilt-affected soils, indicating similar levels of bacterial species richness among the soil samples (Figures 2a,b; Supplementary Table S1). However, the Shannon and Simpson diversity indices for the wilt-affected soil were observed to be lower in comparison to the unplanted and non-wilt-affected soils, with the Simpson index of the diseased ginger soil (0.9726) being significantly lower than that of the non-wilt-affected soil (0.9954, p < 0.05, KW test) (Figures 2c,d; Supplementary Table S1). These findings quantitatively demonstrated a reduced diversity within the rhizosphere soil of wilted ginger, reflecting a measurable impact on the bacterial communities present.
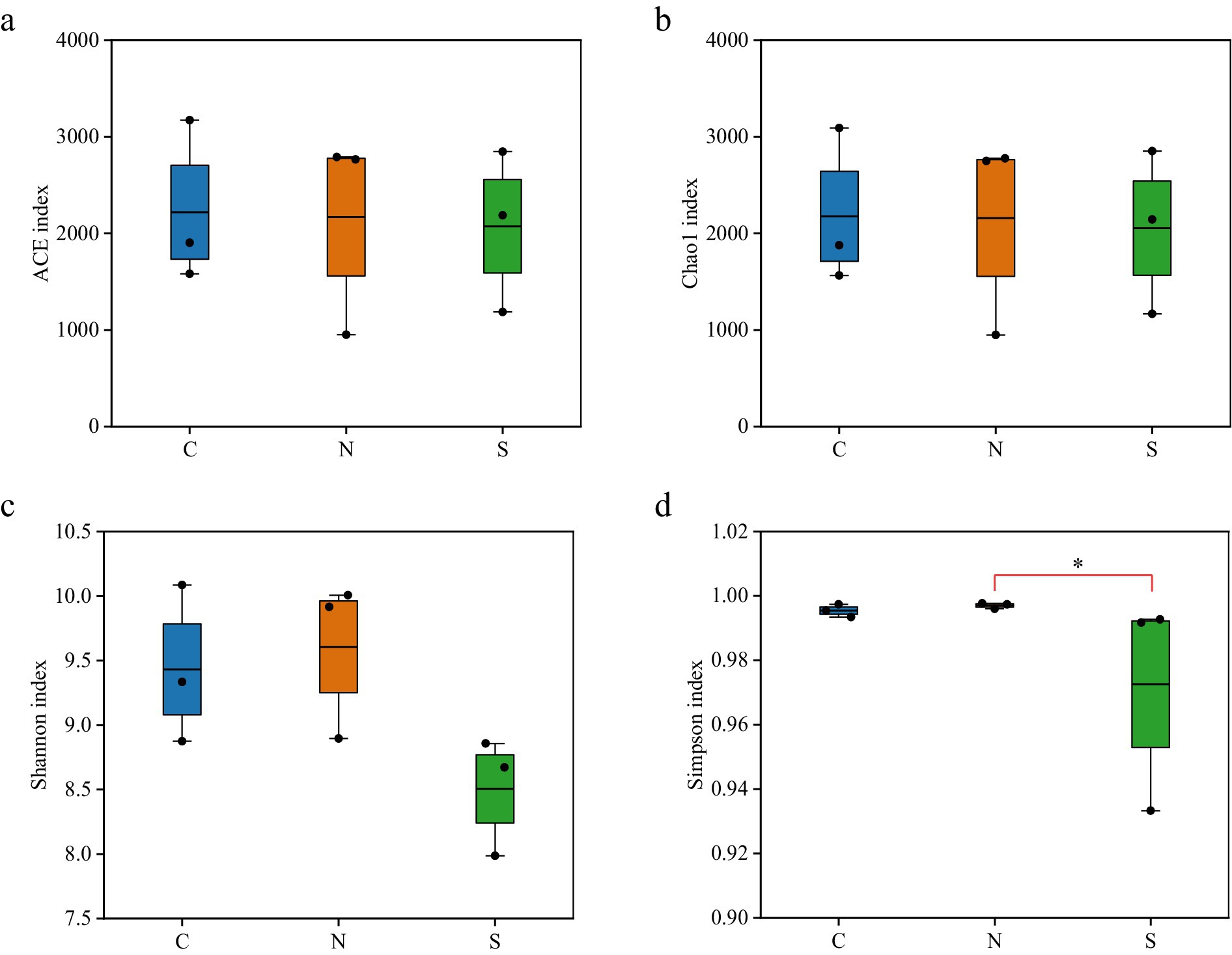
Figure 2. Boxplot of alpha-diversity indices. (a) Ace and (b) Chao1 indices reflect the abundance of ASVs in samples. (c) Shannon and (d) Simpson indices reflect the diversity of ASVs in samples. * represents significant difference (p < 0.05, KW test).
3.4 Beta-diversity and clustered abundance heatmaps
To assess differences in microbial community structure among groups, beta-diversity was analyzed based on Bray–Curtis dissimilarity (Gu et al., 2023). PCoA revealed clear separation between groups, with samples from the same condition clustering together and showing no inter-group overlap (Figure 3a), indicating distinct community structures. PERMANOVA further confirmed significant differences in community composition among groups (R2 = 0.523, p < 0.01, Figure 3b). The between-group dissimilarity substantially exceeded within-group variation, supporting the strong separation observed in ordination plots. Consistent with these findings, NMDS analysis yielded similar clustering results (Supplementary Figure S2). The stress value from the NMDS analysis was 0.0013, well below 0.05, indicating a high representativeness of the results (Dexter et al., 2018).
Figure 3. PCoA (a) and PERMANOVA (b) analysis of bacterial community structure based on the Bray–Curtis distance metric in all soil samples.
The beta-diversity results were further visually corroborated by a clustered abundance heatmap generated using row Z-score normalization of the top 100 bacterial genera (Euclidean distance, complete linkage; Supplementary Figure S3). Group S soils exhibited apparent heterogeneity in genus enrichment among replicates, whereas Group N soils demonstrated consistent enrichment patterns. Group C soils displayed intermediate variability. Despite intra-group variations, samples within each group maintained higher overall similarity in bacterial community composition compared to those between groups.
3.5 Ternary plot analysis
The ternary plot illustrates the distribution of bacterial genera across three sample groups, focusing on the top five most abundant phyla (indicated by color in the legend; Figure 4). The position of a symbol indicates the relative proportional abundance of that genus within each group: proximity to a vertex signifies higher relative abundance in the corresponding group. The visualization reveals that the majority of bacterial genera within the phyla Acidobacteriota and Gemmatimonadota are markedly more abundant in rhizospheric soils from control (Group C) and healthy ginger plants (Group N) than in soils from wilted ginger plants (Group S). Genera classified within Proteobacteria and Bacteroidota are notably more abundant in Group S soils. In Group S soils, an increased abundance of certain bacterial genera within Proteobacteria is observed, particularly among genera positioned close to the Group S vertex. Notably, one genus exhibits particularly high abundance in this region, which is identified as the genus Ralstonia through inspection of the raw data from the ternary plot. This indicates that the significant increase in Proteobacteria abundance observed in Group S soils is predominantly driven by a substantial enrichment of the genus Ralstonia rather than by proportional increases in other Proteobacterial genera.
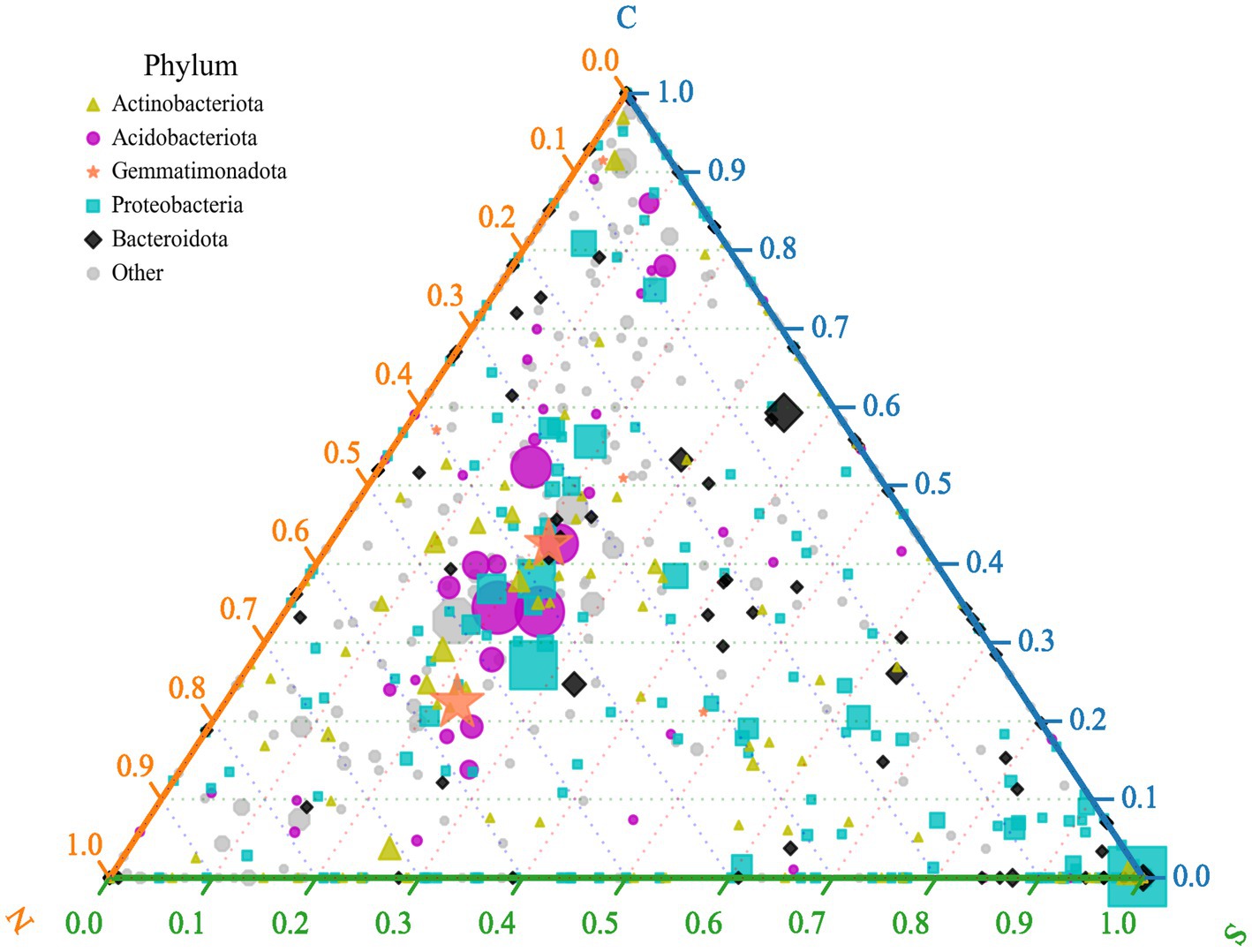
Figure 4. Ternary plot depicts different relative abundances of the genus across three sample groups (groups C, N, and S). Symbols represent individual genera, with size proportional to mean relative abundance across samples and color denoting phylum-level taxonomy.
3.6 Comparative assessment of microbial biomarkers
The LEfSe analysis, executed with an LDA score threshold of 4, revealed distinct biomarker taxa across the groups (Figure 5). Specifically, Figure 5a presents the cladogram (phylogenetic tree) generated by LEfSe, illustrating the taxonomic hierarchy of the features and highlighting those with significant differential abundance. The cladogram identified an unidentified species within the genus Ralstonia of the family Burkholderiaceae as a distinctive biomarker for Group S soils. Figure 5b depicts the LDA scores for the most discriminative features (potential biomarkers), quantifying the effect size of their abundance differences.
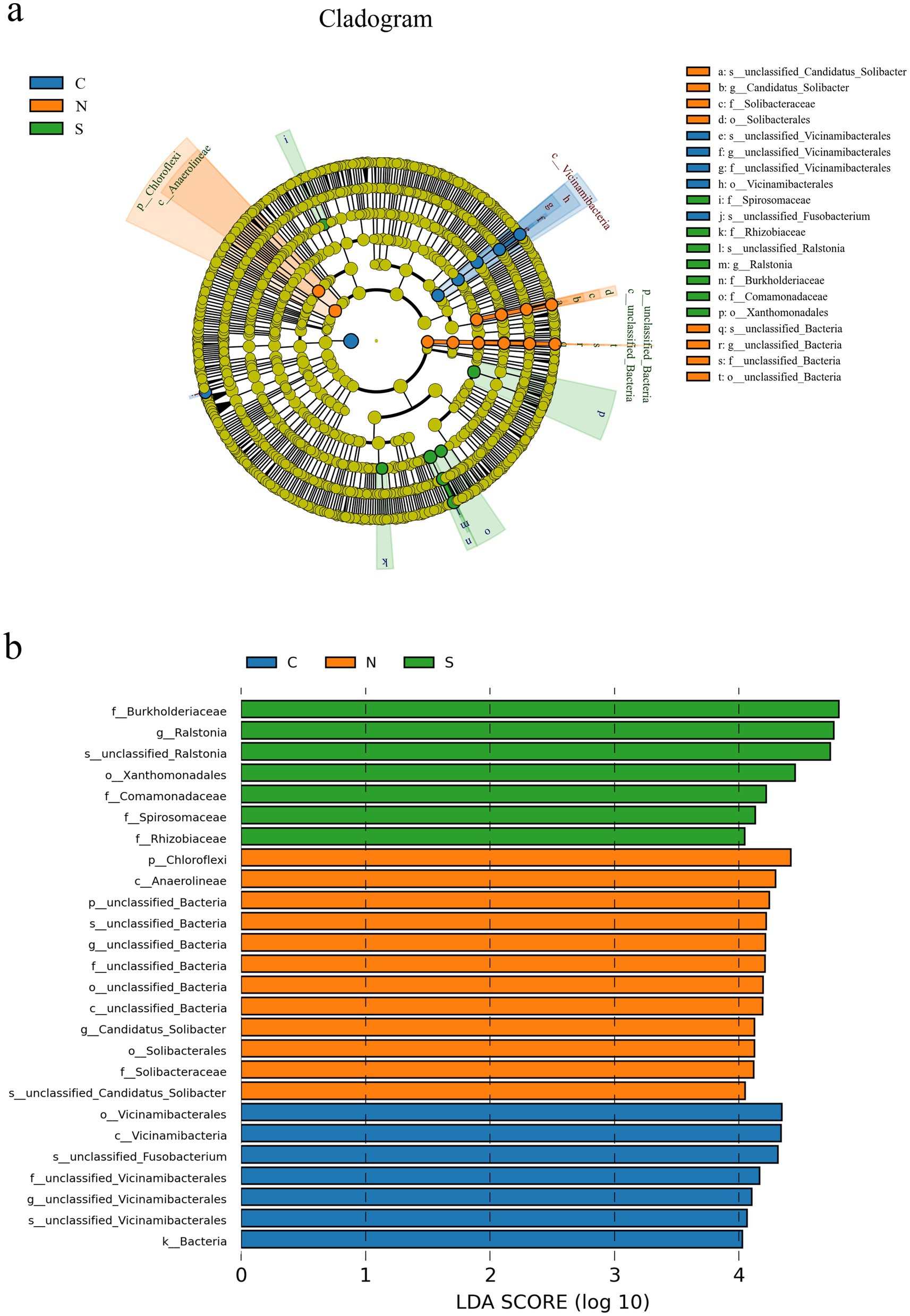
Figure 5. (a) LEfSe cladogram illustrating differentially abundant taxa across groups. (b) Bar plot of LDA effect sizes for significant microbial biomarkers (LDA score >4). (a) Circles from the center to outside layers represent taxonomic levels from phylum to species. Each node represents a specific taxon on the corresponding taxonomic level. The size of the dots is proportional to the relative abundance of that taxon in the group. Nodes colored yellow represent taxa that did not show statistically significant differential abundance among the groups. Otherwise, the nodes were colored according to the group with the highest relative abundance (LDA score >4). Different colors indicate different experimental groups; (b) Each bar represents a taxon identified as a biomarker with significant differential abundance and high discriminative power between groups. The length of the bar corresponds to the LDA score (effect size), indicating the magnitude of the difference in abundance. Different colors correspond to the group in which the taxon is most significantly enriched.
Beyond the primary Ralstonia (LDA = 4.69) biomarker for Group S soils, the LEfSe analysis also revealed that bacteria belonging to the families Comamonadaceae (LDA = 4.26), Spirosomaceae (LDA = 4.17), Rhizobiaceae (LDA = 4.06), and order Xanthomonadales (LDA = 4.49) exhibited higher relative abundance in Group S soils. Conversely, an unidentified bacterium (LDA = 4.39) within the order Vicinamibacterales and an unclassified Fusobacterium (LDA = 4.24) were more prevalent in Group C soils. In Group N soils, a higher relative abundance was observed for the class Anaerolineae (LDA = 4.30) of the phylum Chloroflexi, an unclassified genus Candidatus Solibacter (LDA = 4.17) of the family Soliacteraceae within the order Solibacterales, along with an unidentified bacterium from the domain Bacteria. Collectively, the LEfSe analysis reveals marked disparities in the biomarker taxa across the three sample groups.
3.7 Microbial community metabolic functions
Although a consistent increasing trend in aerobic abundance was noted from Group C to N to S, no statistically significant differences were observed among groups (Figure 6a). In contrast, anaerobic abundance was significantly reduced in the wilt-affected ginger soils (Group S) compared to the control soils (Group C) (p < 0.05, ANOVA; Figure 6a). While no significant difference was detected between the healthy ginger rhizosphere (Group N) and the control, pronounced shifts in anaerobic community composition occurred in both cultivated groups. Specifically, diverse anaerobic taxa prevalent in the uncultivated control soil (e.g., Faecalibacterium, Coprococcus, and Blautia) were largely absent in Groups N and S (Figure 6b). No significant differences in Gram-positive/Gram-negative ratios were detected among groups (p > 0.05, ANOVA). More importantly, BugBase predicted significantly higher relative abundance of mobile elements, oxidative stress tolerance, and pathogenic potential in Group S microbiota compared to other groups (p < 0.05, ANOVA; Figures 6c–e). A deeper investigation into the genera contributing substantially to these phenotypes identified Ralstonia, Rhodanobacter, Dokdonella, and Devosia as prominent. Notably, Ralstonia was the dominant taxon contributing to these predictions in Group S soils (Figures 6c–e).
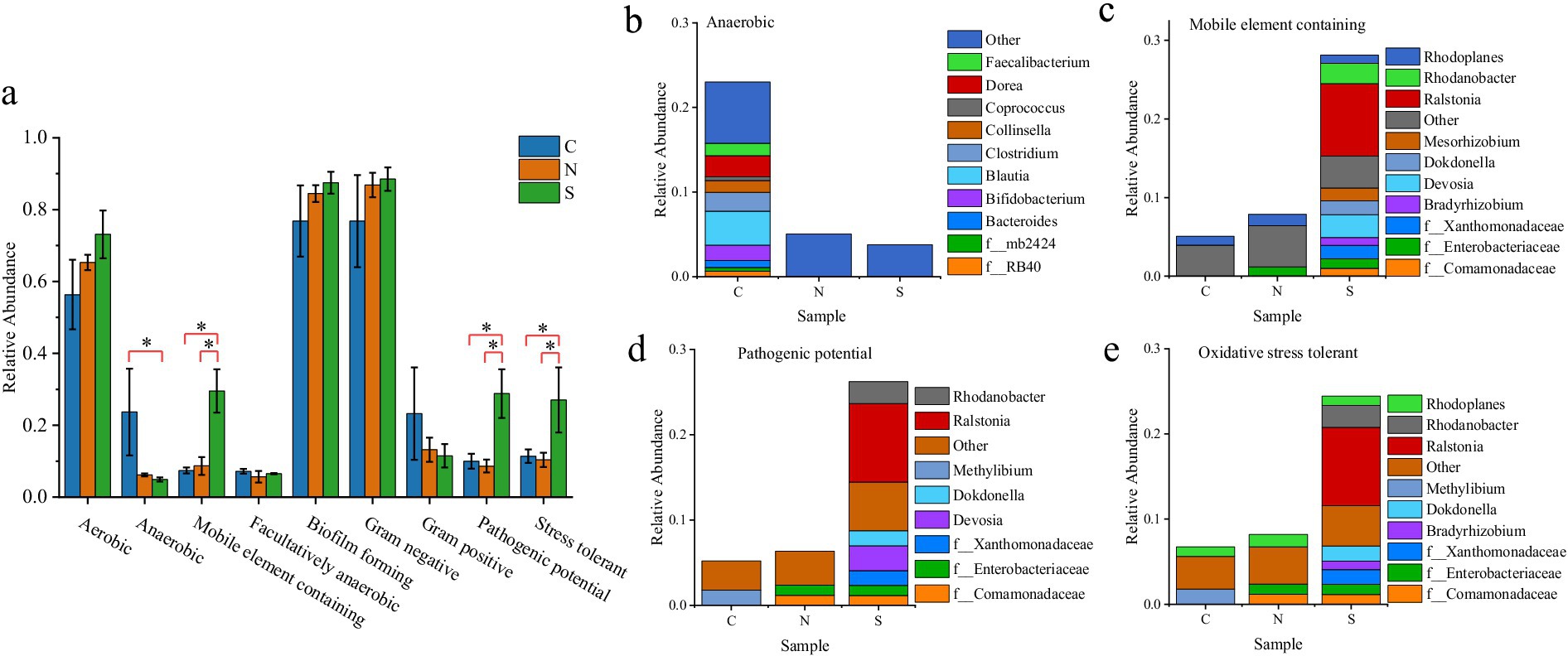
Figure 6. (a) Summary of BugBase-predicted bacterial phenotypes. * represents a significant difference. (b–e) Genus-level composition of bacteria associated with specific phenotypes per group: (b) aerobic, (c) mobile element-containing, (d) pathogenic potential, and (e) oxidative stress-tolerant.
4 Discussion
The structure and composition of soil microbial communities are intricately linked to the soil environment and are significantly influenced by crop cultivation and disease occurrence. In agricultural soils, bacteria and fungi are the predominant microbial groups. Our comprehensive analysis targeted both bacterial and fungal communities. Notably, despite the potential ability of certain fungi, such as the genus Fusarium, to induce wilt diseases (Lal et al., 2024), our data revealed no substantial increase in the genus Fusarium within wilt-affected soils (Supplementary Figure S9). Furthermore, no fungal genera exhibited significant proliferation, leading to the conclusion that fungi are unlikely to be the pathogens responsible for the ginger wilt disease in Rucheng County (Supplementary Figures S9, S13, and S14). Thus, our research focused on elucidating bacterial community dynamics.
In agricultural soils, the phyla Proteobacteria, Actinobacteriota, Bacteroidota, and Acidobacteriota constitute the predominant bacterial groups (Wang et al., 2023). Our study revealed that Proteobacteria and Acidobacteriota were the most abundant in the studied soils, with relative abundances ranging from 35.3 to 65.7%, aligning with previous findings (Yuan et al., 2021; Luo et al., 2023). Abundance analysis indicated that the cultivation of small yellow ginger and the incidence of wilt disease significantly altered the relative abundance of key bacterial phyla, specifically Proteobacteria, Acidobacteriota, Firmicutes, and Chloroflexi. Soils affected by wilt disease exhibited a notable increase in Proteobacteria and a corresponding decrease in Acidobacteriota, Firmicutes, and Chloroflexi. This shift aligns with a previous study on alterations in soil bacterial composition after ginger infection by Ralstonia solanacearum (Dang et al., 2023).
The observed changes in the soil microbial community suggest that both cultivation practices and disease occurrence exert substantial impacts on bacterial composition and overall community structure. Crucially, ternary plot analysis revealed that the significant increase in Proteobacteria abundance in wilt-affected soils was driven primarily by the enrichment of the genus Ralstonia (a known phytopathogen), rather than a broad increase across other Proteobacterial genera. This finding strongly indicates that the shift toward Proteobacteria dominance is specifically linked to the proliferation of this key pathogen. While we acknowledge the inherent functional diversity within Proteobacteria (including beneficial taxa such as Pseudomonas fluorescens) (Maheshwari and Sankar, 2023) and the existence of non-phytopathogenic Ralstonia strains in other contexts, the specific and dominant enrichment of Ralstonia observed here, coinciding with disease symptoms, points decisively to its pathogenic role. Conversely, the decrease in Acidobacteriota, Firmicutes, and Chloroflexi could signify a reduction in microbial functions associated with these groups, such as organic matter decomposition and nutrient cycling (Acidobacteriota) (Kalam et al., 2020), plant growth promotion and biocontrol (Firmicutes, notably Bacillus spp.) (Shen et al., 2023; Jabran et al., 2024), and participation in specific carbon cycling pathways (Chloroflexi) (Freches and Fradinho, 2024).
The relationship between wilted ginger plants and root bacterial diversity appears critical. Previous studies and our own diversity analysis consistently showed that ginger disease led to a decline in soil bacterial diversity (Liu et al., 2017; Wang et al., 2022a; Dang et al., 2023). Our data revealed that the biodiversity of wilt-affected soils was lower compared to both control soils and rhizosphere soils of healthy plants. This suggests that pathogens may disrupt the equilibrium of the rhizosphere microbiota. A less diverse community may be less capable of suppressing pathogen invasion or maintaining plant health, potentially creating conditions more favorable for the proliferation of Ralstonia and contributing to rhizosphere microecological imbalances (Dang et al., 2023). Further analyses (PCoA, NMDS, and heatmap) confirmed significant alterations in microbial community structure and composition in ginger-cultivated soils and wilt-affected soils. Notably, the rhizosphere microbiota of Rucheng’s small yellow ginger exhibits distinctive features when compared to previous studies on ginger wilt in other regions. This discrepancy suggests that the microbial consortia associated with ginger wilt may be strongly influenced by local edaphic conditions, agricultural practices, or host cultivar specificity. The distinct secondary dominant taxa observed here underscore that ginger wilt is not a monolithic disease but a context-dependent dysbiosis, highlighting the need for region-specific management strategies.
The potential mechanisms by which Ralstonia contributes to ginger wilt disease are well-documented (Jiang et al., 2017; Prameela and Suseela Bhai, 2020; Vailleau and Genin, 2023). Ralstonia solanacearum typically invades plant roots through wounds or natural openings, colonizes the xylem vessels, and rapidly multiplies. It produces exopolysaccharides (EPS) that occlude xylem vessels, blocking water transport and causing wilting. Additionally, it secretes cell wall-degrading enzymes and effector proteins that disrupt host cell function and suppress plant defenses. While the genus Ralstonia exhibits genetic diversity and includes non-pathogenic environmental strains, the dominant enrichment of Ralstonia in wilted ginger (supported by ternary plot and LEfSe analyses), coupled with its prominence in pathogenic phenotype predictions (Figure 6e), strongly suggests a pathogenic strain adapted to ginger. Further isolation and pathogenicity testing (e.g., Koch’s postulates) are warranted for confirmation. Beyond Ralstonia, LEfSe analysis (Figure 5) also identified other bacteria enriched in wilt-affected soils (Spirosomaceae, Rhizobiaceae, Comamonadaceae, and Xanthomonadales). While their roles in ginger wilt are less defined, they may contribute to disease dynamics; for example, some Comamonadaceae or Xanthomonadales members might thrive on decaying root material. Conversely, bacteria enriched in healthy plant soils (Anaerolineae and Candidatus Solibacter) may represent beneficial groups whose decline contributes to disease susceptibility.
Functional trait prediction via BugBase suggested that the diseased rhizosphere microbiome harbored a genomic potential indicative of a more pathogenic and resilient state. Crucially, the enhanced pathogenic potential, oxidative stress tolerance, and mobile element content were collectively driven by a network of co-enriched taxa. While Ralstonia was a key player, the genera Rhodanobacter, Dokdonella, and Devosia were also prominent contributors to these predicted traits. This collective enhancement suggests that these taxa are not merely bystanders but may actively facilitate the disease process. Thus, the severity of ginger wilt likely stems from the concerted action of a multigenus pathogenic alliance rather than from Ralstonia alone. It is important to note that these functional predictions are derived from 16S rRNA data and are constrained by limited sample size (n = 3 per group); they should therefore be interpreted as exploratory and hypothesis-generating. Despite these limitations, the predicted genomic traits provide a plausible mechanistic hypothesis for the severity and persistence of ginger wilt under field conditions. However, validation of in situ gene expression is required, warranting further investigation through metagenomic or metatranscriptomic approaches.
These findings have direct agricultural implications for managing ginger wilt in Rucheng County. First, pathogen-specific crop rotations should avoid planting ginger after known Ralstonia solanacearum hosts (e.g., Solanaceae crops like tomato, potato, pepper, and eggplant); instead, implementing extended rotations with non-host species, such as corn (maize), leafy vegetables, or legumes, is recommended to reduce pathogen load in the soil. Second, the observed reduction in beneficial phyla such as Acidobacteriota directly signals a degradation of soil health, impacting its capacity for organic matter decomposition. Therefore, soil health management strategies should prioritize practices that restore Acidobacteriota-mediated decomposition functions through organic amendments (compost, biochar), cover cropping, and reduced tillage to rebuild a suppressive microbiome. Third, the inferred functional synergy within the pathogenic consortium, including enhanced oxidative stress tolerance, suggests that biocontrol requires a multipronged approach. Introducing single antagonistic strains (e.g., Bacillus or Pseudomonas) may be insufficient. Instead, consortia of beneficial microbes should be developed to compete with, and disrupt the functionality of, the entire pathogenic network. Fourth, screening existing small yellow ginger landraces or developing new varieties with enhanced resistance to the local Ralstonia strain represents a sustainable solution. Finally, strict sanitation protocols must be implemented to prevent dissemination via contaminated tools, irrigation water, or soil movement.
5 Conclusion
This study demonstrates that ginger wilt in continuously cropped small yellow ginger significantly alters the composition and functional potential of soil microbial communities. We revealed a clear structural shift in the rhizospheric microbial community of diseased plants, which was significantly distinct from that of healthy plants. This dysbiosis was marked by a pronounced decline in beneficial phyla such as Acidobacteriota and Chloroflexi, concurrently with a significant increase in the abundance of the phylum Proteobacteria. Crucially, this expansion of Proteobacteria was primarily driven by the striking enrichment of a single genus, Ralstonia, which strongly implicates it as a key pathogen and diagnostic biomarker for ginger wilt disease. Beyond these taxonomic changes, functional prediction indicated that the diseased microbiome exhibits an enhanced genetic potential for pathogenicity and stress tolerance, and that this functional shift is contributed to not only by Ralstonia but also by other co-enriched taxa (e.g., Rhodanobacter, Dokdonella), suggesting disease severity may be mediated by a multigenus pathogenic alliance with Ralstonia as a keystone pathogen. These findings provide an ecological perspective for managing ginger wilt, emphasizing the need for strategies that rehabilitate overall microbiome function and resilience—through soil amendments and customized biocontrol—over those that target the pathogen alone. Future research should focus on isolating the ginger-adapted Ralstonia strain for pathogenicity validation and on empirically validating the predicted functional traits through molecular approaches.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA030785, PRJCA030785.
Author contributions
QS: Conceptualization, Data curation, Formal analysis, Methodology, Visualization, Writing – original draft. SF: Formal analysis, Funding acquisition, Resources, Writing – original draft. SH: Formal analysis, Investigation, Writing – original draft. RT: Formal analysis, Writing – original draft. HC: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Supervision, Visualization, Writing – review & editing. SS: Funding acquisition, Project administration, Supervision, Writing – review & editing. BW: Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was financially supported by the Chenzhou National Sustainable Development Agenda Innovation Demonstration Areas Construction Provincial Special Funding (2022sfq06 and 2022sfq51), and General Scientific Research Project of Hunan Provincial Department of Education (24C0488).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1670956/full#supplementary-material
References
Aysanew, E., and Alemayehu, D. (2022). Integrated management of ginger bacterial wilt (Ralstonia solanacearum) in Southwest Ethiopia. Cogent Food Agric. 8:2125033. doi: 10.1080/23311932.2022.2125033
Bai, B., Liu, W., Qiu, X., Zhang, J., Zhang, J., and Bai, Y. (2022). The root microbiome: community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 64, 230–243. doi: 10.1111/jipb.13226
Benzie, I. F. F., and Wachtel-Galor, S. (Eds.) (2011). Herbal medicine: biomolecular and clinical aspects. 2nd Edn. Boca Raton, FL: CRC Press/Taylor & Francis.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Dang, K., Hou, J., Liu, H., Peng, J., Sun, Y., Li, J., et al. (2023). Root exudates of ginger induced by Ralstonia solanacearum infection could inhibit bacterial wilt. J. Agric. Food Chem. 71, 1957–1969. doi: 10.1021/acs.jafc.2c06708
Dexter, E., Rollwagen-Bollens, G., and Bollens, S. M. (2018). The trouble with stress: a flexible method for the evaluation of nonmetric multidimensional scaling. Limnol. Oceanogr. Methods 16, 434–443. doi: 10.1002/lom3.10257
Freches, A., and Fradinho, J. C. (2024). The biotechnological potential of the Chloroflexota phylum. Appl. Environ. Microbiol. 90:e0175623. doi: 10.1128/aem.01756-23
Gu, Y., Liang, W., Li, Z., Liu, S., Liang, S., Lei, P., et al. (2023). The biocontrol agent Bacillus velezensis T-5 changes the soil bacterial community composition by affecting the tomato root exudate profile. Plant Soil 490, 669–680. doi: 10.1007/s11104-023-06114-3
Guji, M. J., Yetayew, H. T., and Kidanu, E. D. (2019). Yield loss of ginger (Zingiber officinale) due to bacterial wilt (Ralstonia solanacearum) in different wilt management systems in Ethiopia. Agric. Food Secur. 8:5. doi: 10.1186/s40066-018-0245-6
Hagerty, S. L., Hutchison, K. E., Lowry, C. A., and Bryan, A. D. (2020). An empirically derived method for measuring human gut microbiome alpha diversity: demonstrated utility in predicting health-related outcomes among a human clinical sample. PLoS One 15:e0229204. doi: 10.1371/journal.pone.0229204
Haq, M., Yu, J., Yao, G., Yang, H., Iqbal, H. A., Tahir, H., et al. (2023). A systematic review on the continuous cropping obstacles and control strategies in medicinal plants. Int. J. Mol. Sci. 24:12470. doi: 10.3390/ijms241512470
Hua, Y., Zheng, Z., Wang, J., Zhu, L., and Zhou, C. (2024). First report of wilt disease of ginger caused by Achromobacter xylosoxidans in China. Plant Dis. 108:778. doi: 10.1094/PDIS-05-23-0891-PDN
Jabran, M., Ali, M. A., Acet, T., Zahoor, A., Abbas, A., Arshad, U., et al. (2024). Growth regulation in bread wheat via novel bioinoculant formulation. BMC Plant Biol. 24:1039. doi: 10.1186/s12870-024-05698-x
Jiang, G., Wei, Z., Xu, J., Chen, H., Zhang, Y., She, X., et al. (2017). Bacterial wilt in China: history, current status, and future perspectives. Front. Plant Sci. 8:1549. doi: 10.3389/fpls.2017.01549
Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., El-Enshasy, H. A., Dailin, D. J., et al. (2020). Recent understanding of soil Acidobacteria and their ecological significance: a critical review. Front. Microbiol. 11:580024. doi: 10.3389/fmicb.2020.580024
Lal, D., Dev, D., Kumari, S., Pandey, S., Aparna,, Sharma, N., et al. (2024). Fusarium wilt pandemic: current understanding and molecular perspectives. Funct. Integr. Genomics 24:41. doi: 10.1007/s10142-024-01319-w
Li, C., Chen, G., Zhang, J., Zhu, P., Bai, X., Hou, Y., et al. (2021). The comprehensive changes in soil properties are continuous cropping obstacles associated with American ginseng (Panax quinquefolius) cultivation. Sci. Rep. 11:5068. doi: 10.1038/s41598-021-84436-x
Li, X., Li, X., Hong, J., Wang, Y., Guo, D., Liu, J., et al. (2023). Comparative analyses of soil bacterial colonies of two types of chinese ginger after a major flood disaster. Microbiol. Spectr. 11, e04355–e04322. doi: 10.1128/spectrum.04355-22
Liu, Q., Cheng, L., Nian, H., Jin, J., and Lian, T. (2023). Linking plant functional genes to rhizosphere microbes: a review. Plant Biotechnol. J. 21, 902–917. doi: 10.1111/pbi.13950
Liu, Y., Wu, L., Wu, X., Li, H., Liao, Q., Zhang, X., et al. (2017). Analysis of microbial diversity in soil under ginger cultivation. Scientifica 2017:8256865. doi: 10.1155/2017/8256865
Liu, M., Xue, R., Yang, C., Han, N., Hu, Y., Gu, K., et al. (2024). Rotation with other crops slow down the fungal process in tobacco-growing soil. Sci. Rep. 14:14160. doi: 10.1038/s41598-024-64735-9
Liu, J., Zhao, Z., and Wang, C. (2021). First report of rhizome rot on ginger (Zingiber officinale) caused by Enterobacter cloacae in Shandong province, China. Plant Dis. 105:210. doi: 10.1094/PDIS-05-20-1108-PDN
Luo, X., Gong, Y., Xu, F., Wang, S., Tao, Y., and Yang, M. (2023). Soil horizons regulate bacterial community structure and functions in Dabie Mountain of the East China. Sci. Rep. 13:15866. doi: 10.1038/s41598-023-42981-7
Maheshwari, P., and Sankar, P. M. (2023). “Chapter 42—culture-independent and culture-dependent approaches in symbiont analysis: in proteobacteria” in Microbial symbionts. ed. D. Dharumadurai (San Diego: Academic Press), 743–763.
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200
Mukhtar, S., Mehnaz, S., and Malik, K. A. (2021). Comparative study of the rhizosphere and root endosphere microbiomes of cholistan desert plants. Front. Microbiol. 12:618742. doi: 10.3389/fmicb.2021.618742
Nishijima, K. A., Alvarez, A. M., Hepperly, P. R., Shintaku, M. H., Keith, L. M., Sato, D. M., et al. (2004). Association of Enterobacter cloacae with rhizome rot of edible ginger in Hawaii. Plant Dis. 88, 1318–1327. doi: 10.1094/PDIS.2004.88.12.1318
Paret, M. L., Silva, A. S.de, Criley, R. A., and Alvarez, A. M. (2008). Ralstonia solanacearum race 4: risk assessment for edible ginger and floricultural ginger industries in Hawaii. HortTechnology, 18: 90–96. doi: 10.21273/HORTTECH.18.1.90
Prameela, T. P., and Suseela Bhai, R. (2020). Bacterial wilt of ginger (Zingiber officinale Rosc.) incited by Ralstonia pseudosolanacearum - a review based on pathogen diversity, diagnostics and management. J. Plant Pathol. 102, 709–719. doi: 10.1007/s42161-020-00487-5
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Saud, S., Jiang, Z., and Fahad, S. (2023). Significance and exploitation of rhizosphere chemical signaling metabolites for enhancing soil nutrient transformation. J. Soil Sci. Plant Nutr. 23, 4827–4842. doi: 10.1007/s42729-023-01526-7
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shen, Y., Yang, H., Lin, Z., Chu, L., Pan, X., Wang, Y., et al. (2023). Screening of compound-formulated Bacillus and its effect on plant growth promotion. Front. Plant Sci. 14:1174583. doi: 10.3389/fpls.2023.1174583
Vailleau, F., and Genin, S. (2023). Ralstonia solanacearum: an arsenal of virulence strategies and prospects for resistance. Annu. Rev. Phytopathol. 61, 25–47. doi: 10.1146/annurev-phyto-021622-104551
Wang, G. F., Praphat, K., Xie, G. L., Zhu, B., Li, B., Liu, B., et al. (2008). Bacterial wilt of mulberry (Morus alba) caused by Enterobacter cloacae in China. Plant Dis. 92:483. doi: 10.1094/PDIS-92-3-0483B
Wang, Q., Song, J., Zhang, J., Qin, X., Kang, Y., Huang, S., et al. (2023). Effects of microbial agent application on the bacterial community in ginger rhizosphere soil under different planting years. Front. Microbiol. 14:1203796. doi: 10.3389/fmicb.2023.1203796
Wang, C.-W., Wong, J.-W. M., Yeh, S.-S., Hsieh, Y. E., Tseng, C.-H., Yang, S.-H., et al. (2022a). Soil bacterial community may offer solutions for ginger cultivation. Microbiol. Spectr. 10, e01803–e01822. doi: 10.1128/spectrum.01803-22
Wang, P., Yan, S., Zhang, W., Xie, X., Li, M., Ren, T., et al. (2022b). Effect of soil management systems on the rhizosphere bacterial community structure of tobacco: continuous cropping vs. paddy-upland rotation. Front. Plant Sci. 13:996858. doi: 10.3389/fpls.2022.996858
Ward, T., Larson, J., Meulemans, J., Hillmann, B., Lynch, J., Sidiropoulos, D., et al. (2017). BugBase predicts organism-level microbiome phenotypes, 133462. bioRxiv. doi: 10.1101/133462
Xu, G., Xing, W., Li, T., Xue, M., Ma, Z., Jiang, N., et al. (2019). Comparative study on the effects of different feeding habits and diets on intestinal microbiota in Acipenser baeri Brandt and Huso huso. BMC Microbiol. 19:297. doi: 10.1186/s12866-019-1673-6
Yang, X., Xiong, J., Du, T., Ju, X., Gan, Y., Li, S., et al. (2024). Diversifying crop rotation increases food production, reduces net greenhouse gas emissions and improves soil health. Nat. Commun. 15:198. doi: 10.1038/s41467-023-44464-9
Yuan, M., Yu, T., Shi, Q., Han, D., Yu, K., Wang, L., et al. (2021). Rhizosphere soil bacterial communities of continuous cropping-tolerant and sensitive soybean genotypes respond differently to long-term continuous cropping in Mollisols. Front. Microbiol. 12:729047. doi: 10.3389/fmicb.2021.729047
Zhang, L. (2024). Effects of mixed biocrusts on soil nutrients and bacterial community structure: a case study from hilly loess plateau, China. Sci. Rep. 14:21265. doi: 10.1038/s41598-024-71927-w
Zhang, R., Xu, K., Li, X., Gao, Y., Sun, Y., and Huang, Q. (2020). Ginger wilt and rot disease caused by Ceratocystis fimbriata in China. Plant Dis. 105:1569. doi: 10.1094/PDIS-08-20-1647-PDN
Zhang, M., Zhao, R., Wang, D., Wang, L., Zhang, Q., Wei, S., et al. (2021). Ginger (Zingiber officinale Rosc.) and its bioactive components are potential resources for health beneficial agents. Phytother. Res. 35, 711–742. doi: 10.1002/ptr.6858
Keywords: ginger, wilt disease, Ralstonia, soil microbiota, microbial diversity
Citation: Sun Q, Fei S, Huang S, Tan R, Chen H, Song S and Wang B (2025) Wilt disease reshapes rhizosphere microbiota in small yellow ginger soils. Front. Microbiol. 16:1670956. doi: 10.3389/fmicb.2025.1670956
Edited by:
Min-Chong Shen, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Jialin Hu, Texas A&M University, United StatesMuhammad Jabran, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2025 Sun, Fei, Huang, Tan, Chen, Song and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haibo Chen, Y2hlbmhhaWJvNTI2QGhvdG1haWwuY29t
 Qing Sun
Qing Sun Sitian Fei1,2,3
Sitian Fei1,2,3 Haibo Chen
Haibo Chen