- 1Hangzhou Academy of Agricultural Sciences, Hangzhou, China
- 2Engineering Research Center of Ministry of Education of China for Food and Medicine, Jilin Agricultural University, Changchun, China
- 3Hangzhou Rural Revitalization Service Center, Hangzhou, China
- 4Lin′an Agriculture and Forestry Technology Extension Center, Hangzhou, China
- 5Tonglu Country Agricultural Technology Promotion Center, Hangzhou, China
Continuous cropping in facility agriculture induces severe soil degradation through acidification, nutrient imbalance, and pathogen accumulation, posing a significant threat to agricultural sustainability; to address this challenge, we developed an innovative Stropharia rugosoannulata-Ornamental Sunflower Rotation System (SR-OS2) incorporating spent mushroom substrate (SMS) and investigated its remediation mechanisms through integrated approaches including soil physicochemical analysis, extracellular enzyme assays, high-throughput sequencing (16S/ITS), co-occurrence network analysis, and Partial Least Squares Path Modeling (PLS-PM). The SR-OS2 system significantly enhanced soil properties by increasing pH (+0.57 units), decreasing electrical conductivity (−37.56%), and boosting available phosphorus (+84.2%), while also shifting microbial communities toward bacterial dominance with a 37.4% increase in bacterial Chao1 diversity and a 39.1% decrease in fungal diversity, alongside strengthened bacterial connectivity (+42%) and reduced fungal modularity in co-occurrence networks. Enzyme stoichiometry further revealed alleviated nitrogen limitation (vector angle: 27.2°–30.9°), and PLS-PM identified dual remediation pathways—a dominant biological pathway (β = 0.92) and a physicochemical pathway (β = −0.501); these improvements collectively demonstrate that the SR-OS2 system synergistically restores soil microecological functions, providing a sustainable paradigm for agricultural waste valorization and effective management of continuous cropping obstacles.
1 Introduction
Continuous cropping obstacles in facility agriculture pose a severe threat to global agricultural sustainability. These obstacles manifest primarily as the deterioration of soil physicochemical properties (acidification, nutrient imbalance) and ecological functional degradation driven by the enrichment of soil-borne pathogens and microbial community dysbiosis, ultimately leading to persistent crop yield reduction (Li Y. et al., 2022). Ornamental sunflower (Helianthus annuus L.), an economically and ecologically valuable crop, faces industrial development constraints due to soil health decline induced by continuous cropping (Puttha et al., 2023). Recent studies demonstrate that incorporating spent mushroom substrate (SMS) into soil is an effective strategy for rehabilitating degraded soils. Its mechanisms involve not only enhancing soil organic matter and activating mineral nutrients (Li et al., 2020) but also significantly altering soil microbial community structure and enzyme activity (Gong et al., 2018; Hao et al., 2024). Specifically, SMS from Stropharia rugosoannulata cultivation—rich in lignin-degrading enzymes (e.g., lignin peroxidase, LiP) and humus precursors (Hao et al., 2024)—exhibits unique potential for mitigating continuous cropping obstacles. To leverage this potential, we innovatively designed the “Stropharia rugosoannulata-Ornamental Sunflower” rotation system (SR-OS2). This system integrates in-situ SMS incorporation with coordinated stubble management to simultaneously achieve agricultural waste valorization and soil health restoration.
Although S. rugosoannulata SMS input has been confirmed to increase soil organic carbon (SOC) and total nitrogen (TN) pools (Gong et al., 2018), critical knowledge gaps remain regarding its deep-layer mechanisms for rehabilitating continuously cropped soils: (1) The regulatory patterns of SMS input and rotation on cross-seasonal C-N-P stoichiometric dynamics—particularly enzyme activity ratios and vector traits—have not yet been quantified (Hu et al., 2020); (2) Microbial community restructuring (e.g., increased bacterial dominance) (Hernandez et al., 2021) has not been effectively linked to topological evolution in microbial co-occurrence networks (e.g., connectivity, modularity); (3) Integrated empirical models are lacking for SMS-driven “physicochemical-biological” multi-level cascade pathways (e.g., trace element activation → microbial community restructuring → enzyme activity response) (Jansson and Baker, 2016).
To address these gaps, this study established a four-stage SR-OS2 treatment using soil from a 5-year continuously cropped sunflower facility: B1: Untreated soil (control), B2: SMS incorporation, B3: First sunflower crop, B4: Second no-till sunflower crop. Prior to the experiment, this facility had documented issues of stunted plant growth, leaf chlorosis, and an estimated yield reduction of over 20% in the most recent cropping season—symptoms characteristic of severe continuous cropping obstacles.
By integrating soil physicochemical analysis, extracellular enzyme activity assays, and high-throughput sequencing, we aimed to: (1) Decipher C-N-P stoichiometric dynamics: Quantify changes in SOC/TN/AP (available phosphorus) content, enzyme activity ratios, and enzymatic traits; (2) Uncover microbial co-occurrence network restructuring: Analyze topological properties (connectivity, modularity) of bacterial/fungal communities and their coupling with key extracellular enzyme activities; (3) Validate multi-level cascade pathways: Test physicochemical (trace elements → metalloenzyme activation) and biological (SOC/TN → microbial community → enzyme balance) cascading effects. This study provides new insights into the ecological mechanisms underpinning continuous cropping obstacle mitigation and supports the optimization of agricultural waste recycling technologies.
2 Materials and methods
2.1 Study region and experimental design
Field experiments were conducted from March 2023 to July 2024 in Lin’an District, Hangzhou, Zhejiang Province, China (119°61′E, 30°20′N), a region characterized by a mid-subtropical monsoon climate with a mean annual temperature of 15.4 °C and annual precipitation of 1,000–1,200 mm. The soil, classified as yellow-brown, had undergone 5 years of continuous ornamental sunflower (Helianthus annuus L.) cultivation, with initial properties of pH 6.82, EC 120 μS cm−1, and SOC 1.25%. We established the SR-OS2 with four treatments: (1) B1: Untreated fallow soil; (2) B2: Soil amended with S. rugosoannulata spent mushroom substrate (SMS) incorporated into the 0–20 cm layer after mushroom harvest. SMS was applied at a rate of 10 t/ha. The SMS had the following basic properties: total organic carbon (TOC) = 35.2%, total nitrogen (TN) = 1.8%, and a C/N ratio of 19.6; (3) B3: Sunflower (H. annuus cv. ‘Jincancan’) planted post-SMS amendment at 6 × 104 plants ha−1 density (90-day growth period); (4) B4: Second sunflower crop planted under no-till conditions after B3 harvest (85-day growth period). A randomized block design with three replicates was implemented, with individual plots (30 m × 20 m) separated by ≥30 m buffers to mitigate edge effects. No additional fertilization was applied during sunflower cultivation. Standard field management practices included drip irrigation to maintain soil moisture at 60–70% of field capacity and manual weeding as necessary.
2.2 Soil sampling and pretreatment
Composite soil samples (0–20 cm depth) were collected at critical stages: B1 (initial soil), B2 (30 days post-SMS incorporation), and B3/B4 (immediately after sunflower harvest). Five subsamples per plot were obtained along an “S”-shaped transect within 1 m × 1 m quadrats and homogenized. Samples were partitioned for: (1) fresh storage (4 °C, enzyme assays within 24 h); (2) flash-freezing in liquid N₂ (−80 °C storage for DNA extraction); (3) air-drying (sieved to 2 mm for physicochemical analysis or 0.15 mm for micronutrient detection).
2.3 Soil physicochemical analysis
Soil pH and EC were determined in 1:2.5 (w/v) soil-water slurry using a PHS-25 pH meter (Leici, Shanghai) and a DDS-307 conductivity meter (Yidian, Shanghai, China), respectively. SOC was quantified via potassium dichromate oxidation (Keming Bio, Suzhou, China), and TN followed the Kjeldahl method (GB 7173–87; Kjeltec 8,400, Foss, Denmark). AP was extracted with 0.5 M NaHCO₃ (pH 8.5) and measured spectrophotometrically (UV-1800, Shimadzu (Shanghai) Global Laboratory Consumables Co., Ltd., Shanghai, China) using the molybdenum blue method. AK (Available Potassium) was extracted with 1 M NH₄OAc and quantified by flame photometry (FP6410, Jingke, Shanghai). Micronutrients (Ca, Mg, Fe, Mn, Cu, Zn, S) were digested with HNO₃-H₂O₂-HF (170 °C, 4 h) and analyzed by ICP-MS (NexION® 1,000, PerkinElmer), with spike recovery rates of 85–110%.
2.4 Extracellular enzyme activity assays
Five extracellular enzymes were assayed colorimetrically: β-glucosidase (BG; p-nitrophenyl-β-D-glucopyranoside substrate, 400 nm) for C mineralization; N-acetylglucosaminidase (NAG; p-nitrophenyl-N-acetyl-β-D-glucosaminide, 400 nm) for fungal N cycling; leucine aminopeptidase (LAP; L-leucine-p-nitroanilide, 405 nm) for bacterial N cycling; alkaline phosphatase (ALP; p-nitrophenyl phosphate, 405 nm) for P mineralization; and polyphenol oxidase (PPO; pyrogallol, 430 nm) for lignin degradation. Fresh soil (0.5 g dry-weight equivalent) was incubated with substrate and modified universal buffer (pH 7.0) at 37 °C for 4 h (BG, NAG, LAP, ALP) or 24 h (PPO). Activities are expressed as μmol product g−1 soil d−1.
2.5 Enzyme stoichiometry analysis
Microbial nutrient limitations were evaluated using enzyme vector analysis. Vector length indicated carbon limitation (higher values), while vector angle (A = arctan [ln(NAG+LAP)/ln(BG)]), identified nitrogen (<45°) or phosphorus (>45°) limitation. Additional ratios included: BG:(NAG + LAP) (C vs. N acquisition investment), NAG:LAP (fungal vs. bacterial N cycling dominance; >1 = fungal), BG:PPO (labile vs. recalcitrant C decomposition), and BG:ALP (C vs. P acquisition strategy).
2.6 Microbial high-throughput sequencing
Soil DNA was extracted using the TIANamp Soil DNA Kit (TIANGEN, Beijing, China). Bacterial 16S rRNA V3–V4 regions were amplified with primers 341F (5’-CCTACGGGNGGCWGCAG-3′)/805R (5’-GACTACHVGGGTATCTAATCC-3′), and fungal ITS1 regions with ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3′)/ITS2R (5’-GCTGCGTTCTTCATCGATGC-3′). PCR conditions included: 98 °C for 30 s; 27 cycles of 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 30 s; and final extension at 72 °C for 5 min. Libraries were validated (Agilent 2100 Bioanalyzer, California, USA) and sequenced on the Illumina NovaSeq 6000 platform (PE250; 60,000 reads/sample target depth).
2.7 Data analysis
2.7.1 Amplicon sequencing data analysis
Raw sequencing data underwent processing with DADA2 (v1.26) for bacterial 16S rRNA amplicons and UNOISE3 (v11) for fungal ITS sequences to generate amplicon sequence variants (ASVs). Taxonomic assignment was performed against the SILVA v138 database for bacteria and UNITE v8.0 for fungi. Alpha diversity was evaluated using Chao1 richness and Simpson’s evenness indices. FUNGuild (v1.2) was used to predict the ecological functions of the fungal communities. Fungal ASVs were assigned to trophic modes and guilds, and those assigned as ‘highly probable’ or ‘probable’ were retained for subsequent analysis. Linear Discriminant Analysis Effect Size (LEfSe) was performed using the online tool1, with an LDA score threshold set to > 3.0 and a significance level of p < 0.05 (Kruskal-Wallis test).
2.7.2 Statistical and multivariate analysis
Treatment effects on soil properties, enzyme activities, and diversity metrics were assessed through one-way ANOVA with Fisher’s LSD post hoc tests (p < 0.05) in R v4.3.2. Microbial community structure variations were visualized via principal coordinates analysis (PCoA) based on Bray-Curtis distances and statistically validated with PERMANOVA (999 permutations; vegan package). Microbial co-occurrence networks were constructed using the SpiecEasi package (v1.1.2) with the MB method (Matrix Bootstrap) for robust association inference. Networks were constructed for each treatment group using all samples within that group. Only robust interactions with |SparCC ρ| > 0.6 and FDR-corrected p < 0.01 were retained. Network topology metrics (nodes, edges, average degree, transitivity) were computed with `igraph` (v1.6.0). Partial Least Squares Path Modeling (PLS-PM) was implemented via the plspm package (v0.4.9) incorporating six latent variables: (1) Rotation treatment (categorical: B1-B4), (2) Soil properties (observed: SOC, TN, AP, pH), (3) Micronutrients (observed: Mg, Fe, Mn, Cu, Zn), (4) Microbial community (observed: bacterial/fungal Chao1 and Simpson indices), (5) Enzyme ratios (observed: BG:(NAG+LAP), BG:PPO, BG:ALP, NAG:LAP), and (6) Enzyme activity (observed: BG, NAG, PPO). Path coefficients were validated with 1,000 bootstraps, and model fit was evaluated using the Goodness-of-Fit (GoF) index, where GoF > 0.6 indicated excellent fit for soil-microbe systems (Rillig et al., 2017).
3 Results
3.1 Soil physicochemical properties
This study systematically investigated the effects of the SR-OS2 on the physicochemical properties of continuous cropping soil. The results (Table 1) demonstrated that the SMS application (B2) significantly increased soil pH and reduced EC, indicating a trend toward mitigating soil acidification and salinization, even though the initial pH was near neutral. A significant increase in soil pH of 0.57 units (p < 0.05) and a 37.56% reduction in EC. This improvement created a more favorable soil environment for subsequent crop growth. Regarding soil nutrients, after the first sunflower crop (B3), SOC and TN contents increased significantly by 20.6 and 23.0%, respectively, compared to the control (B1) (p < 0.05), while AP content increased significantly by 48.1% (p < 0.05). Following the harvest of the second sunflower crop (B4), SOC and TN contents remained at elevated levels, and AP content further increased to 111.16 mg/kg, representing a significant 84.2% increase over the control. These results indicate that the combination of SMS application and ornamental sunflower cultivation significantly promoted the accumulation of SOC and nitrogen, and enhanced phosphorus availability. Correlation analysis (Supplementary Table S1) revealed significant positive correlations between SOC and both TN (r = 0.66*) and AP (r = 0.70*), suggesting that increased SOC contributes to nitrogen and phosphorus accumulation. Concurrently, a highly significant negative correlation was observed between EC and pH (r = −0.87**), indicating that the SMS application effectively mitigated soil acidification by reducing EC.
Figure 1 illustrates the dynamics of soil trace element contents. B2 had no significant effect on Fe, Mg, or Ca contents. However, under B3 and B4 treatments, Ca content increased significantly by 26 and 24%, respectively, compared to B2 (p < 0.05, Figures 1A–C). Mn, S, Zn, and Cu contents decreased after SMS application but rebounded significantly following sunflower cultivation (Figures 1D–G). Notably, in the B4 treatment, Mn, S, and Zn contents were significantly higher than in B1, with increases of 83, 13, and 12%, respectively (p < 0.05). Further correlation analysis indicated positive relationships of Ca and Zn with SOC and TN, while Cu showed a negative correlation with EC (Figure 1H). This suggests that the rotation system significantly influenced the availability of meso- and micro-elements by regulating organic matter decomposition.
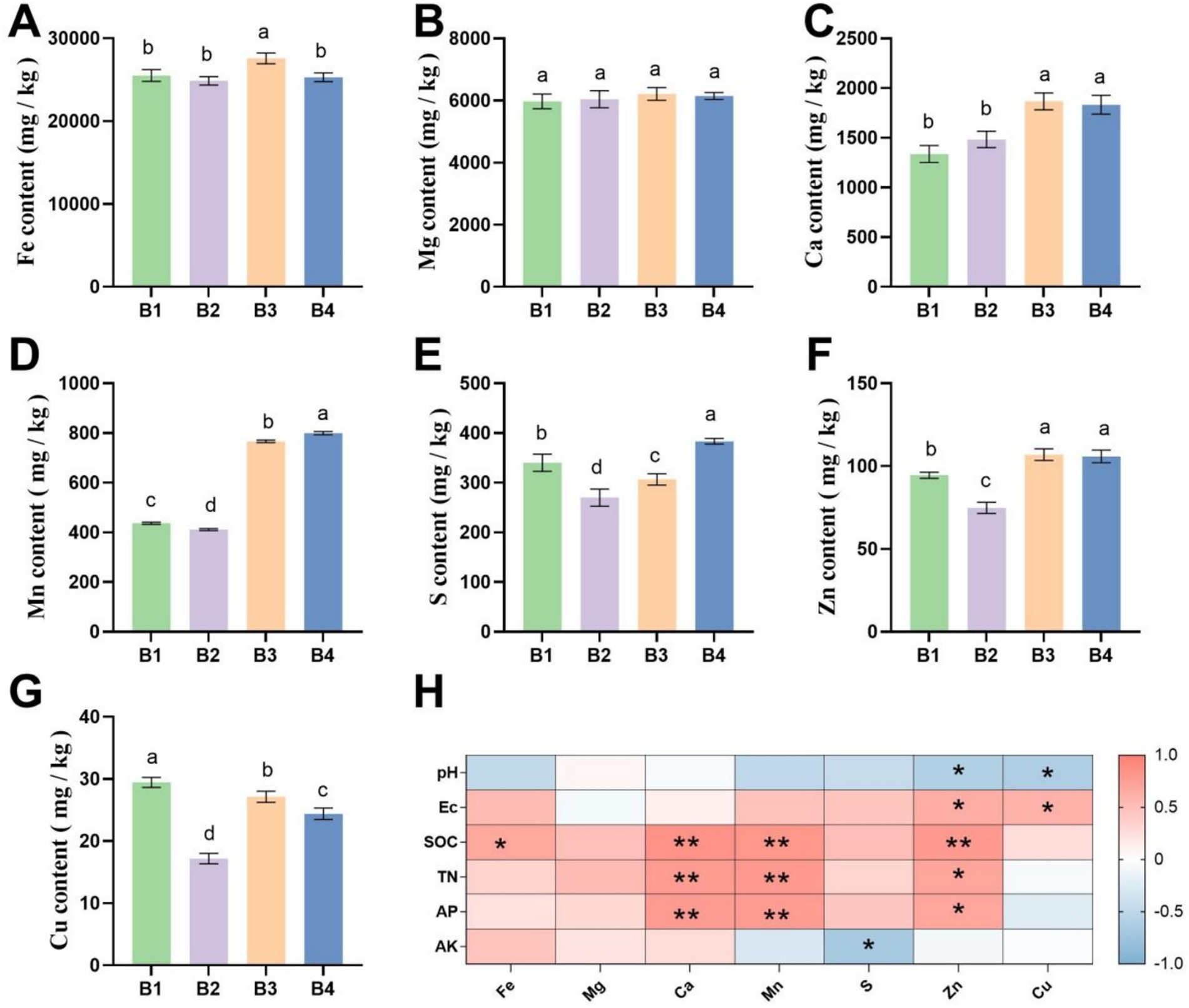
Figure 1. Differences in soil meso- and micro-element contents under SR-OS2 system treatments. (A–G) Contents of Fe, Mg, Ca, Mn, S, Zn, and Cu. Different lowercase letters indicate significant differences among treatments (p < 0.05, LSD test). (H) Correlation between soil basic nutrients and meso−/micro-elements. * denotes p < 0.05; ** denotes p < 0.01; Fe, Iron; Mg, Magnesium; Ca, Calcium; Mn, Manganese; S, Sulfur; Zn, Zinc; Cu, Copper; pH, potential of hydrogen; EC, Electrical Conductivity; SOC, soil organic carbon; TN, total nitrogen; AP, available phosphorus; AK, Available Potassium.
In conclusion, the SR-OS2 system, through the synergistic effect of SMS application and sunflower cultivation, significantly improved the physicochemical properties of continuous cropping soil, thereby creating more favorable conditions for crop growth. These findings provide an important theoretical basis for utilizing agricultural waste like SMS to improve soil quality and promote the sustainable development of protected agriculture.
3.2 Changes in extracellular enzyme activities and stoichiometric ratios
This study comprehensively analyzed the effects of the SR-OS2 on soil extracellular enzyme activities and their stoichiometric ratios. The results demonstrated significant treatment-dependent variations in soil enzyme activities. As shown in Table 2, the SMS application (B2) increased the activities of the nitrogen (N)-cycling enzyme LAP and phosphorus (P)-cycling enzyme ALP by 42.1 and 46.0%, respectively (p < 0.05), while reducing BG activity (C-cycling) by 17.2% (p < 0.05). This shift indicates enhanced microbial demand for N and P acquisition following SMS amendment. After sunflower cultivation (B3), PPO activity peaked at 67.8% above the control (B1) (p < 0.05), suggesting accelerated plant residue decomposition. By the second crop (B4), LAP activity further increased by 103.1% compared to B1, whereas NAG activity remained unchanged. This reflects persistent N-cycling enhancement, potentially constrained by fungal metabolism, since NAG is primarily fungal-derived, while LAP is bacterial-dominated (Xing et al., 2024).
Enzyme stoichiometric ratio analysis (Figure 2) further elucidated microbial nutrient acquisition strategies. The SMS application significantly reduced the BG:(NAG + LAP) ratio, indicating that the increase in N-cycling enzymes surpassed that of C-cycling BG (Figure 2A), thus relatively enhancing soil N-cycling capacity. Concurrently, decreased BG:ALP and BG:PPO ratios suggested improved P-cycling efficiency (Figures 2B,C). Shifts in the NAG:LAP ratio revealed altered fungal versus bacterial contributions to N-cycling (Figure 2D). Ecological enzyme stoichiometry vector analysis showed that SMS amendment significantly decreased vector length (Vector L), indicating alleviated microbial C limitation. The vector angle (Vector A) increased from 27.2° to 30.9° (remaining <45°) (Figures 2E,F), demonstrating mitigated yet persistent N limitation.
![Graphical representation of six bar charts labeled A to F. Each chart compares different biochemical ratios or vector lengths across four groups, B1 to B4. Chart A shows [BG:(NAG+LAP)] ratio with B1 highest. Chart B displays [BG:ALP] ratio, B1 also highest. Chart C presents [BG:PPO] ratio, again B1 is highest. Chart D shows [NAG:LAP] ratio with B1 leading. Chart E represents vector length with B1 highest, while Chart F depicts vector length in degrees with B2 leading. Error bars indicate variability, and statistically significant differences are marked by different letters.](https://www.frontiersin.org/files/Articles/1671484/fmicb-16-1671484-HTML/image_m/fmicb-16-1671484-g002.jpg)
Figure 2. Soil enzyme stoichiometric indices under the SR-OS2 system; (A) BG:(NAG + LAP); (B) BG:ALP; (C) BG:PPO; (D) NAG:LAP; (E) Vector length; (F) Vector angle; BG, β-glucosidase; NAG, N-acetylglucosaminidase; LAP, leucine aminopeptidase; PPO, polyphenol oxidase.
In summary, the SR-OS2 system significantly modulated soil enzyme activities and stoichiometry through SMS amendment and sunflower cultivation. The SMS application stimulated microbial demand for N and P, elevating corresponding enzyme activities, while sunflower cultivation further intensified N-cycling enzyme activity. These dynamic responses reflect microbial adaptations to soil nutrient alterations, providing critical insights into the mechanisms through which rotation systems regulate soil microecology.
3.3 Soil microbial diversity and community structure
The effects of the SR-OS on soil microbial diversity and community structure were analyzed using high-throughput sequencing. The results revealed significant alterations in bacterial and fungal diversity and composition across treatments. As shown in Figures 3A,B, the bacterial Chao1 index decreased by 7.5% (p < 0.05) in the SMS-amended treatment (B2) compared to the control (B1). However, it increased significantly by 37.3 and 37.4% (p < 0.05) following the first (B3) and second sunflower crops (B4), respectively. Conversely, the fungal Chao1 index declined by 23.0% (p < 0.05) in B2 and decreased progressively to 39.1% below B1 (p < 0.05) by B4 (Figure 3B). Simpson index analysis (Figures 3C,D) indicated peak bacterial diversity in B3, while fungal diversity in B2 was significantly lower than in B1 (p < 0.05). This demonstrates that the rotation system enhanced bacterial diversity and evenness but suppressed fungal diversity and evenness post-sunflower cultivation.
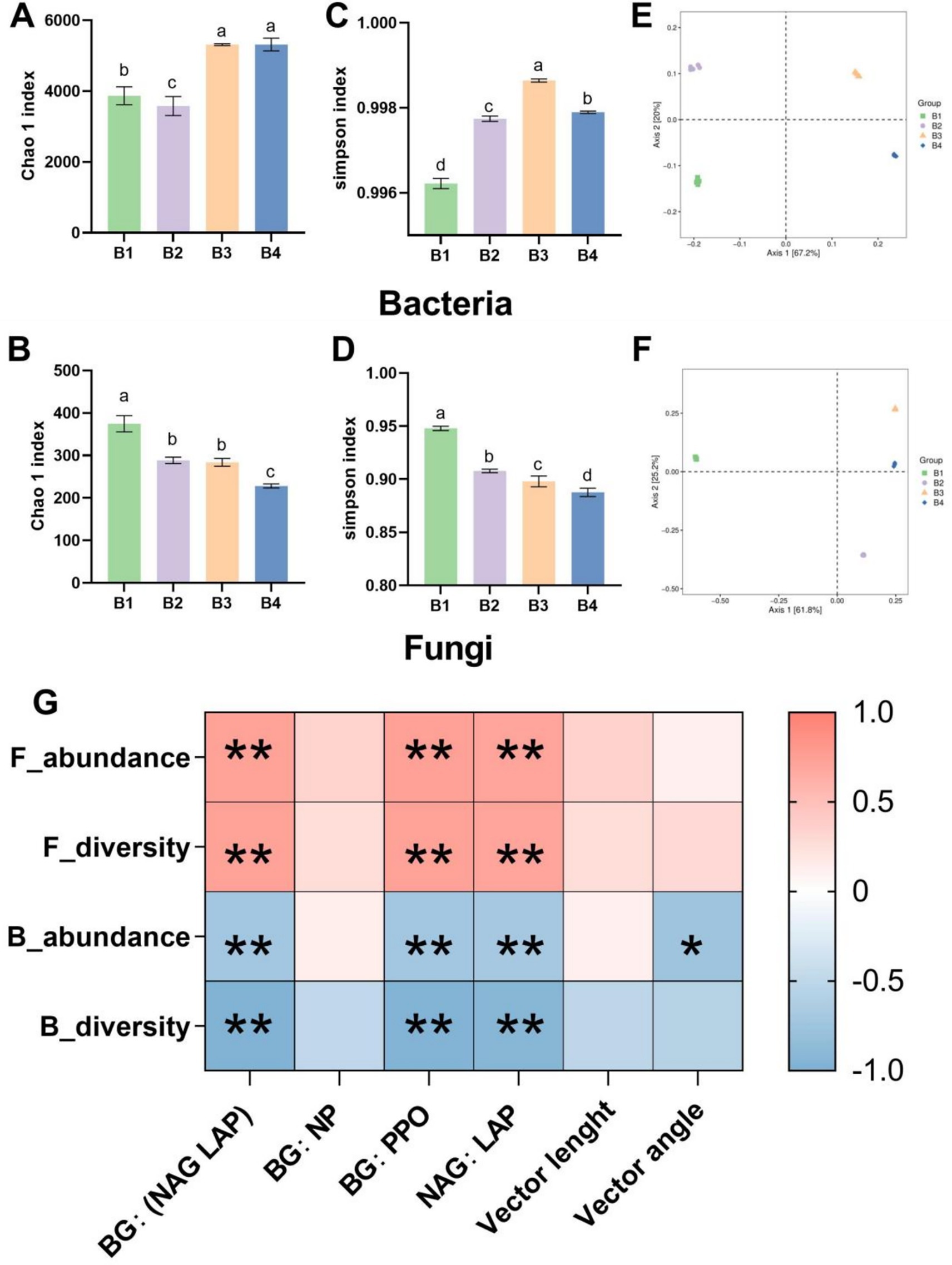
Figure 3. Microbial community abundance, diversity, and enzyme activity correlations under the SR-OS2 system; (A) Bacterial Chao1 index; (B) Bacterial Simpson index; (C) Fungal Chao1 index; (D) Fungal Simpson index; (E) Principal Coordinate Analysis (PCoA) of bacterial communities; (F) PCoA of fungal communities; (G) Correlation heatmap between microbial diversity indices and enzyme activities. Different lowercase letters indicate significant differences among treatments (p < 0.05, ANOVA with LSD test). * denotes p < 0.05; F_abundance, fungal abundance; F_diversity, fungal diversity; B_abundance, bacterial abundance; B_diversity, bacterial diversity; BG, β-glucosidase; NAG, N-acetylglucosaminidase; LAP, leucine aminopeptidase; PPO, polyphenol oxidase.
Principal Coordinates Analysis (PCoA) based on Bray-Curtis distances revealed significant divergence in microbial community composition among treatments. Bacterial PCoA (Figure 3E) showed clear separation between B1/B2 and B3/B4 clusters along the first principal component, indicating that SMS amendment and sunflower cultivation substantially restructured bacterial communities. Fungal PCoA (Figure 3F) exhibited similar clustering patterns, confirming treatment-dependent shifts in microbial assemblages.
Correlation analysis (Figure 3G) identified significant associations between bacterial diversity and enzyme stoichiometric ratios. Increased BG:(NAG+LAP), BG:PPO, and NAG:LAP ratios correlated with enhanced fungal diversity/richness but reduced bacterial diversity/richness (p ≤ 0.05). Additionally, negative correlations between bacterial abundance and vector angles underscored linkages between community structure and enzymatic activity dynamics.
In summary, the SR-OS2 system significantly reshaped soil microbial diversity and community structure through SMS amendment and sunflower cultivation. Initial suppression of fungal diversity was followed by robust bacterial diversification during rotation progression, demonstrating the system’s capacity to restore bacterial communities. These shifts provide critical insights into the mechanisms through which rotation systems regulate soil microecology.
3.4 Impact of the rotation system on soil microbial co-occurrence networks
The SR-OS2 significantly restructured soil microbial community composition and co-occurrence networks through SMS amendment and sunflower cultivation. In bacterial communities, the SMS application (B2) markedly increased the relative abundance of Proteobacteria and Acidobacteriota while reducing Chloroflexi, Actinobacteriota, and Gemmatimonadota. Subsequent sunflower cultivation (B3/B4) induced a progressive decline in Proteobacteria, with Acidobacteriota and Chloroflexi showing initial reduction followed by rebound, while Gemmatimonadota demonstrated an increasing trend (Figure 4A). For fungi, Ascomycota remained dominant, with SMS amendment elevating Ascomycota and Mortierellomycota but suppressing Rozellomycota, Basidiomycota, and Chytridiomycota. By B3/B4, Ascomycota increased by 9.4 and 11.1% versus B1 (p < 0.05), contrasting with drastic declines in Basidiomycota (−78%/−96%), Mortierellomycota (−87%/−88%), Rozellomycota (−95%/−100%), and Chytridiomycota (−100%/−100%) (Figure 4C).
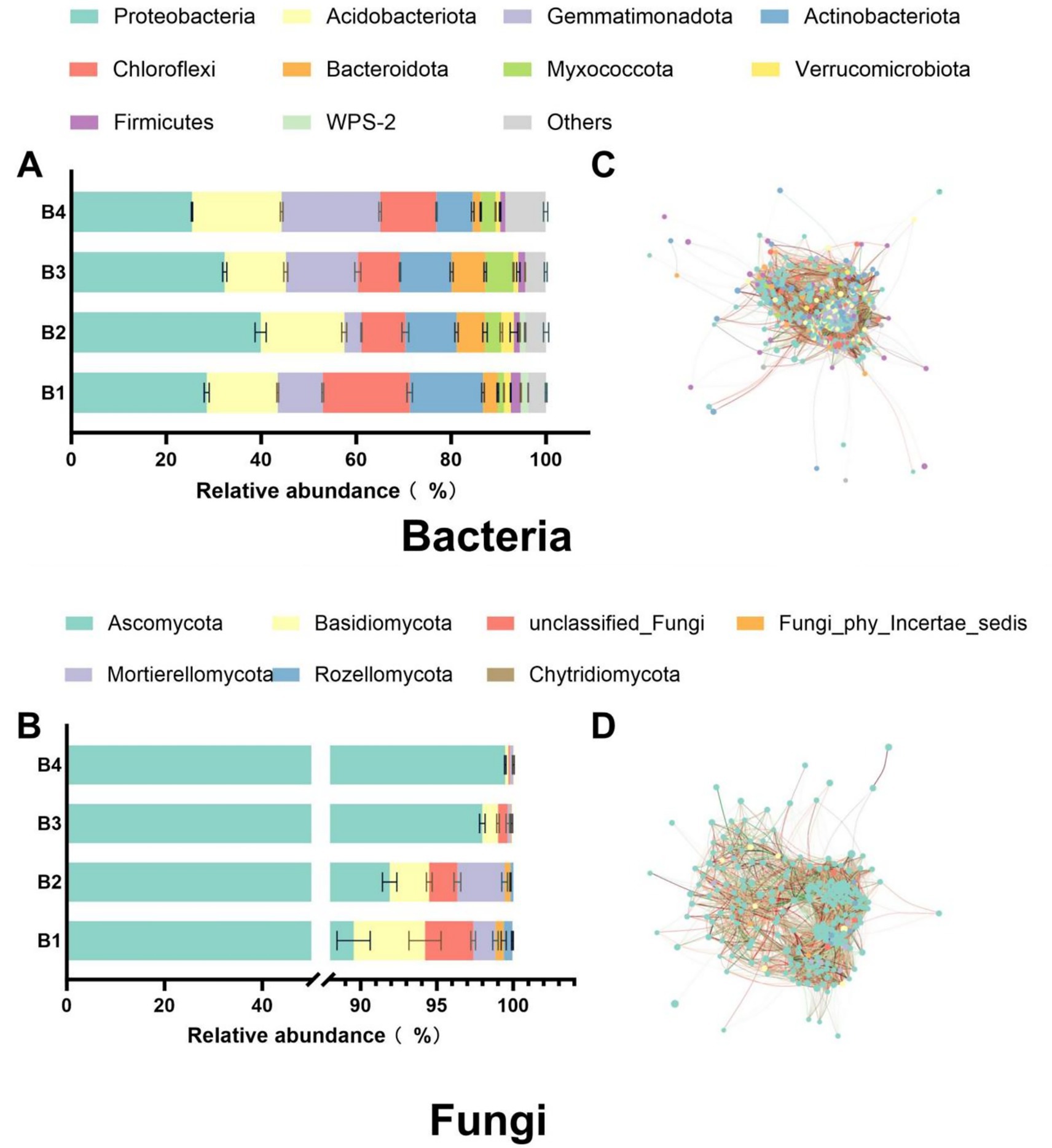
Figure 4. Phylogenetic profiling and co-occurrence network analysis of dominant bacterial and fungal phyla under the SR-OS2 system; (A) Relative abundance of top 10 bacterial phyla; (B) Relative abundance of dominant fungal phyla; (C) Co-occurrence network of top 10 bacterial phyla; (D) Co-occurrence network of dominant fungal phyla. Network edges represent significant species correlations (p < 0.05), with line thickness proportional to correlation coefficient (r). Red lines: positive correlations; Green lines: negative correlations. Node size reflects connectivity degree.
Network topology analysis revealed that SMS amendment and sunflower cultivation substantially modified microbial interactions. Bacterial networks exhibited enhanced scale and connectivity in B2-B4, evidenced by increased node counts and average degrees, with peak complexity in B3 despite slightly reduced transitivity (Figure 4B). Fungal networks showed strengthened diversity and interactions under SMS (B2), but no-till sunflower cultivation (B3/B4) reduced node counts, links, and average degrees, indicating diminished connectivity (Figure 4D). Collectively, these findings demonstrate that SMS amendment enhanced network complexity in both domains, while subsequent sunflower cultivation exerted divergent effects, further complexifying bacterial networks but simplifying fungal networks. This restructuring provides critical insights for designing soil management strategies to optimize ecological functioning.
3.5 Cluster and redundancy analyses integrating environmental factors and microbial communities
Cluster analysis and RDA, integrating environmental factors and microbial data, further elucidated the regulatory mechanisms of soil physicochemical properties on microbial community structure. Cluster analysis based on bacterial (Figure 5A) and fungal abundance (Figure 5B) with soil properties revealed phylum-specific responses to environmental factors. Proteobacteria were inhibited by TN, preferring low-N environments. In contrast, Gemmatimonadota showed significant positive correlations with S, Ca, Mn, Zn, TN, and soil SOC, indicating a competitive advantage in nutrient-rich soils. Actinobacteriota exhibited negative correlations with AK and pH, thriving in acidic, low-K conditions. Bacteroidota dominated in low-S/high-P soils, negatively correlating with S but positively with AP. These results demonstrate that soil physicochemical properties fundamentally shape microbial community assembly.
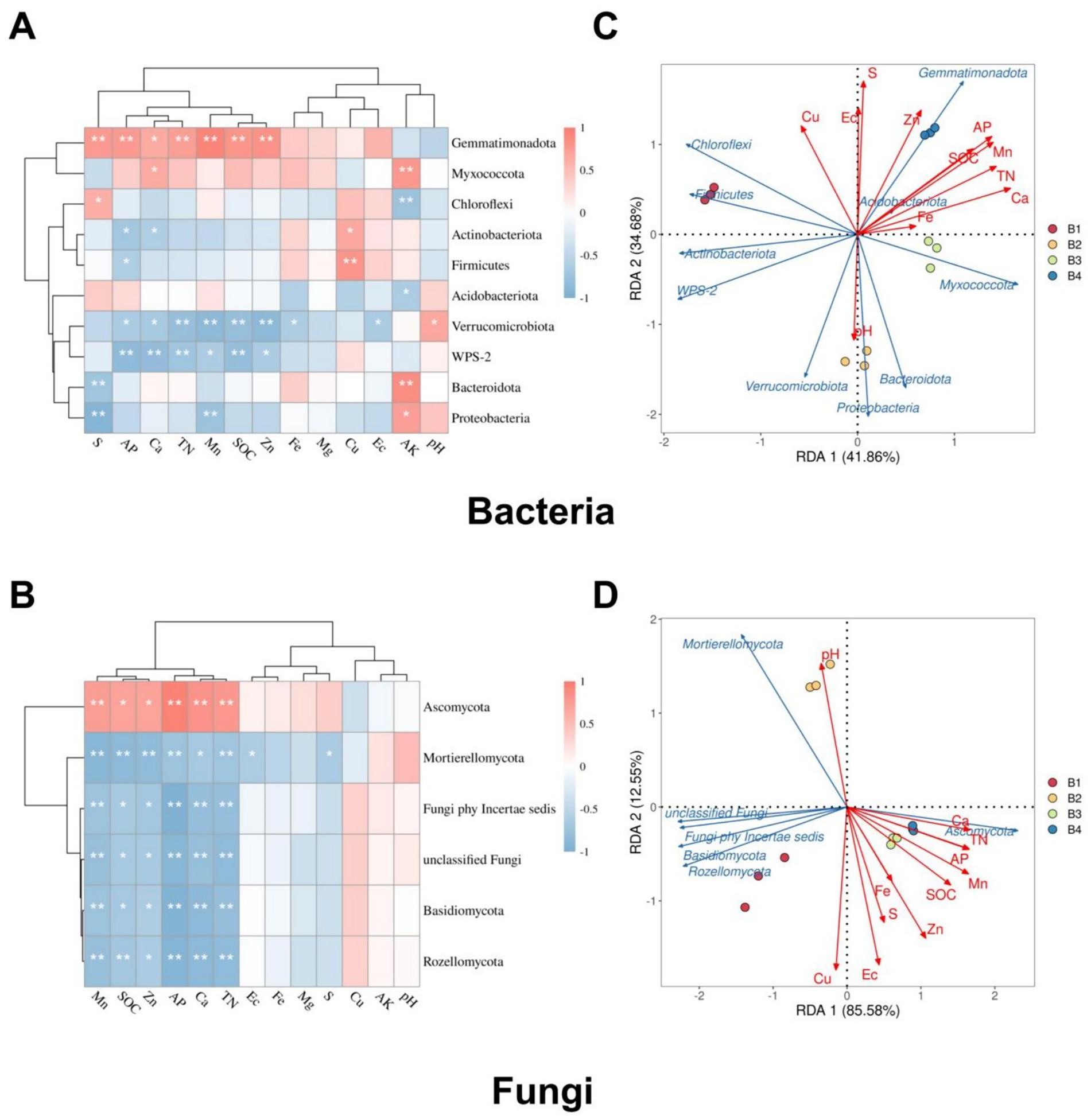
Figure 5. Integrated cluster and redundancy analyses of soil microbial phyla with environmental factors under the SR-OS2 system; (A) Cluster analysis of bacterial phyla based on soil physicochemical properties; (B) Cluster analysis of fungal phyla based on soil physicochemical properties; (C) Redundancy analysis (RDA) of bacterial phyla-environmental relationships (RDA1 = 41.86%, RDA2 = 34.68%); (D) RDA of fungal phyla-environmental relationships (RDA1 = 85.58%, RDA2 = 12.55%). Green arrows represent microbial taxa; Solid red arrows represent soil physicochemical properties; Fe, Iron; Mg, Magnesium; Ca, Calcium; Mn, Manganese; S, Sulfur; Zn, Zinc; Cu, Copper; pH, potential of hydrogen; EC, Electrical Conductivity; SOC, soil organic carbon; TN, total nitrogen; AP, available phosphorus; AK, Available Potassium.
RDA quantified environmental drivers of community variation. For bacteria, RDA1 and RDA2 explained 41.86 and 34.68% of abundance variation (cumulative 76.54%; Figure 5C). Actinobacteriota and Proteobacteria positively correlated with SOC, TN, and Ca, highlighting their competitiveness in organically enriched soils. Conversely, Acidobacteriota and Gemmatimonadota showed negative correlations with S, EC, Zn, AP, and Cu, indicating a preference for low-S, low-EC, and low-Zn environments. For fungi, RDA1 captured 85.58% of the variation with RDA2 explaining 12.55% (cumulative 98.13%; Figure 5D). Ascomycota and Basidiomycota are positively associated with TN and Ca, dominating high-N/high-Ca soils. Mortierellomycota are closely linked with SOC, iron (Fe), and Mn, while unclassified fungi and Rozellomycota were negatively regulated by S, Zn, EC, Cu, and AP, indicating competitiveness in low-S, low-Zn, low-EC, low-Cu, and low-P environments.
Collectively, these analyses demonstrate that key elements (N, Ca, S, Zn, Cu) critically influence microbial distribution patterns through physicochemical regulation of community structure, providing fundamental insights into soil microecological functionality and its governing mechanisms.
3.6 Microbial succession and functional shifts
The SR-OS2 system induced a stage-specific succession of soil microbial communities. LEfSe analysis showed that the untreated soil (B1) was dominated by generalist decomposers (e.g., Sphingomonas, Penicillium). SMS amendment (B2) enriched specialized decomposers, including the bacteria Dyella and Rhodanobacter, and the nematode-trapping fungus Arthrobotrys. Sunflower cultivation (B3/B4) further shifted the community toward nutrient-mobilizing and antipathogenic taxa, such as the phosphate solubilizer Gemmatimonas and the antipathogenic bacterium Lysobacter (Supplementary Table S3).
FUNGuild analysis revealed a functional transition in fungal trophic modes. From B1 to B4, the pure saprotrophic and parasitic-saprotrophic-symbiotic guilds increased dramatically (22.2-fold and 9.1-fold, respectively), while the parasitic-saprotrophic guild decreased (Supplementary Table S4). Compared with B1, the relative abundance of potential fungal pathogens (e.g., Fusarium) was significantly reduced in B3 and B4 treatments. This indicates a systematic shift toward saprotrophy and multifunctionality, reducing pathogen pressure.
3.7 Partial least squares path modeling (PLS-PM)
Figure 6 presents the PLS-PM elucidating the hierarchical regulatory mechanisms through which the SR-OS2 restores soil microecological functions. The model demonstrated good overall fit (GoF = 0.748), with all latent variables exhibiting average variance extracted (AVE) > 0.7, validating measurement model reliability. Two primary pathways mediated the system’s effects: (1) The biological pathway (Rotation → Soil properties → Microbial Community → Enzyme Ratios) showed rotation treatments (B1-B4) strongly driving soil physicochemical alterations (β = 0.92, p < 0.001; R2 = 0.84). Elevated SOC and TN in B3/B4 versus B1/B2 confirmed that SMS amendment, coupled with sunflower cultivation, enhanced soil C-N levels. These changes reduced fungal abundance in high-SOC treatments (B3: 286.6 vs. B1: 361.9), likely through carbon-triggered microbial metabolic compensatory mechanisms. Ultimately, bacterial diversity positively regulated enzyme stoichiometric ratios (β = 0.53, p = 0.003), evidenced by higher BG:PPO ratios in high-diversity B2 (1.42) versus B3 (0.80). (2) The physicochemical pathway (Rotation → Micronutrients → Enzyme Activity → Enzyme Ratios) revealed micronutrient-mediated enzyme activity shifts, where increased BG and PPO activities significantly reduced enzyme balance ratios (β = −0.501, p = 0.004). This was manifested by 45% greater PPO activity in B4 (45.9 μmol g−1 soil d−1) than in B1 (31.6 μmol g−1 soil d−1), alongside decreased BG:(NAG+LAP) from 1.88 to 1.24. Collectively, PLS-PM uncovered how rotation systems cascade through soil-microbe-enzyme hierarchies to restore ecological functions, providing mechanistic foundations for optimizing agricultural management.
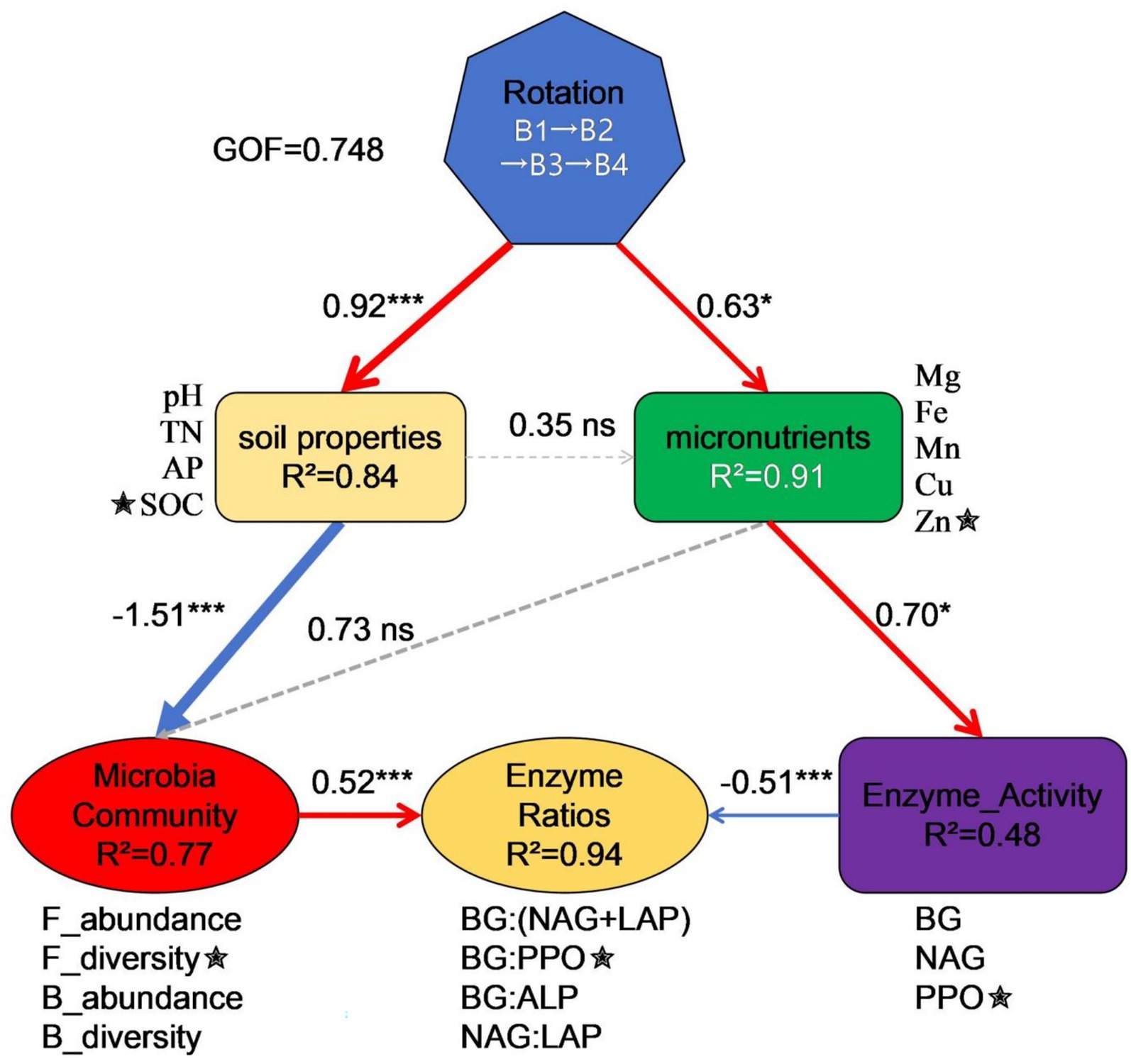
Figure 6. PLS-PM of microecological restoration mechanisms in the SR-OS2 rotation system, Solid red lines: Positive paths, Solid blue lines: Negative paths, Dashed lines: Non-significant paths, Numerical values: Path coefficients, ★: Strongest loadings, ***p < 0.001; *p < 0.05; ns p > 0.05, R2 indicates model explanatory power; GoF = 0.748; pH, potential of hydrogen; SOC, soil organic carbon; TN, total nitrogen; AP, available phosphorus; Fe, Iron; Mg, Magnesium; Mn, Manganese; Zn, Zinc; Cu, Copper; F_abundance, fungal abundance; F_diversity, fungal diversity; B_abundance, bacterial abundance; B_diversity, bacterial diversity; BG, β-glucosidase; NAG, N-acetylglucosaminidase; LAP, leucine aminopeptidase; PPO, polyphenol oxidase.
4 Discussion
4.1 Remediation effects of SR-OS2 on physicochemical properties of continuous cropping soil
This study demonstrates that the SR-OS2 significantly ameliorates physicochemical degradation in continuous cropping soils. SMS application (B2) substantially increased soil pH while reducing EC, effectively mitigating soil acidification and salinization. These findings align with Hu et al. (2020), who reported similar pH improvements through edible mushroom residue amendment (Hu et al., 2020). The pH elevation may be attributed to carbonic anhydrase in SMS catalyzing CO₂ hydration to HCO₃−, thereby buffering soil acidity (Occhipinti and Boron, 2019), while SMS’s porous structure likely immobilized free salt ions through physical adsorption (Wu et al., 2019). Regarding soil nutrients, sunflower cultivation (B3/B4) significantly enhanced SOC, TN, and AP contents. SOC accumulation likely resulted from covalent bonding of lignocellulosic components forming stable organo-mineral complexes (Kleber et al., 2021) and microbial-mediated ammonification of fungal mycelia-derived amino sugars, elevating TN (Li T. et al., 2022). AP increase may stem from SMS phosphorus mineralization and phosphate-solubilizing rhizobacteria secreting acid phosphatases (Tao and Gao, 2023). Trace element dynamics revealed Ca2+ and Zn2+ enrichment via organic acid-mediated mineral weathering (Schmalenberger et al., 2015), contrasting with Cu2+ reduction potentially involving humus chelation (Qin et al., 2023). Significant positive correlations between SOC and both TN and AP suggest SOC accumulation facilitates nutrient retention, implying enhanced SOC inputs can improve soil fertility (Abay et al., 2024). Collectively, SR-OS2 synergistically enhanced soil properties through SMS amendment and sunflower cultivation, providing critical foundations for utilizing agricultural waste in sustainable protected agriculture.
4.2 Microbial nutrient acquisition strategies revealed by extracellular enzyme activity dynamics
The SR-OS2 significantly modulated soil extracellular enzyme activities and stoichiometric ratios, revealing microbial adaptive strategies to nutrient availability shifts. Following SMS amendment (B2), LAP (nitrogen-cycling) and ALP (phosphorus-cycling) activities increased substantially while BG (carbon-cycling) activity decreased. This prioritization of N and P acquisition enzymes under nutrient constraints reflects microbial optimization of substrate capture efficiency, consistent with Sinsabaugh et al.’s framework of nutrient-dependent enzyme allocation (Sinsabaugh et al., 2009). Post-sunflower cultivation (B3), peak polyphenol oxidase (PPO) activity indicated accelerated plant residue decomposition and altered carbon utilization strategies. Sunflower root exudates rich in lignin and cellulose likely stimulated microbial production of ligninolytic enzymes, driving priming effects on recalcitrant carbon decomposition as documented by Baldrian (2008) (Baldrian, 2008). SMS amendment significantly reduced the BG:(NAG+LAP) ratio, demonstrating enhanced nitrogen-cycling capacity relative to carbon acquisition, while decreased BG:ALP and BG:PPO ratios suggested improved phosphorus-cycling efficiency. Vector analysis showed reduced vector length (indicating alleviated carbon limitation) and increased vector angle from 27.2° to 30.9° (persistent nitrogen limitation despite mitigation), aligning with Moorhead’s ecological stoichiometry principles (Moorhead et al., 2016). Collectively, these enzyme dynamics demonstrate how SR-OS2 triggers microbial metabolic reprogramming in response to nutrient shifts, providing a scientific basis for optimizing agricultural management through rotation systems.
4.3 Microbial community restructuring and alleviation mechanisms for continuous cropping obstacles
SMS amendment induced microbial community restructuring characterized by bacterial dominance: Bacterial α-diversity increased progressively during rotation, while fungal diversity declined significantly. This divergence is consistent with the observed increase in oligotrophic taxa such as Acidobacteriota and Chloroflexi (Figure 4A), which are known for their ability to degrade complex aromatic compounds present in SMS (Fierer et al., 2007). Co-occurrence network analysis revealed decreased modularity but increased connectivity density in bacterial networks, indicating SMS input enhanced functional guild cooperation (Banerjee et al., 2016). Conversely, reduced keystone species in fungal networks suggest SMS effectively disrupted pathogenic niches.
Collectively, the SR-OS2 system restructured soil microbial diversity and composition through SMS amendment and sunflower cultivation. Initial fungal suppression was succeeded by significant bacterial diversification during rotation progression, demonstrating the system’s capacity to restore bacterial communities. These shifts elucidate rotation-mediated regulatory mechanisms in soil microecology and provide scientific foundations for optimizing agricultural management.
4.4 Microbial succession as a driver of soil restoration
The SR-OS2 system drove a deterministic microbial succession that directly remediated continuous cropping obstacles. The initial community of generalist decomposers (B1) transitioned to substrate-activated specialists (B2), initiating the breakdown of complex organics and biocontrol of nematodes. Subsequent sunflower cultivation (B3/B4) enriched taxa critical for nutrient mobilization (e.g., Gemmatimonas) and pathogen suppression (e.g., Lysobacter), establishing a resilient soil state.
The concomitant functional shift in fungi—from pathotrophic to saprotrophic dominance—reflects a systematic reduction in pathogen pressure. Thus, the system’s efficacy stems from its ability to orchestrate a phased functional succession within the microbiome, sequentially activating processes that restore soil health.
4.5 Driving effects of nutrient limitation on microbial communities and regulatory roles of the rotation system
Partial Least Squares Path Modeling (PLS-PM; GoF = 0.748) revealed dual cascading pathways through which the Stropharia rugosoannulata-Ornamental Sunflower Rotation System (SR-OS2) restores soil microecological function. In the biological pathway, SMS amendment significantly elevated soil carbon and nitrogen levels (SOC/TN, β = 0.92, p < 0.001; R2 = 0.84), consistent with organic amendments’ role in soil fertility reconstruction (Lal, 2004). High carbon inputs triggered microbial metabolic compensatory mechanisms (Fontaine et al., 2003), suppressing fungal abundance by 20.8% in B3 versus B1, while bacterial diversity (B_diversity) positively regulated enzyme balance (β = 0.53, p = 0.003) via functional redundancy (Wagg et al., 2019). In the physicochemical pathway, rotation altered micronutrient bioavailability, activating metalloenzyme-dependent polyphenol oxidase (PPO) (45% activity increase in B4 vs. B1) (Sinsabaugh, 2010). Coordinated enhancement of β-glucosidase (BG) and PPO activities induced microbial resource allocation trade-offs (Moorhead et al., 2016), significantly reducing enzyme balance ratios (β = −0.501, p = 0.004) and shifting soil organic matter transformation toward recalcitrant carbon decomposition (Cotrufo et al., 2013). Collectively, SR-OS2 achieves waste valorization and fertility enhancement through these synergistic pathways, with enzyme activity ratios (e.g., BG:PPO) serving as sensitive biological indicators for optimizing rotation design.
It is important to note that the primary objective of this study was to decipher the soil-mediated mechanisms (physicochemical and microecological) through which the SR-OS2 system alleviates continuous cropping obstacles. Therefore, we focused our experimental design and resources on intensive soil sampling and analysis. While plant productivity and health are the ultimate agronomic goals, they are lagging indicators that result from the complex interplay of soil conditions remediated by our system. The significant improvements in soil health parameters reported here—such as the alleviation of nutrient limitations, the restructuring of the microbial community away from pathogenic dominance, and the enhancement of beneficial bacterial networks—are well-established precursors to improved plant growth and yield. Future studies will explicitly measure the transgenerational effects of these soil improvements on sunflower agronomic traits to provide a complete assessment of the SR-OS2 system’s benefits.
5 Conclusion
The SR-OS2 system synergistically improved physicochemical properties of continuous cropping soils through integrated SMS application and rotational cultivation. It enhanced nitrogen/phosphorus-cycling enzyme activities, restructured microbial communities, effectively alleviated nutrient limitations, and suppressed pathogen enrichment. These findings establish a technical framework for ecologically controlling sunflower continuous cropping obstacles while providing novel approaches for agricultural waste valorization and sustainable protected agriculture. Future research should elucidate mechanisms of functional microorganisms in SMS, optimize rotation cycles and amendment rates, and advance precision management of this system.
Data availability statement
The data of this study have been deposited in the Genome Sequence Archive database, and the accession number is PRJCA049688.
Author contributions
JL: Data curation, Writing – original draft, Investigation. YC: Writing – original draft, Investigation. JY: Investigation, Writing – original draft. HY: Investigation, Writing – original draft. XY: Writing – review & editing, Investigation. XZ: Writing – review & editing, Investigation. FY: Investigation, Writing – review & editing. ZZ: Investigation, Funding acquisition, Writing – review & editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the China Agriculture Research System (No. CARS-20).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1671484/full#supplementary-material
Footnotes
References
Abay, P., Gong, L., Luo, Y., Zhu, H., and Ding, Z. (2024). Soil extracellular enzyme stoichiometry reveals the nutrient limitations in soil microbial metabolism under different carbon input manipulations. Sci. Total Environ. 913:169793. doi: 10.1016/j.scitotenv.2023.169793
Baldrian, P. (2008). “Enzymes of saprotrophic basidiomycetes” in British mycological society symposia series. eds. L. Boddy, J. C. Frankland, and P. West (Cambridge: Academic Press), 19–41.
Banerjee, S., Kirkby, C. A., Schmutter, D., Bissett, A., Kirkegaard, J. A., and Richardson, A. E. (2016). Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 97, 188–198. doi: 10.1016/j.soilbio.2016.03.017
Cotrufo, M. F., Wallenstein, M. D., Boot, C. M., Denef, K., and Paul, E. (2013). The microbial efficiency-matrix stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Glob. Chang. Biol. 19, 988–995. doi: 10.1111/gcb.12113
Fierer, N., Breitbart, M., Nulton, J., Salamon, P., Lozupone, C., Jones, R., et al. (2007). Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73, 7059–7066. doi: 10.1128/AEM.00358-07
Fontaine, S., Mariotti, A., and Abbadie, L. (2003). The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 35, 837–843. doi: 10.1016/S0038-0717(03)00123-8
Gong, S., Chen, C., Zhu, J., Qi, G., and Jiang, S. (2018). Effects of wine-cap Stropharia cultivation on soil nutrients and bacterial communities in forestlands of northern China. PeerJ 6:e5741. doi: 10.7717/peerj.5741
Hao, H., Yue, Y., Chen, Q., Yang, Y., Kuai, B., Wang, Q., et al. (2024). Effects of an efficient straw decomposition system mediated by Stropharia rugosoannulata on soil properties and microbial communities in forestland. Sci. Total Environ. 916:170226. doi: 10.1016/j.scitotenv.2024.170226
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A., and Afkhami, M. E. (2021). Environmental stress destabilizes microbial networks. ISME J. 15, 1722–1734. doi: 10.1038/s41396-020-00882-x
Hu, L., Li, Y., Tian, S., Wei, J., Liao, D., Feng, M., et al. (2020). Fungi residue returning: effects on soil physicochemical characters, microorganisms and enzyme activity in vegetable field. Chin. Agric. Sci. Bull. 36, 98–104. doi: 10.11924/j.issn.1000-6850.casb18080068
Jansson, J. K., and Baker, E. S. (2016). A multi-omic future for microbiome studies. Nat. Microbiol. 1:16049. doi: 10.1038/nmicrobiol.2016.49
Kleber, M., Bourg, I. C., Coward, E. K., Hansel, C. M., Myneni, S. C. B., and Nunan, N. (2021). Dynamic interactions at the mineral–organic matter interface. Nat. Rev. Earth Environ. 2, 402–421. doi: 10.1038/s43017-021-00162-y
Lal, R. (2004). Soil carbon sequestration to mitigate climate change. Geoderma 123, 1–22. doi: 10.1016/j.geoderma.2004.01.032
Li, F., Kong, Q., Zhang, Q., Wang, H., Wang, L., and Luo, T. (2020). Spent mushroom substrates affect soil humus composition, microbial biomass and functional diversity in paddy fields. Appl. Soil Ecol. 149:103489. doi: 10.1016/j.apsoil.2019.103489
Li, Y., Shi, C., Wei, D., Gu, X., Wang, Y., Sun, L., et al. (2022). Soybean continuous cropping affects yield by changing soil chemical properties and microbial community richness. Front. Microbiol. 13:1083736. doi: 10.3389/fmicb.2022.1083736
Li, T., Zhang, J., Wang, X., Hartley, I. P., Zhang, J., and Zhang, Y. (2022). Fungal necromass contributes more to soil organic carbon and more sensitive to land use intensity than bacterial necromass. Appl. Soil Ecol. 176:104492. doi: 10.1016/j.apsoil.2022.104492
Moorhead, D. L., Sinsabaugh, R. L., Hill, B. H., and Weintraub, M. N. (2016). Vector analysis of ecoenzyme activities reveal constraints on coupled C, N and P dynamics. Soil Biol. Biochem. 93, 1–7. doi: 10.1016/j.soilbio.2015.10.019
Occhipinti, R., and Boron, W. F. (2019). Role of carbonic anhydrases and inhibitors in Acid-Base physiology: insights from mathematical Modeling. Int. J. Mol. Sci. 20:3841. doi: 10.3390/ijms20153841
Puttha, R., Venkatachalam, K., Hanpakdeesakul, S., Wongsa, J., Parametthanuwat, T., Srean, P., et al. (2023). Exploring the potential of sunflowers: agronomy, applications, and opportunities within bio-circular-green economy. Horticulturae 9:1079. doi: 10.3390/horticulturae9101079
Qin, Q., Zhang, Z., and Song, Z. (2023). Research progress on sources, characteristics of humus and mechanism of interaction with heavy metals in soil. Environ. Chem. 42, 1899–1910. doi: 10.7524/j.issn.0254-6108.2022010801
Rillig, M. C., Muller, L. A. H., and Lehmann, A. (2017). Soil aggregates as massively concurrent evolutionary incubators. ISME J. 11, 1943–1948. doi: 10.1038/ismej.2017.56
Schmalenberger, A., Duran, A. L., Bray, A. W., Bridge, J., Bonneville, S., Benning, L. G., et al. (2015). Oxalate secretion by ectomycorrhizal Paxillus involutus is mineral-specific and controls calcium weathering from minerals. Sci. Rep. 5:12187. doi: 10.1038/srep12187
Sinsabaugh, R. L. (2010). Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 42, 391–404. doi: 10.1016/j.soilbio.2009.10.014
Sinsabaugh, R. L., Hill, B. H., and Follstad Shah, J. J. (2009). Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 462, 795–798. doi: 10.1038/nature08632
Tao, D., and Gao, Y. (2023). Advances on the strategies of soil phosphate solubilizing microorganisms to promote plant phosphorus uptake. Acta Ecol. Sin. 43, 4390–4399. doi: 10.5846/stxb202111193253
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G. A. (2019). Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10:4841. doi: 10.1038/s41467-019-12798-y
Wu, Q., Xian, Y., He, Z., Zhang, Q., Wu, J., Yang, G., et al. (2019). Adsorption characteristics of pb(II) using biochar derived from spent mushroom substrate. Sci. Rep. 9:15999. doi: 10.1038/s41598-019-52554-2
Keywords: continuous cropping obstacles, spent mushroom substrate, microbial co-occurrence networks, enzyme stoichiometry, Stropharia rugosoannulata, soil microecological restoration
Citation: Lu J, Chen Y, Yan J, Ye H, Ying X, Zhou X, Yao F and Zhou Z (2025) Synergistic remediation of continuous cropping obstacles in facility agriculture: insights from the Stropharia rugosoannulata-Ornamental Sunflower Rotation System. Front. Microbiol. 16:1671484. doi: 10.3389/fmicb.2025.1671484
Edited by:
Jincai Ma, College of New Energy and Environment/Jilin Unversity, ChinaReviewed by:
WuPing Yan, Jiangxi Agricultural University, ChinaYuanyuan Yan, Nanjing Normal University, China
Copyright © 2025 Lu, Chen, Yan, Ye, Ying, Zhou, Yao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zufa Zhou, emhvdXp1ZmEyMDI1QDE2My5jb20=
 Jia Lu
Jia Lu Yan Chen3
Yan Chen3 Zufa Zhou
Zufa Zhou