- 1College of Animal Science and Veterinary Medicine, Henan Institute of Science and Technology, Xinxiang, China
- 2Postdoctoral Research Base, Henan Institute of Science and Technology, Xinxiang, China
- 3Postdoctoral Research Base, College of Veterinary Medicine, Henan Agricultural University, Zhengzhou, China
- 4Ministry of Education Key Laboratory for Animal Pathogens and Biosafety, Zhengzhou, China
- 5Laboratory of Functional Microbiology and Animal Health, Henan University of Science and Technology, Luoyang, China
Introduction: Carbapenem-resistant Acinetobacter baumannii (CRAB) poses a significant global health challenge owing to its high mortality rates and widespread antibiotic resistance. While the clinical utility of last-resort antibiotics, such as colistin, remains limited. Consequently, developing novel antimicrobial agents is imperative. Antimicrobial peptides have emerged as promising candidates against multidrug-resistant pathogens. Animal venom constitutes a rich reservoir of bioactive peptides.
Methods: In this study, in vitro experiments were conducted to assess the antibacterial activity of the scorpion-derived peptide AaeAP2a against CRAB, its inhibition of biofilm formation, as well as its stability and biocompatibility. Additionally, the antibacterial mechanism was investigated, and in vivo efficacy was evaluated using a mouse model of peritonitis-associated sepsis.
Results: AaeAP2a exhibits potent antibacterial activity against CRAB and a significant inhibitory effect on biofilm formation. Moreover, AaeAP2a maintains high stability under a broad range of stressful physicochemical conditions and exhibits promising biocompatibility in vitro. Mechanistically, AaeAP2a disrupts bacterial membrane integrity, increases membrane permeability, reduces the NAD+/NADH ratio, dissipates the proton motive force, decreases ATP production, and induces reactive oxygen species and hydroxyl radical accumulation. Moreover, in a mouse model of peritonitis-associated sepsis, AaeAP2a treatment enhanced survival rates and reduced bacterial burdens in key organs.
Discussion: These findings underscore the potential of AaeAP2a as a promising therapeutic agent for CRAB infections, offering novel strategies for addressing antimicrobial resistance.
1 Introduction
The escalating antibiotic resistance crisis has extended beyond the healthcare sector, posing a systemic threat within the One Health framework (Aslam et al., 2021; de la Rocque et al., 2023). The World Health Organization (WHO) named carbapenem-resistant Acinetobacter baumannii (CRAB) as the pathogen with the highest alert status in 2017 (Yadav et al., 2025). The emergence of CRAB has further heightened morbidity and mortality rates related to A. baumannii infections, with these rates ranging from 33 to 86% (Monteiro et al., 2025). Notably, studies have observed that the resistance rate of Acinetobacter baumannii to carbapenems surged from 4.7 to 62.8% between 2016 and 2020 (Ju et al., 2023). This alarming status is attributable not only to the overuse of antibiotics but also to the rapid dissemination of drug-resistant genes across various hosts (e.g., humans, companion animals) and environmental settings (e.g., healthcare facilities, pet hospitals, soil, and water ecosystems) (Castañeda-Barba et al., 2024). According to growing data, companion animals, due to their shared close-contact environments with humans (such as households and veterinary clinics), facilitate the exchange of drug-resistant bacteria and genes, thus serving as key vectors for the transmission of antimicrobial resistance (AMR) (Zhao et al., 2022; Price et al., 2024). Such cross-species transmission complicates traditional infection control measures and underscores the necessity of shifting from a “human-centered” approach to a holistic One Health perspective. Colistin is the current last-line defense antibiotic for CRAB; nonetheless, in recent years, there has been a growing number of reports on colistin-resistant Acinetobacter baumannii (Liu et al., 2022). Moreover, its clinical utility is limited due to its nephrotoxicity and neurotoxicity (Guzman et al., 2025a, 2025b). Additionally, colistin overuse has led to the emergence of the mcr-1 resistance gene, first identified in 2015 and spread through plasmids. This accelerates colistin resistance and further jeopardizes its clinical effectiveness (Dalmasso et al., 2023). In light of these challenges, there is an urgent scientific imperative to develop safe and effective alternative antimicrobial agents.
Due to their unique mechanisms of action and broad-spectrum efficacy (Bucataru and Ciobanasu, 2024), antimicrobial peptides (AMPs), which are essential components of the innate immune system, are a promising treatment for multidrug-resistant (MDR) infections (Gao et al., 2024). In addition to its primary neurotoxins, scorpion venom also contains a wide range of bioactive molecules, including AMPs that exhibit antibacterial, antiviral, immunosuppressive, analgesic, and anticancer effects (Rabea et al., 2025; Xin et al., 2025). This study introduces AaeAP2a, an optimized analog of the naturally derived AaeAP2, originally isolated from scorpion venom. Although AaeAP2 exhibits broad-spectrum antimicrobial activity, its efficacy against Gram-negative bacteria is limited (MIC for Escherichia coli >512 μg/mL). Conversely, the modified and synthesized analog AaeAP2a significantly reduces the MIC for E. coli to 16 μg/mL and demonstrates enhanced inhibitory effects on the proliferation of cancer cells, including lung (H460), breast (MB435S, MCF-7), and prostate (PC-3) cancer cell lines (Du et al., 2015). Nevertheless, the antibacterial activity and underlying mechanisms of AaeAP2a against MDR Gram-negative pathogens, particularly CRAB, remain insufficiently characterized.
The objective of this study was to investigate the antimicrobial activity and mechanisms of AaeAP2a against CRAB. Using a CRAB strain isolated from a pet dog as the target pathogen, we observed that AaeAP2a exhibited significant antibacterial activity and dose-dependent inhibition of biofilm formation. Moreover, AaeAP2a demonstrated excellent stability, promising biocompatibility in vitro, and a low propensity for inducing resistance. Mechanistically, AaeAP2a disrupted both inner and outer bacterial membranes, increased membrane permeability, dissipated membrane potential, inhibited ATP synthesis, and induced oxidative stress, cumulatively leading to bacterial death. Notably, AaeAP2a also proved effective in a mouse model of peritonitis-associated sepsis.
2 Materials and methods
2.1 Antimicrobial peptide
The antimicrobial peptide AaeAP2a (FLFKLIPKAIKGLVKAIRK; molecular weight: 2183.84 Da) was synthesized by GL Biochemistry (Shanghai, China) via solid-phase chemical synthesis. The purity of the peptide (>95%) was confirmed by reversed-phase high-performance liquid chromatography (RP-HPLC), and its sequence was validated using electrospray ionization mass spectrometry (ESI-MS).
2.2 Bacterial strains and gene detection
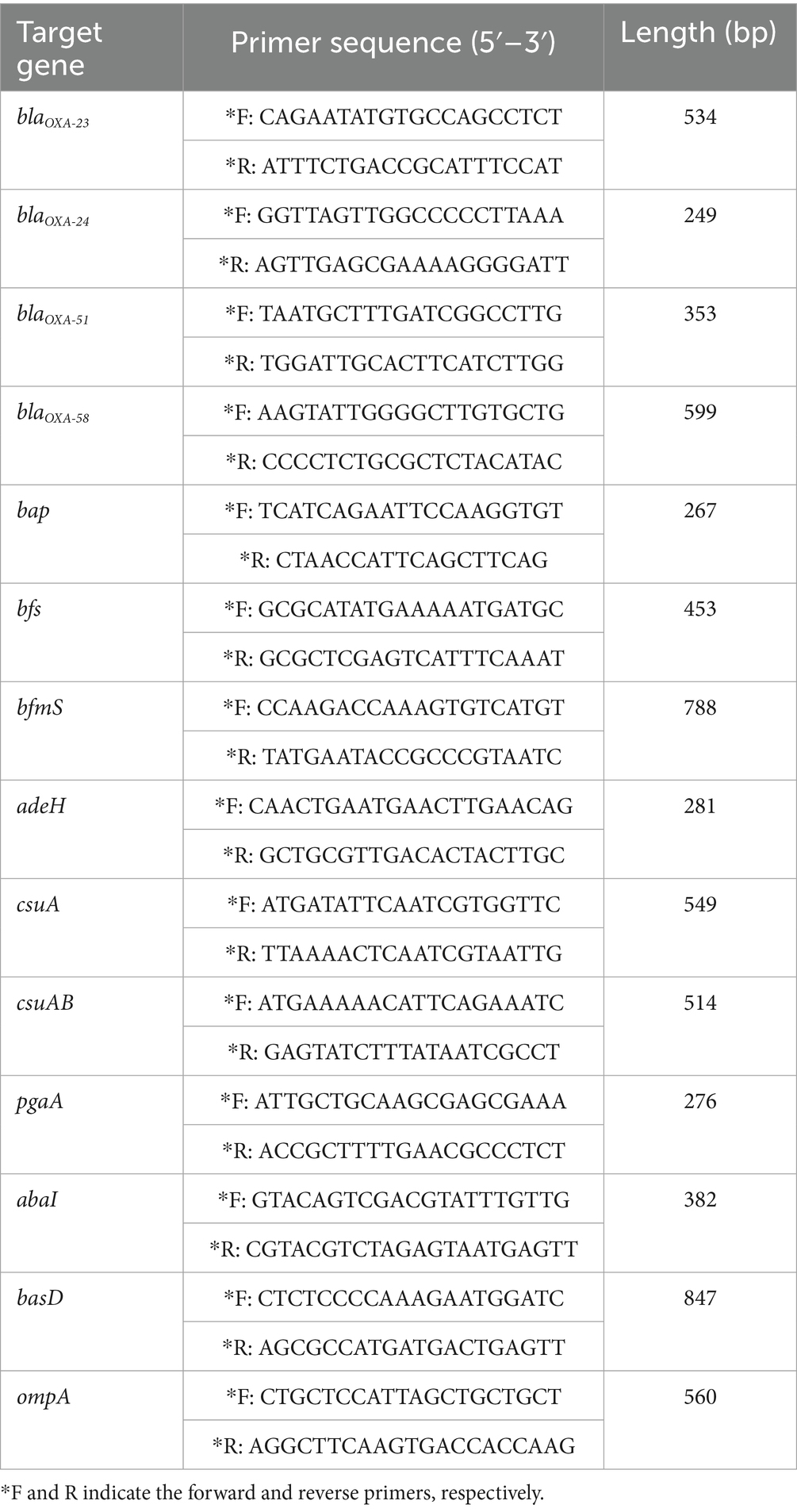
The A. baumannii strain ATCC 19606 was obtained from the American Type Culture Collection (ATCC). The CRAB 236 antibiotic-resistant clinical strain maintained in our laboratory was originally isolated from a pet dog. Polymerase chain reaction (PCR) was used to amplify resistance genes, including blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-58. Additionally, the virulence genes bap, bfs, bfmS, adeH, csuA, csuAB, pgaA, abaI, basD, and ompA were detected (Table 1).
2.3 Evaluation of inhibitory effect (minimum inhibitory concentration, MIC)
The inhibitory activity of the antimicrobial peptide was assessed using the broth microdilution method following the CLSI M100-ED34 standard (Humphries et al., 2021). A single colony of A. baumannii was isolated and inoculated into Mueller-Hinton Broth (MHB), then incubated overnight at 37 °C with agitation at 180 rpm. The overnight cultures were adjusted to a 0.5 McFarland turbidity standard, after which it was diluted 1:100 in MHB medium. The MICs of tetracycline, ciprofloxacin, gentamicin, ceftriaxone, colistin, meropenem, and the antimicrobial peptide AaeAP2a for the CRAB 236 strain were subsequently determined using the broth microdilution method. A. baumannii ATCC 19606 served as the control strain, and the experiments were conducted in triplicate. The Mueller-Hinton Broth medium and the antibiotics used (tetracycline, ciprofloxacin, gentamicin, ceftriaxone, colistin, meropenem) were all purchased from Macklin (China).
2.4 Time-kill curve
The bactericidal potential of AaeAP2a was evaluated using time-kill curve assays on A. baumannii strains ATCC 19606 and CRAB 236 (Yi et al., 2022). After the bacterial cultures were diluted in MHB at a 1:100 ratio, the AaeAP2a was applied in final concentrations corresponding to 0.5, 1, 2, and 4 × MIC, with a control group without the AMPs. Samples were analyzed using 10-fold serial dilutions at 0, 2, 4, 6, 8, 10, and 12 h, and 100 μL aliquots were placed on Mueller-Hinton Agar (MHA) plates. The plates were incubated at 37 °C for 24 h before colony enumeration. Triplicate assays were performed for each condition. Mueller-Hinton Agar (MHA) medium was purchased from Macklin (China); and the incubator was from Thermo Fisher Scientific (USA).
2.5 Determination of biofilm inhibitory activity
In this study, the crystal violet staining method was employed to evaluate the inhibitory efficacy of the antimicrobial peptide AaeAP2a on A. baumannii biofilm formation. An A. baumannii overnight culture was diluted in MHB to yield a bacterial suspension of 1 × 107 CFU/mL; this suspension was prepared from cultures incubated at 37 °C with agitation at 180 rpm. Subsequently, 150 μL of the suspension was dispensed into each well of a sterile 96-well plate. AaeAP2a was added at final concentrations ranging from 0.5 × to 4 × the MIC. A negative control group, which received no AMP, was established, while colistin at 1 × MIC served as the positive control. After incubation at 37 °C for 24 h, the supernatant was removed, and the remaining biofilm was subjected to staining with crystal violet, followed by washing, and then solubilization. The absorbance was then measured at 570 nm to quantify the biofilm mass (Grasteau et al., 2011). All experiments were performed in triplicate. All reagents used in this experiment were obtained from Macklin (China). The incubator and microplate reader were from Thermo Fisher Scientific (United States); and the sterile 96-well microplates were purchased from Corning (United States).
2.6 Stability analysis
We thoroughly examined the effects of storage at room temperature, repeated freeze–thaw cycles, temperature variations, pH levels, salt ions, urea, and protease treatment on the antimicrobial activity of peptide AaeAP2a (Zhu et al., 2022). Through 0 to 12 freeze–thaw iterations, AaeAP2a was incubated for 1 h at various temperatures (0, 25, 50, 75, and 100 °C) and pH values (3 to 11). Additionally, the peptide was incubated with four salt ions—NaCl, KCl, CaCl2, and MgCl2—at concentrations of 0, 10, 50, 100, 150, and 200 mmol/mL, as well as with urea at concentrations of 0, 2, 4, 6, 8, and 10 mmol/mL for 1 h. Furthermore, AaeAP2a was treated with proteinase K or trypsin at concentrations ranging from 0 to 100 μg/mL (i.e., 0, 20, 40, 60, 80, and 100 μg/mL) for 1 h at 37 °C. The changes in its antimicrobial activity were quantitatively assessed using the MIC method. All assays were conducted in triplicate. Notably, for this section, only the highly representative standard strain ATCC 19606 was specifically selected as the experimental strain. Proteinase K, Trypsin, and Urea were obtained from Beyotime Biotechnology (China), while all other reagents were purchased from Macklin (China).
2.7 Biosafety assay
To systematically evaluate the biosafety of the antimicrobial peptide AaeAP2a, we assessed its hemolytic activity and cytotoxicity (Liu et al., 2020a; Zhou F. et al., 2022). A 4% (v/v) erythrocyte suspension was prepared from freshly collected anticoagulated mouse blood by repeatedly resuspending and washing the cells three times with phosphate-buffered saline (PBS), following the established protocol. This suspension was incubated with a series of AaeAP2a portions (0, 2, 4, 8, 16, 32, and 64 × MIC) at 37 °C for 1 h. In this test, 1% (v/v) Triton X-100 was used as the positive control. Three replicates per treatment group were used to study OD at 540 nm using a microplate reader. The cytotoxicity of AaeAP2a on human embryonic kidney 293 T cells was tested using the Cell Counting Kit - 8 (CCK-8). In this experiment, different AaeAP2a concentrations (0, 0.5, 1, 2, 4, 8, 16, and 32 MIC) were added to DMEM-containing wells containing 1 × 104 cells/well and 10% (v/v) fetal bovine serum, followed by 24-h incubation at 37 °C. The CCK-8 assay was thus performed, with three replicates recorded for each concentration.
2.8 Assessment of resistance development
A. baumannii ATCC 19606 in the exponential growth phase was inoculated into MHB supplemented with a sub-inhibitory concentration of AaeAP2a (0.5 × MIC). Colistin served as the positive control at a 0.5 × MIC, while an untreated group was included as the negative control. Following incubation at 37 °C with shaking at 180 rpm for 24 h, the MIC for each group was determined. This incubation and MIC determination procedure was repeated 20 times, after which the fold change in MIC relative to the initial baseline was calculated (Pollard et al., 2012). Sources of key reagents used in the experiment are as follows: Phosphate-Buffered Saline (PBS), 1% Triton X-100 solution, and Cell Counting Kit-8 (CCK-8) were all purchased from Beyotime Biotechnology (China); Dulbecco’s Modified Eagle Medium (DMEM) and Fetal Bovine Serum (FBS) were both sourced from Gibco (United States).
2.9 Outer membrane permeability assay
Bacterial cells from an overnight culture of activated A. baumannii were collected by centrifugation, washed twice with 5 mM HEPES, and resuspended to an OD600 of 0.5 (approximately 1.5 × 108 CFU/mL). The fluorescent probe N-phenylnaphthylamine (Merck; 1-N-phenyl-naphthylamine, NPN) was then added to a final concentration of 10 μM, and the mixture was incubated at 37 °C for 30 min (Zhu et al., 2022). Subsequently, varying concentrations (0.5–4 × MIC) of AaeAP2a or colistin (1 × MIC) were introduced, and the samples were further incubated for 1 h. The fluorescence intensity was measured at an excitation wavelength of 350 nm and an emission wavelength of 420 nm.
2.10 Assessment of cytoplasmic membrane integrity
A cell suspension was prepared as described in section 2.9, and propidium iodide (PI) was added to a final concentration of 0.5 μM. AaeAP2a was administered at concentrations ranging from 0.5 to 4 × MIC, with colistin (1 × MIC) as a positive control after the initial incubation at 37 °C for 30 min. After an additional hour of incubation, a microplate reader with excitation and emission frequencies of 535 and 615 nm was used to determine the light strength (Zhu et al., 2022). Propidium Iodide (PI) and colistin were purchased from Macklin (China).
2.11 Scanning electron microscopy analysis
Scanning Electron Microscopy (SEM) was used to evaluate the effects of the antimicrobial peptide AaeAP2a on the morphology of A. baumannii cells. Overnight-cultured A. baumannii cells were washed with PBS and resuspended to an OD600 of 0.5. AaeAP2a was added to a final concentration of 4 × MIC, and the samples were incubated at 37 °C for 2 and 4 h, respectively. Following incubation, the samples were fixed in a 2.5% glutaraldehyde solution at 4 °C overnight. The fixed samples were subsequently dehydrated using a graded ethanol series (30, 50, 70, 90, and 100%), dried, sputter-coated with a conductive metal, and finally examined by SEM. SEM analysis was performed with a chamber vacuum of ≤2 × 10−4 Pa, an acceleration voltage of 3.00 kV, a probe current of 100 pA, and a working distance of 7.00 mm. The 2.5% glutaraldehyde fixative was obtained from Solarbio (China); and scanning electron microscopy (SEM) was performed using a ZEISS Sigma 300 instrument from Oberkochen, Germany.
2.12 Determination of cell membrane potential
The bacterial suspension was prepared as described in section 2.9, and 3,3’-Dipropylthiadicarbocyanine iodide [DiSC3(5)] was added to a final concentration of 0.5 μM, and the mixture was incubated at 37 °C for 30 min. Subsequently, AaeAP2a (0.5–4 × MIC) and colistin (at 1 × MIC, serving as the positive control) were added, whereas an untreated control group was maintained without drug administration. The bacterial suspension was incubated at 37 °C for an additional hour, and the fluorescence intensity was measured using a microplate reader at excitation/emission wavelengths of 622/670 nm (Dombach et al., 2020). The DiSC3(5) probe was obtained from Thermo Fisher Scientific (United States).
2.13 Determination of cell pH gradient
A bacterial suspension was prepared as described in Section 2.9, and the pH-sensitive fluorescent probe 2′,7′-bis(carboxyethyl)-5(6)-carboxyfluorescein acetate methyl ester (BCECF-AM, 20 μM) was added for incubation. After that, AaeAP2a (0.5–4 × MIC) and colistin (1 × MIC) were introduced, and the cells were kept at 37 °C for 1 h. Each sample was analyzed sequentially using a microplate reader with 500 nm and 522 nm excitation and emission wavelengths. The pH-sensitive fluorescent probe BCECF-AM was purchased from Invitrogen (Thermo Fisher Scientific, United States).
2.14 ATP assay
According to the reference’s instructions (Yi et al., 2022), an ATP assay kit measured intracellular ATP levels in A. baumannii cells (Beyotime Biotechnology, China). Bacterial cultures were rinsed with PBS (pH 7.4) and adjusted to an OD600 of 0.5. The cells were then exposed to AaeAP2a (0.5–4 × MIC) or colistin (1 × MIC) for 1 h. Following incubation, the supernatant was removed after centrifugation, and the bacterial pellet was lysed using the ATP experiment lysis buffer. The resulting supernatant was subsequently used to identify intracellular ATP levels by a microplate reader.
2.15 Determination of total NAD+/NADH content
As previously described, the total NAD+/NADH content in A. baumannii was determined using the Beyotime NAD+/NADH Assay Kit based on the WST − 8 process (Li et al., 2019). Bacterial cultures, adjusted to an OD600 of 0.5, were incubated with various concentrations (0.5–4 × MIC) of AaeAP2a or colistin (1 × MIC) for 1 h. After centrifugation, the supernatant was removed, and the bacterial pellet was lysed using lysozyme. The lysate was soon centrifuged to obtain the supernatant, which was then incubated in the dark. At 450 nm, the absorbance was measured using a microplate reader. The NAD+/NADH Assay Kit (WST-8 method) was obtained from Beyotime Biotechnology (China); lysozyme was purchased from Sangon Biotech (China).
2.16 Determination of ROS content
Log-phase A. baumannii cultures adjusted to an OD600 of 0.5 were incubated with 10 μM 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) for 30 min, followed by treatment with varying concentrations (0.5–4 × MIC) of AaeAP2a or colistin (1 × MIC) for 1 h. Fluorescence strength was determined using a microplate reader at excitation and emission wavelengths of 488 nm and 525 nm, respectively (Liu et al., 2020b). The Reactive Oxygen Species Assay Kit containing the DCFH-DA fluorescent probe was obtained from Beyotime Biotechnology (China).
2.17 Measurement of hydroxyl radicals (•OH)
In the logarithmic growth phase, bacteria were washed and adjusted to an OD600 of 0.5 before 30 min of incubation with the hydroxyphenyl fluorescein (HPF) probe. The cells were then exposed to a range of AaeAP2a (0.5–4 × MIC) or colistin (1 × MIC, serving as a positive control). The treated cells were kept in the dark for an additional hour. The fluorescence intensity at 495 and 515, both at excitation and emission wavelengths, was determined using a microplate reader (Sugimoto et al., 2021). The fluorescent probe hydroxyphenyl fluorescein (HPF) and colistin were purchased from Macklin (China).
2.18 In vivo efficacy evaluation of AaeAP2a
To evaluate the antimicrobial efficacy of AaeAP2a in vivo, specific pathogen-free (SPF) grade BALB/c mice, aged 6 to 8 weeks and weighing 20 ± 2 g, with an equal sex distribution, were used to establish a mouse peritonitis-associated sepsis model. The mice were given 6.75 × 106 CFU of CRAB 236 via intraperitoneal injection. Animals were randomly assigned to five groups (n = 10 per group): a PBS control group, a CRAB 236-infected group, and three treatment groups receiving low (5 mg/kg), medium (10 mg/kg), or high (20 mg/kg) doses of AaeAP2a. Treatment was administered via oral gavage at 2, 24, and 48 h post-infection, and the survival rate was monitored over 72 h. SPF-level BALB/c mice were obtained from Vital River Laboratory Animal Technology Co., Ltd. (China).
In a subsequent experiment under similar conditions, mice were administered AaeAP2a by gavage 2 h post-infection. Twelve hours post-drug administration, the mice were euthanized via cervical dislocation. Blood, liver, and spleen tissues were harvested and homogenized in sterile PBS, and serial dilutions were prepared. An aliquot of 0.1 mL from each dilution was evenly spread on LB agar plates, and bacterial counts in the target tissues were determined after overnight incubation. Each dilution was plated in triplicate. It is important to highlight that in our study, no protective measures (e.g., encapsulation or chemical modification) were applied to AaeAP2a for enhancing its stability. Oral gavage needles were acquired from Servicebio (China), and a tissue homogenizer was procured from Ningbo Scientz Biotechnology Co., Ltd. (SCIENTZ, China).
2.19 Data analysis
Statistical analyses were performed using GraphPad Prism version 9.5.0. Data, derived from a minimum of three biological replicates, are presented as the mean ± standard deviation (SD). Intergroup comparisons were conducted using a nonparametric one-way analysis of variance (ANOVA), with statistical significance defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3 Results
3.1 CRAB 236 exhibits resistance and virulence-associated genes
PCR amplification and subsequent gel electrophoresis were employed to screen for key genes associated with clinically significant traits in A. baumannii. Analysis revealed that CRAB 236 carried the carbapenem-resistance determinants blaOXA-23, blaOXA-51, and blaOXA-58 (Figure 1A), which collectively encode OXA-type carbapenemases, the primary molecular mediators of resistance to carbapenems (Lupo et al., 2017). Virulence gene amplification confirmed that CRAB 236 carries bap, bfs, bfmS, adeH, csuA, csuAB, pgaA, basD, and ompA (Figure 1B), which mediate diverse pathogenic functions including biofilm formation, adhesion, and iron acquisition (Lukovic et al., 2025; Ali et al., 2025). These findings indicate that the strain exhibits a high level of carbapenem resistance, possesses robust biofilm-forming capabilities, and demonstrates significant pathogenic potential.
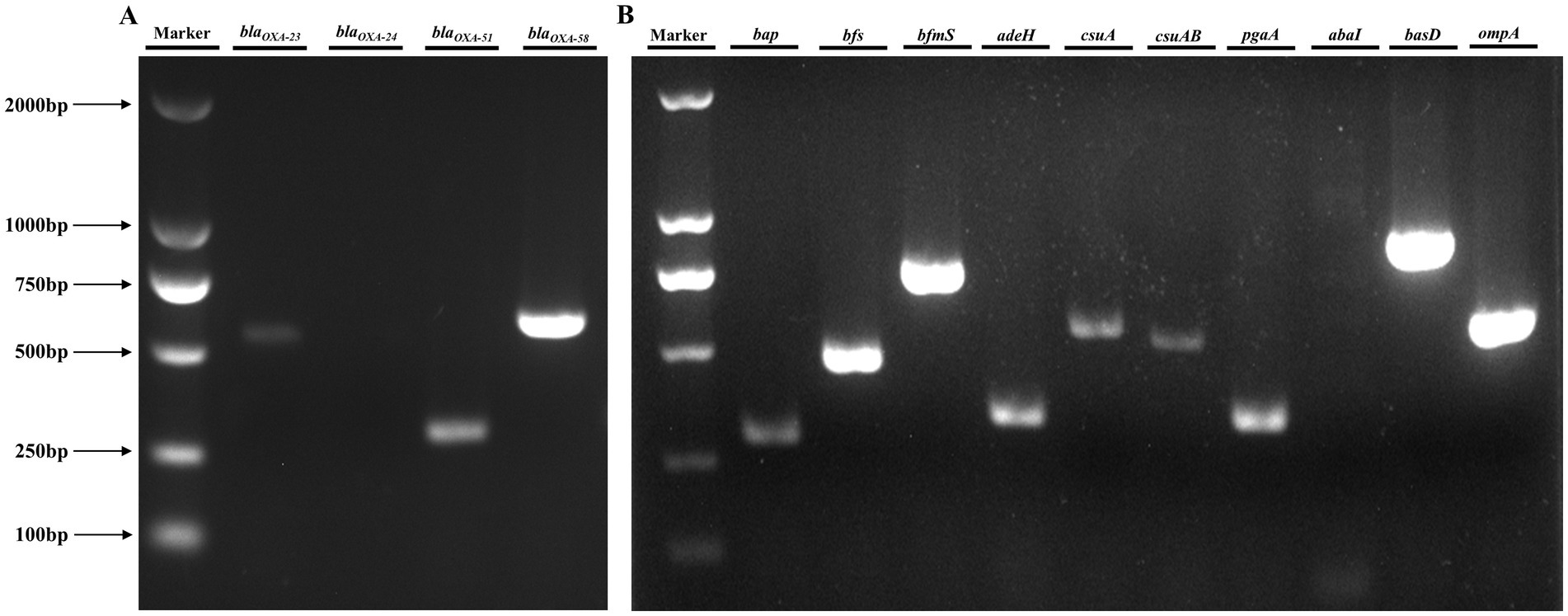
Figure 1. Analysis of resistance and virulence genes in the CRAB 236 strain. (A) Resistance gene detection demonstrated that CRAB 236 successfully amplified blaOXA-23 (534 bp), blaOXA-51 (353 bp), and blaOXA-58 (599 bp). (B) Analysis of virulence genes showed that CRAB 236 tested positive for bap, bfs, bfmS, adeH, csuA, csuAB, pgaA, basD, and ompA. Lane Marker corresponds to the DL2000 DNA marker.
3.2 AaeAP2a exhibits complete bactericidal activity against CRAB
The MIC assay results indicated that isolate CRAB 236 exhibited resistance to a range of antibiotics, including tetracyclines, quinolones, aminoglycosides, and carbapenems (CLSI), thereby confirming its MDR profile. Notably, AaeAP2a demonstrated significant inhibitory activity against this resistant strain (MIC = 6.25 μg/mL) and exhibited even greater efficacy against the sensitive reference strain ATCC 19606 (MIC = 3.125 μg/mL) (Table 2). These findings suggest that AaeAP2a may overcome conventional antibiotic resistance mechanisms.
Further assessment of the dynamic bactericidal efficacy of AaeAP2a was performed using a time-kill curve analysis. AaeAP2a exhibited a potent, concentration-dependent bactericidal effect. Specifically, treatment with 4 × MIC (25 μg/mL) of AaeAP2a for 10 or 6 h resulted in complete eradication of the clinical isolate (CRAB 236) and the standard strain (ATCC 19606), respectively. Furthermore, treatment at a lower concentration of 2 × MIC fully eradicated the strain ATCC 19606 within 12 h (Figures 2A,B). These findings underscore the promise of AaeAP2a as a potential bactericidal antibiotic candidate against CRAB.
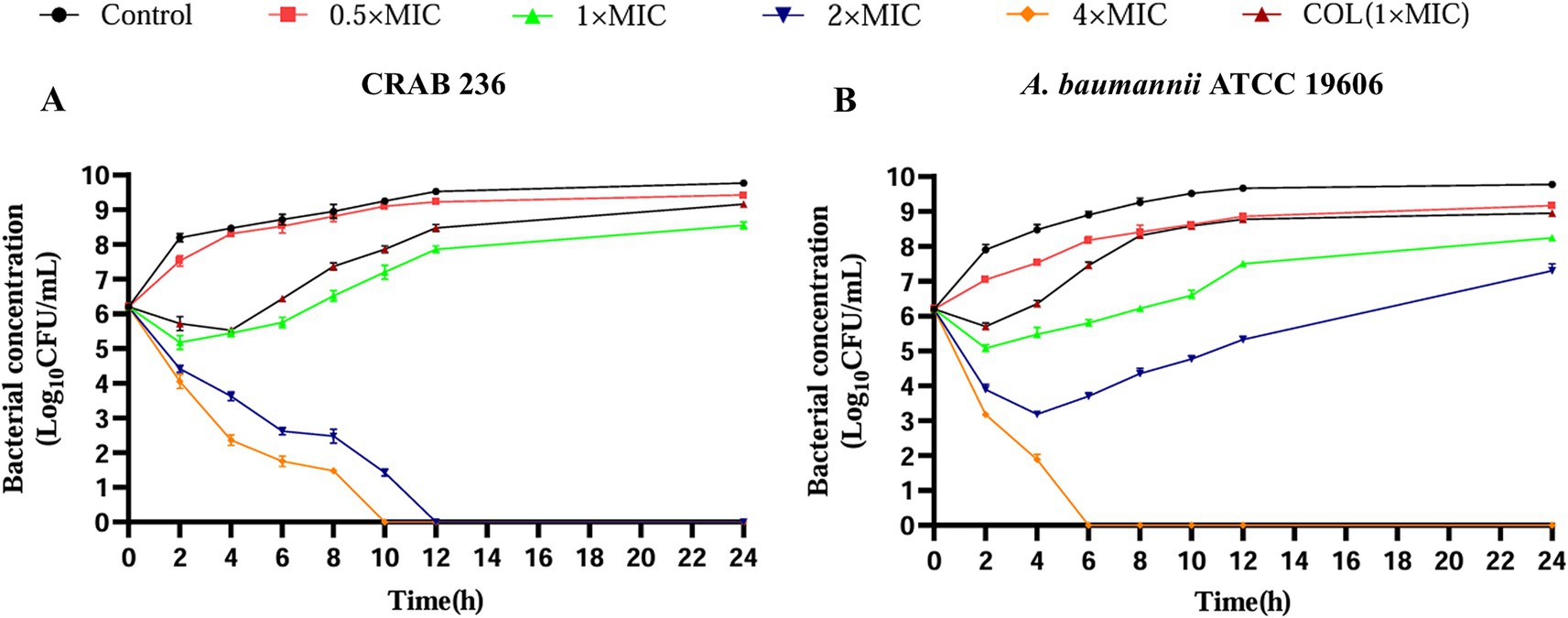
Figure 2. AaeAP2a demonstrates complete bactericidal activity against CRAB. The time-dependent bactericidal effects of AaeAP2a on strains CRAB 236 (A) and ATCC 19606 (B), respectively. Data are presented as the mean ± SD (n = 3).
3.3 AaeAP2a exhibits low propensity for inducing drug resistance and effectively inhibits biofilm formation in vitro
To assess the propensity of AaeAP2a to induce bacterial resistance, A. baumannii ATCC 19606 was repeatedly exposed to 0.5 × MIC (1.5625 μg/mL) of AaeAP2a over 20 consecutive days through serial passaging. The results showed a 4-fold increase in the AaeAP2a MIC (Figure 3A), which is considerably lower than the 32-fold increase observed with the colistin control (0.5 × MIC, 2 μg/mL). Meanwhile, the MIC of the negative control exhibited no significant changes, which thereby rules out the interference of “MIC alterations caused by spontaneous mutations of the strain itself.” This comparison indicates a minimal risk of resistance development with AaeAP2a, supporting its potential for long-term clinical application.
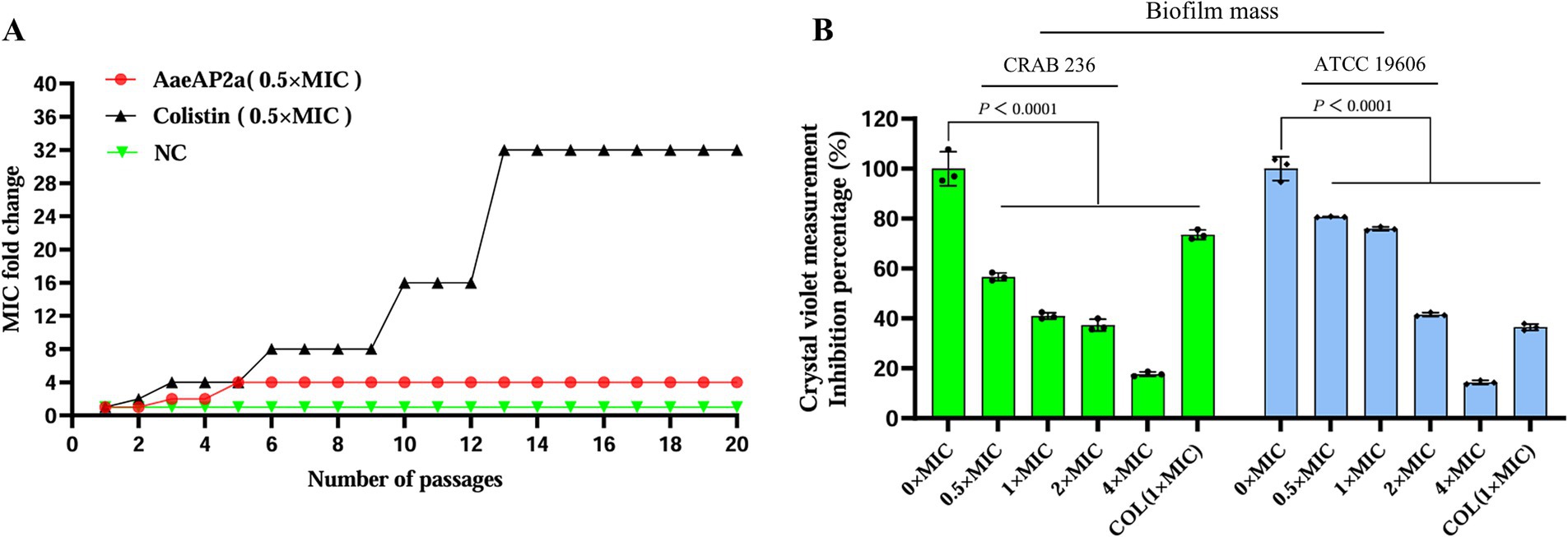
Figure 3. AaeAP2a exhibits low susceptibility to resistance and effectively inhibits biofilm formation. (A) In the resistance development experiment, the MIC of the AaeAP2a group (red) increased 4-fold over 20 generations, compared to a 32-fold increase in the colistin group (black), the negative control group (green) exhibited no changes. These values reflect the multiplicative change in MIC relative to the initial generation. (B) In the biofilm inhibition assay, AaeAP2a significantly reduced biofilm absorbance across concentrations ranging from 0.5 × MIC to 4 × MIC, achieving an inhibition rate exceeding 80% for both strains at 4 × MIC.
Additionally, bacterial biofilm formation significantly enhances pathogenicity and contributes to drug resistance, leading to chronic infections and treatment challenges (Vishwakarma et al., 2021; Ma et al., 2024; Olanrewaju et al., 2024). Notably, AaeAP2a was found to effectively inhibit biofilm formation in a concentration-dependent manner. As demonstrated in Figure 3B, treatment with AaeAP2a resulted in an 85% reduction in biofilm absorbance for ATCC 19606 and an 82% reduction for CRAB 236 at 4 × MIC (relative to the 0 × MIC group, p < 0.001). Notably, at an identical concentration of 1 × MIC, AaeAP2a demonstrated enhanced biofilm inhibitory activity against CRAB 236 compared to colistin. These findings suggest that AaeAP2a not only exhibits potent antibacterial activity but also diminishes bacterial pathogenicity by inhibiting biofilm formation, thereby underscoring its potential clinical utility.
3.4 AaeAP2a exhibits promising biocompatibility and stability in vitro
Ensuring the safety and stability of a pharmaceutical agent is essential for its clinical application. In this study, we thoroughly evaluated the hemolytic activity, cytotoxicity, and overall stability of AaeAP2a under a range of conditions. At 16 × MIC, AaeAP2a induced < 5% hemolysis in mouse erythrocytes and maintained > 90% viability in 293 T cells. Even at 64 × MIC (200 μg/mL), hemolysis remained below 5% (Figures 4A,B). Moreover, regarding physical stability, AaeAP2a maintained full activity after undergoing 12 freeze–thaw cycles (from −80 to 25 °C), exposure to 100 °C for 1 h, or incubation across a pH range of 4–10 for 1 h (Figures 4C,D,F). Even under extreme pH conditions (pH 3 and pH 11), the MIC increased only twofold. Notably, its antibacterial efficacy remained undiminished after storage at room temperature for 4 weeks (Figure 4E). Concerning chemical stability, AaeAP2a demonstrated substantial resistance to urea and proteinase K, although its activity was markedly reduced at trypsin concentrations exceeding 40 μg/mL (Figures 4G,H). While the peptide was not affected by the presence of monovalent cations (Na+/K+), a 10-fold increase in the MIC was observed with 100 μM Ca2+ or Mg2+ (Figure 4I).

Figure 4. AaeAP2a demonstrates favorable biosafety and stability. (A) Hemolysis of mouse erythrocytes. (B) Cytotoxicity in 293 T cells. (C) Stability after freeze–thaw cycles (−80 to 25 °C). (D) Thermal stability at different temperatures. (E) Long-term stability during room temperature storage (25°C). (F) Stability across a range of pH values. (G) Urea tolerance. (H) Protease resistance. (I) Effect of metal ions.
3.5 AaeAP2a disrupts the integrity of inner and outer membranes
To elucidate the bactericidal mechanism of AaeAP2a, we evaluated its impact on the membrane structure of A. baumannii using fluorescent probe techniques and SEM. Fluorescence tracer experiments using 1-N-phenylnaphthylamine (NPN), an outer membrane probe, and propidium iodide (PI), an inner membrane probe, demonstrated that AaeAP2a significantly increased the permeability of both bacterial membranes in a dose-dependent manner. Specifically, CRAB 236 treated with a 0.5 × MIC of AaeAP2a exhibited greater fluorescence intensity for both NPN and PI compared with colistin at 1 × MIC (Figures 5A,B). At 1 × MIC, treatment with AaeAP2a resulted in a 2.3-fold increase in PI fluorescence intensity compared to colistin in the canine-derived CRAB 236 isolate (Figure 5B), indicating marked disruption of both membranes. These findings were further corroborated by SEM observations (Figure 6). After 2 h of treatment with a 4 × MIC of AaeAP2a, the bacterial membranes exhibited severe wrinkling and structural damage, with the deterioration becoming more pronounced over time and resulting in cytoplasmic leakage at 4 h. Consistent effects were observed in the reference strain ATCC 19606 (Figures 6G–L), mirroring the response seen in CRAB 236 (Figures 6A–F).
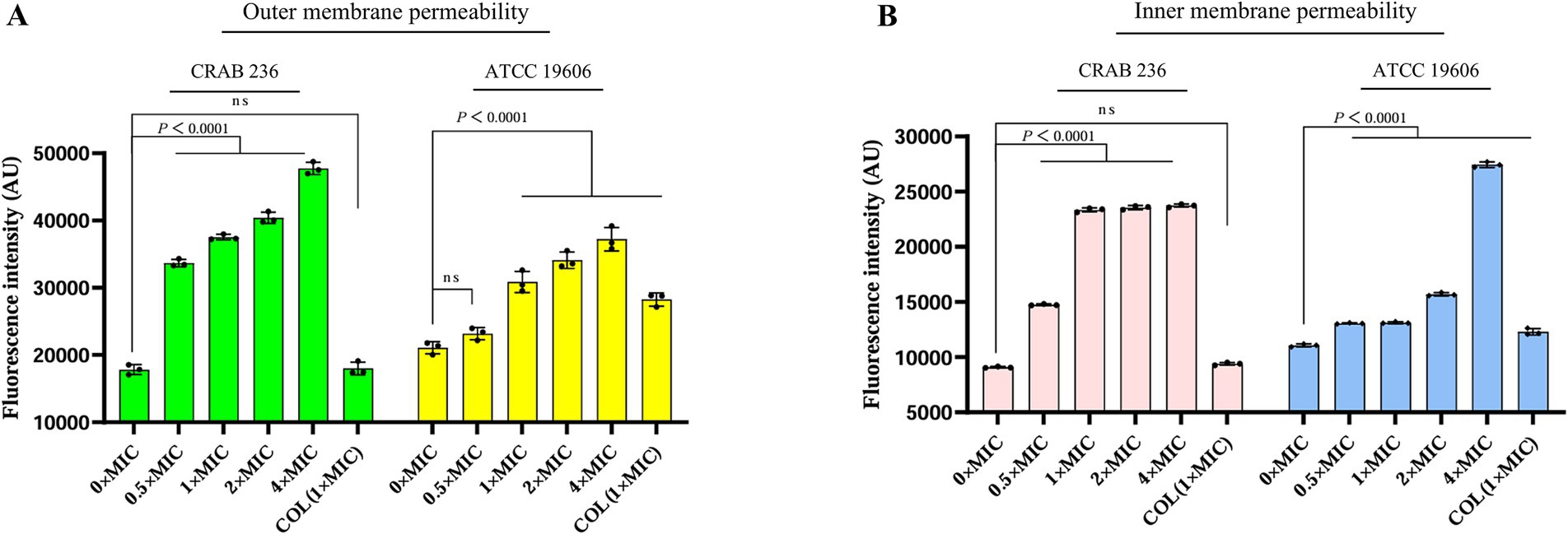
Figure 5. AaeAP2a influences the permeability of the outer and inner membranes of A. baumannii. (A,B) The assessment of membrane permeability after co-incubating AaeAP2a, at concentrations ranging from 0 to 4 × MIC, with either strain ATCC 19606 or CRAB 236 for 1 h. The assays employed 10 μM NPN or 0.5 μM PI as fluorescent indicators. All experiments were conducted in triplicate, with error bars representing the standard deviation (SD). Statistical significance was evaluated using a nonparametric one-way analysis of variance (ANOVA), and “AU” denotes arbitrary units.
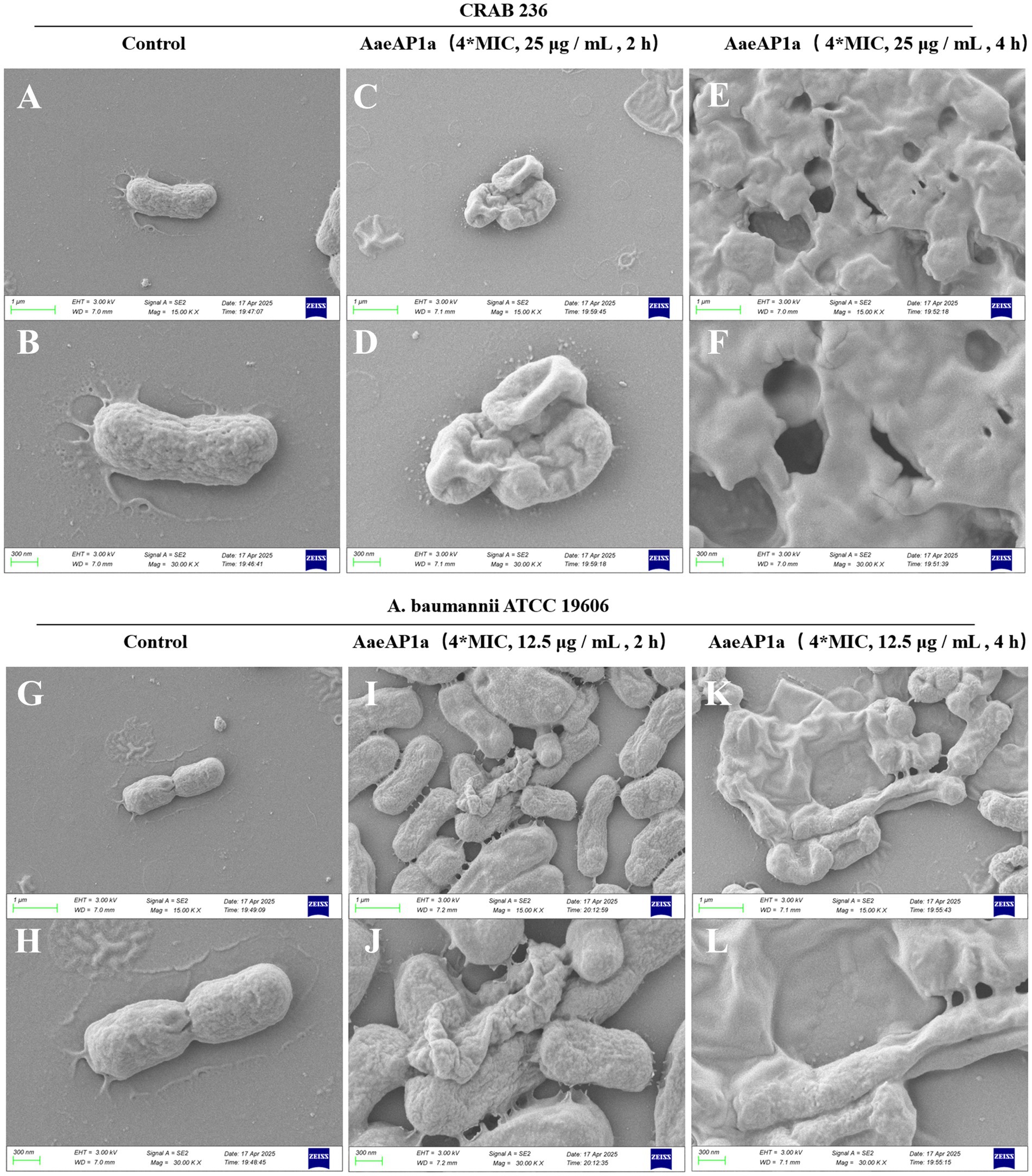
Figure 6. AaeAP2a disrupts the structure of the bacterial membrane. (A–F) SEM images of A. baumannii CRAB 236 post-treatment with 4 × MIC AaeAP2a (25 μg/mL) for either 2 or 4 h. (G–L) SEM images of A. baumannii ATCC 19606 after exposure to 4 × MIC AaeAP2a (12.5 μg/mL) for the same time intervals.
3.6 Multiple mechanisms synergistically enhance the anti-CRAB activity of AaeAP2a
Building on its excellent membrane permeability, we further elucidated the mechanism by which AaeAP2a synergistically disrupts the metabolic homeostasis of A. baumannii through multiple pathways. The NAD+/NADH ratio, an indicator of cellular energy metabolism and redox balance (Xu et al., 2023), was significantly reduced in a dose-dependent manner following treatment with AaeAP2a (Figure 7A). The bacterial PMF, which comprises an electrical potential (Δψ) and a transmembrane proton gradient (ΔpH), is an essential energy mechanism embedded in bacterial membranes. It is crucial for several physiological processes, including ATP creation and the movement of active substances. Because of its significance for bacterial survival, the PMF is a promising target for antimicrobial treatments (Yang et al., 2023). We assessed the impact of AaeAP2a on Δψ and ΔpH using DiSC3(5) and BCECF-AM, respectively. The results demonstrated that AaeAP2a induced dose-dependent membrane depolarization and potential dissipation (Figure 7C), alongside an elevation in intramembrane alkalinization (Figure 7D). Our assays revealed a significant drop in intracellular ATP levels (Figure 7B), which is consistent with the dissipation of PMF and the decrease in NAD+/NADH ratio, ultimately causing a collapse in energy metabolism. Additional investigations showed a rapid accumulation of reactive oxygen species (ROS) and hydroxyl radicals (•OH) (Figures 7E,F), which resulted in multiple oxidative stress-induced injuries.
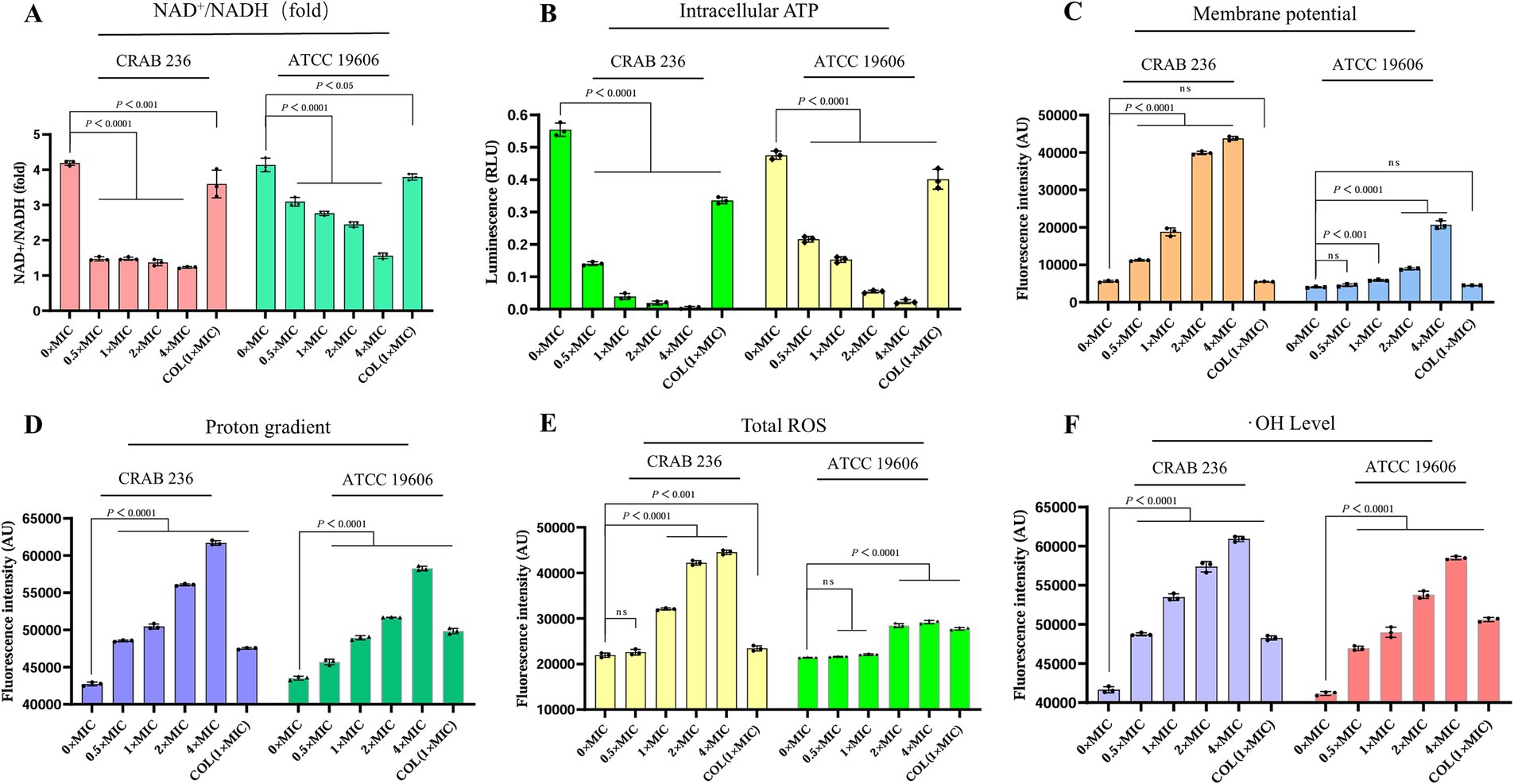
Figure 7. AaeAP2a modulates the NAD+/NADH ratio, disrupts the PMF, reduces ATP levels, and induces oxidative stress damage. (A) NAD+/NADH ratio. (B) Intracellular ATP levels. (C) Δψ measured using DiSC3(5). (D) ΔpH assessed with BCECF-AM. (E) ROS levels. (F) Hydroxyl radical accumulation. All experiments were conducted in triplicate, and the data are presented as mean ± standard deviation (mean ± SD).
3.7 AaeAP2a exhibits significant antimicrobial activity in vivo
To evaluate the in vivo therapeutic efficacy of AaeAP2a, a peritonitis-associated sepsis model was established in BALB/c mice infected with CRAB 236 at a dose of 6.75 × 106 CFU. The results demonstrated that all mice treated with PBS alone succumbed within 48 h post-infection. In contrast, increasing the dosage of AaeAP2a from 5 to 20 mg/kg led to a marked enhancement in the 72-h survival rate, rising from 20 to 80% (Figure 8A). Moreover, AaeAP2a significantly reduced bacterial loads in the blood, liver, and spleen in a dose-dependent manner 12 h after administration (Figures 8B–D), a finding that correlated with reduced symptom severity and improved survival.
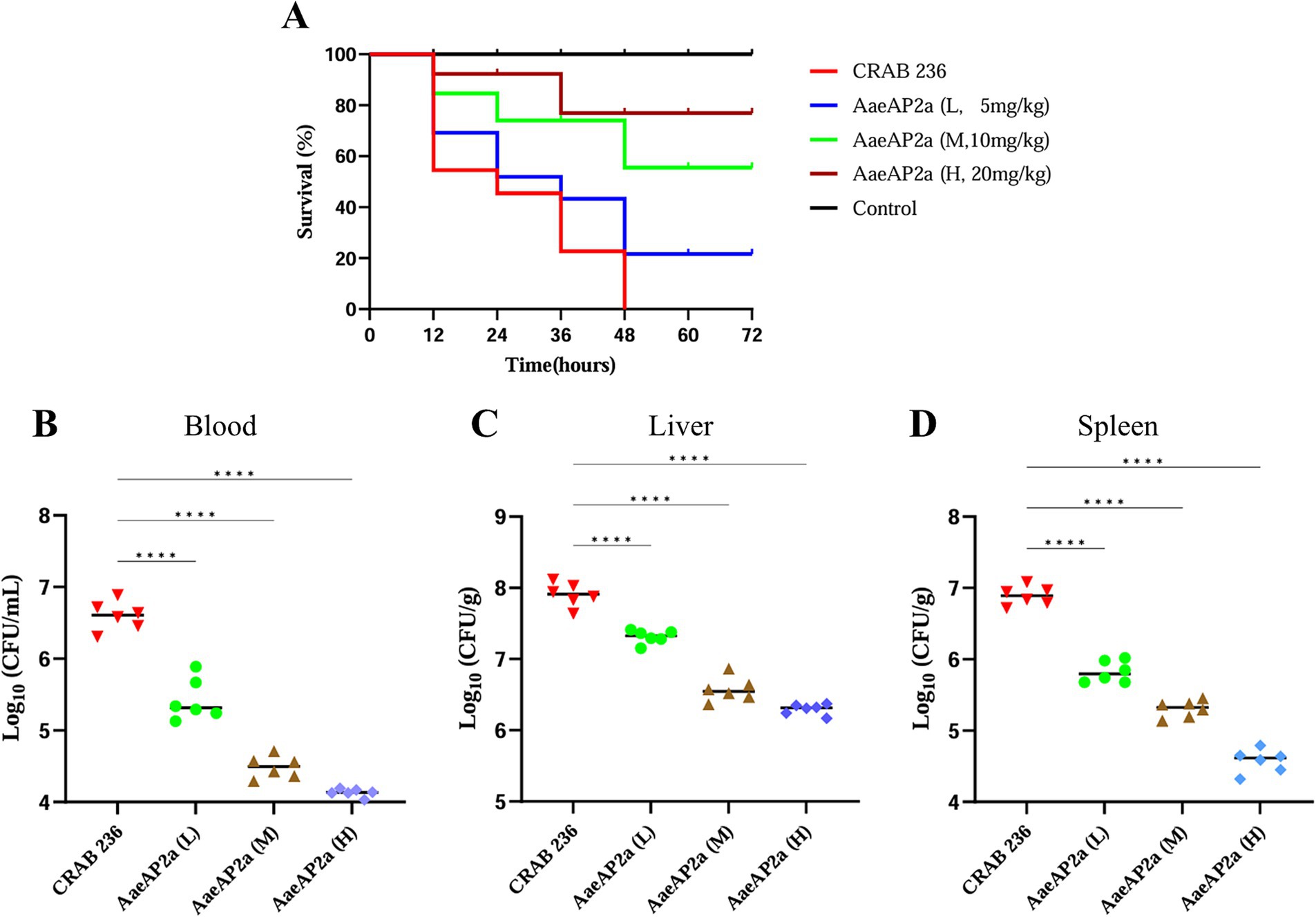
Figure 8. AaeAP2a demonstrates dose-dependent antimicrobial efficacy in a murine peritonitis-associated sepsis model. (A) Survival curves of infected mice (n = 10) treated with various doses of AaeAP2a (5–20 mg/kg) compared to the PBS control group. (B–D) Bacterial loads in the blood, liver, and spleen of infected mice 12 h post-administration of different doses of AaeAP2a. The symbol “****” indicates a statistically significant difference with a P-value less than 0.0001, meaning there is an extremely significant statistical difference between the relevant groups.
4 Discussion
The evolutionary complexity of CRAB undermines conventional therapies through rapid resistance evolution, leading to high mortality rates among infected patients—particularly in cases of bloodstream infections (Luo et al., 2025), COVID-19-related complications (Russo et al., 2022), and post-craniotomy infections (Lan et al., 2024). Recent epidemiological investigations have revealed a concerning trend: companion animals have emerged as potential reservoirs for MDR pathogens, thereby facilitating the establishment of a human–animal–environment transmission network that poses an ongoing threat to public health and safety (Lupo et al., 2017). The co-occurrence of resistance, biofilm-associated, and virulence genes in the canine-derived CRAB 236 strain demonstrates carbapenem resistance, robust biofilm-forming capacity, and significant pathogenic potential. These findings are corroborated by phenotypic evidence in Table 2 (MIC assays), Figure 3B (biofilm quantification), and Figure 8A (murine virulence model), respectively. Based on these findings, we hypothesize that the CRAB 236 strain effectively overcomes host defenses and antimicrobial barriers through these polygenic co-evolutionary mechanisms. Consequently, traditional antibiotics relying on single-target mechanisms face increasing limitations in treating CRAB infections. Moreover, last-resort antibiotics like colistin remain clinically suboptimal due to constraints such as nephrotoxicity concerns (Guzman et al., 2025a, 2025b). Thus, developing innovative approaches to overcome multidrug resistance constraints is imperative.
The scientific community has recognized antimicrobial peptides—natural immune molecules that synergistically eradicate bacteria through multiple mechanisms and are less likely to induce resistance—as a promising solution against MDR bacteria (Song et al., 2021). Scorpion venoms are a powerful natural reservoir of AMPs, offering a foundation for the development of novel agents against a broad spectrum of pathogens, including MDR microbes, fungi, viruses, parasites, and malignant cells (Rabea et al., 2025). In this study, we systematically evaluated the antimicrobial activity, stability, security, and protection against CRAB of AaeAP2a. We also elucidated its antimicrobial mechanism and assessed its therapeutic efficacy in vivo.
In addition to increasing the permeability of antimicrobial agents, bacterial biofilms, which rely on a complex matrix of extracellular polymeric substances, support the evolution of bacterial drug resistance, creating a protective barrier against drug-induced killing (Verderosa et al., 2019). AMPs have shown dual potential in meeting biofilm-related chronic infections: they can inhibit the initial stages of biofilm formation and eradicate mature biofilms. In this study, AaeAP2a, at a 4 × MIC, obtained an inhibition rate approaching 80% against biofilms formed by two strains of Acinetobacter baumannii (Figure 3B). Consistent with previous findings, these results verify the high efficacy of AMPs in preventing biofilm formation (Jung et al., 2021; Wang et al., 2022). However, the recalcitrance of mature biofilms is the main obstacle to treating chronic infections. To tackle this clinical dilemma, several studies have shown that SR25, at a concentration of 1 × MIC, accomplishes a 56% eradication efficiency against Escherichia coli biofilms, profoundly surpassing colistin (35%) (Luo et al., 2024). Moreover, the helical peptide G3 possesses stage-specific activity, initially preventing the adhesion and colonization of Streptococcus mutans, and subsequently targeting extracellular DNA to interfere with mature biofilm structures (Zhang et al., 2020). Likewise, Cec4 shows both inhibitory and eradicative effects on Klebsiella pneumoniae biofilms and can eliminate biofilms generated by CRAB, albeit at increasingly high concentrations (256–512 μg/mL) (Peng et al., 2023; Li et al., 2025). Nevertheless, AMPs that combine biofilm inhibition and eradication functions remain scarce. To solve the limitations of single AMPs in biofilm eradication, researchers have explored innovative strategies. For instance, KKd-11 can self-assemble into hydrogels, facilitating dual intervention through extended proteolytic degradation resistance and prolonged antimicrobial activity (Guo et al., 2021). In another approach, the combined application of melimine or Mel4 with ciprofloxacin has shown synergistic effects in eradicating Pseudomonas aeruginosa biofilms (Yasir et al., 2020). Unfortunately, our study found that AaeAP2a did not exhibit a significant eradicative effect on mature Acinetobacter baumannii biofilms at 4 × MIC (data not shown). Future studies may use combined strategies such as AMP + antibiotics, AMP + quorum sensing inhibitors, or nanocarrier-based delivery systems to explore AaeAP2a biofilm eradication (Abdelhamid and Yousef, 2023; Liu et al., 2025).
After 20 generations of sub-MIC exposure, AaeAP2a exhibited a remarkably low resistance induction rate (Figure 3A), which is consistent with the low abundance and limited transmission of resistance genes (Chen et al., 2023), the multiple antimicrobial mechanisms (Bucataru and Ciobanasu, 2024), as well as rapid bactericidal effect of AMPs (Zhang et al., 2022). Safety and stability are essential prerequisites for the clinical application of pharmaceutical agents. Assessments of hemolytic activity and cytotoxicity have preliminarily confirmed that AaeAP2a exhibits promising biosafety in vitro (Figures 4A,B). However, to truly advance its clinical translation, more comprehensive safety data are needed, including long-term toxicity, toxicokinetics, and tissue compatibility. Notably, the hemolytic activity of AaeAP2a in our study is considerably lower than previously reported. Du et al. observed complete lysis of a 2% erythrocyte suspension at 64 μg/mL, whereas our experiments showed that even at 200 μg/mL (with a 4% suspension), the hemolysis rate remained below 5%. This discrepancy may be attributed to differences in red blood cell sources; our study utilized mouse erythrocytes, in contrast to the horse erythrocytes used by Du et al. The compound also displayed considerable stability, retaining its efficacy after exposure to high temperatures (100 °C), a wide pH range (4–10), and multiple freeze–thaw cycles. Notably, its activity was diminished only at elevated concentrations of divalent cations (Ca2+, Mg2+ > 50 mM) or in the presence of trypsin (> 40 μg/mL), reflecting a competitive displacement effect of Mg2+/Ca2+ ions on lipopolysaccharides (LPS), as previously reported (Sun et al., 2025). Furthermore, its exceptional thermal and freeze–thaw stability, combined with robust long-term storage stability at ambient temperature, underscores its potential for formulation development. Nevertheless, strategies such as altering administration routes (e.g., intravenous, inhalation, or topical), enhancing intrinsic stability (e.g., through D-amino acid substitution, terminal closure, cyclization, or polymerization), and optimizing delivery systems (e.g., liposomes, nanoparticles, or hydrogels) may further mitigate the identified vulnerabilities (Zheng et al., 2025).
Given that AaeAP2a does not readily induce bacterial resistance, we hypothesized that it can offer distinct advantages over the single-target mechanism typical of conventional antimicrobial agents. Indeed, previous studies have demonstrated that AMPs can exert broad-spectrum bactericidal effects through a multimodal synergistic mechanism (Xuan et al., 2023). Numerous AMPs, such as the modified peptide 2K4L (Ji et al., 2023) and the novel peptide LL-1 (Zhou L. et al., 2022) derived from the Linnaeus toadstool, achieve sterilization by disrupting cell membranes. These peptides markedly increase the permeability of MDRAB and E. coli cell membranes, leading to intracellular leakage and cell death. Similarly, our team members Zhang et al. (2015), Wang et al. (2019), and Wang et al. (2019) demonstrated that the antimicrobial peptide JH-3 inhibits Salmonella and E. coli by disrupting the integrity of their cell walls and membranes. In this study, we find that AaeAP2a significantly enhances the permeability of the outer membrane in A. baumannii while compromising the integrity of its inner membrane (Figures 5, 6). Moreover, AaeAP2a disrupts the bacterial membrane potential, accompanied by a disciplinary change of ΔpH, and subsequently perturbs key bacterial metabolic processes. This perturbation resulting in the decomposition of the electron transport chain, ATP loss, ROS, and •OH formation (Figure 7). These findings corroborate previous studies indicating that AMPs can provoke membrane destabilization, PMF collapse, energy metabolism impairment, and oxidative stress (Xia et al., 2021; Shi et al., 2022). This multi-target synergistic bactericidal mechanism successfully overcomes the limitations of conventional antibiotics and provides a persuasive medical justification for AaeAP2a’s potent antibiotic effects.
In conclusion, we conducted a thorough investigation of AaeAP2a, which shows strong in vitro bactericidal activity and effective in vivo therapeutic potential against CRAB. AaeAP2a also demonstrates favorable biosafety and stability. Mechanistic studies indicate that its mode of action involves increased permeability of both inner and outer bacterial membranes, membrane depolarization and potential dissipation, impaired ATP synthesis, and oxidative damage. These findings position AaeAP2a as a promising candidate in the fight against antibiotic resistance. Nonetheless, the crystal structure, in vivo half-life, and metabolic pathway of AaeAP2a remain to be fully elucidated. Further research is warranted to address translational challenges through yield optimization and cost reduction.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Henan Institute of Science and Technology Ethics Committee/Affiliated with Henan Institute of Science and Technology. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
WL: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing. LiZ: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft. HG: Formal analysis, Investigation, Methodology, Software, Writing – original draft. HL: Formal analysis, Investigation, Software, Writing – original draft. XiL: Formal analysis, Investigation, Software, Writing – original draft. YW: Data curation, Resources, Writing – review & editing. HS: Methodology, Software, Writing – review & editing. BH: Formal analysis, Supervision, Writing – review & editing. LoZ: Methodology, Writing – review & editing. WZ: Investigation, Methodology, Writing – review & editing. XuL: Data curation, Validation, Writing – review & editing. RW: Methodology, Writing – review & editing. BW: Formal analysis, Supervision, Writing – original draft. JS: Project administration, Resources, Writing – original draft. CZ: Methodology, Software, Writing – review & editing. YB: Data curation, Funding acquisition, Investigation, Methodology, Software, Supervision, Validation, Writing – review & editing. LW: Writing – review & editing. KD: Conceptualization, Project administration, Supervision, Writing – review & editing. JH: Conceptualization, Funding acquisition, Investigation, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Third-class postdoctoral research grant in Henan Province (202003074), the National Natural Science Foundation of China (32402883), Key Research and Development Project of Henan Province (241111110100), National Natural Science Foundation of China (32172862, 32473037), National Key R&D Program of China (2021YFD1301200), Science and Technology Innovative Research Team in Higher Educational Institutions of Henan Province (24IRTSTHN035), the Joint Fund of Science and Technology Research and Development Plan in Henan province (225200810044), and Outstanding Youth Foundation of Henan Scientific Committee (222300420043).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelhamid, A. G., and Yousef, A. E. (2023). Combating bacterial biofilms: current and emerging Antibiofilm strategies for treating persistent infections. Antibiotics 12:1005. doi: 10.3390/antibiotics12061005
Ali, S., Stavropoulos, A., Jenkins, B., Graves, S., Ahmadi, A., Marzbanrad, V., et al. (2025). Comparative proteomics of biofilm development in Pseudoalteromonas tunicata discovers a distinct family of Ca2+−dependent adhesins. MBio 16:e01069-25. doi: 10.1128/mbio.01069-25
Aslam, B., Khurshid, M., Arshad, M. I., Muzammil, S., Rasool, M., Yasmeen, N., et al. (2021). Antibiotic resistance: one health one world outlook. Front. Cell. Infect. Microbiol. 11:771510. doi: 10.3389/fcimb.2021.771510
Bucataru, C., and Ciobanasu, C. (2024). Antimicrobial peptides: opportunities and challenges in overcoming resistance. Microbiol. Res. 286:127822. doi: 10.1016/j.micres.2024.127822
Castañeda-Barba, S., Top, E. M., and Stalder, T. (2024). Plasmids, a molecular cornerstone of antimicrobial resistance in the one health era. Nat. Rev. Microbiol. 22, 18–32. doi: 10.1038/s41579-023-00926-x
Chen, B., Zhang, Z., Zhang, Q., Xu, N., Lu, T., Wang, T., et al. (2023). Antimicrobial peptides in the global microbiome: biosynthetic genes and resistance determinants. Environ. Sci. Technol. 57, 7698–7708. doi: 10.1021/acs.est.3c01664
Dalmasso, G., Beyrouthy, R., Brugiroux, S., Ruppé, E., Guillouard, L., Bonnin, V., et al. (2023). Genes mcr improve the intestinal fitness of pathogenic E. coli and balance their lifestyle to commensalism. Microbiome 11:12. doi: 10.1186/s40168-022-01457-y
de la Rocque, S., Errecaborde, K. M. M., Belot, G., Brand, T., Shadomy, S., von Dobschuetz, S., et al. (2023). One health systems strengthening in countries: tripartite tools and approaches at the human-animal-environment interface. BMJ Glob. Health 8:e011236. doi: 10.1136/bmjgh-2022-011236
Dombach, J. L., Quintana, J. L. J., Nagy, T. A., Wan, C., Crooks, A. L., Yu, H., et al. (2020). A small molecule that mitigates bacterial infection disrupts gram-negative cell membranes and is inhibited by cholesterol and neutral lipids. PLoS Pathog. 16:e1009119. doi: 10.1371/journal.ppat.1009119
Du, Q., Hou, X., Wang, L., Zhang, Y., Xi, X., Wang, H., et al. (2015). AaeAP1 and AaeAP2: novel antimicrobial peptides from the venom of the scorpion, Androctonus aeneas: structural characterisation, molecular cloning of biosynthetic precursor-encoding cDNAs and engineering of analogues with enhanced antimicrobial and anticancer activities. Toxins 7, 219–237. doi: 10.3390/toxins7020219
Gao, N., Wang, J., Fang, C., Bai, P., Sun, Y., Wu, W., et al. (2024). Combating bacterial infections with host defense peptides: shifting focus from bacteria to host immunity. Drug Resist. Updat. 72:101030. doi: 10.1016/j.drup.2023.101030
Grasteau, A., Tremblay, Y. D. N., Labrie, J., and Jacques, M. (2011). Novel genes associated with biofilm formation of Actinobacillus pleuropneumoniae. Vet. Microbiol. 153, 134–143. doi: 10.1016/j.vetmic.2011.03.029
Guo, Z., Wang, Y., Tan, T., Ji, Y., Hu, J., and Zhang, Y. (2021). Antimicrobial d-peptide hydrogels. ACS Biomater Sci. Eng. 7, 1703–1712. doi: 10.1021/acsbiomaterials.1c00187
Guzman, L., Parcerisas, A., Cano, A., Sánchez-López, E., Verdaguer, E., Auladell, C., et al. (2025a). Colistin treatment causes neuronal loss and cognitive impairment via ros accumulation and neuronal plasticity alterations. Biomed. Pharmacother. 183:117839. doi: 10.1016/j.biopha.2025.117839
Guzman, L., Rabanal, F., Garcia, J., Marqués, A. M., Sánchez, E., Cano, A., et al. (2025b). B-rich colistin and B-pure colistin as novel strategies to increase the therapeutic window of polymyxin antibiotic therapy. Biomed. Pharmacother. 190:118366. doi: 10.1016/j.biopha.2025.118366
Humphries, R., Bobenchik, A. M., Hindler, J. A., and Schuetz, A. N. (2021). Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J. Clin. Microbiol. 59:e0021321. doi: 10.1128/JCM.00213-21
Ji, F., Tian, G., Shang, D., and Jiang, F. (2023). Antimicrobial peptide 2K4L disrupts the membrane of multidrug-resistant Acinetobacter baumannii and protects mice against sepsis. Front. Microbiol. 14:1258469. doi: 10.3389/fmicb.2023.1258469
Ju, Y., Liu, K., Ma, G., Zhu, B., Wang, H., Hu, Z., et al. (2023). Bacterial antibiotic resistance among cancer inpatients in China: 2016–20. QJM 116, 213–220. doi: 10.1093/qjmed/hcac244
Jung, C.-J., Liao, Y.-D., Hsu, C.-C., Huang, T.-Y., Chuang, Y.-C., Chen, J.-W., et al. (2021). Identification of potential therapeutic antimicrobial peptides against Acinetobacter baumannii in a mouse model of pneumonia. Sci. Rep. 11:7318. doi: 10.1038/s41598-021-86844-5
Lan, X., Qin, S., Liu, H., Guo, M., Zhang, Y., Jin, X., et al. (2024). Dual-targeting tigecycline nanoparticles for treating intracranial infections caused by multidrug-resistant Acinetobacter baumannii. J. Nanobiotechnol. 22:138. doi: 10.1186/s12951-024-02373-z
Li, C.-G., Zeng, Q.-Z., Chen, M.-Y., Xu, L.-H., Zhang, C.-C., Mai, F.-Y., et al. (2019). Evodiamine augments NLRP3 Inflammasome activation and anti-bacterial responses through inducing α-tubulin acetylation. Front. Pharmacol. 10:290. doi: 10.3389/fphar.2019.00290
Li, L., Zeng, Y., Tian, M., Cao, H., Qiu, Z., Guo, G., et al. (2025). The antimicrobial peptide Cec4 has therapeutic potential against clinical carbapenem-resistant Klebsiella pneumoniae. Microbiol. Spectr. 13, e02738–e02724. doi: 10.1128/spectrum.02738-24
Liu, C., Chen, K., Wu, Y., Huang, L., Fang, Y., Lu, J., et al. (2022). Epidemiological and genetic characteristics of clinical carbapenem-resistant Acinetobacter baumannii strains collected countrywide from hospital intensive care units (ICUs) in China. Emerg. Microbes Infect. 11, 1730–1741. doi: 10.1080/22221751.2022.2093134
Liu, Y., Jia, Y., Yang, K., Li, R., Xiao, X., Zhu, K., et al. (2020a). Metformin restores Tetracyclines susceptibility against multidrug resistant Bacteria. Adv. Sci. 7:1902227. doi: 10.1002/advs.201902227
Liu, Y., Shi, J., Tong, Z., Jia, Y., Yang, K., and Wang, Z. (2020b). Potent broad-Spectrum antibacterial activity of amphiphilic peptides against multidrug-resistant Bacteria. Microorganisms 8:1398. doi: 10.3390/microorganisms8091398
Liu, H., Song, Z., Zhang, Y., Wu, B., Chen, D., Zhou, Z., et al. (2025). De novo design of self-assembling peptides with antimicrobial activity guided by deep learning. Nat. Mater. 24, 1295–1306. doi: 10.1038/s41563-025-02164-3
Lukovic, B., Kabic, J., Dragicevic, M., Kuljanin, S., Dimkic, I., Jovcic, B., et al. (2025). Genetic basis of antimicrobial resistance, virulence features and phylogenomics of carbapenem-resistant Acinetobacter baumannii clinical isolates. Infection 53, 39–50. doi: 10.1007/s15010-024-02316-8
Luo, Q., Chang, M., Lu, P., Guo, Q., Jiang, X., Xiao, T., et al. (2025). Genomic epidemiology and phylodynamics of Acinetobacter baumannii bloodstream isolates in China. Nat. Commun. 16:3536. doi: 10.1038/s41467-025-58772-9
Luo, X., Hu, C., Yin, Q., Zhang, X., Liu, Z., Zhou, C., et al. (2024). Dual-mechanism peptide SR25 has broad antimicrobial activity and potential application for healing bacteria-infected diabetic wounds. Adv. Sci. 11:2401793. doi: 10.1002/advs.202401793
Lupo, A., Châtre, P., Ponsin, C., Saras, E., Boulouis, H.-J., Keck, N., et al. (2017). Clonal spread of Acinetobacter baumannii sequence type 25 carrying blaOXA-23 in companion animals in France. Antimicrob. Agents Chemother. 61, e01881–e01816. doi: 10.1128/AAC.01881-16
Ma, Y., Tang, W. S., Liu, S. Y., Khoo, B. L., and Chua, S. L. (2024). Juglone as a natural quorum sensing inhibitor against Pseudomonas aeruginosa pqs-mediated virulence and biofilms. ACS Pharmacol. Transl. Sci. 7, 533–543. doi: 10.1021/acsptsci.3c00354
Monteiro, R., Alcantud, B. S., Piersma, S., Hendrickx, A. P. A., Maaß, S., Becher, D., et al. (2025). Outer membrane vesicles of carbapenem-resistant clinical Acinetobacter baumannii isolates protect both the vesicle-producing bacteria and non-resistant bacteria against carbapenems. Microbiol. Res. 297:128175. doi: 10.1016/j.micres.2025.128175
Olanrewaju, R. O., Lee, J.-H., Kim, Y.-G., and Lee, J. (2024). Antimicrobial and antibiofilm activities of halogenated phenols against Staphylococcus aureus and other microbes. Chemosphere 367:143646. doi: 10.1016/j.chemosphere.2024.143646
Peng, J., Wang, Y., Wu, Z., Mao, C., Li, L., Cao, H., et al. (2023). Antimicrobial peptide Cec4 eradicates multidrug-resistant Acinetobacter baumannii in vitro and in vivo. Drug Des. Devel. Ther. 17, 977–992. doi: 10.2147/DDDT.S405579
Pollard, J. E., Snarr, J., Chaudhary, V., Jennings, J. D., Shaw, H., Christiansen, B., et al. (2012). In vitro evaluation of the potential for resistance development to ceragenin CSA-13. J. Antimicrob. Chemother. 67, 2665–2672. doi: 10.1093/jac/dks276
Price, E. R., McDermott, D., Sherman, A., Kelley, L., Mehr, J., Greeley, R., et al. (2024). Canine multidrug-resistant Pseudomonas aeruginosa cases linked to human artificial tears–related outbreak. Emerg. Infect. Dis. 30, 2689–2691. doi: 10.3201/eid3012.240085
Rabea, E. Y., Mahmoud, E. D., Mohamed, N. K., Ansary, E. R., Alrouby, M. R., Shehata, R. R., et al. (2025). Potential of venom-derived compounds for the development of new antimicrobial agents. Toxins 17:238. doi: 10.3390/toxins17050238
Russo, A., Gavaruzzi, F., Ceccarelli, G., Borrazzo, C., Oliva, A., Alessandri, F., et al. (2022). Multidrug-resistant Acinetobacter baumannii infections in COVID-19 patients hospitalized in intensive care unit. Infection 50, 83–92. doi: 10.1007/s15010-021-01643-4
Shi, J., Chen, C., Wang, D., Wang, Z., and Liu, Y. (2022). The antimicrobial peptide LI14 combats multidrug-resistant bacterial infections. Commun. Biol. 5:926. doi: 10.1038/s42003-022-03899-4
Song, X., Liu, P., Liu, X., Wang, Y., Wei, H., Zhang, J., et al. (2021). Dealing with MDR bacteria and biofilm in the post-antibiotic era: application of antimicrobial peptides-based nano-formulation. Mater. Sci. Eng. C 128:112318. doi: 10.1016/j.msec.2021.112318
Sugimoto, W., Miyoshi, D., and Kawauchi, K. (2021). Detection of intracellular reactive oxidative species using the fluorescent probe Hydroxyphenyl fluorescein. Methods Mol. Biol. 2274, 207–215. doi: 10.1007/978-1-0716-1258-3_18
Sun, H., Zhang, J., Li, X., Gao, H., Liao, C., Wang, R., et al. (2025). Fish-derived cationic antimicrobial peptide piscidin 1 exerts broad-spectrum bactericidal activities against multidrug-resistant gram-negative pathogens. LWT 215:117189. doi: 10.1016/j.lwt.2024.117189
Verderosa, A. D., Totsika, M., and Fairfull-Smith, K. E. (2019). Bacterial biofilm eradication agents: a current review. Front. Chem. 7:824. doi: 10.3389/fchem.2019.00824
Vishwakarma, A., Dang, F., Ferrell, A., Barton, H. A., and Joy, A. (2021). Peptidomimetic polyurethanes inhibit bacterial biofilm formation and disrupt surface established biofilms. J. Am. Chem. Soc. 143, 9440–9449. doi: 10.1021/jacs.1c02324
Wang, X., Hong, X., Chen, F., and Wang, K.-J. (2022). A truncated peptide Spgillcin177-189 derived from mud crab Scylla paramamosain exerting multiple antibacterial activities. Front. Cell. Infect. Microbiol. 12:928220. doi: 10.3389/fcimb.2022.928220
Wang, L., Zhao, X., Xia, X., Zhu, C., Qin, W., Xu, Y., et al. (2019). Antimicrobial peptide JH-3 effectively kills Salmonella enterica Serovar Typhimurium strain CVCC541 and reduces its pathogenicity in mice. Probiot. Antimicrob. Proteins 11, 1379–1390. doi: 10.1007/s12602-019-09533-w
Xia, Y., Cebrián, R., Xu, C., Jong, A.de, Wu, W., and Kuipers, O. P. (2021). Elucidating the mechanism by which synthetic helper peptides sensitize Pseudomonas aeruginosa to multiple antibiotics PLoS Pathog. 17:e1009909 doi: 10.1371/journal.ppat.1009909
Xin, K., Sun, R., Xiao, W., Lu, W., Sun, C., Lou, J., et al. (2025). Short peptides from Asian scorpions: bioactive molecules with promising therapeutic potential. Toxins 17:114. doi: 10.3390/toxins17030114
Xu, Y., Ehrt, S., Schnappinger, D., and Beites, T. (2023). Synthetic lethality of Mycobacterium tuberculosis NADH dehydrogenases is due to impaired NADH oxidation. MBio 14:e01045-23. doi: 10.1128/mbio.01045-23
Xuan, J., Feng, W., Wang, J., Wang, R., Zhang, B., Bo, L., et al. (2023). Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resist. Updat. 68:100954. doi: 10.1016/j.drup.2023.100954
Yadav, B., Karad, D. D., Kharat, K. R., Makwana, N., Jaiswal, A., Chawla, R., et al. (2025). Environmental and clinical impacts of antibiotics’ sub-minimum inhibitory concentrations on the development of resistance in acinetobacter baumannii. Sci. Total Environ. 979:179521. doi: 10.1016/j.scitotenv.2025.179521
Yang, B., Tong, Z., Shi, J., Wang, Z., and Liu, Y. (2023). Bacterial proton motive force as an unprecedented target to control antimicrobial resistance. Med. Res. Rev. 43, 1068–1090. doi: 10.1002/med.21946
Yasir, M., Dutta, D., and Willcox, M. D. P. (2020). Activity of antimicrobial peptides and ciprofloxacin against Pseudomonas aeruginosa biofilms. Molecules 25:3843. doi: 10.3390/molecules25173843
Yi, K., Liu, S., Liu, P., Luo, X., Zhao, J., Yan, F., et al. (2022). Synergistic antibacterial activity of tetrandrine combined with colistin against MCR-mediated colistin-resistant Salmonella. Biomed. Pharmacother. 149:112873. doi: 10.1016/j.biopha.2022.112873
Zhang, J., Chen, C., Chen, J., Zhou, S., Zhao, Y., Xu, M., et al. (2020). Dual mode of anti-biofilm action of G3 against Streptococcus mutans. ACS Appl. Mater. Interfaces 12, 27866–27875. doi: 10.1021/acsami.0c00771
Zhang, M., Ouyang, J., Fu, L., Xu, C., Ge, Y., Sun, S., et al. (2022). Hydrophobicity determines the bacterial killing rate of α-helical antimicrobial peptides and influences the bacterial resistance development. J. Med. Chem. 65, 14701–14720. doi: 10.1021/acs.jmedchem.2c01238
Zhang, Q., Xu, Y., Wang, Q., Hang, B., Sun, Y., Wei, X., et al. (2015). Potential of novel antimicrobial peptide P3 from bovine erythrocytes and its analogs to disrupt bacterial membranes in vitro and display activity against drug-resistant bacteria in a mouse model. Antimicrob. Agents Chemother. 59, 2835–2841. doi: 10.1128/AAC.04932-14
Zhao, R., Hao, J., Yang, J., Tong, C., Xie, L., Xiao, D., et al. (2022). The co-occurrence of antibiotic resistance genes between dogs and their owners in families. iMeta 1:e21. doi: 10.1002/imt2.21
Zheng, S., Tu, Y., Li, B., Qu, G., Li, A., Peng, X., et al. (2025). Antimicrobial peptide biological activity, delivery systems and clinical translation status and challenges. J. Transl. Med. 23:292. doi: 10.1186/s12967-025-06321-9
Zhou, F., Kostantin, E., Yang, D.-Q., and Sacher, E. (2022). Cytotoxicity and antibacterial efficacy of AgCu and AgFe NanoAlloys: a comparative study. Antibiotics 11:1737. doi: 10.3390/antibiotics11121737
Zhou, L., Lian, K., Wang, M., Jing, X., Zhang, Y., and Cao, J. (2022). The antimicrobial effect of a novel peptide LL-1 on Escherichia coli by increasing membrane permeability. BMC Microbiol. 22:220. doi: 10.1186/s12866-022-02621-y
Keywords: Crab, antimicrobial peptide, AaeAP2a, bactericidal mechanism, peritonitis-associated sepsis
Citation: Luo W, Zhang L, Gao H, Li H, Li X, Wen Y, Sun H, Hang B, Zhang L, Zhang W, Liu X, Wang R, Wen B, Shen J, Zhu C, Bai Y, Wang L, Ding K and Hu J (2025) AaeAP2a, a scorpion-derived antimicrobial peptide, combats carbapenem-resistant Acinetobacter baumannii via membrane disruption and triggered metabolic collapse. Front. Microbiol. 16:1673333. doi: 10.3389/fmicb.2025.1673333
Edited by:
Nagendran Tharmalingam, Houston Methodist Research Institute, United StatesReviewed by:
Cristian Salinas-Restrepo, University of Antioquia, ColombiaPedro Alejandro Fong Coronado, University of Mississippi Medical Center, United States
Copyright © 2025 Luo, Zhang, Gao, Li, Li, Wen, Sun, Hang, Zhang, Zhang, Liu, Wang, Wen, Shen, Zhu, Bai, Wang, Ding and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhe Hu, eHhqaWFuaGVAMTI2LmNvbQ==; Ke Ding, a2VkaW5nMTlAMTYzLmNvbQ==; Lei Wang, d2xlaV8wMDdAMTYzLmNvbQ==
†These authors share first authorship
 Weiyu Luo
Weiyu Luo Liuwei Zhang
Liuwei Zhang Haolei Gao1,4
Haolei Gao1,4 Lei Wang
Lei Wang