- 1Center for Excellence in Post-Harvest Technologies, North Carolina Agricultural and Technical State University, Kannapolis, NC, United States
- 2Department of Biology, North Carolina Agricultural and Technical State University, Greensboro, NC, United States
Introduction: Food allergy is an increasing health concern worldwide. Microbes, food allergies, and polyphenols are found to be interrelated. However, studies relating polyphenols’ effect on food allergy via microbiome modulation are scarce, and there is a lack of common signature microbiome modulation patterns. Thus, this review aims to summarize the effect of polyphenols on food allergy via microbiome modulation.
Methods: Research articles were searched from Scopus, PubMed, ScienceDirect, and Web of Science database. The in vivo and in vitro studies were assessed via SYRCLE risk of bias and modified CONSORT checklist, respectively. The population characteristics and experimental details were extracted, and the data were synthesized narratively.
Results: The included studies were free of selective reporting of results. The allergy of egg (ovalbumin), milk (𝛽-lactoglobulin), soybean (𝛽-conglycinin), and shrimp allergy contributed to 54%, 23%, 15%, and 8% of the total included studies, respectively. The used compounds were a different source or types of polyphenols such as cocoa, cyanidin-3-O-glucoside (C3G), avenanthramide's (AVA), rosmarinic acid (RA), neohesperidin, and fermented apple juice for egg allergy, luteolin, and green tea polyphenol (GTP) for soybean allergy, and flavonoids (Luteolin, myricetin and hyperoside), ferulic acid, and luteolin for milk allergy. Allergies of milk, egg, wheat, and shrimp occurred with the reduction of Lactobacillus, Alistipes, Odaribactor, Akkermansia, Bacteroides, and Lachnospiraceae_NK4A136_group and an increase of Prevotella, Alloprevotella, Faecalibaculum, Helicobactor, Blautia, Clostridium, and Staphylococcus. The polyphenols modulated these microbes in order to attenuate the food allergies.
Discussion: The types of polyphenols, food allergies, animal model used, and taxonomic resolution of the microbiome studies lead to variation in the results. Thus, by increasing the studies on effect of polyphenols on individual food allergies, and combining with higher taxonomic resolution techniques such as shotgun metagenomics along with metabolomics would increase reliability of the results of the future studies.
1 Introduction
Food allergy is a serious health concern (Camps-Bossacoma et al., 2017; Yang et al., 2023) and about 5% of adults and 8% of children suffer food allergy worldwide (Li et al., 2022; Loh and Tang, 2018). Studies show the relationship among microbiota, polyphenolic compounds, and food allergies (Camps-Bossacoma et al., 2017; Yang et al., 2023; Li et al., 2022; Liu et al., 2023; Wang et al., 2022; Zhou et al., 2023) or cytokine-induced inflammations (Liu et al., 2021). The commensal gut microbes help the breakdown of dietary foods, produce short-chain fatty acid (SCFA), protect intestinal epithelial cells (IEC), modulate protective barriers, promote mucosal immunity system by developing tolerogenic CD103+ dendritic cells (DC) which influence regulatory T cells (T-reg) and IgA production from B cell, and metabolites production (Chen et al., 2016; Goldberg et al., 2020; Gu et al., 2022; Kourosh et al., 2018).
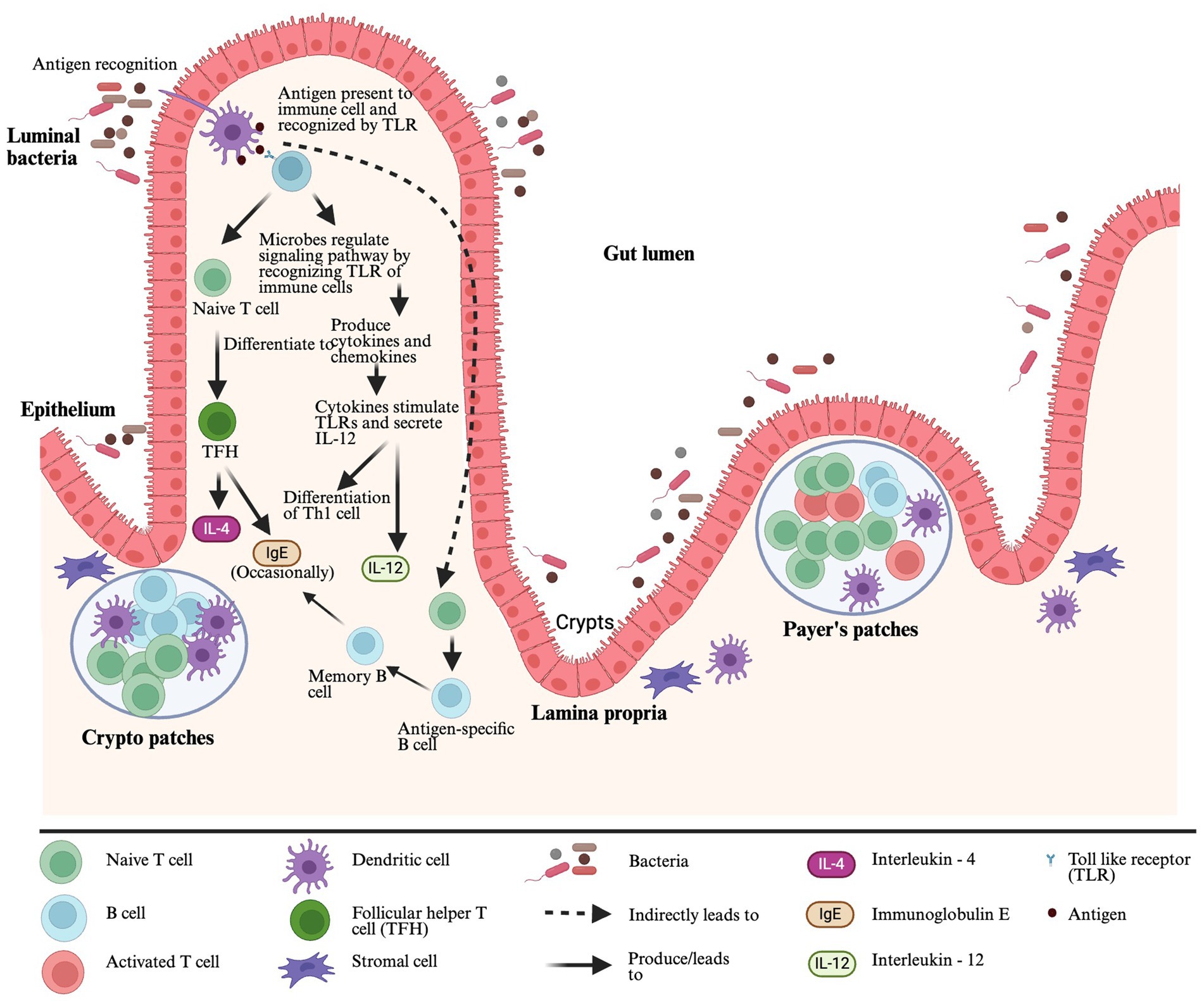
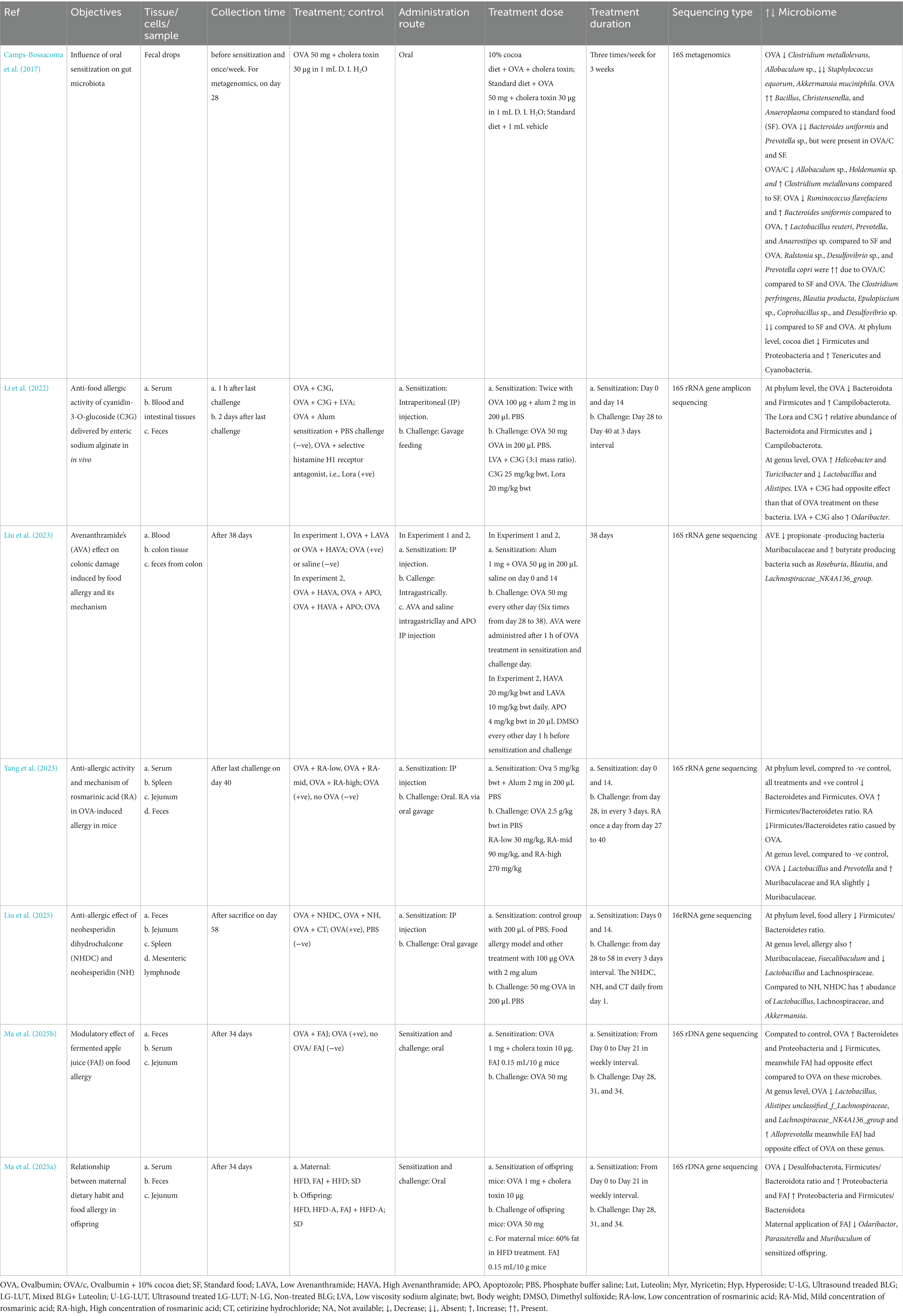
Gut-associated lymphoid tissue (GALT) is a mucosal immune system in the gut and the largest lymphoid tissue in the human body. It helps to promote oral tolerance to specific food allergens by distinguishing allergic and non-allergic food antigens (Ganesh and Versalovic, 2015). The GALT comprises of lymphoid structures and scattered lymphocytes with specialized functions (e.g., Natural killer cells, B cells, and T cells) in epithelium and lamina propria (LP). The lymphoid structures include Payer’s patches (PPs), crypto patches (CPs), DC, stromal cells around the crypts of the small intestine, intraepithelial lymphocytes (IELs), intestinal epithelial cells (IECs) and mesenteric lymph nodes (MLNs) (Sajdel-Sulkowska and Sajdel-Sulkowska, 2019) (Figure 1). The DC is one of the most important antigen-presenting cells (APC), and after getting antigens from the gut lumen, DCs present them to toll-like receptors (TLRs). The TLRs are pattern recognition receptors expressed in the immune cells and recognized by gut microbiota. After recognition of TLRs, microbes can regulate signaling pathways to communicate with a host producing pro- or anti-inflammatory cytokines and chemokines (Liu et al., 2022). When cytokines stimulate TLRs, they secrete Interleukin 12 (IL-12), and IL-12 helps in Type 1 helper CD4+ T cell differentiation in the presence of antigens (Ganesh and Versalovic, 2015). After interacting with antigen-containing DCs, naïve CD4+ T cells differentiate into T follicular helper (TFH) cells. These TFH cells produce IL-4 in response to allergens, and they can also occasionally generate Immunoglobulin E (IgE), replacing the role of Th2 cells in generating IgE (Hong et al., 2019). The DCs present antigens with T cells and can share that information with B cells separately. In that case, naïve B cells will turn into antigen-specific B cells, further differentiating into memory B cells or ultimately turning into plasma cells that produce IgE specific to an antigen (Shi et al., 2015) (Figure 1).
Polyphenol’s inhibitory or stimulatory effects on microbes depend upon the polyphenol’s structure and bacterial species/strains (Makarewicz et al., 2021). Polyphenols have antimicrobial properties with various mechanisms. Polyphenols interact with bacterial proteins on the cell wall, cell membrane, and with those proteins involved in the fundamental metabolism, inhibit DNA synthesis or cause DNA cleavage, disturb membrane permeability, antibiotic resistance, and enzyme formation. Moreover, polyphenols also inhibit ATP synthase and ATPase function, biofilm formation, and quorum sensing activities (Makarewicz et al., 2021; Rodríguez-Daza et al., 2021; Ashwin et al., 2021; Plamada and Vodnar, 2021).
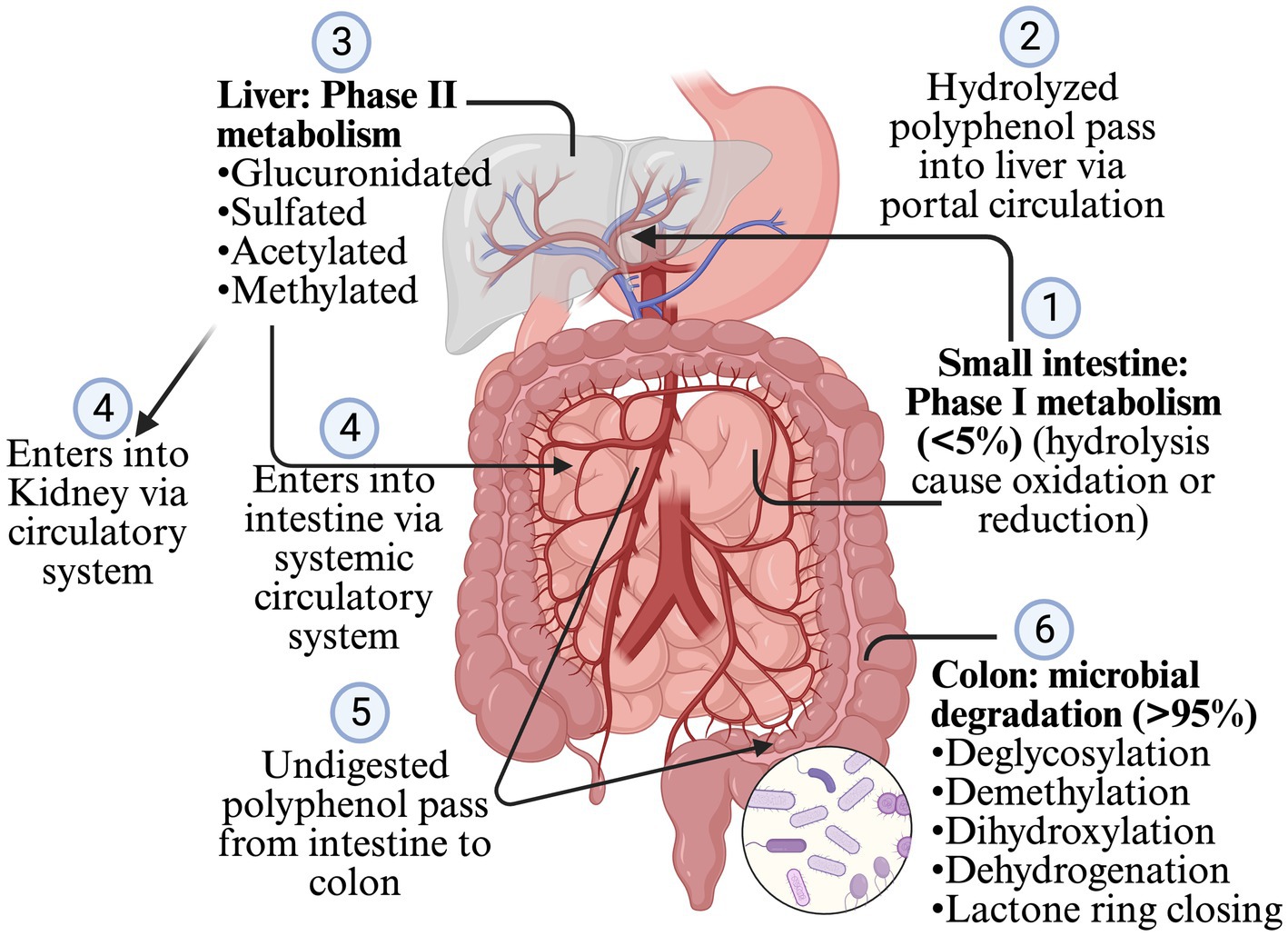
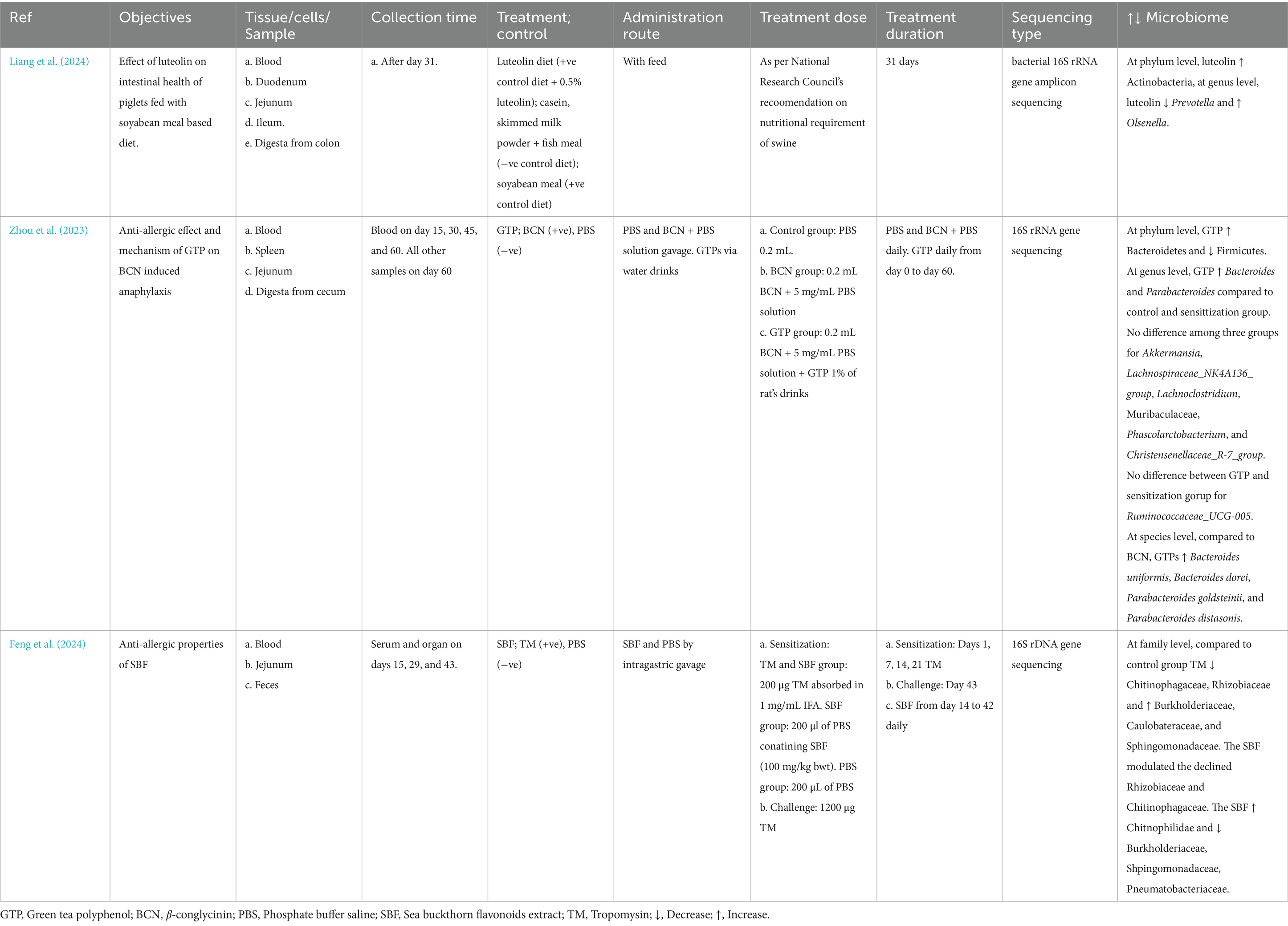
Less than 5% of polyphenols consumed are absorbed in the stomach and intestine, and >95% are undigested and reach the colon and interact with gut microbiota (Makarewicz et al., 2021; Rodríguez-Daza et al., 2021; Ashwin et al., 2021; Loo et al., 2020; Rowland et al., 2018). Most of the polyphenols are found as glycosides or in the polymers, so they need to be converted into aglycone and simple compounds so that enterocytes can absorb them. However, some glycosides, such as anthocyanins, can be absorbed without processing (Ashwin et al., 2021; Plamada and Vodnar, 2021; Loo et al., 2020; Rowland et al., 2018). The polymerization, and types of polyphenols such as flavonoids and non-flavonoids, affect the microbial conversion of the polyphenols. Flavonoids are composed of two benzene rings (A and B rings) linked to a heterocyclic pyrone C-ring. Simple phenolics derived from the A and B rings are released after the gut microbiota breaks down the C-ring in different positions. The hydroxylation pattern and the position of the B-ring determines the types of resulting phenotypes (Plamada and Vodnar, 2021; Ozdal et al., 2016). After absorption from the intestine, the polyphenol is mildly oxidized or reduced by hydrolysis (phage I metabolism, which increases the polarity of polyphenols) when it passes through enterocytes (Figure 2). The resulting simpler forms of polyphenols are transferred to the liver via the portal circulation, where they are glucuronidated, sulfated, acetylated or methylated (phage II metabolism that adds the chemical radicals into polyphenol) (Ashwin et al., 2021; Plamada and Vodnar, 2021; Rowland et al., 2018; Mithul Aravind et al., 2021). The resulting metabolites after phage II metabolism enter different organs via the systemic circulatory system. The undigested polyphenols in the intestine pass into the colon and are further metabolized into simpler forms by GM. The processes involved during this transformation are deglycosylation, demethylation, dihydroxylation, dehydrogenation, and closing of the lactone ring in the lower colon (Ashwin et al., 2021; Plamada and Vodnar, 2021; Rowland et al., 2018). After the resulting metabolites or polyphenols are absorbed from the colon, they go to phase II metabolism in the intestinal tissue and liver. Enterohepatic circulation helps to excrete the conjugated compounds back to the gut, which are again deconjugated by microbes and reabsorbed (Plamada and Vodnar, 2021; Loo et al., 2020) (Figure 2). The Daidzein, ellagitannins, lignans, proanthocyanidins converts primarily (80–90%) into O-desmethylangolensin or S-equol (30–50%), urolithins, enterolactones (and ultimately into enterodiol in human), and isomers of valerolactones (ultimately into phenolic acid), respectively. Similarly, isoflavone converts into propanoic acid or equol, anthocyanin into phenolic acid or phloroglucinol acid, quercetin into hippuric acid or benzoic acid, neochlorogenic acid into caffeic acid and quinoic acid, trans-resveratrol into piceid and resveratrolozide, and curcumin into ferulic acid and dihyroferulic acid (Makarewicz et al., 2021; Rodríguez-Daza et al., 2021; Plamada and Vodnar, 2021; Rowland et al., 2018; Ozdal et al., 2016; Mithul Aravind et al., 2021). The resulting polyphenol metabolites are more bioactive (bioaccessible and absorbed) than the parent polyphenol (Rowland et al., 2018).
Although studies had proven the association between food allergy and microbiome (Bunyavanich and Berin, 2019; Iweala and Nagler, 2023; Nance et al., 2020; Zhao et al., 2020), polyphenol and microbiome (Catalkaya et al., 2020; De Rossi et al., 2025; Kumar Singh et al., 2019; Piekarska-Radzik and Klewicka, 2020), and polyphenol and allergies (Zeng et al., 2022; Wu et al., 2018; Zhang et al., 2024; Wu et al., 2023), studies investigating the effect of polyphenol on food allergies via microbiome modulation are scarce and are in the initial stage (Li et al., 2022). Thus, this systematic review aims to determine common microbiome modulation pattern of polyphenols to mitigate food allergy.
2 Methodology
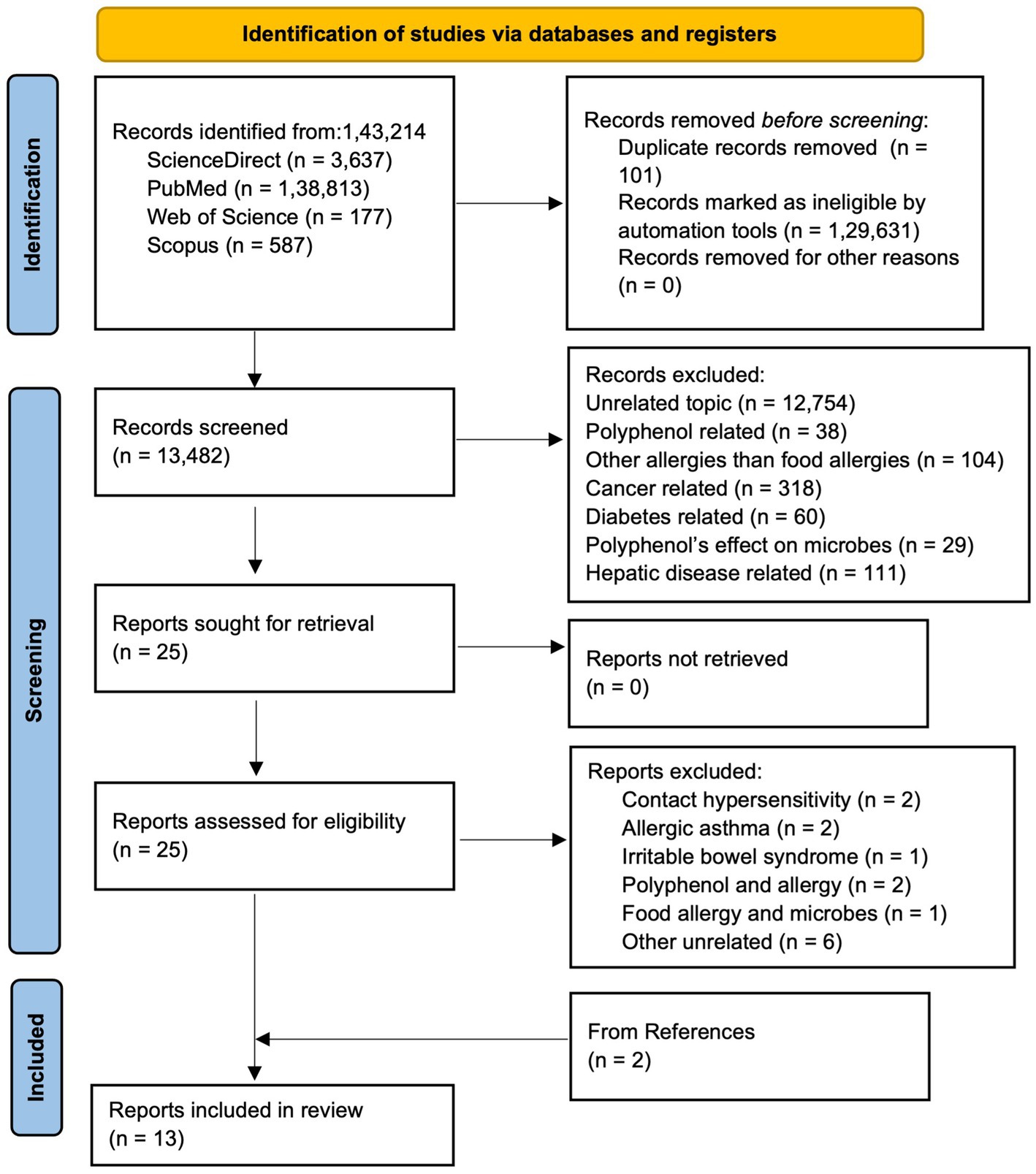
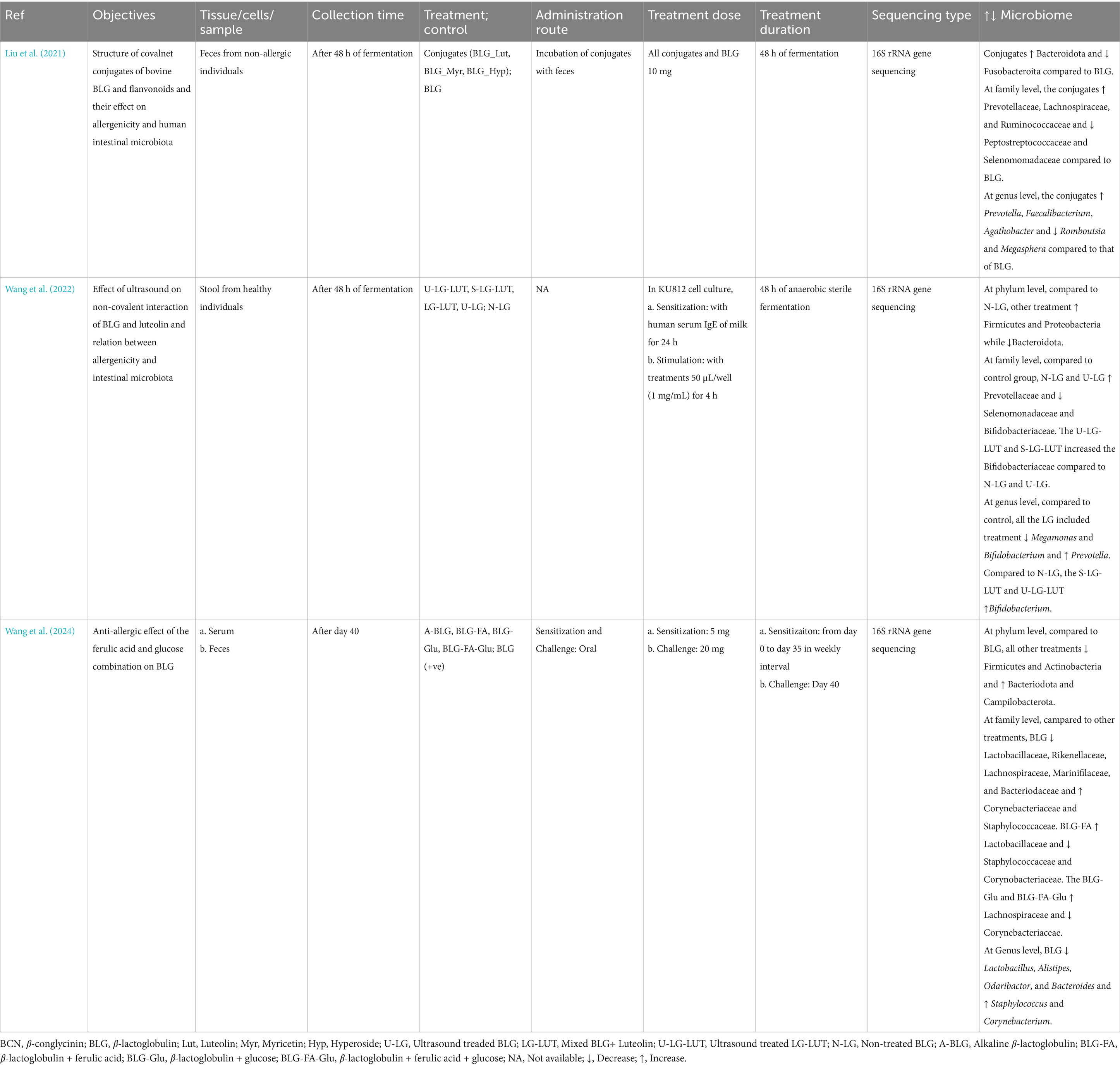
The Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) guideline was followed to conduct the systematic review (Page et al., 2021). Articles published until 2025, were searched from various databases: ScienceDirect, PubMed, Web of Science, and Scopus. The articles were searched in different databases using following combination of keywords: (a)In Scopus and Web of Science, (“hypersensi*” OR “food allerg*” OR “allerg*” OR “anaphyla*”) AND (“microbio*” OR “microorga*”) AND (“proanthocya*” OR “anthocya*” OR “procyan*"OR “flav*” OR “polypheno*”); (b) In ScienceDirect, (“hypersensitivity” OR “food allergy” OR “anaphylaxis”) AND (“microbiome” OR “microorganism”) AND (“procyanidins”OR “flavonoids” OR “polyphenols”); and (c) In PubMed, ((((((“Food Hypersensitivity”[Mesh]) OR “Hypersensitivity”[Mesh]) AND “Gastrointestinal Microbiome”[Mesh]) OR “Microbiota”[Mesh]) AND “Polyphenols”[Mesh]) OR “Flavonoids”[Mesh]) OR “Proanthocyanidins”[Mesh]. As shown in PRISMA flow diagram (Figure 3), from a total of 1,43,214 articles obtained using aforementioned keywords from different databases, 101 were removed in deduplication, 1,29,631 were removed using automation process by search tools as they were one of the following categories: other than original research articles, and published in other than English. Out of 13,482 remaining screened articles, 13,457 were excluded during title and abstract screening. Out of the 25 remaining articles, two were related to contact hypersensitivity, two articles were related allergic asthma, one was related to irritable bowel syndrome, two were related to polyphenol and allergy, one was related to food allergy and microbes only, and six were other unrelated articles. Thus, 11 articles were obtained from screening and two articles were obtained from reference and citation of the 11 articles leading to 13 total articles for this systematic review. Two authors (TR and JR) agreed upon search criteria, searched and screened articles, and discussed and resolved disagreements with the third author (RB). Three authors (TR, RB, and JR) independently extracted data on objectives, tissue or sample type, treatment, treatment administration route, treatment dose, and duration, sequencing type, and microbiome change, and population characteristics (animal type, age, weight, sex, total number). The evidence in articles was determined with the PICO framework as follows: population: animal (mice, rats, pigs) and human, intervention: polyphenol, comparison: food allergen, output: change in microbiome.
Three authors (TR, RB, and JR) performed the risk of bias. The risk of bias evaluation of in vivo studies was performed according to SYRCLE’s risk of bias tool (Hooijmans et al., 2014). Comprehension and unbiases on abstract, background and rationale, objectives, hypothesis, intervention, outcome, statistical method, outcome, limitation, and funding were evaluated. For in vitro studies, the risk of bias was calculated based on a modified CONSORT checklist (Faggion, 2012; Lam et al., 2024). The process of complete randomization, blinding, unbiased and complete reporting of the articles was evaluated. Due to differences in treatment, objective and overall design, and outcomes among selected articles, we narratively synthesized the articles. The authors agreed upon the synthesis process.
3 Results
3.1 Risk of bias
The in vitro studies did report background, objectives, outcomes, and limitations. However, they did not mention the hypothesis, and one of the studies did not clearly mention the statistical method used to analyse the data (Supplementary Table 1). All the in vivo studies did not explain the study’s randomization process and blinding steps. Two of the studies did not mention reasons for incomplete outcomes. All of them were free from selective outcome reporting (Supplementary Table 2).
3.2 Population/study characteristics
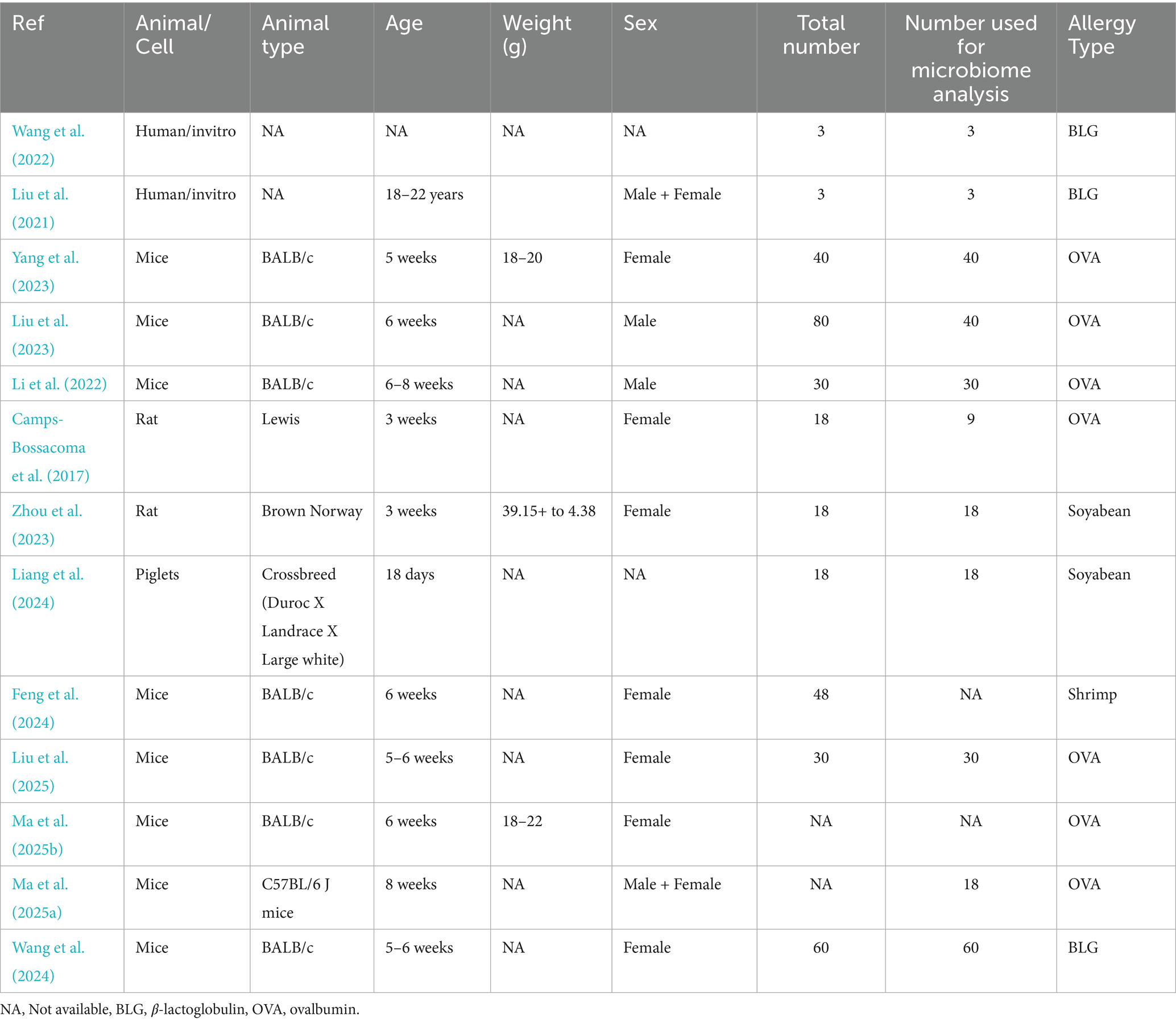
Two studies were done on 3-week-old Lewis and Brown Norway rats, seven on 5–8 weeks-old BALB/c mice, one on C57BL/6 J mice, one on 18-day-old piglets, and two on incubation of human fecal matter in the artificial chamber (Table 1). All animal studies used either female or male animals. The allergy on ovalbumin, beta-lactoglobulin (milk) and soybean, and shrimp contributed 54, 23, 15, and 8% of the included studies, respectively (Table 1).
3.3 Microbiome and food allergies
3.3.1 Egg allergies
In general, ovalbumin (OVA) seem to reduce Lactobacillus (Yang et al., 2023; Li et al., 2022; Ma et al., 2025b; Liu et al., 2025), Alistipes (Li et al., 2022; Ma et al., 2025b), Prevotella (Yang et al., 2023), Akkermansia (Camps-Bossacoma et al., 2017), and members of lachnospiraceae such as Unclassified_f_lachnospiraceae, and Lachnospiraceae_NK4A136_group (Ma et al., 2025b; Liu et al., 2025). The OVA promoted the Bacteroides (Camps-Bossacoma et al., 2017; Ma et al., 2025b), Helicobactor (Li et al., 2022), Faecalibaculum (Liu et al., 2025), and Alloprevotella (Ma et al., 2025b). At higher level of classification, OVA promotes muribaculaceae (Yang et al., 2023; Liu et al., 2025), campylobacteria (Li et al., 2022), and proteobacteria (Ma et al., 2025a), meanwhile the OVA reduced firmicutes (Li et al., 2022; Ma et al., 2025a, 2025b; Liu et al., 2025). Use of various polyphenol helped to alleviate allergy via reducing the microbes promoted by OVA and promoting those microbes reduced by OVA. However, due to variation on the polyphenol and animal model used, diverse effect of polyphenol were found on microbes. For example, cocoa diet promoted Lactobacillus, Provotella, Anastipes, and it reduced Clostridium and Blautia. On the other hand, cyanidin-3-O-Glucoside promoted Rosburia, Blautia, and Lachnospiraceae_NK4A136_group (Table 2).
3.3.2 Soyabean and shrimp allergies
Although soyabean’s effect on microbes were not clearly mentioned, polyphenols such as luteolin reduced Prevotella and increased Olsenella (Liang et al., 2024) and. Unlike in ova allergy, 𝛽-conglycinin or green tea polyphenol (GTP) did not affect the Akkermansia, Lachnospiraceae_NK4A136_group, and Muribaculaceae. However, GTP promoted the Bacteroides and Parabacteroides (Zhou et al., 2023) (Table 2). The shrimp allergen reduced chitinophagaceae, rhizobiaceae, and increased burkholdariaceae, caulobactereaceae and sphingomonadaceae while the polyphenol extract had opposite effect compared to the allergen on these microbes (Feng et al., 2024) (Table 3).
3.3.3 Milk allergies
Milk allergy reduce firmicutes and increases bactereodota (Wang et al., 2022; Wang et al., 2024). The allergy also increases Staphylococcus, Campilobacterota and reduces Lactobacillus, Alistipes, Odaribactor, and Bacteroides (Wang et al., 2024). Use of various polyphenols such as flavonoids, luteolin, ferulic acids increases bifidobactereaceae, lactobacillaceae, Faecalibacterium, and Agathobactor. The polyphenol use reduced the staphylococceae, corynebactereaceae, and Ramboustia (Table 4).
4 Discussion and future perspectives
In general, food allergy is related to reduction of Lactobacillus, Alistipes, Odaribactor, Akkermansia, Bacteroides, and Lachnospiraceae_NK4A136_group and an increase of Prevotella, Alloprevotella, Faecalibaculum, Helicobactor, Blautia, Clostridium, and Staphylococcus (Tables 2–4). Previous studies also found that food allergy is related to reduced Bacteroides, Alistipes, Lachnospriaceae_NK4A136_group, Akkermansia, and Lactobacillus and abundance of Prevotella, Helicobacter and Clostridium (Chen et al., 2016; Gu et al., 2022; Tanaka et al., 2024; Liu et al., 2019; Huang et al., 2025; Hara et al., 2024; Chang et al., 2018; E et al., 2024; Xu et al., 2025; Qiao et al., 2024).
Very few common microbial species were identified across different food allergy studies. Moreover, the effect of polyphenols on food allergies varied according to the type of polyphenol used and type of food allergies. For example, cocoa (Camps-Bossacoma et al., 2017), flavonoids (Liu et al., 2021), and Luteolin (Wang et al., 2022) increased Prevotella compared to allergen alone in milk and egg allergies. However, the Prevotella was reduced by the Luteolin compared to that of soybean allergen (Liang et al., 2024). This implies that the microbiome’s role also varies based on food allergy type (Goldberg et al., 2020; De Filippis et al., 2021). Similar, variation was also found in previous studies where Prevotella copri was increased in milk allergy while it was decreased in peanut allergy (Goldberg et al., 2020). Futhermore, Blautia was increased by cyanidin-3-O-glucoside (C3G) (Li et al., 2022) and Avenanthramide’s (AVA) (Liu et al., 2023) but it was reduced by cocoa diet (Camps-Bossacoma et al., 2017) compared to that of allergens alone. Furthermore, the C3G (Li et al., 2022) increased Lactobacillus alistipes and while cocoa diet (Camps-Bossacoma et al., 2017) increased the Lactobacillus reuteri compared to allergen alone. At phylum level, cocoa diet (Camps-Bossacoma et al., 2017), green tea polyphenol (GTP) (Zhou et al., 2023), or rosmarinic acid (Yang et al., 2023; Zhou et al., 2023) decreased firmicutes, but C3G (Li et al., 2022; Zhou et al., 2023) increased the firmicutes in comparison to the allergens. Other studies investigating the effect of different polyphenols in microbes have also reported the different in effect of various polyphenols on the same genus/species of microbes (Loo et al., 2020; Mithul Aravind et al., 2021). Thus, more studies are needed on the effect of polyphenols on a specific types of food allergies via microbiome modulation in order to identify signature microbiome modulation pattern of the specific types of allergy before determining the pattern for food allergies in general.
Besides the types of polyphenols or food allergens, the taxonomic resolution of the microbiome in a study may also affect the results. For example, effects of both the C3G and cocoa diet were measured at species level, i.e., they both increased the Lactobacillus alistipes and Lactobacillus reuteri, respectively. However, the cocoa diet increased Lactobacillus reuteri but decreased Ruminococcus flavefaciens compared to allergen. Both of these bacterial species are firmicutes, but represent different classes, orders, or families (Camps-Bossacoma et al., 2017; Zhou et al., 2023). Similarly, the C3G (Li et al., 2022) or AVA (Liu et al., 2023) increased Blautia, but cocoa diet (Camps-Bossacoma et al., 2017), reduced Blautia producta compared to the allergen treatment. Furthermore, cocoa diet increased the Clostridium metallovans, but it caused a disappearance of the Clostridium perfringens compared to standard food. Similarly, in other studies, Clostridium senso stricto1 found in healthy children while Clostridium innnocuum were higher in wheat allergic children (Kanchongkittiphon et al., 2024) Not only species but also strains of a species vary in their presence and function (Mennini et al., 2021). These results indicate that it is important to study the higher level of taxonomic resolution of the microbiome in order to accurately determine the effect of polyphenols in food allergy via microbiome modulation.
5 Limitation
This study included the articles published in English. Thus, it may cause omission of important articles in other languages. The included studies also had diverse polyphenol forms and animal models (rats, mice, and piglets). These cause variation in the results, making it hard to find common microbiome signature and their modulation pattern by polyphenol. All included studies used 16S rRNA gene sequencing to investigate the microbiome changes due to the polyphenols. The 16S rRNA sequencing is not rigorous enough to study at species or strain level of microbes. Moreover, some of the studies only reported the results at phylum and genus level, which increased the variability of the microbiome results. Most of the other food allergies and gut microbiome studies also reported microbiome diversity and functional prediction using 16S rRNA sequencing techniques (Chen et al., 2016; Goldberg et al., 2020; Gu et al., 2022; Kourosh et al., 2018; Bunyavanich and Berin, 2019; Tanaka et al., 2024; Mennini et al., 2021; Fazlollahi et al., 2019; Tulyeu et al., 2019). Very few studies have used combined approaches such as the 16S rRNA gene sequencing and metabolomics (Xu et al., 2022) or shutgun metagenomics (De Filippis et al., 2021) to determine microbial signature and their potential functional in various food allergies. Furthermore, out of other important food allergies, only four types were covered by the included studies (egg, milk, shrimp, and soybean). Other important food allergies such as allergies related to peanut, wheat, and tree nuts are yet to be studied in terms of polyphenol’s effect on these allergies via microbiome modification. Thus, finding in this study is limited to modulation of polyphenols on egg, milk, soyabean and shrimp allergy. For modulatory effect of polyphenol on the other important food allergies including peanut, wheat, and nuts, further studies are necessary in the future.
6 Conclusion
Higher level of variation in polyphenol used and animal model used along with lower taxonomic resolution of microbiome in the included studies in this review led to lack of common microbiome modulation pattern of polyphenols in the reduction of food allergy. High-resolution taxonomic level investigation (Jovel et al., 2016) or microbiomes-and-metabolomics approach (Xu et al., 2022) have been proven effective in getting the signature gut microbiome in food allergy studies. Given that 16S rRNA sequencing technique would not provide the species or strain level resolution which is critical for identification of signature microbiome and their functional potential in food allergy. Moreover, shotgun sequencing approach provide higher taxonomic resolution and opportunity to direct assessment of functional potential of the microbiomes (Jovel et al., 2016). Thus, use of shotgun metagenomics combined with metabolomics could provide reliable food allergy microbiome signature and their potential function as well as reliable measure of polyphenol’s effect on food allergy via microbiome modulation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
TR: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. RB: Investigation, Supervision, Writing – review & editing. JR: Investigation, Visualization, Writing – review & editing. LW: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by 1890 Capacity Building Grant Program from the United States Department of Agriculture and National Institute of Food and Agriculture (Project award No. 2023-38821-39979).
Conflict of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1673472/full#supplementary-material
References
Ashwin, K., Pattanaik, A. K., and Howarth, G. S. (2021). Polyphenolic bioactives as an emerging group of nutraceuticals for promotion of gut health: a review. Food Biosci. 44:101376. doi: 10.1016/j.fbio.2021.101376
Bunyavanich, S., and Berin, M. C. (2019). Food allergy and the microbiome: current understandings and future directions. J. Allergy Clin. Immunol. 144, 1468–1477. doi: 10.1016/j.jaci.2019.10.019
Bunyavanich, S., Shen, N., Grishin, A., Wood, R., Burks, W., Dawson, P., et al. (2016). Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 138, 1122–1130. doi: 10.1016/j.jaci.2016.03.041
Camps-Bossacoma, M., Pérez-Cano, F. J., Franch, À., and Castell, M. (2017). Gut microbiota in a rat oral sensitization model: effect of a cocoa-enriched diet. Oxidative Med. Cell. Longev. 2017:7417505. doi: 10.1155/2017/7417505
Catalkaya, G., Venema, K., Lucini, L., Rocchetti, G., Delmas, D., Daglia, M., et al. (2020). Interaction of dietary polyphenols and gut microbiota: microbial metabolism of polyphenols, influence on the gut microbiota, and implications on host health. Food Front. 1, 109–133. doi: 10.1002/fft2.25
Chang, M., Zhao, Y., Qin, G., and Zhang, X. (2018). Fructo-oligosaccharide alleviates soybean-induced anaphylaxis in piglets by modulating gut microbes. Front. Microbiol. 9:9. doi: 10.3389/fmicb.2018.02769
Chen, C., Chen, K., Kong, M., Chang, H., and Huang, J. (2016). Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr. Allergy Immunol. 27, 254–262. doi: 10.1111/pai.12522
De Filippis, F., Paparo, L., Nocerino, R., Della Gatta, G., Carucci, L., Russo, R., et al. (2021). Specific gut microbiome signatures and the associated pro-inflamatory functions are linked to pediatric allergy and acquisition of immune tolerance. Nat. Commun. 12:5958. doi: 10.1038/s41467-021-26266-z
De Rossi, L., Rocchetti, G., Lucini, L., and Rebecchi, A. (2025). Antimicrobial potential of polyphenols: mechanisms of action and microbial responses—a narrative review. Antioxidants 14:14. doi: 10.3390/antiox14020200
E, D. p., Plekhova, V., Vangeenderhuysen, P., Baeck, N., Bullens, D., Claeys, T., et al. (2024). Integrated gut metabolome and microbiome fingerprinting reveals that dysbiosis precedes allergic inflammation in IgE-mediated pediatric cow's milk allergy. Allergy 79, 949–963. doi: 10.1111/all.16005
Faggion, C. M. (2012). Guidelines for reporting pre-clinical in vitro studies on dental materials. J. Evid.Based Dent. Pract. 12, 182–189. doi: 10.1016/j.jebdp.2012.10.001
Fazlollahi, M., Chun, Y., Grishin, A., Wood, R. A., Burks, A. W., Dawson, P., et al. (2019). Early-life gut microbiome and egg allergy. Allergy 73, 1515–1524. doi: 10.1111/all.13389
Feng, X., Yan, Z., Ren, X., Jia, Y., Sun, J., Guo, J., et al. (2024). Sea buckthorn flavonoid extracted with high hydrostatic pressure alleviated shrimp allergy in mice through the microbiota and metabolism. J. Agric. Food Chem. 72, 25094–25102. doi: 10.1021/acs.jafc.4c06928
Ganesh, B. P., and Versalovic, J. (2015). Luminal conversion and immunoregulation by probiotics. Front. Pharmacol. 6:6. doi: 10.3389/fphar.2015.00269
Goldberg, M. R., Mor, H., Magid Neriya, D., Magzal, F., Muller, E., Appel, M. Y., et al. (2020). Microbial signature in IgE-mediated food allergies. Genome Med. 12:92. doi: 10.1186/s13073-020-00789-4
Gu, S., Xie, Q., Chen, C., Liu, C., and Xue, W. (2022). Gut microbial signatures associated with peanut allergy in a BALB/c mouse model. Foods 11:11. doi: 10.3390/foods11101395
Hara, M., Suzuki, H., Hayashi, D., Morii, W., Nakamura, T., Kiyoki, K., et al. (2024). Gut microbiota of one-and-a-half-year-old food-allergic and healthy children. Allergol. Int. 73, 550–555. doi: 10.1016/j.alit.2024.03.004
Hong, S., Lee, J. Y., Lee, M., Han, D., Ko, H., Sprent, J., et al. (2019). Food antigens drive spontaneous IgE elevation in the absence of commensal microbiota. Sci. Adv. 5:eaaw1507. doi: 10.1126/sciadv.aaw1507
Hooijmans, C. R., Rovers, M. M., de Vries, R. B., Leenaars, M., Ritskes-Hoitinga, M., and Langendam, M. W. (2014). SYRCLE’S risk of bias tool for animal studies. BMC Med. Res. Methodol. 14:43. doi: 10.1186/1471-2288-14-43
Huang, J., Qiao, H., Li, Q., Zhang, Y., Zhang, C., Su, H., et al. (2025). Osteopontin protects from ovalbumin-induced asthma by preserving the microbiome and the intestinal barrier function. mSystems 10:e00389-25. doi: 10.1128/msystems.00389-25
Iweala, O. I., and Nagler, C. R. (2023). The microbiome and food allergy. Annu. Rev. Immunol. 37, 377–403. doi: 10.1146/annurev-immunol-042718-041621
(63) Jovel, J., Patterson, J., Wang, W., Hotte, N., O’keefe, S., Mitchel, T., et al. (2016). Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 7:7. doi: 10.3389/fmicb.2016.00459
Kanchongkittiphon, W., Nopnipa, S., Mathuranyanon, R., Nonthabenjawan, N., Sritournok, S., Manuyakorn, W., et al. (2024). Characterization of gut microbiome profile in children with confirmed wheat allergy. Asian Pac. J. Allergy Immunol. doi: 10.12932/ap-080623-1626
Kourosh, A., Luna, R. A., Balderas, M., Nance, C., Anagnostou, A., Devaraj, S., et al. (2018). Fecal microbiome signatures are different in food-allergic children compared to siblings and healthy children. Pediatric Allergy Immunol. 29, 545–554. doi: 10.1111/pai.12904
Kumar Singh, A., Cabral, C., Kumar, R., Ganguly, R., Kumar Rana, H., Gupta, A., et al. (2019). Beneficial effects of dietary polyphenols on gut microbiota and strategies to improve delivery efficiency. Nutrients 11:11. doi: 10.3390/nu11092216
Lam, T., Tran, N. N., Pham, L. D., Lai, N. V., Dang, B. N., Truong, N. N., et al. (2024). Flavonoids as dual-target inhibitors against α-glucosidase and α-amylase: a systematic review of in vitro studies. Nat. Prod. Bioprospect. 14:4. doi: 10.1007/s13659-023-00424-w
Li, J., Zou, C., and Liu, Y. (2022). Amelioration of ovalbumin-induced food allergy in mice by targeted rectal and colonic delivery of cyanidin-3-O-glucoside. Foods 11:11. doi: 10.3390/foods11111542
Liang, X., Zheng, S., Zhou, Y., Li, J., and Zhang, Z. (2024). Luteolin, a natural flavonoid, exhibits a protective effect on intestinal injury induced by soybean meal in early-weaned piglets. J. Anim. Sci. 102:102. doi: 10.1093/jas/skae214
Liu, X., Wang, C., Tang, S., Wang, G., Huang, Y., Yang, F., et al. (2025). Comparative study on the alleviating effect of neohesperidin dihydrochalcones and its synthetic precursor neohesperidin on ovalbumin-induced food allergy. Food Res. Int. 212:116436. doi: 10.1016/j.foodres.2025.116436
Liu, J., Wang, Y., Tu, Z., Chen, W., and Yuan, T. (2021). Bovine β-Lactoglobulin covalent modification by flavonoids: effect on the Allergenicity and human intestinal microbiota. J. Agric. Food Chem. 69, 6820–6828. doi: 10.1021/acs.jafc.1c02482
Liu, Y., Wang, J., and Wu, C. (2022). Modulation of gut microbiota and immune system by probiotics, pre-biotics, and post-biotics. Front. Nutr. 8:8. doi: 10.3389/fnut.2021.634897
Liu, S., Yang, B., Yang, P., and Liu, Z. (2019). Herbal formula-3 ameliorates OVA-induced food allergy in mice may via modulating the gut microbiota. Am. J. Transl. Res. 11:5812.
Liu, P., Zhang, M., Liu, T., Mo, R., Wang, H., Zhang, G., et al. (2023). Avenanthramide improves colonic damage induced by food allergies in mice through altering gut microbiota and regulating Hsp70-NF-κB signaling. Nutrients 15:15. doi: 10.3390/nu15040992
Loh, W., and Tang, M. L. K. (2018). The epidemiology of food allergy in the global context. IJERPH 15:15. doi: 10.3390/ijerph15092043
Loo, Y. T., Howell, K., Chan, M., Zhang, P., and Ng, K. (2020). Modulation of the human gut microbiota by phenolics and phenolic fiber-rich foods. Compr. Rev. Food Sci. Food Saf. 19, 1268–1298. doi: 10.1111/1541-4337.12563
Ma, J., Yu, J., Jia, Y., Luo, Z., Yang, X., Li, H., et al. (2025a). Fermented apple juice reduces the susceptibility of offspring mice to food allergy exacerbated by maternal high-fat diet. Nutrients 17:17. doi: 10.3390/nu17111927
Ma, J., Zhang, L., Ren, X., Luo, Z., Zhao, M., Tong, P., et al. (2025b). High hydrostatic pressure pretreated fermented apple juice attenuated anaphylaxis by improving gut microbiota and metabolic regulation. Food Biosci. 64:105844. doi: 10.1016/j.fbio.2025.105844
Makarewicz, M., Drożdż, I., Tarko, T., and Duda-Chodak, A. (2021). The interactions between polyphenols and microorganisms, especially gut microbiota. Antioxidants 10:10. doi: 10.3390/antiox10020188
Mennini, M., Reddel, S., Del Chierico, F., Gardini, S., Quagliariello, A., Vernocchi, P., et al. (2021). Gut microbiota profile in children with IgE-mediated cow’s Milk allergy and cow’s Milk sensitization and probiotic intestinal persistence evaluation. IJMS 22:22. doi: 10.3390/ijms22041649
Mithul Aravind, S., Wichienchot, S., Tsao, R., Ramakrishnan, S., and Chakkaravarthi, S. (2021). Role of dietary polyphenols on gut microbiota, their metabolites and health benefits. Food Res. Int. 142:110189. doi: 10.1016/j.foodres.2021.110189
Nance, C. L., Deniskin, R., Diaz, V. C., Paul, M., Anvari, S., and Anagnostou, A. (2020). The role of the microbiome in food allergy: a review. Children 7:7. doi: 10.3390/children7060050
Ozdal, T., Sela, D. A., Xiao, J., Boyacioglu, D., Chen, F., and Capanoglu, E. (2016). The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 8:8. doi: 10.3390/nu8020078
Page, M. J., Moher, D., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ 372:n160. doi: 10.1136/bmj.n160
Piekarska-Radzik, L., and Klewicka, E. (2020). Mutual influence of polyphenols and Lactobacillus spp. bacteria in food: a review. Eur. Food Res. Technol. 247, 9–24. doi: 10.1007/s00217-020-03603-y
Plamada, D., and Vodnar, D. C. (2021). Polyphenols—gut microbiota interrelationship: a transition to a new generation of prebiotics. Nutrients 14:137. doi: 10.3390/nu14010137
Qiao, C., Bian, S., Huang, H., Xiao, H., Ma, L., and Han, R. (2024). Impact of ovalbumin allergy on oral and gut microbiome dynamics in 6-week-old BALB/c mice. Front. Microbiol. 15:15. doi: 10.3389/fmicb.2024.1439452
Rodríguez-Daza, M. C., Pulido-Mateos, E. C., Lupien-Meilleur, J., Guyonnet, D., Desjardins, Y., and Roy, D. (2021). Polyphenol-mediated gut microbiota modulation: toward prebiotics and further. Front. Nutr. 8:689456. doi: 10.3389/fnut.2021.689456
Rowland, I., Gibson, G., Heinken, A., Scott, K., Swann, J., Thiele, I., et al. (2018). Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 57, 1–24. doi: 10.1007/s00394-017-1445-8
Sajdel-Sulkowska, E. M., and Sajdel-Sulkowska, E. M. (2019). Altered microbiota-GALT communication in IBD and ASD: changes in IELs and AhR/ARNT gene polymorphism. J. Biotechnol. Biomed. 2:2. doi: 10.26502/jbb.2642-91280018
Shi, Y., Xu, L., Peng, K., Wu, W., Wu, R., Liu, Z., et al. (2015). Specific immunotherapy in combination with Clostridium butyricum inhibits allergic inflammation in the mouse intestine. Sci. Rep. 5:5. doi: 10.1038/srep17651
Tanaka, M., Korenori, Y., Washio, M., Kobayashi, T., Momoda, R., Kiyohara, C., et al. (2024). Signatures in the gut microbiota of Japanese infants who developed food allergies in early childhood. FEMS Microbiol. Ecol. 93:93. doi: 10.1093/femsec/fix099
Tulyeu, J., Kumagai, H., Jimbo, E., Watanabe, S., Yokoyama, K., Cui, L., et al. (2019). Probiotics prevents sensitization to Oral antigen and subsequent increases in intestinal tight junction permeability in juvenile–young adult rats. Microorganisms 7:7. doi: 10.3390/microorganisms7100463
Wang, T., Chen, W., Shao, Y., Liu, J., and Tu, Z. (2022). Ultrasound improved the non-covalent interaction of β-Lactoglobulin with luteolin: regulating human intestinal microbiota and conformational epitopes reduced allergy risks. Foods 11:11. doi: 10.3390/foods11070988
Wang, Y., Zhang, K., Chen, W., Mao, J., Wang, X., Shao, Y., et al. (2024). Gut microbiome-serum metabolism revealed the allergenicity of ferulic acid combined with glucose-modified β-Lactoglobulin. J. Agric. Food Chem. 72, 11746–11758. doi: 10.1021/acs.jafc.4c01545
Wu, T., Li, Z., Wu, Y., Yang, X., Li, L., Chen, S., et al. (2023). Exploring plant polyphenols as anti-allergic functional products to manage the growing incidence of food allergy. Front. Nutr. 10:10. doi: 10.3389/fnut.2023.1102225
Wu, X., Lu, Y., Xu, H., Lin, D., He, Z., Wu, H., et al. (2018). Reducing the allergenic capacity of β-lactoglobulin by covalent conjugation with dietary polyphenols. Food Chem. 256, 427–434. doi: 10.1016/j.foodchem.2018.02.158
Xu, J., Sheikh, T. M. M., Shafiq, M., Khan, M. N., Wang, M., Guo, X., et al. (2025). Exploring the gut microbiota landscape in cow milk protein allergy: clinical insights and diagnostic implications in pediatric patients. J. Dairy Sci. 108, 73–89. doi: 10.3168/jds.2024-25455
Xu, J., Ye, Y., Ji, J., Sun, J., Wang, J., and Sun, X. (2022). Untargeted Metabolomic profiling reveals changes in gut microbiota and mechanisms of its regulation of allergy in OVA-sensitive BALB/c mice. J. Agric. Food Chem. 70, 3344–3356. doi: 10.1021/acs.jafc.1c07482
Yang, Q., Jia, B., Shang, J., Wang, X., Xu, L., Liu, X., et al. (2023). Effects of rosmarinic acid on immune response and intestinal microbiota in ovalbumin-induced intestinal allergy mice. J. Sci. Food Agric. 104, 3002–3012. doi: 10.1002/jsfa.13192
Zeng, B., Jiang, T., Xiong, W., Che, H., and Sun, S. (2022). Protective properties of polyphenols in food allergy: a review. Allergy 78, 1654–1656. doi: 10.1111/all.15459
Zhang, C., Zhang, Q., Li, H., Cheng, Z., Fan, S., Xie, H., et al. (2024). Dietary polyphenols reduced the allergenicity of β-lactoglobulin via non-covalent interactions: a study on the structure-allergenicity relationship. Food Sci. Human Wellness 13, 2617–2628. doi: 10.26599/fshw.2022.9250210
Zhao, W., Ho, H., and Bunyavanich, S. (2020). The gut microbiome in food allergy. Ann. Allergy Asthma Immunol. 122, 276–282. doi: 10.1016/j.anai.2018.12.012
Keywords: polyphenol, food allergy, microbiome, 16s rRNA sequencing, egg allergy, milk allergy, soyabean allergy, shrimp allergy
Citation: Rana TS, Bansode RR, Rana JP and Williams LL (2025) A systematic review: polyphenol’s effect on food allergy via microbiome modulation. Front. Microbiol. 16:1673472. doi: 10.3389/fmicb.2025.1673472
Edited by:
Wenchao Cai, Shihezi University, ChinaReviewed by:
Iftikhar Younis Mallhi, Minhaj University Lahore, PakistanChangqi Liu, San Diego State University, United States
Copyright © 2025 Rana, Bansode, Rana and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonard L. Williams, bGx3QG5jYXQuZWR1
 Tekan Singh Rana
Tekan Singh Rana Rishipal Rastrapal Bansode
Rishipal Rastrapal Bansode Jenny Pakhrin Rana
Jenny Pakhrin Rana Leonard L. Williams
Leonard L. Williams