- 1Department of Medical Laboratory, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Hubei Provincial Engineering Research Center of Intestinal Microecological Diagnostics, Therapeutics, and Clinical Translation, Wuhan, China
- 3National Key Laboratory of Intelligent Tracking and Forecasting for Infectious Diseases, Chinese Center for Disease Control and Prevention, Beijing, China
- 4Cancer Research Institute of Wuhan, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Introduction: A novel Erwinia strain, BC051422T, was isolated from the blood of a patient at the Central Hospital of Wuhan, Wuhan, PR China, in 2022. The strain was identified as gram-negative, facultatively anaerobic, motile, and rod-shaped.
Methods: Preliminary analysis based on the 16S rRNA gene sequence and multilocus sequence analysis of the atpD, infB, rpoB, and gyrB genes unveiled that this strain is closely related to Erwinia members. Whole-genome sequencing was performed, and the average nucleotide identity (ANI) and in silico DNA–DNA hybridization (isDDH) values between strain BC051422T and type strains of all known Erwiniaceae species ranged from 68.8% to 83.4% and 19.2% to 35.5%, respectively, which below the accepted species delineation thresholds of 95% for ANI and 70% for isDDH. The major cellular fatty acids of strain BC051422T were C16:0, C16:1 ω7c/C16:1 ω6c, and C17:0 cyclo, consistent with profiles similar to those observed in other Erwinia species. The genomic DNA G+C content was 55.95 mol%. Strain BC051422T can be differentiated from other Erwinia species by its ability to ferment sucrose, but its inability to metabolize mannose, rhamnose, melibiose, and sorbitol.
Results: Genotypic and phenotypic characteristics together support the classification of strain BC051422T as a novel species of the genus Erwinia, for which the name Erwinia wuhanensis sp. nov. is proposed. The type strain of E. wuhanensis sp. nov. is BC051422T (=GDMCC 1.4074T = JCM 36319T).
Introduction
The genus Erwinia was first described by (Winslow et al. 1920) in 1920 and is currently classified under the family Erwiniaceae, which was proposed to refine the taxonomy within the order Enterobacterales (Adeolu et al., 2016). According to the List of Prokaryotic Names with Standing in Nomenclature (https://lpsn.dsmz.de/genus/erwinia) (Parte et al., 2020), 20 Erwinia species have been validly published to date. Members of the Erwinia genus are widely distributed and primarily isolated from plants, including fruit trees, potato stems, pomelo, and olive (Geider et al., 2006; Ming-Kai et al., 2022; Moretti et al., 2011; Ramirez-Bahena et al., 2016). Some species, such as E. aphidicola, E. typographi, and E. teleogrylli, have been isolated from insects like pea aphid, bark beetles, and crickets, respectively (Harada et al., 1997; Liu et al., 2016; Skrodenyte-Arbaciauskiene et al., 2012). Several Erwinia species are known plant pathogens. E. pyrifoliae, E. piriflorinigrans, E. uzenensis, and E. amylovora are causative agents of fire blight and blossom necrosis in pome fruit trees (Kharadi et al., 2019; Kim et al., 1999; López et al., 2011; Matsuura et al., 2012). A recently described phytopathogen, E. sorbitola, has demonstrated potential pathogenicity to both plants and animals (Tao et al., 2023). Despite this, human infections caused by Erwinia species remain rare. E. persicina CDC 4073-83 was isolated from the urine of an elderly woman with a urinary tract infection and also from bile fluid cultures of another patient with perihilar cholangiocarcinoma (Ceylan and Özden, 2022; O'Hara et al., 1998). E. billingiae has been implicated in cutaneous infections, bacteremia, and septic arthritis, with cases linked to environmental exposure to plants (Bonnet et al., 2019; Prod'homme et al., 2017). E. tasmaniensis was recovered from the severely necrotic tissue of a patient with cervical lymphadenitis and reported as an Erwinia-like organism (Shin et al., 2008). In this study, we report the taxonomic characterization of strain BC051422T, isolated from the blood of a patient with chronic renal failure following hemodialysis. Comprehensive genomic and phenotypic analyses indicate that this strain represents a novel species of the genus Erwinia, for which we propose the name Erwinia wuhanensis sp. nov.
Materials and methods
Isolation and ecology
Strain BC051422T was recovered from the blood culture of a 64-year-old female patient presenting with the symptom of fever after hemodialysis at the Central Hospital of Wuhan (Wuhan; 30 °35′ N, 114 °19′ E; PR China) in 2022. The patient developed fever 2 h after regular hemodialysis, with a body temperature of 38.3 °C. After the consultation, the patient was found to have a history of hypertension chronic heart failure, chronic renal failure, maintenance hemodialysis, and renal anemia. She also had a history of type 2 diabetes for more than 10 years, diabetic nephropathy, diabetic retinopathy, fatty liver, gallstones, gastric ulcers, and had undergone appendectomy and amputation of both hands. Laboratory tests revealed a white blood cell (WBC) count of 12.0 × 109/L (94.1% neutrophils) (normal 3.59.5 × 109/L, 40%−75% neutrophils), hemoglobin of 102 g/L (normal 115–150 g/L), and C-reactive protein of 4.54 mg/dL (normal 0–0.6 mg/dL), and procalcitonin of 3.98ng/mL (normal 0–0.05 ng/mL). The patient tested negative for the novel coronavirus. A computerized tomography scan of Lung and abdomen showed bronchitis, multiple small nodules in both lungs, splenomegaly, bilateral kidney atrophy and stones, proximal ureteral stones on the right side, and sclerosis of the abdominal aorta and its branches and bilateral renal blood vessels. Due to the incresed indicators of inflamation, two sets of blood culture were performed. One of the cultures was positive for gram-negative rods after 18 h of incubation at 37 °C in the presence of 5% CO2 on Columbia blood agar and MacConkey agar plates (Guangzhou Dijing Microbial Technology Co., Ltd., Guangzhou, China). The antimicrobial susceptibility of strain BC051422T was conducted on VITEK 2 platform (bioMérieux) according to the manufacturer's guidelines, and drug sensitivity was determined according to the Clinical and Laboratory Standards Institute standard (Supplementary Table S1). The isolate is sensitive to all antibiotics tested. The patient's condition improved after receiving piperacillin-tazobactam sodium (4.5 g/12 h) and discharged 6 days after admission.
16S rRNA gene sequence analysis
The genomic DNA of purified bacteria was extracted using the TIANamp Bacteria DNA Kit (TIANGEN Biotech, Co., Ltd, Beijing, China). 16S rRNA gene sequencing was performed with universal primers (27F: 5′-AGTTTGATCMTGGCTCAG-3′; 1492R: 5′-GGTTACCTTGTTACGACTT-3′). The amplification was performed on the C1000 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA), and the product was sequenced on the Applied Biosystems 3730XL platform (Thermo Fisher Scientific Inc., MA, USA). The nearly complete 16S rRNA sequence of the strain BC051422T was analyzed with reference to the EzBioCloud Database (Yoon et al., 2017a). The phylogenetic tree was constructed using the neighbor-joining (NJ) and maximum-likelihood (ML) algorithms with Kimura's two-parameter model as well as maximum parsimony algorithm by using the MEGA software version 11 (Tamura et al., 2021).
Multilocus sequence analysis
The internal sequence fragments of partial atpD, infB, rpoB, and gyrB of strain BC051422T were retrieved from their whole-genome sequences. Phylogenetic tree based on concatenated partial atpD, infB, rpoB, and gyrB sequences was constructed using the PhyloSuite platform (Zhang et al., 2020). The best partitioning scheme and evolutionary models were selected using PartitionFinder2 v2.1.1 (Lanfear et al., 2017), with the rcluster algorithm and AICc criterion. Bayesian Inference phylogenies were inferred using MrBayes v3.2.7 (Ronquist et al., 2012) under the partition model (2 parallel runs, 200,000 generations), wherein the initial 25% of the sampled data were discarded as burn-in.
Genome analysis
The draft genome sequencing of the strain BC051422T was performed using the Illumina HiSeq platform by generating paired-end libraries. Fragmented genomic DNA with an average size of 300 bp was selected for sequencing. The raw data of sequencing were evaluated by FastQC v0.11.2 and cut by Trimmomatic v0.36 (Bolger et al., 2014) to obtain relatively accurate and effective data. The filtered reads were assembled into contigs using SPAdes v3.5.0 (Bankevich et al., 2012), and GapFiller v1.11 (Boetzer and Pirovano, 2012) is used to fill the GAP between contigs. The genetic elements were predicted by Prokka v1.10 (Seemann, 2014). The ANI was calculated using the OrthoANIu algorithm (Yoon et al., 2017b). The in silico DNA-DNA hybridization (isDDH) value between BC051422T and its related type strain was analyzed by Genome-to-Genome Distance Calculator 3.0 (Meier-Kolthoff et al., 2022). All other genome sequences of closely related type strains were obtained from the GenBank database, and the genome sequence of strain BC051422T has been deposited in GenBank/EMBL/DDBJ/PIR (accession number: JAUJUH010000000).
Physiological and chemotaxonomic analyses
Growth tests were performed at 30 °C on different media, including Luria–Bertani agar, Columbia blood agar, MacConkey agar, Chocolate agar, and Brain Heart Infusion agar (Guangzhou Dijing Microbial Technology Co., Ltd., Guangzhou, China). The growth temperature range was tested at 4, 10, 25, 28, 30, 35, 37, 42, and 50 °C on Columbia blood agar for 48 h. Tolerance to NaCl and pH was detected in test tubes containing 2 ml of LB broth after incubation for 48 h in a thermostatically controlled incubator at different NaCl concentrations (0–10%, w/v, at intervals of 1%) and pH values (4.0–10.0, at intervals of 1.0 pH unit), respectively. Anaerobic growth was performed by incubating cultures on Columbia blood agar for 7 days in an anaerobic bag (bioMerieux). Gram staining was performed using Rapid Gram Stain (Baso Diagnostics, Inc., Zhuhai, China) as per the manufacturer's instructions. The mass spectrometry was conducted on a matrix-assisted laser desorption ionization/time-of-flight mass spectrometry (MALDI-TOF MS; Bruker Daltonik GmbH, Germany) platform using a direct smear of a fresh bacterial colony on the target plate. The bacterial suspension was fixed in glutaraldehyde phosphate buffer overnight and then stained with sodium phosphotungstate. The cell morphology was observed by using a transmission electron microscope (Hitachi TEM System; Tokyo, Japan). The biochemical characterization of strain BC051422T was performed in a microbiochemical tube as per the manufacturer's instructions (Hangzhou Microbial Reagent, Co., Ltd, Hangzhou, China). E. persicina GDMCC1.331T was used as a positive control, with 3% (v/v) H2O2 used to detect the catalytic activity of catalase. The oxidase activity was measured using filter paper soaked with the reagent tetramethyl-p-phenylenediamine. Erwinia persicina GDMCC1.331T and Erwinia sorbitola J780T were used as control strains for the biochemical analyses. Data for species other than E. wuhanensis, E. persicina, and E. sorbitola were obtained from the Bacterial Diversity Metadatabase (BacDive) (Reimer et al., 2022). Chemotaxonomic analysis of the BC051422T strain and the reference strain was performed by using the GC platform (Agilent GC 6890), flame ionization detector (FID), and Sherlock Microbial Identification System (Sasser, 1990).
Results and Discussion
16S rRNA gene phylogeny
In total, 1,427 contiguous nucleotides of strain BC051422T were sequenced and deposited in GenBank under the accession number OQ938606. The 16S rRNA gene sequence showed the highest similarity to E. tasmaniensis Et1/99T (98.74%), followed by E. piriflorinigrans CFBP 5888T (98.59%), E. endophytica BSTT30T (98.31%), E. billingiae CIP 106121T (98.18%), and Pantoea eucrina LMG 2781T (97.98%). Similarity to other species of the genera Erwinia and Pantoea was below 98.00%. A phylogenetic tree constructed using the neighbor-joining method with 1,000 bootstrap replicates showed that strain BC051422T is distinct from previously described species, including the recently published E. sorbitola J780T (Figure 1). Additionally, a phylogenetic tree constructed using the maximum likelihood algorithm placed strain BC051422T within the family Erwiniaceae, forming a separate lineage (Supplementary Figure S1). To further validate the evolutionary relationships of the strain BC051422T and closely related species, phylogenetic tree with the method of maximum parsimony was constructed (Supplementary Figure S2).
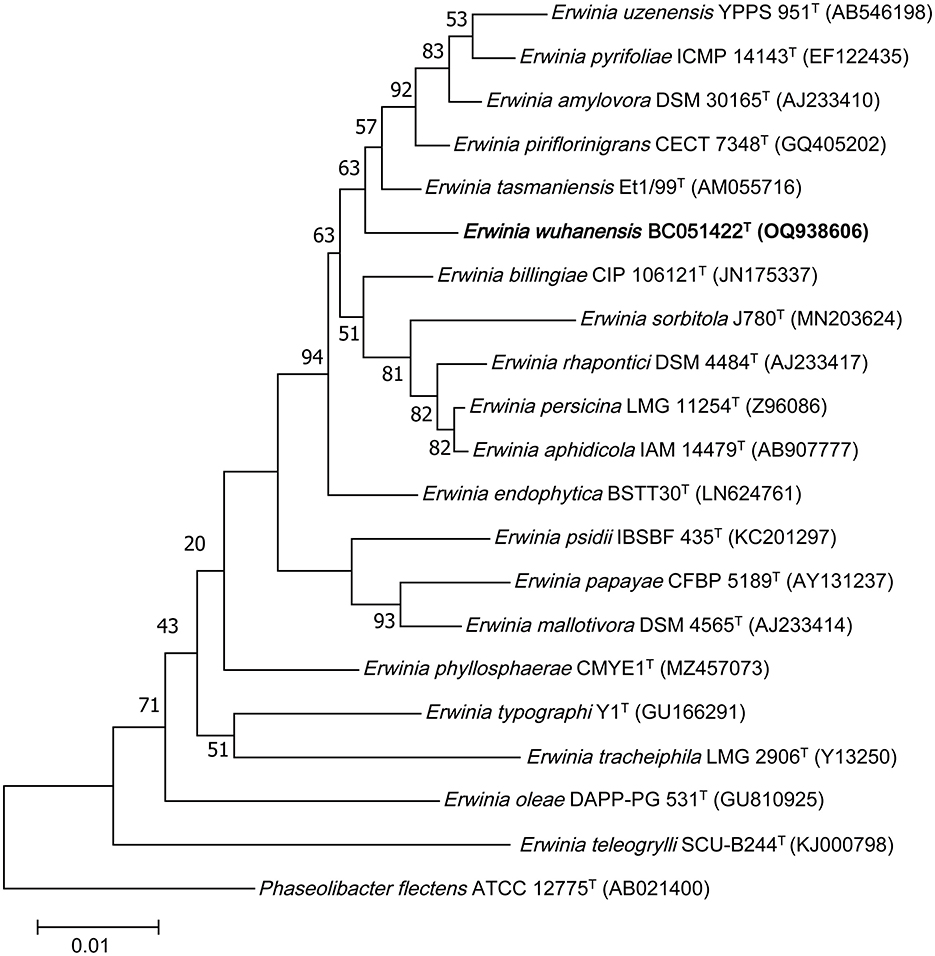
Figure 1. Phylogenetic tree based on the 16S rRNA gene sequences showing the relationship of novel strain BC051422T (bold) and members within genus Erwinia. The tree was reconstructed by the neighbor-joining method, and Phaseolibacter flectens ATCC 12775T (AB021400) was used as an outgroup. Bootstrap values (>50 %) based on 1,000 replicates are shown at branch nodes. T, type strain.
Multilocus sequence analysis
Multilocus sequence analysis (MLSA) is essential for distinguishing closely related genera such as Erwinia, Pantoea, and Tatumella (Brady et al., 2008). Phylogenetic analysis based on the concatenated sequences of partial atpD, infB, rpoB, and gyrB genes revealed that strain BC051422T clusters with type strains of E. amylovora LMG2024T, E. aphidicola JCM21242T, E. tansmaniensis NCPPB 4358T, E. persicina LMG11254T, and E. rhapontici (Figure 2). Sequence accession numbers for all strains used in the MLSA are listed in Supplementary Table S2.
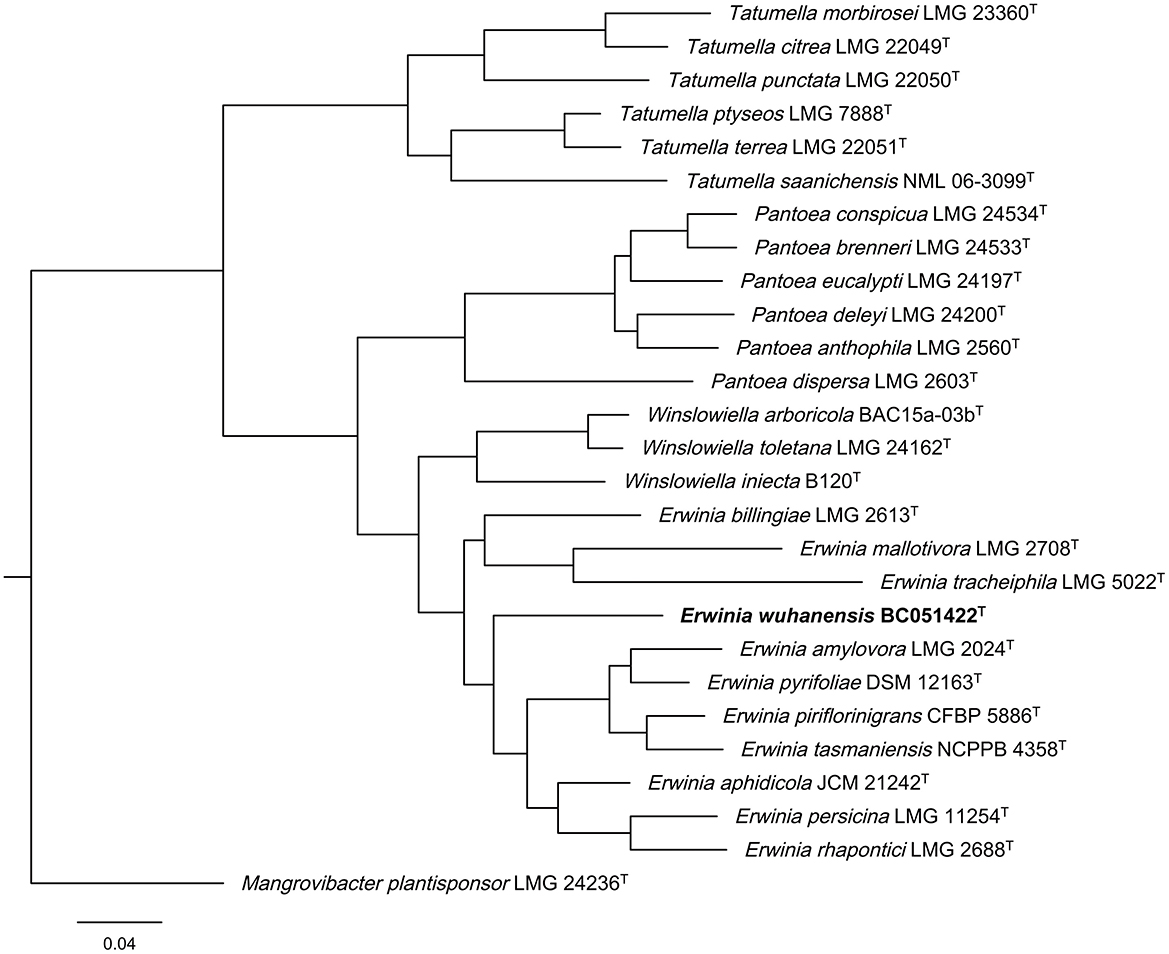
Figure 2. Phylogenetic tree based on concatenated partial atpD, infB, rpoB, and gyrB gene sequences from the genera Erwinia, Pantoea, Tatumella, and Winslowiella. The partial genes of strain BC051422T (bold) were retrieved from their whole genome sequences, and the genes of other reference strains were obtained from the GenBank database. The sequences were subjected to PhyloSuite platform according to the guidelines as described. Mangrovibacter plantisponsor LMG 24236T was used as outgroup strain.
Genomic characterization
The draft genome of strain BC051422T is 4,596,380 bp in length, assembled into 26 contigs, with an N50 value of 489,674 bp, 200 × coverage, and a GC content of 55.95 mol%. ANI and isDDH values between strain BC051422T and closely related type strains in Erwiniaceae ranged 68.75%−83.44% (ANI) and 19.2%−35.5% (isDDH), both well below the species delineation thresholds of 95%−96% (ANI) and 70% (isDDH) (Chun et al., 2018) (Table 1). These results support the classification of strain BC051422T as a novel species within the genus Erwinia. A comparative analysis of genomic features among closely related species identified in the phylogenetic tree was performed. The genome size of these species ranges from 3.8Mb to 5.1Mb, with relatively little variation in GC content and gene mumber of tRNA (Supplementary Table S3).
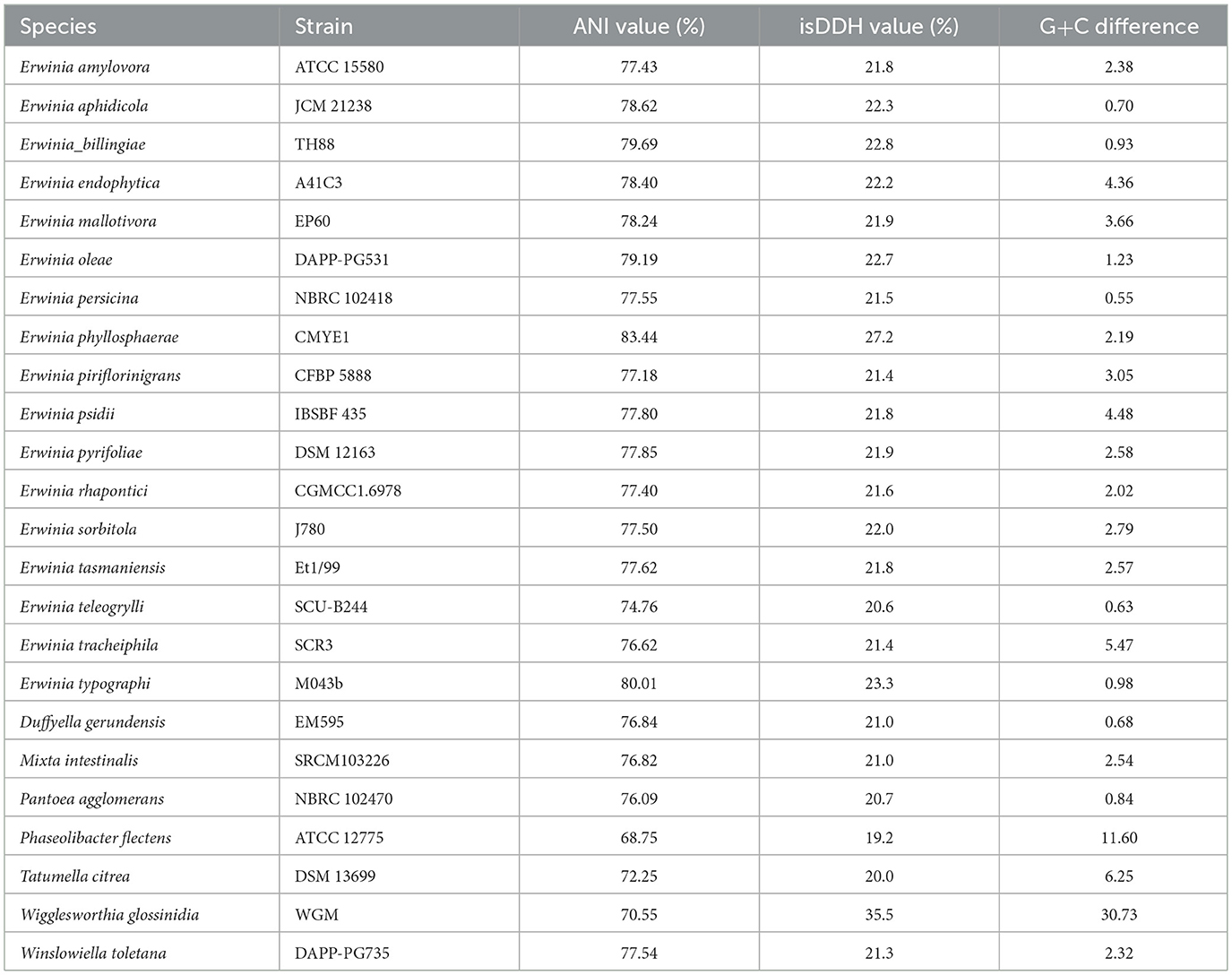
Table 1. Average nucleotide identity (ANI), in silico DNA–DNA hybridization (isDDH), and G+C difference values between strain BC051422T and strains of other members of the family Erwiniaceae.
Prediction of pathogenicity
To further explore the pathogenicity of the novel species, an advanced annotation regarding to virulence factors and antibiotic resisrance was conducted. A total of 121 virulence factor terms were predicted through the core dataset of the Virulence Factors of Pathogenic Bacteria (VFDB) (Liu et al., 2022), which contains 342 predicted proteins, accounting for 8.3% of the total proteins (Figure 3A). The database predicted the most nutritional/Metabolic factors, followed by motility and immune modulation related proteins. Adherence and invasion are critical for initial infection of pathogenic organisms, but the predicted invasion-related proteins are in low proportion, suggesting that the novel species may serve as opportunistic pathogen. Additionally, the strain BC051422T was analyzed by the Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al., 2013) and 75 proteins related to antibiotic resistance were predicted (Figure 3B). The antibiotic resistance ontology (ARO) rsmA and CRP, which belong to resistance-nodulation-division (RND) family, were annotated with high identity of matching region (85.25% and 98.57%, respectively).

Figure 3. Prediction of pathogenicity of the strain BC051422T through database. (A) The virulence factors were predicted through the core dataset of the VFDB. (B) The antibiotic resistance was predicted by CARD.
Physiology and chemotaxonomy
Strain BC051422T grows well on all tested media, forming visible colonies within 48 h. It grows at 10°C−42°C (optimum: 30°C−35°C), in 0%−8% (w/v) NaCl (optimum: 0%−1% NaCl), and at pH 4.0–9.0 (optimum: pH 7.0), including under anaerobic conditions. Colonies are light yellow, smooth, circular, and 1–2 mm in diameter after 24 h on Columbia blood agar at 30 C (Supplementary Figure S1). Cells are gram-negative rods with peripheral flagella, as observed by microscopy and transmission electron microscopy. A representative mass spectrum was acquired through MALDI-TOF MS (Supplementary Figure S3). Strain BC051422T tested positive for β-galactosidase and nitrate reduction, and negative for arginine dihydrolase, ornithine decarboxylase, lysine decarboxylase, urease, Voges-Proskauer reaction, cytochrome oxidase, and H2S production. Acid was produced from glucose, arabinose, and sucrose, but not from mannose, rhamnose, melibiose, or sorbitol (Table 2). These traits differentiate BC051422T from other Erwinia species, particularly its ability to ferment sucrose and arabinose, but not mannose, rhamnose, or sorbitol. Detailed results of the aforementioned tests are presented in the species description.
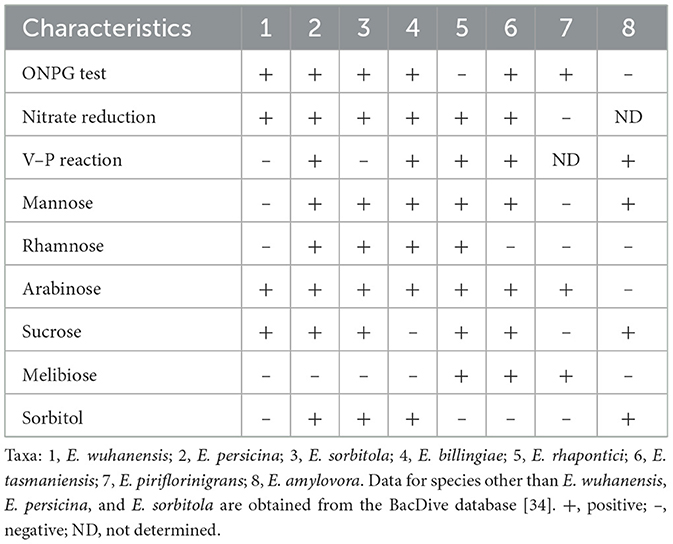
Table 2. Biochemical characteristics of strain E. wuhanensis BC051422T and type strains of other Erwinia species.
The major cellular fatty acids of BC051422T were C16:0 (31.2%), C17:0 cyclo (11.4%), summed feature 3 (C16:1 ω7c/C16:1 ω6c, 23.9%), and summed feature 8 (C18:1 ω7c and/or C18:1 ω6c, 10.8%). The fatty acid profile of E. wuhanensis and strains of closely related species is presented in Table 3.

Table 3. Cellular fatty acid composition (as a percentage of the total) of Erwinia wuhanensis and strains of the most closely related species.
Conclusions
Based on phylogenetic, genomic, phenotypic, and chemotaxonomic evidence, strain BC051422T represents a novel species of the genus Erwinia, for which the name E. wuhanensis sp. nov. is proposed.
Description of E. wuhanensis sp. nov.
E. wuhanensis (wu.han.en'sis. N.L. fem. adj. wuhanensis, referring to Wuhan city, Hubei Province, China, where the type strain was isolated).
Cells are gram-negative, non-spore-forming, facultatively anaerobic rods (1.5–2.5μm), motile with peripheral flagella. Colonies are light yellow, smooth, circular, and 1–2 mm in diameter after 24 h at 30 °C on Columbia blood agar. It grows on LB agar, Columbia blood agar, MacConkey agar, chocolate agar, and brain heart infusion agar. Growth occurs at 10°C−42°C (optimum: 30 C−35 C), in 0%−8% (w/v) NaCl (optimum: 0%−1% NaCl) in brain heart infusion agar, and at pH 4.0–9.0 (optimum: pH 7.0). Positive for catalase, β-galactosidase, aesculin hydrolysis, and nitrate reduction, but negative for cytochrome oxidase, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, urease, Voges-Proskauer reaction, and H2S production. Ferments glucose, arabinose, and sucrose, but not mannose, rhamnose, melibiose, raffinose, sorbitol, or adonitol. Major cellular fatty acids are C16:0, C17:0 cyclo, summed feature 3 (C16:1 ω7c/C16:1 ω6c), and summed feature 8 (C18:1 ω7c and/or C18:1 ω6c).
The type strain is BC051422T, isolated from the blood of a patient at the Central Hospital of Wuhan, Wuhan city, Hubei Province, China, in 2022. The draft genome is 4.6 Mb with a DNA G+C content of 55.95 mol%. Genome and 16S rRNA sequences are available in GenBank/EMBL/DDBJ/PIR under accession numbers JAUJUH010000000 and OQ938606, respectively. The strain is deposited at the Guangdong Microbiology Culture Centre as GDMCC 1.4074T and at the Japan Collection of Microorganisms as JCM 36319T.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving humans were approved by Ethical Committee of the Central Hospital of Wuhan. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
YMZ: Conceptualization, Funding acquisition, Methodology, Writing – original draft. YZ: Formal analysis, Investigation, Writing – original draft. JY: Conceptualization, Supervision, Writing – review & editing. ZXL: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the Wuhan Municipal Health Commission (Project No. WX21Q42) and the Contral Hospital of Wuhan (Project No. 330032) to YMZ.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1675452/full#supplementary-material
References
Adeolu, M., Alnajar, S., Naushad, S., and S Gupta, R. (2016). Genome-based phylogeny and taxonomy of the 'Enterobacteriales': proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Genome Biol. 13:R56. doi: 10.1186/gb-2012-13-6-r56
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonnet, I., Bozzi, B., Fourniols, E., Mitrovic, S., Soulier-Escrihuela, O., Brossier, F., et al. (2019). Erwinia billingiae as unusual cause of septic arthritis, France, 2017. Emerg. Infect. Dis. 25, 1587–1589. doi: 10.3201/eid2508.181073
Brady, C., Cleenwerck, I., Venter, S., Vancanneyt, M., Swings, J., and Coutinho, T. (2008). Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 31, 447–460. doi: 10.1016/j.syapm.2008.09.004
Ceylan, A. N., and Özden, I. (2022). A rare bacterial pathogen in a patient with perihilar cholangiocarcinoma: Erwinia persicina; first case from Turkey. Mikrobiyol. Bul. 56, 574–579. doi: 10.5578/mb.20229716
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Geider, K., Auling, G., Du, Z., Jakovljevic, V., Jock, S., and Volksch, B. (2006). Erwinia tasmaniensis sp. nov., a non-phytopathogenic bacterium from apple and pear trees. Int. J. Syst. Evol. Microbiol. 56, 2937–2943. doi: 10.1099/ijs.0.64032-0
Harada, H., Oyaizu, H., Kosako, Y., and Ishikawa, H. (1997). Erwinia aphidicola, a new species isolated from pea aphid, Acyrthosiphon pisum. J. Gen. Appl. Microbiol. 43, 349–354. doi: 10.2323/jgam.43.349
Kharadi, R. R., Castiblanco, L. F., Waters, C. M., and Sundin, G. W. (2019). Phosphodiesterase genes regulate amylovoran production, biofilm formation, and virulence in Erwinia amylovora. Appl. Environ. Microbiol. 85:e02233-18. doi: 10.1128/AEM.02233-18
Kim, W. S., Gardan, L., Rhim, S. L., and Geider, K. (1999). Erwinia pyrifoliae sp. nov., a novel pathogen that affects Asian pear trees (Pyrus pyrifolia Nakai). Int. J. Syst. Bacteriol. 49, 899–905. doi: 10.1099/00207713-49-2-899
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Liu, B., Luo, J., Li, W., Long, X. F., Zhang, Y. Q., Zeng, Z. G., et al. (2016). Erwinia teleogrylli sp. nov., a bacterial isolate associated with a Chinese cricket. PLoS ONE 11:e0146596. doi: 10.1371/journal.pone.0146596
Liu, B., Zheng, D., Zhou, S., Chen, L., Yang, J., and Liu, C. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
López, M. M., Roselló, M., Llop, P., Ferrer, S., Christen, R., and Gardan, L. (2011). Erwinia piriflorinigrans sp. nov., a novel pathogen that causes necrosis of pear blossoms. Int. J. Syst. Evol. Microbiol. 61, 561–567. doi: 10.1099/ijs.0.020479-0
Matsuura, T., Mizuno, A., Tsukamoto, T., Shimizu, Y., Saito, N., Sato, S., et al. (2012). Erwinia uzenensis sp. nov., a novel pathogen that affects European pear trees (Pyrus communis L.). Int. J. Syst. Evol. Microbiol. 62, 1799–1803. doi: 10.1099/ijs.0.032011-0
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Ming-Kai, P., Guo-Dong-Fang, Yao, Q., Li, J., Liu, C., and Zhu, H. (2022). Erwinia phyllosphaerae sp. nov., a novel bacterium isolated from phyllosphere of pomelo (Citrus maxima). Int. J. Syst. Evol. Microbiol. 72. doi: 10.1099/ijsem.0.005316
Moretti, C., Hosni, T., Vandemeulebroecke, K., Brady, C., De Vos, P., Buonaurio, R., et al. (2011). Erwinia oleae sp. nov., isolated from olive knots caused by Pseudomonas savastanoi pv. savastanoi. Int. J. Syst. Evol. Microbiol. 61, 2745–2752. doi: 10.1099/ijs.0.026336-0
O'Hara, C. M., Steigerwalt, A. G., Hill, B. C., Miller, J. M., and Brenner, D. J. (1998). First report of a human isolate of Erwinia persicinus. J. Clin. Microbiol. 36, 248–250. doi: 10.1128/JCM.36.1.248-250.1998
Parte, A. C., Sardà Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Prod'homme, M., Micol, L. A., Weitsch, S., Gassend, J. L., Martinet, O., and Bellini, C. (2017). Cutaneous infection and bactaeremia caused by Erwinia billingiae: a case report. New Microbes New Infect. 19, 134–136. doi: 10.1016/j.nmni.2017.07.006
Ramirez-Bahena, M. H., Salazar, S., Cuesta, M. J., Tejedor, C., Igual, J. M., Fernandez-Pascual, M., et al. (2016). Erwinia endophytica sp. nov., isolated from potato (Solanum tuberosum L.) stems. Int. J. Syst. Evol. Microbiol. 66, 975–981. doi: 10.1099/ijsem.0.000820
Reimer, L. C., Sardà Carbasse, J., Koblitz, J., Ebeling, C., Podstawka, A., and Overmann, J. (2022). BacDive in 2022: the knowledge base for standardized bacterial and archaeal data. Nucleic Acids Res. 50, D741–D746. doi: 10.1093/nar/gkab961
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sasser, M. (1990). Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. Newark, DE: MIDI Inc.
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shin, S. Y., Lee, M. Y., Song, J. H., and Ko, K. S. (2008). New Erwinia-like organism causing cervical lymphadenitis. J. Clin. Microbiol. 46, 3156–3158. doi: 10.1128/JCM.00716-08
Skrodenyte-Arbaciauskiene, V., Radziute, S., Stunzenas, V., and Buda, V. (2012). Erwinia typographi sp. nov., isolated from bark beetle (Ips typographus) gut. Int. J. Syst. Evol. Microbiol. 62, 942–948. doi: 10.1099/ijs.0.030304-0
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tao, Y., Ge, Y., Yang, J., Song, W., Jin, D., Lin, H., et al. (2023). A novel phytopathogen Erwinia sorbitola sp. nov., isolated from the feces of ruddy shelducks. Front. Cell. Infect. Microbiol. 13:1109634. doi: 10.3389/fcimb.2023.1109634
Winslow, C. E., Broadhurst, J., Buchanan, R. E., Krumwiede, C., Rogers, L. A., and Smith, G. H. (1920). The families and genera of the bacteria: final report of the Committee of the Society of American Bacteriologists on characterization and classification of bacterial types. J. Bacteriol. 5, 191–229. doi: 10.1128/jb.5.3.191-229.1920
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017a). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017b). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie 4Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Keywords: Erwinia wuhanensis, novel species, bacteria, genome, 16S rRNA
Citation: Zhang Y, Zhan Y, Yang J and Lu Z (2025) Erwinia wuhanensis sp. nov. isolated from human blood. Front. Microbiol. 16:1675452. doi: 10.3389/fmicb.2025.1675452
Received: 29 July 2025; Accepted: 20 August 2025;
Published: 04 September 2025.
Edited by:
Axel Cloeckaert, Institut National de recherche pour l'agriculture, l'alimentation et l'environnement (INRAE), FranceReviewed by:
Miloud Sabri, Istituto Agronomico Mediterraneo di Bari, ItalyTao Yuanmeihui, Chinese Center for Disease Control and Prevention, China
Copyright © 2025 Zhang, Zhan, Yang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Yang, eWFuZ2ppbmdAaWNkYy5jbg==; Zhongxin Lu, bHV6aG9uZ3hpbkB6eGhvc3BpdGFsLmNvbQ==
 Yingmiao Zhang
Yingmiao Zhang Yu Zhan1
Yu Zhan1 Jing Yang
Jing Yang Zhongxin Lu
Zhongxin Lu