- Engineering Technology Research Center for Aquatic Organism Conservation and Water Ecosystem Restoration in University of Anhui Province, College of Life Science, Anqing Normal University, Anqing, China
Introduction: Efficient and safe governance of soil contaminated with organophosphate pesticides is of crucial significance for the protection of the ecosystem. This study focuses on soils from typical riparian zones along the project of water diversion from the Yangtze River to Chaohu Lake, aiming to screen acephate-degrading microorganisms and to systematically evaluate their degradation efficiency.
Methods: Acephate-degrading bacteria were isolated from soil via enrichment culture with acephate as the sole carbon source, and their degradation efficiency was subsequently evaluated. Subsequently, a pot experiment was designed to investigate the efficiency of the combined remediation of soil acephate through the synergistic action of the isolated bacteria and plants.
Results: Five acephate-degrading strains were isolated and identified via 16S rDNA sequencing as Enterobacter cloacae, Enterobacter hormaechei, Bacillus badius, Sphingobacterium spiritivorum, and Serratia nematodiphila. Although all strains degraded acephate, their efficiencies differed significantly. Except for the 50 mg L−1 acephate condition with added glucose, B. badius consistently exhibited higher degradation efficiency across all tested conditions. Furthermore, increasing acephate concentration in the culture medium from 10 to 50 mg L−1 reduced degradation efficiency across strains. However, adding 0.1 g L−1 glucose enhanced degradation rates for all strains, with B. badius achieving the highest degradation efficiency (76.17% at 10 mg L−1 acephate). For combined experiments, we paired B. badius (with superior in vitro degradation performance) with Persicaria hydropiper, and S. spiritivorum with Carex dimorpholepis. At both 200 μg kg−1 and 1,000 μg kg−1 soil acephate concentrations, combined remediation efficiencies exceeded those of microbes or plants alone. The combination of B. badius and P. hydropiper achieved the highest removal rate of 91.27% at the 1,000 μg kg−1 acephate concentration.
Conclusion: These findings significantly enrich the repository of acephate-degrading bacteria and demonstrate that combined remediation with B. badius and P. hydropiper is an effective strategy for the bioremediation of acephate-contaminated soils within the project of water diversion from the Yangtze River to Chaohu Lake.
1 Introduction
Acephate has been widely applied in the management of pests across vegetable, fruit, and other crop species, attributable to its high insecticidal efficacy and relatively low toxicity (Hou, 2018). Following the comprehensive prohibition of the highly toxic pesticide methamidophos in China, acephate became the predominant alternative, exhibiting consistently increasing annual usage (Hua, 2014). However, the utilization efficiency of acephate remains low, with only approximately 0.1% of the active ingredient reaching the target organisms, whereas up to 99.9% disperses into ecological ecosystems. For instance, Raj and Krishnan (2023) identified acephate in wastewater, and Omwenga et al. (2021) documented concerning residue levels in common vegetable cultivars. Yan et al. (2024) evaluated the potential risks of acephate to bees and earthworms and found that acephate products had a significant impact on the body weight of earthworms. Dhanushka and Peiris (2017) found that long-term exposure to acephate can lead to alterations in human sperm structure and function, and reduce semen quality, posing potential risks to both the ecological environment and human health (Xie et al., 2008; Tao, 2022). Therefore, effective remediation of acephate-contaminated environments is critical for protecting ecological security and public health.
Currently, remediation strategies for organophosphorus pesticides primarily involve physical, chemical, and biological degradation approaches. Conventional physicochemical methods, though widely employed, frequently result in incomplete degradation, leading to the transformation of soil pesticide residues into secondary pollutants that threaten ecological safety (Mohamed et al., 1998; Tu, 1993). In contrast, biodegradation—particularly microbial degradation—demonstrates greater potential in efficiency and environmental compatibility. Studies indicate microbial degradation achieves removal rates exceeding 90%, with efficiencies over tenfold higher than those of physical or chemical treatments (Giri et al., 2021; Kumar et al., 2018). In natural environments, microbial metabolic activity predominantly mediates the degradation of most organophosphorus pesticides (Sun, 2007). Consequently, applying microorganisms with high degradation capacities has become a focal strategy for remediating soils contaminated with organophosphorus pesticides, especially acephate. For example, Ren et al. (2020) reported that Bacillus paramycoides degraded 500 mg L−1 acephate at a rate of 76% within 48 h. Wang et al. (2013) demonstrated that the bacterial strain Hyphomicrobium sp. achieved complete degradation of 100 mg kg−1 acephate within 9 days at 30 °C and pH 6.8. Similarly, Yu et al. (2010) isolated Stenotrophomonas sp. and Pseudomonas sp. from pesticide-contaminated soils; under conditions of 500–1,000 mg L−1 acephate, 30 °C, pH 8, and 2.5% inoculum, these strains mineralized acephate into phosphate within 1 week, achieving degradation rates approaching 80%.
The principal mechanism for microbial degradation of organophosphorus pesticides entails enzymatic reactions, wherein cleavage of P–O, P–S, and P–N bonds mediates breakdown (Wei et al., 2022). For instance, Lourthuraj et al. (2022) confirmed that E. aerogenes and S. pyogenes secrete organophosphorus hydrolase extracellularly, enhancing chlorpyrifos degradation. However, microbial degradation exhibits high sensitivity to environmental conditions and is influenced by factors including indigenous microorganism abundance and functionality, alongside soil plant community composition (Bu et al., 2023). Previous studies indicate that laboratory-screened strains frequently underperform in field applications, attributable to divergent soil physicochemical properties, competition with native microbes for ecological niches, and insufficient environmental adaptability (Sun et al., 2019). Plant–microbe combined remediation represents a promising strategy, as plant root exudates provide energy and nutrients that enhance microbial activity and degradation efficiency (Oram et al., 2025). For instance, Ma isolated two bacterial strains (Enterobacter sp. and Achromobacter sp.) and a fungal strain (Scedosporium sp.), applying them with Lolium multiflorum to remediate carbendazim-contaminated soil. After 21 days, carbendazim removal efficiencies reached 57.66–78.90%, significantly exceeding microbial-only remediation (41.77–62.9%) (Ma, 2022). Similarly, Chang et al. (2025) confirmed that the combination of Acinetobacter seifertii and Carex dimorpholepis achieved a 93.27% degradation rate for Ethoprophos within 30 days, and this degradation efficiency significantly outperformed that of individual microbial or plant treatments. Lin and You (2009) demonstrated that the combination of Arthrobacter sp. and the plants Sorghum drummondii, Medicago sativa, and Lolium perenne significantly enhanced chlorpyrifos degradation in contaminated soil, surpassing those of individual plants. Consequently, plant–microbe combined remediation demonstrates substantial potential for acephate-contaminated soil remediation, although relevant studies remain limited.
As a strategic water resource allocation project in Anhui Province, the project of water diversion from the Yangtze River to Chaohu Lake (YC-project) conveys water through Caizi Lake, Kongcheng River, Luobu River, and Baishi River before discharging into Chaohu Lake. Recent studies report acephate accumulation in both water bodies and sediments along the YC-project route, exhibiting variable enrichment levels with moderate environmental risk (Song et al., 2022). Literature documents numerous high-efficiency acephate-degrading microorganisms, including Acinetobacter sp., Pseudomonas sp., Exiguobacterium sp., and Rhodococcus sp. (Phugare et al., 2012; Pinjari et al., 2012), predominantly isolated from pesticide manufacturing sites or pesticide-contaminated soils. Nevertheless, riparian zones constitute ecotones between terrestrial and aquatic systems that differ fundamentally from agroecosystems, raising unresolved questions regarding the presence of efficient acephate-degrading strains in these transitional environments.
To address tacephate contamination in soils along the YC-project, this study sampled rhizosphere soils from riparian zones, isolated acephate-degrading strains, and conducted pot experiments to evaluate the efficacy of combined plant–microbe remediation. We hypothesize that combining acephate-degrading bacteria with plant-assisted remediation can enhance pesticide removal from soil. Our findings will enrich the repository of acephate-degrading microbial resources and establish a scientific foundation for the remediation of contaminated soils along the project’s route.
2 Materials and methods
2.1 Soil sample collection
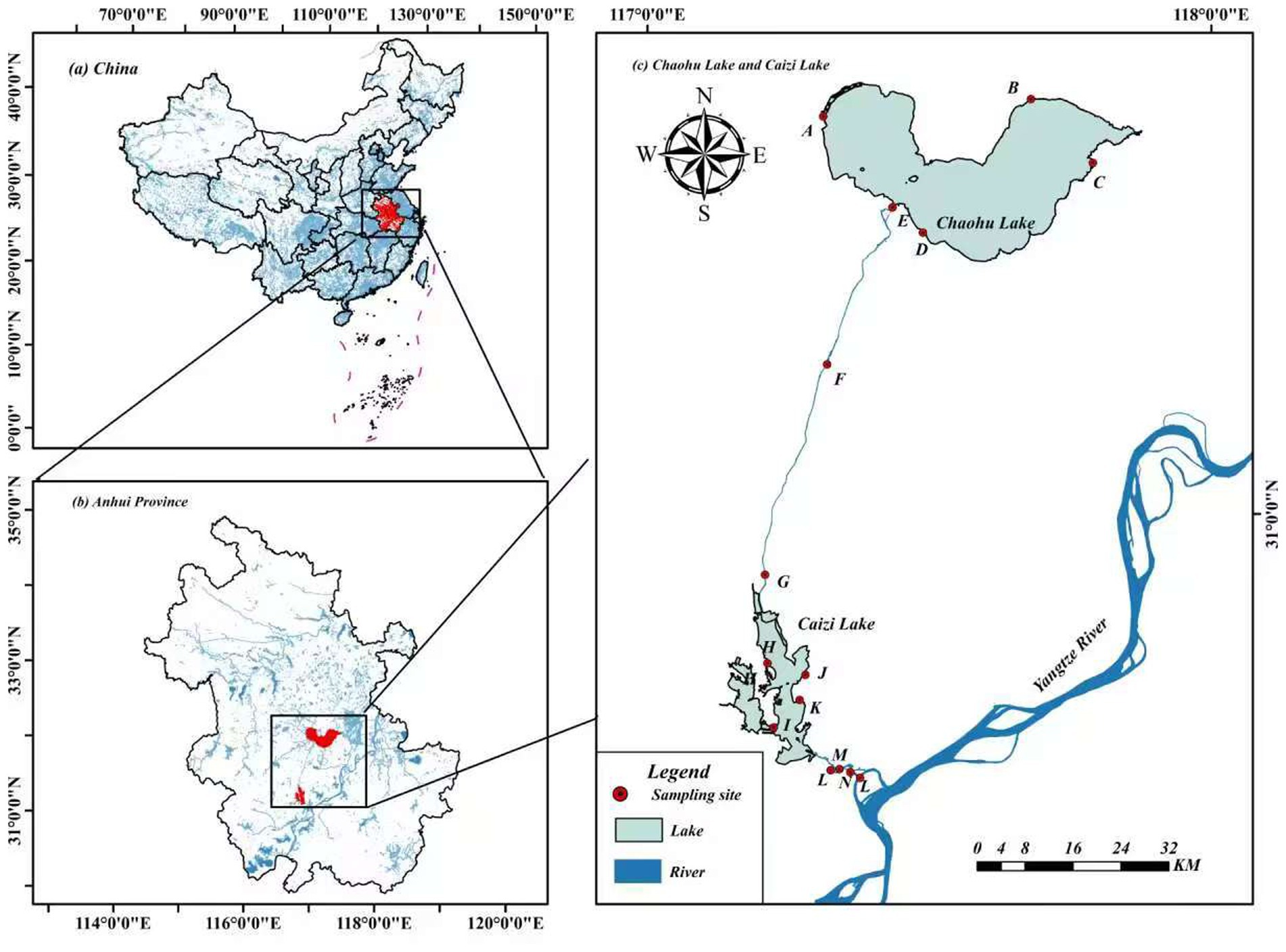
In October 2022, 15 sampling sites were systematically established along the riparian zones of the YC-project (Figure 1). These sites were selected to represent regional typical vegetation types and were evenly distributed along the YC-project route. At each location, 3–6 quadrats (1 m × 1 m) were positioned perpendicular to the riverbank or lakeshore. Rhizosphere soil samples were collected from densely vegetation areas using a soil auger (1.0 m × 50 mm) at 20 cm depth. Samples were immediately sealed in polyethylene bags, stored in dry ice containers, and transported to the laboratory for preservation at −4 °C.
2.2 Isolation of acephate-degrading Bacteria
Acephate-degrading bacteria were isolated from soil using acephate (99.6% purity) as the sole carbon source following the method described by Zhao et al. (2014), involving enrichment, acclimatization, and purification:
1. Ten grams of soil were vortexed with 50 mL of sterile water in a centrifuge tube. The supernatant was transferred to 100 mL nutrient medium (beef extract 3 g, NaCl 5 g, peptone 10 g, deionized water 1,000 mL; pH 6.8) and incubated at 30 °C with shaking at 150 rpm for 24 h;
2. An inoculum was streaked onto nutrient agar plates supplemented with 30 mg L−1 acephate and incubated at 30 °C for 48 h. Colonies were subsequently transferred to plates containing 60 mg L−1 and 120 mg L−1 acephate;
3. Acclimated colonies were streaked onto inorganic salt agar plates containing 240 mg L−1 and 500 mg L−1 acephate as the sole carbon source (Na2HPO4 6.34 g, KH2PO4 1.33 g, (NH4)2SO4 1 g, MgSO4·7H2O 0.2 g, FeSO4 0.001 g, CaCl2 0.04 g, agar 15 g, deionized water 1,000 mL; pH 6.8) for 10 days incubated at 30 °C;
4. Colonies were subcultured, and bacterial suspensions were serially diluted (10−5 to 10−9) with sterile water for single-colony isolation. Purified strains were cryopreserved in 20% glycerol at −60 °C (Koch, 2001).
2.3 Physiological characterization and 16S rDNA sequencing of bacterial strains
Following isolation, the bacterial strains were streaked onto solid media and incubated at 30 °C for 24 h. Colony morphology, including shape, edge, and pigmentation, was recorded. Gram staining was performed to determine cell wall type. Physiological tests included the methyl red test (MR), Voges–Proskauer test (VP), gelatin hydrolysis, starch hydrolysis, nitrate reduction, and catalase assays (McDevitt, 2009). For temperature tolerance, overnight cultures were inoculated into nutrient medium (pH 6.8) and incubated at 20 °C, 25 °C, 30 °C, 35 °C, and 40 °C with shaking at 150 rpm. Optical density at 600 nm (OD600) was measured after 12 h. For pH response, the pH of the nutrient medium was adjusted to 5.0, 6.0, 7.0, 8.0, and 9.0. Inoculated cultures were incubated at 30 °C and 150 rpm, and OD600 values were recorded every 4 h intervals.
Genomic DNA was extracted for 16S rDNA sequencing. PCR amplification was performed using primers B341F (5′-CCTACGGGNGGCWGCAG-3′) and B785R (5′-GACTACHVGGGTATCTAAT-3′). The thermal cycling protocol comprised: initial denaturation at 95 °C for 3 min; 25 cycles of denaturation (95 °C, 30 s), annealing (54 °C, 30 s), and extension (72 °C, 30 s); followed by final extension at 72 °C for 5 min. PCR products were verified through 2% agarose gel electrophoresis and submitted to Sangon Biotech Co., Ltd. (Jiangsu, China) for bidirectional sequencing. Sequences were analyzed via the BLAST algorithm against the NCBI database for homology analysis, and phylogenetic trees were reconstructed using MEGA11 software.
2.4 Measurement of acephate degradation efficiency
Inorganic salt media containing acephate concentrations of 10, 20, and 50 mg L−1 were prepared in 150 mL Erlenmeyer flasks. Each flask was inoculated with bacterial suspension at a 4% inoculum. A sterile filter membrane was secured on flask necks to enable aeration and prevent contamination. Sterile water served as blank controls. Each treatment included three replicates. Flasks were incubated at 30 °C and 150 rpm for 7 days. After incubation, 5 mL aliquots were centrifuged at 4,000 rpm for 3 min. One milliliter of supernatant was filtered through a 0.22 μm membrane, extracted with 10 mL acetonitrile, shaken for 2 min, and sonicated for 20 min. After adding 1 g anhydrous NaCl, the mixture stood for 30 min for phase separation (St-Amand and Girard, 2004). The organic phase was collected for acephate quantification. An additional experiment supplemented with 0.1 g L−1 glucose evaluated external carbon source effects on degradation efficiency.
2.5 Determination of acephate concentration
Acephate concentration was quantified using gas chromatography–mass spectrometry (GC–MS, GCMS-TQ8040, Shimadzu) equipped with a polar SH-Rxi-17Sil MS column (30 m × 0.25 mm × 0.25 μm). Analytical parameters were as follows: injector temperature 250 °C, interface temperature 280 °C, ion source temperature 240 °C, carrier gas helium (99.999%) at 1.97 mL min−1 flow rate, injection volume 1 μL, and selective ion monitoring (SIM) mode. The temperature program was: 65 °C (1 min hold), ramp to 130 °C at 20 °C min−1, then to 280 °C at 10 °C min−1 (10 min hold), and finally to 300 °C at 10 °C min−1 (10 min hold) (Alder et al., 2006).
The standard curve was established by plotting acephate concentration (x-axis) against corresponding peak area (y-axis), yielding a linear regression equation of y = 670,146x − 11,6701. The curve exhibited excellent linearity across the concentration range of 0.1, 0.5, 2, 5, and 10 mg L−1, as confirmed by a correlation coefficient (R2) of 0.9995. Method sensitivity was characterized by detection of 0.01 mg L−1 (liquid matrices) and 0.6 μg kg−1 (soil samples). Validation in an inorganic salt medium spiked with acephate demonstrated acceptable recoveries (93.49–98.73%) and relative standard deviations of 2.47–5.18%, meeting established criteria for pesticide residue analysis.
2.6 Combined plant–microbe degradation of acephate
The experimental soil was prepared by blending peat and riparian zone soil (1:1 v/v ratio). The mixture was sterilized by autoclaving and filled into plastic pots (9.8 cm height × 9.8 cm diameter; 300 g/pot), with soil moisture adjusted to 35% water-holding capacity. Two microbial–plant combinations were tested: Bacillus badius (DA-3) was paired with its host Persicaria hydropiper (30–40 cm height), and S. spiritivorum (DA-4) with its host C. dimorpholepis (10–15 cm height). Bacterial strains were activated and propagated in nutrient broth. Acephate concentrations in soil were set at 200 μg kg−1 and 1,000 μg kg−1. Four treatment groups were established: microbial mono-treatment, plant mono-treatment, plant–microbe combination, and blank control (CK) – all with quintuplicate replicates. Incubation proceeded for 30 d in a climate-controlled growth chamber (30 °C). Bacterial inoculum (10 mL) was supplemented weekly, and deionized water was added triweekly to maintain soil moisture.
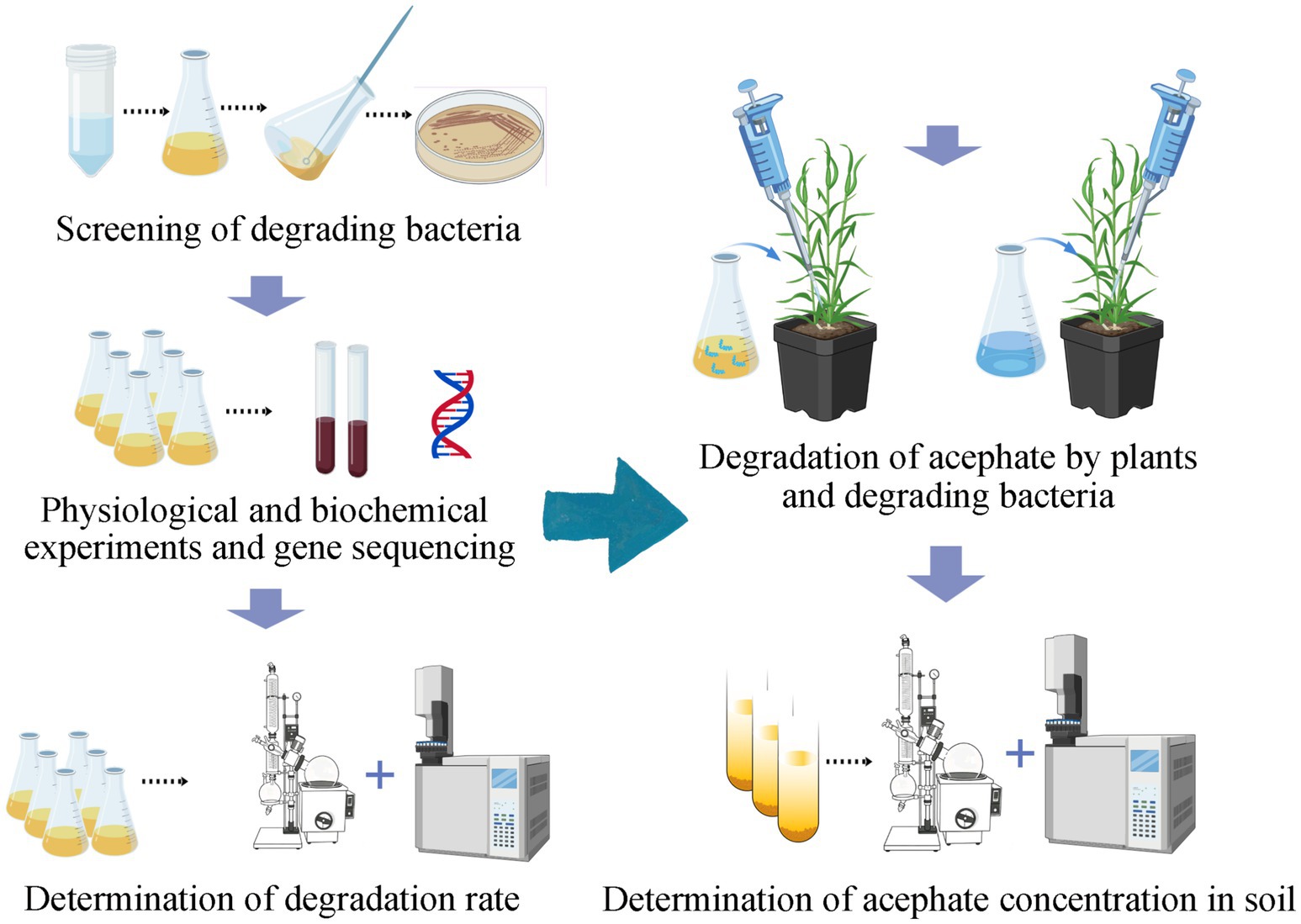
After 30 days, soil samples were collected, freeze-dried, and sieved through a 0.25 mm mesh. Five grams of soil were extracted with 40 mL acetonitrile in 100 mL centrifuge tubes, shaken vigorously, and mixed with 5 g anhydrous NaCl. After centrifugation at 8,000 rpm for 5 min, 10 mL supernatant was transferred to a 100 mL pear-shaped flask and concentrated to ~1 mL a 40 °C. The residue was reconstituted in 3 mL acetonitrile–toluene (1:1), loaded onto a solid-phase extraction (SPE) cartridge, and rinsed twice with 2 mL acetonitrile–toluene. The cartridge was eluted with 25 mL acetonitrile–toluene, and eluents were combined, concentrated to dryness at 40 °C, redissolved in 1 mL ethyl acetate, filtered through an organic microporous membrane, and analyzed via GC–MS. The experimental workflow is depicted in Figure 2.
2.7 Data analysis
All data were compiled and preprocessed in Microsoft Excel 2016. Two-way analysis of variance (ANOVA) was performed using SPSS 24.0. When significant differences were identified (p < 0.05), Tukey’s HSD test was employed for post hoc comparisons. Figures were generated using Python 3.8.
3 Results
3.1 Morphology and physiological characteristics of acephate-degrading strains
Through enrichment, acclimatization, and purification, five target strains were isolated from soil (Figure 3) and designated DA-1, DA-2, DA-3, DA-4, and DA-5, respectively. Gram staining confirmed that DA-3 was Gram-positive, whereas the other four strains were Gram-negative, with all five strains exhibiting entire colony margins (Table 1). Physiological and biochemical characterization are presented in Table 1. MR tests showed DA-4 and DA-5 negative, and the other three positive; VP tests showed DA-4 and DA-5 positive and the other three negative; all strains were gelatin hydrolysis-positive; starch hydrolysis was positive for DA-3 and negative for others; all were nitrate reduction-positive; and all were oxidase-positive.
Temperature significantly influenced the growth of all five strains. Within 20–40 °C, growth initially increased before stabilizing or declining; optimum temperatures for DA-1 through DA-5 were 30, 35, 35, 30, and 35 °C, respectively (Figure 4). pH variations substantially affected DA-4 and DA-5 growth, with significantly higher OD600 values at pH 7 versus other pH levels. All strains entered the stationary phase after 16–20 h, with optimal pH values for DA-1 to DA-5 being 7, 7, 6, 7, and 7, respectively (Figure 5).
3.2 Molecular identification of the degrading strains
The 16S rDNA sequences of the five strains were analyzed via the BLAST algorithm against the NCBI GenBank database. Combined with their physiological and biochemical characteristics, strains DA-1 and DA-2 were identified as Enterobacter cloacae and E. hormaechei, respectively. While DA-2 showed the closest phylogenetic proximity to Pseudotuberculosis, it was not classified as such. Strain DA-3 was identified as B. badius and strain DA-4 as S. spiritivorum (with DA-3 and DA-4 being phylogenetically affiliated). Strain DA-5 was identified as Serratia nematodiphila (Figure 6). DA-3 belongs to the phylum Firmicutes, DA-4 to Bacteroidota, and the other three strains to Proteobacteria (class γ-Proteobacteria, order Enterobacterales, family Enterobacteriaceae). All five strains share close phylogenetic relatedness, with the highest 16S rDNA sequence similarity to Klebsiella quasipneumoniae.

Figure 6. Phylogenetic tree based on 16S rDNA sequences. (DA-1, DA-2, DA-3, DA-4, and DA-5 are the acephate-degrading strains isolated in this study; other microorganisms shown are those known to be most closely related to acephate-degrading bacteria in the NCBI database. The phylogenetic tree was constructed using the neighbor-joining method).
3.3 Degradation efficiency of acephate-degrading strains
The acephate degradation capabilities of the five strains are shown in Figure 7. All strains exhibited measurable acephate degradation ability, though efficiencies varied significantly. The degradation rates decreased with increasing acephate concentration. At 10 mg L−1, DA-2, DA-3, and DA-4 exceeded 50% degradation, with DA-3 achieving the highest rate at 73.69%, which was significantly higher than the other four strains. At 20 mg L−1, DA-3 again exhibited the highest degradation rate (64.36%), significantly higher than the other strains. At 50 mg L−1, DA-3 maintained the highest degradation rate (33.24%), being significantly greater than DA-1 and DA-5 (p < 0.05) but not significantly different from DA-2 and DA-4.

Figure 7. Degradation efficiency of each strain under different acephate concentrations within 7 days. (Different capital letters indicate significant differences in degradation performance among strains at the same acephate concentration; different lowercase letters indicate significant differences in degradation by the same strain at different concentrations. Differences were considered significant at p < 0.05).
Following supplementation of the inorganic salt medium with 0.1 g L−1 glucose, strain degradation efficiencies improved overall (Figure 8), particularly at high acephate concentrations—except for DA-3 at 50 mg L−1, which showed no enhancement. The most significant improvement occurred in DA-2 at 10 mg L−1, with the degradation rate increasing to 69.47% (a 17.13% increase). At 10 mg L−1 acephate, DA-3 maintained the highest degradation rate at 76.17%, significantly higher than DA-1, DA-4, and DA-5, but not significantly different from DA-2 (69.47%). At 20 mg L−1, DA-3 achieved the highest degradation rate of 67.22%, significantly surpassing the other strains. At 50 mg L−1, DA-4 showed peak degradation (34.97%), significantly higher than DA-3 and DA-5 (p < 0.05), but not significantly different from DA-1 and DA-2.

Figure 8. Degradation efficiency of each strain after glucose addition under different acephate concentrations within 7 days. (Different capital letters indicate significant differences among strains at the same acephate concentration; different lowercase letters indicate significant differences within the same strain across different concentrations. Differences were considered significant at p < 0.05).
3.4 Combined degradation of acephate by plants and degrading strains
Plant-microbe combined treatments demonstrated significantly higher acephate degradation efficiency than mono-treatments across concentrations after 30 days (Figure 8). At 200 μg kg−1 acephate, the P. hydropiper-DA-3 combination achieved a degradation efficiency of 89.15%, which was significantly higher than that of DA-3 alone (44.20%) but not significantly different from that of P. hydropiper alone (84.45%, Figure 9a). At 1000 μg kg−1, the combination showed peak degradation efficiency (91.27%), significantly higher than both DA-3 (61.11%) and P. hydropiper (69.85%).

Figure 9. Combined degradation of acephate by different plants and degrading strains. (a) DA-3 and Persicaria hydropiper. (b) DA-4 and Carex dimorpholepis. (PH: P. hydropiper; CD: C. dimorpholepis. Different capital letters indicate significant differences between treatments under the same acephate concentration. Different lowercase letters indicate significant differences among treatments at the same concentration. Differences were considered significant at p < 0.05).
For the C. dimorpholepis-DA-4 combination, the degradation efficiency at 200 μg kg−1 reached 87.97%, significantly higher than that of C. dimorpholepis alone (36.10%) but not significantly different from that of DA-4 alone (86.40%) (Figure 9b). At 1,000 μg kg−1, the combined degradation efficiency was 68.74%, significantly higher than that of both C. dimorpholepis alone (20.78%) and DA-4 alone (59.38%). The P. hydropiper-DA-3 combination showed no significant difference in degradation between 200 μg kg−1 (89.15%) and 1,000 μg kg−1 (91.27%) (Figure 9a). Conversely, the C. dimorpholepis-DA-4 combination exhibited significantly higher degradation at 200 μg kg−1 (87.97%) than at 1,000 μg kg−1 (68.74%) (p < 0.05; Figure 9B).
4 Discussion
4.1 Acephate-degrading microorganisms in riparian zones and their degradation capacity
In this study, five acephate-degrading strains were isolated from riparian zone soils along the YC-project. Among them, DA-4 (S. spiritivorum) is an oligotrophic bacterium, while the other four strains are eutrophic microorganisms (Koch, 2001). Previous studies have suggested that the nutritional type of microorganisms may influence their biodegradation capacity (Sun et al., 2015). Reported high-efficiency acephate-degrading bacteria are mostly isolated from contaminated sites or industrial effluents. For instance, Ramya et al. (2016) found that Enterobacter aerogenes could degrade acephate, which belongs to the same genus as E. cloacae and E. hormaechei identified in our study. Mohan and Naveena (2015) discovered that Lysinibacillus could degrade acephate concentrations up to 500 mg L−1. Lin et al. (2016) reported enhanced acephate degradation by Bacillus subtilis under Pb2+ stress, which is consistent with the genus of B. badius (DA-3) in our study. Dong et al. (2024) found that Serratia marcescens could degrade acetochlor, which belongs to the same genus as S. nematodiphila (DA-5) isolated in our study. Other studies also reported acephate degradation by Pseudomonas aeruginosa (Ramu and Seetharaman, 2014) and Burkholderia sp. (Wu et al., 2023), which were not observed in this study. This indicates that diverse microbial species can degrade acephate, and the types of microorganisms capable of its degradation vary across ecosystems.
We further assessed the degradation performance of the five isolated strains and found that DA-3 exhibited significantly higher degradation efficiency than the other four strains (Figure 7). This may be attributed to interspecies differences in microbial degradation capacity (Bose et al., 2021). For example, Singh et al. (2020) compared the degradation of acephate by P. aeruginosa, Pseudomonas putida, and Pseudomonas azotoformans under identical conditions, finding P. aeruginosa had the highest efficiency, suggesting substantial inter-species variability, consistent with our observations for E. cloacae and E. hormaechei. Lin et al. (2022) reported that a microbial community degraded acephate most efficiently under conditions of 34.1 °C and pH 8.9, achieving 89.5% at an acephate concentration of 200 mg L−1. The lower degradation rate observed in our study may result from differences in microbial species. Numerous studies show that under pure culture conditions, microbial degradation efficiency of organophosphorus pesticides can exceed 90%. For example, Micromonospora sp., Pseudomonas sp., and Enterobacter sp. achieve over 95% degradation or complete mineralization of acephate or methamidophos (Wang et al., 2010; Li et al., 2014; Singh et al., 2017). In this study, B. badius (DA-3) showed a maximum degradation rate of 76.17%, which is potentially due to the influence of culture time or pesticide type (Guerrero Ramírez et al., 2023). However, our findings align with Phugare et al. (2012), who found that Exiguobacterium sp. degraded acephate with a rate of 75.85%, possibly reflecting shared Bacillaceae phylogeny. These findings further support our speculation that variations in acephate degradation efficiency are due to strain-specific characteristics.
We also observed reduced microbial degradation efficiency with increasing acephate concentration, mitigated by glucose supplementation, especially at high acephate concentrations. This may be due to the stimulatory effect of additional carbon sources on microbial growth (Li et al., 2021), which enhanced degradation activity. This suggests that nutrient stress limits acephate biodegradation.
4.2 Combined plant–microbe remediation of acephate
Under natural conditions, plants secrete extracellular enzymes such as esterases and phosphatases through their roots to promote pesticide degradation. Esterases hydrolyze ester bonds, while phosphatases cleave phosphate ester bonds in organophosphorus pesticides, reducing pesticide concentrations (Sun et al., 2010). However, plant root degradation of organophosphorus compounds is often slow and subject to low tolerance (Li and Fantke, 2023). Microorganisms, although efficient degraders, are often limited by environmental factors and nutrient availability. In nutrient-deficient soils, microbial activity may be severely inhibited (Zhou et al., 2020). Plants can offset this limitation by releasing carbon and nitrogen sources through fine root turnover and root exudates, thus promoting microbial growth (Yu et al., 2022). Therefore, plant–microbe combined remediation has emerged as an effective strategy for enhancing pesticide degradation. For example, Vaishnavi and Osborne (2024) reported that co-remediation using Proteus myxofaciens and Chrysopogon zizanioides achieved over 90% degradation of monocrotophos after 45 days. Jabeen et al. (2016) found that combining Lolium perenne with endophytic rhizobia promoted bacterial rhizosphere colonization, aiding chlorpyrifos removal. Our results demonstrated that combining strains with the plant under different concentrations enhanced acephate degradation, consistent with these studies (Xie et al., 2018). Consequently, our findings confirm that plant–microbe interactions accelerate acephate degradation in contaminated soils.
Notably, under 200 μg kg−1 acephate, the P. hydropiper and DA-3 combination achieved 89.15% degradation. When acephate concentration increased to 1,000 μg kg−1, the degradation rate rose to 91.27% (though statistically insignificant, p > 0.05). This unexpected result may stem from DA-3 increasing its degradation from 44.20 to 61.11% under higher concentrations. We speculate that elevated pesticide levels induced DA-3 to secrete broad-spectrum enzymes (phosphatases and sulfur oxidases), enabling co-metabolism of acephate and root-derived carbon sources, thereby mitigating toxicity stress (Ke et al., 2023). For instance, Yu et al. (2010) reported that their acephate-degrading strain Y1 exhibited increased degradation at 100–500 mg L−1. Furthermore, the P. hydropiper-DA-3 combination at 1,000 μg kg−1 (91.27%) outperformed the C. dimorpholepis-DA-4 combination. This may be attributed to the extensive rhizosphere of P. hydropiper in moist soils, forming dense root networks that enhance microbial colonization. Previous studies indicate that flavonoids in P. hydropiper root exudates induce oph gene expression in Enterobacter, facilitating cleavage of P–O alkyl bonds in organophosphates (Li et al., 2020). We thus speculated that these exudates potentially promoted B. badius enrichment and oph gene expression, contributing to the high degradation. In contrast, phenolic compounds such as ferulic acid secreted by C. dimorpholepis may inhibit enzyme activity in Sphingobacterium sp. (Güsewell and Schroth, 2017). Moreover, P. hydropiper is a dicotyledonous plant, while C. dimorpholepis is monocotyledonous, and differences in plant morphology and physiology may also influence degradation capacity (Sreenivasulu and Wobus, 2013; Wang et al., 2023).
Numerous studies demonstrate that hydrolysis, dehalogenation, and oxidation are enzymatic reactions driving organophosphorus pesticide degradation (Bosu et al., 2024; Bosu et al., 2024). Consequently, this degradation proceeds through distributed pathways, yielding diverse intermediate metabolites and terminal products. For example, Kumari et al. (2025) identified acetamide and trimethyl phosphate as intermediate metabolites of monocrotophos. Hou et al. (2021) reported that dialkyl phosphates are the predominant terminal metabolites in bacterial degradation of nonhalogenated organophosphate esters. Sur and Sathiavelu (2024) identified methyl diethanolamine and aspartyl glycine ethyl ester as intermediate metabolites in dimethoate degradation, with O, O, S-trimethyl phosphorothioate as the terminal metabolite. Consequently, terminal metabolites of organophosphorus pesticides are typically non-toxic phosphate derivatives (Das et al., 2024). Therefore, although degradation products of acephate were undetected in this study, we infer its mineralization to non-toxic phosphate, thereby achieving the purpose of pollution remediation. In summary, this work lies not only in the first-time isolation of an acephate-degrading microbe from the riparian zones but also in demonstrating its synergistic potential with plants—achieving over 90% degradation and thus opening avenues for in situ bioremediation.
5 Conclusion
In this study, five acephate-degrading microbial strains were isolated from riparian zone soils along the project of water diversion from the Yangtze River to Chaohu Lake. These strains were identified as E. cloacae, E. hormaechei, B. badius, S. spiritivorum, and S. nematodiphila. Culture experiments demonstrated that B. badius exhibited the highest degradation efficiency for acephate. Increasing acephate concentrations generally reduced microbial degradation efficiency, whereas the addition of glucose alleviated this inhibitory effect (except for B. badius at 50 mg L−1 acephate). Combined degradation experiments showed that at 1,000 μg kg−1 acephate, the co-remediation by B. badius and P. hydropiper achieved optimal degradation, with a removal rate of 91.27%. These findings provide a scientific basis for the bioremediation of acephate-contaminated soils.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
HW: Data curation, Methodology, Writing – original draft, Writing – review & editing. JC: Data curation, Formal analysis, Project administration, Writing – original draft, Writing – review & editing. CP: Formal analysis, Writing – original draft, Writing – review & editing. DJ: Formal analysis, Methodology, Data curation, Writing – review & editing. YW: Methodology, Software, Writing – review & editing. QY: Investigation, Software, Writing – review & editing. XC: Data curation, Writing – review & editing. XL: Methodology, Writing – review & editing. ML: Data curation, Writing – review & editing. XZ: Formal analysis, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Key Research and Development Program of Anhui Province [grant number 2022L07020002], the Key Project of Natural Science foundation for universities of Anhui Province [grant number 2023AH050477 and 2024AH051090]. National Natural Science Foundation of China (32471706).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alder, L., Greulich, K., Kempe, G., and Vieth, B. (2006). Residue analysis of 500 high priority pesticides: better by GC-MS or LC-MS/MS? Mass Spectrom. Rev. 25, 838–865. doi: 10.1002/mas.20091
Bose, S., Kumar, P. S., Vo, D. V. N., Rajamohan, N., and Saravanan, R. (2021). Microbial degradation of recalcitrant pesticides: a review. Environ. Chem. Lett. 19, 3209–3228. doi: 10.1007/s10311-021-01236-5
Bosu, S., Rajamohan, N., Al Salti, S., Rajasimman, M., and Das, P. (2024). Biodegradation of chlorpyrifos pollution from contaminated environment - a review on operating variables and mechanism. Environ Res 248:118212.
Bu, L. Y., Peng, Z. H., Tian, J., Bu, L., Peng, Z., Zhang, X., et al. (2023). Core autotrophic microbes drive functional stability of soil cbbL-containing autotrophic microbes during desertification. Appl. Soil Ecol. 190:105027. doi: 10.1016/j.apsoil.2023.105027
Chang, J. L., Wang, H. L., Pan, C., Shen, J. J., Jiang, D. S., Wang, Y. M., et al. (2025). Isolation of ethoprophos-degrading bacteria and phytoremediation–microbial remediation from soil for the project of water diversion from the Yangtze River to Chaohu Lake. Appl. Ecol. Environ. Res. 23, 4457–4472.
Das, R., Saikia, K., Sarma, P. P., Devi, R., and Thakur, D. (2024). Evaluating the potential of bacillus isolates for chlorpyrifos degradation and their role in tea growth promotion and suppression of pathogens. Curr. Microbiol. 81:332. doi: 10.1007/s00284-024-03859-7
Dhanushka, M. A. T., and Peiris, L. D. C. (2017). Cytotoxic and genotoxic effects of acephate on human sperm. J. Toxicol. 2017:3874817.
Dong, M., Xiao, Y., Yang, B., Wang, S., Sun, L., Han, Z., et al. (2024). Serratia marcescens AB1: a rhizosphere bacterium mitigating the acetochlor stress on the soil environment. Rhizosphere 30:100898. doi: 10.1016/j.rhisph.2024.100898
Giri, B. S., Geed, S., Vikrant, K., Lee, S. S., Kim, K., Kailasa, S. K., et al. (2021). Progress in bioremediation of pesticide residues in the environment. Environ. Eng. Res. 26:2004. 46
Guerrero Ramírez, J. R., Ibarra Muñoz, L. A., Balagurusamy, N., Frías Ramírez, J. E., Alfaro Hernández, L., and Carrillo Campos, J. (2023). Microbiology and biochemistry of pesticides biodegradation. Int. J. Mol. Sci. 24:15969. doi: 10.3390/ijms242115969
Güsewell, S., and Schroth, M. H. (2017). How functional is a trait? Phosphorus mobilization through root exudates differs little between Carex species with and without specialized dauciform roots. New Phytol. 215, 1438–1450. doi: 10.1111/nph.14674
Hou, J. H. (2018). Isolation, identification and characterization of an acephate-degrading Bacillus strain. Biotechnology 28, 286–289, 254. (in Chinese)
Hou, R., Wang, Y., Zhou, S., Zhou, L., Yuan, Y., and Xu, Y. (2021). Aerobic degradation of nonhalogenated organophosphate flame esters (OPEs) by enriched cultures from sludge: kinetics, pathways, bacterial community evolution, and toxicity evaluation. Sci. Total Environ. 760:143385. doi: 10.1016/j.scitotenv.2020.143385
Hua, N. Z. (2014). Advances and applications of the organophosphorus insecticide acephate. World Pesticides 36, 11–16. (in Chinese)
Jabeen, H., Iqbal, S., Ahmad, F., Afzal, M., and Firdous, S. (2016). Enhanced remediation of chlorpyrifos by ryegrass (Lolium multiflorum) and a chlorpyrifos degrading bacterial endophyte Mezorhizobium sp. HN3. Int. J. Phytoremed. 18, 126–133. doi: 10.1080/15226514.2015.1073666
Ke, D., Jiang, Y., Hong, Z. F., Zhang, N. C., Yang, J. W., Lin, Q. Q., et al. (2023). Community structure variation of bacteria and fungi in the Phragmites australis rhizosphere and their environmental limiting factors by the co-metabolism between root exudates and alkylphenols. Acta Sci. Circumst. 43, 362–371. (in Chinese).
Kumar, S., Kaushik, G., Dar, A. M., Nimesh, S., Lopez, J. U., and Villarreal, F. J. (2018). Microbial degradation of organophosphate pesticides: a review. Pedosphere 28, 190–208.
Kumari, A., Ghosh, C., Kannan, N., and Balaji, S. (2025). Lactiplantibacillus plantarum as a sustainable solution for monocrotophos degradation and plant growth enhancement. Int. Microbiol., 1–13.
Li, Z., and Fantke, P. (2023). Considering degradation kinetics of pesticides in plant uptake models: proof of concept for potato. Pest Manag. Sci. 79, 1154–1163. doi: 10.1002/ps.7288
Li, T., Wang, R., Cai, J., Meng, Y., Wang, Z., Feng, X., et al. (2021). Enhanced carbon acquisition and use efficiency alleviate microbial carbon relative to nitrogen limitation under soil acidification. Ecol. Process. 10:32. doi: 10.1186/s13717-021-00309-1
Li, Y., Wang, B. S., Huang, Y. Y., Yao, Y. W., Lin, J. M., Liu, K. H., et al. (2020). Mechanism study on the phytoremediation of cadmium- and arsenic-contaminated soil by polygonaceae plants with Enterobacter sp. J. Agro-environ. Sci. 39, 304–312. (in Chinese)
Li, D. D., Yuan, Y. Z., Chen, Y. L., Li, N., Li, J. L., Zheng, Y. L., et al. (2014). Isolation and characterization of three bacterium strains for biodegradation of methamidophos. Adv. Mater. Res. 955, 235–238.
Lin, W., Huang, Z., Li, X., Liu, M., and Cheng, Y. (2016). Bio-remediation of acephate–pb (II) compound contaminants by Bacillus subtilis FZUL-33. J. Environ. Sci. 45, 94–99. doi: 10.1016/j.jes.2015.12.010
Lin, Z., Pang, S., Zhou, Z., Wu, X., Li, J., Huang, Y., et al. (2022). Novel pathway of acephate degradation by the microbial consortium ZQ01 and its potential for environmental bioremediation. J. Hazard. Mater. 426:127841. doi: 10.1016/j.jhazmat.2021.127841
Lin, C., and You, M. S. (2009). Plant-microorganism combined bioremediation of chlorpyrifos contaminated soil. Entomol. J. East China 18, 081–087. (In Chinese)
Lourthuraj, A. A., Hatshan, M. R., and Hussein, D. S. (2022). Biocatalytic degradation of organophosphate pesticide from the wastewater and hydrolytic enzyme properties of consortium isolated from the pesticide contaminated water. Environ. Res. 205:112553. doi: 10.1016/j.envres.2021.112553
Ma, Y. C. (2022). Isolation and screening of carbendazim-degrading bacteria and their combined use with ryegrass to remediate contaminated soil (In Chinese). Gansu: Gansu Agricultural University.
McDevitt, S. (2009). Methyl red and voges-proskauer test protocols. Washington, D.C.: American Society for Microbiology, 8.
Mohamed, T. A., Saad, M. M. I., and Mabrouk, S. S. (1998). Residues of some chlorinated hydrocarb pesticide in rain water, soil and ground water, and their influence on some soil microorganisms. Environ. Int. 24, 665–670.
Mohan, N., and Naveena, L. (2015). Isolation and determination of efficacy of acephate degrading bacteria from agricultural soil. J. Environ. Sci. Toxicol. Food Technol. 9, 10–20.
Omwenga, I., Kanja, L., Zomer, P., Louisse, J., Rietjens, I. M. C. M., and Mol, H. (2021). Organophosphate and carbamate pesticide residues and accompanying risks in commonly consumed vegetables in Kenya. Food Addit. Contaminan. Part B 14, 48–58. doi: 10.1080/19393210.2020.1861661
Oram, N. J., Brennan, F., Praeg, N., Bardgett, R. D., Illmer, P., Ingrisch, J., et al. (2025). Plant community composition and traits modulate the impacts of drought intensity on soil microbial community composition and function. Soil Biol. Biochem. 200:109644. doi: 10.1016/j.soilbio.2024.109644
Phugare, S. S., Gaikwad, Y. B., and Jadhav, J. P. (2012). Biodegradation of acephate using a developed bacterial consortium and toxicological analysis using earthworms (Lumbricus terrestris) as a model animal. Int. Biodeterior. Biodegrad. 69, 1–9. doi: 10.1016/j.ibiod.2011.11.013
Pinjari, A. B., Novikov, B., Rezenom, Y. H., Russell, D. H., Wales, M. E., and Siddavattam, D. (2012). Mineralization of acephate, a recalcitrant organophosphate insecticide is initiated by a pseudomonad in environmental samples. PLoS One 7:e31963. doi: 10.1371/journal.pone.0031963
Raj, R. S., and Krishnan, K. A. (2023). A comprehensive review on the impact of emerging organophosphorous pesticides and their remedial measures: special focus on acephate. Environ. Nanotechnol. Monit. Manag. 20:100813.
Ramu, S., and Seetharaman, B. (2014). Biodegradation of acephate and methamidophos by a soil bacterium Pseudomonas aeruginosa strain Is-6. J. Environ. Sci. Health B 49, 23–34. doi: 10.1080/03601234.2013.836868
Ramya, S. L., Venkatesan, T., Murthy, K. S., Jalali, S. K., and Varghese, A. (2016). Degradation of acephate by Enterobacter asburiae, Bacillus cereus and Pantoea agglomerans isolated from diamondback moth Plutella xylostella (L), a pest of cruciferous crops. J. Environ. Biol. 37:611.
Ren, J., Wang, C., Huhetaoli,, Li, C., Fan, B., and Niu, D. (2020). Biodegradation of acephate by Bacillus paramycoides NDZ and its degradation pathway. World J. Microbiol. Biotechnol. 36, 1–11.
Singh, S., Kumar, V., Singla, S., Sharma, M., Singh, D. P., Prasad, R., et al. (2020). Kinetic study of the biodegradation of acephate by indigenous soil bacterial isolates in the presence of humic acid and metal ions. Biomolecules 10:433. doi: 10.3390/biom10030433
Singh, S., Kumar, V., Upadhyay, N., Singh, J., Singla, S., and Datta, S.. (2017). Efficient biodegradation of acephate 566 by Pseudomonas pseudoalcaligenes PS-5 in the presence and absence of heavy metal ions [cu(II) and Fe(III)], and 568 humic acid. Biotech 7:262.
Song, J., Lü, D., Ding, R., and Zhang, X. K. (2022). Distribution characteristics and risk assessment of organophosphorus pesticides in caizihu lake line of the leading water project from Yangtze River to Huaihe River. J. Shanghai Ocean Univ. 31, 1502–1513. (in Chinese)
Sreenivasulu, N., and Wobus, U. (2013). Seed-development programs: a systems biology–based comparison between dicots and monocots. Annu. Rev. Plant Biol. 64, 189–217. doi: 10.1146/annurev-arplant-050312-120215
St-Amand, A. D., and Girard, L. (2004). Determination of acephate and its degradation product methamidophos in soil and water by solid-phase extraction (SPE) and GC-MS. Int. J. Environ. Anal. Chem. 84, 739–748. doi: 10.1080/03067310410001729600
Sun, X. H. (2007). Investigation of the degradation mechanisms of pesticides in soil. J. Anhui Agric. Sci. 31, 10036–10037. (in Chinese)
Sun, T. R., Cang, L., Wang, Q. Y., Zhou, D. M., Cheng, J. M., and Xu, H. (2010). Roles of abiotic losses, microbes, plant roots, and root exudates on phytoremediation of PAHs in a barren soil. J. Hazard. Mater. 176, 919–925. doi: 10.1016/j.jhazmat.2009.11.124
Sun, C. C., Ding, W., Pan, X. B., and Run, F. F. (2019). Analysis of factors affecting the application stability of microbial inoculants. Plant Doctor 32, 19–23. (in Chinese)
Sun, R., Han, D. D., Ma, D., and Wang, Y. Y. (2015). Biodegradation of phthalates by ultramicrobacteria under oligotrophic conditions. J. Saf. Environ. 15, 260–264. (in Chinese)
Sur, S., and Sathiavelu, M. (2024). Functional profiling of the rhizospheric Exiguobacterium sp. for dimethoate degradation, PGPR activity, biofilm development, and ecotoxicological risk. Sci. Rep. 14:29361. doi: 10.1038/s41598-024-80559-z
Tao, K. L. (2022). The ecological effect of parathion pollution on soil FDA hydrolase and its mechanism. Xianyang: Northwest A&F University (in Chinese).
Tu, C. M. (1993). Influence of ten herbicides on activities of microorganisms and enzymes in soil. Bull. Environ. Contam. Toxicol. 51, 30–39.
Vaishnavi, J., and Osborne, J. W. (2024). Biodegradation of monocrotophos, cypermethrin & fipronil by Proteus myxofaciens VITVJ1: a plant-microbe based remediation. Heliyon 10:e37384. doi: 10.1016/j.heliyon.2024.e37384
Wang, L., Ling, Q., Wu, C. N., Li, S. P., Jiang, J. D., and Wang, M. (2013). Bioremediation on the soil contaminated by methamidophos, acephate and isocarbophos by Hyphomicrobium sp. MAP-1. J. Agro-environ. Sci. 32, 81–87. (in Chinese)
Wang, L., Wen, Y., Guo, X., Wang, G., Li, S., and Jiang, J. (2010). Degradation of methamidophos by Hyphomicrobium species MAP-1 and the biochemical degradation pathway. Biodegradation 21, 513–523. doi: 10.1007/s10532-009-9320-9
Wang, H., Zhang, X., Xu, Y., Wang, H., Song, M., and Shen, Y. (2023). Ecological regulation of water level should be combined with seed supplementation for lakeshore Carex community restoration in Yangtze-disconnected lakes. Sci. Total Environ. 897:165358. doi: 10.1016/j.scitotenv.2023.165358
Wei, Y. N., Liu, C. G., Fu, H. Y., Wu, T., Song, F. Q., Ma, Y. K., et al. (2022). Advance in microbial remediation of organophosphorus pesticide pollution. Chin Agric. Sci. Bull. 38, 131–137. (in Chinese)
Wu, X., Chen, W. J., Lin, Z., Huang, Y., el Sebai, T. N. M., Alansary, N., et al. (2023). Rapid biodegradation of the organophosphorus insecticide acephate by a novel strain Burkholderia sp. A11 and its impact on the structure of the indigenous microbial community. J. Agric. Food Chem. 71, 5261–5274. doi: 10.1021/acs.jafc.2c07861
Xie, K. Z., Xu, P. Z., Chen, J. S., Tang, S. H., Zhang, F. B., Yang, S. H., et al. (2008). Isolation and identification acephate-degrading bacteria XP-3 and studies on its physiological characterization. Guangdong Acad. Agric. Sci. 8, 79–82. (in Chinese)
Xie, H., Zhu, L., and Wang, J. (2018). Combined treatment of contaminated soil with a bacterial Stenotrophomonas strain DXZ9 and ryegrass (Lolium perenne) enhances DDT and DDE remediation. Environ. Sci. Pollut. Res. 25, 31895–31905. doi: 10.1007/s11356-018-1236-7
Yan, W., Zheng, Q., Zhu, S., Miao, X., Yang, L., Wu, J., et al. (2024). Coating of maize seeds with acephate for precision agriculture: safety assessment in earthworms, bees, and soil microorganisms. Sci. Total Environ. 943:173761. doi: 10.1016/j.scitotenv.2024.173761
Yu, C. H., Zhang, X. T., Song, Y. N., Lu, X. B., Sun, H. W., Zhang, S., et al. (2010). Isolation, identification and degradation characteristics of two acephate-degrading bacteria. Chin. J. Environ. Eng. 4, 2623–2630. (in Chinese)
Yu, L., Zi, H., Zhu, H., Liao, Y., Xu, X., and Li, X. (2022). Rhizosphere microbiome of forest trees is connected to their resistance to soil-borne pathogens. Plant Soil 479, 143–158. doi: 10.1007/s11104-022-05505-2
Zhao, Y., Zhao, P., Wang, Y., and Qi, W. J. (2014). Isolation, identification, and characterization of an organophosphorous pesticide degrading bacterium, Enterobacter ludwigii M2. Adv. Mater. Res. 1051, 398–403. doi: 10.4028/www.scientific.net/AMR.1051.398
Keywords: acephate, riparian zones, bioremediation, pesticide degradation, soil contamination
Citation: Wang H, Chang J, Pan C, Jiang D, Wang Y, Yin Q, Chen X, Liao X, Li M and Zhang X (2025) Isolation of acephate-degrading bacteria and phytoremediation–microbial remediation from soil for the project of water diversion from the Yangtze River to Chaohu Lake. Front. Microbiol. 16:1675842. doi: 10.3389/fmicb.2025.1675842
Edited by:
Gaurav Saxena, Mandsaur University, IndiaReviewed by:
Long Zou, Jiangxi Normal University, ChinaSheel Ratna, Indian Institute of Technology (BHU), India
Copyright © 2025 Wang, Chang, Pan, Jiang, Wang, Yin, Chen, Liao, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Pan, cGFuY2hhbmcyMDIwQDE2My5jb20=; Xiaoke Zhang, enhrc2dzZ0AxNjMuY29t
 Huili Wang
Huili Wang Chang Pan
Chang Pan Xiaoke Zhang
Xiaoke Zhang