- Huzhou Traditional Chinese Medicine Hospital Affiliated to Zhejiang Chinese Medical University (Huzhou Hospital of Traditional Chinese Medicine), Huzhou, China
Asthma is a multifactorial inflammatory airway disease shaped by complex interactions among host genetics, environmental exposures, and the microbiota. The human body hosts a highly diverse microbial ecosystem, comprising more than 10,000 species that profoundly influence host physiology through the regulation of nutrient metabolism and immune homeostasis. Disruption of this balance, or dysbiosis, contributes to the onset and progression of immune-mediated diseases, including asthma. Asthma is a multifactorial disorder driven by the complex interaction of genetic susceptibility and environmental exposures, and its heterogeneous phenotypes and severity are increasingly associated with alterations in the microbiota. In particular, the gut–lung axis represents a critical bidirectional pathway through which microbial communities and their metabolites in the gut and airways shape immune responses and respiratory health. This review summarizes current evidence on microbiota-driven mechanisms underlying asthma pathogenesis, highlights the role of the gut–lung axis in immune regulation, and discusses emerging microbiota-targeted therapeutic strategies, emphasizing their potential for clinical translation in asthma treatment.
1 Introduction
Asthma is a chronic and heterogeneous respiratory disorder characterized by persistent airway inflammation and variable expiratory airflow obstruction (Global Initiative for Asthma, 2025). Clinically, it manifests as recurrent wheezing, dyspnea, chest tightness, and cough, affecting more than 300 million people globally and imposing a substantial socioeconomic burden on healthcare systems and society (Wang et al., 2023). The pathogenesis of asthma involves multiple interrelated processes, including airway hyperresponsiveness (AHR), structural remodeling, T helper (Th) cell imbalance, eosinophilic infiltration, and goblet cell hyperplasia (Maia et al., 2024). Together, these alterations underlie the chronic inflammatory state and phenotypic variability observed in patients.
In recent years, increasing attention has focused on the role of the microbiota as a critical modulator of immune development and respiratory health. The human gastrointestinal tract harbors a dense and diverse microbial community that contributes to nutrient metabolism, epithelial barrier integrity, and the regulation of local and systemic immune responses (Kandari et al., 2024). Although the microbial biomass of the lung is relatively low, its community structure is shaped by inhalation, mucosal dispersion, and host–microbe interactions (Druszczynska et al., 2024). Communication between the gut and lung occurs through lymphatic circulation, hematogenous dissemination, and micro-aspiration, forming a bidirectional regulatory network known as the gut–lung axis. In this review, we define the gut–lung axis as the bidirectional communication between intestinal and respiratory systems mediated by microbial communities, metabolites, and immune signaling pathways. Dysbiosis refers to a state of microbial imbalance characterized by reduced diversity and altered functional capacity, whereas immune regulation encompasses microbial modulation of both innate and adaptive immune responses.
Accumulating evidence indicates that alterations in gut microbial composition are associated with asthma susceptibility, phenotypic expression, and disease progression. Characteristic changes include reduced abundance of beneficial commensals such as Bifidobacterium and Bacteroides fragilis, as well as expansion of potentially pro-inflammatory taxa (Enaud et al., 2020). These microbial imbalances disrupt immune homeostasis, enhance Th2 and Th17 polarization, and promote airway hyperresponsiveness and inflammation. Furthermore, gut dysbiosis has been linked to broader systemic effects through the gut–lung axis, shaping pulmonary immune responses via metabolite signaling and immune regulation. Such disturbances are increasingly recognized as key contributors to asthma heterogeneity, influencing not only disease mechanisms but also clinical phenotypes and therapeutic outcomes.
To improve transparency and reduce citation bias, we searched PubMed and Web of Science for relevant studies published between 2000 and 2025 using combinations of the keywords “asthma,” “gut–lung axis,” “microbiota,” and “immune regulation.” Additional articles were identified through manual screening of reference lists.
Unlike previous reviews that have primarily focused on individual mechanisms or clinical associations, this article proposes a hierarchical framework that classifies microbiota–host interactions into primary drivers, intermediate modulators, and downstream immune effectors. This structure integrates recent advances in metabolite signaling, epigenetic regulation, and neuro–immune crosstalk, providing a more unified mechanistic perspective on the gut–lung axis in asthma. In this review, we synthesize current evidence on microbiota–immune interactions in asthma, focusing on the gut–lung axis as an integrative framework. We further discuss recent progress in microbiota-targeted therapeutic strategies and highlight remaining knowledge gaps to inform future mechanistic and clinical research.
2 Microorganisms and asthma pathogenesis
The development and progression of asthma are shaped by intricate host–microbe interactions, which influence immune maturation, epithelial barrier integrity, and inflammatory responses. Early conceptual frameworks, such as the hygiene hypothesis, proposed that reduced exposure to pathogens during early childhood impairs immune tolerance and increases the risk of allergic diseases. This idea has since evolved into the biodiversity hypothesis, which emphasizes that limited contact with diverse natural environments and environmental microbiota diminishes immune regulatory capacity and heightens susceptibility to asthma and other allergic disorders (Haahtela et al., 2013). Supporting this view, decreased diversity of commensal taxa—including Lactobacillus, Bifidobacterium, and Bacteroides—has been associated with impaired induction of regulatory T (Treg) cells and disrupted immune homeostasis, ultimately enhancing vulnerability to airway inflammation. Current evidence indicates that the influence of the microbiota on asthma involves multiple, interconnected mechanisms that operate across different biological levels. These mechanisms encompass innate immune sensing through pattern recognition receptors (PRRs), modulation of epithelial barrier structure and function, epigenetic regulation, and neuro–immune signaling. Collectively, they can be broadly classified into primary drivers, intermediate modulators, and downstream immune effectors, reflecting the hierarchical organization of microbiota–host interactions in asthma. Figure 1 provides an integrative schematic of these pathways, illustrating how gut microbial communities act as upstream drivers, epithelial and epigenetic processes function as intermediate modulators, and neuro–immune interactions operate as downstream regulatory mechanisms. This framework contextualizes the subsequent sections within a unified model of asthma pathogenesis.
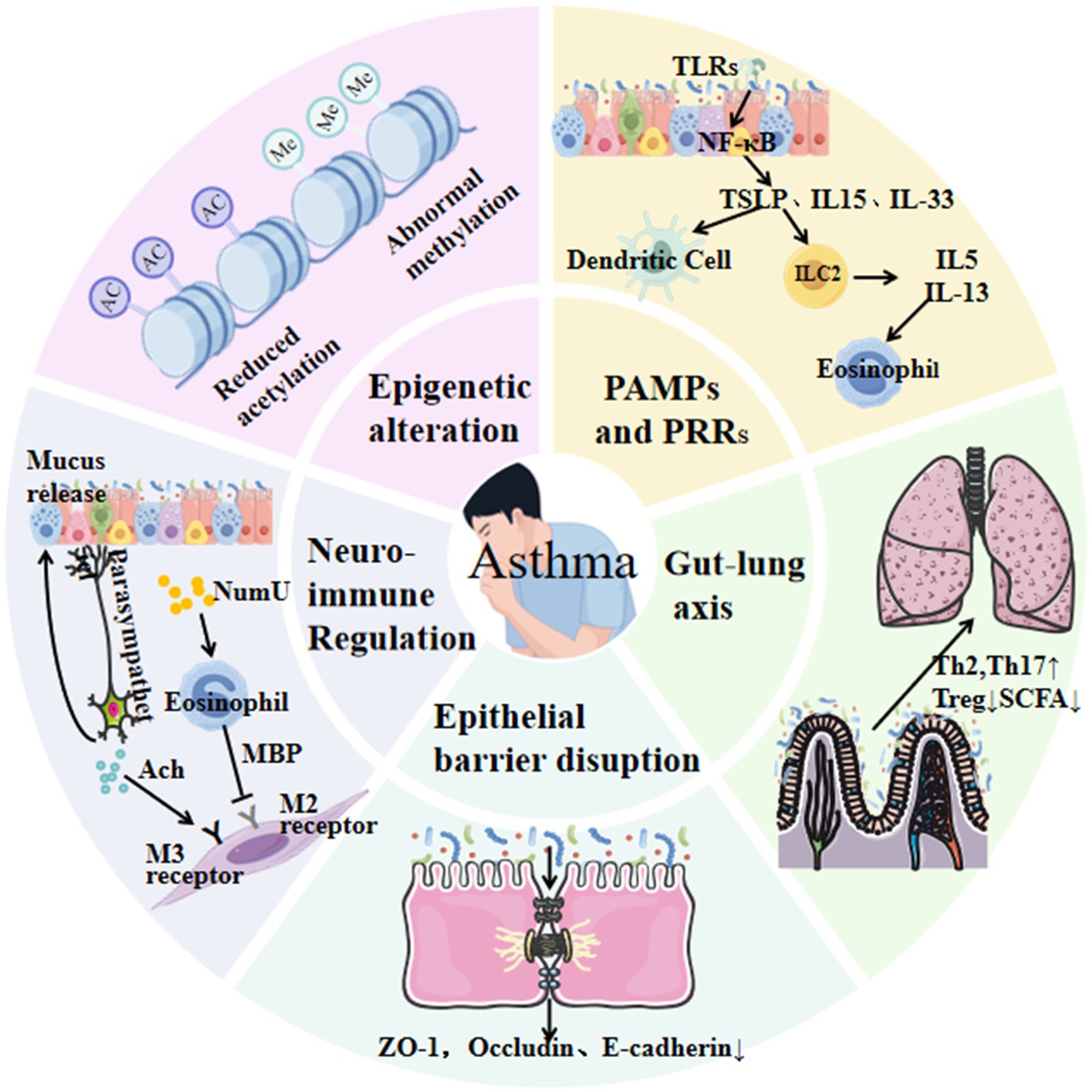
Figure 1. This schematic illustrates key pathways linking microbiota to asthma, including innate immune activation (e.g., TLRs/NF-κB), epithelial barrier disruption, gut–lung axis interactions via reduced short-chain fatty acid (SCFA) production, Th2/Th17 polarization, and neuro-immune crosstalk.
2.1 Activation of pathogen-associated molecular patterns and pattern recognition receptors
Airway epithelial cells serve as the first line of defense by recognizing microbial threats through pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs). Upon detection of pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs), these cells initiate downstream signaling cascades—most notably NF-κB—which drive the production of cytokines and chemokines that shape both innate and adaptive immune responses in asthma (Komlósi et al., 2022). PRR activation exhibits marked pathogen specificity. In the gut, bacterial components are primarily sensed through TLR2, TLR4 (lipopolysaccharide, LPS), and NOD1/NOD2 (peptidoglycan), whereas viral signals are detected by TLR3, TLR7/8, RIG-I, and MDA-5, which recognize viral RNA. Although the pulmonary microbiome contributes to PRR-mediated signaling, its impact is generally secondary to that of gut-derived stimuli, reflecting the higher microbial biomass and metabolic activity of the gastrointestinal tract (Komlósi et al., 2022). Importantly, the consequences of PRR engagement in asthma are highly context dependent. Zakeri et al. demonstrated that the effects of TLR agonists vary according to cell type (hematopoietic vs. non-hematopoietic), allergen characteristics (e.g., house dust mite (HDM), ovalbumin), and the route of administration, collectively determining whether PRR signaling dampens, exacerbates, or promotes disease development. Headland et al. further showed that gut microbiota–derived LPS induces elevated oncostatin M (OSM) expression through the TLR4–MyD88 pathway, driving airway inflammation and mucus hypersecretion characteristic of severe asthma. Consistently, OSM levels are markedly increased in the airways of patients with non–type 2 severe asthma (Headland et al., 2022). Moreover, Porsbjerg and colleagues reported that bronchial epithelial responses to viral mimetics differ across asthma phenotypes and severities, highlighting the heterogeneity of PRR-mediated pathways (Porsbjerg et al., 2022). Collectively, these studies indicate that PRR signaling exerts bidirectional immunomodulatory effects in asthma: gut microbial signals act as central upstream regulators, whereas their downstream consequences are shaped by microbial type, host genetic background, and the local immune microenvironment.
2.2 Epithelial barrier disruption and aberrant repair mechanisms
Under physiological conditions, epithelial cells in both the gut and respiratory tract maintain barrier integrity by producing immunoglobulin A (IgA), defensins, and lysozyme, thereby preventing microbial invasion and regulating immune homeostasis (Losol et al., 2023). Repeated allergen exposure in individuals with asthma leads to structural and functional barrier disruption, which profoundly alters local immune responses. Upon injury, epithelial cells release alarmins such as interleukin (IL) -25, IL-33, and thymic stromal lymphopoietin (TSLP), which activate group 2 innate lymphoid cells (ILC2s) and initiate type 2 inflammatory cascades (Losol et al., 2023). In parallel, epithelial cells interact with dendritic cells (DCs) and other immune cells, amplifying cytokine secretion and promoting airway inflammation.
A variety of microbial pathogens exploit these vulnerabilities to disrupt epithelial homeostasis. For example, the fungal allergen alkaline protease 1 from Aspergillus directly damages bronchiolar cell–cell junctions in murine models by inducing calcium influx via the calcineurin pathway, thereby promoting Th cell–mediated pulmonary eosinophilia (Wiesner et al., 2020). Similarly, respiratory viral infections exert potent barrier-modulating effects. Respiratory syncytial virus (RSV) downregulates tight junction proteins zonula occludens-1 (ZO-1) and E-cadherin through the IL-33/ST2/MyD88 pathway (Miao et al., 2025), while human rhinovirus (HRV) remodels epithelial junctions through IL-15–dependent antiviral responses, leading to enhanced permeability and exacerbated Th2 inflammation (Looi et al., 2016). These microbial insults act synergistically with impaired epithelial repair mechanisms and heightened immune activation to produce sustained barrier dysfunction in both the gut and lungs. Importantly, accumulating evidence indicates that gut dysbiosis serves as a critical upstream driver of these processes, linking microbial community alterations to airway inflammation through barrier disruption and alarmin release. Collectively, epithelial barrier dysfunction represents a key intermediate mechanism connecting gut microbial disturbances to asthma pathogenesis.
2.3 Microbiota-host interactions
The gut and lung are connected through the gut–lung axis, a bidirectional communication network shaped by their shared embryological origin and mucosal immune system. In healthy individuals, the gut microbiota is dominated by the phyla Firmicutes, Bacteroidetes, and Actinobacteria, along with less abundant taxa such as Fusobacteria. This microbial community plays a pivotal role in immune education, metabolic regulation, and epithelial barrier maintenance. Among gut-derived metabolites, SCFAs are key mediators of protective effects. Reduced fecal SCFA concentrations in pediatric atopic disorders support their role in maintaining immune tolerance and suppressing inflammation (Sun et al., 2017; Kumari and Kozyrskyj, 2017). SCFAs modulate epithelial cytokine production—including IL-6 and IL-8—through TLR and NF-κB signaling pathways, thereby exerting broad anti-inflammatory functions. Specifically, butyrate alleviates asthma by suppressing GATA3 expression and reducing IL-5 and IL-13 secretion from ILC2s (Lewis et al., 2019), whereas propionate acts through free fatty acid receptor 3 to promote DC and macrophage differentiation, attenuating allergic inflammation (Zhou et al., 2021). Maternal microbial exposure also influences offspring immunity through epigenetic and metabolic pathways. Perdijk et al. demonstrated that maternal high-fiber or acetate-enriched diets protect offspring against allergic airway disease by inhibiting histone deacetylase 9 (HDAC9) and promoting Foxp3+ Treg cell differentiation (Perdijk et al., 2024). Beyond SCFAs, other metabolite classes such as bile acids and polyamines also modulate airway inflammation. For instance, tauroursodeoxycholic acid (TUDCA) significantly reduces HDM-induced IL-4, IL-5, IL-13, and IgE levels, whereas β-muricholic acid suppresses IL-6 and IL-33 production, underscoring the diverse metabolic pathways through which gut microbes influence asthma pathogenesis (Nakada et al., 2019).
Although recent research has focused on the gut microbiota, the respiratory microbiome also contributes to disease heterogeneity. In healthy adults, the lower airways are typically dominated by Streptococcus, Veillonella, and Prevotella. Phenotype-specific shifts have been observed in asthma: eosinophilic asthma is associated with enrichment of Actinobacteria (Simpson et al., 2016), whereas neutrophilic asthma is characterized by expansion of Proteobacteria, particularly Haemophilus influenzae. Versi et al. showed that H. influenzae dominance correlates with neutrophilic inflammation, prolonged disease duration, frequent exacerbations, high corticosteroid use, and elevated serum IL-8, while fractional exhaled nitric oxide (FeNO) and eosinophilic inflammation are increased only in a subset of patients (Versi et al., 2023). These observations indicate that H. influenzae enrichment reflects neutrophil-driven, rather than T2-high eosinophilic, inflammation.
Regional and longitudinal studies further highlight the systemic impact of gut microbes on asthma phenotypes. Cross-sectional research in South China revealed distinct gut microbial signatures across asthma endotypes, linking altered abundances of Bacteroides, Bifidobacterium, and SCFA-producing taxa with specific clinical manifestations (Zou et al., 2021). Similarly, data from a Chinese birth cohort showed that early-life dynamics of Bifidobacterium, Faecalibacterium, and fecal SCFA levels were associated with subsequent development of eczema and wheezing (Cheung et al., 2023). A recent systematic review confirmed that children with asthma or recurrent wheeze consistently exhibit reduced fecal SCFA concentrations (Sasaki et al., 2024). Collectively, these findings highlight the central role of gut microbial diversity and metabolite production in shaping airway immune responses, supporting the gut–lung axis as a key determinant of asthma pathogenesis.
Finally, certain commensal respiratory taxa confer protective effects. Genera such as Corynebacterium and Dolosigranulum can inhibit pathogenic Streptococcus colonization by producing antimicrobial compounds, thereby lowering the risk of poorly controlled asthma in children (Zhou et al., 2019). These protective interactions emphasize that the microbiome—particularly gut microbial metabolites—modulates asthma pathophysiology through systemic immune regulation and inter-organ communication, while respiratory communities add an additional layer of modulation that contributes to disease heterogeneity.
2.4 Epigenetic modifications
Epigenetic mechanisms regulate gene expression without altering the underlying DNA sequence and include DNA methylation, histone modifications, and non-coding RNA regulation (Wu et al., 2021). These modifications shape immune development and airway inflammation by modulating the accessibility of key regulatory loci. Host epigenetic programs are highly sensitive to microbial and metabolic signals, positioning the microbiota as an upstream driver of epigenetic remodeling in asthma.
Microbial metabolites influence host epigenetics through several complementary pathways. One of the best-characterized examples is butyrate, a major SCFA produced by commensal gut bacteria. Schulthess et al. demonstrated that butyrate promotes monocyte differentiation into macrophages by inhibiting histone deacetylase 3 (HDAC3), thereby reducing mammalian target of rapamycin kinase activity, enhancing LC3-mediated antimicrobial responses, and inducing antimicrobial peptide production (Schulthess et al., 2019). These epigenetic changes augment host defense and modulate immune activation, providing a mechanistic link between microbial metabolites and asthma susceptibility.
Microbiota-mediated epigenetic reprogramming can also occur through community-level alterations. Ramar et al. showed that gut microbiota transplantation (GMT) from asthma-susceptible donors induced dysregulated butyrate metabolism in recipient mice, leading to altered DNA methylation patterns in DCs (Ramar et al., 2025). This reprogramming enhanced antigen-presenting capacity and T cell proliferation, epigenetically amplifying allergic inflammation. These findings highlight epigenetic modification as a mid-level mechanism that integrates upstream microbial signals with downstream immune responses.
Epigenetic programming during early life represents another critical determinant of asthma susceptibility. DeVries and colleagues reported that maternal IFN-γ to IL-13 ratios shape DNA methylation profiles in cord blood mononuclear cells (CBMCs), altering innate immune training and nasal microbiota succession in offspring (DeVries et al., 2022). Maternal diet, microbial exposure, and metabolite availability thereby exert long-term influences on immune trajectories through epigenetic pathways, potentially predisposing individuals to asthma.
Epigenetic regulation also modulates therapeutic responses. Clinical research has shown that inhaled corticosteroids influence epigenetic regulators—including IL-2, TNF-α, NF-κB, and CCAAT/enhancer-binding protein pathways—in parallel with microbiome remodeling (Perez-Garcia et al., 2025). These results suggest that epigenetic states not only reflect disease susceptibility but also determine treatment efficacy, providing a mechanistic rationale for combining microbiota-targeted interventions with epigenetic modulation in future precision therapies.
Collectively, these studies position epigenetic modifications as a critical intermediate layer linking microbial signals, immune programming, and clinical outcomes in asthma. Future research should prioritize clarifying the causal directionality of these interactions, identifying key microbial–epigenetic–immune nodes, and standardizing longitudinal epigenomic profiling strategies to facilitate clinical translation.
2.5 Neuro-immune regulation
The pathogenesis of asthma involves dynamic interactions among the nervous system, immune system, and the microbiota. Neurotransmitters and neuropeptides released by nerve cells rapidly activate immune effector pathways, inducing the production of inflammatory mediators while simultaneously shaping local microbial communities (Joos et al., 2003). Conversely, immune cell–derived mediators such as histamine and neurotrophic factors stimulate sensory neurons, promoting bronchoconstriction and cough reflexes. This bidirectional neuro–immune communication contributes to asthma pathology by regulating type 2 inflammatory processes (Voisin et al., 2017). Recent studies have highlighted the role of the gut–brain–lung axis as a key pathway linking microbial signals to neural and immune responses. In murine models, postprandial acetylcholine release by the parasympathetic nervous system activates ILC2s through muscarinic receptor M4 (Chrm4), leading to increased secretion of IL-5 and IL-13 and exacerbated airway inflammation (Chen et al., 2025). Conversely, genetic deletion of Chrm4 significantly attenuates ILC2-driven airway pathology. These findings indicate that gut microbiota–derived metabolites, particularly butyrate, can modulate neural circuits via the dorsal vagal nucleus, thereby amplifying Th2 cytokine responses and downstream inflammatory cascades.
Under conditions of persistent Th2 inflammation, neutrophils release chromatin and granule proteins to form neutrophil extracellular traps (NETs). While NETs facilitate antifungal defense—for example, against Candida albicans—they also promote dysbiosis and sustain chronic inflammation (Daniel et al., 2019). This reciprocal interaction between neural pathways, immune responses, and microbial communities contributes to disease persistence and heterogeneity in asthma.
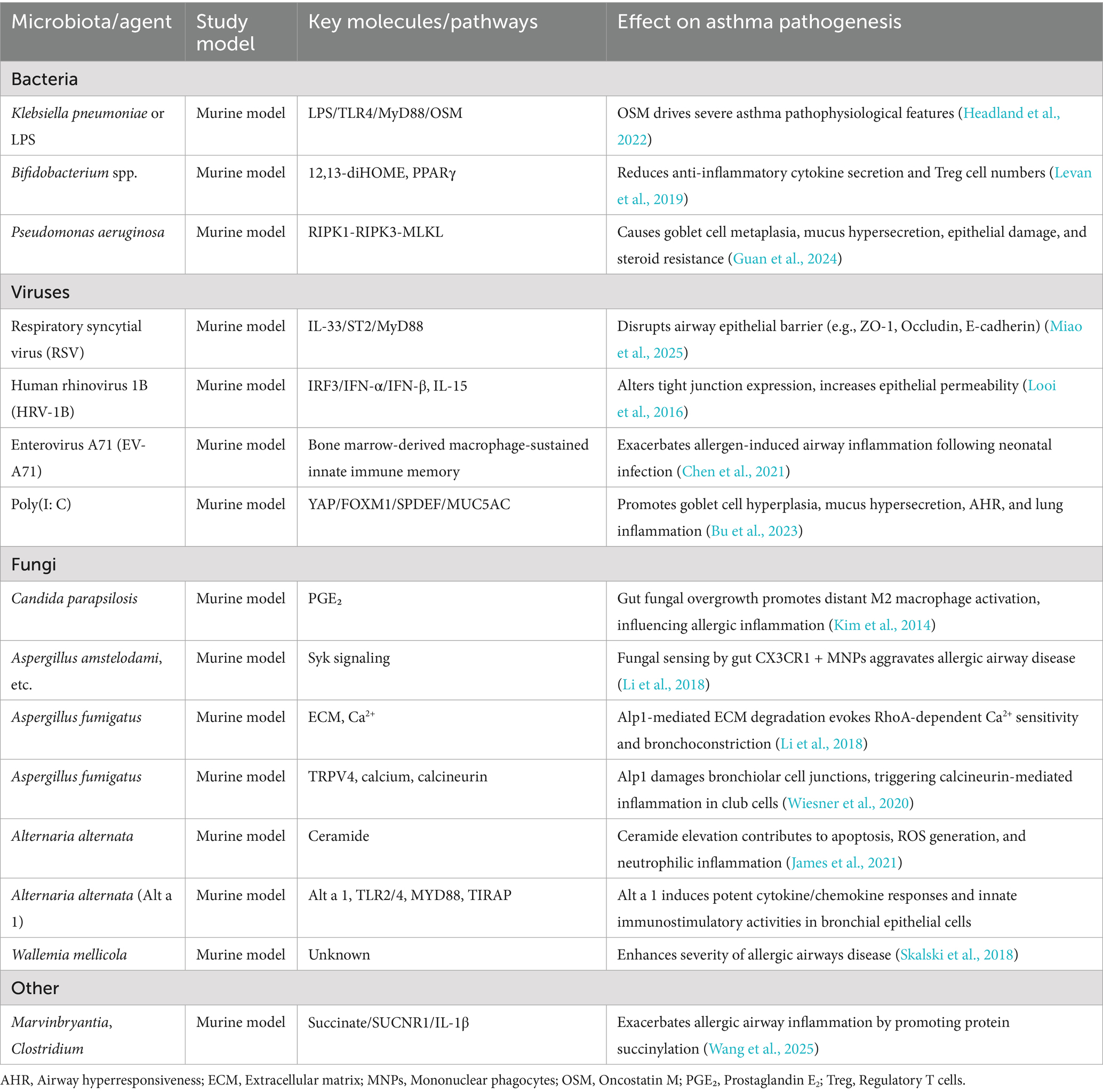
Collectively, microorganisms across different kingdoms—including bacteria, viruses, and fungi—shape asthma pathogenesis through multiple mechanisms, including disruption of epithelial barriers, modulation of innate and adaptive immunity, epigenetic remodeling, and neuro–immune signaling. These neuro–immune interactions represent a downstream regulatory layer that integrates microbial and immune signals, contributing to both acute responses and chronic disease trajectories. Table 1 provides an overview of key microorganisms and their mechanistic contributions to asthma pathogenesis based on experimental and clinical studies.
3 The clinical correlation between microorganisms and asthma
3.1 Microbial diversity and asthma phenotypes
Asthma exhibits marked clinical heterogeneity, posing significant challenges for disease management. Based on predominant inflammatory cells identified in induced sputum or bronchial biopsy—such as eosinophils, neutrophils, and lymphocytes—patients are commonly categorized into distinct inflammatory phenotypes. Among these, the Th2-high phenotype (typically associated with eosinophilic inflammation) and the Th2-low or non-Th2 phenotype (often linked to neutrophilic or lymphocytic inflammation) are the most widely recognized subgroups (Ojanguren et al., 2025). Increasing evidence has indicated that dysbiosis of the human microbiota contributes to the development of these phenotypes, with specific microbial signatures associated with distinct immunological profiles.
In the Th2-high phenotype, characteristic alterations of the respiratory microbiota have been observed. Clinical studies demonstrated that patients with severe eosinophilic asthma exhibited significantly greater abundances of Haemophilus and Streptococcus in bronchial specimens compared with healthy controls or non-eosinophilic asthma patients, correlating positively with sputum eosinophil counts (Versi et al., 2024). Metagenomic sequencing of induced sputum from 55 asthma patients and 12 healthy individuals revealed that the α-diversity of the lower respiratory tract microbiota was significantly higher in Th2-high asthma compared to non-Th2 asthma, accompanied by a reduction in Proteobacteria abundance. Further analysis showed a denser microbial network structure in the Th2-high group, while the relative abundance of Actinobacteria correlated positively with eosinophil counts (Wang W. et al., 2024).
Notably, differences in microbial communities across asthma phenotypes extend beyond the respiratory tract to the gut. Metagenomic sequencing of fecal samples revealed that patients with eosinophilic asthma displayed a significant increase in gut fungal communities, particularly within the yeast phylum. In contrast, individuals with neutrophilic asthma exhibited increased levels of Bacillales (Yan et al., 2024). These findings suggest that both bacterial and fungal communities may serve as candidate biomarkers for distinguishing asthma phenotypes. Additional multi-cohort studies further supported these associations: reduced airway bacterial diversity and enrichment of Proteobacteria were linked to corticosteroid resistance and neutrophilic inflammation (Durack et al., 2020), whereas early-life depletion of gut microbial diversity increased the risk of persistent wheeze and subsequent asthma (Kim and Bunyavanich, 2025). Moreover, loss of butyrate-producing taxa such as Faecalibacterium prausnitzii correlated with impaired SCFA production and poor asthma control (Fujimura et al., 2016).
To illustrate these links, Figure 2 summarizes how specific microbial exposures shape asthma endotypes. Th2-high/eosinophilic inflammation is primarily driven by epithelial cytokines (IL-25, IL-33, TSLP) that amplify Th2 responses, whereas Th2-low/neutrophilic inflammation is more closely associated with TLR-mediated activation of Th1/Th17 pathways. Collectively, these findings provide a mechanistic framework connecting microbial dysbiosis with the heterogeneity of asthma phenotypes.
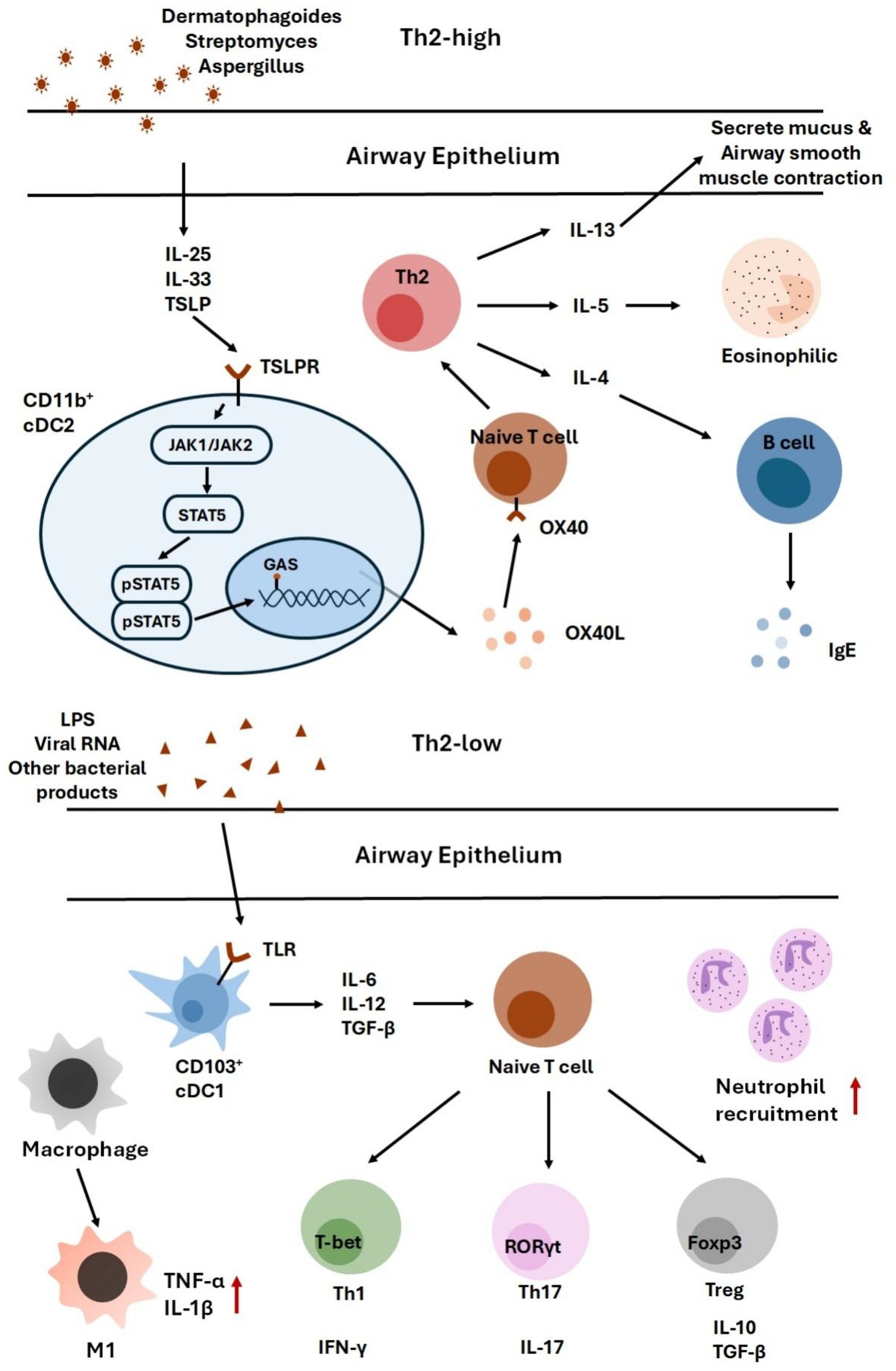
Figure 2. This figure summarizes how distinct microbial exposures contribute to different asthma endotypes. Th2-high/eosinophilic inflammation is driven by epithelial cytokines such as IL-25, IL-33, and TSLP, which amplify Th2 responses. In contrast, Th2-low/neutrophilic inflammation arises from TLR activation, leading to Th1/Th17-mediated immune responses.
3.2 Immunoregulatory functions of microbiota-derived metabolites
Microbial metabolites represented key mediators through which the gut microbiota influenced host immune regulation and contributed to asthma pathogenesis (Ge et al., 2025). Among these, short-chain fatty acids (SCFAs)—including acetate, propionate, and butyrate—were the most extensively studied. Produced in the intestinal lumen via dietary fiber fermentation, SCFAs entered systemic and lymphatic circulation and reached the lungs, where they modulated airway inflammation and immune balance.
Experimental studies demonstrated that butyrate bound to G protein–coupled receptor 43 (GPR43), inhibited the p38 MAPK/NF-κB signaling pathway, and suppressed IL-13 expression. This mechanism reduced Tfh13 cell–mediated production of high-affinity IgE and alleviated allergic airway responses (Yu et al., 2025). Similarly, propionate acted on GPR41 in dendritic cells, promoting the migration of intestinal Tregs to the lungs and limiting excessive Th2 inflammation (Mann et al., 2024). Beyond SCFAs, tryptophan-derived metabolites such as indole, serotonin, and indole-3-aldehyde were found to be enriched in low asthma-risk environments. Network analyses indicated that these protective metabolites were associated with cohabiting microorganisms such as Actinobacteria, suggesting that environmental microbial exposure could shape immune tolerance through metabolite production (Sun et al., 2022).
Not all microbial metabolites exerted protective effects. Animal studies revealed that 4-methylpentanoic acid, generated via branched-chain amino acid metabolism, was elevated in asthma models and positively correlated with IL-5 and IL-13 expression (Liu Y. H. et al., 2025). In addition, LPS released during gut dysbiosis could translocate into the bloodstream, activate TLR4 signaling in airway epithelial and stromal cells, and induce OSM expression. OSM drove mucus hypersecretion and promoted Th2/Th17 inflammation, leading to eosinophil and neutrophil infiltration (Headland et al., 2022; Zhang et al., 2025). However, previous investigations of LPS exposure and asthma yielded inconsistent outcomes, likely reflecting differences in experimental models, treatment timing, and dosage (Zakeri and Russo, 2018). Collectively, these findings suggested that LPS exerted a bidirectional influence on immune responses in asthma, with effects depending on exposure context.
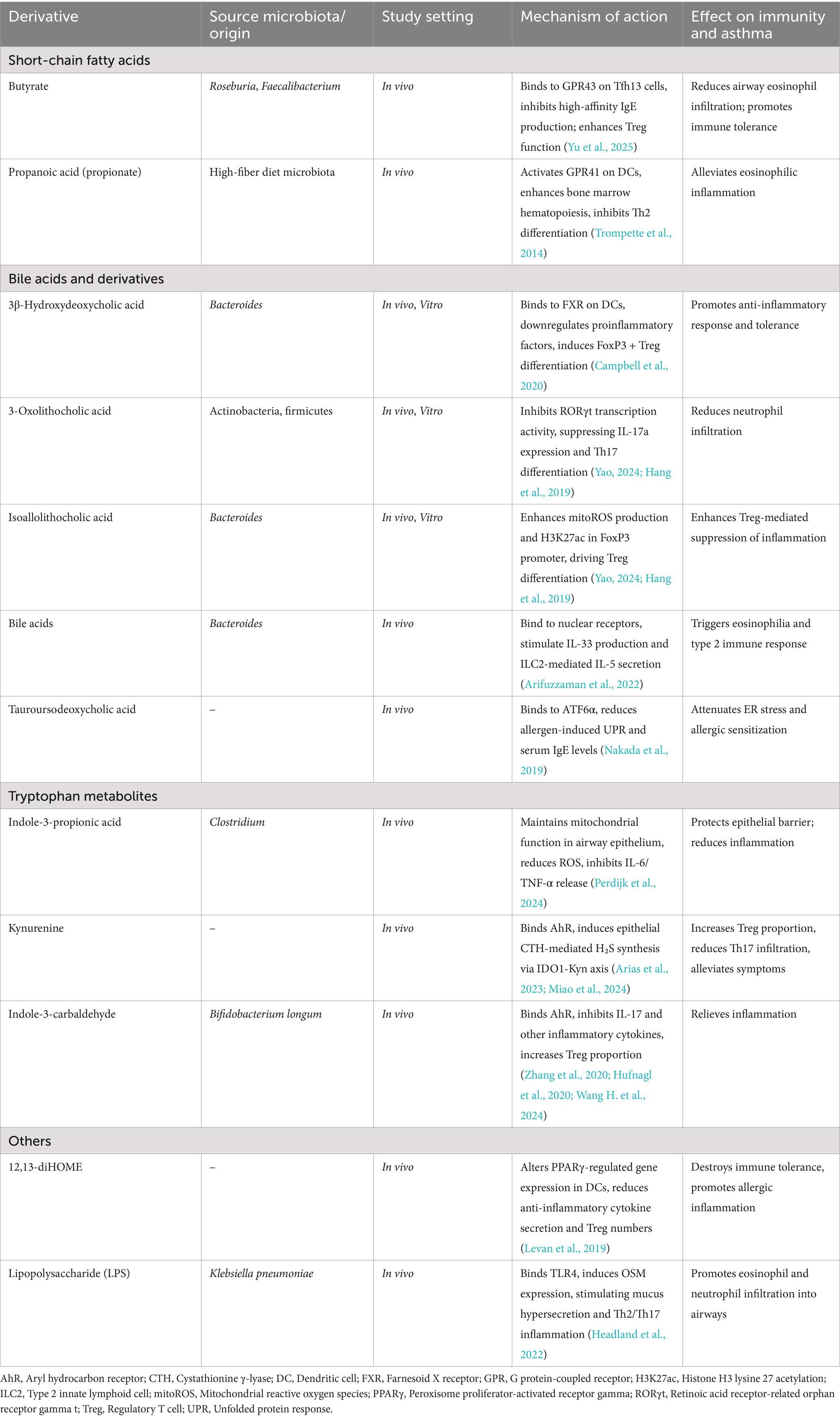
In summary, microbial metabolites exerted diverse immunoregulatory effects in asthma, ranging from protective activities such as maintaining epithelial integrity and promoting Treg differentiation, to pathogenic actions including mucus hypersecretion, eosinophilic infiltration, and Th17-driven inflammation. These effects appeared to depend on metabolite type, concentration, and cellular targets, highlighting the complexity of metabolite-mediated immune modulation. Table 2 provides a comprehensive summary of the immunoregulatory mechanisms of major microbiota-derived metabolites in asthma pathogenesis.
3.3 Microbial biomarkers in asthma diagnostics and prognostication
Conventional prediction of acute asthma exacerbations had relied largely on clinical symptoms and lung function tests. However, these indicators often lag behind inflammatory progression, limiting their utility for early detection. Recent studies suggested that alterations in the airway and gut microbiota contributed to both asthma pathogenesis and acute exacerbations, with microbial composition changes emerging as sensitive predictive biomarkers. For example, elevated abundances of Haemophilus influenzae and Moraxella in sputum from patients with severe asthma correlate with increased sputum eosinophil counts and can predict exacerbation events (Heaney et al., 2016), Within the gut, Ligilactobacillus murinus (LGG) suppressed excessive inflammation through the TLR2/IL-10 pathway, and its fecal abundance was proposed as a marker of systemic immune status and a predictor of post-infection exacerbations (Hu et al., 2022). In addition to compositional markers, microbiota-derived metabolites provided functional insights: short-chain fatty acids (SCFAs) reduced airway eosinophil infiltration by binding to GPR41/43 and inhibiting histone deacetylase (HDAC), thereby reflecting the anti-inflammatory potential of the microbiota-host interaction. Furthermore, integrating microbial markers with established clinical indicators improved prognostic accuracy. A combined evaluation of eosinophil counts (EOS) and FeNO demonstrated greater predictive value for exacerbation events when both metrics were elevated, compared to either alone (Meulmeester et al., 2025). In summary, evidence indicated that the gut microbiota and its metabolites represented promising biomarkers for asthma diagnostics and prognostication. Their incorporation into clinical practice may enhance risk stratification and facilitate precision management of asthma.
4 Therapeutic role of microbiota in asthma treatment
Corticosteroids remain the first-line therapy for suppressing airway inflammation in asthma. However, a subset of patients exhibits suboptimal responses, and systemic administration is associated with adverse effects including hoarseness, oral candidiasis, growth suppression, and hypothalamic–pituitary–adrenal axis suppression (Voo et al., 2022). These limitations have prompted growing interest in complementary therapeutic strategies that target the gut–lung axis.
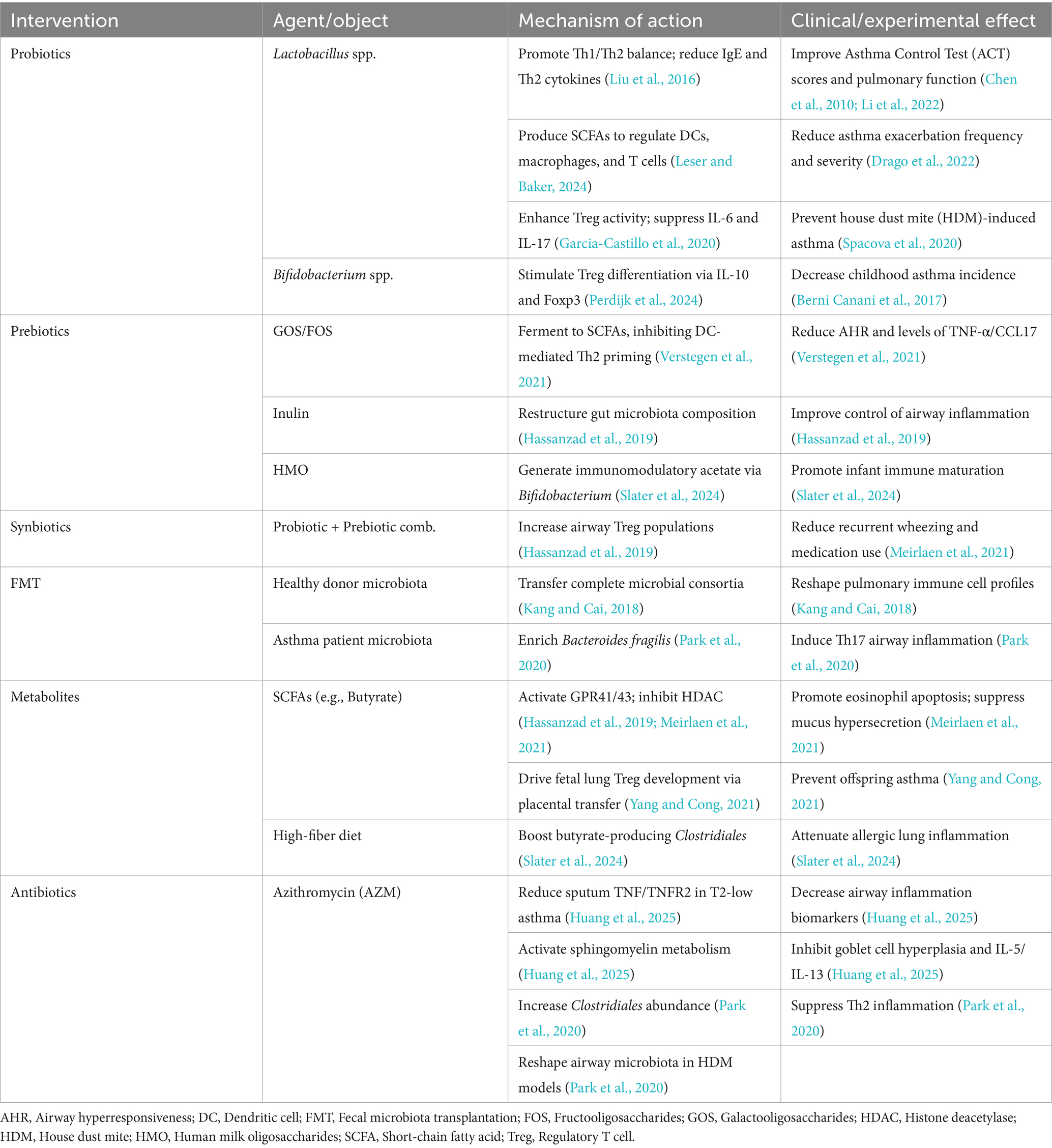
Accumulating evidence indicates that intestinal dysbiosis is a modifiable risk factor contributing to asthma pathogenesis (Liu Y. et al., 2025). Accordingly, microbiota-targeted interventions have emerged as promising approaches to restore immune homeostasis and improve clinical outcomes. These strategies include probiotics, prebiotics, synbiotics, fecal microbiota transplantation (FMT), precision antibiotic regimens, and targeted modulation of microbial metabolites. Their proposed mechanisms and available evidence are summarized in Table 3. Mechanistically, these interventions modulate disease processes by restoring microbial balance, increasing SCFA production, and downregulating type 2 inflammatory pathways, including the suppression of IL-5 and IL-13 as well as eosinophilic activity. Figure 3 illustrates the major pathways through which microbiota-based interventions may alleviate airway inflammation and clinical symptoms.
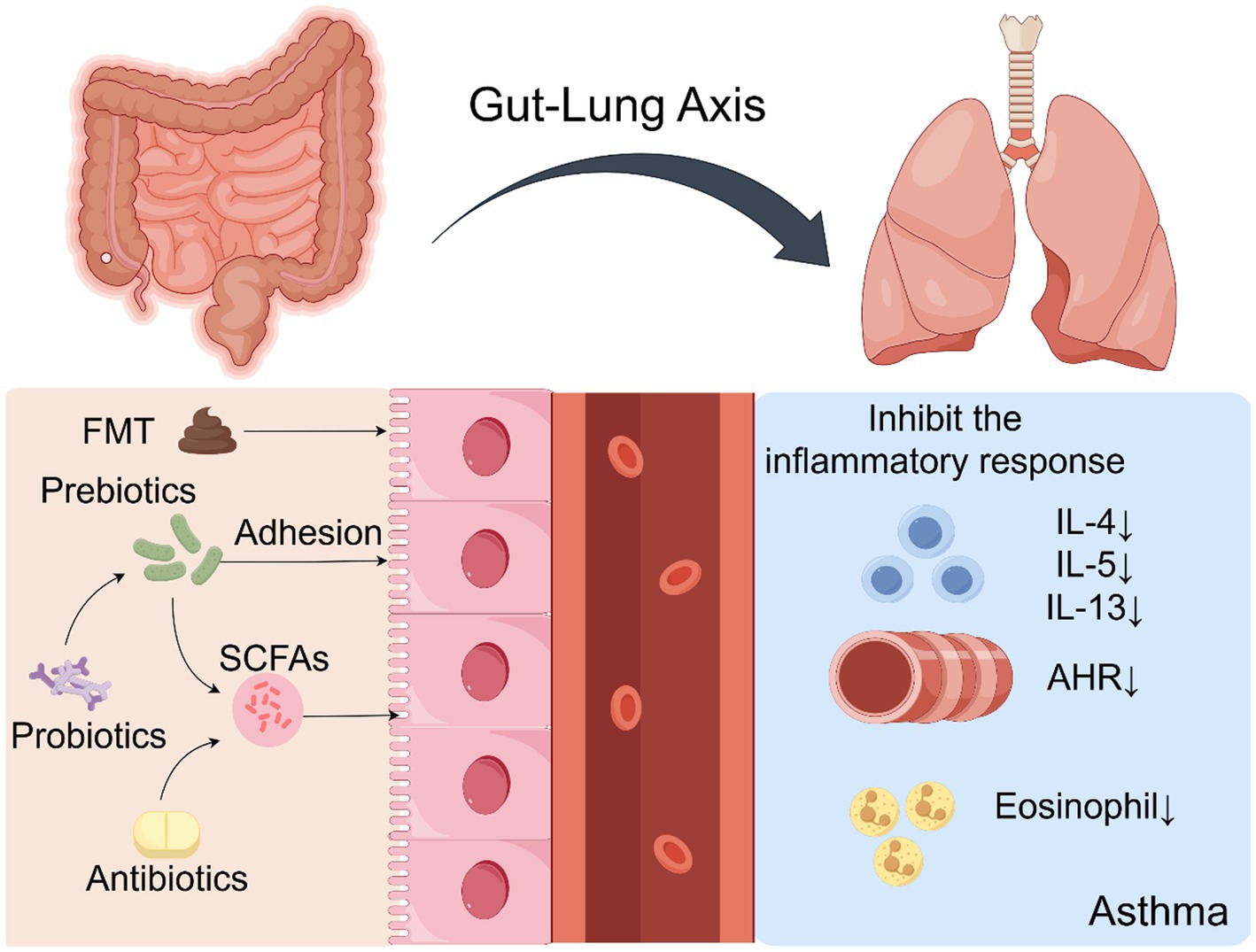
Figure 3. This schematic illustrates microbiota-targeted therapeutic approaches, including probiotics, prebiotics, and FMT. These interventions alleviate airway inflammation and symptoms by restoring microbial balance, enhancing SCFA production, and suppressing pro-inflammatory cytokines (e.g., IL-5, IL-13) and eosinophilic activity.
4.1 Probiotics
Probiotics, defined as “live microorganisms” that confer health benefits when administered in adequate amounts (Peng et al., 2024). Emerging evidence from both preclinical and clinical studies demonstrates that probiotic supplementation exerts beneficial immunomodulatory effects in asthma, acting primarily through the gut–lung axis as a primary upstream driver of host–microbe interactions (Tashiro et al., 2025). As summarized in Figure 3, probiotic interventions restore gut microbial homeostasis, promote anti-inflammatory metabolite production, and modulate systemic immune signaling, thereby influencing pulmonary immunity.
Mechanistically, probiotic-derived signals are first sensed by gut mucosal immune cells via PRRs, leading to the activation of innate immune pathways and subsequent trafficking of immune cells to the lungs. For example, oral probiotic supplementation enhances intestinal Th1 responses and facilitates the migration of CD4+ IFN-γ+ T cells to the airways, rebalancing the Th1/Th2 axis and attenuating allergen-induced sensitization. This process is accompanied by reduced serum levels of IgE and Th2-associated cytokines, including IL-5, IL-6, IL-8, and IL-17 (Garcia-Castillo et al., 2020). In parallel, probiotic-derived SCFAs act as critical intermediate modulators by regulating the activity of DCs, macrophages, and T cells, enhancing Treg differentiation and suppressing pro-inflammatory cytokine production (Leser and Baker, 2024). These interconnected pathways illustrate how probiotics influence asthma pathogenesis through a hierarchical mechanism: gut microbial modulation as the upstream driver, epithelial and metabolic signaling as intermediate modulators, and immune cell reprogramming as downstream effectors.
The therapeutic potential of probiotics in asthma has been investigated through a combination of preclinical studies and randomized controlled trials (RCTs), although the strength of evidence remains heterogeneous (Drago et al., 2022; Li et al., 2020). Preclinical models consistently demonstrate that oral or inhaled administration of Lactobacillus strains can reduce allergen-induced airway inflammation, suppress AHR, and modulate Th2-driven immune responses (Spacova et al., 2020). These findings provide essential mechanistic insights but require careful translation to human disease contexts.
In clinical settings, several RCTs have evaluated the efficacy of probiotics as adjunctive interventions in asthma management, with mixed but generally favorable outcomes. In a double-blind RCT, Li et al. reported that 8-week supplementation with Lactobacillus reuteri CCFM1040 significantly improved respiratory symptoms in patients with allergic rhinitis and asthma, as reflected by higher Asthma Control Test (ACT) scores compared to placebo (Li et al., 2022). Two additional RCTs in pediatric populations demonstrated that daily intake of Lactobacillus strains reduced FeNO levels and improved lung function parameters, suggesting attenuation of airway inflammation (Chen et al., 2010). Beyond symptom control, probiotic supplementation has shown promise in preventing asthma development. An RCT involving 422 children found that administration of Lactobacillus salivarius DSM 22775 significantly reduced the frequency and severity of asthma exacerbations (Drago et al., 2022). Moreover, a three-year longitudinal RCT demonstrated that Lactobacillus rhamnosus GG (LGG)-fortified formula lowered asthma incidence among children with cow’s milk allergy (Berni Canani et al., 2017). These clinical findings collectively support a moderate but growing evidence base for probiotic use in asthma, while also underscoring the need for larger, well-controlled trials to optimize strain selection, dosage, and intervention timing.
In addition to their standalone effects, probiotics can act as adjuvant therapies that enhance the efficacy of conventional asthma treatments. Preclinical studies have shown that co-administration of Bifidobacterium and LGG with glucocorticoids exerts synergistic anti-inflammatory effects, leading to greater suppression of AHR and airway inflammation than glucocorticoid monotherapy (Voo et al., 2022). These findings suggest that probiotic co-therapy may permit lower glucocorticoid dosages while maintaining clinical efficacy, potentially minimizing treatment-related adverse effects.
Probiotics have also been investigated as adjuncts to allergen-specific immunotherapy (AIT). Clinical studies indicate that the combination of probiotics with AIT significantly improves asthma symptom control compared to AIT alone (Liu et al., 2016). Mechanistically, this effect involves enhanced induction of antigen-specific regulatory B10 cells and reduced serum levels of allergen-specific IgE, thereby augmenting immune tolerance. Given the emerging role of the lung microbiota in asthma pathogenesis, inhalation-based probiotic delivery has been proposed as a novel strategy to directly modulate the airway microbial community. By targeting the respiratory tract, this approach may enhance local immune regulation and reduce inflammation, complementing the systemic effects of oral supplementation. Although clinical data remain limited (Glieca et al., 2024; Nicola et al., 2024), early findings suggest that inhaled probiotics could represent a promising avenue for targeted microbiota modulation in asthma management.
Collectively, current evidence supports the therapeutic potential of probiotics as both preventive and adjunctive interventions in asthma management. Preclinical studies consistently demonstrate mechanistic efficacy through modulation of the gut–lung axis, while clinical trials provide emerging, though heterogeneous, support for improvements in airway inflammation and symptom control. Importantly, probiotics are generally well tolerated and carry a favorable safety profile, making them attractive candidates for long-term disease modulation. Nevertheless, the clinical evidence base remains limited by small sample sizes, short follow-up durations, and substantial heterogeneity in probiotic strains, dosages, and patient populations. To advance probiotic therapy toward clinical translation, future research should prioritize large, well-designed randomized controlled trials to determine optimal microbial strains and combinations, standardized dosing regimens, and critical intervention windows (e.g., perinatal versus postnatal periods). Integrating microbiome profiling and immunophenotyping into clinical trial design will be essential for identifying patient subgroups most likely to benefit and for elucidating causal mechanisms. Overall, while probiotics represent a promising and biologically plausible therapeutic avenue, their incorporation into routine asthma care will require more rigorous evidence to define efficacy, durability of effects, and patient-specific therapeutic strategies.
4.2 Prebiotics and synbiotics
Prebiotics are defined as indigestible organic substrates that selectively stimulate the growth and metabolic activity of beneficial commensal microorganisms, thereby contributing to host health by restoring microbial homeostasis (Fan et al., 2025). In the context of asthma prevention and management, commonly studied prebiotics—including inulin, fructooligosaccharides (FOS), and galactooligosaccharides (GOS)—are fermented by the gut microbiota to produce SCFAs. SCFAs act as key intermediate immunomodulators, inhibiting DC–mediated Th2 immune priming and attenuating allergic airway inflammation (Blanco-Pérez et al., 2021). Beyond these synthetic substrates, human milk oligosaccharides (HMOs)—major components of breast milk—play a pivotal role in shaping the neonatal gut microbiota. Metabolism of HMOs by Bifidobacterium species generates high concentrations of acetate, which are critical for early-life immune programming (Slater et al., 2024). Clinically, synbiotic formulations combining prebiotics and probiotics have been shown to enhance gut microbial diversity and modulate immune responses in high-risk infants. RCTs indicate that such supplementation significantly reduces the incidence of wheezing episodes, suggesting potential for early-life asthma prevention (Hassanzad et al., 2019).
The therapeutic potential of prebiotic supplementation in asthma is increasingly supported by both mechanistic and clinical evidence. RCTs have shown that administration of GOS and inulin significantly reduces AHR and inflammatory biomarkers such as TNF-α and C-C motif CCL17, while concurrently improving lung function in patients with stable asthma (Verstegen et al., 2021). Inulin supplementation has been further associated with gut microbiota remodeling and enhanced disease control (Hassanzad et al., 2019). These clinical effects are closely linked to SCFA-mediated immune regulation, which promotes anti-inflammatory responses and fosters colonization of beneficial microbial taxa.
Experimental studies provide complementary mechanistic insights: mice maintained on high-fiber diets exhibit increased SCFA production and display significantly attenuated airway inflammation in experimental asthma models, largely through enhanced Treg responses (Slater et al., 2024). Evidence for synbiotics remains more limited, but early clinical trials indicate promising preventive effects. In a 12-week multicenter RCT involving 90 infants with atopic dermatitis, 7-month synbiotic supplementation reduced the incidence of recurrent wheezing and respiratory symptoms, along with decreased asthma medication use, although no significant differences in total IgE levels were observed (Meirlaen et al., 2021).
Collectively, while current findings highlight the mechanistic plausibility and early clinical benefits of prebiotics and synbiotics in asthma, the evidence base remains fragmentary and heterogeneous. Future research should focus on large-scale, longitudinal RCTs to define optimal substrate types, dosages, and intervention windows, and to identify patient subgroups most likely to benefit.
4.3 Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) involves the transfer of functional microbial communities from healthy donors to recipients with the aim of restoring intestinal microbial homeostasis (Kang and Cai, 2018). Unlike probiotics, which typically introduce single or limited strains, FMT delivers a complex and stable microbial consortium, thereby enabling more durable engraftment and niche restoration. Through these mechanisms, FMT has shown therapeutic potential across several extraintestinal conditions—including metabolic, neuropsychiatric, autoimmune, and allergic disorders—by re-establishing colonization resistance and modulating immune responses. Within the framework of the gut–lung axis, FMT is hypothesized to act as an upstream microbial intervention capable of influencing pulmonary immunity via metabolic and immunological signaling.
Although no clinical trials have yet evaluated FMT specifically for asthma, accumulating preclinical evidence supports its immunomodulatory capacity. In murine models, transplantation of fecal microbiota from patients with asthma—characterized by enrichment of Bacteroides fragilis—into germ-free recipients induced T helper 17 (Th17)–polarized airway inflammation (Park et al., 2020). In antibiotic-treated respiratory disease models, FMT has been shown to reshape pulmonary immune cell landscapes, highlighting the bidirectional communication between gut and lung microbial communities. Compared with probiotic interventions, FMT provides superior microbial biomass and engraftment stability, potentially enabling more profound reconfiguration of host–microbiota interactions.
Despite its promise, the clinical translation of FMT for asthma faces several technical, economic, and safety challenges. Standardized fecal processing protocols and targeted delivery systems remain underdeveloped, and the procedure is currently 3–5 times more expensive per capita than conventional therapies (Hou et al., 2022). Safety concerns—including the risk of pathogen transmission and uncertain outcomes in immunocompromised or pregnant individuals—must be carefully addressed before clinical implementation. Future research should prioritize the development of selective microbial transplantation strategies, ideally supported by large-scale, well-controlled clinical trials. Establishing donor–recipient matching frameworks and optimizing microbial consortia composition will be essential for achieving precise immune modulation in asthma. Furthermore, integration of multi-omics approaches and immune phenotyping may enable better prediction of FMT outcomes and identification of responsive patient subgroups.
4.4 Targeting microbial metabolites
Microbial metabolites serve as critical intermediates linking intestinal microbial composition to systemic and pulmonary immune regulation. Among these, SCFAs—such as acetate, propionate, and butyrate—have been most extensively studied. In animal models, dietary fiber supplementation significantly increases SCFA production, which in turn suppresses allergic airway inflammation by promoting the differentiation and function of Tregs (Yang and Cong, 2021). SCFA-mediated induction of Tregs provides early protection against Th2-driven allergic phenotypes through microbiota-dependent immune education following intestinal colonization.
Early-life exposure represents a critical immunological window for SCFA-mediated effects. Experimental studies have demonstrated that microbial exposure after weaning fails to reverse asthma-related immune and phenotypic abnormalities, underscoring the importance of early colonization for immune programming. Notably, acetate can cross the placental barrier and promote fetal lung Treg development, thereby attenuating postnatal asthma susceptibility (Lee-Sarwar et al., 2020). These findings highlight the potential of maternal diet modulation and early-life microbial interventions as preventive strategies for asthma.
Beyond SCFAs, other microbial metabolite classes—including polyamines, tryptophan derivatives, and bile acids—play emerging roles in modulating immune homeostasis. Polyamines regulate epithelial integrity and inflammatory gene expression, while tryptophan metabolites act via the aryl hydrocarbon receptor to shape innate lymphoid and T cell responses. Secondary bile acids, such as TUDCA and β-muricholic acid, have been shown to suppress Th2 cytokine production and IgE levels, thereby attenuating allergic airway inflammation (Nakada et al., 2019). Although these pathways are less extensively characterized than SCFAs, they provide complementary mechanisms through which the microbiota can influence asthma pathogenesis.
Looking forward, future research should prioritize integrated metabolomics–immunology approaches to define the temporal dynamics and causal relevance of these metabolite classes in asthma. Clinical translation will require well-controlled dietary and microbial interventions aimed at selectively modulating metabolite production during critical developmental periods. Such strategies may ultimately enable precise, metabolite-targeted modulation of the gut–lung axis for asthma prevention and therapy.
4.5 Precision antibiotic therapy
Antibiotic-induced alterations of the gut microbiota exert profound effects on host immune regulation, particularly through disruption of the gut–lung axis. While broad-spectrum antibiotics are effective in controlling infections, their nonspecific antimicrobial activity perturbs intestinal microbial homeostasis, leading to decreased production of SCFAs and impaired Treg function (Donald and Finlay, 2024). In murine models, early-life vancomycin exposure selectively depletes butyrate-producing Clostridium populations, resulting in exacerbated allergic airway inflammation (Chen et al., 2022).
Among available antimicrobials, azithromycin (AZM) displays distinctive dual immunomodulatory properties. Clinical investigations have shown that prolonged AZM therapy significantly reduces sputum TNF/TNFR2 concentrations in non-eosinophilic, Th2-low asthma endotypes. Preclinical studies further demonstrate that AZM mitigates FMT–induced allergic airway inflammation by reducing BALF IL-5 levels and AHR, while concurrently remodeling airway microbial communities in HDM–challenged asthma models (Park et al., 2020). Mechanistically, AZM activates the sphingomyelin (SM) metabolic pathway, leading to reduced goblet cell proliferation and inhibition of Th2 cytokines, including IL-5 and IL-13, thereby attenuating type 2 airway inflammation (Huang et al., 2025). Experimental studies further reveal that antibiotic-naïve HDM-sensitized mice spontaneously develop heightened AHR and elevated Th2 cytokines (IL-4, IL-5, IL-13) accompanied by pronounced eosinophilic infiltration in BALF. 16S rRNA sequencing demonstrates that this inflammatory state is associated with significant gut dysbiosis, despite minimal changes in airway microbiota composition (Park et al., 2020). Collectively, these data underscore the bidirectional interactions between gut microbial communities and pulmonary immune responses and highlight the potential of AZM as both an antimicrobial and immunomodulatory agent.
Future research should aim to define precision antibiotic strategies that minimize collateral microbiota disruption while harnessing the immunomodulatory benefits of specific agents such as AZM. Integrating microbiome profiling with clinical phenotyping may enable individualized antimicrobial interventions that align with microbiota-targeted therapies for allergic asthma.
5 Summary and future perspectives
Microbial dysbiosis disrupts host homeostasis and plays a pivotal role in the pathogenesis of immune-mediated diseases, including asthma. As a multifactorial disorder shaped by both genetic and environmental influences, asthma pathogenesis, clinical phenotypes, and disease severity are closely linked to dynamic fluctuations in microbial communities within the respiratory and gastrointestinal systems. Although the pulmonary microbiota has a relatively low biomass, it engages in continuous crosstalk with the intestinal microbiota through lymphatic circulation, hematogenous dissemination, and micro-aspiration, establishing the bidirectional gut–lung axis. Through this axis, distal microbial communities exert context-dependent regulatory effects on pulmonary immune responses, thereby contributing to the inflammatory and heterogeneous nature of asthma.
Future research should prioritize several interrelated areas to advance mechanistic understanding and clinical translation. First, delineating phenotype–microbiota correlations represents a critical step toward precision medicine in asthma. Longitudinal cohort studies with larger sample sizes are needed to clarify how microbial composition and function relate to inflammatory phenotypes and disease severity in both pediatric and adult populations. Second, establishing causal links between microbial metabolites and asthma pathophysiology remains a key challenge. Integrating single-cell multi-omics with targeted metabolomics will enable high-resolution mapping of host–microbe interactions, identify functional microbial signatures, and uncover novel therapeutic targets. Third, standardizing interventional strategies and clinical trial protocols is essential for translating microbiota research into therapeutic applications. Microbial consortia engineering and rationally designed probiotics offer promising approaches to stably modulate host immunity, but their efficacy must be validated in well-controlled randomized clinical trials. Finally, future efforts should aim to integrate inter-kingdom microbial dynamics—including bacterial, viral, and fungal interactions—into holistic models of asthma pathogenesis. Such integrative frameworks will be crucial for developing individualized microbiota-targeted therapies that account for disease heterogeneity across phenotypes and endotypes.
Collectively, addressing these research priorities will deepen our understanding of the gut–lung axis in asthma and accelerate the development of mechanism-based, microbiota-informed strategies for disease prevention and treatment.
Author contributions
ZY: Conceptualization, Visualization, Writing – original draft. WM: Conceptualization, Funding acquisition, Supervision, Visualization, Writing – original draft, Writing – review & editing. JW: Visualization, Writing – original draft. LY: Visualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Basic Public Welfare Technology Research Projects of Zhejiang Province (No. LGF22H290002).
Acknowledgments
The figures presented in this article were created with FigDraw (accessed on 4 July 2025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arias, C., Sepúlveda, P., Castillo, R. L., and Salazar, L. A. (2023). Relationship between hypoxic and immune pathways activation in the progression of Neuroinflammation: role of HIF-1α and Th17 cells. Int. J. Mol. Sci. 24:3073. doi: 10.3390/ijms24043073
Arifuzzaman, M., Won, T. H., Li, T. T., Yano, H., Digumarthi, S., Heras, A. F., et al. (2022). Inulin fibre promotes microbiota-derived bile acids and type 2 inflammation. Nature 611, 578–584. doi: 10.1038/s41586-022-05380-y
Berni Canani, R., Di Costanzo, M., Bedogni, G., Amoroso, A., Cosenza, L., Di Scala, C., et al. (2017). Extensively hydrolyzed casein formula containing Lactobacillus rhamnosus GG reduces the occurrence of other allergic manifestations in children with cow’s milk allergy: 3-year randomized controlled trial. J. Allergy Clin. Immunol. 139, 1906–1913.e4. doi: 10.1016/j.jaci.2016.10.050
Blanco-Pérez, F., Steigerwald, H., Schülke, S., Vieths, S., Toda, M., and Scheurer, S. (2021). The dietary Fiber pectin: health benefits and potential for the treatment of allergies by modulation of gut microbiota. Curr Allergy Asthma Rep 21:43. doi: 10.1007/s11882-021-01020-z
Bu, M. L., Li, M. H., Feng, M., Wang, J. R., and Sun, L. (2023). Poly(I:C) exacerbates airway goblet cell hyperplasia and lung inflammation in HDM-exposed Balb/C mice by YAP/FOXM1 pathway. Int. Arch. Allergy Immunol. 184, 707–719. doi: 10.1159/000529109
Campbell, C., McKenney, P. T., Konstantinovsky, D., Isaeva, O. I., Schizas, M., Verter, J., et al. (2020). Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479. doi: 10.1038/s41586-020-2193-0
Chen, Y. S., Jan, R. L., Lin, Y. L., Chen, H. H., and Wang, J. Y. (2010). Randomized placebo-controlled trial of lactobacillus on asthmatic children with allergic rhinitis. Pediatr. Pulmonol. 45, 1111–1120. doi: 10.1002/ppul.21296
Chen, P. C., Shao, Y. T., Hsieh, M. H., Kao, H. F., Kuo, W. S., Wang, S. M., et al. (2021). Early-life EV-A71 infection augments allergen-induced airway inflammation in asthma through trained macrophage immunity. Cell. Mol. Immunol. 18, 472–483. doi: 10.1038/s41423-020-00621-4
Chen, Z., Xu, Q., Liu, Y., Wei, Y., He, S., Lin, W., et al. (2022). Vancomycin-induced gut microbiota dysbiosis aggravates allergic rhinitis in mice by altered short-chain fatty acids. Front. Microbiol. 13:1002084. doi: 10.3389/fmicb.2022.1002084
Chen, H., Zhou, X., Liu, T., Liu, J., Wu, D., Xu, X., et al. (2025). Postprandial parasympathetic signals promote lung type 2 immunity. Neuron 113, 670–683.e7. doi: 10.1016/j.neuron.2024.12.020
Cheung, M. K., Leung, T. F., Tam, W. H., Leung, A. S. Y., Chan, O. M., Ng, R. W. Y., et al. (2023). Development of the early-life gut microbiome and associations with eczema in a prospective Chinese cohort. mSystems 8:e0052123. doi: 10.1128/msystems.00521-23
Daniel, C., Leppkes, M., Muñoz, L. E., Schley, G., Schett, G., and Herrmann, M. (2019). Extracellular DNA traps in inflammation, injury and healing. Nat. Rev. Nephrol. 15, 559–575. doi: 10.1038/s41581-019-0163-2
DeVries, A., McCauley, K., Fadrosh, D., Fujimura, K. E., Stern, D. A., Lynch, S. V., et al. (2022). Maternal prenatal immunity, neonatal trained immunity, and early airway microbiota shape childhood asthma development. Allergy 77, 3617–3628. doi: 10.1111/all.15442
Donald, K., and Finlay, B. B. (2024). Mechanisms of microbe-mediated immune development in the context of antibiotics and asthma. Front. Allergy 5:1469426. doi: 10.3389/falgy.2024.1469426
Drago, L., Cioffi, L., Giuliano, M., Pane, M., Amoruso, A., Schiavetti, I., et al. (2022). The probiotics in pediatric asthma management (PROPAM) Study in the primary care setting: a randomized, controlled, double-blind trial with Ligilactobacillus salivarius LS01 (DSM 22775) and Bifidobacterium breve B632 (DSM 24706). J Immunol Res 2022, 1–7. doi: 10.1155/2022/3837418
Druszczynska, M., Sadowska, B., Kulesza, J., Gąsienica-Gliwa, N., Kulesza, E., and Fol, M. (2024). The intriguing connection between the gut and lung microbiomes. Pathogens 13:1005. doi: 10.3390/pathogens13111005
Durack, J., Christian, L. S., Nariya, S., Gonzalez, J., Bhakta, N. R., Ansel, K. M., et al. (2020). Distinct associations of sputum and oral microbiota with atopic, immunologic, and clinical features in mild asthma. J. Allergy Clin. Immunol. 146, 1016–1026. doi: 10.1016/j.jaci.2020.03.028
Enaud, R., Prevel, R., Ciarlo, E., Beaufils, F., Wieërs, G., Guery, B., et al. (2020). The gut-lung Axis in health and respiratory diseases: a place for inter-organ and inter-kingdom Crosstalks. Front. Cell. Infect. Microbiol. 10:9. doi: 10.3389/fcimb.2020.00009
Fan, D., Hu, J., and Lin, N. (2025). Effects of probiotics, prebiotics, synbiotics and postbiotics on pediatric asthma: a systematic review. Front. Nutr. 12:1586129. doi: 10.3389/fnut.2025.1586129
Fujimura, K. E., Sitarik, A. R., Havstad, S., Lin, D. L., Levan, S., Fadrosh, D., et al. (2016). Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191. doi: 10.1038/nm.4176
Garcia-Castillo, V., Tomokiyo, M., Raya Tonetti, F., Islam, M. A., Takahashi, H., Kitazawa, H., et al. (2020). Alveolar macrophages are key players in the modulation of the respiratory antiviral immunity induced by orally administered Lacticaseibacillus rhamnosus CRL 1505. Front. Immunol. 11:568636. doi: 10.3389/fimmu.2020.568636
Ge, Y., Tang, G., Fu, Y., Deng, P., and Yao, R. (2025). The impact of environmental factors on respiratory tract microbiome and respiratory system diseases. Eur. J. Med. Res. 30:236. doi: 10.1186/s40001-025-02517-3
Glieca, S., Quarta, E., Bottari, B., Lal, V. C., Sonvico, F., and Buttini, F. (2024). The role of airways microbiota on local and systemic diseases: a rationale for probiotics delivery to the respiratory tract. Expert Opin. Drug Deliv. 21, 991–1005. doi: 10.1080/17425247.2024.2380334
Global Initiative for Asthma. Global strategy for asthma management and prevention. (2025). Available online at: http://www.ginasthma.org.
Guan, J., Yao, W., Zhang, L., Xie, H., Li, L., Wen, Y., et al. (2024). Contribution of Pseudomonas aeruginosa - mediated club cell necroptosis to the bias of type 17 inflammation and steroid insensitivity in asthma. J. Adv. Res. 75, 137–149. doi: 10.1016/j.jare.2024.10.020
Haahtela, T., Holgate, S., Pawankar, R., Akdis, C. A., Benjaponpitak, S., Caraballo, L., et al. (2013). The biodiversity hypothesis and allergic disease: world allergy organization position statement. World Allergy Organ J. 6:3. doi: 10.1186/1939-4551-6-3
Hang, S., Paik, D., Yao, L., Kim, E., Trinath, J., Lu, J., et al. (2019). Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148. doi: 10.1038/s41586-019-1785-z
Hassanzad, M., Maleki Mostashari, K., Ghaffaripour, H., Emami, H., Rahimi Limouei, S., and Velayati, A. A. (2019). Synbiotics and treatment of asthma: a double-blinded, randomized, placebo-controlled clinical trial. Galen Med. J. 8:e1350. doi: 10.31661/gmj.v8i0.1350
Headland, S. E., Dengler, H. S., Xu, D., Teng, G., Everett, C., Ratsimandresy, R. A., et al. (2022). Oncostatin M expression induced by bacterial triggers drives airway inflammatory and mucus secretion in severe asthma. Sci. Transl. Med. 14:eabf8188. doi: 10.1126/scitranslmed.abf8188
Heaney, L. G., Djukanovic, R., Woodcock, A., Walker, S., Matthews, J. G., Pavord, I. D., et al. (2016). Research in progress: Medical Research Council United Kingdom refractory asthma stratification Programme (RASP-UK). Thorax 71, 187–189. doi: 10.1136/thoraxjnl-2015-207326
Hou, K., Wu, Z. X., Chen, X. Y., Wang, J. Q., Zhang, D., Xiao, C., et al. (2022). Microbiota in health and diseases. Signal Transduct. Target. Ther. 7:135. doi: 10.1038/s41392-022-00974-4
Hu, J., Deng, F., Zhao, B., Lin, Z., Sun, Q., Yang, X., et al. (2022). Lactobacillus murinus alleviate intestinal ischemia/reperfusion injury through promoting the release of interleukin-10 from M2 macrophages via toll-like receptor 2 signaling. Microbiome 10:38. doi: 10.1186/s40168-022-01227-w
Huang, D., Xie, L., Luo, T., Lin, L., Ren, Q., Zeng, Z., et al. (2025). Effects of azithromycin on alleviating airway inflammation in asthmatic mice by regulating airway microbiota and metabolites. Microbiol. Spectr. 13, e02217–e02224. doi: 10.1128/spectrum.02217-24
Hufnagl, K., Pali-Schöll, I., Roth-Walter, F., and Jensen-Jarolim, E. (2020). Dysbiosis of the gut and lung microbiome has a role in asthma. Semin. Immunopathol. 42, 75–93. doi: 10.1007/s00281-019-00775-y
James, B. N., Oyeniran, C., Sturgill, J. L., Newton, J., Martin, R. K., Bieberich, E., et al. (2021). Ceramide in apoptosis and oxidative stress in allergic inflammation and asthma. J. Allergy Clin. Immunol. 147, 1936–1948.e9. doi: 10.1016/j.jaci.2020.10.024
Joos, G. F., De Swert, K. O., Schelfhout, V., and Pauwels, R. A. (2003). The role of neural inflammation in asthma and chronic obstructive pulmonary disease. Ann. N. Y. Acad. Sci. 992, 218–230. doi: 10.1111/j.1749-6632.2003.tb03152.x
Kandari, A., Odat, M. A., Alzaid, F., and Scott, K. P. (2024). Biotics and bacterial function: impact on gut and host health. ISME J. 18:wrae226. doi: 10.1093/ismejo/wrae226
Kang, Y., and Cai, Y. (2018). Future prospect of faecal microbiota transplantation as a potential therapy in asthma. Allergol Immunopathol (Madr) 46, 307–309. doi: 10.1016/j.aller.2017.04.008
Kim, Y. J., and Bunyavanich, S. (2025). Microbial influencers: the airway microbiome’s role in asthma. J. Clin. Invest. 135:e184316. doi: 10.1172/JCI184316
Kim, Y. G., Udayanga, K. G. S., Totsuka, N., Weinberg, J. B., Núñez, G., and Shibuya, A. (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE₂. Cell Host Microbe 15, 95–102. doi: 10.1016/j.chom.2013.12.010
Komlósi, Z. I., van de Veen, W., Kovács, N., Szűcs, G., Sokolowska, M., O'Mahony, L., et al. (2022). Cellular and molecular mechanisms of allergic asthma. Mol. Asp. Med. 85:100995. doi: 10.1016/j.mam.2021.100995
Kumari, M., and Kozyrskyj, A. L. (2017). Gut microbial metabolism defines host metabolism: an emerging perspective in obesity and allergic inflammation. Obes. Rev. 18, 18–31. doi: 10.1111/obr.12484
Lee-Sarwar, K. A., Kelly, R. S., Lasky-Su, J., Zeiger, R. S., O'Connor, G. T., Sandel, M. T., et al. (2020). Fecal short-chain fatty acids in pregnancy and offspring asthma and allergic outcomes. J Allergy Clin Immunol Pract 8, 1100–1102.e13. doi: 10.1016/j.jaip.2019.08.036
Leser, T., and Baker, A. (2024). Molecular mechanisms of Lacticaseibacillus rhamnosus, LGG probiotic function. Microorganisms 12:794. doi: 10.3390/microorganisms12040794
Levan, S. R., Stamnes, K. A., Lin, D. L., Panzer, A. R., Fukui, E., McCauley, K., et al. (2019). Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 4, 1851–1861. doi: 10.1038/s41564-019-0498-2
Lewis, G., Wang, B., Shafiei Jahani, P., Hurrell, B. P., Banie, H., Aleman Muench, G. R., et al. (2019). Dietary Fiber-induced microbial Short chain fatty acids suppress ILC2-dependent airway inflammation. Front. Immunol. 10:2051. doi: 10.3389/fimmu.2019.02051
Li, L., Fang, Z., Lee, Y. K., Zhao, J., Zhang, H., Lu, W., et al. (2020). Prophylactic effects of oral administration of Lactobacillus casei on house dust mite-induced asthma in mice. Food Funct. 11, 9272–9284. doi: 10.1039/d0fo01363c
Li, L., Fang, Z., Lee, Y. K., Zhao, J., Zhang, H., Peng, H., et al. (2022). Efficacy and safety of Lactobacillus reuteri CCFM1040 in allergic rhinitis and asthma: a randomized, placebo-controlled trial. Front. Nutr. 9:862934. doi: 10.3389/fnut.2022.862934
Li, X., Leonardi, I., Semon, A., Doron, I., Gao, I. H., Putzel, G. G., et al. (2018). Response to fungal Dysbiosis by gut-resident CX3CR1+ mononuclear phagocytes aggravates allergic airway disease. Cell Host Microbe 24, 847–856.e4. doi: 10.1016/j.chom.2018.11.003
Liu, J., Chen, F. H., Qiu, S. Q., Yang, L. T., Zhang, H. P., Liu, J. Q., et al. (2016). Probiotics enhance the effect of allergy immunotherapy on regulating antigen specific B cell activity in asthma patients. Am. J. Transl. Res. 8, 5256–5270.
Liu, Y., Dai, J., Zhou, G., Chen, R., Bai, C., and Shi, F. (2025). Innovative therapeutic strategies for asthma: the role of gut microbiome in airway immunity. J. Asthma Allergy 18, 257–267. doi: 10.2147/JAA.S504571
Liu, Y. H., Lee, Y. L., Han, C. L., Lo, Y. C., Liao, Z. A., Shih, Y. S., et al. (2025). ITIH4 alleviates OVA-induced asthma by regulating lung-gut microbiota. Mol. Med. 31:204. doi: 10.1186/s10020-025-01270-x
Looi, K., Troy, N. M., Garratt, L. W., Iosifidis, T., Bosco, A., Buckley, A. G., et al. (2016). Effect of human rhinovirus infection on airway epithelium tight junction protein disassembly and transepithelial permeability. Exp. Lung Res. 42, 380–395. doi: 10.1080/01902148.2016.1235237
Losol, P., Sokolowska, M., Hwang, Y. K., Ogulur, I., Mitamura, Y., Yazici, D., et al. (2023). Epithelial barrier theory: the role of exposome, microbiome, and barrier function in allergic diseases. Allergy Asthma Immunol. Res. 15, 705–724. doi: 10.4168/aair.2023.15.6.705
Maia, L. P., Cunha, T. M., Santos, P. S., Martins, M. M., Briza, P., Ferreira, F., et al. (2024). Exploring inflammatory asthma phenotypes: proteomic signatures in serum and induced sputum. Int. J. Mol. Sci. 25:3501. doi: 10.3390/ijms25063501
Mann, E. R., Lam, Y. K., and Uhlig, H. H. (2024). Short-chain fatty acids: linking diet, the microbiome and immunity. Nat. Rev. Immunol. 24, 577–595. doi: 10.1038/s41577-024-01014-8
Meirlaen, L., Levy, E. I., and Vandenplas, Y. (2021). Prevention and management with pro-, pre and Synbiotics in children with asthma and allergic rhinitis: a narrative review. Nutrients 13:934. doi: 10.3390/nu13030934
Meulmeester, F. L., Mailhot-Larouche, S., Celis-Preciado, C., Lemaire-Paquette, S., Ramakrishnan, S., Wechsler, M. E., et al. (2025). Inflammatory and clinical risk factors for asthma attacks (ORACLE2): a patient-level meta-analysis of control groups of 22 randomised trials. Lancet Respir. Med. 13, 505–516. doi: 10.1016/S2213-2600(25)00037-2
Miao, Q., Yu, R., Shi, F., Li, K., du, X., Gao, Y., et al. (2025). Respiratory syncytial virus infection disrupts airway epithelial barriers via IL-33/ST2/MyD88 signaling Axis. J. Med. Virol. 97:e70432. doi: 10.1002/jmv.70432
Miao, Y., Zhong, C., Bao, S., Wei, K., Wang, W., Li, N., et al. (2024). Impaired tryptophan metabolism by type 2 inflammation in epithelium worsening asthma. iScience 27:109923. doi: 10.1016/j.isci.2024.109923
Nakada, E. M., Bhakta, N. R., Korwin-Mihavics, B. R., Kumar, A., Chamberlain, N., Bruno, S. R., et al. (2019). Conjugated bile acids attenuate allergen-induced airway inflammation and hyperresponsiveness by inhibiting UPR transducers. JCI Insight 4:e98101. doi: 10.1172/jci.insight.98101
Nicola, T., Wenger, N., Xu, X., Evans, M., Qiao, L., Rezonzew, G., et al. (2024). A lactobacilli-based inhaled live biotherapeutic product attenuates pulmonary neutrophilic inflammation. Nat. Commun. 15:7113. doi: 10.1038/s41467-024-51169-0
Ojanguren, I., Quirce, S., Bobolea, I., Pérez de Llano, L., and Del Pozo, V. (2025). Phenotyping asthma exacerbations: one step further in the Management of Severe Asthma. J Investig Allergol Clin Immunol 35, 1–11. doi: 10.18176/jiaci.1043
Park, H. K., Choi, Y., Lee, D. H., Kim, S., Lee, J. M., Choi, S. W., et al. (2020). Altered gut microbiota by azithromycin attenuates airway inflammation in allergic asthma. J. Allergy Clin. Immunol. 145, 1466–1469.e8. doi: 10.1016/j.jaci.2020.01.044
Peng, X., Li, Z., Pei, Y., Zheng, S., Liu, J., Wang, J., et al. (2024). Streptococcus salivarius K12 alleviates Oral Mucositis in patients undergoing radiotherapy for malignant head and neck tumors: a randomized controlled trial. J. Clin. Oncol. 42, 1426–1435. doi: 10.1200/JCO.23.00837
Perdijk, O., Butler, A., Macowan, M., Chatzis, R., Bulanda, E., Grant, R. D., et al. (2024). Antibiotic-driven dysbiosis in early life disrupts indole-3-propionic acid production and exacerbates allergic airway inflammation in adulthood. Immunity 57, 1939–1954.e7. doi: 10.1016/j.immuni.2024.06.010
Perez-Garcia, J., Cardenas, A., Lorenzo-Diaz, F., and Pino-Yanes, M. (2025). Precision medicine for asthma treatment: unlocking the potential of the epigenome and microbiome. J. Allergy Clin. Immunol. 155, 298–315. doi: 10.1016/j.jaci.2024.06.010
Porsbjerg, C., Nieto-Fontarigo, J. J., Cerps, S., Ramu, S., Menzel, M., Hvidtfeldt, M., et al. (2022). Phenotype and severity of asthma determines bronchial epithelial immune responses to a viral mimic. Eur. Respir. J. 60:2102333. doi: 10.1183/13993003.02333-2021
Ramar, M., Wiscovitch-Russo, R., Yano, N., Singh, H., Lamere, E., Short, M., et al. (2025). Live bacteria in gut microbiome dictate asthma onset triggered by environmental particles via modulation of DNA methylation in dendritic cells. Cell Rep. 44:115684. doi: 10.1016/j.celrep.2025.115684
Sasaki, M., Suaini, N. H. A., Afghani, J., Heye, K. N., O'Mahony, L., Venter, C., et al. (2024). Systematic review of the association between short-chain fatty acids and allergic diseases. Allergy 79, 1789–1811. doi: 10.1111/all.16065
Schulthess, J., Pandey, S., Capitani, M., Rue-Albrecht, K. C., Arnold, I., Franchini, F., et al. (2019). The Short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7. doi: 10.1016/j.immuni.2018.12.018
Simpson, J. L., Daly, J., Baines, K. J., Yang, I. A., Upham, J. W., Reynolds, P. N., et al. (2016). Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur. Respir. J. 47, 792–800. doi: 10.1183/13993003.00405-2015
Skalski, J. H., Limon, J. J., Sharma, P., Gargus, M. D., Nguyen, C., Tang, J., et al. (2018). Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 14:e1007260. doi: 10.1371/journal.ppat.1007260
Slater, A. S., Hickey, R. M., and Davey, G. P. (2024). Interactions of human milk oligosaccharides with the immune system. Front. Immunol. 15:1523829. doi: 10.3389/fimmu.2024.1523829
Spacova, I., Van Beeck, W., Seys, S., Devos, F., Vanoirbeek, J., Vanderleyden, J., et al. (2020). Lactobacillus rhamnosus probiotic prevents airway function deterioration and promotes gut microbiome resilience in a murine asthma model. Gut Microbes 11, 1729–1744. doi: 10.1080/19490976.2020.1766345
Sun, M., Wu, W., Liu, Z., and Cong, Y. (2017). Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 52, 1–8. doi: 10.1007/s00535-016-1242-9
Sun, Y., Zhang, M., Ou, Z., Meng, Y., Chen, Y., Lin, R., et al. (2022). Indoor microbiome, microbial and plant metabolites, chemical compounds, and asthma symptoms in junior high school students: a multicentre association study in Malaysia. Eur. Respir. J. 60:2200260. doi: 10.1183/13993003.00260-2022
Tashiro, H., Kuwahara, Y., and Takahashi, K. (2025). Gut-lung axis in asthma and obesity: role of the gut microbiome. Front. Allergy 6:1618466. doi: 10.3389/falgy.2025.1618466
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Versi, A., Azim, A., Ivan, F. X., Abdel-Aziz, M. I., Bates, S., Riley, J., et al. (2024). A severe asthma phenotype of excessive airway Haemophilus influenzae relative abundance associated with sputum neutrophilia. Clin. Transl. Med. 14:e70007. doi: 10.1002/ctm2.70007
Versi, A., Ivan, F. X., Abdel-Aziz, M. I., Bates, S., Riley, J., Baribaud, F., et al. (2023). Haemophilus influenzae and Moraxella catarrhalis in sputum of severe asthma with inflammasome and neutrophil activation. Allergy 78, 2906–2920. doi: 10.1111/all.15776
Verstegen, R. E. M., Kostadinova, A. I., Merenciana, Z., Garssen, J., Folkerts, G., Hendriks, R. W., et al. (2021). Dietary fibers: effects, underlying mechanisms and possible role in allergic asthma management. Nutrients 13:4153. doi: 10.3390/nu13114153
Voisin, T., Bouvier, A., and Chiu, I. M. (2017). Neuro-immune interactions in allergic diseases: novel targets for therapeutics. Int. Immunol. 29, 247–261. doi: 10.1093/intimm/dxx040
Voo, P. Y., Wu, C. T., Sun, H. L., Ko, J. L., and Lue, K. H. (2022). Effect of combination treatment with Lactobacillus rhamnosus and corticosteroid in reducing airway inflammation in a mouse asthma model. J. Microbiol. Immunol. Infect. 55, 766–776. doi: 10.1016/j.jmii.2022.03.006
Wang, H., He, Y., Dang, D., Zhao, Y., Zhao, J., and Lu, W. (2024). Gut microbiota-derived tryptophan metabolites alleviate allergic asthma inflammation in ovalbumin-induced mice. Foods 13:1336. doi: 10.3390/foods13091336
Wang, Z., Li, Y., Gao, Y., Fu, Y., Lin, J., Lei, X., et al. (2023). Global, regional, and national burden of asthma and its attributable risk factors from 1990 to 2019: a systematic analysis for the global burden of disease Study 2019. Respir. Res. 24:169. doi: 10.1186/s12931-023-02475-6
Wang, W., Wang, F. R., Guo, Y., Zhang, H. B., and Jiang, F. F. (2024). Characteristics of airway microbiome co-occurrence network in patients with type 2 and non-type 2 asthma. Zhonghua Jie He He Hu Xi Za Zhi 47, 1121–1129. doi: 10.3760/cma.j.cn112147-20241015-00611
Wang, C., Yu, X., Yu, X., Xiao, H., Song, Y., Wang, X., et al. (2025). Gut flora-derived succinate exacerbates allergic airway inflammation by promoting protein succinylation. Redox Biol. 82:103623. doi: 10.1016/j.redox.2025.103623
Wiesner, D. L., Merkhofer, R. M., Ober, C., Kujoth, G. C., Niu, M., Keller, N. P., et al. (2020). Club cell TRPV4 serves as a damage sensor driving lung allergic inflammation. Cell Host Microbe 27, 614–628.e6. doi: 10.1016/j.chom.2020.02.006
Wu, Y., Wang, C. Z., Wan, J. Y., Yao, H., and Yuan, C. S. (2021). Dissecting the interplay mechanism between epigenetics and gut microbiota: health maintenance and disease prevention. Int. J. Mol. Sci. 22:6933. doi: 10.3390/ijms22136933
Yan, W., Li, X. Q., Liu, B. B., Sun, X. Y., Wu, W. Y., and Shen, N. (2024). Exploratory analysis of gut microbiota differences in patients with bronchial asthma of different inflammatory types. Zhonghua Nei Ke Za Zhi 63, 605–612. doi: 10.3760/cma.j.cn112138-20240207-00103
Yang, W., and Cong, Y. (2021). Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell. Mol. Immunol. 18, 866–877. doi: 10.1038/s41423-021-00661-4
Yao, L. (2024). Breaking boundaries: Bacteria act as architects of host T cell modulators using bile acids. Science 385:37. doi: 10.1126/science.adq2341
Yu, B., Pei, C., Peng, W., Zheng, Y., Fu, Y., Wang, X., et al. (2025). Microbiota-derived butyrate alleviates asthma via inhibiting Tfh13-mediated IgE production. Signal Transduct. Target. Ther. 10:181. doi: 10.1038/s41392-025-02263-2
Zakeri, A., and Russo, M. (2018). Dual role of toll-like receptors in human and experimental asthma models. Front. Immunol. 9:1027. doi: 10.3389/fimmu.2018.01027
Zhang, D., Li, S., Wang, N., Tan, H. Y., Zhang, Z., and Feng, Y. (2020). The Cross-talk between gut microbiota and lungs in common lung diseases. Front. Microbiol. 11:301. doi: 10.3389/fmicb.2020.00301
Zhang, M., Qin, Z., Huang, C., Liang, B., Zhang, X., and Sun, W. (2025). The gut microbiota modulates airway inflammation in allergic asthma through the gut-lung axis related immune modulation: a review. Biomol. Biomed. 25, 727–738. doi: 10.17305/bb.2024.11280
Zhou, Y., Jackson, D., Bacharier, L. B., Mauger, D., Boushey, H., Castro, M., et al. (2019). The upper-airway microbiota and loss of asthma control among asthmatic children. Nat. Commun. 10:5714. doi: 10.1038/s41467-019-13698-x
Zhou, A., Lei, Y., Tang, L., Hu, S., Yang, M., Wu, L., et al. (2021). Gut microbiota: the emerging link to lung homeostasis and disease. J. Bacteriol. 203, e00454–e00420. doi: 10.1128/JB.00454-20
Zou, X. L., Wu, J. J., Ye, H. X., Feng, D. Y., Meng, P., Yang, H. L., et al. (2021). Associations between gut microbiota and asthma Endotypes: a Cross-sectional Study in South China based on patients with newly diagnosed asthma. J. Asthma Allergy 14, 981–992. doi: 10.2147/JAA.S320088
Glossary
AHR - Airway hyperresponsiveness
Th - T helper cell
PRRs - Pattern recognition receptors
PAMPs - Pathogen-associated molecular patterns
DAMPs - Damage-associated molecular patterns
TLRs - Toll-like receptors
Treg - Regulatory T cell
ILC2s - Group 2 innate lymphoid cells
SCFAs - Short-chain fatty acids
IgA - Immunoglobulin A
IgE - Immunoglobulin E
IL - Interleukin
TSLP - Thymic stromal lymphopoietin
GMT - Gut microbiota transplantation
OSM - Oncostatin M
TUDCA - Tauroursodeoxycholic acid
TNF-α - Tumor necrosis factor-alpha
IFN-γ - Interferon-gamma
HDM - House dust mite
BALF - Bronchoalveolar lavage fluid
ACT - Asthma control test
AZM - Azithromycin
DC - Dendritic cell
FeNO - Fractional exhaled nitric oxide
FMT - Fecal microbiota transplantation
FOS - Fructooligosaccharides
GOS - Galactooligosaccharides
HDAC - Histone deacetylase
HMO - Human milk oligosaccharides
LPS - Lipopolysaccharide
Chrm4 - Muscarinic receptor M4
NETs - Neutrophil extracellular traps
RCT - Randomized controlled trial
EOS - Eosinophil counts
LGG - Ligilactobacillus murinus
AIT - Allergen-specific immunotherapy
Keywords: gut microbiota, asthma, gut-lung axis, immunomodulation, probiotics
Citation: Yang Z, Mao W, Wang J and Yin L (2025) The gut–lung axis in asthma: microbiota-driven mechanisms and therapeutic perspectives. Front. Microbiol. 16:1680521. doi: 10.3389/fmicb.2025.1680521
Edited by:
Moez Rhimi, INRAE Centre Jouy-en-Josas, FranceReviewed by:
Francesca Buttini, University of Parma, ItalyBianca Sampaio Dotto Fiuza, Federal University of Bahia, Brazil
Yen-Chu Huang, Taichung Veterans General Hospital, Taiwan
Zhang Jingjing, Qilu Aerospace Information Research Institute, China
Amar Garg, Swami Vivekanand Subharti University, India
Copyright © 2025 Yang, Mao, Wang and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Mao, bXc5ODg2LWh6QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Zihan Yang
Zihan Yang Wei Mao
Wei Mao Junyang Wang
Junyang Wang