- 1Department of Plant Pathology, Centre for Plant Protection Studies, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 2Horticultural Research Station, Tamil Nadu Agricultural University, Udagamandalam, Tamil Nadu, India
- 3Department of Medicine, Houston Methodist Research Institute, Houston, TX, United States
1 Introduction
Microbes (fungi, bacteria, and viruses) are the major cause of plant diseases and are responsible for devastating yield reductions that translate into enormous economic burdens. Global annual losses with regard to plant diseases account for $220 billion (Savary et al., 2019), posing a significant threat to global food security (Sharma et al., 2020). Various strategies have been used to address these losses. For example, traditional breeding approaches help to provide crops with durable resistance, yet it is constrained by the rapid breakdown of resistance and the limited availability of resistant genes (R genes) in the host plant. However, pathogens can overcome that resistance over time. Additionally, chemical pesticides may be used, but most pathogens gain resistance through repeated and often widespread application (Meade et al., 2021). At the molecular level, pathogens, including bacteria, fungi, and viruses, produce effector molecules, which are proteinaceous biological molecules that act as mediators of interaction with the host plant. Effector molecules are released into the apoplast or host cell, thereby helping the pathogen subvert the host's immune response (Liu et al., 2014). These molecules are critical virulence determinants, found mainly in the secretion system of bacteria, haustoria of fungi, and salivary secretions of insects that transmit diseases caused by viruses and phytoplasmas (Gonzalez et al., 2016).
Biotechnology tools have been leveraged to target effectors for plant disease management. These approaches offer specificity and provide long-term resistance to the host (Belete and Boyraz, 2019). In this paper, we highlight the potential of effector binding sites as molecular targets that can be leveraged using techniques such as CRISPR/Cas-based genome editing, RNA interference, decoy engineering, and effectoromics approaches. These approaches involve identifying genes that will accelerate resistance breeding and ultimately contributing to sustainable disease management and food security.
2 Discussion
2.1 Effectors as key components in disease development
Plant pathogenic effectors play a crucial role in the interaction between host and pathogens. These specialized molecules facilitate pathogen colonization and nutrient extraction by modulating host cellular processes (Harris et al., 2023). They modify levels of various phytohormones to promote pathogenicity and evade plant immunity (Han and Kahmann, 2019). Effectors are classified as intracellular or extracellular based on their site of localization. Intracellular effectors are released into the cytoplasm or nucleus, where they suppress plant immunity. Extracellular effectors operate outside the cell, in the apoplast, breaching the physical and chemical barriers of plant defense (De Wit, 2016). Translocated cytoplasmic effectors, primarily produced by bacteria, influence plant responses and disease symptoms (Todd et al., 2022). They achieve this by interfering with gene transcription and targeting susceptible factors, which facilitates pathogen growth. One such group of cytoplasmic effectors is the transcription activator-like effectors (TAL) from Xanthomonas, which alter plant transcription factors. TAL effectors are secreted by the type III secretion system. The RxLR effector, produced by Phytophthora, exhibits pathogenicity and suppresses host defense (Jiang et al., 2008). Some effectors hijack the host cell machinery by mimicking host cell proteins. Phytoplasmas produce effector molecules, such as SAP (secreted aster yellows witches' broom proteins), which target host transcription factors like TCPs (teosinte branched/cycloidea/proliferating cell factor) and RAD23, thereby altering host development and immunity (Janik et al., 2017).
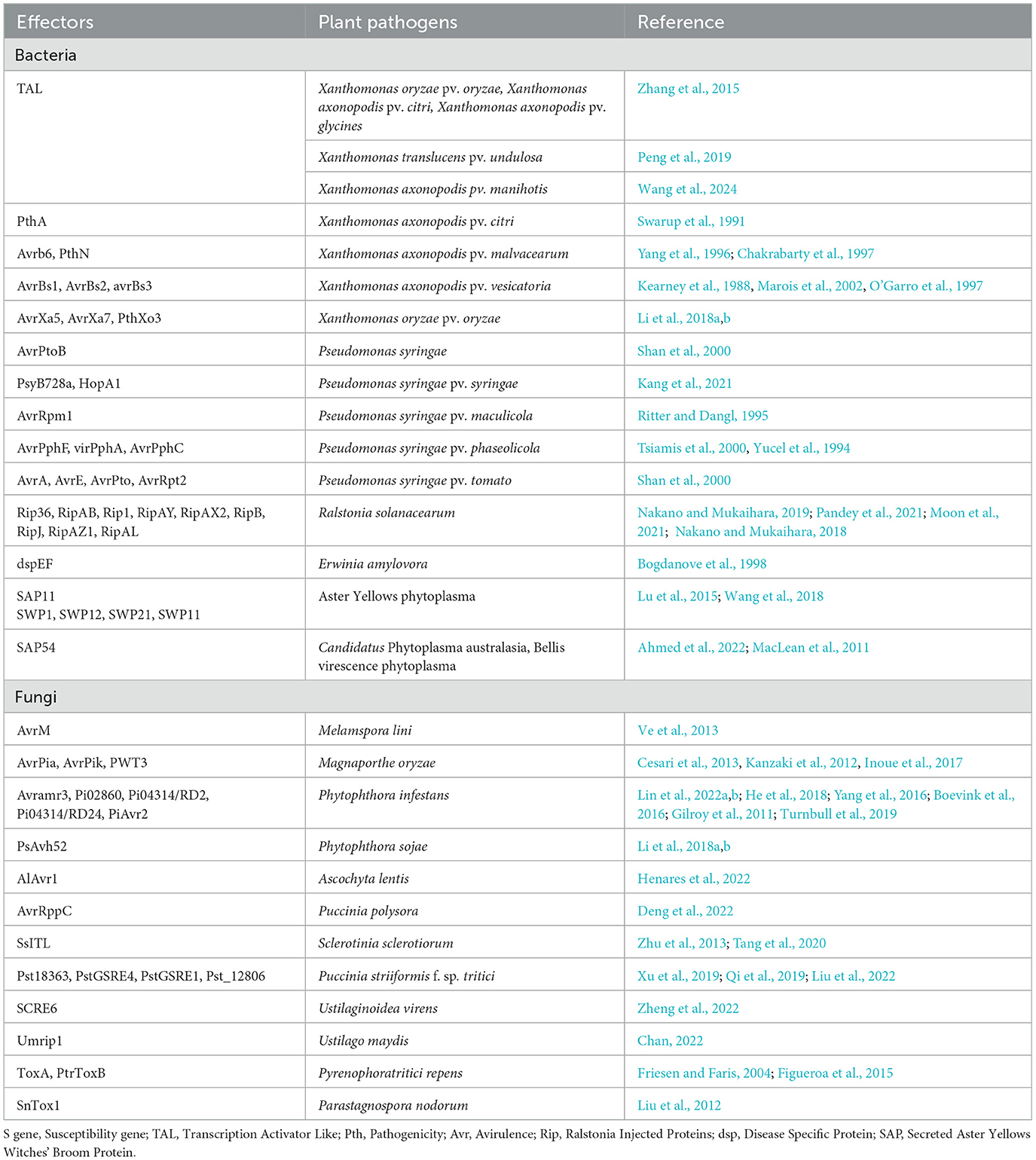
Apoplastic effectors, which are produced by fungi, insects, and nematodes, are characterized by their secretory nature. One such effector is Ecp20-2 produced by Cladosporium fulvum, (Stergiopoulos et al., 2010; Westerink et al., 2004; Van Esse et al., 2007) which inhibits the production of plant enzymes, detoxifies reactive oxygen species, and suppresses PAMP-triggered immunity (Chen et al., 2023). Table 1 provides a list of effector molecules that can be identified and targeted for innovative and improved disease management strategies.
2.2 Improving plant disease management through effector-directed interventions
Resistance achieved through conventional breeding methods can be overcome by pathogens, which generate new, more virulent strains (Shang et al., 2023). In contrast, strategies for targeting effectors for plant disease management offer several promising advantages. First, these strategies can alter pathogenicity and affect the virulence of the pathogen to some extent (Todd et al., 2022). Second, most of the effectors are conserved among multiple pathogenic strains, making them an ideal target for broad-spectrum activity (Sha and Li, 2023). For instance, Avr (avirulence) and RxLR effectors are conserved across various pathogens offering durable resistance to varied pathogens in most crops. Third, targeting site-specific effectors could reduce off-target effects on beneficial microbes in ecosystems. Finally, strategies for effector targeting are compatible with other disease management methods, which could lead to a sustainable, multi-pronged approach in the future.
2.3 Methods of targeting effector proteins
Effectors can be targeted using various biotechnological approaches, such as genome editing tools, RNA interference, effector decoy strategies, and effector breeding and diagnostics. Various -omics approaches can be used to understand the molecular level of these effectors and improve precision management, as shown in Figure 1.
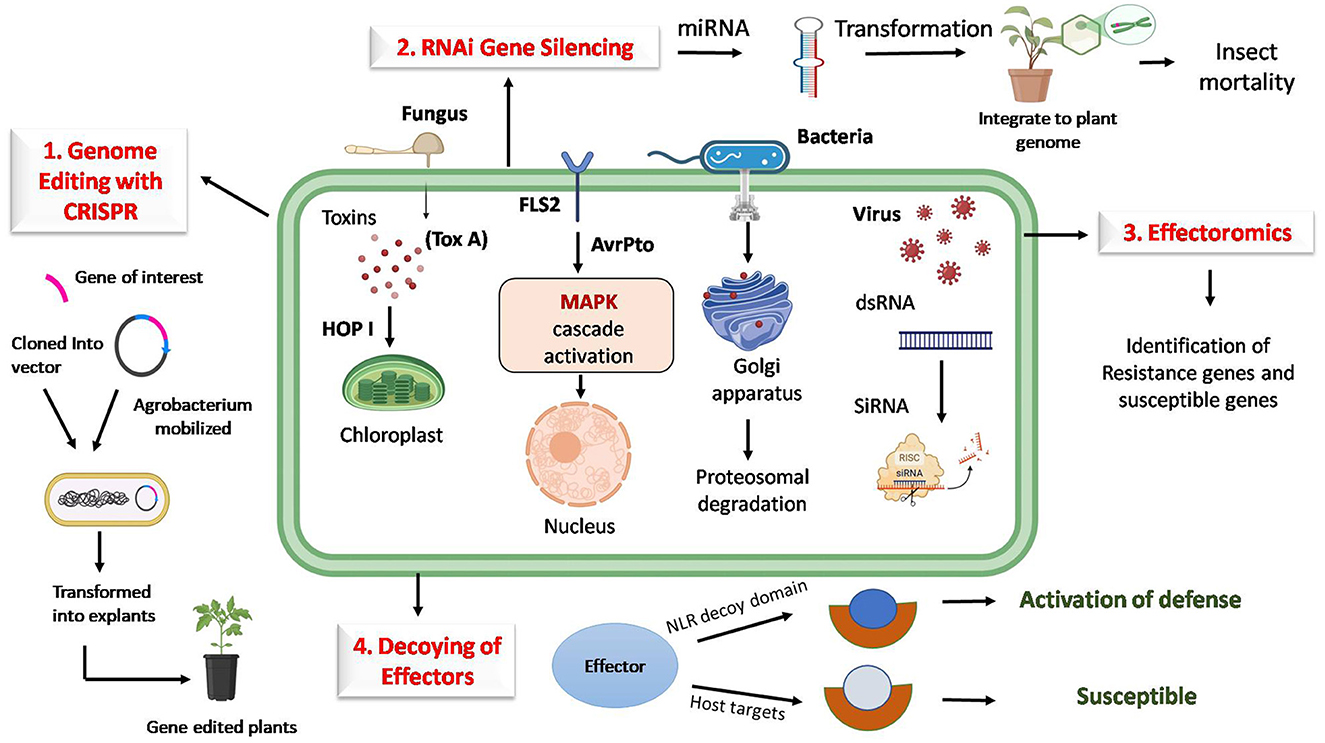
Figure 1. Strategies for targeting host factors to manipulate pathogen effectors for durable plant disease resistance. (1) Genome editing with CRISPR/Cas targets plant genes by impairing effector targets, which disrupts pathogen compatibility. (2) RNA interference (RNAi) enables host-induced gene silencing of effector genes. (3) Engineering effector decoys mimics effector targets and intercepts pathogen effectors. (4) Effectoromics-based identification of gene pathways and networks manipulated by effectors, providing precise intervention points, thereby providing resistance. Together, these approaches offer a layered defense strategy that interrupts the pathogen's effectors and provides durable resistance to host plants.
2.3.1 Genome editing with CRISPR/Cas
Genome editing offers two complementary approaches: disruption of effector binding elements (EBEs) in the promoter regions of host susceptible genes and knocking out negative regulators. By modifying EBEs through mutation, for instance, we can prevent effector binding and subsequent activation of the target site, thereby inhibiting pathogenicity, virulence, recognition, and colonization by the pathogen. For example, the SWEET (sugar will eventually be exported transporter) genes are known susceptibility genes (S genes) to which TAL effectors bind at specific EBEs in the promoters of these genes, leading to their overexpression. Sugar efflux into the apoplast provides the pathogen with nutrients, thereby enhancing infection and disease progression. CRISPR/Cas can be used to edit SWEET genes (OsSWEET11, 13, and 14) to disrupt EBEs in their promoters confers resistance against bacterial leaf blight in rice (Zhou et al., 2015). In cassava, the SWEET10a gene targets host genes that increase the resistance toward Xanthomonas axonopodis pv. manihotis (Wang et al., 2024). This prevents TAL effector-mediated activation and confers resistance to bacterial blight in elite rice cultivars (IR64, Ciherang-Sub1, and Kitaake). Disrupting TAL-EBEs blocks the pathogen-induced gene activation and enhances blight resistance without affecting plant development (Li et al., 2025). Second, the Mildew Locus O (MLO) gene family encodes membrane-associated proteins that negatively regulate plant defense responses. These genes are well-characterized susceptibility genes in both monocots and dicots, as loss-of-function mutations in MLO result in broad-spectrum resistance to powdery mildew pathogens. Using CRISPR/Cas9, targeted knockouts or frameshift mutations in MLO genes have been achieved in species such as wheat, tomato, and grapevine. This reduces or eliminates functional MLO protein activity and thereby confers resistance without significant developmental penalties (Nekrasov, 2019). In both banana and tomato plants, knocking out the DMR6 gene led to increased resistance to Xanthomonas (Tripathi et al., 2021; Thomazella et al., 2021). Similarly, the transgenic expression of the Bs2 gene from pepper detects the effectors produced by Xanthomonas, thereby providing resistance.
Although targeted genome editing can provide durable resistance, identifying S genes is challenging because they are often recessive and have multiple copies, unlike resistance genes. Identification methods are thus time-consuming and labor-intensive, often relying on wild cultivars to achieve optimal results. Furthermore, targeting S genes is known to have pleiotropic effects, including negative effects on plant growth and yield. This is undesirable for disease management in agriculture. Validating these effectors as S genes highlights the need to balance pathogen specificity with agronomic performance.
2.3.2 RNA interference and gene silencing
RNAi-mediated silencing enables the direct targeting of pathogen effector molecules either through host-induced gene silencing (HIGS) or spray-induced gene silencing (SIGS). HIGS is durable and can silence multiple effectors simultaneously, but it relies on stable transgenics, which pose regulatory challenges. On the contrary, SIGS provides a non-transgenic and eco-friendly alternative, but it depends on the stability and delivery efficiency of dsRNA. Compared to CRISPR, RNAi offers greater flexibility in targeting multiple effectors, but it lacks the long-term durability of genetic modifications, making RNAi a suitable option as an interim strategy. There are reports that RNAi is successful in silencing the effector genes of plant-parasitic nematodes, such as Meloidogyne incognita, leading to reduced infectivity (Shivakumara et al., 2016). In M. incognita, RNAi targets and suppresses genes such as msp-18, msp-20, msp-24, msp-33, and msp-16. These genes interact with host transcription factors by altering the expression of cell wall-degrading enzymes (Shivakumara et al., 2016). Putative effectors in the nematode, Pratylenchus thornei were identified, and upon introducing RNAi, they exhibited severe effects on phenotype, behavior, gene expression, and the reproductive system (Khot, 2018). Similar effects were observed using RNAi in the fungal pathogens, such as Fusarium, Verticilium, and Rhizoctonia (Foroud et al., 2014), as well as in insect vectors, including whiteflies and aphids (Feng et al., 2023). Host plants adopt a mechanism of host-induced gene silencing when they use RNAi molecules. This mechanism targets and silences specific effectors, thereby reducing the pathogen's virulence and inhibiting colonization. This reduces pest and disease incidence and provides better management strategies.
2.3.3 Decoying of effectors
Decoy engineering converts susceptible nature into resistance by providing plants with engineered proteins that mimic natural effector targets, sequestering effectors before they interact with host proteins. When the pathogens bind to the decoys, they are prevented from reaching their actual targets within the host, thereby suppressing pathogen infection. This approach is highly specific once the effector-target interaction is well-established. These decoys prevent the effectors from reaching their EBEs, a mechanism that has been well-documented in R genes, which provide host plants with broad-spectrum resistance. In the future, synthesizing such decoys could provide an opportunity to design novel resistance strategies based on specific EBEs.
2.3.4 Effectoromics
Effectoromics is a potentially powerful approach for quickly and efficiently identifying novel R genes. Pathogen effectors act as tools that identify resistance genes across germplasm collections through immune response screening (Domazakis et al., 2017). They also differentiate functional redundancy and specificity. These R genes form the basis for breeding methods that increase resistance and incorporate effector-triggered immunity into crop improvement programs. Similarly, R genes such as Rpi-amr4, Rpi-amr16, and Rpi-amr17 were identified in potatoes in response to the late blight pathogen, Phytophthora infestans effector RxLR genes Avramr4, Avramr16, and Avramr17 (Lin et al., 2022a,b). These genes act as resistance genes in the host plant and are used for effective disease management. However, this approach is data-intensive and functional validation of candidate susceptible genes remains time-consuming; it does not confer resistance, but serves as an indispensable backbone that informs and strengthens effector targeting strategies.
3 Conclusion
The major current and future challenges in agriculture on a global level are emerging plant diseases, pathogen resistance, and climate change. Hence an urgent need for innovative, cost-effective and sustainable solutions is critical. Targeting effectors is durable and eco-friendly, disabling the limitations of chemical-based management, such as emerging pathogen resistance and harm to the beneficial microbiome within the ecosystem. Targeting effectors disarms the pathogen at the molecular level, modifying the strategy toward an ecologically based approach to crop protection. Leveraging new technologies such as genome editing, RNA interference (RNAi), decoying of effectors, and effectoromics can advance plant disease management results, which face uncertainties in durability, delivery efficiency and environmental stability. An effector-based approach could be the future technology, transforming plant pathology into a science driven by prediction and precision rather than reaction. However, biosafety and ecological considerations such as unintended impacts on beneficial microbes or non-target organisms must be critically evaluated. This shift would help to secure global food security by enabling the development of disease resistant varieties. Further, to translate these approaches into practical crop improvement, it requires integration of effectoromics into breeding pipeline, their validation under field conditions, incorporating with integrated plant disease management provides a path forward, ensuring that effector targeting strategies can make a meaningful contribution to global food security.
Author contributions
GS: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MS: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. EP: Data curation, Formal analysis, Methodology, Resources, Validation, Visualization, Writing – review & editing. TA: Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – review & editing. MK: Methodology, Software, Validation, Visualization, Writing – review & editing. NT: Funding acquisition, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. We used to proofread the language.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, E. A., Farrag, A. A., Kheder, A. A., and Shaaban, A. (2022). Effect of phytoplasma associated with sesame phyllody on ultrastructural modification, physio-biochemical traits, productivity and oil quality. Plants 11:477. doi: 10.3390/plants11040477
Belete, T., and Boyraz, N. (2019). Biotechnological tools for detection, identification and management of plant diseases. Afr. J. Biotechnol. 18, 797–807. doi: 10.5897/AJB2018.16591
Boevink, P. C., Wang, X., McLellan, H., He, Q., Naqvi, S., Armstrong, M. R., et al. (2016). A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat. Commun. 7:10311. doi: 10.1038/ncomms10311
Bogdanove, A. J., Kim, J. F., Wei, Z., Kolchinsky, P., Charkowski, A. O., Conlin, A. K., et al. (1998). Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc. Nat. Acad. Sci USA. 95, 1325–1330. doi: 10.1073/pnas.95.3.1325
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Chakrabarty, P. K., Duan, Y. P., and Gabriel, D. W. (1997). Cloning and characterization of a member of the Xanthomonas avr/pth gene family that evades all commercially utilized cotton R genes in the United States. Phytopathology 87, 1160–1167. doi: 10.1094/PHYTO.1997.87.11.1160
Chan, C. (2022). The intricate dance between Ustilago effector and maize defense. Plant Cell 34, 2586–2587. doi: 10.1093/plcell/koac109
Chen, X. R., Wang, Y., Kale, S. D., Fang, Y., and Srivastava, V. (2023). Apoplastic effectors—what roles do they play in plant-pathogen interactions? Front. Microbiol. 14:1149771. doi: 10.3389/fmicb.2023.1149771
De Wit, P. J. (2016). Apoplastic fungal effectors in historic perspective; a personal view. New Phytol. 212, 805–813. doi: 10.1111/nph.14144
Deng, C., Leonard, A., Cahill, J., Lv, M., Li, Y., Thatcher, S., et al. (2022). The RppC-AvrRppC NLR-effector interaction mediates the resistance to southern corn rust in maize. Mol. Plant 15, 904–912. doi: 10.1016/j.molp.2022.01.007
Domazakis, E., Lin, X., Aguilera-Galvez, C., Wouters, D., Bijsterbosch, G., Wolters, P. J., et al. (2017). Effectoromics-Based Identification of Cell Surface Receptors in Potato. New York, NY: Humana Press, 337–353. doi: 10.1007/978-1-4939-6859-6_29
Feng, H., Chen, W., Hussain, S., Shakir, S., Tzin, V., Adegbayi, F., et al. (2023). Horizontally transferred genes as RNA interference targets for aphid and whitefly control. Plant Biotechnol. J. 21, 754–768. doi: 10.1111/pbi.13992
Figueroa, M., Manning, V. A., Pandelova, I., and Ciuffetti, L. M. (2015). Persistence of the host-selective toxin Ptr ToxB in the apoplast. Mol. Plant-Microbe Interact. 28, 1082–1090. doi: 10.1094/MPMI-05-15-0097-R
Foroud, N. A., Chatterton, S., Reid, L. M., Turkington, T. K., Tittlemier, S. A., and Gräfenhan, T. (2014). “Fusarium diseases of Canadian grain crops: impact and disease management strategies,” in Future Challenges in Crop Protection Against Fungal Pathogens, eds A. Goyal, and C. Manoharachary (New York, NY: Springer New York), 267–316. doi: 10.1007/978-1-4939-1188-2_10
Friesen, T. L., and Faris, J. D. (2004). Molecular mapping of resistance to Pyrenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. Theor. Appl. Genet. 109, 464–471. doi: 10.1007/s00122-004-1678-9
Gilroy, E. M., Breen, S., Whisson, S. C., Squires, J., Hein, I., Kaczmarek, M., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191, 763–776. doi: 10.1111/j.1469-8137.2011.03736.x
Gonzalez, C., Brito, N., and Sharon, A. (2016). “Infection process and fungal virulence factors.” in Botrytis-The Fungus, The Pathogen and its Management in Agricultural Systems, eds Y. Elad, B. Williamson, P. Tudzynski, and N. Delen (Cham, Switzerland: Springer International Publishing), 229–246. doi: 10.1007/978-3-319-23371-0_12
Han, X., and Kahmann, R. (2019). Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 10:822. doi: 10.3389/fpls.2019.00822
Harris, W. J., Kim, S., V?lz, R., and Lee, Y.-H. (2023). Nuclear effectors of plant pathogens: distinct strategies to be one step ahead. Mol. Plant Pathol. 24, 637–650. doi: 10.1111/mpp.13315
He, Q., Naqvi, S., McLellan, H., Boevink, P. C., Champouret, N., Hein, I., et al. (2018). Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proc. Natl. Acad. Sci. USA. 115, E7834–E7843. doi: 10.1073/pnas.1808585115
Henares, B. M., Debler, J. W., Farfan-Caceres, L. M., Grime, C. R., Syme, R. A., Blake, S. N., et al. (2022). The novel avirulence effector AlAvr1 from Ascochyta lentis mediates host cultivar specificity of ascochyta blight in lentil. Mol. Plant Pathol. 23, 984–996. doi: 10.1111/mpp.13203
Inoue, Y., Vy, T. T. P., Yoshida, K., Asano, H., Mitsuoka, C., Asuke, S., et al. (2017). Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 357, 80–83. doi: 10.1126/science.aam9654
Janik, K., Mithöfer, A., Raffeiner, M., Stellmach, H., Hause, B., and Schlink, K. (2017). An effector of apple proliferation phytoplasma targets TCP transcription factors-a generalized virulence strategy of phytoplasma? Mol. Plant Pathol. 18, 435–442. doi: 10.1111/mpp.12409
Jiang, R. H., Tripathy, S., Govers, F., and Tyler, B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA. 105, 4874–4879. doi: 10.1073/pnas.0709303105
Kang, H., Nguyen, Q.-M., Iswanto, A. B. B., Hong, J. C., Bhattacharjee, S., Gassmann, W., et al. (2021). Nuclear localization of HopA1Pss61 is required for effector-triggered immunity. Plants 10:888. doi: 10.3390/plants10050888
Kanzaki, H., Yoshida, K., Saitoh, H., Fujisaki, K., Hirabuchi, A., Alaux, L., et al. (2012). Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907. doi: 10.1111/j.1365-313X.2012.05110.x
Kearney, B., Ronald, P. C., Dahlbeck, D., and Staskawicz, B. J. (1988). Molecular basis for evasion of plant host defence in bacterial spot disease of pepper. Nature 332, 541–543. doi: 10.1038/332541a0
Khot, S. D. (2018). Silencing Parasitism Effectors Of the Root Lesion Nematode, Pratylenchus thornei. Murdoch University Research Repository. Available online at: https://researchrepository.murdoch.edu.au/id/eprint/41078/ (Accessed November 6, 2025).
Li, C., Liu, B., Dong, H., and Yang, B. (2025). Enhancing resistance to bacterial blight in rice using CRISPR-based base editing technology. Crop J. 13, 115–124. doi: 10.1016/j.cj.2024.09.003
Li, H., Wang, H., Jing, M., Zhu, J., Guo, B., Wang, Y., et al. (2018a). A phytophthora effector recruits a host cytoplasmic transacetylase into nuclear speckles to enhance plant susceptibility. Elife 7:e40039. doi: 10.7554/eLife.40039
Li, R., Wang, S., Sun, R., He, X., Liu, Y., and Song, C. (2018b). Xanthomonas oryzae Pv. oryzae type III effector PthXo3JXOV suppresses innate immunity, induces susceptibility and binds to multiple targets in rice. FEMS Microbiol. Lett. 365:fny037. doi: 10.1093/femsle/fny037
Lin, X., Jia, Y., Heal, R., Prokchorchik, M., Sindalovskaya, M. L., Olave-Achury, A., et al. (2022a). The Solanum americanum pangenome and effectoromics reveal new resistance genes against potato late blight. bioRxiv [preprint]. doi: 10.1101/2022.08.11.503608
Lin, X., Olave-Achury, A., Heal, R., Pais, M., Witek, K., Ahn, H.-K., et al. (2022b). A potato late blight resistance gene protects against multiple Phytophthora species by recognizing a broadly conserved RXLR-WY effector. Mol. Plant 15, 1457–1469. doi: 10.1016/j.molp.2022.07.012
Liu, C., Wang, Y., Wang, Y., Du, Y., Song, C., Song, P., et al. (2022). Glycine-serine-rich effector PstGSRE4 in Puccinia striiformis f. sp. tritici inhibits the activity of copper zinc superoxide dismutase to modulate immunity in wheat. PLoS Pathog. 18:e1010702. doi: 10.1371/journal.ppat.1010702
Liu, T., Song, T., Zhang, X., Yuan, H., Su, L., Li, W., et al. (2014). Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5:4686. doi: 10.1038/ncomms5686
Liu, Z., Zhang, Z., Faris, J. D., Oliver, R. P., Syme, R., McDonald, M. C., et al. (2012). The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 8:e1002467. doi: 10.1371/journal.ppat.1002467
Lu, Y. T., Li, M. Y., Cheng, K. T., Tan, C. M., Su, L. W., and Yang, J. Y. (2015). Phytoplasma effector SAP11 altered phosphate starvation responses and root architecture in Arabidopsis. Phytopathogen. Mollicut. 5, S125–S126. doi: 10.5958/2249-4677.2015.00054.7
MacLean, A. M., Sugio, A., Makarova, O. V., Findlay, K. C., Grieve, V. M., Tóth, R., et al. (2011). Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 157, 831–841. doi: 10.1104/pp.111.181586
Marois, E., Van den Ackerveken, G., and Bonas, U. (2002). The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant-Microbe Interact. 15, 637–646. doi: 10.1094/MPMI.2002.15.7.637
Meade, E., Slattery, M. A., and Garvey, M. (2021). Biocidal resistance in clinically relevant microbial species: a major public health risk. Pathogens 10:598. doi: 10.3390/pathogens10050598
Moon, H., Pandey, A., Yoon, H., Choi, S., Jeon, H., Prokchorchik, M., et al. (2021). Identification of RipAZ1 as an avirulence determinant of Ralstonia solanacearum in Solanum americanum. Mol. Plant Pathol. 22, 317–333. doi: 10.1111/mpp.13030
Nakano, M., and Mukaihara, T. (2018). Ralstonia solanacearum type III effector RipAL targets chloroplasts and induces jasmonic acid production to suppress salicylic acid-mediated defense responses in plants. Plant Cell Physiol. 59, 2576–2589. doi: 10.1093/pcp/pcy177
Nakano, M., and Mukaihara, T. (2019). The type III effector RipB from Ralstonia solanacearum RS1000 acts as a major avirulence factor in Nicotiana benthamiana and other nicotiana species. Mol. Plant Pathol. 20, 1237–1251. doi: 10.1111/mpp.12824
Nekrasov, V. (2019). Sequence-specific nucleases as tools for enhancing disease resistance in crops. Transgenic Res. 28, 75–80. doi: 10.1007/s11248-019-00137-2
O'Garro, L. W., Gibbs, H., and Newton, A. (1997). Mutation in the avrBs1 avirulence gene of Xanthomonas campestris pv. vesicatoria influences survival of the bacterium in soil and detached leaf tissue. Phytopathology 87, 960–966. doi: 10.1094/PHYTO.1997.87.9.960
Pandey, A., Moon, H., Choi, S., Yoon, H., Prokchorchik, M., Jayaraman, J., et al. (2021). Ralstonia solanacearum type III effector RipJ triggers bacterial wilt resistance in Solanum pimpinellifolium. Mol. Plant-Microbe Interact. 34, 962–972. doi: 10.1094/MPMI-09-20-0256-R
Peng, Z., Hu, Y., Zhang, J., Huguet-Tapia, J. C., Block, A. K., Park, S., et al. (2019). Xanthomonas translucens commandeers the host rate-limiting step in ABA biosynthesis for disease susceptibility. Proc. Natl. Acad. Sci. USA. 116, 20938–20946. doi: 10.1073/pnas.1911660116
Qi, T., Guo, J., Liu, P., He, F., Wan, C., Islam, M. A., et al. (2019). Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant 12, 1624–1638. doi: 10.1016/j.molp.2019.09.010
Ritter, C., and Dangl, J. L. (1995). The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol. Plant-Microbe Interact. 8, 444–453. doi: 10.1094/MPMI-8-0444
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Sha, G., and Li, G. (2023). Effector translocation and rational design of disease resistance. Trends Microbiol. 31, 1202–1205. doi: 10.1016/j.tim.2023.09.007
Shan, L., He, P., Zhou, J. M., and Tang, X. (2000). A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol. Plant-Microbe Interact. 13, 592–598. doi: 10.1094/MPMI.2000.13.6.592
Shang, S., Liu, G., Zhang, S., Liang, X., Zhang, R., and Sun, G. (2023). A fungal CFEM-containing effector targets NPR1 regulator NIMIN2 to suppress plant immunity. Plant Biotechnol J. 22:82–97. doi: 10.1111/pbi.14166
Sharma, P., Sharma, M. M. M., Anamika Kapoor, D., Rani, K., Singh, D., and Barkodia, M. (2020). “Role of microbes for attaining enhanced food crop production,” in Microbial Biotechnology: Basic Research and Applications eds, Singh, J., Gehlot, P., and Narula, A. (Singapore: Springer), 55–78. doi: 10.1007/978-981-15-2817-0_3
Shivakumara, T. N., Papolu, P. K., Dutta, T. K., Kamaraju, D., Chaudhary, S., and Rao, U. (2016). RNAi-induced silencing of an effector confers transcriptional oscillation in another group of effectors in the root-knot nematode, Meloidogyne incognita. Nematology 18, 857–870. doi: 10.1163/15685411-00003003
Stergiopoulos, I., Van Den Burg, H. A., Ökmen, B., Beenen, H. G., van Liere, S., Kema, G. H., et al. (2010). Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA. 107, 7610–7615. doi: 10.1073/pnas.1002910107
Swarup, S., De Feyter, R., Brlansky, R. H., and Gabriel, D. W. (1991). A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of X. campestris to elicit cankerlike lesions on citrus. Phytopathology 81, 802–809. doi: 10.1094/Phyto-81-802
Tang, L., Yang, G., Ma, M., Liu, X., Li, B., Xie, J., et al. (2020). An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 21, 686–701. doi: 10.1111/mpp.12922
Thomazella, D. P. T., Seong, K., Mackelprang, R., Dahlbeck, D., Geng, Y., Gill, U. S., et al. (2021). Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA. 118:e2026152118. doi: 10.1073/pnas.2026152118
Todd, J. N. A., Carreón-Anguiano, K. G., Islas-Flores, I., and Canto-Canché, B. (2022). Microbial effectors: key determinants in plant health and disease. Microorganisms 10:1980. doi: 10.3390/microorganisms10101980
Tripathi, J. N., Ntui, V. O., Shah, T., and Tripathi, L. (2021). CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease. Plant Biotechnol. J. 19, 1291–1293. doi: 10.1111/pbi.13614
Tsiamis, G., Mansfield, J. W., Hockenhull, R., Jackson, R. W., Sesma, A., Athanassopoulos, E., et al. (2000). Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 19, 3204–3213. doi: 10.1093/emboj/19.13.3204
Turnbull, D., Wang, H., Breen, S., Malec, M., Naqvi, S., Yang, L., et al. (2019). AVR2 targets BSL family members, which act as susceptibility factors to suppress host immunity. Plant Physiol. 180, 571–581. doi: 10.1104/pp.18.01143
Van Esse, H. P., Bolton, M. D., Stergiopoulos, I., de Wit, P. J., and Thomma, B. P. (2007). The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant-Microbe Interact. 20, 1092–1101. doi: 10.1094/MPMI-20-9-1092
Ve, T., Williams, S. J., Catanzariti, A. M., Rafiqi, M., Rahman, M., Ellis, J. G., et al. (2013). Structures of the flax-rust effector AvrM reveal insights into the molecular basis of plant-cell entry and effector-triggered immunity. Proc. Natl. Acad. Sci. USA 110, 17594–17599. doi: 10.1073/pnas.1307614110
Wang, N., Yang, H., Yin, Z., Liu, W., Sun, L., and Wu, Y. (2018). Phytoplasma effector SWP1 induces witches' broom symptom by destabilizing the TCP transcription factor BRANCHED1. Mol. Plant Pathol. 19, 2623–2634. doi: 10.1111/mpp.12733
Wang, Y., Geng, M., Pan, R., Zhang, T., Lu, X., Zhen, X., et al. (2024). Editing of the MeSWEET10a promoter yields bacterial blight resistance in cassava cultivar SC8. Mol. Plant Pathol. 25:e70010. doi: 10.1111/mpp.70010
Westerink, N., Brandwagt, B. F., De Wit, P. J., and Joosten, M. H. (2004). Cladosporium fulvum circumvents the second functional resistance gene homologue at the Cf-4 locus (Hcr9-4E) by secretion of a stable avr4E isoform. Mol. Microbiol. 54, 533–545. doi: 10.1111/j.1365-2958.2004.04288.x
Xu, Q., Tang, C., Wang, X., Sun, S., Zhao, J., Kang, Z., et al. (2019). An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 10:5571. doi: 10.1038/s41467-019-13487-6
Yang, L., McLellan, H., Naqvi, S., He, Q., Boevink, P. C., Armstrong, M., et al. (2016). Potato NPH3/RPT2-like protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiol. 171, 645–657. doi: 10.1104/pp.16.00178
Yang, Y., Yuan, Q., and Gabriel, D. W. (1996). Watersoaking function (s) of XcmH1005 are redundantly encoded by members of the Xanthomonas avr/pth gene family. Mol. Plant Microbe Interact. 9, 105–113. doi: 10.1094/MPMI-9-0105
Yucel, I., Slaymaker, D., Boyd, C., Murillo, J., Buzzell, R. I., and Keen, N. T. (1994). Avirulence gene avrPphC from Pseudomonas syringae pv. phaseolicola 3121: a plasmid-borne homologue of avrC closely linked to an avrD allele. Mol. Plant-Microbe Interact. 7, 677–679. doi: 10.1094/MPMI-7-0677
Zhang, J., Yin, Z., and White, F. (2015). TAL effectors and the executor R genes. Front. Plant Sci. 6:641. doi: 10.3389/fpls.2015.00641
Zheng, X., Fang, A., Qiu, S., Zhao, G., Wang, J., Wang, S., et al. (2022). Ustilaginoidea virens secretes a family of phosphatases that stabilize the negative immune regulator OsMPK6 and suppress plant immunity. Plant Cell 34, 3088–3109. doi: 10.1093/plcell/koac154
Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J. S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. doi: 10.1111/tpj.12838
Keywords: effectors, S genes, CRiSPR/Cas, RNAi, decoys, plant immunity, durable resistance, food security
Citation: Senthilraja G, Sandhya M, Priyadharshini E, Anand T, Kavitha M and Tharmalingam N (2025) Targeting effector proteins of plant pathogens as a strategy for durable plant disease resistance. Front. Microbiol. 16:1681047. doi: 10.3389/fmicb.2025.1681047
Received: 13 August 2025; Accepted: 27 October 2025;
Published: 14 November 2025.
Edited by:
Assefa Sintayehu, University of Gondar, EthiopiaReviewed by:
Jaindra Nath Tripathi, International Institute of Tropical Agriculture (IITA), KenyaTesfaye Alemu, Addis Ababa University, Ethiopia
Umesh Goutam, Lovely Professional University, India
Copyright © 2025 Senthilraja, Sandhya, Priyadharshini, Anand, Kavitha and Tharmalingam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Govindasamy Senthilraja, Z3NyLnBhdGhAZ21haWwuY29t; Nagendran Tharmalingam, bnRoYXJtYWxpbmdhbUBob3VzdG9ubWV0aG9kaXN0Lm9yZw==
 Govindasamy Senthilraja
Govindasamy Senthilraja Maddi Sandhya
Maddi Sandhya Eswaran Priyadharshini1
Eswaran Priyadharshini1 Nagendran Tharmalingam
Nagendran Tharmalingam