- Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (CAAS), Shanghai, China
Background: Pathogens employ a variety of effectors to modulate key host signaling pathways, thereby facilitating bacterial survival and enhancing pathogenicity. Despite lacking a complete ubiquitin system of their own, bacterial effectors frequently function as ubiquitin ligases or deubiquitinases (DUBs) to disrupt the eukaryotic ubiquitin machinery. DUBs have been found in a variety of bacteria, including ElaD, which has recently been recognized as a DUB in Escherichia coli (E. coli). However, the distribution and evolutionary analyses of ElaD in different E. coli remains largely unknown.
Methods: We retrieved and analyzed the elaD gene sequences of 530 E. coli strains. Then, molecular characterization of each strain was determined. According to all the statistical information, the distribution of elaD gene in E. coli was comprehensively investigated, and the relationship between elaD and E. coli pathotypes, serotypes, phylogenetic groups and MLSTs was analyzed. Phylogenetic tree was also constructed to analyze the evolutionary relationships between different ElaD.
Results: Our findings demonstrate that the elaD gene was present in 66.60% (353/530) of both pathogenic and nonpathogenic E. coli strains. elaD gene is predominantly found in the O157, O26, O139 and O8 serotypes. The majority of elaD-positive strains belonged to phylogenetic groups B1, A, E and D, with the predominant sequence types being ST11, ST21, ST10, ST1 and ST69. ElaD from different strains clustered in the phylogenetic tree in a correlation with O serotypes and phylogenetic groups. In addition, ElaD of some branches showed premature translation termination.
Conclusion: The widespread occurrence of the elaD gene among various E. coli strains suggests its potential significance in E. coli, although its precise functional role remains to be elucidated.
Introduction
Pathogenic Escherichia coli (E. coli) capable of causing severe diseases from gastroenteritis to extraintestinal infections according to the acquisition of a mixture of comprehensive mobile genetic elements, which encode virulence factors (Frost et al., 2005; Pakbin et al., 2021). According to the different pathogenesis and lesion location, it can be divided into intestinal pathogenic E. coli (IPEC) and extraintestinal pathogenic E. coli (ExPEC) (Russo and Johnson, 2003). Among them, IPEC includes enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), enterohaemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), diffusely adherent E. coli (DAEC), as well as a new pathotype, adherent invasive E. coli (AIEC), which mainly causes diarrhea and intestinal diseases; newborn meningitis E. coli (NMEC), uropathogenic E. coli (UPEC), avian pathogenic E. coli (APEC), which can cause human urinary tract infections, neonatal meningitis, avian respiratory tract or systemic infections, are considered ExPEC (Sora et al., 2021). E. coli are also characterized by the serotype, phylogenetic group and MLST (multilocus sequence typing) to which they belong (Riley, 2020). Some pathogenic E. coli infecting humans can cause serious morbidity and mortality worldwide (Clements et al., 2012). Animal pathogenic E. coli (such as APEC), in addition to bringing huge economic losses to farms, may also be transmitted to humans through feces, drinking water or undercooked meat, posing a potential health threat, which has a tremendous burden on public health (Hu et al., 2022; Tapader et al., 2019).
The binding of ubiquitin molecules to eukaryotic proteins helps to regulate post-translational modifications, thus influencing and participating in the majority of cellular processes such as cell cycle, apoptosis and cell signal transduction (Kerscher et al., 2006; Song and Luo, 2019). Ubiquitination is a strictly regulated and reversible process (Liu et al., 2005). On the one hand, ubiquitin-activating enzyme, ubiquitin-conjugating enzyme and ubiquitin ligase catalyze a sophisticated three-step enzymatic cascade to add one ubiquitin (Ub) or a chain of Ubs to the substrate, which helps to complete the process of post-translational modification of proteins; on the other hand, deubiquitinases (DUBs) can remove Ub from the substrate proteins (Di Gregorio et al., 2023; Fang et al., 2023; Wolberger, 2014). DUBs are involved in counterbalancing and proofreading ubiquitin processes, as well as recycling ubiquitins (Snyder and Silva, 2021). The dynamic balance between DUBs and ubiquitin ligases is essential for many aspects of cellular processes (Komander et al., 2009; Li et al., 2023).
Increasing studies highlight bacterial pathogens also encode effectors with DUB activity (Sheedlo et al., 2015). These bacterial DUBs subvert the immune response by targeting the host’s ubiquitin system, thereby contributing to bacterial survival in host cells and enhancing bacterial pathogenicity (Pruneda et al., 2016; Qiu and Luo, 2017). For example, the Salmonella T3SS effector SseL represents the first reported bacterial DUB within the CE family. It exhibits the ability to hydrolyze both K48- and K63-linked polyubiquitin chains. Studies have demonstrated that SseL enhances Salmonella intracellular survival by suppressing the NF-κB signaling pathway and inflammatory responses, cleaving polyubiquitin chains on the Salmonella-containing vacuole (SCV) to inhibit autophagy, and reducing host cell cytotoxicity (Geng et al., 2019; Mesquita et al., 2012; Rytkönen et al., 2007). Similarly, Chlamydia trachomatis effectors ChlaDUB1 and ChlaDUB2 both possess DUB activity. Beyond deubiquitinating IκBα to inhibit NF-κB activation, ChlaDUBs also stabilize the anti-apoptotic protein Mcl-1, thereby modulating host cell apoptosis (Fischer et al., 2017). Chlamydia pneumoniae encodes an OTU-family DUB, ChlaOTU, which reduces polyubiquitin accumulation by binding to the autophagy receptor NDP52 (Fischer et al., 2017; Schubert et al., 2020). Furthermore, the Legionella pneumophila effector RavD specifically cleaves M1-linked linear ubiquitin chains on the Legionella-containing vacuole (LCV) within infected macrophages. This process attenuates NF-κB-mediated inflammatory signaling and promotes intracellular bacterial survival (Wan et al., 2019). However, it is predicted that E. coli also has a DUB called ElaD. Genome comparison results showed that ElaD is only present in all IPEC, but its function is unknown (Catic et al., 2007).
Here, we comprehensively describe the distribution of elaD in E. coli, and then connect elaD with the pathotypes, serotypes, phylogenetic groups and MLSTs of E. coli. We analyze the genetic and evolutionary relationships of ElaD in different E. coli strains, and elucidate the importance of ElaD in E. coli from the perspective of genomic analysis. Our findings provide insight into the distribution of ElaD and the basis for further research.
Materials and methods
Sequence information of Escherichia coli strains
The whole-genome of all E. coli strains were accessed from the database of the National Center for Biotechnology Information,1 from which 522 strains with known pathotypes were selected. Beside 522 strains from NCBI, we included 8 E. coli strains from different clinical tissue samples collected in our laboratory. These 530 strains were subjected to genome download or whole genome sequencing for bioinformatics analysis.
Determination of elaD distribution by bioinformatics analysis
The complete genome sequences of 530 E. coli strains were used to create a local BLAST database, and the distribution rate of elaD in the genomes of the 530 strains was investigated by running BLAST. The gene was described as present if the result match three parameters simultaneously: identity>92%, query cover>90%, and E-value = 0.
Analyses of pathotypes, serotypes, phylogenetic groups and MLSTs of Escherichia coli strains
To further elucidate the relationship between elaD distribution and E. coli pathotypes, serotypes, phylogenetic groups and MLSTs, each E. coli was determined: pathotypes were identified during the download of the E. coli whole-genome file by searching literatures and NCBI website details; serotypes were identified by comparison with EcOH database (Ingle et al., 2017) using ABRicate v.0.8;2 phylogenetic groups were interfered using the Clermon Typing method in silico (Beghain et al., 2018) and sequence typing (ST) was performed with the MLST scheme of Achtman (Jolley and Maiden, 2010) using mlst v.2.11 software.3
Phylogenomic analysis of ElaD
Based on the ElaD amino acid sequences of all 353 E. coli strains, phylogeny was used for sequence analysis. After using ClustalW to match sequences under default parameters, a phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap values in MEGA 11 software. Phylogenetic tree was annotated and visualized using the Interactive Tree of Life (iTOLs) tool. Pathotypes, serotypes, phylogenetic groups and MLSTs of E. coli to which ElaD belongs were displayed alongside the phylogenetic tree.
Statistical analysis
The chi-square test was used to identify differences in the prevalence of elaD gene of E. coli strains. All analyses were performed with SPSS version 16.
Results
Distribution of elaD genes in Escherichia coli
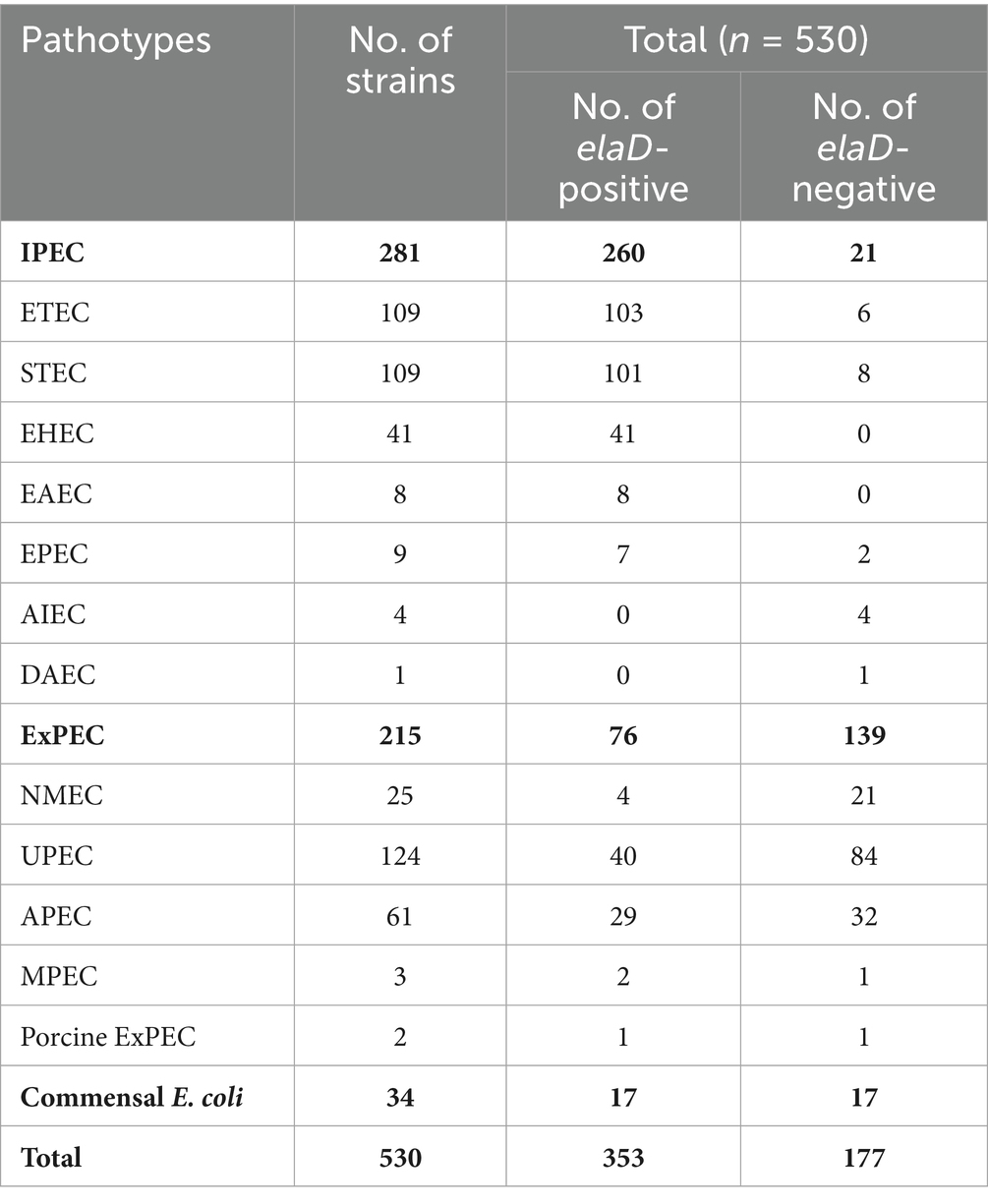
To examine the prevalence of the elaD sequence among E. coli strains, BLASTn was performed. The elaD gene was detected in 66.60% (353/530) of the E. coli strains. These elaD sequences shared >92% identity and were generally full length. Notably, further analysis of these elaD sequences revealed that 16.60% (88/530) of them matched the full-length reference gene, yet contained premature stop codons. These mutations prevent the translation of a full-length protein, suggesting that these variants are likely non-functional. Meanwhile, 2.26% (12/530) elaD sequences cannot encode full-length proteins but contain functionally relevant structural domains and could be considered functional. In addition, 33.40% (177/530) E. coli genomes did not match the elaD sequence at all (Figure 1). Therefore, ElaD is widely present in E. coli strains.
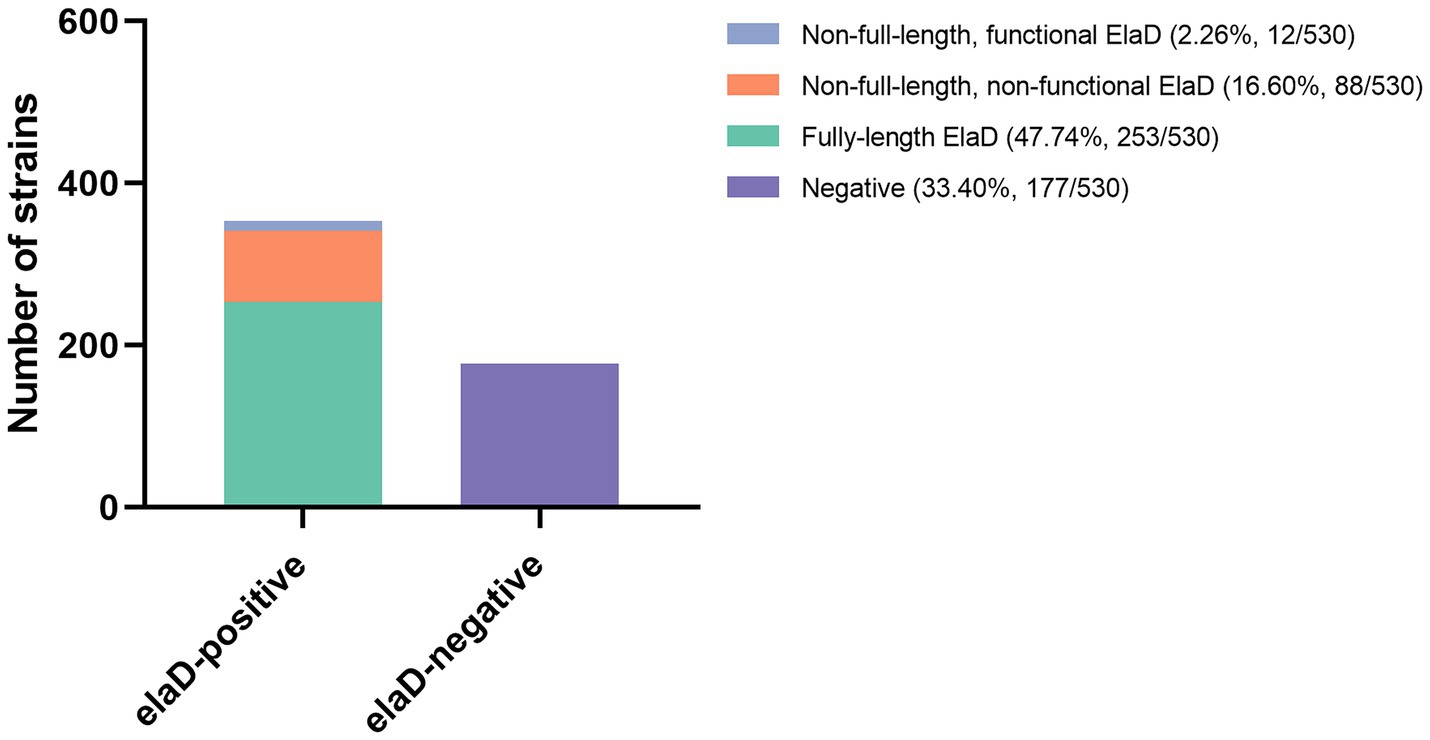
Figure 1. The distribution of elaD in E. coli. The distribution of the elaD gene in E. coli was confirmed using BLASTn, while the integrity of the protein encoded by the elaD gene was clarified by amino acid sequence analysis.
The prevalence of ElaD in various Escherichia coli pathotypes
In order to assign the E. coli strains to the different pathotypes, we summarized the details of all the strains (Table 1). As shown in Figure 2A, 13 pathotypes were identified among the 530 E. coli strains. 23.40% (124/530) of the strains were grouped as UPEC, 20.57% (109/530) as ETEC, 20.57% (109/530) as STEC, 11.51% (61/530) as APEC, 7.74% (41/530) as EHEC, 6.42% (34/530) as commensal E. coli and 4.72% (25/530) as NMEC; the remaining 27 strains (5.09%), including EPEC, EAEC, AIEC, MPEC, porcine ExPEC and DAEC, accounted for only a small percentage. Next, we determined the pathotype in the elaD-positive E. coli strains. Correspondingly, of the 353 E. coli strains positive for elaD, 29.18% (103/353) were ETEC, 28.61% (101/353) STEC, 11.61% (41/353) EHEC, 11.33% (40/353) UPEC, 8.22% (29/353) APEC, 4.82% (17/353) commensal E. coli and 6.23% others (Figure 2B).
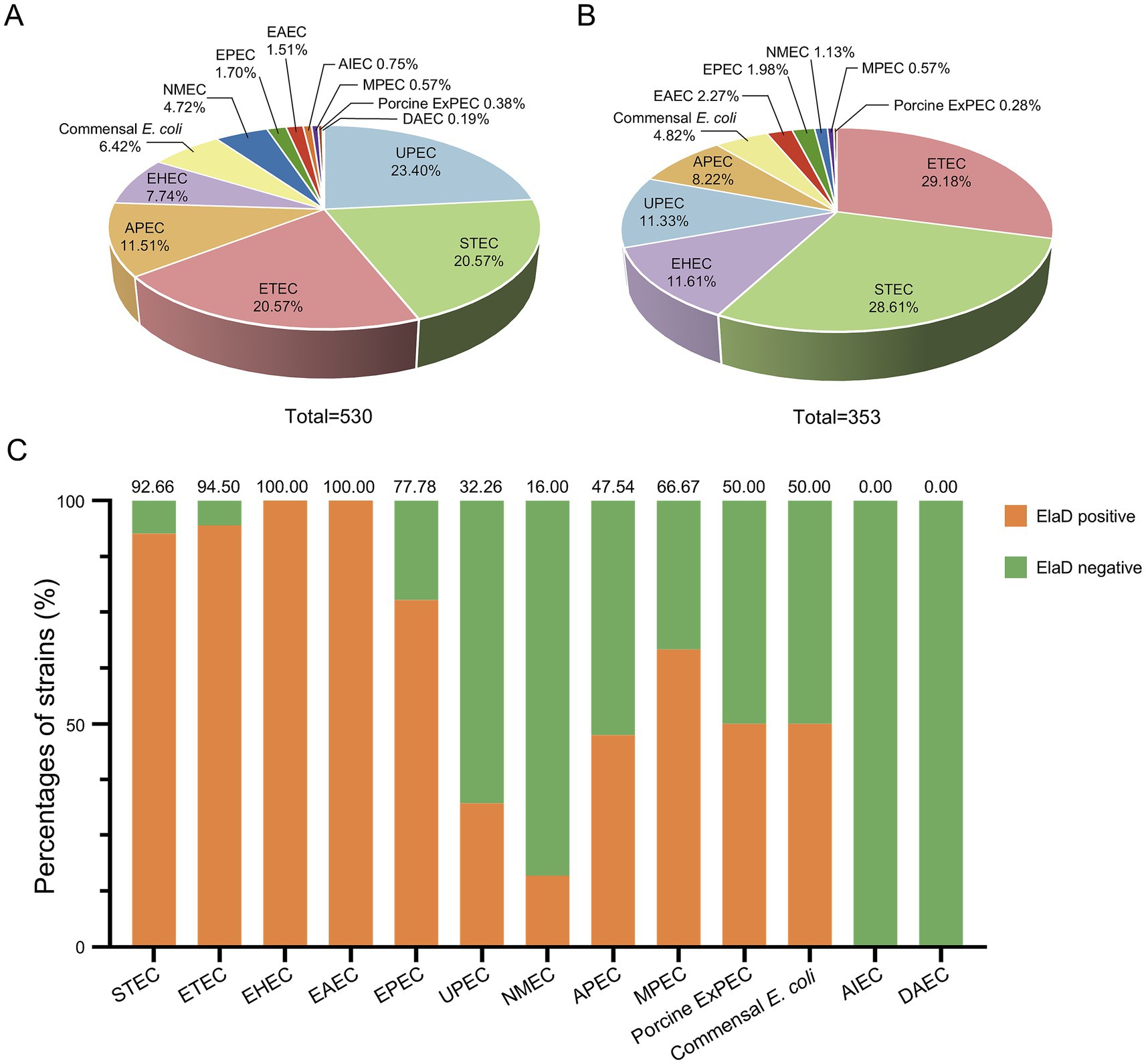
Figure 2. The prevalence of elaD in various pathotypes of E. coli. (A) The distribution of different pathotypes of all 530 E. coli. (B) The distribution of different pathotypes of 353 elaD-positive E. coli strains. (C) The elaD positivity for individual pathotypes in elaD-positive E. coli strains. The orange color represents the positive rate of elaD gene, while the green represents the negative rate.
However, the elaD was present in most of the STEC (92.66%, 101/109), ETEC (94.50%, 103/109), EHEC (100.00%, 41/41), EAEC (100.00%, 8/8) and EPEC (77.78%, 7/9). A feature clearly distinguishing ExPEC from IPEC was the lower occurrence of elaD, which was detected in only 66.67% of MPEC (2/3), 50.00% of porcine ExPEC (1/2), 32.26% of UPEC (40/124), 47.54% of APEC (29/61) and 16.00% of NMEC (4/25). Further categorical statistics have also confirmed this viewpoint, which is that elaD is distributed in 92.53% (260/281) of IPEC, but only exists in 35.35% (76/215) of ExPEC. The prevalence of elaD in IPEC strains was significantly higher than ExPEC (92.53% vs. 35.35%, p: <0.0001). But the gene was completely absent in AIEC and DAEC strains. It even presents in the 50.00% (17/34) commensal E. coli, which refers to harmless members of the normal intestinal bacterial microflora in humans and animals (Figure 2C).
The prevalence of ElaD in various O serotypes
To compare the prevalence of elaD in various O serotypes, 530 strains were characterized using ABRicate v.0.8 software. Of these, 123 O serotypes were successfully identified, with Onovel31 (7.55%, 40/530) being the most prevalent one, followed by O157 (6.98%, 37/530), O2 (5.85%, 31/530), O6 (4.72%, 25/530), O78 (3.02%, 16/530), O1 (2.83%, 15/530), O75 (2.83%, 15/530), O139 (2.64%, 14/530), O18 (2.64%, 14/530), O26 (2.26%, 12/530), O8 (2.26%, 12/530), O111 (2.08%, 11/530) and O25 (2.08%, 11/530), while 52.26% were distributed among an additional 110 O-types (Figure 3A). elaD gene is found in the O157 (10.48%, 37/353), O139 (3.97%, 14/353), O26 (3.40%, 12/353), O8 (3.40%, 12/353), O78 (2.83%, 10/353), O111 (2.83%, 10/353), O104 (2.55%, 9/353), O103 (2.27%, 8/353), O115 (1.98%, 7/353), O9 (1.98%, 7/353), O113 (1.98%, 7/353), O148 (1.98%, 7/353) and strains from the other 111 O-types (Figure 3B).
Figure 3. The prevalence of elaD in various O serotypes of E. coli. (A) The distribution of different O serotype of all 530 E. coli. (B) The distribution of different O serotype of 353 elaD-positive E. coli strains. (C) The elaD positivity for individual O serotype in elaD-positive E. coli strains. The orange color represents the positive rate of elaD gene, while the green represents the negative rate.
The distribution of elaD genes for each O serotype strains are shown in Figure 3C. The elaD positivity rate was 100.00% across all 89 O serotypes (including O8, O9, O26, O103, O104, O113, O115, O139, O148 and O157). However, elaD was rarely present in all Onovel31, O75 and O18 serotype strains detected. These results suggest that there is a correlation between E. coli elaD and O serotypes, especially the O157, O139 and O26 serotypes.
The prevalence of ElaD in various phylogenetic groups
The distribution of the 530 collected E. coli strains among the phylogenetic groups was determined using the Clermon Typing method and is shown in Figure 4A. Most of the strains could be categorized into groups B1 (31.13%, 165/530) and B2 (28.87%, 153/530), while other strains belonged to groups A (14.91%, 79/530), E (9.06%, 48/530), D (7.17%, 38/530), C (3.40%, 18/530), G (2.64%, 14/530) and F (2.26%, 12/530). 3 strains (0.57%) were not assigned to any group. A comparison of the distribution of the phylogenetic groups among elaD-positive E. coli revealed that 46.74% (165/353), 20.40% (72/353), 13.60% (48/353), 10.48% (38/353), 4.82% (17/353) and 3.40% (12/353) of the elaD-positive strains were assigned to groups B1, A, E, D, C and F, respectively (Figure 4B).
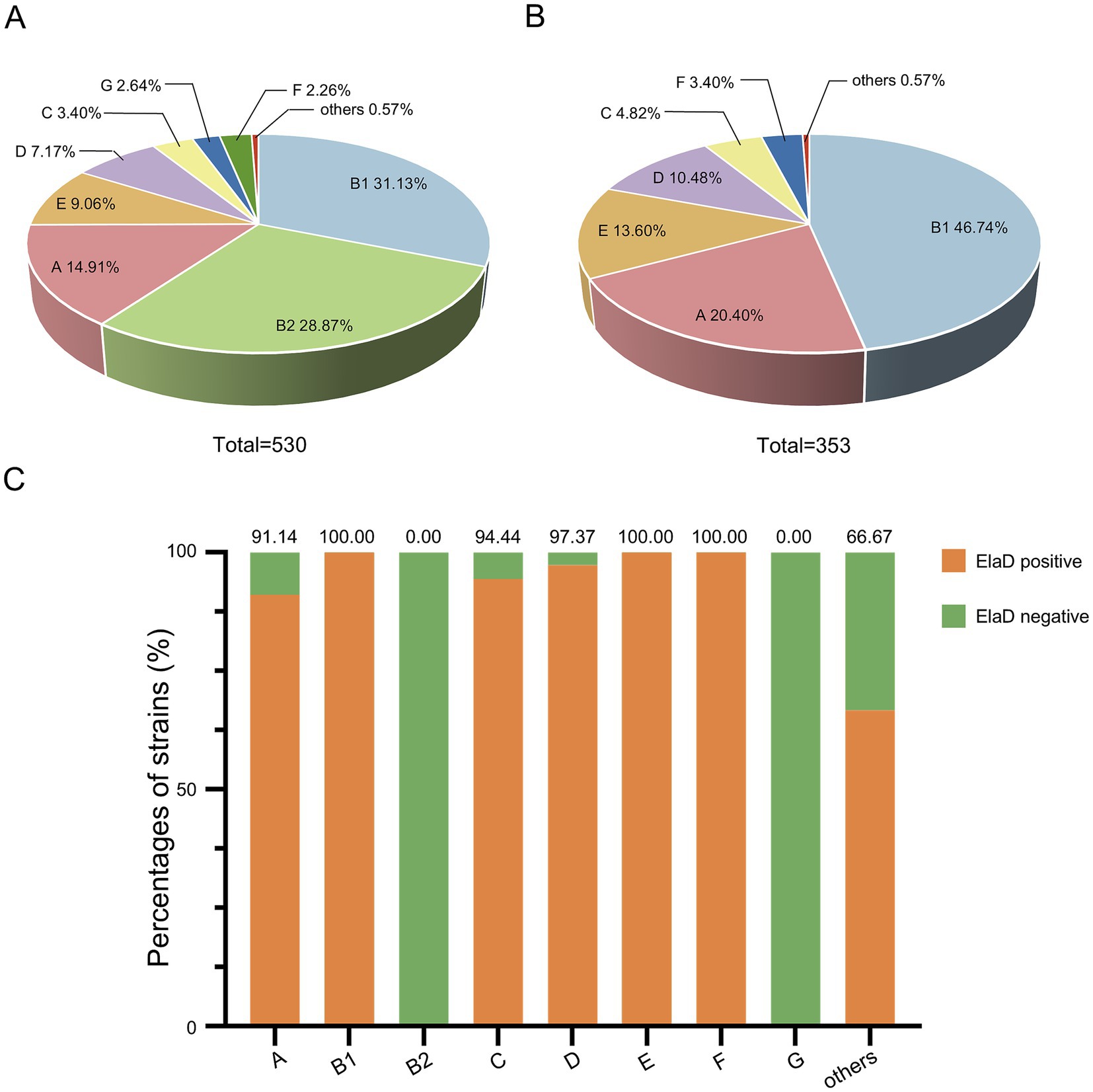
Figure 4. The prevalence of elaD in various phylogenetic groups of E. coli. (A) The distribution of different phylogenetic group of all 530 E. coli. (B) The distribution of different phylogenetic group of 353 elaD-positive E. coli strains. (C) The elaD positivity for individual phylogenetic group in elaD-positive E. coli strains. The orange color represents the positive rate of elaD gene, while the green represents the negative rate.
The analyses showed a clear correlation between the distribution of the elaD gene in E. coli and the phylogenetic group. The elaD gene was found in all B1, E and F examined in our E. coli database. Notably, our results showed that elaD was found in none of the groups B2 and G. In addition, elaD gene was present in more than 90% of the A, C and D E. coli (Figure 4C).
The prevalence of ElaD and association of MLST
The conventional seven-gene MLST analysis revealed that the strains were relatively diverse with a total of 174 STs in the 530 E. coli strains, among which ST131 comprised 7.74% (41/530) of the strains in this study, followed by ST95 (6.60%, 35/530), ST11 (6.60%, 35/530), ST10 (3.40%, 18/530), ST21 (2.64%, 14/530), ST1193 (2.64%, 14/530), ST1 (2.26%, 12/530), ST69 (2.08%, 11/530) and ST73 (1.89%, 10/530), while 64.15% were distributed among an additional 165 STs (Figure 5A). In contrast, the distribution of STs of 353 elaD-positive E. coli strains was 9.92% ST11 (35/353), 4.53% ST10 (18/353), 3.97% ST21 (14/353), 3.40% ST1 (12/353), 3.12% ST69 (11/353), 1.98% ST678 (7/353), 1.98% ST443 (7/353), 1.98% ST223 (7/353), 1.98% ST17 (7/353), 1.98% ST16 (7/353) and 65.16% others, which are almost the same STs mentioned earlier to predominate among 530 E. coli strains (Figure 5B).
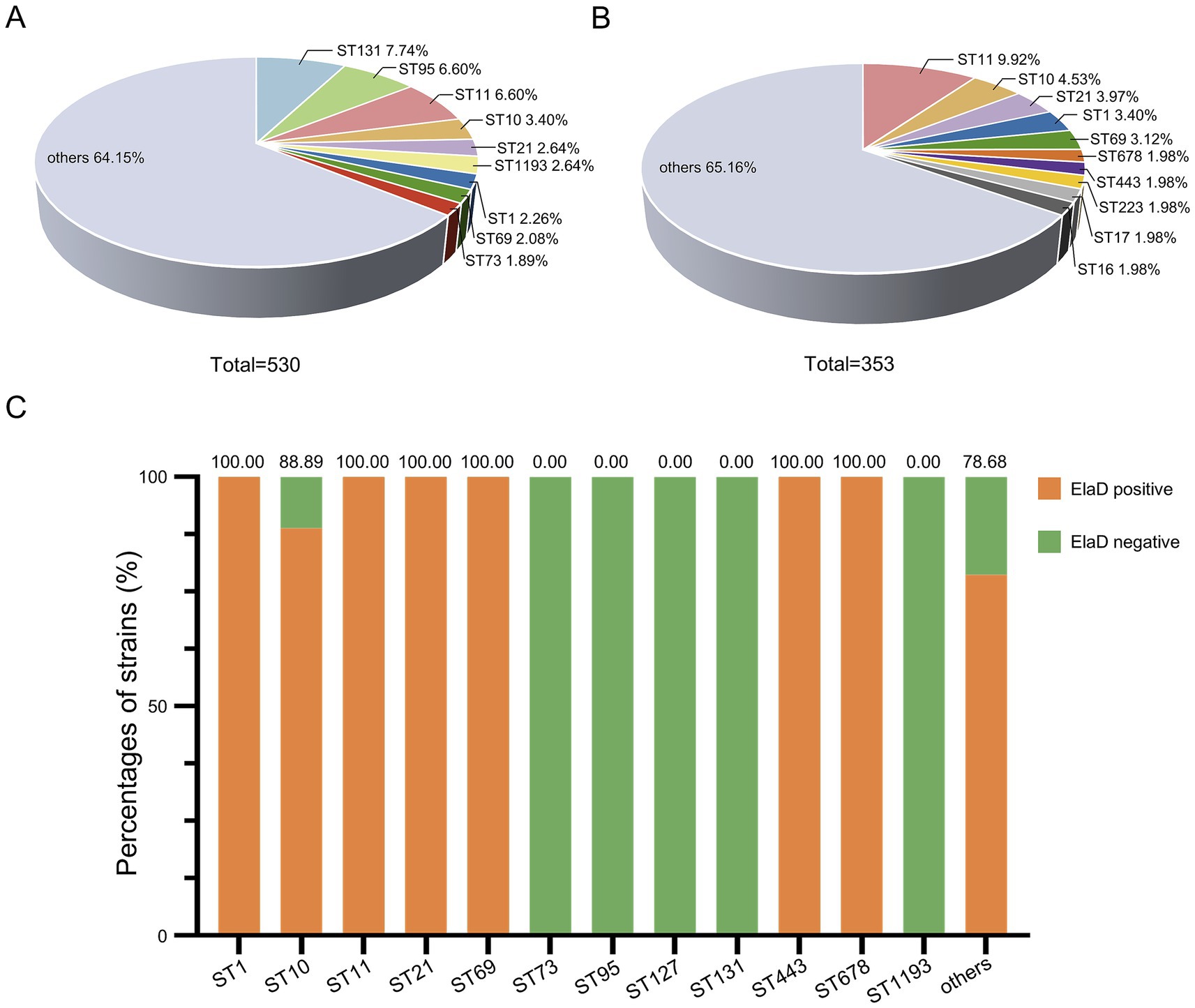
Figure 5. The prevalence of elaD in various MLSTs of E. coli. (A) The distribution of different MLSTs of all 530 E. coli. (B) The distribution of different MLSTs of 353 elaD-positive E. coli strains. (C) The elaD positivity for individual MLSTs in elaD-positive E. coli strains. The orange color represents the positive rate of elaD gene, while the green represents the negative rate.
The elaD gene was present in all tested strains of ST1, ST11, ST21, ST69, ST443 and ST678 (including most of the other STs identified), demonstrating a strong association between elaD and E. coli MLST. Surprisingly, ST73, ST95, ST127, ST131 and ST1193 E. coli appear to completely lack the elaD gene (Figure 5C).
Phylogenetic analysis of the ElaD of Escherichia coli
To further elucidate the distribution of the elaD in E. coli, we constructed a phylogenetic tree using the ElaD amino acid sequence of 353 strains. The pathotypes, serotypes, phylogenetic groups and MLSTs of the 353 strains were presented along with the phylogenetic tree and displayed with distinguishable colors and stripes.
The ElaD amino acid sequences of 353 strains were extensively distributed across the phylogenetic tree, of which 53 and 22 ElaD showed premature translation termination and were clustered in two regions, respectively. This phenomenon suggested that the elaD gene may have mutated during evolution leading to premature termination of translation. However, the remaining 13 ElaD proteins with premature translation termination were scattered in other clades. There were no major clusters of STs on the tree, except for ST11. Likewise, overlaying information on the pathotype from which these E. coli strains were derived revealed no clear relevance between placement within the phylogeny and bacterial pathotype. The O antigens, such as O157, O148, O111 and O139, corresponded with the phylogenomic results to a certain extent, however, other O antigens are interspersed in the tree. Furthermore, by analyzing the correlation between the bacterial phylogenetic group and the ElaD, we found that the ElaD from the same phylogenetic group are generally on the same clade, such as A, B1 and E (Figure 6).
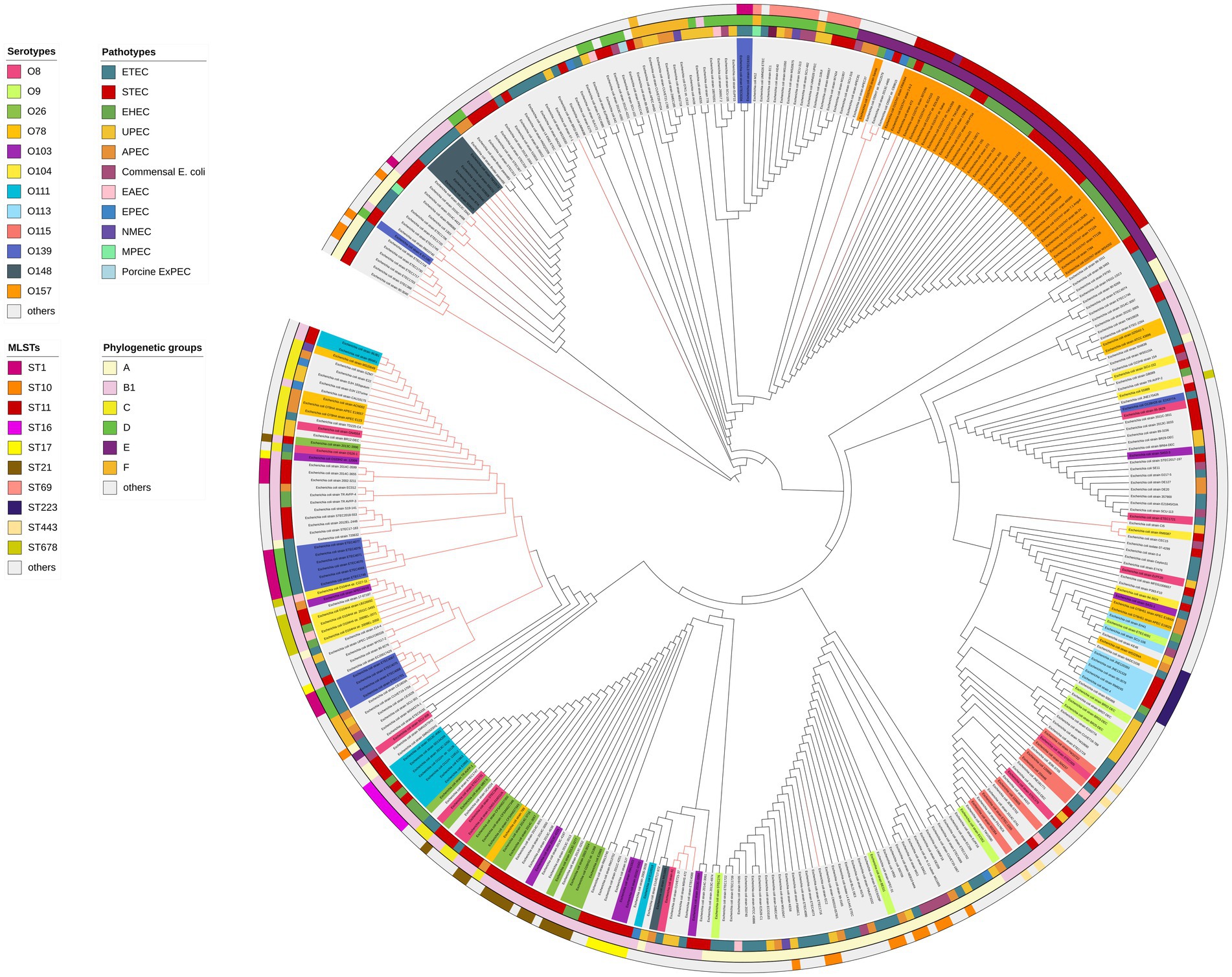
Figure 6. ElaD amino acid sequences based phylogenetic tree of 353 E. coli strains. The color range of the ring shows the names of the individual strains and their O serotypes. Then, the color strips from the innermost to the outermost represent the pathotypes, phylogenetic groups and MLSTs of E. coli strains, respectively. Diverse type is marked in different colors. In addition, the red branches represent the non-full-length amino acid sequence translated by the gene.
Discussion
Pathogenic E. coli can cause a wide range of diseases, from gastroenteritis and diarrhea to extraintestinal infections, threatening worldwide human health and the development of the livestock farming (Croxen and Finlay, 2010). During infection, the secretion system can deliver effectors to host cells that interfere with specific cellular and host immune responses (Green and Mecsas, 2016). A number of bacterial pathogens have been shown to encode and deliver effectors with DUB activity, which promote bacterial survival and enhance pathogenicity by targeting and disrupting the host ubiquitination system. Recently, ElaD has been identified as a DUB in E. coli. However, the distribution and evolution of this gene in E. coli remains unknown.
This study showed that 66.60% of the 530 E. coli strains analyzed contained the elaD gene. Meanwhile, the detection rate of elaD exceeded 70% in all five collected IPEC pathotypes—namely EHEC, STEC, ETEC, EAEC and EPEC. These pathotypes of E. coli can cause acute and persistent diarrhea in children, adults and other mammals (calves, piglets, etc.), and in severe cases, lead to hemorrhagic colitis or lethal hemolytic uremic syndrome, which is a health hazard (Bai et al., 2016; Kolenda et al., 2015; Kolodziejek et al., 2022). IPEC primarily relies on intestinal adhesion, colonization and the modulation of inflammatory responses to promote bacterial survival and replication, which ultimately contribute to disease development (Jesser and Levy, 2020). This process relates to various adhesins, toxins and effectors. The type III secretion system (T3SS) or E. coli type III secretion system 2 (ETT2) has been identified in various IPEC, which can directly deliver effectors into host cells (Gaytán et al., 2016; Wang et al., 2024). Furthermore, previous studies have compared ElaD with deubiquitinases from other bacteria in terms of evolution, structure, and function. Phylogenetic analyses indicate that E. coli ElaD within the same clade as Legionella pneumophila effector and Salmonella Typhimurium SseL, showing high sequence homology. It has been demonstrated that ElaD possesses deubiquitinating enzyme activity (Catic et al., 2007). Given that SseL has been confirmed as a T3SS effector that inhibit host inflammatory responses via its deubiquitination activity, we hypothesize that the high detection rate of elaD in IPEC might also indicate that the presence of elaD contributes to the infection process of IPEC.
A previous study showed that elaD was only present in commensal E. coli K12 and all IPEC in 16 sequenced E. coli strains. However, our results found that ExPEC also possessed elaD but its detection rate was relatively low, about 35.35%. In addition, elaD was also present in other commensal E. coli. The wide distribution of elaD in different pathogenic E. coli demonstrates its significance.
Our results revealed a high distribution of elaD among the O157, O139, O26, O8, O111 and O78 serotypes. These six serotypes are known to be closely associated with human and animal disease, with O157 considered to be the major serotype responsible for serious foodborne disease caused by STEC and EHEC infections worldwide, while the non-O157 serotypes, including O26 and O111, are equally important in causing outbreaks of intestinal infections (Mathusa et al., 2010; Wang et al., 2013). ETEC serotypes O8 and O139 are considered to be the main diarrheal pathogens affecting pigs below four weeks of age (Wyrsch et al., 2015). Meanwhile, O78 is the predominant serotype in APEC strains. The detection rate of the elaD gene was 100% for all 4 O serotypes, except for O111 (90.91%) and O78 (62.50%). The correlation between the distribution of elaD and bacterial serotypes also demonstrates the importance of ElaD.
A close relationship between phylogenetic group and virulence factors (VFs) of the pathogens was reported. EHEC, ETEC and STEC/EIEC and their specific VFs were found only in A, B1, C or E groups. In contrast, ExPEC strains and their VFs preferentially belonged to B2 and D group (Escobar-Páramo et al., 2004). Our study also demonstrated the relationship between the distribution of the elaD gene and the phylogenetic group of the pathogens. The positivity rates of elaD in E. coli was more than 90.00% in A, B1, C, D, E and F groups. Notably, in all B2 and G groups E. coli detected, the elaD gene was absent. It further suggests the notion that the presence of the elaD gene on IPEC or ExPEC is correlated with its pathogenic potential.
Furthermore, the MLST results offer a crucial reference for our analysis. ST11 represents a lineage of E. coli that is primarily associated with the O157: H7 serotype (Pugh et al., 2023). Our findings revealed that the elaD gene was present in all examined ST11 E. coli strains, all of which belonged to the O157 serotype and the E group. This is also evidenced in the phylogenetic tree.
ST131, initially recognized for its association with the carriage of extended-spectrum β-lactamase genes, has emerged as the predominant ST among global ExPEC isolates, belonging to phylogenetic group B2 (Nicolas-Chanoine et al., 2014). Notably, the elaD gene was completely absent in all examined ST131 strains, which corresponds to a 100% elaD negativity rate within the B2 group. Likewise, ST95 strains belonging to phylogroup B2 do not carry the elaD gene (Xia et al., 2022). During the long-term evolution, E. coli genome exhibits frequent alterations by increased rates of homologous recombination or horizontal gene transfer (Mageiros et al., 2021). These genes stably gained or lost, contribute to the fitness of the group B2 strains (Touchon et al., 2009). Therefore, the absence of elaD in the B2 group ExPEC may reflect the loss of adaptive genes, which may be caused by environmental pressure or niche specialization. Besides, ExPEC strains usually translocate from the gut to colonization sites, evade the host’s defense system (such as complement and phagocytosis) and establish persistent infection. ExPEC can even spread in bloodstream and cause fatal multisystemic infection (Biran et al., 2021). During this process, ExPEC employs various virulence factors, including adhesins, invasins, iron uptake factors, protectines, and toxins, which can cooperate and contribute to the pathogenic potential (Sora et al., 2021). Therefore, we hypothesize that the pathogenic process of these B2 group ExPEC may not primarily depend on ElaD. Further research is essential to comprehensively investigate the distribution and function of ElaD across E. coli.
In this study, E. coli strains showed a high prevalence of the elaD gene. Its presence correlating significantly with specific bacterial pathotypes, serotypes, phylogenetic groups, and MLSTs. Our results tentatively suggest that ElaD is strongly associated with bacterial pathogenicity, especially IPEC, but the specific regulatory mechanism by which the ElaD will act as a DUB has not yet been clarified. Further studies are still needed to evaluate the role of ElaD in the pathogenic E. coli to help us resolve the mechanism of E. coli infections and provide a reference for the prevention of E. coli disease and potential human and animal infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The manuscript presents research on bacteria isolation that does not require ethical approval.
Author contributions
XW: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. WG: Data curation, Software, Writing – review & editing. JH: Funding acquisition, Supervision, Writing – review & editing. BZ: Supervision, Writing – review & editing. JQ: Supervision, Writing – review & editing. MT: Supervision, Writing – review & editing. YB: Supervision, Writing – review & editing. LD: Supervision, Writing – review & editing. SW: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32172856, 32302881), National Key Research and Development Program of China (2021YFD1800402), the Natural Science Foundation of Shanghai (22ZR1476100, 23ZR1476600), and the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2021-SHVRI-07).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1681308/full#supplementary-material
Footnotes
References
Bai, X., Hu, B., Xu, Y., Sun, H., Zhao, A., Ba, P., et al. (2016). Molecular and phylogenetic characterization of non-O157 Shiga toxin-producing Escherichia coli strains in China. Front. Cell. Infect. Microbiol. 6:143. doi: 10.3389/fcimb.2016.00143
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4:e000192. doi: 10.1099/mgen.0.000192
Biran, D., Sura, T., Otto, A., Yair, Y., Becher, D., and Ron, E. Z. (2021). Surviving serum: the Escherichia coli iss gene of extraintestinal pathogenic E. coli is required for the synthesis of group 4 capsule. Infect. Immun. 89:e0031621. doi: 10.1128/iai.00316-21
Catic, A., Misaghi, S., Korbel, G. A., and Ploegh, H. L. (2007). ElaD, a deubiquitinating protease expressed by E. coli. PLoS One 2:e381. doi: 10.1371/journal.pone.0000381
Clements, A., Young, J. C., Constantinou, N., and Frankel, G. (2012). Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 3, 71–87. doi: 10.4161/gmic.19182
Croxen, M. A., and Finlay, B. B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38. doi: 10.1038/nrmicro2265
Di Gregorio, J., Appignani, M., and Flati, V. (2023). Role of the mitochondrial E3 ubiquitin ligases as possible therapeutic targets in cancer therapy. Int. J. Mol. Sci. 24:17176. doi: 10.3390/ijms242417176
Escobar-Páramo, P., Clermont, O., Blanc-Potard, A. B., Bui, H., Le Bouguénec, C., and Denamur, E. (2004). A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21, 1085–1094. doi: 10.1093/molbev/msh118
Fang, Y. Z., Jiang, L., He, Q., Cao, J., and Yang, B. (2023). Deubiquitination complex platform: a plausible mechanism for regulating the substrate specificity of deubiquitinating enzymes. Acta Pharm. Sin. B 13, 2955–2962. doi: 10.1016/j.apsb.2023.02.019
Fischer, A., Harrison, K. S., Ramirez, Y., Auer, D., Chowdhury, S. R., Prusty, B. K., et al. (2017). Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense. eLife 6:e21465. doi: 10.7554/eLife.21465
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Gaytán, M. O., Martínez-Santos, V. I., Soto, E., and González-Pedrajo, B. (2016). Type three secretion system in attaching and effacing pathogens. Front. Cell. Infect. Microbiol. 6:129. doi: 10.3389/fcimb.2016.00129
Geng, S., Wang, Y., Xue, Y., Wang, H., Cai, Y., Zhang, J., et al. (2019). The SseL protein inhibits the intracellular NF-κB pathway to enhance the virulence of Salmonella pullorum in a chicken model. Microb. Pathog. 129, 1–6. doi: 10.1016/j.micpath.2019.01.035
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4:19. doi: 10.1128/microbiolspec.VMBF-0012-2015
Hu, J., Afayibo, D. J. A., Zhang, B., Zhu, H., Yao, L., Guo, W., et al. (2022). Characteristics, pathogenic mechanism, zoonotic potential, drug resistance, and prevention of avian pathogenic Escherichia coli (APEC). Front. Microbiol. 13:1049391. doi: 10.3389/fmicb.2022.1049391
Ingle, D. J., Valcanis, M., Kuzevski, A., Tauschek, M., Inouye, M., Stinear, T., et al. (2017). Corrigendum: in silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 3:e000109. doi: 10.1099/mgen.0.000109
Jesser, K. J., and Levy, K. (2020). Updates on defining and detecting diarrheagenic Escherichia coli pathotypes. Curr. Opin. Infect. Dis. 33, 372–380. doi: 10.1097/qco.0000000000000665
Jolley, K. A., and Maiden, M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595
Kerscher, O., Felberbaum, R., and Hochstrasser, M. (2006). Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180. doi: 10.1146/annurev.cellbio.22.010605.093503
Kolenda, R., Burdukiewicz, M., and Schierack, P. (2015). A systematic review and meta-analysis of the epidemiology of pathogenic Escherichia coli of calves and the role of calves as reservoirs for human pathogenic E. coli. Front. Cell. Infect. Microbiol. 5:23. doi: 10.3389/fcimb.2015.00023
Kolodziejek, A. M., Minnich, S. A., and Hovde, C. J. (2022). Escherichia coli O157:H7 virulence factors and the ruminant reservoir. Curr. Opin. Infect. Dis. 35, 205–214. doi: 10.1097/qco.0000000000000834
Komander, D., Clague, M. J., and Urbé, S. (2009). Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563. doi: 10.1038/nrm2731
Li, S., Song, Y., Wang, K., Liu, G., Dong, X., Yang, F., et al. (2023). USP32 deubiquitinase: cellular functions, regulatory mechanisms, and potential as a cancer therapy target. Cell Death Discov. 9:338. doi: 10.1038/s41420-023-01629-1
Liu, Y. C., Penninger, J., and Karin, M. (2005). Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5, 941–952. doi: 10.1038/nri1731
Mageiros, L., Méric, G., Bayliss, S. C., Pensar, J., Pascoe, B., Mourkas, E., et al. (2021). Genome evolution and the emergence of pathogenicity in avian Escherichia coli. Nat. Commun. 12:765. doi: 10.1038/s41467-021-20988-w
Mathusa, E. C., Chen, Y., Enache, E., and Hontz, L. (2010). Non-O157 Shiga toxin-producing Escherichia coli in foods. J. Food Prot. 73, 1721–1736. doi: 10.4315/0362-028x-73.9.1721
Mesquita, F. S., Thomas, M., Sachse, M., Santos, A. J., Figueira, R., and Holden, D. W. (2012). The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 8:e1002743. doi: 10.1371/journal.ppat.1002743
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/cmr.00125-13
Pakbin, B., Brück, W. M., and Rossen, J. W. A. (2021). Virulence factors of enteric pathogenic Escherichia coli: a review. Int. J. Mol. Sci. 22:9922. doi: 10.3390/ijms22189922
Pruneda, J. N., Durkin, C. H., Geurink, P. P., Ovaa, H., Santhanam, B., Holden, D. W., et al. (2016). The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol. Cell 63, 261–276. doi: 10.1016/j.molcel.2016.06.015
Pugh, H. L., Connor, C., Siasat, P., McNally, A., and Blair, J. M. A. (2023). E. coli ST11 (O157:H7) does not encode a functional AcrF efflux pump. Microbiology 169:001324. doi: 10.1099/mic.0.001324
Qiu, J., and Luo, Z. Q. (2017). Hijacking of the host ubiquitin network by Legionella pneumophila. Front. Cell. Infect. Microbiol. 7:487. doi: 10.3389/fcimb.2017.00487
Riley, L. W. (2020). Distinguishing pathovars from nonpathovars: Escherichia coli. Microbiol. Spectr. 8:10.1128. doi: 10.1128/microbiolspec.AME-0014-2020
Russo, T. A., and Johnson, J. R. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5, 449–456. doi: 10.1016/s1286-4579(03)00049-2
Rytkönen, A., Poh, J., Garmendia, J., Boyle, C., Thompson, A., Liu, M., et al. (2007). Ssel, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. USA 104, 3502–3507. doi: 10.1073/pnas.0610095104
Schubert, A. F., Nguyen, J. V., Franklin, T. G., Geurink, P. P., Roberts, C. G., Sanderson, D. J., et al. (2020). Identification and characterization of diverse OTU deubiquitinases in bacteria. EMBO J. 39:e105127. doi: 10.15252/embj.2020105127
Sheedlo, M. J., Qiu, J., Tan, Y., Paul, L. N., Luo, Z. Q., and Das, C. (2015). Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc. Natl. Acad. Sci. USA 112, 15090–15095. doi: 10.1073/pnas.1514568112
Snyder, N. A., and Silva, G. M. (2021). Deubiquitinating enzymes (DUBs): regulation, homeostasis, and oxidative stress response. J. Biol. Chem. 297:101077. doi: 10.1016/j.jbc.2021.101077
Song, L., and Luo, Z. Q. (2019). Post-translational regulation of ubiquitin signaling. J. Cell Biol. 218, 1776–1786. doi: 10.1083/jcb.201902074
Sora, V. M., Meroni, G., Martino, P. A., Soggiu, A., Bonizzi, L., and Zecconi, A. (2021). Extraintestinal pathogenic Escherichia coli: virulence factors and antibiotic resistance. Pathogens 10:1355. doi: 10.3390/pathogens10111355
Tapader, R., Basu, S., and Pal, A. (2019). Secreted proteases: a new insight in the pathogenesis of extraintestinal pathogenic Escherichia coli. Int. J. Med. Microbiol. 309, 159–168. doi: 10.1016/j.ijmm.2019.03.002
Touchon, M., Hoede, C., Tenaillon, O., Barbe, V., Baeriswyl, S., Bidet, P., et al. (2009). Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. doi: 10.1371/journal.pgen.1000344
Wan, M., Wang, X., Huang, C., Xu, D., Wang, Z., Zhou, Y., et al. (2019). A bacterial effector deubiquitinase specifically hydrolyses linear ubiquitin chains to inhibit host inflammatory signalling. Nat. Microbiol. 4, 1282–1293. doi: 10.1038/s41564-019-0454-1
Wang, F., Yang, Q., Kase, J. A., Meng, J., Clotilde, L. M., Lin, A., et al. (2013). Current trends in detecting non-O157 Shiga toxin-producing Escherichia coli in food. Foodborne Pathog. Dis. 10, 665–677. doi: 10.1089/fpd.2012.1448
Wang, X., Zhu, H., Hu, J., Zhang, B., Guo, W., Wang, Z., et al. (2024). Genetic distribution, characterization, and function of Escherichia coli type III secretion system. iScience 27:109763. doi: 10.1016/j.isci.2024.109763
Wolberger, C. (2014). Mechanisms for regulating deubiquitinating enzymes. Protein Sci. 23, 344–353. doi: 10.1002/pro.2415
Wyrsch, E., Roy Chowdhury, P., Abraham, S., Santos, J., Darling, A. E., Charles, I. G., et al. (2015). Comparative genomic analysis of a multiple antimicrobial resistant enterotoxigenic E. coli O157 lineage from Australian pigs. BMC Genomics 16:165. doi: 10.1186/s12864-015-1382-y
Xia, F., Cheng, J., Jiang, M., Wang, Z., Wen, Z., Wang, M., et al. (2022). Genomics analysis to identify multiple genetic determinants that drive the global transmission of the pandemic ST95 lineage of extraintestinal pathogenic Escherichia coli (ExPEC). Pathogens 11:1489. doi: 10.3390/pathogens11121489
Keywords: Escherichia coli , deubiquitinase, ElaD, distribution, pathotype
Citation: Wang X, Guo W, Hu J, Zhang B, Qi J, Tian M, Bao Y, Deng L and Wang S (2025) The deubiquitinase ElaD is present in the majority of Escherichia coli strains. Front. Microbiol. 16:1681308. doi: 10.3389/fmicb.2025.1681308
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Wentong Cai, Chinese Academy of Agricultural Sciences, ChinaKushal Grakh, Lala Lajpat Rai University of Veterinary and Animal Sciences, India
Xutong Xue, Boston Children's Hospital and Harvard Medical School, United States
Copyright © 2025 Wang, Guo, Hu, Zhang, Qi, Tian, Bao, Deng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaohui Wang, c2h3YW5nMDgyN0AxMjYuY29t
 Xinyu Wang
Xinyu Wang Weiqi Guo
Weiqi Guo Jiangang Hu
Jiangang Hu Beibei Zhang
Beibei Zhang Jingjing Qi
Jingjing Qi Mingxing Tian
Mingxing Tian Yanqing Bao
Yanqing Bao Lei Deng
Lei Deng Shaohui Wang
Shaohui Wang