- 1Department of Epidemiology, The First Affiliated Hospital of Shandong First Medical University, Jinan, China
- 2College of Biomedical Sciences, Key Lab for Genetic Engineering and Synthetic Biology of Shandong Province, Shandong First Medical University, Shandong Academy of Medical Sciences, Jinan, China
- 3School of Municipal and Environmental Engineering, Shandong Jianzhu University, Jinan, China
Ansamitocin P-3 (AP-3), a maytansinoid antibiotic produced by Actinosynnema pretiosum, exhibits potent anticancer activity. However, its biosynthetic regulation in A. pretiosum remains largely unknown. Two-component systems (TCSs) are ubiquitous in actinomycetes and primarily regulate the biosynthesis of secondary metabolites. In this study, we identified a novel TCS, designated CrsRK, in A. pretiosum X47 through sequence analysis. Deletion of the response regulator gene crsR drastically decreased AP-3 production. RNA-seq revealed CrsR’s global regulatory role, significantly altering transcription of primary metabolic genes, especially those in purine metabolism. Crucially, the deletion of crsR also significantly downregulated transcription of the AP-3 biosynthetic genes, including asm7, asm10–15, asm21, asm23–24, asmAB, and asm43–47, which encode enzymes for multiple steps in AP-3 biosynthesis. Electrophoretic mobility shift assays confirmed direct binding of CrsR to promoters of asm21, asm43–44, and asm45–47 operons, indicating direct transcriptional control. Our results demonstrate that CrsR positively regulates AP-3 biosynthesis by directly and indirectly controlling transcription within the AP-3 biosynthetic gene cluster. In conclusion, this study elucidates the critical role of CrsR in AP-3 biosynthesis and expands our understanding of AP-3 regulatory mechanisms and TCS functions in A. pretiosum.
Introduction
Maytansinoid antibiotics are widely used as the cytotoxic “warhead” in antibody–drug conjugates (ADCs) due to their potent microtubule-depolymerizing activity (Lopus et al., 2010; Zafar et al., 2023). Ansamitocins were first isolated from Nocardia sp. C-15003 (now reclassified as Actinosynnema pretiosum), which has been established as its primary producing strain (Hasegawa et al., 1983; Higashide et al., 1977). Based on structural differences in the R substituent at the C-3 position, ansamitocins are classified into six derivatives: ansamitocin P-0, P-1, P-2, P-3′, P-3, and P-4. Among the ansamitocin derivatives, ansamitocin P-3 (AP-3) is the most abundant derivative in the fermentation yield and exhibits the highest biological activity, and its derivatives can be converted in vitro into the clinically used maytansinoid antibiotics DM1 and DM4 (Prota et al., 2014; Venghateri et al., 2013). These payloads are utilized in the FDA-approved targeted anti-tumor drugs, Kadcyla and Elahere, for treating metastatic breast cancer and platinum-resistant epithelial ovarian cancer, respectively (Modi et al., 2022; Narayan et al., 2021; Wedam et al., 2020). Therefore, microbial fermentation of AP-3 attracts considerable attention.
AP-3 biosynthesis is controlled by two genetic biosynthetic clusters (asm BGC) (Carroll et al., 2002; Yu et al., 2002). The pathway initiates with uridine diphosphate (UDP)-glucose, which is converted into 3-amino-5-hydroxybenzoic acid (AHBA) through the aminoshikimate pathway (Floss et al., 2011; Ghisalba and Nuesch, 1981; Kibby and Rickards, 1981; Yu et al., 2002). Increasing intracellular concentrations of UDP-glucose or the precursor methylmalonyl-ACP significantly enhances AP-3 production in A. pretiosum (Fan et al., 2016; Zhao et al., 2017). Subsequently, AHBA is condensed with three propionate units, three acetate units, and one glycosyl unit under the catalysis of type I polyketide synthases (PKSs) to form the proansamitocin. Following multiple post-modification steps, the proansamitocin is converted into AP-3 (Kubota et al., 2006; Li et al., 2011; Moss et al., 2002; Spiteller et al., 2003; Wu et al., 2011; Zhao et al., 2008). Furthermore, altering the fermentation medium or conditions significantly impacts AP-3 yield in A. pretiosum (Fan et al., 2014; Jia and Zhong, 2011; Lin et al., 2011).
Bacterial responses to environmental signals are generally regulated by sigma factors and two-component systems (TCSs). In TCSs, a membrane-localized kinase senses specific signals, autophosphorylates, and transfers the phosphate to a response regulator (Zschiedrich et al., 2016). The activation of the regulator influences target gene expression. TCS functions are well-studied in Actinobacteria, where multiple TCSs are involved in antibiotic biosynthesis. In Streptomyces lincolnensis, the TCS AflQ1/AflQ2 acts as a repressor of lincomycin biosynthesis, exerting its control through multiple downstream regulatory cascades (Wang et al., 2023). RspA1/RspA2 is directly involved in regulating the production of the polyether antibiotic salinomycin and primary metabolism in Streptomyces albus (Zhang et al., 2021; Zhang et al., 2020). MtrAB, a TCS in actinomycetes, plays a vital role in regulating antibiotic production. In Streptomyces coelicolor, deletion of MtrA resulted in a significant reduction in the biosynthesis of actinorhodin (ACT), undecylprodigiosin (RED), calcium-dependent antibiotic (CDA), and the yellow polyketide compound (yCPK) (Som et al., 2017b; Zhu et al., 2020). Similarly, in Streptomyces venezuelae, loss of MtrA function impaired the production of chloramphenicol (CHL) and jadomycin (JAD) (Som et al., 2017a). In S. coelicolor, single or double mutations in MacR/S largely inhibited ACT production while promoting aerial mycelium formation (Liu et al., 2021, 2019). In Streptomyces gilvosporeus F607, MacRS positively regulates natamycin biosynthesis and sporulation processes (Zong et al., 2022). DraR/K, another TCS in S. coelicolor, exhibits differential regulation, activating ACT biosynthesis while suppressing RED and yCPK production (Yu et al., 2012, 2014). Similarly, in Streptomyces bingchenggensis, AtcR/K functions as a global regulator coordinating multiple secondary metabolites (Yan et al., 2024). In the A. pretiosum X47 strain, TCS CNX_RS34865/CNX_RS34870 was found to regulate AP-3 biosynthesis, and the response regulator CNX_RS34870 positively modulates the expression of biosynthetic cluster genes and primary metabolic genes to enhance AP-3 production (Zhang K. et al., 2022). Furthermore, characterization of the PhoP/PhoR system in this strain reveals that PhoP acts as a negative regulator of morphological development, repressing the transcription of differentiation-associated genes, but does not affect AP-3 biosynthesis (Zhang P. et al., 2022).
In this study, we identified a novel TCS CrsR/CrsK in the genome of A. pretiosum X47 and generated a CrsR (response regulator) deletion mutant. Our findings demonstrate that CrsR deletion significantly impaired AP-3 biosynthesis. Furthermore, we demonstrated that CrsR directly activates the transcription of AP-3 biosynthetic genes (asm43–44, asm45, and asm21) to promote antibiotic production. These results elucidate CrsR-mediated regulation, providing a framework to improve AP-3 yields.
Materials and methods
Strains, plasmids, and culture conditions
For spore production, conjugation, and phenotype analysis, A. pretiosum X47 and derivatives were cultured at 30 °C on solid International Streptomyces Project-2 (ISP2) medium, Mannitol Soya Flour (MS) medium, and BSCA medium, respectively (Qin et al., 2017; Ma et al., 2007). The seed culture medium for A. pretiosum strains contained (w/v): glucose, 2%; soluble starch, 4%; soybean meal, 1%; polypeptone, 0.5%; NaCl, 0.3%; and CaCl₂, 0.5% (pH 7.0). The fermentation medium consisted of (w/v): maltodextrin, 3%; soluble starch, 3%; malt extract, 1%; polypeptone, 0.5%; and CaCl₂, 1% (pH 7.0). Escherichia coli DH5α (for cloning), BL21 (DE3) (for heterologous protein expression), and ET12567 (pUZ8002) (for conjugation) (Kieser et al., 2000) are cultured in Luria-Bertani (LB) or LB agar (LA) medium at 37 °C supplemented with appropriate antibiotics. The plasmids pET-15b (for in vitro expression of CrsR protein), pMD-18 T (subcloning vector for knockout or complementation plasmid construction), pJTU1278 (for gene knockout plasmid construction), pMS82 (for gene complementation plasmid construction), and pSET152 (template for the apramycin resistance cassette) (Bierman et al., 1992) were used in this study.
crsR knockout in Actinosynnema pretiosum X47
To construct the crsR deletion mutant (ΔcrsR) in A. pretiosum X47, approximately 1.5 kb flanking regions upstream and downstream of the crsR gene were amplified from X47 genomic DNA using primer pairs crsR-L-F/L-R and crsR-R-F/R-R (Supplementary Table S1). The apramycin resistance cassette (aac(3)IV) was amplified from plasmid pSET152 using primers Apra-F/Apra-R (Bierman et al., 1992). The upstream flanking region, apramycin resistance cassette, and downstream flanking region were directionally assembled and ligated into pMD18-T using the ClonExpress II One Step Cloning Kit (Vazyme). The assembled fragment was subsequently excised from pMD18-T by digestion with XbaI and HindIII and cloned into pJTU1278, generating the deletion plasmid pM-crsR. For conjugal transfer, pM-crsR was introduced into E. coli ET12567(pUZ8002). This donor strain was then conjugated with A. pretiosum X47 as described (Kieser et al., 2000). Apramycin-resistant exconjugants were selected, and successful deletion of crsR was confirmed by polymerase chain reaction (PCR) using primers crsR-V-F/R.
Genetic complementation of the crsR deletion mutant
For ΔcrsR complementation, a 2,344-bp fragment spanning the crsRK coding region and the native promoter of crsK was amplified from X47 genomic DNA using primers crsR-Com-F/R (Supplementary Table S1). The PCR product was cloned into pMD18-T and then subcloned as a HindIII fragment into pMS82 to generate plasmid pC-crsR. The resulting plasmid was transformed into E. coli ET12567 (pUZ8002), and the hygromycin B-resistant transformants were conjugated with the ΔcrsR strain. Exconjugants selected for hygromycin B were confirmed by PCR verification using primers V-F/R.
HPLC quantification of AP-3 yield
The spore suspensions of A. pretiosum X47 and derivatives were inoculated into seed medium. Cultures were incubated at 28 °C with shaking at 220 rpm for 48 h. The seed cultures were transferred to fermentation medium and incubated at 28 °C and 220 rpm for 144 h. Culture supernatants of A. pretiosum strains were extracted with ethyl acetate. AP-3 was quantified by high-performance liquid chromatography (HPLC) using a Diamonsil C18 column (250 mm × 4.6 mm), an acetonitrile–water gradient mobile phase, and detection at 254 nm (Zhong et al., 2019). The dry cell weight (DCW) of A. pretiosum strains was determined by collecting 1 mL of culture supernatant, followed by centrifugation, removal of the supernatant, and drying the mycelia at 80 °C to a constant weight. The AP-3 production and DCW data for A. pretiosum strains were derived from three independent biological replicates, and the results are presented as the mean ± standard deviation.
Total RNA isolation, RNA-Seq, and qRT-PCR assays
Spore suspensions of A. pretiosum strains were first cultivated in seed medium and then transferred to fermentation medium for an additional 48 h. Mycelia were harvested and processed simultaneously in a single batch. Total RNA was extracted from mycelia using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA integrity and total RNA quantity were assessed using an Agilent 2,100 Bioanalyzer. Transcriptome sequencing was performed using NovoGene (Beijing, China) on the Illumina NovaSeq platform with 150 bp paired-end reads. Raw reads were processed using in-house Perl scripts to remove adapter sequences, reads containing N bases, and low-quality reads to obtain clean data. The quality of the clean data was evaluated by calculating Q20, Q30 scores, and GC content. All downstream analyses were performed using high-quality clean data. The reference genome was indexed using Bowtie2 v2.2.3, and the clean reads were aligned with the reference genome using the same tool. Differential gene expression between the X47 and the ΔcrsR mutant strains was analyzed using the DESeq2 R package. Genes with an adjusted p-value of <0.05 and |log₂(Fold Change)| ≥ 1 were considered significantly differentially expressed. Subsequent bioinformatic analysis followed the standard computational pipeline.
For qRT-PCR analysis, the X47 and ΔcrsR strains were cultured in fermentation medium for 48 h. Total RNA was then extracted using TRIzol reagent (Invitrogen) and reverse-transcribed into cDNA after genomic DNA removal using the PrimeScript FAST RT Reagent Kit with gDNA Eraser (RR092A, Takara). Amplification was performed on a LightCycler 480 instrument (Roche) using TB Green® Premix Ex Taq™ (Tli RNaseH Plus) (RR420A, Takara) and gene-specific primers (Supplementary Table S1). The hrdB gene was used for normalization. Relative gene expression in the ΔcrsR mutant, presented as fold change compared to the X47 strain, was calculated from three independent biological replicates and is expressed as mean ± standard deviation.
Recombinant protein expression and purification
The crsR gene was amplified and ligated into linearized pET-15b by homologous recombination using the ClonExpress II One Step Cloning Kit (Vazyme), yielding the His₆-tagged CrsR expression plasmid. This plasmid was transformed into E. coli BL21(DE3) competent cells. His-tagged CrsR expression was induced with 0.5 mM IPTG at 28 °C for 4 h. The protein was purified using Ni-NTA Sepharose 6FF resin (Sangon Biotech). Bacterial cells were resuspended in a lysis buffer supplemented with 20 mM imidazole and lysed via ultrasonication on ice. The clarified supernatant was applied to a Ni-NTA Sepharose 6FF column (Sangon Biotech). The column was initially washed with a buffer containing 100 mM imidazole to remove weakly bound and non-specific proteins. His-tagged CrsR was subsequently eluted in a buffer containing 250 mM imidazole. The purity of the eluted protein was evaluated by SDS-PAGE. The protein was then dialyzed in a buffer containing 20 mM Tris–HCl, 50 mM NaCl, and 10% glycerol (pH 8.0) and concentrated using centrifugal filters (Amicon® Ultra). Protein concentration was determined using a Bradford Protein Assay Kit (Sangon Biotech) according to the manufacturer’s instructions. A standard curve was generated using bovine serum albumin (BSA) provided in the kit, and the absorbance was measured at 595 nm.
Electrophoretic mobility shift assays
The upstream regions of genes or operons were amplified and 5′-labeled with biotin to generate probes. A total of 50–100 fmol probes were incubated with His₆-tagged CrsR protein in a binding buffer containing poly(dI-dC) for 20 min at room temperature. A 100-fold molar excess of either an unlabeled specific probe (identical to the labeled probe) or unlabeled non-specific competitor poly(dI-dC) was used in binding assays to assess the specificity of protein–probe interactions. To validate concentration-dependent binding, the assays were conducted using two concentrations of CrsR protein (0.5 and 1 μg). Protein–DNA complexes were then resolved on 8% non-denaturing polyacrylamide gels, and the band patterns were transferred to nylon membranes and UV cross-linked. Following blocking, membranes were incubated with HRP-conjugated streptavidin (Beyotime) in blocking buffer. After two washes, biotinylated probes were detected using the ECL Western Blotting Detection System (Thermo Fisher Scientific).
Bioinformatic analysis
Sequence and conserved domain analysis of CrsR and CrsK were performed using BLAST.1 The three-dimensional structures of CrsR and CrsK were predicted by AlphaFold 3, with the putative binding sites of CrsR also being analyzed by the same platform2 (Abramson et al., 2024). The transmembrane helices of CrsK were predicted using TMHMM-2.03 (Krogh et al., 2001; Sonnhammer et al., 1998).
Results
Bioinformatic identification of the CrsRK TCS (encoded by CNX_RS21345/CNX_RS21350) in the Actinosynnema pretiosum X47 genome
Genomic sequencing and comparative analysis revealed the presence of dozens of TCSs in the genome of the A. pretiosum X47 strain, most of which lack functional characterization. To systematically identify TCSs influencing AP-3 biosynthesis, we generated knockout mutants targeting the response regulator genes of multiple TCSs. The mutation in CNX_RS21345 led to a pronounced reduction in the AP-3 titer, prompting its identification as a key candidate.
Sequence alignment analysis identified the adjacent genes CNX_RS21345 and CNX_RS21350 in the X47 genome, predicted to form an operon based on their 24-bp intergenic spacer. Bioinformatic analysis identified CNX_RS21345 and CNX_RS21350 as a TCS pair. CNX_RS21345 encodes a 217-amino acid protein belonging to the NarL family of response regulators, which comprises a phosphoacceptor receiver (REC) domain and a helix-turn-helix (HTH) DNA-binding domain (Figures 1a,b). CNX_RS21350 encodes a 429-amino acid sensor histidine kinase, featuring a C-terminal histidine kinase (HK) domain and an N-terminal sensor domain (Figures 1a,c). In this study, we designated CNX_RS21345 and CNX_RS21350 as CrsR and CrsK, respectively.
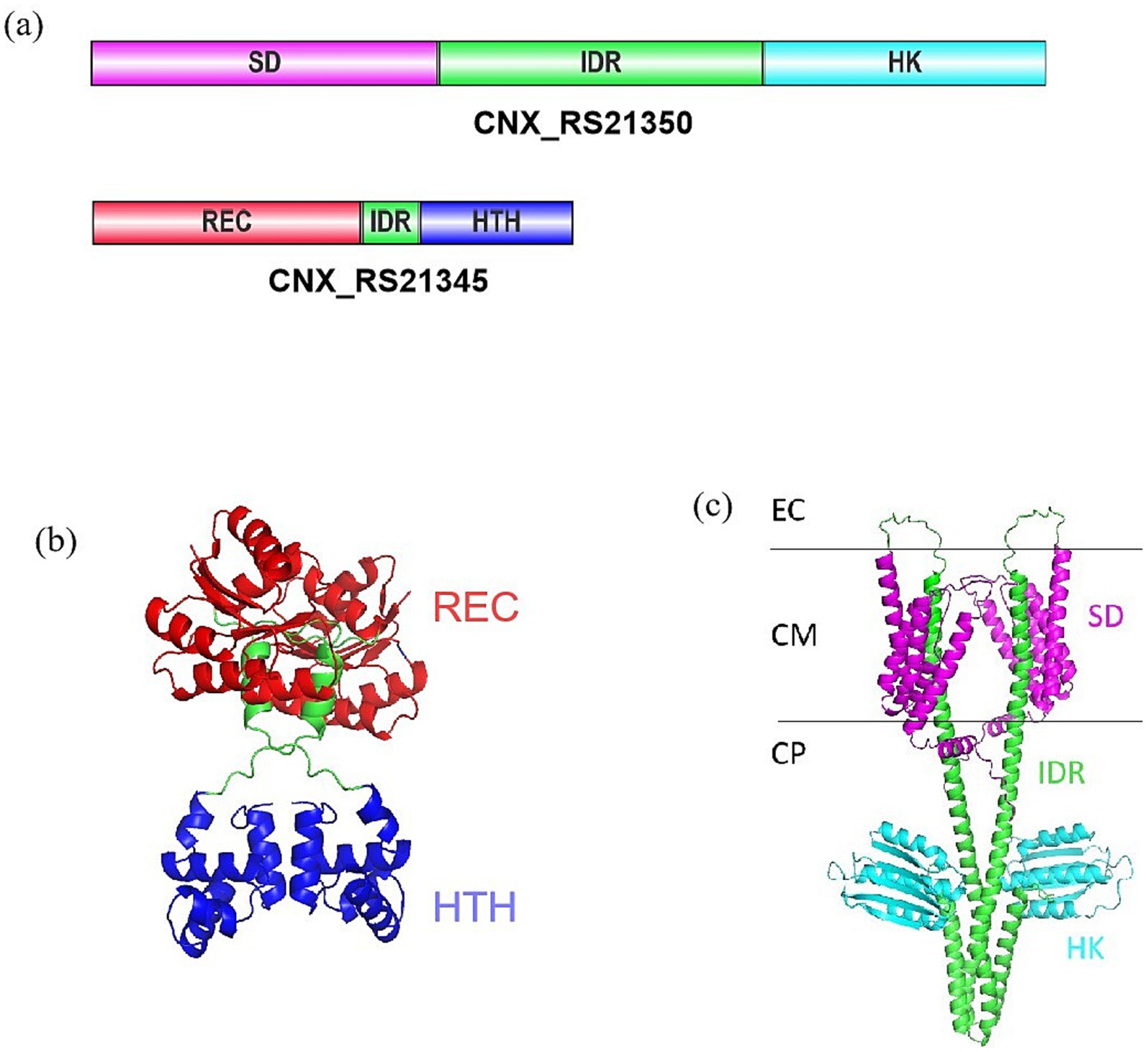
Figure 1. Protein structure prediction of CNX_RS21345 and CNX_RS21350. (a) Domain architecture of CNX_RS21345 and CNX_RS21350. (b) Computationally predicted the tertiary structure of CNX_RS21345. (c) Computationally predicted tertiary structure of CNX_RS21350. EC, extracellular; CM, cell membrane; CP, cytoplasm. Schematic representations depict conserved domains: SD, sensor domain; IDR, intrinsically disordered region; HK, histidine kinase; REC, phosphoacceptor receiver; HTH, helix-turn-helix motif.
crsR deletion led to a significant decrease in AP-3 biosynthesis in Actinosynnema pretiosum X47
To investigate the function of CrsR, we constructed a deletion mutant of the gene crsR in the X47 strain. The mutant strain was verified by PCR using primer V-F/R, which produced a 727 bp amplicon in the X47 strain and a 1,310 bp fragment in the mutant strain (Figures 2a,b). Phenotypic comparison of A. pretiosum X47, ∆crsR, and C-∆crsR grown on BSCA and ISP2 media revealed no significant morphological differences between the ∆crsR mutant and the wild-type strain (Figure 2c). The AP-3 production in both the X47 and ΔcrsR strains was analyzed. The mutant exhibited markedly lower production levels than X47 as early as 48 h (Figure 2d). At 48 h, the X47 strain produced 2.67 ± 0.87 mg/L of AP-3, while the ΔcrsR mutant yielded only 0.82 ± 0.14 mg/L. By 144 h, AP-3 titers in X47 reached 18.36 ± 2.21 mg/L, whereas the ΔcrsR strain showed a 64% reduction (6.67 ± 1.54 mg/L) relative to the wild-type strain (Supplementary Table S2). The complemented strain showed restored AP-3 production, reaching yields similar to those of the X47 strain. Under fermentation conditions, the biomass of all strains remained similar across different time points (Supplementary Figure S1), demonstrating that the deletion of the crsR gene does not impact growth, which indicates that the significant differences in AP-3 production were not caused by variations in biomass.
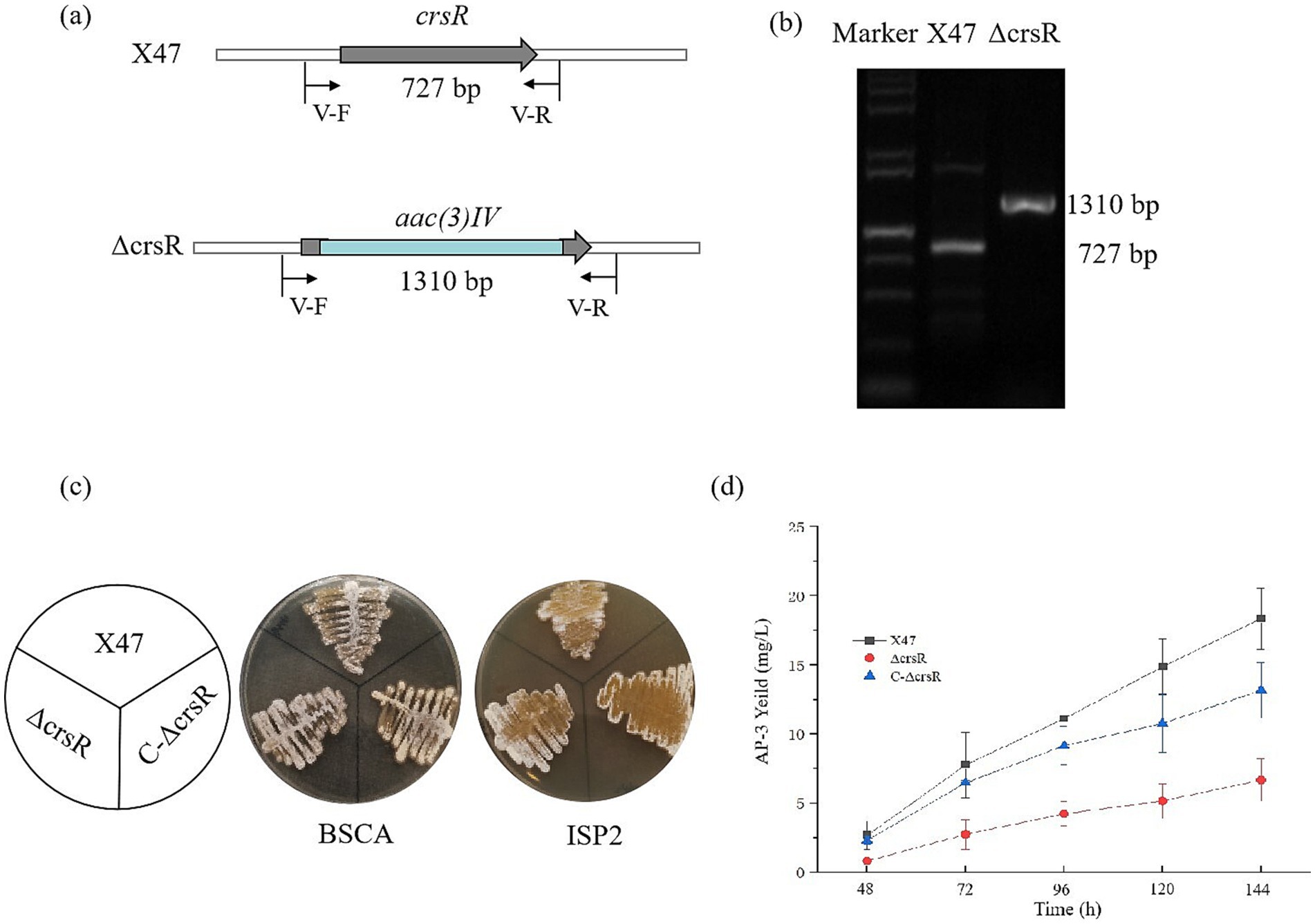
Figure 2. Deletion of crsR leads to reduced AP-3 production in A. pretiosum X47. (a) Schematic of crsR internal deletion (468 bp) replaced by an apramycin resistance cassette. (b) PCR confirmation of the crsR deletion using primers flanking the deletion site. (c) Phenotypes of A. pretiosum X47, ∆crsR, and C-∆crsR strains grown on BSCA and ISP2 media for 72 h. (d) Comparative AP-3 production in A. pretiosum X47, ∆crsR, and C-∆crsR strains. The results represent the mean ± SD of three independent biological replicates.
To assess the effect of CrsR overexpression, we integrated a native crsRS cassette into the X47 genome, creating the X47:pMS82-crsRS strain, with an empty vector integrant as a control. The overexpressed strain exhibited approximately 60% enhancement in AP-3 production at 144 h relative to the control strains. Furthermore, no significant differences in biomass were observed among the three strains at either 48 h or 144 h (Supplementary Figure S2). Collectively, these results demonstrate that CrsR acts as a crucial positive regulator of AP-3 biosynthesis in A. pretiosum.
crsR deletion causes genome-wide transcriptional changes
To determine how CrsR regulates AP-3 synthesis, we analyzed gene transcription in the wild-type X47 strain and the ΔcrsR mutant under fermentation conditions. The results showed that crsR deletion caused altered expression levels of numerous genes. At 48 h of fermentation, compared to the wild-type strain, 684 genes were upregulated and 948 genes were downregulated in the ΔcrsR mutant. GO enrichment analysis revealed that the differentially expressed genes (DEGs) were significantly associated with pathways related to purine nucleotide metabolism/biosynthesis, carbohydrate metabolic process, and nutrient catabolism (Figure 3a), suggesting that CrsR is a global regulator involved in core metabolic homeostasis.
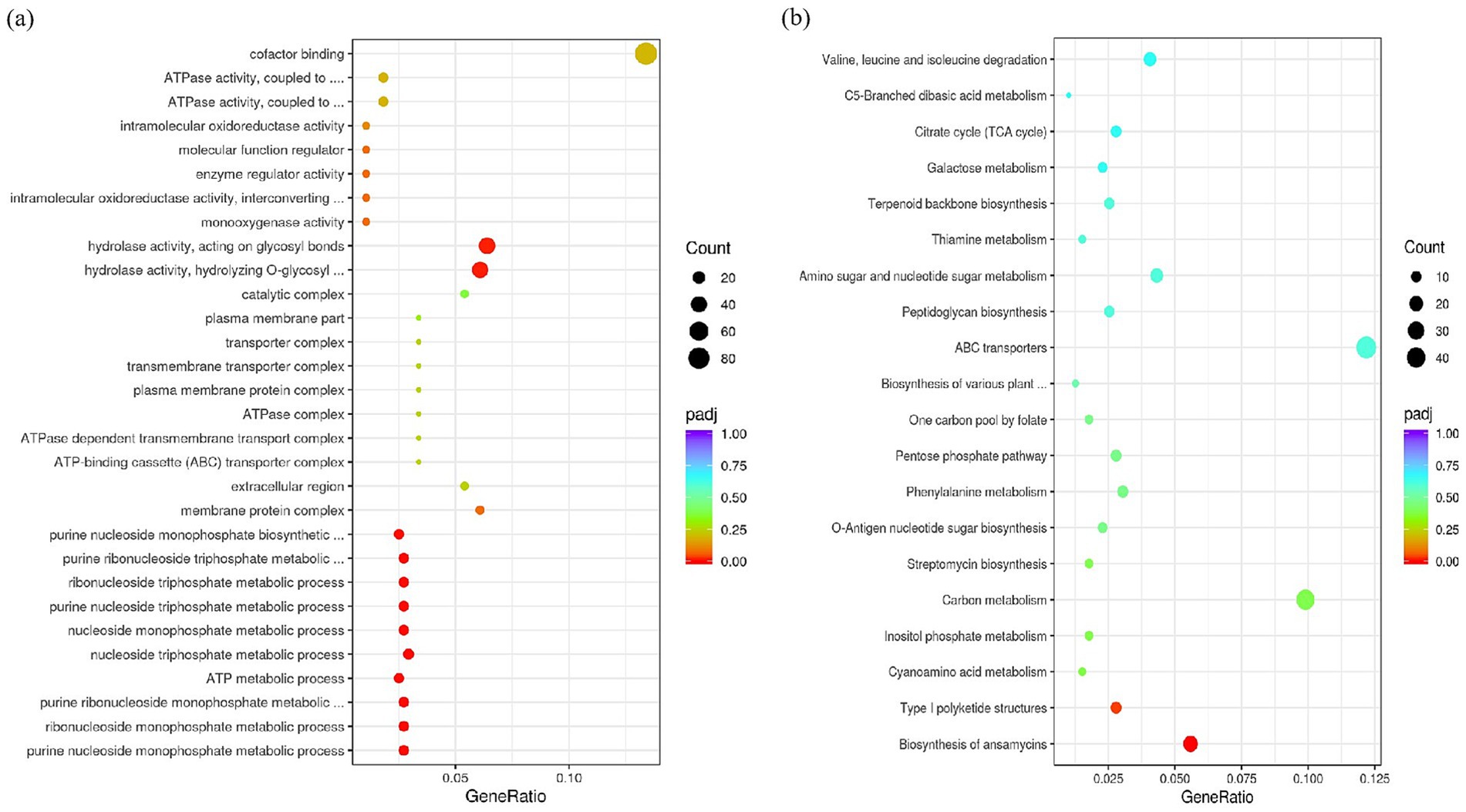
Figure 3. GO and KEGG enrichment analysis of DEGs in the crsR mutant. (a) Dot plot shows significantly enriched GO terms. (b) Dot plot shows significantly enriched KEGG pathways. The x-axis represents the gene ratio. The y-axis represents the enriched GO terms (a) or KEGG pathways (b). Dot size corresponds to the gene count. Dot color indicates the statistical significance.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis identified ansamitocin biosynthesis as the most significantly enriched pathway (Figure 3b), consistent with altered AP-3 production in the ΔcrsR mutant. The transcriptional levels of genes involved in the AP-3 biosynthesis pathway were quantitatively assessed in the ΔcrsR mutant relative to the wild-type strain (Supplementary Table S3). Uridine Diphosphate Glucose (UDPG) serves as the starting material and is converted to the starter unit AHBA (Yu et al., 2002). The genes encoding key enzymes involved in this process, asm23–24 and asm43–47, were significantly downregulated in the ΔcrsR mutant. In the proansamitocin biosynthetic pathway of the mutant strain, the transcriptional levels of the polyketide synthase (PKS) genes (asmA, asmB) and the post-PKS modification genes (asm13–15, asm17) were significantly downregulated (Figure 4a).
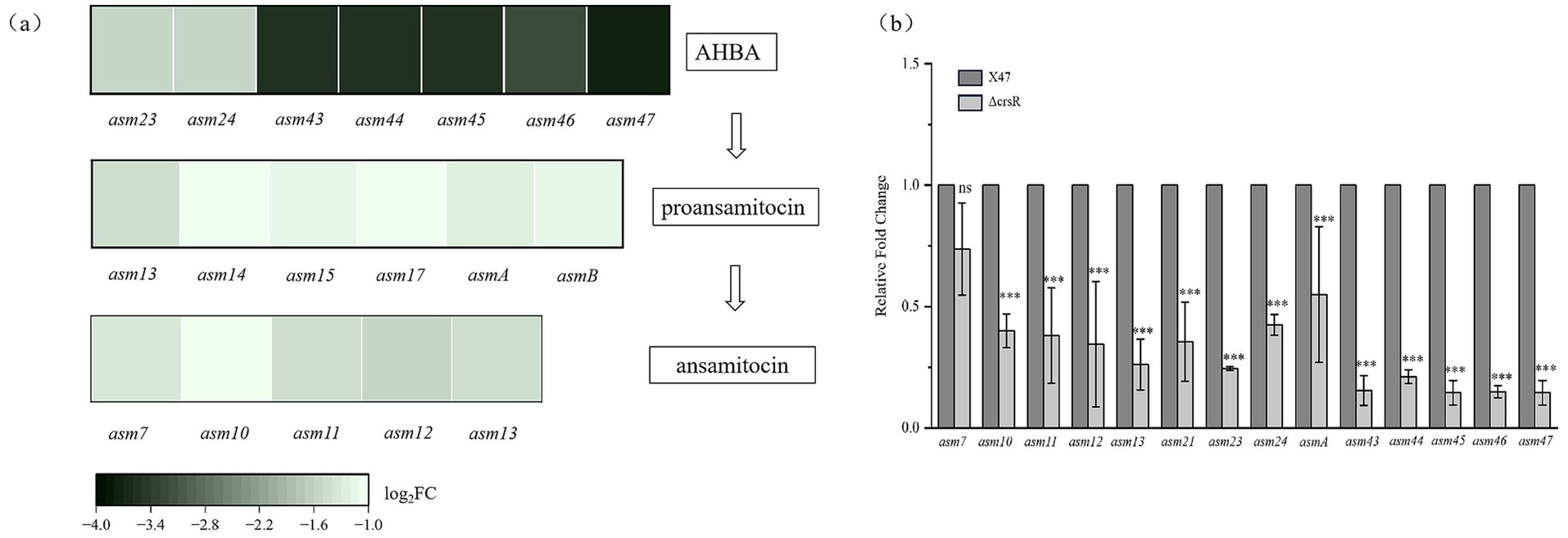
Figure 4. CrsR regulates the expression of AP-3 biosynthetic genes. (a) Heatmap depicts the significantly altered genes (p < 0.05) involved in the biosynthesis of AP-3. The analysis encompasses genes for AHBA biosynthesis, PKS assembly, post-assembly modifications, and tailoring steps. Each column represents a gene, and each row represents the ΔcrsR strain. The color gradient corresponds to the Log2 fold change (Log2 FC) in gene expression of the ΔcrsR strain relative to the X47 strain, with negative values indicating downregulation in the ΔcrsR mutant. (b) qRT-PCR validation of DEGs from the RNA-seq analysis. Expression levels were normalized to hrdB, and data are presented as mean ± SD from three biological replicates. Statistical significance is denoted as: ***, p ≤ 0.001; **, p ≤ 0.01; *, and p ≤ 0.05; ns, not significant.
To validate the transcriptome sequencing results, we quantified the expression of asm genes (asm7, asm10–13, asmA, asm23–asm24, and asm43–asm47) in strains X47 and ΔcrsR using qRT-PCR. The results were consistent with the RNA-seq data, except for asm7, which showed no significant difference in expression between the ΔcrsR and X47 strains (Figure 4b). These findings collectively suggest that CrsR acts as a global regulatory factor that positively modulates the transcription of AP-3 biosynthetic genes, thereby influencing AP-3 production.
CrsR binds to the promoters of asm21, asm43–44, and asm45–47 genes
EMSAs were performed to assess whether CrsR directly binds to AP-3 biosynthetic gene promoters. Based on operon organization determined from gene structure and RNA-seq analysis, promoter regions of asm7, asm13–15 (asm10–12), asm21, asm24, asm43–44, asm45–47, and asmA-B were amplified and biotin-labeled as probes (Figure 5a). Our results demonstrate that the His₆-tagged CrsR protein does not bind to the promoters of asm7, asm13–15 (asm10–12), asm24, or asmA (Figure 5b). Instead, it specifically binds to the promoters of asm21, asm43–44, and asm45–47 (Figures 5c–e). The concentration dependence of CrsR binding was evaluated by comparing its binding activity at 0.5 μg and 1.0 μg of protein (Figures 5c–e). Compared to the control lane with no CrsR, the addition of 0.5 μg CrsR resulted in a clearly visible shifted band, indicating that binding occurred, while a significant amount of free probe remained unbound. When the CrsR concentration was increased to 1.0 μg, the signal of the shifted band intensified significantly. The results revealed a marked increase in the DNA–protein complex signal at the higher CrsR concentration, confirming that binding is concentration-dependent. In competitive binding assays, a 100-fold excess of unlabeled specific DNA fragment significantly reduced binding to the asm21 promoter, whereas an equivalent excess of the non-specific competitor poly(dI-dC) had no effect (Figure 5c). Similar binding patterns were observed for the asm43 and asm45 promoters (Figures 5d,e). These findings indicate that His₆-tagged CrsR specifically interacts with the asm21, asm43–44, and asm45–47 promoters, suggesting that it directly regulates their transcription.
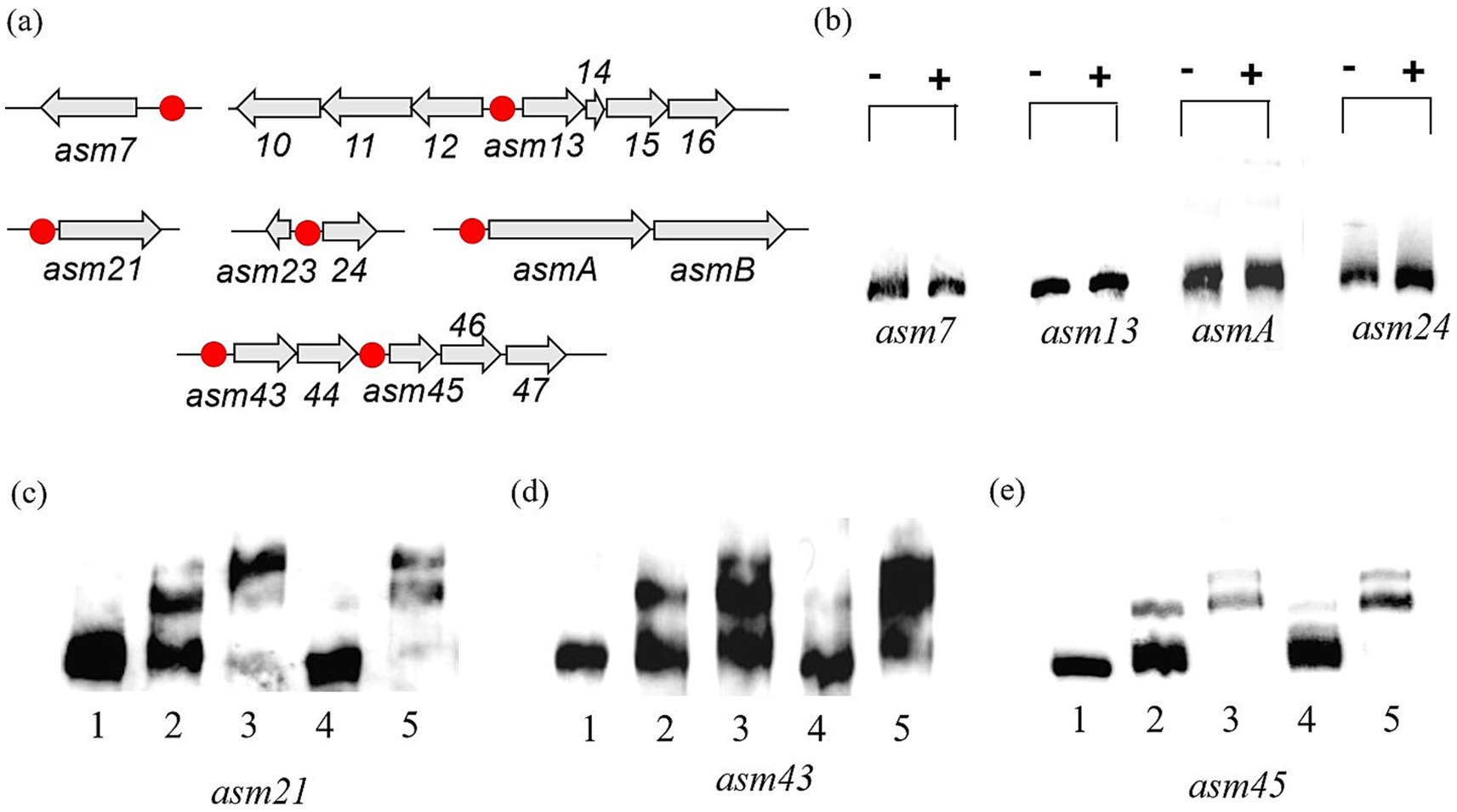
Figure 5. EMSAs with the His₆-tagged CrsR with target probes. (a) Transcriptional organization of CrsR-affected genes or operons within the asm cluster. Probe locations are marked with red dots. (b) Binding of CrsR to the asm7, asm13, asm24, and asmA promoters. (−): no protein; (+): 1 μg CrsR. (b–d) Binding of CrsR to the asm21 (c), asm43 (d), and asm45 (e) promoters. Lane 1: probe only (no protein); Lane 2: probe + 0.5 μg CrsR; Lane 3: probe + 1 μg CrsR; Lane 4: probe + 1 μg CrsR + 100-fold molar excess of unlabeled specific probe; and Lane 5: probe + 1 μg CrsR + 100-fold molar excess of unlabeled non-specific probe.
We used AlphaFold 3 to predict the precise binding sites of CrsR in the promoter regions of its target genes (Supplementary Table S5). The resulting models exhibited low confidence scores, with ipTM <0.6 and pTM < 0.5. Furthermore, no clearly conserved binding motif was identified among the predicted sequences.
Discussion
The A. pretiosum X47 genome encodes dozens of putative TCSs, most of which are uncharacterized. Notably, we functionally characterized CrsR, the response regulator of TCS CrsRK, and proposed a CrsR-dependent regulatory model for AP-3 biosynthesis (Figure 6). KEGG pathway analysis indicated that the CrsR mutation primarily disrupted the AP-3 biosynthetic pathway. The transcriptional levels of the majority of genes (asm23–asm24 and asm43–asm47) in the cluster were downregulated to varying degrees, with the most pronounced changes observed for asm43–asm47 (Yu et al., 2002). EMSA results demonstrated that CrsR directly binds to the promoters of asm43–asm47, which regulates the transcription of these genes, ultimately affecting the metabolic flux from UDP-glucose toward AHBA synthesis. Furthermore, reduced transcription of genes (asm10–asm15, asm21, and asmA–asmB) involved in the PKS pathway and post-modification steps further impaired the production of AP-3 (Spiteller et al., 2003). CrsR directly regulates asm21, as evidenced by its binding to the asm21 promoter region in vitro. In contrast, CrsR does not directly regulate other genes, including asm10–15, asmA, asmB, and asm23–24. EMSAs confirmed the absence of CrsR binding to their promoter regions, indicating an indirect regulatory mechanism. Although the pathway-specific regulators within the asm BGC were not controlled by CrsR (Supplementary Table S4), we proposed that CrsR modulates the expression of asm10–15, asmA, and asm23–24 through other pleiotropic regulators. However, the precise mechanism remains to be elucidated. Collectively, our results demonstrate that the response regulator CrsR acts as a global transcriptional activator of AP-3 biosynthesis, directly or indirectly regulating the asm BGC.
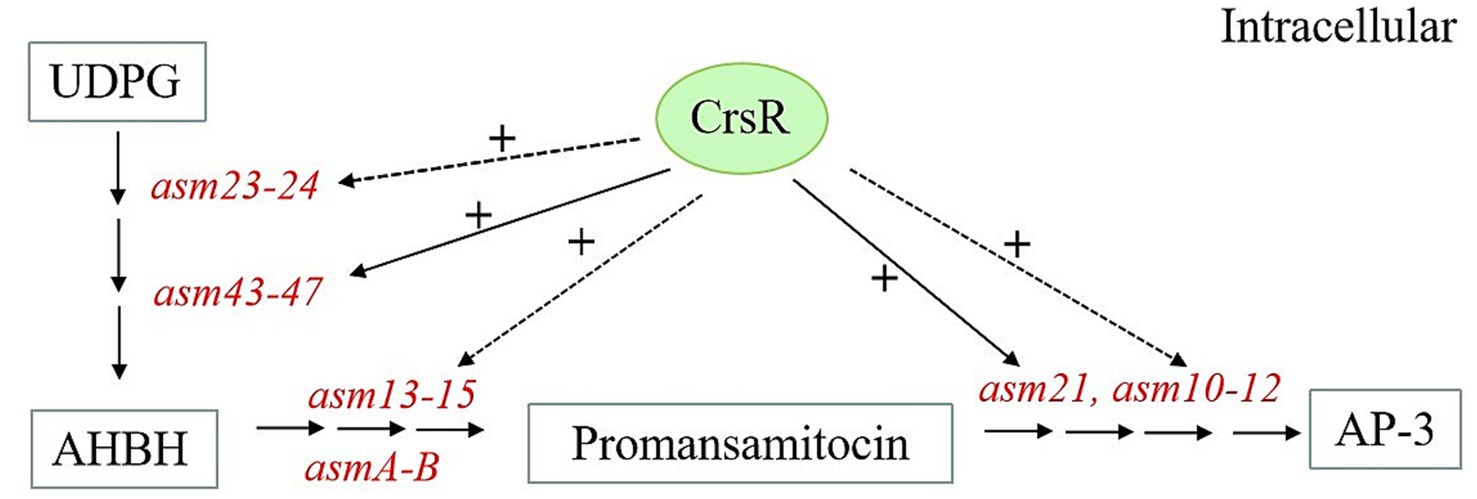
Figure 6. Proposed model of CrsR regulation in asm BGC. CrsR positively regulates AP-3 biosynthetic genes. (+) Denotes positive regulation. Direct and indirect regulations are represented by solid and dashed lines, respectively.
In a typical TCS, the kinase senses environmental signals, undergoes autophosphorylation, and subsequently phosphorylates its cognate response regulator (Zschiedrich et al., 2016). CrsK is the histidine kinase component of the CrsRK TCSs, equipped with a signal perception domain and a histidine kinase domain. Its proposed role is to sense external signals, autophosphorylate, and then phosphorylate the response regulator CrsR to activate downstream gene regulation. Although we speculate that the kinase domain mediates the interaction with CrsR, the exact environmental signals that CrsK detects cannot be inferred due to a lack of functional data on homologous proteins. A critical next step to fully decipher the regulatory logic of this pathway will be to systematically investigate the signals perceived by CrsK and the precise mechanism of its interaction with CrsR.
Actinomycetes are the predominant reservoir of bioactive natural products, accounting for most of the clinically used antibiotics (Barka et al., 2016). Antibiotic biosynthetic genes are typically organized in clusters within the genome. Antibiotic biosynthesis is governed by a hierarchical regulatory framework comprising pathway-specific control mediated by cluster-situated regulators (CSRs) and global regulation orchestrated by pleiotropic regulators, which collectively constitute an integrated network that dynamically coordinates antibiotic production (Wei et al., 2018). Knowledge of antibiotic biosynthesis regulation is crucial for discovering novel antibiotics and enhancing the yields of known compounds. For instance, in S. coelicolor, ACT biosynthesis is governed by the CSR ActII-ORF4 while simultaneously being regulated by global regulators including AdpA, MtrA, MacR, WblA, and DraR (Kang et al., 2007; Lee et al., 2013; Liu et al., 2021; Som et al., 2017b; Yu et al., 2012). Few research has elucidated the CSRs governing AP-3 biosynthesis in A. pretiosum, offering diverse strategies for yield improvement. Within the asm BGCs, the LuxR family regulator Asm8 directly activates the AHBA formation, while others, such as Asm18, not only increase ansamitocin production but also enhance the chemical diversity of the metabolites produced (Li et al., 2016; Pan et al., 2013). Asm2 and Asm39 function as positive regulators of ansamitocin biosynthesis, and constitutive overexpression of these genes significantly enhances AP-3 production (Ng et al., 2009). Additionally, global regulators governing AP-3 biosynthesis have been characterized. In A. pretiosum subsp. auranticum ATCC 31565, the AdpA family regulator AdpA_1075 pleiotropically links morphological differentiation to ansamitocin biosynthesis (Guo et al., 2022). AdpA_1075 positively regulates ansamitocin biosynthesis by directly controlling asm28 expression. CrsR indirectly regulates the expression of genes such as asmAB and asm10–15 but has no effect on the transcription of regulatory proteins within the gene cluster (Supplementary Table S4), suggesting that other regulatory pathways control the expression of the ansamitocin gene cluster. In A. pretiosum X47, knockout of the response regulator in the CNX_RS34865/CNX_RS34870 TCS led to reduced AP-3 biosynthesis, accompanied by significant downregulation of several asm genes, including asm1, asm2, asm30, asm32–asm35, and asm37 (Zhang P. et al., 2022). Our study reveals that CrsR, the response regulator of the CrsRK TCS, acts as another key regulator in the AP-3 biosynthetic network. Notably, these two regulatory systems appear to operate independently, as RNA-seq analysis revealed that CrsR does not affect the expression of CNX_RS34865/CNX_RS34870, and each system regulates a distinct subset of asm genes. This clear segregation of target genes suggests that AP-3 synthesis is finely tuned through parallel signaling pathways, allowing the integration of different environmental or physiological cues for precise metabolic control.
Our RNA-seq data support that CrsR functions as a global regulator, affecting not only AP-3 biosynthesis but also primary metabolic processes such as purine synthesis. Although the deletion of crsR led to transcriptional upregulation of these metabolic genes (e.g., guaA, atpA, atpD, atpF, atpG, and atpH), AP-3 production decreased, suggesting that the enhanced metabolic flux may have been redirected toward other cellular processes rather than being channeled into AP-3 biosynthesis.
As a response regulator, CrsR is expected to specifically bind to conserved DNA motifs within the promoters of its target genes. Our data confirm that CrsR does bind directly to the promoters of asm21, asm43, and asm45. However, AlphaFold-based predictions failed to reveal a consistently conserved binding motif among these regions, and the resulting models exhibited low confidence scores. This apparent discrepancy may reflect inherent limitations of current structure prediction tools in accurately modeling protein–DNA interactions, particularly when binding involves flexible regions or non-canonical interfaces (Abramson et al., 2024). Therefore, in future studies, combining ChIP-seq with EMSAs and bioinformatic analyses will be essential to define the conserved binding motif of CrsR and comprehensively elucidate its regulatory mechanism.
In conclusion, this study identifies and functionally characterizes the novel TCS CrsRK in A. pretiosum X47, revealing its global regulatory role in AP-3 biosynthesis. We demonstrate that CrsR directly targets key biosynthetic genes (asm21 and asm43–47) through promoter binding, as confirmed by EMSAs. However, the mechanistic basis of CrsR-mediated regulation, including its conserved DNA-binding motifs, potential co-regulatory partners, and the environmental signals sensed by its cognate kinase CrsK, requires further investigation. Elucidating these mechanisms will advance the fundamental understanding of how TCSs orchestrate complex secondary metabolism in actinomycetes and enable rational engineering of high-yield AP-3 strains.
Data availability statement
RNA-Seq data has been deposited in the Sequence Read Archive422 (SRA) under the accession number PRJNA1301260.
Author contributions
PZ: Conceptualization, Writing – original draft. GZ: Formal analysis, Writing – review & editing. TW: Investigation, Software, Writing – review & editing. SZ: Investigation, Writing – review & editing. RS: Investigation, Writing – review & editing. JF: Validation, Software, Writing – review & editing. ML: Conceptualization, Writing – original draft. GC: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grants No. 82204255 and 32200071) and the Shandong Provincial Natural Science Foundation (Grants No. ZR2021QC169 and ZR2023MC068). Grants No. 82204255, 32200071, and ZR2021QC169 provided funding for the research, while Grant No. ZR2023MC068 specifically supported data collection and analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1684526/full#supplementary-material
Footnotes
References
Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., et al. (2024). Accurate structure prediction of biomolecular interactions with alphafold 3. Nature 630, 493–500. doi: 10.1038/s41586-024-07487-w
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Bierman, M., Logan, R., O'Brien, K., Seno, E. T., Rao, R. N., and Schoner, B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49. doi: 10.1016/0378-1119(92)90627-2
Carroll, B. J., Moss, S. J., Bai, L., Kato, Y., Toelzer, S., Yu, T. W., et al. (2002). Identification of a set of genes involved in the formation of the substrate for the incorporation of the unusual "glycolate" chain extension unit in ansamitocin biosynthesis. J. Am. Chem. Soc. 124, 4176–4177. doi: 10.1021/ja0124764
Fan, Y., Gao, Y., Zhou, J., Wei, L., Chen, J., and Hua, Q. (2014). Process optimization with alternative carbon sources and modulation of secondary metabolism for enhanced ansamitocin p-3 production in actinosynnema pretiosum. J. Biotechnol. 192 Pt A, 1–10. doi: 10.1016/j.jbiotec.2014.10.020
Fan, Y., Zhao, M., Wei, L., Hu, F., Imanaka, T., Bai, L., et al. (2016). Enhancement of udpg synthetic pathway improves ansamitocin production in actinosynnem pretiosum. Appl. Microbiol. Biotechnol. 100, 2651–2662. doi: 10.1007/s00253-015-7148-2
Floss, H. G., Yu, T. W., and Arakawa, K. (2011). The biosynthesis of 3-amino-5-hydroxybenzoic acid (ahba), the precursor of mc7n units in ansamycin and mitomycin antibiotics: a review. J. Antibiot. (Tokyo) 64, 35–44. doi: 10.1038/ja.2010.139
Ghisalba, O., and Nuesch, J. (1981). A genetic approach to the biosynthesis of the rifamycin-chromophore in nocardia mediterranei. Iv. Identification of 3-amino-5-hydroxybenzoic acid as a direct precursor of the seven-carbon amino starter-unit. J. Antibiot. (Tokyo) 34, 64–71. doi: 10.7164/antibiotics.34.64
Guo, S., Leng, T., Sun, X., Zheng, J., Li, R., Chen, J., et al. (2022). Global regulator adpa_1075 regulates morphological differentiation and ansamitocin production in actinosynnema pretiosum subsp. auranticum. Bioengineering (Basel) 9:719. doi: 10.3390/bioengineering9110719
Hasegawa, T., S, T., and Hatano, K. (1983). Motile actinomycetes: Actinosynnema pretiosum sp. nov., subsp. nov., and Actinosynnema pretiosum subsp. pretiosum subsp. auranticum subsp. nov. Int. J. Syst. Bacteriol. 33, 314–320.
Higashide, E., Asai, M., Ootsu, K., Tanida, S., Kozai, Y., Hasegawa, T., et al. (1977). Ansamitocin, a group of novel maytansinoid antibiotics with antitumour properties from nocardia. Nature 270, 721–722. doi: 10.1038/270721a0
Jia, Y., and Zhong, J. J. (2011). Enhanced production of ansamitocin p-3 by addition of mg2+ in fermentation of actinosynnema pretiosum. Bioresour. Technol. 102, 10147–10150. doi: 10.1016/j.biortech.2011.08.031
Kang, S. H., Huang, J., Lee, H. N., Hur, Y. A., Cohen, S. N., and Kim, E. S. (2007). Interspecies dna microarray analysis identifies wbla as a pleiotropic down-regulator of antibiotic biosynthesis in streptomyces. J. Bacteriol. 189, 4315–4319. doi: 10.1128/JB.01789-06
Kibby, J. J., and Rickards, R. W. (1981). The identification of 3-amino-5-hydroxybenzoic acid as a new natural aromatic amino acid. J. Antibiot. (Tokyo) 34, 605–607. doi: 10.7164/antibiotics.34.605
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical streptomyces genetics, Norwich: John Innes Foundation.
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kubota, T., Brunjes, M., Frenzel, T., Xu, J., Kirschning, A., and Floss, H. G. (2006). Determination of the cryptic stereochemistry of the first pks chain-extension step in ansamitocin biosynthesis by actinosynnema pretiosum. Chembiochem 7, 1221–1225. doi: 10.1002/cbic.200500506
Lee, H. N., Kim, J. S., Kim, P., Lee, H. S., and Kim, E. S. (2013). Repression of antibiotic downregulator wbla by adpa in streptomyces coelicolor. Appl. Environ. Microbiol. 79, 4159–4163. doi: 10.1128/AEM.00546-13
Li, S., Lu, C., Chang, X., and Shen, Y. (2016). Constitutive overexpression of asm18 increases the production and diversity of maytansinoids in actinosynnema pretiosum. Appl. Microbiol. Biotechnol. 100, 2641–2649. doi: 10.1007/s00253-015-7127-7
Li, Y., Zhao, P., Kang, Q., Ma, J., Bai, L., and Deng, Z. (2011). Dual carbamoylations on the polyketide and glycosyl moiety by asm21 result in extended ansamitocin biosynthesis. Chem. Biol. 18, 1571–1580. doi: 10.1016/j.chembiol.2011.11.007
Lin, J., Bai, L., Deng, Z., and Zhong, J. J. (2011). Enhanced production of ansamitocin p-3 by addition of isobutanol in fermentation of actinosynnema pretiosum. Bioresour. Technol. 102, 1863–1868. doi: 10.1016/j.biortech.2010.09.102
Liu, M., Xu, W., Zhu, Y., Cui, X., and Pang, X. (2021). The response regulator macr and its potential in improvement of antibiotic production in streptomyces coelicolor. Curr. Microbiol. 78, 3696–3707. doi: 10.1007/s00284-021-02633-3
Liu, M., Zhang, P., Zhu, Y., Lu, T., Wang, Y., Cao, G., et al. (2019). Novel two-component system macrs is a pleiotropic regulator that controls multiple morphogenic membrane protein genes in streptomyces coelicolor. Appl. Environ. Microbiol. 85:e02178-18. doi: 10.1128/AEM.02178-18
Lopus, M., Oroudjev, E., Wilson, L., Wilhelm, S., Widdison, W., Chari, R., et al. (2010). Maytansine and cellular metabolites of antibody-maytansinoid conjugates strongly suppress microtubule dynamics by binding to microtubules. Mol. Cancer Ther. 9, 2689–2699. doi: 10.1158/1535-7163.MCT-10-0644
Ma, J., Zhao, P. J., and Shen, Y. M. (2007). New amide n-glycosides of ansamitocins identified from actinosynnema pretiosum. Arch. Pharm. Res. 30, 670–673. doi: 10.1007/BF02977625
Modi, S., Jacot, W., Yamashita, T., Sohn, J., Vidal, M., Tokunaga, E., et al. (2022). Trastuzumab deruxtecan in previously treated her2-low advanced breast cancer. N. Engl. J. Med. 387, 9–20. doi: 10.1056/NEJMoa2203690
Moss, S. J., Bai, L., Toelzer, S., Carroll, B. J., Mahmud, T., Yu, T. W., et al. (2002). Identification of asm19 as an acyltransferase attaching the biologically essential ester side chain of ansamitocins using n-desmethyl-4,5-desepoxymaytansinol, not maytansinol, as its substrate. J. Am. Chem. Soc. 124, 6544–6545. doi: 10.1021/ja020214b
Narayan, P., Osgood, C. L., Singh, H., Chiu, H. J., Ricks, T. K., Chiu, Y. C. E., et al. (2021). Fda approval summary: fam-trastuzumab deruxtecan-nxki for the treatment of unresectable or metastatic her2-positive breast cancer. Clin. Cancer Res. 27, 4478–4485. doi: 10.1158/1078-0432.CCR-20-4557
Ng, D., Chin, H. K., and Wong, V. V. (2009). Constitutive overexpression of asm2 and asm39 increases ap-3 production in the actinomycete actinosynnema pretiosum. J. Ind. Microbiol. Biotechnol. 36, 1345–1351. doi: 10.1007/s10295-009-0619-7
Pan, W., Kang, Q., Wang, L., Bai, L., and Deng, Z. (2013). Asm8, a specific lal-type activator of 3-amino-5-hydroxybenzoate biosynthesis in ansamitocin production. Sci. China Life Sci. 56, 601–608. doi: 10.1007/s11427-013-4502-4
Prota, A. E., Bargsten, K., Diaz, J. F., Marsh, M., Cuevas, C., Liniger, M., et al. (2014). A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs. Proc. Natl. Acad. Sci. USA 111, 13817–13821. doi: 10.1073/pnas.1408124111
Qin, R., Zhong, C., Zong, G., Fu, J., Pang, X., and Cao, G. (2017). Improvement of clavulanic acid production in Streptomyces clavuligerus F613-1 by using a claR-neo reporter strategy. Electron. J. Biotechnol. 28, 41–46. doi: 10.1016/j.ejbt.2017.05.002
Som, N. F., Heine, D., Holmes, N., Knowles, F., Chandra, G., Seipke, R. F., et al. (2017b). The mtrab two-component system controls antibiotic production in streptomyces coelicolor a3(2). Microbiology (Reading) 163, 1415–1419. doi: 10.1099/mic.0.000524
Som, N. F., Heine, D., Holmes, N. A., Munnoch, J. T., Chandra, G., Seipke, R. F., et al. (2017a). The conserved actinobacterial two-component system mtrab coordinates chloramphenicol production with sporulation in streptomyces venezuelae nrrl b-65442. Front. Microbiol. 8:1145. doi: 10.3389/fmicb.2017.01145
Sonnhammer, E. L., von Heijne, G., and Krogh, A. (1998). A hidden markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182.
Spiteller, P., Bai, L., Shang, G., Carroll, B. J., Yu, T. W., and Floss, H. G. (2003). The post-polyketide synthase modification steps in the biosynthesis of the antitumor agent ansamitocin by Actinosynnema pretiosum. J. Am. Chem. Soc. 125, 14236–14237. doi: 10.1021/ja038166y
Venghateri, J. B., Gupta, T. K., Verma, P. J., Kunwar, A., and Panda, D. (2013). Ansamitocin p3 depolymerizes microtubules and induces apoptosis by binding to tubulin at the vinblastine site. PLoS One 8:e75182. doi: 10.1371/journal.pone.0075182
Wang, R., Zhou, T., Kong, F., Hou, B., Ye, J., Wu, H., et al. (2023). Aflq1-q2 represses lincomycin biosynthesis via multiple cascades in streptomyces lincolnensis. Appl. Microbiol. Biotechnol. 107, 2933–2945. doi: 10.1007/s00253-023-12429-z
Wedam, S., Fashoyin-Aje, L., Gao, X., Bloomquist, E., Tang, S., Sridhara, R., et al. (2020). Fda approval summary: ado-trastuzumab emtansine for the adjuvant treatment of her2-positive early breast cancer. Clin. Cancer Res. 26, 4180–4185. doi: 10.1158/1078-0432.CCR-19-3980
Wei, J., He, L., and Niu, G. (2018). Regulation of antibiotic biosynthesis in actinomycetes: perspectives and challenges. Synth. Syst. Biotechnol. 3, 229–235. doi: 10.1016/j.synbio.2018.10.005
Wu, Y., Kang, Q., Shang, G., Spiteller, P., Carroll, B., Yu, T. W., et al. (2011). N-methylation of the amide bond by methyltransferase Asm10 in ansamitocin biosynthesis. Chembiochem 12, 1759–1766. doi: 10.1002/cbic.201100062
Yan, Y., Zou, L., Wei, H., Yang, M., Yang, Y., Li, X., et al. (2024). An atypical two-component system, Atcr/Atck, simultaneously regulates the biosynthesis of multiple secondary metabolites in Streptomyces bingchenggensis. Appl. Environ. Microbiol. 90:e130023. doi: 10.1128/aem.01300-23
Yu, T. W., Bai, L., Clade, D., Hoffmann, D., Toelzer, S., Trinh, K. Q., et al. (2002). The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA 99, 7968–7973. doi: 10.1073/pnas.092697199
Yu, Z., Zhu, H., Dang, F., Zhang, W., Qin, Z., Yang, S., et al. (2012). Differential regulation of antibiotic biosynthesis by drar-k, a novel two-component system in streptomyces coelicolor. Mol. Microbiol. 85, 535–556. doi: 10.1111/j.1365-2958.2012.08126.x
Yu, Z., Zhu, H., Zheng, G., Jiang, W., and Lu, Y. (2014). A genome-wide transcriptomic analysis reveals diverse roles of the two-component system drar-k in the physiological and morphological differentiation of streptomyces coelicolor. Appl. Microbiol. Biotechnol. 98, 9351–9363. doi: 10.1007/s00253-014-6102-z
Zafar, S., Armaghan, M., Khan, K., Hassan, N., Sharifi-Rad, J., Habtemariam, S., et al. (2023). New insights into the anticancer therapeutic potential of maytansine and its derivatives. Biomed. Pharmacother. 165:115039. doi: 10.1016/j.biopha.2023.115039
Zhang, K., Mohsin, A., Dai, Y., Ali, M. F., Chen, Z., Zhuang, Y., et al. (2021). Role of a two-component signal transduction system rspa1/a2 in regulating the biosynthesis of salinomycin in streptomyces albus. Appl. Biochem. Biotechnol. 193, 1296–1310. doi: 10.1007/s12010-020-03357-z
Zhang, K., Mohsin, A., Yu, J., Hu, Y., Ali, M. F., Chen, Z., et al. (2020). Two-component-system rspa1/a2-dependent regulation on primary metabolism in streptomyces albus a30 cultivated with glutamate as the sole nitrogen source. Front. Microbiol. 11:1658. doi: 10.3389/fmicb.2020.01658
Zhang, K., Zhang, P., Fu, J., Kang, N., Zong, G., Ma, X., et al. (2022). Characterization of the two-component transduction system cnx_rs34865/cnx_rs34870 in regulating the biosynthesis of ansamitocin. Chin. J. Antibiot. 4, 354–360. doi: 10.3969/j.issn.1001-8689.2022.04.006
Zhang, P., Zhang, K., Liu, Y., Fu, J., Zong, G., Ma, X., et al. (2022). Deletion of the response regulator phop accelerates the formation of aerial mycelium and spores in actinosynnema pretiosum. Front. Microbiol. 13:845620. doi: 10.3389/fmicb.2022.845620
Zhao, P., Bai, L., Ma, J., Zeng, Y., Li, L., Zhang, Y., et al. (2008). Amide n-glycosylation by asm25, an n-glycosyltransferase of ansamitocins. Chem. Biol. 15, 863–874. doi: 10.1016/j.chembiol.2008.06.007
Zhao, M., Fan, Y., Wei, L., Hu, F., and Hua, Q. (2017). Effects of the methylmalonyl-coa metabolic pathway on ansamitocin production in actinosynnema pretiosum. Appl. Biochem. Biotechnol. 181, 1167–1178. doi: 10.1007/s12010-016-2276-4
Zhong, C., Zong, G., Qian, S., Liu, M., Fu, J., Zhang, P., et al. (2019). Complete genome sequence of actinosynnema pretiosum x47, an industrial strain that produces the antibiotic ansamitocin ap-3. Curr. Microbiol. 76, 954–958. doi: 10.1007/s00284-018-1521-1
Zhu, Y., Zhang, P., Zhang, J., Wang, J., Lu, Y., and Pang, X. (2020). Impact on multiple antibiotic pathways reveals mtrA as a master regulator of antibiotic production in Streptomyces spp. and potentially in other actinobacteria. Appl. Environ. Microbiol. 86:e01201-20. doi: 10.1128/AEM.01201-20
Zong, G., Cao, G., Fu, J., Zhang, P., Chen, X., Yan, W., et al. (2022). Macrs controls morphological differentiation and natamycin biosynthesis in streptomyces gilvosporeus f607. Microbiol. Res. 262:127077. doi: 10.1016/j.micres.2022.127077
Keywords: Actinosynnema pretiosum, ansamitocin, two-component system, biosynthesis, response regulator
Citation: Zhang P, Zong G, Wang T, Zhao S, Sun R, Fu J, Liu M and Cao G (2025) The response regulator CrsR positively regulates ansamitocin P-3 biosynthesis in Actinosynnema pretiosum. Front. Microbiol. 16:1684526. doi: 10.3389/fmicb.2025.1684526
Edited by:
Yixin Shi, Arizona State University, United StatesReviewed by:
Xin Li, Shanghai Academy of Agricultural Sciences, ChinaYanyan Zhang, Chinese Academy of Agricultural Sciences, China
Copyright © 2025 Zhang, Zong, Wang, Zhao, Sun, Fu, Liu and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Liu, MTQwMzVAc2RqenUuZWR1LmNu; Guangxiang Cao, Y2FvZ3Vhbmd4aWFuZ0BzZGZtdS5lZHUuY24=
†These authors have contributed equally to this work
 Peipei Zhang
Peipei Zhang Gongli Zong
Gongli Zong Tengfei Wang2
Tengfei Wang2 Jiafang Fu
Jiafang Fu Meng Liu
Meng Liu