- 1Christian Medical College, Vellore, Tamil Nadu, India
- 2The Tamil Nadu Dr. M.G.R. Medical University, Chennai, Tamil Nadu, India
- 3Indira Gandhi Institute of Child Health, Bengaluru, Karnataka, India
- 4St. John’s Medical College Hospital, Bengaluru, Karnataka, India
- 5Baby Memorial Hospital, Kozhikode, Kerala, India
- 6KIMSHEALTH Hospital, Thiruvananthapuram, Kerala, India
- 7Agilus Diagnostic Labs, Mumbai, Maharashtra, India
- 8Sahyadri Speciality Labs, Pune, Maharashtra, India
- 9Fortis Hospital, Bengaluru, Karnataka, India
- 10Lilavati Hospital & Research Centre, Mumbai, India
- 11P. D. Hinduja Hospital & Medical Research Centre, Mumbai, India
- 12Descriptive Research Division, Indian Council of Medical Research, New Delhi, India
Background: The rise of antimicrobial resistance (AMR) in Salmonella enterica serovar Typhi poses a serious threat to global enteric fever control. In particular, the emergence of resistance to third-generation cephalosporins and azithromycin critically undermines available treatment options. Sustained genomic surveillance of high-risk S. Typhi lineages and resistance determinants is essential for informing antibiotic policy and optimizing typhoid conjugate vaccine (TCV) introduction in endemic regions. In this study, we report a multicenter outbreak of carbapenem-resistant S. Typhi in India and investigate its genomic epidemiology, resistance mechanisms, and evolutionary origins.
Methods: A total of 31 carbapenem-resistant S. Typhi isolates collected from multiple tertiary care hospitals were subjected to phenotypic antimicrobial susceptibility testing and whole-genome sequencing (WGS). Short-read WGS data were used to analyze core-genome SNPs, infer phylogenetic relationships, and investigate AMR determinants. Two representative isolates underwent long-read Oxford Nanopore sequencing for plasmid reconstruction and comparative genomic analysis with Enterobacterales.
Results: Antimicrobial susceptibility testing of isolates revealed resistance to ampicillin, ciprofloxacin, ceftriaxone, and carbapenems while retaining susceptibility to chloramphenicol, cotrimoxazole, and azithromycin. The genomic analysis identified the presence of two plasmids: IncFIB(K) harboring blaCTX-M-15, qnrS1, tetA, and IncX3, carrying the blaNDM-5 gene. Phylogenetic analysis classified the isolates within a novel genotype, 4.3.1.1.1, belonging to genotype 4.3.1.1 (H58 lineage I). Notably, plasmid comparison revealed high similarity to resistance plasmids circulating in co-endemic Escherichia coli and Klebsiella pneumoniae, indicating recent horizontal gene transfer.
Conclusion: This is the first documented outbreak of blaNDM-mediated carbapenem-resistant S. Typhi, highlighting a new stage in the evolution of drug-resistant typhoid. The acquisition of high-risk plasmids by S. Typhi and their integration into successful epidemic lineages underscores the urgent need for strengthened genomic surveillance and inter-species AMR tracking. Our findings have direct implications for treatment guidelines, TCV implementation strategies, and efforts to prevent global dissemination of carbapenem-resistant S. Typhi.
Introduction
Enteric fever, primarily caused by Salmonella enterica serovars Typhi and Paratyphi A, remains a major public health challenge, particularly in low- and middle-income countries (LMICs) (Parry et al., 2002; Crump et al., 2015). This disease is primarily transmitted through the ingestion of food or water contaminated with fecal matter, potentially leading to severe complications, such as intestinal perforation, if untreated (Radhakrishnan et al., 2018; Crump, 2019). Recent estimates indicate a global burden of approximately 9.24 million enteric fever cases in 2021,1 with South Asia accounting for 62% of the total incidence. Within this region, India bears the highest burden, contributing 58% of global cases, followed by Pakistan and Bangladesh (Piovani et al., 2024). Notably, children under 5 years of age are disproportionately affected, experiencing a significant share of the disease’s morbidity and mortality, particularly in endemic areas (John et al., 2023).
The management of typhoid fever has historically relied on antimicrobial therapy; however, the emergence of antimicrobial-resistant (AMR) strains has significantly compromised treatment efficacy (Browne et al., 2024). In the late 20th century, multidrug-resistant (MDR) S. Typhi strains emerged, exhibiting resistance to first-line antibiotics such as ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole (Crump et al., 2015). This situation prompted a shift towards fluoroquinolones as the primary treatment option (Marchello et al., 2020). Unfortunately, the subsequent rise of fluoroquinolone non-susceptible (FQNS) strains necessitated the exploration of alternative therapies, including azithromycin and third-generation cephalosporins (Kirchhelle et al., 2019). In recent years, the emergence and spread of ceftriaxone-resistant S. Typhi strains in Pakistan and India has further complicated the treatment strategies. In Pakistan, the outbreak of extensively drug-resistant (XDR) S. Typhi, has spread across the country, with ~5,274 cases reported by December 2018 (Akram et al., 2020). These XDR strains exhibit resistance to first-line antibiotics, fluoroquinolones, and third-generation cephalosporins, leaving azithromycin as the primary treatment options (Akram et al., 2020; Klemm et al., 2018). Similarly, ceftriaxone-resistant S. Typhi outbreaks have been documented in India, including in Mumbai and Vadodara (Jacob et al., 2021; Thirumoorthy et al., 2025). Furthermore, azithromycin-resistant strains, driven by mutations in the acrB gene, threaten one of the few remaining oral treatment options, highlighting the urgent need for new therapeutic strategies (Carey et al., 2021).
Genomic surveillance is a key tool for monitoring AMR transmission and tracking outbreaks of S. Typhi (Baker et al., 2018). The GenoTyphi scheme provides a robust framework for classifying S. Typhi into four major lineages and over 75 genotypes, enabling the identification of distinct transmission pathways on a global scale (Wong et al., 2016; Dyson and Holt, 2021). Among these, haplotype 58 (H58, genotype 4.3.1) has become the dominant strain worldwide, primarily due to its enhanced transmissibility and MDR (Wong et al., 2015; Pragasam et al., 2020; Carey et al., 2024). Within this lineage, key subclades include 4.3.1.1, associated with MDR, 4.3.1.2, linked to FQNS, and 4.3.1.3, which predominates in Bangladesh, providing critical epidemiological insights into the evolution and spread of drug-resistant typhoid (Carey et al., 2023). Further the GenoTyphi framework has proven effective in managing outbreaks of drug-resistant S. Typhi. For instance, the XDR S. Typhi outbreak in Pakistan, attributed to subclade 4.3.1.1.P1, was tracked using this scheme (Klemm et al., 2018). Similarly, ceftriaxone-resistant S. Typhi outbreaks in India, including in Mumbai (subclade 4.3.1.2.1) and Vadodara (4.3.1.2.2) were effectively tracked and characterized (Jacob et al., 2021; Thirumoorthy et al., 2025; Argimón et al., 2022).
Recently, sporadic reports have highlighted the emergence of carbapenem-resistant S. Typhi (CRST) in Pakistan, raising serious concerns about the evolving landscape of antimicrobial resistance (AMR) in this pathogen (Ain et al., 2022; Nizamuddin et al., 2023). More alarmingly, similar cases have been reported in various cities across southern and western India, indicating a potential regional spread of this resistance profile in a region where typhoid fever remains endemic. Despite these developments, the genomic epidemiology and evolutionary trajectory of CRST strains in India remain poorly understood. In this study, we aimed to investigate the genomic characteristics and evolutionary dynamics of CRST isolates in India, with particular focus on their rising prevalence and the role of plasmids in mediating resistance (Tharani Priya et al., 2025). A deeper understanding of these factors is critical to inform public health strategies and guide targeted interventions to curb the spread of CRST in endemic settings.
Methods
Study settings
A total of 32 Salmonella Typhi isolates were obtained between August 2024 and May 2025 from enteric fever patients treated at hospitals in various cities in South and West India and from individuals with recent travel exposure to these regions. Among the isolates, sixteen originated from St. John’s Medical College Hospital, Bengaluru, five from Fortis Hospital, Bengaluru, four from Indira Gandhi Institute of Child Health, Bengaluru, two from Baby Memorial Hospital, Kozhikode and one each from Agilus Diagnostics Lab, Mumbai, KIMS Health Hospital, Trivandrum, P. D. Hinduja Hospital and Medical Research Centre, Mumbai, Christian Medical College, Chittoor campus, Andhra Pradesh and Sahyadri Speciality Labs, Pune, Maharashtra. Participating institutions flagged these isolates due to atypical antimicrobial resistance profiles detected via phenotypic susceptibility testing and/or automated VITEK 2 systems. Detailed patient information, including clinical affiliations and travel histories, is provided in Supplementary Table S1. For in-depth analysis of resistance mechanisms, all isolates were transferred to the Department of Clinical Microbiology at Christian Medical College (CMC), Vellore, for whole-genome sequencing and genomic characterization.
Bacterial isolates and phenotypic testing
The isolates, once received at CMC Vellore, was cultured on blood and MacConkey agar plates to ensure purity. The isolates were confirmed S. Typhi by conventional biochemical tests, serotyping (Kauffmann–White scheme), and qPCR (Nair et al., 2019). Antimicrobial susceptibility (AST) of the study isolates were evaluated by Kirby–Bauer disk diffusion technique, with inhibition zone measurements and interpretations adhering to the Clinical and Laboratory Standards Institute criteria (Clinical and Laboratory Standards Institute, 2024). The tested antibiotics included ampicillin (10 μg), chloramphenicol (30 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), ciprofloxacin (5 μg), pefloxacin (5 μg), ceftriaxone (30 μg), cefixime (5 μg), azithromycin (15 μg), meropenem (10 μg), and ertapenem (10 μg). To complement these findings, the broth microdilution (BMD) method was employed to assess the minimum inhibitory concentrations (MICs) of additional antibiotics, namely cefepime, aztreonam, piperacillin-tazobactam (fixed 4 μg/mL), ceftazidime-avibactam (fixed 4 μg/mL), aztreonam-avibactam (fixed 4 μg/mL), and colistin. Colistin MIC was interpreted according to EUCAST guidelines (The European Committee on Antimicrobial Susceptibility Testing, 2024).
DNA extraction and whole genome sequencing (WGS)
Genomic DNA was isolated from samples using the QIAamp® Mini Kit (250) (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. DNA purity and concentration were quantified using a Nanodrop One spectrophotometer (Thermo Fisher Scientific, Waltham, United States) and a Qubit Fluorometer with the dsDNA HS Assay Kit (Life Technologies, Carlsbad, United States). The presence of carbapenemase and ESBL genes were identified by multiplex PCR with an in-house gene panel, adapted from the methodology described by Poirel et al. (2011).
For short-read sequencing, DNA was fragmented, and paired-end libraries were prepared using the Illumina Nextera DNA Flex Library Kit and Nextera DNA CD Indexes (Illumina, Massachusetts, United States). Equimolar library pools were sequenced on the Illumina NovaSeq 6000 platform (available at Unipath Specialty Laboratory Limited, Ahmedabad, India), generating 2 × 150 bp paired-end reads. All steps, including tagmentation, library amplification, and purification, were performed as specified by the manufacturer.
To complement the short-read data, long-read sequencing was performed on two isolates using Oxford Nanopore Technology (ONT). For each sample, approximately 200 ng of DNA was processed with the Nanopore Rapid Barcoding Kit 96 V14 (SQK-RBK114.96; Oxford Nanopore Technologies, Oxford, United Kingdom) following the manufacturer’s protocol. Sequencing was carried out on a PromethION P2 Solo platform with real-time base-calling enabled during the run. Basecalling of POD5 files was executed using Dorado v0.8.3 (Oxford Nanopore Technologies) with the dna_r10.4.1_e8.2_400bps_sup@v5.0.0 model to ensure high-accuracy sequence reconstruction.
Quality control, assembly and annotation
Quality assessment of Illumina reads was performed using FastQC v0.12.12 followed by adapter and index trimming with Trimmomatic v0.39 (Bolger et al., 2014). Contaminant screening and filtering were executed with Kraken v1.1.1 (Wood et al., 2019),3 followed by sequence coverage analysis. High-quality reads (Phred score >30) were assembled into draft genomes using SKESA (Souvorov et al., 2018).4 For hybrid assembly, the Hybracter pipeline v0.8.0 (Bouras et al., 2024)5 was employed to process Oxford Nanopore (ONT) long reads. This workflow first improved the read quality with Filtlong, assembled long reads de novo using Flye, and polished the assemblies iteratively with Medaka. Further refinement was achieved by polishing Illumina short reads via Polypolish and PyPolca. Final genome completeness and accuracy were evaluated using QUAST v5.2.0 (Gurevich et al., 2013). Genome annotations were performed using Bakta v1.10.1 (Schwengers et al., 2021).6 Unless specified, default parameters were applied throughout all analytical steps.
Comparative genome analysis
Draft genome assemblies were analyzed with SeqSero v2.0 (Zhang et al., 2019)7 to verify the antigenic composition of the serotype. In silico multilocus sequence typing (MLST) (Larsen et al., 2012) was performed on all isolates using the pipeline provided by the Center for Genomic Epidemiology.8 Antimicrobial resistance (AMR) genes were identified using NCBI AMRFinderPlus v4.0.3 (Feldgarden et al., 2021).9 Plasmid content was determined by querying genome sequences against the PlasmidFinder database (Carattoli et al., 2014).10 Plasmids were compared using the Basic Local Alignment Search Tool (BLAST), and circular maps were prepared using the Proksee server11 (Grant et al., 2023).
Genotyping and phylogeny
The isolates were assigned to previously defined genotypes using the GenoTyphi pipeline (available at: https://github.com/katholt/genotyphi). Unique single nucleotide polymorphisms (SNPs) characterizing the novel sub-lineage were and subsequently incorporated into the GenoTyphi framework to track the outbreak in future investigations.
For phylogenetic analysis, genome assemblies of S. Typhi (n = 412) representing all major genotypes were obtained from the curated global collection provided by the Global Typhoid Genomics Consortium12 (Carey et al., 2023), with data sourced from NCBI (Supplementary Table S2). The sequencing reads were aligned to the reference genome of S. Typhi CT18 (GenBank: AL513382.1) using Snippy v4.6.0.13 The resulting full alignment was first filtered to exclude SNPs located within the 354 kb of repetitive regions in the S. Typhi CT18 reference chromosome, using previously established coordinates (Wong et al., 2015) processed using Gubbins v3.3.3 (Croucher et al., 2015). After masking these regions, the alignment was processed with Gubbins14 to remove recombination sites (Wood et al., 2019). SNPs were extracted using SNP-sites v2.5.115 (Page et al., 2016). A maximum-likelihood phylogeny was then reconstructed from the filtered alignment using the TVM + F + ASC + G4 model with 1,000 bootstraps using IQ-TREE v.2.4.0 (Nguyen et al., 2015) integrated with ModelFinder (Kalyaanamoorthy et al., 2017). The resulting phylogenetic tree was visualized and annotated using the Interactive Tree of Life software (iTOL v.5) (Letunic and Bork, 2021).
Results
Identification of CRST isolates and resistance profile
A total of 32 non-duplicated S. Typhi isolates were included in this study, collected from patients diagnosed with typhoid fever across multiple healthcare institutions in southern and western India, between 2024 and 2025. All isolates were confirmed as S. Typhi through standard biochemical tests, conventional serotyping methods, and qPCR. AST by disk diffusion revealed a consistent resistance profile across all Salmonella Typhi isolates except one, with resistance to ampicillin, ciprofloxacin, ceftriaxone, and carbapenems. One isolate (ERR15187853) was resistant to ampicillin, ciprofloxacin, and ceftriaxone but susceptible to carbapenems. Conversely, all isolates remained susceptible to chloramphenicol, trimethoprim-sulfamethoxazole, and azithromycin.
MIC testing confirmed high-level resistance to ciprofloxacin, ceftriaxone, ertapenem, and meropenem, with retained susceptibility to chloramphenicol, trimethoprim-sulfamethoxazole, azithromycin, and aztreonam-avibactam. MIC values for all antibiotics tested, including β-lactam/β-lactamase inhibitor combinations and colistin, are provided in Table 1. The multiplex PCR analysis confirmed the presence of both blaNDM and blaCTX-M genes in 31 CRST isolates, while the remaining isolate carried blaCTX-M alone. This indicates the predominance of co-occurring carbapenemase (blaNDM) and extended-spectrum β-lactamase (blaCTX-M) genes among CRST strains.
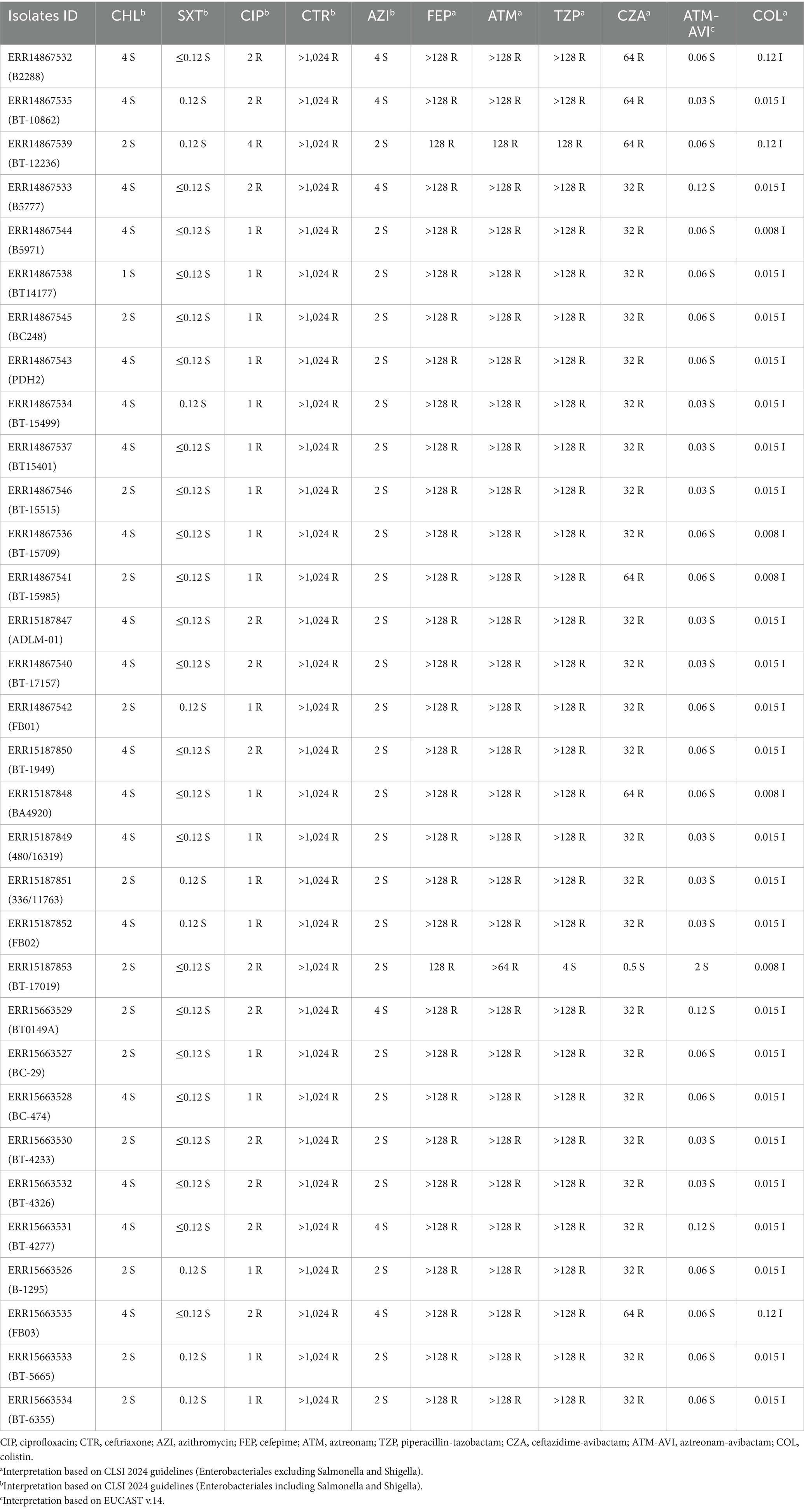
Table 1. Antimicrobial susceptibility profile and minimum inhibitory concentration (MIC) in μg/mL of different antibiotics against S. Typhi isolates.
Genotyping and comparative genome analysis
The S. Typhi genomes (n = 32) were characterized using the GenoTyphi genotyping scheme (Wong et al., 2016; Dyson and Holt, 2021). All study isolates were identified as belonging to the H58 haplotype (genotype 4.3.1), specifically falling within the 4.3.1.1 genotype (H58 Lineage I) (Supplementary Figure S1). Based on their shared genomic features and epidemiological significance related to carbapenem resistance, these isolates have been assigned to a novel sub-genotype within 4.3.1.1, designated as 4.3.1.1.1.
AMR gene profiling identified the presence of blaNDM-5, blaCTX-M-15, qnrS, and tetA among the isolates. In addition, resistance-associated point mutation analysis revealed an S83Y substitution in gyrA within the quinolone resistance-determining region (QRDR). The presence of blaNDM-5 correlates with resistance to carbapenems, blaCTX-M-15 confers resistance to third-generation cephalosporins (3GCs), tetA is linked to tetracycline resistance, while both the S83Y mutation in gyrA and qnrS contribute to fluoroquinolone resistance. Among the two plasmids identified IncFIB(K) carried blaCTX-M-15, qnrS, and tetA, while blaNDM-5 was harbored by IncX3 plasmid.
Population structure of CRST isolates from India
A core genome SNP-based phylogenetic analysis, incorporating the 31 CRST study isolates and 311 global reference isolates, revealed that the CRST isolates formed a distinct subclade within the H58 lineage I (genotype 4.3.1.1) (Figure 1). This CRST subclade was distinguished from its parent clade by seven unique SNPs, underscoring its genetic distinctiveness (Supplementary Table S3). Notably, the CRST isolates exhibited the closest genetic relatedness (10 SNP difference) to a previously sequenced cluster of six S. Typhi isolates from India, identified through the Surveillance for Enteric Fever in India (SEFI) study. These six isolates were collected from two geographic locations Anantapur, Andhra Pradesh (ERR4790795, ERR5200930, ERR4790761, ERR5201325, ERR5201273) and Bengaluru, Karnataka (ERR5200874). All isolates were sequenced between 2018 and 2020 as part of the SEFI initiative (Supplementary Figure S2). Isolates closely related to the CRST clone but lacking resistance plasmids have been circulating in India since at least 2018, indicating a pre-existing susceptible lineage that subsequently acquired plasmid-mediated resistance determinants. This finding suggests that the CRST clone likely evolved locally from these endemic lineages through the recent acquisition of resistance plasmids. Furthermore, the CRST isolates sequenced in this study were phylogenetically distinct from both the previously reported CRST isolate from Pakistan (SRR22801806; 35 SNPs difference) and the ceftriaxone-resistant outbreak isolates recently identified in Gujarat, India (31 SNPs difference).
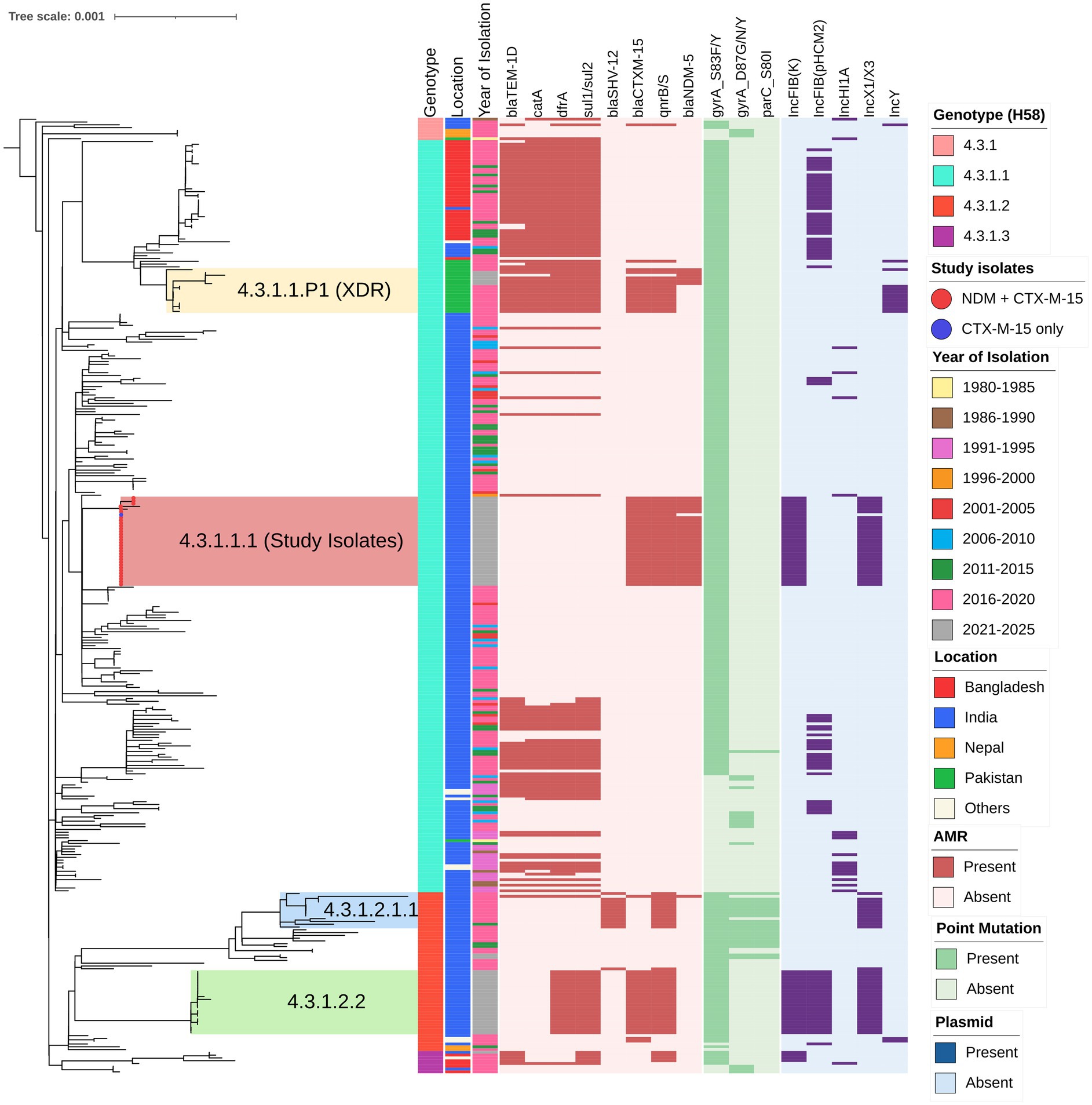
Figure 1. Phylogenetic relationship and genetic characteristics of 32 S. Typhi strains from India. A maximum likelihood phylogenetic tree was constructed using core genome SNPs from the study isolates (marked with red circles at the branch tips) along with 311 global H58 isolates, revealing an overall SNP difference of 1,149 among the H58 isolates. The tree is rooted against outgroup isolate belonging to genotype 4.3.1 (ERR1764570) and inferred using IQ-TREE2 (http://www.iqtree.org/), with bootstrap support values calculated from 1,000 replicates. Previously reported high-risk clones of cephalosporin-resistant clusters in the tree, including 4.3.1.2.1.1 (blue), 4.3.1.1.P1 (yellow), and 4.3.1.2.2 (green). The color-coded metadata strips adjacent to the tree represent: strip 1 denotes genotype, strip 2 indicates the country location, and strip 3 shows the year of isolation of each isolate. The heatmap alongside the tree depicts the distribution of antimicrobial resistance (AMR) genes, point mutations, and plasmid replicons, which are predominantly concentrated within the H58 cluster. The scale bar indicates substitutions per site. The tree was visualized and annotated using iTOL (https://itol.embl.de/).
Characterization of plasmids
All CRST isolates carried AMR genes located on both IncFIB(K) and IncX3 plasmids. AMR gene analysis revealed that the ~73 kb IncFIB(K) plasmid harbored blaCTX-M-15, qnrS1, and tet(A), while the ~47 kb IncX3 plasmid carried blaNDM-5 gene. To investigate the origin of these plasmids, complete circular sequences of IncFIB(K) and IncX3 plasmids from the study isolates were BLAST-compared with previously reported plasmids in the NCBI database. The IncFIB(K) plasmid (Accession No. CP189855) showed 100% sequence identity to an E. coli IncFIB(K) plasmid (CP116920), as well as to plasmids previously reported in ceftriaxone-resistant S. Typhi isolates from Gujarat, India (CP173298 and CP168964). However, the sequence coverage was 98% for E. coli and 93% for S. Typhi, indicating that while highly similar, the plasmids are not identical. Consistent with previous reports (Thirumoorthy et al., 2025), blaCTX-M-15 gene in the IncFIB(K) plasmid was located downstream of an IS1380 family ISEcp1 element, suggesting a potential association with its mobilization (Figure 2A).
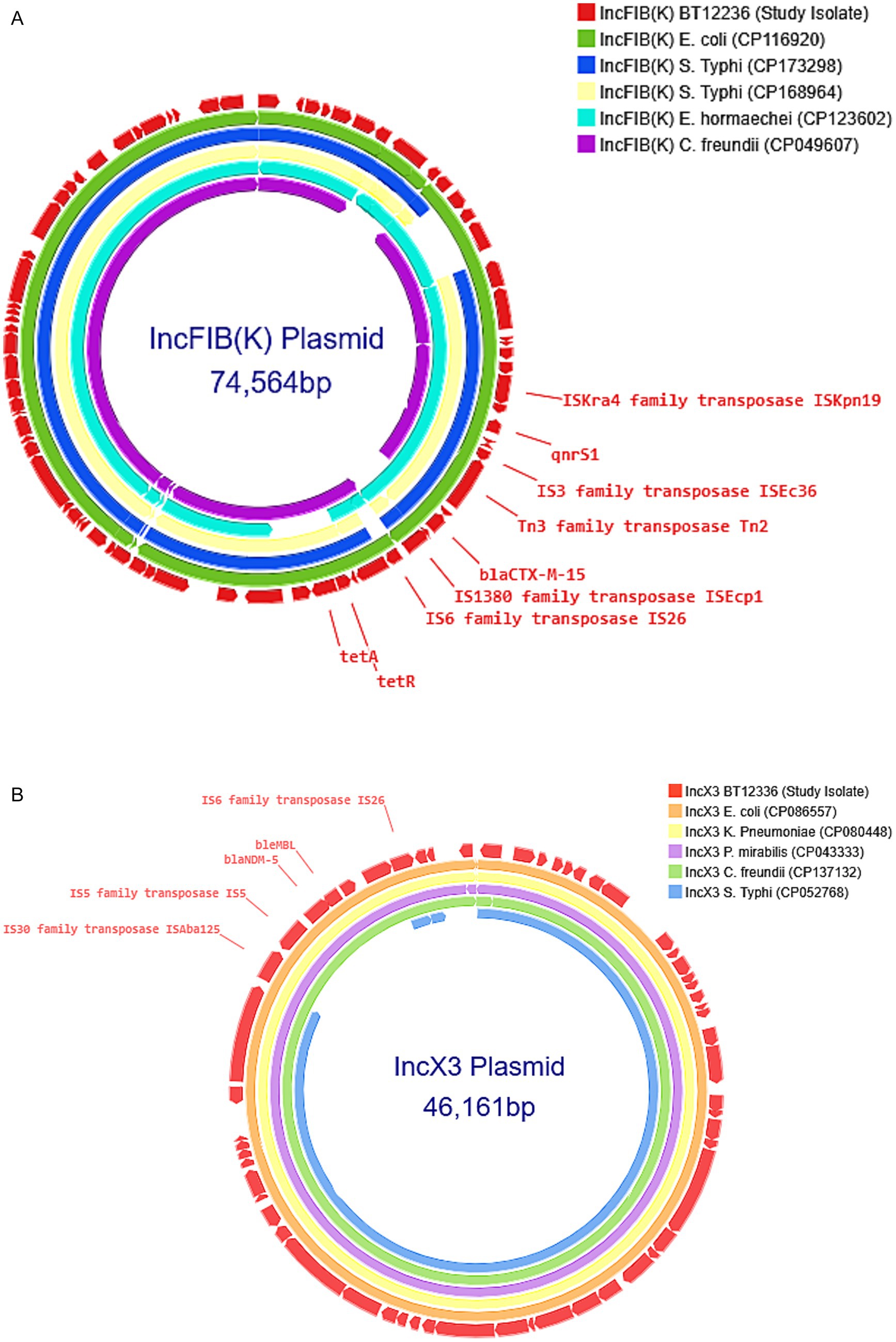
Figure 2. (A) Comparison of IncFIB(K) plasmid: The IncFIB(K) plasmid from the carbapenem-resistant Salmonella Typhi (CRST) isolate was BLAST-searched against the NCBI BLASTn database and compared with Enterobacteriales carrying plasmids of the same incompatibility group [(CP116920), (CP173298), (CP168964), (CP123602), (CP049607)] with 90–100% identity. The selected plasmids were then annotated and visualized using Proksee. (B) Comparison of IncX3 plasmid: The IncX3 plasmid from the carbapenem-resistant Salmonella Typhi (CRST) isolate was BLAST-searched against the NCBI BLASTn database and compared with Enterobacteriales carrying plasmids of the same incompatibility group [(CP086557), (CP080448), (CP043333), (CP137132), (CP052768)] with 90–100% identity. The selected plasmids were then annotated and visualized using Proksee.
A similar BLAST analysis of the IncX3 plasmid (Accession No. CP189856) carrying blaNDM-5 revealed 100% sequence identity and query coverage with IncX3 plasmids previously reported in E. coli (CP086557) and K. pneumoniae (CP080448). Notably, the IncX3 plasmid from this study showed only 83% query coverage with the SHV-carrying IncX3 plasmid (CP052768) identified from the S. Typhi outbreak in Mumbai, suggesting it is genetically distinct. The blaNDM-5 was located within the characteristic genetic structure ISAba125-IS5-blaNDM–5-bleMBL-trpF-dsbC-IS26 found in IncX3 type plasmids (Figure 2B).
Discussion
The emergence of CRST signifies a critical evolutionary escalation, building on the shift from MDR to XDR strains observed in Pakistan by 2016 (Klemm et al., 2018). Before 2016, resistance in S. Typhi was primarily limited to first-line antibiotics and fluoroquinolones (FQNS), with the H58 lineage dominating across South Asia and Africa (Carey et al., 2024). Within this lineage, subclades 4.3.1.1 (MDR) and 4.3.1.2 (FQNS) were distinguishable via GenoTyphi genotyping (Wong et al., 2016). Subsequent emergence of ceftriaxone-resistant and XDR S. Typhi in India and Pakistan, respectively, was further mapped through finer subclade resolution (Dyson and Holt, 2021; Carey et al., 2023). For example, the Sindh XDR outbreak (4.3.1.1-P1) diverged by six SNPs from its nearest contemporaries (Klemm et al., 2018), while the Vadodara outbreak in India (4.3.1.2.1) exhibited 21 SNPs from its closest relative (Thirumoorthy et al., 2025). Notably, the CRST isolates in this study have accumulated seven unique chromosomal mutations (Supplementary Table S3) while acquiring a blaNDM-5 harboring plasmid. Given the low mutation rate of S. Typhi (0.63 SNPs per genome per year; Wong et al., 2016), this shift appears to be a plasmid-driven phenotypic leap rather than gradual chromosomal adaptation, underscoring the role of horizontal gene transfer under selective antibiotic pressure (Rodríguez-Beltrán et al., 2021). These findings highlight unregulated antibiotic use in South Asia as a key driver of resistance evolution, emphasizing the urgent need for antimicrobial stewardship interventions.
The acquisition of diverse plasmids has played a pivotal role in the evolution of antimicrobial resistance in S. Typhi, driving the transition from MDR strains to emerging CRST variants. MDR S. Typhi strains harbored IncHI1 pST6 plasmids, almost exclusively within the H58 lineage, which became the dominant clade in South Asia and Africa (Holt et al., 2011). Resistance escalated further with the acquisition of blaCTX-M-15-carrying IncY plasmids, likely originating from E. coli, which facilitated the emergence of third-generation cephalosporin-resistant strains (Klemm et al., 2018). Subsequent ceftriaxone-resistant S. Typhi outbreaks in Mumbai and Vadodara, India, were linked to the horizontal acquisition of distinct plasmids, specifically IncX3 and IncFIB(K), respectively, likely sourced from co-circulating Enterobacteriaceae (Jacob et al., 2021; Thirumoorthy et al., 2025). This pattern indicates that S. Typhi has repeatedly leveraged plasmid pools from E. coli and Klebsiella spp., allowing it to bypass previously effective antibiotic therapies. The CRST isolates in this study represent the next stage in this evolutionary trajectory, having acquired an IncX3 plasmid harboring blaNDM-5 and an IncFIB(K) plasmid carrying blaCTX-M-15, both likely derived from Enterobacteriaceae (Figure 2). Notably, the IncFIB(K) plasmid in the Vadodara outbreak differs from that found in CRST isolates, suggesting independent acquisition events rather than clonal dissemination. Similarly, the first reported carbapenem-resistant S. Typhi case in Peshawar, Pakistan (July 2022), carried blaNDM-5 on an IncN plasmid, again resembling plasmids found in other Enterobacteriaceae (Nizamuddin et al., 2023). The presence of multiple plasmid types (IncX3 and IncN) conferring carbapenem resistance suggests that CRST has arisen independently across different settings, rather than spreading from a single source. These findings highlight a rapidly evolving resistance landscape, where S. Typhi continues to integrate plasmid-borne resistance genes from Enterobacteriaceae, facilitating stepwise antibiotic resistance escalation.
In South Asia, MDR and FQNS S. Typhi infections are primarily treated with oral cefixime or azithromycin for outpatient cases, while intravenous ceftriaxone and azithromycin are used in combination for severe infections (Dolecek et al., 2019; Kuehn et al., 2022). For XDR or ceftriaxone resistant strains azithromycin remains the primary oral therapy for uncomplicated typhoid fever, whereas intravenous meropenem and azithromycin are recommended for severe cases (Qureshi et al., 2020; Parry et al., 2023). However, the emergence of CRST in India, co-producing blaNDM and blaCTX-M enzymes, renders both ceftriaxone and meropenem ineffective (Park et al., 2024). If CRST isolates remain susceptible to azithromycin (MIC ≤ 16 μg/mL), azithromycin remains the preferred oral treatment, leveraging its intracellular efficacy. Alarmingly, the first report of CRST from Peshawar, Pakistan, identified co-carriage of the mphA gene, conferring phenotypic resistance to azithromycin (Nizamuddin et al., 2023). In such cases, ceftazidime-avibactam + aztreonam combination therapy is recommended for severe infections (Tamma et al., 2021). Where aztreonam-avibactam is unavailable, colistin, fosfomycin, or tigecycline monotherapy may serve as alternative options (Parry et al., 2019). Given the limited clinical evidence for treating CRST, treatment decisions should be guided by individual patient factors, local resistance patterns, and expert consultation.
The rapid evolution of S. Typhi resistance, culminating in CRST, signals an urgent public health crisis in South Asia, where the failure of ceftriaxone and meropenem leaves severely limited treatment options for typhoid fever (Nabarro et al., 2022). Given the increasing ineffectiveness of antibiotics, preventive strategies must take priority to reduce disease burden and slow resistance evolution. One of the most effective interventions is the widespread adoption of the typhoid conjugate vaccine (TCV), which successfully curbed the Sindh XDR outbreak in Pakistan by reducing case numbers and lowering antibiotic selective pressure (Nampota-Nkomba et al., 2023; Qamar et al., 2024). Expanding TCV coverage across India and South Asia is critical to prevent CRST from becoming endemic (Mogasale et al., 2024). Beyond vaccination, strengthening water, sanitation, and hygiene (WASH) infrastructure is essential, as poor sanitation fuels S. Typhi’s fecal-oral transmission, sustaining high infection rates and increasing exposure to resistant strains (Luby et al., 2018). Enhanced genomic surveillance is also crucial for tracking CRST’s plasmid-driven spread and emerging resistance mutations—particularly acrB-R717Q/L mutations and mphA-mediated azithromycin resistance, as seen in Peshawar’s first CRST case (Nizamuddin et al., 2023). Finally, urgent antibiotic stewardship reforms are needed to curb the unregulated use of broad-spectrum antibiotics, a key driver of resistance escalation. Preserving azithromycin’s efficacy and ensuring restricted use of last-resort drugs like aztreonam-avibactam will be essential to maintaining effective treatment options for future cases.
In conclusion, the emergence of high-risk S. Typhi clones, particularly those harboring carbapenem resistance, underscores the urgent need for a comprehensive, multi-pronged strategy to contain their spread. Robust genomic surveillance is critical for tracking resistance trends, deciphering genetic evolution, and understanding transmission dynamics. Strengthening national antibiotic stewardship policies can help curb selective pressure and slow resistance escalation. Additionally, widespread typhoid conjugate vaccine (TCV) deployment can significantly reduce disease burden and limit antibiotic exposure. A coordinated global response, integrating surveillance, stewardship, and vaccination, is essential to mitigate the growing threat posed by carbapenem-resistant S. Typhi.
Author’s note
This publication is part of the Ph.D. thesis of The Tamil Nadu Dr. M.G.R. Medical University.
Data availability statement
The datasets presented in this study are available in online repositories. They can be retrieved under the repository name “Genomic Insights into the Emergence of Carbapenem-Resistant Salmonella Typhi harboring blaNDM-5 in Bengaluru, India” with the accession number PRJEB88734, and the plasmid sequences are available under the BioProject number PRJNA1212325.
Ethics statement
This study was approved by the Institutional Review Board (IRB) of Christian Medical College, Vellore (IRB Min No.10393 dated 30.11.2016 & IRB Min No.15247 dated 22.03.2023).
Author contributions
TT: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft. JJa: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft. MT: Investigation, Project administration, Data curation, Writing – review & editing. SMah: Investigation, Project administration, Writing – review & editing. BJ: Investigation, Project administration, Writing – review & editing. SMan: Investigation, Project administration, Writing – review & editing. SN: Investigation, Project administration, Writing – review & editing. JSa: Investigation, Project administration, Writing – review & editing. PP: Investigation, Project administration, Writing – review & editing. JSu: Investigation, Project administration, Writing – review & editing. AN: Investigation, Project administration, Writing – review & editing. SV: Investigation, Project administration, Writing – review & editing. RG: Investigation, Project administration, Writing – review & editing. DJ: Investigation, Project administration, Writing – review & editing. VN: Investigation, Project administration, Writing – review & editing. CR: Investigation, Project administration, Writing – review & editing. PN: Investigation, Data curation, Methodology, Writing – review & editing. AV: Project administration, Data curation, Investigation, Writing – review & editing. KS: Investigation, Data curation, Methodology, Writing – review & editing. JJo: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Validation. KW: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. BV: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Validation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The Bill & Melinda Gates Foundation (Investment ID INV-009497 OPP1159351) supported this study’s phenotypic work under the project “National Surveillance System for Enteric Fever in India.” BV and JJ received support from the same grant. The genomic sequencing component was funded by the Indian Council of Medical Research (ICMR) through the Expression of Interest titled “To establish ICMR’s Genomic Surveillance for Antimicrobial Resistance” (Grant ID: AMR/DX/TYP/3/2024-CD).”
Acknowledgments
We gratefully acknowledge Drs. Duncan Steele and Supriya Kumar from the Bill & Melinda Gates Foundation for their technical guidance throughout the study, provided on behalf of the SEFI consortium. We extend our appreciation to all the members of the SEFI reference laboratory team at CMC Vellore for their dedicated efforts, with special thanks to Ms. P. Agila Kumari, Ms. S. Baby Abirami, Mr. N. Ayyanraj, Ms. Sowmya Murugan, Ms. Yamini Umashankar, and Mr. Praveen Thilagan for their contributions to phenotypic testing and maintenance of stock cultures. We sincerely thank Dr. Anton Spadar (London School of Hygiene & Tropical Medicine, United Kingdom) for his significant contribution in updating the GenoTyphi genotyping scheme, enabling the assignment of new genotypes in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1685068/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Phylogenetic relationship of 31 Carbapenem-resistant S. Typhi isolates from India against 412 global S. Typhi strains. The maximum likelihood phylogenetic tree was constructed based on single nucleotide polymorphisms (SNPs) and mapped against the Salmonella Typhi CT18 reference strain, with an overall SNP count of 6,780. The genotypes of S. Typhi strains are represented as gradient-colored strips, while the study isolates are highlighted as red dots.
SUPPLEMENTARY FIGURE S2 | Phylogenetic tree depicting the CRST subclade and its closely related isolates (n = 70) This figure illustrates the phylogenetic relationships among the CRST subclade and closely related Salmonella Typhi isolates sequenced between 2018 and 2020 as part of the SEFI initiative to enhance clarity and visualization of the genomic relationships.
Footnotes
1. ^https://www.cdc.gov/mmwr/volumes/72/wr/mm7207a2.htm#:~:text=In%202019%2C%20an%20updated%20modeling%20study%20estimated,Eastern%20Mediterranean%20(187)%2C%20and%20African%20(111)%20regions
12. ^https://bridges.monash.edu/articles/dataset/Global_Typhoid_Genomics_Consortium_2022_-_Genome_Assemblies/21431883
References
Ain, Q., Tahir, M., Sadaqat, A., Ayub, A., Awan, A. B., Wajid, M., et al. (2022). First detection of extensively drug-resistant Salmonella Typhi isolates harboring VIM and GES genes for carbapenem resistance from Faisalabad, Pakistan. Microb. Drug Resist. 28, 1087–1098. doi: 10.1089/mdr.2022.0094
Akram, J., Khan, A. S., Khan, H. A., Gilani, S. A., Akram, S. J., Ahmad, F. J., et al. (2020). Extensively drug-resistant (XDR) typhoid: evolution, prevention, and its management. Biomed. Res. Int. 2020:6432580. doi: 10.1155/2020/6432580
Argimón, S., Nagaraj, G., Shamanna, V., Sravani, D., Vasanth, A. K., Prasanna, A., et al. (2022). Circulation of third-generation cephalosporin resistant Salmonella Typhi in Mumbai, India. Clin. Infect. Dis. 74, 2234–2237. doi: 10.1093/cid/ciab897
Baker, S., Thomson, N., Weill, F. X., and Holt, K. E. (2018). Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 360, 733–738. doi: 10.1126/science.aar3777
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouras, G., Houtak, G., Wick, R. R., Mallawaarachchi, V., Roach, M. J., Papudeshi, B., et al. (2024). Hybracter: enabling scalable, automated, complete and accurate bacterial genome assemblies. Microb. Genom. 10:001244. doi: 10.1099/mgen.0.001244
Browne, A. J., Chipeta, M. G., Fell, F. J., Haines-Woodhouse, G., Kashef Hamadani, B. H., Kumaran, E. A. P., et al. (2024). Estimating the subnational prevalence of antimicrobial resistant Salmonella enterica serovars Typhi and Paratyphi A infections in 75 endemic countries, 1990–2019: a modelling study. Lancet Glob. Health 12, e406–e418. doi: 10.1016/S2214-109X(23)00585-5
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carey, M. E., Dyson, Z. A., Ingle, D. J., Amir, A., Aworh, M. K., Chattaway, M. A., et al. (2023). Global diversity and antimicrobial resistance of typhoid fever pathogens: insights from a meta-analysis of 13,000 Salmonella Typhi genomes. eLife 12:e85867. doi: 10.7554/eLife.85867
Carey, M. E., Jain, R., Yousuf, M., Maes, M., Dyson, Z. A., Thu, T. N. H., et al. (2021). Spontaneous emergence of azithromycin resistance in independent lineages of Salmonella Typhi in northern India. Clin. Infect. Dis. 72, e120–e127. doi: 10.1093/cid/ciaa1773
Carey, M. E., Thi Nguyen, T. N., Tran, D. H., Tran, D. H. N., Dyson, Z. A., Keane, J. A., et al. (2024). The origins of haplotype 58 (H58) Salmonella enterica serovar Typhi. Commun. Biol. 7:775. doi: 10.1038/s42003-024-06451-8
Clinical and Laboratory Standards Institute (2024). Performance Standards for Antimicrobial Susceptibility Testing Guideline M100. 34th Edn. Malvern, PA, USA: Clinical and Laboratory Standards Institute.
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Crump, J. A. (2019). Progress in typhoid fever epidemiology. Clin. Infect. Dis. 68, S4–S9. doi: 10.1093/cid/ciy846
Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A., and Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
Dolecek, C., Pokharel, S., Basnyat, B., and Olliaro, P. (2019). “Antibiotics for typhoid fever” in Selection and Use of Essential Medicines. WHO Technical Report Series (Geneva: World Health Organization), 19–26.
Dyson, Z. A., and Holt, K. E. (2021). Five years of GenoTyphi: updates to the global Salmonella Typhi genotyping framework. J. Infect. Dis. 224, S775–S780. doi: 10.1093/infdis/jiab414
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Grant, J. R., Enns, E., Marinier, E., Mandal, A., Herman, E. K., Chen, C. Y., et al. (2023). Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492. doi: 10.1093/nar/gkad326
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Holt, K. E., Phan, M. D., Baker, S., Duy, P. T., Nga, T. V. T., Nair, S., et al. (2011). Emergence of a globally dominant IncHI1 plasmid type associated with multiple drug resistant typhoid. PLoS Negl. Trop. Dis. 5:e1245. doi: 10.1371/journal.pntd.0001245
Jacob, J. J., Pragasam, A. K., Vasudevan, K., Veeraraghavan, B., Kang, G., John, J., et al. (2021). Salmonella Typhi acquires diverse plasmids from other Enterobacteriaceae to develop cephalosporin resistance. Genomics 113, 2171–2176. doi: 10.1016/j.ygeno.2021.05.003
John, J., Bavdekar, A., Rongsen-Chandola, T., Dutta, S., Gupta, M., Kanungo, S., et al. (2023). Burden of typhoid and paratyphoid fever in India. N. Engl. J. Med. 388, 1491–1500. doi: 10.1056/NEJMoa2209449
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., Von Haeseler, A., and Jermiin, L. S. (2017). Modelfinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kirchhelle, C., Dyson, Z. A., and Dougan, G. (2019). A biohistorical perspective of typhoid and antimicrobial resistance. Clin. Infect. Dis. 69, S388–S394. doi: 10.1093/cid/ciz556
Klemm, E. J., Shakoor, S., Page, A. J., Qamar, F. N., Judge, K., Saeed, D. K., et al. (2018). Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio 9, 10–128. doi: 10.1128/mBio.00105-18
Kuehn, R., Stoesser, N., Eyre, D., Darton, T. C., Basnyat, B., and Parry, C. M. (2022). Treatment of enteric fever (typhoid and paratyphoid fever) with cephalosporins. Cochrane Database Syst. Rev. 11:CD010452. doi: 10.1002/14651858.CD010452.pub2
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Luby, S. P., Rahman, M., Arnold, B. F., Unicomb, L., Ashraf, S., Winch, P. J., et al. (2018). Effects of water quality, sanitation, handwashing, and nutritional interventions on diarrhoea and child growth in rural Bangladesh: a cluster randomised controlled trial. Lancet Glob. Health 6, e302–e315. doi: 10.1016/S2214-109X(17)30490-4
Marchello, C. S., Carr, S. D., and Crump, J. A. (2020). A systematic review on antimicrobial resistance among Salmonella Typhi worldwide. Am. J. Trop. Med. Hyg. 103, 2518–2527. doi: 10.4269/ajtmh.20-0258
Mogasale, V. V., Sinha, A., John, J., Hasan Farooqui, H., Ray, A., Chantler, T., et al. (2024). Typhoid conjugate vaccine implementation in India: a review of supportive evidence. Vaccine X 21:100568. doi: 10.1016/j.jvacx.2024.100568
Nabarro, L. E., McCann, N., Herdman, M. T., Dugan, C., Ladhani, S., Patel, D., et al. (2022). British Infection Association guidelines for the diagnosis and management of enteric fever in England. J. Infect. 84, 469–489. doi: 10.1016/j.jinf.2022.01.014
Nair, S., Patel, V., Hickey, T., Maguire, C., Greig, D. R., Lee, W., et al. (2019). Real-time PCR assay for differentiation of typhoidal and nontyphoidal Salmonella. J. Clin. Microbiol. 57, 10–128. doi: 10.1128/JCM.00167-19
Nampota-Nkomba, N., Carey, M. E., Jamka, L. P., Fecteau, N., and Neuzil, K. M. (2023). Using typhoid conjugate vaccines to prevent disease, promote health equity, and counter drug-resistant typhoid fever. Open Forum Infect. Dis. 10, S6–S12. doi: 10.1093/ofid/ofad022
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nizamuddin, S., Khan, E. A., Chattaway, M. A., and Godbole, G. (2023). Case of carbapenem-resistant Salmonella Typhi infection, Pakistan, 2022. Emerg. Infect. Dis. 29, 2395–2397. doi: 10.3201/eid2911.230499
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2:e000056. doi: 10.1099/mgen.0.000056
Park, S. Y., Baek, Y. J., Kim, J. H., Seong, H., Kim, B., Kim, Y. C., et al. (2024). Guidelines for antibacterial treatment of carbapenem-resistant Enterobacterales infections. Infect. Chemother. 56, 308–328. doi: 10.3947/ic.2024.0038
Parry, C. M., Hien, T. T., Dougan, G., White, N. J., and Farrar, J. J. (2002). Typhoid fever. N. Engl. J. Med. 347, 1770–1782. doi: 10.1056/NEJMra020201
Parry, C. M., Qamar, F. N., Rijal, S., McCann, N., Baker, S., and Basnyat, B. (2023). What should we be recommending for the treatment of enteric fever? Open Forum Infect. Dis. 10, S26–S31. doi: 10.1093/ofid/ofad179
Parry, C. M., Ribeiro, I., Walia, K., Rupali, P., Baker, S., and Basnyat, B. (2019). Multidrug resistant enteric fever in South Asia: unmet medical needs and opportunities. BMJ 364:k5322. doi: 10.1136/bmj.k5322
Piovani, D., Figlioli, G., Nikolopoulos, G. K., and Bonovas, S. (2024). The global burden of enteric fever, 2017–2021: a systematic analysis from the global burden of disease study 2021. EClinicalMedicine 77:102883. doi: 10.1016/j.eclinm.2024.102883
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Pragasam, A. K., Pickard, D., Wong, V., Dougan, G., Kang, G., Thompson, A., et al. (2020). Phylogenetic analysis indicates a longer term presence of the globally distributed H58 haplotype of Salmonella Typhi in southern India. Clin. Infect. Dis. 71, 1856–1863. doi: 10.1093/cid/ciz1112
Qamar, F. N., Qureshi, S., Haq, Z., Yousafzai, T., Qazi, I., Irfan, S., et al. (2024). Longevity of immune response after a single dose of typhoid conjugate vaccine against Salmonella Typhi among children in Hyderabad, Pakistan. Int. J. Infect. Dis. 147:107187. doi: 10.1016/j.ijid.2024.107187
Qureshi, S., Naveed, A. B., Yousafzai, M. T., Ahmad, K., Ansari, S., Lohana, H., et al. (2020). Response of extensively drug resistant Salmonella Typhi to treatment with meropenem and azithromycin, in Pakistan. PLoS Negl. Trop. Dis. 14:e0008682. doi: 10.1371/journal.pntd.0008682
Radhakrishnan, A., Als, D., Mintz, E. D., Crump, J. A., Stanaway, J., Breiman, R. F., et al. (2018). Introductory article on global burden and epidemiology of typhoid fever. Am. J. Trop. Med. Hyg. 99, 4–9. doi: 10.4269/ajtmh.18-0032
Rodríguez-Beltrán, J., DelaFuente, J., León-Sampedro, R., MacLean, R. C., and San Millán, Á. (2021). Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 19, 347–359. doi: 10.1038/s41579-020-00497-1
Schwengers, O., Jelonek, L., Dieckmann, M. A., Beyvers, S., Blom, J., and Goesmann, A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 7:000685. doi: 10.1099/mgen.0.000685
Souvorov, A., Agarwala, R., and Lipman, D. J. (2018). SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19:153. doi: 10.1186/s13059-018-1540-z
Tamma, P. D., Aitken, S. L., Bonomo, R. A., Mathers, A. J., van Duin, D., and Clancy, C. J. (2021). Infectious Diseases Society of America guidance on the treatment of extended-spectrum β-lactamase producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 72, e169–e183. doi: 10.1093/cid/ciaa1478
Tharani Priya, T., Jacob, J. J., Monisha Priya, T., Mahantesh,, Bhavana,, Suhani,, et al. (2025). Genomic insights into the emergence of carbapenem-resistant Salmonella Typhi harboring blaNDM-5 in India. medRxiv. Available online at: https://doi.org/10.1101/2025.05.20.25327816. [Epub ahead of preprint]
The European Committee on Antimicrobial Susceptibility Testing. (2024). Breakpoint tables for interpretation of MICs and Zone diameters, version 14.0. Available online at: http://www.eucast.org/clinical_breakpoints (Accessed June 8. 2024).
Thirumoorthy, T. P., Jacob, J. J., Velmurugan, A., Teekaraman, M. P., Shah, B., Iyer, V., et al. (2025). Recent emergence of cephalosporin-resistant Salmonella Typhi in India due to the endemic clone acquiring IncFIB (K) plasmid encoding blaCTX-M-15 gene. Microbiol. Spectr. 13:e0087524. doi: 10.1128/spectrum.00875-24
Wong, V. K., Baker, S., Connor, T. R., Pickard, D., Page, A. J., Dave, J., et al. (2016). An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat. Commun. 7:12827. doi: 10.1038/ncomms12827
Wong, V. K., Baker, S., Pickard, D. J., Parkhill, J., Page, A. J., Feasey, N. A., et al. (2015). Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter-and intracontinental transmission events. Nat. Genet. 47, 632–639. doi: 10.1038/ng.3281
Wood, D. E., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0
Keywords: Salmonella Typhi, enteric fever, carbapenem-resistant, whole genome sequencing, outbreak investigation
Citation: Thirumoorthy TP, Jacob JJ, Teekaraman MP, Mahantesh S, Jagannatha B, Manasa S, Nagaraj S, Savio J, Padaki PA, Sudarsana J, Nair A, Verma S, Gaikwad R, Joshi D, Nagvekar VC, Rodrigues C, Narayanan PS, Velmurugan A, Santhosh KB, John J, Walia K and Veeraraghavan B (2025) Emergence of carbapenem-resistant Salmonella Typhi harboring blaNDM-5 in India: genomic evidence from a multicenter study. Front. Microbiol. 16:1685068. doi: 10.3389/fmicb.2025.1685068
Edited by:
Jess Vergis, ICAR Indian Veterinary Research Institute, IndiaReviewed by:
Chien-Shun Chiou, Taiwan Centers for Disease Control (CDC), TaiwanElli Mylona, University of Cambridge, United Kingdom
Copyright © 2025 Thirumoorthy, Jacob, Teekaraman, Mahantesh, Jagannatha, Manasa, Nagaraj, Savio, Padaki, Sudarsana, Nair, Verma, Gaikwad, Joshi, Nagvekar, Rodrigues, Narayanan, Velmurugan, Santhosh, John, Walia and Veeraraghavan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Balaji Veeraraghavan, dmJhbGFqaUBjbWN2ZWxsb3JlLmFjLmlu
 Tharani Priya Thirumoorthy
Tharani Priya Thirumoorthy Jobin John Jacob
Jobin John Jacob Monisha Priya Teekaraman
Monisha Priya Teekaraman S. Mahantesh3
S. Mahantesh3 Bhavana Jagannatha
Bhavana Jagannatha Suhani Manasa
Suhani Manasa Savitha Nagaraj
Savitha Nagaraj Jayanthi Savio
Jayanthi Savio Priyadarshini A. Padaki
Priyadarshini A. Padaki J. Sudarsana
J. Sudarsana Ashalatha Nair
Ashalatha Nair Sukanya Verma
Sukanya Verma Raman Gaikwad
Raman Gaikwad Divya Joshi
Divya Joshi Camilla Rodrigues
Camilla Rodrigues Pavithra Sathya Narayanan
Pavithra Sathya Narayanan K. B. Santhosh
K. B. Santhosh Kamini Walia
Kamini Walia Balaji Veeraraghavan
Balaji Veeraraghavan