- 1Microbiology Laboratory, Quanzhou Women's and Children's Hospital, Quanzhou, China
- 2The Affiliated Women’s and Children's Hospital of Huaqiao University, Quanzhou, China
- 3Respiratory Department, Quanzhou Women’s and Children’s Hospital, Quanzhou, China
- 4Research Administration Office, Quanzhou Women's and Children's Hospital, Quanzhou, China
Background: Phytobacter diazotrophicus (P. diazotrophicus) is an emerging opportunistic pathogen responsible for various human infections. However, it remains unclear whether aldehyde dehydrogenase B (aldB) mediates antibiotic resistance in P. diazotrophicus, and whether acetoin catabolism operon regulator (acoR) gene is associated with antibiotics. This study aims to explore the association between the aldB gene and bacterial susceptibility to β-lactam antibiotics, investigate the potential mechanism by which aldB mediates antibiotic resistance, and clarify the regulatory mechanism of aldB by the upstream adjacent gene acoR.
Methods: Gene knockout was performed using homologous recombination. The minimum inhibitory concentrations (MICs) of 12 β-lactam antibiotics were determined using the MH agar plate dilution method. RNA transcriptome analysis was performed on the wild-type and aldB knockout strains of P. diazotrophicus Pd1. mRNA expression levels were measured using real-time quantitative polymerase chain reaction (qPCR). The binding of the purified AcoR-HTH protein to PaldB was analyzed using electrophoretic mobility shift assays (EMSA). Ceftazidime was used for antibiotic stimulation tests.
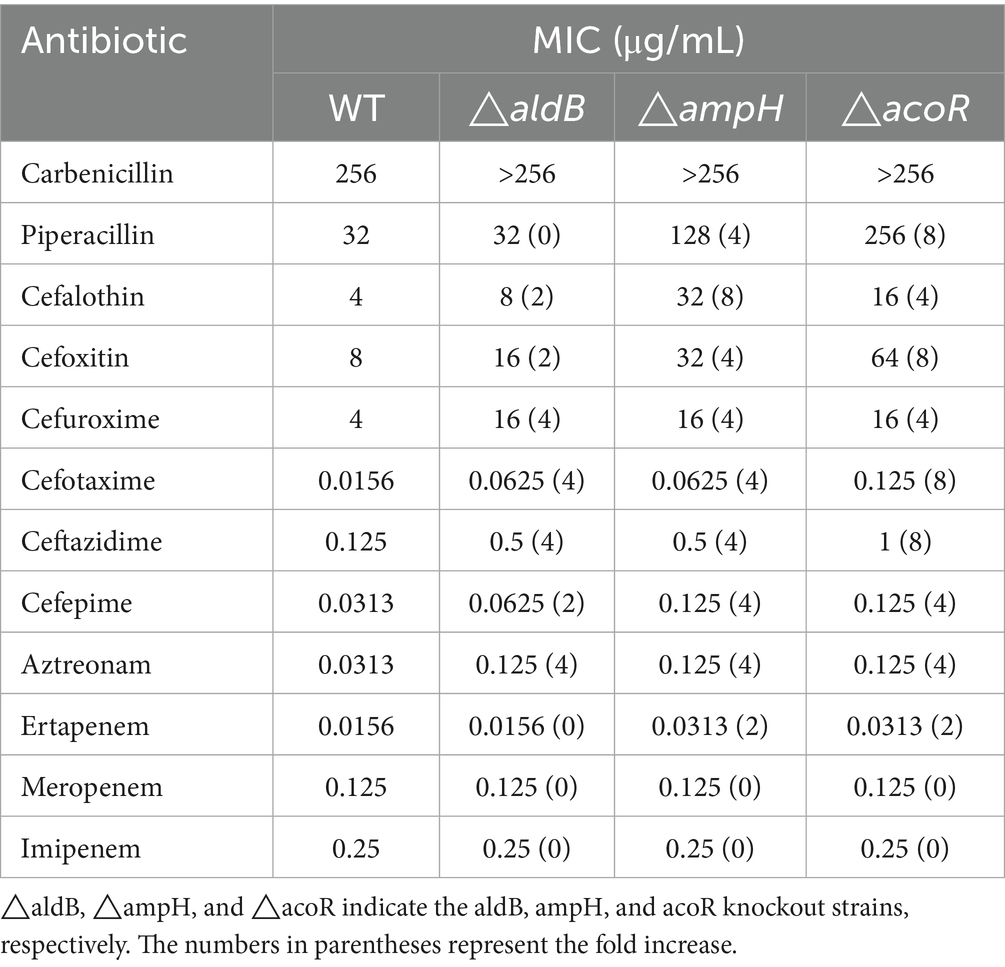
Results: Compared with the wild-type strain, the aldB knockout strain exhibited significantly increased MICs for carbenicillin, cefalotin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, and aztreonam by at least 2-, 2-, 2-, 4-, 4-, 4-, 2-, and 4-fold, respectively. RNA transcriptome sequencing revealed that ampH was most significantly downregulated in the aldB knockout strain, which was confirmed by qPCR. Compared with the wild-type strain, the ampH-knockout strain exhibited significantly increased MICs for carbenicillin, piperacillin, cefalotin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, aztreonam, and ertapenem by at least 2-, 4-, 8-, 4-, 4-, 4-, 4-, 4-, 4-, and 2-fold, respectively. Compared with the wild-type strain, the acoR knockout strain exhibited significantly increased MICs for carbenicillin, piperacillin, cefalotin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, aztreonam, and ertapenem by at least 2-, 4-, 4-, 8-, 4-, 8-, 8-, 4-, 4-, and 2-fold, respectively. Compared with the wild-type strain, the acoR knockout strain significantly downregulates the mRNA expression of aldB. The sequence ACGACACAGTTCGCGAA was identified as a recognition site for AcoR in P. diazotrophicus through software alignment and EMSA experiments. Compared with the untreated wild-type strain, aldB mRNA expression levels in the ceftazidime-stimulated wild-type strain reduced significantly.
Conclusion: In P. diazotrophicus, aldB and acoR reduced some β-lactam resistance by facilitating ampH and aldB expression, respectively. This is the first report that links acoR to β-lactam antibiotics and demonstrates that AcoR positively regulates aldB. Under ceftazidime stress, P. diazotrophicus reduced aldB expression to increase its tolerance to the antibiotic. The discovery of the mechanism by which AcoR regulates aldB expression provides preliminary evidence for subsequent research on drug resistance mechanisms.
Introduction
Phytobacter diazotrophicus (P. diazotrophicus), belonging to the family Enterobacteriaceae, is an emerging opportunistic pathogen associated with various human infections (sepsis and biliary tract infections) (Lin et al., 2024; Smits et al., 2022) and occasionally causes fatal outbreaks (Pillonetto et al., 2018). The clinical significance of P. diazotrophicus has likely been underestimated due to frequent misidentification of clinical isolates as other Enterobacteriaceae. β-lactam antibiotics are commonly used to treat bacterium-induced infections. Over the past 2 years, clinical reports have confirmed the emergence of multidrug-resistant P. diazotrophicus strains (Hon et al., 2023; Almuzara et al., 2024). Therefore, assessing drug resistance mechanisms in P. diazotrophicus is crucial for the treatment and prevention of clinical infections.
The intergenic region between aldehyde dehydrogenase B (aldB) and yiaW may contribute to the tolerance of Escherichia coli to antimicrobial agents rather than the aldB itself (Hu and Coates, 2005). However, aldB gene reduces the resistance of Lactobacillus reuteri (L. reuteri) to chloramphenicol but not to β-lactam antibiotics (Wang et al., 2024). This study did not assess why aldB reduced chloramphenicol resistance. The presence of aldB increases the number of persisters under non-stress conditions. However, the effect of aldB on persister counts is environmentally dependent. For example, under amino acid limitation, aldB knockdown unexpectedly increased persister numbers. Because this increase is associated with glycolytic metabolites, metabolic flux may play a crucial role in aldB-mediated persister formation (Kawai et al., 2018). Therefore, it can be concluded that the aldB regulates antibiotic resistance in certain bacteria (but not all) and is involved in persister formation.
In E. coli, low-molecular-mass penicillin-binding proteins (LMM PBPs) are crucial for the proper formation of cell morphology. These enzymes possess DD-carboxypeptidase and/or DD-endopeptidase activities that are associated with the maturation and remodeling of peptidoglycan (PG) (Sauvage et al., 2008). AmpH is an AmpH-type class C LMM PBP (Sauvage et al., 2008) involved in PG recycling as a bifunctional enzyme with both DD-endopeptidase and DD-carboxypeptidase activities (González-Leiza et al., 2011). Although AmpH is closely associated with ampicillin class C β-lactamase (AmpC) and other class C β-lactamases, it did not exhibit β-lactamase activity toward nitrocefin in previous studies (Henderson et al., 1997). However, AmpH can strongly bind to penicillin G, cefoxitin, and cephalosporin C (Henderson et al., 1997). Additionally, AmpH exhibits weak β-lactamase activity against nitrocefin and is a penicillin-binding protein owing to its affinity for β-lactam substances (such as the fluorescent penicillin Bocillin FL and cefmetazole) (González-Leiza et al., 2011). It is demonstrated that AmpH consistently binds to certain β-lactam antibiotics. Genetically, ampH has not yet been knocked out to assess its association with β-lactam antibiotics.
In the genome of P. diazotrophicus Pd1, acetoin catabolism regulator (acoR) gene is located directly upstream of aldB and is divergently transcribed relative to it. acoR encodes a sigma-54-dependent transcriptional regulatory protein of factor for inversion stimulation (Fis) family. Fis is a versatile DNA-binding protein that plays a crucial role in coordinating global bacterial gene expression in response to growth phase and environmental stress. It inhibits aldB expression (Xu and Johnson, 1995) and enhances bacterial resistance to ciprofloxacin by downregulating genes involved in pyocin biosynthesis (FLong et al., 2020). Additionally, Fis acts as an accessory transcriptional activator at the promoter of multiple antibiotic-resistant marRAB operon (Martin and Rosner, 1997). The acetoin catabolic pathway, which is significant in numerous microorganisms, is transcriptionally regulated by acoR (Peng et al., 2020). It remains unclear whether aldB mediates antibiotic resistance in P. diazotrophicus, and whether acoR is associated with antibiotics.
In this study, we report that aldB mediates antibiotic resistance in P. diazotrophicus by affecting ampH expression. Additionally, we demonstrated the association between acoR and antibiotics, its regulatory role in aldB-mediated antibiotic resistance, and the role of aldB under ceftazidime stimulation. This study aims to elucidate the mechanism by which AcoR regulates aldB in P. diazotrophicus, thereby providing a theoretical basis for the prevention and control of related infections.
Materials and methods
Bacterial strains and culture conditions
Phytobacter diazotrophicus Pd1, which was isolated from the blood of a newborn with D-galactosemia complicated by sepsis, was preserved in our laboratory (Lin et al., 2024). The recombinant pKAS32 plasmid carrying the tetracycline resistance gene cassette was purchased from Baosai Biotechnology Co., Ltd., Hangzhou, China. The pDM4 plasmid was purchased from Wuhan Miaoling Biotechnology Co., Ltd. and used for constructing suicide plasmid. E. coli S17-1 (λpir) was purchased from Beyotime Biotechnology and provides a host environment for the pDM4 plasmid. P. diazotrophicus Pd1 was cultured on a blood agar plate at 37 °C for 24 h.
Construction of deletion mutant strains
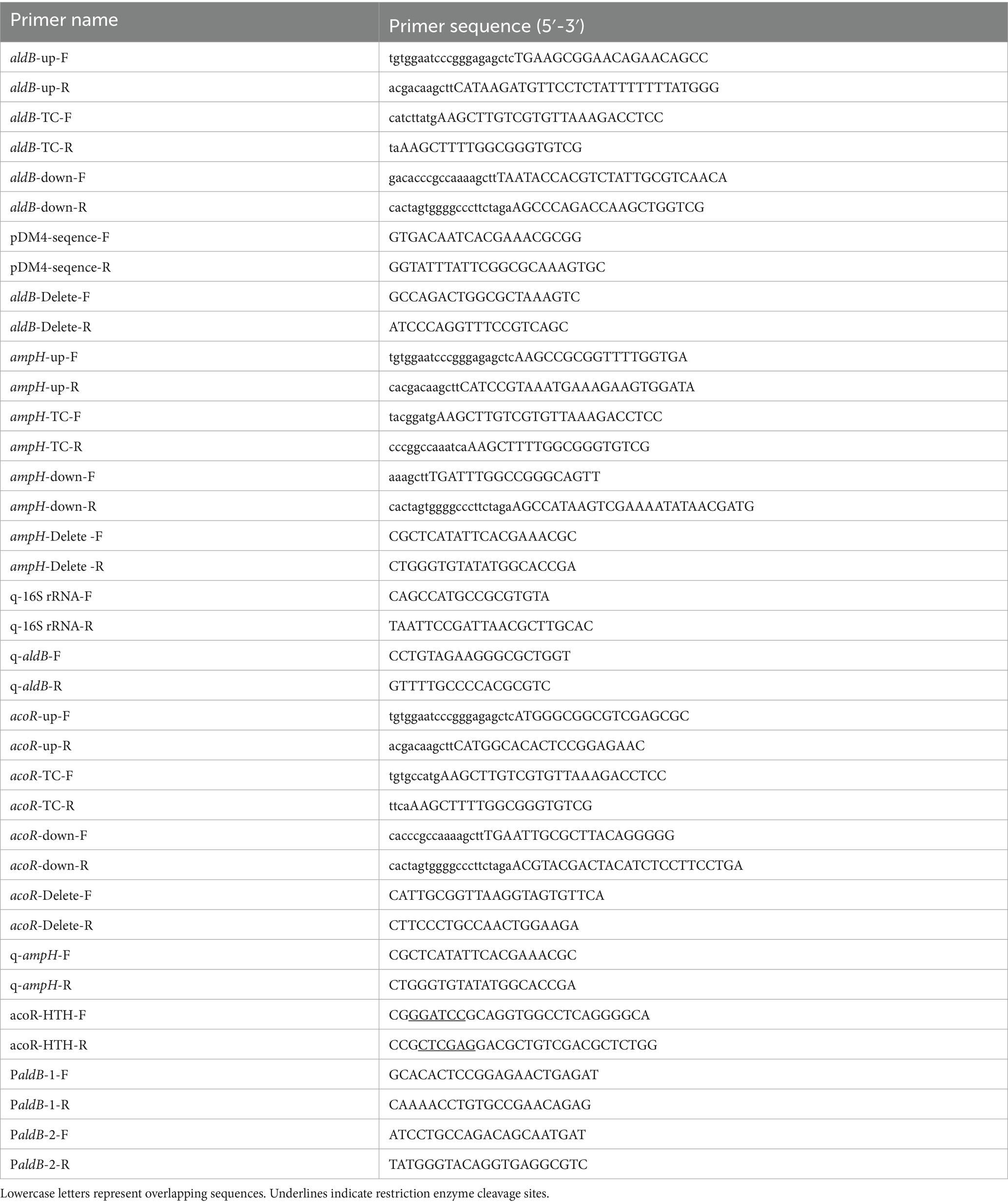
Construction of aldB gene knockout based on homologous recombination method (Lin et al., 2025). The genome of P. diazotrophicus Pd1 was extracted in accordance with the requirements of TIANamp Bacteria DNA Kit from Tiangen Biochemical Technology Co., Ltd. The upstream and downstream arms of aldB were amplified from P. diazotrophicus Pd1 genomic DNA by high-fidelity PCR using the primer pairs aldB-up-F/aldB-up-R and aldB-down-F/aldB-down-R, respectively. The tetracycline resistance gene was amplified from the pKAS32 plasmid by high-fidelity PCR using the primer pair aldB-TC-F and aldB-TC-R. High-fidelity PCR was purchased from Vazyme. The total reaction volume of 50 μL consisted of 20 μL ddH2O, 25 μL 2 × Phanta Max Master Mix, 2 μL primer F, 2 μL primer R, and 1 μL template DNA. The thermal cycle was programmed for 3 min at 95 °C for pre-denaturation, followed by 32 cycles of 15 s at 95 °C for denaturation, 15 s at 60 °C for annealing, 60 s at 72 °C for extension; a final extension was conducted for 5 min at 72 °C (Lin et al., 2025). The above three fragments were ligated into the Suicide pDM4 plasmid using the ClonExpress MultiS One Step Cloning Kit (Vazyme Biotech Co., Ltd) with LIC (Ligation-Independent Cloning). The recombinant pDM4 plasmid was verified for ligation by Sanger sequencing using the primer pairs pDM4-sequence-F and pDM4-sequence-R. The recombinant pDM4 plasmid was electroporated into competent P. diazotrophicus Pd1 using an electroporation instrument (1.8 kV, 400 Ω). Subsequently, the bacterial suspension was spread on LB agar plates containing tetracycline (20 μg/mL) and piperacillin (24 μg/mL). Single colonies were picked and subcultured for 5 generations. The knockout of the aldB gene was verified by PCR using the primer pair aldB-Delete-F and aldB-Delete-R. The primer sequences are listed in Table 1.
The construction process of the ampH knockout strain is consistent with that of the aldB knockout strain. High-fidelity PCR technology was used to amplify the upstream and downstream homologous arms and the tetracycline resistance gene cassette using the primer pairs ampH-up-F/ampH-up-R, ampH-down-F/ampH-down-R, and ampH-TC-F/ampH-TC-R, respectively. PCR verification was performed with the primer pair ampH-Delete-F/ampH-Delete-R to confirm the successful knockout of the ampH gene. All primer sequences are detailed in Table 1.
The construction of the acoR gene knockout strain also followed the method used for the aldB knockout strain. Amplification of the upstream homologous arm, downstream homologous arm, and tetracycline resistance gene cassette was accomplished using high-fidelity PCR with the primer pairs acoR-up-F/acoR-up-R, acoR-down-F/acoR-down-R, and acoR-TC-F/acoR-TC-R, respectively. The knockout efficiency of the acoR gene was confirmed by PCR verification using the primer pair acoR-Delete-F/acoR-Delete-R. The sequences of all related primers are detailed in Table 1.
Antibiotic susceptibility test
The minimum inhibitory concentration (MIC) values of the strains against 12 β-lactam antibiotics, including carbenicillin, piperacillin, cefalotin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, aztreonam, ertapenem, meropenem, and imipenem, were determined using the MH agar plate dilution method. The specific procedure was as follows: first, the strains were revived on blood plates for 24 h, and single colonies were picked and subcultured for 24 h. Subsequently, each antibiotic with a serial dilution concentration was added to empty plates, and then 25 mL of MH agar medium at approximately 50 °C was poured into each plate. Before conducting the experiment, mark the regions for the wild-type strain and the knockout mutant strain on the bottom of each MH agar plate. Next, the concentration of the bacterial suspension was adjusted to the 0.5 McFarland turbidity standard using a McFarland turbidimeter with sterile 0.65% physiological saline, and 1 μL of the bacterial suspension was titrated onto the surface of the MH agar plates. After air-drying, the plates were placed in an incubator at 37 °C, and the MIC values of each strain were observed after 20 h. E. coli ATCC8739 was selected as the quality control bacterium. The MIC determination results of this reference strain fell within the range specified in the Clinical and Laboratory Standards Institute (CLSI) document M100-Ed35. This biological experiment was repeated three times.
RNA transcriptome analysis
RNA transcriptome analysis was conducted on the aldB knockout strain and wild-type strain of P. diazotrophicus Pd1. Total RNA was extracted from the strains using the RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions, and the concentration and purity of the extracted RNA were determined using a Nanodrop 2000 spectrophotometer. RNA-seq paired-end sequencing was performed on the Illumina NovaSeq 6000 platform. Bioinformatics analysis of the sequencing data generated by the Illumina platform was carried out using the cloud computing platform (cloud.majorbio.com) provided by Shanghai Majorbio Bio-pharm Technology Co., Ltd. After obtaining the read counts for each gene, differential expression analysis was performed using DESeq2 software with reference to the P. diazotrophicus Pd1 genome. The criteria for screening differentially expressed genes were as follows: an adjusted p-value < 0.05, and a fold change (FC) in gene expression between samples of ≥2 (up-regulated) or ≤0.5 (down-regulated).
RNA isolation and quantitative PCR
Total RNA was extracted using FreeZol Reagent (Vazyme Biotech Co., Ltd., Nanjing, China), with operations strictly following the manufacturer’s instructions. The specific steps were as follows: 1 mL of bacterial suspension was placed in an EP tube and centrifuged at 11,200 rpm for 3 min. After discarding the supernatant, 100 μL of lysozyme reagent (30 mg/mL) was added to resuspend the bacterial pellet, followed by pipetting to mix thoroughly, and incubation at 37 °C for 30 min. After this treatment, 500 μL of FreeZol Reagent was added, and the mixture was vortexed thoroughly to mix well, then left to stand at room temperature for 5 min. Subsequently, it was centrifuged at 11,200 rpm for 15 min at room temperature, and the upper aqueous phase was carefully aspirated. An equal volume of isopropanol was added, and the mixture was gently inverted to mix well, then allowed to stand at room temperature for 10 min. It was centrifuged again at 11,200 rpm for 10 min at room temperature, and the supernatant was discarded. One milliliter of 75% ethanol was added, the EP tube was inverted 5 times, and centrifugation was performed at 9,100 rpm for 3 min at room temperature. The supernatant was discarded, and the steps of ethanol washing, centrifugation, and supernatant discarding were repeated once. The tube was left at room temperature for 3 min to allow the precipitate to air-dry naturally, and finally, 20 μL of RNase-free double-distilled water was added to dissolve the precipitate. After extraction, the RNA concentration was measured using a Nano-300 micro-spectrophotometer.
Five hundred nanogram of total RNA was taken, and cDNA was synthesized via reverse transcription using the HiScript III RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme Biotech Co., Ltd., Nanjing, China). The specific steps were as follows: First, to remove genomic DNA, a 16 μL reaction system was prepared, containing 0.5 μL of RNA, 4 μL of 4 × gDNA Wiper Mix, and 11.5 μL of RNase-free double-distilled water. After mixing, it was incubated at 42 °C for 2 min. Subsequently, 5 μL of HiScript III RT SuperMix was added, and reverse transcription was performed under the following conditions: incubation at 37 °C for 15 min, followed by treatment at 85 °C for 5 s.
The real-time fluorescent quantitative PCR (qPCR) reaction was performed using 2.5 ng of cDNA as the template, with ChamQ Blue Universal SYBR PCR Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China) on the Gentier 96 fully automated real-time fluorescent quantitative PCR analysis system (Tianlong Technology Co., Ltd., Xi’an, China). The 20 μL reaction system consisted of 10 μL of 2 × ChamQ Blue Universal SYBR qPCR Master Mix, 0.4 μL of forward primer (10 μM), 0.4 μL of reverse primer (10 μM), 1 μL of cDNA, and 8.2 μL of double-distilled water. The qPCR reaction conditions were: pre-denaturation at 95 °C for 30 s; 40 cycles of denaturation at 95 °C for 10 s and annealing-extension at 60 °C for 10 s; finally, melting curve analysis was performed according to the instrument’s default program. Detailed information on the primers used is provided in Table 1. To validate qPCR primer efficiency, the efficiency (E) is calculated using the formula E = [10^(−1/slope) - 1] × 100%. A primer is considered to have qualified efficiency if 90% ≤ E ≤ 110%; if the E value falls outside this range, the primer needs to be optimized. Data normalization was performed using the housekeeping gene 16S rRNA as an internal reference, and the relative expression levels of the ampH and aldB genes were calculated using the 2^(−ΔΔCt) method. For the qPCR experiment, five biological replicates were selected for each group, and the Cohen’s d value was set to > 0.8.
Analysis of AcoR binding sites
In this study, the sequence upstream of the translation initiation site of the aldB gene in P. diazotrophicus was aligned with the reported promoter sequence bound by the AcoR protein using DNAMAN software to analyze the potential binding site of AcoR.
Expression and purification of the helix-turn-helix domain of AcoR
The DNA fragment of the acoR gene, which encompasses its helix-turn-helix (HTH) domain, was amplified via PCR from the genomic DNA of P. diazotrophicus Pd1. The PCR primers used for this amplification were acoR-HTH-F and acoR-HTH-R. Following amplification, the PCR product was subjected to restriction enzyme digestion and subsequently ligated into the expression vector pET21b. The resulting recombinant plasmid, designated as pET-HTH, was transformed into E. coli strain BL21 (DE3) to generate the recombinant strain BL21 (pET-HTH). Finally, the BL21 (pET-HTH) strain was cultured in LB medium until it reached the exponential growth phase. Expression and purification of the HTH-His protein was performed as previously described (Lin et al., 2025). Before purification, the inclusion body-borne HTH-His protein was solubilized in a 50 mM Tris–HCl buffer (pH 8.0) containing 8 M urea, following 20 min of ultrasonic disruption. The lysate was centrifuged at 12,000 rpm for 20 min, and the supernatant was collected. After purification, dialysis bags filled with the protein-containing supernatant were placed in the following dialysates, respectively, for dialysis: 50 mM Tris–HCl buffer (pH 8.0) containing 100 mM NaCl, 1 mM DTT, 0.02% Tween 80 and with urea concentrations of 6 M, 4 M, 2 M respectively, as well as PBS buffer (pH 7.35). Dialysis was performed at 4 °C, and the dialysate needed to be replaced after 6 h of dialysis each time.
Electrophoresis mobility shift assays
Promoter region PaldB-1 was designed to contain the potential AcoR-binding site (sequence: ACGACACAGTTCGCGAA). To further verify whether this sequence functions as a binding site, promoter region PaldB-2 was constructed to lack this specific motif. Promoter regions PaldB-1 and PaldB-2 from P. diazotrophicus Pd1 genome were PCR amplified using primer pairs PaldB-1-F/PaldB-1-R and PaldB-2-F/PaldB-2-R, respectively. The concentrations of PaldB-1 and PaldB-2 were determined using a Nano-300 micro-spectrophotometer. Electrophoretic mobility shift assays (EMSA) were performed as previously described (Peng et al., 2014), with minor modifications to analyze binding of the purified AcoR-HTH protein to PaldB. Finally, the non-denaturing electrophoresis gel was stained with SYBR™ Green for 1 h and imaged using a CLINX GenoSens 1,660 instrument.
Ceftazidime stimulation test
Twenty-five microliter of bacterial suspension (either the wild-type strain or the acoR knockout strain) at a 0.5 McFarland standard was inoculated into 25 mL of Brain Heart Infusion (BHI) medium and incubated for 24 h at 37 °C. For the dose-dependent experiment, ceftazidime should be individually added to each bacterial suspension in the experimental groups to adjust the final concentration to 0.125, 0.25, 0.5, and 1 μg/mL, respectively, (corresponding to 1×, 2×, 4×, and 8 × MIC). After mixing well, the suspensions are incubated statically at 37 °C for 1 h. For the time kinetic experiment, ceftazidime should be individually added to each bacterial suspension in the experimental groups to adjust the final concentration to 0.25 μg/mL (equivalent to 2 × MIC). After mixing well, the suspensions are incubated statically at 37 °C for 0.5, 1, 2, and 3 h, respectively. Meanwhile, under the same conditions, the untreated wild-type strain and untreated acoR knockout strain (without ceftazidime exposure) served as control groups. Since ceftazidime was dissolved in sterile double-distilled water, and the volume of ceftazidime added to the experimental group was extremely small, which is negligible relative to the 25 mL culture medium, it is unnecessary to set up a solvent control group.
Next, 1 mL aliquots of each bacterial suspension (both experimental and control groups) were centrifuged at 6,000 rpm for 3 min. The supernatant was carefully aspirated, and the bacterial pellet was washed three times with PBS buffer (pH 7.35) to remove residual medium and antibiotics. Finally, bacterial RNA isolation and qPCR were performed following the procedures described above. This biological experiment was independently replicated five times to validate the reproducibility of the results.
Statistical analysis
In this study, the data of all sample groups were verified to exhibit a normal distribution via the Shapiro–Wilk test, and the homogeneity of variances between groups was confirmed through the Levene test. Based on the above prerequisite conditions, Student’s t-test was used to evaluate the statistical significance of differences between groups. A p-value < 0.05 was considered to indicate a statistically significant difference.
Results
aldB is associated with antibiotic susceptibility
To demonstrate the association between aldB and β-lactam antibiotics, we constructed an aldB deletion strain (Supplementary Figure S1) and determined the minimum inhibitory concentrations (MICs) of 12 β-lactam antibiotics in the wild-type and aldB knockout strains. Compared with wild-type P. diazotrophicus, the aldB knockout strain exhibited increased MICs for carbenicillin, cefalotin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, and aztreonam by at least 2-, 2-, 2-, 4-, 4-, 4-, 2-, and 4-fold, respectively (Table 2). These results indicate that aldB is associated with some β-lactam susceptibility and reduces P. diazotrophicus resistance to these antibiotics.
aldB reduces antibiotic resistance by upregulating ampH
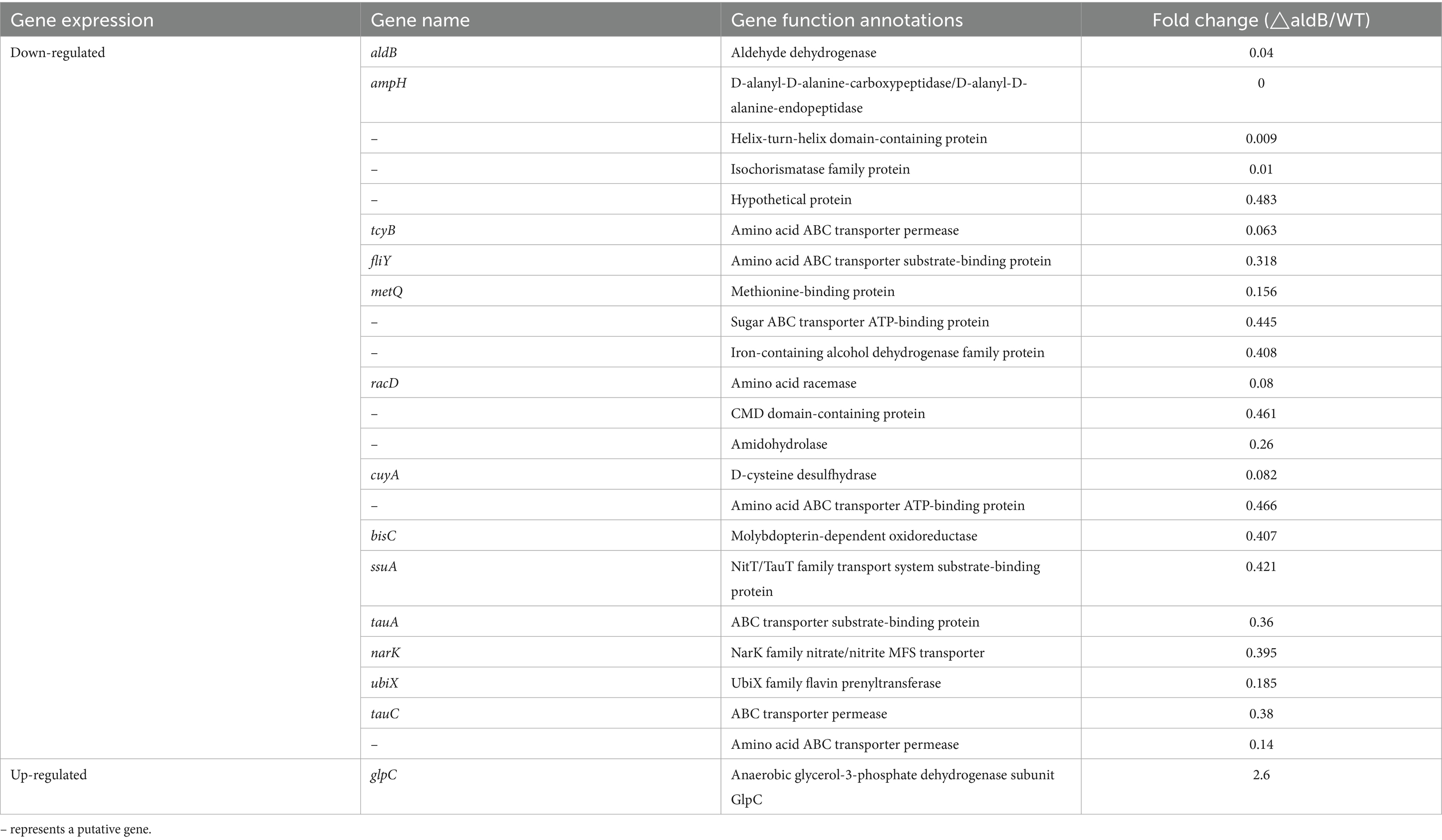
To assess how aldB reduces resistance to β-lactam antibiotics in P. diazotrophicus, transcriptome sequencing was conducted on both the wild-type and aldB knockout strains. Compared to wild-type P. diazotrophicus, the aldB knockout strain upregulated one gene and downregulated 22 genes (Table 3). Transcriptome analysis demonstrated that aldB was most significantly downregulated, confirming its successful knockout. Excluding aldB, ampH was most significantly down-regulated (Figure 1; Table 3). Real-time quantitative polymerase chain reaction (qPCR) was performed to confirm transcriptome sequencing results. The E values of 16S rRNA and ampH are 100 and 107% respectively, both falling within the ideal range (90–110%) for qPCR primer efficiency (Supplementary Figures S2, S3). Both the melting curves of 16S rRNA and ampH show a single peak, which indicates that the primers for 16S rRNA and ampH have good specificity (Supplementary Figure S4). ampH expression in the aldB knockout strain was reduced by approximately 100-fold than that in the wild-type strain (Supplementary Figure S5; Figure 2). This indicates that aldB upregulated ampH expression.
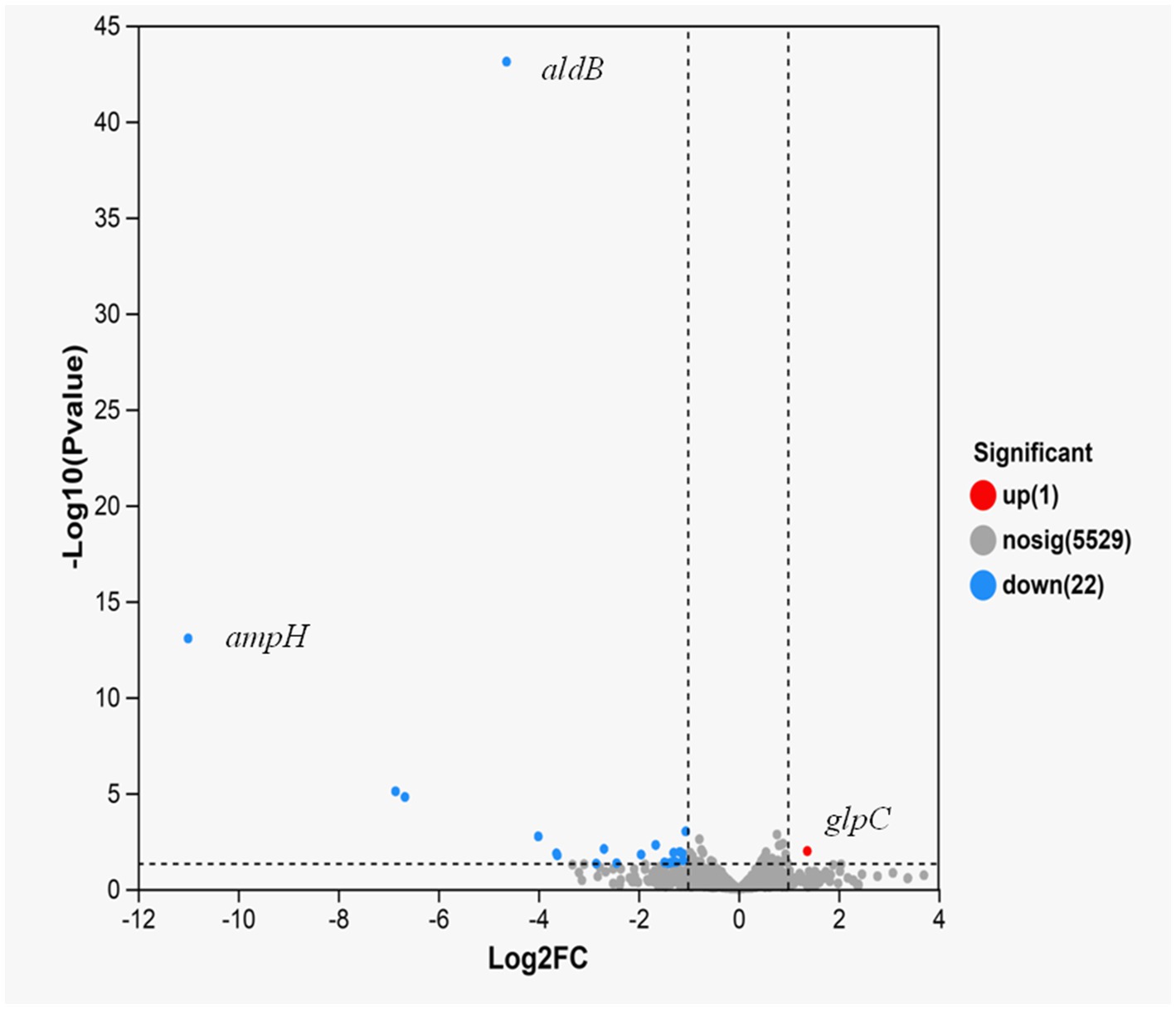
Figure 1. Volcano plot of expression difference for aldB knockout strain VS wild-type strain. The abscissa represents the fold change value of gene expression difference between the two groups of samples, i.e., the FC value. The ordinate represents the statistical test value of the difference in gene expression changes, i.e., the p-value. A higher p-value indicates a more significant expression difference, and both the abscissa and ordinate values have undergone logarithmic processing. Each point in the figure represents a specific gene: red points indicate significantly up-regulated genes, green points indicate significantly down-regulated genes, and gray points indicate genes with non-significant differences. After mapping all genes, it can be seen that the points on the left represent genes with down-regulated expression differences, and the points on the right represent genes with up-regulated expression differences. The points closer to the two sides and the top have more significant expression differences.
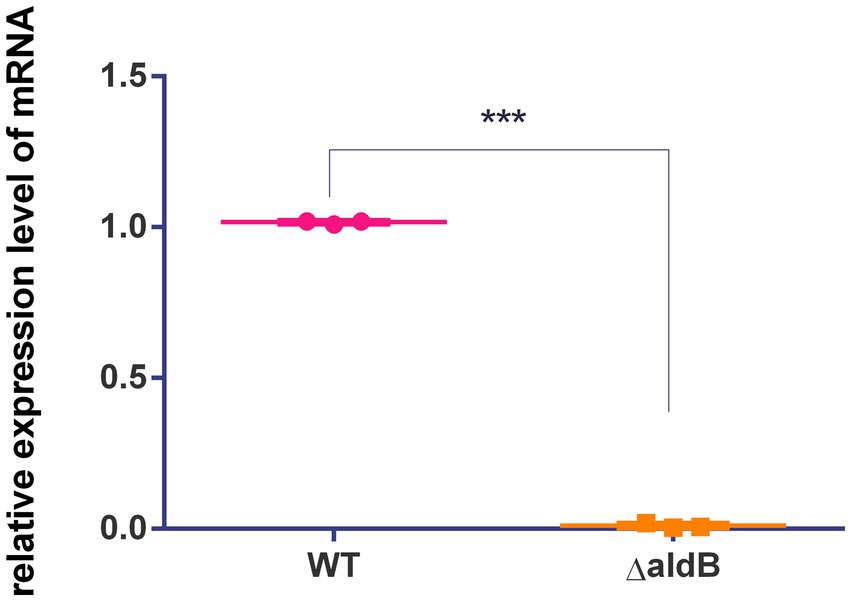
Figure 2. Comparison of relative expression levels of ampH between the wild-type strain and aldB knockout strain of P. diazotrophicus. Data represent the mean ± standard deviation of five sample results. *** indicates p < 0.001.
ampH binds to β-lactam antibiotics (González-Leiza et al., 2011; Henderson et al., 1997). We inferred that ampH may play an important role in the bactericidal effect of β-lactam antibiotics. To demonstrate the association between ampH and β-lactam antibiotics, we constructed an ampH deletion strain (Supplementary Figure S6) and determined the MICs of 12 β-lactam antibiotics for the wild-type and ampH knockout strains. Compared with wild-type P. diazotrophicus, the ampH knockout strain exhibited increased MICs for carbenicillin, piperacillin, cefalotin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, aztreonam, and ertapenem by at least 2-, 4-, 8-, 4-, 4-, 4-, 4-, 4-, 4-, and 2-fold, respectively (Table 2). This indicates that aldB upregulated ampH expression and reduced P. diazotrophicus resistance to these antibiotics.
acoR is associated with antibiotic susceptibility
Since the acoR gene is located directly upstream of the aldB gene in the genome, we hypothesize that these two genes are associated. To demonstrate the association between acoR and β-lactam antibiotics, we constructed an acoR deletion strain (Supplementary Figure S1) and determined the MICs of 12 β-lactam antibiotics for the wild-type and acoR knockout strains. Compared with wild-type P. diazotrophicus, the acoR knockout strain exhibited increased MICs for carbenicillin, piperacillin, cefalotin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, aztreonam, and ertapenem by at least 2-, 4-, 4-, 8-, 4-, 8-, 8-, 4-, 4-, and 2-fold, respectively (Table 2). This indicates that acoR is associated with some β-lactam antibiotics and reduces P. diazotrophicus resistance to these antibiotics.
acoR reduces antibiotic resistance by upregulating aldB
To demonstrate that acoR regulates aldB expression, qPCR was used to measure aldB mRNA levels in the wild-type and acoR knockout strains. The E value of aldB is 101.5%, which falls within the ideal range (90–110%) for qPCR primer efficiency (Supplementary Figure S7). The melting curve of aldB shows a single peak, indicating that the primer for aldB has good specificity (Supplementary Figure S4). Compared with wild-type P. diazotrophicus, aldB mRNA levels in the acoR knockout strain were significantly reduced (Figure 3). This indicates that acoR upregulated aldB expression and reduced P. diazotrophicus resistance to these antibiotics.
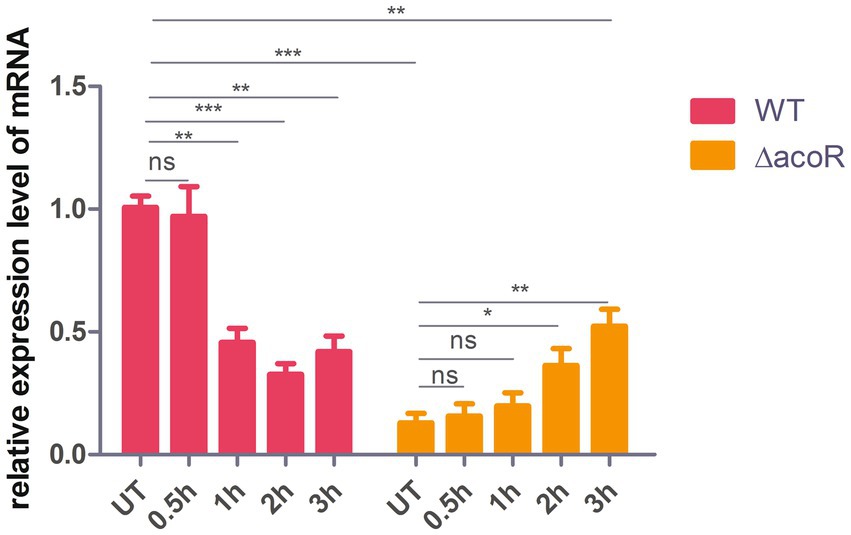
Figure 3. Comparison of relative aldB expression levels in wild-type and acoR knockout strains of P. diazotrophicus following simultaneous stimulation with 2 × MIC ceftazidime for 0.5 h, 1 h, 2 h, and 3 h. WT represents the wild-type strain, △acoR represents the acoR knockout strain, and UT indicates no treatment. Data represent the mean ± standard deviation of five sample results. * indicates p < 0.05; ** indicates p < 0.01; *** indicates p < 0.001; ns indicates no statistically significant difference.
The sequence where AcoR binds to the aldB promoter
The upstream activating sequences (UAS) sequence (CAGTTTGAGAC) of the aco operon is the most likely recognition and binding site for the AcoR protein (Ali et al., 2001). In this study, we aligned this UAS with the sequence upstream of the aldB translation start site (ATG) and observed that a similar sequence (CAGTTCGCGAA) was present at position +24 downstream of the aldB transcription start site (Figure 4). Additionally, we aligned the sequence upstream of ATG with the AcoR-binding site (GACAAAACGAGACAGATGTCTCATTTTGTC) reported by Peng et al. (2020) and identified a similar sequence (ACGACAC) (Figure 4). The terminal C of this sequence overlaps with the initial C of CAGTTCGCGAA, indicating that ACGACACAGTTCGCGAA may be a recognition site for AcoR in P. diazotrophicus. The potential AcoR-binding motifs is ACNANACAGTTNGNGAN.
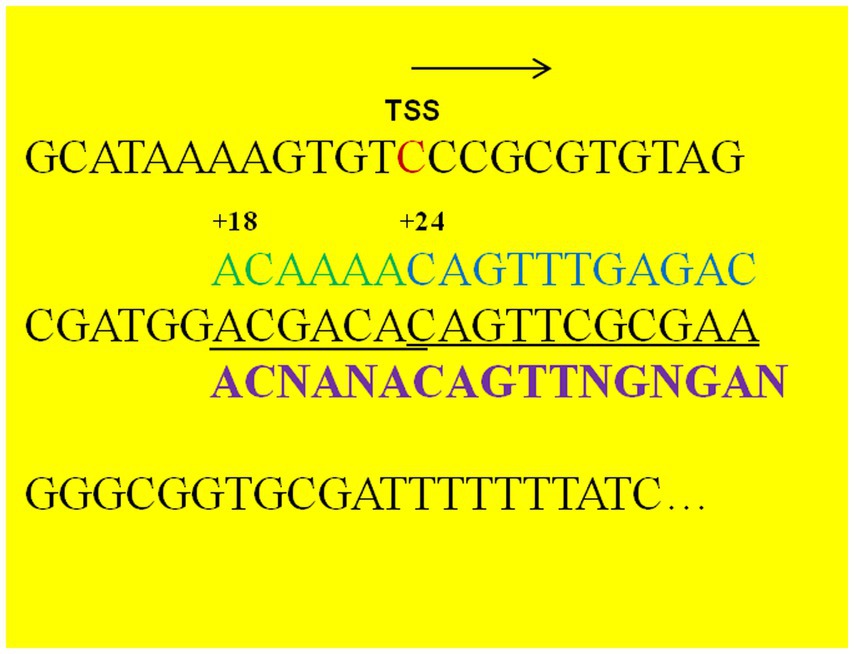
Figure 4. Nucleotide sequence downstream of the aldB transcription start site. TSS refers to the transcription start site identified by transcriptomic analysis. Arrows indicate the direction of transcription. The nucleotide sequences in black font (including the TSS) represent the upstream sequence of the translation initiation codon of P. diazotrophicus aldB, the nucleotide sequences in green and blue fonts represent the reported binding sequences of the AcoR protein from other bacterial species, the underlined part indicates the potential binding region of AcoR protein in P. diazotrophicus, and the sequences in purple font represent the binding motifs of the AcoR protein.
AcoR directly binds to the aldB promoter
To further confirm that AcoR directly binds to the aldB promoter and regulates aldB expression, we performed EMSA experiments. As shown in the right panel of Figure 5, for the PaldB-1 promoter containing the predicted binding site, distinct shifted bands were clearly observed in lanes 2–5. In contrast, no shifted bands were detected in lanes 2 and 3 of the left panel of Figure 5, which corresponds to the PaldB-2 promoter lacking the predicted binding site. These results indicate that the predicted binding site is highly likely to be the recognition and binding site of AcoR, which strongly supports the conclusion that AcoR directly binds to the aldB promoter to regulate aldB expression.
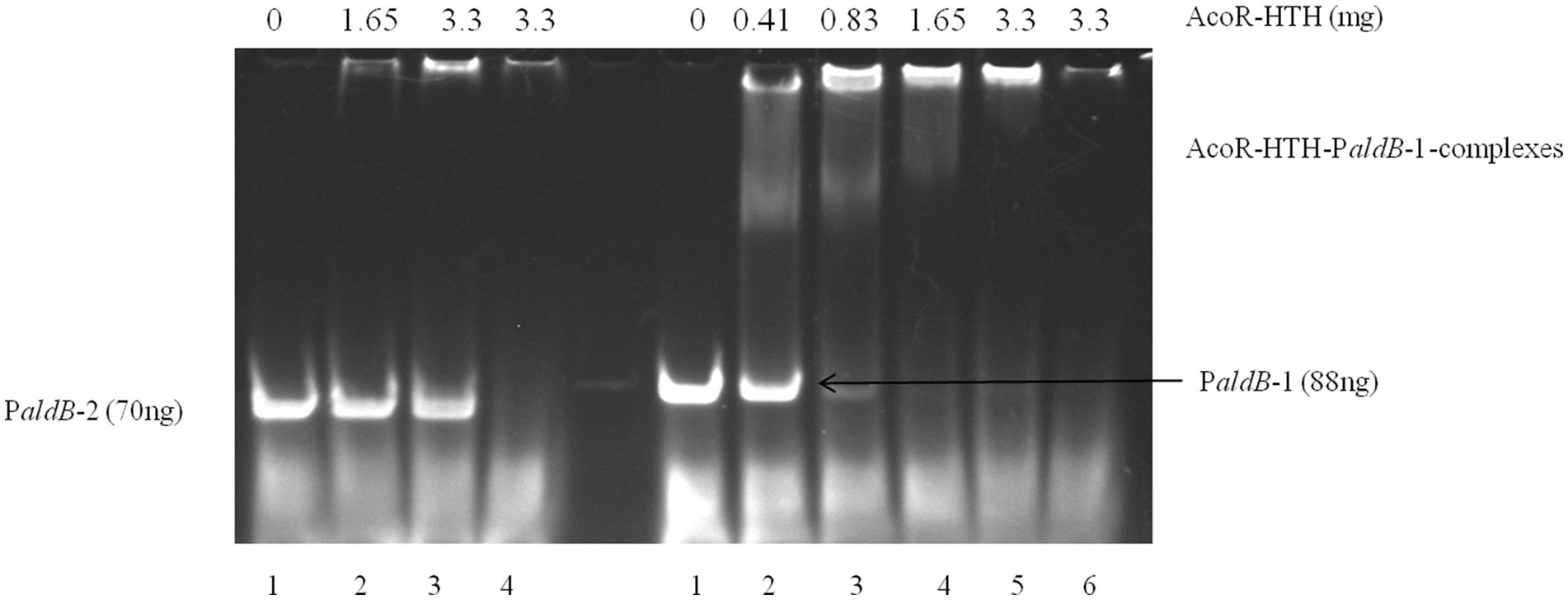
Figure 5. EMSA of AcoR-HTH protein with the aldB promoter region. In the left panel of the figure, only probe PaldB-2 was added to Lane 1 with no AcoR protein included. Different concentrations of AcoR protein were added to probe PaldB-2 in Lanes 2–3. The highest amount of AcoR protein was added to Lane 4, while no probe PaldB-2 was included. In the right panel of the figure, only probe PaldB-1 was added to Lane 1 with no AcoR protein included. Different concentrations of AcoR protein were added to probe PaldB-1 in Lanes 2–5. The highest amount of AcoR protein was added to Lane 6, while no probe PaldB-1 was included.
aldB enhances ceftazidime resistance
To assess the role of acoR-regulated aldB pathway in P. diazotrophicus in response to ceftazidime stress, we treated both the wild-type and acoR knockout strains with ceftazidime for the dose-dependent and the time kinetic experiment. qPCR was performed to measure aldB mRNA levels in the untreated wild-type, untreated acoR knockout, ceftazidime-stimulated wild-type, and ceftazidime-stimulated acoR knockout strains. First, to assess the alterations in aldB under ceftazidime stimulation, we compared aldB expression levels between the untreated and ceftazidime-stimulated wild-type strains. Compared with the untreated wild-type strain, the expression level of aldB in the ceftazidime-stimulated wild-type strain was significantly reduced under the following two sets of conditions: first, at 2 × MIC with incubation times of 1 h, 2 h, and 3 h; second, at an incubation time of 1 h with concentrations of 4 × MIC, and 8 × MIC (Figures 3, 6). The result showed that under ceftazidime stimulation, the expression of aldB decreased, which suggests that aldB is involved in the tolerance to ceftazidime.
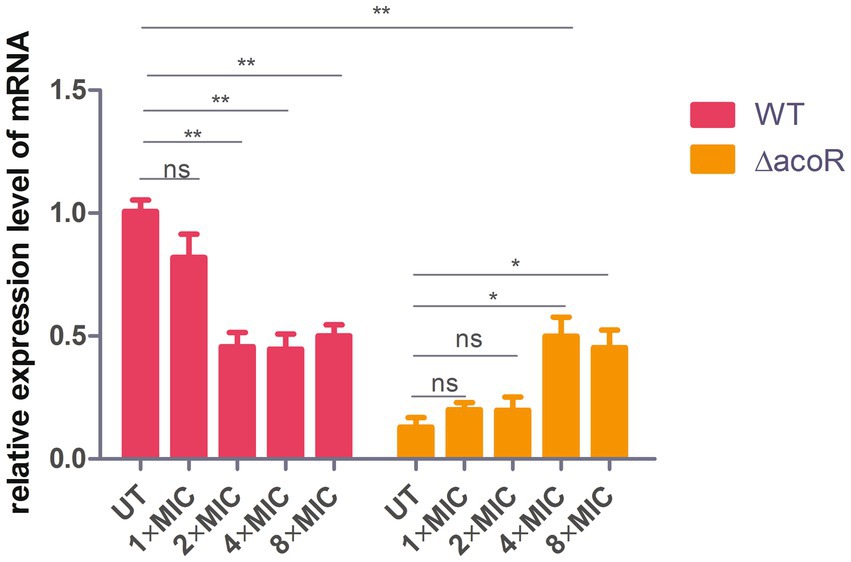
Figure 6. Comparison of relative aldB expression levels in wild-type and acoR knockout strains of P. diazotrophicus following simultaneous stimulation with ceftazidime at different MIC concentrations for 1 h. WT represents the wild-type strain, △acoR represents the acoR knockout strain, and UT indicates no treatment. Data represent the mean ± standard deviation of five sample results. * indicates p < 0.05; ** indicates p < 0.01; ns indicates no statistically significant difference.
To further evaluate whether acoR is involved in ceftazidime-induced aldB downregulation, we compared aldB expression levels between the untreated acoR knockout and ceftazidime-stimulated acoR knockout strains. Compared with the untreated acoR knockout strain, the ceftazidime-stimulated acoR knockout strain did not significantly upregulate the expression of aldB at 2 × MIC for 0.5 h and 1 h, but showed significant upregulation at 2 h and 3 h (Figure 3). Compared with the untreated acoR knockout strain, the ceftazidime-stimulated acoR knockout strain did not significantly upregulate the expression of aldB at 1 × MIC for 1 h, but exhibited significant upregulation at 4 × MIC and 8 × MIC (Figure 6). The result indicates that acoR may not be involved in ceftazidime-induced aldB downregulation.
Discussion
AmpH exhibits weak β-lactamase activity against nitrocefin (González-Leiza et al., 2011). If AmpH is a β-lactamase, aldB-mediated ampH expression may enhance β-lactam antibiotic resistance, in contrast to aldB-induced reduction in resistance in P. diazotrophicus. Therefore, we concluded that ampH does not function as a β-lactamase in P. diazotrophicus. ampH may serve as an auxiliary drug target for certain β-lactam antibiotics. The aldB knockout reduced ampH expression, thereby reducing the number of binding sites for β-lactam antibiotics. This reduces the bactericidal effect and increases the resistance of the aldB knockout strain to these antibiotics. However, the mechanism by which ampH reduces resistance to β-lactam antibiotics requires further exploration.
This study indicates that aldB in P. diazotrophicus is associated with some β-lactam antibiotics, inconsistent with the report by Wang et al. (2024), who observed no such association. We observed that the complete genomes of L. reuteri TD1 and JCM 1112 lack ampH, indicating that its absence may have caused this discrepancy.
AcoR upregulates aldB, unlike Fis that downregulates aldB (Xu and Johnson, 1995), indicating that the members of the Fis family of transcriptional regulators can regulate aldB through different mechanisms. Central-domain activators interact with σ54 holoenzyme through DNA looping after binding to the enhancer, known as UAS or downstream activating sequences (DAS). Unlike other σ54-dependent genes, rocG lacks the UAS; instead, its expression depends on DAS that is located downstream of the rocG transcription start site (Belitsky and Sonenshein, 1999). Similarly, AcoR enhanced aldB expression by binding to DAS.
In the majority of cases, mutation-mediated resistance to ceftazidime is because of the overproduction of chromosomally encoded inducible AmpC β-lactamase (Livermore, 2002). In this study, ampC was not detected in P. diazotrophicus Pd1, indicating that this bacterium does not resist ceftazidime through induced ampC expression but through other pathways. Exposure of bacteria to ceftazidime stress reduced intracellular lipid levels, indicating altered membrane permeability (Peng et al., 2019) a finding supported by our research. In P. diazotrophicus, ceftazidime exposure inhibited aldB expression, thereby reducing ampH expression that affects peptidoglycan synthesis and may alter cell membrane permeability to ceftazidime.
In conclusion, in P. diazotrophicus, aldB reduced resistance to some β-lactam antibiotics by upregulating ampH, whereas AcoR reduced resistance by facilitating aldB expression. Under ceftazidime stress, P. diazotrophicus downregulated aldB, increasing its antibiotic tolerance. The elucidation of the mechanism by which AcoR regulates aldB expression provides new clues and directions for the study of bacterial drug resistance.
Data availability statement
The data presented in this study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/sra, accession PRJNA1298522.
Ethics statement
This study followed the principles of the Declaration of Helsinki and was approved by the Ethics Committee of the Quanzhou Women’s and Children’s Hospital (Approval No. 2024109), confirming that all methods were performed in accordance with relevant guidelines and regulations.
Author contributions
JL: Methodology, Data curation, Writing – original draft, Conceptualization, Funding acquisition. JZ: Writing – review & editing, Data curation. GW: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Joint Funds for the Innovation of Science and Technology, Fujian Province (no: 2024Y9466).
Acknowledgments
We express our gratitude for the assistance provided by Xiaoyun You.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1687374/full#supplementary-material
References
Ali, N. O., Bignon, J., Rapoport, G., and Debarbouille, M. (2001). Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. J. Bacteriol. 183, 2497–2504. doi: 10.1128/JB.183.8.2497-2504.2001
Almuzara, M., Cittadini, R., Traglia, G., Haim, M. S., De Belder, D., Alvarez, C., et al. (2024). Phytobacter spp: the emergence of a new genus of healthcare-associated Enterobacterales encoding carbapenemases in Argentina: a case series. Infect. Prev. Pract. 6:100379. doi: 10.1016/j.infpip.2024.100379
Belitsky, B. R., and Sonenshein, A. L. (1999). An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96, 10290–10295. doi: 10.1073/pnas.96.18.10290
FLong, Y., Fu, W., Wang, S., Deng, X., Jin, Y., Bai, F., et al. (2020). Fis contributes to resistance of Pseudomonas aeruginosa to ciprofloxacin by regulating pyocin synthesis. J. Bacteriol. 202:e00064-20. doi: 10.1128/JB.00064-20
González-Leiza, S. M., dPM, S., and Ayala, J. A. (2011). AmpH, a bifunctional DD-endopeptidase and DD-carboxypeptidase of Escherichia coli. J. Bacteriol. 193, 6887–6894. doi: 10.1128/JB.05764-11
Henderson, T. A., Young, K. D., Denome, S. A., and Elf, P. K. (1997). AmpC and AmpH, proteins related to the class C-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J. Bacteriol. 179, 6112–6121.
Hon, P., Ko, K. K. K., Zhong, J. C. W., De, P. P., Smits, T. H. M., Low, J., et al. (2023). Genomic identification of two Phytobacter diazotrophicus isolates from a neonatal intensive care unit in Singapore. Microbiol. Resour. Announc. 12:e0016723. doi: 10.1128/mra.00167-23
Hu, Y., and Coates, A. R. (2005). Transposon mutagenesis identifies genes which control antimicrobial drug tolerance in stationary-phase Escherichia coli. FEMS Microbiol. Lett. 243, 117–124. doi: 10.1016/j.femsle.2004.11.049
Kawai, Y., Matsumoto, S., Ling, Y., Okuda, S., and Tsuneda, S. (2018). AldB controls persister formation in Escherichia coli depending on environmental stress. Microbiol. Immunol. 62, 299–309. doi: 10.1111/1348-0421.12587
Lin, J., Lin, C., and Zheng, J. (2025). LysR-type transcriptional regulator CARR represses the expression of blaCAR-2 and reduces P. Diazotrophicus resistance to cefalothin, cefuroxime and cefotaxime. Front. Cell. Infect. Microbiol. 15:1616646. doi: 10.3389/fcimb.2025.1616646
Lin, J., Wu, J., Gong, L., Li, X., and Wang, G. (2024). Sepsis caused by Phytobacter diazotrophicus complicated with galactosemia type 1 in China: a case report. BMC Infect. Dis. 24:599. doi: 10.1186/s12879-024-09458-y
Livermore, D. M. (2002). Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34, 634–640. doi: 10.1086/338782
Martin, R. G., and Rosner, J. L. (1997). Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or rob. J. Bacteriol. 179, 7410–7419. doi: 10.1128/jb.179.23.7410-7419.1997
Peng, M. W., Wei, X. Y., Yu, Q., Yan, P., Chen, Y. P., and Guo, J. S. (2019). Identification of ceftazidime interaction with bacteria in wastewater treatment by Raman spectroscopic mapping. RSC Adv. 9, 32744–32752. doi: 10.1039/C9RA06006E
Peng, Q., Yang, M., Wang, W., Han, L., Wang, G., Wang, P., et al. (2014). Activation of gab cluster transcription in Bacillus thuringiensis by γ-aminobutyric acid or succinic semialdehyde is mediated by the sigma 54-dependent transcriptional activator GabR. BMC Microbiol. 14:306. doi: 10.1186/s12866-014-0306-3
Peng, Q., Zhao, X., Wen, J., Huang, M., Zhang, J., and Song, F. (2020). Transcription in the acetoin catabolic pathway is regulated by AcoR and CcpA in Bacillus thuringiensis. Microbiol. Res. 235:126438. doi: 10.1016/j.micres.2020.126438
Pillonetto, M., Arend, L., Gomes, S. M. T., Oliveira, M. A. A., Timm, L. N., Martins, A. F., et al. (2018). Molecular investigation of isolates from a multistate polymicrobial outbreak associated with contaminated total parenteral nutrition in Brazil. BMC Infect. Dis. 18:397. doi: 10.1186/s12879-018-3287-2
Sauvage, E., Kerff, F., Terrak, M., Ayala, J. A., and Charlier, P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258. doi: 10.1111/j.1574-6976.2008.00105.x
Smits, T. H. M., Arend, L. N. V. S., Cardew, S., Tång-Hallbäck, E., Mira, M. T., Moore, E. R. B., et al. (2022). Resolving taxonomic confusion: establishin.G the genus Phytobacter on the list of clinicallyrelevant Enterobacteriaceae. Eur. J. Clin. Microbiol. Infect. Dis. 41, 547–558. doi: 10.1007/s10096-022-04413-8
Wang, X., Chen, P., Wang, J., Wang, Y., Miao, Y., Wang, X., et al. (2024). Acetolactate decarboxylase as an important regulator of intracellular acidification, morphological features, and antagonism properties in the probiotic Lactobacillus reuteri. Mol. Nutr. Food Res. 68:e2300337. doi: 10.1002/mnfr.202300337
Keywords: Phytobacter diazotrophicus , aldB , ampH , acoR , antibiotic resistance
Citation: Lin J, Zheng J and Wang G (2025) Regulatory relationships among aldB, ampH, and acoR and their impact on β-lactam susceptibility in Phytobacter diazotrophicus. Front. Microbiol. 16:1687374. doi: 10.3389/fmicb.2025.1687374
Edited by:
Atte Johannes Von Wright, University of Eastern Finland, FinlandReviewed by:
Alexey V. Rakov, Central Research Institute of Epidemiology (CRIE), RussiaEsaú De La Vega Camarillo, National Polytechnic Institute (IPN), Mexico
Copyright © 2025 Lin, Zheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingyang Zheng, Mjc5Mjc3NDgxQHFxLmNvbQ==; Gaoxiong Wang, d2FuZ2dhb3hpb25nMjAxM0AxNjMuY29t
 Jiansheng Lin
Jiansheng Lin Jingyang Zheng
Jingyang Zheng Gaoxiong Wang
Gaoxiong Wang