- College of Forestry Engineering, Shandong Agriculture and Engineering University, Jinan, Shandong, China
Introduction: This study elucidated how different plantation types in limestone mountains shape the community structure and co-occurrence networks of soil fungal habitatspecialization groups, offering a habitat-adaptation perspective on the assembly mechanisms of soil microbial diversity during ecological restoration.
Methods: In this study, we grouped soil fungi from plantations of coniferous forests (CF), mixed forests (MF) and broad-leaved forests (BF) into habitat-generalists, specialists and opportunists based on niche breadth, and examined how forest type shapes their diversity, community structure and co-occurrence networks along a habitat-specialization gradient.
Results: We found that: (1) The number and abundance of habitat-specialists significantly exceeded those of generalists. Habitat-generalists exhibited the highest abundance and ecological niche width in BF, whereas habitat-specialists were most abundant in CF. (2) The diversity index was the highest in BF across all habitat specialization groups. Differences in community structure among forest types increased with habitat specialization, and the composition of dominant tree species significantly influenced the community structure of each group. Soil properties primarily affected the community structure of habitat generalists and opportunists between BF and other forest types. (3) The network structure of habitat specialists exhibited high modularity, while habitat generalists formed independent subnetworks with more fragile structures. CF and BF exhibited strong intra-module connections and high modularity, whereas MF displayed high intermodule connectivity, which reduced their modularity. Both within-module (Zi) and between-module connectivity (Pi) of increased with habitat specialization and the proportion of broadleaved tree species.
Discussion: We concluded that broad-leaved plantations, by increasing soil environmental heterogeneity, promoted the diversity of habitat-specialists and enhancing their network hub roles, representing the optimal strategy for optimizing below-ground biodiversity and stability in limestone mountain forest restoration.
1 Introduction
The biological and abiotic components in forest soil interact with various environmental factors, interconnecting through material cycling and energy flow, thereby facilitating their turnover and stability, and forming a soil ecosystem (Zhu et al., 2022). Soil microorganisms, as the most important biological components of soil ecosystems, typically exhibited rapid responses and high turnover rates to environmental changes. The ecological linkages between aboveground and belowground biota during forest succession have been considered an important mechanism in forest development and succession (Guo et al., 2024). For example, changes in plant species strongly influenced the community structure and function of soil microorganisms (Nagati et al., 2018). The key factor is that litter input alters soil abiotic properties such as pH and nutrient availability (Deng et al., 2016). Soil fungi, acting as decomposers, symbionts, and pathogens, are essential functional components of the soil. Extracellular enzymes produced by fungi participate in the cycling and metabolism of soil C, N, and P, significantly affecting the decomposition of plant litter (Lin et al., 2018; Li et al., 2018). Fungal biomass is sensitive to changes in soil properties, and generally higher in broad-leaved forests than in coniferous forests (Noll et al., 2016). Saprophytic soil fungi can decompose soil substrates and enhance soil nutrient cycling (Nagati et al., 2018; Liu et al., 2018), while ectomycorrhizal fungi (Preetisri et al., 2017), as symbionts, are known to improve plant nutrition status (Becquer et al., 2018; Clasen et al., 2018).
Community assembly refers to the mechanisms of its diversity formation and maintenance and mainly comprises deterministic processes and stochastic processes. The former involves environmental filtering and biological interactions, while the latter includes dispersal limitation and random variation. According to Peng et al. (2021), it is a necessary factor for coupling microbial community structure with ecosystem function (Wan et al., 2021). The construction of soil fungal communities has been proved to be influenced by complex interactions between plant community changes and various environmental factors (Canini et al., 2019; Geng et al., 2023). Vegetation type significantly affected soil fungal community structures (Porazinska et al., 2018), which might be achieved by altering the chemical composition and content of litter and root exudates or by mediating through the regulation of soil physical and chemical properties (Yang and Zhu, 2015; Ortiz et al., 2022). Studies have shown significant differences in soil fungal community structures between coniferous forests and broadleaved forests (Vuong et al., 2020). Based on ecological niche width and distribution range (Abdullah Al et al., 2022), soil fungal species exhibit varying degrees of adaptability to environmental changes, with the community including habitat generalists that can adapt to substantial changes in environmental quality and habitat-specialists that can only survive in relatively narrow habitat ranges (Sriswasdi et al., 2017). Previous studies have indicated that habitat generalists and habitat specialists among soil fungi adapt to environmental changes differently, suggesting variations in their community assembly mechanisms (Zheng et al., 2023; Mo et al., 2021; Yan et al., 2022; Zuo et al., 2023). However, how different plantation types filter soil fungal communities along a habitat-specialization gradient in limestone forest remains poorly understood.
Intraspecific and interspecific interactions of soil fungi were key drivers in community assembly. Co-occurrence network analysis effectively quantified interactions by identifying potential associations between species (Faust and Raes, 2012). Network topology parameters (such as average degree, closeness centrality) can identify the key species that have the greatest impact on community stability (Banerjee et al., 2018; Li et al., 2020). Furthermore, the modular structure of complex co-occurrence networks is crucial for maintaining overall community stability and functional differentiation (Xue et al., 2018; Liu et al., 2019; Ratzke et al., 2020). Worth noting that, recent studies indicated that habitat specialists contribute more significantly to maintaining network structure than habitat generalists. (Abdullah Al et al., 2022; Yan et al., 2022).
Shandong Province is located in the North Temperate Zone, where agriculture and forestry were originally suitable for habitation. Historical deforestation has led to the degradation of most mountainous regions into barren rock (Wu, 2017; Tan et al., 2020), Severely damaged the original diversity and service functions of the forest ecosystem (Wang et al., 2025). It was not until the 1960s that fast-growing and drought-resistant tree species such as Pinus tabulaeformis, P. thunbergii, and Robinia pseudoacacia were intentionally introduced on thin limestone slopes, gradually establishing the existing artificial forests (Du et al., 2020). Our study area, the Fanggan Ecological Restoration Area, is located in the northern foothills of the Taishan Mountain Range in Shandong Province. Since 1970, villagers have planted over three million trees for ecological restoration, forming diverse plantation types including coniferous, broad-leaved, and mixed forests. Currently, the ecological restoration area evolved into a thriving community with a well-defined vertical structure and dense canopy, supporting the healthy growth of various forest types (Qiu et al., 2022). Consistent soil conditions before planting, due to soil transfer for vegetation recovery on barren mountains, enable a more accurate assessment of the impact of different plant types on the soil ecosystem. Previous reports have detailed the effects of plantation types on soil properties, bacterial community structure and diversity (Qiu et al., 2022) and analyzed the characteristics and mechanisms of bacterial community structure changes in response to plantation types from both habitat specialization and abundance dimensions (Qiu et al., 2023). However, studies on how the community structure and co-occurrence networks of soil fungi respond to different afforestation types along a habitat specialization gradient were still particularly lacking.
This study focused on soil fungi in coniferous forests, mixed forests, and broad-leaved forests developed in the Fanggan Ecological Restoration Area. We explored the characteristics and environmental driving mechanisms of diversity and structure responses of different habitat specialization groups to different afforest types, and conducted analyses of fungal co-occurrence network features, topological parameters, and key groups. Our research aimed to reveal: (1) differences in the response of abundance, ecological niche width, diversity, and community structure of different habitat specialization groups to afforestation types; (2) differences in the community assembly characteristics of different habitat specialization groups among afforestation types; (3) differences in network structure characteristics and key groups of different habitat specialization groups among afforestation types.
2 Materials and methods
2.1 Study site and plot sets
The Fanggan Ecological Restoration Area is located in the northern region of the Shandong Central Mountain Range, with geographical coordinates spanning from 117°24′45″ to 117°28′5″E and 36°24′23″ to 36°26′44″N. This area is characterized by mountainous and hilly terrain, reaching a maximum altitude of 860 m and featuring over 30 peaks exceeding 400 m in height. The climate of restoration zone is classified as a continental monsoon climate, with the average annual temperature at 12.4 °C, and the lowest extreme temperature record of −22.5 °C. The average value of annual precipitation exceeded 830 mm, predominantly occurred from July to September. The predominant soil type in the region is mountain brown soil, with an average litter layer of 4 cm. The vegetation community in the area displays a distinct vertical structure, but species diversity is low. The tree layer primarily comprises Pinus densiflora Siebold & Zucc., P. tabulaeformis Carrière, Populus davidiana Dode, Robinia pseudoacacia L., and Diospyros lotus L. The shrub layer is mainly composed of Vitex negundo var. heterophylla (Franch.) Rehd., Ziziphus jujuba var. spinosa (Bunge) Hu ex H. F. Chow., Grewia biloba G. Don, along with seedlings regenerating from arbor layer. The herbaceous layer predominantly consists of species from the Gramineae, Cyperaceae, and Compositae. All plantations within the restoration area were established between 1975 and 1985, resulting in a relatively uniform forest age.
Based on a review of relevant literature and field surveys, the forests in the restoration area are primarily located between 300 and 750 m in altitude. Consequently, an investigation into the vegetation communities was conducted in regions with dense forest canopies within this altitude range. 24 sample plots with each area of 10 m × 10 m were established, covering three plantation forest types: six coniferous forest (CF) plots, six mixed coniferous and broad-leaved forest (MF) plots, and twelve broad-leaved forest (BF) plots. To minimize the impact of topographical factors, the design of each plot aimed to have as similar topographical conditions as possible. Additionally, the geographical coordinates, elevation, aspect, and slope of each plot were recorded.
2.2 Vegetation community survey and soil physicochemical property analysis
Measurements were taken for each tree with DBH ≥ 3 cm. α-diversity indices of the vegetation community, including the Shannon-Wiener index (Equation 1) (Shannon and Weaver, 1949), Simpson diversity index (Equation 2) (Simpson, 1949), Shannon evenness index (Equation 3), Simpson evenness index (Equation 4), Pielou evenness index (Equation 5) (Pielou, 2011), and species richness were calculated.
In each plot, topsoil samples within the 0–10 cm range were collected using the five-point sampling method. The sampling points were arranged at the center and four corners (3 m from the vertex of the plot). Then the samples at the five points in each plot were thoroughly mixed and then divided into two parts. As a result, there were a total of 48 soil samples. One part was stored in liquid nitrogen for molecular biology extraction and analysis, while the other was kept at room temperature and promptly brought back to the laboratory for physicochemical analysis.
Soil pH was determined by potentiometric method with 2.5:1 water-soil ratio. Organic carbon content (SOC) was determined by potassium dichromate oxidation-spectrophotometry (Ministry of Environmental Protection, PRC, 2011), and available phosphorus (AP) was determined by molybdenum-antimony anti-color spectrophotometry (Ministry of Agriculture, PRC, 2012). We used potassium chloride solution extraction-spectrophotometry and ultraviolet spectrophotometry to determine ammonium nitrogen (NH4+-N) and the content of Rapid-acting potassium (SK) was determined using the ammonium acetate extraction and flame photometric method.
2.3 DNA extraction, amplification, and sequencing
Total genomic DNA was extracted from each soil sample using the TGuide S96 Magnetic Soil/Stool DNA Kit [Tiangen Biotech (Beijing) Co., Ltd.]. The quality and quantity of the extracted DNA were examined using electrophoresis on a 1.8% agarose gel, and DNA concentration and purity were determined with Nano Drop 2000 UV–Vis spectrophotometer (Thermo Scientific, Wilmington, USA). The ITS1 sequence were amplified with primer pairs F: CTTGGTCATTTAGAGGAAGTAA and R: GCTGCGTTCTTCATCGATGC for 25 cycles. The amplification reaction system was as follows: 25 μL of Taq enzyme, 1 μL of 10 mM forward and reverse primers, 3 μL of template DNA (20 ng μL−1), and ddH2O added up to 50 μL. The thermal cycling conditions were: initial denaturation at 95 °C for 5 min; denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, extension at 72 °C for 40 s, repeated for 25 cycles; final extension at 72 °C for 7 min. PCR amplicons were purified with VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Qubit dsDNA HS Assay Kit and Qubit 3.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Oregon, USA). After the individual quantification step, amplicons were pooled in equal amounts. SMRTbell libraries were prepared from the amplified DNA by SMRTbell Express Template Prep Kit 2.0 according to the manufacturer’s instructions (Pacific Biosciences). Purified SMRTbell libraries from the pooled and barcoded samples were sequenced on a Illumina platform (Beijing Biomarker Technologies Co., Ltd., Beijing, China) using Sequel II binding kit 2.0.
The bioinformatics analysis of this study was performed with the aid of the BMK Cloud.1 The raw reads generated from sequencing were filtered and demultiplexed using the SMRT Link software (version 8.0) with the min Passes ≥5 and min Predicted Accuracy ≥0.9, in order to obtain the circular consensus sequencing (CCS) reads. Subsequently, the lima (version 1.7.0) was employed to assign the CCS sequences to the corresponding samples based on their barcodes. CCS reads containing no primers and those reads beyond the length range (1,200–1,650 bp) were discarded through the recognition of forward and reverse primers and quality filtering using the Cut adapt (version 2.7) quality control process (Bolger et al., 2014). UCHIME algorithm (v8.1) was used in detecting and removing chimera sequences to obtain the clean reads (Büchi and Vuilleumier, 2014). Clean reads then were conducted on feature classification to output an ASVs (amplicon sequence variants) by DADA2 (Callahan et al., 2016), and the ASVs counts less than 2 in all samples were filtered. Taxonomy annotation of the ASVs was performed based on the Naive Bayes classifier in QIIME2 (Bolyen et al., 2019) using the SILVA database (Quast et al., 2012) (release 138.1) with a confidence threshold of 70%.
2.4 Identification of habitat generalists, specialists, and opportunists of soil fungi
Levins niche width (Equation 6) (Levins, 1968) was calculated using the ‘spaa’ package of R v 4.0.0 (Zhang and Ma, 2014). The frequency of occurrence of each ASV annotate to species level was randomized and rearranged 1,000 times using the replacement method of the ‘EcolUtilis’ package (Salazar, 2019), respectively. Then the zero distribution of the niche width index was calculated for each ASV annotate to species level. Based on the consensus that habitat-generalists owe a wider niche width than the habitat-specialists (Wilson and Hayek, 2015), ASV species were classified into habitat-generalists or specialists based on whether the frequency of observed occurrence exceeded the upper 95% confidence interval or fell below the lower 5% confidence interval, and the others with the observed niche width within in the 5 to 95% confidence interval range were distributed to the habitat-opportunists (Wu et al., 2022).
2.5 Co-occurrence network and key taxa analysis
The co-occurrence patterns of soil fungi communities among the different forest types were determined using network analysis. Co-occurring networks based on Spearman correlation analysis in this study were conducted using the “Hmisc” package in R program. The co-occurrence patterns of soil fungal communities were studied based on strong correlations (r > 0.7) and significant correlations (p < 0.05). Network analysis was performed using “igraph” package (Csárdi et al., 2024) and then visualized in Gephi 0.9.
Moreover, the “igraph” package was employed to calculate the within-module connectivity (Zi) and between-module connectivity (Pi) of the constructed network, aiming to identify key species. Based on within-module connectivity (Zi) and among-module connectivity (Pi), ASVs were classified into network hubs (Zi ≥ 2.5; Pi ≥ 0.62), module hubs (Zi ≥ 2.5; Pi < 0.62), connectors (Zi < 2.5; Pi ≥ 0.62), and peripherals (Zi < 2.5; Pi < 0.62) Generally, the other three types of nodes except peripherals were classified as key nodes (Deng et al., 2012), and these nodes played a crucial role in maintaining the stability of the network structure (Hu et al., 2024).
2.6 Data analysis
Based on the ASVs abundance matrix, the Shannon-Wiener diversity indices and ASVs species richness for each habitat-specialists were computed. After testing for normality and homogeneity of variances, one-way ANOVA (SPSS 19.0) was used to analyze the significance of soil physicochemical properties, niche width, abundance, and α—diversity indices among different afforestation types. Principal Coordinates Analysis (PCoA) function from the ‘vegan’ package was utilized to examine the differences in community structure of each habitat-specialization group among afforestation types. The significance of these structural differences was assessed through a Permanova test (using the adonis function in the vegan package of R software, with permutations = 999 and the Bray-Curtis distance metric). The RDA function was applied to analyze the impacts of tree composition, tree diversity, and soil physicochemical properties on the microbial subgroup community structure. The significance of the RDA model was tested using ANOVA, and the Monte Carlo permutation test was employed to evaluate the significance of the influencing factors of each explanatory variable in the RDA model (via the anova.cca and envfit functions in the vegan package of R software, with permutations = 999). The graph was created using the ggplot2 package in R version 4.2.2.
3 Results and analysis
3.1 Response characteristics in the abundance of ASVs and niche width of each habitat specialization group to afforestation types
Soil pH and available potassium content of broadleaved forest soil were significantly higher than those of other forest types (p < 0.05), indicating that establishing broadleaved forests can more effectively increase soil pH and available potassium content compared to other forest types. The effective phosphorus content in mixed forests and broadleaved forests was higher than that in coniferous forests, but the difference was not significant (p > 0.05), suggesting that both mixed forests and broadleaved forests positively contribute to increasing soil effective phosphorus content. However, ammonium nitrogen was significantly higher in coniferous forests and mixed forests compared to broadleaved forests (p < 0.05). The organic carbon content showed no significant variation with different forest types (Table 1). The number of sequences obtained from sample sequencing ranged from 35,076 to 61,484, with ASVs varying between 304 and 1,359. Among them, 1,189 ASVs were annotated to the species level. 90 of habitat generalist accounted for 7.57%, 534 of habitat opportunists accounted for 44.91%, and 564 of habitat specialists accounted for 47.43% were identified.
The abundance of soil fungi in each type of plantation forests increased with the degree of habitat specialization. In broadleaved forests, the abundance of habitat-generalist and opportunities was higher compared to coniferous and mixed forests, while the abundance of habitat-specialist showed the opposite trend (Figure 1), suggesting a higher degree of habitat specialization in coniferous forest soil environments. No significant difference in niche widths was found among each habitat-specialization group. Among them, habitat generalists showed a significantly higher niche width in broadleaved forests compared to other forest types, while habitat-opportunities and specialists have a higher niche width in coniferous forests (Figure 2).
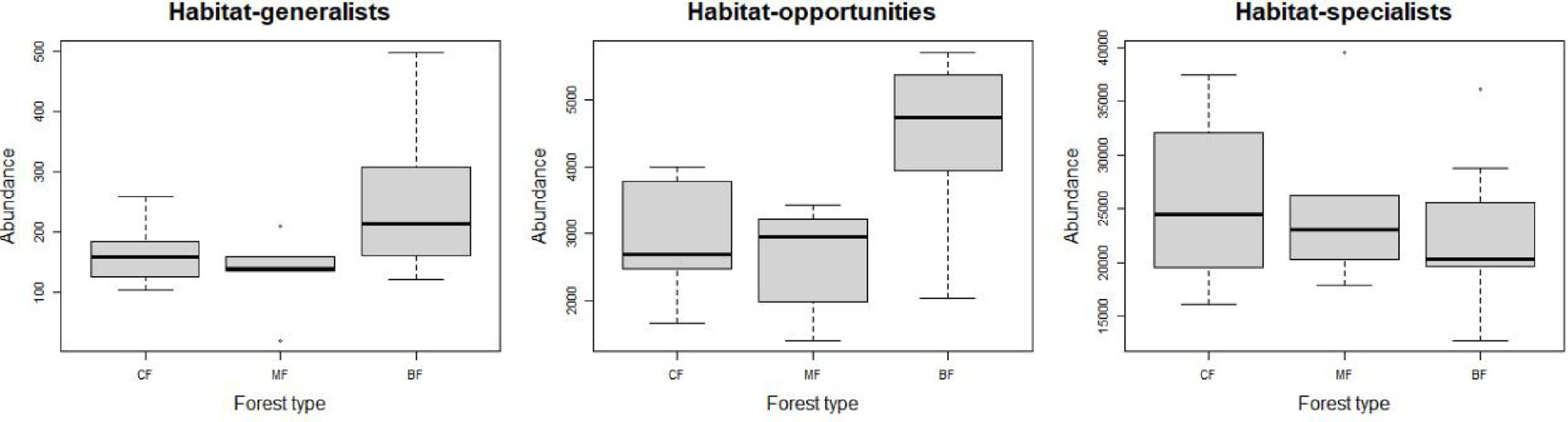
Figure 1. Response of fungal abundance with each habitat specialization group to afforestation types (CF, coniferous forest; MF, mixed forest; BF, broadleaved forest).
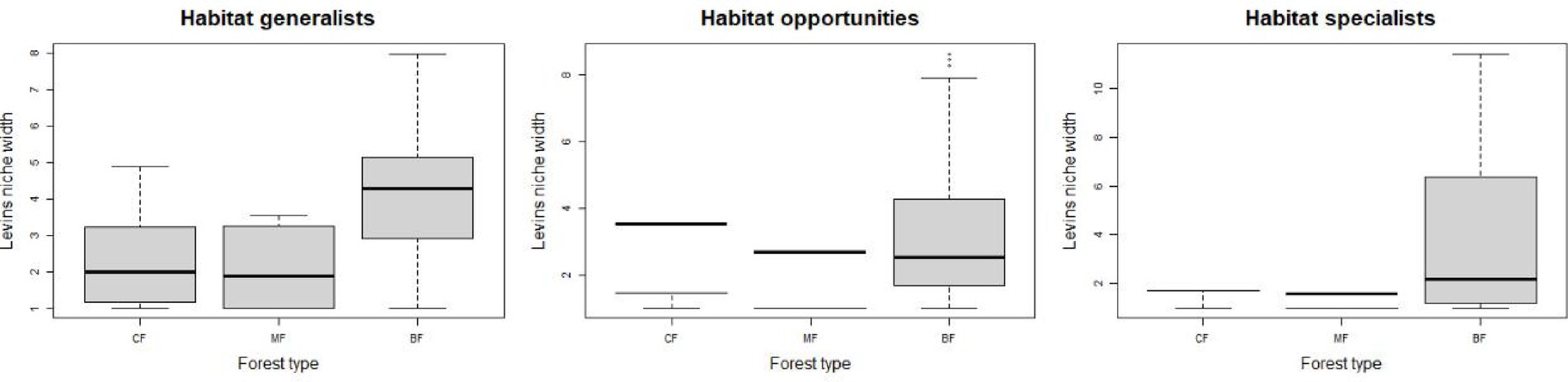
Figure 2. Response of fungal niche breadth among each habitat-specialization group to afforestation types (CF, coniferous forest; MF, mixed forest; BF, broadleaved forest).
3.2 Response characteristics of α-diversity and community structure of each habitat-specialization group of soil fungi to afforestation types
Our research indicated that the Shannon-Winner diversity index and species richness of each habitat-specialization group of soil fungi in broadleaved forests significantly higher than other afforestation types (Figures 3–5). However, the situation differs between coniferous and mixed forests: The diversity indexes were higher in coniferous for habitat-generalists and specialists, while they were higher in mixed forests for habitat-opportunities (Figures 3–5).
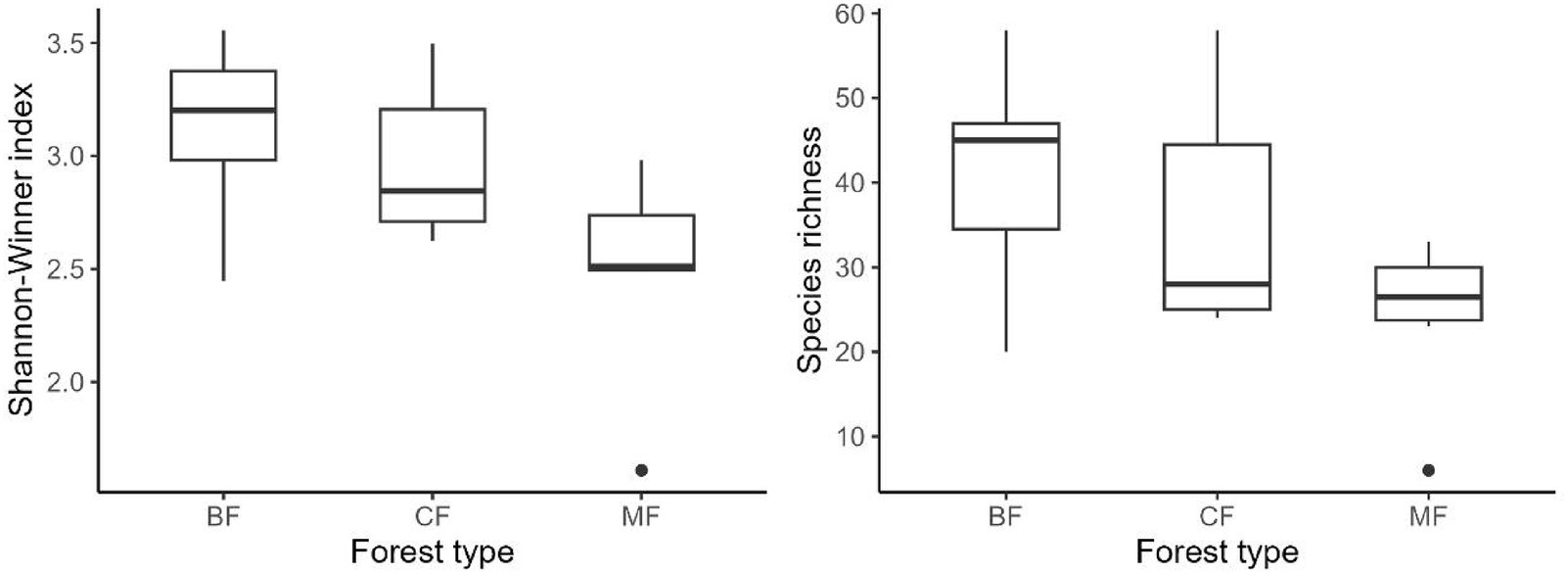
Figure 3. Response of species richness and Shannon-Winner index of habitat generalist to forest stands (CF, coniferous forest; MF, mixed forest; BF, broadleaf forest).
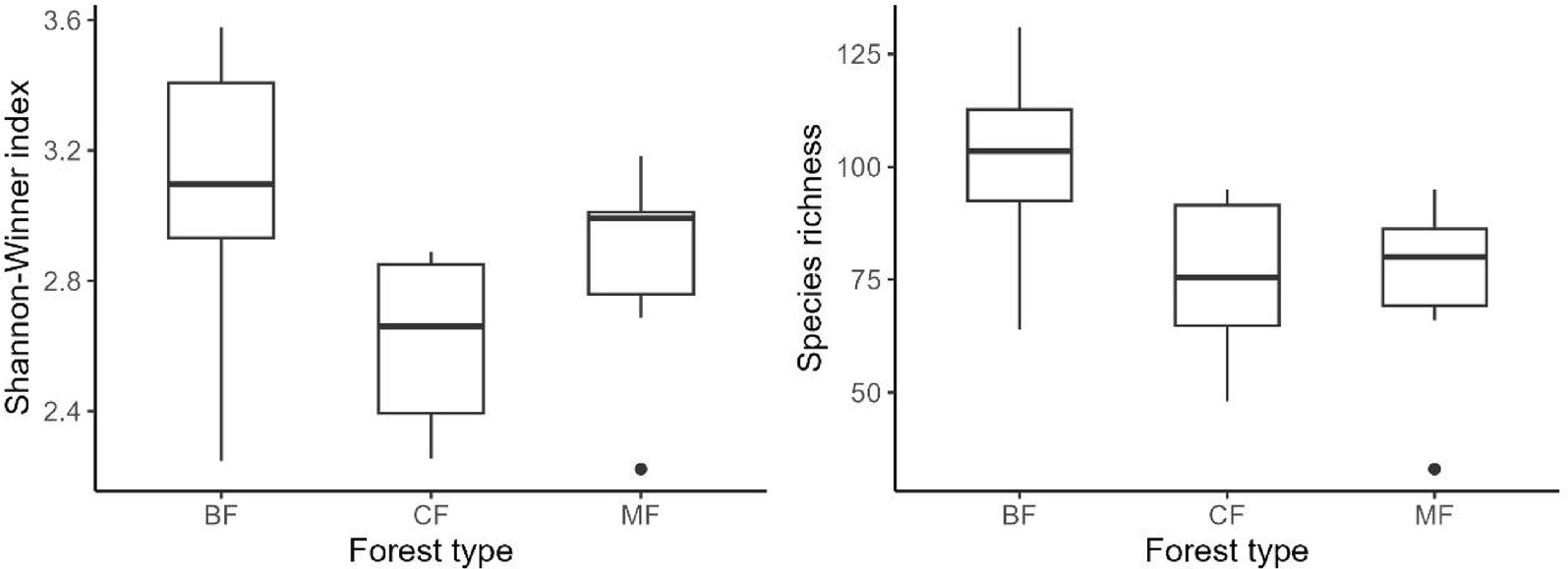
Figure 4. Response of species richness and Shannon-Winner index of habitat opportunists to forest stands (CF, coniferous forest; MF, mixed forest; BF, broadleaved forest).
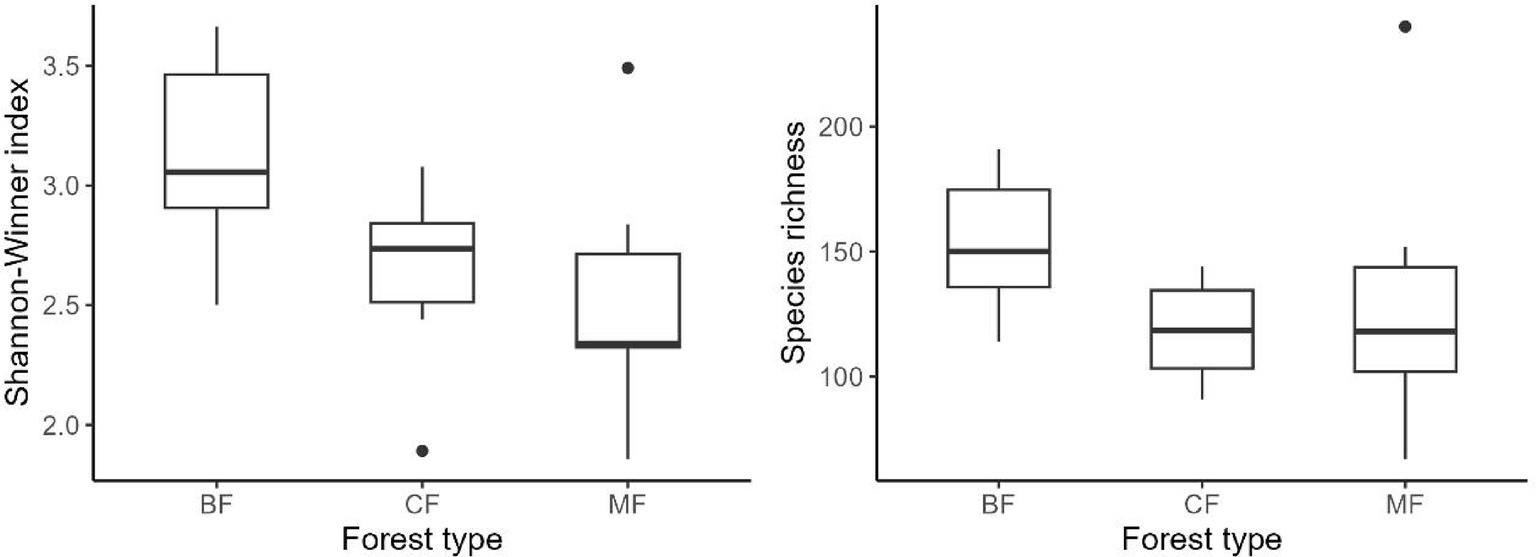
Figure 5. Response of species richness and Shannon-Winner index of habitat specialists to forest stands (CF, coniferous forest; MF, mixed forest; BF, broadleaved forest).
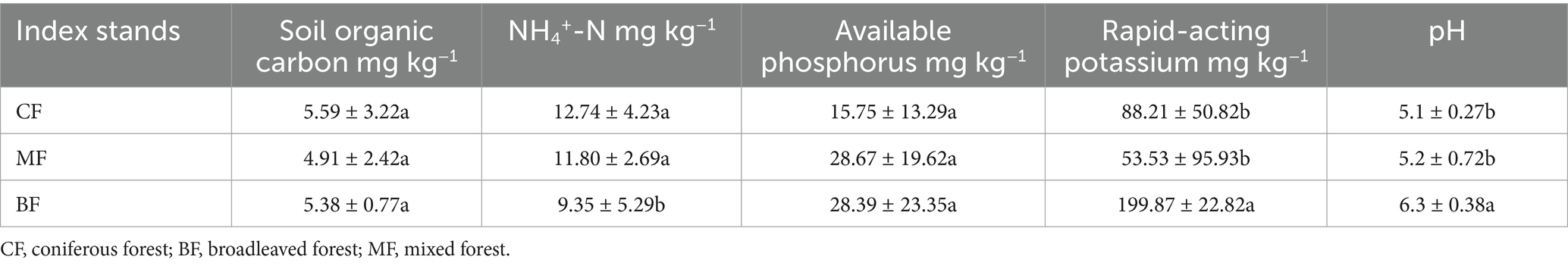
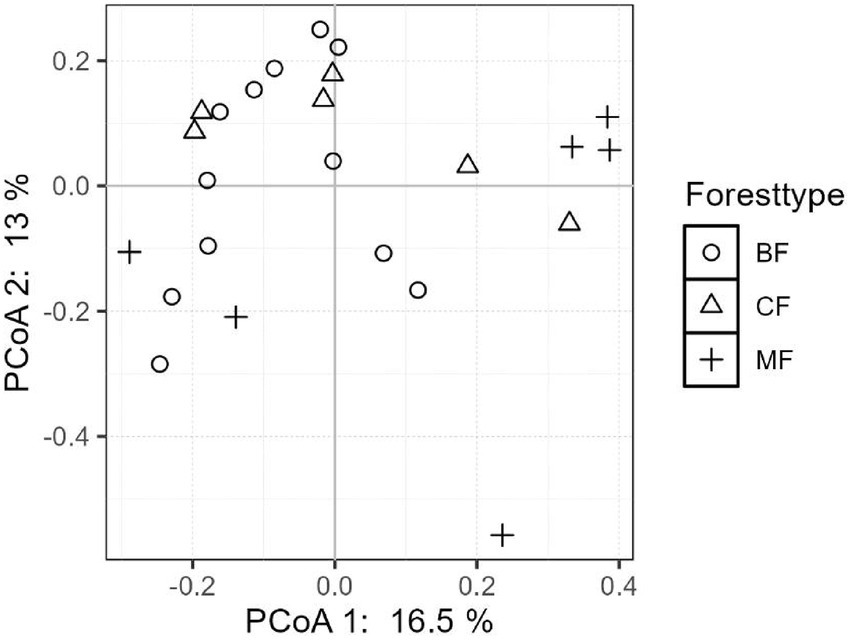
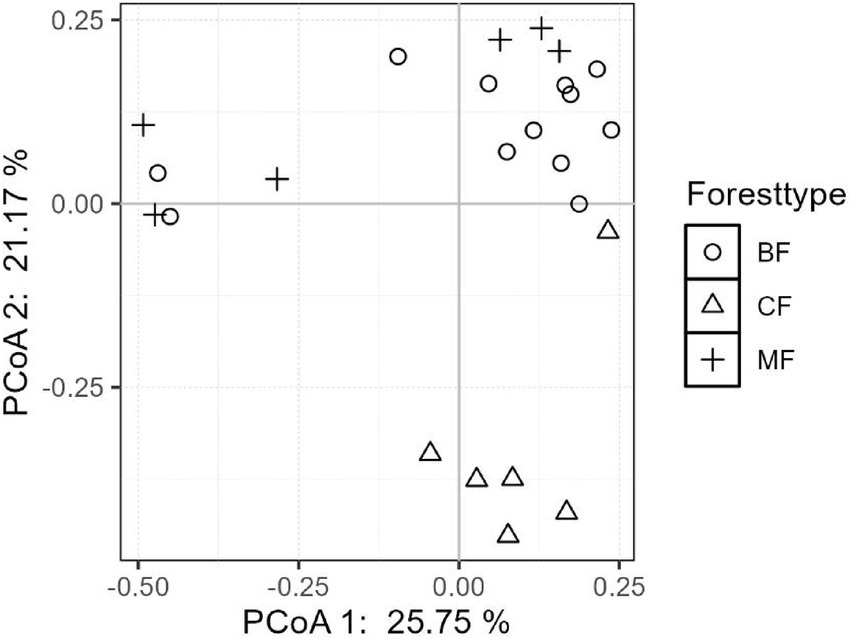
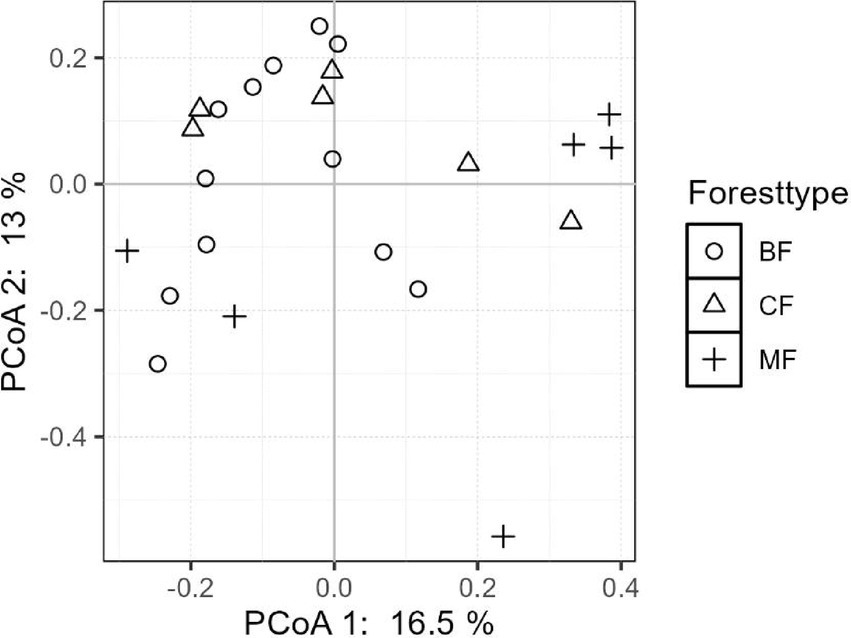
The differentiation of community structure among afforestation types increased with the degree of habitat specialization. The community structure of habitat generalist varied greatly within forest type but showed direct mixing between forest types, indicated no significant difference in community structure among afforestation types (Figure 6). In the case of habitat opportunists, the coordinate points of coniferous forests were already clearly separated from those of mixed and broadleaved forests, suggested a clear difference in community structure. The coordinate points of mixed forests were mostly mixed with those of broadleaved forests, suggested no significant difference in the structure of habitat opportunities (Figure 7). For habitat specialists, the coordinate points of different forest stands had distinct aggregation ranges in a two-dimensional graph, indicated a significant difference in community structure among afforestation types (Figure 8).
3.3 Environmental driving mechanisms of community structure for each habitat-specialization group among afforestation type
3.3.1 The driving mechanisms of plant-diversity on community structure
Research results indicated the structure of habitat opportunists was more significantly influenced by plant community diversity indices. All diversity indices collectively explain the structure of habitat opportunities at an extremely significant level (p < 0.01). In contrast, habitat specialists were not significantly affected by plant diversity indices (Figure 9, Table 2).
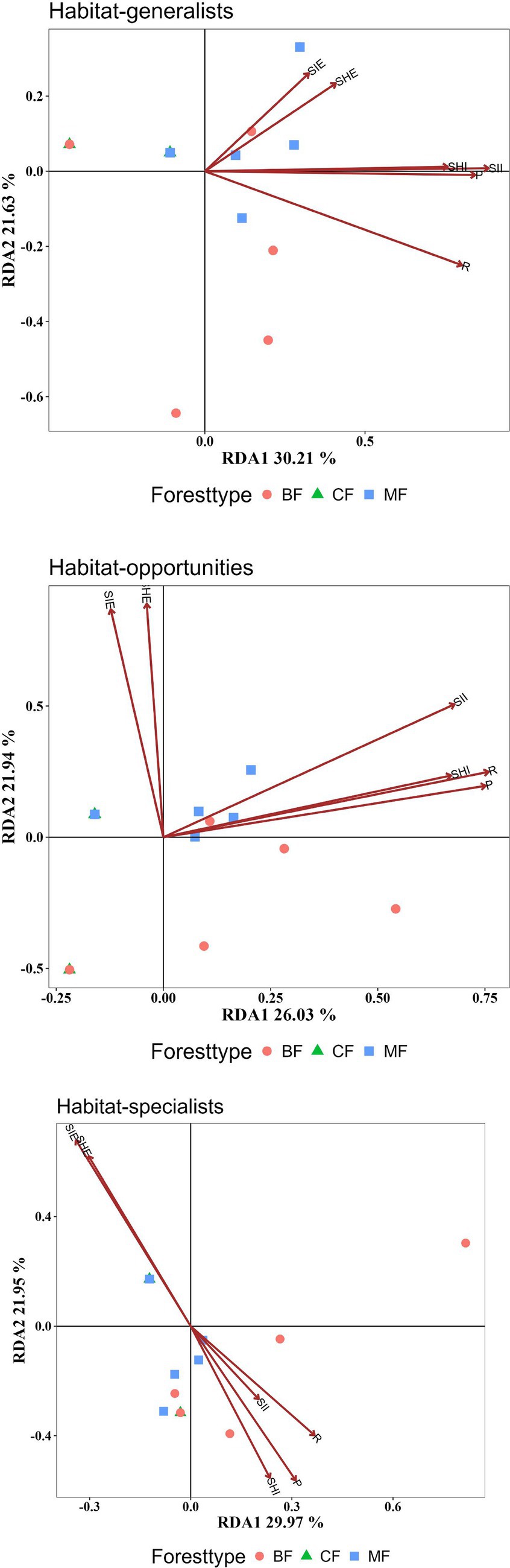
Figure 9. RDA analysis of the impact of plant diversity indices on the structure of each habitat-specialization group (CF, coniferous forest; MF, mixed forest; BF, broadleaved forest; R, Species Richness; SHI, Shannon-Winner Index; SII, Simpson Index; SHE, Shannon Evenness Index; SIE, Simpson Evenness Index; P, Pielou Evenness Index).
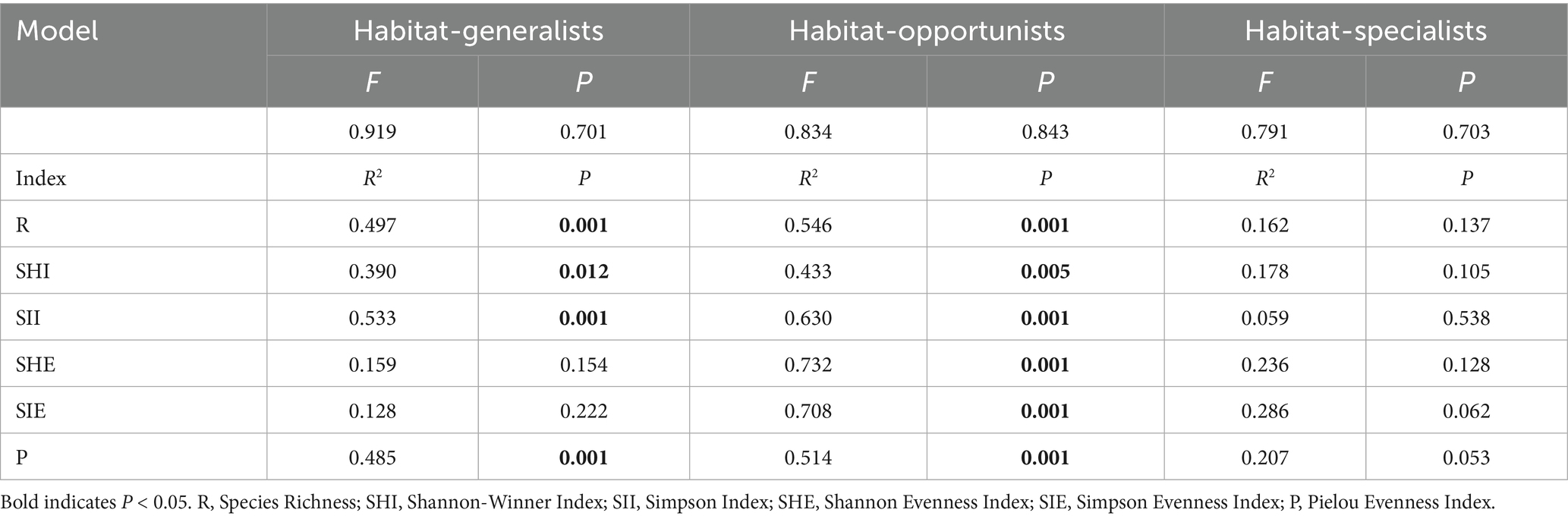
Table 2. Results of Monte Carlo test for RDA redundancy analysis of plant diversity on each habitat-specialization group.
3.3.2 The driving mechanisms of tree species composition on community structure
Studies have shown that the differences in the community structure of habitat-specialists between afforestation types were all influenced by the dominant tree species within the communities. The structural differences of habitat generalists among afforestation types were significantly influenced by the importance values of Populus davidiana and Platycladus orientalis. Habitat-opportunities were significantly affected by the importance values of Pinus tabulaeformis, Robinia pseudoacacia, and Populus davidiana. The habitat-specialists were significantly influenced by the importance values of Robinia pseudoacacia and Populus davidiana. The structural characteristics of each habitat-specialists vary from coniferous forests, mixed forests to broad-leaved forests, showing a positive correlation with the increase of Populus davidiana importance values and the decrease of Pinus tabulaeformis and Platycladus orientalis importance values (Figure 10, Table 3).
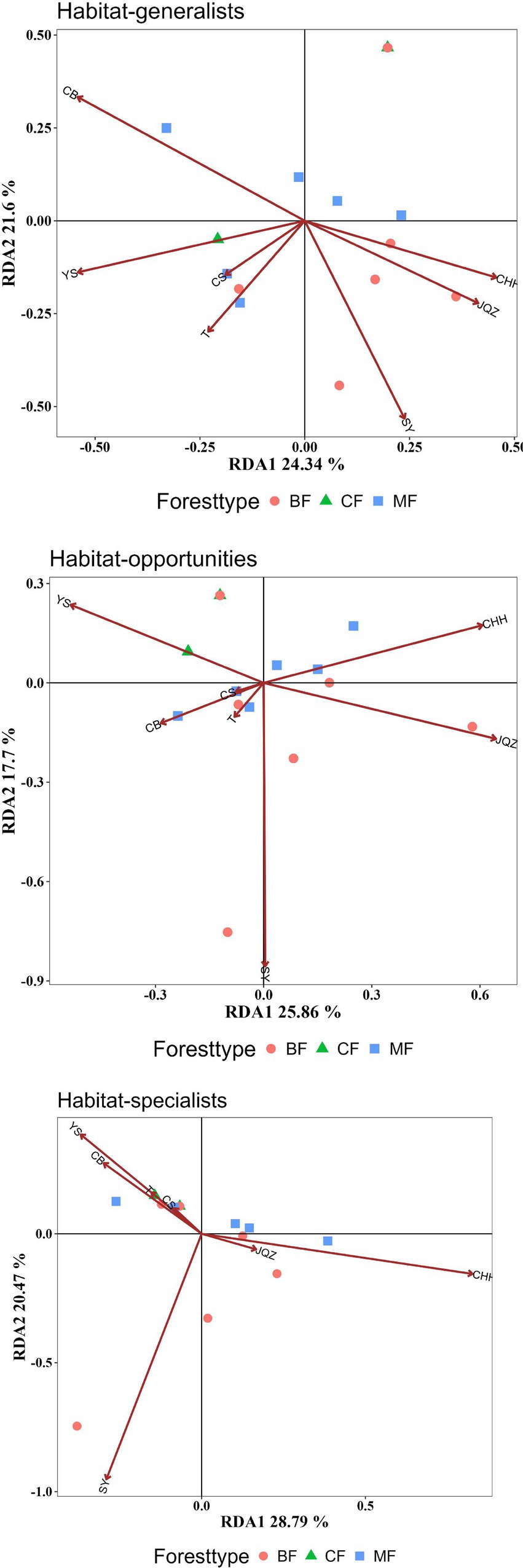
Figure 10. RDA analysis of the impact of plant composition on the structure of each habitat-specialization group (CF, coniferous forest; MF, mixed forest; BF, broadleaved forest; YS: Pinus tabulaeformis; CH: Robinia pseudoacacia; JQZ: Diospyros lotus; SY: Populus davidiana; CB: Platycladus orientalis; T: Prunus persica; CHH: Pinus densiflora).
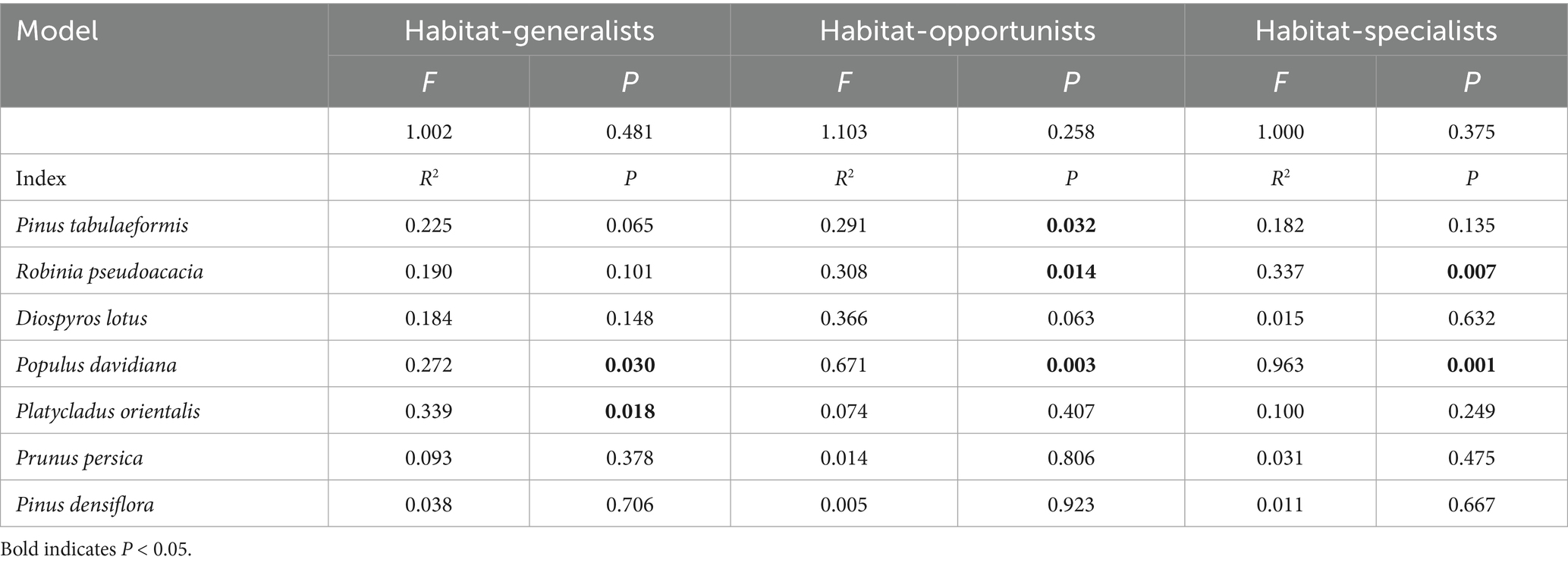
Table 3. Results of Monte Carlo test for Redundancy Analysis (RDA) of plant composition on each habitat-specialization group.
3.3.3 The driving mechanisms of soil properties on community structure
Habitat-generalists and opportunities tended to be significantly influenced by more soil indicators than specialists. AN and pH significantly affected on the structure of all habitat-specialization groups. Habitat generalist and opportunities of broadleaved forests could be distinctly separated on the RDA1 axis from coniferous and mixed forests, while habitat specialists showed overlapping coordinates across different forest types, indicated that soil properties significantly influence the community structure of generalist and opportunities broadleaved forests. The increase in SK and pH positively correlated with the differences in community structure between broadleaved forests and coniferous forests for each habitat specialization group, whereas AN content was negatively correlated with structural changes in habitat generalists and opportunities and (Figure 11, Table 4).
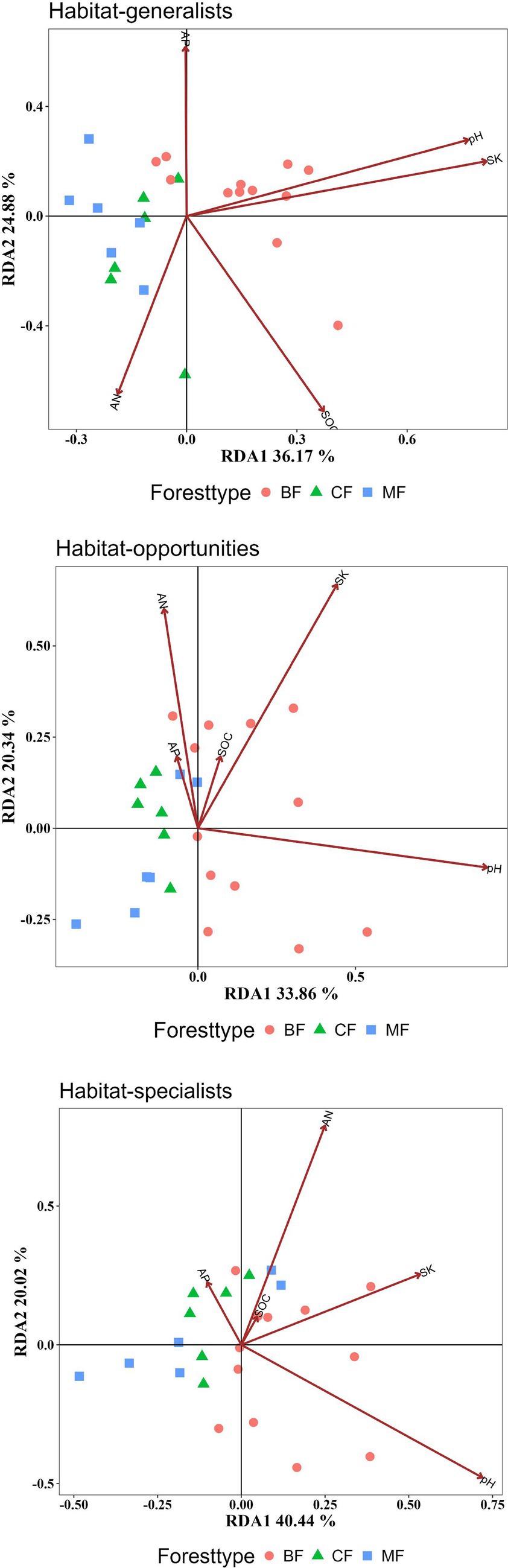
Figure 11. RDA analysis of the impact of soil properties on the structure of each habitat-specialization group (CF, coniferous forest; MF, mixed forest; BF, broadleaved forest).
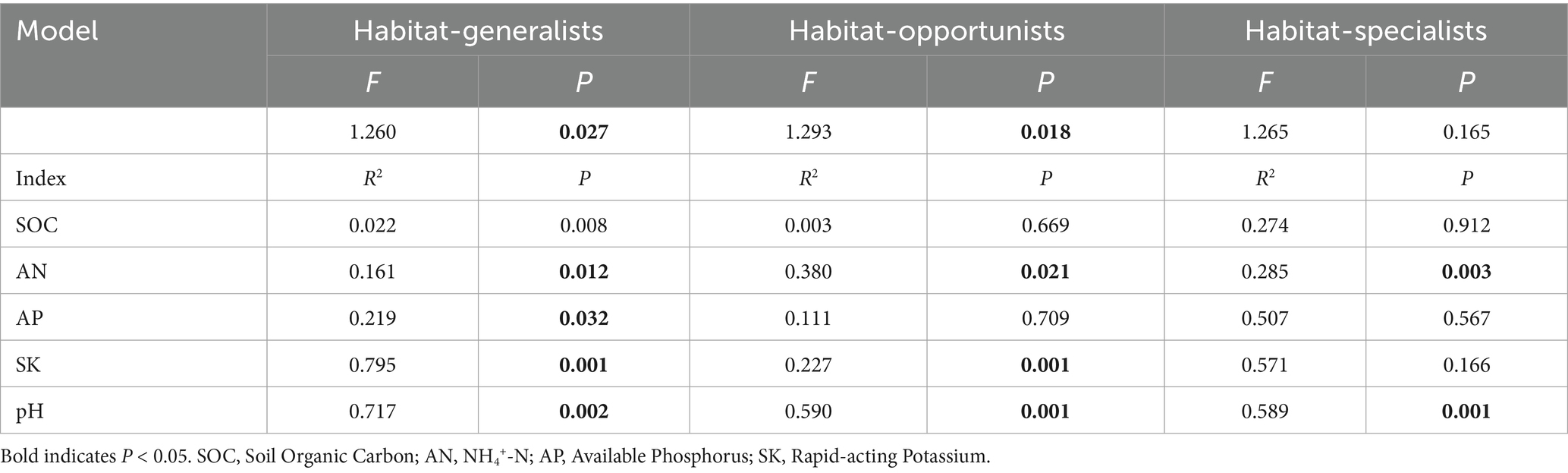
Table 4. Results of Monte Carlo test for Redundancy Analysis (RDA) of soil properties on each habitat-specialization group.
3.4 Response characteristics of network structure and key groups of each habitat specialization group to afforestation types
3.4.1 Modularity and topological characteristics of network structure of habitat-specialization groups
Compared to habitat generalists, the network structures of habitat opportunities and habitat specialists were more complex and have higher modularity (Supplementary Figure). Firstly, the connectivity between nodes of habitat specialists was higher, which can be reflected by the higher node degree, hub index, clustering coefficient, eigenvector centrality, and average path length compared to those of habitat opportunities and habitat specialists. These suggested that specialists and opportunities have higher internal connectivity within the network and were more likely to form modular structures. On the contrary, Habitat generalists formed more independent subnetworks, which were structurally more fragile, as evidenced by their higher eccentricity, betweenness centrality, module categories, PageRank values, greater number of components, and shorter average path lengths (see Table 5).
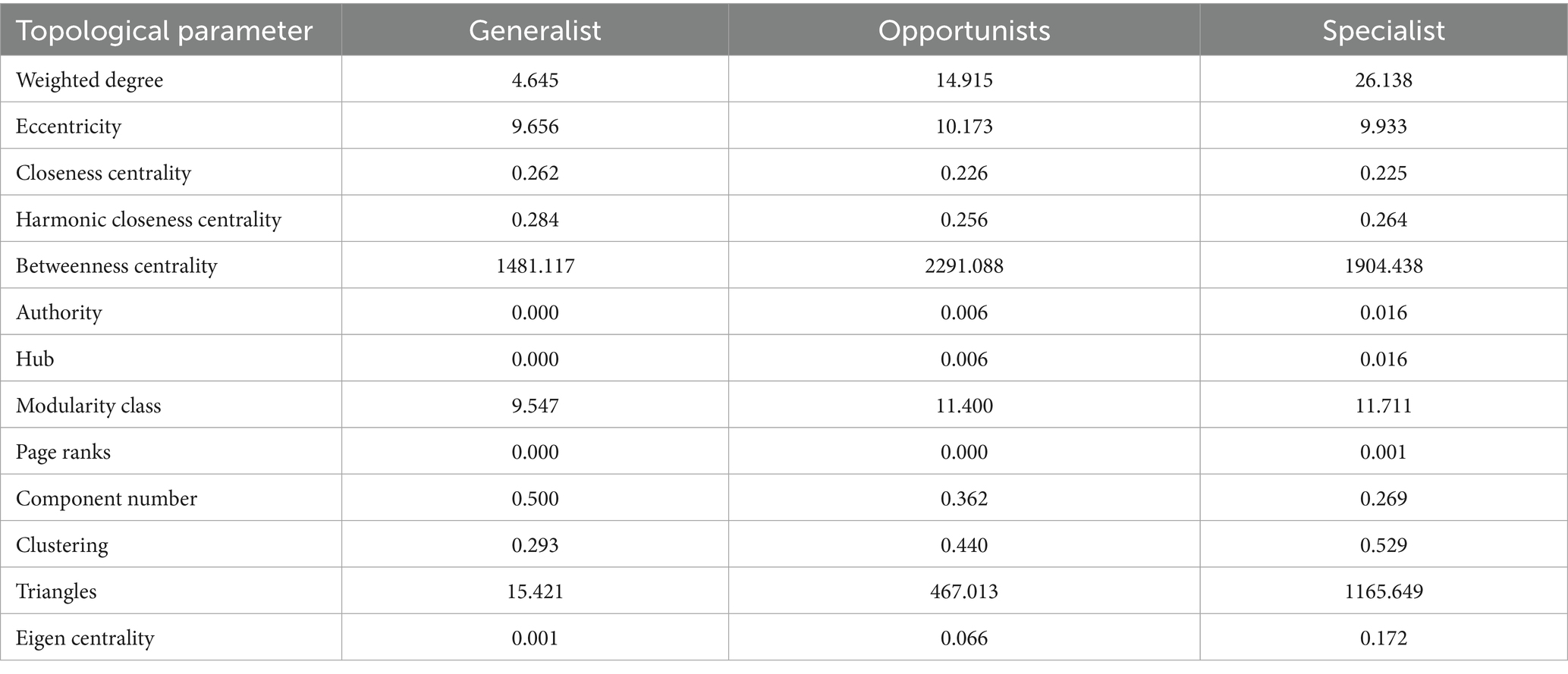
Table 5. Comparison of topological parameters of network structures for each habitat specialization group.
3.4.2 Response of network structures of habitat-specialization groups to forest type
Results indicated that the topological parameters of network structure of each habitat specialization group responded consistently to changes in forest stands. The average degree and graph density were the highest in mixed forests (Figures 12A,B), while modularity and the number of connected components were the lowest (Figures 12E,F). This can be interpreted as the complex soil environment of mixed forests generated more frequent and complex interactions among fungi, lead to higher average degree and graph density. This disrupted the clear boundaries between modules, thereby reducing the modularity index. In broad-leaved forests, the average path length and network diameter were the highest (Figures 12C,D), while the average clustering coefficient were the lowest (Figure 12G), indicating that the fungal network structure in broad-leaved forests is loose with weak node connections. This was also consistent with low graph density and high modularity index in broad-leaved forests.
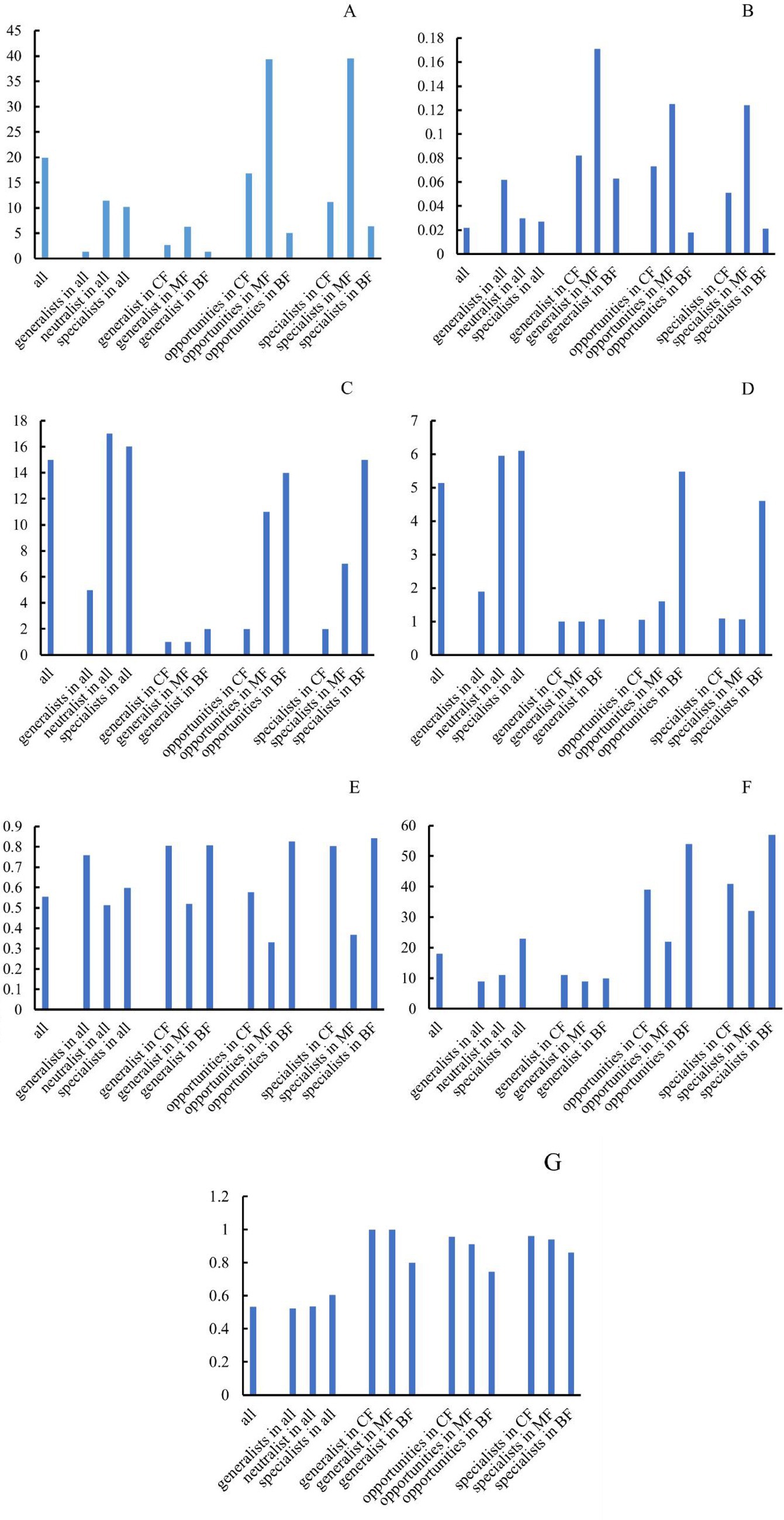
Figure 12. Difference of network topological index among forest stands for each habitat-specialization group (A: average weighted degree; B: density of graph; C: network diameter; D: average path length; E: modular; F: connective component; G: average clustering coefficient; CF, coniferous forest; MF, mixed forest; BF, broadleaved forest).
3.4.3 Key taxa of network structure
Studies have shown that the number of high-value Zi and Pi increased with the degree of habitat specialization. In the network structure of habitat generalists, only peripheral species were found (Figures 13A–D). For habitat opportunities, 4 species were identified as module hubs, with 1 in mixed forests, and 3 in the overall forest type. Additionally, 3 species were identified as connectors (Figures 13E–H). For habitat specialists, 4 species were identified as module hubs, and 1 was connector (Figures 13I–L). In different types of forest stands, the number of high-value Zi and Pi showed a trend of increasing from coniferous forests to mixed forests and then to broad-leaved forests. Nodes with Pi>0 were only present in broad-leaved forests, and their numbers were significantly higher in habitat opportunities and habitat specialists than in habitat generalists.
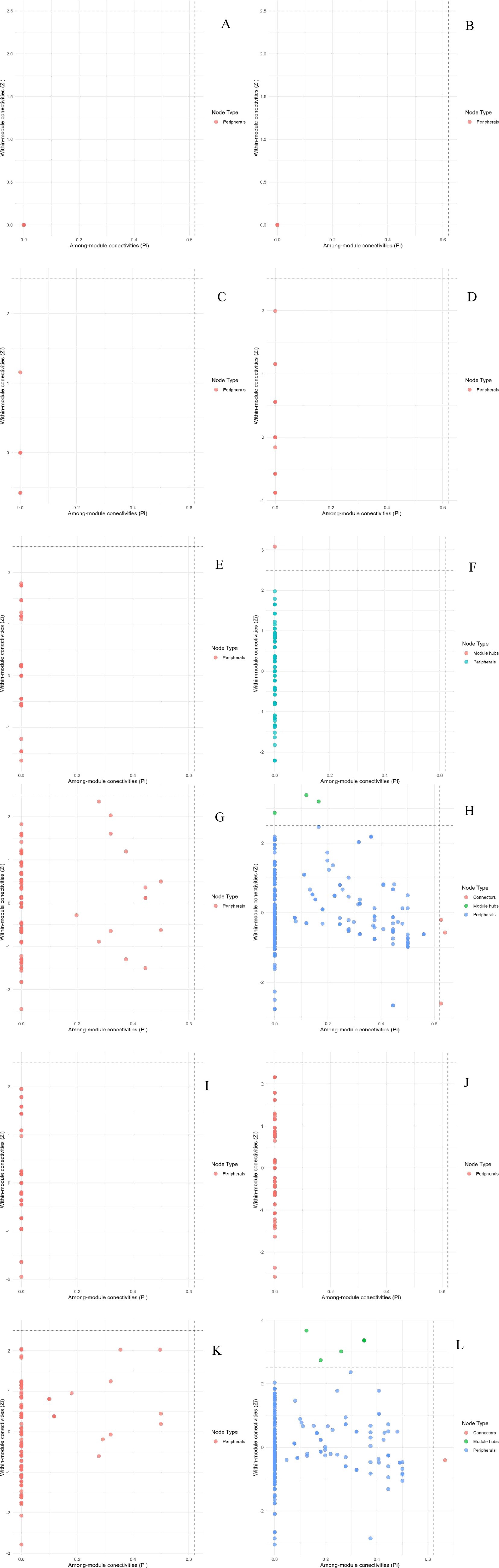
Figure 13. Differences of Zi and Pi of each habitat-specialization group between forest stands (A: Generalists of CF; B: Generalists of MF; C: Generalists of BF; D: Generalists of all stands; E: Opportunists of CF; F: Opportunists of MF; G: Opportunists of BF; H: Opportunists of all stands; I: Specialists of CF; J: Specialists of MF; K: Specialists of BF; L: Specialists of all stands).
The distribution characteristics of Zi and Pi values revealed that planting broad-leaved tree species enhanced the within-module and between-module connections of habitat opportunities and habitat specialists. First of all, the number of negative Zi values for habitat opportunities and habitat specialists was greater than the number of positive Zi values in coniferous forests, while in mixed forests and broad-leaved forests, the number of positive Zi values was greater than the number of negative Zi values (Figure 14). In addition, in coniferous forests, the positive mean of Zi was greater than the negative mean for habitat opportunities and habitat specialists, while in mixed forests and broad-leaved forests was the opposite trend, with negative mean of Zi greater than the positive mean (Figure 15). The connectivity between modules in all habitat specialization groups was poor, with most Pi values being 0. Only a few nodes with positive Pi values are present in the broad-leaved forest for habitat opportunities and habitat specialists (Figures 16, 17).
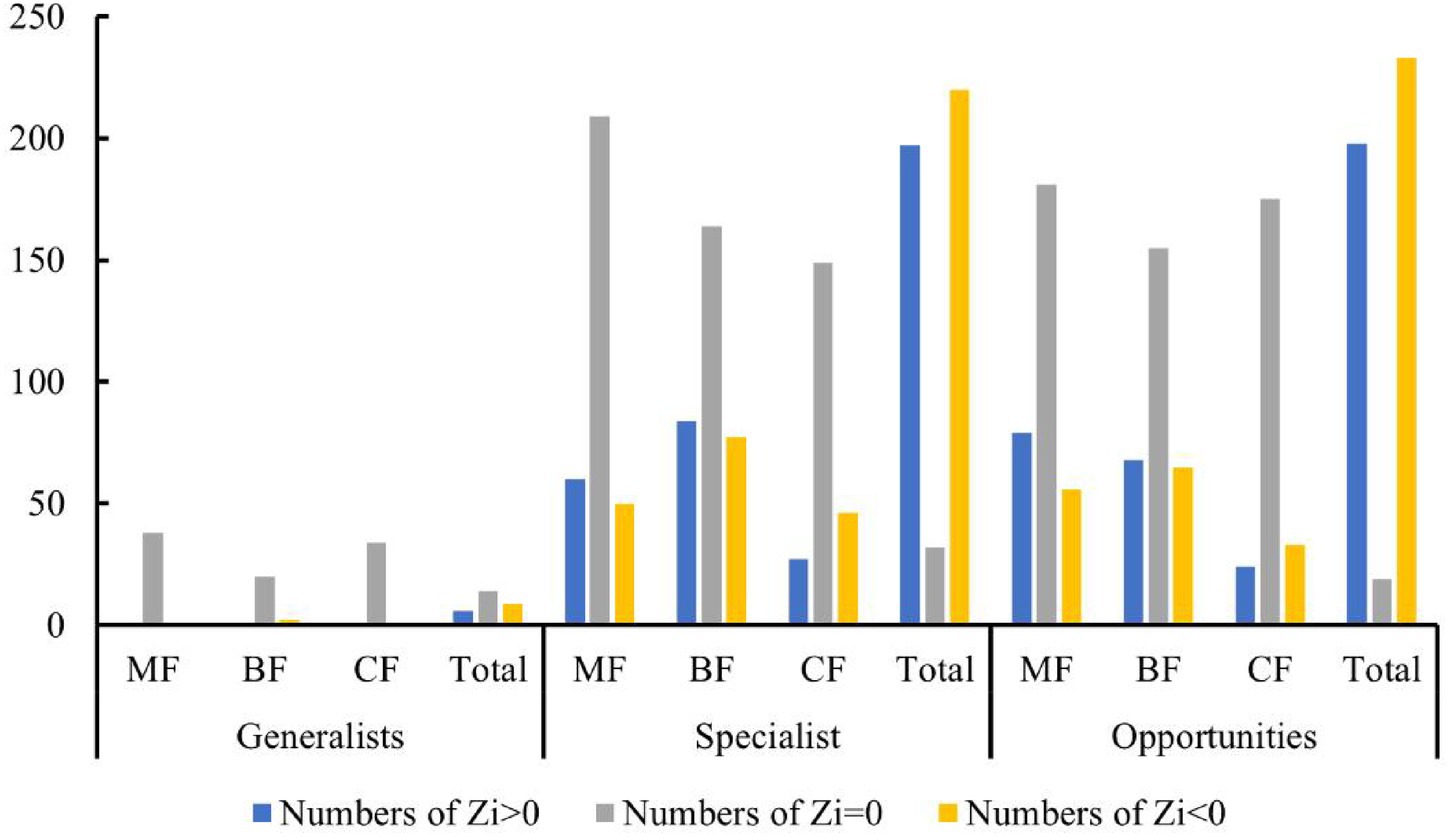
Figure 14. Differences of numbers of positive and negative Zi values among forest types of each habitat-specialization group.
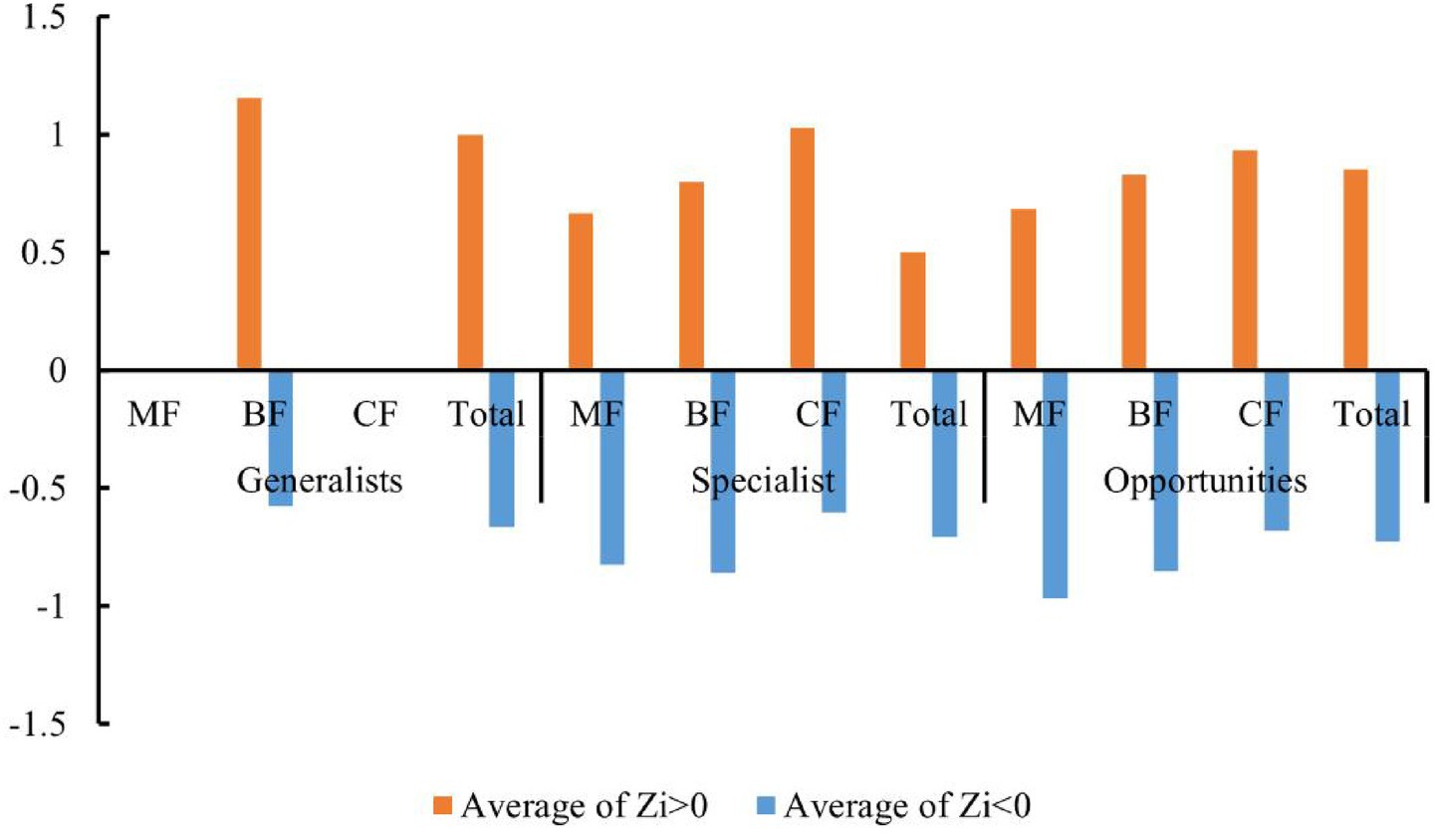
Figure 15. Differences of average positive and negative Zi values among forest types of each habitat-specialization group (CF, coniferous forest; MF, mixed forest; BF, broadleaf forest).
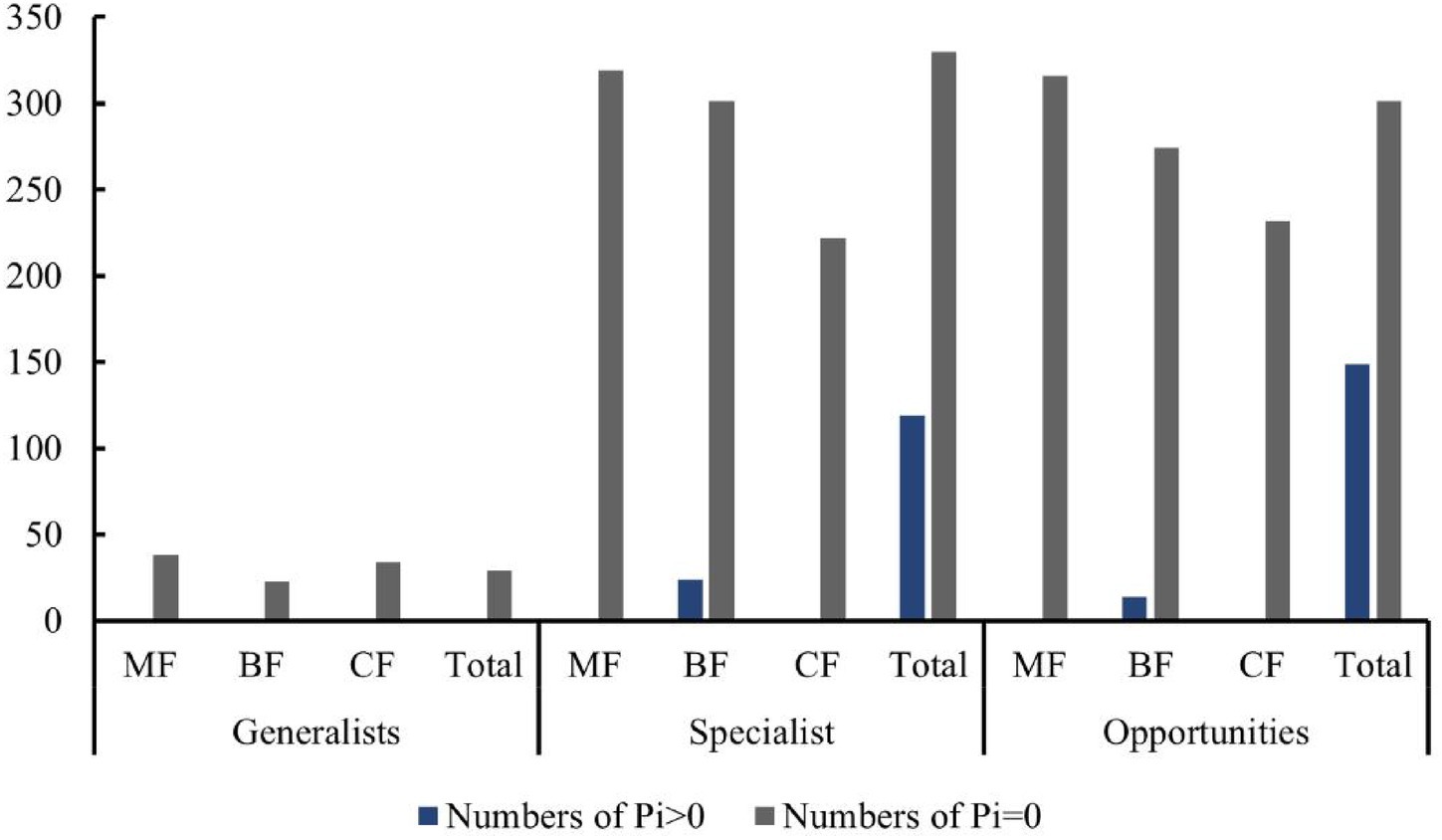
Figure 16. Differences of numbers of positive and 0 Pi values among forest types of each habitat-specialization group (CF, coniferous forest; MF, mixed forest; BF, broadleaf forest).
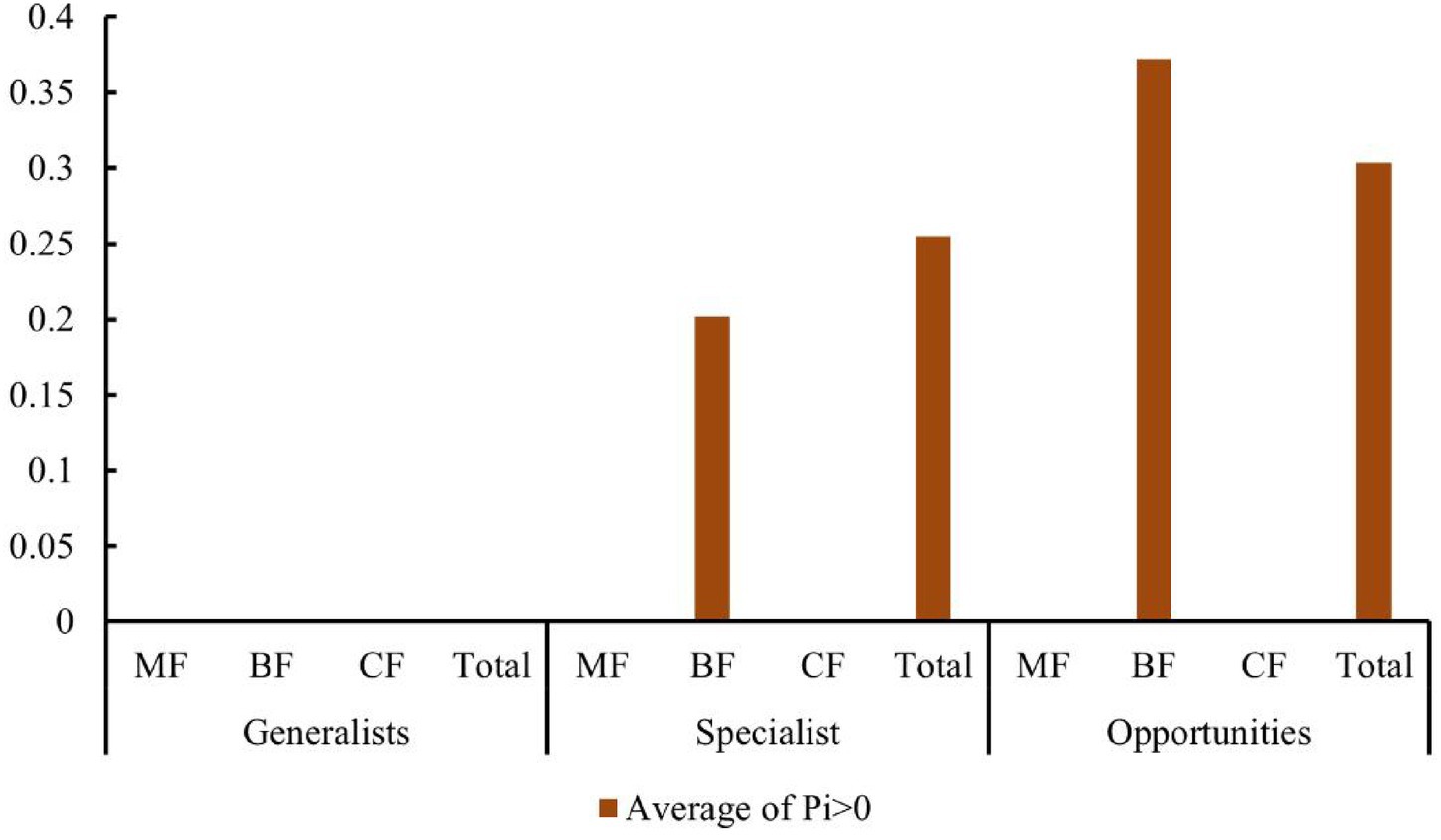
Figure 17. Differences of average Pi values among forest types of each habitat-specialization group (CF, coniferous forest; MF, mixed forest; BF, broadleaf forest).
4 Discussion
4.1 Impact of plantation types on the abundance and α-diversity of soil fungi with different habitat specialization
This study categorized soil fungi based on the confidence interval distribution of niche width, into three major groups: habitat generalists, specialists, and neutrals. Furthermore, the response characteristics of soil fungal community composition and structure, network relationships, and assembly mechanisms to different types of artificial forests were explored along this habitat specialization gradient. Our research results strongly supported the highest heterogeneity of soil environments in broad-leaved forest. Firstly, the abundance of habitat-generalists was the highest in broad-leaved forests, this group demonstrated a higher tolerance to environmental changes, effectively utilizes diverse resources, and thrives across various habitats (Wan et al., 2021; Jia et al., 2018; Xu et al., 2021). Secondly, the niche breadth of habitat generalists was higher in broad-leaved forests than in other forest types, and it is higher than that of habitat specialist species in all forest types. Finally, the diversity of similarly habitat-specialized groups showed a trend of broad-leaved forests > mixed forests > coniferous forests, indicating that the soil environment of broad-leaved forests is suitable for the coexistence of more fungal species, which verifies its high environmental heterogeneity. In addition, the study also found the number and abundance of specialist and opportunities significantly higher than generalists, indicated that most fungal members had a limited distribution range, while only a small portion were highly distributed in space and time (Sriswasdi et al., 2017; Delgado-Baquerizo et al., 2018). This finding aligned with the conclusions of Li et al. (2025), and could also be expected that the different afforestation types studied exert strong selection pressure on fungi (Logares et al., 2013). The abundance of habitat-specialists was the highest in coniferous forests, indicating that the soil environment in coniferous forests has the highest degree of specialization and exerts the strongest filtering effect on fungal community assembly. The reason was why the specialists rely heavily on specific resource conditions, showing stronger competitiveness in more stable environments and playing key hub and regulatory roles in maintaining community structural stability (Liu et al., 2023).
4.2 Impact of plantation types on the β-diversity and environmental driving mechanisms of soil fungi with different habitat specialization
Soil fungal community was influenced by both deterministic and stochastic processes, with the former mainly including interspecific interactions such as coexistence, competition, and exclusion caused by environmental selection, and the latter mainly including random diffusion effects (Peng et al., 2021; Wan et al., 2021). Both processes play an equally important role in driving community assembly of habitat-specialized clades (Büchi and Vuilleumier, 2014). This study showed that the structural turnover rate of habitat-specialists was higher than generalists and opportunities, consistent with the view that the community assembly of habitat specialists was mainly driven by deterministic processes because their narrow environmental tolerance range, making them easily replaced by other species (Lindh et al., 2016), and they were less affected by random diffusion (Liao et al., 2016). The distribution pattern of habitat generalists was more neutral, attributable to their wider ecological adaptability, reduced impact from environmental protection, and increased influence from stochastic factors (FAO, 2022). In coniferous forests, the homogeneity of structures of habitat-specialists was higher than generalists, whereas in broadleaved forests, there was no significant difference in the heterogeneity of each habitat-specialization group structures, confirmed the specialized soil environments in coniferous forests and high heterogeneity of soil environments in broad-leaved forests. These differences in the structures of habitat specialists among afforestation types reflected that the balance between deterministic and stochastic processes can be regulated by environmental factors (Zhang et al., 2019). RDA analysis indicated that changes in community dominant tree species significantly affect the community structure of each habitat-specialized group, consistent with the view of Urbanová et al. (2015). Porazinska et al. (2018) also proposed that particular plant species, rather than plant diversity, more effectively influence the structure of soil microbial communities. Changes in soil properties resulted in significant differences in the community structure of generalist and opportunities between broadleaved forests and other forest types, while little impacted on specialists. This could be attributed to the high environmental heterogeneity of broadleaved forest soils, which weakens the response strength of specialists to environmental changes and reduces their susceptibility to deterministic processes (Mo et al., 2021; Gad et al., 2020). Among these, SK and pH were identified positively correlated to the difference of structure between broadleaved forest and other forests for each habitat specialization group and AN was proved to be negatively correlated to that for generalists and opportunities (Barcenas-Moreno et al., 2016; Dawud et al., 2016).
4.3 Impact of plantation types on network structural features of soil fungi with different habitat specialization
Co-occurrence network analysis served as a crucial basis for determining community assembly mechanisms (Xue et al., 2017; Zhou et al., 2021), it also provided powerful tools for interpreting microbial interactions, identifying key species, and calculating topological features (Guan et al., 2021). Relevant studies indicated the key taxa an important role in maintaining network structure, primarily existing as module hubs, and their disappearance may disrupt the network structure (Sun et al., 2021). Our study concluded that the habitat-specialists were more crucial in preserving network structure and stability than habitat generalists and opportunities. Firstly, habitat specialists exhibited higher node degree, centrality and modularity in network topology compared to habitat generalists and opportunities, aligning with the finding of Mo et al.’s (2021). Secondly, the number of key taxa of network in habitat-specialists was notably higher compared to habitat generalist and opportunities. The Key taxa in habitat-specialists mainly served as module hubs than connectors, underscoring their critical role in maintaining network structure, aligning with the findings of Li et al. (2024). In addition, habitat specialists exhibited the highest node degree and graph density and the lowest modularity in mixed forests, whereas the showed higher modularity indices in coniferous and broad-leaved forests. This could be explained as the connectivity of soil fungi in mixed forests increased interactions among species, breaking down module boundaries and thus lowering the modularity index. In mixed and broad-leaved forests, the positive interactions between habitat specialists and opportunities exceeded the negative ones, with opposite in coniferous forests. This suggested that coexistence and symbiosis dominate among habitat specialists and opportunities in mixed and broad-leaved forests, while competition and exclusion prevail in coniferous forests (Liu et al., 2024).
5 Conclusion
From the perspective of habitat specialization, this study systematically revealed the impact characteristics of different afforestation types in limestone mountainous areas on soil fungal diversity, community structure, and co-occurrence networks. It concluded that broad-leaved forests can enhance the diversity of habitat-specialists and strengthen the hub function of their network by increasing soil environmental heterogeneity. This research provided a clear theoretical basis and strategic reference for optimizing underground biodiversity and enhancing ecosystem stability through tree species configuration in the ecological restoration of plantations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
ZQ: Data curation, Visualization, Methodology, Investigation, Writing – review & editing, Writing – original draft. JL: Investigation, Writing – review & editing, Data curation. TP: Methodology, Investigation, Writing – review & editing, Visualization. ZW: Data curation, Writing – review & editing, Investigation. YS: Data curation, Writing – review & editing, Investigation. XW: Data curation, Writing – review & editing, Investigation. JS: Investigation, Resources, Methodology, Writing – review & editing, Visualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This project is funded by the high-level talent project funds of Shandong Agricultural Engineering University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1691167/full#supplementary-material
Footnotes
References
Abdullah Al, M., Xue, Y., Xiao, P., Xu, J., Chen, H., Mo, Y., et al. (2022). Community assembly of microbial habitat generalists and specialists in urban aquatic ecosystems explained more by habitat type than pollution gradient. Water Res. 220:118693. doi: 10.1016/J.WATRES.2022.118693
Banerjee, S., Schlaeppi, K., and Van Der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Barcenas-Moreno, G., Bååth, E., and Rousk, J. (2016). Functional implications of the pH-trait distribution of the microbial community in a re-inoculation experiment across a pH gradient. Soil Biol. Biochem. 93, 69–78. doi: 10.1016/j.soilbio.2015.10.024
Becquer, A., Garcia, K., Amenc, L., Rivard, C., Doré, J., Trives-Segura, C., et al. (2018). The Hebeloma cylindrosporum HcPT2 pi transporter plays a key role in ectomycorrhizal symbiosis. New Phytol. 220, 1185–1199. doi: 10.1111/nph.15281
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Büchi, L., and Vuilleumier, S. (2014). Coexistence of specialist and generalist species is shaped by dispersal and environmental factors. Am. Nat. 183, 612–624. doi: 10.1086/675756
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Canini, F., Zucconi, L., Pacelli, C., Selbmann, L., Onofri, S., and Geml, J. (2019). Vegetation pH and water content as main factors for shaping fungal richness, community composition and functional guilds distribution in soils of Western Greenland. Front. Microbiol. 10:2348. doi: 10.3389/fmicb.2019.02348
Clasen, B. E., Silveira, A. D., Baldoni, D. B., Montagner, D. F., Jacques, R. J. S., and Antoniolli, Z. I. (2018). Characterization of ectomycorrhizal species through molecular biology tools and morphotyping. Sci. Agric. 75, 246–254. doi: 10.1590/1678-992X-2016-0419
Csárdi, G., Nepusz, T., Müller, K., Horvát, S., Traag, V., Zanini, F., et al. (2024). igraph for R: R interface of the igraph library for graph theory and network analysis (v2.1.2). Zenodo. doi: 10.5281/zenodo.14347716
Dawud, S. M., Raulund-Rasmussen, K., Domisch, T., Finér, L., Jaroszewicz, B., and Vesterdal, L. (2016). Is tree species diversity or species identity the more important driver of soil carbon stocks, C/N ratio and pH. Ecosystems 19, 645–660. doi: 10.1007/s10021-016-9958-1
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-González, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Deng, Q., Cheng, X. L., Hui, D. F., Zhang, Q., Li, M., and Zhang, Q. F. (2016). Soil microbial community and its interaction with soil carbon and nitrogen dynamics following afforestation in Central China. Scitotenv 541, 230–237. doi: 10.1016/j.scitotenv.2015.09.080
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13, 1–20. doi: 10.3389/FEVO.2020.588430
Du, Z. Y., Lian, Y., Ge, Z. Q., Li, Z. T., Li, Y. T., Lv, L. C., et al. (2020). Soil quality characteristics of Platycladus orientalis plantations with different densities in central mountainous area of Shandong Province. South Univ. For. Technol. 40, 104–112. doi: 10.14067/j.cnki.1673-923x.2020.09.012
FAO. (2022) Forest pathways for green recovery and building inclusive, resilient and sustainable economies. Rome, Italy. doi: 10.4060/cb9360en
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Gad, M., Li, Y. H., Jiang, W. L., Yang, W., Rashid, A., Neng, W. C., et al. (2020). Distinct mechanisms underlying the assembly of microeukaryotic generalists and specialists in an anthropogenically impacted river. Sci. Total Environ. 748:141434. doi: 10.1016/j.scitotenv.2020.141434
Geng, X. Z., Zuo, J. C., Meng, Y. H., Zhuge, Y. H., Zhu, P., Wu, N., et al. (2023). Changes in nitrogen and phosphorus availability driven by secondary succession in temperate forests shape soil fungal communities and function. Ecol. Evol. 13:12. doi: 10.1002/ece3.10593
Guan, Y. P., Jiang, N. N., Wu, Y. X., Yang, Z. Z., Bello, A., and Yang, W. (2021). Disentangling the role of salinity-sodicity in shaping soil microbiome along a natural saline-sodic gradient. Sci. Total Environ. 765:142738. doi: 10.1016/j.scitotenv.2020.142738
Guo, X. F., Wang, S. J., Wang, C., Lan, M. J., Yang, S. Q., Luo, S., et al. (2024). The changes, aggregation processes, and driving factors for soil fungal communities during tropical forest restoration. J. Fungi 10:27. doi: 10.3390/jof10010027
Hu, H., Wei, X. Y., Liu, L., Wang, Y. B., Bu, L. K., Jia, H. J., et al. (2024). Biogeographic patterns of meio-and micro-eukaryotic communities in dam-induced river-reservoir systems. Appl. Microbiol. Biotechnol. 108:130. doi: 10.1007/s00253-023-12993-4
Jia, X., Andreote, F. D., and Salles, J. F. (2018). Community assembly processes of the microbial rare biosphere. Trends Microbiol. 26, 738–747. doi: 10.1016/j.tim.2018.02.011
Levins, R. (1968). Evolution of diversity, efficiency and community stability. Am. Zool. 10, 17–25. doi: 10.1093/icb/10.1.17
Li, J. B., Li, C. N., Kou, Y. P., Yao, M. J., He, Z. L., and Li, X. Z. (2020). Distinct mechanisms shape soil bacterial and fungal co-occurrence networks in a mountain ecosystem. FEMS Microbiol. Ecol. 96:fiaa030. doi: 10.1093/femsec/fiaa030
Li, Y. J., Li, R. Y., Li, Q., Zhao, X. D., Zhao, P. Y., Yan, P. M., et al. (2025). Study on the synergistic mechanisms of fungal biodiversity and ecosystem multifunctionality across vegetation diversity gradients. Sci. Total Environ. :964178563. doi: 10.1016/J.SCITOTENV.2025.178563
Li, H. L., Ostermann, A., Karunarathna, S. C., Xu, J. C., Hyde, K. D., and Mortimer, P. E. (2018). The importance of plot size and the number of sampling seasons on capturing macrofungal species richness. Fungal Biol. 122, 692–700. doi: 10.1016/j.funbio.2018.03.004
Li, S. Z., Yan, X., Abdullah, M., Ren, K. X., Rensing, C., Hu, A. Y., et al. (2024). Ecological and evolutionary processes involved in shaping microbial habitat generalists and specialists in urban park ecosystems. mSystems 9:e0046924. doi: 10.1128/MSYSTEMS.00469-24
Liao, J. Q., Cao, X. F., Zhao, L., Wang, J., Gao, J., Wang, M. C., et al. (2016). The importance of neutral and niche processes for bacterial community assembly differs between habitat generalists and specialists. FEMS Microbiol. Ecol. 92:fiw174. doi: 10.1093/femsec/fiw174
Lin, Z. W., Li, Y. F., Tang, C. X., Luo, Y., Fu, W. J., Cai, X. Q., et al. (2018). Converting natural evergreen broadleaved forests to intensively managed moso bamboo plantations affects the pool size and stability of soil organic carbon and enzyme activities. Biol. Fertil. Soils 54, 467–480. doi: 10.1007/s00374-018-1275-8
Lindh, M. V., Sjöstedt, J., Casini, M., Andersson, A., Legrand, C., and Pinhassi, J. (2016). Local environ-mental conditions shape generalist but not specialist components of microbial metacommunities in the Baltic Sea. Front. Microbiol. 7:2078. doi: 10.3389/fmicb.2016.02078
Liu, L. M., Chen, H. H., Liu, M., Yang, J. R., Xiao, P., Wilkinson, D. M., et al. (2019). Response of the eukaryotic plankton community to the cyanobacteria biomass cycle over 6 years in two subtropical reservoirs. ISME J. 13, 2196–2208. doi: 10.1038/s41396-019-0417-9
Liu, X., Chu, H. Y., Godoy, O., Fan, K. K., Gao, G. F., Yang, T., et al. (2024). Positive associations fuel soil biodiversity and ecological networks worldwide. Proc. Natl. Acad. Sci. USA 121:e2308769121. doi: 10.1073/PNAS.2308769121
Liu, H. T., Hu, T. L., Wang, H., Zhang, Y. H., Guo, S. W., and Xie, Z. B. (2023). Community construction and functional potential of typical rice soil bacterial subgroups: generalized species and specialized species. Acta Pedol. Sin. 60, 546–557. (in Chinese). doi: 10.11766/trxb202105310284
Liu, Y. P., Sun, Q. B., Li, J., and Lian, B. (2018). Bacterial diversity among the fruit bodies of ectomycorrhizal and saprophytic fungi and their corresponding hyphosphere soils. Sci. Rep. 8:11672. doi: 10.1038/s41598-018-30120-6
Logares, R., Lindstrom, E. S., Langenheder, S., Logu, J. B., Paterson, H., LaybournParry, J., et al. (2013). Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J. 7, 937–948. doi: 10.1038/ismej.2012.168
Ministry of Agriculture, PRC (2012). Soil testing-method for determination of available phosphorus in soil. NY/T 1121.7-2014. Beijing, China: Ministry of Agriculture, PRC (in Chinese).
Ministry of Environmental Protection, PRC (2011). Soil determination of organic carbon-potassium dichromate oxidation Spectro photometric method. HJ615-2011. Beijing, China: Ministry of Environmental Protection, PRC (in Chinese).
Mo, Y., Zhang, W., Wilkinson, D. M., Yu, Z., Xiao, P., and Yang, J. (2021). Biogeography and co-occurrence patterns of bacterial generalists and specialists in three subtropical marine bays. Limnol. Oceanogr. 66, 793–806. doi: 10.1002/lno.11643
Nagati, M., Roy, M., Manzi, S., Richard, F., Desrochers, A., Gardes, M., et al. (2018). Impact of local forest composition on soil fungal communities in a mixed boreal forest. Plant Soil 432, 345–357. doi: 10.1007/s11104-018-3806-3
Noll, L., Leonhardt, S., Arnstadt, T., Hoppe, B., Poll, C., Matzner, E., et al. (2016). Fungal biomass and extracellular enzyme activities in coarse woody debris of 13 tree species in the early phase of decomposition. For. Ecol. Manag. 378, 181–192. doi: 10.1016/j.foreco.2016.07.035
Ortiz, D. C., Souza, T. A. F. D., Pech, T. M., Bartz, M. L. C., Baretta, D., Siminski, A., et al. (2022). Soil ecosystem changes by vegetation on old-field sites over five decades in the Brazilian Atlantic forest. J. For. Res. 33, 667–677. doi: 10.1007/S11676-021-01368-Y
Peng, Z. H., Wang, Z. F., Liu, Y., Yang, T. Y., Chen, W. M., Wei, G. H., et al. (2021). Soil phosphorus determines the distinct assembly strategies for abundant and rare bacterial com munities during successional reforestation. Soil Ecol. Lett. 3, 342–355. doi: 10.1007/S42832-021-0109-Z
Porazinska, D. L., Farrer, E. C., Spasojevic, M. J., Bueno, d. M. C. P., Sartwell, S. A., Smith, J. G., et al. (2018). Plant diversity and density predict belowground diversity and function in an early successional alpine ecosystem. Ecology 99, 1942–1952. doi: 10.1002/ecy.2420
Preetisri, B., Riitta, H., Linnea, B. S., Clemmensen, K. E., Ågren, G. I., Lindahl, B. D., et al. (2017). Modelling the influence of ectomycorrhizal decomposition on plant nutrition and soil carbon sequestration in boreal forest ecosystems. New Phytol. 213, 1452–1465. doi: 10.1111/nph.14213
Qiu, Z. L., Li, J., Wang, P., Wang, D., Han, L., Gao, X. J., et al. (2023). Response of soil bacteria on habitat-specialization and abundance gradient to different afforestation types. Sci. Rep. 13:18181. doi: 10.1038/S41598-023-44468-X
Qiu, Z. L., Shi, C., Zhao, M. Y., Wang, K. F., Zhang, M., Wang, T. T., et al. (2022). Improving effects of afforestation with different forest types on soil nutrients and bacterial community in barren hills of North China. Sustainability 14:1202-1202. doi: 10.3390/SU14031202
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ratzke, C., Barrere, J., and Gore, J. (2020). Strength of species interactions determines biodiversity and stability in microbial communities. Nat. Ecol. Evol. 4, 376–383. doi: 10.1038/s41559-020-1099-4
Salazar, G. (2019). EcolUtils: utilities for community ecology analysis. R package version 0.1. Available online at: https://github.com/GuillemSalazar/EcolUtils
Shannon, C. E., and Weaver, W. (1949). The mathematical theory of communication. Urbana: University of Illinois Press.
Sriswasdi, S., Yang, C., and Iwasaki, W. (2017). Generalist species drive microbial dispersion and evolution. Nat. Commun. 8:1162. doi: 10.1038/s41467-017-01265-1
Sun, C. X., Zhang, B., Ning, D., Zhang, Y., Dai, T. J., Wu, L. W., et al. (2021). Seasonal dynamics of the microbial community in two full-scale wastewater treatment plants: diversity, composition, phylogenetic group based assembly and cooccurrence pattern. Water Res. 200:117295. doi: 10.1016/J.WATRES.2021.117295
Tan, L. C., Liu, W., Wang, T. L., Cheng, P., Zang, J. J., Wang, X. Q., et al. (2020). A multiple-proxy stalagmite record reveals historical deforestation in Central Shandong, northern China. Sci. Sinica 50, 1643–1654. doi: 10.1007/s11430-019-9649-1
Urbanová, M., Šnajdr, J., and Baldrian, P. (2015). Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 84, 53–64. doi: 10.1016/j.soilbio.2015.02.011
Vuong, T. M. D., Zeng, J. Y., and Man, X. L. (2020). Soil fungal and bacterial communities in southern boreal forests of the Greater Khingan Mountains and their relationship with soil properties. Sci. Rep. 10:22025. doi: 10.1038/s41598-020-79206-0
Wan, W. J., Gadd, G. M., Yang, Y. Y., Yuan, W. K., Gu, J. D., Ye, L. P., et al. (2021). Environmental adaptation is stronger for abundant rather than rare microorganisms in wetland soils from the Qinghai-Tibet plateau. Mol. Ecol. 30, 2390–2403. doi: 10.1111/mec.15882
Wang, H. Y., Li, J. L., Ke, Y. M., Zhang, Y. H., Zhou, C. H., Wei, C. X., et al. (2025). The patterns of the Populus euphratica forest and cropland changes upstream of the Tarim River under the policy-driven human activities during 1960–2023. Int. J. Remote Sens. 46, 5087–5106. doi: 10.1080/01431161.2025.2513565
Wilson, B., and Hayek, L. A. C. (2015). Distinguishing relative specialist and generalist species in the fossil record. Mar. Micropaleontol. 119, 7–16. doi: 10.1016/j.marmicro.2015.07.001
Wu, J. Z. (2017). The development of handicraft industry and forest change in Shandong area of Tang dynasty. J. Shandong Agric. Univ. (Soc. Sci. Ed.) 19, 14–21. (in Chinsese)
Wu, W., Wang, X., Ren, Z., Zhou, X., and Du, G. (2022). N-induced species loss dampened by clipping mainly through suppressing dominant species in an alpine meadow. Front. Plant Sci. 13:815011. doi: 10.3389/fpls.2022.815011
Xu, Q., Vandenkoornhuyse, P., Li, L., Guo, J., Zhu, C., Guo, S., et al. (2021). Microbial generalists and specialists differently contribute to the community diversity in farmland soils. J. Adv. Res. 40, 17–27. doi: 10.1016/J.JARE.2021.12.003
Xue, Y. Y., Chen, H. H., Yang, J. R., Liu, M., Huang, B. Q., and Yang, J. (2018). Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 12, 2263–2277. doi: 10.1038/s41396-018-0159-0
Xue, L., Ren, H., Li, S., Leng, X., and Yao, X. (2017). Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Front. Microbiol. 8:2377. doi: 10.3389/fmicb.2017.02377
Yan, Q., Liu, Y., Hu, A., Wan, W., Zhang, Z., and Liu, K. (2022). Distinct strategies of the habitat generalists and specialists in sediment of Tibetan lakes. Environ. Microbiol. 24, 4153–4166. doi: 10.1111/1462-2920.16044
Yang, K., and Zhu, J. J. (2015). The effects of N and P additions on soil microbial properties in paired stands of temperate secondary forests and adjacent larch plantations in Northeast China. Soil Biol. Biochem. 90, 80–86. doi: 10.1016/j.soilbio.2015.08.002
Zhang, J. L., and Ma, K. P. (2014) spaa: An R package for computing species association and niche overlap. Research progress of biodiversity conservation in China, 165-174 (in Chinese).
Zhang, K. P., Shi, Y., Cui, X. Q., Yue, P., Li, K. H., Liu, X. J., et al. (2019). Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems 4, e00225–e00218. doi: 10.1128/mSystems.00225-18
Zheng, W., Feng, G., Liang, K., Wang, T., Lv, F., Zhang, X., et al. (2023). SOC mediates the contribution of generalists and specialists to changes in soil nirK bacterial diversity: evidence from apple orchards in main production areas of China. Appl. Soil Ecol. 182:104713. doi: 10.1016/J.APSOIL.2022.104713
Zhou, L., Zhou, Y. Q., Tang, X. M., Zhang, Y. L., Zhu, G. W., Székely, A. J., et al. (2021). Eutrophication alters bacterial co-occurrence networks and increases the importance of chromophoric dissolved organic matter composition. Limnol. Oceanogr. 66, 2319–2332. doi: 10.1002/LNO.11756
Zhu, Y. G., Chen, B. D., and Fu, W. (2022). Research frontiers in soil ecology. Sci. Technol. Rev. 40, 25–31(in Chinese). doi: 10.3981/j.issn.1000-7857.2022.03.002
Keywords: habitat-generalists, habitat-specialists, network, afforestation, soil fungi
Citation: Qiu Z, Liu J, Pan T, Wang Z, Shen Y, Wang X and Shu J (2025) Response of soil fungal community structure and co-occurrence network features on plantations of limestone mountains along habitat specialization gradient. Front. Microbiol. 16:1691167. doi: 10.3389/fmicb.2025.1691167
Edited by:
Fred O. Asiegbu, University of Helsinki, FinlandReviewed by:
Mingmin Zhao, Inner Mongolia Agricultural University, ChinaYonglong Wang, Baotou Teachers’ College, China
Copyright © 2025 Qiu, Liu, Pan, Wang, Shen, Wang and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Shu, MTU2NjY3NzU2NzdAMTYzLmNvbQ==
 Zhenlu Qiu
Zhenlu Qiu Jin Liu
Jin Liu