- 1College of Life Science and Technology, Harbin Normal University, Harbin, China
- 2Key Laboratory of Photoelectric Conversion and Utilization of Solar Energy, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, China
- 3Shandong Energy Institute, Qingdao, China
- 4Laboratory for Marine Biology and Biotechnology, Qingdao Marine Science and Technology Center, Qingdao, China
Due to the robust capabilities in hydrocarbon/herbicide degradation, biopolymer/compatible solute synthesis, steroid bioconversion, and zinc salt solubilization, Pseudomonas oleovorans has shown considerable potential for industrial, agricultural, and environmental applications. However, the poor availability of genetic tools for this bacterium hinders genetic, biochemical, metabolic, and engineering studies. In the present study, a genetic manipulation system that is based on electroporation was established for P. oleovorans strain T9AD. Antibiotic susceptibility profiling demonstrated that aminoglycoside-type antibiotics, such as kanamycin and gentamycin, are suitable selective markers. Optimization of electroporation parameters, including processing temperature for competent cell preparation, DNA concentration, DNA-cell pre-incubation, and post-pulse recovery, yielded stable electroporation efficiencies at levels of 104 CFU/μg DNA. Among five candidate genomic neutral sites, two were experimentally verified and exhibited favorable suitability for gene integration. Integration of reporter genes at these sites did not affect cell growth, salt tolerance, and compatible solute anabolism. Using these neutral sites or the broad-host-range plasmid pBBR1MCS-5, regulated gene expression via the genome- or plasmid-based strategies was successfully achieved. All together, these tools, in combination with established conjugation methods, set up a robust technological platform to facilitate fundamental and application research in P. oleovorans.
1 Introduction
Pseudomonas spp., comprising one of the largest genera of Gram-negative bacteria, thrive in a broad spectrum of environments from soils and waters to extreme niches (Lalucat et al., 2022; Saati-Santamaría et al., 2022). Given their metabolic diversity, these bacteria exhibit remarkable potential in industrial, agricultural, and environmental applications (Craig et al., 2021; Mehmood et al., 2023). As a member with rarely reported pathogenic associations, Pseudomonas oleovorans displays robust capabilities in alkane degradation and biopolymer biosynthesis, such as poly (3-hydroxyalkanotes), thereby attracting considerable biotechnological interest (Huisman et al., 1991; Fiedler et al., 2002; Nisar et al., 2025). Subsequent studies have unveiled additional functions of this species, including degradation of halogenated herbicides (e.g., acetochlor; Xu et al., 2006; Chen et al., 2024), bioconversion of steroids (e.g., hydrocortisone to prednisolone; Abd El-Hady and Abd El-Rehim, 2004), solubilization of insoluble zinc salts (Rehman et al., 2021), and compatible solute synthesis (e.g., glucosylglycerol [GG]). These discoveries further expand the biotechnological relevance of P. oleovorans to herbicide bioremediation, biofertilizer development, pharmaceutical synthesis, and moisturizer production.
A convenient and stable genetic manipulation system in P. oleovorans is essential for in-depth investigations into genetic, biochemical, and regulatory mechanisms of the aforementioned functions, as well as for advancing genetic engineering studies. However, reports on specialized methods and genetic tools for P. oleovorans remain extremely limited. Most of these studies rely on approaches (e.g., conjugation) developed for well-characterized species such as P. putida (Klinke et al., 2000; Nandy et al., 2021; Guo et al., 2022). In the present study, an electroporation-based genetic manipulation system for P. oleovorans was established by using a marine-derived strain, T9AD (Wang et al., 2021).
2 Materials and methods
2.1 Bacterial strains and cultivation conditions
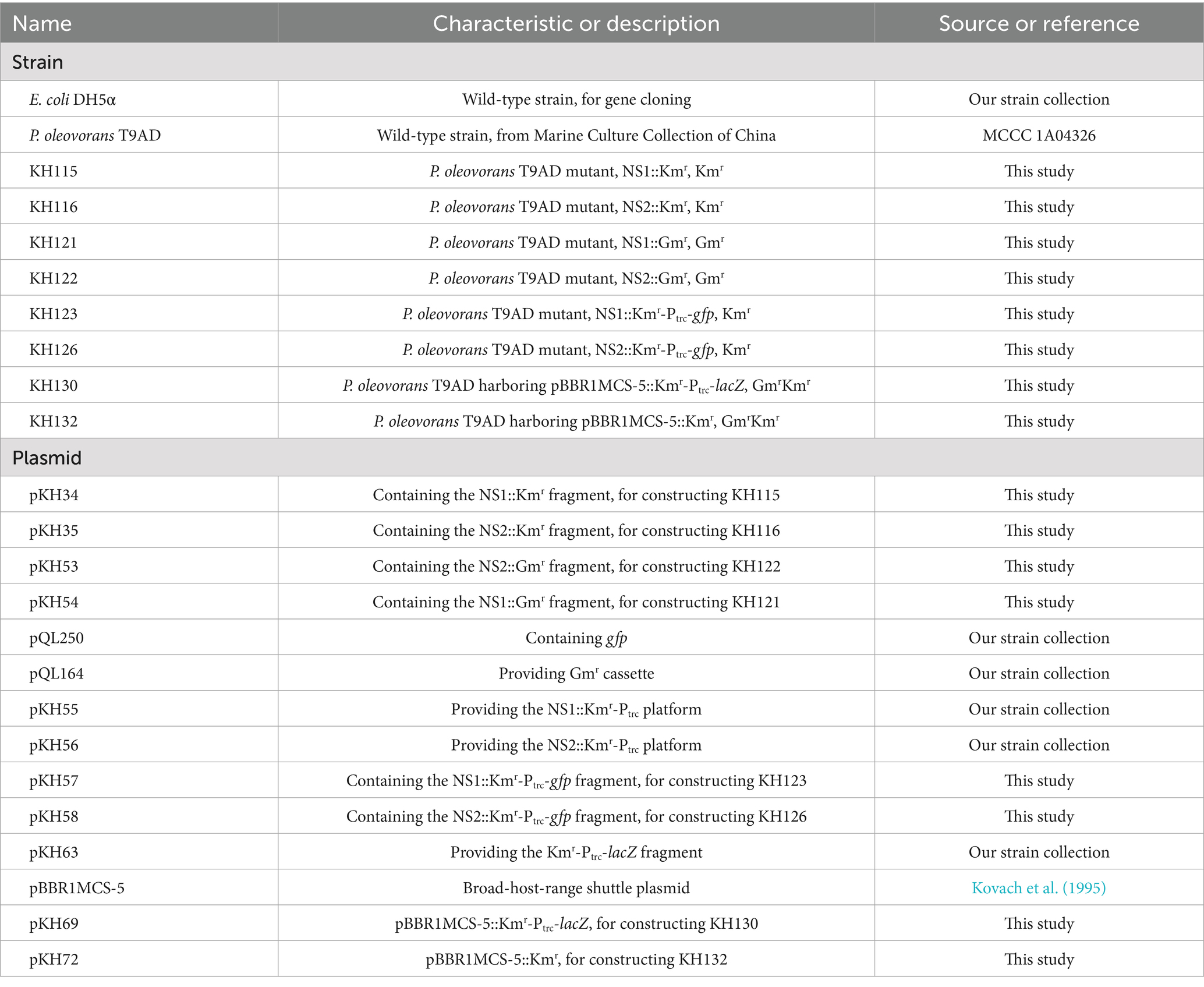
The bacterial strains used in the present study are listed in Table 1. P. oleovorans T9AD was grown in LB medium (for growth experiment) or M9 minimal medium (containing 0.5% wt/vol L-lactic acid as the sole carbon source, for GG production) at 30 °C. Escherichia coli was grown in LB medium at 37 °C. Cell growth was monitored by measuring the optical density at a wavelength of 600 nm (OD600). Solid media were prepared by adding 1.5% (wt/vol) agar. Kanamycin (Km) of 50 μg/mL and/or gentamycin (Gm) of 10 μg/mL were added when required.
In the test of antibiotic sensitivity, P. oleovorans T9AD was inoculated into 96-well plates containing LB medium supplemented with different concentrations (0, 1, 2, 5, 10, 20, 50, and 100 μg/mL) of Km, ampicillin (Amp), chloramphenicol (Cm), spectinomycin (Spe), apramycin (Apr), Gm, neomycin (Neo), and streptomycin (Stp). A negative control was included with no cell inoculation. After 20 h cultivation on a horizontal shaker (MB100-2A, Hangzhou Aosheng, China) at 500 rpm, cell growth was determined by measuring the OD₆₀₀ on a microplate reader (SpectraMax M3, Molecular Devices, United States).
2.2 Sequence analysis
The P. oleovorans T9AD genome under the GenBank accession number LR130779.2 was used for sequence analysis. The manual examination of open reading frames (ORFs) was conducted using the ARTEMIS program (Rutherford et al., 2000).
2.3 Competent cells of Pseudomonas oleovorans and electroporation
The optimized procedure for competent cell preparation and electroporation was as follows: P. oleovorans strain T9AD was grown in 100 mL of LB medium in 250 mL flasks. Overnight cultures were harvested by centrifugation at room temperature (RT), washed twice with 0.3 M sucrose solution, and resuspended in the same solution. Electroporation was performed with the following procedure: a proper amount of plasmid DNA was added into 100 μL of competent cells to a final concentration of 0.02 μg/mL. An incubation step was not necessary. The mixtures were immediately transferred into electrocuvettes, which have a gap of 2 mm, and electropulse treated by an electroporator (Bio-Rad Micropulser, United States) with the following parameters, 12.5 kV/cm, 25 μF, 200 Ω, and a pulse duration of 5 ms. Immediately after pulses, cells were supplemented with 900 μL of SOC medium (Russell and Sambrook, 2001) and incubated at 30 °C for 1 h recovery. Then cells were plated onto LB agar plates containing antibiotics for selection. Generation of Gm-resistant (Gmr) and/or Km-resistant (Kmr) transformants was examined after 20 h of cultivation. During the optimization of the electroporation method, different processing temperatures for competent cell preparation, such as 4 °C and RT, DNA concentrations (0–5 μg/mL), durations for DNA-cell incubation (0–2 h), and times for cell recovery (0–24 h) were tested.
2.4 DNA manipulation
The plasmids and primers used in the present study were listed in Table 1 and Supplementary Table S1, respectively. To construct mutant KH115, the upstream (0.90 kb) and downstream (0.98 kb) flanking regions of the neutral site 1 (NS1) were amplified from the total DNA of P. oleovorans T9AD using primer pairs NS1-up-F/NS1-up-R(Km) and NS1-dn-F(Km)/NS1-dn-R, respectively. A 0.93 kb Kmr cassette was amplified from pCE-Zero (Vazyme Biotech, China) using primers Km-F(NS1) and Km-R(NS1). The three fragments were assembled and cloned into pUC19 using the ClonExpress® Ultra One-Step Cloning Kit (Vazyme Biotech, China), resulting in plasmid pKH34. After sequence confirmation by sequencing, pKH34 was electrotransformed into P. oleovorans T9AD. After 20 h of cultivation for homologous double crossover, Kmr transformants were obtained on LB agar plates containing Km, and the genotypes of the transformants were confirmed by PCR. The other recombinant strains of P. oleovorans, including KH116 (using primers NS2-up-F/NS2-up-R(Km) and NS2-dn-F(Km)/NS2-dn-R for flanking regions), KH121 (using primers NS1-up-F/NS1-up-R(Gm) and NS1-dn-F(Gm)/NS1-dn-R for flanking regions), and KH122 (using primers NS2-up-F/NS2-up-R(Gm) and NS2-dn-F(Gm)/NS2-dn-R for flanking regions), were constructed following a similar procedure. The 1.19 kb Gmr cassette was obtained from pQL164 using primers Gm-F(NS1)/Gm-R(NS1) or Gm-F(NS2)/Gm-R(NS2). For constructing KH123 and KH126, the ORF of the gfp gene (coding for green fluorescent protein, 0.72 kb) was amplified from pQL250 using primer pair gfp-F/gfp-R(NS1) or gfp-F/gfp-R(NS2) and cloned into pKH55 and pKH56 at the sites downstream of the trc promoter (Ptrc), generating plasmids pKH57 and pKH58 for electroporation. For constructing KH130, the Kmr-Ptrc-lacZ cassette, amplified from pKH63 using primers KPZ-F and KPZ-R, was cloned into pBBR1MCS-5, generating pKH69 for electroporation.
2.5 Phenotype analyses
To determine GG production, P. oleovorans cells grown in M9 minimal medium were harvested by centrifugation and inoculated into 100 mL of the same medium supplemented with 3% (wt/vol) NaCl at an initial OD600 of 0.8. After 6 h of cultivation, GG was extracted from P. oleovorans cells for determination. GG extraction and quantification were performed as previously described (Qiao et al., 2018; Qiao et al., 2019).
To analyze GFP fluorescence, strains KH123 and KH126 were grown in LB medium and an isopropyl-D-1-thiogalactopyranoside (IPTG) concentration ranging from 0 to 0.25 mM was applied to induce gfp expression. After 6 or 20 h of induction, cells were harvested and resuspended in M9 minimal medium with an OD₆₀₀ of ~1.0. 200 μL of cell suspensions was transferred to 96-well microplates, and fluorescence intensity was measured using a microplate reader (SpectraMax M3, Molecular Devices, United States) with an excitation wavelength of 488 nm and an emission wavelength of 525 nm. Visualization of GFP fluorescence was accomplished using a fluorescence microscope (Axio Imager 2, Zeiss, Germany).
To analyze β-galactosidase (LacZ) activity, strains KH130 and KH132 were grown in LB medium and induced by adding IPTG (final concentrations of 0–0.25 mM). After 6 or 20 h of induction, β-galactosidase activity was determined following a reported protocol (Miller, 1972; Jaishankar and Srivastava, 2020; Fitzgerald et al., 2024).
3 Results and discussion
3.1 Antibiotic sensitivity profiling of Pseudomonas oleovorans T9AD
A clear profiling of antibiotic susceptibility forms the foundation for developing genetic manipulation tools in a given microbe, as it guides rational selection of selective markers and tool vectors (Aparicio et al., 2015). Due to the generally low outer membrane permeability and the presence of multiple efflux pumps and modification enzymes (e.g., aminoglycoside-modifying enzymes and 16S rRNA methylase) in the cell, Pseudomonas species generally exhibit intrinsic resistance to many antibiotics (Burns et al., 1989; Ramos et al., 2002; Daniels and Ramos, 2009; Fernández et al., 2012; Aghazadeh et al., 2013). Additionally, Pseudomonas can also acquire antibiotic resistance through chromosomal mutations and horizontal gene transfer (Botelho et al., 2019). For example, while P. aeruginosa is generally sensitive to carbapenems, some clinic isolates of this species acquire carbapenemase-encoding genes and exhibit resistance to these antibiotics (Botelho et al., 2015; Oliver et al., 2015). Therefore, for a given strain, the antibiotic resistance profile can be species- or strain- specific and requires clear determination on a case-by-case basis.
To determine the antibiotic susceptibility profile of P. oleovorans T9AD, eight commonly used antibiotics, namely Km, Amp, Cm, Spe, Apr, Gm, Neo, and Stp, at final concentrations of up to 100 μg/mL, were employed to apply selective pressure (Figure 1A; Supplementary Table S2). In liquid culture, Km and Gm exhibited the most potent inhibitory effects on cell growth. A concentration of 5 μg/mL completely arrested the growth of P. oleovorans T9AD, indicating a minimal inhibitory concentration (MIC) of ≤ 5 μg/mL. By contrast, Stp, Apr, Neo and Spe, showed moderate inhibition. Complete suppression of cell growth was observed at antibiotic concentrations of 20 μg/mL (for Apr, Neo, and Stp) or 50 μg/mL (for Spe). For Amp and Cm, no inhibitory effect was detected under the tested conditions. A similar inhibitory pattern was also observed in solid culture (Supplementary Figure S1). These results agreed well with the established understanding that Pseudomonas species are generally susceptible to aminoglycoside-type antibiotics while exhibiting resistance to β-lactam- and amphenicol-type antibiotics (Kadurugamuwa and Beveridge, 1997; Fernández et al., 2012; Hujer et al., 2023). For example, P. aeruginosa, P. putida, and P. fluorescens are sensitive to Gm, Km, Stp, and tobramycin. These antibiotics were frequently used in the genetic studies of Pseudomonas (Shivaji et al., 1989; Ghiglione et al., 1999; Wu et al., 2015; Calero et al., 2018). A key factor underlying the efficacy of these antibiotics is their strong ability to penetrate bacterial cell membranes (Lang et al., 2023). On the other hand, many Pseudomonas bacteria (e.g., P. cepacia, P. loganensis, P. aeruginosa, and P. putida) also exhibit natural resistance to Cm, carbapenems, and penicillins (Burns et al., 1989; Fernández et al., 2012; Horna et al., 2018; Karaman et al., 2025). The wide presence of RND family multidrug efflux pumps (such as MexAB-OprM and MexXY-OprM) and hyperexpression of modifying enzymes (such as AmpC β-lactamases) in the cells contribute considerably to their intrinsic antibiotic resistance by reducing intracellular drug accumulation and inactivating β-lactam compounds, respectively (Barceló et al., 2022; Lorusso et al., 2022).
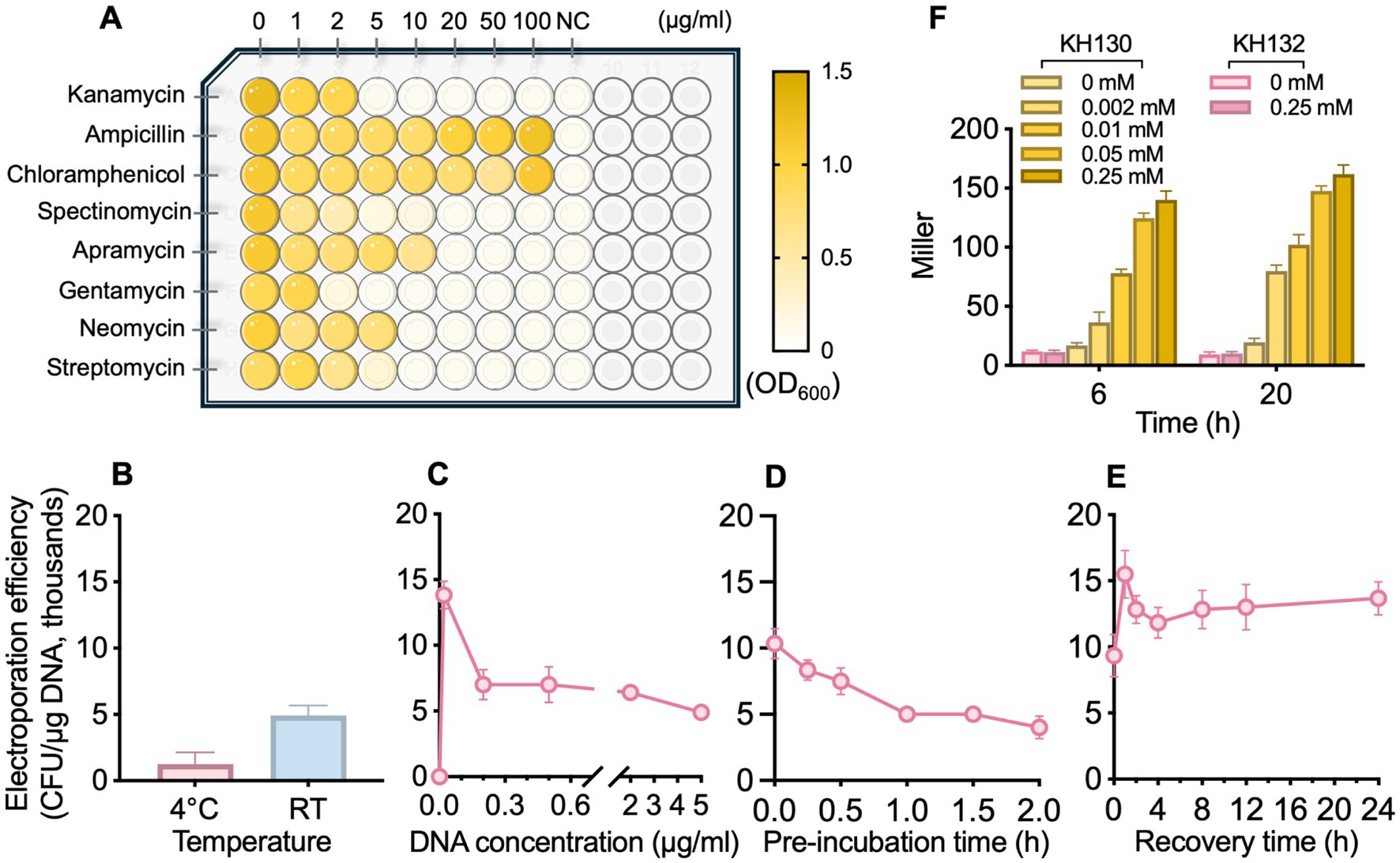
Figure 1. Determination of the antibiotic susceptibility profile of P. oleovorans T9AD (A), optimization of its electroporation efficiency (B–E), and assessment of plasmid-based gene expression in P. oleovorans T9AD (F). In (A), P. oleovorans T9AD was grown in liquid LB medium supplemented with different antibiotics (as indicated) at an initial optimal density at 600 nm (OD₆₀₀) of 0.05. After 20 h of cultivation, cell growth was monitored by measuring OD600. Uninoculated LB medium was used as the negative control (NC). In (B–E), the effects of processing temperature for cell preparation and electropulse (B, 4 °C and RT), DNA concentration (C, 0–5 μg/mL), pre-incubation time for DNA-cell mixture (D, 0–2 h), and post-pulse cell recovery (E, 0–24 h) on the electroporation efficiency were analyzed. Each parameter was altered independently, whereas the others remained constant. In (F), the β-galactosidase activity of mutants KH130 (expressing lacZ in pBBR1MCS-5) and KH132 (blank control) was examined. Cells were grown in LB medium, and IPTG of 0–0.25 mM was supplemented to induce lacZ expression. The data are presented as means from three independent replicates with standard deviations.
3.2 Electroporation optimization of Pseudomonas oleovorans T9AD
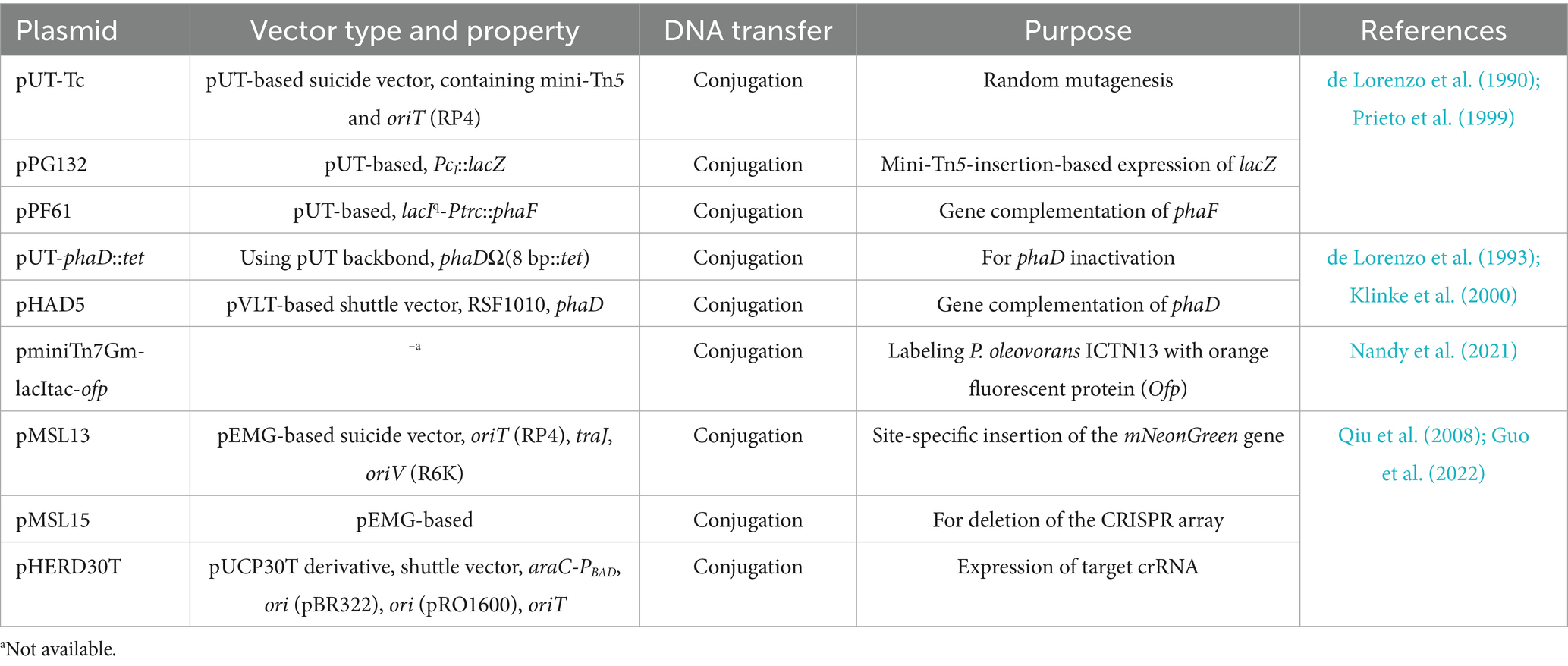
To introduce foreign DNA into Pseudomonas cells, conjugation and electroporation are the most commonly used methods. The former enables the transfer of large DNA fragments but is generally more time-consuming and less efficient for small fragments compared with electroporation (Harris et al., 2016; Bai et al., 2024). To the best of our knowledge, the few genetic investigations in P. oleovorans have all relied on conjugation. By employing suicide vectors (e.g., pUT- and pEGM-series) and shuttle vectors (e.g., pVLT- and pHERD-series), genetic constructs have been delivered into P. oleovorans cells, enabling successful gene inactivation (e.g., phaD), chromosomal disruption (e.g., CRISPR array), expression of reporter genes (e.g., lacZ), and gene complementation (e.g., phaD and phaF; Table 2; Prieto et al., 1999; Klinke et al., 2000; Nandy et al., 2021; Guo et al., 2022). In comparison with conjugation, electroporation offers greater flexibility with respect to the types and lengths of foreign DNA. For example, based on this strategy, a rapid all-in-one plasmid-based CRISPR/Cas9 system was established for genome editing in P. putida. This allowed one edit in less than 1.5 days. Although being fast and simple for gene transfer, electroporation generally suffers from low efficiency in Pseudomonas species at initial attempts. Reducing nonessential genetic components in genomes and optimizing the electroporation procedure can substantially improve transformation efficiency (Choi et al., 2006; Fan et al., 2024; Wen et al., 2024). Here, the feasibility of electroporation in P. oleovorans T9AD was examined using the broad-host-range plasmid pBBR1MCS-5 (Kovach et al., 1995).
Our initial attempt of electrotransforming P. oleovorans T9AD yielded an efficiency of only 66.7 CFU/μg DNA, a level that was too low for efficient genetic manipulation. Therefore, a systematically optimization of electroporation efficiency was conducted. Optimization began with the following settings: (i) preparing electrocompetent cells at 4 °C, (ii) using 1.5 μg/mL plasmid DNA, (iii) no pre-incubation of DNA with cells, and (iv) 4 h-recovery after pulses. Each parameter was modified individually while the others remained constant. In many cases, low temperatures were used for competent cell preparation and electroporation pulses (Diver et al., 1990; Qin et al., 2022). However, some studies have also reported improved electroporation efficiencies at RT for Pseudomonas species (Choi et al., 2006; Tu et al., 2016). Here, a similar result was observed in P. oleovorans T9AD (Figure 1B). When competent cells were prepared and pulsed at RT, the electroporation efficiency increased 3.8-fold compared with the level of 4 °C. The value raised from 1.3 × 103 to 4.9 × 103 CFU/μg DNA. Therefore the RT condition was applied in the following experiments. In the assessment of the relationship between electroporation efficiency and DNA concentration, the highest efficiency (1.4 × 104 CFU/μg DNA) was observed at a concentration of 0.02 μg/mL, followed by a sharp decline at 0.2 μg/mL (Figure 1C). At higher concentrations (≥ 0.2 μg/mL), the value continued to decrease gradually. Thus, low DNA concentrations favor efficient electroporation in P. oleovorans. Similar findings have been reported in Rhodobacter sphaeroides, Nocardia nova, and Bacillus subtilis (Luo et al., 2013; Zhang et al., 2015; Lee et al., 2024).
The effects of DNA-cell pre-incubation and cell recovery were further examined. With an increase of incubation time, the number of positive transformants exhibited a decreasing tendency (Figure 1D). Compared with the non-incubated control (0 h), a 2 h pre-incubation reduced the efficiency by half. Therefore, a pre-incubation of competent cells with DNA prior to electroporation is unnecessary. Regarding the process after electropulses, a recovery of pulse-treated cells in antibiotic-free medium seems helpful to increase efficiency (Figure 1E). In all recovery attempts with different times (0–24 h), increased numbers of transformants were observed in comparison with the control (0 h). The highest level (1.6 × 104 CFU/μg DNA) was achieved after 1 h recovery. From five randomly selected transformants, plasmids were extracted and examined by restriction digestion. They all exhibited identical pattern to the parent plasmid in the electroporation analysis (Supplementary Figure S2). Taken together, these optimizations yielded stable electroporation efficiencies of 104 CFU/μg DNA in P. oleovorans, sufficient for genetic manipulations such as gene inactivation or expression.
3.3 Controllable gene expression on a plasmid platform
For genetic and metabolic engineering studies in Pseudomonas, stable and controllable gene expression platforms are required due to the needs of gene complementation, heterologous protein expression, pathway construction, metabolic flux regulation, and other applications. These are usually accomplished by plasmid-based systems and genomic integration.
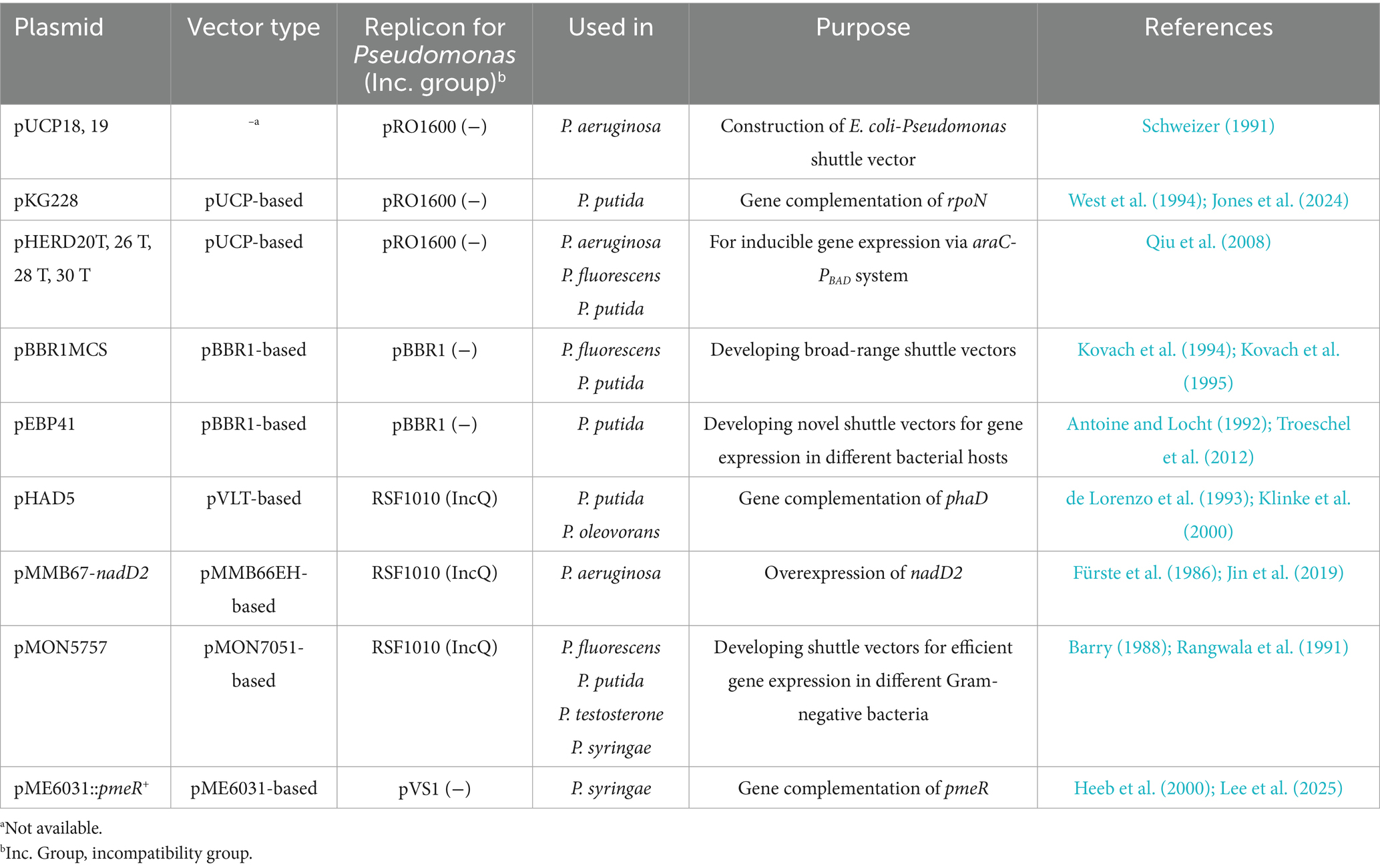
Plasmid-based systems usually offer advantages in expression flexibility (e.g., temporal control and intensity regulation) and ease of manipulation (Rangwala et al., 1991; Blatny et al., 1997). Several types of shuttle vectors have been validated for use in Pseudomonas (Table 3). The pBBR1MCS-series plasmids, known for their broad-host-range, compact size, moderate copy number, and multiple cloning sites, have been validated for stable heterologous gene expression in many Gram-negative bacteria including Pseudomonas species (Elzer et al., 1995; Battisti and Minnick, 1999; Takahashi et al., 2016). They could be ideal candidates for gene or pathway expression in P. oleovorans. During electroporation optimization, the stable and autonomous replication of pBBR1MCS-5, a Gmr version of pBBR1MCS, in P. oleovorans has been demonstrated. Thus, this plasmid was further employed to drive inducible expression of the lacZ reporter gene via the commonly used E. coli trc promoter (Ptrc) in strain T9AD. Upon plasmid transformation, a dose-dependent induction of lacZ expression was observed in the recombinant strain KH130 (Figure 1F). It displayed β-galactosidase activity in the presence of IPTG, with enzyme activity increasing progressively along with elevated IPTG concentrations. In contrast, the control strain KH132, which harbored a blank plasmid, exhibited only basal activity regardless of IPTG concentration. These results validated the utility of pBBR1MCS-based vectors and the trc promoter for controllable expression of non-native genes in P. oleovorans. Considering that P. oleovorans exhibits sensitivity to Km, Apr, Spe, Stp, and tetracycline (Figure 1A; Prieto et al., 1999), other pBBR1MCS variants harboring appropriate resistant cassettes (e.g., pBBR1MCS-2 and pBBR1MCS-3; Kovach et al., 1995) could be also applied. In addition to the broad-host-range capability, pBBR1MCS plasmids also demonstrate good compatibility with other broad-host-range plasmids from the IncP, IncW, and IncQ groups. This property is particular useful for synthetic biology investigations, as it enables flexible co-expression of multiple genes or pathways using plasmids from different incompatibility groups (such as those listed in Table 3; Antoine and Locht, 1992; Kovach et al., 1994; Li et al., 2020).
3.4 Development of genomic neutral platforms in Pseudomonas oleovorans
Genomic NSs are useful tools for bacterial genetic and metabolic engineering studies. Insertion of genes in these chromosomal loci does not cause detectable phenotypic changes of cells. Since no NSs have been identified in P. oleovorans to date, the genome of strain T9AD was analyzed for this purpose and four criteria were considered: (i) chromosomal regions lacking predicted ORFs; (ii) region of 250–300 bp in length to minimize the presence of other possible functional elements, (iii) flanking genes with opposite transcriptional directions facing each other, and (iv) the absence of similar sequences to the target NS and its flanking genes elsewhere in the genome. Five candidate sites of 256–292 bp in length were identified (Supplementary Figure S3), and two of them with the longest flanking genes, NS1 (POT9AD_2766–2,767) and NS2 (POT9AD_5113–5,114), were selected to investigate their neutrality. A Kmr gene and a Gmr gene were independently introduced into these sites via homologous recombination (Figure 2A). The resulting mutants KH115, KH116, KH121, and KH122 exhibited the expected antibiotic-resistances. Regardless of whether the medium contained 5% NaCl or not, the mutant strains showed growths comparable to that of the wild-type strain (Figures 2B,C). Thus, integration of foreign genes into NS1 and NS2 did not affect the basic growth of P. oleovorans T9AD as well as its salt-tolerance. During genome analysis, it was found that P. oleovorans T9AD harbors a putative glucosylglycerol (GG) synthetic pathway encoded by the GG-phosphate phosphatase/synthase gene ggpPS (Cheng et al., 2024), suggesting this strain might synthesize GG as compatible solute for salt acclimation. The GG-synthesizing capability of T9AD was examined (Figure 2D). Under the condition of 3% NaCl, salt-induced GG production was observed in the wild-type and mutant strains. The mutant strains accumulated GG levels comparable to the wild-type. Thus gene insertion in NS1 and NS2 did not influence GG anabolism of P. oleovorans.
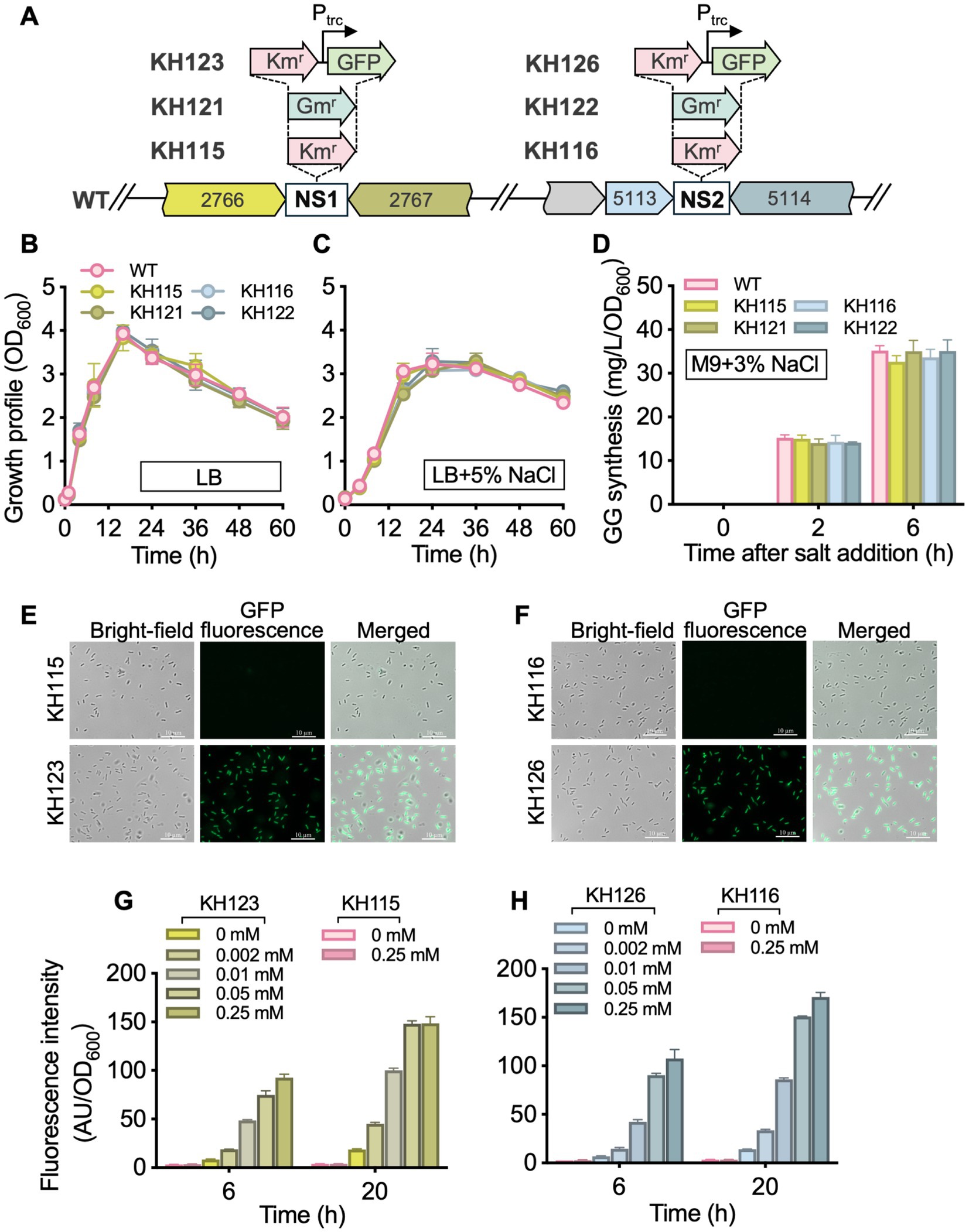
Figure 2. Development of genomic NSs in P. oleovorans T9AD. (A) Illustrates the genomic loci of NS1 and NS2 in the wild-type (WT) strain and the insertion of foreign genes in these sites. KH115, KH116, KH121, KH122, KH123, and KH126 are the resulting insertion mutants. The prefix “POT9AD_” of locus tags POT9AD_2766, POT9AD_2,767, POT9AD_5113, and POT9AD_5,114 is omitted. Growth profiles of the WT and mutant strains in the absence and presence of 5% salinity are demonstrated in (B,C), respectively. Pseudomonas cells were grown in LB medium with or without 5% (wt/vol) NaCl addition. (D) Demonstrates the GG-synthesizing ability of the WT and mutant strains of P. oleovorans T9AD. Cells were grown in M9 minimal medium and 3% (wt/vol) NaCl was added to induce GG production. In (E–H), GFP fluorescence of mutants KH123 and KH126 were examined. Cells were grown in LB medium, and IPTG of 0–0.25 mM was supplemented to induce gfp expression. The data of (B–D) and (G,H) are presented as means from three independent replicates with standard deviations. Kmr, Km-resistant; Gmr, Gm-resistant; Ptrc, the trc promoter.
Compared to plasmid-based gene expression, the genome-integrated system eliminates the need to consider plasmid loss, plasmid incompatibility, and host range limitations. It also allows cultivation without the addition of antibiotics (Friehs, 2004; Bassalo et al., 2016; Li et al., 2020; Kazi et al., 2022). To investigate the feasibility of NS1 and NS2, the green fluorescent protein reporter gene (gfp) was integrated into these sites under the control of the trc promoter (Figure 2A). Under fluorescence microscopy, the recombinant strains KH123 and KH126 showed strong fluorescence, indicating successful gfp expression (Figures 2E,F). The fluorescence intensity correlated positively with increasing IPTG concentrations (0 to 0.25 mM) and induction durations (6 to 20 h; Figures 2G,H). Under the same conditions, no GFP signal was detected in the control strains (KH115 and KH116; Figures 2E–H). These results demonstrated that stable and dose-dependent expression of non-native genes or pathways could be achieved via the trc regulatory system within the NS1 and NS2 platforms. Previously, chromosome-based gene integration in Pseudomonas, such as P. fluorescens and P. putida, has mainly been achieved employing the miniTn7-transposition system (Koch et al., 2001; Lambertsen et al., 2004). This requires a miniTn7 vector and relies on the presence of the attTn7 site, a neutral intergenic region located immediately downstream of the glucosamine-6-phosphate synthetase gene (glmS). In P. oleovorans, an attTn7-like site is detected in the genome, and the newly developed NS1 and NS2 sites offer alternative options alongside attTn7, making them suitable for multi-module expression.
4 Conclusion
In summary, this study established an electroporation-based genetic system for P. oleovorans, a species with biotechnological potential. Aminoglycoside antibiotics, especially Km and Gm, showed potent inhibitory effects on cell growth, providing a rational basis for selective marker design. By optimizing the temperature for cell preparation and electropulse, DNA concentration, and post-pulse cell recovery, the electroporation efficiency was improved to stably achieve the level of ~104 CFU/μg DNA. Employing the broad-host-range vector pBBR1MCS-5, plasmid-based inducible expression of foreign genes could be achieved. Additionally, two genomic NSs (NS1 and NS2) were developed and validated for gene integration without major phenotypic influence. Collectively, these tools, along with established conjugation method, set up a robust technological platform to facilitate further fundamental and application studies in P. oleovorans.
Data availability statement
The data presented in this work are available upon request from the authors.
Author contributions
HK: Writing – original draft, Validation, Data curation, Formal analysis, Visualization, Investigation. ZZ: Writing – review & editing, Data curation. YL: Conceptualization, Writing – review & editing. QL: Funding acquisition, Conceptualization, Writing – original draft, Data curation. XL: Writing – review & editing, Funding acquisition, Resources, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study is supported by Natural Science Foundation of China (32470040), Taishan Scholarship of Shandong Province, and Natural Science Foundation of Shandong Province (ZR2023MC175).
Acknowledgments
Thanks to Marine Culture Collection of China for providing Pseudomonas oleovorans T9AD.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1691967/full#supplementary-material
References
Abd El-Hady, A., and Abd El-Rehim, H. A. (2004). Production of prednisolone by Pseudomonas oleovorans cells incorporated into PVP/PEO radiation crosslinked hydrogels. J. Biomed. Biotechnol. 2004, 219–226. doi: 10.1155/S1110724304308053
Aghazadeh, M., Rezaee, M. A., Nahaei, M. R., Mahdian, R., Pajand, O., Saffari, F., et al. (2013). Dissemination of aminoglycoside-modifying enzymes and 16S rRNA methylases among Acinetobacter baumannii and Pseudomonas aeruginosa isolates. Microb. Drug Resist. 19, 282–288. doi: 10.1089/mdr.2012.0223
Antoine, R., and Locht, C. (1992). Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6, 1785–1799.
Aparicio, T., de Lorenzo, V., and Martínez-García, E. (2015). “Broadening the SEVA plasmid repertoire to facilitate genomic editing of gram-negative bacteria” in Hydrocarbon and lipid microbiology protocols. Springer Protocols Handbooks. eds. T. McGenity, K. Timmis, and B. Nogales (Berlin, Heidelberg: Springer).
Bai, S., Luo, H., Tong, H., Wu, Y., and Yuan, Y. (2024). Advances on transfer and maintenance of large DNA in bacteria, fungi, and mammalian cells. Biotechnol. Adv. 76:108421. doi: 10.1016/j.biotechadv.2024.108421
Barceló, I. M., Jordana-Lluch, E., Escobar-Salom, M., Torrens, G., Fraile-Ribot, P. A., Cabot, G., et al. (2022). Role of enzymatic activity in the biological cost associated with the production of AmpC β-lactamases in Pseudomonas aeruginosa. Microbiol. Spectr. 10:e0270022. doi: 10.1128/spectrum.02700-22
Barry, G. F. (1988). A broad-host-range shuttle system for gene insertion into the chromosomes of gram-negative bacteria. Gene 71, 75–84.
Bassalo, M. C., Liu, R., and Gill, R. T. (2016). Directed evolution and synthetic biology applications to microbial systems. Curr. Opin. Biotechnol. 39, 126–133. doi: 10.1016/j.copbio.2016.03.016
Battisti, J. M., and Minnick, M. F. (1999). Development of a system for genetic manipulation of Bartonella bacilliformis. Appl. Environ. Microbiol. 65, 3441–3448.
Blatny, J. M., Brautaset, T., Winther-Larsen, H. C., Karunakaran, P., and Valla, S. (1997). Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38, 35–51.
Botelho, J., Grosso, F., and Peixe, L. (2019). Antibiotic resistance in Pseudomonas aeruginosa - mechanisms, epidemiology and evolution. Drug Resist. Updat. 44:100640. doi: 10.1016/j.drup.2019.07.002
Botelho, J., Grosso, F., Sousa, C., and Peixe, L. (2015). Characterization of a new genetic environment associated with GES-6 carbapenemase from a Pseudomonas aeruginosa isolate belonging to the high-risk clone ST235. J. Antimicrob. Chemother. 70, 615–617. doi: 10.1093/jac/dku391
Burns, J. L., Hedin, L. A., and Lien, D. M. (1989). Chloramphenicol resistance in Pseudomonas cepacia because of decreased permeability. Antimicrob. Agents Chemother. 33, 136–141.
Calero, P., Jensen, S. I., Bojanovič, K., Lennen, R. M., Koza, A., and Nielsen, A. T. (2018). Genome-wide identification of tolerance mechanisms toward p-coumaric acid in Pseudomonas putida. Biotechnol. Bioeng. 115, 762–774. doi: 10.1002/bit.26495
Chen, W. J., Chen, S. F., Song, H., Li, Z., Luo, X., Zhang, X., et al. (2024). Current insights into environmental acetochlor toxicity and remediation strategies. Environ. Geochem. Health 46:356. doi: 10.1007/s10653-024-02136-7
Cheng, L., Zhang, Z., Zhu, D., Luo, Q., and Lu, X. (2024). Glucosylglycerol phosphorylase, a potential novel pathway of microbial glucosylglycerol catabolism. Appl. Microbiol. Biotechnol. 108:214. doi: 10.1007/s00253-024-13035-3
Choi, K. H., Kumar, A., and Schweizer, H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397. doi: 10.1016/j.mimet.2005.06.001
Craig, K., Johnson, B. R., and Grunden, A. (2021). Leveraging Pseudomonas stress response mechanisms for industrial applications. Front. Microbiol. 12:660134. doi: 10.3389/fmicb.2021.660134
Daniels, C., and Ramos, J. L. (2009). Adaptive drug resistance mediated by root-nodulation-cell division efflux pumps. Clin. Microbiol. Infect. 15, 32–36. doi: 10.1111/j.1469-0691.2008.02693.x
de Lorenzo, V., Eltis, L., Kessler, B., and Timmis, K. N. (1993). Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123, 17–24.
de Lorenzo, V., Herrero, M., Jakubzik, U., and Timmis, K. N. (1990). Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172, 6568–6572.
Diver, J. M., Bryan, L. E., and Sokol, P. A. (1990). Transformation of Pseudomonas aeruginosa by electroporation. Anal. Biochem. 189, 75–79.
Elzer, P. H., Kovach, M. E., Phillips, R. W., Robertson, G. T., and Peterson, K. M. (1995). In vivo and in vitro stability of the broad-host-range cloning vector pBBR1MCS in six Brucella species. Plasmid 33, 51–57.
Fan, S., Ren, H., Fu, X., Kong, X., Wu, H., and Lu, Z. (2024). Genome streamlining of Pseudomonas putida B6-2 for bioremediation. mSystems 9:e0084524. doi: 10.1128/msystems.00845-24
Fernández, M., Conde, S., de la Torre, J., Molina-Santiago, C., Ramos, J. L., and Duque, E. (2012). Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440. Antimicrob. Agents Chemother. 56, 1001–1009. doi: 10.1128/AAC.05398-11
Fiedler, S., Steinbüchel, A., and Rehm, B. H. (2002). The role of the fatty acid β-oxidation multienzyme complex from Pseudomonas oleovorans in polyhydroxyalkanoate biosynthesis: molecular characterization of the fadBA operon from P. Oleovorans and of the enoyl-CoA hydratase genes phaJ from P. Oleovorans and Pseudomonas putida. Arch. Microbiol. 178, 149–160. doi: 10.1007/s00203-002-0444-0
Fitzgerald, M. J., Scavone, A., Moolaveesala, C., Pearson, M. M., and Mobley, H. L. T. (2024). Development of a suite of Proteus mirabilis-derived urea-inducible promoters. Appl. Environ. Microbiol. 90:e0127324. doi: 10.1128/aem.01273-24
Friehs, K. (2004). Plasmid copy number and plasmid stability. Adv. Biochem. Engin/Biotechnol. 86, 47–82. doi: 10.1007/b12440C
Fürste, J. P., Pansegrau, W., Frank, R., Blöcker, H., Scholz, P., Bagdasarian, M., et al. (1986). Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48, 119–131.
Ghiglione, J. F., Philippot, L., Normand, P., Lensi, R., and Potier, P. (1999). Disruption of narG, the gene encoding the catalytic subunit of respiratory nitrate reductase, also affects nitrite respiration in Pseudomonas fluorescens YT101. J. Bacteriol. 181, 5099–5102.
Guo, X., Sanchez-Londono, M., Gomes-Filho, J. V., Hernandez-Tamayo, R., Rust, S., Immelmann, L. M., et al. (2022). Characterization of the self-targeting type IV CRISPR interference system in Pseudomonas oleovorans. Nat. Microbiol. 7, 1870–1878. doi: 10.1038/s41564-022-01229-2
Harris, J. R., Lundgren, B. R., Grzeskowiak, B. R., Mizuno, K., and Nomura, C. T. (2016). A rapid and efficient electroporation method for transformation of Halomonas sp. O-1. J. Microbiol. Methods 129, 127–132. doi: 10.1016/j.mimet.2016.08.009
Heeb, S., Itoh, Y., Nishijyo, T., Schnider, U., Keel, C., Wade, J., et al. (2000). Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13, 232–237. doi: 10.1094/MPMI.2000.13.2.232
Horna, G., López, M., Guerra, H., Saénz, Y., and Ruiz, J. (2018). Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 8:16463. doi: 10.1038/s41598-018-34694-z
Huisman, G. W., Wonink, E., Meima, R., Kazemier, B., Terpstra, P., and Witholt, B. (1991). Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J. Biol. Chem. 266, 2191–2198. doi: 10.1016/S0021-9258(18)52227-4
Hujer, A. M., Marshall, S. H., Mack, A. R., Hujer, K. M., Bakthavatchalam, Y. D., Umarkar, K., et al. (2023). Transcending the challenge of evolving resistance mechanisms in Pseudomonas aeruginosa through β-lactam-enhancer-mechanism-based cefepime/zidebactam. MBio 14:e0111823. doi: 10.1128/mbio.01118-23
Jaishankar, J., and Srivastava, P. (2020). Strong synthetic stationary phase promoter-based gene expression system for Escherichia coli. Plasmid 109:102491. doi: 10.1016/j.plasmid.2020.102491
Jin, Y., Zhang, M., Zhu, F., Peng, Q., Weng, Y., Zhao, Q., et al. (2019). NrtR regulates the type III secretion system through cAMP/Vfr pathway in Pseudomonas aeruginosa. Front. Microbiol. 10:85. doi: 10.3389/fmicb.2019.00085
Jones, I., Vermillion, D., Tracy, C., Denton, R., Davis, R., and Geszvain, K. (2024). Isolation, characterization, and genetic manipulation of cold-tolerant, manganese-oxidizing Pseudomonas sp. strains. Appl. Environ. Microbiol. 90:e0051024. doi: 10.1128/aem.00510-24
Kadurugamuwa, J. L., and Beveridge, T. J. (1997). Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40, 615–621.
Karaman, M. Z., Yetiman, A. E., Zhan, J., and Fidan, O. (2025). Biochemical characterization and genome analysis of Pseudomonas loganensis sp. nov., a novel endophytic bacterium. Microbiology 14:e70051. doi: 10.1002/mbo3.70051
Kazi, T. A., Acharya, A., Mukhopadhyay, B. C., Mandal, S., Arukha, A. P., Nayak, S., et al. (2022). Plasmid-based gene expression systems for lactic acid bacteria: a review. Microorganisms 10:1132. doi: 10.3390/microorganisms10061132
Klinke, S., de Roo, G., Witholt, B., and Kessler, B. (2000). Role of phaD in accumulation of medium-chain-length poly(3-hydroxyalkanoates) in Pseudomonas oleovorans. Appl. Environ. Microbiol. 66, 3705–3710. doi: 10.1128/AEM.66.9.3705-3710.2000
Koch, B., Jensen, L. E., and Nybroe, O. (2001). A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45, 187–195. doi: 10.1016/s0167-7012(01)00246-9
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M. II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176.
Kovach, M. E., Phillips, R. W., Elzer, P. H., Roop, R. M. II, and Peterson, K. M. (1994). pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16, 800–802.
Lalucat, J., Gomila, M., Mulet, M., Zaruma, A., and García-Valdés, E. (2022). Past, present and future of the boundaries of the Pseudomonas genus: proposal of Stutzerimonas gen. Nov. Syst. Appl. Microbiol. 45:126289. doi: 10.1016/j.syapm.2021.126289
Lambertsen, L., Sternberg, C., and Molin, S. (2004). Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6, 726–732. doi: 10.1111/j.1462-2920.2004.00605.x
Lang, M., Carvalho, A., Baharoglu, Z., and Mazel, D. (2023). Aminoglycoside uptake, stress, and potentiation in gram-negative bacteria: new therapies with old molecules. Microbiol. Mol. Biol. Rev. 87:e0003622. doi: 10.1128/mmbr.00036-22
Lee, C. Y., Irvine, M., and Kunkel, B. (2025). PmeR, a TetR-like transcriptional regulator, is involved in both auxin signaling and virulence in the plant pathogen Pseudomonas syringae strain PtoDC3000. MBio 16:e0115225. doi: 10.1128/mbio.01152-25
Lee, Y. R., Lee, J., Hong, S., Lee, S. Y., Lee, W. H., Koh, M., et al. (2024). Optimization of electroporation conditions for introducing heterologous DNA into Rhodobacter sphaeroides. J. Microbiol. Biotechnol. 34, 2347–2352. doi: 10.4014/jmb.2408.08044
Li, Z., Cai, Z., Cai, Z., Zhang, Y., Fu, T., Jin, Y., et al. (2020). Molecular genetic analysis of an XDR Pseudomonas aeruginosa ST664 clone carrying multiple conjugal plasmids. J. Antimicrob. Chemother. 75, 1443–1452. doi: 10.1093/jac/dkaa063
Lorusso, A. B., Carrara, J. A., Barroso, C. D. N., Tuon, F. F., and Faoro, H. (2022). Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 23:15779. doi: 10.3390/ijms232415779
Luo, Q., Hiessl, S., Poehlein, A., and Steinbüchel, A. (2013). Microbial gutta-percha degradation shares common steps with rubber degradation by Nocardia nova SH22a. Appl. Environ. Microbiol. 79, 1140–1149. doi: 10.1128/AEM.03016-12
Mehmood, N., Saeed, M., Zafarullah, S., Hyder, S., Rizvi, Z. F., Gondal, A. S., et al. (2023). Multifaceted impacts of plant-beneficial Pseudomonas spp. in managing various plant diseases and crop yield improvement. ACS Omega 8, 22296–22315. doi: 10.1021/acsomega.3c00870
Miller, J. H. (1972). Experiments in molecular genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory.
Nandy, S., Arora, U., Tarar, P., Viggor, S., Jõesaar, M., Kivisaar, M., et al. (2021). Monitoring the growth, survival and phenol utilization of the fluorescent-tagged Pseudomonas oleovorans immobilized and free cells. Bioresour. Technol. 338:125568. doi: 10.1016/j.biortech.2021.125568
Nisar, N., Fareed, A., Naqvi, S. T. A., Zeb, B. S., Amin, B. A. Z., Khurshid, G., et al. (2025). Biodegradation study of used engine oil by free and immobilized cells of the Pseudomonas oleovorans strain NMA and their growth kinetics. ACS Omega 10, 541–549. doi: 10.1021/acsomega.4c06964
Oliver, A., Mulet, X., López-Causapé, C., and Juan, C. (2015). The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 21-22, 41–59. doi: 10.1016/j.drup.2015.08.002
Prieto, M. A., Bühler, B., Jung, K., Witholt, B., and Kessler, B. (1999). PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J. Bacteriol. 181, 858–868.
Qiao, C., Duan, Y., Zhang, M., Hagemann, M., Luo, Q., and Lu, X. (2018). Effects of reduced and enhanced glycogen pools on salt-induced sucrose production in a sucrose-secreting strain of Synechococcus elongatus PCC 7942. Appl. Environ. Microbiol. 84:e02023-17. doi: 10.1128/AEM.02023-17
Qiao, C., Zhang, M., Luo, Q., and Lu, X. (2019). Identification of two two-component signal transduction mutants with enhanced sucrose biosynthesis in Synechococcus elongatus PCC 7942. J. Basic Microbiol. 59, 465–476. doi: 10.1002/jobm.201800676
Qin, J., Hong, Y., Pullela, K., Morona, R., Henderson, I. R., and Totsika, M. (2022). A method for increasing electroporation competence of gram-negative clinical isolates by polymyxin B nonapeptide. Sci. Rep. 12:11629. doi: 10.1038/s41598-022-15997-8
Qiu, D., Damron, F. H., Mima, T., Schweizer, H. P., and Yu, H. D. (2008). PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74, 7422–7426. doi: 10.1128/AEM.01369-08
Ramos, J. L., Duque, E., Gallegos, M. T., Godoy, P., Ramos-Gonzalez, M. I., Rojas, A., et al. (2002). Mechanisms of solvent tolerance in gram-negative bacteria. Ann. Rev. Microbiol. 56, 743–768. doi: 10.1146/annurev.micro.56.012302.161038
Rangwala, S. H., Fuchs, R. L., Drahos, D. J., and Olins, P. O. (1991). Broad host-range vector for efficient expression of foreign genes in gram-negative bacteria. Biotechnology (N.Y) 9, 477–479.
Rehman, H. F., Ashraf, A., Muzammil, S., Siddique, M. H., and Ali, T. (2021). Assessment of zinc solubilization potential of zinc-resistant Pseudomonas oleovorans strain ZSB13 isolated from contaminated soil. Braz. J. Biol. 83:e240015. doi: 10.1590/1519-6984.240015
Russell, D., and Sambrook, J. (2001). Molecular cloning: A laboratory manual. third Edn. New York: Cold Spring Harbor Laboratory.
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Saati-Santamaría, Z., Baroncelli, R., Rivas, R., and García -Fraile, P. (2022). Comparative genomics of the genus Pseudomonas reveals host- and environment-specific evolution. Microbiol. Spectr. 10:e0237022. doi: 10.1128/spectrum.02370-22
Schweizer, H. P. (1991). Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97, 109–112.
Shivaji, S., Rao, N. S., Saisree, L., Sheth, V., Reddy, G. S., and Bhargava, P. M. (1989). Isolation and identification of Pseudomonas spp. from Schirmacher oasis, Antarctica. Appl. Environ. Microbiol. 55, 767–770.
Takahashi, E., Lee, J. M., Mon, H., Chieda, Y., Yasunaga-Aoki, C., Kusakabe, T., et al. (2016). Effect of antibiotics on extracellular protein level in Pseudomonas aeruginosa. Plasmid 84-85, 44–50. doi: 10.1016/j.plasmid.2016.03.001
Troeschel, S. C., Thies, S., Link, O., Real, C. I., Knops, K., Wilhelm, S., et al. (2012). Novel broad host range shuttle vectors for expression in Escherichia coli, Bacillus subtilis and Pseudomonas putida. J. Biotechnol. 161, 71–79. doi: 10.1016/j.jbiotec.2012.02.020
Tu, Q., Yin, J., Fu, J., Herrmann, J., Li, Y., Yin, Y., et al. (2016). Room temperature electrocompetent bacterial cells improve DNA transformation and recombineering efficiency. Sci. Rep. 6:24648. doi: 10.1038/srep24648
Wang, S. Z., Cruaud, C., Aury, J. M., Vallenet, D., Poulain, J., Vacherie, B., et al. (2021). Complete genome sequences of two Pseudomonas species isolated from marine environments of the Pacific Ocean. Microbiol. Resour. Announc. 10, e01062–e01019. doi: 10.1128/MRA.01062-19
Wen, Q., Chen, J., Li, J., Dharmasiddhi, I. P. W., Yang, M., Xing, J., et al. (2024). A single-plasmid-based, easily curable CRISPR/Cas9 system for rapid, iterative genome editing in Pseudomonas putida KT2440. Microb. Cell Factories 23:349. doi: 10.1186/s12934-024-02634-4
West, S. E., Schweizer, H. P., Dall, C., Sample, A. K., and Runyen-Janecky, L. J. (1994). Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86. doi: 10.1016/0378-1119(94)90237-2
Wu, X., Held, K., Zheng, C., Staudinger, B. J., Chavez, J. D., Weisbrod, C. R., et al. (2015). Dynamic proteome response of Pseudomonas aeruginosa to tobramycin antibiotic treatment. Mol. Cell. Proteomics 14, 2126–2137. doi: 10.1074/mcp.M115.050161
Xu, J., Qiu, X., Dai, J., Cao, H., Yang, M., Zhang, J., et al. (2006). Isolation and characterization of a Pseudomonas oleovorans degrading the chloroacetamide herbicide acetochlor. Biodegradation 17, 219–225. doi: 10.1007/s10532-005-4220-0
Keywords: Pseudomonas, genetic manipulation, antibiotic susceptibility, electroporation, neutral site
Citation: Ke H, Zhang Z, Liu Y, Luo Q and Lu X (2025) Development of genetic manipulation tools for Pseudomonas oleovorans. Front. Microbiol. 16:1691967. doi: 10.3389/fmicb.2025.1691967
Edited by:
Rongming Liu, Dalian University of Technology, ChinaReviewed by:
Liya Liang, Dalian University of Technology, ChinaAngelica Severino, University of Naples Federico II, Italy
Copyright © 2025 Ke, Zhang, Liu, Luo and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuefeng Lu, bHZ4ZkBxaWJlYnQuYWMuY24=; Quan Luo, bHVvcXVhbkBxaWJlYnQuYWMuY24=; Yan Liu, eWFubGl1aHJiQGhvdG1haWwuY29t
 Hongjiao Ke
Hongjiao Ke Zhichao Zhang2,3
Zhichao Zhang2,3 Yan Liu
Yan Liu Quan Luo
Quan Luo Xuefeng Lu
Xuefeng Lu