- Society-Environment Research Group, Institute of Geography, Friedrich-Alexander-Universität Erlangen-Nüremberg, Erlangen, Germany
This minireview focuses on recent developments regarding mobile genetic elements (MGEs) and horizontal gene transfer (HGT) in wastewater treatment plants (WWTPs) and proximal environments. WWTPs are often discussed as hotspots and bioreactors for the evolution of MGEs and ARGs and their horizontal transfer. Firstly, the article reviews the effects of emerging contaminants on HGT and MGEs with a specific focus on microplastics and per- and polyfluoroalkyl substances (PFAS). Secondly, the review focuses on how extreme weather and climate change can overwhelm WWTPs, increase the input of diverse genetic elements, and alter the dynamics of HGT. Finally, the trophic connections between the WWTP microbiota and external ecosystems underscore the potential for wider transmission of MGEs. Here, the focus is on transfer of MGEs to larger organisms in the vicinity of WWTPs. In sum, the review focuses on emerging areas of research that refine our understanding of the WWTP environment as a hotspot for HGT and dissemination of MGEs with potentially deleterious implications for human and wider ecosystem health.
1 Introduction
Horizontal genetic transfer (HGT) refers to the sharing of genetic material between microorganisms (Soucy et al., 2015). HGT events are mediated via several distinct mechanisms including conjugation, transduction, and transformation, and via gene transfer agents including plasmids and phages. HGT is a key driver of microbial genomic plasticity and is central to microbes’ ability to adapt to changing and/or stressful environmental conditions (Arnold et al., 2022). Along with the activity of transposons and integron/integrases which drive recombination events within the genome, HGT is an important source of genetic innovation and novelty in microorganisms (Carscadden et al., 2023). Increasingly complex and reticulated genomic structures such as plasmids with multiple drug resistances (Mbanga et al., 2021) and large integron cassettes are often the end products of successive HGT events (Ghaly et al., 2021). The transfer of MGEs is catalyzed by anthropogenic pressures (Gillings, 2017; Groussin et al., 2021; Bradshaw, 2024a) and is particularly intense within wastewater treatment plant (WWTP) environments. Due to their unique function and position in relation anthropogenic activities, WWTPs and their surrounding environments can be seen, on the one hand, as a rescaled microcosm (or model) of the wider socio-environmental dynamics that characterize the Anthropocene (see Achmon et al., 2018). On the other hand, attention to WWTPs can unearth key dynamics of microbial evolution and interaction, particularly HGT, as it adapts to an era of unprecedented environmental change. These interrelated processes are two sides of the same coin that foreground the entangled complexity of the WWTP and its connections to wider environments.
This minireview summarizes recent research focusing on HGT and MGEs in WWTPs. The aim is not to provide a systematic overview of the variables influencing HGT in these environments but to focus on areas that are currently emerging in the literature and which represent important areas for future study. Specifically, the focus is on the role of emerging contaminants, the impact of environmental volatility, and on identifying HGT events to the microbiomes of other ecosystems and species that live and feed in and around WWTPs. These three areas correspond to emerging hazards posed by WWTPs and their interfacing with an increasingly chemically diverse human society and anthropogenic climate change. They represent key sites for ongoing research and are schematically represented in Figure 1.
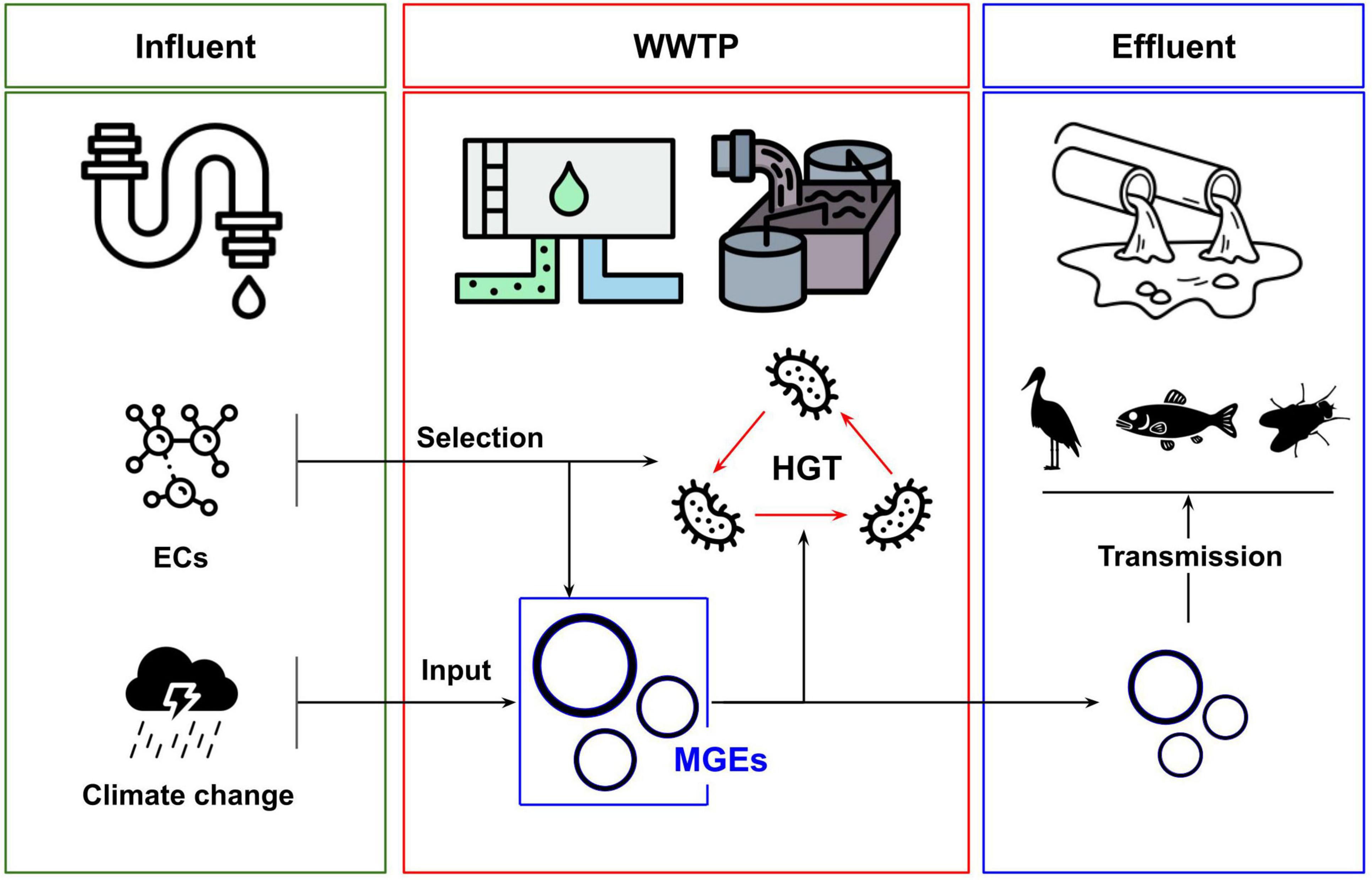
Figure 1. Overview of relationships between emerging contaminants (ECs), climate change and transmission of mobile genetic elements (MGEs) with wastewater treatment plant environments (WWTPs). ECs can select for increased rates of HGT and the abundance of MGEs. Strong weather events can influence the input of MGEs into WWTPs. The dissemination of MGEs in WWTP effluent can promote transmission to downstream ecologies, including larger organisms.
2 Wastewater treatment plants and horizontal gene transfer
Wastewater treatment plants (WWTPs) are unique ecological environments that collect and treat the effluent from human societies. Depending on the precise organization of the WWTP, this can include municipal/domestic wastewater and/or more specialized wastewater streams (hospital, agricultural, and/or industrial) as well as stormwater runoff. The primary method of wastewater treatment is the activated sludge process (AS) in which microbial aerobic digestion is enrolled to reduce the biological oxygen demand (BOD) of influent (McCabe and Eckenfelder, 1961) allowing the treated water to be returned to watersheds (oceans or rivers). The AS microbiome is a complex community that is characterized by a set of core microorganisms (Saunders et al., 2016), dominated by denitrifying bacteria (Yu and Zhang, 2012) and exhibits seasonal variation (Ju et al., 2014). In addition to the core functions of nutrient removal, the AS microbiome contains biodegradation genes targeted toward specific pollutants (e.g., Cai et al., 2013; Fang et al., 2013, 2018) although their function tends to be variable (Douziech et al., 2018).
The WWTP environment exposes a highly dense and diverse microbial community to various hard selection pressures including antibiotics, heavy metals, and disinfectants, along with emerging contaminants of concern (see below). This in turn promotes bacterial evolution and the sharing of genetic material (Brown et al., 2024; Fang et al., 2024). Because of the nature of these selective agents and their effects on bacterial communities, WWTPs are increasingly recognized as “hotspots” (Guo et al., 2017) that foster the emergence and evolution of antimicrobial resistance (AMR; see Bradshaw, 2024b). Firstly, the WWTP community can itself harbor ARGs (on MGEs), thereby acting as an AMR “reservoir” (Yoo et al., 2020; Yin et al., 2022; Zhang et al., 2022). Secondly, the release of MGEs from WWTPs can promote the transfer of ARGs to downstream ecosystems and environments (Sambaza and Naicker, 2023). These processes underscore the framing of WWTPs as risky environments that both concentrate and disperse various hazards.
Over the last century or so HGT has contributed to the reprogramming of bacterial genetic and phenotypic identity on a planetary scale (Gillings, 2017; Gillings and Paulsen, 2014). This evolution is both tightly coupled to anthropogenic activities on the one hand and tends to promote the emergence of drug-resistant and pathogenic microbes, on the other. Further, MGEs with origins that can be traced to single bacterial strains are now observed dispersed throughout the global microbial population, a phenomenon that emerges from the interaction between microbial HGT and human infrastructures (Haraoui, 2022). Hence, although MGEs and HGT events have been critical in evolutionary flexibility, genomic innovation, and the evolution of complex life forms (Jain et al., 2003; Daubin and Szöllősi, 2016), contemporary concerns often focus on these processes as vectors for the dissemination of antimicrobial resistance genes (ARGs) and the emergence of novel pathogens (Emamalipour et al., 2020; Vos, 2020) in an era of unprecedented socio-ecological connectivity.
3 Contaminants of emerging concern in WWTPs and HGT events
A wide variety of emerging contaminants have been implicated in the selection and evolution of ARGs and MGEs, and their transmission. Many of these contaminants concentrate in WWTPs, including pharmaceuticals, personal care products, fungicides, biocides, and herbicides (Feng et al., 2021; Alderton et al., 2021). Disinfectants (Tan et al., 2021), heavy metals (Lin et al., 2019), preservatives (Cen et al., 2020), and organic pollutants are also implicated in the HGT of ARGs (Sharma et al., 2025) and in the evolution of metabolic genes related to pollutant metabolism and detoxification (Top and Springael, 2003; Miguel et al., 2020). Here, the focus here is on the effect of two contaminants of emerging concern on HGT and MGEs: microplastics and per- and polyfluoroalkyl substances. These contaminants are discussed due to the small but growing body of research elucidating their effects and the need for further investigation.
3.1 Microplastics
Microplastics (MPs), small (<5 mm) plastic particles of various polymer composition and structure, are emerging contaminants of concern in wastewater streams and aquatic, marine, estuarine and terrestrial ecosystems across the globe. MPs act as a physical substrate for microbial biofilm formation and have been demonstrated to facilitate HGT under various experimental conditions (Arias-Andres et al., 2018; Liu et al., 2022). MPs of different polymer type have been associated with increased prevalence of ARGs, MGEs, and HGT in mariculture (Liu et al., 2025) estuarine (Zhou et al., 2024), and constructed wetland (Zhao et al., 2023) environments, to name a few. This effect may be associated with the induction of oxidative stress, ROS production, and the formation of a specific “plastisphere” community that enriches MGE-harboring microorganisms (Luo et al., 2023; Liu et al., 2025).
Microplastics are increasingly shed into waste streams and produced in situ through the degradation of larger pieces of plastic (Zhang et al., 2021). These particles are dispersed widely in hydrological systems, bypass initial filtering in WWTPs, and may be released in effluent (Ziajahromi et al., 2016; Mason et al., 2016; Edo et al., 2020). In addition to the environments noted above, researchers have investigated the relationships between MPs and MGEs, ARGs, and HGT events in WWTP systems. For instance, Pham et al. (2021) observed that intI1 was enriched up to 4.5-fold on microplastics incubated in a WWTP in the USA. From a functional perspective, Feng et al. (2023) demonstrated that the ARGs in MP biofilms can be horizontally transferred to free cells, demonstrating the potential correlation between gene abundance and functional effects.
Certain pollutants present in wastewater can be adsorbed onto MP surfaces resulting in localized increases in concentration and leading to bacterial stress responses, increased eDNA uptake, and lateral sharing of genetic information (Shen et al., 2023; Wang et al., 2023). For instance, Wang et al. (2021b) investigated the interactions between MPs isolated from wastewater and different pharmaceuticals. They hypothesized that the colocalization of MPs with multiple pollutants may have synergistic effects on the selection of MGEs. In agreement, their study demonstrated that microorganisms cultured with MPs adsorbed with tetracycline, triclosan, or ampicillin had significantly increased MGEs and ARGs as compared to those cultured with pharmaceuticals alone. Their findings further contribute to the model of MPs as hotspots for the selection, concentration and the transfer of MGEs. On the other hand, however, Xu M. et al.’s (2024) study suggested that MPs effectively reduced ARGs and MGEs in anaerobic digestion environments. Further, MPs were associated with a reduction in genes functionally linked to HGT which was subsequently reduced on MP surfaces. Different polymer types were linked to different effects on MGEs, with polypropylene and polyethylene having a greater effect than polyamide. The effect of MP polymeric identity on HGT and the microbial community involved (e.g., aerobic vs. anaerobic) are important areas for future research. WWTPs should continue to be a key site for these studies (Junaid et al., 2022; Syranidou and Kalogerakis, 2022).
3.2 Per- and polyfluoroalkyl substances
Per- and polyfluoroalkyl substances (PFAS) are a large family (>4700 in commercial use; Wang et al., 2021a) of chemically related compounds characterized by the presence of carbon-fluoride bonds and environmental and biological stability. PFAS are ecotoxic, bioaccumulative, and highly persistent in aquatic and terrestrial environments (Brunn et al., 2023). Exposure to PFAS is associated with changes to the function and structure of the human gut-associated microbiome and various environmental microbiomes (Sen et al., 2024; Beale et al., 2022; Laue et al., 2023). They are contaminants of emerging concern and are linked to health issues in humans and other species. PFAS also concentrate in WWTPs where they are removed with varying degrees of efficiency (Ilieva et al., 2024; Barisci and Suri, 2021).
A small number of studies have begun investigating the roles of PFAS substances on HGT events. For instance, Liu et al. (2023) demonstrated that low levels of two PFAS chemicals, perfluorooctanoic acid (PFOA), perfluorododecanoic acid (PFDoA) and ammonium perfluoro (2-methyl-3-oxahexanoate) (GenX) (at 0.01 and 0.1 mg/L) promoted the conjugative transfer of plasmid RP4 between Escherichia coli. However, higher levels (1.0 and 10 mg/L) inhibited transfer suggesting a non-linear dose response (Xu Z. et al., 2024). Mechanistic studies demonstrated that conjugative transfer was linked to oxidative stress, and higher levels of PFAS reduced ATP thereby inhibiting transfer. Another study demonstrated that PFOA can increase transmission of plasmid-encoded ARGs by up to 3.5-fold, although this was to a soil bacterial community (Yin L. et al., 2023).
Perfluorooctanoic acid also increased the risk of horizontal ARG transmission in groundwater ecosystems by selecting for denitrifying bacterial communities that harbor ARGs associated with MGEs (Chen et al., 2023). On the other hand, Chen et al. (2022) demonstrated that PFOA-induced microbial stress reduced the expression of plasmid-mediated horizontally transmissible ARGs (also in a groundwater model), demonstrating that the effects of PFAS on HGT are complex and likely depend on the microbial community involved and environmental factors. Another study demonstrates that in water distribution systems, PFAS and phthalate esters may act additively to increase MGE levels in biofilms, an effect that was also dependent upon pipe material (Yin H. et al., 2023). Furthermore, a recent study by Wang et al. (2025) demonstrated that in the presence of the quaternary ammonium compound diallyl dimethylammonium chloride, PFOA, PFHxA, and PFBS all increased the level of antimicrobial resistance genes in a nitrification community. This increase was correlated with elevated MGEs, suggesting that PFAS was involved in HGT in the community.
Collectively, these studies demonstrate that the effects of PFAS on MGEs and HGT are complex and environment dependent. A recent molecular dynamics and machine learning study by Xiao et al. (2025) predicted that PFAS increase bacterial HGT, an observation that was influenced by the electronegativity of PFAS molecules. A synergistic effect on HGT was predicted in the co-presence of PFAS and MPs. However, given the variability discussed above, it is important to validate these observations experimentally and with various microbial communities. Moreover, questions remain as to how those results obtained in model systems with controlled levels of PFAS might translate to the in situ activated sludge process with variable and dynamic PFAS concentrations, compositions, and distributions. Further research is also required to assess how PFAS may interact with other pollutants, stressors, and physical substrates in complex and potentially synergistic ways to affect HGT in wastewater operations.
4 HGT events from WWTPs associated with environmental volatility
There are complex connections between anthropogenic climate change and the selection of AMR and its transmission via MGEs and HGT. Climate change may exacerbate AMR via numerous mechanisms, including increased proximity between humans and animals (Magnano San Lio et al., 2023) altered spatial range of vectors and pathogens, and increased usage of antibiotics in humans and domesticated animals (Reverter et al., 2020). Moreover, higher temperatures are directly associated with increased rates of AMR (Meinen et al., 2023) although it has varying effects on the spread of resistance (Bagra et al., 2024a). In the Yellow river in China, for instance, ARG and MGE levels correlated with temperature (Yu et al., 2023). Other studies demonstrate that the spatial distribution (sediment or water) and mobilization potential of ARGs via MGEs differs depending on the time of year, with more MGEs in sediment during the Spring drought in Asian and European contexts (Guo et al., 2023). In the context of this review, the focus is on how climate change significantly impacts the hydrological cycle and how this influences the resistome profile of WWTPs and the spread of MGEs.
Zhou et al. (2022) identified a carbapenem-resistant Citrobacter sedlakii strain isolated from aerosols sampled outside a WWTP in China. Resistance was related to the presence of the blaNDM-5 gene, which was located on the IncX3 plasmid (pCSNDM-5). The plasmid could be transferred to the Escherichia coli recipient J53, demonstrating a possible transmission pathway for the spread of clinically relevant ABR and MGEs from WWTPs. Given that WWTP bio-aerosol generation is related to environmental conditions, including temperature, rainfall, and wind (Tian et al., 2022), Zhou et al.’s (2022) findings demonstrate the complex and interconnected pathways of MGE transmission in anthropogenic environments stemming from WWTPs. In this respect, Hou et al. (2022) investigated the effects of a severe precipitation event on the resistome of lagoon surface waters in China. Their study demonstrated that heavy rain events were associated with the promotion of HGT events by increasing the input of MGEs from diverse urban sources, such as road and agricultural runoff as well as WWTP effluent. Indeed, MGE levels were highest immediately following the storm, followed by a reduction to baseline levels that was associated with environmental controls such as pumping and floodgate opening. Urban stormwater often contains high levels of MGEs as well as selective pressures such as heavy metals, which are transported to WWTPs in sediment from roads (Zuo et al., 2022). The effects of floods and monsoons on MGE-mediated ARG dissemination is exacerbated in regions where wastewater treatment is suboptimal and untreated effluent contaminates river environments (Bagra et al., 2024b).
Collectively, these observations demonstrate the highly dynamic nature of MGE dissemination and HGT events, and highlight the temporal distribution of risk following extreme weather events. However, MGE distribution is geographically specific and dissemination depends on factors including host phylogeny (Johansson et al., 2023), suggesting that potential dissemination may be curbed by local ecology and climate. Hurricanes (Davis et al., 2020) have been associated with altered MGE profiles in watersheds, whilst a recent study has demonstrated that following a mining-associated tsunami in Brazil, MGEs were significantly increased in the disturbed river environment, among them the highly mobile and promiscuous ARG blaOXA (Suhadolnik et al., 2022). Less research has focused on the effects of droughts on the abundance of MGEs and rates of HGTs in WWTPs, but given that drought can significantly impact pollutant load in WWTPs and receiving environments (Köck-Schulmeyer et al., 2011) this is an important area for future study. Collectively, these findings demonstrate that climatic conditions are significant variables in modulating the landscape of MGEs and HGT events from WWTPs. As extreme weather events continue to increase in the modern world, their effects on the quantity, distribution, and transmission of MGEs and associated risks should become a specific focal point of research.
5 Transmission of genes from WWTPs to downstream organisms
The environment immediately surrounding WWTPs, and particularly the receiving waters of discharged effluent are under increased pressures. WWTPs exist in a gradient, along which the impact of anthropogenic activities can be detected, often via an increased prevalence of MGEs (Gillings et al., 2015) and ARGs. From a One Health perspective (Larsson et al., 2023), it is important to understand the connections between the MGEs discharged from WWTPs and recipient ecosystems, including environmental microbiomes, aquatic species, birds, and terrestrial organisms such as insects. In turn, these organisms can act as vectors further promoting the transmission of MGEs beyond the immediate WWTP environment.
5.1 Transmission to microbiomes
In the first instance, studies demonstrate that microbiomes downstream of WWTP effluent have altered community structure and dynamics (Price et al., 2018). However, it is uncertain whether these effects are related to selective chemicals acting on the receiving population, the transmission of MGEs from incoming effluent, or a combination of both. Using a nanopore approach, a study by Wu et al. (2022) demonstrated that receiving seawater contained a 10-fold enrichment of ARGs compared to clean seawater. The resistome profile matched that of the WWTP and was primarily due to the revival of microbes in the effluent that bypassed disinfection. Their results further suggested that plasmids and class I integrons were key players in the dissemination and persistence of ARGs in receiving waters. In agreement, Zhang et al. (2021) demonstrated that river microbiomes downstream of sewage treatment plants contain increased resistome diversity, due to invasion by resistance elements from WWTP effluents. These studies collectively suggest that the impacts of WWTP on HGT extend beyond the confines of the engineered ecosystem and readily infiltrate neighboring microbial communities. However, recent evidence suggests it is not only the treatment efficiency and quality of the final effluent that contributes to resistome infiltration, but also the background contamination of the river environment (Ferreira et al., 2022). This indicates that multiple stressors interact in complex ways to sculpt the mobilome of recipient ecosystems.
5.2 Transmission to larger organisms
The majority of studies investigating the effects of WWTP effluent on the downstream ecosystem health have involved taxonomic metagenomic studies. Such studies have demonstrated effects on the microbiomes of fish (Restivo et al., 2021), birds (Marcelino et al., 2018), mussels (Millar et al., 2022a), and insects (Millar et al., 2022b) living near WWTPs and exposed to their effluent. A comparatively smaller number of studies, however, have studied the presence and potential origins of mobile genetic elements in downstream microbial biofilms and in host microbiomes.
Insects are considered to act as vectors for the transmission of ARBs and ARGs (Gwenzi et al., 2021; Rawat et al., 2023). The feeding behavior and environmental niches of specific insects such as “filth flies” (Onwugamba et al., 2018) favors their contact with enteric pathogens potentially containing ARGs. In terms of HGT, events Doud et al. (2014) observed that house flies (Musca domestica) isolated from WWTPs and neighboring urban areas contain Enterococcus faecalis clones resistant to several antibiotics. Resistance was horizontally transferable between species as demonstrated by in vitro conjugation experiments. Genetic experiments (PFGE) suggested that the isolates were from an agricultural source, but higher resolution sequencing studies would be necessary to investigate this hypothesis. Future experiments should continue to use high resolution metagenomic sequencing to trace the origins of AMR genes in insect species living near WWTPs. On the other hand, Lee et al. (2023) demonstrated that WWTP-originating ARGs were not enriched in amphipods in receiving environments, thereby demonstrating the species and ecosystem-dependence of HGT in the vicinity of WWTPs.
In terms of larger organisms, the microbiomes of Hemiculter leucisculus living downstream of WWTPs demonstrated increased co-occurence of MGEs and ARGs (Xue et al., 2021), suggesting that the MGEs from WWTP effluent infiltrated fish microbiota (Guan et al., 2022). Specifically, the findings suggested that plasmids were key vectors for the transmission of ARGs along an anthropogenic gradient from WWTP to pristine river environments. Gulls are considered a key organism involved in the wide-range transmission of ARGs between different species due to their intimate connections across human and aquatic environments (Wyrsch et al., 2022; Lin et al., 2020; Zeballos-Gross et al., 2021; Marcelino et al., 2018). Indeed, gulls living in proximity to anthropogenic environments hosted Escherichia coli and Klebsiella pneumoniae strains resistant to clinically important antibiotics, with some determinants carried on plasmids (Mukerji et al., 2023). Further, Woksepp et al. (2023) detected carbapenemase-producing Enterobacterales (CPE) in Swedish wastewater and gull feces. Four different carbapenemases were identified (blaGES-5, blaIMI-3, blaOXA-181 and blaOXA-244) which were carried on plasmids [IncP and IncFII(Yp)]. Those MGEs detected in the gull feces were strongly suggested to have been acquired from the WWTP where the authors had observed the birds feeding. These results are concerning, especially given that certain carbapenemases and their MGEs were detected in gull feces 2 km from the WWTP.
6 Conclusion
This minireview has focused on three scales and domains of HGT in and around WWTPs that are emerging in the scientific literature. However, it is important to consider the inseparability and connectivity between these processes. Researchers should continue to analyze the role of specific factors, such as micropollutants, in the dynamics of HGT but more integrated perspectives are also necessary, such as “One Health,” “Global Health” or “Planetary Health” approaches (Horvat and Kovačević, 2025; Ramsamy et al., 2022). WWTPs are themselves sites of integration for municipal, industrial, and agricultural effluent, microbial evolution, and increasingly volatile climate regimes. On the one hand, the studies summarized here point to general trends in which WWTPs act as catalysts for HGT and the selective accumulation of MGEs. On the other hand, the growing research also points to important differences that are site, context, and microbial community-dependent. These elements of variability underscore the need to understand bacterial dynamics on a case-by-case basis, even as their effects reverberate through larger systems. Thus, whilst an interconnected picture begins to link WWTPs and HGT with novel and emerging contaminants, volatile weather, and trophic links to wider ecosystems, future research is necessary to explore these connections across specific environments and socio-ecological systems.
Author contributions
AB: Funding acquisition, Writing – original draft, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by the Deutsche Forschungsgemeinschaft (DFG) Project number 556660619: “MicroFlows: Socio-Microbial Relations and Urban Water Metabolism in the Bacterial City.”
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achmon, Y., Achmon, M., Dowdy, F. R., Spiegel, O., Claypool, J. T., Toniato, J., et al. (2018). Understanding the Anthropocene through the lens of landfill microbiomes. Front. Ecol. Environ. 16:354–360. doi: 10.1002/fee.1819
Alderton, I., Palmer, B. R., Heinemann, J. A., Pattis, I., Weaver, L., Gutiérrez-Ginés, M. J., et al. (2021). The role of emerging organic contaminants in the development of antimicrobial resistance. Emerg. Contam. 7, 160–171. doi: 10.1016/j.emcon.2021.07.001
Arias-Andres, M., Klümper, U., Rojas-Jimenez, K., and Grossart, H.-P. (2018). Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 237, 253–261. doi: 10.1016/j.envpol.2018.02.058
Arnold, B. J., Huang, I.-T., and Hanage, W. P. (2022). Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 20, 206–218. doi: 10.1038/s41579-021-00650-4
Bagra, K., Kneis, D., Padfield, D., Szekeres, E., Teban-Man, A., Coman, C., et al. (2024a). Contrary effects of increasing temperatures on the spread of antimicrobial resistance in river biofilms. Msphere 9:e00573-23. doi: 10.1128/msphere.00573-23
Bagra, K., Singh, H., Klümper, U., and Singh, G. (2024b). Drivers of antibiotic resistance in two monsoon-impacted Indian urban rivers receiving untreated wastewater. bioRxiv [Preprint] doi: 10.1101/2024.10.31.621109
Barisci, S., and Suri, R. (2021). Occurrence and removal of poly/perfluoroalkyl substances (PFAS) in municipal and industrial wastewater treatment plants. Water Sci. Technol. 84, 3442–3468. doi: 10.2166/wst.2021.484
Beale, D. J., Bissett, A., Nilsson, S., Bose, U., Nelis, J. L. D., Nahar, A., et al. (2022). Perturbation of the gut microbiome in wild-caught freshwater turtles (Emydura macquarii macquarii) exposed to elevated PFAS levels. Sci. Total Environ. 838:156324.
Bradshaw, A. (2024a). Abundance and absence: Human-microbial co-evolution in the Anthropocene. Anthropocene Rev. 11, 26–48. doi: 10.1177/20530196231153925
Bradshaw, A. (2024b). The invisible city: The mundane biogeographies of urban microbial ecologies. Geo Geogr. Environ. 11:e00148. doi: 10.1002/geo2.148
Brown, C. L., Maile-Moskowitz, A., Lopatkin, A. J., Xia, K., Logan, L. K., Davis, B. C., et al. (2024). Selection and horizontal gene transfer underlie microdiversity-level heterogeneity in resistance gene fate during wastewater treatment. Nat. Commun. 15:5412. doi: 10.1038/s41467-024-49742-8
Brunn, H., Arnold, G., Körner, W., Rippen, G., Steinhäuser, K. G., and Valentin, I. (2023) PFAS: forever chemicals—persistent, bioaccumulative and mobile. Reviewing the status and the need for their phase out and remediation of contaminated sites. Environ. Sci. Eur. 35, 1–50. doi: 10.1186/s12302-023-00721-8
Cai, L., Yu, K., Yang, Y., Chen, B.-W., Li, X.-D., and Zhang, T. (2013). Metagenomic exploration reveals high levels of microbial arsenic metabolism genes in activated sludge and coastal sediments. Appl. Microbiol. Biotechnol. 97, 9579–9588. doi: 10.1007/s00253-012-4678-8
Carscadden, K. A., Batstone, R. T., and Hauser, F. E. (2023). Origins and evolution of biological novelty. Biol. Rev. 98, 1472–1491. doi: 10.1111/brv.12963
Cen, T., Zhang, X., Xie, S., and Li, D. (2020). Preservatives accelerate the horizontal transfer of plasmid-mediated antimicrobial resistance genes via differential mechanisms. Environ. Int. 138:105544. doi: 10.1016/j.envint.2020.105544
Chen, C., Fang, Y., and Zhou, D. (2023). Selective pressure of PFOA on microbial community: Enrichment of denitrifiers harboring ARGs and the transfer of ferric-electrons. Water Res. 233:119813.
Chen, C., Fang, Y., Cui, X., and Zhou, D. (2022). Effects of trace PFOA on microbial community and metabolisms: microbial selectivity, regulations and risks. Water Res. 226:119273.
Daubin, V., and Szöllősi, G. J. (2016). Horizontal gene transfer and the history of life. Cold Spring Harbor Perspect. Biol. 8:a018036. doi: 10.1101/cshperspect.a018036
Davis, B. C., Riquelme, M. V., Ramirez-Toro, G., Bandaragoda, C., Garner, E., Rhoads, W. J., et al. (2020). Demonstrating an integrated antibiotic resistance gene surveillance approach in Puerto Rican watersheds post-hurricane Maria. Environ. Sci. Technol. 54, 15108–15119. doi: 10.1021/acs.est.0c05567
Doud, C. W., Morgan, H., Scott, L., and Zurek, J. (2014). Role of house flies in the ecology of Enterococcus faecalis from wastewater treatment facilities. Microb. Ecol. 67, 380–391. doi: 10.1007/s00248-013-0337-6
Douziech, M., Conesa, I. R., Benítez-López, A., Franco, A., Huijbregts, M., and van Zelm, R. (2018). Quantifying variability in removal efficiencies of chemicals in activated sludge wastewater treatment plants–a meta-analytical approach. Environ. Sci. Processes Impacts 20, 171–182. doi: 10.1039/C7EM00493A
Edo, C., González-Pleiter, M., Leganés, F., Fernández-Pinas, F., and Rosal, R. (2020). Fate of microplastics in wastewater treatment plants and their environmental dispersion with effluent and sludge. Environ. Pollut. 259:113837. doi: 10.1016/j.envpol.2019.113837
Emamalipour, M., Seidi, K., Vahed, S. Z., Jahanban-Esfahlan, A., Jaymand, M., Majdi, H., et al. (2020). Horizontal gene transfer: From evolutionary flexibility to disease progression. Front. Cell Dev. Biol. 8:229. doi: 10.3389/fcell.2020.00229
Fang, G.-Y., Liu, X.-Q., Jiang, Y.-J., Mu, X.-J., and Huang, B.-W. (2024). Horizontal gene transfer in activated sludge enhances microbial antimicrobial resistance and virulence. Sci. Total Environ. 912:168908. doi: 10.1016/j.scitotenv.2023.168908
Fang, H., Cai, L., Yu, Y., and Zhang, T. (2013). Metagenomic analysis reveals the prevalence of biodegradation genes for organic pollutants in activated sludge. Bioresource Technol. 129, 209–218. doi: 10.1016/j.biortech.2012.11.054
Fang, H., Zhang, H., Han, L., Mei, J., Ge, Q., Long, Z., et al. (2018). Exploring bacterial communities and biodegradation genes in activated sludge from pesticide wastewater treatment plants via metagenomic analysis. Environ. Pollut. 243, 1206–1216. doi: 10.1016/j.envpol.2018.09.080
Feng, G., Huang, H., and Chen, Y. (2021). Effects of emerging pollutants on the occurrence and transfer of antibiotic resistance genes: A review. J. Hazard. Mater. 420:126602. doi: 10.1016/j.jhazmat.2021.126602
Feng, Y., Sun, J.-W., Shi, W.-W., Duan, J.-L., Sun, X.-D., Feng, L.-J., et al. (2023). Microplastics exhibit accumulation and horizontal transfer of antibiotic resistance genes. J. Environ. Manage. 336:117632. doi: 10.1016/j.jenvman.2023.117632
Ferreira, C., Abreu-Silva, J., and Manaia, C. M. (2022). The balance between treatment efficiency and receptor quality determines wastewater impacts on the dissemination of antibiotic resistance. J. Hazard. Mater. 434:128933. doi: 10.1016/j.jhazmat.2022.128933
Ghaly, T. M., Gillings, M. R., Penesyan, A., Qi, Q., Rajabal, V., and Tetu, S. G. (2021). The natural history of integrons. Microorganisms 9:2212. doi: 10.3390/microorganisms9112212
Gillings, M. R. (2017). Lateral gene transfer, bacterial genome evolution, and the Anthropocene. Ann. N. Y. Acad. Sci. 1389, 20–36. doi: 10.1111/nyas.13213
Gillings, M. R., and Paulsen, I. T. (2014). Microbiology of the Anthropocene. Anthropocene 5, 1–8. doi: 10.1016/j.ancene.2014.06.004
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y.-G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Groussin, M., Poyet, M., Sistiaga, A., Kearney, S. M., Moniz, K., Noel, M., et al. (2021). Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184, 2053–2067. doi: 10.1016/j.cell.2021.02.052
Guan, Y., Xue, X., Jia, J., Li, X., Xing, H., and Wang, Z. (2022). Metagenomic assembly and binning analyses the prevalence and spread of antibiotic resistome in water and fish gut microbiomes along an environmental gradient. J. Environ. Manage. 318:115521. doi: 10.1016/j.jenvman.2022.115521
Guo, J., Li, J., Chen, H., Bond, P. L., and Yuan, Z. (2017). Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 123, 468–478. doi: 10.1016/j.watres.2017.07.002
Guo, Z.-F., Boeing, W. J., Xu, Y.-Y., Borgomeo, E., Liu, D., and Zhu, Y.-G. (2023). Data-driven discoveries on widespread contamination of freshwater reservoirs by dominant antibiotic resistance genes. Water Res. 229:119466. doi: 10.1016/j.watres.2022.119466
Gwenzi, W., Chaukura, N., Muisa-Zikali, N., Teta, C., Musvuugwa, T., Rzymski, P., et al. (2021). Insects, rodents, and pets as reservoirs, vectors, and sentinels of antimicrobial resistance. Antibiotics 10:68. doi: 10.3390/antibiotics10010068
Haraoui, L.-P. (2022). Networked collective microbiomes and the rise of subcellular ‘units of life’. Trends Microbiol. 30, 112–119. doi: 10.1016/j.tim.2021.09.011
Horvat, O., and Kovačević, Z. (2025). Human and veterinary medicine collaboration: Synergistic approach to address antimicrobial resistance through the lens of planetary health. Antibiotics 14:38. doi: 10.3390/antibiotics14010038
Hou, L., Li, J., Wang, H., Chen, Q., Su, J.-Q., Gad, M., et al. (2022). Storm promotes the dissemination of antibiotic resistome in an urban lagoon through enhancing bio-interactions. Environ. Int. 168:107457. doi: 10.1016/j.envint.2022.107457
Ilieva, Z., Hamza, R. A., and Suehring, R. (2024). The significance of fluorinated compound chain length, treatment technology, and influent composition on per-and polyfluoroalkyl substances removal in worldwide wastewater treatment plants. Integr. Environ. Assess. Manag. 20, 59–69. doi: 10.1002/ieam.4778
Jain, R., Rivera, M. C., Moore, J. E., and Lake, J. A. (2003). Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20, 1598–1602.
Johansson, M. H. K., Aarestrup, F. M., and Petersen, T. N. (2023). Importance of mobile genetic elements for dissemination of antimicrobial resistance in metagenomic sewage samples across the world. PLoS One 18:e0293169. doi: 10.1371/journal.pone.0293169
Ju, F., Guo, F., Ye, L., Xia, Y., and Zhang, T. (2014). Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ. Microbiol. Rep. 6, 80–89.
Junaid, M., Liu, S., Liao, H., Liu, X., Wu, Y., and Wang, J. (2022). Wastewater plastisphere enhances antibiotic resistant elements, bacterial pathogens, and toxicological impacts in the environment. Sci. Total Environ. 841:156805. doi: 10.1016/j.scitotenv.2022.156805
Köck-Schulmeyer, M., Ginebreda, A., Postigo, C., López-Serna, R., Pérez, S., Brix, R., et al. (2011). Wastewater reuse in Mediterranean semi-arid areas: The impact of discharges of tertiary treated sewage on the load of polar micro pollutants in the Llobregat river (NE Spain). Chemosphere 82, 670–678. doi: 10.1016/j.chemosphere.2010.11.005
Larsson, D. G. J., Gaze, W. H., Laxminarayan, R., and Topp, E. (2023). AMR, one health and the environment. Nat. Microbiol. 8, 754–755. doi: 10.1038/s41564-023-01351-9
Laue, H. E., Moroishi, Y., Palys, T. J., Christensen, B. C., Criswell, R. L., Peterson, L. A., et al. (2023). Early-life exposure to per-and polyfluoroalkyl substances and infant gut microbial composition. Environ. Epidemiol. 7:e238. doi: 10.1097/EE9.0000000000000238
Lee, J., Ju, F., Beck, K., and Bürgmann, H. (2023). Differential effects of wastewater treatment plant effluents on the antibiotic resistomes of diverse river habitats. ISME J. 17, 1993–2002. doi: 10.1038/s41396-023-01506-w
Lin, H., Jiang, L., Li, B., Dong, Y., He, Y., and Qiu, Y. (2019). Screening and evaluation of heavy metals facilitating antibiotic resistance gene transfer in a sludge bacterial community. Sci. Total Environ. 695:133862. doi: 10.1016/j.scitotenv.2019.133862
Lin, Y., Dong, X., Sun, R., Wu, J., Tian, L., Rao, D., et al. (2020). Migratory birds-one major source of environmental antibiotic resistance around Qinghai Lake, China. Sci. Total Environ. 739:139758. doi: 10.1016/j.scitotenv.2020.139758
Liu, C., Zhu, X., You, L., Gin, K. Y.-H., Chen, H., and Chen, B. (2023). Per/polyfluoroalkyl substances modulate plasmid transfer of antibiotic resistance genes: A balance between oxidative stress and energy support. Water Res. 240:120086. doi: 10.1016/j.watres.2023.120086
Liu, X., Wang, H., Li, L., Deng, C., Chen, Y., Ding, H., et al. (2022). Do microplastic biofilms promote the evolution and co-selection of antibiotic and metal resistance genes and their associations with bacterial communities under antibiotic and metal pressures? J. Hazard. Mater. 424:127285. doi: 10.1016/j.jhazmat.2021.127285
Liu, Y., Liu, L., Wang, X., Shao, M., Wei, Z., Wang, L., et al. (2025). Microplastics enhance the prevalence of antibiotic resistance genes in mariculture sediments by enriching host bacteria and promoting horizontal gene transfer. Eco-Environ. Health 4:100136. doi: 10.1016/j.eehl.2025.100136
Luo, T., Dai, X., Chen, Z., Wu, L., Wei, W., Xu, Q., et al. (2023). Different microplastics distinctively enriched the antibiotic resistance genes in anaerobic sludge digestion through shifting specific hosts and promoting horizontal gene flow. Water Res. 228:119356. doi: 10.1016/j.watres.2022.119356
Magnano San Lio, R., Favara, G., Maugeri, A., Barchitta, M., and Agodi, A. (2023). How antimicrobial resistance is linked to climate change: An overview of two intertwined global challenges. Int. J. Environ. Res. Public Health 20:1681. doi: 10.3390/ijerph20031681
Marcelino, V. R., Wille, M., Hurt, A. C., González-Acuña, D., Klaassen, M., Eden, J.-S., et al. (2018). High levels of antibiotic resistance gene expression among birds living in a wastewater treatment plant. bioRxiv [Preprint] doi: 10.1101/462366
Mason, S. A., Garneau, D., Sutton, R., Chu, Y., Ehmann, K., Barnes, J., et al. (2016). Microplastic pollution is widely detected in US municipal wastewater treatment plant effluent. Environ. Pollut. 218, 1045–1054. doi: 10.1016/j.envpol.2016.08.056
Mbanga, J., Amoako, D. G., Abia, A. L. K., Allam, M., Ismail, A., and Essack, S. Y. (2021). Genomic insights of multidrug-resistant Escherichia coli from wastewater sources and their association with clinical pathogens in South Africa. Front. Vet. Sci. 8:636715. doi: 10.3389/fvets.2021.636715
McCabe, B. J., and Eckenfelder, W. W. (1961). BOD removal and sludge growth in the activated sludge process. J. Water Pollut. Control Fed. 33, 258–271.
Meinen, A., Tomczyk, S., Wiegand, F. N., Abu Sin, M., Eckmanns, T., and Haller, S. (2023). Antimicrobial resistance in Germany and Europe-A systematic review on the increasing threat accelerated by climate change. J. Health Monit. 8, 93–108. doi: 10.25646/11404
Miguel, A. B. R., Jetten, M. S. M., and Welte, C. U. (2020). The role of mobile genetic elements in organic micropollutant degradation during biological wastewater treatment. Water Res. X 9:100065. doi: 10.1016/j.wroa.2020.100065
Millar, E. N., Kidd, K. A., Surette, M. G., Bennett, C. J., Salerno, J., and Gillis, P. L. (2022a). Effects of municipal wastewater effluents on the digestive gland microbiome of wild freshwater mussels (Lasmigona costata). Ecotoxicol. Environ. Saf. 241:113774. doi: 10.1016/j.ecoenv.2022.113774
Millar, E. N., Surette, M. G., and Kidd, K. A. (2022b). Altered microbiomes of aquatic macroinvertebrates and riparian spiders downstream of municipal wastewater effluents. Sci. Total Environ. 809:151156.
Mukerji, S., Sahibzada, S., Abraham, R., Stegger, M., Jordan, D., Hampson, D. J., et al. (2023). Proximity to human settlement is directly related to carriage of critically important antimicrobial-resistant Escherichia coli and Klebsiella pneumoniae in Silver Gulls. Vet. Microbiol. 280:109702. doi: 10.1016/j.vetmic.2023.109702
Onwugamba, F. C., Fitzgerald, J. R., Rochon, K., Guardabassi, L., Alabi, A., Kühne, S., et al. (2018). The role of ‘filth flies’ in the spread of antimicrobial resistance. Travel Med. Infect. Dis. 22, 8–17. doi: 10.1016/j.tmaid.2018.02.007
Pham, D. N., Clark, L., and Li, M. (2021). Microplastics as hubs enriching antibiotic-resistant bacteria and pathogens in municipal activated sludge. J. Hazard. Mater. Lett. 2:100014. doi: 10.1016/j.hazl.2021.100014
Price, J. R., Ledford, S. H., Ryan, M. O., Toran, L., and Sales, C. M. (2018). Wastewater treatment plant effluent introduces recoverable shifts in microbial community composition in receiving streams. Sci. Total Environ. 613, 1104–1116. doi: 10.1016/j.scitotenv.2017.09.162
Ramsamy, Y., Mlisana, K. P., Amoako, D. G., Abia, A. L. K., Ismail, A., Allam, M., et al. (2022). Mobile genetic elements-mediated Enterobacterales-associated carbapenemase antibiotic resistance genes propagation between the environment and humans: A one health South African study. Sci. Total Environ. 806:150641. doi: 10.1016/j.scitotenv.2021.150641
Rawat, N., Sabu, B., Jamwal, R., Devi, P. P., Yadav, K., Raina, H. S., et al. (2023). Understanding the role of insects in the acquisition and transmission of antibiotic resistance. Sci. Total Environ. 858:159805. doi: 10.1016/j.scitotenv.2022.159805
Restivo, V. E., Kidd, K. A., Surette, M. G., Servos, M. R., and Wilson, J. Y. (2021). Rainbow darter (Etheostoma caeruleum) from a river impacted by municipal wastewater effluents have altered gut content microbiomes. Sci. Total Environ. 751:141724. doi: 10.1016/j.scitotenv.2020.141724
Reverter, M., Sarter, S., Caruso, D., Avarre, J.-C., Combe, M., Pepey, E., et al. (2020). Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 11:1870. doi: 10.1038/s41467-020-15735-6
Sambaza, S. S., and Naicker, N. (2023). Contribution of wastewater to antimicrobial resistance: A review article. J. Glob. Antimicrob. Resist. 34, 23–29. doi: 10.1016/j.jgar.2023.05.010
Saunders, A. M., Albertsen, M., Vollertsen, J., and Nielsen, P. H. (2016). The activated sludge ecosystem contains a core community of abundant organisms. ISME J. 10, 11–20. doi: 10.1038/ismej.2015.117
Sen, P., Fan, Y., Schlezinger, J. J., Ehrlich, S. D., Webster, T. F., Hyötyläinen, T., et al. (2024). Exposure to environmental toxicants is associated with gut microbiome dysbiosis, insulin resistance and obesity. Environ. Int. 186:108569. doi: 10.1016/j.envint.2024.108569
Sharma, P., Pal, N., Kumawat, M., Singh, S., Das, D., Tilwari, A., et al. (2025). Investigating the antibiotic resistance genes and mobile genetic elements in water systems impacted with anthropogenic pollutants. Environ. Res. 269:120814. doi: 10.1016/j.envres.2025.120814
Shen, H., Yang, M., Yin, K., Wang, J., Tang, L., Lei, B., et al. (2023). Size-and surface charge-dependent hormetic effects of microplastics on bacterial resistance and their interactive effects with quinolone antibiotic. Sci. Total Environ. 903:166580. doi: 10.1016/j.scitotenv.2023.166580
Soucy, S. M., Huang, J., and Gogarten, J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482. doi: 10.1038/nrg3962
Suhadolnik, M. L. S., Costa, P. S., Paiva, M. C., Salim, A. C. M., Barbosa, F. A. R., Lobo, F. P., et al. (2022). Spatiotemporal dynamics of the resistome and virulome of riverine microbiomes disturbed by a mining mud tsunami. Sci. Total Environ. 806:150936. doi: 10.1016/j.scitotenv.2021.150936
Syranidou, E., and Kalogerakis, N. (2022). Interactions of microplastics, antibiotics and antibiotic resistant genes within WWTPs. Sci. Total Environ. 804:150141. doi: 10.1016/j.scitotenv.2021.150141
Tan, Q., Chen, J., Chu, Y., Liu, W., Yang, L., Ma, L., et al. (2021). Triclosan weakens the nitrification process of activated sludge and increases the risk of the spread of antibiotic resistance genes. J. Hazard. Mater. 416:126085. doi: 10.1016/j.jhazmat.2021.126085
Tian, J., Yan, C., Alcega, S. G., Hassard, F., Tyrrel, S., Coulon, F., et al. (2022). Detection and characterization of bioaerosol emissions from wastewater treatment plants: Challenges and opportunities. Front. Microbiol. 13:958514. doi: 10.3389/fmicb.2022.958514
Top, E. M., and Springael, D. (2003). The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14, 262–269. doi: 10.1016/S0958-1669(03)00066-1
Vos, M. (2020). The evolution of bacterial pathogens in the Anthropocene. Infect. Genet. Evol. 86:104611. doi: 10.1016/j.meegid.2020.104611
Wang, H., Gao, J., Cui, Y., Wang, Y., Guo, Y., and Chen, H. (2025). Short-chain per/polyfluoroalkyl substances alternatives enhance horizontal gene transfer risks in nitrification systems under quaternary ammonia compounds antimicrobials co-stress despite lower acute toxicity than perfluorooctanoic acid. Water Res. 287:124274. doi: 10.1016/j.watres.2025.124274
Wang, H., Xu, K., Wang, J., Feng, C., Chen, Y., Shi, J., et al. (2023). Microplastic biofilm: An important microniche that may accelerate the spread of antibiotic resistance genes via natural transformation. J. Hazard. Mater. 459:132085. doi: 10.1016/j.jhazmat.2023.132085
Wang, Z., Buser, A. M., Cousins, I. T., Demattio, S., Drost, W., Johansson, O., et al. (2021a). A new OECD definition for per-and polyfluoroalkyl substances. Environ. Sci. Technol. 55, 15575–15578. doi: 10.1021/acs.est.1c06896
Wang, Z., Gao, J., Zhao, Y., Dai, H., Jia, J., and Zhang, D. (2021b). Plastisphere enrich antibiotic resistance genes and potential pathogenic bacteria in sewage with pharmaceuticals. Sci. Total Environ. 768:144663. doi: 10.1016/j.scitotenv.2020.144663
Woksepp, H., Karlsson, K., Borjesson, S., Karlsson Lindsjö, O., Söderlund, R., and Bonnedahl, J. (2023). Dissemination of carbapenemase-producing Enterobacterales through wastewater and gulls at a wastewater treatment plant in Sweden. Sci. Total Environ. 886:163997. doi: 10.1016/j.scitotenv.2023.163997
Wu, Z., Che, Y., Dang, C., Zhang, M., Zhang, X., Sun, Y., et al. (2022). Nanopore-based long-read metagenomics uncover the resistome intrusion by antibiotic resistant bacteria from treated wastewater in receiving water body. Water Res. 226:119282. doi: 10.1016/j.watres.2022.119282
Wyrsch, E. R., Nesporova, K., Tarabai, H., Jamborova, I., Bitar, I., Literak, I., et al. (2022). Urban wildlife crisis: Australian silver gull is a bystander host to widespread clinical antibiotic resistance. mSystems 7:e00158-22. doi: 10.1128/msystems.00158-22
Xiao, B., Pu, Q., Ding, G., Wang, Z., Li, Y., and Hou, J. (2025). Synergistic effect of horizontal transfer of antibiotic resistance genes between bacteria exposed to microplastics and per/polyfluoroalkyl substances: An explanation from theoretical methods. J. Hazardous Mater. 492:138208.
Xu, M., Gao, P., Gao, Y., Xiong, S.-J., Chen, H.-Q., and Shen, X.-X. (2024). Impacts of microplastic type on the fate of antibiotic resistance genes and horizontal gene transfer mechanism during anaerobic digestion. J. Environ. Manage. 360:121090. doi: 10.1016/j.jenvman.2024.121090
Xu, Z., Xiong, J., Li, C., Hu, S., Li, Z., Ma, Y., et al. (2024). Environmental concentrations of per/polyfluoroalkyl substances promote the conjugative transfer of antibiotic resistance genes. Chem. Eng. J. 498:155500.
Xue, X., Jia, J., Yue, X., Guan, Y., Zhu, L., and Wang, Z. (2021). River contamination shapes the microbiome and antibiotic resistance in sharpbelly (Hemiculter leucisculus). Environ. Pollut. 268:115796. doi: 10.1016/j.envpol.2020.115796
Yin, H., Chen, R., Wang, H., Schwarz, C., Hu, H., Shi, B., et al. (2023). Co-occurrence of phthalate esters and perfluoroalkyl substances affected bacterial community and pathogenic bacteria growth in rural drinking water distribution systems. Sci. Total Environ. 856:158943. doi: 10.1016/j.scitotenv.2022.158943
Yin, L., Wang, X., Xu, H., Yin, B., Wang, X., Zhang, Y., et al. (2023). Unrecognized risk of perfluorooctane sulfonate in promoting conjugative transfers of bacterial antibiotic resistance genes. Appl. Environ. Microbiol. 89:e00533-23. doi: 10.1128/aem.00533-23
Yin, X., Yang, Y., Deng, Y., Huang, Y., Li, L., Chan, L. Y. L., et al. (2022). An assessment of resistome and mobilome in wastewater treatment plants through temporal and spatial metagenomic analysis. Water Res. 209:117885. doi: 10.1016/j.watres.2021.117885
Yoo, K., Yoo, H., Lee, J., Choi, E. J., and Park, J. (2020). Exploring the antibiotic resistome in activated sludge and anaerobic digestion sludge in an urban wastewater treatment plant via metagenomic analysis. J. Microbiol. 58, 123–130. doi: 10.1007/s12275-020-9309-y
Yu, K., and Zhang, T. (2012). Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS One 7:e38183. doi: 10.1371/journal.pone.0038183
Yu, Q., Han, Q., Shi, S., Sun, X., Wang, X., Wang, S., et al. (2023). Metagenomics reveals the response of antibiotic resistance genes to elevated temperature in the Yellow River. Sci. Total Environ. 859:160324. doi: 10.1016/j.scitotenv.2022.160324
Zeballos-Gross, D., Rojas-Sereno, Z., Salgado-Caxito, M., Poeta, P., Torres, C., and Benavides, J. A. (2021). The role of gulls as reservoirs of antibiotic resistance in aquatic environments: A scoping review. Front. Microbiol. 12:703886. doi: 10.3389/fmicb.2021.703886
Zhang, K., Hamidian, A. H., Tubić, A., Zhang, Y., Fang, J. K. H., Wu, C., et al. (2021). Understanding plastic degradation and microplastic formation in the environment: A review. Environ. Pollut. 274:116554. doi: 10.1016/j.envpol.2021.116554
Zhang, Y., Liu, C., Chen, H., Chen, J., Li, J., and Teng, Y. (2022). Metagenomic insights into resistome coalescence in an urban sewage treatment plant-river system. Water Res. 224:119061. doi: 10.1016/j.watres.2022.119061
Zhao, Y., Hu, Z., Xie, H., Wu, H., Wang, Y., Xu, H., et al. (2023). Size-dependent promotion of micro (nano) plastics on the horizontal gene transfer of antibiotic resistance genes in constructed wetlands. Water Res. 244:120520. doi: 10.1016/j.watres.2023.120520
Zhou, Y., Zhang, G., Zhang, D., Zhu, N., Bo, J., Meng, X., et al. (2024). Microplastic biofilms promote the horizontal transfer of antibiotic resistance genes in estuarine environments. Mar. Environ. Res. 202:106777. doi: 10.1016/j.marenvres.2024.106777
Zhou, Z., Berglund, B., Liu, J., Zhao, L., Xia, H., Zou, H., et al. (2022). Emergence of IncX3 plasmid-harboring blaNDM-5 in a Citrobacter sedlakii isolated from outdoor aerosol in wastewater treatment plant. Microb. Drug Resist. 28, 199–204. doi: 10.1089/mdr.2021.0057
Ziajahromi, S., Neale, P. A., and Leusch, F. D. L. (2016). Wastewater treatment plant effluent as a source of microplastics: Review of the fate, chemical interactions and potential risks to aquatic organisms. Water Sci. Technol. 74, 2253–2269. doi: 10.2166/wst.2016.414
Keywords: MGE, HGT, PFAS, microplastics, contaminants of emerging concern, WWTP, AMR
Citation: Bradshaw A (2025) Mobile genetic elements and wastewater treatment: contaminants of emerging concern, climate change, and trophic transmission. Front. Microbiol. 16:1699325. doi: 10.3389/fmicb.2025.1699325
Received: 04 September 2025; Accepted: 20 October 2025;
Published: 10 November 2025.
Edited by:
Yanchu Ke, Fujian Agriculture and Forestry University, ChinaReviewed by:
Shahbaz Raza, Sungkyunkwan University, Republic of KoreaCopyright © 2025 Bradshaw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aaron Bradshaw, YWFyb24uYnJhZHNoYXdAZmF1LmRl
 Aaron Bradshaw
Aaron Bradshaw