- 1Clinical Research Center (CRC), Medical Pathology Center (MPC), Cancer Early Detection and Treatment Center (CEDTC) and Translational Medicine Research Center (TMRC), Chongqing University Three Gorges Hospital, Chongqing University, Chongqing, China
- 2CQU-Ferenc Krausz Nobel Laureate Scientific Workstation, Chongqing University Three Gorges Hospital and Academy for Advanced Interdisciplinary Technology, Chongqing, China
- 3Chongqing Technical Innovation Center for Quality Evaluation and Identification of Authentic Medicinal Herbs, Chongqing, China
- 4School of Medicine, Chongqing University, Chongqing, China
- 5State Key Laboratory of Digital Medical Engineering, Key Laboratory of Biomedical Engineering of Hainan Province, School of Biomedical Engineering, Hainan University, Sanya, Hainan, China
- 6West China School of Basic Medical Sciences and Forensic Medicine, Institute of Biomedical Engineering, Sichuan University, Chengdu, China
Colorectal cancer (CRC) is closely linked to gut microbiota dysbiosis. We synthesize evidence that carcinogenic microbes promote CRC through chronic inflammation, bacterial genotoxins, and metabolic imbalance, highlighting key pathways involving Fusobacterium nucleatum, pks+Escherichia coli, and enterotoxigenic Bacteroides fragilis (ETBF). Building on these mechanisms, we propose a minimal diagnostic signature that integrates multi-omics with targeted qPCR, and a pathway–therapy–microbiome matching framework to guide individualized treatment. Probiotics, fecal microbiota transplantation (FMT), and bacteriophage therapy show promise as adjunctive strategies; however, standardization, safety monitoring, and regulatory readiness remain central hurdles. We advocate a three-step path to clinical implementation—stratified diagnosis, therapy matching, and longitudinal monitoring—supported by spatial multi-omics and AI-driven analytics. This approach aims to operationalize microbiome biology into deployable tools for risk stratification, treatment selection, and surveillance, advancing toward microbiome-informed precision oncology in CRC.
1 Introduction
Colorectal cancer (CRC) is one of the most prevalent malignant tumors worldwide. According to the latest data released by the International Agency for Research on Cancer (IARC), the global incidence of CRC is expected to exceed 3.2 million new cases in 2040, with nearly 1.6 million deaths, ranking third among all cancers after breast and lung cancer (Morgan et al., 2022). While early detection rates are relatively high in some developed countries, such as the United States and European nations, due to well-established screening programs, the situation remains critical in developing regions including India and Africa, where screening coverage is limited and over 60% of cases are diagnosed at advanced stages (Lee and Holmes, 2023). This “high-incidence and high-mortality” pattern not only poses a significant threat to public health but also imposes a considerable burden on global healthcare systems.
With the rapid development of high-throughput sequencing, metagenomics, and metabolomics, the role of the gut microbiota in human health and disease has drawn increasing attention (Fan and Pedersen, 2020). Gut microbes maintain intestinal homeostasis and host immunity. They also contribute to CRC via chronic inflammation, bacterial genotoxins, oxidative stress, and dysregulated microbial metabolites (Dougherty and Jobin, 2023; White and Sears, 2023). Given that the colon and rectum harbor a highly dense microbial ecosystem, gut microbiota dysbiosis is now considered a pivotal environmental factor contributing to CRC onset and progression.
Throughout the multistage development of CRC, the gut microbiota interacts dynamically with the host (Kim and Lee, 2022). On one hand, specific bacterial taxa, including Fusobacterium nucleatum (F. nucleatum), Escherichia coli (E. coli), and Bacteroides fragilis (B. fragilis), are enriched in tumor tissues and can promote tumorigenesis by activating pro-inflammatory pathways, inducing DNA damage, and modulating oncogenic signaling (Wong and Yu, 2023; Ternes et al., 2020). On the other hand, strategies aimed at modulating the gut microbiome hold promise for early detection, therapeutic synergy, and even prevention of CRC (Chen and Chen, 2021).
In this review, we comprehensively summarize current advances in understanding the relationship between the gut microbiota and CRC. We focus on microbial mechanisms of tumor initiation and progression, key bacterial species with carcinogenic potential, cutting-edge microbiome detection technologies, emerging microbiota-targeted therapeutic strategies, and translational landscape, providing theoretical and translational insights for CRC prevention, early diagnosis, and precision therapy.
2 Mechanisms linking gut microbiota to colorectal carcinogenesis
CRC is driven by a multifactorial interplay of genetic susceptibility, environmental exposures, and gut microbiota dysbiosis. Among these factors, the gut microbiota has emerged as a pivotal environmental contributor that participates in tumor initiation, progression, and metastasis through both direct and indirect mechanisms (Cheng Y. et al., 2020). Accumulating evidence indicates that microbial imbalance disrupts host immune homeostasis and epithelial barrier integrity, while triggering chronic inflammation, genotoxic stress, oxidative damage, and metabolic dysregulation-collectively fostering a microenvironment conducive to malignant transformation (Hanus et al., 2021).
Pathogenic bacteria drive chronic inflammation that fuels precancerous growth (Zhang et al., 2023); Genotoxins damage DNA and foster driver mutations (Lai et al., 2021); Oxidative stress promotes chromosomal aberrations and perturbs key signaling pathways (Shi et al., 2021); Metabolites such as secondary bile acids, acetaldehyde, and TMAO further promote malignant transformation via immune and signaling effects (You et al., 2023).
Additionally, the gut microbiota can influence the metabolic activation or inactivation of exogenous carcinogens and chemotherapeutic agents, further modulating host tumor susceptibility. These mechanistic insights not only highlight the central role of gut microbiota dysbiosis in CRC pathogenesis but also provide a foundation for identifying microbial biomarkers and developing microbiota-targeted interventions.
In the following sections, we delineate four core mechanistic pathways underlying the contribution of gut microbes to colorectal tumorigenesis: (i) chronic inflammation driven by pathogenic bacteria; (ii) DNA damage induced by bacterial genotoxins; (iii) oxidative stress-mediated chromosomal instability; and (iv) the oncogenic influence of microbial metabolites.
2.1 Pathogenic bacteria and chronic inflammation
Chronic inflammation is one of the most prominent pro-tumorigenic factors within the colorectal tumor microenvironment. Various pathogenic gut bacteria initiate and sustain mucosal inflammatory responses through multiple mechanisms, including epithelial adhesion, toxin secretion, and immune activation (Lee et al., 2022; Jergens et al., 2021). Among them, F. nucleatum is the most extensively studied pro-inflammatory bacterium in CRC. Its FadA adhesin binds to E-cadherin on host intestinal epithelial cells, triggering β-catenin signaling and inducing the expression of inflammatory cytokines such as IL-6 and IL-8, thereby establishing a localized inflammatory niche (Li et al., 2022). Beyond β-catenin pathway engagement via FadA–E-cadherin, downstream nuclear β-catenin activity upregulates prototypical targets including MYC and CCND1 (Cyclin D1), which promote proliferation and cell-cycle progression. In parallel, TLR4/MYD88/NF-κB signaling augments pro-inflammatory cytokines and cooperates with Wnt pathway, together reinforcing epithelial proliferation and a tumor-permissive niche (Figure 1A) (Hu et al., 2021).
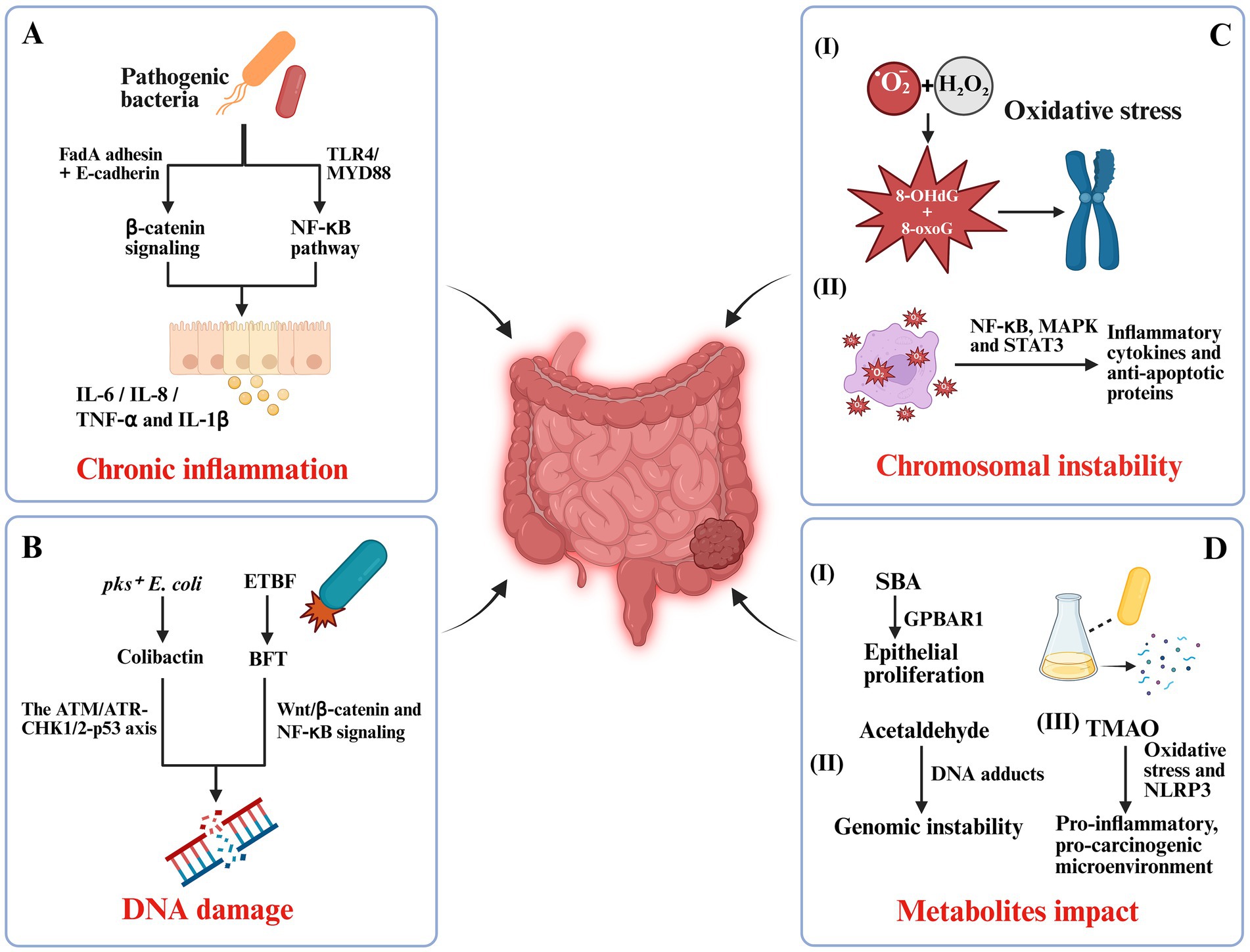
Figure 1. Mechanistic pathways linking gut microbiota dysbiosis to colorectal carcinogenesis. (A) Pathogenic bacteria-induced chronic inflammation: Fusobacterium nucleatum and adherent-invasive Escherichia coli (AIEC) trigger pro-inflammatory cascades via FadA adhesin–E-cadherin binding and TLR4/MYD88/NF-κB signaling, promoting IL-6, IL-8, TNF-α, and IL-1β secretion. (B) Bacterial genotoxins and DNA damage: pks+ E. coli produces colibactin that induces double-strand breaks through the ATM/ATR-CHK1/2-p53 axis, while enterotoxigenic Bacteroides fragilis (ETBF) secretes BFT toxin, activating Wnt/β-catenin and NF-κB pathways. (C) Oxidative stress and chromosomal instability: Reactive oxygen species (ROS) from Enterococcus faecalis cause oxidative DNA lesions (e.g., 8-OHdG, 8-oxoG) and activate NF-κB, MAPK, and STAT3, enhancing inflammatory and anti-apoptotic signaling. (D) Microbial metabolite effects: Secondary bile acids (SBA), acetaldehyde, and trimethylamine N-oxide (TMAO) drive epithelial proliferation, genomic instability, oxidative stress, and a pro-carcinogenic microenvironment.
Certain E. coli strains, including adherent-invasive E. coli (AIEC), are also frequently enriched in patients with inflammatory bowel disease and CRC. These bacteria can invade epithelial cells, evade immune clearance, and continuously stimulate the activation of T cells and dendritic cells, leading to the release of pro-inflammatory mediators such as TNF-α and IL-1β (Figure 1A) (Viladomiu et al., 2021). This persistent, low-grade inflammation compromises epithelial barrier integrity, promotes abnormal epithelial proliferation, and increases the likelihood of mutational accumulation.
Sustained chronic inflammation further promotes tumorigenesis by reshaping the immune microenvironment. It recruits myeloid-derived suppressor cells (MDSCs) and polarizes tumor-associated macrophages (TAMs), thereby facilitating immune evasion (Siddiqui and Glauben, 2022; Li et al., 2020). Moreover, inflammatory cytokines upregulate cyclooxygenase-2 (COX-2) expression and enhance prostaglandin E2 (PGE2) production, which collectively stimulate angiogenesis and extracellular matrix degradation (Finetti et al., 2020). These processes generate a supportive “fertile soil” for tumor invasion and distant metastasis.
2.2 Genotoxins and DNA damage
Certain gut bacterial strains contribute to colorectal carcinogenesis by producing genotoxic compounds that directly compromise genomic integrity, representing a key early event in tumor initiation (Cao et al., 2022). A prime example is E. coli strains harboring the pks pathogenicity island, which encodes colibactin. Colibactin is an alkylating genotoxin that binds the DNA minor groove. It induces double-strand breaks and activates DDR pathways, including the ATM/ATR–CHK1/2–p53 axis (Figure 1B) (Gerstberger et al., 2025). In murine models, colonization with pks+ E. coli significantly increases γ-H2AX foci formation and the incidence of microadenomas in the intestinal epithelium, confirming its carcinogenic potential (Iftekhar et al., 2021).
Enterotoxigenic B. fragilis (ETBF) secretes BFT, a zinc-dependent metalloprotease that cleaves epithelial E-cadherin. This proteolysis disrupts the cadherin–catenin complex, releasing β-catenin from the adherens junction and permitting its nuclear translocation, where it partners with TCF/LEF to drive transcription of canonical Wnt targets (MYC, CCND1/Cyclin D1) (Lee et al., 2022). Thus, the pathway is not a direct ligand-like “activation” of Wnt; rather, E-cadherin cleavage is the proximal event that enables β-catenin–dependent transcription, alongside BFT-associated NF-κB signaling and barrier disruption (Figure 1B) (Curti and Campaner, 2021).
Importantly, these genotoxins often act synergistically with chronic inflammation, creating a mutagenic microenvironment. By promoting oxidative stress, epigenetic alterations, and chromosomal instability, they drive epithelial cells into a “high-variability” state that greatly increases the likelihood of malignant transformation (Wang and Fu, 2023).
2.3 Oxidative stress and chromosomal abnormalities
Oxidative stress, resulting from the excessive accumulation of reactive oxygen species (ROS) and reactive nitrogen species (RNS), is a critical molecular driver in colorectal carcinogenesis. Gut microbiota dysbiosis can markedly alter the redox balance of the intestinal microenvironment, promoting DNA damage and genomic instability (Hamamah et al., 2024). In host–microbe interactions, Enterococcus faecalis (E. faecalis) generates ROS, primarily superoxide anions (O₂−) and hydrogen peroxide (H₂O₂), which induce oxidative DNA damage in the host. This damage is characterized by the formation of 8-hydroxy-2′-deoxyguanosine (8-OHdG), along with oxidized bases (e.g., 8-oxoguanine, 8-oxoG) and strand breaks that challenge DNA repair pathways. Under mismatch repair (MMR) deficiency, the processing of oxidative mismatches—such as 8-oxoG: A mispairs—becomes error-prone and inadequately corrected, thereby increasing mutational burden and promoting microsatellite instability (MSI). Thus, crosstalk between ROS and MMR mechanistically links E. faecalis–driven oxidative stress to genomic instability and may account for the heightened susceptibility of dMMR/MSI-high contexts to microbe-induced mutagenesis (Figure 1C) (Tang et al., 2022; Fang et al., 2024). Such oxidative base modifications and strand breaks, particularly when they occur in tumor suppressor genes (e.g., TP53, APC) or proto-oncogene regions, can lead to point mutations, misrepaired double-strand breaks, and structural chromosomal abnormalities such as translocations, deletions, or amplifications (Kavec et al., 2022). These alterations are detectable even at early adenoma stages, highlighting oxidative stress as a pivotal tumor-initiating factor.
Beyond direct genotoxicity, oxidative stress activates multiple pro-tumorigenic signaling pathways, including NF-κB, MAPK, and STAT3, which upregulate inflammatory cytokines and anti-apoptotic proteins, thereby facilitating the survival of genetically damaged cells (Figure 1C) (Alanazi et al., 2024; Mukherjee et al., 2022). A persistent positive feedback loop between oxidative stress and inflammation forms a “pro-carcinogenic ecosystem,” particularly evident in colitis-associated colorectal cancer (CAC) (Bardelčíková et al., 2023).
Collectively, microbially induced oxidative stress not only drives DNA damage and chromosomal instability but also shapes an inflammatory and anti-apoptotic tumor microenvironment that favors malignant progression (Neganova et al., 2021).
2.4 Microbial metabolites and their impact
Microbial metabolites serve as critical mediators of host–microbe interactions and exert profound effects on intestinal epithelial homeostasis, immune regulation, and tumorigenesis. Among these, secondary bile acids (SBAs), acetaldehyde, and TMAO are strongly implicated in CRC development (Figure 1D) (Zhang et al., 2021).
SBAs, such as deoxycholic acid (DCA) and lithocholic acid (LCA), are generated through the microbial transformation of primary bile acids, primarily by Clostridium species. Elevated levels promote epithelial proliferation, inhibit apoptosis, and enhance adhesion and migration via GPBAR1/TGR5 (Qi Y. et al., 2022). High concentrations of DCA can also trigger endoplasmic reticulum (ER) stress and activate the unfolded protein response (UPR), shifting the balance toward cell survival and facilitating malignant transformation (Oakes, 2020).
Acetaldehyde, a highly reactive and mutagenic metabolite generated by microbial alcohol dehydrogenase during ethanol metabolism, directly forms DNA adducts, induces interstrand crosslinks, and interferes with base excision repair, cumulatively driving mutational burden and genomic instability (Figure 1D) (Oakes, 2020).
TMAO, a gut microbiota-derived metabolite of choline, L-carnitine, and phosphatidylcholine, has emerged as a systemic tumor-promoting factor. It can disrupt cellular energy metabolism, induce oxidative stress, and activate the NLRP3 inflammasome, thereby facilitating a pro-inflammatory, pro-carcinogenic microenvironment (Figure 1D) (Lei et al., 2023).
Additionally, β-glucuronidase (GUS) activity is markedly increased in the fecal samples of CRC patients. This enzyme hydrolyzes glucuronide conjugates of carcinogens excreted in bile, reactivating these compounds within the intestinal lumen and enhancing DNA damage (Hillege et al., 2024). Collectively, these microbial metabolites function not only as mechanistic effectors of tumor initiation and progression but also as potential biomarkers and therapeutic targets for metabolic interventions in CRC.
3 Key microbial species and their CRC-specific mechanisms
Comparative analyses show a marked microbial imbalance in CRC. Pro-carcinogenic species are enriched, while commensal or beneficial bacteria are depleted. Pathogenic taxa such as F. nucleatum, toxigenic E. coli, and enterotoxigenic B. fragilis are significantly enriched in CRC patients, whereas beneficial microbes including Bifidobacterium and Lactobacillus species are markedly reduced (Senthakumaran et al., 2023; Qu et al., 2023). This gut microbiota dysbiosis not only disrupts intestinal homeostasis but also impairs epithelial barrier integrity and weakens immune surveillance, creating a permissive niche for microbial colonization and tumor initiation.
High-resolution analyses of tumor-associated microbiota demonstrate that some pathogens exhibit striking tissue tropism. For example, F. nucleatum can accumulate within adenomas and carcinoma tissues at levels several-fold to hundreds of times higher than in adjacent normal mucosa (Wang and Fang, 2022). Concomitantly, CRC patients typically show reduced microbial diversity, which diminishes ecological resilience and heightens susceptibility to environmental perturbations such as high-fat diets, antibiotic exposure, or chemotherapy (Yang et al., 2020; Kenneth et al., 2025). This dysbiotic state favors the expansion of pathobionts and triggers pro-carcinogenic processes through chronic inflammation, genotoxic insult, and metabolite-driven signaling (Rossi et al., 2020; Finetti et al., 2020).
In the following sections, we highlight six key microbial species or genera with well-characterized contributions to CRC pathogenesis-F. nucleatum, E. coli, B. fragilis, E. faecalis, Streptococcus bovis (S. bovis), and Peptostreptococcus anaerobius (P. anaerobius). We summarize their molecular mechanisms, associated host signaling pathways, and clinical implications, providing a foundation for the development of microbiota-based biomarkers and targeted interventions.
3.1 Fusobacterium nucleatum
F. nucleatum is an anaerobic Gram-negative bacterium that is commonly found in the oral cavity but is consistently enriched in the colorectal tumors of patients with CRC. Its abundance is particularly elevated in adenomas and carcinoma tissues compared with adjacent normal mucosa, suggesting an active role in early tumorigenesis and progression. Mechanistically, F. nucleatum contributes to CRC through three interconnected processes: inflammation activation, signaling pathway modulation, and immune evasion (Wang and Fang, 2022).
First, F. nucleatum promotes a pro-inflammatory tumor microenvironment. Its surface lipopolysaccharide (LPS) interacts with Toll-like receptor 4 (TLR4) on colonic epithelial cells, activating the TLR4/MYD88/NF-κB signaling axis (Luo et al., 2025). This leads to the transcription of pro-inflammatory cytokines and upregulation of oncogenic microRNA-21 (miR-21) (Sun et al., 2021). miR-21 downregulates RASA1, releasing suppression of the RAS-MAPK pathway, thereby enhancing tumor cell proliferation, invasion, and resistance to apoptosis (Bhere et al., 2020).
Second, the bacterial adhesin FadA binds to E-cadherin on epithelial cells, triggering β-catenin nuclear translocation and activation of the Wnt/β-catenin pathway (Jiang et al., 2024). This signaling cascade accelerates cell cycle progression and abnormal epithelial proliferation, key events in the adenoma-to-carcinoma sequence. Additionally, F. nucleatum modulates host lipid metabolism by promoting the production of linoleic acid-derived 12,13-epoxyoctadecenoic acid (12,13-EpOME), which facilitates epithelial-mesenchymal transition (EMT) and enhances metastatic potential (Kong et al., 2021).
Third, F. nucleatum exerts dual effects on the immune microenvironment. While it can activate the STING pathway to enhance dendritic cell antigen presentation, it simultaneously suppresses natural killer (NK) cell cytotoxicity and elevates ROS levels, thereby creating a tumor-permissive niche (Gao et al., 2021). This immunomodulatory balance underlies its capacity to promote tumor progression and immune evasion.
Collectively, F. nucleatum acts as a multifaceted driver of CRC through the induction of chronic inflammation, activation of oncogenic signaling pathways, and remodeling of the immune microenvironment. Its tissue enrichment and mechanistic links to tumorigenesis position it as a promising biomarker for early CRC detection and as a potential target for microbiota-based therapeutic interventions.
3.2 Escherichia coli
E. coli is a facultative anaerobic Gram-negative bacterium that exists as both a commensal and an opportunistic pathogen in the human gut. Certain pathogenic strains, particularly those harboring the pks genomic island, have been strongly implicated in colorectal carcinogenesis. The pks island encodes colibactin, a genotoxic secondary metabolite capable of alkylating DNA and inducing interstrand crosslinks and double-strand breaks (Pleguezuelos-Manzano et al., 2020; Gerstberger et al., 2025). These lesions activate canonical DDR pathways, including the ATM/ATR-CHK1/2-p53 axis, and lead to mutational accumulation and genomic instability if repair is incomplete (Pleguezuelos-Manzano et al., 2020; Huber et al., 2024). Animal studies have demonstrated that colonization with pks+ E. coli elevates γ-H2AX levels and promotes early adenoma formation, directly linking colibactin activity to tumor initiation.
Beyond its direct genotoxicity, E. coli contributes to CRC through inflammatory and signaling mechanisms. The bacterium can activate Wnt/β-catenin and STAT3 pathways, driving epithelial proliferation and anti-apoptotic signaling (Trejo-Solís et al., 2021; Zhou et al., 2021). Other virulence factors, such as Shiga toxins and hemolysins, exacerbate epithelial barrier injury, enhance ROS production, and amplify chronic inflammation (Warr et al., 2020). Interaction with host immune cells further accelerates tumor-promoting inflammation: E. coli engages the TLR4/NF-κB signaling axis to induce pro-inflammatory cytokines, including IL-6, IL-8, and TNF-α, which sustain tumor-promoting microenvironments and facilitate angiogenesis (Lan et al., 2022; Zhuang et al., 2020).
High abundance of these strains has been associated with worse overall survival and higher recurrence rates, highlighting their potential as prognostic biomarkers. Given their multifaceted roles in DNA damage, signaling activation, and immune modulation, pathogenic E. coli strains represent not only key drivers of CRC but also compelling targets for microbiome-focused diagnostic and therapeutic strategies.
3.3 Bacteroides fragilis
B. fragilis is a common anaerobic bacterium in the human gut, but its pathogenic potential is largely associated with toxigenic strains, collectively known as ETBF. ETBF secretes BFT, a zinc-dependent metalloprotease that cleaves E-cadherin at the epithelial junctions, compromising barrier integrity and facilitating inflammatory infiltration (Lukiw, 2020). This early disruption of epithelial homeostasis is a critical initiating event in microbe-driven tumorigenesis.
Mechanistically, BFT drives CRC progression through both inflammatory and oncogenic signaling pathways. First, BFT induces COX-2 expression and promotes PGE2 production, fostering a state of chronic inflammation that supports epithelial proliferation and survival (Wei et al., 2022). Second, BFT activates the STAT3 signaling cascade, enhancing anti-apoptotic gene expression and promoting the maintenance of cancer stem-like properties (Yang J. et al., 2024). ETBF colonization also reshapes local immune responses: it suppresses IL-2 expression in regulatory T cells, promotes the differentiation of Th17 cells, and elevates IL-17 levels, which in turn induces IL-6 production (Jo et al., 2023). This creates a self-sustaining IL-6/STAT3 positive feedback loop that amplifies tumor-promoting inflammation (Pandey et al., 2024).
Evidence from animal models further supports the carcinogenic potential of ETBF. Colonization with ETBF accelerates colonic epithelial proliferation, induces sustained mucosal inflammation, and promotes adenoma formation (Yang J. et al., 2024). Given its well-defined role at the intersection of inflammation and oncogenesis, ETBF is increasingly recognized as both a potential microbial biomarker for CRC risk and a candidate target for microbiome-focused preventive or therapeutic interventions (Zamani et al., 2020).
3.4 Enterococcus faecalis
E. faecalis is a Gram-positive, facultative anaerobe that commonly inhabits the human gut and oral cavity (Madani et al., 2024). Although long regarded as a commensal—and in some contexts explored as a probiotic candidate—accumulating evidence indicates a strain-dependent, dual role in CRC (Elnar and Kim, 2025; Daca and Jarzembowski, 2024). Beneficial or food-derived strains can support barrier function and immune homeostasis, whereas pathogenic or clinical isolates harbor virulence and antimicrobial-resistance determinants and are capable of driving pro-carcinogenic biology (Zhang L. et al., 2024).
Mechanistically, E. faecalis contributes to tumor promotion through intertwined inflammatory, oxidative, and genotoxic processes. Certain isolates generate high levels of reactive oxygen species (ROS; superoxide and hydrogen peroxide), producing oxidized DNA bases (e.g., 8-oxoG/8-OHdG) and strand breaks that challenge canonical repair pathways (Kouhzad et al., 2025). This oxidative stress operates alongside mucosal inflammation to create a microenvironment favorable to malignant transformation and progression (Catalano et al., 2025). Importantly, MMR status modulates the genomic consequences of E. faecalis–derived ROS: in MMR-deficient settings, error-prone processing of oxidative mismatches (such as 8-oxoG: A mispairs) is inadequately corrected, amplifying mutational burden and microsatellite instability, and thereby linking microbial ROS directly to genomic instability in susceptible hosts (Sun et al., 2025).
Beyond oxidative injury, E. faecalis can influence epithelial signaling and cell-cycle control. ROS-driven kinase activation and inflammatory transcriptional programs converge on pathways that enhance proliferation and survival, and may cooperate with other microbe-host axes present in CRC lesions (Hong et al., 2024). The net effect is a genotoxic, pro-inflammatory niche that lowers the threshold for oncogenic evolution and can attenuate responses to cytotoxic therapy (Catalano et al., 2025).
These features have practical implications. First, because oncogenic potential varies by strain, future translational work should incorporate genomic and virulence profiling (including toxin gene content and antimicrobial-resistance genes) when E. faecalis is considered in diagnostics or as a therapeutic adjunct. Second, in mechanistic and clinical studies, MMR context should be specified a priori, as dMMR/MSI-high tumors may be particularly vulnerable to ROS-mediated mutagenesis (Sun et al., 2025). Third, where E. faecalis is implicated in gut microbiota dysbiosis, narrow-spectrum or sequencing-guided antimicrobial strategies, coupled with restoration of barrier function and short-chain fatty acid production, may mitigate collateral damage while addressing the offending strains. Finally, targeted qPCR panels and metagenomic readouts that resolve E. faecalis at the strain level can support peri-operative monitoring and help relate microbial dynamics to treatment tolerance and outcome.
In sum, E. faecalis exemplifies how a common commensal can become a context- and strain-specific contributor to CRC pathogenesis. Recognizing its dual identity—and explicitly accounting for strain heterogeneity and host MMR status—will be essential for accurate risk attribution and for the rational design of microbiome-informed prevention and therapeutic strategies.
3.5 Streptococcus bovis
Streptococcus bovis/Streptococcus equinus complex (SBSEC) remains strongly associated with colorectal neoplasia, but recent evidence favors risk estimates from meta-analyses over broad prevalence ranges. A 2023 systematic review reported that patients with SBSEC bacteremia were 3.73-fold more likely to harbor underlying CRC than those without bacteremia (RR 3.73, 95% CI 2.79–5.01) (Ouranos et al., 2023). In pooled case–control data, CRC cases were ~2.27-fold more likely to demonstrate SBSEC colonization or anti–S. gallolyticus IgG responses than controls. The association appears genospecies-dependent, being stronger for S. gallolyticus subsp. gallolyticus than for other SBSEC members. These data support the clinical recommendation that patients with SBSEC bacteremia or infective endocarditis undergo routine colonoscopic evaluation to detect CRC and guide early management (Ouranos et al., 2023).
The tumor-promoting mechanisms of S. bovis are multifaceted, involving both inflammatory and proliferative pathways (Karpiński et al., 2022). The bacterium can stimulate the NF-κB pathway, inducing the secretion of pro-inflammatory cytokines such as IL-6, IL-8, and TNF-α, thereby fostering a chronic inflammatory microenvironment that facilitates tumor initiation and progression (Neumann et al., 2021). In parallel, S. bovis can engage TLR2 and TLR4 signaling, driving the recruitment of CD11b+ TLR4+ monocytes to the tumor site, which contributes to local immune suppression and facilitates immune evasion by cancer cells (Huang et al., 2020).
Beyond immune modulation, S. bovis can directly influence epithelial cell proliferation by activates the Ras–Raf–MEK–ERK and JNK/p38 MAPK pathways, accelerating cell-cycle progression and enhancing oncogenic signaling (Xiao et al., 2020). This dual action-chronic inflammation coupled with aberrant epithelial proliferation-creates a pro-tumorigenic niche.
Notably, S. bovis has also been detected in the bloodstream of CRC patients, suggesting a potential role in tumor-associated bacteremia. Enhanced bacterial adhesion to endothelial surfaces may promote vascular permeability, support tumor-associated angiogenesis, and facilitate distant metastasis (Laupland et al., 2023). Owing to its early enrichment during CRC development and its detectability via blood cultures or serological testing, S. bovis has been proposed as a candidate biomarker for high-risk CRC screening (Thind et al., 2021).
Translationally, these data support a genospecies-aware, oncology-aligned approach. SBSEC bacteremia—or infective endocarditis—should trigger colonoscopic evaluation to detect occult CRC and enable early management, consistent with recent meta-analytic risk estimates (Ouranos et al., 2023). Diagnostic workups should aim for lineage-level resolution (e.g., S. gallolyticus subsp. gallolyticus) and, where feasible, integrate virulence profiling into future qPCR panels for risk stratification. Within oncology pathways, SBSEC positivity should prompt assessment of metastatic risk and source control (mucosal lesions, dental foci, biliary tract) alongside guideline-concordant antimicrobials, reflecting modern evidence on genospecies-specific risk.
Looking ahead, embedding SBSEC diagnostics and management within oncology workflows enables earlier case finding, molecularly informed risk tiers, and longitudinal surveillance that links infection control to CRC prevention, staging, and treatment planning. Priorities for future research include high-resolution characterization of virulence factors, host–receptor interactions, and strain-specific pathogenicity, together with the co-development of deployable qPCR-based SBSEC panels that can be integrated into perioperative and adjuvant care pathways.
3.6 Peptostreptococcus anaerobius
P. anaerobius is a Gram-positive anaerobic coccus that commonly resides in the human oral cavity and gastrointestinal tract. In recent years, this bacterium has gained attention for its potential role in CRC progression. Multiple independent metagenomic and tissue-based studies have demonstrated its significant enrichment in CRC patients, particularly within tumor-adjacent mucosa and intratumoral regions, suggesting a spatially specific association with tumorigenesis (Conde-Pérez et al., 2024).
Mechanistically, P. anaerobius promotes CRC through several interrelated pathways. It activates TLR2 and TLR4 mediated signaling, triggering the NF-κB pathway and upregulating pro-inflammatory cytokines such as IL-1β, TNF-α, and IL-6, thereby fostering a chronic inflammatory microenvironment conducive to malignant transformation (Shen et al., 2024). Concurrently, the bacterium interacts with membrane cholesterol to form cholesterol-rich microdomains that facilitate the activation of Ras–Raf–MEK–ERK and JNK/p38 MAPK signaling cascades (Bahar et al., 2023). These pathways enhance epithelial cell proliferation, migration, and survival, supporting tumor progression.
In addition to its pro-inflammatory and pro-proliferative effects, P. anaerobius can exacerbate genomic instability by increasing ROS production and downregulating DNA repair enzymes, resulting in oxidative DNA damage and accelerated cell-cycle progression via upregulation of Cyclin D1 (Gupta et al., 2023). This genotoxic and proliferative environment establishes a fertile ground for carcinogenesis. Furthermore, emerging evidence suggests that P. anaerobius may impair antitumor immunity by reducing CD8+ T-cell infiltration and inducing PD-L1 expression on tumor cells, thereby facilitating immune evasion (Wang et al., 2020).
These mechanisms have direct therapeutic relevance. Associations between P. anaerobius abundance and attenuated responses to immune checkpoint blockade or cytotoxic chemotherapy point to a microbiome-linked axis of resistance mediated by inflammatory remodeling of the tumor microenvironment and the expansion of myeloid-derived suppressor cells. Where resources allow, baseline microbial profiling should be incorporated into treatment planning—particularly for immunotherapy or irinotecan-based regimens—to anticipate resistance trajectories and to guide supportive measures.
The organism’s dependency on cholesterol-enriched membrane domains also highlights tractable points of intervention. Approaches under investigation include membrane-lipid remodeling and small-molecule blockade of microdomain formation to disrupt adhesion, signaling, and biofilm stability, with the aim of restoring sensitivity to systemic therapy. In clinical practice, when a high P. anaerobius signal is detected by tissue or fecal qPCR/metagenomics, it is reasonable to prioritize regimens that pair anticancer therapy with anti-inflammatory measures and targeted antimicrobials. Sequencing-guided, narrow-spectrum strategies may limit gut microbiota dysbiosis; longitudinal qPCR can then be used to track organismal clearance and relate microbial dynamics to immune activation and clinical response.
From a prognostic perspective, P. anaerobius clusters within chemoresistance-associated microbiome profiles, suggesting value as a predictive biomarker for treatment response and outcome (Xing et al., 2025). Future work should define strain-level virulence factors and host–pathogen interactions, test mechanism-anchored adjuncts in biomarker-selected cohorts, and embed microbial–immune monitoring into peri-operative and adjuvant pathways to establish causal links between microbial modulation and restoration of antitumor immunity. This integrated view positions P. anaerobius alongside F. nucleatum and SBSEC as a microbiome target with clear diagnostic and therapeutic implications.
4 Gut microbiome detection technologies
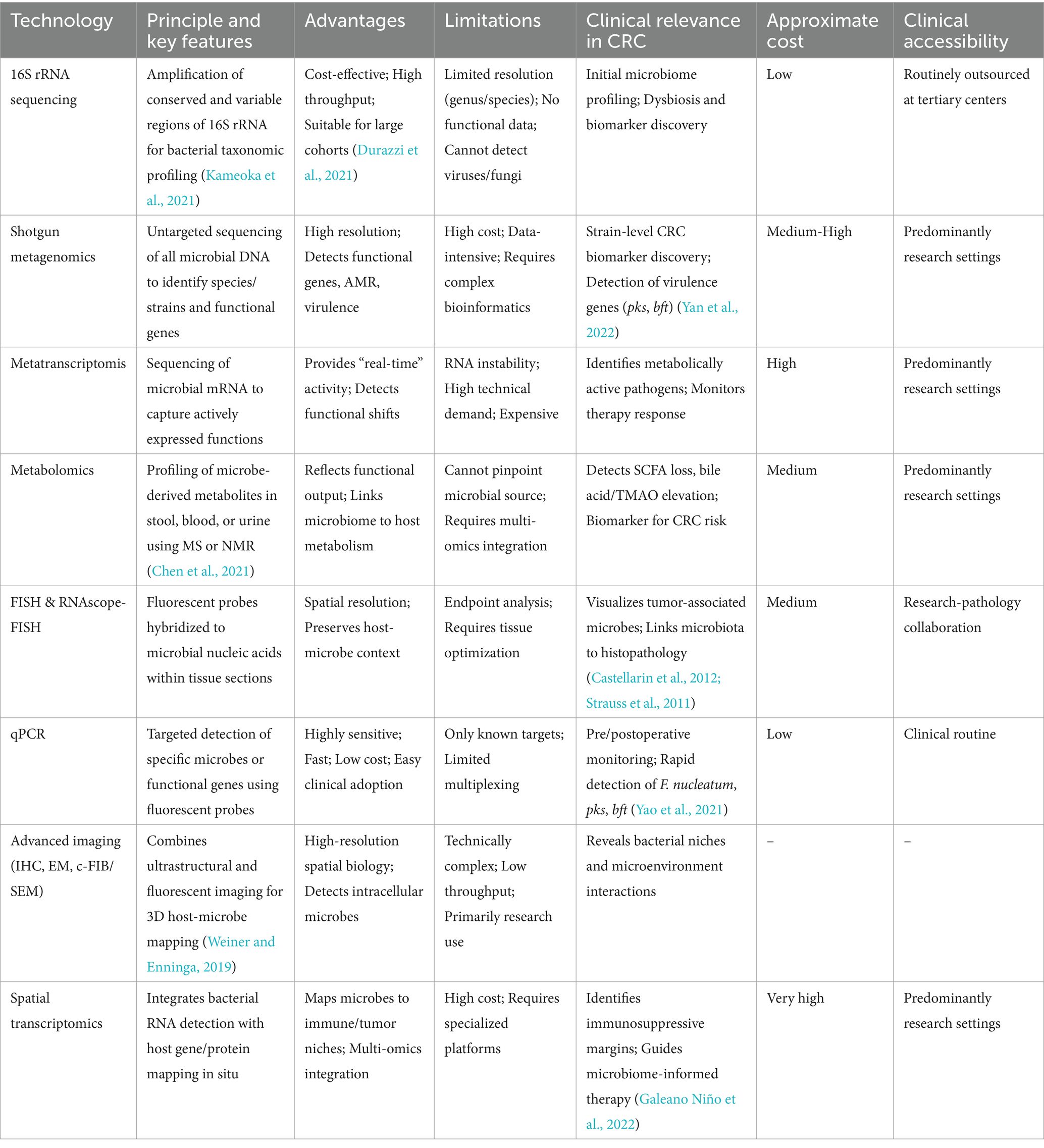
Advances in molecular biology and multi-omics have dramatically expanded our ability to characterize the gut microbiome, enabling both taxonomic and functional profiling of microbial communities (Deissova et al., 2023). These methods have not only deepened our understanding of microbial contributions to CRC but also opened avenues for early detection, therapeutic monitoring, and precision interventions. Contemporary microbiome detection strategies can be broadly categorized into nucleic acid-based sequencing, functional and metabolic analyses, and imaging-based spatial profiling (Table 1).
4.1 16S rRNA and metagenomic sequencing
16S rRNA gene sequencing remains the most widely used approach for initial gut microbiota profiling. By amplifying conserved and variable regions of the 16S rRNA gene, this method provides insights into microbial community structure, diversity, and composition (Kameoka et al., 2021). It is cost-effective and suitable for large-scale population studies or preliminary CRC microbiome screening (Durazzi et al., 2021). Several studies have demonstrated enrichment of F. nucleatum and other pathobionts, alongside depletion of Bifidobacterium and Lactobacillus, in CRC patients (Yuan et al., 2022; Qi Z. et al., 2022). However, 16S sequencing is limited in taxonomic resolution (typically to genus or species level), cannot detect viruses or fungi, and does not provide direct functional information (Matchado et al., 2023).
Shotgun metagenomic sequencing overcomes these limitations by performing untargeted, whole-genome sequencing of all microbial DNA in a sample. This approach enables species and strain-level identification and captures functional gene content, including virulence factors, antibiotic resistance genes, and key metabolic pathways (Durazzi et al., 2021; Xie et al., 2023). In CRC research, metagenomics has been instrumental in linking specific microbial genes-such as the pks island encoding colibactin and bft encoding B. fragilis toxin-to tumorigenesis (Yan et al., 2022). Despite its strengths, metagenomics is data-intensive, costly, and requires rigorous sample processing and bioinformatics pipelines.
4.2 Metatranscriptomics and metabolomics
Beyond static DNA-based analyses, metatranscriptomics captures the active transcriptional state of the microbiome, providing a real-time snapshot of microbial activity. This approach reveals which genes are actively expressed in specific clinical contexts, such as chemotherapy exposure or post-surgical recovery, and can identify functionally active pathogens or beneficial commensals (Uehara et al., 2022). For instance, increased GUS expression during irinotecan treatment has been linked to drug toxicity, while Bifidobacterium longum activity correlates with anti-inflammatory effects (Chung et al., 2020).
Metabolomics, using mass spectrometry or nuclear magnetic resonance, interrogates the small-molecule metabolites produced by the gut microbiome (Chen et al., 2021). This provides critical insights into host-microbe metabolic crosstalk, including alterations in short-chain fatty acids (SCFAs), secondary bile acids, and TMAO associated with CRC progression (Coker et al., 2022). Integrating metagenomics and metabolomics enables the construction of microbiome-metabolite-disease networks, offering a robust framework for biomarker discovery and therapeutic monitoring (Gao et al., 2022).
4.3 Quantitative PCR
Quantitative PCR (qPCR) serves as a targeted, rapid, and highly sensitive technique for detecting known microbial species or virulence genes. In CRC-related applications, qPCR is commonly employed to detect F. nucleatum, toxigenic E. coli, and B. fragilis, as well as pks and bft genes (Yao et al., 2021). This method is well-suited for dynamic monitoring using fecal or tissue samples, supporting preoperative screening, postoperative surveillance, and evaluation of microbiota-targeted interventions. Although qPCR is limited to known targets, its low cost, high sensitivity, and rapid turnaround make it a valuable complement to high-throughput sequencing, especially in clinical workflows (Senthakumaran et al., 2024).
Despite its utility, qPCR is susceptible to false-positive results arising from (i) sample contamination or carryover amplicons; (ii) non-specific amplification/primer–dimer artifacts; (iii) homology-driven cross-reactivity with related taxa; (iv) detection of DNA from non-viable cells or extracellular DNA (especially in low-biomass matrices); and (v) thresholding bias (liberal Ct cutoffs) and multiple-target testing without correction. To reduce these risks in CRC workflows, we recommend strict pre-analytical controls tailored for low-biomass work (e.g., negative extraction/NTCs, unidirectional workflow) together with transparent contamination reporting, as emphasized in recent guidance for low-biomass microbiome studies (Fierer et al., 2025). Carryover prevention with uracil-N-glycosylase systems remains a best practice to limit amplicon contamination between runs (Mizumoto-Teramura et al., 2024). Assay design should rely on validated primer–probe sets with in-silico specificity checks and empirical verification (melt curves/amplicon sequencing); recent tools and updates facilitate rigorous, scalable in-silico screening (Collatz et al., 2025). Standard curves with explicit limits of detection/quantification, appropriate multiple-testing control, and conservative Ct thresholds are mandated by the updated MIQE 2.0 recommendations (Bustin et al., 2025). Finally, interpretation should be context-aware: where viability is uncertain, note that PMA/EMA-qPCR has important limitations and may only qualitatively distinguish live/dead under constrained conditions, warranting orthogonal confirmation (e.g., duplex targets or sequencing of representative positives) and, when possible, integration with metagenomic/spatial evidence and clinical phenotype (Kaur et al., 2024).
4.4 Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) is a spatially resolved detection method that uses fluorescently labeled oligonucleotide probes to hybridize with microbial RNA or DNA within tissue samples. FISH allows direct visualization of microbial localization in CRC tissues, preserving spatial context and enabling co-staining with host markers (Yincharoen et al., 2025). Its integration with confocal microscopy or 3D imaging provides high-resolution insights into bacteria-host interactions, especially for low-biomass or intracellular organisms that may be underrepresented in sequencing data.
Recent adaptations, such as RNAscope-FISH, offer enhanced signal amplification and single-cell resolution, enabling the visualization of metabolically active tumor-associated bacteria (Castellarin et al., 2012; Strauss et al., 2011). However, this approach is largely limited to endpoint analyses and requires careful optimization of tissue processing and probe design.
4.5 Advanced imaging techniques
Modern imaging-based microbial profiling has further expanded the ability to study host–microbe interactions at high spatial resolution. Techniques such as immunohistochemistry (IHC), high-resolution electron microscopy (EM), and correlative focused ion beam/scanning electron microscopy (c-FIB/SEM) combine ultrastructural imaging with fluorescent microbial labeling, revealing 3D bacterial niches within tumor tissue (Weiner and Enninga, 2019).
Use of fluorochrome-conjugated, bacteria-specific antibodies and bacterial metabolic labeling, such as fluorescent D-alanine incorporation into bacterial cell walls, allows selective imaging of live, metabolically active bacteria in fresh tumor samples (Casasanta et al., 2020; Puschhof et al., 2021; Xue et al., 2023). These methods are particularly valuable for linking microbial spatial distribution to functional interactions, though they remain largely limited to research settings due to technical complexity and endpoint constraints.
4.6 Spatial transcriptomics and multi-omics profiling
Spatial transcriptomics and multi-omics integration now provide unprecedented resolution in mapping tumor-associated microbial niches. RNAscope-FISH allows single-cell bacterial RNA localization, while digital spatial profiling (GeoMX) simultaneously quantifies dozens of immune-related proteins, correlating microbial presence with local immune contexture.
The 10x Visium spatial transcriptomics platform enables host gene expression mapping in situ, linking microbial colonization to tumor margin characteristics, including hypovascular and immunosuppressive microenvironments (Galeano Niño et al., 2022). These integrated spatial approaches bridge microbial detection and functional tumor biology, revealing how localized microbial communities influence immune evasion, tumor progression, and therapy response.
5 Gut microbiota-based interventions in CRC therapy
With growing recognition of the gut microbiome as a pivotal modulator of host health, microbiota-targeted interventions have emerged as promising adjunct strategies in CRC management. Mounting evidence indicates that the gut microbial community not only contributes to tumor initiation and progression but also modulates responses to chemotherapy, immunotherapy, and other treatment modalities through its impact on drug metabolism, host immunity, and inflammatory signaling (Sánchez-Alcoholado et al., 2020; Kim and Lee, 2022; Kalasabail et al., 2021). Altered microbial profiles in CRC, characterized by enrichment of pro-carcinogenic species such as F. nucleatum and depletion of beneficial taxa like Bifidobacterium, have been associated with poor treatment responses and increased therapy-related toxicity (Fong et al., 2020). Conversely, specific beneficial microbes, including members of the Bifidobacterium genus, have been shown to enhance the efficacy of oxaliplatin and other chemotherapeutics, likely through immune modulation and reduction of intestinal inflammation (Chawrylak et al., 2024). In contrast, broad-spectrum antibiotic-induced dysbiosis can attenuate therapeutic efficacy and exacerbate adverse effects (Van Dingenen et al., 2023).
These observations underscore the concept that the “microbial status” of the gut is a determinant of therapeutic outcomes, providing a rationale for microbiota-targeted interventions. Currently, three major strategies have been investigated in preclinical and clinical settings: (i) probiotics and prebiotics, which support the growth and function of beneficial microbes; (ii) fecal microbiota transplantation (FMT), which reconstructs a balanced microbial ecosystem; and (iii) bacteriophage therapy, which selectively eliminates pro-carcinogenic or antibiotic-resistant bacteria (Masheghati et al., 2024; Zuo et al., 2024; Song et al., 2020).
5.1 Microbiota modulation of therapeutic responses
The gut microbiome influences CRC therapy through multiple mechanisms, including regulation of drug metabolism, modulation of tumor immunity, and alteration of the inflammatory tumor microenvironment. For example, irinotecan (CPT-11), a widely used chemotherapeutic, requires conversion to its active metabolite SN-38, which is subsequently inactivated by glucuronidation in the liver. Gut microbial GUS can reactivate SN-38 in the colon, enhancing its local antitumor effect but simultaneously causing dose-limiting gastrointestinal toxicity (Bell et al., 2021; Yang Q. et al., 2023). Similarly, ROS generation induced by microbial modulation of NADPH oxidase activity is essential for the cytotoxic efficacy of platinum-based chemotherapies such as oxaliplatin (Figure 2A) (Cheng W. et al., 2020).
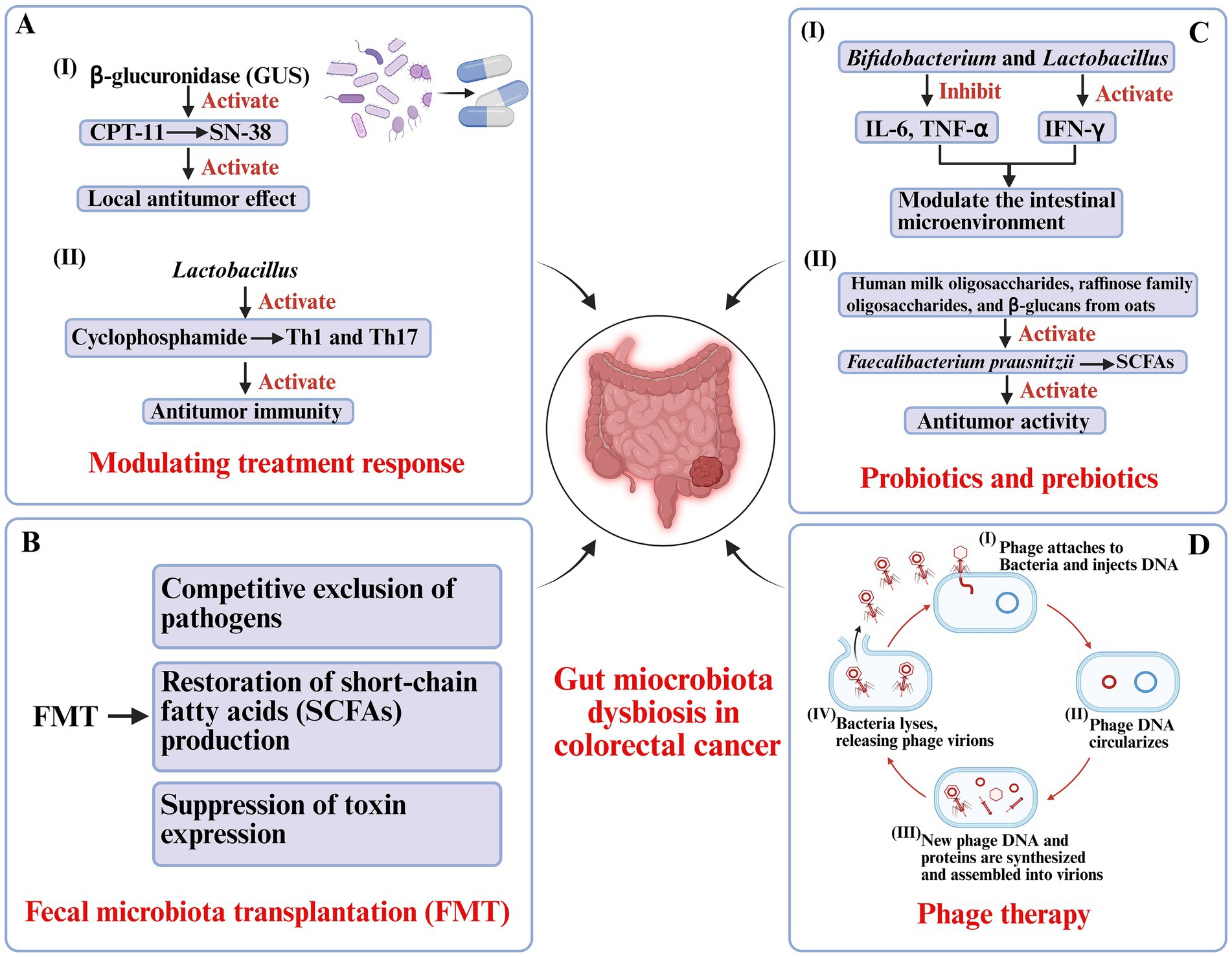
Figure 2. Microbiota-based therapeutic strategies in colorectal cancer. (A) Modulation of treatment responses: Gut microbial β-glucuronidase (GUS) reactivates irinotecan metabolite SN-38, enhancing local antitumor effects but contributing to toxicity. Lactobacillus augments cyclophosphamide-induced Th1/Th17 responses and antitumor immunity. (B) Fecal microbiota transplantation (FMT): Restores gut homeostasis by excluding pathogens, reestablishing short-chain fatty acid (SCFA) production, and suppressing toxin expression. (C) Probiotics and prebiotics: Bifidobacterium and Lactobacillus modulate cytokine expression, strengthen intestinal barrier function, and promote SCFA-producing bacteria (Faecalibacterium prausnitzii), supporting antitumor immunity. (D) Phage therapy: Bacteriophages selectively lyse pathogenic bacteria through infection cycles, reducing tumor-promoting microbes while sparing commensals.
Microbiota-driven immune modulation also plays a crucial role. Certain commensals, such as Lactobacillus species, facilitate cyclophosphamide-induced Th1 and Th17 immune responses, enhancing antitumor immunity. Barnesiella intestinihominis has been shown to promote interferon-γ (IFN-γ) release and reduce regulatory T cell activity, creating a tumor-suppressive immune milieu (Aghamajidi and Vareki, 2022; Rad et al., 2024). Conversely, gut microbiota dysbiosis may attenuate therapeutic sensitivity and even contribute to treatment-related complications (Mohseni et al., 2023). These findings suggest that restoring or optimizing the gut microbial ecosystem could serve as a strategy to improve both the efficacy and safety of CRC therapy (Figure 2A).
5.2 Fecal microbiota transplantation
FMT represents a more comprehensive approach to restoring gut microbial homeostasis by transferring processed stool from healthy donors to recipients. This method allows for ecosystem-wide reconstruction of microbial networks, potentially reestablishing metabolic and immune functions disrupted in CRC (Andary et al., 2024). FMT has already achieved high cure rates in recurrent Clostridioides difficile infection, with mechanisms including competitive exclusion of pathogens, restoration of SCFA production, and suppression of toxin expression (Figure 2B) (Yadegar et al., 2024).
In oncology, preclinical studies have shown that antibiotic-induced microbiome disruption diminishes the efficacy of chemotherapeutic agents like oxaliplatin and cyclophosphamide, while FMT can reverse this effect and restore antitumor immunity (Yang Y. et al., 2024). Early clinical studies suggest that FMT may improve postoperative gut function, enhance epithelial barrier integrity, and modulate immune signaling pathways such as IL-10 and IFN-γ, potentially contributing to a less inflammatory tumor microenvironment (Xu et al., 2022).
For oncology applications, patient safety remains paramount. Key risks include pathogen transmission (e.g., multidrug-resistant organisms [MDROs]), bacteremia/sepsis, transmission of opportunistic or emerging agents, and horizontal gene transfer of antimicrobial-resistance genes. Additional concerns involve long-term engraftment stability, metabolic effects (e.g., unintended weight/metabolic shifts), and exacerbation of inflammation in vulnerable hosts (immunocompromised, mucosal injury) (Peery et al., 2024). Best practices therefore include (i) rigorous donor screening (travel/behavioral risks, comprehensive serology and stool pathogen panels with MDRO screening); (ii) standardized processing (closed systems, defined storage, traceability, batch QC/endotoxin testing); (iii) clear contraindications and informed consent; (iv) post-procedure pharmacovigilance with predefined adverse event (AE)/serious adverse event (SAE) reporting windows; and (v) registry-based long-term follow-up. For interventional trials, we recommend protocolized lot release criteria, recipient risk stratification, and co-primary safety endpoints (e.g., serious infection rate, MDRO colonization), alongside efficacy readouts.
5.3 Probiotics and prebiotics
Probiotics-live microorganisms that confer health benefits to the host-have demonstrated promising roles in CRC prevention and adjunct therapy. Species such as Bifidobacterium and Lactobacillus modulate the intestinal microenvironment by enhancing barrier integrity, downregulating pro-inflammatory cytokines (e.g., IL-6, TNF-α), and upregulating antitumor cytokines (e.g., IFN-γ) (Li et al., 2024). Preclinical studies using Bifidobacterium longum have shown delayed tumor growth, reduced inflammation, and restoration of epithelial barrier function in murine CRC models (Shang et al., 2024). Clinically, supplementation with Lactobacillus rhamnosus GG (LGG) for four weeks postoperatively reduced infection rates, decreased intestinal inflammation, and improved gut microbial diversity in CRC patients (Rafter et al., 2007). Mechanistically, these effects involve enhanced expression of tight junction proteins, promotion of epithelial repair, inhibition of pathogen adhesion, and stimulation of antimicrobial peptide secretion (Figure 2C) (Si et al., 2021).
Prebiotics-non-digestible substrates that selectively promote beneficial microbial growth-serve as “metabolic enhancers” for probiotics (Park et al., 2020). Compounds such as human milk oligosaccharides, raffinose family oligosaccharides, and β-glucans from oats have been shown to increase populations of SCFA-producing bacteria like Faecalibacterium prausnitzii. The SCFAs, particularly butyrate, maintain mucosal homeostasis, regulate immune responses, and suppress inflammation-driven tumorigenesis (Kok et al., 2020). Epidemiological data further support that diets rich in prebiotic fibers (e.g., whole grains, fruits, and vegetables) are associated with a reduced risk of CRC (Turati et al., 2022). Emerging “synbiotic” strategies that combine probiotics and prebiotics have demonstrated synergistic effects, enhancing microbial resilience and antitumor activity (Figure 2C) (Oh et al., 2020).
5.4 Bacteriophage therapy
Bacteriophages are viruses that specifically infect bacteria and can selectively target pro-carcinogenic or multidrug-resistant species in the gut (Danis-Wlodarczyk et al., 2021). Lytic phages bind bacterial surface receptors, replicate within the host, and induce bacterial lysis, thereby reducing pathogen burden and associated inflammatory signaling (Shahin et al., 2021). Compared with broad-spectrum antibiotics, phages offer precision targeting with minimal disruption to commensal microbes.
In CRC–relevant gut microbiota dysbiosis, phages can be engineered or selected to target defined taxa while preserving commensals. For example, phages directed against F. nucleatum or E. coli harboring the pks island (pks+ E. coli) may lower inflammatory signaling, curtail genotoxic potential, and mitigate therapy-interfering microbial functions. Such approaches can be integrated with chemotherapy or immunotherapy in biomarker-defined contexts to enhance therapeutic efficacy.
Successful translation must also account for phage–host coevolution and polymicrobial ecology. Bacteria can rapidly evolve receptor modifications, deploy restriction–modification or abortive-infection systems, and activate CRISPR–Cas defenses, which can shorten phage durability. Recent work underscores these arms-race dynamics and multi-defense synergies in natural and engineered settings (Chen et al., 2024). Countermeasures include rational phage cocktails that diversify receptor usage, adjuvants such as depolymerases and biofilm disruptors, and adaptive reformulation guided by longitudinal microbiome surveillance; contemporary reviews also highlight CRC-relevant opportunities and constraints for phage strategies (Mayorga-Ramos et al., 2024). In the polymicrobial, biofilm-rich microenvironments typical of CRC lesions, community interactions may impede phage adsorption or metabolically shield targets; phage-enabled and nanocomposite approaches are being explored to improve biofilm penetration (Mayorga-Ramos et al., 2024). Practical implications include (i) testing efficacy in community/biofilm models, (ii) optimizing dosing routes and pharmacokinetics for mucosal delivery (e.g., encapsulation, mucoadhesive matrices), (iii) monitoring neutralizing antibodies and mucin binding, and (iv) implementing rigorous quality control for manufacturing (potency, purity/endotoxin, stability); emerging clinical and pharmaceutical data emphasize antibody-mediated neutralization and formulation/stability requirements (Sawa et al., 2024). Thoughtful antibiotic–phage sequencing (to harness synergy and avoid antagonism) and real-time resistance surveillance should be embedded in trial designs to preserve activity and ecological balance; recent overviews of phage therapy for multidrug-resistant (MDR) infections and gastrointestinal (GI) contexts reinforce these design principles (Kim M. K. et al., 2025).
Although preclinical studies have shown encouraging antitumor potential, clinical translation remains constrained by host immune clearance, the emergence of bacterial resistance, and manufacturing standardization (Khalid et al., 2021). A deeper understanding of phage–microbiome–host interactions, together with integration of phage therapy into existing microbiota-based strategies, may open new therapeutic avenues for CRC (Figure 2D).
6 Translational landscape and challenges
Diagnostic tools: Multi-omics has matured from exploratory discovery to clinically actionable workflows. 16S rRNA sequencing and shotgun metagenomics are best positioned for initial screening and feature extraction, enabling strain-level detection of carcinogenic determinants (e.g., pks, bft) and cross-cohort reproducibility. Targeted qPCR panels provide rapid, low-cost surveillance for perioperative and longitudinal follow-up—well suited to monitoring F. nucleatum, pks+ E. coli, and enterotoxigenic Bacteroides fragilis in feces or tissue. For difficult or refractory cases, FISH and spatially resolved profiling (e.g., RNAscope, digital spatial proteomics) localize microbes within tumor–immune niches and link presence to pathway activity. Practically, we recommend high-throughput sequencing for discovery and risk stratification, qPCR for routine monitoring, and tissue imaging when spatial context will change management (Table 1).
Interventional measures: Probiotics and prebiotics can reinforce barrier integrity, temper mucosal inflammation, and restore SCFA production; they are suitable as adjuncts around surgery or during chemotherapy to reduce gastrointestinal toxicity. FMT shows signals for postoperative functional recovery and immune modulation, but translation hinges on stringent donor screening, standardized processing, and pharmacovigilance for long-term safety. Bacteriophage (phage) therapy offers precision removal of pro-carcinogenic taxa (e.g., F. nucleatum or pks+ E. coli) with minimal off-target disruption, yet faces hurdles including host immune clearance, resistance, and scalable manufacturing. A pragmatic clinical algorithm is to prioritize (i) anti-inflammatory and antimicrobial adjuncts when F. nucleatum burden is high; (ii) β-glucuronidase (GUS) inhibition and toxicity management when pks/colibactin signatures are present; and (iii) consider phage or targeted antimicrobial strategies in biomarker-defined, refractory microbiome states.
Challenges and proposed solutions: (1) Causality and trial design—move beyond association by embedding microbial mechanisms into eligibility and randomization: enroll by “microbe–pathway–therapy” matches (e.g., F. nucleatum-high → antibiotic/phage + chemotherapy; pks+ → irinotecan-GUS mitigation bundles) and power trials for DFS/OS as well as toxicity endpoints. (2) Standardization—harmonize biospecimen handling, sequencing/qPCR pipelines, and reporting standards to enable multi-center validation and regulatory review. (3) Safety and durability—establish long-term surveillance for microbiome interventions (FMT, phage, high-dose probiotics), including resistance and horizontal gene transfer monitoring. (4) Minimal, deployable multi-omics panels—co-develop small, cost-effective signatures (metagenomics + metabolomics + qPCR) that track with clinical endpoints (DFS, OS, adverse events) and integrate into perioperative and adjuvant treatment pathways. Together, these steps outline a feasible road map toward microbiome-informed precision oncology.
7 Conclusions and future perspectives
The intricate relationship between the gut microbiota and CRC has gained considerable attention in recent years, revealing profound implications for tumor biology, diagnosis, and therapy. Accumulating evidence indicates that the gut microbiome contributes to CRC initiation, progression, and prognosis through multiple interrelated mechanisms, including chronic inflammation, bacterial genotoxin production, oxidative stress, and aberrant microbial metabolism (Png et al., 2022). Specific taxa, such as F. nucleatum, E. coli, and B. fragilis, are frequently enriched within tumor tissues, and their high abundance has been correlated with poor clinical outcomes, highlighting their potential roles as biomarkers and therapeutic targets (Alexander et al., 2023). At the same time, heterogeneity across studies—driven by differences in sampling, sequencing, and analysis pipelines, as well as confounding influences of diet, antibiotic exposure, and host genetics—complicates the interpretation of associations and underscores the need for methodological rigor and standardization (Diacova et al., 2025).
Advances in high-throughput sequencing, multi-omics integration, and bioinformatics have greatly enhanced our ability to characterize gut microbial composition and function with increasing precision, enabling earlier detection of dysbiosis, improved prediction of therapeutic responses, and movement toward individualized interventions in CRC. Recent multi-omics studies and reviews illustrate prognostic modeling and subtype discovery, as well as clinical screening potential, when metagenomics is integrated with metabolomics/proteomics and other layers (Xu et al., 2024). However, variability in pre-analytical procedures (e.g., stool collection/stabilization/storage) and platform effects (targeted amplicon vs. shotgun metagenomics) can introduce batch effects and limit cross-cohort comparability. Comparative work from 2024 to 2025 underscores method-dependent differences between 16S and shotgun approaches, while studies on FIT-derived material, domestic freezer storage, and field-collection protocols highlight how handling choices shape profiles (Bars-Cortina et al., 2024). Downstream bioinformatics also contributes to between-study variability, motivating updated computational best practices (Pita-Galeana et al., 2025). Addressing these issues will require harmonized protocols, shared reference materials, and transparent reporting standards (e.g., STORMS and related community efforts), alongside systematic capture of key covariates—particularly detailed diet and medication histories and host genomic data—to enable robust adjustment for confounding; recent large-scale analyses further emphasize microbial load, diet, and medications as major drivers of variability (Forry et al., 2024).
Microbiota-targeted interventions—including probiotics, prebiotics, FMT, and bacteriophage therapy—are emerging as promising strategies for both CRC prevention and adjunctive treatment (Lei et al., 2025). These approaches aim to restore microbial homeostasis, reshape the tumor microenvironment, and enhance the efficacy of existing therapeutic modalities, providing new opportunities for precision oncology. Nonetheless, considerable interindividual variability in treatment response is consistently observed. Sources of this variability likely include baseline community structure and function, host immune tone, mucosal ecology (e.g., biofilms), diet, recent antibiotic exposure, and host genetic variation in pathways mediating microbe–host interactions (Kim K. et al., 2025). Prospective trials should therefore incorporate responder/non-responder stratification, dietary control or standardized counseling, pre-specified antibiotic washout periods where feasible, and integration of host multi-omics to identify predictive biomarkers and guide patient selection.
To make these advances clinically actionable, we outline a multi-omics–to-clinic framework. We prioritize a minimal diagnostic signature that pairs metagenomics (with optional metabolomics) and targeted qPCR to deliver reproducible, cost-aware risk stratification. We then implement a pathway–therapy–microbiome matching schema that aligns dominant microbial mechanisms (genotoxin production, chronic inflammation, metabolite dysregulation, biofilm ecology) with tailored interventions (e.g., β-glucuronidase mitigation, anti-inflammatory adjuncts, probiotics/prebiotics, FMT, or phage targeting of F. nucleatum and pks+ E. coli). Finally, we recommend a three-step clinical path—stratified diagnosis → therapy matching → longitudinal monitoring—to support prospective validation, safety/quality oversight, and real-time adaptation based on microbiome dynamics.
Several critical knowledge gaps remain. First, establishing causal relationships between gut microbes and CRC is essential, necessitating well-designed in vivo and ex vivo models to move beyond correlative studies (Liu et al., 2023). Complementary causal-inference approaches (e.g., longitudinal designs, mediation analyses) may help disentangle confounding by diet, antibiotics, and host genetics. Second, there is an urgent need for highly sensitive, specific, and cost-effective diagnostic tools to facilitate the clinical implementation of microbiome-based biomarkers (Kværner et al., 2021). Such tools should be validated across centers using standardized workflows, external quality controls, and comprehensive metadata capture to ensure generalizability. Third, the safety, efficacy, and durability of microbiota-targeted interventions require validation in large, multicenter clinical trials, with careful attention to interindividual variability, host–microbiome interactions, and the potential ecological trade-offs of therapy (Huang et al., 2022). Pragmatic trial designs, real-time resistance/ecology surveillance (for antibiotics and phages), and consensus manufacturing/quality criteria (potency, purity, stability, endotoxin burden) will be important for translation.
Looking forward, the integration of advanced technologies such as artificial intelligence (AI) and machine learning with multi-omics datasets—encompassing microbiome, metabolome, genome, and single-cell transcriptome—offers an unprecedented opportunity to build predictive models for CRC risk, prognosis, and therapeutic response (Fusco et al., 2024; Yang L. et al., 2023). To realize this potential, models must be trained on well-annotated, harmonized datasets that include standardized laboratory and computational pipelines and rich covariate metadata (diet, medications, host genetics). AI-driven feature extraction can accelerate the identification of microbial signatures for precision diagnostics, while deep learning approaches may elucidate complex host–microbiota network interactions, informing personalized treatment strategies (Zhang J. et al., 2024; Zhou et al., 2022). Equally important are prospective external validation, assessment of model transportability across populations and diets, and interpretable frameworks that link features to mechanism and actionability.
Overall, gut microbiome research is transitioning from descriptive studies toward precision applications in oncology. Realizing the promise of “microbiome-informed precision oncology” will depend not only on mechanistic insight and therapeutic innovation but also on field-wide standardization, rigorous control of confounding, and deliberate accommodation of interindividual variability in trial design and clinical implementation. By coupling methodological best practices with mechanistic and translational advances, microbiota-based diagnostics and therapeutics are poised to become integral components of comprehensive CRC management.
Author contributions
BB: Writing – original draft, Writing – review & editing. JM: Writing – original draft. WX: Writing – review & editing. XiC: Writing – review & editing. XuC: Writing – review & editing. CL: Writing – review & editing. WS: Writing – review & editing. YL: Writing – review & editing. HS: Writing – review & editing. BZ: Writing – review & editing. DX: Writing – review & editing. ZL: Writing – review & editing. YW: Writing – review & editing. JS: Writing – review & editing. MY: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the grant of the China Postdoctoral Science Foundation (Grant No. 2025M772548), the Chongqing Wanzhou Municipal Science and Health Joint Medical Research Project Youth Program (Grant No. wzwjw-kw2024005), Youth Project of the Scientific and Technological Research Program of Chongqing Municipal Education Commission (Grant No. KJQN202500113), Natural Science Foundation of Chongqing, China (Grant No. CSTB2025NSCQ-GPX0109), Science and Technology Innovation Key R&D Program of Chongqing (Grant No. CSTB2023TIAD-STX0011), and The National Key R&D Programmes (NKPs) of China (Grant No. 2022YFC3601802).
Acknowledgments
We gratefully acknowledge all the researchers for taking part of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aghamajidi, A., and Vareki, S. M. (2022). The effect of the gut microbiota on systemic and anti-tumor immunity and response to systemic therapy against cancer. Cancer 14:3563. doi: 10.3390/cancers14153563
Alanazi, S., Salama, S., Althobaiti, M., Alotaibi, R., Alabdullatif, A., Musa, A., et al. (2024). Alleviation of copper-induced hepatotoxicity by bergenin: diminution of oxidative stress, inflammation, and apoptosis via targeting SIRT1/FOXO3a/NF-κB axes and p 38 MAPK signaling. Biol. Trace Elem. Res. 203, 3195–3207. doi: 10.1007/s12011-024-04401-3
Alexander, J., Posma, J., Scott, A., Poynter, L., Mason, S., Doria, M., et al. (2023). Pathobionts in the tumour microbiota predict survival following resection for colorectal cancer. Microbiome 11:100. doi: 10.1186/s40168-023-01518-w
Andary, C., Al, K., Chmiel, J. A., Gibbons, S., Daisley, B., Parvathy, S., et al. (2024). Dissecting mechanisms of fecal microbiota transplantation efficacy in disease. Trends Mol. Med. 30, 209–222. doi: 10.1016/j.molmed.2023.12.005
Bahar, M. E., Kim, H. J., and Kim, D. (2023). Targeting the RAS/RAF/MAPK pathway for cancer therapy: from mechanism to clinical studies. Signal Transduct. Target. Ther. 8:455. doi: 10.1038/s41392-023-01705-z
Bardelčíková, A., Šoltys, J., and Mojžiš, J. (2023). Oxidative stress, inflammation and colorectal cancer: An overview. Antioxidants 12:901. doi: 10.3390/antiox12040901
Bars-cortina, D., Ramon, E., Rius-sansalvador, B., Guinó, E., Garcia-serrano, A., Mach, N., et al. (2024). Comparison between 16S rRNA and shotgun sequencing in colorectal cancer, advanced colorectal lesions, and healthy human gut microbiota. BMC Genomics 25:730. doi: 10.1186/s12864-024-10621-7
Bell, H., Rebernick, R., Goyert, J., Singhal, R., Kuljanin, M., Kerk, S., et al. (2021). Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell 40, 185–200.e6. doi: 10.1016/j.ccell.2021.12.001
Bhere, D., Arghiani, N., Lechtich, E. R., Yao, Y., Alsaab, S., Bei, F., et al. (2020). Simultaneous downregulation of miR-21 and upregulation of miR-7 has anti-tumor efficacy. Sci. Rep. 10:1779. doi: 10.1038/s41598-020-58072-w
Bustin, S. A., Ruijter, J. M., Van den hoff, M. J. B., Kubista, M., Pfaffl, M. W., Shipley, G. L., et al. (2025). MIQE 2.0: revision of the minimum information for publication of quantitative real-time PCR experiments guidelines. Clin. Chem. 71, 634–651. doi: 10.1093/clinchem/hvaf043
Cao, Y., Oh, J., Xue, M., Huh, W., Wang, J., González-Hernández, J., et al. (2022). Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science 378:eabm3233. doi: 10.1126/science.abm3233
Casasanta, M. A., Yoo, C. C., Udayasuryan, B., Sanders, B. E., Umaña, A., Zhang, Y., et al. (2020). Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 13:eaba9157. doi: 10.1126/scisignal.aba9157
Castellarin, M., Warren, R. L., Freeman, J. D., Dreolini, L., Krzywinski, M., Strauss, J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306. doi: 10.1101/gr.126516.111
Catalano, T., Selvaggi, F., Cdtellese, R., and Aceto, G. M. (2025). The role of reactive oxygen species in colorectal cancer initiation and progression: perspectives on theranostic approaches. Cancers (Basel) 17:752. doi: 10.3390/cancers17050752
Chawrylak, K., Leśniewska, M., Mielniczek, K., Sędłak, K., Pelc, Z., Pawlik, T., et al. (2024). Gut microbiota—adversary or ally? Its role and significance in colorectal cancer pathogenesis, progression, and treatment. Cancer 16:2236. doi: 10.3390/cancers16122236
Chen, Y., and Chen, Y.-X. (2021). Microbiota-associated metabolites and related immunoregulation in colorectal cancer. Cancer 13:4054. doi: 10.3390/cancers13164054
Chen, F., Dai, X., Zhou, C.-C., Li, K., Zhang, Y.-J., Lou, X., et al. (2021). Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut 71, 1315–1325. doi: 10.1136/gutjnl-2020-323476
Chen, L., Zhao, X., Wongso, S., Lin, Z., and Wang, S. (2024). Trade-offs between receptor modification and fitness drive host-bacteriophage co-evolution leading to phage extinction or co-existence. ISME J. 18:wrae214. doi: 10.1093/ismejo/wrae214
Cheng, Y., Ling, Z., and Li, L. (2020). The intestinal microbiota and colorectal cancer. Front. Immunol. 11:615056. doi: 10.3389/fimmu.2020.615056
Cheng, W., Wu, C.-Y., and Yu, J. (2020). The role of gut microbiota in cancer treatment: friend or foe? Gut 69, 1867–1876. doi: 10.1136/gutjnl-2020-321153
Chung, Y. W., Gwak, H.-J., Moon, S., Rho, M., and Ryu, J.-H. (2020). Functional dynamics of bacterial species in the mouse gut microbiome revealed by metagenomic and metatranscriptomic analyses. PLoS One 15:e0227886. doi: 10.1371/journal.pone.0227886
Coker, O., Liu, C., Wu, W., Wong, S., Jia, W., Sung, J., et al. (2022). Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome 10:35. doi: 10.1186/s40168-021-01208-5
Collatz, M., Braun, S. D., Reinicke, M., Müller, E., Monecke, S., and Ehricht, R. (2025). AssayBLAST: a bioinformatic tool for in silico analysis of molecular multiparameter assays. Appl. Biosci. 4:18. doi: 10.3390/applbiosci4020018
Conde-Pérez, K., Aja-Macaya, P., Buetas, E., Trigo-Tasende, N., Nasser-Ali, M., Rumbo-Feal, S., et al. (2024). The multispecies microbial cluster of fusobacterium, parvimonas, bacteroides and faecalibacterium as a precision biomarker for colorectal cancer diagnosis. Mol. Oncol. 18, 1093–1122. doi: 10.1002/1878-0261.13604
Curti, L., and Campaner, S. (2021). MYC-induced replicative stress: a double-edged sword for cancer development and treatment. Int. J. Mol. Sci. 22:6168. doi: 10.3390/ijms22126168
Daca, A., and Jarzembowski, T. (2024). From the friend to the foe-enterococcus faecalis diverse impact on the human immune system. Int. J. Mol. Sci. 25:2422. doi: 10.3390/ijms25042422
Danis-Wlodarczyk, K., Dąbrowska, K., and Abedon, S. T. (2021). Phage therapy: the pharmacology of antibacterial viruses. Curr. Issues Mol. Biol. 81-164, 81–164. doi: 10.21775/cimb.040.081
Deissova, T., Zapletalová, M., Kunovský, L., Kroupa, R., Grolich, T., Kala, Z., et al. (2023). 16S rRNA gene primer choice impacts off-target amplification in human gastrointestinal tract biopsies and microbiome profiling. Sci. Rep. 13:12577. doi: 10.1038/s41598-023-39575-8
Diacova, T., Cifelli, C. J., Davis, C. D., Holscher, H. D., Kable, M. E., Lampe, J. W., et al. (2025). Best practices and considerations for conducting research on diet-gut microbiome interactions and their impact on health in adult populations: An umbrella review. Adv Nutr 16:100419. doi: 10.1016/j.advnut.2025.100419
Dougherty, M., and Jobin, C. (2023). Intestinal bacteria and colorectal cancer: etiology and treatment. Gut Microbes 15:2185028. doi: 10.1080/19490976.2023.2185028
Durazzi, F., Sala, C., Castellani, G., Manfreda, G., Remondini, D., and De Cesare, A. (2021). Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 11:3030. doi: 10.1038/s41598-021-82726-y
Elnar, A. G., and Kim, G. B. (2025). Probiotic potential and safety assessment of bacteriocinogenic Enterococcus faecalis CAUM157. Front. Microbiol. 16:1563444. doi: 10.3389/fmicb.2025.1563444
Fan, Y., and Pedersen, O. (2020). Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71. doi: 10.1038/s41579-020-0433-9
Fang, Y., Fu, M., Li, X., Zhang, B., and Wan, C. (2024). Enterohemorrhagic escherichia coli effector espF triggers oxidative DNA lesions in intestinal epithelial cells. Infect. Immun. 92:e0000124. doi: 10.1128/iai.00001-24
Fierer, N., Leung, P. M., Lappan, R., Eisenhofer, R., Ricci, F., Holland, S. I., et al. (2025). Guidelines for preventing and reporting contamination in low-biomass microbiome studies. Nat. Microbiol. 10, 1570–1580. doi: 10.1038/s41564-025-02035-2
Finetti, F., Travelli, C., Ercoli, J., Colombo, G., Buoso, E., and Trabalzini, L. (2020). Prostaglandin E2 and cancer: insight into tumor progression and immunity. Biology 9:434. doi: 10.3390/biology9120434
Fong, W., Li, Q., and Yu, J. (2020). Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene, 39, 4925–4943. doi: 10.1038/s41388-020-1341-1
Forry, S. P., Servetas, S. L., Kralj, J. G., Soh, K., Hadjithomas, M., Cano, R., et al. (2024). Variability and bias in microbiome metagenomic sequencing: an interlaboratory study comparing experimental protocols. Sci. Rep. 14:9785. doi: 10.1038/s41598-024-57981-4
Fusco, W., Bricca, L., Kaitsas, F., Tartaglia, M. F., Venturini, I., Rugge, M., et al. (2024). Gut microbiota in colorectal cancer: from pathogenesis to clinic. Best Pract. Res. Clin. Gastroenterol. 72:101941. doi: 10.1016/j.bpg.2024.101941
Galeano Niño, J. L., Wu, H., Lacourse, K. D., Kempchinsky, A. G., Baryiames, A., Barber, B., et al. (2022). Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817. doi: 10.1038/s41586-022-05435-0
Gao, Y., Bi, D., Xie, R.-T., Li, M., Guo, J., Liu, H., et al. (2021). Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct. Target. Ther. 6:398. doi: 10.1038/s41392-021-00795-x
Gao, R., Wu, C., Zhu, Y., Kong, C., Zhu, Y., Gao, Y., et al. (2022). Integrated analysis of colorectal cancer reveals cross-cohort gut microbial signatures and associated serum metabolites. Gastroenterology 163, 1024–1037.e9. doi: 10.1053/j.gastro.2022.06.069
Gerstberger, S., Lumish, M., Hartner, S., Shah, F., Choi, S., Lum, K., et al. (2025). Abstract 2856: Pks+ E. coli trigger intestinal stem cell plasticity and early onset colorectal cancer. Cancer Res. 85:2856. doi: 10.1158/1538-7445.AM2025-2856
Gupta, S. V., Campos, L., and Schmidt, K. (2023). Mitochondrial superoxide dismutase Sod2 suppresses nuclear genome instability during oxidative stress. Genetics 225:iyad147. doi: 10.1093/genetics/iyad147
Hamamah, S., Lobiuc, A., and Covașă, M. (2024). Antioxidant role of probiotics in inflammation-induced colorectal cancer. Int. J. Mol. Sci. 25:9026. doi: 10.3390/ijms25169026
Hanus, M., Parada-Venegas, D., Landskron, G., Wielandt, A., Hurtado, C., Álvarez, K., et al. (2021). Immune system, microbiota, and microbial metabolites: the unresolved triad in colorectal cancer microenvironment. Front. Immunol. 12:612826. doi: 10.3389/fimmu.2021.612826
Hillege, L., Stevens, M., Kristen, P., De Vos-Geelen, J., Penders, J., Redinbo, M., et al. (2024). The role of gut microbial β-glucuronidases in carcinogenesis and cancer treatment: a scoping review. J. Cancer Res. Clin. Oncol. 150:495. doi: 10.1007/s00432-024-06028-2
Hong, Y., Boiti, A., Vallone, D., and Foulkes, N. S. (2024). Reactive oxygen species signaling and oxidative stress: transcriptional regulation and evolution. Antioxidants (Basel) 13:312. doi: 10.3390/antiox13030312
Hu, L.-J., Liu, Y., Kong, X., Wu, R., Peng, Q., Zhang, Y., et al. (2021). Fusobacterium nucleatum facilitates M2 macrophage polarization and colorectal carcinoma progression by activating TLR4/NF-κB/S100A9 cascade. Front. Immunol. 12:658681. doi: 10.3389/fimmu.2021.658681
Huang, H.-C., Cai, B.-H., Suen, C.-S., Lee, H.-Y., Hwang, M., Liu, F.-T., et al. (2020). BGN/TLR4/NF-κB mediates epigenetic silencing of immunosuppressive siglec ligands in colon cancer cells. Cells 9:397. doi: 10.3390/cells9020397
Huang, J., Liu, D., Wang, Y., Liu, L., Li, J., Yuan, J., et al. (2022). Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut 71, 734–745. doi: 10.1136/gutjnl-2020-321031
Huber, A. R., Pleguezuelos-Manzano, C., Puschhof, J., Ubels, J., Boot, C., Saftien, A., et al. (2024). Improved detection of colibactin-induced mutations by genotoxic E. coli in organoids and colorectal cancer. Cancer Cell 42, 487–496. doi: 10.1016/j.ccell.2024.02.009
Iftekhar, A., Berger, H., Bouznad, N., Heuberger, J., Boccellato, F., Dobrindt, U., et al. (2021). Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat. Commun. 12:1003. doi: 10.1038/s41467-021-21162-y
Jergens, A., Parvinroo, S., Kopper, J., and Wannemuehler, M. (2021). Rules of engagement: epithelial-microbe interactions and inflammatory bowel disease. Front. Med. 8:669913. doi: 10.3389/fmed.2021.669913
Jiang, Y., Huang, Y., Hu, Y., Yang, Y., You, F., Hu, Q., et al. (2024). Banxia xiexin decoction delays colitis-to-cancer transition by inhibiting E-cadherin/β-catenin pathway via fusobacterium nucleatum fadA. J. Ethnopharmacol. 328:117932. doi: 10.1016/j.jep.2024.117932
Jo, M., Hwang, S., Lee, C., Hong, J., Kang, D.-H., Yoo, S.-H., et al. (2023). Promotion of colitis in B cell-deficient C57BL/6 mice infected with enterotoxigenic bacteroides fragilis. Int. J. Mol. Sci. 25:364. doi: 10.3390/ijms25010364
Kalasabail, S., Engelman, J., Zhang, L. Y., El-Omar, E., and Yim, H. (2021). A perspective on the role of microbiome for colorectal cancer treatment. Cancer 13:4623. doi: 10.3390/cancers13184623
Kameoka, S., Motooka, D., Watanabe, S., Kubo, R., Jung, N., Midorikawa, Y., et al. (2021). Benchmark of 16S rRNA gene amplicon sequencing using japanese gut microbiome data from the V1-V2 and V3-V4 primer sets. BMC Genomics 22:527. doi: 10.1186/s12864-021-07746-4
Karpiński, T., Ożarowski, M., and Stasiewicz, M. (2022). Carcinogenic microbiota and its role in colorectal cancer development. Semin. Cancer Biol. 86, 420–430. doi: 10.1016/j.semcancer.2022.01.004
Kaur, S., Ortiz, L. B., Rudakov, G., and Verma, M. S. (2024). Propidium monoazide is unreliable for quantitative live-dead molecular assays. bioRxiv, 2024.06.05.597603. doi: 10.1101/2024.06.05.597603
Kavec, M., Urbanova, M., Makovicky, P., Opattova, A., Tomasova, K., Kroupa, M., et al. (2022). Oxidative damage in sporadic colorectal cancer: molecular mapping of base excision repair glycosylases MUTYH and hOGG1 in colorectal cancer patients. Int. J. Mol. Sci. 23:5704. doi: 10.3390/ijms23105704
Kenneth, M. J., Wu, C.-C., Fang, C.-Y., Hsu, T. K., Lin, I. C., Huang, S.-W., et al. (2025). Exploring the impact of chemotherapy on the emergence of antibiotic resistance in the gut microbiota of colorectal cancer patients. Antibiotics 14:264. doi: 10.3390/antibiotics14030264
Khalid, A., Lin, R. C. Y., and Iredell, J. R. (2021). A phage therapy guide for clinicians and basic scientists: background and highlighting applications for developing countries. Front. Microbiol. 11:599906. doi: 10.3389/fmicb.2020.599906
Kim, J., and Lee, H. (2022). Potential role of the gut microbiome in colorectal cancer progression. Front. Immunol. 12:807648. doi: 10.3389/fimmu.2021.807648
Kim, K., Lee, M., Shin, Y., Lee, Y., and Kim, T. J. (2025). Optimizing cancer treatment through gut microbiome modulation. Cancers (Basel) 17:1252. doi: 10.3390/cancers17071252
Kim, M. K., Suh, G. A., Cullen, G. D., Perez rodriguez, S., Dharmaraj, T., Chang, T. H. W., et al. (2025). Bacteriophage therapy for multidrug-resistant infections: current technologies and therapeutic approaches. J. Clin. Invest. 135:e187996. doi: 10.1172/jci187996
Kok, C. R., Brabec, B., Chichlowski, M., Harris, C. L., Moore, N., Wampler, J. L., et al. (2020). Stool microbiome, pH and short/branched chain fatty acids in infants receiving extensively hydrolyzed formula, amino acid formula, or human milk through two months of age. BMC Microbiol. 20:337. doi: 10.1186/s12866-020-01991-5
Kong, C., Yan, X., Zhu, Y., Zhu, H., Luo, Y., Liu, P., et al. (2021). Fusobacterium nucleatum promotes the development of colorectal cancer by activating a cytochrome P450/Epoxyoctadecenoic acid axis via TLR4/Keap1/NRF2 signaling. Cancer Res. 81, 4485–4498. doi: 10.1158/0008-5472.CAN-21-0453
Kouhzad, M., Götz, F., Navidifar, T., Taki, E., Ghamari, M., Mohammadzadeh, R., et al. (2025). Carcinogenic and anticancer activities of microbiota-derived secondary bile acids. Front. Oncol. 15:1514872. doi: 10.3389/fonc.2025.1514872
Kværner, A. S., Birkeland, E., Bucher-Johannessen, C., Vinberg, E., Nordby, J. I., Kangas, H., et al. (2021). The CRCbiome study: a large prospective cohort study examining the role of lifestyle and the gut microbiome in colorectal cancer screening participants. BMC Cancer 21:930. doi: 10.1186/s12885-021-08640-8
Lai, Y.-R., Chang, Y. F., Ma, J., Chiu, C. H., Kuo, M., and Lai, C.-H. (2021). From DNA damage to cancer progression: potential effects of cytolethal distending toxin. Front. Immunol. 12:760451. doi: 10.3389/fimmu.2021.760451
Lan, C., Chen, S., Jiang, S., Lei, H., Cai, Z., and Huang, X. (2022). Different expression patterns of inflammatory cytokines induced by lipopolysaccharides from escherichia coli or porphyromonas gingivalis in human dental pulp stem cells. BMC Oral Health 22:121. doi: 10.1186/s12903-022-02161-x
Laupland, K., Edwards, F., Furuya-Kanamori, L., Paterson, D., and Harris, P. (2023). Bloodstream infection and colorectal cancer risk in Queensland Australia, 2000-2019. Am. J. Med. 136, 896–901. doi: 10.1016/j.amjmed.2023.05.003
Lee, R., and Holmes, D. (2023). Barriers and recommendations for colorectal cancer screening in Africa. Glob. Health Action 16:2181920. doi: 10.1080/16549716.2023.2181920
Lee, C., Hwang, S., Gwon, S.-Y., Park, C., Jo, M., Hong, J., et al. (2022). Bacteroides fragilis toxin induces intestinal epithelial cell secretion of interleukin-8 by the E-cadherin/β-catenin/NF-κB dependent pathway. Biomedicine 10:827. doi: 10.3390/biomedicines10040827
Lei, D., Yu, W., Liu, Y., Jiang, Y., Li, X.-H., Lv, J., et al. (2023). Trimethylamine N-oxide (TMAO) inducing endothelial injury: UPLC-MS/MS-based quantification and the activation of cathepsin B-mediated NLRP3 inflammasome. Molecules 28:3817. doi: 10.3390/molecules28093817
Lei, W., Zhou, K., Lei, Y., Li, Q., and Zhu, H. (2025). Gut microbiota shapes cancer immunotherapy responses. NPJ Biofilms Microbiomes 11:143. doi: 10.1038/s41522-025-00786-8
Li, Y., Li, Q., Yuan, R., Wang, Y., Guo, C., and Wang, L. (2024). Bifidobacterium breve-derived indole-3-lactic acid ameliorates colitis-associated tumorigenesis by directing the differentiation of immature colonic macrophages. Theranostics 14, 2719–2735. doi: 10.7150/thno.92350
Li, R., Shen, J., and Xu, Y. (2022). Fusobacterium nucleatum and colorectal cancer. Infect. Drug Resist. 15, 1115–1120. doi: 10.2147/IDR.S357922
Li, L., Yu, R., Cai, T., Chen, Z., Lan, M., Zou, T., et al. (2020). Effects of immune cells and cytokines on inflammation and immunosuppression in the tumor microenvironment. Int. Immunopharmacol. 88:106939. doi: 10.1016/j.intimp.2020.106939
Liu, X., Tang, H., Zhou, Q., Zeng, Y., Lu, B., Chen, D., et al. (2023). Gut microbiota composition in patients with advanced malignancies experiencing immune-related adverse events. Front. Immunol. 14:1109281. doi: 10.3389/fimmu.2023.1109281
Lukiw, W. (2020). Gastrointestinal (GI) tract microbiome-derived neurotoxins—potent neuro-inflammatory signals from the GI tract via the systemic circulation into the brain. Front. Cell. Infect. Microbiol. 10:22. doi: 10.3389/fcimb.2020.00022
Luo, R., Yao, Y., Chen, Z., and Sun, X. (2025). An examination of the LPS-TLR4 immune response through the analysis of molecular structures and protein-protein interactions. Cell Commun. Signal 23:142. doi: 10.1186/s12964-025-02149-4
Madani, W. A. M., Ramos, Y., Cubillos-ruiz, J. R., and Morales, D. K. (2024). Enterococcal-host interactions in the gastrointestinal tract and beyond. FEMS Microbes 5:xtae027. doi: 10.1093/femsmc/xtae027
Masheghati, F., Asgharzadeh, M., Jafari, A., Masoudi, N., and Maleki-Kakelar, H. (2024). The role of gut microbiota and probiotics in preventing, treating, and boosting the immune system in colorectal cancer. Life Sci. 344:122529. doi: 10.1016/j.lfs.2024.122529
Matchado, M., Rühlemann, M., Reimeiter, S., Kacprowski, T., Frost, F., Haller, D., et al. (2023). On the limits of 16S rRNA gene-based metagenome prediction and functional profiling. Microb. Genom. 10:001203. doi: 10.1099/mgen.0.001203
Mayorga-ramos, A., Carrera-pacheco, S. E., Barba-ostria, C., and Guamán, L. P. (2024). Bacteriophage-mediated approaches for biofilm control. Front. Cell. Infect. Microbiol. 14:1428637. doi: 10.3389/fcimb.2024.1428637
Mizumoto-teramura, Y., Kamogashira, T., Kondo, K., and Yamasoba, T. (2024). Heterochronous multiplex real-time PCR with intercalating dye using uracil-DNA N-glycosylase (UNG) and multiple primer pairs to revaluate post PCR product. MethodsX 13:102818. doi: 10.1016/j.mex.2024.102818
Mohseni, A., Taghinezhad-S, S., Casolaro, V., Lv, Z., and Li, D. (2023). Potential links between the microbiota and T cell immunity determine the tumor cell fate. Cell Death Dis. 14:154. doi: 10.1038/s41419-023-05560-2
Morgan, E., Arnold, M., Gini, A., Lorenzoni, V., Cabasag, C., Laversanne, M., et al. (2022). Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut 72, 338–344. doi: 10.1136/gutjnl-2022-327736
Mukherjee, S., Dutta, A., and Chakraborty, A. (2022). The interaction of oxidative stress with MAPK, PI3/AKT, NF-κB, and DNA damage kinases influences the fate of γ-radiation-induced bystander cells. Arch. Biochem. Biophys. 725:109302. doi: 10.1016/j.abb.2022.109302
Neganova, M., Liu, J., Aleksandrova, Y., Klochkov, S., and Fan, R. (2021). Therapeutic influence on important targets associated with chronic inflammation and oxidative stress in cancer treatment. Cancer 13:6062. doi: 10.3390/cancers13236062
Neumann, A., Happonen, L., Karlsson, C., Bahnan, W., Frick, I., and Björck, L. (2021). Streptococcal protein SIC activates monocytes and induces inflammation. iScience 24:102339. doi: 10.1016/j.isci.2021.102339
Oakes, S. (2020). Endoplasmic reticulum stress signaling in cancer cells. Am. J. Pathol. 190, 934–946. doi: 10.1016/j.ajpath.2020.01.010
Oh, N., Lee, J., Kim, Y.-T., Kim, S., and Lee, J.-H. (2020). Cancer-protective effect of a synbiotic combination between Lactobacillus gasseri 505 and a Cudrania tricuspidata leaf extract on colitis-associated colorectal cancer. Gut Microbes 12:1785803. doi: 10.1080/19490976.2020.1785803
Ouranos, K., Gardikioti, A., Bakaloudi, D. R., Mylona, E. K., Shehadeh, F., and Mylonakis, E. (2023). Association of the streptococcus bovis/streptococcus equinus complex with colorectal neoplasia: a systematic review and meta-analysis. Open Forum Infect. Dis. 10:ofad547. doi: 10.1093/ofid/ofad547
Pandey, V., Premkumar, K., Kundu, P., and Shankar, B. (2024). PGE2 induced miR365/IL-6/STAT3 signaling mediates dendritic cell dysfunction in cancer. Life Sci. 350:122751. doi: 10.1016/j.lfs.2024.122751
Park, I., Lee, J.-H., Kye, B.-H., Oh, H.-K., Cho, Y., Kim, Y.-T., et al. (2020). Effects of PrObiotics on the symptoms and surgical ouTComes after anterior REsection of colon cancer (POSTCARE): a randomized, double-blind, placebo-controlled trial. J. Clin. Med. 9:2181. doi: 10.3390/jcm9072181
Peery, A. F., Kelly, C. R., Kao, D., Vaughn, B. P., Lebwohl, B., Singh, S., et al. (2024). AGA clinical practice guideline on fecal microbiota-based therapies for select gastrointestinal diseases. Gastroenterology 166, 409–434. doi: 10.1053/j.gastro.2024.01.008
Pita-galeana, M. A., Ruhle, M., López-vázquea, L., De anda-jáuregui, G., and Hernández-lemus, E. (2025). Computational metagenomics: state of the art. Int. J. Mol. Sci. 26:9206. doi: 10.3390/ijms26189206
Pleguezuelos-Manzano, C., Puschhof, J., Huber, A. R., Van Boxtel, R., Rosendahl Huber, A., van Hoeck, A., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic pks+E. coli. Nature 580, 269–273. doi: 10.1038/s41586-020-2080-8
Png, C., Chua, Y.-K., Law, J., Zhang, Y., and Tan, K.-K. (2022). Alterations in co-abundant bacteriome in colorectal cancer and its persistence after surgery: a pilot study. Sci. Rep. 12:9829. doi: 10.1038/s41598-022-14203-z
Puschhof, J., Pleguezuelos-Manzano, C., Martinez-Silgado, A., Akkerman, N., Saftien, A., Boot, C., et al. (2021). Intestinal organoid cocultures with microbes. Nat. Protoc. 16, 4633–4649. doi: 10.1038/s41596-021-00589-z
Qi, Y., Duan, G., Wei, D., Zhao, C., and Yonggui, M. (2022). The bile acid membrane receptor TGR5 in cancer: friend or foe? Molecules 27:5292. doi: 10.3390/molecules27165292
Qi, Z., Zhibo, Z., Jing, Z., Zhanbo, Q., Shugao, H., Weili, J., et al. (2022). Prediction model of poorly differentiated colorectal cancer (CRC) based on gut bacteria. BMC Microbiol. 22:312. doi: 10.1186/s12866-022-02712-w
Qu, R., Zhang, Y., Yanpeng, M., Zhou, X., Sun, L., Jiang, C., et al. (2023). Role of the gut microbiota and its metabolites in tumorigenesis or development of colorectal cancer. Adv. Sci. 10:e2205563. doi: 10.1002/advs.202205563
Rad, H. F., Tahmasebi, H., Javani, S., Hemati, M., Zakerhamidi, D., Hosseini, M., et al. (2024). Microbiota and cytokine modulation: innovations in enhancing anticancer immunity and personalized cancer therapies. Biomedicine 12:2776. doi: 10.3390/biomedicines12122776
Rafter, J., Bennett, M., Caderni, G., Clune, Y., Hughes, R., Karlsson, P. C., et al. (2007). Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 85, 488–496. doi: 10.1093/ajcn/85.2.488
Rossi, T., Vergara, D., Fanini, F., Maffia, M., Bravaccini, S., and Pirini, F. (2020). Microbiota-derived metabolites in tumor progression and metastasis. Int. J. Mol. Sci. 21:5786. doi: 10.3390/ijms21165786
Sánchez-Alcoholado, L., Ramos-Molina, B., Otero, A., Laborda-Illanes, A., Ordoñez, R., Medina, J., et al. (2020). The role of the gut microbiome in colorectal cancer development and therapy response. Cancer 12:1406. doi: 10.3390/cancers12061406
Sawa, T., Moriyama, K., and Kinoshita, M. (2024). Current status of bacteriophage therapy for severe bacterial infections. J. Intensive Care 12:44. doi: 10.1186/s40560-024-00759-7
Senthakumaran, T., Moen, A., Tannæs, T., Endres, A., Brackmann, S., Rounge, T., et al. (2023). Microbial dynamics with CRC progression: a study of the mucosal microbiota at multiple sites in cancers, adenomatous polyps, and healthy controls. Eur. J. Clin. Microbiol. Infect. Dis. 42, 305–322. doi: 10.1007/s10096-023-04551-7
Senthakumaran, T., Tannæs, T., Moen, A., Brackmann, S., Jahanlu, D., Rounge, T., et al. (2024). Detection of colorectal-cancer-associated bacterial taxa in fecal samples using next-generation sequencing and 19 newly established qPCR assays. Mol. Oncol. 19, 412–429. doi: 10.1002/1878-0261.13700
Shahin, K., Barazandeh, M., Zhang, L., Hedayatkhah, A., He, T., Bao, H., et al. (2021). Biodiversity of new lytic bacteriophages infecting shigella spp. in freshwater environment. Front. Microbiol. 12:619323. doi: 10.3389/fmicb.2021.619323
Shang, F., Jiang, X., Wang, H., Guo, S., Kang, S., Xu, B., et al. (2024). Bifidobacterium longum suppresses colorectal cancer through the modulation of intestinal microbes and immune function. Front. Microbiol. 15:1327464. doi: 10.3389/fmicb.2024.1327464
Shen, X.-H., Guan, J., Lu, D.-P., Hong, S.-C., Yu, L., and Chen, X. (2024). Peptostreptococcus anaerobius enhances dextran sulfate sodium-induced colitis by promoting nf-κB-NLRP3-dependent macrophage pyroptosis. Virulence 15:2435391. doi: 10.1080/21505594.2024.2435391
Shi, T., Van Soest, D., Polderman, P., Burgering, B., and Dansen, T. (2021). DNA damage and oxidant stress activate p53 through differential upstream signaling pathways. Free Radic. Biol. Med. 172, 298–311. doi: 10.1016/j.freeradbiomed.2021.06.013
Si, W., Liang, H., Bugno, J., Xu, Q., Ding, X.-C., Yang, K., et al. (2021). Lactobacillus rhamnosus GG induces cGAS/STING-dependent type I interferon and improves response to immune checkpoint blockade. Gut 71, 521–533. doi: 10.1136/gutjnl-2020-323426
Siddiqui, S., and Glauben, R. (2022). Fatty acid metabolism in myeloid-derived suppressor cells and tumor-associated macrophages: key factor in cancer immune evasion. Cancer 14:250. doi: 10.3390/cancers14010250
Song, M., Chan, A., and Sun, J. (2020). Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology 158, 322–340. doi: 10.1053/j.gastro.2019.06.048
Strauss, J., Kaplan, G. G., Beck, P. L., Rioux, K., Panaccione, R., Devinney, R., et al. (2011). Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17, 1971–1978. doi: 10.1002/ibd.21606
Sun, M., Monahan, K., Moquet, J., and Barnard, S. (2025). Colorectal cancers associated with mismatch repair deficiency. Front. Med. (Lausanne) 12:1649565. doi: 10.3389/fmed.2025.1649565
Sun, Y., Zhang, L., Hong, L., Zheng, W., Cui, J., Liu, X., et al. (2021). MicroRNA-181b-2 and MicroRNA-21-1 negatively regulate NF-κB and IRF3-mediated innate immune responses via targeting TRIF in teleost. Front. Immunol. 12:734520. doi: 10.3389/fimmu.2021.734520
Tang, J., Song, X., Zhao, M., Chen, H., Wang, Y., Zhao, B., et al. (2022). Oral administration of live combined bacillus subtilis and enterococcus faecium alleviates colonic oxidative stress and inflammation in osteoarthritic rats by improving fecal microbiome metabolism and enhancing the colonic barrier. Front. Microbiol. 13:1005842. doi: 10.3389/fmicb.2022.1005842
Ternes, D., Karta, J., Tsenkova, M., Wilmes, P., Haan, S., and Letellier, E. (2020). Microbiome in colorectal cancer: how to get from meta-omics to mechanism? Trends Microbiol. 28, 401–423. doi: 10.1016/j.tim.2020.01.001
Thind, S., Shibib, D., and Gentry, C. (2021). The effect of nomenclature revision of streptococcus bovis to streptococcus gallolyticus on subsequent colon cancer screening. Open Forum Infect. Dis. 8:ofab426. doi: 10.1093/ofid/ofab426
Trejo-Solís, C., Escamilla-Ramírez, A., Jimenez-Farfan, D., Castillo-Rodríguez, R., Flores-Nájera, A., and Cruz-Salgado, A. (2021). Crosstalk of the Wnt/β-catenin signaling pathway in the induction of apoptosis on cancer cells. Pharmaceuticals 14:871. doi: 10.3390/ph14090871
Turati, F., Concina, F., Rossi, M., Fiori, F., Parpinel, M., Taborelli, M., et al. (2022). Association of prebiotic fiber intake with colorectal cancer risk: the PrebiotiCa study. Eur. J. Nutr. 62, 455–464. doi: 10.1007/s00394-022-02984-y
Uehara, M., Inoue, T., Kominato, M., Hase, S., Sasaki, E., Toyoda, A., et al. (2022). Intraintestinal analysis of the functional activity of microbiomes and its application to the common marmoset intestine. mSystems 7:e0052022. doi: 10.1128/msystems.00520-22
Van Dingenen, L., Segers, C., Wouters, S., Mysara, M., Leys, N., Kumar-Singh, S., et al. (2023). Dissecting the role of the gut microbiome and fecal microbiota transplantation in radio- and immunotherapy treatment of colorectal cancer. Front. Cell. Infect. Microbiol. 13:1298264. doi: 10.3389/fcimb.2023.1298264
Viladomiu, M., Metz, M., Lima, S., Jin, W.-B., Chou, L., Guo, C.-J., et al. (2021). Adherent-invasive E. coli metabolism of propanediol in Crohn's disease regulates phagocytes to drive intestinal inflammation. Cell Host Microbe 29, 607–619.e8. doi: 10.1016/j.chom.2021.01.002
Wang, N., and Fang, J. (2022). Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 31, 159–172. doi: 10.1016/j.tim.2022.08.010
Wang, Y., and Fu, K. (2023). Genotoxins: the mechanistic links between escherichia coli and colorectal cancer. Cancer 15:1152. doi: 10.3390/cancers15041152
Wang, J., Zhang, R.-G., Lin, Z., Zhang, S., Chen, Y., Tang, J., et al. (2020). CDK7 inhibitor THZ1 enhances antiPD-1 therapy efficacy via the p38α/MYC/PD-L1 signaling in non-small cell lung cancer. J. Hematol. Oncol. 13:99. doi: 10.1186/s13045-020-00926-x
Warr, A., Kuehl, C., and Waldor, M. (2020). Shiga toxin remodels the intestinal epithelial transcriptional response to enterohemorrhagic Escherichia coli. PLoS Pathog. 17:e1009290. doi: 10.1371/journal.ppat.1009290
Wei, J., Zhang, J., Wang, D., Cen, B., Lang, J., and Dubois, R. (2022). The COX-2-PGE2 pathway promotes tumor evasion in colorectal adenomas. Cancer Prev. Res. 15, 285–296. doi: 10.1158/1940-6207.CAPR-21-0572
Weiner, A., and Enninga, J. (2019). The pathogen-host interface in three dimensions: correlative FIB/SEM applications. Trends Microbiol. 27, 426–439. doi: 10.1016/j.tim.2018.11.011
White, M., and Sears, C. (2023). The microbial landscape of colorectal cancer. Nat. Rev. Microbiol. 22, 240–254. doi: 10.1038/s41579-023-00973-4
Wong, C., and Yu, J. (2023). Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 1–24. doi: 10.1038/s41571-023-00766-x
Xiao, K., Liu, C., Tu, Z., Xu, Q., Chen, S., Zhang, Y., et al. (2020). Activation of the NF-κB and MAPK signaling pathways contributes to the inflammatory responses, but not cell injury, in IPEC-1 cells challenged with hydrogen peroxide. Oxidative Med. Cell. Longev. 2020, 1–14. doi: 10.1155/2020/5803639
Xie, Z., Canalda-Baltrons, A., D’Enfert, C., and Manichanh, C. (2023). Shotgun metagenomics reveals interkingdom association between intestinal bacteria and fungi involving competition for nutrients. Microbiome 11:275. doi: 10.1186/s40168-023-01693-w
Xing, G., Cui, Y., Guo, Z., Han, B., and Zhao, G. (2025). Progress on the mechanism of intestinal microbiota against colorectal cancer. Front. Cell. Infect. Microbiol. 15:1565103. doi: 10.3389/fcimb.2025.1565103
Xu, H.-X., Cao, C., Ren, Y., Weng, S., Liu, L., Guo, C. C., et al. (2022). Antitumor effects of fecal microbiota transplantation: implications for microbiome modulation in cancer treatment. Front. Immunol. 13:949490. doi: 10.3389/fimmu.2022.949490
Xu, Y. J., He, Y., Chen, C., Shi, J., He, M., Liu, Y., et al. (2024). Multiomics analysis revealed colorectal cancer pathogenesis. J. Proteome Res. 23, 2100–2111. doi: 10.1021/acs.jproteome.3c00894
Xue, C., Chu, Q., Zheng, Q., Yuan, X., Su, Y., Bao, Z., et al. (2023). Current understanding of the intratumoral microbiome in various tumors. Cell Rep Med 4:100884. doi: 10.1016/j.xcrm.2022.100884
Yadegar, A., Bar-Yoseph, H., Monaghan, T., Pakpour, S., Severino, A., Kuijper, E., et al. (2024). Fecal microbiota transplantation: current challenges and future landscapes. Clin. Microbiol. Rev. 37:e0006022. doi: 10.1128/cmr.00060-22
Yan, J., Liao, C., Taylor, B., Fontana, E., Amoretti, L., Wright, R., et al. (2022). A compilation of fecal microbiome shotgun metagenomics from hematopoietic cell transplantation patients. Sci Data 9:219. doi: 10.1038/s41597-022-01302-9
Yang, Y., An, Y., Dong, Y., Chu, Q., Wei, J.-Q., Wang, B., et al. (2024). Fecal microbiota transplantation: no longer cinderella in tumour immunotherapy. EBioMedicine 100:104967. doi: 10.1016/j.ebiom.2024.104967
Yang, L., Li, A., Wang, Y., and Zhang, Y. (2023). Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct. Target. Ther. 8:35. doi: 10.1038/s41392-022-01304-4
Yang, J., Wang, X., Hu, T., Huang, H., Chen, G., Jin, B., et al. (2024). Entero-toxigenic bacteroides fragilis contributes to intestinal barrier injury and colorectal cancer progression by mediating the BFT/STAT3/ZEB2 pathway. Cell Cycle 23, 70–82. doi: 10.1080/15384101.2024.2309005
Yang, Q., Wang, B., Zheng, Q., Li, H., Meng, X., Zhou, F., et al. (2023). A review of gut microbiota-derived metabolites in tumor progression and cancer therapy. Adv. Sci. 10:e2207366. doi: 10.1002/advs.202207366
Yang, J., Wei, H., Zhou, Y., Szeto, C., Li, C., Lin, Y., et al. (2020). High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology 162, 135–149.e2. doi: 10.1053/J.GASTRO.2021.08.041
Yao, Y., Ni, H.-T., Wang, X., Xu, Q., Zhang, J., Jiang, L., et al. (2021). A new biomarker of fecal bacteria for non-invasive diagnosis of colorectal cancer. Front. Cell. Infect. Microbiol. 11:744049. doi: 10.3389/fcimb.2021.744049
Yincharoen, P., Mordmuang, A., Techarang, T., Tangngamsakul, P., Kaewubon, P., Atipairin, P., et al. (2025). Microbiome and biofilm insights from normal vs tumor tissues in thai colorectal cancer patients. NPJ Precis Oncol. 9:98. doi: 10.1038/s41698-025-00873-1
You, M., Xie, Z., Zhang, N., Zhang, Y., Xiao, D., Liu, S., et al. (2023). Signaling pathways in cancer metabolism: mechanisms and therapeutic targets. Signal Transduct. Target. Ther. 8:196. doi: 10.1038/s41392-023-01442-3
Yuan, D., Tao, Y., Wang, H., Wang, J., Cao, Y., Cao, W., et al. (2022). A comprehensive analysis of the microbiota composition and host driver gene mutations in colorectal cancer. Investig. New Drugs 40, 884–894. doi: 10.1007/s10637-022-01263-1
Zamani, S., Taslimi, R., Sarabi, A., Jasemi, S., Sechi, L., and Feizabadi, M. (2020). Enterotoxigenic bacteroides fragilis: a possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front. Cell. Infect. Microbiol. 9:449. doi: 10.3389/fcimb.2019.00449
Zhang, W., An, Y., Qin, X., Wu, X., Wang, X., Hou, H., et al. (2021). Gut microbiota-derived metabolites in colorectal cancer: the bad and the challenges. Front. Oncol. 11:739648. doi: 10.3389/fonc.2021.739648
Zhang, L., Deng, M., Liu, J., Zhang, J., Wang, F., and Yu, W. (2024). The pathogenicity of vancomycin-resistant Enterococcus faecalis to colon cancer cells. BMC Infect. Dis. 24:230. doi: 10.1186/s12879-024-09133-2
Zhang, S., Shen, Y., Liu, H., Zhu, D., Fang, J., Pan, H., et al. (2023). Inflammatory microenvironment in gastric premalignant lesions: implication and application. Front. Immunol. 14:1297101. doi: 10.3389/fimmu.2023.1297101
Zhang, J., Wang, P., Wang, J., Wei, X., and Wang, M. (2024). Unveiling intratumoral microbiota: An emerging force for colorectal cancer diagnosis and therapy. Pharmacol. Res. 203:107185. doi: 10.1016/j.phrs.2024.107185
Zhou, Y., Xu, J., Luo, H., Meng, J., Chen, M., and Zhu, D. (2021). Wnt signaling pathway in cancer immunotherapy. Cancer Lett. 525, 84–96. doi: 10.1016/j.canlet.2021.10.034
Zhou, P., Yang, D., Sun, D., and Zhou, Y. (2022). Gut microbiome: new biomarkers in early screening of colorectal cancer. J. Clin. Lab. Anal. 36:e24359. doi: 10.1002/jcla.24359
Zhuang, C., Liu, G., Barkema, H., Zhou, M., Xu, S., Rahman, U., et al. (2020). Selenomethionine suppressed TLR4/NF-κB pathway by activating aelenoprotein S to alleviate ESBL Escherichia coli-induced inflammation in bovine mammary epithelial cells and macrophages. Front. Microbiol. 11:1461. doi: 10.3389/fmicb.2020.01461
Keywords: gut microbiota dysbiosis, carcinogenic mechanisms, key microbial species, multi-omics technologies, microbiota-based therapeutics
Citation: Bai B, Ma J, Xu W, Chen X, Chen X, Lv C, Su W, Li Y, Sun H, Zhang B, Xiang D, Li Z, Wu Y, Sun J and Yin M (2025) Gut microbiota and colorectal cancer: mechanistic insights, diagnostic advances, and microbiome-based therapeutic strategies. Front. Microbiol. 16:1699893. doi: 10.3389/fmicb.2025.1699893
Edited by:
Roshan Kumar, Medical College of Wisconsin, United StatesReviewed by:
Zhengrui Li, Shanghai Jiao Tong University, ChinaCopyright © 2025 Bai, Ma, Xu, Chen, Chen, Lv, Su, Li, Sun, Zhang, Xiang, Li, Wu, Sun and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingbing Bai, YmFpYmJAY3F1LmVkdS5jbg==; Jian Sun, c3VuamlhbkBjcXUuZWR1LmNu; Mingzhu Yin, eWlubWluZ3podUBjcXUuZWR1LmNu
†These authors have contributed equally to this work
 Bingbing Bai
Bingbing Bai Jianing Ma
Jianing Ma Wenlong Xu1,2,3,4†
Wenlong Xu1,2,3,4† Chao Lv
Chao Lv Wei Su
Wei Su Yaoxu Li
Yaoxu Li Hongyin Sun
Hongyin Sun Zhongsha Li
Zhongsha Li Yuesong Wu
Yuesong Wu Jian Sun
Jian Sun