- 1School of Forestry and Protoculture, Ningxia University, Yinchuan, China
- 2School of Agriculture, Ningxia University, Yinchuan, China
- 3Department of Pharmacotherapy and Translational Research, University of Florida, Gainesville, FL, United States
Introduction: Arbuscular mycorrhizal fungi (AMF) are symbiotic microorganisms that exert positive effects on their host plants. However, their colonization and community diversity in wine grapes remain unclear.
Methods: This study investigated roots and rhizosphere soils from Cabernet Sauvignon grapevines in vineyards in seven ecological regions at the eastern foot of the Helan Mountains in Ningxia, China. We employed Illumina MiSeq high-throughput sequencing to analyze AMF community composition and diversity in the rhizosphere soil, and examined the effects of soil factors on AMF communities.
Results: The results showed that the grapevine root system was colonized by AMF, with significant spatial heterogeneity in colonization rates and spore densities across the sample plots. Differences in the diversity of the AMF communities in the rhizosphere soil of wine grapes in the different sample plots were observed, and these AMF communities were further divided into three groups. In total, 168 operational taxonomic units were detected in the rhizosphere soil, corresponding to 40 AMF species from five orders, seven families, and seven genera. The Glomus and Glomus melanosporum were the dominant genus and species, respectively. Claroideoglomus and Glomus were identified as biomarkers. Soil pH and organic matter were key factors influencing AMF colonization, abundance, diversity, and community composition.
Discussion: The grape rhizosphere in this region hosts a rich diversity of AMF. This finding provides a reference for the protection and commercial cultivation of AMF in wine grape rhizospheres.
1 Introduction
Arbuscular mycorrhizal fungi (AMF), obligate symbiotic fungi belonging to the phylum Glomeromycota and Mucoromycota (Lutz et al., 2025), are a crucial group of soil microorganisms that are widespread in natural ecosystems (Liang et al., 2022; Xu et al., 2022). AMF make symbiotic associations with more than 80% of terrestrial plant roots (Luginbuehl et al., 2017; Liao et al., 2018; Ma et al., 2022). In this relationship, AMF obtain photosynthetic metabolites from plants and improve water and mineral nutrient uptake (Mickan et al., 2016; Xu et al., 2022) enhancing plant adaptability (Shaul-Keinan et al., 2002). Considering their ecological significance and the pervasive occurrence of their symbiotic associations, elucidating the factors that modulate AMF community composition is pivotal to understanding their distribution patterns and optimizing their ecological functions.
AMF community composition is affected by geographical area (Zhang et al., 2021), environmental conditions (Jansa et al., 2014; Yang et al., 2016), management measures (Geel et al., 2015), host plants (Vályi et al., 2014) and other factors. At local scales, host plants and environmental factors are the primary drivers of AMF community formation. While differences in host plants at the species or genotypes influence the structure of AMF communities (Zeng et al., 2023), studies have found that environmental factors have a greater impact on root AMF community structure. Among these, soil pH is considered one of the factors most closely related to the root AMF community (Han et al., 2023; Xu et al., 2023). In addition, soil chemical factors such as total nitrogen (Han et al., 2020; Lu et al., 2022), organic carbon (Garo et al., 2022), total phosphorus (Zhang M. et al., 2022), available potassium (Wang et al., 2024), available phosphorus (Garo et al., 2022; Xu et al., 2023); climatic characteristic factors such as precipitation (Li et al., 2021); and geographical factors, such as altitude (Rasmussen et al., 2022) and slope direction (Wang et al., 2022) have been reported to affect the AMF community structure of roots. These studies underscore the complexity of the influence of soil factors on the formation of rooted AMF communities, highlighting the need to distinguish between ecosystems for in-depth analysis.
Grapes (Vitis vinifera L.) are a globally cultivated perennial fruit crop prized for their high nutritional value and extensive history of cultivation (Massa et al., 2020). Investigations on grapes are primarily concentrated on postharvest technology (Mencarelli and Bellincontro, 2020), disease and pest management (Girardello et al., 2019), terpenoids (Mele et al., 2021), variety breeding (Fernández-López et al., 2022) and wine aroma (Liu et al., 2017). However, the exploration of beneficial microbial resources, particularly the diversity of symbiotic AMF, remains limited. Schreiner and Mihara (2009) demonstrated the stability of V. vinifera root AMF diversity in Oregon vineyards across seasons, with observed variations based on soil type and vine age. Subsequent research further elucidated the influence of soil depth, management zones, and timing of the growing season on the grape root AMF community (Schreiner, 2020). However, studies specifically targeting AMF community diversity associated with Chinese wine grapes remain notably scarce.
The eastern foot of the Helan Mountains (EFHM) in Ningxia is a prominent geographical indication product protection area for wine in China (Zhang Z. et al., 2022). Located in the global “golden zone” for grape cultivation, this region boasts significant international acclaim (Hu et al., 2021). Fang et al. (2011) investigated the spatial distribution of root AMF in V. vinifera cv. Cabernet Sauvignon across five distinct vineyards in northwest China. Their findings confirmed the beneficial symbiotic relationship between grapes and AMF, with Glomus species being dominant. Common species such as G. constictum, G. intraradices, G. mosseae, and G. versiforme and rare species such as G. claroideum, G. coronatum, G. etunicatum, G. luteum, A. rehmii, and A. rugosa haved been identified in the rhizosphere of grapes in the EFHM. Zhang et al. (2025) observed differences in the diversity and composition of rhizosphere soil AMF communities among different wine grape varieties in the EFHM. Specifically, they observed the richest AMF community composition for Cabernet Sauvignon. Nevertheless, comprehensive assessments of AMF colonization and community composition in the root system and rhizosphere soil across different wine grape ecotopes in this region remain unexplored. Our study applied high-throughput sequencing to investigate the rhizosphere soils of V. vinifera cv. Cabernet Sauvignon grapes in various ecological regions within the EFHM. We classified and identified the isolated AMF spores based on their morphological characteristics. The acetic acid-ink staining method was used to observe the AMF infection status in grapevine roots. Specifically, the objectives were to explore the colonization patterns of AMF in wine grape roots, analyze AMF diversity in wine grape rhizosphere soil, elucidate the community composition and distribution of AMF in wine grape rhizosphere soil, and examine the relationships between AMF community diversity and soil factors. This study lays the groundwork for maximizing the utilization of AMF resources and the development of specialized microbial fertilizers tailored to wine grapes.
2 Materials and methods
2.1 Study region
The study was conducted in a grape-producing area in the EFHM in the Ningxia Hui Autonomous Region of China. This region, located at 37°43′–39°23′N and 105°45′–106°47′E on the western edge of the Yinchuan Plain and with an average altitude of 1,118–1,333 m, is one of China’s most important grape-producing areas (Figure 1). The area experiences a temperate continental arid and semi-arid climate, with daily temperature variations of 12 to 15 °C and annual precipitation of 150–240 mm. The soil in this region is primarily composed of sand, silt, and gravel, with low organic matter content and good aeration and permeability. These unique geographical and environmental conditions are conducive to the growth of wine grapes.
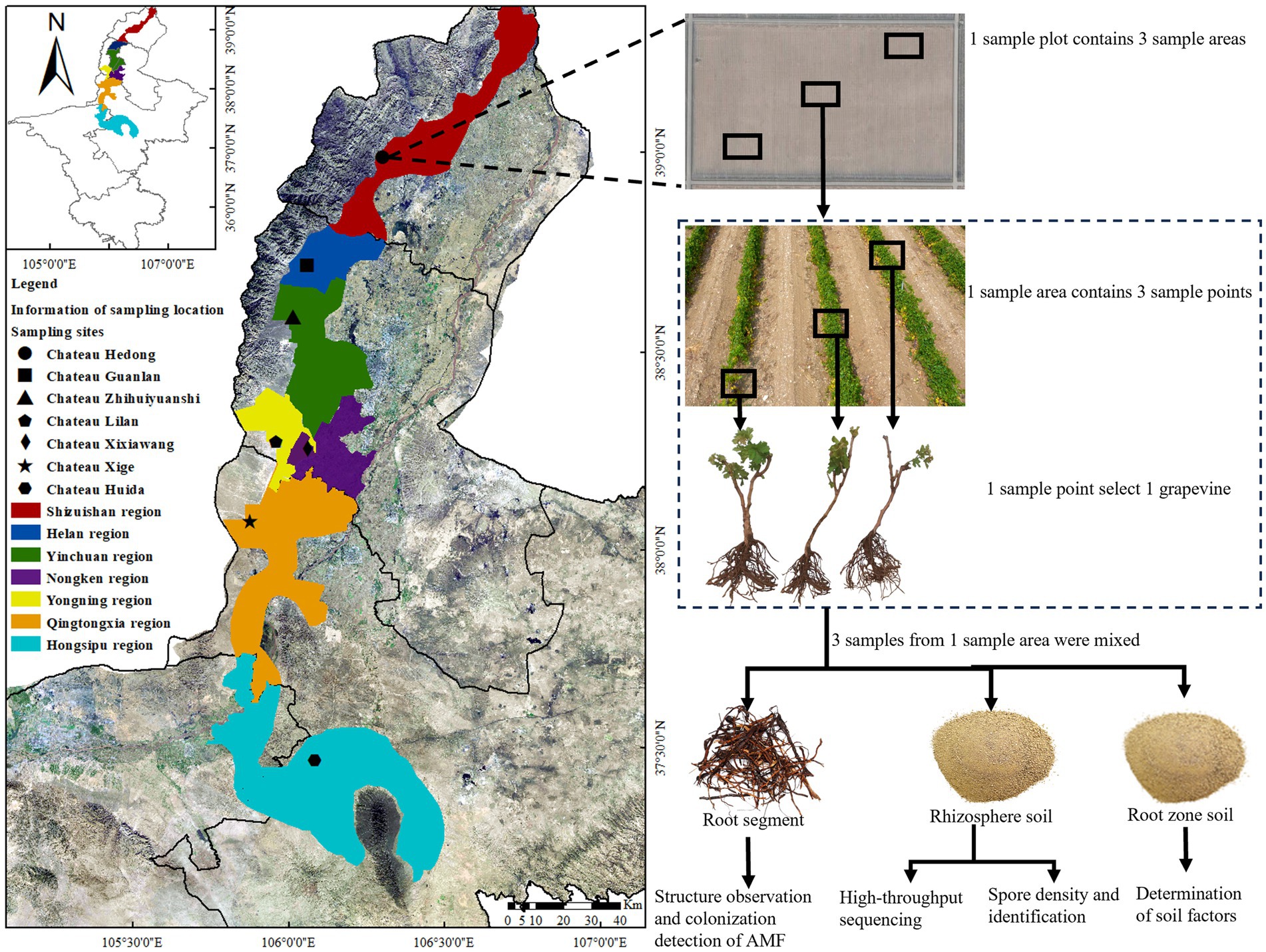
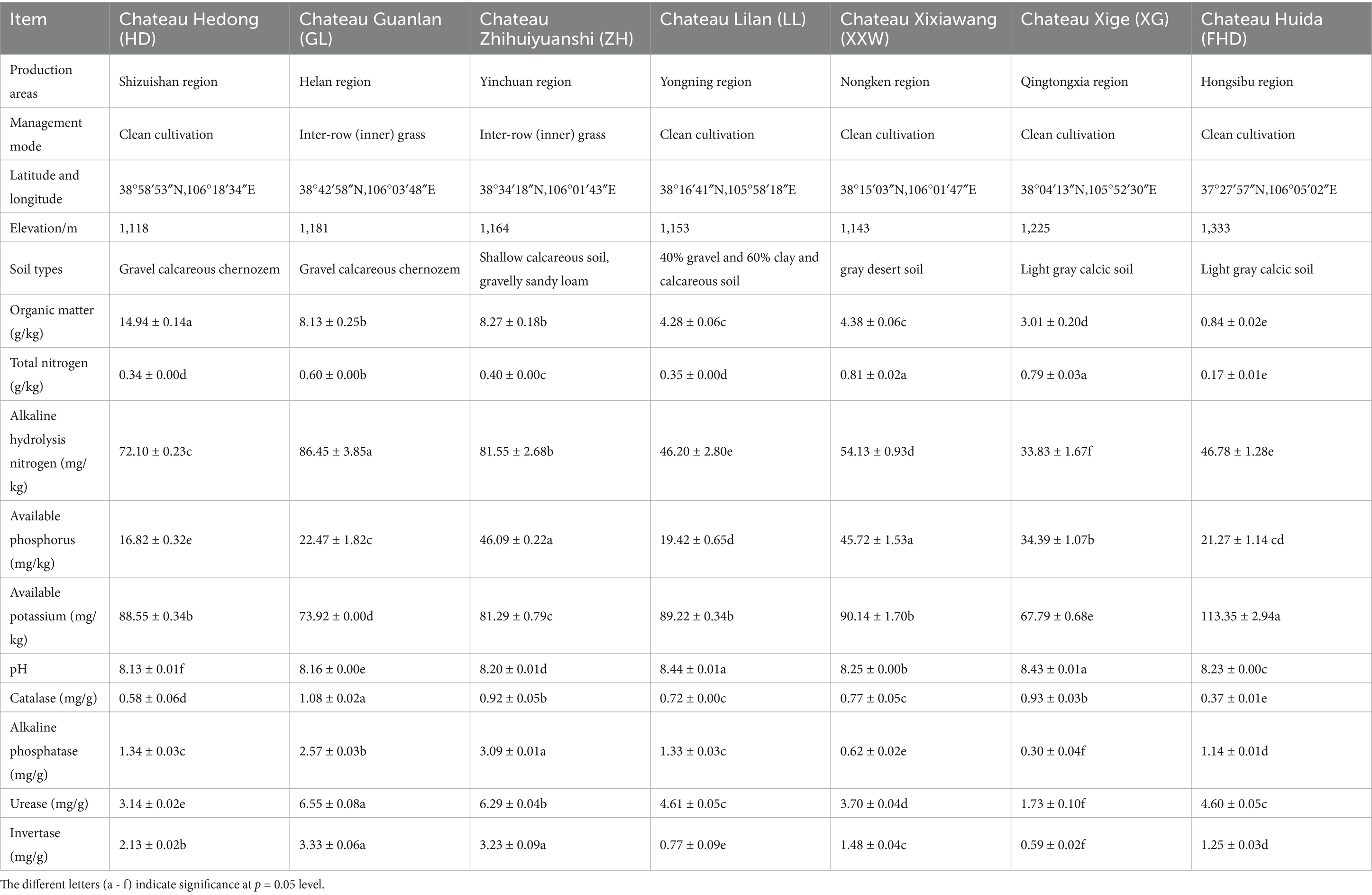
Unlike most wine-producing regions globally, in the western part of China, including the EFHM, the temperature in winter reaches as low as −20 °C. Therefore, to mitigate the risk of grapevine damage due to freezing, it is necessary to prune vines and bury them in the soil in November. The grapevines in this region predominantly consist of self-rooted seedlings, as opposed to grafted rootstocks, which is a key factor in the successful implementation of this burying method. These regions of western China are designated as “buried cold regions”. Based on different factors such as climate, soil type, and altitude, the grape-producing area was divided into seven ecological regions, predominantly cultivating V. vinifera cv. Cabernet Sauvignon, Merlot, Syrah, Chardonnay, and Riesling varieties. In this study, representative winery vineyard plots were selected as sample collection points for seven ecological regions. To reduce experimental error, seven Cabernet Sauvignon vineyards with vines aged >8 years were selected. The vineyard locations were Chateau Hedong (HD) in the Shizuishan production area, Chateau Guanlan (GL) in the Helan production area, Chateau Zhihuiyuanshi (ZH) in the Yinchuan production area, Chateau Lilan (LL) in the Yongning production area, Chateau Xixiawang (XXW) in the Nongken production area, Chateau Xige (XG) in the Qingtongxia production area, and Chateau Huida (FHD) in the Hongsipu-production area. The general characteristics of the seven winery plots are shown in Figure 1 and Table 1.
2.2 Experimental design and sampling strategy
In August 2022, we collected root segments and rhizosphere soil samples from V. vinifera cv. Cabernet Sauvignon wine grapes from seven vineyard plots (HD, GL, ZH, LL, XXW, XG, and FHD). Each plot covered approximately 6.67–8 hm2 and the same cultivation and management practices were followed. Before sampling, we conducted preliminary observations by excavating the roots of grape. We found that the roots were primarily distributed within 0–60 cm of the soil surface. Consequently, the sampling range was set at this depth.
At each plot, we established three sampling points (20 × 20 m) using the diagonal sampling method, ensuring each point was more than 100 m apart. After removing the surface gravel and dead weeds, we randomly selected three healthy grapevines at each sampling point. Lateral root segments (diameter: 0.1–0.5 mm and length: 10–100 mm) and rhizosphere soil samples were collected from the 0–60 cm soil layer, within a 0–30 cm radius around the main stem of the grapevines. Nine grape trees were collected from each plot. The soil adhering to the roots was defined as the rhizosphere soil (Zhou et al., 2019). The sampling method was as described by Liu et al. (2022). We used a sterile brush to brush the soil tightly attached to the root surface and then passed it through a 1 mm soil sieve to prevent excess dead root bark litter from entering the sample. After the rhizosphere soil of three vines in each sample point was fully mixed, we retained 5 g soil samples using the quartering method, placed them into 2 mL sterile centrifuge tubes, transported them to the laboratory in a cooling box, and froze them at −80 °C for high-throughput sequencing. The remaining rhizosphere soil was stored at 4 °C for AMF spore identification and spore density detection. Grape root segments were cut using sterile scissors, washed with sterile water, and stored in formalin-aceto-alcohol (FAA) fixative solution (V (38% formaldehyde): V (glacial acetic acid): V (75% alcohol) = 1:1:18) for morphological structure observation and AMF colonization detection. The soil in the plant growth area was defined as the root zone soil (Shi et al., 2019), and 500 g of root zone soil was collected at each sample point for the determination of soil physical and chemical factors. A total of 21 root segment samples, 21 rhizosphere soil samples (HTS), 21 rhizosphere soil samples (spore identification and spore density detection of AMF) and 21 root zone soil samples were collected from 63 grape trees in seven plots, with three replicates for each treatment (Figure 1).
2.3 Observation of AMF structure and infection in wine grape roots
Wine grape root AMF colonization was stained according to the methods described previously by Phillips and Hayman (1970). The root segments were taken out from the FAA and cut into 1 cm. For each sample, 100 roots were selected, transferred into 10% KOH at 90 °C for 1 h, acidified with lactic acid for 5 to 10 min, dyed with 0.05% (v/v) acetic acid-ink staining solution (95 mL of white vinegar, 5 mL of Quink ink) for 5 min, and decolored with lactic acid glycerol (the volume ratio of lactic acid and glycerol was 1:1) for 12 h. After decolorization, the samples were observed under an Olympus BX53 microscope (Olympus, Japan) at 200 × magnification, and the root AMF colonization status was assessed using the intersection method (McGonigle et al., 1990)—30 visual fields were examined for each root segment to ensure data representativeness. The root AMF colonization rate and colonization intensity were calculated as follows (Equations 1, 2) (Li et al., 2011; Yan et al., 2014):
2.4 Isolation and identification of AMF in wine grape rhizosphere soil
After the rhizosphere soil samples were dried in the shade, AMF spores were isolated through wet sieve precipitation-sucrose centrifugation (Song et al., 2019). The number of spores was observed and recorded under an Olympus BX53 microscope (Olympus, Japan) at 200 × magnification, and the number of AMF spores per 50 g of air-dried soil was counted as the spore density. Morphological features of spores and the phenotypic and histochemical characters of spore wall layers were determined as described by Omar et al. (1978), using a mixture of polyvinyl-lactic acid-glycerin (PVLG) and Melzer’s reagent (1:1, v/v). The preparation of spores for study, determination of color, and photography of spores and mycorrhizal structures were performed as previously described (Blaszkowski et al., 2012). Species identification was performed according to “Manual for the Identification of Va Mycorrhizal Fungi” (Schenck and Perez, 1990) and the latest taxonomic description and pictures provided by the International Culture Collection of (vesicular) Arbuscular Mycorrhizal Fungi (INVAM) on https://invam.ku.edu/ (Sturmer et al., 2021).
2.5 Determination of soil factors
Root zone soil samples were sieved through a 1 mm mesh for the determination of soil physicochemical properties, following the method described by Bao (2000): soil organic matter (SOM) was determined via potassium dichromate-ferrous sulfate titration; total nitrogen (TN) via Kjeldahl digestion; alkali-hydrolyzable nitrogen (AN) via alkali diffusion; pH via a PHS-25 (Shanghai Leici, China) precision pH meter; available phosphorus (AP) via molybdenum-antimony anti-colorimetry; and available potassium (AK) via flame photometry.
Reserved root zone soil samples were sieved through a 0.25 mm mesh for soil enzyme activity analysis, following the method described by Guan (1986). Sucrase activity was determined using 3,5-dinitrosalicylic acid colorimetry, expressed as mg glucose per gram of soil after 24 h; urease activity via indophenol blue colorimetry, expressed as mg NH₃-N per 5 grams of soil after 24 h; catalase activity via potassium permanganate titration, expressed as mL of 0.1 N potassium permanganate per gram of soil after 20 min; and alkaline phosphatase activity via sodium phenyl phosphate colorimetry, expressed as mg phenol released per gram of soil after 24 h.
2.6 Fungal community identification with Illumina MiSeq sequencing
2.6.1 DNA extraction and PCR amplification
For DNA extraction, microbial DNA was isolated from the prepared soil samples (0.25 g soil per sample) using the Fast DNA® SPIN Kit for Soil (QBIOgene Inc., Carlsbad, CA, United States), following the manufacturer’s protocols with strict adherence to ensure high-quality DNA recovery. For the PCR amplification, nested PCR was conducted to amplify the AMF 18S rRNA. The primer pairs AML1 (5′-ATCAACTTTCGATGGTAGGATAGA-3′) and AML2 (5’-GAACCCAAACACTTTGGTTTCC-3′) (Lee et al., 2008) were used in the first run. The primer pairs AMV4.5NF (5’-AAGCTCGTAGTTGAATTTCG-3′) and AMDGR (5’-CCCAACTATCCCTATTAATCAT-3′) were used in the second run (Van Geel et al., 2014). The reaction volume was 20 μL, consisting of 4 μL 5 × TransStart FastPfu Buffer, 2 μL of dNTPs (2.5 mmol/L), 0.8 μL each forward and reverse primers (5 μmol/L), 0.4 μL TransStart FastPfu Polymerase (2.5 U/μL), 10 ng/L DNA template, and ddH2O to 20 μL. The PCR procedure was as follows: 95 °C for 3 min; 95 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s; and 72 °C for 10 min and 10 °C for holding. The annealing temperature was 55 °C in the first run for 32 cycles and 55 °C in the first run for 20 cycles. DNA quality was checked using 2% agarose gel electrophoresis.
2.6.2 Illumina sequencing
Purified amplicons were pooled in equimolar amounts, and paired-end sequencing (2 × 300 bp) was conducted using an Illumina Nextseq2000 platform (Illumina, San Diego, CA, United States) according to the standard protocols of Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Sequence read processing was performed using QIIME v.1.9.1 (Wassermann et al., 2019). The raw reads were stored in the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA966782) https://www.ncbi.nlm.nih.gov/sra/PRJNA966782.
2.6.3 Bioinformatics analysis
Raw FASTQ files were demultiplexed, quality-filtered using Trimmomatic (Bolger et al., 2014), and merged using FLASH with the following criteria: reads were subjected to truncation at any site that received an average quality score of less than 20 over a 50-base pair sliding window. Primers were matched exactly, with a maximum of two nucleotide mismatches permitted, and reads containing ambiguous bases were removed. Sequences exhibiting a base-pair overlap greater than 10 were amalgamated based on the overlapping sequences. Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (v.7.1; http://drive5.com/uparse/) (Edgar, 2013), and chimeric sequences were identified and removed using UCHIME (Edgar et al., 2011). The taxonomy of each 18S gene sequence was analyzed using RDP Classifier (v.2.2, http://sourceforge.net/projects/rdp-classifier/) and the maarjam20220506/AM1 18S database, with a confidence threshold of 95% (Luo et al., 2020).
2.7 Data analysis
The alpha diversity indices of the samples, including Simpson even (reflecting community uniformity), Shannon (reflecting community diversity), Chao (reflecting community richness), and PD (reflecting the pedigree diversity of species), were calculated using Mothur software (version v.1.30.2) (Schloss et al., 2009). SPSS 23.0 Duncan’s multiple range test was used for the inter-group test of AMF total colonization rate, colonization intensity, and alpha diversity indexes (Ju et al., 2022). Based on the Qiime software (version 1.9.1) (Bolyen et al., 2019), the beta diversity distance matrix was calculated and the Adonis inter-group difference test results were integrated to construct the principal coordinate analysis (PCoA) diagram (Yan et al., 2022) to determine the differences in AMF among samples. Permutational analysis of variance (PERMANOVA) (Ziegler et al., 2019) was used to decompose the total variance using a semi-metric (Bray-Curtis), analyze the explanatory power of different grouping factors on sample differences, and use a substitution test to analyze the statistical significance of the division. The species composition and relative abundance of each species in each sample were determined at different taxonomic levels and the composition of the dominant species in the different samples was visualized using Pie and Bar diagrams. A Venn diagram (Yan et al., 2022), was used to show the number of shared and unique OTUs of AMF in different groups. LEfSe multi-level species difference discriminant analysis (Segata et al., 2011) was used to evaluate the flora that had a significant impact on sample differences. The free online platform Majorbio I-Sanger Cloud Platform was used to analyze microbiological data.2 Soil factors were screened using Variance Inflation Factor (VIF) analysis in the R software vegan package (vsesion2.4.3). VIF analysis was performed on soil factors across different sampling sites, filtering out those with VIF > 10. This screening process was repeated until all selected environmental factors had a VIF < 10. The correlations between soil factors and AMF colonization were analyzed using origin2022, while correlation analysis and mapping between soil factors and AMF communities were performed using RDA in the R software vegan package (version 2.4.3).
3 Results
3.1 AMF colonization and identification of soil spores of wine grape roots in the EFHM
AMF invaded the roots of wine grapes, formed a typical AMF structure (arum-type structure), and established a good plant symbiotic relationship (Figure 2). AMF hyphae spread and grew outside the root system of host wine grapes to form extra root hyphae (Figure 2A). It also infected the root system of wine grapes and formed intercellular hyphae in the intercellular spaces of cortical cells. Some intercellular hyphae were straight (Figure 2B), some formed binary branches (Figure 2C), and some were entangled to form mycelial circles (Figure 2D) and intracellular arbuscular structures (Figures 2B–F). Most of the hyphae were non-septate hyphae, although and septate hyphae were formed (Figure 2C). The tops of the hyphae expanded to form vesicles of varying sizes and shapes (Figures 2F–H). The vesicles were mostly round, oval, or irregular in shape and some had inclusions (Figures 2G,H).
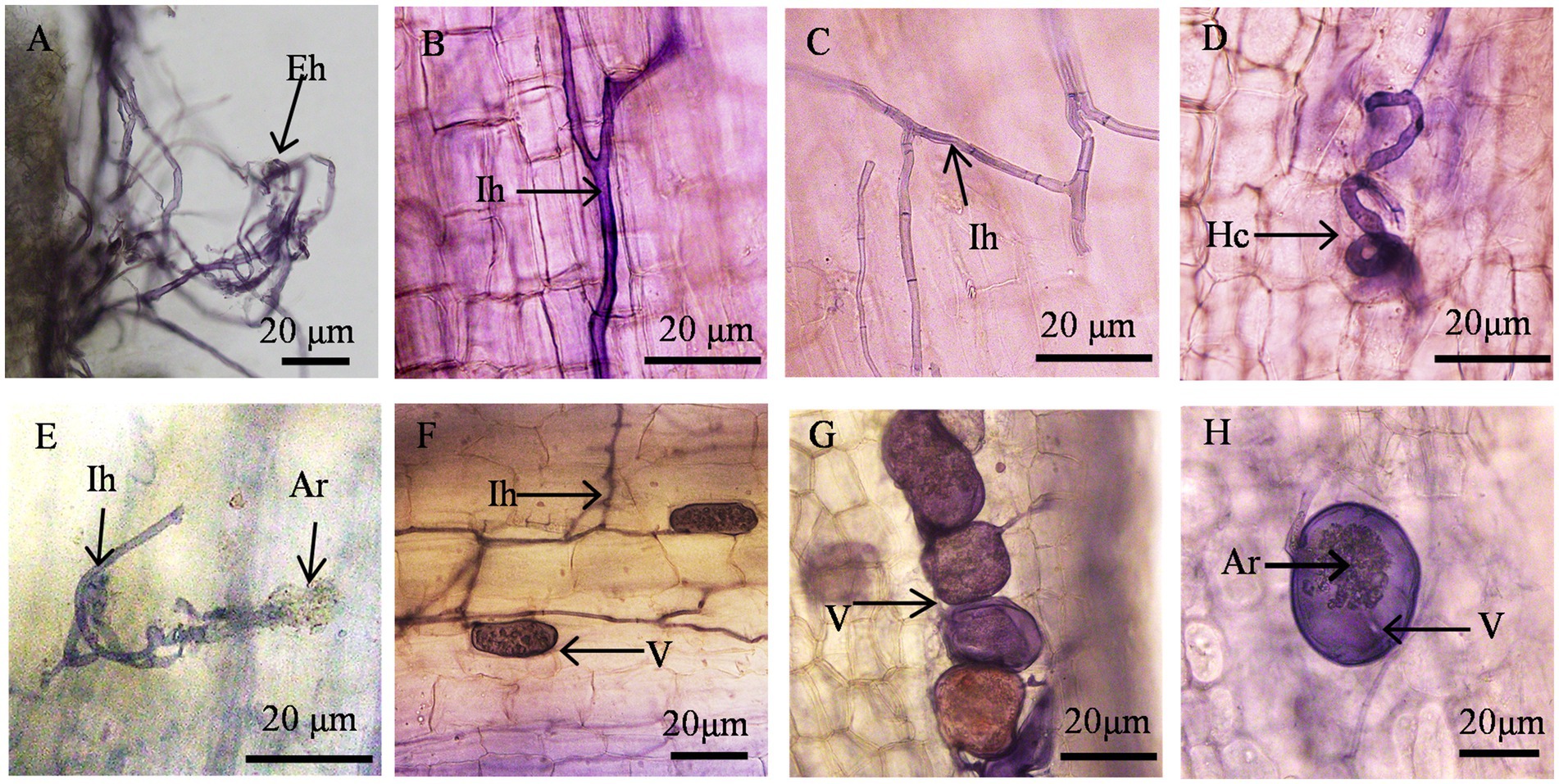
Figure 2. Structure of AMF Colonization in the Root Systems of Wine Grapes. (A) Hyphae (Eh); (B, C) Internal hyphae (Ih); (D) Circular hyphae (Hc); (E) Internal hyphae (Ih) and arbuscule (Ar); (F) Vesicle (V) and internal hyphae (Ih); (G, H) Vesicle (V) and arbuscule (Ar).
As shown in Figure 3A and Supplementary Table S1, the spore density of AMF in the rhizosphere differed by sample plot. The average spore density of AMF was the highest at ZH, at 252 per 50 g, followed by LL, at 199.63 per 50 g. XXW had the lowest, at 63 per 50 g. The root samples of wine grapes in each sample plot were infected with AMF, with colonization rates ranging from 6.28 to 70.04%. Among them, the AMF colonization rate of ZH was the highest (70.04%) and that of HD was the lowest (6.28%; Figure 3B). The AMF colonization intensity in ZH was significantly higher than that observed in the other plots (p < 0.05), followed by XG and HD, which were 75.52, 67.41, and 25.14%, respectively (Figure 3C). These data indicate that wine grapes are susceptible to AMF infection.
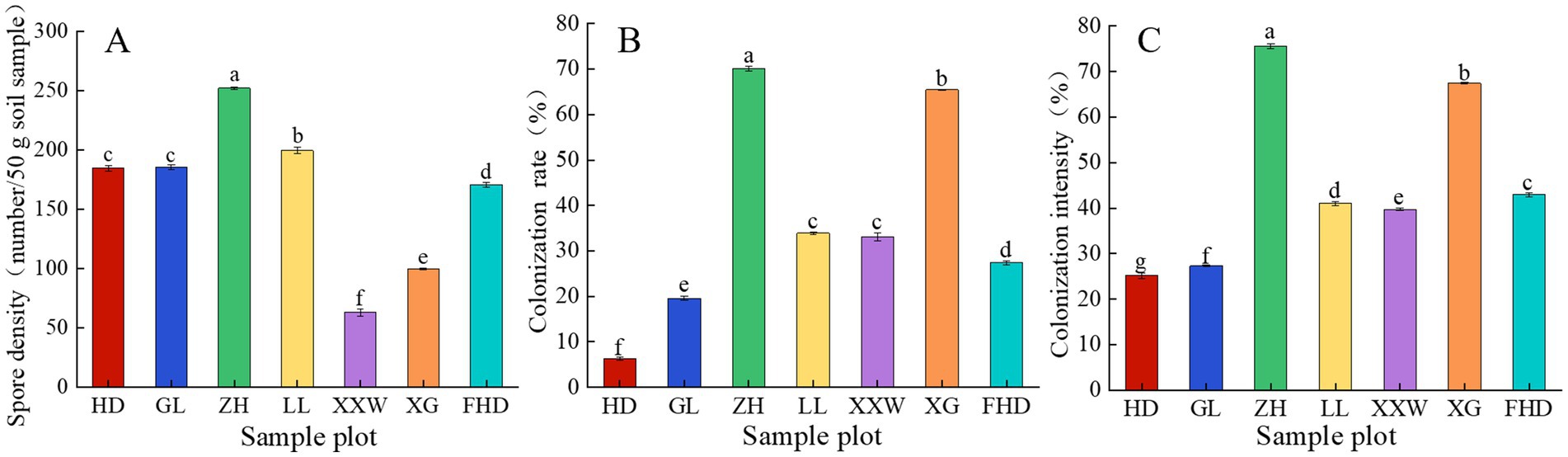
Figure 3. AMF spore density (A), colonization rate (B) and colonization intensity (C) of grapevines at different sample plots.
The morphological characteristics of AMF spores were meticulously documented through microscopic observation, and identified by VA mycorrhizal identification manual. A total of 16 species of AMF belonging to 6 genera were identified (Figure 4; Supplementary Table S2). There were ten species of Glomus, two species of Claroideoglomus, one species of Diversispora, one species of Paraglomus, one species of Acaulospora and one species of Scutellospora. Glomus was the dominant genus, and G. melanosporum was the dominant species (Supplementary Table S2). Claroideoglomus, Diversispora and Paraglomus were first discovered in grape plants in Northwest China.
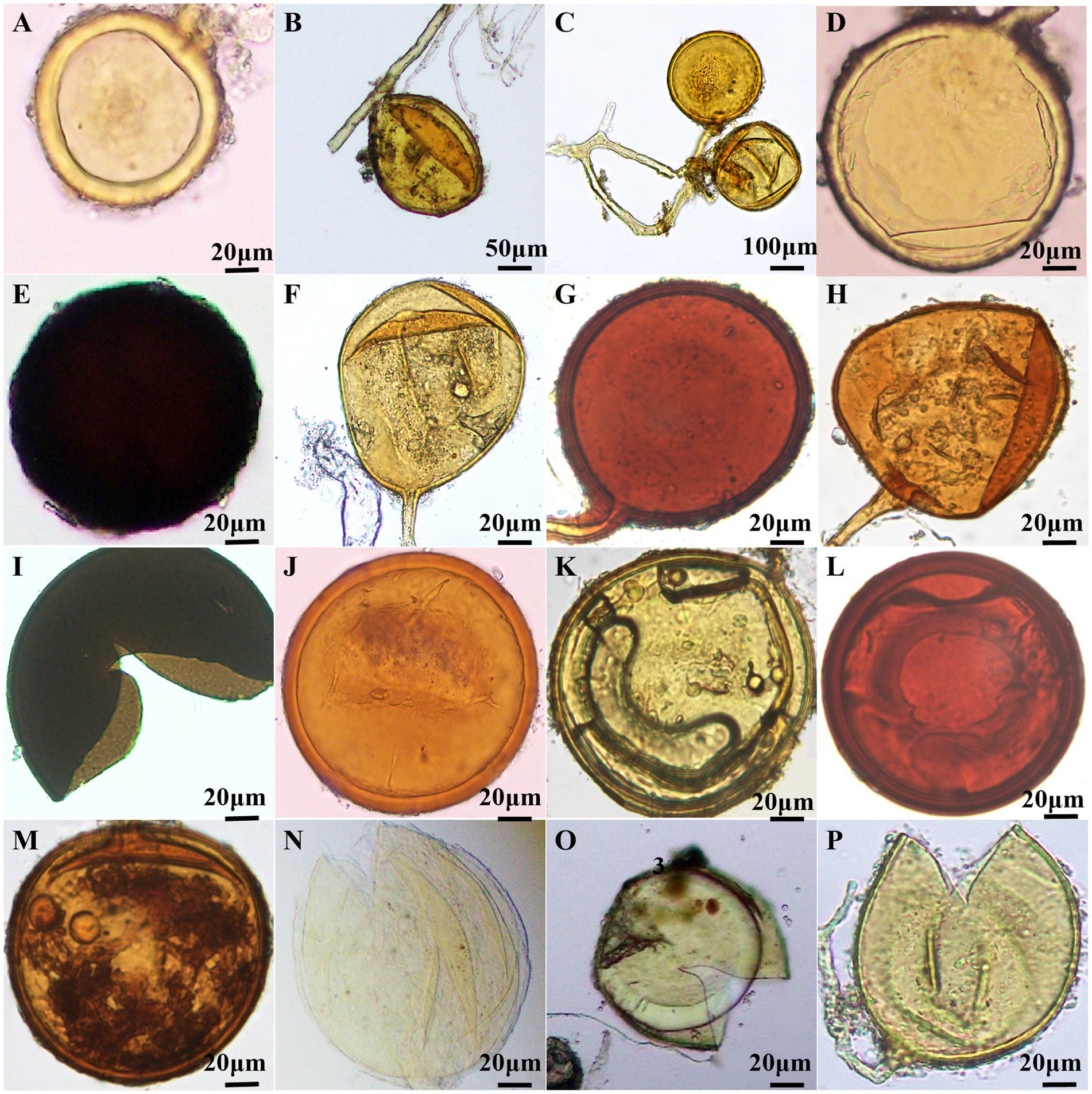
Figure 4. Morphology of AMF spores in wine grape roots. (A) C. claroideum; (B) C. etunicatum; (C) Glomus aggregatumv; (D) G. glomerulatum; (E) G. melanosporum; (F) G. pansihalos; (G) G. reticulatum; (H) G. Dolichosporm; (I) G. constrictum; (J) G. multiforum; (K) G. clarum; (L) G. halonatum; (M) D. etunicatum; (N) P. brasilianum; (O) A. dilatata; (P) S. calospora.
3.2 Analysis of AMF community diversity in rhizosphere soil of wine grape in EFHM in Ningxia
3.2.1 AMF α diversity in wine grape rhizosphere soil
High-throughput sequencing was performed on the rhizosphere soil from different sample plots and 1,864,896 raw reads were obtained, with a total base number of 559,468,800. After optimizing the data, 1,258,505 valid sequences were obtained, with an average sequence length of 216 bp and base number of 271,975,386 (Supplementary Table S3). Based on a classification level of 97% similarity, a total of 168 OTUs were annotated. The dilution curve (Supplementary Figure S1) showed that, with an increase in the number of sequencing samples, the OTU dilution curve of the rhizosphere soil samples of wine grapes gradually tended to be gentle, indicating that the amount of sequencing data in this experiment was sufficient to fully reflect the AMF species diversity in the rhizosphere soil of different wine grape varieties.
As shown in Figure 5, variations in AMF α-diversity were observed across plots. The Chao index of LL was significantly higher than that of the other plots (p < 0.05). The ZH plot (17.67) and XXW plot (17.67) ranked second, while the FHD plot had the lowest Chao index (10.33). This indicates that the LL plot had the highest AMF community richness, whereas the FHD plot had the lowest. The Shannon index of the LL plot (2.06) was significantly higher than that of the other plots (p < 0.05), with the GL plot (1.58) ranking second and the FHD plot (0.77) having the lowest, indicating that the community diversity of the LL plot was the highest and that of the FHD plot was the lowest. The Simpson-even index (0.36) of the LL plot was the largest, and the Simpson-even index (0.09) of the ZH plot was the smallest, indicating that the uniformity of the LL plot was the highest and that of the ZH plot was the lowest. The highest PD index was observed in the LL plot (2.10), followed by the ZH plot (3.07), whereas the lowest was observed in the HD plot (1.63). The diversity of the AMF lineages in the rhizosphere soil of wine grapes in the seven sample plots was not significantly different (Supplementary Table S4).
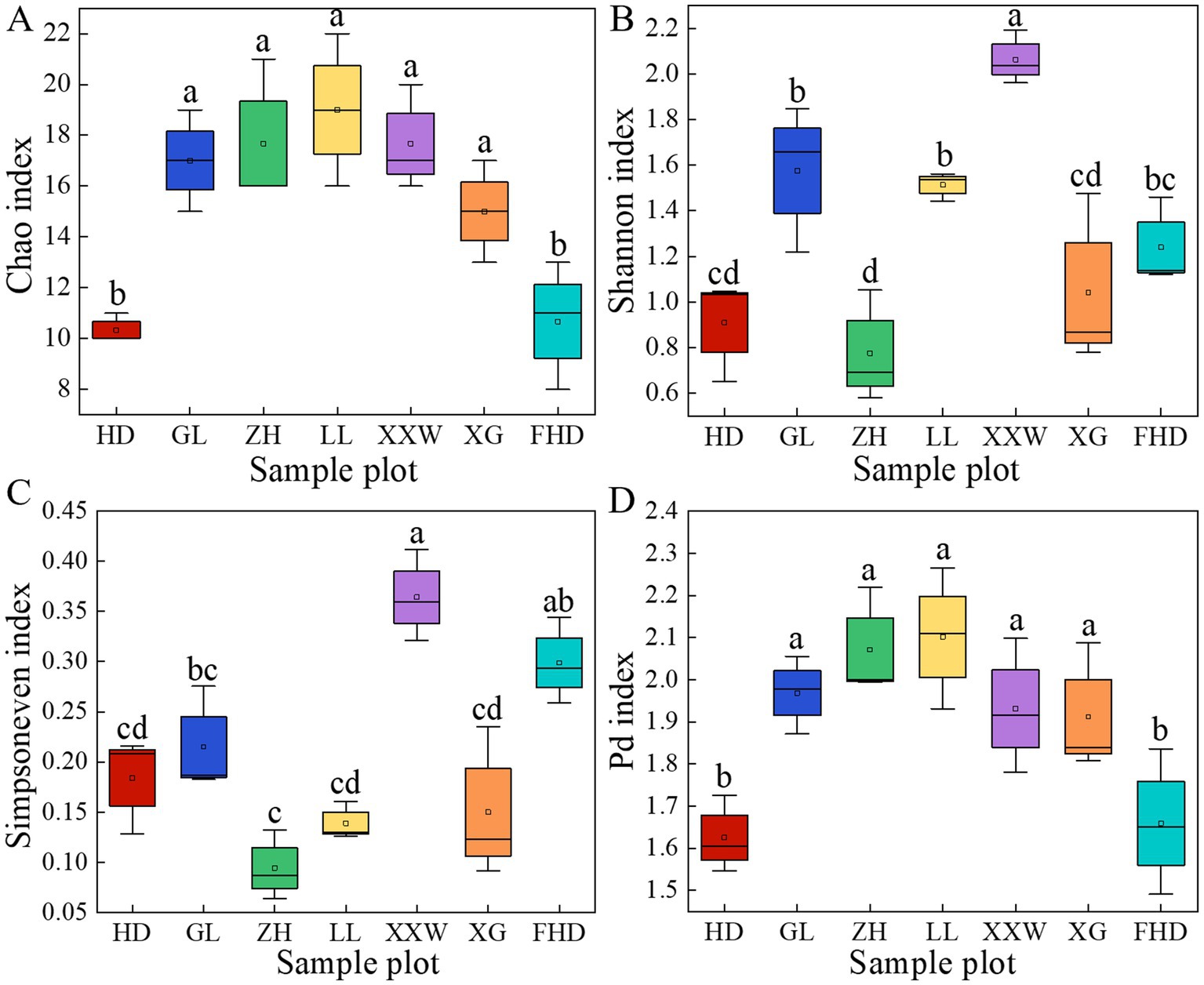
Figure 5. The α diversity index of AMF community in rhizosphere soil of wine grape at species level in different plots. (A) The Chao index represents community richness; (B) The Shannon index represents community diversity; (C) The Simpson evenness index represents community evenness; (D) The Pd index (phylogenetic diversity index) represents phylogenetic diversity. The different letters (a - f) indicate significance at p = 0.05 level.
3.2.2 Analysis of AMF β diversity in wine grape rhizosphere soil
The β diversity of AMF communities in the rhizosphere soil of wine grapes in different plots was evaluated at the species level, and the analysis results are shown in Figure 6. In the ANOSIM analysis based on the Bray-Curtis distance algorithm (Figure 6A), Clustering was performed on the AMF communities in the rhizosphere soil of wine grapes in the seven sample plots. The variation among samples was significantly higher than that within groups (p = 0.001), indicating that the selection of sampling points in this experiment was statistically reliable. Furthermore, PCoA revealed distinct differences in the rhizosphere AMF community of wine grapes from different regions (R = 0.7202, p = 0.001; Figure 6B). The PC1 axis explained 33.53% of the data, and the PC2 axis explained 19.54%. The AMF community in the rhizosphere soil of wine grapes in the seven sample plots was divided into three groups. Group I consisted of HD, LL, ZH, and XG. The distribution of samples in these four sample plots was relatively compact, indicating that the similarity of the bacterial community structure was high and the difference in AMF community composition was small. Group II was GL and XXW, and the distance between the two groups was close, indicating that the difference in AMF community structure between the two groups was small. Group III was FHD, which was far from the other six groups, indicating that the similarity of the AMF community structure between FHD and these six groups was low, and the difference was large (Figure 6B).
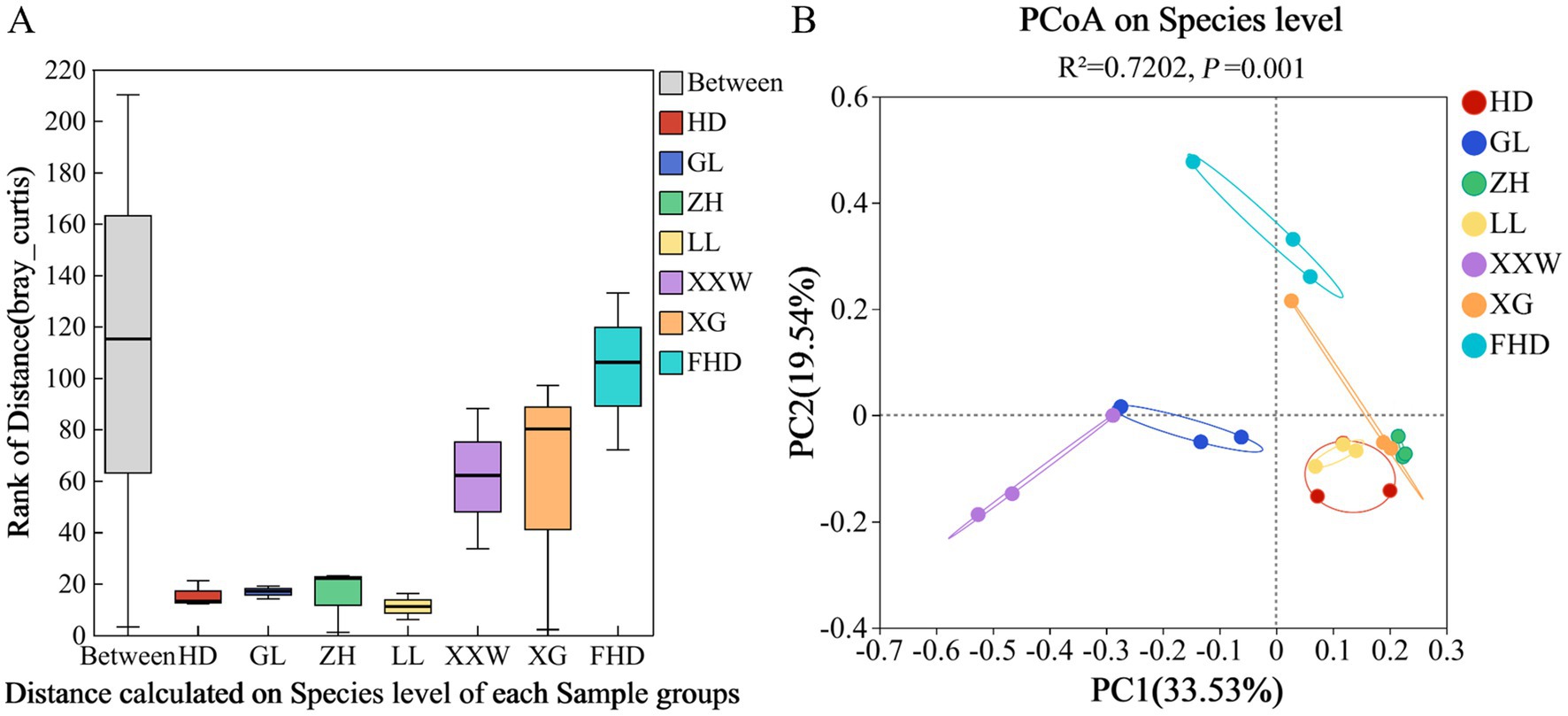
Figure 6. PCoA analysis diagram of soil AMF beta diversity analysis based on the chord algorithm of grapes for different sample plots. (A) The X-axis is the distance value within or between groups, the box corresponding to “Between” represents the distance value of the difference between the groups, and the rest of the boxes represent the distance value of the difference within the group; the Y-axis scale represents the size of the distance value. (B) The X-axis and Y-axis represent the two selected main coordinate axes, and the percentage represents the interpretation value of the main coordinate axis for the difference in sample composition; The scales of the X-axis and Y-axis are relative distances and have no practical significance; Points with different colors or shapes represent different groups of samples, and the closer the two sample points are, the more similar the species composition of the two samples is.
PERMANOVA analysis (Supplementary Table S5) revealed notably high explanatory power (R2 = 0.72) and statistical significance (p = 0.001) for variations in the microbial community of wine grapes across different sample plots. Furthermore, PERMANOVA on low (HD: 1118 m), medium (XG: 1225 m), and high (FHD: 1333 m) altitudes also demonstrated substantial explanatory power (R2 = 0.53) and significance (p = 0.006) for such variations. Collectively, these results indicate that different sample plots and altitudes are important factors contributing to variations in AMF community within the rhizosphere soil of wine grapes.
3.3 AMF community composition of wine grape roots in EFHM in Ningxia
Presently, the membership method is used to define core microbiomes. Membership determines the core microbiome by determining whether there is a common OTU in the host microbiome (Dong et al., 2021). The Venn diagram intuitively reflects the differences in the OTU composition of the AMF community in the rhizosphere soil of different wine grapes and common species. Each sample contained unique OTUs (Figure 7A). The specific OTUs for HD, GL, ZH, LL, XXW, XG, and FHD were 3, 8, 5, 13, 9, 4, and 5, respectively. Fourteen OTUs in the seven plots represented the core microbial group of the soil layer. The shared OTUs were diverse in classification (Supplementary Table S6) but most belonged to Glomus and unclassified Glomeromycetes, indicating that the proportion of the same species in the rhizosphere soil samples of wine grapes in the different sample plots was small.
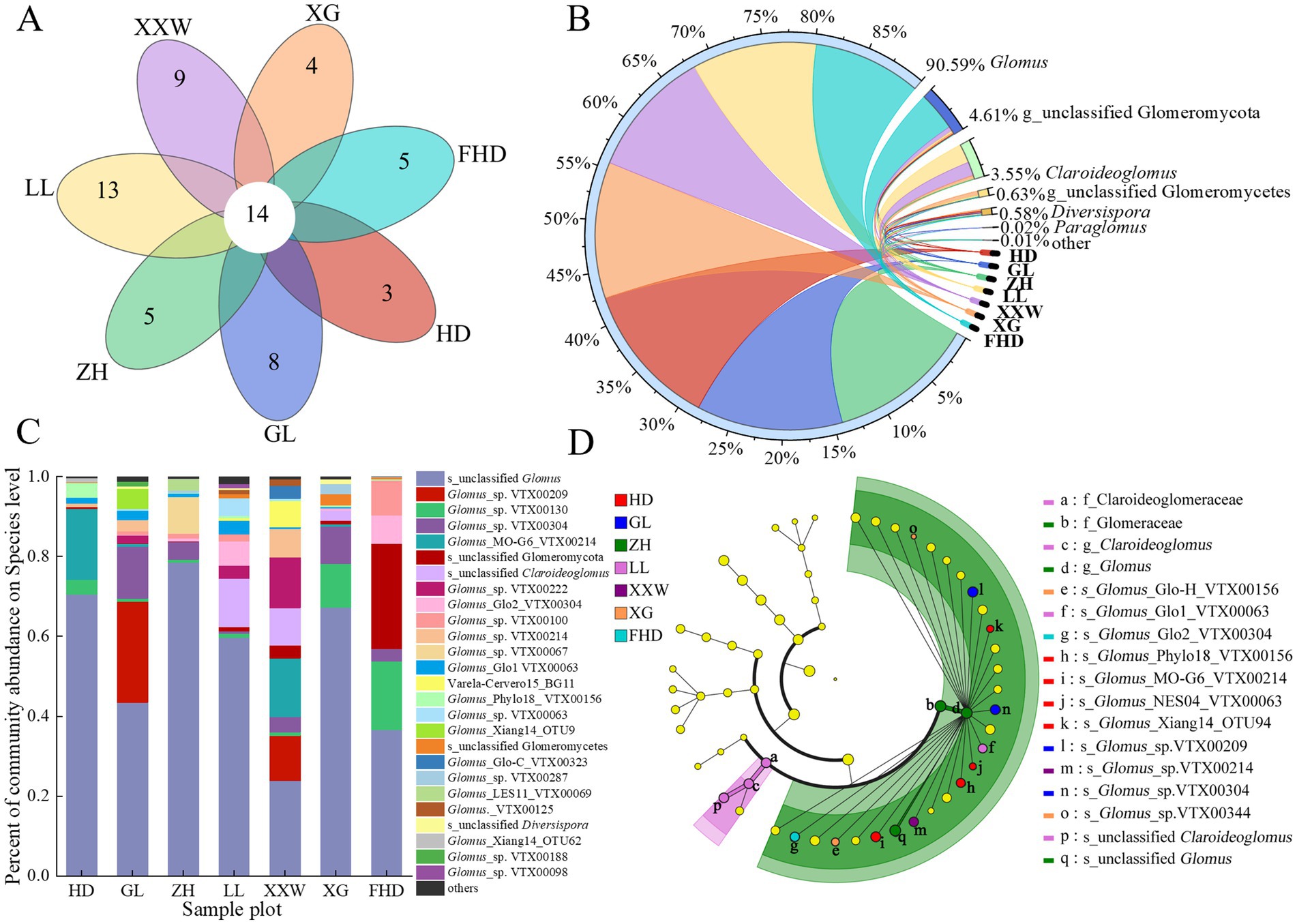
Figure 7. AMF species composition in rhizosphere soil of wine grape in different sample plots. (A) Venn diagram of AMF OTU numbers in inter-root soils of “Cabernet Sauvignon” grapes for different sample plots. (B) Circos map of horizontal species relationship with AMF genus in the rhizosphere soil of wine grapes (the color of the outer ribbon represents the species, the color of the inner ribbon represents the group from which the sample is distributed, and the length represents the distribution proportion of the sample in a certain species). (C) Differences in species composition at the species levels of AMF in inter-root soils of wine grapes at different sample sites (the ordinate is the name of the sample, the abscissa is the proportion of the species in the sample, the columns of different colors represent different species, and the length of the columns represents the proportion of the species). (D) LEfSe hierarchical tree analysis of AMF community composition of wine grapes in different plots (the six large circles are phylum, class, order, family and genus from the inside out. Small circle size represents the relative abundance of the species at the taxonomic level. Species with no significant difference are marked in yellow, while species with significant difference are marked in color).
One phylum, three classes, five orders, seven families, seven genera, and 40 species of AMF were identified in the rhizosphere soil of wine grapes from the seven sample plots. At the genus level, the combined proportion outside this genus was relatively low (p < 0.01). The five genera with the highest relative abundances were Glomus (78.78–99.96%), unclassified Glomeromycota (0.05–26.35%), Claroideoglomus (0.20–12.80%), unclassified Glomeromycetes (0.05–2.81%), and Diversipora (0.18–1.11%) (Figure 7B). At the species level, unclassified Glomus was the dominant species of AMF in the rhizosphere soil of wine grape, with a relative abundance of 23.85–78.55% (Figure 7C). There were substantial disparities in the composition of AMF communities and the prevalence of dominant species in the rhizosphere soil of wine grapes across the diverse sample plots.
LEfSe analysis results indicate that at the species level, the relative abundance of 13 AMFs in the soil of the seven plots showed significant differences (p < 0.05) (Figure 7D), mainly belonging to Claroideoglomus and Glomus. The AMF Claroideoglomus was significantly enriched only in LL (p < 0.05). The AMF of Glomus identified biomarkers in all seven plots, of which HD had the most biomarkers (four biomarkers), followed by XG (two biomarkers) and GL (two biomarkers), while LL, XXW, FHD, and ZH sites each had one Glomus AMF identified as a biomarker.
3.4 Correlation analysis between soil factors and AMF in different sample plots
All plots had alkaline pH, while other soil nutrients showed some variations (Table 1): HD had the highest organic matter (OM) content (14.94 g/kg, p < 0.05). GL exhibited significantly higher contents of alkaline hydrolyzable nitrogen (AN, 86.45 mg/kg), catalase (CAT, 1.08 mg/g), urease (URE, 6.55 mg/g), and sucrase (3.33 mg/g) than other plots (p < 0.05). ZH had the significantly highest contents of available phosphorus (AP, 46.09 mg/kg) and alkaline phosphatase (ALP, 3.09 mg/g) (p < 0.05). Regarding AN content, XXW ranked the highest (0.81 mg/kg). FHD had the significantly highest available potassium (AK) content (p < 0.05). Among these, GL and ZH sites had relatively higher nutrient contents, with more fertile soils. After screening via VIF analysis, the retained soil factors were: soil OM, total nitrogen (TN), AK, AP, pH, and URE—were subjected to correlation analysis (Supplementary Table S7).
Correlation analysis (Figure 8A) revealed the following relationships: AMF colonization rate was significantly positively correlated with soil pH (p < 0.05) and highly significantly positively correlated with AP (p < 0.001), whereas it was significantly negatively correlated with SOM (p < 0.05); colonization intensity showed a highly significant positive correlation with AP (p < 0.001); spore density was significantly positively correlated with SOM (p < 0.05) and urease (highly significant, p < 0.001), but highly significantly negatively correlated with TN (p < 0.001).
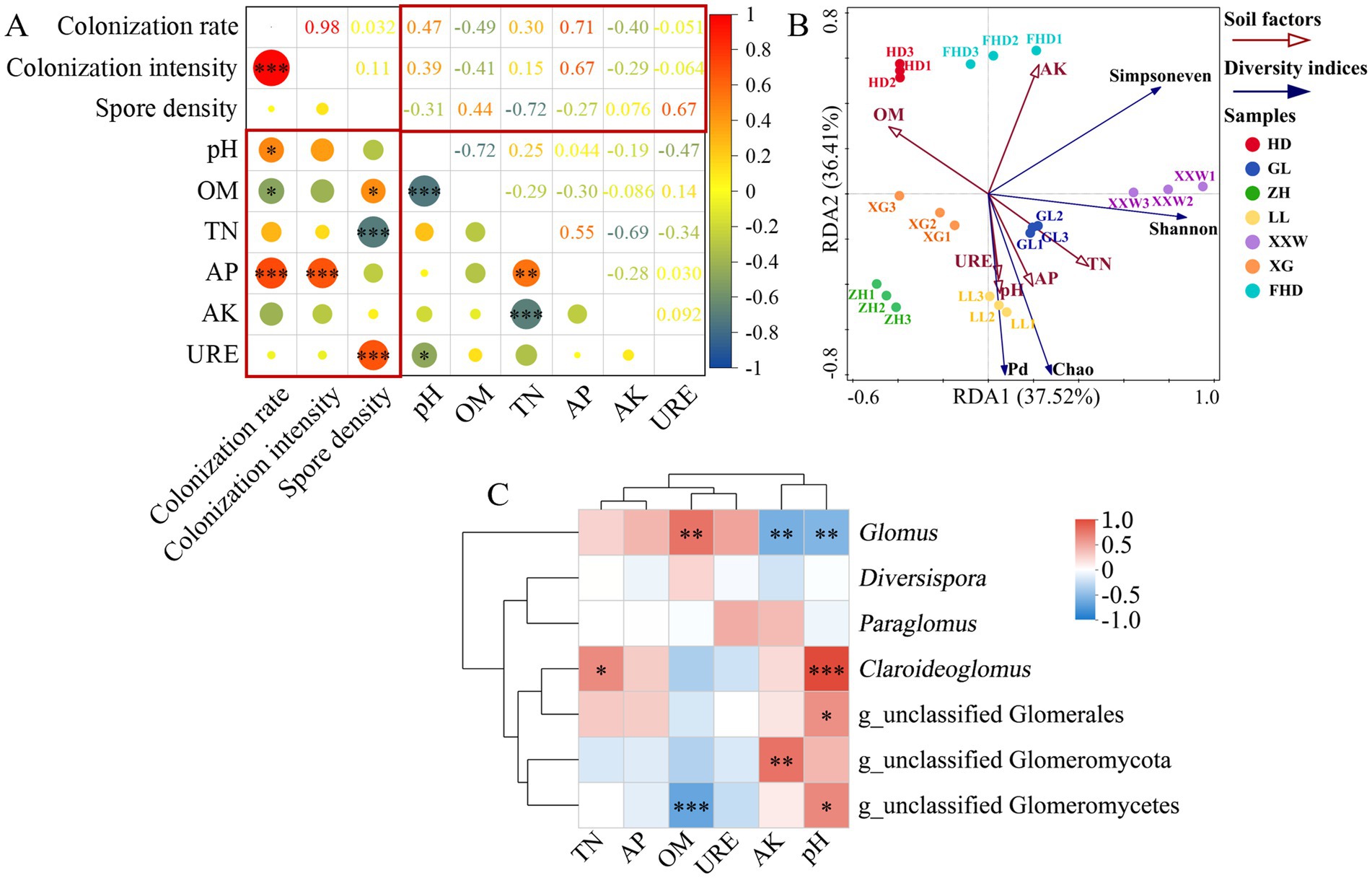
Figure 8. Correlation analysis between environmental factors and AMF. (A) Correlation analysis of soil factors and AMF colonization status in wine grapes. (* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, OM, organic matter; TN, total nitrogen; AP, available potassium; AK, available phosphorus; URE, urease). (B) Analysis of correlation between soil factors and AMF diversity index (species level) in different places (the Angle between the environmental factor arrows represents a positive and negative correlation, acute Angle: positive correlation; Obtuse Angle: negative correlation; Right Angle: no correlation; Projected from the sample point to the arrow of the quantitative environmental factor, the distance from the projection point to the origin represents the relative influence of environmental factors on the distribution of the sample community). (C) Spearman’s correlation heat map of AMF community composition (genus level) and soil environmental factors.
Redundancy analysis was conducted between the soil factors retained after the VIF analysis and the AMF diversity index (species level) in the wine grape rhizosphere soil. Total nitrogen, available phosphorus, pH, and urease levels positively correlated with AMF diversity (Shannon index), richness (Chao index), and phylogenetic diversity (Pd index). Organic matter was negatively correlated with AMF diversity, richness, evenness (Simpson’s index), and phylogenetic diversity. Available potassium was positively correlated with AMF diversity and evenness and negatively correlated with richness and phylogenetic diversity (Figure 8B). In summary, the principal factors influencing the diversity and abundance of AMF in all environmental variables were soil pH, total nitrogen, available phosphorus, urease, and organic matter.
The perspective of AMF genus Glomus was significantly positively correlated with organic matter (p < 0.01) and significantly negatively correlated with pH and available potassium (p < 0.01). Claroideoglomus was significantly positively correlated with pH (p < 0.001) and total nitrogen (p < 0.05). g_nclassified Glomeromycota was significantly negatively correlated with available potassium (p < 0.01). g_unclassified glomerulales was significantly positively correlated with pH (p < 0.05). g_nclassified Glomeromycetes was significantly positively correlated with pH (p < 0.001) and significantly negatively correlated with organic matter (p < 0.05) (Figure 8C).
4 Discussion
AMF constitute essential components of the rhizosphere microbial community, forming symbiotic associations with approximately 80% of plants in terrestrial ecosystems (Aseel et al., 2019). In our study, AMF colonized wine grape roots, appearing as clumped mycorrhizae, with total colonization rates ranging from 25.14 to 75.52% and colonization intensities from 6.28 to 70.04%. These findings underscore the ability of AMF to establish symbiotic relationships with wine grape roots. Arbuscular mycorrhiza exhibits major structural classes, including Arum, Paris, and intermediate types (Smith and Smith, 1997). Conversely, A-types are frequently observed in cultivated plants, distinguished by extensive intercellular hyphal growth within the root cortex and the development of terminal arbuscules on intracellular hyphal branches (Smith and Smith, 1997). Our findings indicated that AMF infection in wine grape roots predominantly forms a characteristic arum-type structure, representing a key feature of AMF symbiosis. Given the longstanding cultivation of wine grapes as economically significant fruit trees, this could be one reason for the dominance of this type of infection among AMF structures in wine grape roots.
The methods for identifying AMF include both morphological identification of AMF spores and molecular analysis (Lee et al., 2006; Bidondo et al., 2018; Stevens et al., 2020). Aguilera et al. (2024) identified 15 AMF species from vineyards along a 1,000-km stretch of Chile via spore morphological analysis. They found that the AMF communities in these vineyards were mainly dominated by the genera Acaulospora, Claroideoglomus, Septoglomus and Simiglomus. Likar et al. (2013) used nested PCR to investigate the colonization and diversity of AMF on grapevine roots in vineyards of karst along the coast of the Adriatic Sea. Their study identified 30 fungal taxa, including the first reported colonization of G. indicum in grapevines. In our study, 16 species from 6 AMF genera were identified in 7 plots using the spore morphology identification method. Among these, Glomus was the dominant genus, G. melanosporum was the dominant species and Claroideoglomus, Diversispora, and Paraglomus were identified for the first time in rhizosphere soils of wine grapes in China. However, spore identification is a time- and energy-intensive process, influenced by regional variations, host plant species, AMF age, and spore morphological variability. Consequently, traditional spore morphological identification poses practical difficulties and fails to truly reflect the natural occurrence of AMF communities (Redecker et al., 2003; Oepik et al., 2009).
High-throughput sequencing provides a more robust and accurate method for studying the community structure and diversity of AMF and can identify AMF in any given root or soil sample without relying on spores (Rodríguez-Echeverría et al., 2017; Rodríguez-Caballero et al., 2018; Faggioli et al., 2019). Moukarzel et al. (2024) used Illumina MiSeq to investigate the composition and abundance of AMF communities in the roots of 12 vineyards in Marburg, New Zealand. The most prevalent AMF genus was Glomus, followed by Entrophospora, Funneliformis, Rhizophagus and Diversispora. In the present study, Illumina MiSeq sequencing identified the fungal communities and divided the AMF OTUs into 1 phylum, 1 class, 5 orders, 7 families, 7 genera, and 40 species. Among them, Claroideoglomus and Glomus were identified as biomarkers and Glomus was the dominant genus in the seven plots, which is consistent with the results of Moukarzel. In addition, Glomus is the core genus of AMF in the rhizosphere of wine grapes and plays an active role in plant biological defense. Ye et al. (2022) studied the effects of G. aggregatum on the growth, photosynthetic activity, and chlorophyll fluorescence of Vitis vinifera L. cv. Ecolly under drought stress, and the results showed that colonization by G. aggregatum increased the dry biomass of shoots and roots, photosynthetic rate, stomatal conductance, and transpiration rate—while decreasing the intercellular CO₂ concentration. Thus, G. aggregatum effectively alleviated the negative effects of drought stress on grapevines. Moukarzel et al. (2022) explored the effect of compound microbial agents with Glomus on grape black foot disease. The study produced evidence that compound microbial agent with Glomus treatments lowered disease incidence at 5 cm and disease severity in vines by 40 to 50% compared to the vines inoculated with the pathogen only. Whether Glomus promotes the growth and development of wine grapes in the EFHM requires further investigation.
The diversity and community structure of soil microorganisms are affected by natural factors such as ecological region, climate, soil properties and vegetation. Among them, the ecological region is the main factor affecting soil AMF diversity (Betancur-Agudelo et al., 2021; Rezaee Danesh et al., 2022). Aguilar-Paredes et al. (2024) conducted a study on the diversity of AMF in vineyards across 9 distinct ecological regions within the Mediterranean climate of Chile, and their findings revealed that AMF exhibited a specific geographic distribution, Acaulospora, Scutellospora sp., C. etunicatum, Pacispora scintillans and Paraglomus sp. were detected only in the southern valleys, while Septoglomus sp. was observed in the Northern and Center Valleys. In the present study, the AMF community structure in the rhizosphere soil of wine grapes from seven ecological regions was divided into three groups. The LEfSe results showed that each ecological region had unique identifying biomarkers. There were some differences in the diversity and abundance of AMF among the different ecological regions. LL and XXW had the highest performance, followed by GL, while FHD had the lowest. We speculate that this may be because LL is located in the Yinchuan Plain, while FHD is located in the Loess Plateau. Changes in topography caused differences in the AMF diversity between the two regions. In addition, the elevation difference of the seven ecological regions in our study exceeded 200 m. PERMANOVA analysis showed that altitude was also a factor that caused AMF diversity and community variation in the rhizosphere soil of wine grapes, which further confirmed that different ecological regions would cause changes in AMF diversity in the ecosystem.
Ecological regions are key macroscale drivers shaping the diversity and community structure of AMF in wine grape rhizospheres. Soil, as the direct habitat for arbuscular mycorrhizal fungi, represents another set of critical regulatory factors. Soil physical and chemical properties and soil enzyme activity, have a direct impact on AMF diversity, community composition, and function (Ren et al., 2021; Song et al., 2021). Correlation analysis showed that soil pH and organic matter were key factors affecting AMF colonization, diversity, and species composition in our study. Soil pH can affect the absorption and utilization of soil elements by plants. An increase in soil pH limits the utilization of soil nutrients and affects the formation and sporulation of AMF (Rillig and Allen, 1999). In our study, the pH values of seven plots ranged from 8.06 to 8.54, with an average of 8.26, indicating that the weakly alkaline soil may be more suitable for the symbiotic relationship between AMF and wine grapes. Correlation analysis showed that pH was significantly positively correlated with the total colonization rate, AMF diversity index and richness index, indicating that the increase of pH was conducive to the formation of AMF, which was consistent with the study of Liu et al. on the correlation between Hedysarum scoparium and pH in the northwest desert (Liu et al., 2018). In addition, pH was significantly positively correlated with Claroideoglomus, and significantly negatively correlated with Glomus, indicating that an alkaline environment was favorable for the growth of Claroideoglomus, whereas Glomus preferred a neutral and acidic environment. Soil organic matter is a crucial source of nutrients and a significant indicator of soil fertility and quality. Its content directly affects soil fertility (Geng et al., 2023). Correlation analysis showed that organic matter was negatively correlated with total colonization rate and AMF diversity, and positively correlated with spore density and Glomus, which was inconsistent with previous studies (Boddington and Dodd, 2000; Wang et al., 2008). A possible reason for this is that AMF spore density and soil organic matter content are only positively correlated within a certain threshold. When the organic matter content exceeded a certain range, this trend stopped or was even reversed, inhibiting the growth of AMF hyphae and vesicle germination.
This study has significant implications for the development and utilization of symbiotic AMF resources in wine grapes in the EFHM of Ningxia, China. Future research could delve deeper into the following areas: identification of beneficial core or key AMF strains to underpin the sustainable cultivation and breeding of grape plants. This entails reducing fertility demands, enhancing nutrient absorption, fortifying resistance to biotic or abiotic stresses, and improving soil structure (Karimi and Noori, 2022). Exploration of how AMF symbiosis influences fruit quality by modulating phenol metabolism, including anthocyanin production, and altering chemical components such as malic acid and tartaric acid. This represents a promising avenue for enhancing grape quality and serves as a potential regulatory resource (Velasquez et al., 2020). Investigation of the impact of AMF on amino acids or volatile compounds in grape plants to enhance the taste, flavor, and aroma of wine. This research can lead to advancements in the wine industry. It is crucial to safeguard the natural mycorrhizal fungal communities in vineyards, emphasizing the importance of preserving and protecting the delicate balance of these beneficial symbionts within the ecosystem.
5 Conclusion
High-throughput sequencing technology was used to comprehensively analyze the composition and diversity of AMF communities in the rhizosphere of wine grapes grown in the EFHM. The roots of wine grapes can be colonized by AMF and form a typical structure. The co-evolution of wine grapes and AMF has produced AMF diversity with ecological and environmental specificity. A total of 168 OTUs were detected in the rhizosphere soil of wine grapes in different locations, belonging to five orders, seven families, seven genera, and 40 species of AMF, among which Glomus was the dominant genus, indicating abundant AMF resources in the rhizosphere of grapes. Correlation analysis showed that pH and organic matter were key factors affecting AMF richness, diversity, and community composition. This information provides a valuable basis for the protection of AMF communities in the rhizosphere of wine grapes and the commercial cultivation of AMF.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA966782.
Author contributions
QiaZ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. SM: Data curation, Investigation, Writing – review & editing. RW: Investigation, Software, Supervision, Writing – review & editing. LL: Data curation, Investigation, Methodology, Software, Writing – review & editing. QinZ: Supervision, Writing – review & editing. MJ: Supervision, Writing – review & editing. PG: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Major R&D project of Ningxia Hui Autonomous Region (2023BCF01026) and the National Key Research and Development project (2019YFD1002502).
Acknowledgments
We thank the Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China) for their important contribution to the use of R software and the preparation for archiving of high-throughput sequencing data. Thanks also to Wiley editing services for improving our manuscript in terms of language.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1700411/full#supplementary-material
Footnotes
References
Aguilar-Paredes, A., Turrini, A., Avio, L., Stuardo, C., Velasquez, A., Becerra, J., et al. (2024). Agricultural managements influence the diversity of arbuscular mycorrhizal Fungi in vineyards from Chilean Mediterranean climate ecosystems. J. Soil Sci. Plant Nutr. 24, 6099–6112. doi: 10.1007/s42729-024-01963-y
Aguilera, P., Silva-Flores, P., Gainza-Cortes, F., Pastenes, C., Castillo, C., Borie, F., et al. (2024). Drivers of arbuscular mycorrhizal fungal diversity across 1,000 km of Chilean vineyards. J. Soil Sci. Plant Nutr. 24, 3675–3686. doi: 10.1007/s42729-024-01787-w
Aseel, D. G., Rashad, Y. M., and Hammad, S. M. (2019). Arbuscular mycorrhizal fungi trigger transcriptional expression of flavonoid and chlorogenic acid biosynthetic pathways genes in tomato against tomato mosaic virus. Sci. Rep. 9:9692. doi: 10.1038/s41598-019-46281-x
Betancur-Agudelo, M., Meyer, E., and Lovato, P. E. (2021). Arbuscular mycorrhizal fungus richness in the soil and root colonization in vineyards of different ages. Rhizosphere 17:100307. doi: 10.1016/j.rhisph.2021.100307
Bidondo, L. F., Colombo, R. P., Recchi, M., Silvani, V. A., Pérgola, M., Martínez, A., et al. (2018). Detection of arbuscular mycorrhizal fungi associated with pecan (Carya illinoinensis) trees by molecular and morphological approaches. MycoKeys 42, 73–88. doi: 10.3897/mycokeys.42.26118
Blaszkowski, J., Kovacs, G. M., Gaspar, B. K., Balazs, T. K., Buscot, F., and Ryszka, P. (2012). The arbuscular mycorrhizal Paraglomus majewskii sp nov represents a distinct basal lineage in Glomeromycota. Mycologia 104, 148–156. doi: 10.3852/10-430
Boddington, C. L., and Dodd, J. C. (2000). The effect of agricultural practices on the development of indigenous arbuscular mycorrhizal fungi. I. Field studies in an Indonesian ultisol. Plant Soil 218, 137–144. doi: 10.1023/a:1014966801446
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2 (vol 37, pg 852, 2019). Nat. Biotechnol. 37:1091. doi: 10.1038/s41587-019-0252-6
Dong, C., Shao, Q., Zhang, Q., Yao, T., Huang, J., Liang, Z., et al. (2021). Preferences for core microbiome composition and function by different definition methods: evidence for the core microbiome of Eucommia ulmoide bark. Sci. Total Environ. 790:148091. doi: 10.1016/j.scitotenv.2021.148091
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Faggioli, V., Menoyo, E., Geml, J., Kemppainen, M., Pardo, A., Salazar, M. J., et al. (2019). Soil lead pollution modifies the structure of arbuscular mycorrhizal fungal communities. Mycorrhiza 29, 363–373. doi: 10.1007/s00572-019-00895-1
Fang, Y., Qu, Y., Zhang, J., Meng, J., Liu, J., Cheng, B., et al. (2011). The screening, identification of arbuscular mycorrhizal fungi in northwestern vineyards. J. Plant Nutr. Fertil. Sci. 17, 124–130. doi: 10.11674/zwyf.2011.0117
Fernández-López, D. J., Fernández-Fernández, J. I., Martínez-Mora, C., Bleda-Sánchez, J. A., and Ruiz-García, L. (2022). Productiveness and berry quality of new wine grape genotypes grown under drought conditions in a semi-arid wine-producing mediterranean region. Plants 11:1363. doi: 10.3390/plants11101363
Garo, G., Van Geel, M., Eshetu, F., Swennen, R., Honnay, O., and Vancampenhout, K. (2022). Arbuscular mycorrhizal fungi community composition, richness and diversity on enset (Ensete ventricosum (Welw.) Cheesman) in Ethiopia is influenced by manure application intensity in low-input farming systems. Plant Soil 478, 409–425. doi: 10.1007/s11104-022-05462-w
Geel, M. V., Ceustermans, A., Hemelrijck, W. V., Lievens, B., and Honnay, O. (2015). Decrease in diversity and changes in community composition of arbuscular mycorrhizal fungi in roots of apple trees with increasing orchard management intensity across a regional scale. Mol. Ecol. 24, 941–952. doi: 10.1111/mec.13079
Geng, H., Wang, X., Shi, S., Ye, Z., and ZHou, W. (2023). Effects of combined application of fungal residue and chemical fertilizer on soil microbial community composition and diversity in paddy soil. Environ. Sci. 44, 2338–2347. doi: 10.13227/j.hjkx.202205202
Girardello, R. C., Rich, V., Smith, R. J., Brenneman, C., Heymanna, H., and Oberholster, A. (2019). The impact of grapevine red blotch disease on Vitis vinifera L. chardonnay grape and wine composition and sensory attributes over three seasons. J. Sci. Food Agric. 100, 1436–1447. doi: 10.1002/jsfa.10147
Han, Y., Feng, J., Han, M., and Zhu, B. (2020). Responses of arbuscular mycorrhizal fungi to nitrogen addition: a meta-analysis. Glob. Chang. Biol. 26, 7229–7241. doi: 10.1111/gcb.15369
Han, S., Wang, X., Cheng, Y., Wu, G., Dong, X., He, X., et al. (2023). Multidimensional analysis reveals environmental factors that affect community dynamics of arbuscular mycorrhizal fungi in poplar roots. Front. Plant Sci. 13:1068527. doi: 10.3389/fpls.2022.1068527
Hu, Z., Zhang, Y., Zhang, X., and Wang, R. (2021). Characteristics and evaluation of soil fertility in different grape production areas at the eastern foot of Helan Mountain, Ningxia. Soil Fertil. Sci. China 6, 35–41. doi: 10.11838/sfsc.1673-6257.20488
Jansa, J., Erb, A., Oberholzer, H.-r., Smilauer, P., and Egli, S. (2014). Soil and geography are more important determinants of indigenous arbuscular mycorrhizal communities than management practices in Swiss agricultural soils. Mol. Ecol. 23, 2118–2135. doi: 10.1111/mec.12706
Ju, M., Zhang, Q., Wang, R., Yan, S., Li, Z., Li, P., et al. (2022). Correlation in endophytic fungi community diversity and bioactive compounds of Sophora alopecuroides. Front. Microbiol. 13:955647. doi: 10.3389/fmicb.2022.955647
Karimi, R., and Noori, A. (2022). Streptomyces rimosus rhizobacteria and Glomus mosseae mycorrhizal fungus inoculation alleviate salinity stress in grapevine through morphophysiological changes and nutritional balance. Sci. Hortic. 305:111433. doi: 10.1016/j.scienta.2022.111433
Lee, J., Lee, S., and Young, J. P. W. (2008). Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 65, 339–349. doi: 10.1111/j.1574-6941.2008.00531.x
Lee, J., Park, S.-H., and Eom, A.-H. (2006). Molecular identification of arbuscular mycorrhizal fungal spores collected in Korea. Mycobiology 34, 7–13. doi: 10.4489/myco.2006.34.1.007
Li, T., Liu, M. J., Zhang, X. T., Zhang, H. B., Sha, T., and Zhao, Z. W. (2011). Improved tolerance of maize (Zea mays L.) to heavy metals by colonization of a dark septate endophyte (DSE) Exophiala pisciphila. Sci. Total Environ. 409, 1069–1074. doi: 10.1016/j.scitotenv.2010.12.012
Li, X., Qi, Z., Yu, X., Xu, M., Liu, Z., Du, G., et al. (2021). Soil pH drives the phylogenetic clustering of the arbuscular mycorrhizal fungal community across subtropical and tropical pepper fields of China. Appl. Soil Ecol. 165:103978. doi: 10.1016/j.apsoil.2021.103978
Liang, J., Zou, R., Huang, Y., Qin, H., Tang, J., Wei, X., et al. (2022). Structure and diversity of mycorrhizal fungi communities of different part of Bulbophyllum tianguii in three terrestrial environments. Front. Plant Sci. 13:992184. doi: 10.3389/fpls.2022.992184
Liao, D., Wang, S., Cui, M., Liu, J., Chen, A., and Xu, G. (2018). Phytohormones regulate the development of arbuscular mycorrhizal symbiosis. Int. J. Mol. Sci. 19:3146. doi: 10.3390/ijms19103146
Likar, M., Hancevic, K., Radic, T., and Regvar, M. (2013). Distribution and diversity of arbuscular mycorrhizal fungi in grapevines from production vineyards along the eastern Adriatic coast. Mycorrhiza 23, 209–219. doi: 10.1007/s00572-012-0463-x
Liu, H. Y., Li, X. M., Zhang, L. L., Wang, J. J., and He, X. L. (2018). Eco-geographical distribution of arbuscular mycorrhizal fungi associated with Hedysarum scoparium in the desert zone of northwestern China Chinese. J. Plant Ecol. 42, 252–260. doi: 10.17521/cjpe.2017.0138
Liu, W., Qiu, K., Xie, Y., Wang, R., Li, H., Meng, W., et al. (2022). Years of sand fixation with Caragana korshinskii drive the enrichment of its rhizosphere functional microbes by accumulating soil N. PeerJ 10:e14271. doi: 10.7717/peerj.14271
Liu, J., Zhu, X.-L., Ullah, N., and Tao, Y.-S. (2017). Aroma glycosides in grapes and wine. J. Food Sci. 82, 248–259. doi: 10.1111/1750-3841.13598
Lu, N., Zhang, P., Wang, P., Wang, X., Ji, B., and Mu, J. (2022). Environmental factors affect the arbuscular mycorrhizal fungal community through the status of host plants in three patterns of Chinese fir in southern China. Glob. Ecol. Conserv. 36:e02121. doi: 10.1016/j.gecco.2022.e02121
Luginbuehl, L. H., Menard, G. N., Kurup, S., Erp, H. V., Radhakrishnan, G. V., Breakspear, A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178. doi: 10.1126/science.aan0081
Luo, Y., Wang, Z., He, Y., Li, G., Lv, X., and Zhuang, L. (2020). High-throughput sequencing analysis of the rhizosphere arbuscular mycorrhizal fungi (AMF) community composition associated with Ferula sinkiangensis. BMC Microbiol. 20:335. doi: 10.1186/s12866-020-02024-x
Lutz, S., Mikryukov, V., Bahram, M., Jones, A., Panagos, P., Delgado-Baquerizo, M., et al. (2025). Global richness of arbuscular mycorrhizal fungi. Fungal Ecol. 74:101407. doi: 10.1016/j.funeco.2024.101407
Ma, J., Xie, Y., Yang, Y., Jing, C., You, X., Yang, J., et al. (2022). AMF colonization affects allelopathic effects of Zea mays L. root exudates and community structure of rhizosphere bacteria. Front. Plant Sci. 13:1050104. doi: 10.3389/fpls.2022.1050104
Massa, N., Bona, E., Novello, G., Todeschini, V., Boatti, L., Mignone, F., et al. (2020). AMF communities associated to Vitis vinifera in an Italian vineyard subjected to integrated pest management at two different phenological stages. Sci. Rep. 10:9197. doi: 10.1038/s41598-020-66067-w
McGonigle, T. P., Miller, M. H., Evans, D. G., Fairchild, G. L., and Swan, J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x
Mele, M. A., Kang, H., Lee, Y., and Islam, M. Z. (2021). Grape terpenoids: flavor importance, genetic regulation, and future potential. Crit. Rev. Food Sci. Nutr. 61, 1429–1447. doi: 10.1080/10408398.2020.1760203
Mencarelli, F., and Bellincontro, A. (2020). Recent advances in postharvest technology of the wine grape to improve the wine aroma. J. Sci. Food Agric. 100, 5046–5055. doi: 10.1002/jsfa.8910
Mickan, B. S., Abbott, L. K., Stefanova, K., and Solaiman, Z. M. (2016). Interactions between biochar and mycorrhizal fungi in a water-stressed agricultural soil. Mycorrhiza 26, 565–574. doi: 10.1007/s00572-016-0693-4
Moukarzel, R., Jones, E. E., Panda, P., Larrouy, J., Ramana, J. V., Guerin-Laguette, A., et al. (2024). Vineyard management systems influence arbuscular mycorrhizal fungi recruitment by grapevine rootstocks in New Zealand. J. Appl. Microbiol. 135:lxae211. doi: 10.1093/jambio/lxae211
Moukarzel, R., Ridgway, H. J., Liu, J., Guerin-Laguette, A., and Jones, E. E. (2022). AMF Community diversity promotes grapevine growth parameters under high black foot disease pressure. J. Fungi 8:250. doi: 10.3390/jof8030250
Oepik, M., Metsis, M., Daniell, T. J., Zobel, M., and Moora, M. (2009). Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol. 184, 424–437. doi: 10.1111/j.1469-8137.2009.02920.x
Omar, M. B., Bolland, L., and Heather, W. A. (1978). A permanent mounting medium for fungi. Stain. Technol. 53, 293–294.
Phillips, J. M., and Hayman, D. S. J. B. M. S. T. (1970). Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55, 158–160. doi: 10.1016/S0007-1536(70)80110-3
Rasmussen, P. U., Abrego, N., Roslin, T., Opik, M., Sepp, S.-K., Blanchet, F. G., et al. (2022). Elevation and plant species identity jointly shape a diverse arbuscular mycorrhizal fungal community in the high Arctic. New Phytol. 236, 671–683. doi: 10.1111/nph.18342
Redecker, D., Hijri, I., and Wiemken, A. (2003). Molecular identification of arbuscular mycorrhizal fungi in roots: perspectives and problems. Folia Geobot. 38, 113–124. doi: 10.1007/bf02803144
Ren, C., Su, W., Pan, L., Han, Z., Wu, D., Li, L., et al. (2021). Fungal community structures and diversities in dhizosphere soils in different growth stages of kiwifruit seedlings based on high-throughput sequencing. Soils 53, 545–554. doi: 10.13758/j.cnki.tr.2021.03.014
Rezaee Danesh, Y., Kariman, K., Keskin, N., and Najafi, S. (2022). Characterization of arbuscular mycorrhizal fungal communities associated with vineyards in northwestern Iran. Turk. J. Agric. For. 46, 271–279. doi: 10.55730/1300-011x.3001
Rillig, M. C., and Allen, M. F. (1999). What is the role of arbuscular mycorrhizal fungi in plant-to-ecosystem responses to elevated atmospheric CO2? Mycorrhiza 9, 1–8. doi: 10.1007/s005720050257
Rodríguez-Caballero, G., Caravaca, F., and Roldán, A. (2018). The unspecificity of the relationships between the invasive Pennisetum setaceum and mycorrhizal fungi may provide advantages during its establishment at semiarid Mediterranean sites. Sci. Total Environ. 630, 1464–1471. doi: 10.1016/j.scitotenv.2018.02.321
Rodríguez-Echeverría, S., Teixeira, H., Correia, M., Timóteo, S., Heleno, R., Öpik, M., et al. (2017). Arbuscular mycorrhizal fungi communities from tropical Africa reveal strong ecological structure. New Phytol. 213, 380–390. doi: 10.1111/nph.14122
Schenck, N., and Perez, Y. (1990). Manual for the identification of Va mycorrhizal Fungi. Gainesville, Florida, USA: Synergistic Publications.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schreiner, R. P. (2020). Depth structures the community of arbuscular mycorrhizal fungi amplified from grapevine (Vitis vinifera L.) roots. Mycorrhiza 30, 149–160. doi: 10.1007/s00572-020-00930-6
Schreiner, R. P., and Mihara, K. L. (2009). The diversity of arbuscular mycorrhizal fungi amplified from grapevine roots (Vitis vinifera L.) in Oregon vineyards is seasonally stable and influenced by soil and vine age. Mycologia 101, 599–611. doi: 10.3852/08-169
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shaul-Keinan, O., Gadkar, V., Ginzberg, I., Grünzweig, J. M., Chet, I., Elad, Y., et al. (2002). Hormone concentrations in tobacco roots change during arbuscular mycorrhizal colonization with Glomus intraradices. New Phytol. 154, 501–507. doi: 10.1046/j.1469-8137.2002.00388.x
Shi, W., Li, M., Wei, G., Tian, R., Li, C., Wang, B., et al. (2019). The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome 7:14. doi: 10.1186/s40168-019-0629-2
Smith, F. A., and Smith, S. E. (1997). Tansley review no. 96 structural diversity in (vesicular)—arbuscular mycorrhizal symbioses. New Phytol. 137, 373–388. doi: 10.1046/j.1469-8137.1997.00848.x
Song, J., Han, Y., Bai, B., Jin, S., He, Q., and Ren, J. (2019). Diversity of arbuscular mycorrhizal fungi in rhizosphere soils of the Chinese medicinal herb Sophora flavescens ait. Soil Tillage Res. 195:104423. doi: 10.1016/j.still.2019.104423
Song, J., Zhang, N., Zhang, Z., Zhou, Z., Liang, J., and Lu, J. (2021). High-throughput sequencing analysis of fungal diversity in agarwood wound locations Chinese journal of tropical. Crops 42, 3358–3368. doi: 10.3969/j.issn.1000-2561.2021.11.039
Stevens, B. M., Propster, J. R., Öpik, M., Wilson, G. W. T., Alloway, S. L., Mayemba, E., et al. (2020). Arbuscular mycorrhizal fungi in roots and soil respond differently to biotic and abiotic factors in the Serengeti. Mycorrhiza 30, 79–95. doi: 10.1007/s00572-020-00931-5
Sturmer, S. L., Bever, J. D., Schultz, P. A., and Bentivenga, S. P. (2021). Celebrating INVAM: 35 years of the largest living culture collection of arbuscular mycorrhizal fungi. Mycorrhiza 31, 117–126. doi: 10.1007/s00572-020-01008-z
Vályi, K., Rillig, M. C., and Hempel, S. (2014). Land-use intensity and host plant identity interactively shape communities of arbuscular mycorrhizal fungi in roots of grassland plants. New Phytol. 205, 1577–1586. doi: 10.1111/nph.13236
Van Geel, M., Busschaert, P., Honnay, O., and Lievens, B. (2014). Evaluation of six primer pairs targeting the nuclear rRNA operon for characterization of arbuscular mycorrhizal fungal (AMF) communities using 454 pyrosequencing. J. Microbiol. Methods 106, 93–100. doi: 10.1016/j.mimet.2014.08.006
Velasquez, A., Valenzuela, M., Carvajal, M., Fiaschi, G., Avio, L., Giovannetti, M., et al. (2020). The arbuscular mycorrhizal fungus Funneliformis mosseae induces changes and increases the concentration of volatile organic compounds in Vitis vinifera cv. Sangiovese leaf tissue. Plant Physiol. Biochem. 155, 437–443. doi: 10.1016/j.plaphy.2020.06.048
Wang, Q., Pan, J., Ke, Y., Yu, S., Murray, P. J. J., Luo, T., et al. (2022). Impact of aspect on arbuscular mycorrhizal fungal diversity and community composition in a natural Toona ciliata var. pubescens forest in subtropical China. Forests 13:2100. doi: 10.3390/f13122100
Wang, S., Tang, M., Niu, Z.-C., and Zhang, H.-Q. (2008). Relationship between AM fungi resources of rare medicinal plants and soil factors in Lishan mountain. Acta Botan. Boreali-Occiden. Sin. 28:7. doi: 10.3321/j.issn:1000-4025.2008.02.023
Wang, Z., Zhao, J., Xiao, D., Chen, M., and He, X. (2024). Higher colonization but lower diversity of root-associated arbuscular mycorrhizal fungi in the topsoil than in deep soil. Appl. Soil Ecol. 194:105195. doi: 10.1016/j.apsoil.2023.105195
Wassermann, B., Cernava, T., Müller, H., Berg, C., and Berg, G. (2019). Seeds of native alpine plants host unique microbial communities embedded in cross-kingdom networks. Microbiome 7:108. doi: 10.1186/s40168-019-0723-5
Xu, X., Qiu, Y., Zhang, K., Yang, F., Chen, M., Luo, X., et al. (2022). Climate warming promotes deterministic assembly of arbuscular mycorrhizal fungal communities. Glob. Chang. Biol. 28, 1147–1161. doi: 10.1111/gcb.15945
Xu, D., Yu, X., Chen, J., Liu, H., Zheng, Y., Qu, H., et al. (2023). Arbuscular mycorrhizae Fungi diversity in the root-rhizosphere-soil of Tetraena mongolica, Sarcozygium xanthoxylon, and Nitraria tangutorum Bobr in Western Ordos, China. Agronomy 13:1485. doi: 10.3390/agronomy13061485
Yan, J., He, X., Zhang, Y., Xu, W., Zhang, J., and Zhao, L. (2014). Colonization of arbuscular mycorrhizal fungi and dark septate endophytes in roots of desert Salix psammophila. Chin. J. Plant Ecol. 38, 949–958. doi: 10.3724/sp.J.1258.2014.00089
Yan, K., Pei, Z., Meng, L., Zheng, Y., Wang, L., Feng, R., et al. (2022). Determination of community structure and diversity of seed-vectored endophytic fungi in Alpinia zerumbet. Front. Microbiol. 13:814864. doi: 10.3389/fmicb.2022.814864
Yang, H., Xiong, Y., Wang, Q., Guo, Y., Dai, Y., and Xu, M. (2016). Arbuscular mycorrhizal fungal species diversity: ecological functioning, determinants and assembling mechanisms. Acta Ecol. Sin. 36, 2826–2832. doi: 10.5846/stxb201410112001
Ye, Q., Wang, H., and Li, H. (2022). Arbuscular mycorrhizal fungi improve growth, photosynthetic activity, and chlorophyll fluorescence of Vitis vinifera L. cv. Ecolly under drought stress. Agronomy 12:1563. doi: 10.3390/agronomy12071563
Zeng, Q., Lebreton, A., Auer, L., Man, X., Jia, L., Wang, G., et al. (2023). Stable functional structure despite high taxonomic variability across fungal communities in soils of old-growth montane forests. Microbiome 11:217. doi: 10.1186/s40168-023-01650-7
Zhang, Q., Li, L., Wang, R., and Gu, P. (2025). Diversity and structure of arbuscular mycorrhizal fungi communities in rhizosphere soil of different wine grape varieties in the eastern foot of Helan Mountain. J. China Agric. Univ. 30, 76–89. doi: 10.11841/j.issn.1007-4333.2025.01.07
Zhang, M., Shi, Z., Xu, X., and Wang, X. (2022). Arbuscular mycorrhizal fungi associated with roots reveal high diversity levels at different elevations in tropical montane rainforests. Diversity 14:587. doi: 10.3390/d14080587
Zhang, M., Shi, Z., Yang, M., Lu, S., Cao, L., and Wang, X. (2021). Molecular diversity and distribution of arbuscular mycorrhizal fungi at different elevations in Mt. Taibai of Qinling Mountain. Front. Microbiol. 12:609386. doi: 10.3389/fmicb.2021.609386
Zhang, Z., Zhang, Q., Yang, H., Sun, L., Xia, H., Sun, W., et al. (2022). Bacterial communities related to aroma formation during spontaneous fermentation of ‘cabernet sauvignon’ wine in Ningxia, China. Foods 11:2775. doi: 10.3390/foods11182775
Zhou, T., Wang, L., Li, S., Gao, Y., Du, Y., Zhao, L., et al. (2019). Interactions between light intensity and phosphorus nutrition affect the P uptake capacity of maize and soybean seedling in a low light intensity area. Front. Plant Sci. 10:183. doi: 10.3389/fpls.2019.00183
Keywords: arbuscular mycorrhizal fungi (AMF), wine grape, colonization status, diversity, soil factors
Citation: Zhang Q, Ma S, Wang R, Li L, Zhang Q, Ju M and Gu P (2025) Diversity and drivers of arbuscular mycorrhizal fungi in the rhizosphere soil of wine grape in the eastern foot of Helan Mountain in Ningxia of China. Front. Microbiol. 16:1700411. doi: 10.3389/fmicb.2025.1700411
Edited by:
Weiwei She, Beijing Forestry University, ChinaReviewed by:
Yangui Qiao, Inner Mongolia University, ChinaKateřina Štůsková, Mendel University in Brno, Czechia
Copyright © 2025 Zhang, Ma, Wang, Li, Zhang, Ju and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peiwen Gu, Z3VwZWl3ZW4yMDE5QG54dS5lZHUuY24=
 Qiangqiang Zhang
Qiangqiang Zhang Siyuan Ma2
Siyuan Ma2 Ruotong Wang
Ruotong Wang Mingxiu Ju
Mingxiu Ju Peiwen Gu
Peiwen Gu