- 1Department of Infection Control, West China Second University Hospital, Sichuan University, Chengdu, China
- 2Department of Laboratory Medicine, Jintang First People’s Hospital, Chengdu, China
- 3Department of Medical Affairs, West China Hospital, Sichuan University, Chengdu, China
- 4Hospital Management Office, West China Hospital Sichuan University Jintang Hospital, Jintang First People’s Hospital, Chengdu, China
- 5Institute of Antibiotics, Huashan Hospital, Fudan University, Shanghai, China
Klebsiella pneumoniae is a significant human pathogen in both hospital and community settings; however, limited data exist regarding its prevalence in district-level hospitals. This study aimed to characterize the drug resistance mechanisms, molecular epidemiology, and virulence profiles of K. pneumoniae in a district hospital in Chengdu, China. A total of 114 clinical isolates were collected between May 2023 and May 2024. Antimicrobial susceptibility testing using the broth microdilution method revealed resistance rates of 14.0–21.1% to third-generation cephalosporins, and 5.3% to carbapenems. Whole-genome sequencing showed that 18.4% (21/114) of the isolates carried ESBL genes, with blaCTX–M–15 (n = 7) being the most common. Six carbapenem-resistant isolates were identified, of which four produced carbapenemases: three harbored blaNDM–5 and one carried blaKPC–2. MLST analysis identified 67 sequence types, with ST23 (n = 9) being the most prevalent. The 21 ESBL-producing isolates were distributed across 15 sequence types, while the six carbapenem-resistant isolates were assigned to four distinct sequence types. Virulence-associated genes were detected at high frequencies, including ybt (39.5%), clb (12.3%), iuc (50.0%), iro (60.5%), rmpA (59.6%), and rmpA2 (36.8%), and were commonly found in ST23 (n = 9), ST412 (n = 6), ST25 (n = 5), ST268 (n = 5), and ST375 (n = 4). In conclusion, K. pneumoniae isolates from this district hospital showed low resistance rates but a worrying high prevalence of virulence genes. This highlights the urgent need for continuous surveillance and early intervention strategies to prevent the emergence of highly virulent and multidrug-resistant strains in healthcare settings.
Introduction
Klebsiella pneumoniae is an important human pathogen both in hospital and community settings, which can cause a series of infections including respiratory tract infections, bloodstream infections, and urinary tract infections (Long et al., 2017; Martin and Bachman, 2018). Based on the data from the China Antimicrobial Surveillance Network (CHINET) in 2024, K. pneumoniae was the clinical isolate with the second highest detection rate, accounting for 13.9%. In the era of antibiotic resistance, K. pneumoniae has been an important multidrug-resistant (MDR) pathogen affecting humans worldwide (Navon-Venezia et al., 2017), and the extended-spectrum beta-lactamase (ESBL)-producing K. pneumoniae constitutes one of the most prevalent MDR bacterial pathogens worldwide (Breurec et al., 2013). Furthermore, increasing resistance to carbapenem in K. pneumoniae raises a global public health concern because of the prevalence of carbapenem-resistant K. pneumoniae (CRKP) and the associated high rate of mortality (Logan and Weinstein, 2017; Navon-Venezia et al., 2017).
Hypervirulent K. pneumoniae (hvKp) is another serious public health threat (Choby et al., 2020). Compared with classical K. pneumoniae (cKp), hvKp usually cause community-acquired infections, such as liver abscess and meningitis, with more severe clinical manifestations (Lee et al., 2006; Siu et al., 2012). Recently, multidrug-resistant hvKp (MDR-hvKp), mainly due to the acquisition of resistance genes by hvKp or the acquisition of virulence plasmids by cKp, has been reported globally (Hao et al., 2020; Shankar et al., 2020). For example, Jin et al. reported a hvKp strain acquired carbapenem resistance by obtaining a plasmid carrying blaKPC–2 (Jin et al., 2021), and Jiang et al. (2025) identified a novel IncFIIK34 blaKPC–2 plasmid that drives the global emergence of carbapenem-resistant hvKp.
Current research on the epidemiology, antimicrobial resistance, and virulence factors of K. pneumoniae primarily focuses on large tertiary teaching hospitals, with less attention given to district-level hospitals (Hu et al., 2020; Lan et al., 2021). Furthermore, most studies have primarily focused on CRKP strains. Therefore, in this study, we investigated the molecular epidemiological characteristics, resistance mechanisms, and virulence profile of K. pneumoniae in a district-level hospital in Chengdu, China. These findings will provide an important basis for formulating effective measures to curb the rapid spread of MDR-Kp and hvKp isolates in district-level hospitals in China and other healthcare settings in developing countries and regions.
Materials and methods
Bacterial isolates
From May 2023 to May 2024, a total of 114 consecutive, non-duplicate clinical K. pneumoniae isolates were obtained from a district comprehensive tertiary hospital in Chengdu, China. All of the isolates were initially identified as K. pneumoniae using a Vitek II system (bioMerieux, Marcy-l’Etoile, France) according to the manufacturer’s recommendations. K. pneumoniae was grown on Luria-Bertani (LB) agar or broth at 37 °C. The isolates were stored at −80 °C in LB broth containing 30% glycerol (v/v) until used.
Antimicrobial susceptibility testing
All of the clinical isolates were tested for susceptibility to 13 antimicrobial agents, including aztreonam, ceftazidime, cefotaxime, cefepime, cefoxitin, ciprofloxacin, gentamicin, amikacin, ertapenem, imipenem, meropenem, tigecycline, and piperacillin-tazobactam. Minimum inhibitory concentrations (MICs) of antimicrobial agents were determined using the broth microdilution method of the Clinical and Laboratory Standards Institute (CLSI, 2019 29th ed. Clinical and Laboratory Standards Institute; Wayne, PA: supplement M100.). The breakpoints of tigecycline were interpreted according to the US Food and Drug Administration (FDA). E. coli ATCC 25922 was used as a quality control strain. The definition of MDR was determined as non-susceptible to ≥1 agent in ≥3 antimicrobial categories according to an international expert proposal for interim standard definitions for acquired resistance (Magiorakos et al., 2012).
Whole-genome sequencing and bioinformatics analysis
A total of 114 K. pneumoniae isolates were sequenced using a HiSeq X10 Sequencer (Illumina, San Diego, CA, USA), with 150 bp paired-end short reads and 200X coverage. Draft genomes were assembled using SPAdes v3.13.0 (Prjibelski et al., 2020). Kleborate v2.3 (Lam et al., 2021) was employed to screen the genome assemblies for the prediction of sequence type, capsular type and the detection of virulence loci, including hypermucoidy genes rmpADC, rmpA2 and siderophore systems, such as yersiniabactin (ybt), colibactin (clb), aerobactin (iuc), and salmochelin (iro). Resistance genes were called by AMRFinderPlus v3.10.20 (Feldgarden et al., 2021). For phylogenetic analyses, core-genome alignments based on the concatenation of core genes were obtained using Panaroo v1.5.0 (Tonkin-Hill et al., 2020) with default parameters, where core genes were defined as those present in more than 98% of the genomes. Maximum-likelihood phylogenetic trees were constructed using FastTree v2.1.11 (model GTR + GAMMA) (Price et al., 2009). The tree was visualized using iTOL v6 (Letunic and Bork, 2024).
Statistical analysis
Statistical significance was assessed using SPSS v.20.0 software (SPSS Inc., Chicago, IL, USA). Categorical variables, expressed as numbers and percentages, were compared by the Chi-square or Fisher’s exact test. The continuous data were expressed as mean ± standard deviation (mean ± SD) appropriately. P-values of <0.05 were considered to be significant.
Results
Clinical characteristics of K. pneumoniae isolates
Table 1 provides a summary of the clinical characteristics. A total of 114 isolates were mainly cultured from sputum (56.1%, 64/114), followed by urine (13.2%, 15/114) and pus (12.3%, 14/114). Most of the patients were male (64.0%, 73/114) with a mean age of 65.3 ± 17.3 years. Of the K. pneumoniae isolates, 36.8% (42/114) were obtained from the intensive care unit (ICU). These findings were consistent with previous studies that older adults are at increased risk of K. pneumoniae infection, and that the ICU remains a major hotspot for K. pneumoniae detection. More than half (50.9%, 58/114) of the patients have undergone invasive procedures, such as surgery, mechanical ventilation, or catheterization, which are closely related to the risk of K. pneumoniae infection. In addition, nearly half (46.5%, 53/114) of the patients have underlying comorbidities (e.g., diabetes, liver/kidney dysfunction), and six patients were in an immunocompromised state with HIV or cancer.
Antimicrobial susceptibility and antimicrobial resistance genes
Among 114 K. pneumoniae isolates, the prevalence of MDR isolates was 12.3% (14/114). As shown in Table 2, ESBL-producing isolates exhibited significantly higher resistance rates to 12 tested antimicrobial agents, including amikacin, aztreonam, cefotaxime, cefoxitin, ceftazidime, cefepime, ciprofloxacin, gentamicin, ertapenem, imipenem, meropenem, and piperacillin/tazobactam, compared to ESBL-negative isolates. The resistance rate to ceftazidime, cefotaxime, and cefepime were 14.0%, 21.1%, and 20.2%, respectively, which is consistent with the finding that 18.4% (21/114) of the K. pneumoniae isolates carried ESBL genes. Among K. pneumoniae isolates carrying ESBLs, the blaCTX–M–15 type (n = 7) was the most prevalent, followed by blaCTX–M–14 (n = 4) and blaCTX–M–3 (n = 4). Of concern, 5.3% (6/114) of the K. pneumoniae isolates were identified as carbapenem-resistant. Resistance to ertapenem was observed in all six isolates, with five also exhibiting non-susceptibility to both meropenem and imipenem. Four of the six carbapenem-resistant isolates were identified as carbapenemase producers, including three harboring blaNDM–5 and one carrying blaKPC–2. Additionally, the resistance rate to ciprofloxacin, amikacin, and gentamicin were 12.3% (14/114), 1.8% (2/114), and 11.4% (13/114), respectively, while fluoroquinolone and aminoglycoside resistance genes were detected in 21.9% (25/114) and 28.1% (32/114) of the isolates, respectively, with qnrS1 (19/25) and strA/B (25/32) being the most prevalent. Notably, all K. pneumoniae isolates remained fully susceptible to tigecycline. The detailed phenotypic profile of antimicrobial resistance was shown in Figure 1, and the detailed genotypic profile of antimicrobial resistance was listed in Supplementary Table 1. Most of the OmpK35 were intact, and only five isolates (4.4%, 5/114) harbored truncated OmpK35 (Figure 1 and Supplementary Table 1).
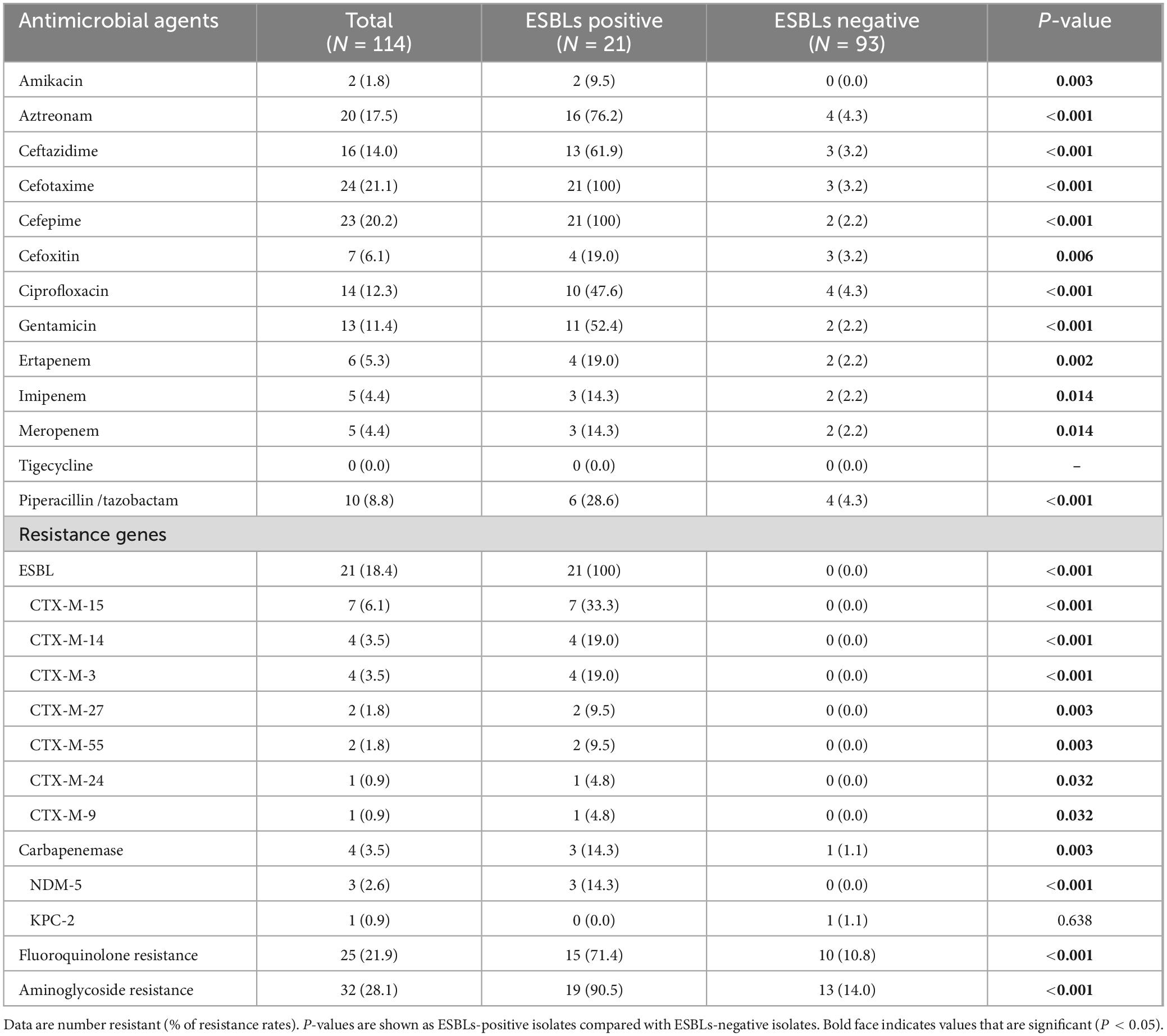
Table 2. Antimicrobial susceptibility and carriage of resistance genes of K. pneumoniae isolates with or without ESBLs.
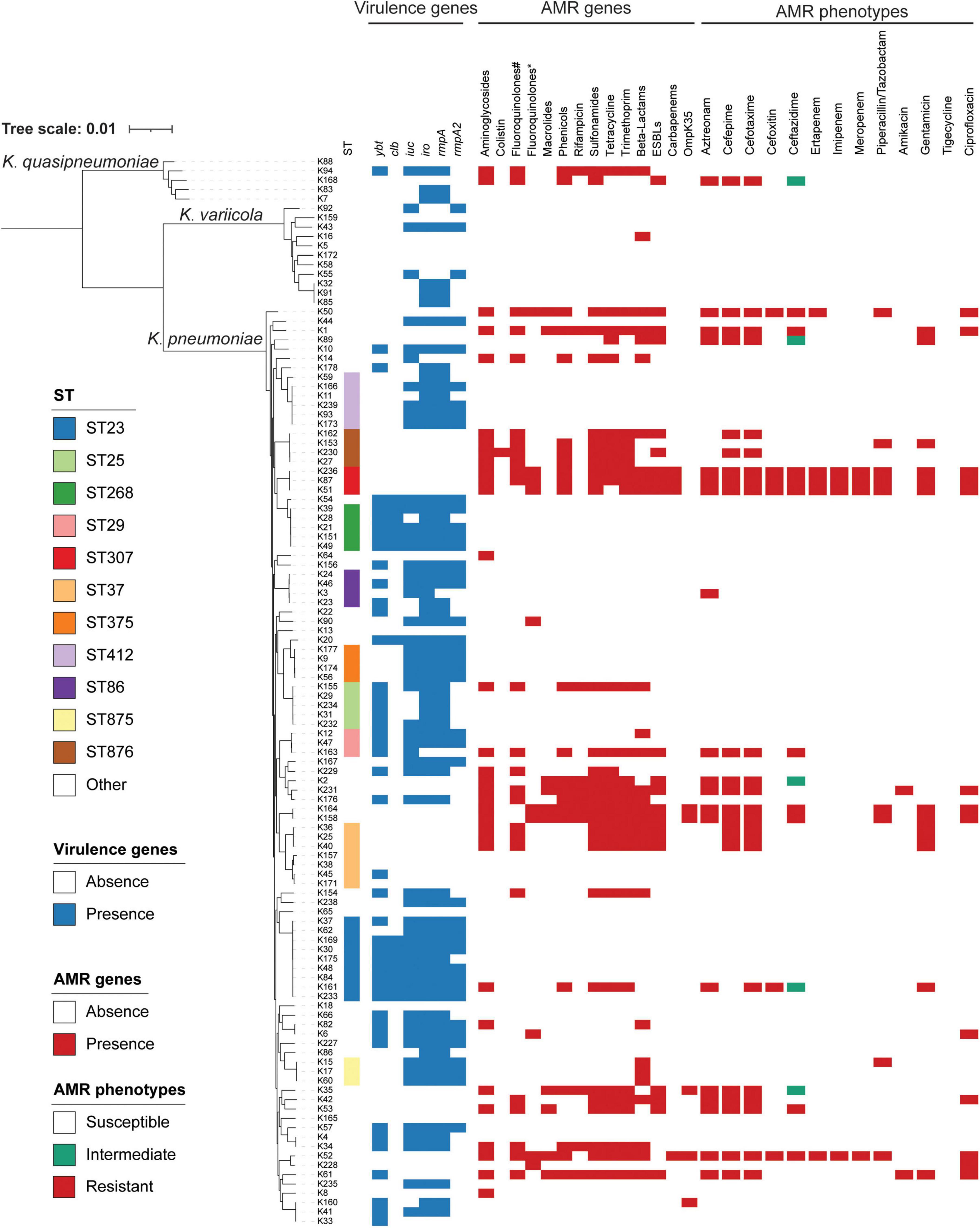
Figure 1. Phylogenetic analysis of 114 isolates. ST, virulence genes, AMR genes, and AMR phenotypes are indicated in different colors. The presence of OmpK35 stands for the mutation of OmpK35. ST, sequence typing; AMR, antimicrobial resistance; #fluoroquinolone resistance genes; *mutations found in the quinolone-resistance determining regions of GyrA and ParC.
Molecular epidemiology of K. pneumoniae isolates
The detailed molecular epidemiological characteristics of the K. pneumoniae isolates are presented in Figure 1. In this study, a total of 114 isolates belonging to the K. pneumoniae complex were identified, comprising three subspecies: K. pneumoniae (n = 98), K. quasipneumoniae subsp. similipneumoniae (n = 5), and K. variicola subsp. variicola (n = 11). MLST classified all isolates into 67 distinct sequence types (STs), with ST23 being the most prevalent (7.9%, 9/114), followed by ST37 (6.1%, 7/114) and ST412 (5.2%, 6/114). The 21 ESBL-positive K. pneumoniae isolates were distributed across 15 sequence types, with ST37 (n = 3) and ST307 (n = 3) being the most common, followed by ST876 (n = 2) and ST6839 (n = 2). The four carbapenemase-producing K. pneumoniae isolates belonged to two distinct sequence types: ST307, which harbored blaNDM–5 (n = 3), and ST709, which carried blaKPC–2 (n = 1).
Capsular types and virulence-associated determinants
Among the 114 K. pneumoniae isolates, 67 were assigned to 19 distinct capsular (K) types, whereas the remaining 47 isolates were non-typeable. Among the 67 K. pneumoniae isolates with identifiable K-types, K2 was the most prevalent serotype (n = 17), followed by KL107 (n = 13) and K1 (n = 12). In addition, 91 of the 114 K. pneumoniae isolates were assigned to eight distinct O-serotypes, with O1ab being the most prevalent (n = 49), followed by O2a (n = 12) and O5 (n = 12); the remaining 23 isolates were O-non-typeable. Details of capsular and LPS serotypes were provided in Supplementary Table 1.
The distribution of virulence-associated genes among the isolates is also presented in Figure 1. Among the 114 clinical K. pneumoniae isolates, the prevalence of virulence-associated genes was as follows: ybt (39.5%, 45/114), clb (12.3%, 14/114), iuc (50.0%, 57/114), iro (60.5%, 69/114), rmpA (59.6%, 68/114), and rmpA2 (36.8%, 42/114). In this study, the six virulence-associated genes were frequently detected in isolates belonging to ST23 (n = 9), ST412 (n = 6), ST25 (n = 5), ST268 (n = 5), and ST375 (n = 4), with ST23 being the most prevalent sequence type.
Although hypervirulent K. pneumoniae (HvKP) isolates are generally susceptible to most antimicrobial agents, the emergence of convergent K. pneumoniae strains that combine features of HvKP and CRKP has raised concern by linking virulence and resistance within single isolates (Chen and Kreiswirth, 2018). We did not identify such convergent isolates in our study; however, among K. pneumoniae isolates carrying the virulence-associated genes ybt, clb, iuc, iro, rmpA, and rmpA2, the proportions of MDR were 4.4% (2/45), 7.1% (1/14), 1.8% (1/57), 2.9% (2/69), 2.9% (2/68), and 2.4% (1/42), respectively. Details for all six virulence-associated genes are provided in Supplementary Table 1.
Discussion
According to the 2024 report from the CHINET1, the resistance rate of K. pneumoniae to cefotaxime was 41.8%, and the prevalence of CRKP reached 22.6%. However, the prevalence of antimicrobial resistance varied across provinces in China. For example, in Sichuan province, the resistance rates to cefotaxime and carbapenems in 2023 were 26.3 and 9.0%, respectively, according to data from the China Antimicrobial Resistance Surveillance System (CARSS)2. Our findings revealed cefotaxime and carbapenem resistance rates of 21.1 and 5.3%, respectively, among K. pneumoniae isolates from a district-level tertiary hospital in Chengdu, Sichuan—both of which were lower than the corresponding averages reported for Sichuan Province.
CRKP is predominantly associated with ST11 in China and South America, whereas ST258 is the dominant lineage in Europe and the United States (Wang M. et al., 2022). ST11 is a single-locus variant of ST258, differing at the tonB gene, and both belong to clonal complex 258 (CC258) (Guo et al., 2022). In China, KPC-2 is the predominant carbapenemase among CRKP isolates, accounting for up to 94% of cases (Wang M. et al., 2022). However, in the present study, three of the four carbapenemase-producing K. pneumoniae isolates harbored blaNDM–5, while only one carried blaKPC–2. Notably, the three NDM-5-positive isolates belonged to ST307, whereas the KPC-2-producing isolate was assigned to ST709. ST307 is recognized as an emerging high-risk antimicrobial-resistant clone with global distribution and increasing prevalence; however, its occurrence in China has remained sporadic and infrequent. For example, a recent study reported an outbreak of ST307 CRKP isolates producing NDM-5 in a teaching hospital in Shanghai, China (Zhu et al., 2024).
In our study, two CRKP isolates did not harbor any carbapenemase genes. Notably, one of these isolates exhibited resistance to ertapenem alone, while remaining susceptible to meropenem and imipenem. A previous study demonstrated that mutations in ramR can lead to overexpression of efflux pumps and suppression of the outer membrane porin OmpK35, a mechanism specifically associated with ertapenem resistance in K. pneumoniae (Wang D. et al., 2022).
Notably, ST23—a prototypical lineage of hvKp—emerged as the most dominant sequence type among the isolates analyzed in this study. In addition, the prevalence of six virulence-associated genes—ybt, clb, iuc, iro, rmpA, and rmpA2—was relatively high, with three of them (iuc, iro, and rmpA) present in over 50% of the 114 K. pneumoniae isolates. A previous study reported that 37.8% K. pneumoniae isolates in China were hvKp defined by aerobactin detection, and the prevalence of hvKp varied among different cities, with the highest rate in Wuhan (73.9%) and the lowest in Zhejiang (8.3%) (Zhang et al., 2016). All six virulence-associated genes were most frequently detected in ST23, followed by ST25, ST268, ST375, and ST412. These sequence types have all been associated with high virulence. For example, previous studies have reported that ST25 K. pneumoniae strains are implicated in hospital-acquired infections (Cienfuegos-Gallet et al., 2019). Moreover, ST25 has also been identified as the predominant sequence type among carbapenem-resistant hvKp strains in a hospital in south-central China (Li et al., 2019). Multiple studies have indicated that ESBL-producing K. pneumoniae ST268 strains are strongly associated with hvKp (Ku et al., 2017; Falgenhauer et al., 2019; Kakuta et al., 2020). K. pneumoniae K2-ST375 is considered one of the most clinically concerning hvKp lineages, capable of causing pyogenic liver abscesses and primary lung abscesses (Hoashi et al., 2019; Hirai et al., 2020; Xie et al., 2021); A previous study of global ST412 K. pneumoniae strains revealed that the earliest strain lacked virulence plasmids; however, within approximately 2 years, they progressively acquired plasmids harboring virulence genes, leading to their evolution into hvKp (Liu et al., 2023). Nonetheless, further studies are needed to assess the virulence and fitness of the isolates characterized in this study.
Limitation
In this study, the K. pneumoniae isolates were obtained from a single regional hospital, which may limit representativeness and generalizability. Although we compared our resistance rates with CHINET and CARSS (Qin et al., 2024) in the discussion, expanding the sample to include hospitals of different tiers would help clarify regional differences in resistance and virulence characteristics in future work.
Conclusion
In summary, our study of K. pneumoniae isolates from a district-level hospital in Chengdu revealed slightly lower resistance rates to cefotaxime and carbapenems compared to the provincial averages in Sichuan. However, a high prevalence of virulence-associated genotypes was observed, warranting further investigation to demonstrate the virulent phenotypes and the pathogenicity. Although the prevalence of MDR-HvKP was extremely low in this study, our findings highlight the urgent need for continuous surveillance and early intervention strategies to prevent the emergence of highly virulent and multidrug-resistant isolates in healthcare settings.
Data availability statement
The genome sequences have been deposited in the DDBJ/ENA/GenBank under the bioproject PRJNA676183.
Ethics statement
The studies involving humans were approved by West China Second University Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
YH: Investigation, Methodology, Writing – original draft, Conceptualization, Data curation. XL: Investigation, Writing – review & editing. YS: Writing – review & editing, Data curation. MY: Data curation, Writing – review & editing. LZ: Data curation, Writing – review & editing. JJ: Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. JL: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Shanghai under Award Number 25ZR1401040, and the Natural Science Foundation of Chengdu under Award Number 2022-YF05-01792-SN.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1700934/full#supplementary-material
Footnotes
References
Breurec, S., Guessennd, N., Timinouni, M., Le, T., Cao, V., Ngandjio, A., et al. (2013). Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: Multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol. Infect. 19, 349–355. doi: 10.1111/j.1469-0691.2012.03805.x
Chen, L., and Kreiswirth, B. (2018). Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect. Dis. 18, 2–3. doi: 10.1016/S1473-3099(17)30517-0
Choby, J., Howard-Anderson, J., and Weiss, D. (2020). Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern Med. 287, 283–300. doi: 10.1111/joim.13007
Cienfuegos-Gallet, A., Ocampo de Los Ríos, A. M., Sierra Viana, P., Ramirez Brinez, F., Restrepo Castro, C., and Roncancio Villamil, G. (2019). Risk factors and survival of patients infected with carbapenem-resistant Klebsiella pneumoniae in a KPC endemic setting: A case-control and cohort study. BMC Infect Dis. 19:830. doi: 10.1186/s12879-019-4461-x
CLSI (2019). Performance standards for antimicrobial susceptibility testing supplement M100, 29th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Falgenhauer, L., Schwengers, O., Schmiedel, J., Baars, C., Lambrecht, O., Heß, S., et al. (2019). Multidrug-resistant and clinically relevant gram-negative bacteria are present in german surface waters. Front. Microbiol. 10:2779. doi: 10.3389/fmicb.2019.02779
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J., Haendiges, J., Haft, D., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Guo, L., Wang, L., Zhao, Q., Ye, L., Ye, K., Ma, Y., et al. (2022). Genomic analysis of KPC-2-producing Klebsiella pneumoniae ST11 isolates at the respiratory department of a tertiary care hospital in Beijing. China. Front. Microbiol. 13:929826. doi: 10.3389/fmicb.2022.929826
Hao, M., Shi, X., Lv, J., Niu, S., Cheng, S., Du, H., et al. (2020). In vitro activity of apramycin against carbapenem-resistant and hypervirulent Klebsiella pneumoniae Isolates. Front. Microbiol. 11:425. doi: 10.3389/fmicb.2020.00425
Hirai, J., Sakanashi, D., Momose, M., Koga, T., Kinjo, T., Haranaga, S., et al. (2020). Case report of primary lung abscesses due to hypervirulent Klebsiella pneumoniae (Serotype K2, Sequence Type 375): An emerging isolate in Okinawa, Japan. Infect. Drug Resist. 13, 1691–1695. doi: 10.2147/IDR.S252251
Hoashi, K., Harada, S., Ishii, Y., Aoki, K., Ishikawa, S., Oshiro, Y., et al. (2019). Community-acquired liver abscess caused by capsular genotype K2-ST375 hypervirulent Klebsiella pneumoniae isolates. IDCases 17:e00577. doi: 10.1016/j.idcr.2019.e00577
Hu, Y., Liu, C., Shen, Z., Zhou, H., Cao, J., Chen, S., et al. (2020). Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg. Microbes Infect. 9, 1771–1779. doi: 10.1080/22221751.2020.1799721
Jiang, J., Wang, L., Hu, Y., Chen, X., Li, P., Zhang, J., et al. (2025). Global emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae driven by an IncFIIK34 KPC-2 plasmid. EBioMedicine 113:105627. doi: 10.1016/j.ebiom.2025.105627
Jin, X., Chen, Q., Shen, F., Jiang, Y., Wu, X., Hua, X., et al. (2021). Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerg. Microbes Infect. 10, 1129–1136. doi: 10.1080/22221751.2021.1937327
Kakuta, N., Nakano, R., Nakano, A., Suzuki, Y., Masui, T., Horiuchi, S., et al. (2020). Molecular characteristics of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in Japan: Predominance of CTX-M-15 and emergence of hypervirulent clones. Int. J. Infect. Dis. 98, 281–286. doi: 10.1016/j.ijid.2020.06.083
Ku, Y., Chuang, Y., Chen, C., Lee, M., Yang, Y., Tang, H., et al. (2017). Klebsiella pneumoniae isolates from meningitis: Epidemiology, virulence and antibiotic resistance. Sci. Rep. 7:6634. doi: 10.1038/s41598-017-06878-6
Lam, M., Wick, R., Watts, S., Cerdeira, L., Wyres, K., and Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12:4188. doi: 10.1038/s41467-021-24448-3
Lan, P., Jiang, Y., Zhou, J., and Yu, Y. (2021). A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 25, 26–34. doi: 10.1016/j.jgar.2021.02.020
Lee, H., Chuang, Y., Yu, W., Lee, N., Chang, C., Ko, N., et al. (2006). Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: Association with invasive syndrome in patients with community-acquired bacteraemia. J. Intern. Med. 259, 606–614. doi: 10.1111/j.1365-2796.2006.01641.x
Letunic, I., and Bork, P. (2024). Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82. doi: 10.1093/nar/gkae268
Li, J., Huang, Z., Yu, T., Tao, X., Hu, Y., Wang, H., et al. (2019). Isolation and characterization of a sequence type 25 carbapenem-resistant hypervirulent Klebsiella pneumoniae from the mid-south region of China. BMC Microbiol. 19:219. doi: 10.1186/s12866-019-1593-5
Liu, R., Xu, H., Zhao, J., Hu, X., Wu, L., Qiao, J., et al. (2023). Emergence of mcr-8.2-harboring hypervirulent ST412 Klebsiella pneumoniae strain from pediatric sepsis: A comparative genomic survey. Virulence 14, 233–245. doi: 10.1080/21505594.2022.2158980
Logan, L., and Weinstein, R. (2017). The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Long, S., Linson, S., Ojeda Saavedra, M., Cantu, C., Davis, J., Brettin, T., et al. (2017). Whole-genome sequencing of human clinical Klebsiella pneumoniae Isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2:e00290-17. doi: 10.1128/mSphereDirect.00290-17
Magiorakos, A., Srinivasan, A., Carey, R., Carmeli, Y., Falagas, M., Giske, C., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Martin, R., and Bachman, M. (2018). Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 8:4. doi: 10.3389/fcimb.2018.00004
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Price, M., Dehal, P., and Arkin, A. (2009). FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., and Korobeynikov, A. (2020). Using SPAdes De Novo assembler. Curr. Protoc. Bioinformatics 70:e102. doi: 10.1002/cpbi.102
Qin, X., Ding, L., Hao, M., Li, P., Hu, F., and Wang, M. (2024). Antimicrobial resistance of clinical bacterial isolates in China: Current status and trends. JAC Antimicrob. Resist. 6:dlae052. doi: 10.1093/jacamr/dlae052
Shankar, C., Jacob, J., Vasudevan, K., Biswas, R., Manesh, A., Sethuvel, D., et al. (2020). Emergence of multidrug resistant Hypervirulent ST23 Klebsiella pneumoniae: Multidrug resistant plasmid acquisition drives evolution. Front. Cell. Infect. Microbiol. 10:575289. doi: 10.3389/fcimb.2020.575289
Siu, L., Yeh, K., Lin, J., Fung, C., and Chang, F. (2012). Klebsiella pneumoniae liver abscess: A new invasive syndrome. Lancet Infect. Dis. 12, 881–887. doi: 10.1016/S1473-3099(12)70205-0
Tonkin-Hill, G., MacAlasdair, N., Ruis, C., Weimann, A., Horesh, G., Lees, J., et al. (2020). Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 21:180. doi: 10.1186/s13059-020-02090-4
Wang, D., Wang, M., He, T., Li, D., Zhang, L., Zhang, D., et al. (2022). Molecular epidemiology and mechanism of Klebsiella pneumoniae resistance to ertapenem but not to other carbapenems in China. Front. Microbiol. 13:974990. doi: 10.3389/fmicb.2022.974990
Wang, M., Earley, M., Chen, L., Hanson, B., Yu, Y., Liu, Z., et al. (2022). Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): A prospective, multicentre, cohort study. Lancet Infect. Dis. 22, 401–412. doi: 10.1016/S1473-3099(21)00399-6
Xie, M., Yang, X., Xu, Q., Ye, L., Chen, K., Zheng, Z., et al. (2021). Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun. Biol. 4:650. doi: 10.1038/s42003-021-02148-4
Zhang, Y., Zhao, C., Wang, Q., Wang, X., Chen, H., Li, H., et al. (2016). High prevalence of hypervirulent Klebsiella pneumoniae infection in China: Geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Chemother. 60, 6115–6120. doi: 10.1128/AAC.01127-16
Keywords: Klebsiella pneumoniae, resistance mechanisms, virulence, epidemiology, genomic analysis
Citation: Hu Y, Lu X, Sun Y, Yu M, Zhang L, Jiang J and Li J (2025) Clinical and genomic characterization of Klebsiella pneumoniae in a district hospital in Chengdu, China. Front. Microbiol. 16:1700934. doi: 10.3389/fmicb.2025.1700934
Received: 16 September 2025; Accepted: 28 October 2025;
Published: 19 November 2025.
Edited by:
Raffaele Zarrilli, University of Naples Federico II, ItalyReviewed by:
Anahit Sedrakyan, Institute of Molecular Biology (IMB), ArmeniaXiuru Guan, Harbin Medical University, China
Copyright © 2025 Hu, Lu, Sun, Yu, Zhang, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianping Jiang, amlhbmdqaWFucGluZ0BmdWRhbi5lZHUuY24=; Juan Li, NzIyMzc3MEBxcS5jb20=
 Yiyi Hu
Yiyi Hu Xingyu Lu1
Xingyu Lu1 Jianping Jiang
Jianping Jiang Juan Li
Juan Li