Abstract
A genetic predisposition to PML is now substantially supported by case reports of patients molecularly diagnosed with an inborn error of immunity (IEI) and progressive multifocal leukoencephalopathy (PML). Over the past 10 years, 4 IEI genes linked to PML has now grown to 26 as of 2025. Of these 26 genes believed to be causal of an IEI and PML, 24 (92%) are also linked with hemophagocytic lymphohistiocytosis/macrophage activation syndrome (HLH/MAS)—a severe hyper-inflammation syndrome associated with several IEI genes, most notably in 4 genes (PRF1, STX11, STXBP2, UNC13D) causing familial forms of the syndrome. Many HLH-linked genes are associated with life-threatening Epstein–Barr virus infections, which analogously suggests JC virus infection plus presence of a pathogenic variant in an HLH-linked IEI gene also increases risk of PML. PML also occurs as a serious adverse event for a subset of immunosuppressive therapies (e.g., natalizumab and rituximab) used to treat patients with immune disorders (e.g., multiple sclerosis and hematological malignancies). Recently, 4 PML risk variants were reported for use in a PML risk test to screen patients who are considering treatment with PML-linked therapies. Interestingly, of the 4 genes with a PML risk variant, 2 (LY9 and STXBP2) cause or are linked to HLH. The aim of our review is two-fold: (1) raise awareness among researchers and clinicians (e.g., neurologists, oncologists, and rheumatologists) that patient genetics are a key risk factor for PML, and (2) further reinforce the rationale for screening at-risk patients for PML risk variants before prescribing a PML-linked drug.
Introduction
Progressive multifocal leukoencephalopathy (PML) is a neurological disorder characterized by progressive white matter degeneration. PML occurs as a secondary and often fatal brain disease in immune-suppressed patients infected with human polyomavirus 2 (HPyV2) (1, 2), commonly known as JC virus (JCV) (3, 4). Immune-linked primary diseases associated with an increased risk of PML include HIV infection, hematological malignancies, and autoimmune disorders (3). Treatment of a patient’s primary disease with an immunosuppressant therapy (or non-compliance with antiretroviral therapy in HIV-infected patients) is often a triggering factor for developing PML. We propose that a patient’s underlying genetics are also a key risk factor for developing PML based on two lines of investigation: (1) our genome-wide study of two large cohorts of PML cases revealed four genes that increase PML risk (5, 6) and (2) our assembly of an updated review of the PML case report literature (Figure 1; Supplementary Table 1) on patients diagnosed with an inborn error of immunity (IEI) (7). We note that a majority (73%) of the cases reported in Supplementary Table 1 had a PML diagnosis of definite or probable (3, 8), but diagnostic criteria were not reported for the other PML case reports. PML is an under-appreciated risk in IEI patients and in the wide range of patients on immunosuppressant therapies. Our principal aims are to raise awareness in the clinical communities and increase the vigilance for PML onset, especially in patients with deleterious genetic variants in PML-linked IEI genes.
Figure 1
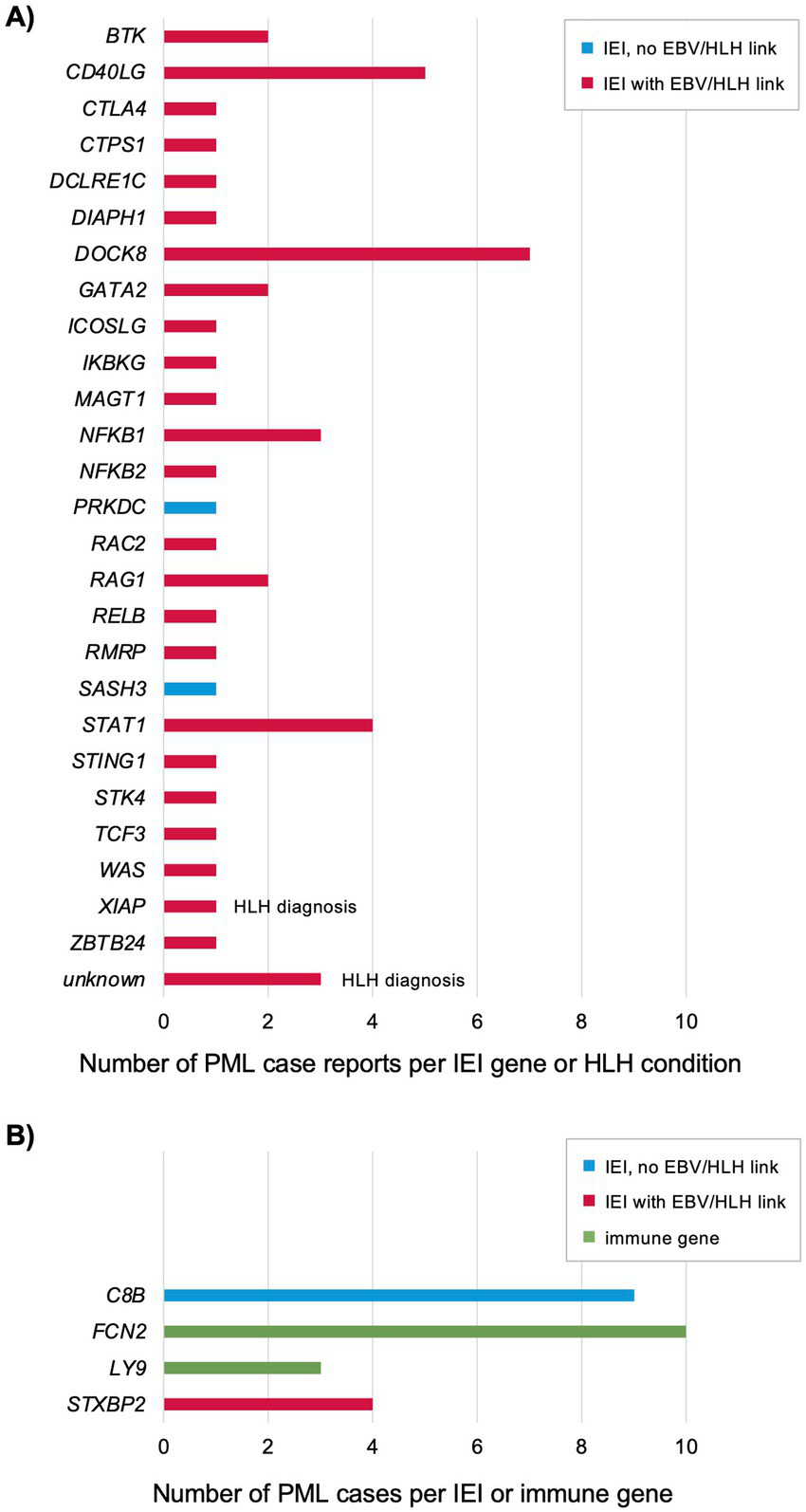
Genes reported for previously published PML cases with an IEI diagnosis, HLH diagnosis, or positive for a PML genetic risk variant (see Supplementary Table 1) (10, 20, 35–38, 96–127). (A) Each patient was genetically diagnosed with an IEI plus developed PML or diagnosed with HLH (indicated) and PML. All other PML cases were not reported to also have HLH, but 19 of 26 IEI genes are linked to an increased risk of EBV and/or HLH as associated features (7, 13, 16–28, 30–34, 128). (B) Each patient tested positive for a PML risk variant (5, 6). Two genes (C8B and FCN2) are in the complement pathway (129–133) and two genes (LY9 and STXBP2) are linked to EBV/HLH (see Figure 2).
Genetic underpinnings of PML—IEI case reports and PML risk variants
Host genetics as an underlying risk factor for PML were first proposed based on a limited number of case reports in patients diagnosed with an IEI and PML (9, 10). The International Union of Immunological Societies (IUIS) Expert Committee has reported there are now 508 IEI genes (7), but none are presently reported to cause an increased risk of PML. In the last 10 years, IEI genes linked to PML based on patients diagnosed with both disorders has increased from 4 to 26 (Figure 1A; Supplementary Table 1). Including PML cases found to have a PML risk variant (Figure 1B; Supplementary Table 1) (6), the total is 28 IEI genes. Notably, a majority of these IEI genes are directly causal or implicated in an increased risk of the hyperinflammation syndrome hemophagocytic lymphohistiocytosis (HLH) (11, 12) and/or severe Epstein–Barr virus (EBV) infections (7, 13), see below for details. Of the four PML risk variant genes (Figure 1B), two are known to cause an IEI, C8B is 1 of 33 IEI genes causing complement deficiencies and STXBP2 is 1 of 7 IEI genes causing familial HLH (FHL) syndromes. Two genes not yet known to cause an IEI are linked to complement and HLH, respectively—FCN2 is 1 of 3 ficolin genes (FCN3 is an IEI gene) and LY9 is linked to the EBV/HLH IEI gene SH2D1A via interaction of their protein products—see below for details. We also note that, like IEI in general, there is extensive genetic and phenotypic heterogeneity reported for IEI plus PML genes, with many (21 of 26, 81%) linked to the broader category of common variable immunodeficiency (CVID) (14). Incomplete penetrance is common and attributed to a number of factors, such as digenic/oligogenic/polygenic inheritance (14) and allele-specific expression (termed transcriptotype) (15). Thus, it should not be surprising that many individuals with an IEI are undiagnosed and PML only emerges upon treatment with immunosuppressive drugs (see below and Supplementary Table 2).
PML and the EBV/HLH connection
Primary HLH (FHL caused by an IEI gene) and secondary HLH (often a complication of rheumatic diseases)—also known as macrophage activation syndrome (MAS) and cytokine storm syndrome (CSS)—are now considered to be a continuum of immune dysfunction (11, 12). The term HLH/MAS has been adopted by experts in the field (12) but, for simplicity, herein will be termed HLH. Since the vast majority of PML case report patients (Figure 1A) were also diagnosed with an IEI linked to EBV/HLH (24 of 26, 92%) (7, 13, 16–34), we also searched the literature and public databases for case reports of patients diagnosed with PML and HLH. We found three cases although genetic information was not reported (unknown genes in Figure 1A; Supplementary Table 1) (35–37). Along with the XIAP plus PML case report (38), there are four patients with a diagnosis of PML and HLH. Given the rarity of PML (39, 40) and HLH (11), this is highly unlikely to be a chance association.
PML risk genes STXBP2 and LY9 further underscore the connection to EBV/HLH. Familial HLH (FHL syndromes) have a high risk for serious EBV infections (13, 23, 29). Of the four FHL genes, only STXBP2 (FHL5) is reported to have an inheritance model of autosomal dominant (AD) or autosomal recessive (AR), while all others are AR only (7). All four PML cases with the same STXBP2 variant were heterozygous (Supplementary Table 1) and in a comparison of natalizumab-treated multiple sclerosis (MS) patients who developed PML (n = 2/86) versus matched controls (natalizumab-treated MS patients who did not develop PML, n = 0/604) there was a 36-fold increased risk of PML (observed positive predictive value of 100%) (6). While LY9 is not known to cause an IEI, it is 1 of 9 signaling lymphocytic activation molecule family (SLAMF) members (41, 42) involved in host defense against pathogens (43, 44). SLAMF proteins interact with the protein product of SH2D1A (gene alias SAP), an IEI gene that causes X-linked lymphoproliferative syndrome (XLP1) characterized by severe EBV infections (13, 23, 29). Another SLAMF gene, CD48 (gene alias SLAMF2), is potentially the first family member linked to an IEI (not yet reported by the IUIS) and HLH based on one case report with a de novo variant (45). SLAMF genes are also implicated in cancers (particularly hematological malignancies) (46, 47) and autoimmune diseases such as rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) (48). Interestingly, the SLAMF locus on chromosome 1 (1q23) harbors all nine SLAMF genes and SLE genetic linkage studies in human (between markers SPTA1 and FCGR3A) (49) and mouse (50) also map to this region. Follow up studies further support the link between SLAMF genes and SLE (51–54).
To highlight the interactions between PML-linked IEI and other immune genes, we performed a protein network analysis using STRING (Figure 2) (55). The analysis included 37 genes: 26 IEI plus PML case report genes (Figure 1A), SH2D1A, PML risk variant genes (STXBP2 and LY9, but not complement pathway genes C8B and FCN2), and other SLAMF genes. All genes had multiple connections except ZBTB24 (Figure 2, upper right). However, 14 of 26 IEI plus PML genes (including ZBTB24) are linked to natural killer (NK) cell deficiency or impaired NK cell function (56, 57). PML risk genes LY9 and STXBP2 are also linked to NK cell function (57–59). Finally, while several complement system genes cause IEI (including PML risk gene C8B) (7), they are not extensively linked to HLH. However, more recent studies do show coexistence of defects in complement and HLH (60), particularly in patients diagnosed with both HLH and thrombotic microangiopathy (TMA) (61, 62).
Figure 2
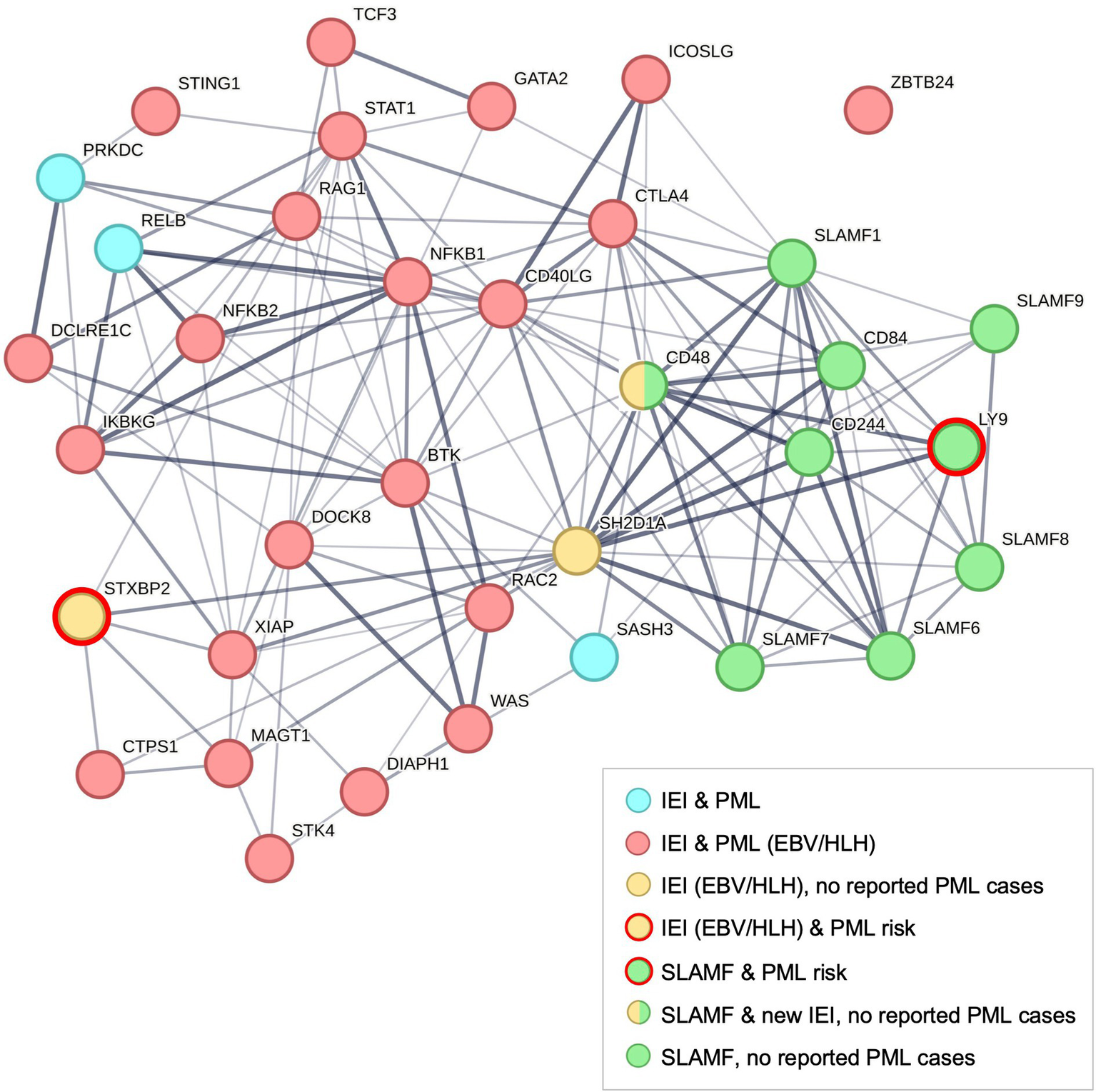
Protein network analysis of PML-linked IEI genes using STRING (55). Default STRING settings were used for 37 genes: 26 IEI + PML case report genes (see Figure 1), 9 SLAMF gene family members (41, 42), SLAMF-interacting gene SH2D1A (an IEI gene that causes XLP1) (7), and 2 PML risk genes (SLAMF gene LY9 and IEI gene STXBP2) (5, 6). The protein–protein interaction (PPI) enrichment p-value is < 1.0E-16. ZBTB24 is the only unconnected IEI & PML gene (upper right), see text. SLAMF gene CD48 (SLAMF2) was recently identified as HLH-linked and a potential new IEI, the first member of this gene family found to cause an immune disorder (45, 134). Both IEI and SLAMF genes have been linked to increased risk of autoimmune diseases (48) and hematological cancers (135, 136).
Functional evidence for PML-linked genes and JCV
For detailed background on JCV biology, see three recent reviews (4, 63, 64). One of the earliest links between JCV and an IEI gene is a study (65) that found JCV’s agnoprotein interacts with the protein product of XRCC6 (gene alias KU70, a DNA repair protein) and impairs function of the protein product of IEI gene PRKDC (66), which causes DNA-PKcs deficiency (7). Subsequent work (67, 68) by this group identified JCV protein links to two other DNA repair genes, IEI genes DCLRE1C (gene alias ARTEMIS) and RAD51 (a cause of Fanconi anemia) (7). PML case reports were found for patients with mutations in DCLRE1C and PRKDC (Figure 1A; Supplementary Table 1). Another group (69) reported an interaction between JCV’s agnoprotein and the protein product of AP3D1, which is an IEI gene that causes an FHL syndrome (Hermansky-Pudlak, type 10) (7). The agnoprotein-AP3D1 interaction was validated in a proteomics study (70). Adapter protein (AP) complexes are comprised of host gene proteins, such as AP3D1, that many viruses hijack for viral propagation and evading host immune responses (71).
Further evidence supporting PML-linked genes comes from a recent study (72) using proteomic and single cell RNA sequencing methods on cerebrospinal fluid (CSF) and serum samples from PML patients. Top genes/proteins from these analyses included chemokines and their receptors (e.g., CCL4, CCL5, CCR2, CCR5, CXCR3, CXCR6) as a key feature in PML versus non-PML samples. This is not surprising given their role in NK cell biology (58, 59), but this study also highlighted a link to PML risk genes LY9 and STXBP2 (6). We observed the following genes in the top quartile (25%, average log2 fold change) of genes in the RNA sequencing data: CD4-PML cluster vs. other CD4 + T cells included PML risk gene STXBP2, 2 SLAMF genes (SLAMF1, SLAMF6), 8 PML plus IEI case report genes (CD40LG, DOCK8, IKBKG, RAC2, RMRP, SASH3, STAT1, WAS), and ITGA4 (target of natalizumab); CD8-PML cluster vs. other CD8 + T cells included SLAMF7, 9 PML plus IEI case report genes (CTPS1, DIAPH1, DOCK8, RAC2, RELB, RMRP, SASH3, STAT1, WAS), and ITGA4.
Like other IEI disorders, severe infection risk is increased for some genes/viruses, although the IUIS has yet to add risk of PML due to JCV infection to a subset of their current list of 508 IEI genes (7), likely because PML from JCV infection is a rarer entity in IEI patients. We note two key parallels between EBV and JCV causing severe, life-threatening infections in IEI patients and those with milder immunodeficiency (e.g., lymphoma and SLE patients): (1) the ubiquitous presence of these viruses in worldwide populations (EBV > 90%, JCV 60–80%) (4, 13) wherein the infection is usually asymptomatic or relatively benign, and (2) both viruses are linked to HLH, which we think the current evidence suggests is due to host genetics (Figure 1; Supplementary Table 1). While co-infection with HIV and JCV leading to PML was common before the era of antiretroviral therapies (3, 4, 73), not much is known about patients co-infected with EBV and JCV. Two interesting observations that will hopefully lead to further research are EBV-JCV recombination leading to increased neurovirulence of JCV (74) and a case report of a HIV-infected patient who developed primary central nervous system lymphoma with tumors infected with both EBV and JCV (75).
Iatrogenic (drug-linked) PML as a serious adverse event (SAE) is not going away
In the first two epochs of reported PML cases, hematological malignancies and acquired immunodeficiency syndrome (AIDS) due to HIV infection were the main risk factors (3, 4). Around 2002–2005, reports of iatrogenic PML (i.e., drug-linked) were emerging (3). PML is now recognized as a significant risk factor in a wide range of patients treated with a wide range of immunosuppressive therapies for their primary disease. The highest number of drug-linked PML cases to date are from natalizumab (used to treat MS and Crohn’s disease) and rituximab, which is primarily used to treat cancers and autoimmune disorders (76–78). To assess the current landscape of drugs with the highest PML risk, we used the FDA Adverse Event Reporting System (FAERS)1 to identify the number of PML cases reported after treatment with a given drug (Supplementary Table 2). The FAERS database is an excellent resource even though underreporting is a limitation of this database (79–81) and not all reported PML cases will have been validated as definite/probable PML (3, 8). Our FAERS analysis focused on the past 5 years (2020–2024) in order to better represent the current situation for older drugs plus highlight newer drugs with an appreciable number of PML cases. To minimize counting duplicate reports of PML cases, we filtered the data using the original manufacturer (Sender) for a given drug, although this may result in an underreporting of PML cases linked to generic drugs. Also, since natalizumab is the highest risk PML-linked drug, when filtering the data we excluded instances for a given drug if natalizumab was also listed (i.e., oftentimes multiple drugs are listed for a given PML case). Drug data in Supplementary Table 2 are grouped according to four main indications (MS, hematological malignancies, non-MS autoimmune diseases such as RA and SLE, and other). We also highlighted PML risk drugs with a boxed warning (the highest warning issued by the FDA) for PML. Finally, we listed some newer drugs that have yet to report a PML case to the FDA if it had the same mechanism of action (MOA) as drugs already linked to PML.
Even after excluding the large number of historical PML cases (i.e., before 2020), the two highest risk drugs continue to be natalizumab and rituximab. We note rituximab PML cases are listed under 3 of 4 subsections of Supplementary Table 2, reflecting its use to treat a wide range of disorders. For MS drugs, natalizumab (n = 231) and fingolimod (n = 51, 58 for all drugs targeting S1P modulators) had the highest number of PML cases but ocrelizumab (n = 28) and other CD20-targeting drugs also have an appreciable number of reported PML cases (44 total in the MS section for all CD20 drugs). Importantly, we noted FAERS now reports instances of natalizumab patients treated with extended interval dosing (EID, also termed Q6W) (82, 83), which is reported under the Reactions column as “Prescribed Underdose.” About a third of natalizumab FAERS PML cases were classified as “prescribed underdose” but this did not reduce the death rate: regular dose 155 PML cases and 17% died vs. 76 underdose and 21% died.
Interestingly, we note that some MS drugs have been linked to cases of HLH (84–88), although we did not find any case reports of patients treated with these drugs who developed HLH and PML. Importantly, several IEI genes are also drug targets (Supplementary Table 2): BTK, C5, CD19, CD20 (gene symbol now MS4A1), CD3D, CD3E, CD3G, CD79B, JAK1, and JAK3 (7). The PML-linked drug belimumab targets TNFSF13B (gene aliases BAFF and BLYS), the ligand of IEI genes TNFRSF13B (gene alias TACI) and TNFRSF13C (gene alias BAFFR) (89). We also note there are several case reports of multiple myeloma (MM) patients diagnosed with PML (90–93). Reported drugs for a subset of these cases included bortezomib, daratumumab, ixazomib, lenalidomide, pomalidomide, and thalidomide. All of these MM drugs have been linked to ≥ 3 PML cases in FAERS (Supplementary Table 2). Elotuzumab, an MM drug that targets HLH-linked gene SLAMF7, has 5 PML cases reported in FAERS and for one case report the MM patient developed PML during treatment with lenalidomide and elotuzumab (93). There are presently limited or no warnings of PML in the prescribing information for these MM drugs (Supplementary Table 2) despite the growing number of PML cases reported to FAERS (e.g., in the last 5 years, there are 27 PML cases reported for daratumumab but still no warning of PML in its prescribing information). These observations, in concert, underscore the delicate balance of the immune system in having too much or too little of a given IEI gene product.
For clinicians and regulators, the key points to consider are: (1) drug-linked cases of PML occur for a wide range of drugs and primary diseases, (2) efforts to mitigate risk for natalizumab (e.g., EID/Q6W treatment regimen and regular JCV antibody testing) are insufficient, (3) additional early detection measures (more frequent brain MRIs, JCV testing, and other biomarkers) could be implemented for higher risk patients (not just MS patients), and (4) preventive testing for PML risk genetic variants/genes (6) may help reduce the number of drug-linked PML cases (94). Given that PML is often life-threatening, up-to-date information should be made available to clinicians, including via prescribing information (i.e., drug labels) (95), to better inform clinicians about recent advances in genetic testing for PML risk.
Constellation of PML risk factors: five recommendations for clinicians and regulators
Based on the continuing increase in PML plus IEI case reports and PML cases that carry a PML risk variant (Figure 1; Supplementary Table 1), a predominant risk factor of PML appears to be host genetics (Supplementary Figure 1). Primary diseases (each with their own predisposing genetic variants in immune-linked genes), immunosuppressant drugs, and infections (JCV is required but co-infection with HIV increases the risk and this may be true for other viruses linked to severe infections in IEI patients) also provide multiple pathways leading to the development of PML. Since there are no approved treatments for PML, prevention is the best defense. Therefore, we propose that experts in the field consider the following recommendations for increased vigilance of PML: (1) add JCV and PML to IUIS tables of IEI, (2) consider HLH/MAS (both primary and secondary) to be a concomitant risk factor of PML, (3) use the PML risk genetic test in all at risk patients (i.e., included but not limited to MS patients prior to treatment with natalizumab), (4) for at risk patients, such as carriers of the PML risk variants, implement more frequent brain MRIs plus more frequent and widespread JCV DNA and antibody testing, and (5) promote PML awareness campaigns to patients and clinicians for other diseases (and immunosuppressant drugs used to treat them) besides the MS community.
Statements
Author contributions
PE: Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. ES: Data curation, Formal analysis, Writing – review & editing. SJ: Data curation, Writing – review & editing. EH: Conceptualization, Investigation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We are grateful to the PML patients and their families who have participated in PML research over the years and our many dedicated and insightful collaborators with a shared goal to advance the field and save lives.
Conflict of interest
EH (UK), ES (USA), PE (USA) and SJ (UK) are employees of Population Bio, Inc.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Correction note
This article has been corrected with minor changes. These changes do not impact the scientific content of the article.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2025.1629581/full#supplementary-material
References
1.
Ehlers B Anoh AE Ben Salem N Broll S Couacy-Hymann E Fischer D et al . Novel polyomaviruses in mammals from multiple orders and reassessment of polyomavirus evolution and taxonomy. Viruses. (2019) 11:930. doi: 10.3390/v11100930
2.
Polyomaviridae Study Group of the International Committee on Taxonomy of Viruses Calvignac-Spencer S Feltkamp MC Daugherty MD Moens U Ramqvist T et al . A taxonomy update for the family Polyomaviridae. Arch Virol. (2016) 161:1739–50. doi: 10.1007/s00705-016-2794-y
3.
Cortese I Reich DS Nath A . Progressive multifocal leukoencephalopathy and the spectrum of JC virus-related disease. Nat Rev Neurol. (2021) 17:37–51. doi: 10.1038/s41582-020-00427-y
4.
Rocchi A Sariyer IK Berger JR . Revisiting JC virus and progressive multifocal leukoencephalopathy. J Neurovirol. (2023) 29:524–37. doi: 10.1007/s13365-023-01164-w
5.
Eis PS Bruno CD Richmond TA Koralnik IJ Hanson BA Major EO et al . Germline genetic risk variants for progressive multifocal leukoencephalopathy. Front Neurol. (2020) 11:186. doi: 10.3389/fneur.2020.00186
6.
Hatchwell E Smith EB III Jalilzadeh S Bruno CD Taoufik Y Hendel-Chavez H et al . Progressive multifocal leukoencephalopathy genetic risk variants for pharmacovigilance of immunosuppressant therapies. Front Neurol. (2022) 13:1016377. doi: 10.3389/fneur.2022.1016377
7.
Poli MC Aksentijevich I Bousfiha AA Cunningham-Rundles C Hambleton S Klein C et al . Human inborn errors of immunity: 2024 update on the classification from the International Union of Immunological Societies Expert Committee. J Hum Immun. (2025) 1:e20250003. doi: 10.70962/jhi.20250003
8.
Berger JR Aksamit AJ Clifford DB Davis L Koralnik IJ Sejvar JJ et al . PML diagnostic criteria: consensus statement from the AAN neuroinfectious disease section. Neurology. (2013) 80:1430–8. doi: 10.1212/WNL.0b013e31828c2fa1
9.
Hatchwell E . Is there a (host) genetic predisposition to progressive multifocal leukoencephalopathy?Front Immunol. (2015) 6:216. doi: 10.3389/fimmu.2015.00216
10.
Zerbe CS Marciano BE Katial RK Santos CB Adamo N Hsu AP et al . Progressive multifocal leukoencephalopathy in primary immune deficiencies: stat 1 gain of function and review of the literature. Clin Infect Dis. (2016) 62:986–94. doi: 10.1093/cid/civ1220
11.
Henter JI . Hemophagocytic Lymphohistiocytosis. N Engl J Med. (2025) 392:584–98. doi: 10.1056/NEJMra2314005
12.
Shakoory B Geerlinks A Wilejto M Kernan K Hines M Romano M et al . The 2022 EULAR/ACR points to consider at the early stages of diagnosis and management of suspected haemophagocytic lymphohistiocytosis/macrophage activation syndrome (HLH/MAS). Ann Rheum Dis. (2023) 82:1271–85. doi: 10.1136/ard-2023-224123
13.
Latour S . Human immune responses to Epstein-Barr virus highlighted by Immunodeficiencies. Annu Rev Immunol. (2025) 43:723–49. doi: 10.1146/annurev-immunol-082323-035455
14.
Peng XP Caballero-Oteyza A Grimbacher B . Common variable immunodeficiency: more pathways than roads to Rome. Annu Rev Pathol. (2023) 18:283–310. doi: 10.1146/annurev-pathmechdis-031521-024229
15.
Stewart O Gruber C Randolph HE Patel R Ramba M Calzoni E et al . Monoallelic expression can govern penetrance of inborn errors of immunity. Nature. (2025) 637:1186–97. doi: 10.1038/s41586-024-08346-4
16.
Boast B Goel S Gonzalez-Granado LI Niemela J Stoddard J Edwards ESJ et al . TCF3 haploinsufficiency defined by immune, clinical, gene-dosage, and murine studies. J Allergy Clin Immunol. (2023) 152:736–47. doi: 10.1016/j.jaci.2023.05.017
17.
Canna SW Marsh RA . Pediatric hemophagocytic lymphohistiocytosis. Blood. (2020) 135:1332–43. doi: 10.1182/blood.2019000936
18.
Fan H Yang Z Wu Y Lu X Li T Lu X et al . Human inborn errors of immunity underlying Talaromyces marneffei infections: a multicenter, retrospective cohort study. Front Immunol. (2025) 16:1492000. doi: 10.3389/fimmu.2025.1492000
19.
Kamae C Imai K Kato T Okano T Honma K Nakagawa N et al . Clinical and immunological characterization of ICF syndrome in Japan. J Clin Immunol. (2018) 38:927–37. doi: 10.1007/s10875-018-0559-y
20.
Kaustio M Nayebzadeh N Hinttala R Tapiainen T Astrom P Mamia K et al . Loss of DIAPH1 causes SCBMS, combined immunodeficiency, and mitochondrial dysfunction. J Allergy Clin Immunol. (2021) 148:599–611. doi: 10.1016/j.jaci.2020.12.656
21.
Klemann C Camacho-Ordonez N Yang L Eskandarian Z Rojas-Restrepo JL Frede N et al . Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front Immunol. (2019) 10:297. doi: 10.3389/fimmu.2019.00297
22.
La Rosée P Horne A Hines M von Bahr Greenwood T Machowicz R Berliner N et al . Recommendations for the management of hemophagocytic lymphohistiocytosis in adults. Blood. (2019) 133:2465–77. doi: 10.1182/blood.2018894618
23.
Latour S Fischer A . Signaling pathways involved in the T-cell-mediated immunity against Epstein-Barr virus: lessons from genetic diseases. Immunol Rev. (2019) 291:174–89. doi: 10.1111/imr.12791
24.
Leopizzi M Mundo L Messina E Campolo F Lazzi S Angeloni A et al . Epstein-Barr virus-encoded EBNA2 downregulates ICOSL by inducing mi R-24 in B-cell lymphoma. Blood. (2024) 143:429–43. doi: 10.1182/blood.2023021346
25.
Münz C . Co-stimulatory molecules during immune control of Epstein Barr virus infection. Biomol Ther. (2021) 12:38. doi: 10.3390/biom12010038
26.
Qiu KY Liao XY Wu RH Huang K Fang JP Zhou DH . X-linked hyper-IgM syndrome: a phenotype of Crohn's disease with Hemophagocytic Lymphohistiocytosis. Pediatr Hematol Oncol. (2017) 34:428–34. doi: 10.1080/08880018.2017.1409301
27.
Sathishkumar D Gach JE Ogboli M Desai M Cole T Hogler W et al . Cartilage hair hypoplasia with cutaneous lymphomatoid granulomatosis. Clin Exp Dermatol. (2018) 43:713–7. doi: 10.1111/ced.13543
28.
Schulert GS Cron RQ . The genetics of macrophage activation syndrome. Genes Immun. (2020) 21:169–81. doi: 10.1038/s41435-020-0098-4
29.
Tangye SG Latour S . Primary immunodeficiencies reveal the molecular requirements for effective host defense against EBV infection. Blood. (2020) 135:644–55. doi: 10.1182/blood.2019000928
30.
Thomsen MM Skouboe MK Mohlenberg M Zhao J de Keukeleere K Heinz JL et al . Impaired STING activation due to a variant in the E3 ubiquitin ligase AMFR in a patient with severe VZV infection and Hemophagocytic Lymphohistiocytosis. J Clin Immunol. (2024) 44:56. doi: 10.1007/s10875-024-01653-5
31.
Yan H Mo Y Liu S Luo X Liu L Zhou L et al . Case report: Hemophagocytic lymphohistiocytosis in a child with primary immunodeficiency infected with Talaromyces marneffei. Front Immunol. (2022) 13:1038354. doi: 10.3389/fimmu.2022.1038354
32.
Zeng Q Jin Y Yin G Yang D Li W Shi T et al . Peripheral immune profile of children with Talaromyces marneffei infections: a retrospective analysis of 21 cases. BMC Infect Dis. (2021) 21:287. doi: 10.1186/s12879-021-05978-z
33.
Zhang L Lv G Peng Y Yang L Chen J An Y et al . A novel RAC2 mutation causing combined immunodeficiency. J Clin Immunol. (2023) 43:229–40. doi: 10.1007/s10875-022-01373-8
34.
Zhang W Chen X Gao G Xing S Zhou L Tang X et al . Clinical relevance of gain- and loss-of-function germline mutations in STAT1: a systematic review. Front Immunol. (2021) 12:654406. doi: 10.3389/fimmu.2021.654406
35.
Ishikawa Y Kasuya T Ishikawa J Fujiwara M Kita Y . A case of developing progressive multifocal leukoencephalopathy while using rituximab and mycophenolate mofetil in refractory systemic lupus erythematosus. Ther Clin Risk Manag. (2018) 14:1149–53. doi: 10.2147/TCRM.S167109
36.
Koop A Zaied A Yataco J . A case of progressive multifocal leukoencephalopathy in a patient with EBV-associated hemophagocytic lymphohistiocytosis. Chest. (2016) 150:374A. doi: 10.1016/j.chest.2016.08.387
37.
Kumar A . The newly available FAERS public dashboard: implications for health care professionals. Hosp Pharm. (2019) 54:75–7. doi: 10.1177/0018578718795271
38.
Seguier J Briantais A Ebbo M Meunier B Aurran T Coze S et al . Late-onset progressive multifocal leukoencephalopathy (PML) and lymphoma in a 65-year-old patient with XIAP deficiency. J Clin Immunol. (2021) 41:1975–8. doi: 10.1007/s10875-021-01139-8
39.
Iacobaeus E Burkill S Bahmanyar S Hakim R Bystrom C Fored M et al . The national incidence of PML in Sweden, 1988-2013. Neurology. (2018) 90:e498–506. doi: 10.1212/WNL.0000000000004926
40.
Joly M Conte C Cazanave C Le Moing V Tattevin P Delobel P et al . Progressive multifocal leukoencephalopathy: epidemiology and spectrum of predisposing conditions. Brain. (2023) 146:349–58. doi: 10.1093/brain/awac237
41.
Engel P Eck MJ Terhorst C . The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat Rev Immunol. (2003) 3:813–21. doi: 10.1038/nri1202
42.
Ma CS Nichols KE Tangye SG . Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu Rev Immunol. (2007) 25:337–79. doi: 10.1146/annurev.immunol.25.022106.141651
43.
Angulo A Cuenca M Martinez-Vicente P Engel P . Viral CD229 (Ly9) homologs as new manipulators of host immunity. J Leukoc Biol. (2019) 105:947–54. doi: 10.1002/JLB.2MR1018-413R
44.
van Driel BJ Liao G Engel P Terhorst C . Responses to microbial challenges by SLAMF receptors. Front Immunol. (2016) 7:4. doi: 10.3389/fimmu.2016.00004
45.
Volkmer B Planas R Gossweiler E Lunemann A Opitz L Mauracher A et al . Recurrent inflammatory disease caused by a heterozygous mutation in CD48. J Allergy Clin Immunol. (2019) 144:1441–1445.e17. doi: 10.1016/j.jaci.2019.07.038
46.
Fouquet G Marcq I Debuysscher V Bayry J Rabbind Singh A Bengrine A et al . Signaling lymphocytic activation molecules Slam and cancers: friends or foes?Oncotarget. (2018) 9:16248–62. doi: 10.18632/oncotarget.24575
47.
Ishibashi M Morita R Tamura H . Immune functions of signaling lymphocytic activation molecule family molecules in multiple myeloma. Cancers (Basel). (2021) 13:279. doi: 10.3390/cancers13020279
48.
Gartshteyn Y Askanase AD Mor A . SLAM associated protein signaling in T cells: tilting the balance toward autoimmunity. Front Immunol. (2021) 12:654839. doi: 10.3389/fimmu.2021.654839
49.
Moser KL Neas BR Salmon JE Yu H Gray-McGuire C Asundi N et al . Genome scan of human systemic lupus erythematosus: evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci USA. (1998) 95:14869–74. doi: 10.1073/pnas.95.25.14869
50.
Wandstrat AE Nguyen C Limaye N Chan AY Subramanian S Tian XH et al . Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. (2004) 21:769–80. doi: 10.1016/j.immuni.2004.10.009
51.
Cunninghame Graham DS Vyse TJ Fortin PR Montpetit A Cai YC Lim S et al . Association of LY9 in UK and Canadian SLE families. Genes Immun. (2008) 9:93–102. doi: 10.1038/sj.gene.6364453
52.
Humbel M Bellanger F Horisberger A Suffiotti M Fluder N Makhmutova M et al . SLAMF receptor expression identifies an immune signature that characterizes systemic lupus erythematosus. Front Immunol. (2022) 13:843059. doi: 10.3389/fimmu.2022.843059
53.
Karampetsou MP Comte D Kis-Toth K Kyttaris VC Tsokos GC . Expression patterns of signaling lymphocytic activation molecule family members in peripheral blood mononuclear cell subsets in patients with systemic lupus erythematosus. PLoS One. (2017) 12:e0186073. doi: 10.1371/journal.pone.0186073
54.
Liu Q Deng Y Liu X Zheng Y Li Q Cai G et al . Transcriptomic analysis of B cells suggests that CD70 and LY9 may be novel features in patients with systemic lupus erythematosus. Heliyon. (2023) 9:e15684. doi: 10.1016/j.heliyon.2023.e15684
55.
Szklarczyk D Nastou K Koutrouli M Kirsch R Mehryary F Hachilif R et al . The STRING database in 2025: protein networks with directionality of regulation. Nucleic Acids Res. (2025) 53:D730–7. doi: 10.1093/nar/gkae1113
56.
Abdalgani M Hernandez ER Pedroza LA Chinn IK Forbes Satter LR Rider NL et al . Clinical, immunologic, and genetic characteristics of 148 patients with natural killer cell deficiency. J Allergy Clin Immunol. (2025) 155:1623–34. doi: 10.1016/j.jaci.2025.01.030
57.
Mace EM Orange JS . Emerging insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol Rev. (2019) 287:202–25. doi: 10.1111/imr.12725
58.
Mace EM . Human natural killer cells: form, function, and development. J Allergy Clin Immunol. (2023) 151:371–85. doi: 10.1016/j.jaci.2022.09.022
59.
Vivier E Raulet DH Moretta A Caligiuri MA Zitvogel L Lanier LL et al . Innate or adaptive immunity? The example of natural killer cells. Science. (2011) 331:44–9. doi: 10.1126/science.1198687
60.
Shu X Gao X Dai Y Wang Y Liu Y Wang D et al . C3 as a predictive and prognostic biomarker in adult hemophagocytic lymphohistiocytosis: a large cohort study in China. Blood Adv. (2025) 9:1836–46. doi: 10.1182/bloodadvances.2024014715
61.
Gloude NJ Dandoy CE Davies SM Myers KC Jordan MB Marsh RA et al . Thinking beyond HLH: clinical features of patients with concurrent presentation of Hemophagocytic Lymphohistiocytosis and thrombotic Microangiopathy. J Clin Immunol. (2020) 40:699–707. doi: 10.1007/s10875-020-00789-4
62.
Minoia F Tibaldi J Muratore V Gallizzi R Bracaglia C Arduini A et al . Thrombotic Microangiopathy associated with macrophage activation syndrome: a multinational study of 23 patients. J Pediatr. (2021) 235:196–202. doi: 10.1016/j.jpeds.2021.04.004
63.
Atkinson AL Atwood WJ . Fifty years of JC polyomavirus: a brief overview and remaining questions. Viruses. (2020) 12:969. doi: 10.3390/v12090969
64.
Butic AB Spencer SA Shaheen SK Lukacher AE . Polyomavirus wakes up and chooses neurovirulence. Viruses. (2023) 15:2112. doi: 10.3390/v15102112
65.
Darbinyan A Siddiqui KM Slonina D Darbinian N Amini S White MK et al . Role of JC virus agnoprotein in DNA repair. J Virol. (2004) 78:8593–600. doi: 10.1128/JVI.78.16.8593-8600.2004
66.
Yue X Bai C Xie D Ma T Zhou PK . DNA-PKcs: a multi-faceted player in DNA damage response. Front Genet. (2020) 11:607428. doi: 10.3389/fgene.2020.607428
67.
Darbinyan A White MK Akan S Radhakrishnan S Del Valle L Amini S et al . Alterations of DNA damage repair pathways resulting from JCV infection. Virology. (2007) 364:73–86. doi: 10.1016/j.virol.2007.02.015
68.
Trojanek J Croul S Ho T Wang JY Darbinyan A Nowicki M et al . T-antigen of the human polyomavirus JC attenuates faithful DNA repair by forcing nuclear interaction between IRS-1 and rad 51. J Cell Physiol. (2006) 206:35–46. doi: 10.1002/jcp.20425
69.
Suzuki T Orba Y Makino Y Okada Y Sunden Y Hasegawa H et al . Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc Natl Acad Sci USA. (2013) 110:18668–73. doi: 10.1073/pnas.1311457110
70.
Saribas AS Datta PK Safak M . A comprehensive proteomics analysis of JC virus Agnoprotein-interacting proteins: Agnoprotein primarily targets the host proteins with coiled-coil motifs. Virology. (2020) 540:104–18. doi: 10.1016/j.virol.2019.10.005
71.
Strazic Geljic I Kucan Brlic P Musak L Karner D Ambriovic-Ristov A Jonjic S et al . Viral interactions with adaptor-protein complexes: a ubiquitous trait among viral species. Int J Mol Sci. (2021) 22:5274. doi: 10.3390/ijms22105274
72.
Deffner M Schneider-Hohendorf T Schulte-Mecklenbeck A Falk S Lu IN Ostkamp P et al . Chemokine-mediated cell migration into the central nervous system in progressive multifocal leukoencephalopathy. Cell Rep Med. (2024) 5:101622. doi: 10.1016/j.xcrm.2024.101622
73.
Schweitzer F Laurent S Cortese I Fink GR Silling S Skripuletz T et al . Progressive multifocal leukoencephalopathy: pathogenesis, diagnostic tools, and potential biomarkers of response to therapy. Neurology. (2023) 101:700–13. doi: 10.1212/WNL.0000000000207622
74.
Wortman MJ Lundberg PS Dagdanova AV Venkataraman P Daniel DC Johnson EM . Opportunistic DNA recombination with Epstein-Barr virus at sites of control region rearrangements mediating JC virus Neurovirulence. J Infect Dis. (2016) 213:1436–43. doi: 10.1093/infdis/jiv755
75.
Barbier MT Del Valle L . Co-detection of EBV and human polyomavirus JCPyV in a case of AIDS-related multifocal primary central nervous system diffuse large B-cell lymphoma. Viruses. (2023) 15:755. doi: 10.3390/v15030755
76.
Bennett CL Focosi D Socal MP Bian JC Nabhan C Hrushesky WJ et al . Progressive multifocal leukoencephalopathy in patients treated with rituximab: a 20-year review from the southern network on adverse reactions. Lancet Haematol. (2021) 8:e593–604. doi: 10.1016/S2352-3026(21)00167-8
77.
Brancati S Gozzo L Longo L Vitale DC Drago F . Rituximab in multiple sclerosis: are we ready for regulatory approval?Front Immunol. (2021) 12:661882. doi: 10.3389/fimmu.2021.661882
78.
Sarsour K Beckley-Kartey S Melega S Odueyungbo A Kirchner P Khalife N et al . Rituximab utilization for approved and off-label nononcology indications and patients' experiences with the patient alert card. Pharmacol Res Perspect. (2020) 8:e00555. doi: 10.1002/prp2.555
79.
Alatawi YM Hansen RA . Empirical estimation of under-reporting in the U.S. Food and Drug Administration adverse event reporting system (FAERS). Expert Opin Drug Saf. (2017) 16:761–7. doi: 10.1080/14740338.2017.1323867
80.
Hazell L Shakir SA . Under-reporting of adverse drug reactions: a systematic review. Drug Saf. (2006) 29:385–96. doi: 10.2165/00002018-200629050-00003
81.
Sakaeda T Tamon A Kadoyama K Okuno Y . Data mining of the public version of the FDA adverse event reporting system. Int J Med Sci. (2013) 10:796–803. doi: 10.7150/ijms.6048
82.
Foley JF Defer G Ryerson LZ Cohen JA Arnold DL Butzkueven H et al . Pharmacokinetics and pharmacodynamics of Natalizumab 6-week dosing vs continued 4-week dosing for relapsing-remitting multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. (2024) 11:e200321. doi: 10.1212/NXI.0000000000200321
83.
Ryerson LZ Foley J Chang I Kister I Cutter G Metzger RR et al . Risk of natalizumab-associated PML in patients with MS is reduced with extended interval dosing. Neurology. (2019) 93:e1452–62. doi: 10.1212/WNL.0000000000008243
84.
Ikumi K Ando T Katano H Katsuno M Sakai Y Yoshida M et al . HSV-2-related hemophagocytic lymphohistiocytosis in a fingolimod-treated patient with MS. Neurol Neuroimmunol Neuroinflamm. (2016) 3:e247. doi: 10.1212/NXI.0000000000000247
85.
Machlańska A Helbig G Chromik K Zapała M Zwiernik B Selmaj K . Hemophagocytic lymphohistiocytosis associated with ocrelizumab treatment in a patient with multiple sclerosis. Mult Scler. (2021) 27:1803–5. doi: 10.1177/1352458521993070
86.
Ošep AB Brecl E Skerget M Savsek L . An unforeseen reality: Hemophagocytic lymphohistiocytosis following alemtuzumab treatment for a multiple sclerosis. Clin Neurol Neurosurg. (2023) 228:107675. doi: 10.1016/j.clineuro.2023.107675
87.
Romero A Midaglia L Salcedo MT Viladomiu L Guillen E Bajana I et al . Hemophagocytic syndrome following alemtuzumab treatment for multiple sclerosis: a case report. Mult Scler Relat Disord. (2020) 40:101973. doi: 10.1016/j.msard.2020.101973
88.
Saarela M Senthil K Jones J Tienari PJ Soilu-Hanninen M Airas L et al . Hemophagocytic lymphohistiocytosis in 2 patients with multiple sclerosis treated with alemtuzumab. Neurology. (2018) 90:849–51. doi: 10.1212/WNL.0000000000005420
89.
Smulski CR Eibel H . BAFF and BAFF-receptor in B cell selection and survival. Front Immunol. (2018) 9:2285. doi: 10.3389/fimmu.2018.02285
90.
Hoeynck BW Cohen AD Stadtmauer EA Susanibar-Adaniya SP Vogl DT Waxman AJ et al . Progressive multifocal leukoencephalopathy in multiple myeloma. Eur J Haematol. (2023) 110:322–9. doi: 10.1111/ejh.13909
91.
Koutsavlis I . Progressive multifocal leukoencephalopathy in multiple myeloma. A literature review and lessons to learn. Ann Hematol. (2021) 100:1–10. doi: 10.1007/s00277-020-04294-x
92.
Usui K Kitazaki Y Enomoto S Morita M Nakamichi K Hamano T . A case of progressive multifocal leukoencephalopathy associated with daratumumab, bortezomib, and dexamethasone for multiple myeloma. Rinsho Shinkeigaku. (2023) 63:513–7. doi: 10.5692/clinicalneurol.cn-001847
93.
Usui Y Nakano H Komatsu J Nakamichi K Saijo M Takano S et al . Progressive multifocal leukoencephalopathy during treatment with lenalidomide and elotuzumab for multiple myeloma. Leuk Lymphoma. (2020) 61:2234–7. doi: 10.1080/10428194.2020.1765237
94.
Berger JR Hartung HP . Commentary: progressive multifocal leukoencephalopathy genetic risk variants for pharmacovigilance of immunosuppressant therapies. Front Neurol. (2023) 14:1146027. doi: 10.3389/fneur.2023.1146027
95.
Pacanowski M Schuck RN . Evidence, in context: a regulatory perspective on Pharmacogenetics. Clin Pharmacol Ther. (2022) 111:1202–4. doi: 10.1002/cpt.2347
96.
Adelon J Abolhassani H Esenboga S Fouyssac F Cagdas D Tezcan I et al . Human DNA-dependent protein kinase catalytic subunit deficiency: a comprehensive review and update. J Allergy Clin Immunol. (2024) 154:1300–12. doi: 10.1016/j.jaci.2024.06.018
97.
Al Shekaili L Sheikh F Al Gazlan S Al Dhekri H Al Mousa H Al Ghonaium A et al . Novel mutation in DOCK8-HIES with severe phenotype and successful transplantation. Clin Immunol. (2017) 178:39–44. doi: 10.1016/j.clim.2016.08.002
98.
Aschermann Z Gomori E Kovacs GG Pal E Simon G Komoly S et al . X-linked hyper-IgM syndrome associated with a rapid course of multifocal leukoencephalopathy. Arch Neurol. (2007) 64:273–6. doi: 10.1001/archneur.64.2.273
99.
Bahrami S Arshi S Nabavi M Bemanian MH Fallahpour M Rezaeifar A et al . Progressive multifocal leukoencephalopathy in a patient with novel mutation in the RAC2 gene: a case report. J Med Case Rep. (2022) 16:235. doi: 10.1186/s13256-022-03333-7
100.
Day-Williams AG Sun C Jelcic I McLaughlin H Harris T Martin R et al . Whole genome sequencing reveals a chromosome 9p deletion causing DOCK8 deficiency in an adult diagnosed with hyper IgE syndrome who developed progressive multifocal leukoencephalopathy. J Clin Immunol. (2015) 35:92–6. doi: 10.1007/s10875-014-0114-4
101.
Delmonte OM Bergerson JRE Kawai T Kuehn HS McDermott DH Cortese I et al . SASH3 variants cause a novel form of X-linked combined immunodeficiency with immune dysregulation. Blood. (2021) 138:1019–33. doi: 10.1182/blood.2020008629
102.
Dhalla F Murray S Sadler R Chaigne-Delalande B Sadaoka T Soilleux E et al . Identification of a novel mutation in MAGT1 and progressive multifocal leucoencephalopathy in a 58-year-old man with XMEN disease. J Clin Immunol. (2015) 35:112–8. doi: 10.1007/s10875-014-0116-2
103.
Dobbs K Tabellini G Calzoni E Patrizi O Martinez P Giliani SC et al . Natural killer cells from patients with recombinase-activating gene and non-homologous end joining gene defects comprise a higher frequency of CD56bright NKG2A+++ cells, and yet display increased degranulation and higher Perforin content. Front Immunol. (2017) 8:798. doi: 10.3389/fimmu.2017.00798
104.
Donadieu J Lamant M Fieschi C de Fontbrune FS Caye A Ouachee M et al . Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica. (2018) 103:1278–87. doi: 10.3324/haematol.2017.181909
105.
Downes SM Black GC Hyman N Simmonds M Morris J Barton C . Visual loss due to progressive multifocal leukoencephalopathy in a congenital immunodeficiency disorder. Arch Ophthalmol. (2001) 119:1376–8. doi: 10.1001/archopht.119.9.1376
106.
Durkee-Shock J Zhang A Liang H Wright H Magnusson J Garabedian E et al . Morbidity, mortality, and therapeutics in combined immunodeficiency: data from the USIDNET registry. J Allergy Clin Immunol Pract. (2022) 10:1334–1341.e6. doi: 10.1016/j.jaip.2022.01.042
107.
Emmanouilidou E Kosmara D Papadaki E Mastorodemos V Constantoulakis P Repa A et al . Progressive multifocal leukoencephalopathy in systemic lupus erythematosus: a consequence of patient-intrinsic or -extrinsic factors?J Clin Med. (2023) 12:6945. doi: 10.3390/jcm12216945
108.
Engelhardt KR Gertz ME Keles S Schaffer AA Sigmund EC Glocker C et al . The extended clinical phenotype of 64 patients with dedicator of cytokinesis 8 deficiency. J Allergy Clin Immunol. (2015) 136:402–12. doi: 10.1016/j.jaci.2014.12.1945
109.
Engelhardt KR McGhee S Winkler S Sassi A Woellner C Lopez-Herrera G et al . Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. (2009) 124:1289–1302.e4. doi: 10.1016/j.jaci.2009.10.038
110.
Hadjadj J Guffroy A Delavaud C Taieb G Meyts I Fresard A et al . Progressive multifocal leukoencephalopathy in primary Immunodeficiencies. J Clin Immunol. (2019) 39:55–64. doi: 10.1007/s10875-018-0578-8
111.
Hernandez-Trujillo V Zhou C Scalchunes C Ochs HD Sullivan KE Cunningham-Rundles C et al . A registry study of 240 patients with X-linked Agammaglobulinemia living in the USA. J Clin Immunol. (2023) 43:1468–77. doi: 10.1007/s10875-023-01502-x
112.
Le Voyer T Maglorius Renkilaraj MRL Moriya K Perez Lorenzo M Nguyen T Gao L et al . Inherited human Rel B deficiency impairs innate and adaptive immunity to infection. Proc Natl Acad Sci USA. (2024) 121:e2321794121. doi: 10.1073/pnas.2321794121
113.
Le Voyer T Parent AV Liu X Cederholm A Gervais A Rosain J et al . Autoantibodies against type I IFNs in humans with alternative NF-kappa B pathway deficiency. Nature. (2023) 623:803–13. doi: 10.1038/s41586-023-06717-x
114.
Li Q Tang C Zhu J Zhang L . A case of progressive multifocal leukoencephalopathy with hypogammaglobulinemia and a TCF3 mutation. J Neurovirol. (2022) 28:616–8. doi: 10.1007/s13365-022-01092-1
115.
Lorenzini T Fliegauf M Klammer N Frede N Proietti M Bulashevska A et al . Characterization of the clinical and immunologic phenotype and management of 157 individuals with 56 distinct heterozygous NFKB1 mutations. J Allergy Clin Immunol. (2020) 146:901–11. doi: 10.1016/j.jaci.2019.11.051
116.
MacDougall M Afzal S Jiang S McGhee S Lewis D . A case of inducible T-cell co-stimulator ligand deficiency with severe viral infection and autoimmune hepatitis. Ann Allergy Asthma Immunol. (2024) 133:S179–80. doi: 10.1016/j.anai.2024.08.698
117.
Maffucci P Filion CA Boisson B Itan Y Shang L Casanova JL et al . Genetic diagnosis using whole exome sequencing in common variable immunodeficiency. Front Immunol. (2016) 7:220. doi: 10.3389/fimmu.2016.00220
118.
Mørup SB Nazaryan-Petersen L Gabrielaite M Reekie J Marquart HV Hartling HJ et al . Added value of reanalysis of whole exome- and whole genome sequencing data from patients suspected of primary immune deficiency using an extended gene panel and structural variation calling. Front Immunol. (2022) 13:906328. doi: 10.3389/fimmu.2022.906328
119.
Nademi Z Wynn RF Slatter M Hughes SM Bonney D Qasim W et al . Hematopoietic stem cell transplantation for cytidine triphosphate synthase 1 (CTPS1) deficiency. Bone Marrow Transplant. (2019) 54:130–3. doi: 10.1038/s41409-018-0246-x
120.
Nitta H Unoki M Ichiyanagi K Kosho T Shigemura T Takahashi H et al . Three novel ZBTB24 mutations identified in Japanese and cape Verdean type 2 ICF syndrome patients. J Hum Genet. (2013) 58:455–60. doi: 10.1038/jhg.2013.56
121.
Sampaio EP Hsu AP Pechacek J Bax HI Dias DL Paulson ML et al . Signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations and disseminated coccidioidomycosis and histoplasmosis. J Allergy Clin Immunol. (2013) 131:1624–1634.e17. doi: 10.1016/j.jaci.2013.01.052
122.
Schröder C Baerlecken NT Pannicke U Dörk T Witte T Jacobs R et al . Evaluation of RAG1 mutations in an adult with combined immunodeficiency and progressive multifocal leukoencephalopathy. Clin Immunol. (2017) 179:1–7. doi: 10.1016/j.clim.2016.12.013
123.
Schwartzmann Y Vaknin-Dembinsky A Gomori JM Elinav H Berkun Y Levin N et al . Tofacitinib-induced progressive multifocal leukoencephalopathy-immune reconstitution inflammatory syndrome. Neurol Sci. (2023) 44:3737–9. doi: 10.1007/s10072-023-06897-4
124.
Suzuki H Takahashi Y Miyajima H . Progressive multifocal leukoencephalopathy complicating X-linked hyper-IgM syndrome in an adult. Intern Med. (2006) 45:1187–8. doi: 10.2169/internalmedicine.45.6023
125.
Teramoto T Kaneko H Funato M Sawa H Nagashima K Hirose Y et al . Progressive multifocal leukoencephalopathy in a patient with X-linked agammaglobulinemia. Scand J Infect Dis. (2003) 35:909–10. doi: 10.1080/00365540310016673
126.
Tuijnenburg P Lango Allen H Burns SO Greene D Jansen MH Staples E et al . Loss-of-function nuclear factor kappa B subunit 1 (NFKB1) variants are the most common monogenic cause of common variable immunodeficiency in Europeans. J Allergy Clin Immunol. (2018) 142:1285–96. doi: 10.1016/j.jaci.2018.01.039
127.
Volk T Warnatz K Marks R Urbach H Schluh G Strohmeier V et al . Pembrolizumab for treatment of progressive multifocal leukoencephalopathy in primary immunodeficiency and/or hematologic malignancy: a case series of five patients. J Neurol. (2022) 269:973–81. doi: 10.1007/s00415-021-10682-8
128.
Almeida DPM Reis B Faccion RS Cunha D Agonigi B Machado L et al . Novel non-coding variant in DCLRE1C segregates with distinct severe combined immunodeficiency phenotypes and Hodgkin lymphoma in consanguineous siblings. Hematol Transfus Cell Ther. (2024) 46:S208–9. doi: 10.1016/j.htct.2024.09.349
129.
Agrawal P Sharma S Pal P Ojha H Mullick J Sahu A . The imitation game: a viral strategy to subvert the complement system. FEBS Lett. (2020) 594:2518–42. doi: 10.1002/1873-3468.13856
130.
Kolev M Le Friec G Kemper C . Complement--tapping into new sites and effector systems. Nat Rev Immunol. (2014) 14:811–20. doi: 10.1038/nri3761
131.
Mayilyan KR . Complement genetics, deficiencies, and disease associations. Protein Cell. (2012) 3:487–96. doi: 10.1007/s13238-012-2924-6
132.
Murugaiah V Varghese PM Beirag N De Cordova S Sim RB Kishore U . Complement proteins as soluble pattern recognition receptors for pathogenic viruses. Viruses. (2021) 13:824. doi: 10.3390/v13050824
133.
Mastellos DC Hajishengallis G Lambris JD . A guide to complement biology, pathology and therapeutic opportunity. Nat Rev Immunol. (2024) 24:118–41. doi: 10.1038/s41577-023-00926-1
134.
Planas R Felber M Vavassori S Pachlopnik Schmid J . The hyperinflammatory spectrum: from defects in cytotoxicity to cytokine control. Front Immunol. (2023) 14:1163316. doi: 10.3389/fimmu.2023.1163316
135.
Mastio J Saeed MB Wurzer H Krecke M Westerberg LS Thomas C . Higher incidence of B cell malignancies in primary Immunodeficiencies: a combination of intrinsic genomic instability and exocytosis defects at the immunological synapse. Front Immunol. (2020) 11:581119. doi: 10.3389/fimmu.2020.581119
136.
Riaz IB Faridi W Patnaik MM Abraham RS . A systematic review on predisposition to lymphoid (B and T cell) Neoplasias in patients with primary Immunodeficiencies and immune Dysregulatory disorders (inborn errors of immunity). Front Immunol. (2019) 10:777. doi: 10.3389/fimmu.2019.00777
Summary
Keywords
hemophagocytic lymphohistiocytosis, inborn error of immunity, immunodeficiency, JC virus, natalizumab, progressive multifocal leukoencephalopathy, rituximab, PML
Citation
Eis PS, Smith EB III, Jalilzadeh S and Hatchwell E (2025) Genetics of progressive multifocal leukoencephalopathy: update on case reports with an inborn error of immunity and risk variants found in drug-linked cases. Front. Neurol. 16:1629581. doi: 10.3389/fneur.2025.1629581
Received
16 May 2025
Accepted
30 June 2025
Published
15 July 2025
Corrected
21 July 2025
Volume
16 - 2025
Edited by
Joseph R. Berger, University of Pennsylvania, United States
Reviewed by
Dennis Kolson, University of Pennsylvania, United States
Updates
Copyright
© 2025 Eis, Smith, Jalilzadeh and Hatchwell.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peggy S. Eis, pegeis@populationbio.comEli Hatchwell, elihatchwell@populationbio.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.