- 1Faculty of Information Technology, Beijing University of Technology, Beijing, China
- 2Beijing Institute of Smart City, Beijing University of Technology, Beijing, China
- 3Engineering Research Center of Digital Community, Beijing University of Technology, Beijing, China
- 4Beijing Key Laboratory of Magnetic Resonance Imaging (MRI) and Brain Informatics, Beijing University of Technology, Beijing, China
- 5Beijing International Collaboration Base on Brain Informatics and Wisdom Services, Beijing University of Technology, Beijing, China
Provenances are a research focus of neuroimaging resources sharing. An amount of work has been done to construct high-quality neuroimaging provenances in a standardized and convenient way. However, besides existing processed-based provenance extraction methods, open research sharing in computational neuroscience still needs one way to extract provenance information from rapidly growing published resources. This paper proposes a literature mining-based approach for research sharing-oriented neuroimaging provenance construction. A group of neuroimaging event-containing attributes are defined to model the whole process of neuroimaging researches, and a joint extraction model based on deep adversarial learning, called AT-NeuroEAE, is proposed to realize the event extraction in a few-shot learning scenario. Finally, a group of experiments were performed on the real data set from the journal PLOS ONE. Experimental results show that the proposed method provides a practical approach to quickly collect research information for neuroimaging provenance construction oriented to open research sharing.
Introduction
Open and FAIR (Findable, Accessible, Interoperable, and Reusable) sharing (Poldrack and Gorgolewski, 2015; Abrams et al., 2021) of neuroimaging data has been widely recognized and has been a concern in computational neuroscience. From fMRIDC (Van Horn et al., 2001) and OpenfMRI (Poldrack and Gorgolewski, 2015) to OpenNeuro (Gorgolewski et al., 2017), a series of public neuroimaging databases have been constructed and have played an important role in brain and intelligence researches. Only taking the INDI database as an example, by March 22, 2017, Google Scholar search results showed that 913 publications clearly indicated the use of INDI data, with a total of 20,697 citations (Milham et al., 2018). Besides sharing raw experimental data, research information sharing1 has also become an important way of neuroimaging data sharing (Poline et al., 2012), which can effectively avoid various challenges of sharing raw experimental data, such as cost, privacy protection, and data ownership. BrainMap2 collects the published information related to brain activated areas from the neuroimaging article to support coordinate based meta-analysis (Laird et al., 2005). Neurosynth3 provides a specialized tool to automatically extract coordinates of activated brain area and research topic information from published neuroimaging articles and shared them by a unified platform (Yarkoni et al., 2011). NeuroVault4 collected non-threshold statistical graphs and related information from different computational neuroscience to support graph based meta-analysis (Gorgolewski et al., 2016). These efforts on sharing neuroimaging research information enrich the content and method of neuroimaging data sharing and effectively support the brain and intelligence studies in computational neuroscience. For example, statistics show that the number of papers based on the neurosynth platform has reached 14,371 by July 2018. Related studies are involved with the brain mechanism of cognitive states (Alcalá-López et al., 2017), functional analysis of brain areas (Genon et al., 2018), large-scale brain structure-function mappings (Bolt et al., 2020), and so on.
Provenances (Moreau et al., 2008) are a research issue of neuroimaging data sharing Poldrack and Gorgolewski (2014) pointed out that sharing task-based fMRI (functional magnetic resonance imaging) data requires the appropriate descriptions of how they were acquired, including not only the MRI acquisition parameters but also the specific order and timing of stimulus presentation during the task, and how they have been transformed. These descriptions are provenances. The special topic “Collaborative efforts for understanding the human brain” of the journal Frontiers in Neuroinformatics emphasized that the key to controlling the re-executability of the publication of computational neuroscience is the generation and reporting, at all stages of the process, machine readable provenance documentation (Kennedy et al., 2019). From the data model (Keator et al., 2013; Maumet et al., 2016), the management tool (Keator et al., 2019) to the acquisition platform (Gorgolewski et al., 2016), an amount of work has been done to construct high-quality neuroimaging provenances in a standardized and convenient way. The neuroimaging provenances have been not only metadata of task-based experimental data but also structured descriptions of research cases in computational neuroscience, which collect massive research information about the whole research process of computational science to support multi-aspect resource integration, fast hypothesis generation, large-scale metadata analysis, strict result evaluation, etc., for realizing open and FAIR neuroscience (Abrams et al., 2021). However, existing studies on neuroimaging construction mainly adopt experts or process recording-based methods to obtain neuroimaging provenances. All methods are centered on the actual experimental data. Besides them, open research sharing in computational neuroscience still needs one new way to automatically and quickly extract provenance information from rapidly growing published sources.
Neuroimaging text mining recognizes important information from neuroimaging texts, especially neuroimaging articles, and provides a practical way to automatically extract provenance information from published sources. In recent years, related studies mainly focused on recognizing neuroimaging entities (Abacha et al., 2017; Shardlow et al., 2018; Riedel et al., 2019), hierarchical terminology system (Huangfu et al., 2020), and span of interests (Zhu et al., 2020). Because extracted information is mainly some words or phrases, and lacks rich relations, especially non-taxonomical relations, these studies can only obtain some fragmented key points of the research process and cannot provide a vivid process description. Hence, it is necessary to develop a new technology of neuroimaging text mining for research sharing-oriented neuroimaging provenance construction.
Based on this observation, this paper proposes a literature mining-based approach for neuroimaging provenance construction. The rest of this paper is organized as follows. Section “Related Work” summarizes previous work-related to neuroimaging provenances, neuroimaging text mining, and biomedical event extraction. Section “Contributions” introduces our contributions. Section “Materials and Methods” describes the proposed method. Experiments and results are presented in section “Results” to validate the effectiveness of the proposed method. Finally, section “Discussion” gives concluding remarks.
Related Work
Neuroimaging Provenances
Provenances can be described in various terms depending on the domains where they are applied (Simmhan et al., 2005). Buneman et al. (2004) defined provenances as the description of the origins of data and the process by which it arrived at the database. Lanter (1991) characterized provenances as information describing materials and transformations applied to derive the data. Greenwood et al. (2003) viewed provenances as metadata recording the process of experiment workflows, annotations, and notes. Simmhan et al. (2005) defined provenances as information that helped determine the derivation history of a data product. Related studies have been widely concerned by many domains. Science magazine pointed out that provenance is an important element of service quality control (Foster, 2005). The NSF (National Science Foundation, Alexandria, VA, United States) Advisory Committee believed that a robust and stable cyberinfrastructure should support provenances (Task Force on Grand Challenges, 2011). The World Wide Web Consortium standard of provenances, called PROV-DM (Provenance Data Model),5 has been released in 2013. A series of international conferences about provenances, such as IPAW (International Provenance and Annotation Workshop, since 2006) and TaPP (Workshop on Theory and Practice of Provenance, since 2009), has attracted a large number of researchers.
Computational neuroscience is an important application domain of provenances. Neuroimaging provenances are the research focus. As early as 2006, the first fMRI data provenance challenge has been held (Moreau et al., 2008). A group of minimum provenance information (MI) guidelines (Gibson et al., 2008; Poldrack et al., 2008; Frishkoff et al., 2011) has been proposed for different types of neuroimaging data. Guided by these MI guidelines, the main neuroimaging data model, such as DICOM (Digital Imaging and Communications in Medicine)6 and XCEDE (XML-based Clinical and Experimental Data Exchange),7 provides the support for representing and storing provenance information. However, the provenance information storage embedded into experimental data models hampers the representation of more complex relationships among data (Ruiz-Olazar et al., 2016). Provenances independent from experimental data have drawn attentions. The representative research work is NIDM (Neuroimaging Data Model), which provides a complete description of provenance for computational neuroscience, from raw data to the final results including all the steps in between (Maumet et al., 2016). NIDM builds upon PROV-DM and extends it with terms found in XCEDE and DICOM headers. It consists of NIDM-Experiment, NIDM-Workflow, and NIDM-Results; NIDM-Experiment and NIDM-Workflow (Keator et al., 2013) model acquisition and processing of experimental data, respectively. NIDM-Results (Maumet et al., 2016) focuses on neuroimaging statistical results along with key image data summarizing the experiment.
In recent years, many studies are working to support the construction of neuroimaging provenances based on NIDM for realizing machine readable descriptions of the data collection, the processing workflow and environment, and the statistical procedures and results. The export and import of NIDM documents have been realized in many widely used neuroimaging tools and platforms, such as SPM, FSL, and NeuroVault. Keator et al. (2019) developed a python-based API (PyNIDM) which followed the simple organizational structure of NIDM-Experiment with functions to create, query, export, import, and transform NIDM-Experiment documents. Kennedy et al. (2019) developed the function to generate NIDM based provenances on ReproNim, which is a center for reproducible neuroimaging computation. Maumet et al. (2019) built a JSON-LD representation for NIDM-Results data and exposed it to the neuroimaging data management tool Datalad. With the growth of scale, the applications of neuroimaging provenances also have broken through traditional data descriptions oriented to data sharing (Van Horn et al., 2001) and process recordings oriented to analysis (Dinov et al., 2009), and are moved toward verification of scientific outputs (Arshad et al., 2019), research artifact sharing (Maumet, 2020), etc. Provenance-based research sharing (Yuan et al., 2018), which uses provenances as “research objects” (Belhajjame et al., 2015; Miksa and Rauber, 2017; That et al., 2017) (i.e., digital research artifacts for knowledge sharing and reproducibility in computational experiments), is becoming a trend for open and FAIR neuroscience.
However, provenance-based research sharing needs large-scale and high-quality neuroimaging provenances. Existing studies of neuroimaging provenances mainly adopt experts or process recordings-based construction methods. Public neuroimaging data or derived data sharing platforms, such as BrainMap and NeuroVault, rely on experts’ manual annotations to collect neuroimaging provenance information. The obtained provenances vary dramatically in quality and grow slowly. Even if Brainspell8 adopts the crowdsourcing-based annotation mode (Badhwar et al., 2016), the situation has not been fundamentally improved. Aiming at this problem, many neuroimaging tools, such as SPM, FSL, and PyNIDM, have provided the supporting for process-based provenance extraction. They can collect provenance information at the time of data acquisition or processing and do not require any extra effort. Process-based provenance extraction has become the most important means to construct neuroimaging provenances for large-scale data sharing. However, this kind of provenance extraction method is centered on actual experimental data. At present, neuroimaging data sharing still faces some important obstacles, such as privacy and data ownership. Public data account for only a small part of the total data. Similarly, many researchers do not actively generate and share neuroimaging provenances during the data acquisition or processing. Therefore, besides collecting provenance information during data acquisition or processing, it is also necessary to develop one new way to extract provenance information from published sources, especially published scientific articles.
Neuroimaging Text Mining
Neuroimaging technologies can non-invasively detect the connection between cognitive states and patterns of brain activity, and have received widespread attentions in computational neuroscience. Neuroimaging texts, especially scientific articles, are growing fast. Taking only fMRI (Wegrzyn et al., 2018) as an example, 307 relevant articles have been published in the journal PLOS ONE in 2020. These neuroimaging texts are valuable knowledge resources for studying human intelligence, pathological mechanism of brain and mental diseases, brain-computer interface, and so on. In the last decade, various neuroimaging meta-analysis (Yarkoni et al., 2011; Neumann et al., 2016) and collaborative analysis (Lei et al., 2020) have made great achievements. The value of neuroimaging texts as knowledge resources is already obvious. How to automatically and continuously extract knowledge from neuroimaging texts has become a key issue for open and FAIR neuroscience.
Neuroimaging text mining is an important branch of biomedical text mining and provides an effective approach to extract knowledge from neuroimaging texts. Early studies mainly focused on extracting neuroimaging knowledge for decoding a wide range of cognitive states Naud and Usui (2008) constructed the neuroscience terminology space by using the Vector Space Model and performed k-means based cluster on the abstract of neuroscience articles for extracting cognitive state-related topics. Neurosynth (Yarkoni et al., 2011) recognized terms based on word frequency and used the Naive Bayesian Classifier to predict the occurrence of specific terms based on the whole-brain activation patterns. Poldrack et al. (2013) adopted the Latent Dirichlet Allocation (LDA) method to identify topics of articles from the Neurosynth database and mapped these topics to brain activation data for discovering mechanisms of cognitive states. French et al. (2012) developed a co-occurrence-based method to extract brain regions and their relations from neuroscience articles. Alhazmi et al. (2018) extracted topic words based on word frequency and constructed relations between semantic spaces of topics and brain-activated regions by using correspondence analysis and hierarchical clustering. In order to decode cognitive states, these studies only extracted cognitive states and activated brain regions and their relations from neuroimaging articles. Main adopted text mining technologies are the frequency-based or probabilistic model-based topic learning methods and the clustering-based relation recognition methods. The obtained knowledge was a group of topic words and their relations. Because of adopting an open-domain task to extract unknown types of topics and relations, many general words were often included in topics, resulting in low-quality results of knowledge extraction. For example, neurosynth topic words include many general words, such as “using,” “repeat,” “asked,” and domain general words, such as “magnetic resonance,” “brain” (Abacha et al., 2017). Poldrack et al. (2013) had to use concepts in the Cognitive Atlas (Poldrack et al., 2011) to further filter neurosynth topic words. Such knowledge extraction oriented to the decoding of cognitive states cannot effectively characterize the whole research process for sharing “research objects.”
In recent years, studies on neuroimaging text mining began to expend the knowledge extraction perspective from decoding cognitive states to characterize the whole research process. The adopted methods also change from topic modeling to named entity recognition. Ben Abacha et al. (2017) adopted the rule-based method and various conditional random field (CRF)-based methods to recognize fifteen functional neuroimaging entity categories, including gross brain anatomy, functional neuroanatomy, medical problem, stimuli, and responses, etc. Shardlow et al. (2018) recognized various entities, including brain regions, experimental values, neuron types, etc., by using active and deep learning, for curating researches in computational neuroscience. Riedel et al. (2019) completed a comprehensive evaluation on recognizing various entities related to the cognitive experiment, which are defined by the Cognitive Paradigm Ontology (Turner and Laird, 2012) and involved with behavioral domain, paradigm class, instruction, stimulus modality, etc., based on multiple corpus features and various classifiers, including Bernoulli naïve Bayes, k-nearest neighbors, logistic regression, and support vector classifier. Huangfu et al. (2020) extracted the term list of the neuroscientific research process from the PubMed Central database, for creating neuroscientific knowledge organization system (i.e., hierarchical terminology system). Zhu et al. (2020) proposed the hierarchical attentive decoding to extract a span of interests (i.e., terms about neuroscientific research process) from neuroscience articles, to predict research species. Whether it is the entity, method term, or research interest, the extracted knowledge in current studies of neuroimaging text mining is no longer limited to cognitive states and brain regions but extended to the whole research process, from the experiment to analysis. Furthermore, most of these studies transformed the open-domain topic modeling task to the close-domain entity recognition task with predefined entity/term/interest types from external domain knowledge, such as Cognitive Paradigm Ontology. By this kind of transformation of task definition, the shortcoming of general word noises in early studies of neuroimaging text mining can be effectively solved. Therefore, rapid growth neuroimaging articles, especially open access articles, and the maturing technologies of neuroimaging text mining provide a practical approach for continuous and rapid collection of neuroimaging provenance information.
However, for provenance-based research sharing, existing studies on neuroimaging text mining still have the following shortcoming. Words or phrases are the main form of extracted knowledge. Because of lacking the relation structure among words, these words or phrases can only show some fragmentary and key points of the research process and cannot provide a vivid process description. Hence, it is necessary to develop a new technology of neuroimaging text mining for research sharing-oriented neuroimaging provenance construction.
Biomedical Event Extraction
In the definition of ACE (Automatic Content Extraction) (Doddington et al., 2004), “event” is described as the occurrence of an action or the change of state. It includes an event trigger and multiple arguments with different roles. Event extraction is to obtain structured representations of events, so as to help answering the “5W1H” questions, including “who, when, where, what, why” and “how,” of real-world events from numerous sources of texts (Xiang and Wang, 2019). Biomedical event extraction is to extract events from biomedical texts and has become one of the most actively researched areas in biomedical text mining (Chung et al., 2020). Since the first biomedical natural language processing shared task (BioNLP-ST) challenge in 2009, the fine-grained biomedical event extraction has received extensive attentions from academia and industry.
Different from news (Abera, 2020), finance (Zheng et al., 2019) and other fields, biomedical event extraction has two main challenges. Firstly, biomedical texts have the higher complexity. They often contain many abbreviations and long sentences with complex structures. Secondly, high-quality annotated corpora are lacking. Most of the existing biomedical event corpora are small scale and have serious class imbalance problems. In recent years, a large number of studies have tried to solve these two problems. Various embedding technologies, such as dependency path embedding (Bjrne and Salakoski, 2018) and entity property embedding from external ontologies (Li et al., 2016), were developed to capture multi-aspect feature information for modeling complex biomedical texts. The hierarchical attention mechanism (Chen, 2019) was proposed to model the global document context for identifying biomedical event triggers from long biomedical sentences. Distance supervision (Araki and Mitamura, 2018), transfer learning (Chen, 2019), and other technologies have also been adopted to solve the problem of lack and imbalance of training samples.
In recent years, the knowledge base-oriented extraction task is becoming the research focus of biomedical event extraction. It evaluates the extraction systems or methods by measuring how much information content can be extracted from corpora (Deléger et al., 2016). Traditional pipeline models divided the event extraction task into multiple independent sub-tasks and resulted in various challenges, such as error propagation and lack of dependencies and interactions among subtasks. Therefore, the joint model becomes the inevitable choice of event extraction that oriented the knowledge base. Nguyen and Nguyen (2019) proposed a novel model to jointly perform predictions for entity mentions, event triggers, and arguments based on the shared hidden representations. Yu et al. (2019) developed an end-to-end model based on LSTM (Long Short-Term Memory) to optimize biomedical event extraction. Trieu et al. (2020) proposed an end-to-end neural nested event extraction model, named DeepEventMine, which can extract multiple overlapping directed acyclic graph structures from a raw sentence. Li et al. (2020) proposed a parallel multi-pooling convolutional neural network model to capture the compositional semantic features of sentences for improving biomedical event extraction on the MLEE (Multi-Level Event Extraction) dataset. Zhao et al. (2021) proposed a novel framework of reinforcement learning for the task of multiple biomedical event extraction, in which trigger identification and argument detection were treated as the main task and the subsidiary task.
Because of rich semantic information, the event is naturally more suitable to represent neuroimaging provenances, which characterize the research process of computational science, than isolated words. However, there are still two challenges in applying existing biomedical event extraction technologies, especially various joint models of biomedical event extraction, on research sharing-oriented neuroimaging provenance construction:
• Modeling the neuroimaging research process and result based on events. Existing biomedical event extraction tasks mainly focus on gene, protein, and disease-related events. As a new field of biomedical event extraction, neuroimaging event extraction lacks referential studies for task definition. Therefore, it is necessary to model the neuroimaging research process and results based on the perspective of events firstly. This is a challenging work because the definition of events needs to take into account both the demands of research sharing and the availability of knowledge in articles. The task type, content, and granularity of event extraction needs to be designed systematically by balancing various factors, including the task complexity, the capability of existing technologies, the importance and completeness of knowledge in research sharing, and the availability of knowledge in articles.
• Constructing the joint model of neuroimaging events in a few-shot learning scenario. Research sharing-oriented neuroimaging provenance construction is a typical task of knowledge base extraction and needs to adopt the joint model of event extraction. However, the complex network structure of joint model brings the higher requirements on the scale of training data. As a new field of event extraction, there are no existing annotated corpora for neuroimaging event extraction. Furthermore, neuroimaging event extraction in this study is involved with multi-domain entities for characterizing the research process of computational neuroscience. This also hinders the use of distance supervision (Araki and Mitamura, 2018) and transfer learning (Chen, 2019). Therefore, this is necessary to develop a new joint model of biomedical event extraction based on a small-scale “gold” corpus set annotated by domain experts.
Contributions
The main contributions can be summarized as follows:
1. Firstly, this paper designs a provenance-guided approach for modeling the neuroimaging research process and result based on events. Considering the requirements of research sharing in open and FAIR neuroscience and availability of knowledge in neuroimaging articles, an improved brain informatics (BI) provenance model is defined based on NIDM. A mapping between the BI model and the definition of events is also proposed to obtain six categories of neuroimaging event-containing attributes for constructing research sharing-oriented neuroimaging provenances.
2. Secondly, this paper proposes a joint extraction model based on deep adversarial learning, called the adversarial training based neuroimaging event and attribute extraction model (AT-NeuroEAE), to extract the defined neuroimaging events. To the best of our knowledge, it is the first event extraction joint model containing attributes. Furthermore, the FreeAT (Free Adversarial Training)-based adversarial learning is introduced into the event joint extraction for improving the accuracy of event extraction in a few-shot learning scenario. This kind of optimization makes the proposed model more practical for computational neuroscience which lacks the “gold” labeled corpora.
3. Thirdly, a group of experiments were performed based on real data from the journal PLOS ONE. Experimental results show that, compared with existing event extraction models, the proposed model can more effectively and completely extract neuroimaging provenance information from neuroimaging articles based on a small-scale corpus set annotated by domain experts. This is a practical and effective approach for large-scale and low-cost neuroimaging provenance construction.
Materials and Methods
Overall Structure
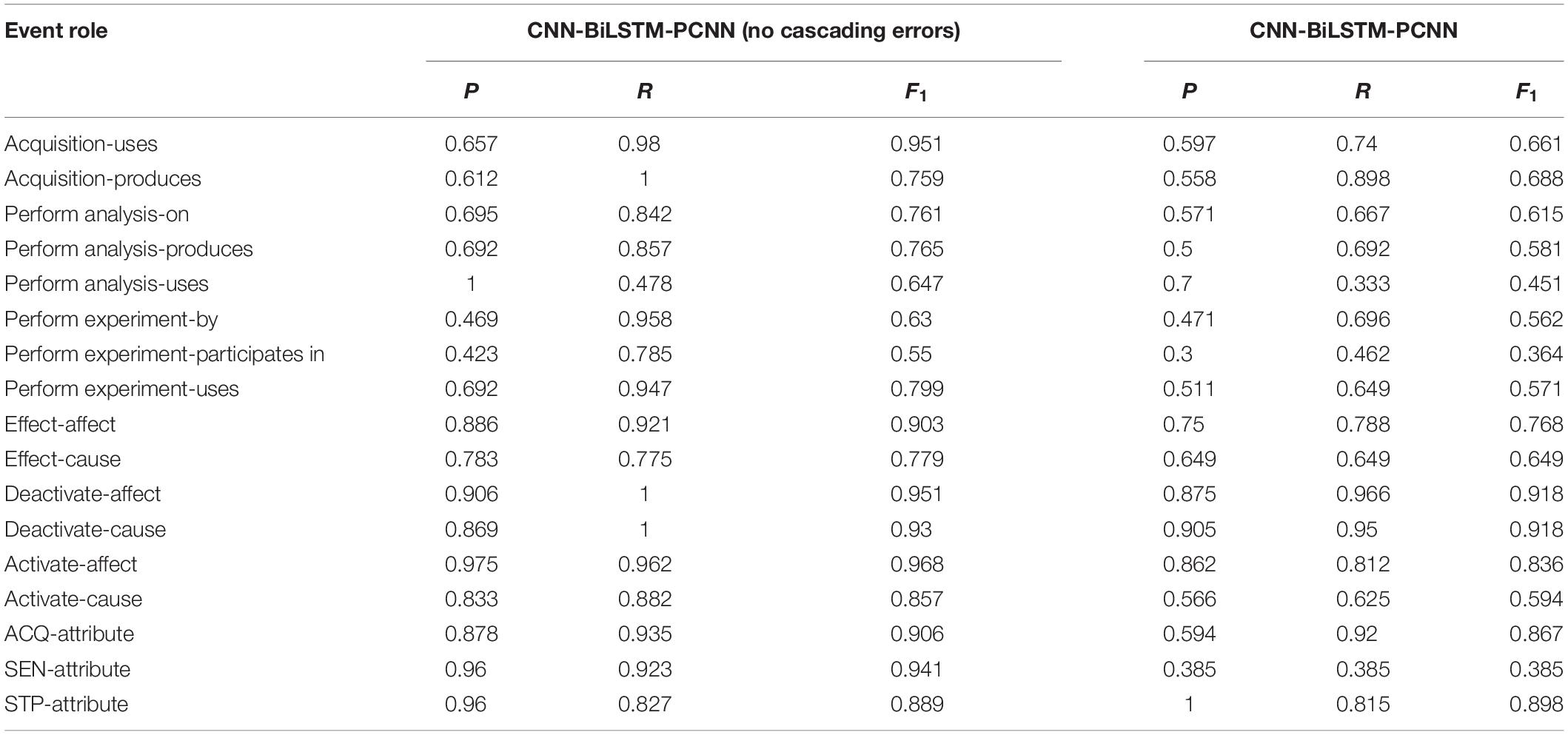
This paper proposes a joint extraction model of events with attributes, called AT-NeuroEAE, for research sharing-oriented neuroimaging provenance construction. By using NeuroEAE, provenance information can be automatically and quickly extracted from published neuroimaging articles, especially open-access scientific articles. As shown in Figure 1, the whole process includes four steps. Details will be described in the subsequent subsections.
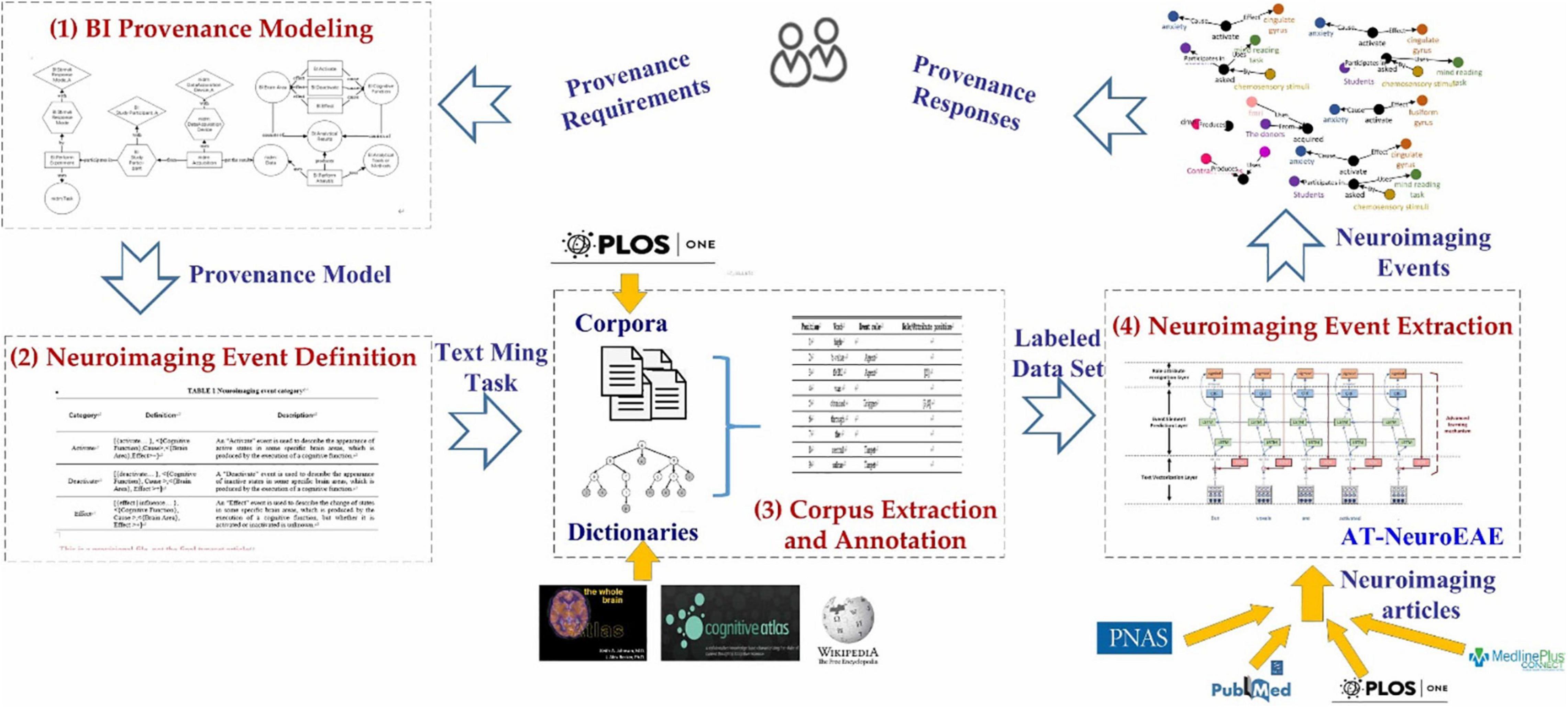
Figure 1. The whole process of provenance information extraction. (1) Brain informatics provenance modeling: construct an improved BI provenance model to capture the provenance requirements of research sharing in open and FAIR neuroscience. (2) Neuroimaging event definition: define a group of neuroimaging events to transform the BI provenance model into text mining tasks. (3) Corpus extraction and annotation: construct a group of labeled corpora for model training and test. (4) Neuroimaging event extraction: develop the NeuroEAE model to extract defined neuroimaging events for meeting the provenance requirements in open and FAIR neuroscience.
Brain Informatics Provenance Modeling
Computational neuroscience is a new application field of biomedical event extraction. In order to construct neuroimaging provenance by using biomedical event extraction, the first task needs to model the neuroimaging research process and result.
At present, NIDM is the most widely used neuroimaging provenance model and the representative work for open and FAIR Neuroscience (Abrams et al., 2021). However, as stated above, its element granularity is still incomplete for neuroimaging research sharing. It also contains a lot of model elements which aren’t available in articles. In our previous studies (Chen et al., 2012; Chen and Ning, 2013), a BI provenance model has been proposed to describe the origin and subsequent processing of various human brain data in systematic BI studies. This study reconstructs the BI provenance model by considering both the importance of knowledge in research sharing and the availability of knowledge in articles.
Firstly, the newest version of BI provenance model (Sheng et al., 2019) is updated based on the widely used NIDM. All entities, activities and agents are replaced by NIDM classes, such as “nidm:AcquisitionObject,” “nidm:Task,” “nidm:Acquisition,” and “nidm:DataAcquisitionDevice.” This update ensures that the new BI provenance model follows the FAIR facets I1: “(meta) data use a formal, accessible, shared, and broadly applicable language for knowledge representation” and I2: “(meta) data use vocabularies that follow FAIR principles.”
Secondly, according to the FAIR facet F2, “data are described with rich metadata” and the deficiency of NIDM in experiment description; the abstract classes of experimental design in NIDM is extended as the activity “BI:PerformExperiment” and three related elements, including the entity “nidm:Task” and the entity “BI:StimuliResponseMode.” The entity “nidm:Task” and “nidm:StudyParticipant” are inherited from NIDM and used to indicate the task design type. The entity “BI:StimuliResponseMode” are used to describe the experiment in more detail. The former describes the sensory stimuli or response of experiment and the latter describes the participant of experiment. They are two important factors for cognitive experiments and need to be considered in multiple key steps of computational neuroscience, including experimental design, data analysis, result interpretation, etc. Neuroimaging articles also often introduce them in the “Experiment” section. Furthermore, both the stimuli mode and the subject are involved with multi-aspect attribute information, such as the perception channel, the stimuli task category, age, gender, and occupation, but it is not sure which and how much attribute information will be described in an article. As shown in Figure 2, different articles may describe subjects by using different attributes, including medication history, age, gender, medical history, and other complex characteristics. Therefore, this study adds the general attribute entity to represent all attributes of the entity. For example, the entity “BI:StudyParticipant_A” is added into the model to represent all possible attribute information of subjects. Similarly, according to the FAIR facet F2 “data are described with rich metadata” and the deficiency of NIDM in result description; this study adds three categories of activities “BI:Activate,” “BI:Deactivate,” “BI:Effect” and two related elements, including the entities “BI:BrainArea” and “BI:CognitiveFunction,” to describe the brain mechanism of cognitive states, which are core research findings in computational neuroscience.

Figure 2. Diverse attributes of subjects in different articles. In example 1 (Daniel et al., 2014), the attributes of subjects include medication history, age, medical history, and gender. In example 2 (Lanting et al., 2014), the attributes of subjects include health condition and medical history.
Thirdly, considering both the importance and the availability of information, this study simplifies the description of NIDM about analytical tools or methods. NIDM contains many classes about parameters of analytical tools or methods. However, many parameters are generally not mentioned in the article. Therefore, this study uses an entity “BI:AnalyticalToolsorMethods,” which is connected to the activity “BI:PerformAnalysis,” to represent analytical tools or methods. Two entities “nidm:AcquisitionObject” and “BI:AnalyticalResults” are also connected to the activity “BI:PerformAnalysis” for representing the analytical process more clearly. Compared with the traditional class or concept tree, this kind of event-based description contains various roles within the event and can better describe the research process than the single subclass relation.
Figure 3 gives the reconstructed BI provenance model. It characterizes experimental design, analytical process, and results by four categories of elements: entity, activity, agent, and attribute.
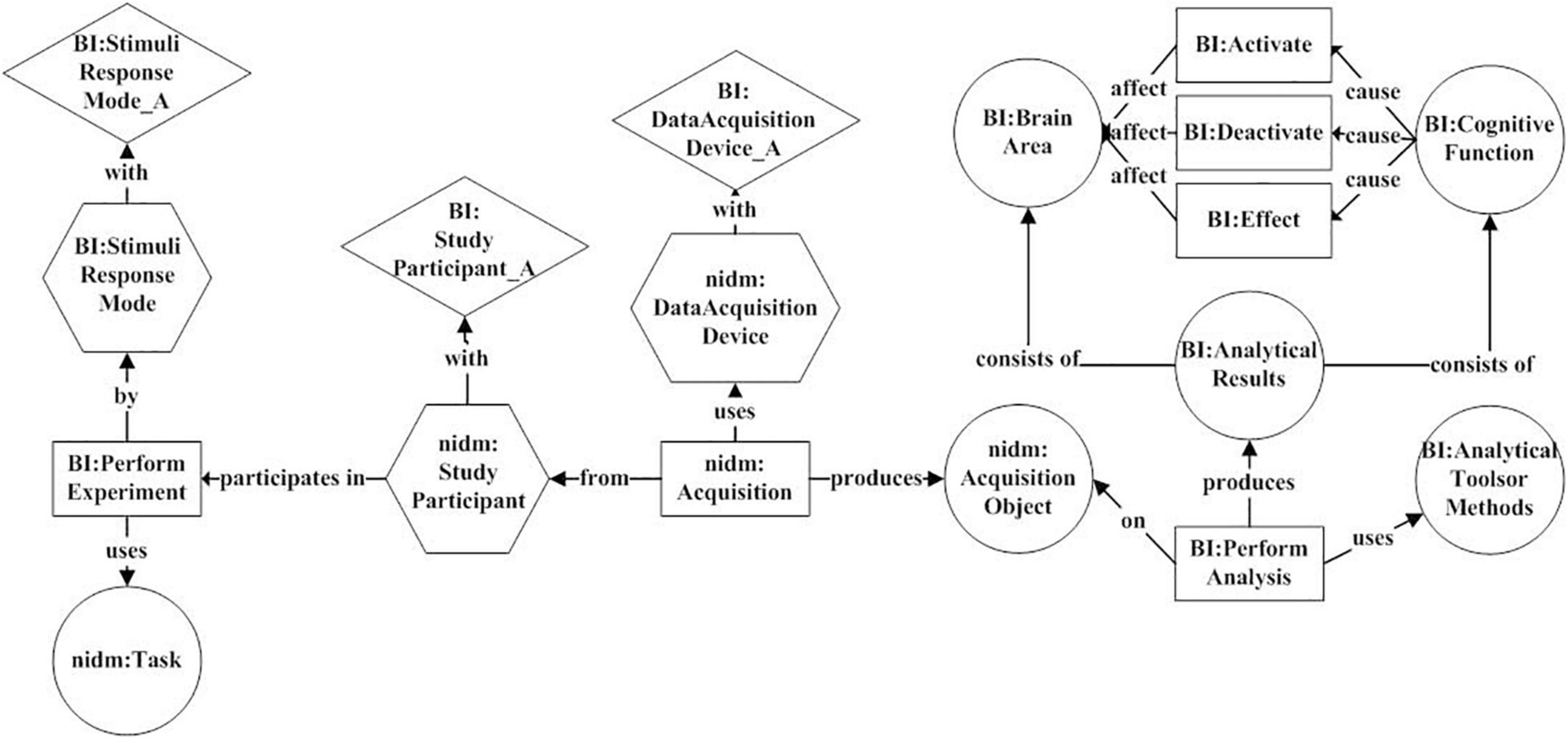
Figure 3. The BI provenance model. It uses six activities (rectangle), including “BI:PerformExperiment,” “nidm:Acquisition,” “BI:PerformAnalysis,” “BI:Activate,” “BI:Deactivate,” and “BI:Effect” to characterize experimental design, analytical process, and results. Six entities (circle), three agents (hexagon), and three attributes (diamond) are connected to these activities for describing various key factors in the research process.
Model-Guided Neuroimaging Event Definition
This study designs a model-guided approach to transform BI provenance model into text mining tasks. A group of neuroimaging events are defined by using the following mapping rules:
• Firstly, each activity and its directly connected elements (entities or agents) are divided into an independent activity unit.
• secondly, each activity unit is mapped to a meta event. The activity is mapped to the trigger words. The entities and agents are mapped to arguments. The relations between activity and elements are mapped to argument roles.
Based on the above rules, six categories of neuroimaging events can be obtained from the BI provenance model. Tables 1, 2 give the category definitions of events and corresponding arguments.
In this study, the definition of events is represented as trigger + parameter structure (Sun et al., 2017). For example, an “Acquisition” event can be represented as follows:
Event Acquisition
= [trigger, < argument1(attribute)#, role1 > #, < argument2(attribute)#, role2 > #…]
= [{assess | acquire | obtain…}, < {Acquisition Object}, produces > *, < {Data Acquisition Device (Data Acquisition Device_A)*}, uses > +, < {Study Participant (Study Participant _A)*}*, from > ]
The above expression shows that an “Acquisition” event consists of a trigger word and three categories of arguments. “*” indicates that the element may occur zero or more times and “+” indicates that the element may occur one or more times. The trigger word may be one of “assess,” “acquire,” “obtain,” etc. Argument 1 belongs to the “Acquisition Object” category and is used to describe the results produced by the “Acquisition” event. Argument 2 belongs to the “Data Acquisition Device” category and is used to describe the device used in the “Acquisition” event. Argument 3 belongs to the “Study Participant” category and is used to describe the object of the “Acquisition” event. Both Argument 2 and Argument 3 have zero or several attributes. Figure 4 gives an example of an “Acquisition” event in a neuroimaging article.
Figure 4. An example of an “Acquisition” event from the article (Mutschler et al., 2016). This event consists of a trigger word “using,” two “Data Acquisition Device” category of arguments “fMRI” and “SCR,” and one “Study Participant” category of argument “infant.”
Corpus Extraction and Annotation
Corpora were crawled from the journal PLOS ONE. We searched the articles based on the keywords “fMRI” or “functional magnetic resonance imaging” or “functional MRI” in their abstracts, and set the published time from July 18, 2014, to July 18, 2019. A total of 677 articles were obtained. Based on these articles, event mentions were extracted by using rule matching. The process can be described as follows.
The first step is dictionary construction. Seven categories of argument dictionaries, including “Brain Area,” “Cognitive Function,” “Study Participant,” “Stimuli Response Mode,” “Task,” “Analytical Tools or Method,” and “Analytical Results” came from our previous study (Sheng et al., 2020). We collected candidate trigger words and arguments based on the co-occurrence with these seven categories of arguments, and then manually filtered them to construct the trigger word dictionaries and other categories of argument dictionaries. Similarly, three categories of argument attribute dictionaries were constructed based on the co-occurrence and manual filtering. Table 3 gives the number of terms in each dictionary.
The second step is mention extraction. Based on the dictionaries and event definitions, event mentions were extracted by using the following rule:
Rule1: If an event trigger word and two arguments appear in the same sentence, this sentence can be marked as a complete event.
However, Rule 1 cannot extract enough the “Deactivate” event. We defined another more relaxed rule:
Rule2: If a “Deactivate” event trigger word and a “BRI” argument appear in the same sentence, this sentence can be marked as a “Deactivate” event.
Based on Rules 1 and 2, a neuroimaging event mention set can be obtained as the experimental data set. It includes 788 “Activate” event mentions, 128 “Deactivate” event mentions, 1169 “Effect” event mentions, 665 “Perform Experiment” event mentions, 266 “Acquisition” event mentions, and 315 “Perform Analysis” event mentions. The distribution of event mentions in the experimental data set is shown in Figure 5.
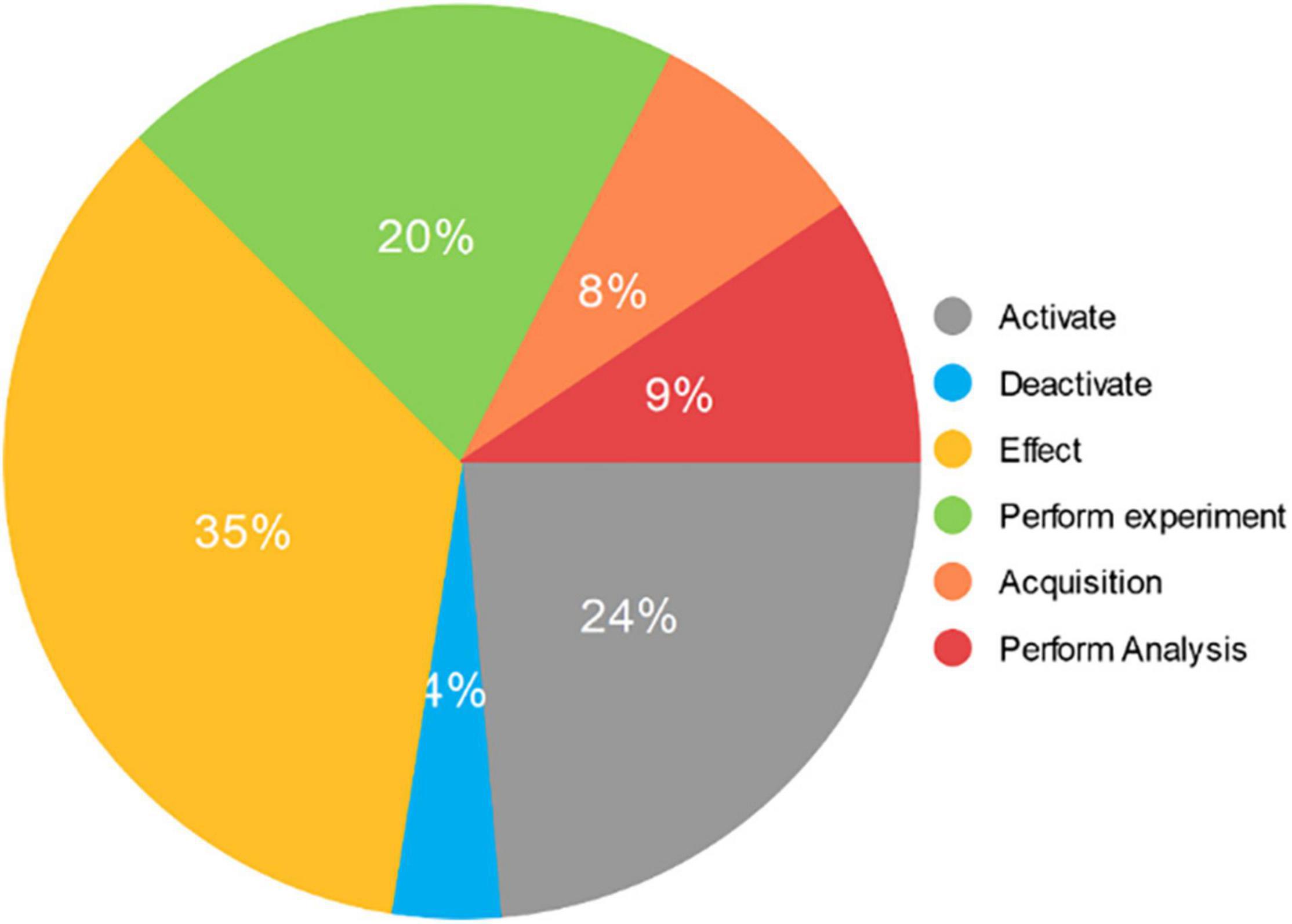
Figure 5. The distribution of event mentions in the experimental data set. It consists of 3331 event mentions extracted from 677 neuroimaging articles. The “Activate” category includes 788 mentions and accounts for 24% of the total. The “Deactivate” category includes 128 mentions and accounts for 4% of the total. The “Effect” category includes 1169 mentions and accounts for 35% of the total. The “Perform experiment” category includes 665 mentions and accounts for 20% of the total. The “Acquisition” category includes 266 mentions and accounts for 8% of the total. The “Perform Analysis” category includes 315 mentions and accounts for 9% of the total.
In the training dataset, the annotation of triggers and arguments adopts the “B/I/O-category abbreviation” mode. The “B/I/O” is the BIO annotation mode (Soomro et al., 2017). “B” indicates the beginning of entity. “I” stands for the middle or end, and “O” stands for other, which is used to mark irrelevant characters. Category abbreviations are involved with argument categories and trigger categories. The entity annotation of attributes adopts the “B/I/O-category abbreviation_A” model. Table 4 gives an annotation example. The labels “BRI” and “AOB” indicate the argument categories “Brain Area” and “Acquisition Object,” respectively. The label “Acq” indicates the trigger category “Acquisition.” The label “O” on the word “high” indicates “high” does not belong to any entity. The label “B-AOB” on the word “fMRI” indicates “fMRI” is the beginning of the “Acquisition Object” argument.
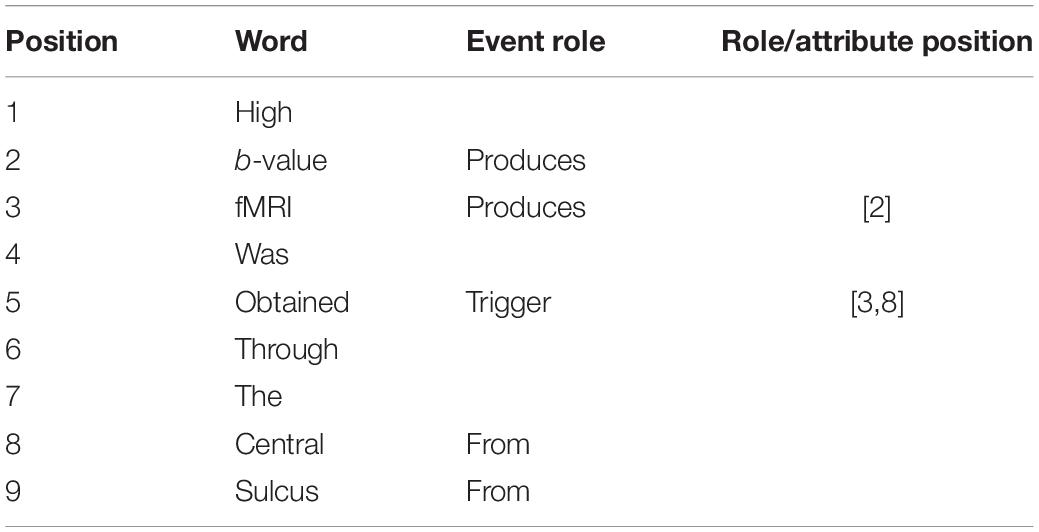
The annotation of event roles and attributes is based on trigger words. As shown in Table 5, the first column is the position and the second column is the corresponding word. If the word is a trigger word, its third column is marked “Trigger” and its fourth column is marked with the form of “[ArgPos1, ArgPos2,…, ArgPosn,…]” in which “ArgPosn” is the starting position of the corresponding argument in rolen. If the word is an argument, its third column is marked by the corresponding role category label. And if this argument has attributes, its fourth column is marked with the form of “[AttPos1, AttPos2,…, AttPosn,…]” in which “AttPosn” is the starting position of its attributen.
Neuroimaging Event Extraction Based on AT-NeuroEAE
In order to realize the event extraction containing attributes, this study proposes a joint event extraction model based on deep adversarial learning, called AT-NeuroEAE. The multi-layer joint extraction model of neuroimaging events with attributes is constructed to synchronously realize the prediction of event elements, including trigger, arguments, and attributes and the extraction of element relations, including argument roles and argument attributes, by the end-to-end mode. The adversarial learning based on FreeAT is combined with this joint model to realize the event extraction in a few-shot learning scenario. Figure 6 gives the whole structure of the model, which consists of the text vectorization layer, the event element prediction layer, the role-attribute recognition layer, and the adversarial learning mechanism.
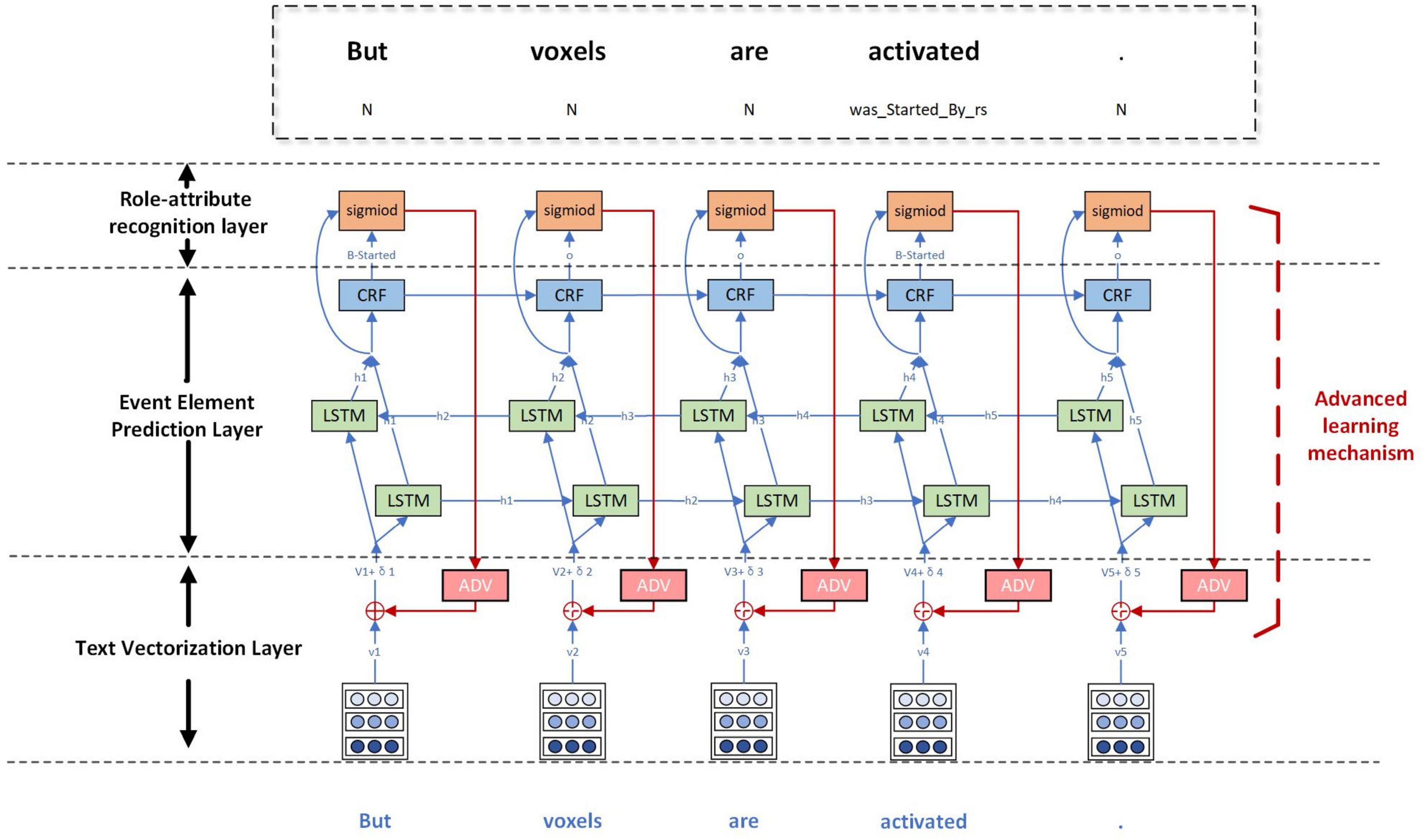
Figure 6. The AT-NeuroEAE model. The text vectorization layer encodes sentences as textual vectors based on lexical units, case features, and domain terminology dictionaries. The event element prediction layer predicts the potential event elements by using the BiLSTM-CRF model. The role-attribute recognition layer identifies the role and attribute of argument by using the sigmoid function. The adversarial learning mechanism adds small and persistent disturbances to the input of joint model for improving the robustness and generalization of the model. BiLSTM: bi-directional long short-term memory; CRF: conditional random fields; adv: adversarial learning.
In this experiment, we set the dimension of the word vector to 25, the number of iterations to 200, the dimension of the LSTM hidden layer to 64, the number of LSTM layers to 3, the dropout to 0.9, the learning rate to 1e-3, the activation function to tanh, and the adversarial disturbance parameter alpha to 0.01. Parameter optimization used the Adam algorithms.
Text Vectorization Layer
The text vectorization layer encodes sentences as textual vectors for the input of model. Suppose that there is a sentence s = [w1, w2, … wn]. For each word wi, three categories of feature vectors are constructed based on lexical units, case features, and domain terminology dictionaries, respectively.
• Word vector. The word vector is learned to contain as much semantic and grammatical information as possible (Pennington et al., 2014). This study adopts the Glove word vector model which was trained on 6 billion words of Wikipedia and web texts (Pennington et al., 2014).
• Case feature vector. Neuroimaging event mentions are involved with a large number of domain terms with capitalized abbreviated names, such as fMRI. In order to capture this kind of corpus features, this study constructs a one-hot case feature vector including six dimensions “numeric,” “allLower,” “allUpper,” “initialUpper,” “mainly_numeric,” “contains_digit,” and “other.”
• Terminology dictionary vector. Neuroimaging event elements involve nine types of important domain entities shown in Table 2. This study constructs the one-dimensional terminology dictionary vector (Xu et al., 2019) to capture this kind of domain corpus features. Nine term dictionaries are collected for these nine categories of domain entities firstly, and then a label list is constructed base on the “B/I/O-entity category abbreviation.” When wi matches any term in the dictionaries, a label index is set as the dimensional value of terminology dictionary vector of wi.
Concatenating these three categories of feature vectors (Zhang et al., 2019), the combined word vector of wi can be obtained as follows:
where vw, vc, and vt are the corresponding word vector, case feature vector, and terminology dictionary vector of wi.
Event Element Prediction Layer
The event element prediction layer predicts the potential event elements (i.e., argument, attribute, or trigger word) by using the BiLSTM-CRF model. BiLSTM is used to model context information of sentences based on input text vectors. The process can be defined as follows:
where, vi is the combination vector of wi, is the forward hidden layer output of LSTM, is the backward hidden layer output of LSTM, and hi is a word representation of each word token.
CRF is used to predict the element category and boundary. Event element prediction is a named entity recognition task which can be formulated as a sequence labeling problem. Based on the BIO annotation mode, each event element often consists of multiple sequential tokens. We calculate the score of each token wi for each event element tag:
where the superscript (e) means event element prediction; f(⋅) is the activation function; V(e) ∈ ℝp = l, U(e) ∈ ℝ2d = l, and l are the width of LSTM layer; and d is the hidden size of LSTM layer. The linear-chain CRF score of the s is defined as:
where yi ∈ Y is the tag of wi, si,yi is the predicted score of wi given a tag yi, and T is the transition matrix.
Then the probability of a given tag sequence on all possible tag sequences of the input sentence sen is defined as:
In the process of training, this study minimizes the cross-entry loss ℒNER:
Role-Attribute Recognition
The role-attribute recognition layer identifies the role and attribute of argument by using the sigmoid function. Recognizing the attribute of argument is the one-to-one relation extraction between the argument and its attribute, but recognizing the role of argument is the one-to-n relations because the trigger word is usually corresponding to multiple arguments in an event. Hence, this paper models the role recognition task as a multi-label head selection problem (Zheng et al., 2017), i.e., the relation extraction between one trigger word and two or more arguments.
As shown in Figure 4, the role-attribute recognition layer receives the hidden state of LSTM from the event element prediction layer. The predication score between tokens wi and wj given a role-attribute tag rk can be calculated as follows:
where the superscript (r) means role-attribute recognition, f(⋅) is the activation function, hi is the output of the hidden state of LSTM corresponding to wi, V(r) ∈ ℝl l is the width of the LSTM layer, U(r) ∈ ℝl*2d, W(r) ∈ ℝl*2d, and d is the hidden size of LSTM layer. The probability of token wj to be selected as the head of token wi with the role-attribute tag rk between them can be calculated as follows:
where θ is a set of parameters and σ(⋅) is the sigmoid function.
In the process of training, this paper minimizes the cross-entry loss ℒRAR:
where yi is the ground truth vector and rk is the role-attribute tag, and yi⊆ sen and rk ⊆ R and m (m < n) are the number of associated heads (i.e., roles or attributes) of wi. During decoding, the most probable heads and roles/attributes are selected using threshold-based prediction.
Finally, for the joint event extraction task, the final objective is calculated as ℒNER + ℒRAR. This study minimizes this objective function in the process of training of joint model.
Adversarial Learning Mechanism
The adversarial learning mechanism adds small and persistent disturbances to the input of the joint model. After the adversarial training with disturbances, the output distribution is consistent with the original distribution, but the robustness and generalization of the model can be improved (Wang et al., 2018).
This study adopts the perturbation strategy of FreeAT proposed by Shafahi et al. (2019). As shown in Figure 5, repeat K times for each sample continuously to find the optimal perturbation. The gradient of the previous step is multiplexed when calculating the perturbation. Finally, the overall iteration is divided by K to greatly reduce the speed problem caused by the inner iteration. The approximation of the perturbation is defined as:
where, radν is the perturbation, is a copy of the current model parameter, t ⊆ K, ∈ is a hyperparameter, α is the factor and D is the dimension of the embeddings vi, and t is the number of inner iterations.
Finally combining the original examples and the confrontation examples for model adversarial training, the final loss of joint model is as follows:
where is the current value of the model parameter.
Evaluation
The precision rate P, the recall rate R, and the F1 value (Curiskis et al., 2019) are adopted to evaluate experimental results. They can be calculated as follows:
In this study, P, R, and F1 values are used to evaluate the results of subtasks, but only F1 values are used to evaluate the overall results of event extraction.
Results
Baseline Methods
The CNN-BiLSTM-PCNN (Convolutional Neural Network-Bi-directional Long Short-Term Memory-Pulse Coupled Neural Network) pipeline model (Zheng et al., 2017) and the GCN (Graph convolution Network) joint model9 are used in the two control experiments. The CNN-BiLSTM-CNN pipeline model uses the CNN-BiLSTM model for the subtask of event element prediction, and the PCNN model for the subtask of role attribute recognition.
In many competitions, these models have been proved to be stable and relatively optimal for various biomedical named entity recognition tasks and relation extraction tasks. The GCN joint model was used on multiple shared tasks of biomedical event extraction, including BB3 (Bacteria Biotope) event data set (Lever and Jones, 2016), and SeeDev (Plant Seed Development) event data set (Agirre et al., 2019), and has obtained good experimental results.
In this study, these two models used their original parameters firstly, and then we adjusted parameters referring to some classic articles (Kip and Welling, 2016; Ma and Hovy, 2016; Zhang et al., 2019). Based on the results, a group of optimal parameters were chosen as follows:
• The CNN-BILSTM-PCNN pipeline model: For the event element prediction subtask, the dimension size of the word vector was set at 100. The epoch number was set at 50, the convolution width at 3, the CNN output size at 30, the dimensional number of LSTM hidden layer at 200, the mini-batch size at 9, and the dropout at 0.5. The Adam algorithm was used to optimize parameters. For the role-attribute recognition subtask, the dimension size of the word vector was set at 50, the convolution width at 3, the dimensions of the two hidden layers at 200 and 100, respectively, and the learning rate at 0.01.
• The GCN joint model: The dimension size of the word vector was set at 300, the dimension size of the Hidden layer at 200, the epoch number at 50, dropout of GCN at 0.5, dropout of word at 0.5, and the batch size at 16.
Results Analysis
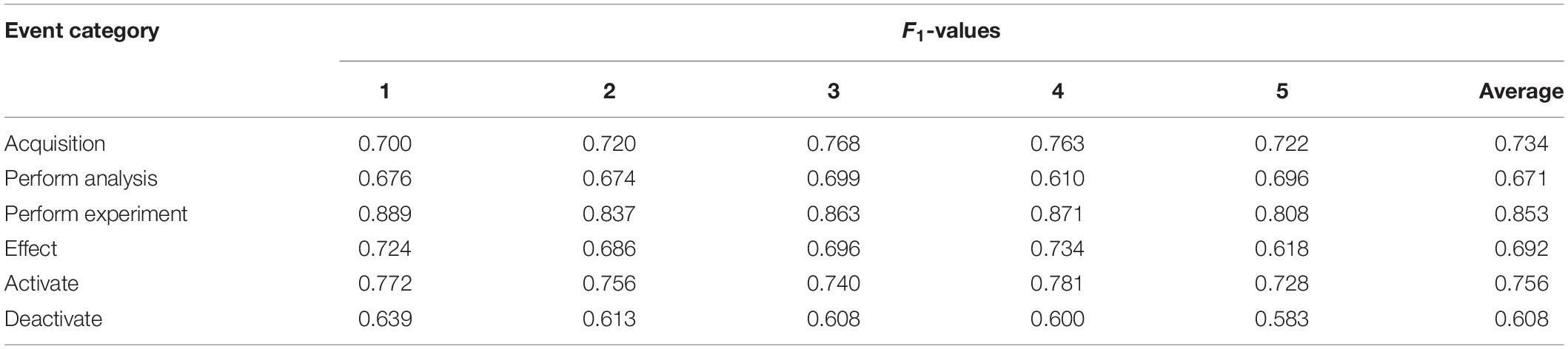
This study adapted fivefold cross validation to improve the objectivity of results. Each category of event mentions was randomly divided into five equal parts. In order to simulate the few-shot learning scenario, each experiment used one part as the training data set and the remaining four parts as the test data set. After five experiments, the average results were taken as the final results.
Table 6 gives F1 values of all models. It can be seen that the CNN-BiLSTM-PCNN pipeline event model lags behind the other three models in five categories of events except “Acquisition.” This shows the necessity of joint model in the complex text mining task, such as event extraction. The proposed NeuroEAE model is superior to the GCN joint model in all event categories except the “Analytical results” event. This proves the validity of proposed model structure for extracting neuroimaging event containing attributes.
The proposed AT-NeuroEAE model introduces adversarial learning into NeuroEAE. It achieved the better results on most of event categories than other models. This shows the validity of adversarial learning in the few-shot learning scenario. However, AT-NeuroEAE loses to NeuroEAE on the “Activate” and “Deactivate” events. This may be due to the fact that its structure and elements are relatively single compared with other event categories. As the conclusion of computational neuroscience research, the “Activate” and “Deactivate” events are often described by using the simple and clear languages in articles. For example, “We previously showed that a common network of brain regions was activated for past and future thinking in healthy older adults” (Rodolphe et al., 2014) and “This cortical area was found to be activated in studies where participants had to perform a task of face vs. non-face recognition” (Prehn-Kristensen et al., 2009). Under the same degree of adversarial disturbance of the model, such a simple event structure is easy to produce the collapse problem, which makes the model unable to further learn. This shows that the increasing disturbance can lead to the reduction of model accuracy under the simple event extraction. For other event categories, whether it is the “Analytical Results” event with the complex structure or the “Effect” event with more trigger word instances, the model has advantages by using adversarial learning. Hence, the proposed AT-NeuroEAE is an effective model to extract neuroimaging events from complex event mentions, which are common in neuroimaging articles. If the event is simpler, the NeuroEAE model is the better choice. Extracting neuroimaging event-containing attributes can be realized by flexibly using these two kinds of models.
Table 7 gives the F1 values of each experiment in fivefold cross validation. As shown in this table, the results are stable in each experiment. Our experiments were performed on the personal computer with CPU Intel Core 9th Generation i7 and Graphics Card NVIDIA GeForce GTX 1650. The epoch number was set at 100. Table 8 gives the running time of each experiment. The average running time of model training is almost 4.8 h and the average running time of model test is almost 6.7 s. This shows that, although the model training takes a long time, the speed of event extraction is still relatively fast and can meet the requirements of rapid extraction of provenances.
Discussion
Results Analysis on Subtasks
Neuroimaging event extraction consists of two subtasks: event element prediction and role-attribute recognition. This section will analyze results of each subtask separately to understand and evaluate the proposed AT-NeuroEAE model in depth.
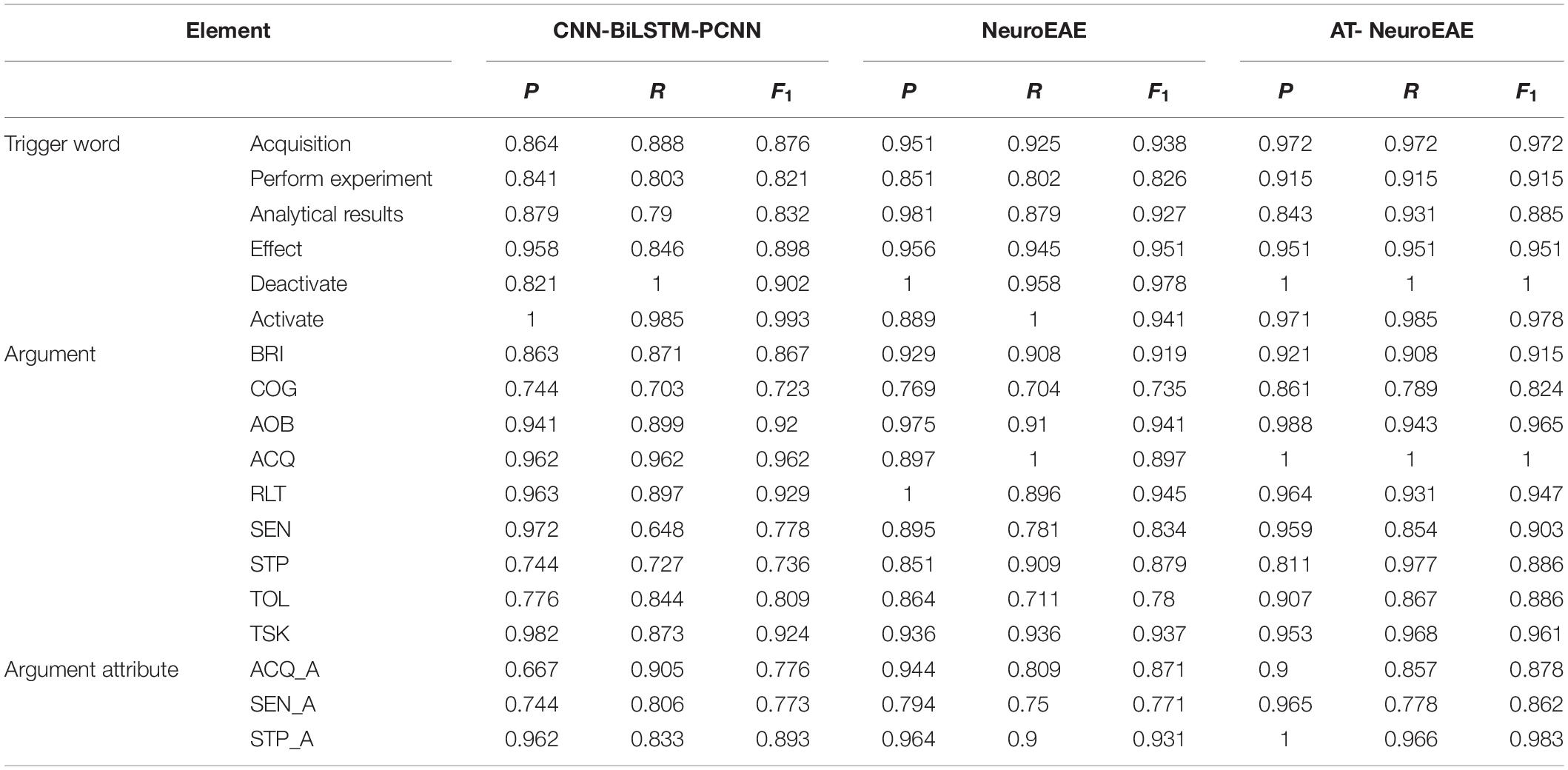
Table 9 gives the results of event element prediction, which are involved with three kinds of elements, i.e., trigger words, arguments, and attributes. The full names of arguments are mentioned in Table 2. The “abbreviation_A,” such as DAT_A, denotes the attribute of the corresponding argument. In this study, the prediction task of attribute is most ambiguous because the general attribute indicates all of possible actual attributes, as stated above. The prediction task of arguments is clearly defined but each argument category is often involved with a large number of entity instances. The prediction task of trigger words is clearest and simplest. Each category of trigger words only involved limited words. Comparing prediction results on these three kinds of elements, we can see that the clearer and simpler the task, the better the result. Generally, all elements are recognized well. F1 values of the AT-NeuroEAE model are almost above 0.9. Even the worst model CNN-BiLSTM-PCNN, its F1 values on all categories of elements are more than 0.7. This shows that the existing deep learning models have been able to effectively solve the task of named entity recognition, as stated in Armelle et al. (2014).
Comparing AT-NeuroEAE and NeuroEAE, most F1 values are improved and proved the effectiveness of adversarial learning. The negative effects of adversarial learning mainly appear in attribute prediction. This shows that the ambiguous task definition may not be suitable for adversarial learning.
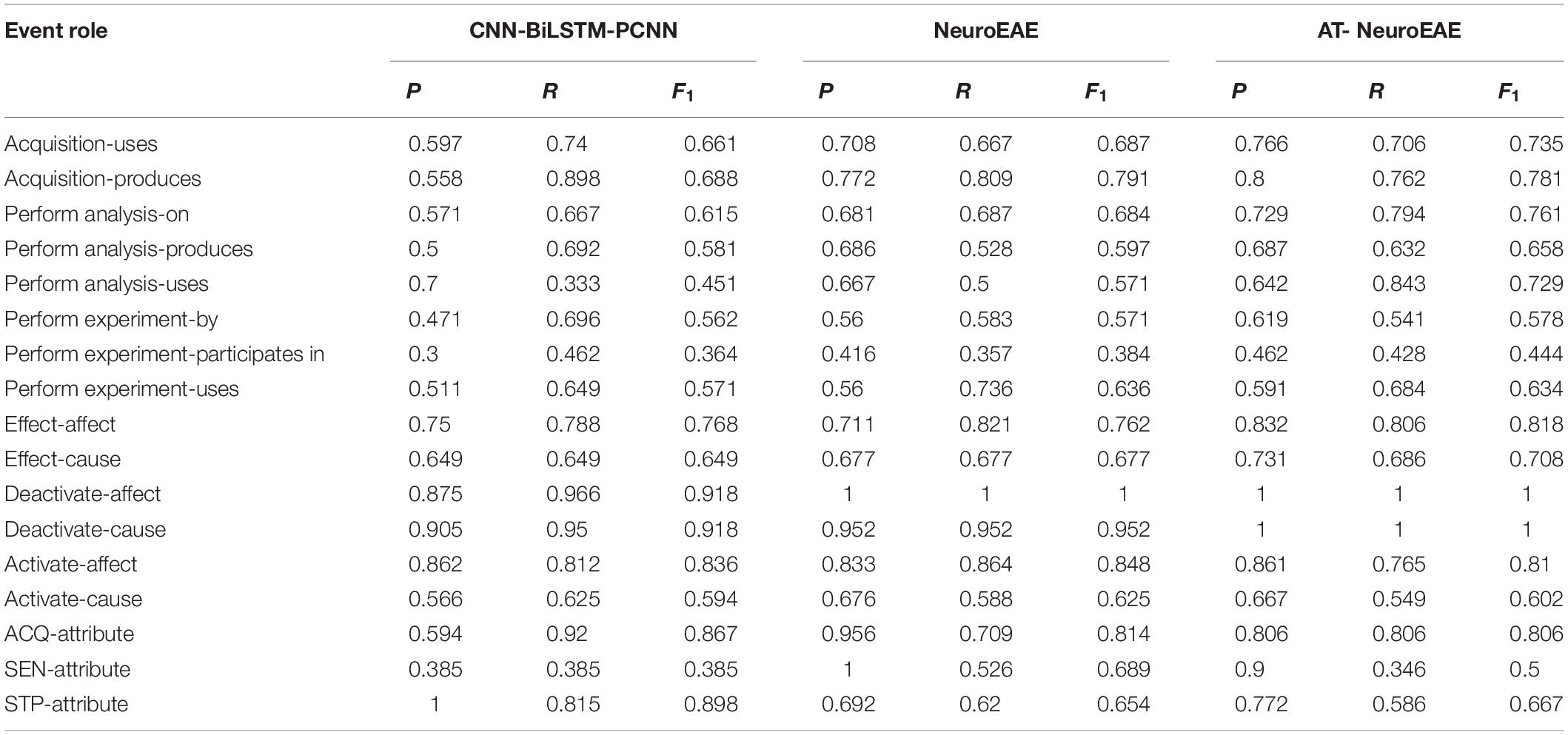
Table 10 gives the results of role-attribute recognition, which are involved with two aspects of relations, i.e., element roles and element-attribute relations. Obviously, adversarial learning improves the F1 values of role-attribute recognition for the majority of roles and attributes. The F1 value of “Analysis-Agent” role is significantly improved after adding adversarial learning. The main reason is that the corresponding “Perform Analysis” event has the complex structure. This confirms the above judgment that the adversarial learning is more suitable for complex tasks.
In addition, we can see that the F1 values of the “Activate-affect” role and “Activate-cause” role in the “Activate” event decrease after adding adversarial learning. As stated above, the “Activate” event often has the simple and clear language structure because it is the research conclusion. Table 9 shows that the F1 values of the element “Brain Area” (BRI) in this event also decrease after adding adversarial learning. This confirms the above judgment again that the adversarial learning is not suitable for simple tasks. The F1 values of another event element “Cognitive Function” (COG) don’t decrease mainly because “Cognitive Function” is most complex element category and has thousands of entity instances (Poldrack et al., 2011) in different forms. The F1 values of two categories of attribute relations “ACQ-Attribute” and “SEN-Attribute” also decrease after adding adversarial learning. This confirms the above judgment again that adversarial learning is not suitable for ambiguous tasks.
Comparing Tables 9, 10, we can see that in the first subtask, the results of CNN-BiLSTM-PCNN pipeline model are similar to our two joint models, but there is a big gap in the second subtask. The reason is the existence of cascading errors. As shown in Table 11, when cascading errors are removed manually, CNN-BiLSTM-PCNN can also achieve good results. This proves the validity of joint models.
Result Comparison With Existing Studies on Neuroimaging Text Mining
As stated in section “Neuroimaging Text Mining,” existing studies on neuroimaging text mining can be divided into neuroimaging topic modeling and neuroimaging named entity recognition. This section will compare the proposed AT-NeuroEAE with these two kinds of neuroimaging text mining, respectively.
LDA is the most commonly used method for neuroimaging topic modeling and has been applied to many previous studies (French et al., 2012; Alhazmi et al., 2018). Different from quantitative analysis based on P, R, and F1, this section compares extracted contents between the proposed AT-NeuroEAE and LDA-based neuroimaging topic modeling by using a specific article. Figure 7 shows the results of AT-NeuroEAE and LDA for the article titled “Induction of Empathy by the Smell of Anxiety” (Prehn-Kristensen et al., 2009). Different colors represent different categories of entities or topics. The random state was set to 1, the number of topics to 15, and alpha to 0.5.
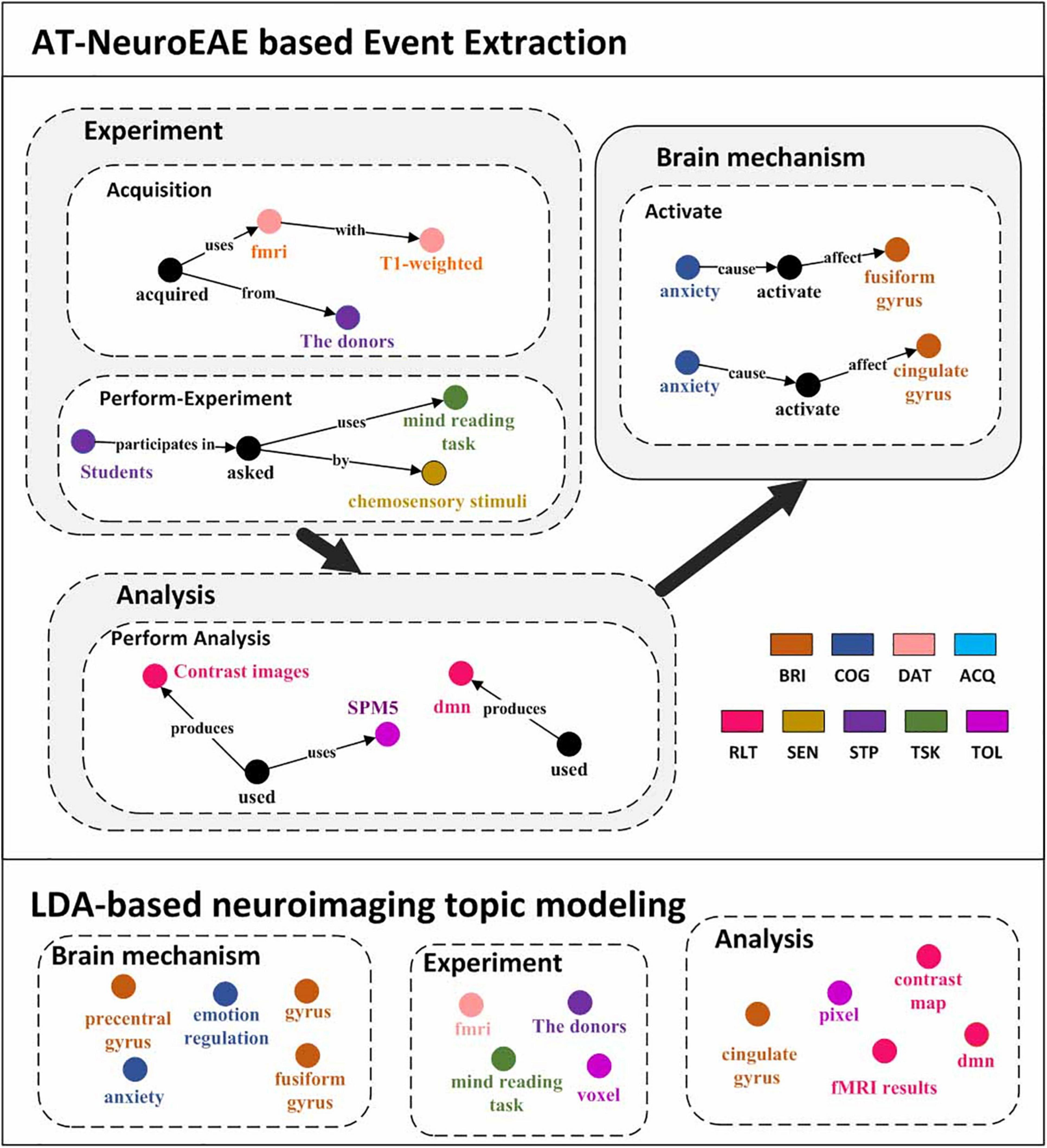
Figure 7. Visualization results of event extraction. The top is five events extracted from the article (Prehn-Kristensen et al., 2009). The bottom is LDA topics extracted from the same article. In order to compare with the results of AT-NeuroEAE, LDA topics are manually divided into three classes, brain mechanism, experiment, and analysis.
As shown in Figure 7, LDA can obtain multi-aspect topics, which are involved with brain mechanism (e.g., anxiety, frontal gyrus), the experiment (e.g., mind reading task), and analysis (e.g., voxel). However, these topics are isolated and cannot effectively characterize the whole of computational neuroscience research. Some general words, such as “conclusion” and “figure,” are also included in topics because a large amount of noise in full-text corpora affects topic recognition based on word distribution.
Different from LDA, the proposed AT-NeuroEAE can extract a group of events with rich semantics for outlining the whole research process:
• Brain mechanisms: The article reveals that “anxiety” activates the brain areas “cingulate gyrus” and “fusiform gyrus.”
• Experiment process: The experiments adopted the “mind reading task” with “chemosensory stimulus.” Researchers used “T1 weighted” “fMRI” device to collect data from “the donors.”
• Analytical process: The study got the “contrast map” by the “SPM5.” “dmn” was also obtained as the analytical result.
Because the extraction is based on pre-defined event categories, those general words can be effectively filtered.
Named entity recognition is the current research focus in neuroimaging text mining. Because related studies (Shardlow et al., 2018; Riedel et al., 2019; Sheng et al., 2019) have different entity or label categories, this study only qualitatively compares their entity/label categories with the proposed AT-NeuroEAE. The result is shown in Figure 8.
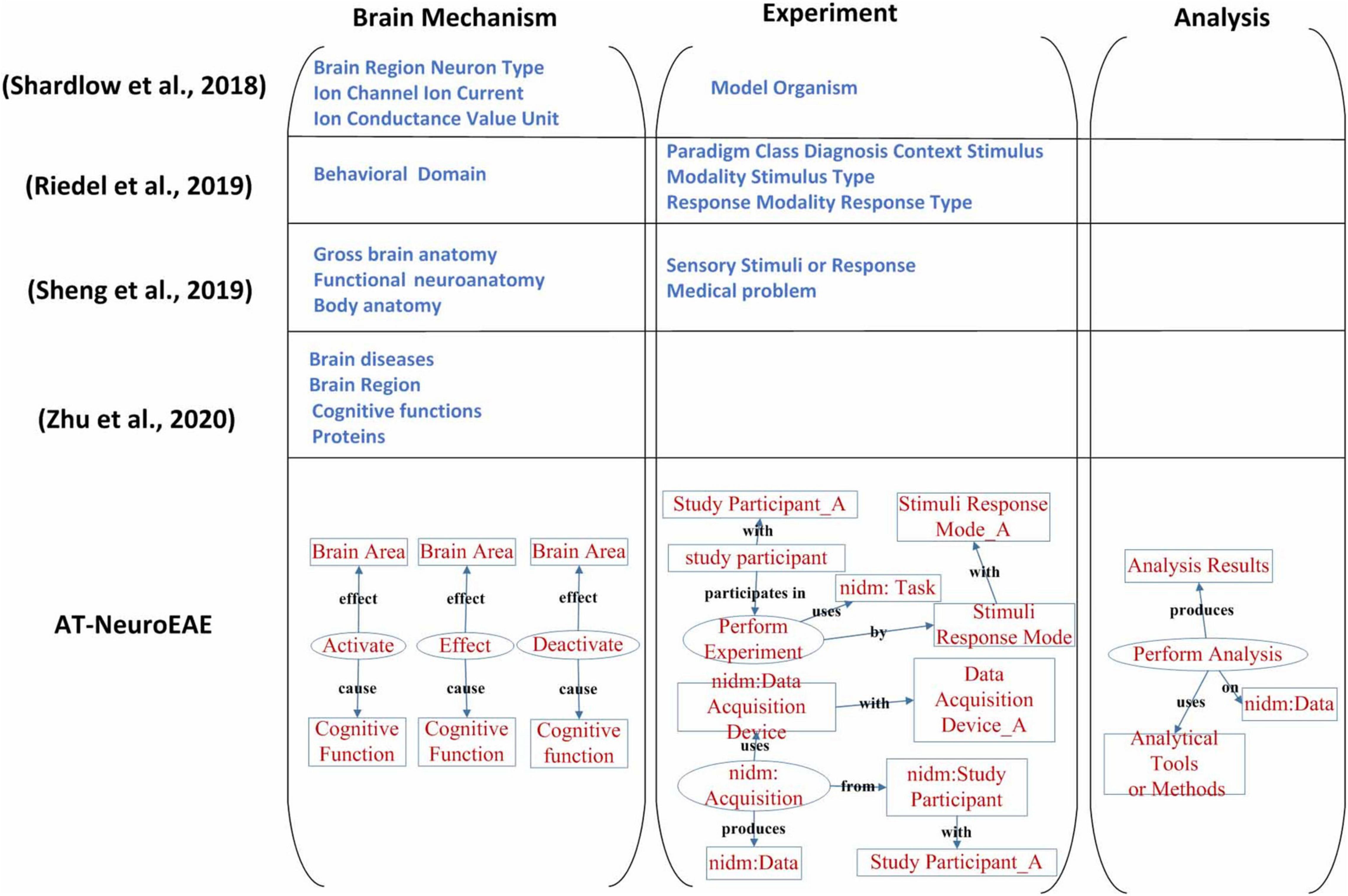
Figure 8. A comparison of neuroimaging entity/label categories/span of interest. The study on Shardlow et al. (2018) mainly focused on brain mechanism, especially multi-level brain structures. The study on Riedel et al. (2019) only took account of the experimental process. The study on Sheng et al. (2019) focused on brain mechanism and two experimental factors, including sensory stimuli or response and study participants’ medical problems. The study on Zhu et al. (2020) paid attention on pathology and mechanism of brain diseases. Our study is involved with the whole research process and extracted information is organized by events with rich semantics.
The study on (Shardlow et al., 2018) mainly focused on brain mechanism, especially multi-level brain structures. The cognitive function and the research process were neglected. In contrast, the study on (Riedel et al., 2019) only took account of the experimental process and neglected brain mechanisms and the analytical process. The study of Sheng et al. (2019) focused on brain mechanism, but neglected the analysis process which is very important to understand and evaluate the conclusion about brain mechanism. About the experiment, only sensory stimuli or response and study participants’ medical problems were extracted. Some key experimental factors, including experimental tasks, data acquisition equipment, etc., have not been paid enough attention. The study of Zhu et al. (2020) paid attention on pathology and mechanism of brain diseases, and its spans of interests are limited to brain diseases, brain regions, cognitive functions, and proteins.
In this study, neuroimaging events related to brain mechanism and the whole research process were extracted. In the aspect of brain mechanism, three event categories “Activate,” “Deactivate,” and “Effect” represent not only location information but also physiological characteristics information such as brain network. In the aspect of experiment, the event category “Perform Experiment” represents as much information as the study on (Riedel et al., 2019) because the representations about stimuli, response, and tasks in neuroimaging articles are flexible and it is not necessary to distinguish the form and type of stimulus, task paradigm, and context in detail. The event category “Acquisition” represents the imaging devices and scanning parameters. In the aspect of analysis, the event category “Perform Analysis” represents three important factors and their relationships during neuroimaging data analysis, including methods/tools, results and brain areas. All extracted information in this study is not an isolated entity, but organized as a systematic information network in the form of events, which can better describe the whole research process.
Conclusion
This study proposed an approach based on event extraction to realize research sharing-oriented neuroimaging provenance construction. Guided by the provenance model, the neuroimaging research process and result are modeled as six categories of neuroimaging event-containing attributes. A joint extraction model based on deep adversarial learning, called AT-NeuroEAE, is proposed to extract the defined neuroimaging events in a few-slot learning scenario. The experimental results on the PLOS ONE data set show that the model can realize the large-scale and low-cost neuroimaging provenance construction for open and FAIR research sharing in computational neuroscience.
Software Dependencies
As described above, the analyses presented in this work rely on the following dependencies: numpy (van der Walt et al., 2011), pandas (McKinney, 2011), statsmodels (Seabold and Perktold, 2010), SciPy (Jones et al., 2001), scikit-learn (Pedregosa et al., 2011), IPython (Pérez and Granger, 2007), nltk (Bird et al., 2009), pdfminer (Shinyama, 2007), seaborn (Waskom et al., 2017), and many core libraries provided with Python 2.7.11. Additionally, the ontological expansion of Cognitive Atlas term weights was influenced by Poldrack et al. (2011).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SL, JC, and ZX developed the study design and performed the coding and analyses. All authors contributed to the implementation and contributed to the manuscript revision, and read and approved the submitted version.
Funding
The work was supported by National Key Research and Development Program of China (Grant No. 2020YFB2104402).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2021.739535/full#supplementary-material
Footnotes
- ^ It is difficult to distinguish information from knowledge clearly because they have varying definitions in different applications. In this study, “research information” refers to all descriptions about computational science in scientific literature. It is involved with not only research findings, such as coordinates of activated brain areas, but also the whole research process, from experimental design to data analysis.
- ^ http://www.brainmap.org/
- ^ http://www.neurosynth.org/
- ^ http://neurovault.org
- ^ http://www.w3.org/TR/prov-dm/
- ^ http://medical.nema.org/dicom/
- ^ https://github.com/incf-nidash/XCEDE
- ^ http://brainspell.org
- ^ https://github.com/wangmm88/BioNLP-ST-2016_BB3-event
References
Abacha, A. B., Herrera, A., Ke, W., Long, L. R., Antani, S., and Demner-Fushman, D. (2017). “Named entity recognition in functional neuroimaging literature,” in Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, 2218–2220. doi: 10.1109/BIBM.2017.8218002
Abera, B. (2020). Event extraction and representation model from news articles. Int. J. Innov. Eng. Technol. 16, 1–8.
Abrams, M. B., Bjaalie, J. G., Das, S., Egan, G. F., and Martone, M. E. (2021). A standards organization for open and fair neuroscience: the international neuroinformatics coordinating facility. Neuroinformatics 1–12. doi: 10.31219/6osf.io/3rt9b
Agirre, A. G., Marimo, M., Intxaurrondo, A., Rabal, O., and Krallinger, M. (2019). “PharmaCoNER: pharmacological substances, compounds and proteins named entity recognition track,” in Proceedings of the 5th Workshop on BioNLP Open Shared Tasks, Hong Kong.
Alcalá-López, D., Smallwood, J., Jefferies, E., Van Overwalle, F., Vogeley, K., Mars, R. B., et al. (2017). Computing the social brain connectome across systems and states. Cereb. Cortex 28, 2207–2232. doi: 10.1093/cercor/bhx121
Alhazmi, F. H., Beaton, D., and Abdi, H. (2018). Semantically defined subdomains of functional neuroimaging literature and their corresponding brain regions. Hum. Brain Mapp. 39, 2764–2776. doi: 10.1002/hbm.24038
Araki, J., and Mitamura, T. (2018). “Open-domain event detection using distant supervision,” in Proceedings of the 27th International Conference on Computational Linguistics, Santa Fe, NM, 878–891.
Armelle, V., Pascale, P., Serge, B., Vincent, D., Béatrice, D., Francis, E., et al. (2014). Episodic future thinking in semantic dementia: a cognitive and fmri study. PLoS One 9:e111046. doi: 10.1371/journal.pone.0111046
Arshad, B., Munir, K., Mcclatchey, R., Shamdasani, J., and Khan, Z. (2019). Neuroprov: provenance data visualisation for neuroimaging analyses. J. Comput. Lang. 52, 72–87. doi: 10.1016/j.cola.2019.04.004
Badhwar, A. P., Kennedy, D., Poline, J. B., and Toro, R. (2016). Distributed collaboration: the case for the enhancement of Brainspell’s interface. Gigascience 5, 1–2.
Belhajjame, K., Zhao, J., Garijo, D., Gamble, M., Hettne, K., Palma, R., et al. (2015). Using a suite of ontologies for preserving workflow-centric research objects. J. Web Semant. 32, 16–42.
Bird, S., Klein, E., and Loper, E. (2009). Natural Language Processing with Python: Analyzing Text with the Natural Language Toolkit. Sebastopol, CA: O’Reilly Media, Inc.
Bjrne, J., and Salakoski, T. (2018). “Biomedical event extraction using convolutional neural networks and dependency parsing,” in Proceedings of the BioNLP 2018 workshop, Melbourne, VC.
Bolt, T., Nomi, J. S., Arens, R., Vij, S. G., Riedel, M., Salo, T., et al. (2020). Ontological dimensions of cognitive-neural mappings. Neuroinformatics 18, 451–463. doi: 10.1007/s12021-020-09454-y
Buneman, P., Khanna, S., and Wang-Chiew, T. (2004). “Why and where: a characterization of data provenance,” in Proceedings of the International Conference on Database Theory, (Berlin: Springer). doi: 10.1109/TVCG.2015.2467551
Chen, J. H., and Ning, Z. (2013). Toward the data-brain driven systematic brain data analysis. IEEE Trans. Syst. Man Cybernet. Syst. 43, 222–228. doi: 10.1109/TSMCA.2012.2199300
Chen, J. H., Ning, Z., and Liang, P. (2012). Data-brain driven systematic human brain data analysis: a case study in numerical inductive reasoning centric investigation. Cogn. Syst. Res. 15–32, 17–32. doi: 10.1016/j.cogsys.2010.12.014
Chen, Y. (2019). Multiple-level biomedical event trigger recognition with transfer learning. BMC Bioinformatics 20:459. doi: 10.1186/s12859-019-3030-z
Chung, J. W., Yang, W., and Park, J. (2020). Unsupervised inference of implicit biomedical events using context triggers. BMC Bioinformatics. 21:29. doi: 10.1186/s12859-020-3341-0
Curiskis, S. A., Drake, B., Osborn, T. R., and Kennedy, P. J. (2019). An evaluation of document clustering and topic modelling in two online social networks: twitter and reddit. Inform. Process. Manag. 57:102034. doi: 10.1016/j.ipm.2019.04.002
Daniel, W., Svenja, T., Anna, B., Münte Thomas, F., Michael, S., Manfred, C., et al. (2014). Tracking functional brain changes in patients with depression under psychodynamic psychotherapy using individualized stimuli. PLoS One 9:e109037. doi: 10.1371/journal.pone.0109037
Deléger, L., Bossy, R., Chaix, E., Ba, M., Ferré, A., Bessières, P., et al. (2016). “Overview of the bacteria biotope task at BioNLP shared task 2016,” in Proceedings of the 4th BioNLP Shared Task Workshop, Berlin, 12–22.
Dinov, I., Van Horn, J., Lozev, K. M., Magsipoc, R., Petrosyan, P., Liu, Z., et al. (2009). Efficient, distributed and interactive neuroimaging data analysis using the loni pipeline. Front. Neuroinform. 3:22. doi: 10.3389/neuro.11.022.2009
Doddington, G., Mitchell, A., Przybocki, M. A., Ramshaw, L., Strassel, S., and Weischedel, R. (2004). “The automatic content extraction (ACE) program - tasks, data, and evaluation,” in Proceedings of the 4th International Conference on Language Resources and Evaluation (LREC’04), Lisbon. doi: 10.1007/s12021-011-9126-x
French, L., Lane, S., Xu, L., Siu, C., Kwok, C., Chen, Y., et al. (2012). Application and evaluation of automated methods to extract neuroanatomical connectivity statements from free text. Bioinformatics 28, 2963–2970. doi: 10.1093/bioinformatics/bts542
Frishkoff, G., Sydes, J., Mueller, K., Frank, R., Curran, T., Connolly, J., et al. (2011). Minimal information for neural electromagnetic ontologies (MINEMO): a standards-compliant method for analysis and integration of eventrelated potentials (ERP) data. Stand. Genom. Sci. 5, 211–223. doi: 10.4056/sigs.2025347
Genon, S., Reid, A., Li, H., Fan, L., Müller, V. I., Cieslik, E. C., et al. (2018). The heterogeneity of the left dorsal premotor cortex evidenced by multimodal connectivity-based parcellation and functional characterization. Neuroimage 170, 400–411. doi: 10.1016/j.neuroimage.2017.02.034
Gibson, F., Overton, P. G., Smulders, T. V., Schultz, S. R., and Lord, P. (2008). Minimum information about a neuroscience investigation (mini): electrophysiology. Nat. Preced. 3. doi: 10.1038/npre.2008.1720.1
Gorgolewski, K. J., Esteban, O., Schaefer, G., Wandell, B. A., and Poldrack, R. A. (2017). OpenNeuro-a free online platform for sharing and analysis of neuroimaging data. Poster at the 23rd Annual Meeting of the Organization for Human Brain Mapping (OHBM) 2017, Vancouver, BC, 1677.
Gorgolewski, K. J., Varoquaux, G., Rivera, G., Schwartz, Y., Sochat, V. V., Ghosh, S. S., et al. (2016). NeuroVault.org: a repository for sharing unthresholded statistical maps, parcellations, and atlases of the human brain. Neuroimage 124, 1242–1244. doi: 10.1016/j.neuroimage.2015.04.016
Greenwood, M., Goble, C., Stevens, R., Zhao, J., and Oinn, T. (2003). “Provenance of e-science experiments-experience from bioinformatics,” in Proceedings of The UK OST e-Science Second All Hands Meeting 2003, Manchester, 223–226.
Huangfu, C., Zeng, Y., and Wang, Y. (2020). Creating neuroscientific knowledge organization system based on word representation and agglomerative clustering algorithm. Front. Neuroinform. 14:38. doi: 10.3389/fninf.2020.00038
Jones, E., Oliphant, T., and Peterson, P. (2001). SciPy: Open Source Scientific Tools for Python. Available online at: http://www.scipy.org/
Keator, D., Helmer, K., Maumet, C., Padhy, S., Jarecka, D., Ghosh, S., et al. (2019). “Tools for FAIR neuroimaging experiment metadata annotation with NIDM experiment,” in Proceedings of the OHBM 2019 – 25th Annual Meeting of the Organization for Human Brain Mapping Jun 2019, Rome, 1–5.
Keator, D., Helmer, K., Steffener, J., Turner, J. A., Van Erp, T. G., Gadde, S., et al. (2013). Towards structured sharing of raw and derived neuroimaging data across existing resources. NeuroImage 82, 647–661. doi: 10.1016/j.neuroimage.2013.05.094
Kennedy, D. N., Abraham, S. A., Bates, J. F., Crowley, A., Ghosh, S., Gillespie, T., et al. (2019). Everything matters: the reproNim perspective on reproducible neuroimaging. Front. Neuroinform. 13:1. doi: 10.3389/fninf.2019.00001
Kip, F. T. N., and Welling, M. (2016). Semi-supervised classification with graph convolutional networks. arXiv [Preprint]. arXiv:1609.02907.
Laird, A. R., Lancaster, J. L., and Fox, P. T. (2005). BrainMap: the social evolution of a human brain mapping database. Neuroinformatics 3, 65–78. doi: 10.1385/ni:3:1:065
Lanter, D. P. (1991). Design of a lineage-based meta-data base for GIS. Cartogr. Geogr. Inform. Syst. 18, 255–261. doi: 10.1559/152304091783786718
Lanting, C. P., Kleine, E. D., Langers, D., and Dijk, P. V. (2014). Unilateral tinnitus: changes in connectivity and response lateralization measured with fmri. PLoS One 9:e110704. doi: 10.1371/journal.pone.0110704
Lei, B., Wu, F., Zhou, J., Xiong, D., and Wu, K. (2020). Neuro-learn: a solution for collaborative pattern analysis of neuroimaging data. Neuroinformatics 19, 79–91. doi: 10.1007/s12021-020-09468-6
Lever, J., and Jones, S. J. (2016). “VERSE: event and relation extraction in the BioNLP 2016 shared task,” in Proceedings of the 4th BioNLP Shared Task Workshop, Berlin.
Li, D., Huang, L., Ji, H., and Han, J. (2016). “Biomedical event extraction based on knowledge-driven Tree-LSTM,” in Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics, vol. 1 (Minneapolis, Minnesota, Hennepin: Association for Computational Linguistics), 1421–1430. doi: 10.18653/v1/N19-1145
Li, L., Liu, Y., and Qin, M. (2020). Extracting biomedical events with parallel multi-pooling convolutional neural networks. IEEE ACM Trans. Comput. Biol. Bioinform. 17, 599–607. doi: 10.1109/TCBB.2018.2868078
Ma, X., and Hovy, E. (2016). “End-to-end sequence labeling via bi-directional lstm-cnns-crf,” in Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics, Berlin. doi: 10.13140/RG.2.1.2182.5685
Maumet, C. (2020). “Open Science in action: doing research as a community!,” in Proceedings of the 26th Annual Meeting of the Organization for Human Brain Mapping OHBM 2020, Montréal, QC, 1–19.
Maumet, C., Auer, T., Bowring, A., Chen, G., Das, S., Flandin, G., et al. (2016). Sharing brain mapping statistical results with the neuroimaging data model. Sci. Data 3:160102. doi: 10.1038/sdata.2016.102
Maumet, C., Ghosh, S., Halchenko, Y., Jarecka, D., Nichols, N., Poline, J. B., et al. (2019). “The best of both worlds: using semantic web with JSON-LD. An example with NIDM-Results and Datalad,” in Proceedings of the OHBM 2019 - 25th Annual Meeting of the Organization for Human Brain Mapping, Rome.
McKinney, W. (2011). pandas: a foundational Python library for data analysis and statistics. Python High Perform. Sci. Comput. 14, 1–9.
Miksa, T., and Rauber, A. (2017). Using ontologies for verification and validation of workflow-based experiments. J. Web Seman. 43, 25–45. doi: 10.1016/j.websem.2017.01.002
Milham, M. P., Cameron, C. R., Son, J. J., Fleischmann, M., Clucas, J., Xu, H., et al. (2018). Assessment of the impact of shared brain imaging data on the scientific literature. Nat. Commun. 9:2818. doi: 10.1038/s41467-018-04976-1
Moreau, L., Ludscher, B., Altintas, I., Barga, R. S., and Zhao, Y. (2008). Special issue: the first provenance challenge. Concurr. Comput. Pract. Exp. 20, 409–418.
Mutschler, I., Ball, T., Kirmse, U., Wieckhorst, B., Pluess, M., Klarhöfer, M. et al. (2016). The role of the subgenual anterior cingulate cortex and amygdala in environmental sensitivity to infant crying. PLoS One 11:e0161181. doi: 10.1371/journal.pone.0161181
Naud, A., and Usui, S. (2008). Exploration of a collection of documents in neuroscience and extraction of topics by clustering. Neural Netw. 21, 1205–1211. doi: 10.1016/j.neunet.2008.05.009
Neumann, N., Lotze, M., and Eickhoff, S. B. (2016). Cognitive expertise: an ale meta-analysis. Hum. Brain Mapp. 37, 262–272. doi: 10.1002/hbm.23028
Nguyen, T. M., and Nguyen, T. H. (2019). “One for all: neural joint modeling of entities and events,” in Proceedings of the 33rd AAAI Conference on Artificial Intelligence, Honolulu, HI, 6851–6858. doi: 10.1371/journal.pone.0235796
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2011). Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830.
Pennington, J., Socher, R., and Manning, C. (2014). “Glove: global vectors for word representation,” in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha.
Pérez, F., and Granger, B. E. (2007). IPython: a system for interactive scientific computing. Comput. Sci. Eng. 9, 21–29. doi: 10.1109/MCSE.2007.53
Poldrack, R. A., and Gorgolewski, K. J. (2014). Making big data open: data sharing in neuroimaging. Nat. Neurosci. 17, 1510–1517. doi: 10.1038/nn.3818
Poldrack, R. A., and Gorgolewski, K. J. (2015). Openfmri: open sharing of task fmri data. NeuroImage 144, 259–261. doi: 10.1016/j.neuroimage.2015.05.073
Poldrack, R. A., Aniket, K., Donald, K., Eric, M., Christian, S., Yolanda, G., et al. (2011). The cognitive atlas: toward a knowledge foundation for cognitive neuroscience. Front. Neuroinform. 5:17. doi: 10.3389/fninf.2011.00017
Poldrack, R. A., Fletcher, P. C., Henson, R. N., Worsley, K. J., Brett, M., and Nichols, T. E. (2008). Guidelines for reporting an fMRI study. Neuroimage 40, 409–414. doi: 10.1016/j.neuroimage.2007.11.048
Poldrack, R. A., Mumford, J. A., Schonberg, T., Kalar, D., Barman, B., Yarkoni, T., et al. (2013). Discovering relations between mind, brain, and mental disorders using topic mapping. PLoS Comput. Biol. 8:e1002707. doi: 10.1371/journal.pcbi.1002707
Poline, J. B., Breeze, J. L., Ghosh, S., Gorgolewski, K., Halchenko, Y. O., Hanke, M., et al. (2012). Data sharing in neuroimaging research. Front. Neuroinform. 6:9. doi: 10.3389/fninf.2012.00009
Prehn-Kristensen, A., Wiesner, C., Bergmann, T. O., Wolff, S., Jansen, O., Mehdorn, H. M., et al. (2009). Induction of empathy by the smell of anxiety. PLoS One 4:e5987. doi: 10.1371/journal.pone.0005987
Riedel, M. C., Salo, T., Hays, J., and Md, Turner, and Laird, A. R. (2019). Automated, efficient, and accelerated knowledge modeling of the cognitive neuroimaging literature using the ATHENA toolkit. Front. Neurosci. 13:494. doi: 10.3389/fnins.2019.00494
Rodolphe, N., Allendorfer, J. B., Szaflarski, J. P., and Yoko, H. (2014). A model for visual memory encoding. PLoS One 9:e107761. doi: 10.1371/journal.pone.0107761
Ruiz-Olazar, M., Rocha, E. S., Rabaa, S. S., Ribas, C. E., and Braghetto, K. R. (2016). “A review of guidelines and models for representation of provenance information from neuroscience experiments,” in Proceedings of the 6th International Workshop on Provenance and Annotation of Data and Processes, (Berlin: Springer-Verlag).
Seabold, S., and Perktold, J. (2010). “Statsmodels: econometric and statistical modeling with python,” in Proceedings of the 9th Python in Science Conference, Vol. 57. (San Francisco, CA: Scipy), 61.
Shafahi, A., Najibi, M., Ghiasi, A., Xu, Z., Dickerson, J., Studer, C., et al. (2019). “Adversarial training for free!,” in Proceedings of the 33rd International Conference on Neural Information Processing Systems, Vol. 302, (Red Hook, NY: Curran Associates Inc.), 3358–3369.
Shardlow, M., Ju, M., Li, M., O’Reilly, C., Iavarone, E., McNaught, J., et al. (2018). A text mining pipeline using active and deep learning aimed at curating information in computational neuroscience. Neuroinformatics 17, 391–406. doi: 10.1007/s12021-018-9404-y
Sheng, Y., Chen, J., He, X., Xu, Z., and Lin, S. (2020). A topic learning pipeline for curating brain cognitive researches. IEEE Access 8, 191758–191774. doi: 10.1109/ACCESS.2020.3032173
Sheng, Y., Lin, S., Gao, J., He, X., and Chen, J. (2019). “Research sharing-oriented functional neuroimaging named entity recognition,” in Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Vol. 2019, (San Diego, CA: IEEE Press), 1629–1632.
Shinyama, Y. (2007). PDFMine-Python PDF Parser. Available online at: http://pypi.python.org/pypi/pdfminer/ (accessed April 29, 2016).
Simmhan, Y. L., Plale, B., and Gannon, D. (2005). A survey of data provenance in e-science. ACM SIGMOD Record. 34, 31–36. doi: 10.1145/1084805.1084812
Soomro, P. D., Kumar, S., Banbhrani, Shaikh, A. A., and Raj, H. (2017). Bio-NER: biomedical named entity recognition using rule-based and statistical learners. Int. J. Adv. Comput. Sci. Appl. 8, 163–170. doi: 10.14569/IJACSA.2017.081220
Sun, R., Guo, S., and Ji, D. H. (2017). Topic representation integrated with event knowledge. Chin. J. Comput. 40, 791–804. doi: 10.11897/SP.J.1016.2017.00791
Task Force on Grand Challenges (2011). NSF Advisory Committee for Cyberinfrastructure. Arlington, VA: National Science Foundation.
That, D., Fils, G., Yuan, Z., and Malik, T. (2017). Sciunits: reusable research objects. arXiv [Preprint]. arXiv:1707.05731v2,Google Scholar
Trieu, H. L., Tran, T. T., Duong, K. N. A., Nguyen, A., Miwa, M., and Ananiadou, S. (2020). DeepEventMine: end-to-end neural nested event extraction from biomedical texts. Bioinformatics 36, 4910–4917. doi: 10.1093/bioinformatics/btaa540
Turner, J. A., and Laird, A. R. (2012). The cognitive paradigm ontology: design and application. Neuroinformatics 10, 57–66. doi: 10.1007/s12021-011-9126-x
van der Walt, S., Colbert, S. C., and Varoquaux, G. (2011). The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 13, 22. doi: 10.1109/MCSE.2011.37
Van Horn, J. D., Grethe, J. S., Kostelec, P., Woodward, J. B., Aslam, J. A., Rus, D., et al. (2001). The functional magnetic resonance imaging data center (fMRIDC): the challenges and rewards of large-scale databasing of neuroimaging studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1412, 1323–1339. doi: 10.1098/rstb.2001.0916
Wang, G., Li, C., Wang, W., Zhang, Y., and Carin, L. (2018). “Joint embedding of words and labels for text classification,” in Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics, Melbourne, VC.
Waskom, M., Botvinnik, O., O’Kane, D., Hobson, P., Lukauskas, S., Gemperline, D. C., et al. (2017). mwaskom/seaborn: v0.8.1 (Version v0.8.1). doi: 10.5281/zenodo.883859
Wegrzyn, M., Aust, J., Barnstorf, L., Gippert, M., Harms, M., Hautum, A., et al. (2018). Thought experiment: decoding cognitive processes from the fmri data of one individual. PLoS One 13:e0204338. doi: 10.1371/journal.pone.0204338
Xiang, W., and Wang, B. (2019). A survey of event extraction from text. IEEE Access 7, 173111–173137. doi: 10.1109/ACCESS.2019.2956831
Xu, K., Yang, Z., Kang, P., Wang, Q., and Liu, W. (2019). Document-level attention-based bilstm-crf incorporating disease dictionary for disease named entity recognition. Comput. Biol. Med. 108, 122–132. doi: 10.1016/j.compbiomed.2019.04.002
Yarkoni, T., Poldrack, R. A., Nichols, T. E., Van Essen, D. C., and Wager, T. D. (2011). Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods 8, 665–670. doi: 10.1038/nmeth.1635
Yu, X., Rong, W., Liu, J., Zhou, D., Ouyang, Y., and Xiong, Z. (2019). LSTM-based end-to-end framework for biomedical event extraction. IEEE/ACM Trans. Comput. Biol. Bioinform. 17, 2029–2039. doi: 10.1109/TCBB.2019.2916346
Yuan, Z., Ton That, D. H., Kothari, S., Fils, G., and Malik, T. (2018). Utilizing provenance in reusable research objects. Informatics 5:14.
Zhang, S., Sheng, Y., Gao, J., Chen, J., Huang, J., and Lin, S. (2019). “A multi-domain named entity recognition method based on part-of-speech attention mechanism,” in Proceedings of the CCF Conference on Computer Supported Cooperative Work and Social Computing, (Singapore: Springer).
Zhao, W. Z., Zhao, Y., Jiang, X. P., He, T., Liu, F., and Li, N. (2021). Efficient multiple biomedical events extraction via reinforcement learning. Bioinformatics 37, 1891–1899. doi: 10.1093/bioinformatics/btab024
Zheng, S., Cao, W., Xu, W., and Bian, J. (2019). “Doc2edag: an end-to-end document-level framework for chinese financial event extraction,” in Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong. doi: 10.18653/v1/D19-1032
Zheng, S., Hao, Y., Lu, D., Bao, H., Xu, J., Hao, H., et al. (2017). Joint entity andrelation extraction based on a hybrid neural network. Neurocomputing 257, 59–66. doi: 10.1016/j.neucom.2016.12.075
Keywords: neuroimaging provenance, neuroimaging text mining, event extraction, attribute extraction, deep adversarial learning
Citation: Lin S, Xu Z, Sheng Y, Chen L and Chen J (2022) AT-NeuroEAE: A Joint Extraction Model of Events With Attributes for Research Sharing-Oriented Neuroimaging Provenance Construction. Front. Neurosci. 15:739535. doi: 10.3389/fnins.2021.739535
Received: 11 July 2021; Accepted: 20 December 2021;
Published: 07 March 2022.
Edited by:
Yuanpeng Zhang, Nantong University, ChinaReviewed by:
Karl Helmer, Massachusetts General Hospital and Harvard Medical School, United StatesYufeng Yao, Changshu Institute of Technology, China
Copyright © 2022 Lin, Xu, Sheng, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhui Chen, Y2hlbmppYW5odWlAYmp1dC5lZHUuY24=
 Shaofu Lin1,2
Shaofu Lin1,2 Zhe Xu
Zhe Xu