- 1Department of Neuroscience II, Research Institute of Environmental Medicine, Nagoya University, Nagoya, Japan
- 2Department of Neural Regulation, Nagoya University Graduate School of Medicine, Nagoya, Japan
Sleep and wakefulness are regulated by both the homeostatic mechanism and circadian clock. In mammals, the central circadian clock, the suprachiasmatic nucleus, in the hypothalamus plays a crucial role in the timing of physiology and behavior. Recently, we found that the circadian regulation of wakefulness was transmitted via corticotropin-releasing factor (CRF) neurons in the paraventricular nucleus of the hypothalamus to orexin neurons in the lateral hypothalamus. However, it is still unclear how the molecular clock in the CRF neurons contributes to the regulation of sleep and wakefulness. In the present study, we established CRF neuron-specific Bmal1-deficient mice and measured locomotor activity or electroencephalography and electromyography. We found that these mice showed normal circadian locomotor activity rhythms in both light–dark cycle and constant darkness. Furthermore, they showed normal daily patterns of sleep and wakefulness. These results suggest that Bmal1 in CRF neurons has no effect on either circadian locomotor activity or sleep and wakefulness.
Introduction
Physiology and behavior, such as sleep and wakefulness, exhibit daily variations that are controlled by the circadian clock. In mammals, almost all cells contain the circadian clock that is thought to generate approximately 24-h rhythms by the transcription–translation feedback loop of several clock genes such as Bmal1, Clock, Period, and Cryptochrome (Reppert and Weaver, 2002). The circadian clocks are present in a variety of tissues and cells. Among them, the central circadian clock is located in the suprachiasmatic nucleus (SCN) of the hypothalamus (Moore and Eichler, 1972; Stephan and Zucker, 1972), which has a role for the coordination of the temporal timing of circadian behavioral rhythms (Sawaki et al., 1984; Silver et al., 1996). The SCN comprises thousands of neurons that are neuroanatomically and functionally heterogenous (Van den Pol, 1980; Abrahamson and Moore, 2001). For instance, several neuropeptides, such as arginine vasopressin, vasoactive intestinal peptides, and gastrin- releasing peptides are involved in the SCN. Importantly, almost all SCN neurons are GABAergic, and this inhibitory signal is suggested to be involved in the regulation of circadian outputs such as sleep and wake cycles (Ono et al., 2019, 2020; Maejima et al., 2021). Circadian period, amplitude, and robustness are different in individual SCN neurons without neuronal networks when studied in a dispersed cell culture (Welsh et al., 1995). On the other hand, the circadian rhythms of SCN cells synchronize with each other via neuronal networks to make stable and robust circadian rhythms in the SCN (Herzog et al., 2004; Honma et al., 2004).
Previous reports show that the SCN neurons innervate several brain areas (Watts et al., 1987; Watts and Swanson, 1987) and play a role in physiology and behavior (Gizowski et al., 2016; Ono et al., 2020; Paul et al., 2020; Todd et al., 2020; Jones et al., 2021). Importantly, the circadian regulation of wakefulness is transmitted via the corticotropin-releasing factor (CRF) neurons in the paraventricular nucleus (PVN) of the hypothalamus to orexin neurons in the lateral hypothalamus (LH) (Ono et al., 2020). Optogenetic activation of CRF neurons in the PVN facilitates wakefulness, and pharmacogenetic suppression and ablation of CRF neurons attenuate wakefulness and locomotor activity, respectively (Ono et al., 2020). These results suggest that circadian information in the SCN transmits through the PVN to the LH pathway.
Although CRF neurons in the PVN are suggested to be involved in circadian-regulated wakefulness, it remains unclear how the molecular circadian clock in the CRF neurons affects sleep and wakefulness. To answer this question, we created CRF neuron-specific Bmal1-deficient mice using the Cre-loxp system. We measured locomotor activity, electroencephalogram (EEG), and electromyogram (EMG) to assess circadian rhythms and sleep/wakefulness in CRF neuron-specific Bmal1-deficient mice. Circadian locomotor activity rhythms were not different between the CRF neuron-specific Bmal1-deficient and control mice. Furthermore, CRF neuron-specific Bmal1 deficient mice did not show differences in sleep and wakefulness. These results suggest that a deficiency of Bmal1 in the CRF neurons has no significant effects on circadian locomotor activity and sleep/wakefulness.
Materials and Methods
Animals
We used CRF-Cre mice (Itoi et al., 2014), Rosa26-LSL-tdTomato (Ai9) mice (Madisen et al., 2010), and Bmal1flox/flox mice (Storch et al., 2007) on C57BL/6J background. All mice were housed in a 12-h dark–light cycle before the experiment (lights on 8:00–20:00). Room temperature (RT) was maintained at 23 ± 2°C, and humidity was maintained between 40 and 70%. Food and water were provided ad libitum. All experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committees at the Research Institute of Environmental Medicine at Nagoya University (approval numbers: R210729, R210730).
Locomotor Activity Recording
Both male and female mice were used for locomotor activity recording. The mice were individually housed in a polycarbonate cage placed in a light-tight and air-conditioned box. Spontaneous movements were measured by a passive infrared sensor that detects changes in animal thermal radiation due to movement. The amount of movement was recorded every minute using a computer software (ClockLab, Actimetrics). Free-running period was calculated using chi-square periodogram.
Electroencephalogram and Electromyogram Surgical Procedure
Male mice were anesthetized with 2% isoflurane (095–06573, FUJIFILM Wako Pure Chemical Corporation), and three EEG electrodes (U-1430–01, Wilco) were implanted on the skull, and two EMG electrodes (AS633, Cooner Wire) were attached through the rhomboid muscle (Tsunematsu et al., 2011). Carprofen (Zoetis Inc., Parsippany-Troy Hills, NJ, United States) 20 mg/kg subcutaneous injection was administered the day of and after surgery because of its anti-inflammatory and analgesic properties. After surgery, the mice were housed for about 7 days for recovery. To acclimate the mice to the recording conditions, they were individually connected to a cable with a slip ring (Kissei Comtec Co., Ltd., Matsumoto, Japan) for 7 days before the start of EEG and EMG recordings.
Sleep/Wake Recordings
Electroencephalogram and EMG signals were filtered at 1.5–30 Hz and 15–300 Hz, respectively, and amplified by an amplifier (AB-610J, Nihon Kohden). Animal behavior was monitored using a charge-coupled device video camera (SPK-E700CHP1, Keiyo Techno) and recorded using an infrared activity sensor (Kissei Comtec). EEG and EMG data were recorded using VitalRecorder with 128 Hz sampling rate (Kissei Comtec) and analyzed with SleepSign software (Kissei Comtec).
Vigilance State Analysis
Fast Fourier transform (FFT) was performed on the EEG signal; δ (1 ≤ δ < 6 Hz) and θ (6 ≤ θ < 11 Hz) bandwidths were calculated from FFT over a 0–10 Hz window with 1 Hz resolution. Then, EEG and EMG were automatically screened in 4 s epochs by SleepSign and classified as reported in previous studies (Tsunematsu et al., 2011; Hung et al., 2020; Ono et al., 2020). In short, wakefulness was categorized by high EMG amplitude or locomotion score combined with low EEG amplitude; nonrapid eye movement (NREM) sleep was identified by low EMG amplitude with high EEG δ power, and rapid eye movement (REM) sleep was characterized by low EMG and low EEG amplitude with over 50% of θ activity.
Immunohistochemistry
Mice were anesthetized with isoflurane, and colchicine (3 μl, 10 μg/μl in saline) was injected in the lateral ventricle. About 48 h later, mice were anesthetized and then sequentially perfused with saline and formalin (066–03847, FUJIFILM Wako). Brains were removed and immersed in formalin solution overnight at 4°C, and then immersed in a 30% sucrose solution in phosphate-buffered saline (PBS) for at least 2 days. The brains were frozen in embedding solution (4583, Sakura Finetek Japan) and stored in a −80°C freezer. Brains were subsequently sectioned into 40 μm thick slices using cryostat (CM3050-S, Leica Microsystems K.K.).
Coronal brain sections were washed three times for 10 min in PBS containing 0.25% Triton X-100 (35501–15, Nacalai Tesque) with 1% bovine serum albumin (A7905-500G, MilliporeSigma) (PBS-BX). The brain sections were incubated in PBS-BX with rabbit anti-Bmal1 antibody (1:2,500, NB100-2288; Novus Biologicals, Centennial, CO, United States) and goat anti-CRF antibody (1:200, sc-1761; Santa Cruz Biotechnology, Dallas, TX, United States) overnight at 4°C. Sections were then washed three times for 10 min in PBS-BX and incubated with CF 488-conjugated anti-goat antibody (1:1,000; Biotium Inc., Hayward, CA, United States) and CF 594-conjugated anti-rabbit antibody (Biotium Inc., Hayward, CA, United States) in PBS-BX for 2 h at RT. Sections were then washed in PBS-BX three times for 10 min and stained with 4′,6-diamidino-2-phenylindole (DAPI). After staining, sections were mounted, and the cover-glass was affixed with 50% glycerol in PBS. Photomicrographs were obtained using a microscope (BZ-X700, Keyence) or confocal microscope (LSM710, Carl Zeiss AG).
Statistical Analyses
Statistical analyses were performed using Statview, Statcel 3, OriginPro 2019 (OriginLab, Northampton, MA, United States) or Excel.
Results
Corticotropin-Releasing Factor Expression Patterns in the Brain
To confirm which brain areas expressed CRF in the brain, we crossed CRF-Cre (Itoi et al., 2014) and Rosa26-LSL-tdTomato mice (Madisen et al., 2010). We observed tdTomato-positive cells in several areas in the brain, including the PVN, bed nucleus of the stria terminalis (BNST), central nucleus of the amygdala (CeA), piriform cortex, interstitial nucleus of the posterior limb of the anterior commissure, laterodorsal tegmental nucleus, and inferior olivary nucleus (Figure 1). Some axons of CRF neurons were also observed. These results were consistent with previous reports (Itoi et al., 2014; Walker et al., 2019).
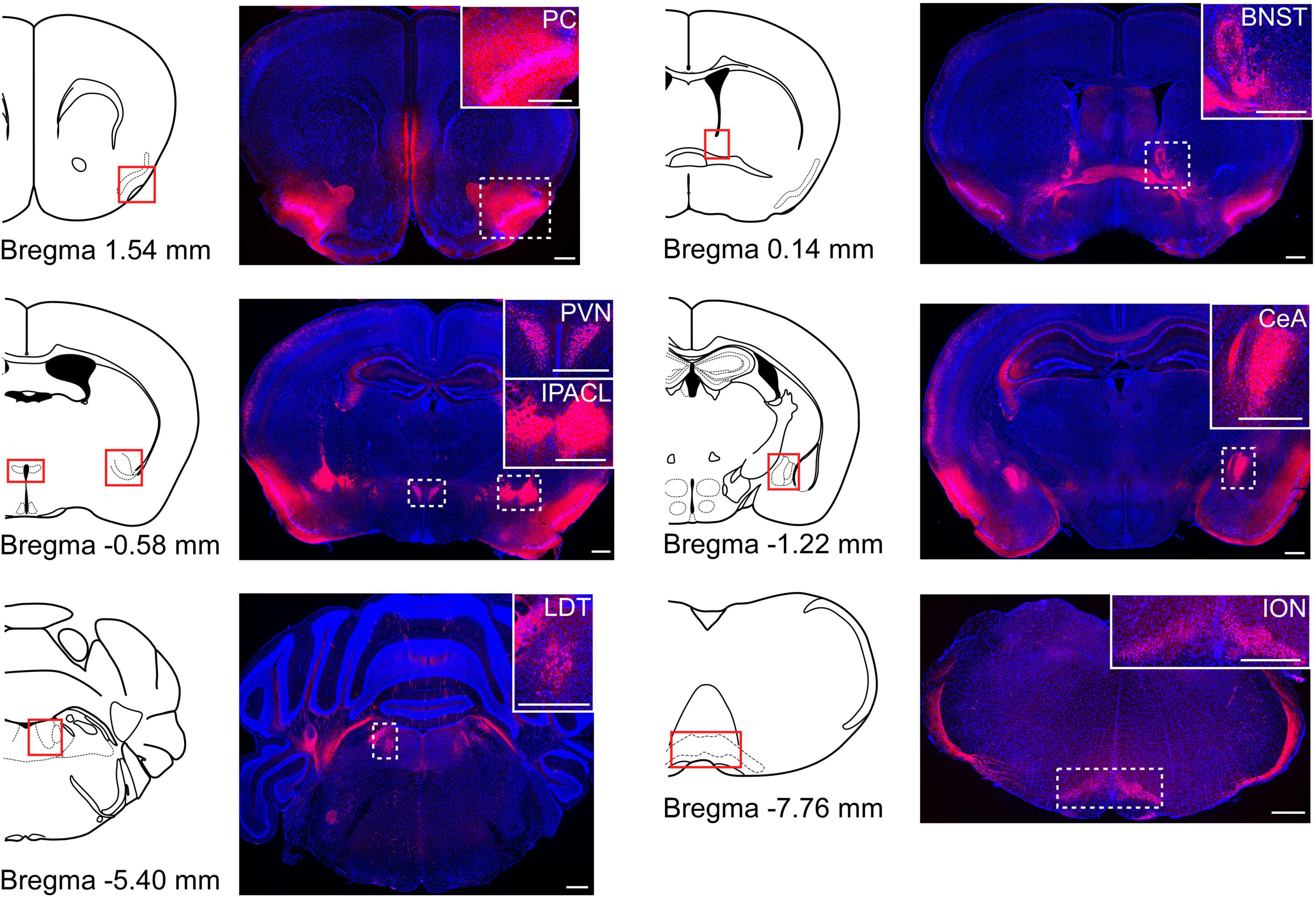
Figure 1. The distribution of CRF neurons in the brain. Cre recombinase specific expression of tdTomato obtained from the CRF-Cre mice crossed with Rosa26-tdTomato reporter mice. Coronal brain sections were stained for DAPI (blue). Schematic drawing of coronal brain slices is shown in the left side of each fluorescence image (modified from Allen brain atlas). Several brain areas expressed tdTomato-positive cells. PC, piriform cortex; BNST, bed nucleus of the stria terminalis; PVN, paraventricular nucleus; IPACL, interstitial nucleus of the posterior limb of the anterior commissure; CeA, central nucleus of the amygdala; LDT, laterodorsal tegmental nucleus; ION, inferior olivary nucleus. White dashed rectangles indicate magnified areas in the panels. Scale bars indicate 500 μm.
Generation of Corticotropin-Releasing Factor Neuron-Specific Bmal1-Deficient Mice
Corticotropin-releasing factor neurons are the key regulators of the hypothalamic–pituitary–adrenal axis, which is involved in the regulation of stress response (Vale et al., 1981). Recently, sleep and wake regulation via CRF neurons in the PVN was reported (Li et al., 2020; Ono et al., 2020). Circadian rhythms in the SCN regulate the neuronal activity in the CRF neurons, and the activation of CRF neurons in the PVN further activates the neuronal activity of orexin neurons in the LH (Ono et al., 2020). Although this neuronal circuit is suggested to be involved in the regulation of sleep and wakefulness, it remains unclear whether the circadian clock in the CRF neurons contributes to the regulation of sleep and wakefulness. To answer this question, we crossed CRF-Cre (Itoi et al., 2014) and Bmal1 flox mice (Storch et al., 2007) to generate CRF neuron-specific Bmal1 knockout mice (CRF-Bmal1flox/flox mice). We obtained CRF-Bmal1flox/flox and Bmal1flox/flox (control) littermate mice and stained for CRF peptides and Bmal1 proteins in the brain using immunohistochemistry. Almost all CRF neurons expressed BMAL1 in the control mice (97.5, 97.3, and 91.7% in the PVN, CeA, and BNST, respectively; n = 2 mice), whereas Bmal1 proteins were rarely observed in the CRF peptide-positive neurons in the CRF-Bmal1flox/flox mice (12.8, 27.8, and 21.2% in the PVN, CeA, and BNST, respectively; n = 2 mice) (Figure 2). The number of BMAL1- positive neurons among non-CRF neurons in the PVN was not different between two genotypes (control mice: 91.4%, CRF-Bmal1flox/flox mice: 90.9%, n = 2 mice). We also counted the number of CRF-positive neurons in the BNST, CeA, and PVN of control (BNST: 490 and 505 cells; CeA: 229 and 480 cells; PVN: 338 and 218 cells, n = 2 mice) and CRF-Bmal1flox/flox (BNST: 242 and 265 cells; CeA: 194 and 138 cells; PVN: 303 and 387 cells, n = 2 mice) mice. These results indicate that Bmal1 expression was deleted in the majority of CRF neurons in the CRF-Bmal1flox/flox mice.
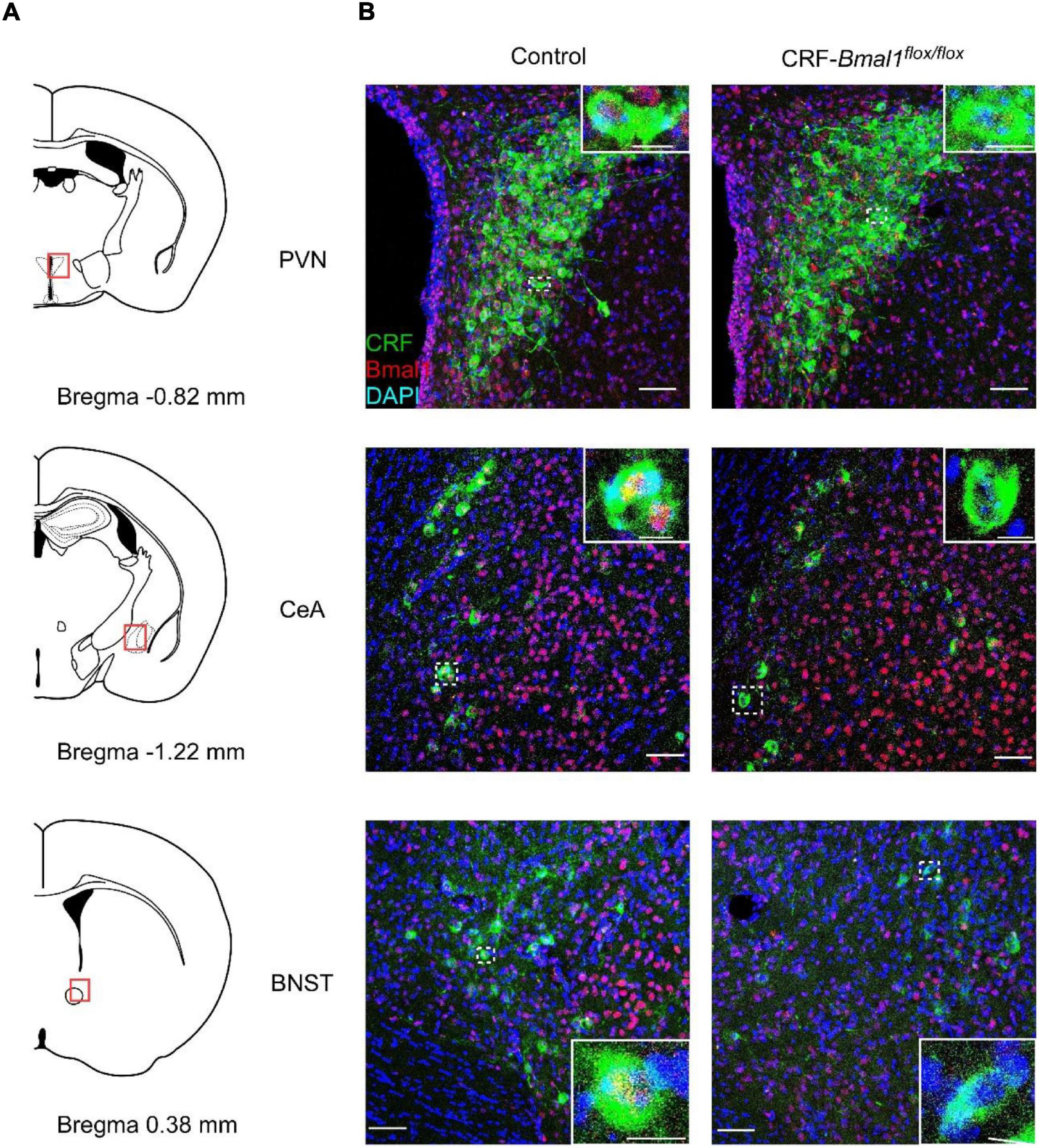
Figure 2. Confirmation of BMAL1 expression in CRF neuron-specific Bmal1-deficient mice. (A) Schematic drawing of coronal brain slices (modified from Allen brain atlas). (B) Fluorescence images of PVN, CeA, and BNST in a Bmal1flox/flox (control) or CRF-Cre/Bmal1flox/flox mice. Coronal brain sections were immunostained with CRF (green), BMAL1 (red), and DAPI (blue). White dashed rectangles indicate magnified areas in the panels. Scale bars indicate 50 and 10 μm in images with low and high magnification, respectively.
Locomotor Activity Rhythm Was Not Affected by Bmal1 Deficiency in the Corticotropin-Releasing Factor Neurons
First, we measured locomotor activity using an infrared sensor from control and CRF-Bmal1flox/flox mice under light–dark (LD) cycle and constant darkness (DD). The daily patterns of circadian locomotor activity rhythms were not different between control and CRF-Bmal1flox/flox mice in both LD and DD conditions (Figures 3A,C). Moreover, the amount of locomotion was not different. The free-running period under DD calculated by chi-square periodogram was not different between the two groups (Figure 3B). These results indicate that Bmal1 deficiency in CRF neurons did not change circadian behavioral rhythms.
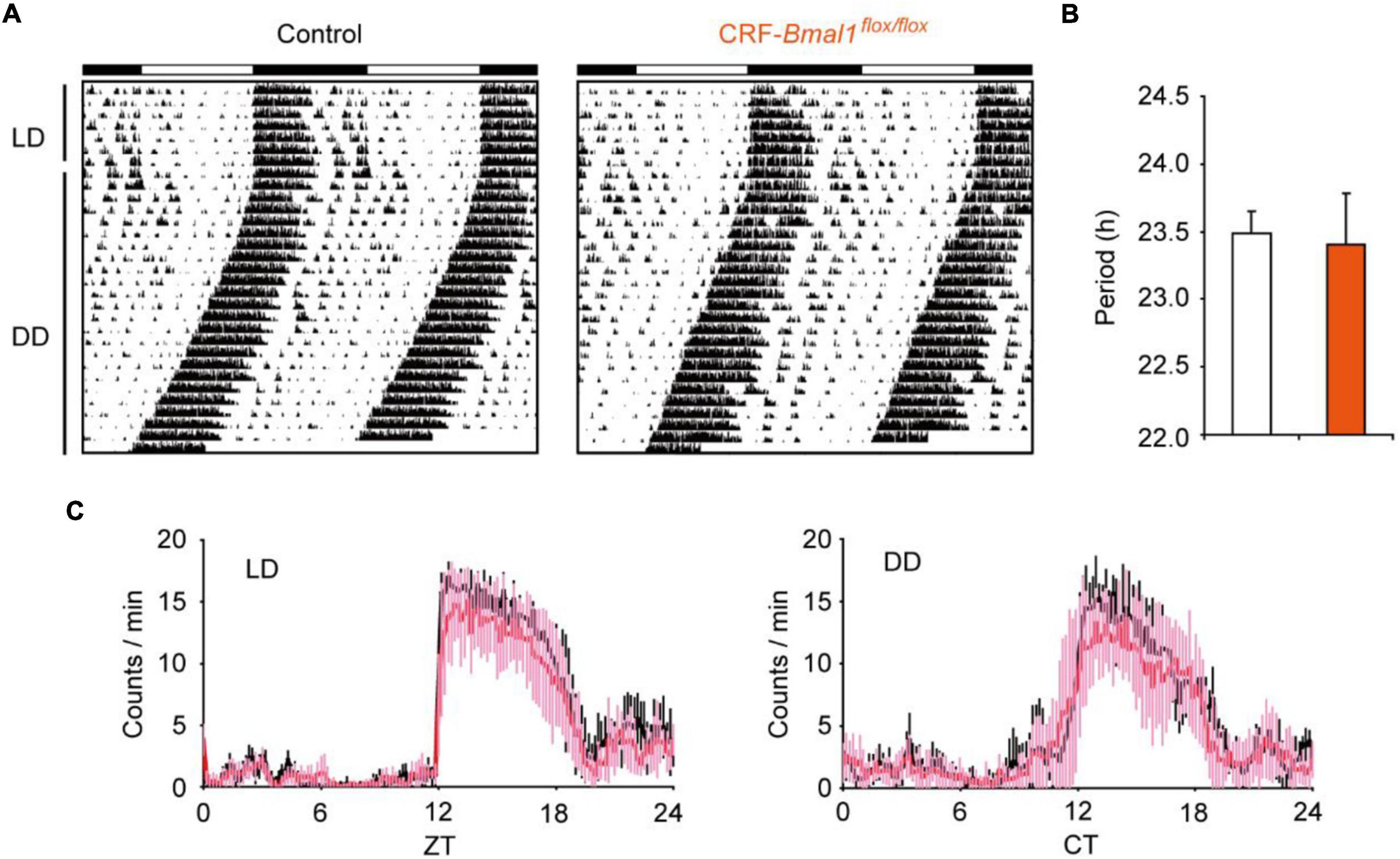
Figure 3. Circadian behavioral rhythms in CRF neuron-specific Bmal1- deficient mice. (A) Representative example of locomotor activity rhythms in control (male, 2; female, 4) or CRF-Cre/Bmal1flox/flox (male, 6; female, 1) mice under LD and DD. (B) Free-running period calculated by chi-square periodogram under DD. (C) Daily patterns’ locomotor activity in both genotypes (black: control mice, Pink: CRF-Cre/Bmal1flox/flox mice). Data are expressed as mean ± SD.
Bmal1 Deficiency in Corticotropin-Releasing Factor Neurons Did Not Have Significant Effects in Sleep and Wakefulness
Next, we measured the sleep and wakefulness from control and CRF-Bmal1flox/flox mice using EEG and EMG recordings under LD and DD. As for the sleep/wakefulness in DD, we analyzed the EEG and EMG data during the second day of DD. Clear daily patterns of wakefulness, NREM, and REM sleep were observed in both groups under LD and DD (Figures 4A,B). There was no significant difference between the two groups in these daily patterns in LD and DD (Figures 4A,B). The total time, number of bouts, and mean duration of bouts were also not different between the two groups (Figures 4C,D). When we compared the EEG power spectrum, no difference was detected (Figure 4E). These results indicate that Bmal1 in CRF neurons have essentially no effect on sleep and wakefulness.
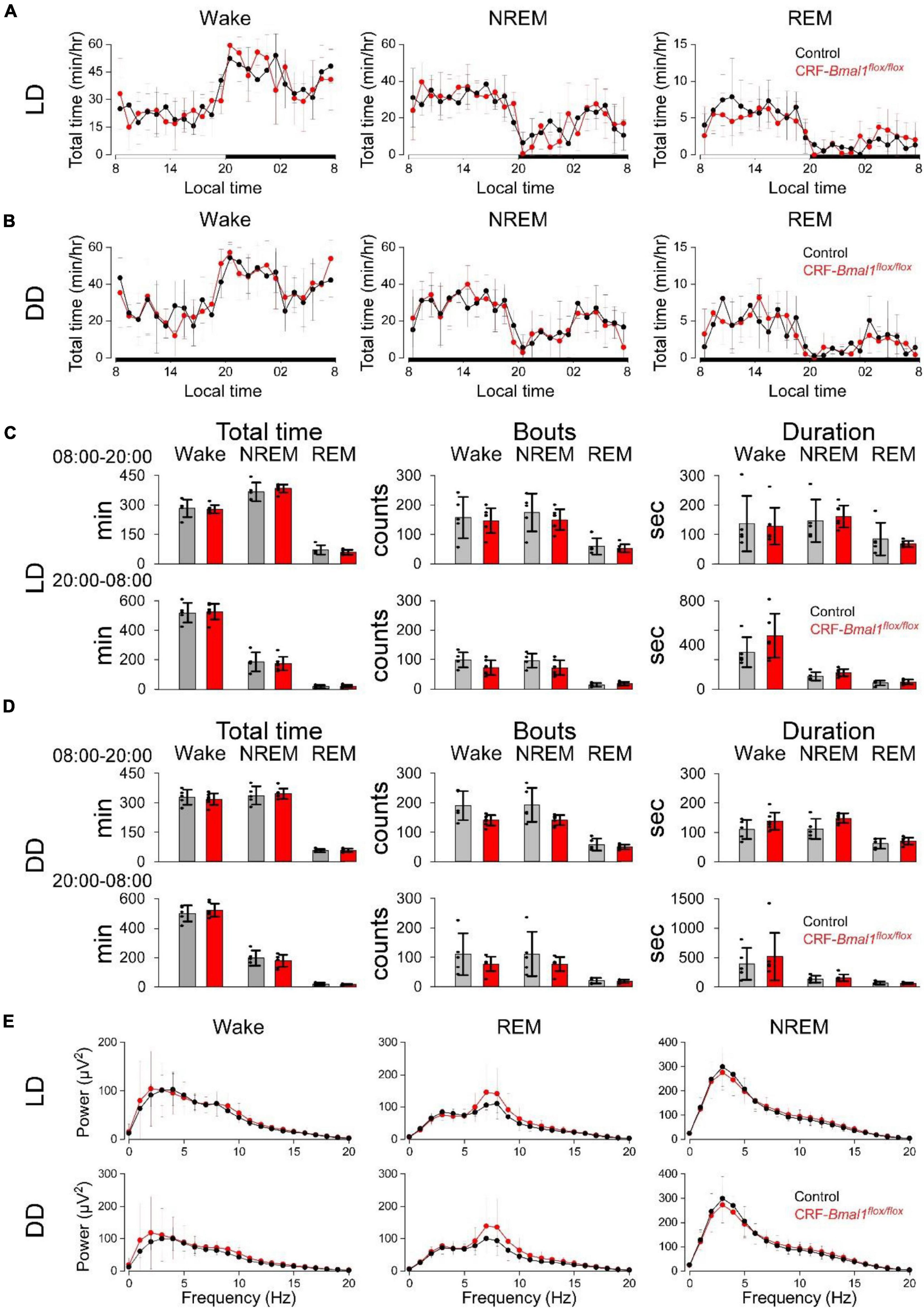
Figure 4. Sleep and wakefulness in CRF neuron-specific Bmal1 deficient mice. (A,B) Daily patterns of wake, NREM, and REM sleep in control (black) (n = 7) or CRF-Cre/Bmal1flox/flox (n = 5) mice (red) under LD (A) and DD (B). (C,D) Total time (left), number of bouts (middle), and duration (right) of wake, NREM, and REM sleep under LD (C) and DD (D). (E) EEG power during wake, NREM, and REM sleep in both genotypes under LD (top) or DD (bottom). Data are expressed as mean ± SD.
Discussion
We found that Bmal1 in the CRF neurons was not involved in the regulation of circadian locomotor activity rhythms and was not related to the circadian clock-regulated wakefulness in mice. Importantly, the mice with the whole-body deletion of Bmal1 exhibit arrhythmic circadian behavior (Bunger et al., 2000) and increases of NREM and REM sleep (Laposky et al., 2005). These results and our findings indicate that Bmal1 plays a role for the regulation of sleep and wakefulness, but these effects were not related to the CRF neurons in mice.
The CRF neurons in the PVN were previously reported to be important in circadian clock-related wakefulness (Ono et al., 2020). However, the CRF neurons in other regions could also have a role in the modulation of sleep and wakefulness. CRF neurons are observed in several brain areas. Among them, CeA contains several important neurons that regulate sleep (Sanford et al., 2002; Ma et al., 2019) and has several functions such as the control of fear, anxiety, and feeding (Ciocchi et al., 2010; Cai et al., 2014; Fadok et al., 2017). The CRF neurons in the CeA might have a functional role in sleep and wake regulation.
Although the brain contains many neurons that are crucial for the regulation of sleep and wakefulness, peripheral tissues also have a potential for the regulation of sleep. For instance, the restoration of Bmal1 specifically into the brain in whole-body Bmal1-deficient mice did not restore sleep abnormality (Ehlen et al., 2017), whereas the restoration of Bmal1 in skeletal muscle completely restored NREM sleep duration to wild-type levels, indicating that Bmal1 in skeletal muscle plays a role in the regulation of sleep and wakefulness. Recently, Jones reported that CRF neuron-specific Bmal1-deficient mice dampened corticosterone rhythms (Jones et al., 2021). Since corticosterone administration or the inhibition of corticosterone synthesis modulates sleep and wakefulness (Vazquez-Palacios et al., 2001; Drouet et al., 2011), sleep and wakefulness in mice might also be affected by corticosterone rhythms.
The functional roles of Bmal1 in sleep and wakefulness have been reported, but other clock genes also play a role in the regulation of sleep. For example, Dec2 is one of the clock genes in mammals (Honma et al., 2002) that is a negative component of the molecular circadian clock. In humans, familial advanced sleep-phase syndrome is a disorder in which the timing of sleep and the peak period of alertness are advanced several hours relative to the social clock (He et al., 2009). This phenotype is due to point mutation in Dec2. Furthermore, this point mutation suppresses prepro-orexin promoter activity (Hirano et al., 2018). Therefore, clock genes in some neurons that are involved in the regulation of sleep and wakefulness have the potential to modulate sleep and wake regulation.
Conclusion
Corticotropin-releasing factor neurons in the PVN are critical for the regulation of circadian clock-derived wakefulness, but Bmal1 in the CRF neurons is not essential for the regulation of sleep and wakefulness in mice.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committees at the Research Institute of Environmental Medicine at Nagoya University (approval numbers: R210729 and R210730).
Author Contributions
DO designed the research. AY contributed in the discussion of the research. DO and CH performed the research, analyzed data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Takeda Science Foundation, SECOM Science and Technology Foundation, Casio Science Promotion Foundation, LOTTE Foundation, Taiju Life Social Welfare Foundation, and JSPS KAKENHI (18H02477, 20KK0177, 21H00422, 21H00307, and 21H02526 to DO, 20J11041 to CH, and 18H02523 and 18KK0223 to AY).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank S. Tsukamoto and S. Nasu for animal care. We also thank K. Itoi for the CRF-Cre mice.
References
Abrahamson, E. E., and Moore, R. Y. (2001). Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 916, 172–191. doi: 10.1016/s0006-8993(01)02890-6
Bunger, M. K., Wilsbacher, L. D., Moran, S. M., Clendenin, C., Radcliffe, L. A., Hogenesch, J. B., et al. (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017. doi: 10.1016/s0092-8674(00)00205-1
Cai, H., Haubensak, W., Anthony, T. E., and Anderson, D. J. (2014). Central amygdala PKC-delta(+) neurons mediate the influence of multiple anorexigenic signals. Nat. Neurosci. 17, 1240–1248. doi: 10.1038/nn.3767
Ciocchi, S., Herry, C., Grenier, F., Wolff, S. B., Letzkus, J. J., Vlachos, I., et al. (2010). Encoding of conditioned fear in central amygdala inhibitory circuits. Nature 468, 277–282. doi: 10.1038/nature09559
Drouet, J. B., Rousset, C., Maury, R., Michel, V., Buguet, A., Cespuglio, R., et al. (2011). Single administration of metyrapone modifies sleep-wake patterns in the rat. Eur. J. Pharmacol. 652, 60–64. doi: 10.1016/j.ejphar.2010.10.071
Ehlen, J. C., Brager, A. J., Baggs, J., Pinckney, L., Gray, C. L., DeBruyne, J. P., et al. (2017). Bmal1 function in skeletal muscle regulates sleep. Elife 6:e26557. doi: 10.7554/eLife.26557
Fadok, J. P., Krabbe, S., Markovic, M., Courtin, J., Xu, C., Massi, L., et al. (2017). A competitive inhibitory circuit for selection of active and passive fear responses. Nature 542, 96–100. doi: 10.1038/nature21047
Gizowski, C., Zaelzer, C., and Bourque, C. W. (2016). Clock-driven vasopressin neurotransmission mediates anticipatory thirst prior to sleep. Nature 537, 685–688. doi: 10.1038/nature19756
He, Y., Jones, C. R., Fujiki, N., Xu, Y., Guo, B., and Holder, J. L. Jr. (2009). The transcriptional repressor DEC2 regulates sleep length in mammals. Science 325, 866–870. doi: 10.1126/science.1174443
Herzog, E. D., Aton, S. J., Numano, R., Sakaki, Y., and Tei, H. (2004). Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J. Biol. Rhythm 19, 35–46. doi: 10.1177/0748730403260776
Hirano, A., Hsu, P. K., Zhang, L., Xing, L., McMahon, T., Yamazaki, M., et al. (2018). DEC2 modulates orexin expression and regulates sleep. Proc. Natl. Acad. Sci. U.S.A. 115, 3434–3439. doi: 10.1073/pnas.1801693115
Honma, S., Kawamoto, T., Takagi, Y., Fujimoto, K., Sato, F., Noshiro, M., et al. (2002). Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419, 841–844.
Honma, S., Nakamura, W., Shirakawa, T., and Honma, K. (2004). Diversity in the circadian periods of single neurons of the rat suprachiasmatic nucleus depends on nuclear structure and intrinsic period. Neurosci. Lett. 358, 173–176. doi: 10.1016/j.neulet.2004.01.022
Hung, C. J., Ono, D., Kilduff, T. S., and Yamanaka, A. (2020). Dual orexin and MCH neuron-ablated mice display severe sleep attacks and cataplexy. Elife 9:e54275. doi: 10.7554/eLife.54275
Itoi, K., Talukder, A. H., Fuse, T., Kaneko, T., Ozawa, R., Sato, T., et al. (2014). Visualization of corticotropin-releasing factor neurons by fluorescent proteins in the mouse brain and characterization of labeled neurons in the paraventricular nucleus of the hypothalamus. Endocrinology 155, 4054–4060. doi: 10.1210/en.2014-1182
Jones, J. R., Chaturvedi, S., Granados-Fuentes, D., and Herzog, E. D. (2021). Circadian neurons in the paraventricular nucleus entrain and sustain daily rhythms in glucocorticoids. Nat. Commun. 12:5763. doi: 10.1038/s41467-021-25959-9
Laposky, A., Easton, A., Dugovic, C., Walisser, J., Bradfield, C., and Turek, F. (2005). Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep 28, 395–409. doi: 10.1093/sleep/28.4.395
Li, S. B., Borniger, J. C., Yamaguchi, H., Hedou, J., Gaudilliere, B., and de Lecea, L. (2020). Hypothalamic circuitry underlying stress-induced insomnia and peripheral immunosuppression. Sci. Adv. 6:eabc2590. doi: 10.1126/sciadv.abc2590
Ma, C., Zhong, P., Liu, D., Barger, Z. K., Zhou, L., Chang, W. C., et al. (2019). Sleep regulation by neurotensinergic neurons in a thalamo-amygdala circuit. Neuron 103, 323–334 e7. doi: 10.1016/j.neuron.2019.05.015
Madisen, L., Zwingman, T. A., Sunkin, S. M., Oh, S. W., Zariwala, H. A., Gu, H., et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. doi: 10.1038/nn.2467
Maejima, T., Tsuno, Y., Miyazaki, S., Tsuneoka, Y., Hasegawa, E., Islam, M. T., et al. (2021). GABA from vasopressin neurons regulates the time at which suprachiasmatic nucleus molecular clocks enable circadian behavior. Proc. Natl. Acad. Sci. U.S.A. 118:e2010168118. doi: 10.1073/pnas.2010168118
Moore, R. Y., and Eichler, V. B. (1972). Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in rat. Brain Res. 42, 201–206. doi: 10.1016/0006-8993(72)90054-6
Ono, D., Honma, K., Yanagawa, Y., Yamanaka, A., and Honma, S. (2019). GABA in the suprachiasmatic nucleus refines circadian output rhythms in mice. Commun. Biol. 2:232. doi: 10.1038/s42003-019-0483-6
Ono, D., Mukai, Y., Hung, C. J., Chowdhury, S., Sugiyama, T., and Yamanaka, A. (2020). The mammalian circadian pacemaker regulates wakefulness via CRF neurons in the paraventricular nucleus of the hypothalamus. Sci. Adv. 6:eabd0384. doi: 10.1126/sciadv.abd0384
Paul, S., Hanna, L., Harding, C., Hayter, E. A., Walmsley, L., Bechtold, D. A., et al. (2020). Output from VIP cells of the mammalian central clock regulates daily physiological rhythms. Nat. Commun. 11:1453. doi: 10.1038/s41467-020-15277-x
Reppert, S. M., and Weaver, D. R. (2002). Coordination of circadian timing in mammals. Nature 418, 935–941. doi: 10.1038/nature00965
Sanford, L. D., Parris, B., and Tang, X. (2002). GABAergic regulation of the central nucleus of the amygdala: implications for sleep control. Brain Res. 956, 276–284. doi: 10.1016/s0006-8993(02)03552-7
Sawaki, Y., Nihonmatsu, I., and Kawamura, H. (1984). Transplantation of the neonatal suprachiasmatic nuclei into rats with complete bilateral suprachiasmatic lesions. Neurosci. Res. 1, 67–72. doi: 10.1016/0168-0102(84)90031-2
Silver, R., LeSauter, J., Tresco, P. A., and Lehman, M. N. (1996). A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature 382, 810–813. doi: 10.1038/382810a0
Stephan, F. K., and Zucker, I. (1972). Circadian-rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic-lesions. Proc. Natl. Acad. Sci. U.S.A. 69, 1583–1586. doi: 10.1073/pnas.69.6.1583
Storch, K. F., Paz, C., Signorovitch, J., Raviola, E., Pawlyk, B., Li, T., et al. (2007). Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130, 730–741. doi: 10.1016/j.cell.2007.06.045
Todd, W. D., Venner, A., Anaclet, C., Broadhurst, R. Y., De Luca, R., Bandaru, S. S., et al. (2020). Suprachiasmatic VIP neurons are required for normal circadian rhythmicity and comprised of molecularly distinct subpopulations. Nat. Commun. 11:4410. doi: 10.1038/s41467-020-17197-2
Tsunematsu, T., Kilduff, T. S., Boyden, E. S., Takahashi, S., Tominaga, M., and Yamanaka, A. (2011). Acute optogenetic silencing of orexin/hypocretin neurons induces slow-wave sleep in mice. J. Neurosci. 31, 10529–10539. doi: 10.1523/JNEUROSCI.0784-11.2011
Vale, W., Spiess, J., Rivier, C., and Rivier, J. (1981). Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science 213, 1394–1397. doi: 10.1126/science.6267699
Van den Pol, A. N. (1980). The hypothalamic suprachiasmatic nucleus of rat: intrinsic anatomy. J. Comp. Neurol. 191, 661–702. doi: 10.1002/cne.901910410
Vazquez-Palacios, G., Retana-Marquez, S., Bonilla-Jaime, H., and Velazquez-Moctezuma, J. (2001). Further definition of the effect of corticosterone on the sleep-wake pattern in the male rat. Pharmacol. Biochem. Behav. 70, 305–310. doi: 10.1016/s0091-3057(01)00620-7
Walker, L. C., Cornish, L. C., Lawrence, A. J., and Campbell, E. J. (2019). The effect of acute or repeated stress on the corticotropin releasing factor system in the CRH-IRES-Cre mouse: a validation study. Neuropharmacology 154, 96–106. doi: 10.1016/j.neuropharm.2018.09.037
Watts, A. G., and Swanson, L. W. (1987). Efferent projections of the suprachiasmatic nucleus: II. Studies using retrograde transport of fluorescent dyes and simultaneous peptide immunohistochemistry in the rat. J. Comp. Neurol. 258, 230–252. doi: 10.1002/cne.902580205
Watts, A. G., Swanson, L. W., and Sanchez-Watts, G. (1987). Efferent projections of the suprachiasmatic nucleus: I. Studies using anterograde transport of Phaseolus vulgaris leucoagglutinin in the rat. J. Comp. Neurol. 258, 204–229. doi: 10.1002/cne.902580204
Keywords: circadian rhythm, locomotor activity, sleep, Bmal1, CRF neurons
Citation: Hung CJ, Yamanaka A and Ono D (2022) Conditional Knockout of Bmal1 in Corticotropin-Releasing Factor Neurons Does Not Alter Sleep–Wake Rhythm in Mice. Front. Neurosci. 15:808754. doi: 10.3389/fnins.2021.808754
Received: 03 November 2021; Accepted: 29 December 2021;
Published: 18 February 2022.
Edited by:
Christopher S. Colwell, University of California, Los Angeles, United StatesReviewed by:
Matt Carter, Williams College, United StatesMichihiro Mieda, Kanazawa University, Japan
Copyright © 2022 Hung, Yamanaka and Ono. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daisuke Ono, ZGFpLW9ub0ByaWVtLm5hZ295YS11LmFjLmpw
 Chi Jung Hung
Chi Jung Hung Akihiro Yamanaka
Akihiro Yamanaka Daisuke Ono
Daisuke Ono