- 1Bacteriology Laboratory, Division of Laboratory Medicine, Department of Diagnostics, Geneva University Hospitals, Geneva, Switzerland
- 2Genomic Research Laboratory, Division of Infectious Diseases, Department of Medicine, Faculty of Medicine, Geneva University Hospitals, Geneva, Switzerland
Background: Multidrug-resistant organisms (MDROs) have become a serious public health concern worldwide. Screening for MDROs can encompass various phenotypic and genotypic approaches. Recently, culture-based MDROs screening has substantially benefitted from laboratory automation and artificial intelligence.
Objectives: To evaluate the performance of PhenoMATRIX® PLUS for an institution-wide culture-based rectal screening for CPOs, ESBL-E, Acinetobacter, and VRE carriage.
Methods: The number of non-duplicated rectal Eswabs specimens used to generate culture plate images for the PhenoMATRIX® PLUS machine learning phase amounted to 2′900 for chromID™ ESBL agar, 2′403 for chromID™ OXA-48 agar, 2′031 for CHROMagar™ Acinetobacter agar, and 9′333 for chromID™ VRE agar. All such plates and images were classified by manual reading as either positive or negative. The validation of the settings derived during the learning phase was achieved on an additional 4′517 non-duplicate specimens. Digital images of 22′585 media plates (i.e., five per specimen) were prospectively analyzed by PhenoMATRIX® PLUS. All media plates were incubated in the WASPLab® and imaged at predefined time points.
Results: According to the manual work-up results, the output agreements of the PhenoMATRIX® PLUS for the three results “no growth,” “negative,” and “Send to Reader” reached 98.5, 99.6%. and 74.8%, respectively. Importantly, no false negative results were returned by PhenoMATRIX® PLUS in the present study.
Conclusion: PhenoMATRIX® PLUS contributes to a significant reduction in the workload. Therefore, technologists’ routine tasks have evolved to the confirmatory tests of the presumptively positive colonies, allowing better management of the laboratory staff and the resources.
Introduction
Over the last decade, multidrug-resistant organisms (MDROs) have become a serious public health concern worldwide. Public health agencies have targeted various MDROs requiring enhanced barrier precautions in the hospital setting (Sartelli et al., 2024). This is especially relevant for carbapenemase-producing organisms (CPOs), Acinetobacter spp., vancomycin-resistant Enterococci (VRE), and extended-spectrum β-lactamase-producing Enterobacterales (ESBL-E). Swift and accurate detection of these MDROs is of paramount importance, since their patient-to-patient transmission is facilitated by their location on mobile genetic elements and the surrounding high antibiotic selective pressure (Mancuso et al., 2023). Detection of carriers triggers infection control procedures at an early stage to prevent the spread of these MDROs within the hospital environment, besides the enforcement of appropriate antibiotic stewardship measures (Durazzi et al., 2023).
Defining a workflow providing simple, rapid, reliable, and affordable routine diagnostic methods to identify carriers of such MDROs constitutes a challenge. Nowadays, several molecular assays including PCR panels or loop-mediated isothermal amplification (LAMP) assays are available (Tenover, 2021; Skally et al., 2025). These assays can be used for the screening, directly from samples (Novazzi et al., 2024). However, their analytical performances vary considerably, not to mention their costs (Kamel et al., 2022). Today, the most widespread approach for the screening of MDROs carriers is typically a combination of molecular assays coupled to culture and phenotypic tests (Dortet et al., 2016; Ciesielczuk et al., 2018; Gelmez et al., 2021).
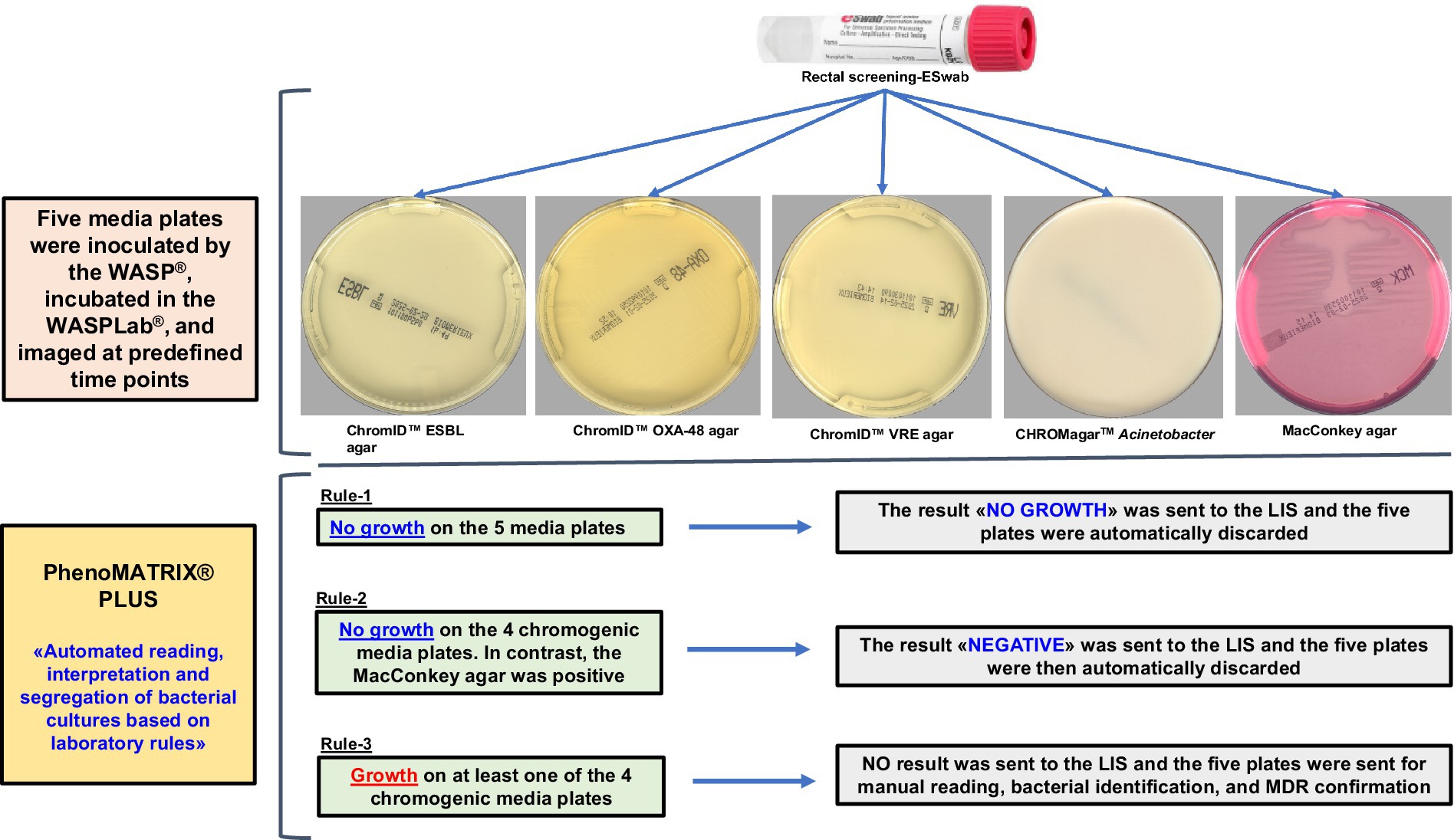
The implementation of chromogenic culture media has considerably improved the performance of screening by cultures (Birlutiu et al., 2025). In addition, culture testing has largely benefitted from laboratory automation and the routine usage of artificial intelligence (AI). PhenoMATRIX® PLUS (Copan, Brescia, Italy) is a software designed to streamline culture-based workflows. It can sort and process culture media plates according to predefined rules. PhenoMATRIX® PLUS enables the automatic release of the negative results to the electronic patients’ file and the automatic discharge of the negative plates from the incubators.
The main objective of the present study is to assess the performance of PhenoMATRIX® PLUS for an institution-wide culture-based rectal screening to detect CPOs, ESBL-E, Acinetobacter, and VRE carriage.
Materials and methods
Setting
Geneva University Hospitals are a Swiss tertiary care center. In 2024, the bacteriology laboratory processed 196′456 specimens, for a total of 423′614 analyses. The screening for carbapenemase-producing organisms (CPOs), extended-spectrum β-lactamase-producing Enterobacterales (ESBL-E), Acinetobacter, and vancomycin-resistant Enterococcus (VRE) carriers was carried out on 13′728 rectal Eswabs (7.0% of our activity, in volume). For the screening of MRSA carriers, we processed 17′407 nasal and inguinal/perineal Eswabs (4.1% of our activity). Current hours of operation of the laboratory span from 7: 30 A.M. to 7: 30 P.M. Monday to Friday; Saturday from 7: 30 A.M. to 5: 00 P.M.; and Sunday from 7: 30 A.M. to 1: 00 P.M.
Culture-based rectal screening of CPOs, ESBL-E, Acinetobacter, and VRE
Rectal CPOs, ESBL-E, Acinetobacter, and VRE screening-ESwabs (490 CE.A, Copan Italia S.p.A. Brescia, Italy) were streaked by the WASP® (Copan) on chromID™ ESBL agar (bioMérieux), chromID™ OXA-48 agar, (bioMérieux), CHROMagar™ Acinetobacter, and chromID™ VRE agar (bioMérieux), in addition to a MacConkey agar (bioMérieux). This last medium was included to assess the quality of the sample. All media plates were incubated in the WASPLab® (Copan) and imaged at predefined time points. We applied the final incubation time points as previously validated for CPOs, ESBL-E and VRE (Cherkaoui et al., 2019; Cherkaoui et al., 2020). Digital images were manually read by the technologists. The identification of screened organisms and their antimicrobial susceptibility testing were performed during the day shift. For all presumptively positive CPOs, ESBL-E, and Acinetobacter colonies on chromogenic media, bacterial identification was performed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF/MS) (MBT Compass 4.1, library version 11.0 (11′410 spectra), Bruker Daltonics, Bremen, Germany) according to the manufacturer’s instructions. The antimicrobial susceptibility testing (AST) was carried out by disk diffusion according to EUCAST recommendations. The ESBL-E was confirmed phenotypically by double-disc synergy tests (DDST20 and DDST30) using Mueller Hinton E agar (MHE) (bioMérieux).
We used the eazyplex® SuperBug complete tests (Amplex Biosystems GmbH, Giessen, Germany) to identify the genes encoding carbapenemases in isolates with reduced susceptibility to carbapenems according to the EUCAST screening breakpoints. This system is based on a loop-mediated isothermal amplification. The carbapenemases detected by the eazyplex® SuperBug complete tests were KPC, NDM, VIM, OXA-23, OXA-40, OXA-48, OXA-58, and OXA-181. In addition, extended-spectrum β-lactamases from the CTX-M-1 and CTX-M-9 groups were also identified. Carbapenemase gene confirmation and sequencing were performed by the national reference center for early detection and monitoring of antibiotic resistance (NARA, Fribourg, Switzerland).
For all presumptively positive VRE colonies on chromID™ VRE agar, their identification by MALDI-TOF/MS and AST were followed by a qPCR assay targeting vanA and vanB (Thermo Fisher Scientific™—QuantStudio™ 5 Real-Time PCR and Absolute™ qPCR MasterMIX Thermo Scientific ABgene®) (Naserpour Farivar et al., 2014).
PhenoMATRIX® PLUS: machine learning phase
The number of non-duplicated rectal Eswabs specimens used to generate culture plate images amounted to 2′900 for chromID™ ESBL agar, 2′403 for chromID™ OXA-48 agar, 2′031 for CHROMagar™ Acinetobacter agar, and 9′333 for chromID™ VRE agar. All such plates and images were classified by manual reading as either positive or negative. These images were then used as the ground truth to train the convolutional neural networks.
PhenoMATRIX® PLUS: validation phase
The validation of the settings derived during the learning phase was conducted on an additional 4′517 non-duplicate specimens assigned to our laboratory for analysis. Digital images of 22′585 media plates (i.e., five per specimen) were prospectively analyzed by PhenoMATRIX® PLUS. Seeking maximum accord, all the media plate images were read manually by the technologists, but they were kept blind of the results dispensed by PhenoMATRIX® PLUS. Discrepancies between PhenoMATRIX® PLUS and manual reading were recorded and reviewed by an independent reader.
Figure 1 depicts the laboratory screening workflow for carbapenemase-producing organisms (CPOs), extended-spectrum β-lactamase-producing Enterobacterales (ESBL-E), Acinetobacter spp., vancomycin-resistant Enterococci (VRE). Rule-2 was applied to remove the bias associated with the non-specific bacterial growth on MacConkey agar. Therefore, all the “no growth” and “negative” results defined by PhenoMATRIX® PLUS were only associated to the specific growth on chromogenic media used in this study.
Results
PhenoMATRIX® PLUS performance for the rectal screening of CPOs, ESBL-E, Acinetobacter, and VRE
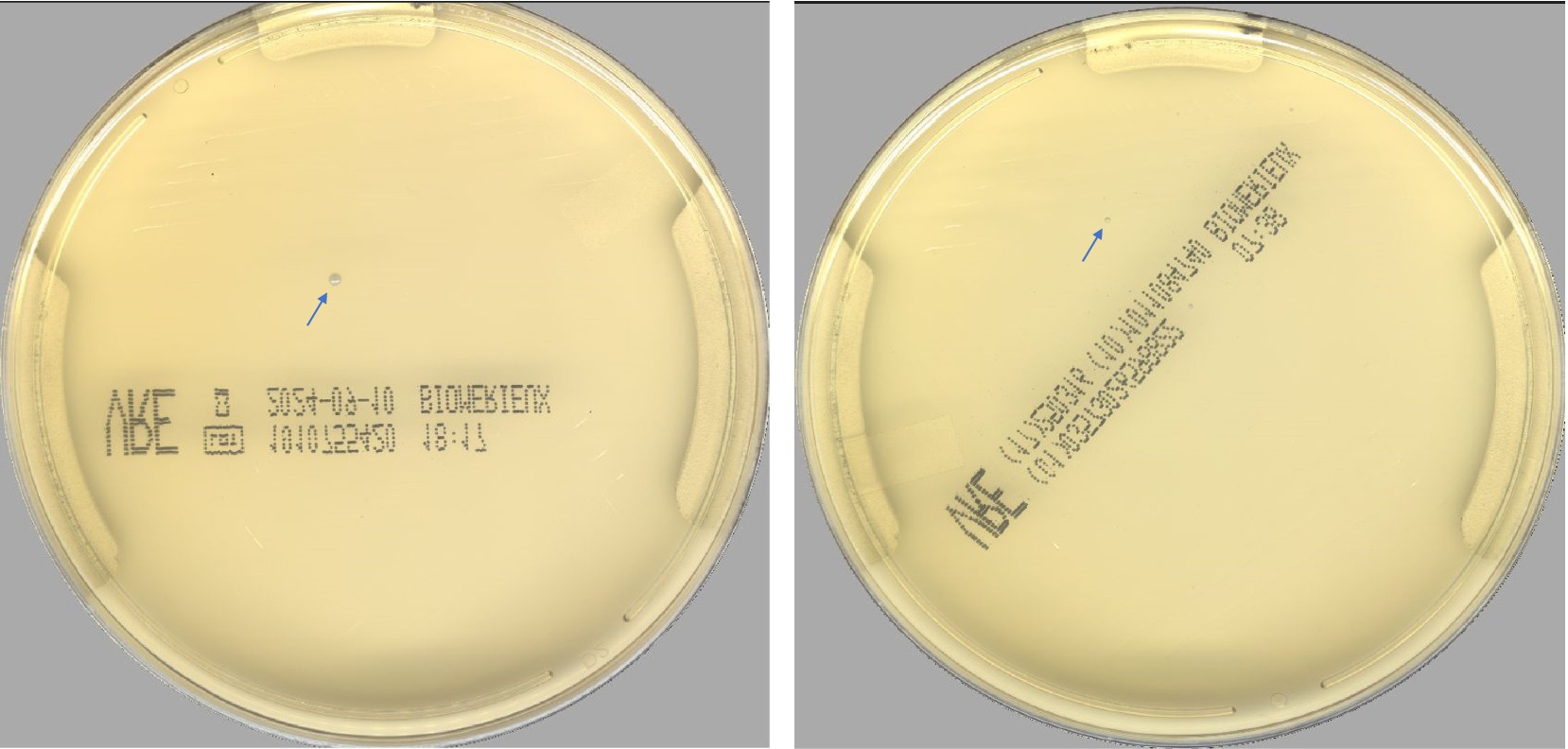
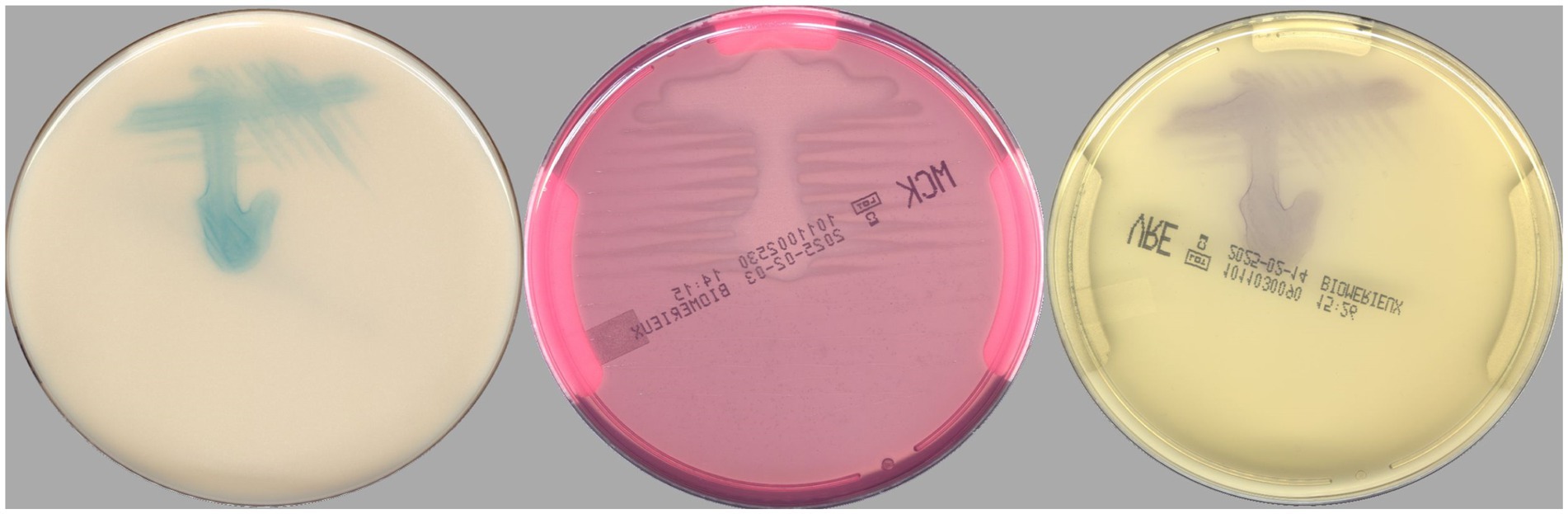
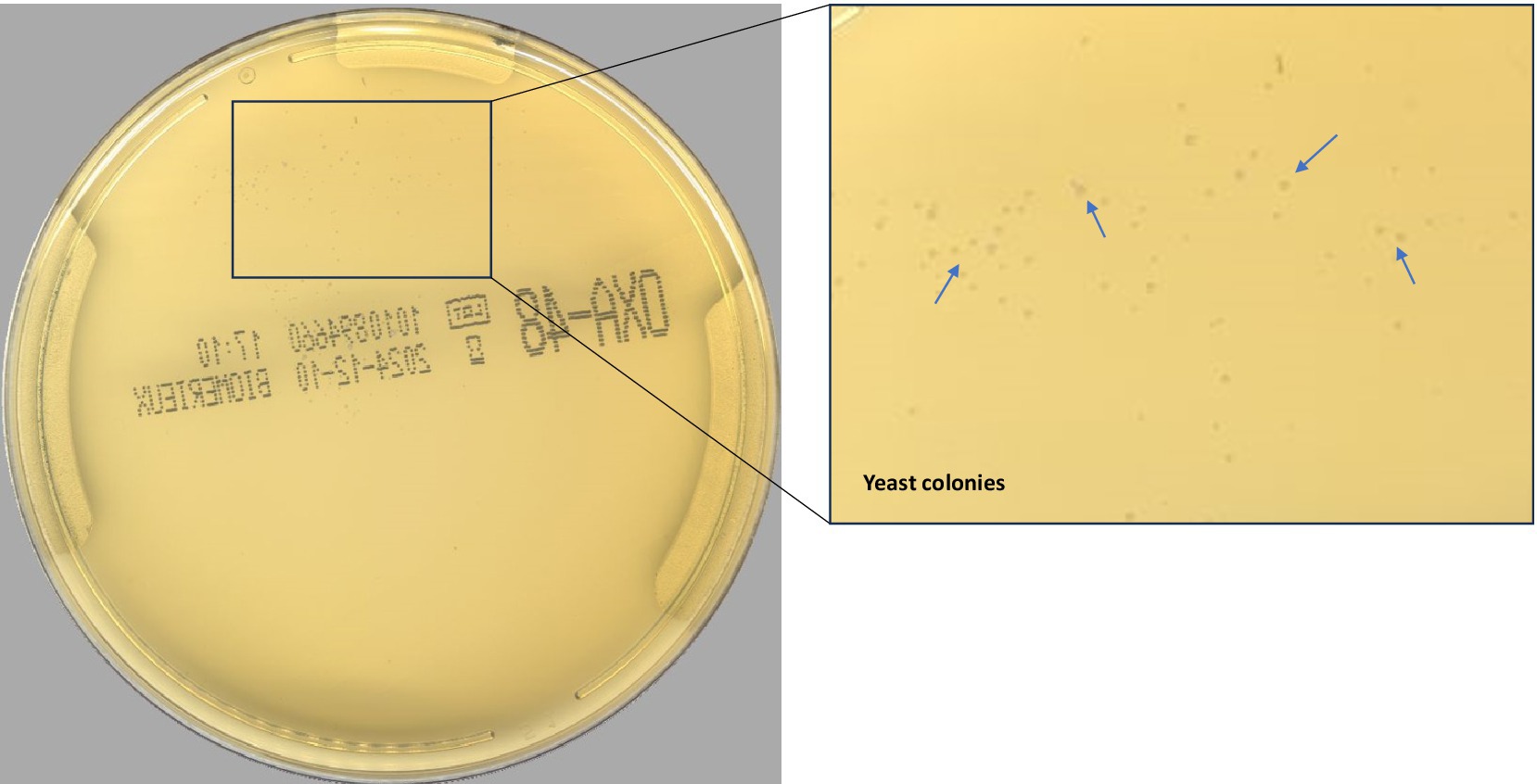
According to the PhenoMATRIX® PLUS workflow depicted in Figure 1, the clinical performances of PhenoMATRIX® PLUS for the screening of CPOs, ESBL-E, Acinetobacter, and VRE were compared to the manual work-up results (Table 1). The results were broken down by media plates (Table 1a) and samples (Table 1b). Among the 4′517 non-duplicated rectal Eswabs specimens analyzed, six were defined by PhenoMATRIX® PLUS as “No growth” but reported as “Negative” by manual reading. The image review of these 30 media plates showed that discordance could be explained by the presence of a few unspecific colonies on the chromID™ VRE agar (Figure 2). Seven specimens were segregated by PhenoMATRIX® PLUS as “Negative” but reported as “No growth” by manual reading. There was no evidence of bacterial growth on the MacConkey agar plates but only sample traces according to the image review of the media (Figure 3). Forty specimens were segregated by PhenoMATRIX® PLUS as “Send to Reader” but reported as “No growth” by manual reading. No evidence of bacterial growth was revealed but there were only sample traces (Figure 3). Finally, 554 specimens were segregated by PhenoMATRIX® PLUS as “Send to Reader” but reported as “negative” by manual reading. Yeast colonies were observed on some chromID™ OXA-48 agar plates (Figure 4). Growth of small colonies of Gram-positive bacteria on chromID™ OXA-48 agar and unspecific colonies on chromID™ VRE agar, explained discordance for most of these specimens. According to the manual work-up results, the output agreements of the PhenoMATRIX® PLUS for the three results “no growth,” “negative,” and “Send to Reader” reached 98.5, 99.6%, and 74.8%, respectively.
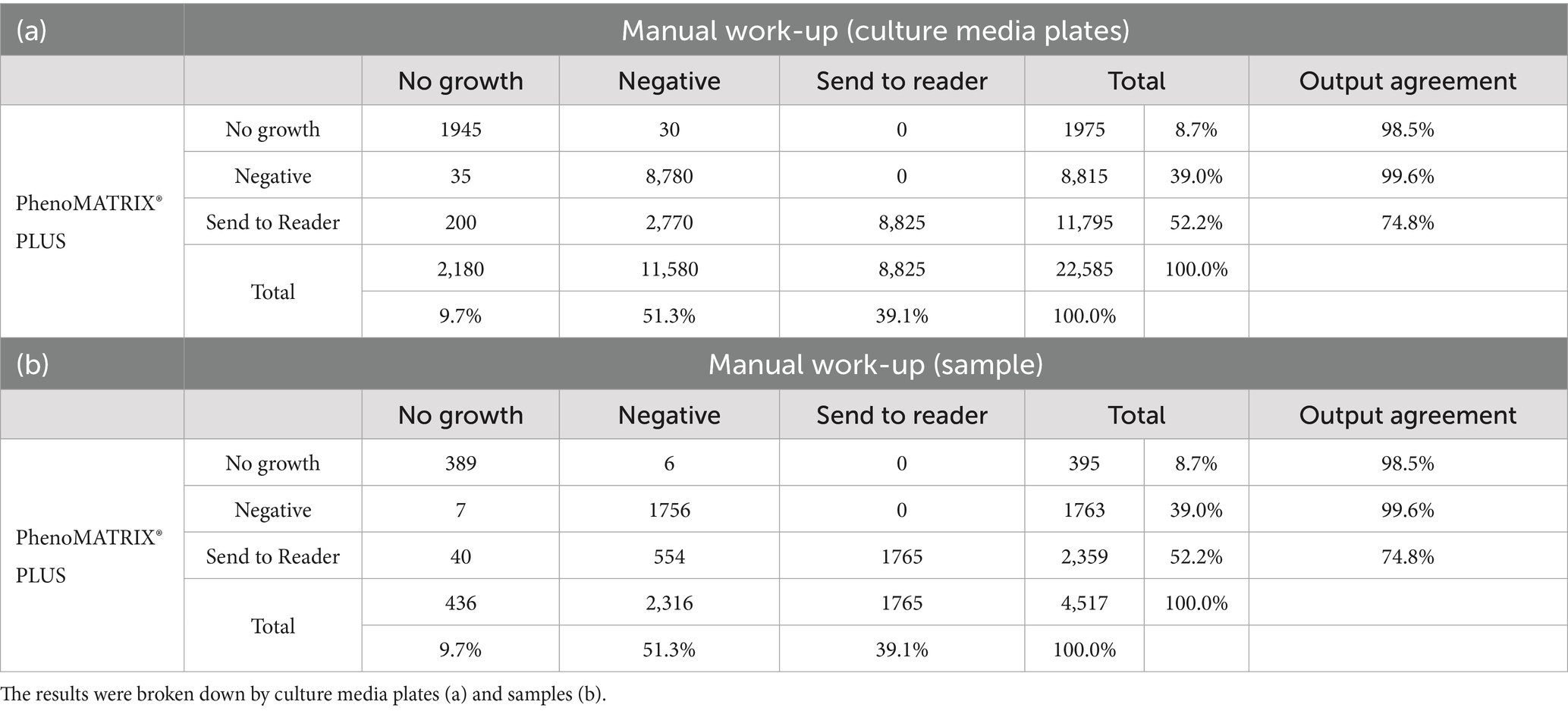
Table 1. Clinical performances of PhenoMATRIX® PLUS for the screening of CPOs, ESBL-E, Acinetobacter, and VRE compared to the manual work-up results.
Discussion
PhenoMATRIX® PLUS enables complete handling of the negative specimens (i.e., assessment of the culture images, automatic release of the results to the LIS, and discarding of the negative plates without technical validation). The accuracy of such software has been previously evaluated for MRSA screening (Cherkaoui et al., 2024). In the present study, we defined a culture-based protocol for rectal Eswab CPOs, ESBL-E, Acinetobacter, and VRE screening. This protocol included four chromogenic media and one MacConkey agar plate, the latter for sampling quality. The settings used for PhenoMATRIX® PLUS during the machine learning phase were not based on the specific color of the chromogenic media but only on the organisms’ growth since no specific color is defined by the manufacturers of CPOs and ESBL-E. Thus, three predefined rules were applied in the validation phase of the PhenoMATRIX® PLUS «No growth», «Negative», and «Send to Reader» as explained above. PhenoMATRIX® PLUS ensured the total and independent management of 48% (10′790/22′585) of the media plates flagged as «No growth» or «Negative» which corresponded to 2′158 negative specimens. The remaining media plates (52%, 11′795/22′585) whose organisms’ growth was segregated as presumptive positive cases still required confirmation by the technologists. In about 8% of the media plates analyzed, PhenoMATRIX® PLUS declared them as positive without evidence of bacterial growth but only due to sample traces. These false positive results should be the subject for upcoming improvement of the PhenoMATRIX® PLUS algorithm. Importantly, no false negative results were returned by PhenoMATRIX® PLUS in the present study.
Currently, PhenoMATRIX® PLUS is coupled to another software (Valab® expert system, Flourens, France) in our lab workflow. After results are automatically sent to the LIS by PhenoMATRIX® PLUS, Valab® automatically validates them and updates immediately the electronic patient file.
The present study has some limitations: (i) PhenoMATRIX® PLUS settings for the chromID™ VRE agar were based on bacterial growth and not on the specific color of the colonies. Settings based on the specific color on VRE chromogenic medium could have reduced the number of false positives, and (ii) sample traces were considered in several cases as bacterial growth by PhenoMATRIX® PLUS.
Conclusion
Facing increasing numbers of specimens dedicated to the screening of MDROs carriage, the implementation of PhenoMATRIX® PLUS contributes to a significant reduction in workload. Therefore, technologists’ routine tasks have evolved to the confirmatory tests of the presumptively positive colonies, allowing better management of the laboratory staff and resources. Some adjustments appear necessary to improve the specificity of PhenoMATRIX® PLUS for the rectal screening of MDROs, but the advantages of this software in terms of sensitivity and workload reduction in the routine lab are already obvious.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
AC: Data curation, Methodology, Supervision, Conceptualization, Writing – original draft, Validation, Formal analysis. GR: Formal analysis, Writing – review & editing. MT-E: Writing – review & editing, Formal analysis. JS: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was performed by using internal funding.
Acknowledgments
The authors thank Copan’s and Ruwag’s teams for the high-quality technical assistance.
Conflict of interest
This research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest or competing interests.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Birlutiu, V., Birlutiu, R. M., Ene, R., and Rusu, D. (2025). Experience in implementing colonization screening in a Multidisciplinary County clinical Hospital in Romania. Microorganisms 13:775. doi: 10.3390/microorganisms13040775
Cherkaoui, A., Renzi, G., Azam, N., Schorderet, D., Vuilleumier, N., and Schrenzel, J. (2020). Rapid identification by MALDI-TOF/MS and antimicrobial disk diffusion susceptibility testing for positive blood cultures after a short incubation on the WASPLab. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1063–1070. doi: 10.1007/s10096-020-03817-8
Cherkaoui, A., Renzi, G., Charretier, Y., Blanc, D. S., Vuilleumier, N., and Schrenzel, J. (2019). Automated incubation and digital image analysis of chromogenic media using Copan WASPLab enables rapid detection of vancomycin-resistant Enterococcus. Front. Cell. Infect. Microbiol. 9:379. doi: 10.3389/fcimb.2019.00379
Cherkaoui, A., Renzi, G., and Schrenzel, J. (2024). Evaluation of PhenoMATRIX and PhenoMATRIX PLUS for the screening of MRSA from nasal and inguinal/perineal swabs using chromogenic media. J. Clin. Microbiol. 62:e0115223. doi: 10.1128/jcm.01152-23
Ciesielczuk, H., Phee, L. M., Dolphin, H., Wilks, M., Cherian, B. P., and Wareham, D. W. (2018). Optimal detection of carbapenemase-producing Enterobacteriaceae from rectal samples: a role for enrichment? J. Hosp. Infect. 98, 270–274. doi: 10.1016/j.jhin.2017.10.012
Dortet, L., Cuzon, G., Plesiat, P., and Naas, T. (2016). Prospective evaluation of an algorithm for the phenotypic screening of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 71, 135–140. doi: 10.1093/jac/dkv308
Durazzi, F., Pezzani, M. D., Arieti, F., Simonetti, O., Canziani, L. M., Carrara, E., et al. (2023). Modelling antimicrobial resistance transmission to guide personalized antimicrobial stewardship interventions and infection control policies in healthcare setting: a pilot study. Sci. Rep. 13:15803. doi: 10.1038/s41598-023-42511-5
Gelmez, G. A., Can, B., Hasdemir, U., and Soyletir, G. (2021). Evaluation of phenotypic tests for detection of carbapenemases: new modifications with new interpretation. J. Infect. Chemother. 27, 226–231. doi: 10.1016/j.jiac.2020.09.021
Kamel, N. A., Tohamy, S. T., Yahia, I. S., and Aboshanab, K. M. (2022). Insights on the performance of phenotypic tests versus genotypic tests for the detection of carbapenemase-producing Gram-negative bacilli in resource-limited settings. BMC Microbiol. 22:248. doi: 10.1186/s12866-022-02660-5
Mancuso, G., De Gaetano, S., Midiri, A., Zummo, S., and Biondo, C. (2023). The challenge of overcoming antibiotic resistance in Carbapenem-resistant Gram-negative Bacteria: "attack on titan". Microorganisms 11:1912. doi: 10.3390/microorganisms11081912
Naserpour Farivar, T., Najafipour, R., Johari, P., Aslanimehr, M., Peymani, A., Jahani Hashemi, H., et al. (2014). Development and evaluation of a quadruplex Taq man real-time PCR assay for simultaneous detection of clinical isolates of Enterococcus faecalis, Enterococcus faecium and their vanA and vanB genotypes. Iran. J. Microbiol. 6, 335–340
Novazzi, F., Arcari, G., Drago Ferrante, F., Boutahar, S., Genoni, A. P., Carcione, D., et al. (2024). Combined use of phenotypic screening and of a novel commercial assay (REALQUALITY carba-screen) for the rapid molecular detection of carbapenemases: a single-center experience. Diagnostics 14:1599. doi: 10.3390/diagnostics14151599
Sartelli, M., Marini, C. P., Mcnelis, J., Coccolini, F., Rizzo, C., Labricciosa, F. M., et al. (2024). Preventing and controlling healthcare-associated infections: the first principle of every antimicrobial stewardship program in hospital settings. Antibiotics 13:896. doi: 10.3390/antibiotics13090896
Skally, M., Cafferkey, J., Russell, M., Mcbrierty, L., Bijoy, R., Burns, K., et al. (2025). Screening for carbapenemase-producing Enterobacterales (CPE)-considering the practical implications of molecular results, the value of culture and deciding criteria for resistance. J. Antimicrob. Chemother. 80, 1402–1406. doi: 10.1093/jac/dkaf088
Keywords: PhenoMATRIX® PLUS, CPOs, MDROs, ESBL-E, Acinetobacter, VRE, chromogenic media, WASPLab®
Citation: Cherkaoui A, Renzi G, Tittel-Elmer M and Schrenzel J (2025) Evaluation of PhenoMATRIX® PLUS for the culture-based rectal screening of CPOs, ESBL-E, Acinetobacter, and VRE carriage. Front. Microbiol. 16:1624411. doi: 10.3389/fmicb.2025.1624411
Edited by:
Yi-Wei Tang, Chongqing Medical University, ChinaReviewed by:
Salome N. Seiffert, Zentrum für Labormedizin (ZLM), SwitzerlandDiego Josa, Clínica Shaio, Colombia
Copyright © 2025 Cherkaoui, Renzi, Tittel-Elmer and Schrenzel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdessalam Cherkaoui, YWJkZXNzYWxhbS5jaGVya2FvdWlAaHVnLmNo
 Abdessalam Cherkaoui
Abdessalam Cherkaoui Gesuele Renzi1
Gesuele Renzi1 Jacques Schrenzel
Jacques Schrenzel