- Department of Clinical Laboratory, Affiliated Jinhua Hospital, Zhejiang University School of Medicine (Jinhua Municipal Central Hospital), Jinhua, Zhejiang, China
Background: Citrobacter portucalensis is an emerging multidrug-resistant (MDR) pathogen within the Citrobacter genus. Although individual occurrences of blaKPC–2 or blaNDM–1 have been sporadically reported, the coexistence of both carbapenemase genes in a single strain remains extremely rare.
Methods: We performed whole-genome sequencing and conjugation assays on a bloodstream isolate of C. portucalensis (JH112) obtained from a critically ill patient. Plasmid structure, resistance determinants, and transferability were comprehensively analyzed using in vitro assays and bioinformatic pipelines.
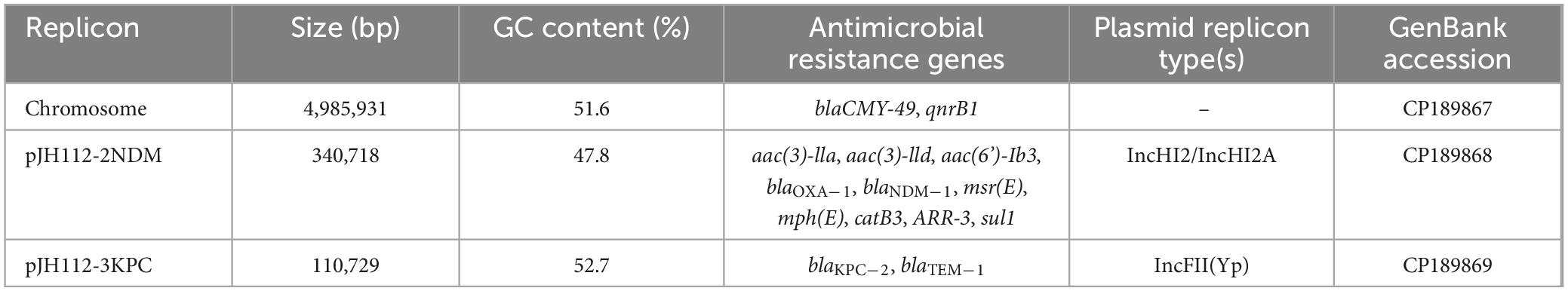
Results: JH112 exhibited an extensively drug-resistant phenotype and carried two major carbapenemase genes, blaKPC–2 and blaNDM–1, located on distinct plasmids. The blaKPC–2 gene resided on an IncFII(Yp)-type plasmid (∼110 kb) with a complete conjugation module and was successfully transferred to a recipient strain. This plasmid also harbored an O-antigen biosynthesis gene cluster, potentially enhancing host adaptation. In contrast, the blaNDM–1 gene was located on a 340 kb IncHI2/HI2A-type megaplasmid with incomplete conjugation machinery and failed to transfer under standard conditions. Both plasmids showed unique structural arrangements compared to known references. The chromosome also carried blaCMY–49 and qnrB1, contributing to broad-spectrum resistance.
Conclusion: We report a rare clinical C. portucalensis isolate co-harboring two carbapenemase genes on genetically distinct plasmids with divergent mobility. This highlights the species’ potential role as a resistance gene reservoir and the need for enhanced molecular surveillance in both clinical and environmental settings.
Introduction
The genus Citrobacter, a member of the Enterobacteriaceae family, comprises common opportunistic pathogens that are widely distributed in human intestines, animal surfaces, food products, aquatic systems, and healthcare environments (Yao et al., 2021). Although traditionally considered to have lower pathogenicity than high-risk pathogens such as Escherichia coli and Klebsiella pneumoniae, Citrobacter species have increasingly been implicated in infections among critically ill, immunocompromised, and long-term hospitalized patients (Lee et al., 2019; Ju et al., 2025). Notably, many isolates have shown high levels of antimicrobial resistance (AMR), particularly to β-lactam antibiotics (Liu et al., 2021). Of concern, certain Citrobacter strains are capable of acquiring and disseminating a broad range of resistance genes, serving as reservoirs and potential mediators for horizontal gene transfer (HGT) (Fonton et al., 2024). Citrobacter portucalensis, a recently redefined species within the genus, is phenotypically indistinguishable from Citrobacter freundii and therefore often misidentified by conventional biochemical methods (Ribeiro et al., 2017). Initially studied in the context of bacterial taxonomy, emerging reports have highlighted its potential to harbor clinically significant resistance determinants, especially carbapenemase genes, suggesting its underestimated role in the evolution of multidrug-resistance (MDR) (Sellera et al., 2023; Cao et al., 2021).
Carbapenems are considered last-line antibiotics against MDR Gram-negative bacteria. Among the carbapenemases, New Delhi metallo-beta-lactamase (NDM) and K. pneumoniae carbapenemase (KPC) are of particular clinical relevance. NDM belongs to the NDM family (Ambler class B), while KPC is a class A serine beta-lactamase; both confer resistance to carbapenems and most other beta-lactam agents. These resistance genes are often located on mobile genetic elements (MGEs) such as plasmids, integrons, and transposons, facilitating their rapid dissemination across different species and ecological boundaries, thus posing a major challenge to infection control (Juhas, 2015). The clinical implications of blaKPC–2 and blaNDM–1 carriage are profound. Infections caused by strains harboring either of these genes are associated with limited treatment options, higher morbidity, prolonged hospital stays, and increased mortality rates, particularly among vulnerable populations such as ICU patients and those with immunosuppression (Logan and Weinstein, 2017; Nordmann et al., 2011). Notably, co-harboring of both blaKPC–2 and blaNDM–1 within the same strain can further exacerbate therapeutic challenges by conferring resistance to most β-lactams, including ceftazidime-avibactam and meropenem-vaborbactam, which are typically effective against either enzyme alone (Papp-Wallace et al., 2020). Such dual-carbapenemase producers often require reliance on last-resort agents like tigecycline, colistin, or combination regimens with limited clinical efficacy and increased toxicity risks. Therefore, strains carrying both blaKPC–2 and blaNDM–1 represent a serious clinical threat, underscoring the need for prompt detection, genomic surveillance, and rigorous infection control measures. Previous studies have shown that C. portucalensis can act as a cryptic intermediate host for these resistance determinants, bridging their transmission between humans, animals, and the environment (Wang et al., 2022).
While sporadic cases of C. portucalensis carrying either blaNDM or blaKPC have been reported, isolates co-harboring both genes remain exceptionally rare. Current knowledge on their resistance mechanisms, plasmid architecture, and transfer dynamics is still limited, warranting further investigation.
In addition, Citrobacter species exhibit strong ecological adaptability, being not only isolated from clinical samples but also stably colonizing hospital effluents, sewage systems, aquatic environments, and the intestinal tracts of animals (Alglave et al., 2025). This cross-host and cross-environment persistence suggests a critical role in the “One Health” framework, where they may serve as reservoirs and recombination platforms for AMR genes (Castañeda-Barba et al., 2024). Several studies have demonstrated that Citrobacter can persist in humid ICU environments for extended periods, acting as a potential source of MDR outbreaks (Fonton et al., 2025; De Geyter et al., 2017).
In this study, we report a clinical bloodstream isolate of C. portucalensis (strain JH112), and conduct whole-genome sequencing and resistance gene profiling. Special emphasis is placed on the genetic characterization of two plasmids respectively harboring blaKPC–2 and blaNDM–1. Through comparative genomic and phylogenetic analyses, we explore their potential evolutionary pathways and transfer mechanisms, aiming to provide molecular insight into the emergence and dissemination of MDR Citrobacter strains and support evidence-based surveillance and control strategies.
Materials and methods
Bacterial strain isolation and species identification
The strain C. portucalensis JH112 was isolated in September 2023 from a blood specimen of a 82-year-old male patient with acute myocardial infarction at Jinhua Central Hospital, Zhejiang Province, China. The sample was cultured on Columbia blood agar plates at 37°C for 24 h, followed by purification. Initial identification was performed using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS, bioMérieux, France), which suggested Citrobacter braakii. To achieve accurate species-level identification, the 16S rRNA gene was sequenced and compared using the EzBioCloud database1 (Chalita et al., 2024). Average Nucleotide Identity (ANI) analysis further confirmed the strain as C. portucalensis. In addition, ribosomal multilocus sequence typing (rMLST) was performed using the PubMLST species identification tool2 (Jolley et al., 2012). This method indexes variation across 53 ribosomal protein subunit genes (rps genes) and confirmed the species identity as C. portucalensis, with 100% match to the reference profile.
Antimicrobial susceptibility testing and carbapenemase gene detection
Minimum inhibitory concentrations (MICs) for multiple antibiotics were determined using the broth microdilution method, including agents from the carbapenem (meropenem and ertapenem), cephalosporin (ceftazidime and cefepime), aminoglycoside, fluoroquinolone, sulfonamide, tigecycline, and colistin classes. MIC interpretations were based on the CLSI 2023 guidelines, except for tigecycline and colistin, which were interpreted according to EUCAST standards. E. coli ATCC 25922 was used as the quality control strain.
The classification of MDR and extensively drug-resistant (XDR) phenotypes was based on the criteria which consider only acquired resistance to antimicrobial agents. Intrinsic resistance mechanisms were excluded from the categorization (Magiorakos et al., 2012).
Carbapenemase production was initially screened using the NG-Test Carba 5 (NG Biotech), which detects KPC, NDM, IMP, VIM, and OXA-type carbapenemases. Positive results were confirmed by PCR amplification and Sanger sequencing for genotype identification.
Conjugation assay and transfer frequency determination
Conjugation assays were performed using a liquid mating protocol. The donor strain JH112 was mixed with sodium azide-resistant E. coli J53 recipient cells at a 1:3 ratio and incubated statically in LB broth at 37°C for 18 h. Transconjugants were selected on LB agar plates containing sodium azide (100 μg/ml) and meropenem (2 μg/ml) to suppress the donor and select for transconjugants. Positive colonies were confirmed by antimicrobial susceptibility testing and PCR amplification of resistance genes. Conjugation frequency was calculated as the number of transconjugant colony-forming units (CFU) per donor CFU.
Transformation assay
Plasmid DNA was extracted from C. portucalensis JH112, using the Qiagen Plasmid Midi Kit. The mixed plasmid preparation was electroporated into E. coli DH5α using standard conditions (2.5 kV, 200 Ω, 25 μF). Transformants were selected on LB agar plates supplemented with meropenem (2 μg/ml). Colonies growing on selective media were screened by PCR for blaNDM–1 and blaKPC–2 to determine which plasmid(s) had been acquired.
Whole-genome sequencing and bioinformatics analysis
Genomic DNA was extracted using the TIANamp Bacterial DNA Kit (Tiangen Biotech, Beijing, China). Whole-genome sequencing of strain JH112 was conducted using a hybrid approach. Long-read sequencing was performed on the Oxford Nanopore MinION platform, and Illumina short-read sequencing was additionally performed for this strain to improve base accuracy. Hybrid genome assembly was carried out using Unicycler v0.4.9 (Wick et al., 2017), which integrates both long and short reads, and SPAdes v3.13.0 (Bankevich et al., 2012). Genome annotation was carried out with Prokka v1.14.6 (Seemann, 2014), and completeness was evaluated using BUSCO v4.1.2 (Simão et al., 2015). Resistance genes were annotated using the ResFinder (Florensa et al., 2022) and CARD databases (Alcock et al., 2020). Plasmid replicon types were identified with PlasmidFinder (Carattoli et al., 2014). Insertion sequences (ISs) were annotated using the ISfinder database (Siguier et al., 2006). Plasmid mobility elements, including oriT, relaxase, and type IV secretion system (T4SS) components, were detected using the oriTfinder tool (Li et al., 2018).
Phylogenetic and MLST analysis
To determine the phylogenetic position of JH112 within the C. portucalensis population, all available C. portucalensis genome sequences were downloaded from the PubMLST database (as of December 2024) (Jolley et al., 2018). MLST was performed using the BIGSdb platform (Jolley and Maiden, 2010). Minimum spanning tree (MST) analysis based on MLST profiles was conducted with GrapeTree to evaluate the genetic relationships among different sequence types (STs). In addition, core genome alignment-based phylogenetic trees were visualized using the Interactive Tree of Life (iTOL)3 to determine the evolutionary clustering of JH112 among global isolates.
Genetic context analysis and visualization
To characterize the genetic environment of blaKPC–2 and blaNDM–1, nucleotide sequences were aligned using BLASTn against representative reference plasmids in the NCBI database. Local synteny and structural variations were visualized using Easyfig v2.2.3. Circular maps of complete plasmid structures were generated using the Proksee platform4 to illustrate homologous regions, structural rearrangements, insertion sequences, and MGEs.
Results
Clinical case overview
An 82-year-old male patient with end-stage renal disease, chronic heart failure, hypertension, and type 2 diabetic nephropathy was admitted to Jinhua Central Hospital in August 2023 following acute cardiovascular decompensation. He was diagnosed with ST-segment elevation myocardial infarction and transferred to the ICU after emergency coronary intervention.
Despite empirical antimicrobial treatment with ceftriaxone, linezolid, and piperacillin/tazobactam, the patient’s condition remained critical. On hospital day 3, blood cultures yielded two Gram-negative bacilli, initially identified by MALDI-TOF MS as C. braakii and Aeromonas caviae. The Citrobacter isolate was later confirmed by 16S rRNA sequencing and whole-genome sequencing as C. portucalensis, while the Aeromonas isolate could not be revived for further characterization.
Antimicrobial susceptibility testing revealed that the C. portucalensis strain exhibited an MDR profile, including resistance to carbapenems, cephalosporins, aminoglycosides, and fluoroquinolones. The A. caviae isolate also showed MDR in preliminary testing. The patient’s condition deteriorated despite intensive care, and following family consent, life-sustaining therapy was withdrawn. The patient died on 5 September 2023.
Antimicrobial susceptibility profile of C. portucalensis JH112
Antimicrobial susceptibility testing indicated that C. portucalensis JH112 exhibited an MDR phenotype. The strain showed high-level resistance to various β-lactams, aminoglycosides, quinolones, and combination agents, with MICs as follows: ceftazidime (>128 μg/ml), cefepime (>128 μg/ml), meropenem (32 μg/ml), ertapenem (128 μg/ml), gentamicin (4 μg/ml), rifampicin (>128 μg/ml), tobramycin (>128 μg/ml), and ceftazidime-avibactam (>128 μg/ml). The isolate remained susceptible only to tigecycline (0.25 μg/ml) and polymyxin B (<0.125 μg/ml) (Table 1).
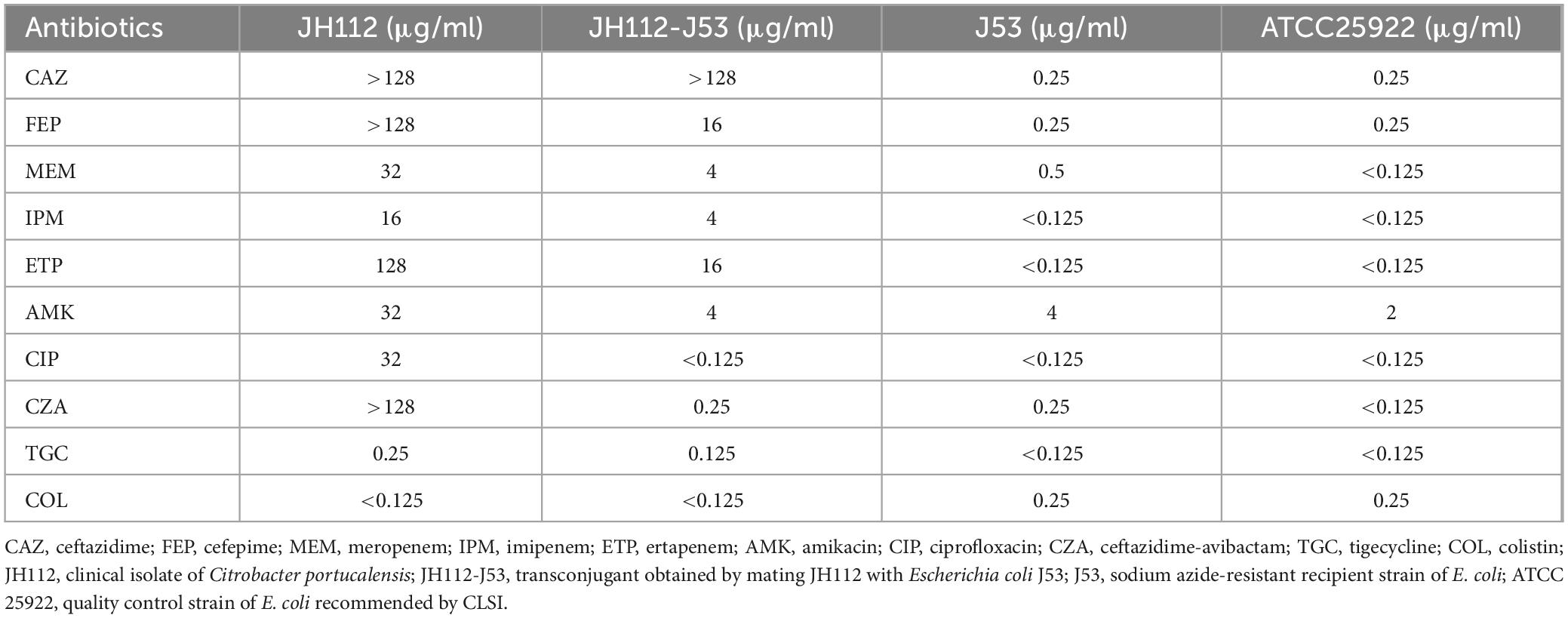
Table 1. Minimum inhibitory concentration values demonstrating resistance transfer from Citrobacter portucalensis JH112 to Escherichia coli J53.
Phylogenetic analysis
To investigate the population structure of C. portucalensis and determine the evolutionary placement of the clinical isolate JH112, we performed two complementary analyses. First, a MST was constructed using all STs available in the PubMLST database as of December 2024. JH112 was classified as ST252, which has only one other recorded isolate in the database. Interestingly, ST252 occupied a central node within the MST, connected to multiple other STs through single-locus variants, suggesting that it may represent an ancestral or intermediary genotype within the current population structure (Figure 1). However, due to the limited number of isolates per ST and the incomplete epidemiological metadata in the database, further inferences about its prevalence or clinical importance remain tentative.
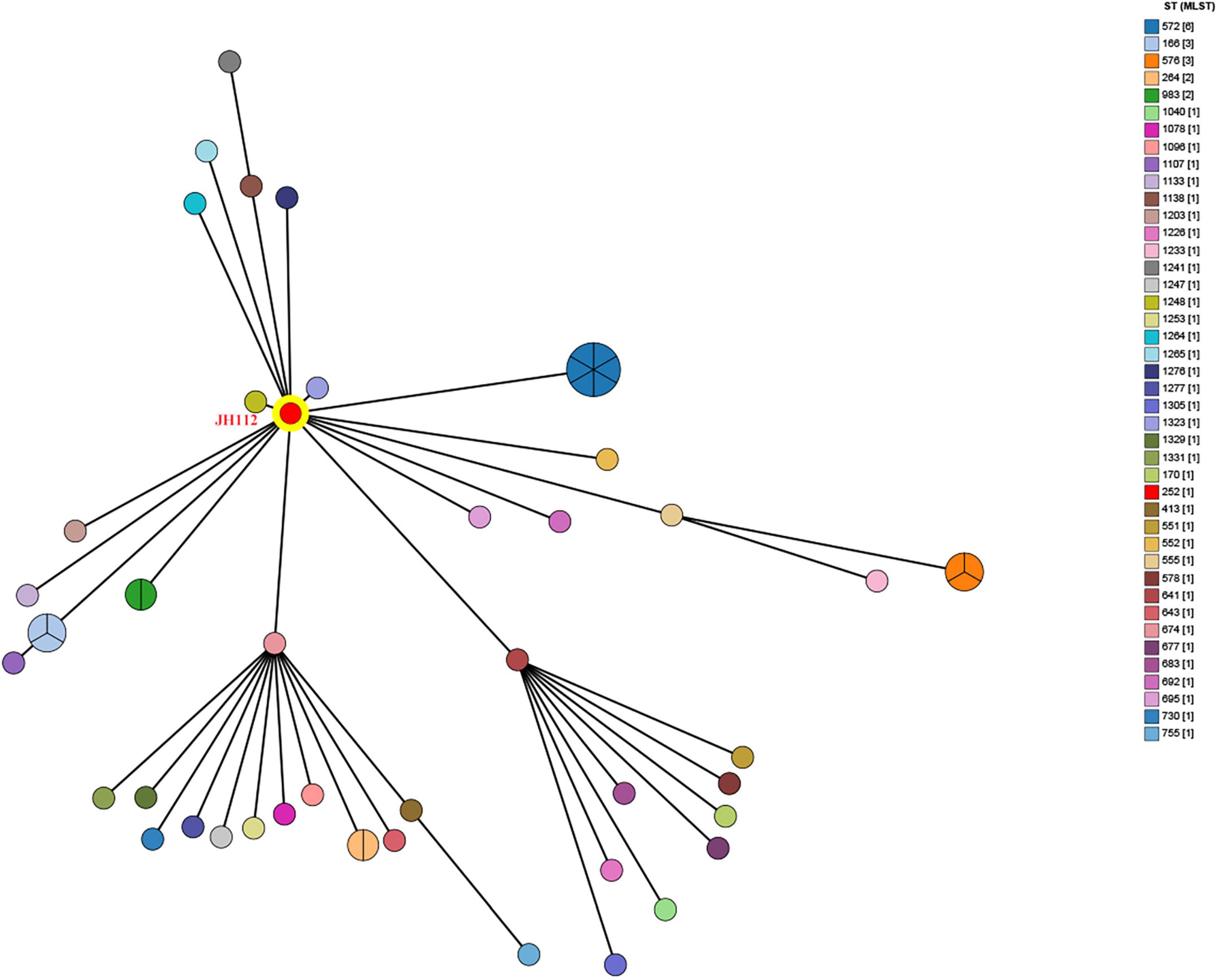
Figure 1. Minimum spanning tree based on multilocus sequence typing (MLST) profiles of Citrobacter portucalensis isolates available in the PubMLST database. Each node represents a unique sequence type (ST), with node size proportional to the number of isolates reported. The clinical isolate JH112, assigned to ST252, is highlighted in red. ST252 lies centrally and connects to multiple STs through single- or double-locus variants, indicating potential allelic proximity. However, most STs are represented by only one isolate, limiting inference of clonal expansion or dominance.
To further explore the genomic relationship of JH112 to other C. portucalensis strains, we constructed a core genome-based maximum likelihood phylogenetic tree using all publicly available genomes from PubMLST. The result showed that JH112 clustered closely with environmental isolates 2SOAct20merA (Czechia) and LACPHL-BACT202400417 (United States), both of which share allelic similarities but belong to distinct STs (ST1248 and ST1323, respectively). Despite their phylogenetic proximity, these STs differ by at least one of the seven canonical MLST loci, underscoring the limitations of classical MLST in capturing fine-scale evolutionary relationships. The absence of defined clonal complexes for these STs in the PubMLST database further reflects the early diversification phase of this species. Overall, the tree supports a model of high genomic heterogeneity within C. portucalensis, with no evidence of dominant epidemic clones or geographic clustering, consistent with sporadic emergence from diverse ecological niches (Figure 2).

Figure 2. Core genome-based phylogenetic tree of Citrobacter portucalensis strains. The tree was constructed using concatenated alignments of core genes from C. portucalensis genomes with assigned STs in the PubMLST database (as of December 2024). JH112 (ST252) is highlighted in red and clusters with ST1248 and ST1323. While these STs appear phylogenetically close in the core genome tree, they remain distinct based on MLST allelic profiles and are not currently assigned to the same clonal complex. The tree shows a broadly dispersed population structure with no evidence of dominant epidemic clones, supporting the view that C. portucalensis is a genetically diverse species undergoing early-stage diversification.
Resistance genotype and plasmid transfer potential of JH112
Whole-genome sequencing of JH112 revealed a 4,985,931 bp chromosome (GC content: 51.6%) harboring two resistance genes: blaCMY–49 and qnrB1. CMY-49 encodes an AmpC-type β-lactamase conferring resistance to various cephalosporins and is not inhibited by commonly used β-lactamase inhibitors. qnrB1 encodes a pentapeptide repeat protein that protects DNA gyrase, contributing to low-level quinolone resistance.
Two plasmids were identified: pJH112-2NDM (IncHI2/IncHI2A, 340,718 bp) and pJH112-3KPC (IncFII(Yp), 110,729 bp). pJH112-2NDM carries multiple resistance genes, including blaNDM–1, two copies of blaOXA–1, aac(3)-IIa, aac(3)-IId, aac(6’)-Ib3, msr(E), mph(E), catB3, ARR-3, and sul1, forming a complex MDR island. Additionally, it harbors tellurite resistance genes (terA, terC, terD, terE, terZ, and terW), facilitating survival under heavy metal stress. The presence of MGEs such as ISAba125, IS26, ISVsa5, and ISYps3 suggests a history of recombination and potential for HGT (Table 2 and Figure 3).
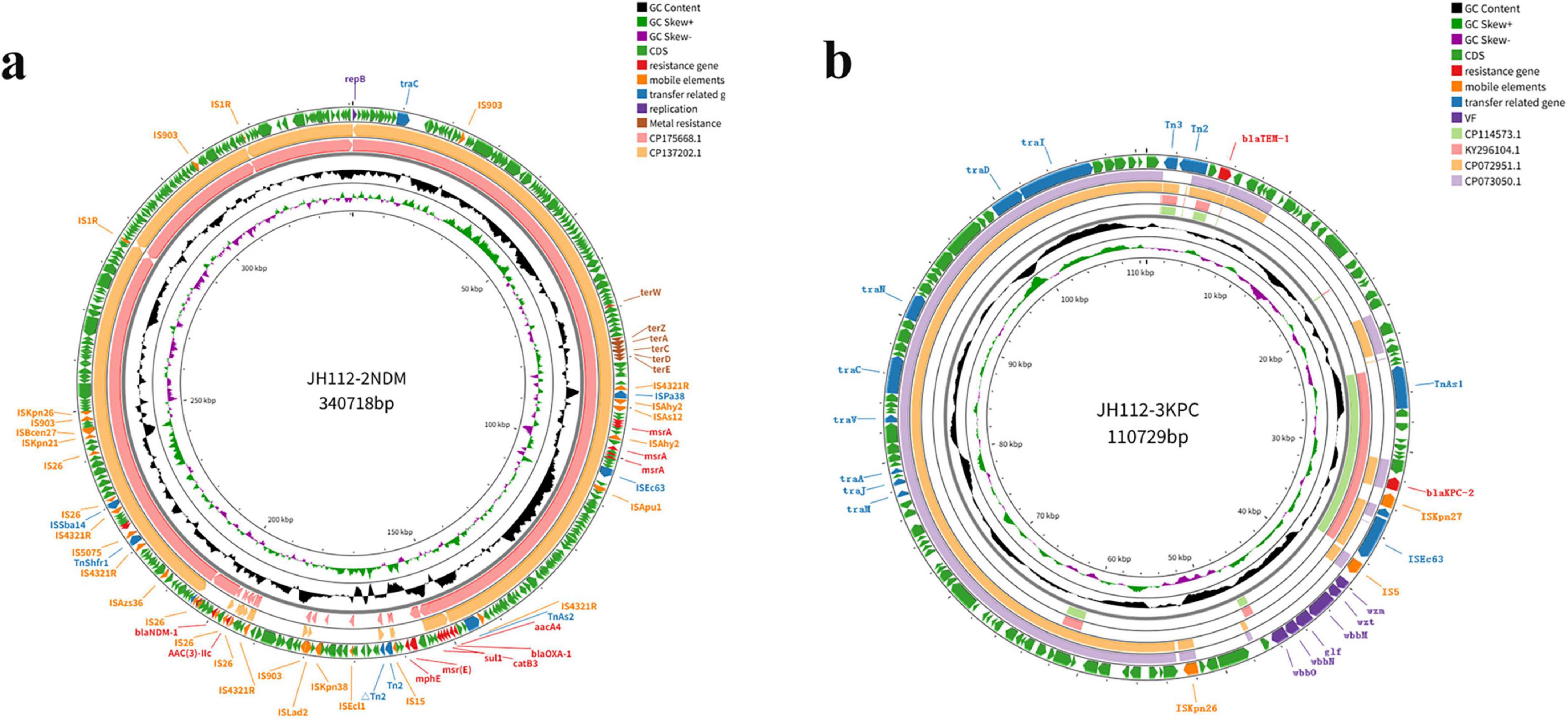
Figure 3. Circular maps of the resistance plasmids carried by Citrobacter portucalensis JH112. (a) pJH112-2NDM (IncHI2/IncHI2A), harboring blaNDM–1, additional resistance genes, tellurite operon, and multiple mobile elements. (b) pJH112-3KPC (IncFII[Yp]), containing blaKPC–2, blaTEM–1, a complete conjugation module, and an O-antigen biosynthesis cluster. Gene functions are color-coded.
To further investigate the evolutionary relationships of the identified plasmids, phylogenetic analyses were performed using representative plasmid sequences retrieved from NCBI. The phylogenetic tree based on the complete plasmid sequences revealed that pJH112-3KPC formed a distinct lineage, separate from other known KPC-2–bearing plasmids, supporting its unique structural configuration and evolutionary divergence. In contrast, pJH112-2NDM clustered closely with pNDM-CRNMS3 and pF3221-NDM, indicating shared ancestry and potential derivation from a common IncHI2 backbone. These findings are consistent with the >90% sequence identity observed in pairwise alignment and highlight differing evolutionary pressures and mobility potential between the two plasmids (Figure 4).
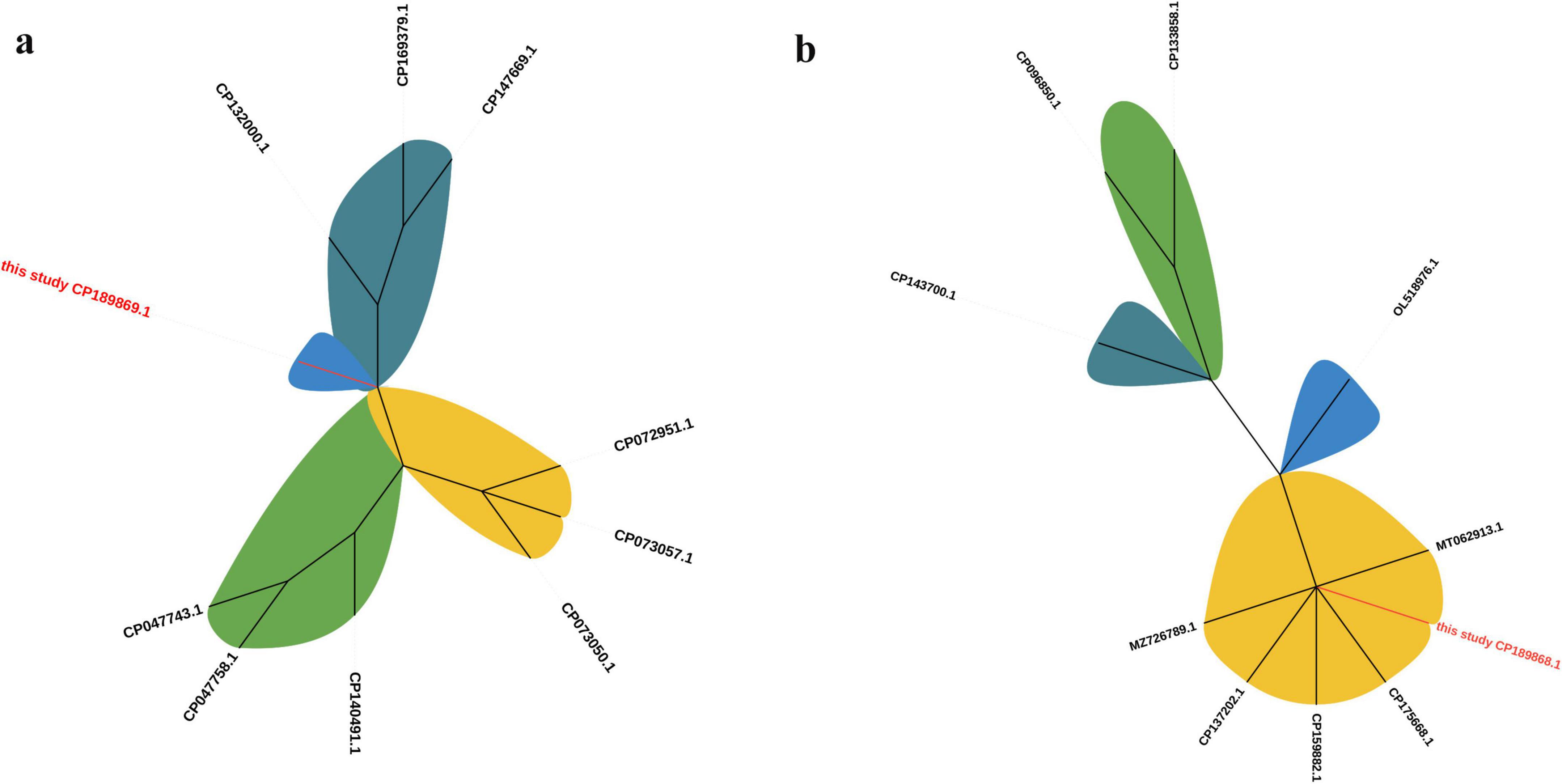
Figure 4. Phylogenetic trees of plasmids pJH112-2NDM and pJH112-3KPC. Maximum likelihood phylogenetic trees were constructed using complete plasmid sequences aligned with related plasmids retrieved from NCBI. (a) pJH112-3KPC (CP189869.1) clusters on an independent branch. (b) pJH112-2NDM (CP189868.1) shows high similarity to pNDM-CRNMS3 (CP175668.1) and pF3221-NDM (CP137202.1).
The genetic context of blaNDM–1 includes an upstream ISAba125–IS26–YokD structure, forming a typical Tn125-like transposon. This structure is incomplete in reference plasmids pNDM-CRNMS3 and pF3221-NDM, indicating that pJH112-2NDM may represent an earlier acquisition state or an independently evolved module. Downstream, a gene cluster composed of FrmB, FrmA, FrmR, ISVsa5, blaOXA–1, IS26, and ISYps3 was identified, with potential mobility mediated by IS26 and ISYps3. Comparative analysis showed structural variations compared to reference plasmids, suggesting divergent evolutionary paths.
Together, these findings indicate that blaNDM–1 is embedded within a complex and modular resistance island resembling a Tn125-like transposon, with extensive flanking insertion sequences (ISAba125, IS26, ISVsa5, and ISYps3) that may enhance environmental recombination and HGT, especially under antibiotic or heavy metal selective pressure. This structure differs substantially from the blaKPC–2 genetic context, reflecting divergent mobility mechanisms.
pJH112-3KPC harbors blaKPC–2 and blaTEM–1, and was the only plasmid successfully transferred in conjugation experiments. It contains a complete conjugation module, including oriT, relaxase, and a fully functional T4SS. Conjugation assays confirmed successful transfer to E. coli J53, and the resulting transconjugants remained susceptible to ceftazidime-avibactam, indicating that the blaNDM–1-carrying plasmid was not co-transferred. PCR confirmed the transfer of blaKPC–2 only. The conjugation frequency of pJH112-3KPC was estimated at approximately 3.6 × 10–3 transconjugants per donor cell, suggesting an efficient horizontal transfer potential under laboratory conditions.
BLAST analysis revealed that no known plasmid shares complete structural identity with pJH112-3KPC, though partial homology (up to 66% coverage and 99.9% nucleotide identity) was observed with plasmids from various Enterobacteriaceae species, indicating a potentially novel plasmid backbone. Upstream of blaKPC–2, the ISKpn27–tnpR–ISE63 module was present; however, typical accessory elements such as hin2, ISKpn19, and ISEc15 were absent and replaced by an IS5 insertion, suggesting IS5-mediated recombination. Downstream, six conserved ORFs, hin1, and TnAs1 were fully retained, indicating a structurally complete transposition region (Figure 5).
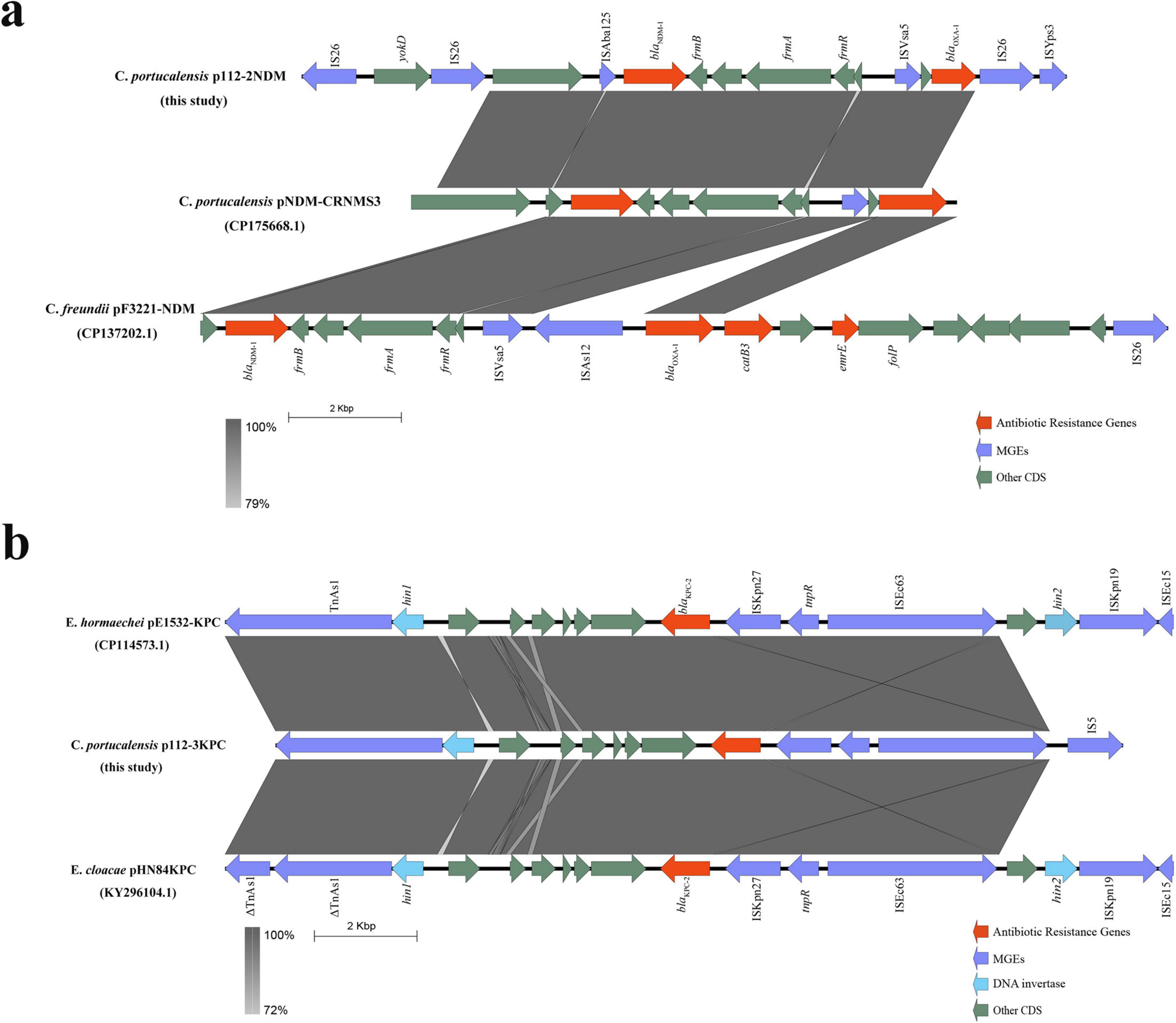
Figure 5. Genetic environment of blaNDM–1 and blaKPC–2 in plasmids from Citrobacter portucalensis JH112. (a) Comparative analysis of blaNDM–1 region on pJH112-2NDM showing a Tn125-like element and a downstream resistance island, relative to reference plasmids. (b) Structure of blaKPC–2 region on pJH112-3KPC, highlighting IS5-mediated rearrangements and a conserved downstream transposition unit. Homologous regions are shaded.
Notably, pJH112-3KPC also carries a complete O-antigen biosynthesis gene cluster, including wzm, wzt, wbbM, wbbN, wbbO, and glf, which is uncommon in Enterobacterales plasmids and may have originated from Klebsiella species. While not classified as traditional virulence factors, these genes are involved in lipopolysaccharide biosynthesis and may enhance immune evasion and environmental fitness, suggesting that the plasmid confers both AMR and potential virulence traits.
In contrast to pJH112-2NDM, the blaKPC–2 locus is integrated within a compact and structurally complete transposition region with functional conjugation machinery, supporting high transferability. The absence of typical accessory elements and the presence of IS5 insertion suggest independent evolution and recombination events. This plasmid thus represents a distinct mobile platform combining resistance and potential virulence features.
Although oriT and relaxase elements were present on pJH112-2NDM, no transconjugants were obtained under standard laboratory conditions, suggesting impaired or inactive conjugation functionality under the tested parameters. To further evaluate its mobility potential, three independent electroporation assays were conducted using purified plasmid DNA from JH112 and E. coli DH5α as the recipient. Despite selection on meropenem-containing media, no transformants harboring blaNDM–1 were recovered, while a small number of colonies carrying blaKPC–2 were detected, indicating that only pJH112-3KPC was successfully introduced. These results further support that pJH112-2NDM was non-transmissible under the tested conditions, likely reflecting impaired conjugation function.
Discussion
In recent years, the prevalence of carbapenem-resistant Citrobacter species has shown a notable upward trend globally, with increasing reports from Europe, the United States, and several Asian countries including China, India, and South Africa (Nobrega et al., 2023; Sommer et al., 2024). The dissemination of resistance genes such as blaNDM and blaVIM has been frequently associated with these isolates. Citrobacter spp. are commonly found in humid hospital environments, sewage systems, and ICU drainage networks. Their exceptional environmental persistence enables them to serve as difficult-to-eradicate reservoirs of AMR genes and act as critical mediators of HGT (Sellera et al., 2022).
This study reports a clinical bloodstream isolate, C. portucalensis JH112, recovered from a patient with multiple comorbidities, including end-stage renal disease, hypertension, diabetes mellitus, and coronary artery disease. Notably, a MDR A. caviae was co-isolated from the same patient, although subsequent recovery failed. The coexistence of multiple MDR pathogens under such critical infection settings may facilitate interspecies gene transfer and the acceleration of resistance evolution through community-level gene exchange. This observation is consistent with previous studies linking Citrobacter bacteremia with polymicrobial infections and chronic disease comorbidities (Hashimoto et al., 2021; Goncalves et al., 2024). Members of the genus Aeromonas possess strong integrative capacities and may act as collectors or transport platforms for resistance genes, promoting the assembly and dissemination of mobile resistance islands in collaboration with species such as Citrobacter (Carusi et al., 2024).
Multilocus sequence typing analysis revealed that JH112 belongs to ST252, a rarely reported ST. Although C. portucalensis exhibits high ST diversity, most isolates display sporadic distribution, with no dominant epidemic clones identified to date, unlike high-risk lineages such as ST11 or ST258 in K. pneumoniae (Li et al., 2021; Zhang et al., 2024). The presence of multiple MDR plasmids in such strains suggests that this species may currently be in a “pre-epidemic” evolutionary phase, with the potential to emerge as a dominant MDR clone under selective pressure in high-risk settings like the ICU.
A major concern highlighted in this study is the coexistence of two distinct carbapenemase-encoding plasmids within a single C. portucalensis isolate. From a clinical and epidemiological perspective, this configuration represents a high-risk resistance profile. The plasmid harboring blaKPC–2 demonstrated successful conjugative transfer under laboratory conditions, underscoring its potential for rapid dissemination within nosocomial environments, particularly in settings with intense antimicrobial selective pressure.
Such dual-carbapenemase producers pose a major therapeutic challenge, as they often exhibit resistance to nearly all β-lactams, including advanced β-lactam/β-lactamase inhibitor combinations like ceftazidime-avibactam and meropenem-vaborbactam, which are typically reserved for either KPC or NDM producers alone. Dual producers such as this strain are associated with poor clinical outcomes and are increasingly recognized as a significant global threat (Lee et al., 2022). This narrows available treatment options to toxic or less effective agents such as tigecycline or colistin, particularly concerning for bloodstream infections or immunocompromised patients. Consequently, infections caused by such strains are associated with increased treatment failure, prolonged hospitalization, and higher mortality.
Remarkably, one of the plasmids (pJH112-3KPC) also carried a complete O-antigen biosynthesis gene cluster, which may have been acquired from Klebsiella spp. Though not conventional virulence determinants, these genes contribute to lipopolysaccharide synthesis and may enhance immune evasion, colonization potential, and host adaptation (Hsieh et al., 2012). The fusion of resistance determinants with putative pathogenicity elements suggests that this plasmid may represent a hybrid resistance-adaptation platform with multifactorial advantages in clinical settings.
Structurally, pJH112-2NDM retained the ISAba125–IS26–YokD module upstream of blaNDM–1, forming a typical Tn125-like transposon not observed in comparator plasmids pNDM-CRNMS3 and pF3221-NDM. This structural configuration may reflect an earlier evolutionary acquisition event or an independent integration pathway, consistent with previous reports highlighting the role of Tn125 and ISAba125 in facilitating the global dissemination of blaNDM (Khan et al., 2017). The downstream presence of FrmA/B/R, blaOXA–1, IS26, and ISYps3 forms a complex resistance locus, with Frm genes potentially enhancing bacterial stress tolerance. The blaOXA–1 gene and associated IS elements may constitute a functional composite transposon, further increasing the plasticity of the resistance region.
From a clinical and epidemiological standpoint, the carriage of multiple resistance determinants—including aac(3)-IIa, aac(3)-IIb, aac(6’)-Ib3, msr(E), mph(E), catB3, ARR-3, sul1—and tellurite resistance genes (terA–terW) greatly expands the bacterium’s potential to survive in hostile environments, such as those exposed to antibiotics or heavy metals in hospital settings. The dense array of MGEs (ISAba125, IS26, ISVsa5, and ISYps3) not only reflects a recombinogenic genome structure but also poses a risk for further HGT events across different species and ecological niches.
Although pJH112-2NDM harbors recognizable oriT and relaxase elements, no conjugative transfer was observed under standard laboratory conditions. This failure is likely multifactorial. The plasmid’s T4SS is incomplete, spanning only 9 kb and lacking auxiliary genes essential for conjugation. Its large size (>340 kb) may also impose a significant metabolic burden on the host, reducing mobilization efficiency. Furthermore, IncHI2 plasmids are known to require low-temperature induction for expression of their conjugation machinery, while our experiments were conducted at 37°C—conditions that may suppress transfer activity (Forns et al., 2005). In addition, recipient incompatibility or entry exclusion systems may contribute to the observed transfer failure. These findings align with prior research by Li et al. (2022), which reported that only 3.1% of dual-carbapenemase-producing isolates were capable of simultaneous plasmid co-transfer, emphasizing the potential role of interplasmid incompatibility and regulatory constraints. While pJH112-2NDM appears non-conjugative under current conditions, its structurally intact resistance region—rich in MGEs—suggests latent mobility potential under selective pressure or future recombination, warranting ongoing genomic surveillance and infection control vigilance.
Additionally, the chromosomal presence of blaCMY–49 and qnrB1 suggests intrinsic resistance to β-lactams and quinolones. While CMY-type β-lactamases are typically plasmid-borne, chromosomally encoded variants, such as blaCMY–37 in C. freundii (Ahmed and Shimamoto, 2008) and blaCMY–190 in Citrobacter youngae (Liu et al., 2025), have been documented. Moreover, both C. freundii and Enterobacter cloacae are known to carry inducible chromosomal AmpC β-lactamase genes (Sader et al., 2021). Chromosomal carriage of AmpC may facilitate inducible expression under β-lactam exposure and contribute to treatment failure (Tebano et al., 2024).
Taken together, C. portucalensis JH112 represents a clinically relevant MDR isolate with dual-carbapenemase production, structural novelty, and potential for dissemination. This study provides a comprehensive “strain–plasmid–gene” analysis of its resistance architecture and highlights the need for enhanced surveillance of cryptic resistance reservoirs in the context of One Health.
Conclusion
In summary, the isolation of C. portucalensis JH112 underscores the growing complexity and clinical relevance of carbapenem resistance within this emerging species. The strain harbors both transferable blaKPC–2 and structurally intact yet non-transmissible blaNDM–1 plasmids—representing the two most concerning carbapenemase platforms in clinical settings. The discovery of an O-antigen biosynthesis gene cluster on the blaKPC-plasmid suggests added potential for host adaptation and immune evasion. In addition to the two carbapenemase genes, the chromosomally encoded blaCMY–49 contributes to third-generation cephalosporin resistance and reduces the efficacy of β-lactam/β-lactamase inhibitor combinations, reflecting a multi-layered β-lactam resistance background. Furthermore, qnrB1 provides low-level quinolone resistance, which may facilitate the selection of higher-level resistance under antibiotic pressure.
These findings not only elucidate the plasmid-level architecture of dual-carbapenemase resistance but also highlight the role of C. portucalensis as a potential reservoir and vector of high-risk resistance determinants in the hospital environment. Continued genomic surveillance, functional assessment of resistance plasmids, and environmental monitoring are essential to mitigate the risk of further dissemination in high-risk populations and healthcare settings.
Data availability statement
The genome sequencing data are publicly available at NCBI GenBank under the BioProject accession number: PRJNA1255133.
Ethics statement
The studies involving humans were approved by Ethics Committee of Jinhua Central Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
JH: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Project administration, Software, Supervision, Visualization, Writing – original draft. KS: Formal Analysis, Funding acquisition, Methodology, Software, Writing – original draft. KC: Supervision, Validation, Visualization, Writing – original draft. JW: Formal Analysis, Resources, Writing – original draft. YZ: Formal Analysis, Methodology, Project administration, Resources, Supervision, Writing – review and editing. JS: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Jinhua City Science and Technology Project (2024-3-055) and Jinhua City Science and Technology Project (2023-4-082).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ahmed, A. M., and Shimamoto, T. (2008). Emergence of a cefepime- and cefpirome-resistant Citrobacter freundii clinical isolate harbouring a novel chromosomally encoded AmpC beta-lactamase, CMY-37. Int. J. Antimicrob. Agents 32, 256–261. doi: 10.1016/j.ijantimicag.2008.04.019
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alglave, L., Faure, K., and Mullié, C. (2025). Plasmid dissemination in multispecies carbapenemase-producing enterobacterales outbreaks involving clinical and environmental strains: A narrative review. Microorganisms 13:810. doi: 10.3390/microorganisms13040810
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Cao, X., Xie, H., Huang, D., Zhou, W., Liu, Y., Shen, H., et al. (2021). Detection of a clinical carbapenem-resistant Citrobacter portucalensis strain and the dissemination of C. portucalensis in clinical settings. J. Glob. Antimicrob. Resist. 27, 79–81. doi: 10.1016/j.jgar.2021.04.027
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Carusi, J., Kabuki, D. Y., de Seixas Pereira, P. M., and Cabral, L. (2024). Aeromonas spp. in drinking water and food: Occurrence, virulence potential and antimicrobial resistance. Food Res. Int. 175:113710. doi: 10.1016/j.foodres.2023.113710
Castañeda-Barba, S., Top, E. M., and Stalder, T. (2024). Plasmids, a molecular cornerstone of antimicrobial resistance in the One Health era. Nat. Rev. Microbiol. 22, 18–32. doi: 10.1038/s41579-023-00926-x
Chalita, M., Kim, Y. O., Park, S., Oh, H. S., Cho, J. H., Moon, J., et al. (2024). EzBioCloud: A genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 74:006421. doi: 10.1099/ijsem.0.006421
De Geyter, D., Blommaert, L., Verbraeken, N., Sevenois, M., Huyghens, L., Martini, H., et al. (2017). The sink as a potential source of transmission of carbapenemase-producing Enterobacteriaceae in the intensive care unit. Antimicrob. Resist. Infect. Control. 6:24. doi: 10.1186/s13756-017-0182-3
Florensa, A. F., Kaas, R. S., Clausen, P. T. L. C., Aytan-Aktug, D., and Aarestrup, F. M. (2022). ResFinder - an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 8:000748. doi: 10.1099/mgen.0.000748
Fonton, P., Grant, R., Gasser, M., Buetti, N., Kronenberg, A., and Harbarth, S. (2025). Incidence and resistance patterns of Citrobacter spp. in Switzerland: A nationwide, retrospective surveillance study (2010-2022). Microorganisms 13:786. doi: 10.3390/microorganisms13040786
Fonton, P., Hassoun-Kheir, N., and Harbarth, S. (2024). Epidemiology of Citrobacter spp. infections among hospitalized patients: A systematic review and meta-analysis. BMC Infect. Dis. 24:662. doi: 10.1186/s12879-024-09575-8
Forns, N., Baños, R. C., Balsalobre, C., Juárez, A., and Madrid, C. (2005). Temperature-dependent conjugative transfer of R27: Role of chromosome- and plasmid-encoded Hha and H-NS proteins. J. Bacteriol. 187, 3950–3959. doi: 10.1128/jb.187.12.3950-3959.2005
Goncalves, H., Sá, R., de Oliveira Simões, F., Domingues, R. M., Oliveira, N., and Pimentel, T. (2024). Purulent pericarditis in end-stage renal disease: A rare case of Citrobacter freundii infection. Cureus 16:e62308. doi: 10.7759/cureus.62308
Hashimoto, M., Mogi, K., Sakurai, M., Sakata, T., Tani, K., and Takahara, Y. (2021). Rupture of a dissecting thoracoabdominal aortic aneurysm due to Citrobacter freundii infection. Clin. Case Rep. 9:e04719. doi: 10.1002/ccr3.4719
Hsieh, P. F., Lin, T. L., Yang, F. L., Wu, M. C., Pan, Y. J., Wu, S. H., et al. (2012). Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS One 7:e33155. doi: 10.1371/journal.pone.0033155
Jolley, K. A., Bliss, C. M., Bennett, J. S., Bratcher, H. B., Brehony, C., Colles, F. M., et al. (2012). Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology (Reading) 158(Pt 4), 1005–1015. doi: 10.1099/mic.0.055459-0
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Jolley, K. A., and Maiden, M. C. (2010). BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 11:595. doi: 10.1186/1471-2105-11-595
Ju, X., Xiong, P., Yan, Z., Chen, G., Cai, C., and Zhang, R. (2025). Emergence of carbapenem-resistant Citrobacter spp. across human, animal, and water environments in China. Int. J. Antimicrob. Agents 65:107463. doi: 10.1016/j.ijantimicag.2025.107463
Juhas, M. (2015). Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 41, 101–108. doi: 10.3109/1040841x.2013.804031
Khan, A. U., Maryam, L., and Zarrilli, R. (2017). Structure, genetics and worldwide spread of New Delhi Metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 17:101. doi: 10.1186/s12866-017-1012-8
Lee, R., Choi, S. M., Jo, S. J., Lee, J., Cho, S. Y., Kim, S. H., et al. (2019). Clinical characteristics and antimicrobial susceptibility trends in citrobacter bacteremia: An 11-Year single-center experience. Infect. Chemother. 51, 1–9. doi: 10.3947/ic.2019.51.1.1
Lee, Y. L., Chen, H. M. I, Hii, M., and Hsueh, P. R. (2022). Carbapenemase-producing Enterobacterales infections: Recent advances in diagnosis and treatment. Int. J. Antimicrob. Agents 59:106528. doi: 10.1016/j.ijantimicag.2022.106528
Li, P., Liang, Q., Liu, W., Zheng, B., Liu, L., Wang, W., et al. (2021). Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: An epidemiological, genomic and functional study. Virulence 12, 377–388. doi: 10.1080/21505594.2020.1867468
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234. doi: 10.1093/nar/gky352
Li, Y., Fang, C., Qiu, Y., Dai, X., and Zhang, L. (2022). Genomic characterization of a carbapenem-resistant Citrobacter freundii cocarrying bla(KPC-2) and bla(NDM-1). J. Glob. Antimicrob. Resist. 29, 289–292. doi: 10.1016/j.jgar.2022.04.014
Liu, L., Zhang, L., Zhou, H., Yuan, M., Hu, D., Wang, Y., et al. (2021). Antimicrobial resistance and molecular characterization of Citrobacter spp. causing extraintestinal infections. Front. Cell Infect. Microbiol. 11:737636. doi: 10.3389/fcimb.2021.737636
Liu, Z., Shen, S., Zhang, X., Lei, J., Tang, C., Wu, S., et al. (2025). Identification of CMY-190, a novel chromosomally encoded AmpC β-lactamase, and plasmid-encoded KPC-2 in a clinical isolate of Citrobacter youngae. Front. Microbiol. 16:1526882. doi: 10.3389/fmicb.2025.1526882
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 215(Suppl. 1), S28–S36. doi: 10.1093/infdis/jiw282
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Nobrega, D., Peirano, G., Matsumura, Y., and Pitout, J. D. D. (2023). Molecular epidemiology of global carbapenemase-producing Citrobacter spp. (2015–2017). Microbiol. Spectr. 11:e0414422. doi: 10.1128/spectrum.04144-22
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Papp-Wallace, K. M., Mack, A. R., Taracila, M. A., and Bonomo, R. A. (2020). Resistance to novel β-Lactam-β-Lactamase inhibitor combinations: The “Price of Progress”. Infect. Dis. Clin. North Am. 34, 773–819. doi: 10.1016/j.idc.2020.05.001
Ribeiro, T. G., Gonçalves, B. R., da Silva, M. S., Novais, Â, Machado, E., Carriço, J. A., et al. (2017). Citrobacter portucalensis sp. nov., isolated from an aquatic sample. Int. J. Syst. Evol. Microbiol. 67, 3513–3517. doi: 10.1099/ijsem.0.002154
Sader, H. S., Mendes, R. E., Doyle, T. B., Davis, A. P., and Castanheira, M. (2021). Characterization of Enterobacter cloacae and Citrobacter freundii species complex isolates with decreased susceptibility to cephalosporins from United States hospitals and activity of ceftazidime/avibactam and comparator agents. JAC Antimicrob. Resist. 3:dlab136. doi: 10.1093/jacamr/dlab136
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sellera, F. P., Fernandes, M. R., Fuga, B., Fontana, H., ásquez-Ponce, F. V., Goldberg, D. W., et al. (2022). Phylogeographical landscape of Citrobacter portucalensis carrying clinically relevant resistomes. Microbiol. Spectr. 10:e0150621. doi: 10.1128/spectrum.01506-21
Sellera, F. P., Fuentes-Castillo, D., Fuga, B., Goldberg, D. W., Kolesnikovas, C. K. M., and Lincopan, N. (2023). New Delhi metallo-β-lactamase-1-producing Citrobacter portucalensis belonging to the novel ST264 causing fatal sepsis in a vulnerable migratory sea turtle. One Health 17:100590. doi: 10.1016/j.onehlt.2023.100590
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Sommer, J., Reiter, H., Sattler, J., Cacace, E., Eisfeld, J., Gatermann, S., et al. (2024). Emergence of OXA-48-like producing Citrobacter species, Germany, 2011 to 2022. Euro Surveill. 29:2300528. doi: 10.2807/1560-7917.es.2024.29.15.2300528
Tebano, G., Zaghi, I., Cricca, M., and Cristini, F. (2024). Antibiotic treatment of infections caused by AmpC-Producing enterobacterales. Pharmacy (Basel) 12:142. doi: 10.3390/pharmacy12050142
Wang, L., Li, Z., Xiao, N., Tang, J., He, Y., Guo, J., et al. (2022). Genetic characterization of bla (NDM-1)-Carrying citrobacter portucalensis sequence type 328 and Citrobacter freundii sequence type 98. Infect. Drug Resist. 15, 2235–2242. doi: 10.2147/idr.s361761
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Yao, Y., Falgenhauer, L., Falgenhauer, J., Hauri, A. M., Heinmüller, P., Domann, E., et al. (2021). Carbapenem-Resistant Citrobacter spp. as an emerging concern in the hospital-setting: Results from a genome-based regional surveillance study. Front. Cell Infect. Microbiol. 11:744431. doi: 10.3389/fcimb.2021.744431
Keywords: Citrobacter portucalensis, blaKPC–2, blaNDM–1, plasmid-mediated resistance, antimicrobial resistance, conjugation
Citation: Huang J, Shen K, Chen K, Wu J, Zhu Y and Shi J (2025) Genomic characterization of a multidrug-resistant Citrobacter portucalensis isolate co-harboring blaKPC–2 and blaNDM–1 on distinct plasmids. Front. Microbiol. 16:1633493. doi: 10.3389/fmicb.2025.1633493
Received: 22 May 2025; Accepted: 01 July 2025;
Published: 16 July 2025.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Vittoria Mattioni Marchetti, University of Pavia, ItalyRaffaele Zarrilli, University of Naples Federico II, Italy
Joana Moreira Da Silva, Instituto Superior de Ciências da Saúde Egas Moniz, Portugal
Copyright © 2025 Huang, Shen, Chen, Wu, Zhu and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingchao Shi, MjAyMTUxNjEwMjlAemNtdS5lZHUuY24=; Yijun Zhu, emh1eWlqdW53ekBzaW5hLmNvbQ==
 Junwei Huang
Junwei Huang Kai Shen
Kai Shen Yijun Zhu
Yijun Zhu Jingchao Shi
Jingchao Shi