Abstract
The influence of environmental factors on the efficacy of the endosymbiont Wolbachia used in mosquito and pathogen control are poorly characterized and may be critical for disease control. We studied the vector mosquito Culex pipiens quinquefasciatus (Say) to determine the effect of temperature on the composition of the relative abundance of Wolbachia spp. and the microbiome, as well as key immune genes of interest in the Toll and IMD pathways. 16S barcode sequencing was used to determine the microbiome composition and qPCR was used to determine the relative abundance of Wolbachia spp. based on the highly utilized marker Wolbachia surface protein (wsp) gene. We found no effect of temperature within a single generation on the relative abundance of Wolbachia or immune gene expression, nor on the alpha or beta diversity of the microbiome. However, there was a significant difference in the abundance of Wolbachia between generations at high temperatures (≥ 28°C), but not at lower temperatures (≤ 23°C). These results support the idea that Wolbachia are reduced at higher temperatures between generations, which has an influence on the establishment of pathogens including West Nile Virus (WNV). Modulation of the Toll or IMD mosquito immune pathways was not indicated. Wolbachia endosymbiosis and trans-generation transmission appears especially sensitive to high temperatures, which may have implications for Wolbachia-based vector control strategies under climate change scenarios.
Introduction
Culex mosquitoes are responsible for spreading numerous arboviruses and parasites, such as West Nile Virus (WNV) and lymphatic filariasis, respectively (1–4). Culex pipiens quinquefasciatus (Say) is part of the overall Culex pipiens complex, and can be found broadly distributed across the world, typically in lower altitude areas between latitudes of 30°N and 30°S (5, 6). In light of global climate change and anticipated changes in temperature particularly, the range of vector mosquitoes has been increasing into areas previously uninhabitable by these insects. The expansion of mosquito ranges globally is, in turn, likely to lead to an increase in vector-borne disease transmission (5, 7–11).
In response to the growing threat of mosquito-borne disease expansion due to climate change, there has been great interest in using microbial methods to control mosquito populations, and subsequently the spread of those vector-borne pathogens and parasites (12–20). The use of Wolbachia has been particularly attractive, as this Rickettsia-like bacterium spreads easily through mosquito populations (due to selective advantage of hosts with infections), induces cytoplasmic incompatibility, and effectively blocks the establishment of some viruses (DENV and CHIKV) in mosquito hosts (17, 18, 21–25). Wolbachia can either be maternally inherited or can be artificially introduced, frequently by transinfection, to insect populations. Wolbachia infection in mosquitoes can lead to cytoplasmic incompatibility, male embryo feminization, selective male killing, and parthenogenesis (26, 27). Cytoplasmic incompatibility (CI) is of particular interest as an incompatible insect technique (IIT) to reduce vector populations. CI occurs when sperm and egg cannot successfully form a viable zygote, which is bidirectional in many mosquitoes. Generally, when male mosquitoes are infected and females are uninfected (or infected with a dissimilar strain), no viable offspring are produced. When Wolbachia-infected females mate with uninfected males, they will produce hybrid offspring that carry and perpetuate the spread of the bacterium (21, 23, 24, 28–30). Wolbachia also acts competitively with and against members of the mosquito microbiome to structure community assemblage and influence pathogen invasion. This influence on pathogen and disease dynamics has led to increased interest in the role of microbiome community dynamics in mosquitoes (21, 18, 23–25, 30–35).
While after nearly a century of research on Wolbachia it is clear that Wolbachia could be immensely advantageous given the correct conditions for disease control, in situ applications remain complicated and require a better understanding of factors influencing efficacy in application (19, 27). For instance, Dodson et al. (36) found that Wolbachia infections in Culex tarsalis did not impede, but actually enhanced, infection of WNV in the mosquito host (37) also found that Wolbachia infection in Culex tarsalis did not inhibit Rift Valley fever virus. Hughes et al. (38) found that introduction of Wolbachia into Anopheles mosquitoes induced higher mortality in hosts and was inhibited by the native microbiome. Novakova et al. (39) found that in situ mosquitoes of the Culex pipiens complex had lower abundances of Wolbachia at higher temperatures, which corresponded with higher infection rates with WNV. Additional studies have reported impacts of temperature on Wolbachia, with several reporting similar reductions of Wolbachia abundances at higher temperatures (40–44). It has become increasingly clear that Wolbachia does not respond equally effectively in terms of control across mosquito species or environmental conditions, indicating that a better understanding of these factors is necessary to gain the maximum benefit from the use of Wolbachia for vector-borne control.
One factor that also can aid in pathogen transmission reduction is Wolbachia’s manipulation of the mosquito host’s immune response. Wolbachia has the ability to alter mosquito innate immune gene expression in response to pathogen invasion of the host (45–50). While the effect of Wolbachia on the mosquito immune system is well-documented in Aedes mosquitoes, less is understood about these effects in Culex mosquitoes (48, 51). The Toll, IMD (Immune Deficiency), and JAK-STAT pathways, in addition to other pathways, play an essential role in pathogen inhibition in mosquitoes (51–55). While gene expression may be altered at many stages within these pathways, two particular genes of interest that are altered with Wolbachia infection are Rel1 (homologue to Drosophila dorsal), an integral part of the Toll pathway that aids in the transcription of innate immune factors (like antimicrobial peptides), and Def1, a gene in both the Toll and IMD pathways (48, 51). Interestingly, Ant et al. (30) analyzed various Wolbachia strains for effective infection and cytoplasmic incompatibility in Culex quinquefasciatus and found that immune genes, including Rel1 and Def1 as well as others associated with the Toll, IMD, and JAK-STAT pathways, were neither up- or down-regulated in relation to any Wolbachia infection. Given the importance of these pathways in mosquito immunity, this lack of change to immune gene expression in Culex quinquefasciatus with Wolbachia infection warrants further investigation.
Environmental conditions affect both mosquito hosts and endosymbiotic bacteria like Wolbachia. Given the implications of global climate change on global temperature and alterations of localized climate patterns, understanding the impact of changing environmental factors on vector control is of the utmost importance (10, 56–58). Boukal et al. (59) found that insects are particularly susceptible to the effects of prolonged increased temperatures at the individual and community levels, due to stress to insect physiological systems, behavior, body size, and, importantly, spatiotemporal distribution. They also found that these changes led to a restructuring of insect communities. Harvey et al. (60) further elaborated on this in their discussion of our current, and very limited, understanding of insect tolerance of temperature extremes due to climate change. Mordecai et al. (58) analyzed the thermal biology of numerous vector mosquito species and found that Culex quinquefasciatus had a thermal optimum for transmission of West Nile Virus (WNV) at 25.2°C, with a range for transmission between 19.0°C to 31.8°C. Several recent studies have indicated that WNV transmission tends to be greater under high temperature field conditions, which may be due to a decrease in naturally-occurring Wolbachia infection efficacy in mosquitoes (39, 42). Indeed, Wolbachia transmission is thermally sensitive in a number of systems including mosquitoes and Drosophila (44, 61–63). In addition, Wolbachia colonization can increase thermal sensitivity of host mosquitoes and other insects (64, 65). To ensure that novel methods of mosquito control will remain functional (or may be adapted to maintain function) under new climate regimes, experiments exploring the impacts of temperature on the survival and efficacy of control microbes, like Wolbachia, are imperative.
To better understand the effects of environmental temperature on host and Wolbachia endosymbionts, we analyzed the relative abundance of native Wolbachia (presumed to be wPip) in Culex quinquefasciatus across upper thermal optima for the transmission of WNV in this species (39, 58). We hypothesized that Wolbachia abundances would be reduced at higher temperatures in line with climate change predictions, and that, similarly to Aedes mosquitoes and in-line with current thought in the field, Wolbachia abundances in Culex quinquefasciatus would influence immune genes, particularly in the Toll and IMD pathways (39, 46–48, 50, 51). The overarching goal of this study was to understand if, across a range of temperatures and across generations, Wolbachia abundance would be altered and if the mosquito host’s immune gene expression would be altered in response to potential differences in Wolbachia abundance.
Methods
Laboratory Mosquito Husbandry
Culex quinquefasciatus were ordered from Benzon Research (Carlisle, PA, USA) as 1st instar larvae and were co-housed at the same 25°C temperature in large 1L sterile glass containers in sterile pond water until 2nd instar. Larvae were fed autoclaved fish food mixed into sterile pond water and added at 2mL to each large 1L larval container until used in the experiment. Second instar larvae were then added along a temperature gradient beam and were used in the transgenerational temperature experiment.
Specifically in the transgenerational temperature experiment, mosquitoes were reared through two generations (with adults collected at generation 0 and 1) and were bloodfed using a membrane-style feeder with pig intestine casing to encase single donor human whole blood in sodium heparin purchased from Innovative Research, Inc (Novi, MI, USA). Mosquitoes had 10% sucrose withheld 24 hours prior to blood feeding. Second generation eggs were hatched by bubbling nitrogen gas through sterilized tubing into water containing eggs to reduce oxygen levels through displacement and trigger hatching. Hatch rate was estimated to be approximately 75%.
Experimental Design-Temperature Gradient Beam
A temperature gradient was designed to house both individual and groups of mosquito larvae along an insulated aluminum beam using Peltier devices to control heating and cooling. Automated temperature measurements along the beam were recorded hourly for the duration of the experiment, and water temperatures of vessels (without larvae) in the beam were measured daily to verify similar temperature measurements. The average temperature for each zone (A-E) was calculated by averaging all temperature measurements for that zone over the duration of the experiment (Figure 1). Temperature range was established based on the thermal optima of West Nile virus transmission in Culex quinquefasciatus presented in Mordecai et al. (58). Primary containment over the glass vessels consisted of sterilized mesh enclosing the top of the vessel with a sterile tie and secondary containment consisted of a mesh enclosure around the aluminum temperature gradient beam.
Figure 1

The experimental design for temperature gradient beam and transgenerational temperature experiments in Culex quinquefasciatus. The temperature gradient beam ran from 33.69-23.23°C with mosquitoes (n = 30 at setup) reared through eclosion at each of the five temperature groups (A-E) along the beam. Adult mosquitoes were harvested within 8 hours of eclosion. The transgenerational experiment was run in two enclosed chambers with several replicate populations (n = ~200) at each temperature (28/30 and 23°C) reared through generation 0 and the eclosion of generation 1 with adult mosquitoes (n=5 for each group for qPCR) sampled from each generation.
Culex larvae were housed individually (5mL water) and in groups of 5 (25mL water) in glass vessels filled with sterile pond water and were fed sterile fish food in sterile pond water per larvae. Water was replenished as needed and was replaced simultaneously from all vessels to maintain relatively consistent levels of oxygenation. The light cycle was 12:12 and larvae were fed every 3-4 days. Larvae were added to the glass vessels simultaneously with all larvae at the second instar stage. Within eight hours of eclosion, mosquito adults were aspirated for collection out of the glass vessels and frozen at -20°C.
Experimental Design-Transgenerational Temperature Experiment
Incubators (Percival I-36VL incubator (Perry, Iowa, USA) and Conviron Gen1000 incubator (Pembina, North Dakota, USA)) were set up in a biosecure insectary with 14:10 light/dark cycles, 65% relative humidity, and set temperatures of 23°C and 30°C, with both incubators reduced 3°C during dark cycles (Figure 1). Temperatures in the 30°C chamber were reduced to 28°C prior to the hatching of generation 1 to reduce the potential for mortality, though 28°C is still considered in the same high range, high potential transmission of WNV for this mosquito species (58).
Mosquitoes were reared over two generations with adults collected from generations 0 and 1. Larvae were co-housed in large glass beakers with the non-sterile water in which they arrived from Benzon Research (Carlisle, Pennsylvania, USA) and had sterile pond water added to reduce density. Culex quinquefasciatus second instar larvae were placed in large 1L glass beakers with sterile pond water in three sterilized mesh cages in each incubator. Larvae were fed sterile fish food ad libitum and upon emergence were fed a 10% sucrose solution using soaked cotton balls. Adults were collected from each generation and were frozen at -20°C. Adult females in generation 0 were blood fed and eggs were hatched as described above. Generation 1 larval housing was covered by sterile mesh to prevent any remote potential for escape into the parent generation prior to the removal of any remaining parents.
Specimen Storage and Extraction
Culex quinquefasciatus samples from both experimental designs were frozen at -20°C and had 100uL of 1X DNA RNA Shield (Zymo Research, Irvine, CA, USA) added after being freeze-killed. Samples were frozen at -20°C until extraction. Samples had wings, legs, and heads removed and carcasses were macerated in DNA RNA Shield. Samples were then extracted using Quick DNA/RNA MagBead kits (Zymo Research, Irvine, CA, USA) and frozen at -80°C. Sample total nucleic acid concentration following extraction was 0.5ug/uL.
qPCR of Relative Wolbachia Abundance (wsp gene) and Immune Genes Rel1 and Def1
Quantitative PCRs were performed in duplicate on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) to analyze the relative abundance of Wolbachia and key immune genes. For Wolbachia qPCRs (n = all surviving adult mosquitoes across groups), the wsp gene was targeted as a proxy for relative abundance and using slightly modified primers forward-5’-AGATAGTGTAACAGCRTTTTCAGGAT- 3’ and reverse-5’-CACCATAAGAACCAAAATAACGAG- 3’ from (66). The reaction mix for the qPCRs was: 5.1uL Milli-Q Ultrapure water, 10uL 2X AzuraQuant Green Fast qPCR Mix LoRox (Azura Genomics, Raynham, MA, USA), 0.8uL of 10uM forward primer, 0.8uL of 10uM reverse primer, 0.3uL 20mg/mL bovine serum albumin, and 3uL template DNA per well. Cycling conditions were: 1 cycle at 95°C, then 40 cycles of 5 seconds at 95°C and 31 seconds of 60°C. Transcript-free negative controls were used in every qPCR run for Wolbachia, 18S, and immune genes.
DNase I (Zymo Research, Irvine, CA, USA) was used to degrade genomic DNA from a subsample of the total nucleic acid extractions. Complementary DNA (cDNA) for use establishing active transcription of immune genes was then created from extracted RNA using SuperScript IV VILO Master Mix (Invitrogen, Carlsbad, CA, USA). Concentrations of cDNA were measured using a Nanodrop-2000 (Thermo Fisher Scientific, Waltham, MA, USA) and were diluted to 2ng/uL in the final reaction mix. For the qPCRs of immune genes and the 18S gene (used here as a housekeeping gene, as done in 30), primers from (30) were used (Table 1). The same PCR reaction mix (though with 8.1uL Milli-Q water and 1uL of template) and cycling conditions as described above were used.
Table 1
| Primer name | 5′–3′ sequence |
|---|---|
| Rel1‐F | GCGACTTTGGCATCAAGCTC |
| Rel1‐R | GTTCGACCGGAGCGTAGTAG |
| Def1‐F | GGTCCAATACTTCGCCAATAC |
| Def1‐R | GATTGGGCGTCAACGATAGT |
| 18S rRNA‐F | CGCGGTAATTCCAGCTCCACTA |
| 18S rRNA‐R | GCATCAAGCGCCACCATATAGG |
Primers from (30) used in qPCRs to analyze the relative abundance of immune genes Rel1 and Def1, as well as the 18S gene utilized for housekeeping and standardization.
Microbiome Sequencing and Preparation
The microbiomes of Culex quinquefasciatus (n = 68; all surviving adults) from the temperature gradient beam experiment were analyzed using 16S V4 rRNA gene region barcoding following the Earth Microbiome Protocol (EMP) (67–72). The 515F (with barcode) and 806R primers were used for amplification, standard 25uL/well reaction PCR recipe and cycling conditions were followed, but 5Prime HotMasterMix (Quantabio, Beverly, MA, USA) was used for PCR in place of the EMP recommended master mix. Post-PCR samples were normalized using a Mag-Bind Pure Library Normalization Kit (Omega Bio-Tek, Norcross, GA, USA). Normalized samples were pooled into a library, quantified using a Qubit 2 Fluorometer (Invitrogen, Carlsbad, CA, USA), and appropriately diluted to follow EMP protocols (73, 74). The library was sequenced using an Illumina MiSeq v2 300-cycle kit (Canton, MA, USA) with the EMP recommended PhiX addition (Illumina, Canton, MA, USA).
Bioinformatic and Statistical Analyses
Mosquito survival across temperature groups in the temperature beam experiment were analyzed using a Chi-Square test in Microsoft Excel. Relative abundances of Wolbachia, Rel1, and Def1 were calculated in Microsoft Excel by standardizing the average qPCR Cq value for each sample and each gene by its corresponding housekeeping 18S Cq value (30). Average Cq values were then scaled by the relative percent change between 18S housekeeping values to adjust and further account for any variance in the starting concentration of DNA or cDNA in each sample. These corrected Cq values were then analyzed in SPSS (IBM SPSS Statistics, v. 26) by testing for normality using Levene’s test and subsequently analyzing the data using either one-way ANOVAs or Kruskal-Wallis tests when data did not meet the criteria for parametric statistics. We used linear regressions to test for correlation of Wolbachia abundance, Rel1, or Def1 with generation, temperature and sex.
Microbiomes of the temperature gradient beam mosquitoes were analyzed using QIIME2. Microbiome data were demultiplexed, deblurred, quality filtered to q-scores of 20, and rarefied to 2500 sequences per sample in QIIME2. Taxonomy was matched using the 2019.7 SILVA pre-trained QIIME2 classifier (75, 76). ANOSIMs and Kruskal-Wallis tests were performed to analyze differences in beta (Bray Curtis dissimilarity matrices) and alpha (sOTU Richness, Shannon Diversity Index) diversity between groups, respectively.
Results
Larval Survival Across the Temperature Gradient Beam
Percentage survival at emergence of Culex quinquefasciatus larvae reared on the temperature gradient beam was determined for each temperature group, as represented by that group’s average temperature over the length of the experiment (Figure 2). Temperature groups with an average temperature over 30°C had markedly lower survival (16.7%) through eclosion when compared to the lower temperatures. The lowest temperature group had the highest survival rate through eclosion, at 83.3% survival. A Chi Square test indicated that there was a significant difference in survival across the temperature groups (X2 = 55.07; df =4; p<0.001).
Figure 2
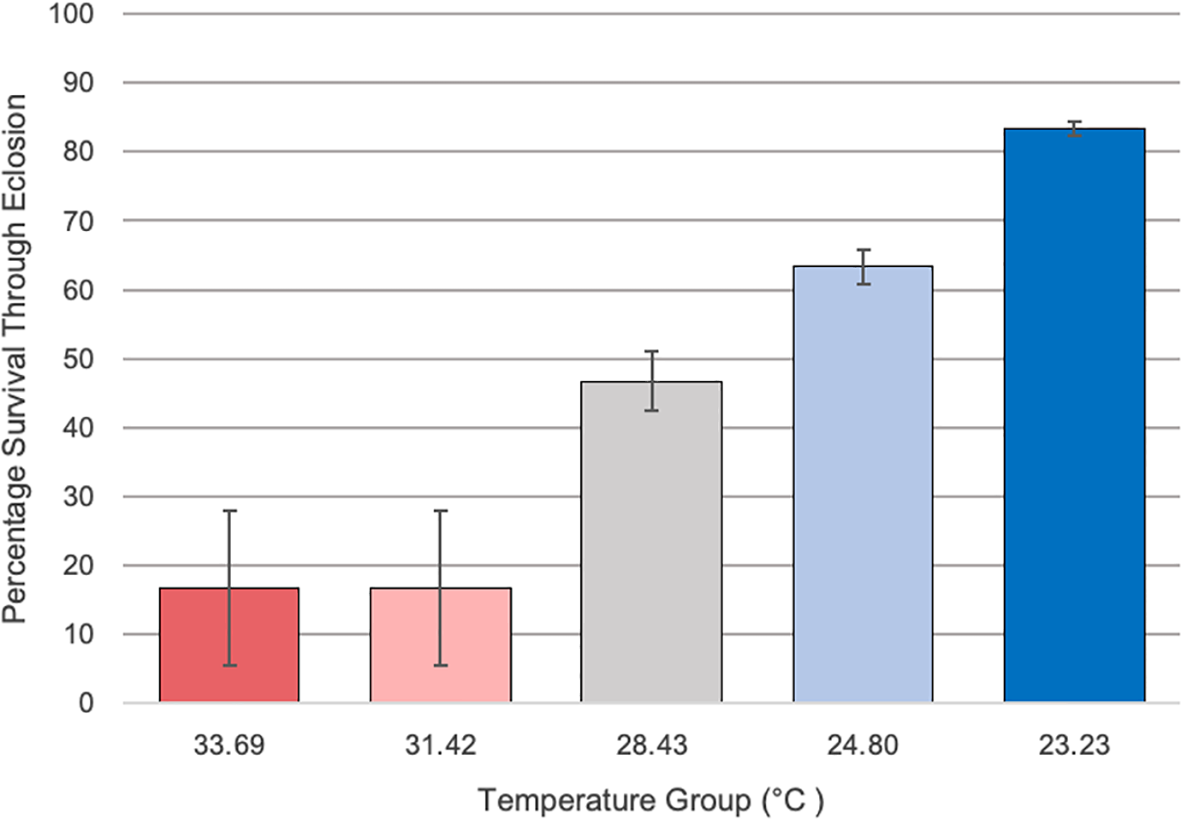
The percentage survival of Culex larvae from second instar through emergence along the various temperature groups on the temperature gradient beam. Temperatures ranged from 33.69 to 23.23°C across sections along the temperature gradient beam. Temperatures over 30°C showed a marked reduction in survival (16.7% survival) through emergence compared to the other groups (83.3% in lowest temperature group). Original experimental setups began with n = 30 and error bars indicate standard error. A Chi Square test indicated that there was a significant difference in survival across the temperature groups (X2 = 55.07; df =4; p<0.001).
Microbiome Diversity Across the Temperature Gradient Beam: sOTU Richness,Shannon Diversity, and Bray Curtis Dissimilarity
Alpha diversity was analyzed using. Kruskal-Wallis tests. There were no significant differences in sOTU richness when analyzed using Kruskal-Wallis tests across temperature groups (n=66, df=4, H=5.057, p= 0.410), sexes (n=65, df=1, H=0.810, p= 0.667), or whether larvae were reared solitary or in groups (n=62, df=1, H= 0.018, p= 0.895). There were also no significant differences in Shannon Diversity index values for the microbiomes between temperature groups (n=66, df=4, H=5.176, p=0.365), sexes (n=65, df=1, H=0.374, p=0.829), or larval grouping conditions (n=62, df=1, H=1.351, p=0.245).
ANOSIMs were run to determine differences between groups in terms of beta diversity. There were no significant differences in Bray Curtis dissimilarity when analyzed using ANOSIMs for temperature groups (n=66, df=4, Test Statistic= 0.067, p= 0.092), sexes (n=65, df=1, Test Statistic= 0.052, p= 0.095), or solitary or grouped larvae (n=62, df= 1, Test Statistic=0.011, p= 0.364). Negative controls (extraction and PCR blanks) were significantly different in pairwise comparisons across all experimental groups and tests than experimental samples, though negative controls were removed prior to running Kruskal-Wallis tests within experimental groups. Prominent genera across all samples, in order of relative frequency, included: Wolbachia, unspecified Enterobacteriaceae, Massilia, Pseudomonas, Bacteriodetes, Aeromonas, unspecified Burkholderiaceae, Elizabethkingia, Pedobacter, and Flectobacillus (Supplementary Figure 1).
Wolbachia and Immune Genes Across Experiments: Temperature Gradient Beam Experiment
Kruskal-Wallis tests were used to analyze differences between Cq values across temperature groups and sexes independently for Wolbachia relative abundance (in the form of the wsp gene), Rel1, and Def1. Levene’s test for homogeneity indicated the need for non-parametric test use. Kruskal-Wallis tests were run in iterations both including Cq values of zero as well as with zeros removed. Tests analyzing temperature groups including the Cq zero values were non-significant overall for Wolbachia (n=68, Test Statistic=1.525, df=4, p=0,822), Rel1 (n=68, Test Statistic=0.283, df=3,p=0.991), and Def1 (n=68, Test Statistic=0.330, df=4, p=0.988). Kruskal-Wallis test iterations with zero values removed also indicated that there was no significant difference between any temperature groups in regard to Wolbachia abundance (n=64, Test Statistic=3.662, df=4, p=0.454; Figure 3), the expression of Rel1 (n=62, Test Statistic=3.496, df=4, p=0.478; Figure 4), or the expression of Def1(n=62, Test Statistic=3.826, df=4, p=0.430; Figure 5). Test iterations with zeros removed also indicated that there was no significant difference between mosquito sex and Wolbachia abundance (n=64, Test Statistic=0.451, df=1, p=0.502), Rel1 expression (n=62, Test Statistic=1.075, df=1, p=0.3), or Def1 expression (n=62, Test Statistic=1.018, df=1, p=0.313). These collectively indicated that there were no differences between temperature groups or sexes for relative Wolbachia abundance, or the relative expression of Rel1 and Def1.
Figure 3
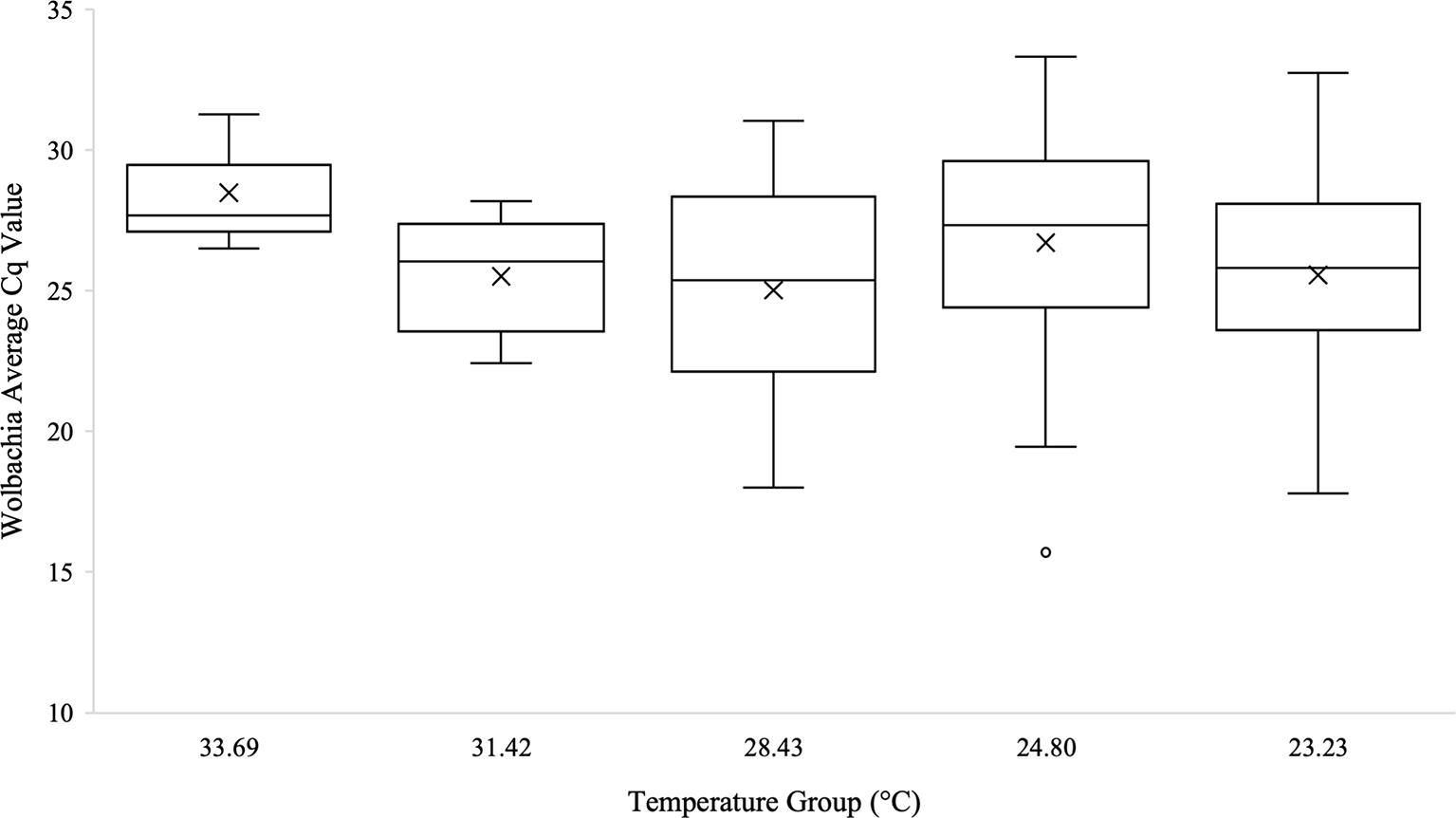
Box and whisker plot of the Wolbachia Cq values from qPCR across temperature groups on the temperature gradient beam. Wolbachia (wsp gene) technical replicates were averaged and all biological replicates that amplified during qPCR are shown across the temperature groups. There was no significant difference in Wolbachia Cq values, which are representative of the Wolbachia abundance, across the temperature beam groups as analyzed by Kruskal-Wallis test (n=64, Test Statistic=3.662, df=4, p=0.454). Mean Cq values for each temperature group are indicated on the plots with an “X”.
Figure 4
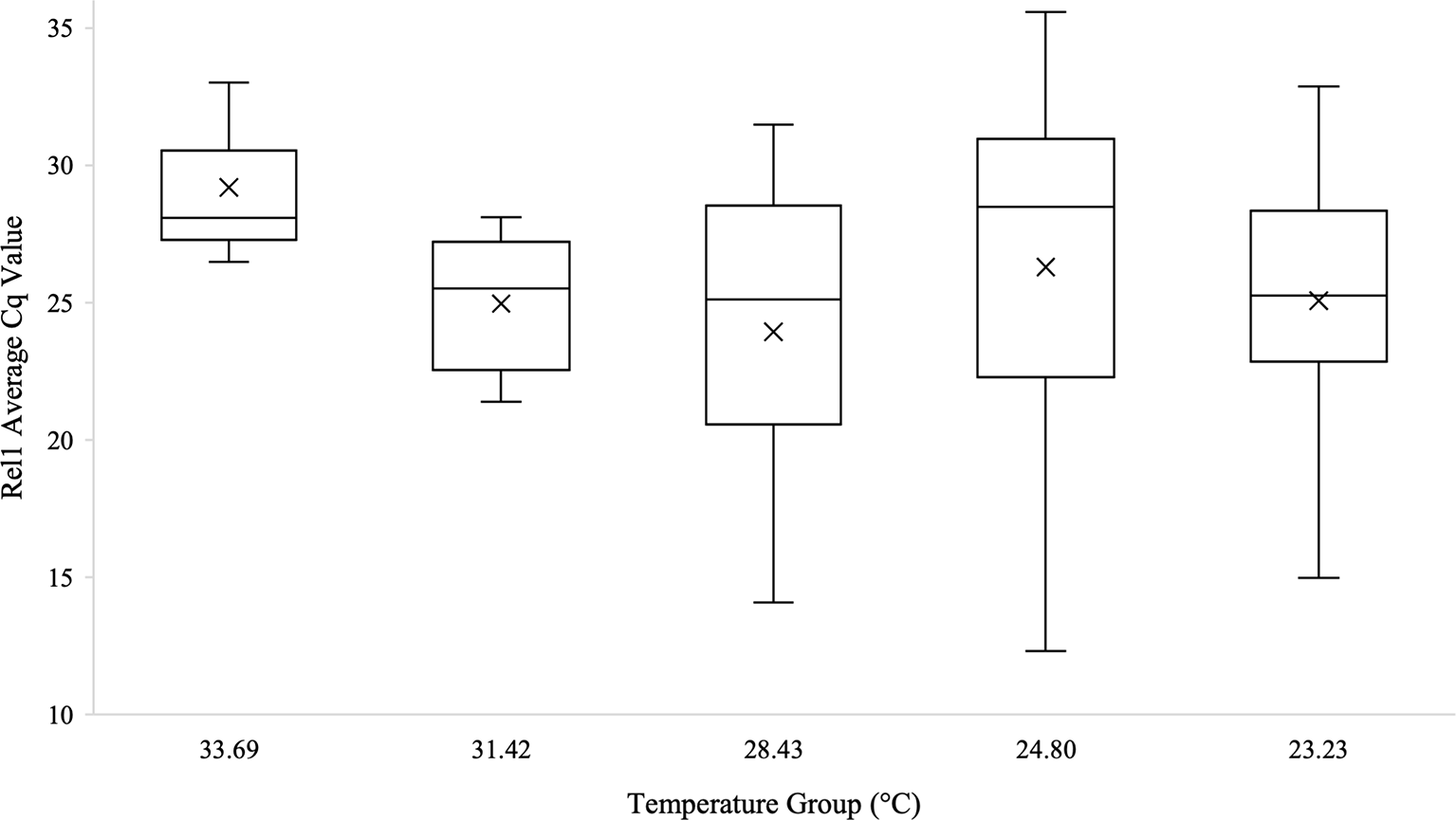
Box and whisker plot of the Rel1 Cq values across temperature groups on the temperature gradient beam. Rel1 expression technical replicates were averaged and all biological replicates that amplified during qPCR are shown across the temperature groups. There was no significant difference in Rel1 Cq values across the temperature beam groups as analyzed by Kruskal-Wallis test (n=62, Test Statistic=3.496, df=4, p=0.478). Mean Cq values for each temperature group are indicated on the plots with an “X”.
Figure 5
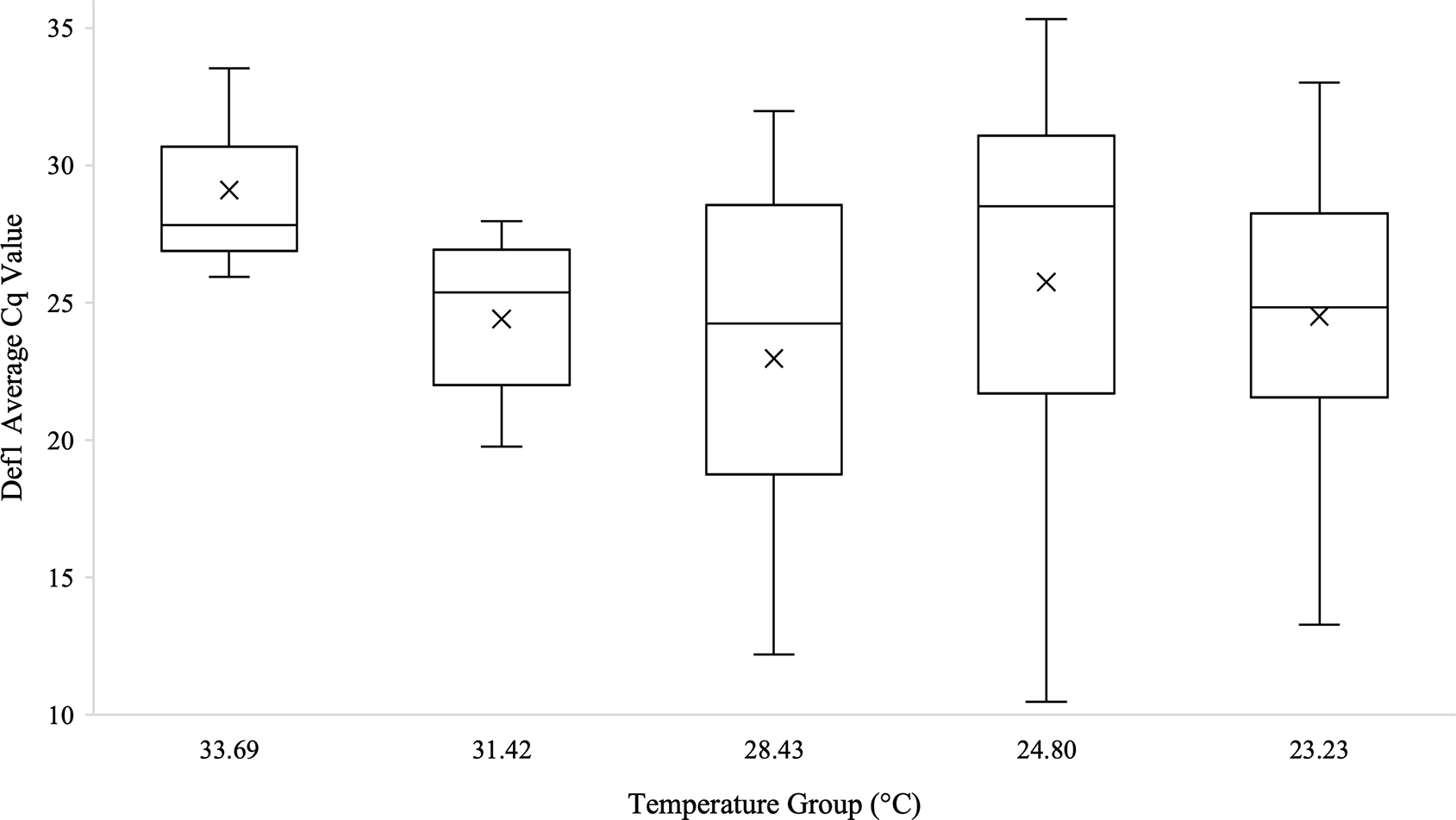
Box and whisker plot of the Def1 Cq values across temperature groups on the temperature gradient beam. Def1 expression technical replicates were averaged and all biological replicates that amplified during qPCR are shown across the temperature groups. There was no significant difference in Def1 Cq values across the temperature beam groups as analyzed by Kruskal-Wallis test (n=62, Test Statistic=3.826, df=4, p=0.430). Mean Cq values for each temperature group are indicated on the plots with an “X”.
Wolbachia and Immune Genes Across Experiments: Transgenerational Temperature Experiment
One-way ANOVAs were run with post-hoc Tukey’s HSD tests for pairwise comparisons (where appropriate) to analyze differences in Wolbachia and immune genes Rel1 and Def1 within and between generations at 23 and 30/28°C (n = 5 randomly and blindly sampled per group). Zero values were removed from analyses, which only removed one sample from Generation 0 at 23 degrees. The ANOVA for Wolbachia abundance (in the form of the wsp gene) was significantly different across temperature groups (n=19, df=2, F=5.110, p=0.019), which the Tukey’s HSD pairwise comparison indicated was driven by the only pairwise significant difference, which was between the 28- and 30-degree temperature groups (n=19, St. Error=0.928, p=0.017, 95% CI=0.5003-5.2877). There was no significant difference between temperature groups for either Rel1 gene expression (n=19, df=2, F=1.022, p=0.382) or Def1 gene expression (n=19, df=2, F=1.072, p=0.366). There was also no significant difference between generations 0 and 1 for Rel1 (n=19, df=18, F=0.525, p=0.479) or Def1 (n=19, df=18, F=0.492, p=0.493), but there was a significant difference in Wolbachia between generations (n=19, df=18, F=5.738, p=0.028).
Additional one-way ANOVAs were run to determine differences between joint temperature groups across generations. Tests indicated that there was no difference between temperature groups and generations for Rel1 expression (n=19, df=3, F=0.680, p=0.578) or Def1 expression (n=19, df=18, F=0.734, p=0.548), but that there was a significant difference across the groups in regard to Wolbachia abundance (n=19, df=18, F=3.326, p=0.048). Interestingly, the Tukey’s HSD Post-Hoc multiple comparisons indicated that there was a significant reduction in Wolbachia wsp gene expression between Generation 0 and Generation 1 at 30/28°C (n=19, St. Error=0.95, p=0.037). There were no significant differences in Wolbachia within generation 0 between temperatures. Linear regression analysis also indicated that Wolbachia abundance is significantly correlated with generation (n=19, R2 = 0.273, F=6.371, p=0.022). However, Rel1 expression (n=19, R2 = 0.048, F=0.401, p=0.676) and Def1 expression (n=19, R2 = 0.044, F=0.372, p=0.695) was not correlated with generation, temperature, or sex.
Moreover, when mosquito sex was analyzed by ANOVA, there was no significant difference for Rel1 (n=19, df=18, F=0.555, p=0.467) or Def1 (n=19, df=18, F=0.538, p=0.473), but there was a significant difference in Wolbachia abundance by sex, with greater abundance occurring in males (n=19, df=18, F=6.017, p=0.025). A linear regression further indicated that there was a correlation between mosquito sex and Wolbachia abundance (n=19, df=18, F=4.170, p=0.035). Additionally, there were no significant differences in Wolbachia abundance (n=14, df=13, F=0.415, p=0.826), Rel1 expression (n=14, df=13, F=0.659, p=0.665), or Def1 expression (n=14, df=13, F= 0.741, p=0.614) among mosquito rearing cage positions in the incubators.
Discussion
Across both the temperature gradient beam and transgenerational experiments in Culex quinquefasciatus, our results indicated that Wolbachia and immune genes Rel1 and Def1 did not differ with temperature within a single generation. Interestingly, Wolbachia relative abundances differed significantly between generations, but only at high temperatures above 28°C, and with no significant change in immune gene expression. Rel1 and Def1 expression did not differ between any experimental groups, but Wolbachia abundance differed by generation and by sex. Differences in Wolbachia abundance by sex were likely due to cytoplasmic compatibility (21, 23, 28, 30). Female mosquitoes and mosquitoes in generation 0 had higher average abundance of Wolbachia. Furthermore, there was no difference in microbiome diversity across mosquitoes in the temperature beam experiment.
Reductions in Wolbachia abundance at high temperatures are fairly well documented in Aedes species (41, 42, 44), particularly in Aedes albopictus where Wolbachia was markedly reduced at 37°C (40). Novakova et al. (39) found that in situ mosquitoes of the Culex pipiens complex had lower abundances of Wolbachia at higher temperatures, and that a reduction in Wolbachia also negatively correlated with West Nile virus (WNV), which is also supported by other works (40, 77, 78). Our results further support both of these findings in Culex pipiens complex mosquitoes, indicating that Wolbachia abundance is generally reduced at high temperatures, similar to findings in a meta-analysis of insect microbiomes (79). Novakova et al. also hypothesized that this reduction of Wolbachia in the Culex pipiens complex differed across generations and may have been influenced mosquito immune function, which is consistent with findings in Aedes mosquitoes (22, 47, 51, 80). Our findings support the reduction of Wolbachia abundance at high temperatures across generations (Figure 6), but do not provide support that Wolbachia influenced immune function in Culex pipiens quinquefasciatus, as we observed no differences in key Toll and IMD pathway genes at varied temperatures.
Figure 6
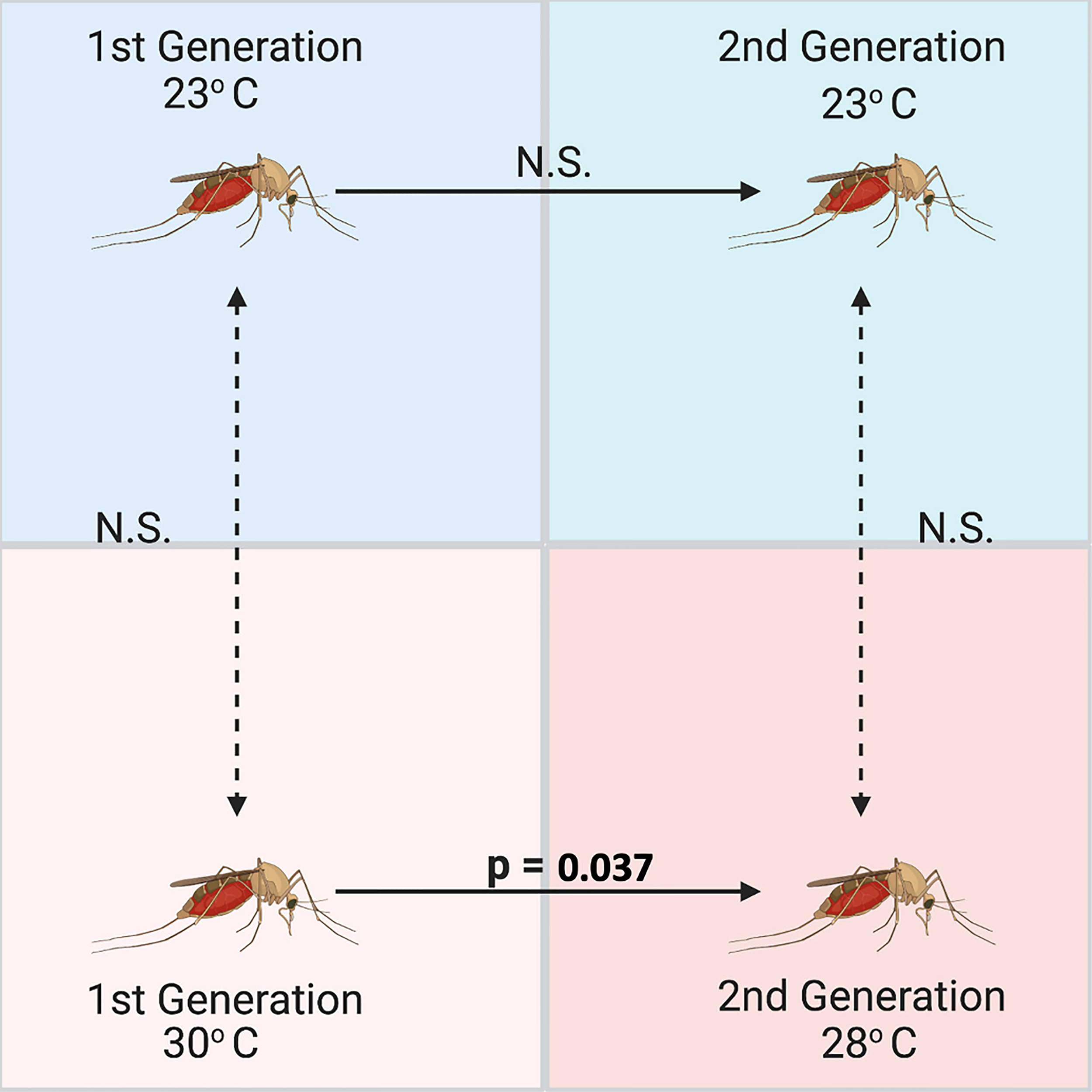
Significant differences across generations between relative abundances of Wolbachia (wsp gene) in Culex quinquefasciatus measured using qPCR and standardized with 18S as a housekeeping gene (n = 5 mosquito adults per group). Culex were reared over two generations at a low (23°C) and high (30/28°C) temperature, but Wolbachia was only significantly different between generations at the high temperature treatment based on an ANOVA comparing the Wolbachia abundances and a subsequent Tukey’s HSD multiple comparisons test (n=19, St. Error=0.95, p=0.037).
Rel1 (homologue to Drosophila dorsal) is an integral part of the Toll pathway that aids in the transcription of innate immune factors (like antimicrobial peptides) and Def1 is part of both the Toll and IMD pathways (48, 51). While no genes of the JAK-STAT pathway were analyzed here, based on the findings of Ant et al. (30), we would not expect to see any changes in this pathway either. They found that immune genes across multiple pathways were neither up- nor down-regulated in relation to Wolbachia infection in Culex quinquefasciatus (30). Their results, taken together with our results and Novakova et al. (39), indicate that temperature has an impact on the abundance of Wolbachia in Culex pipiens quinquefasciatus (Say) over generations but Wolbachia appears unlikely to be altering immune gene expression in turn.
We propose further study of the tripartite interactions between mosquito host, microbiome, and pathogens to help elucidate the mechanism by which high temperatures alter the abundance of Wolbachia (and subsequently, infection with pathogens like WNV). We also propose further investigation into the potential impacts of temperature variations (i.e. seasonal, daily) on the abundance of Wolbachia in Culex mosquitoes. Additionally, we anticipate that there would be interactions between other host factors that have not been accounted for in this study, including competition within the microbiome at varied temperatures, influence of other immune pathways, and potential changes in other signal pathways due to changes in environmental factors (13, 38, 39, 81). Furthermore, we recommend that temperature, weather, and climate conditions be considered when timing the deployment of Wolbachia-based control methods in the field.
Funding
We would like to thank our funders for supporting this work: National Science Foundation grant DGE 1249946, Integrative Graduate Education and Research Traineeship (IGERT): Coasts and Communities – Natural and Human Systems in Urbanizing Environments, a generous donation from Dr. Charles Robertson and Patricia Robertson, the University of Massachusetts Sanofi-Genzyme Doctoral Fellowship, Craig R. Bollinger Memorial Research Grant, National Science Foundation Research Experiences for Undergraduates: Research Experiences in Integrative and Evolutionary Biology Award Number 1950051, US Department of Education Ronald E. McNair Postbaccalaureate Achievement Scholars Program at University of Massachusetts Boston, and the Nancy Goranson Endowment Fund. No funders had a role in the planning, execution, or conclusions of this study.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA809017.
Author contributions
AT-P and DCW conceived, planned, and acquired funding for this study. AT-P, JJ, MF, and SL carried out the experiments. AT-P and JP analyzed the data. AT-P wrote the manuscript with input from the other authors. All authors contributed to the article and approved the submitted version.
Acknowledgments
We would like to thank our collaborators Michael Pollard and Dr. Rob Stevenson for building the temperature gradient beam and training us on operating the beam for this study. Content of this manuscript was previously included in AT-P’s dissertation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2022.762132/full#supplementary-material
Supplementary Figure 1Taxa plot of adult Culex quinquefasciatus microbiomes following emergence across the temperature gradient beam temperature groups. Group A is 33.69°C, Group B is 31.42°C, Group C is 28.43°C, Group D is 24.80°C, and Group E is 23.23°C. The taxa legend includes the twenty highest relative frequency sOTUs at the Genus level. There were no significant differences in sOTU richness by Kruskal-Wallis test (n=66, df=4, H=5.057, p= 0.410), in Shannon diversity by Kruskal-Wallis test (n=66, df=4, H=5.176, p=0.365), or in beta diversity based on Bray-Curtis dissimilarity tested using ANOSIM (n=62, df=4, p= 0.092) between temperature groups.
References
1
Sudomo M Chayabejara S Duong S Hernandez L Wu WP Bergquist R . Elimination of Lymphatic Filariasis in Southeast Asia. Adv Parasitol (2010), 205–33. doi: 10.1016/S0065-308X(10)72008-X
2
Ramaiah KD Das PK Michael E Guyatt HL et al . The Economic Burden of Lymphatic Filariasis in India. Parasitol Today (2000) 16(6):251–3. doi: 10.1016/S0169-4758(00)01643-4
3
Sardelis MR Turell MJ Dohm DJ O’Guinn ML . Vector Competence of Selected North American Culex and Coquillettidia Mosquitoes for West Nile Virus. Emerg Inf Dis (2001) 7:1018–22. doi: 10.3201/eid0706.010617
4
Ciota AT Ehrbar DJ Matacchiero AC Van Slyke GA Kramer LD . The Evolution of Virulence of West Nile Virus in a Mosquito Vector: Implications for Arbovirus Adaptation and Evolution. BMC Evol Biol (2013) 13(1):71. doi: 10.1186/1471-2148-13-71
5
Samy AM Elaagip AH Kenawy MA Ayres CFJ Peterson AT Soliman DE . Climate Change Influences on the Global Potential Distribution of the Mosquito Culex Quinquefasciatus, Vector of West Nile Virus and Lymphatic Filariasis. PLoS One (2016) 11(10):e0163863. doi: 10.1371/JOURNAL.PONE.0163863
6
Gorris ME Bartlow AW Temple SD Romero-Alvarez D Shutt DP Fair JM et al . Updated Distribution Maps of Predominant Culex Mosquitoes Across the Americas. Parasit Vectors (2021) 14(1):1–13. doi: 10.1186/S13071-021-05051-3
7
Lee SH Nam KW Jeong JY Yoo SJ Koh Y-S Lee S et al . The Effects of Climate Change and Globalization on Mosquito Vectors: Evidence From Jeju Island, South Korea on the Potential for Asian Tiger Mosquito (Aedes Albopictus) Influxes and Survival From Vietnam Rather Than Japan. PLoS One (2013) 8(7):e68512. doi: 10.1371/journal.pone.0068512
8
Tjaden NB Caminade C Beierkuhnlein C Thomas SM . Mosquito-Borne Diseases: Advances in Modelling Climate-Change Impacts. Trends Parasitol (2017) 34:227–45. doi: 10.1016/j.pt.2017.11.006
9
Shearer FM Longbottom J Browne AJ Pigott DM Brady OJ Kraemer MUG et al . Existing and Potential Infection Risk Zones of Yellow Fever Worldwide: A Modelling Analysis. Lancet Global Health (2018) 6(3):e270–8. doi: 10.1016/S2214-109X(18)30024-X
10
Kraemer MUG Reiner RC Brady OJ Messina JP Gilbert M Pigott DM et al . Past and Future Spread of the Arbovirus Vectors Aedes Aegypti and Aedes Albopictus. Nat Microbiol (2019) 4(5):854–63. doi: 10.1038/s41564-019-0376-y
11
Liu B Gao X Zheng K Ma J Jiao Z Xiao J et al . The Potential Distribution and Dynamics of Important Vectors Culex Pipiens Pallens and Culex Pipiens Quinquefasciatus in China Under Climate Change Scenarios: An Ecological Niche Modelling Approach. Pest Manag Sci (2020) 76(9):3096–107. doi: 10.1002/PS.5861
12
McMeniman CJ Lane R Cass BN Fong AWC Sidhu M Wang Y-F et al . Stable Introduction of a Life-Shortening Wolbachia Infection Into the Mosquito Aedes Aegypti. Science (New York NY) (2009) 323(5910):141–4. doi: 10.1126/science.1165326
13
Moreira LA Iturbe-Ormaetxe I Jeffery JA Lu G Pyke AT Hedges LM et al . A Wolbachia Symbiont in Aedes Aegypti Limits Infection With Dengue, Chikungunya, and Plasmodium. Cell (2009) 139(7):1268–78. doi: 10.1016/J.CELL.2009.11.042
14
Bian G Joshi D Dong Y Lu P Zhou G Pan X et al . Wolbachia Invades Anopheles Stephensi Populations and Induces Refractoriness to Plasmodium Infection. Science (New York NY) (2013) 340(6133):748–51. doi: 10.1126/science.1236192
15
Hancock PA Sinkins SP Godfray HCJ . Strategies for Introducing Wolbachia to Reduce Transmission of Mosquito-Borne Diseases. PLoS Negl Trop Dis (2011) 5(4):e1024. doi: 10.1371/JOURNAL.PNTD.0001024
16
Hoffmann AA Montgomery BL Popovici J Iturbe-Ormaetxe I Johnson PH Muzzi F et al . Successful Establishment of Wolbachia in Aedes Populations to Suppress Dengue Transmission. Nature (2011) 476(7361):454. doi: 10.1038/nature10356
17
Walker T Johnson PH Moreira LA Iturbe-Ormaetxe I Frentiu FD McMeniman CJ et al . The Wmel Wolbachia Strain Blocks Dengue and Invades Caged Aedes Aegypti Populations. Nature (2011) 476(7361):450–3. doi: 10.1038/nature10355
18
Kamtchum-Tatuene J Makepeace BL Benjamin L Baylis M Solomon T . The Potential Role of Wolbachia in Controlling the Transmission of Emerging Human Arboviral Infections. Curr Opin Infect Dis (2017) 30:108–16. doi: 10.1097/QCO.0000000000000342
19
O’Neill SL . The Use of Wolbachia by the World Mosquito Program to Interrupt Transmission of Aedes Aegypti Transmitted Viruses. In: Dengue and Zika: Control and Antiviral Treatment Strategies. Singapore: Springer (2018). p. 355–60. doi: 10.1007/978-981-10-8727-1_24
20
Tokash-Peters AG Tokash IW Campos AJ Woodhams DC . Developing Effective Mosquito Control Strategies by Utilizing Vector Mosquito Life Histories and Ecology. Case Stud Environ (2019) 3:1–12. doi: 10.1525/cse.2018.001743
21
Zabalou S Riegler M Theodorakopoulou M Stauffer C Savakis C Bourtzis K . Wolbachia-Induced Cytoplasmic Incompatibility as a Means for Insect Pest Population Control. Proc Natl Acad Sci USA (2004) 101(42):15042–5. doi: 10.1073/pnas.0403853101
22
Bian G Xu Y Lu P Xie Y Xi Z . The Endosymbiotic Bacterium Wolbachia Induces Resistance to Dengue Virus in Aedes Aegypti. PLoS Pathog (2010) 6(4):e1000833. doi: 10.1371/journal.ppat.1000833
23
Atyame CM Pasteur N Dumas E Tortosa P Tantely ML Pocquet N et al . Cytoplasmic Incompatibility as a Means of Controlling Culex Pipiens Quinquefasciatus Mosquito in the Islands of the South-Western Indian Ocean. PLoS Negl Trop Dis (2011) 5(12). doi: 10.1371/journal.pntd.0001440
24
Blagrove MSC Arias-Goeta C Failloux AB Sinkins SP . Wolbachia Strain Wmel Induces Cytoplasmic Incompatibility and Blocks Dengue Transmission in Aedes Albopictus. Proc Natl Acad Sci USA (2012) 109(1):255–60. doi: 10.1073/pnas.1112021108
25
Beckmann JF Ronau JA Hochstrasser M . A Wolbachia Deubiquitylating Enzyme Induces Cytoplasmic Incompatibility. Nat Microbiol (2017) 2. doi: 10.1038/nmicrobiol.2017.7
26
Laven H . Eradication of Culex Pipiens Fatigans Through Cytoplasmic Incompatibility. Nature (1967) 216(5113):383–4. doi: 10.1038/216383a0
27
Kaur R Shropshire JD Cross KL Leigh B Mansueto AJ Stewart V et al . Living in the Endosymbiotic World of Wolbachia: A Centennial Review. Cell Host Microbe (2021) 29(6):879–93. doi: 10.1016/J.CHOM.2021.03.006
28
Calvitti M Moretti R Skidmore AR Dobson SL . Wolbachia Strain Wpip Yields a Pattern of Cytoplasmic Incompatibility Enhancing a Wolbachia-Based Suppression Strategy Against the Disease Vector Aedes Albopictus. Parasit Vectors (2012) 5(1):1–9. doi: 10.1186/1756-3305-5-254/FIGURES/2
29
Chen L Zhu C Zhang D . Naturally Occurring Incompatibilities Between Different Culex Pipiens Pallens Populations as the Basis of Potential Mosquito Control Measures. PLoS Negl Trop Dis (2013) 7(1):e2030. doi: 10.1371/JOURNAL.PNTD.0002030
30
Ant TH Herd C Louis F Failloux AB Sinkins SP . Wolbachia Transinfections in Culex Quinquefasciatus Generate Cytoplasmic Incompatibility. Insect Mol Biol (2019) 29(1):1–8. doi: 10.1111/imb.12604
31
Barr AR . Cytoplasmic Incompatibility in Natural Populations of a Mosquito, Culex Pipiens L. [17]. Nature (1980) 71–2. doi: 10.1038/283071a0
32
Sinkins SP Walker T Lynd AR Steven AR Makepeace BL Godfray HCJ et al . Wolbachia Variability and Host Effects on Crossing Type in Culex Mosquitoes. Nature (2005) 436(7048):257–60. doi: 10.1038/nature03629
33
Dumas E Atyame CM Malcolm CA Le Goff G Unal S Makoundou P et al . Molecular Data Reveal a Cryptic Species Within the Culex Pipiens Mosquito Complex. Insect Mol Biol (2016) 25(6):800–9. doi: 10.1111/imb.12264
34
Hegde S Khanipov K Albayrak L Golovko G Pimenova M Saldaña MA et al . Microbiome Interaction Networks and Community Structure From Laboratory-Reared and Field-Collected Aedes Aegypti, Aedes Albopictus, and Culex Quinquefasciatus Mosquito Vectors. Front Microbiol (2018) 9:2160. doi: 10.3389/fmicb.2018.02160
35
Cansado-Utrilla C Zhao SY McCall PJ Coon KL Hughes GL . The Microbiome and Mosquito Vectorial Capacity: Rich Potential for Discovery and Translation. Microbiome (2021) 9(1):1–11. doi: 10.1186/S40168-021-01073-2
36
Dodson BL Andrews ES Turell MJ Rasgon JL . Wolbachia Enhances West Nile Virus (WNV) Infection in the Mosquito Culex Tarsalis. PLoS Negl Trop Dis (2014) 8(7):e2965. doi: 10.1371/journal.pntd.0002965
37
Dodson BL Hughes GL Paul O Matacchiero AC Kramer LD Rasgon JL . Wolbachia Effects on Rift Valley Fever Virus Infection in Culex Tarsalis Mosquitoes. PLoS Negl Trop Dis (2017) 11(10):e0006050. doi: 10.1371/journal.pntd.0006050
38
Hughes GL Dodson BL Johnson RM Murdock CC Tsujimoto H Suzuki Y et al . Native Microbiome Impedes Vertical Transmission of Wolbachia in Anopheles Mosquitoes. Proc Natl Acad Sci (2014) 111(34):12498–503. doi: 10.1073/pnas.1408888111
39
Novakova E Woodhams DC Rodríguez-Ruano SM Brucker RM Leff JW Maharaj A et al . Mosquito Microbiome Dynamics, a Background for Prevalence and Seasonality of West Nile Virus. Front Microbiol (2017) 8:526. doi: 10.3389/fmicb.2017.00526
40
Wiwatanaratanabutr I Kittayapong P . Effects of Crowding and Temperature on Wolbachia Infection Density Among Life Cycle Stages of Aedes Albopictus. J Invertebr Pathol (2009) 102(3):220–4. doi: 10.1016/j.jip.2009.08.009
41
Ross PA Wiwatanaratanabutr I Axford J White V Endersby-Harshman N Hoffmann A . Wolbachia Infections in Aedes Aegypti Differ Markedly in Their Response to Cyclical Heat Stress. PLoS Pathog (2017) 13(1):e1006006. doi: 10.1371/JOURNAL.PPAT.1006006
42
Ross PA Axford JK Yang Q Staunton KM Ritchie SA Richardson KM et al . Heatwaves Cause Fluctuations in Wmel Wolbachia Densities and Frequencies in Aedes Aegypti. PLoS Negl Trop Dis (2020) 14(1):1–18. doi: 10.1371/journal.pntd.0007958
43
Ulrich JN Beier JC Devine GJ Hugo LE . Heat Sensitivity of Wmel Wolbachia During Aedes Aegypti Development. PLoS Negl Trop Dis (2016) 10(7):e0004873. doi: 10.1371/JOURNAL.PNTD.0004873
44
Mancini MV Ant TH Herd CS Martinez J Murdochy SM Gingell DD et al . High Temperature Cycles Result in Maternal Transmission and Dengue Infection Differences Between Wolbachia Strains in Aedes Aegypti. mBio (2021) 12(6):e0025021. doi: 10.1128/MBIO.00250-21
45
Kambris Z Cook PE Phuc HK Sinkins SP . Immune Activation by Life-Shortening Wolbachia and Reduced Filarial Competence in Mosquitoes. Science (2009) 326(5949):134–6. doi: 10.1126/science.1177531
46
Pinto SB Mariconti M Bazzocchi C Bandi C Sinkins SP . Wolbachia Surface Protein Induces Innate Immune Responses in Mosquito Cells. BMC Microbiol (2012) 12(SUPPL. 1):1–6. doi: 10.1186/1471-2180-12-S1-S11
47
Rancès E Ye YH Woolfit M McGraw EA O’Neill SL . The Relative Importance of Innate Immune Priming in Wolbachia-Mediated Dengue Interference. PLoS Pathog (2012) 8(2):e1002548. doi: 10.1371/journal.ppat.1002548
48
Sim S Jupatanakul N Dimopoulos G . Mosquito Immunity Against Arboviruses. Viruses (2014), 4479–504. doi: 10.3390/v6114479
49
Pan X Pike A Joshi D Bian G McFadden MJ Lu P et al . The Bacterium Wolbachia Exploits Host Innate Immunity to Establish a Symbiotic Relationship With the Dengue Vector Mosquito Aedes Aegypti. ISME J (2018) 12(1):277. doi: 10.1038/ISMEJ.2017.174
50
Zhang D Wang Y He K Yang Q Gong M Ji M et al . Wolbachia Limits Pathogen Infections Through Induction of Host Innate Immune Responses. PLoS One (2020) 15(2):e0226736. doi: 10.1371/JOURNAL.PONE.0226736
51
Xi Z Ramirez JL Dimopoulos G . The Aedes Aegypti Toll Pathway Controls Dengue Virus Infection. PLoS Pathog (2008) 4(7):e1000098. doi: 10.1371/journal.ppat.1000098
52
Lemaitre B Nicolas E Michaut L Reichhart JM Hoffmann JA . The Dorsoventral Regulatory Gene Cassette Spatzle/Toll/Cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell (1996) 86(6):973–83. doi: 10.1016/S0092-8674(00)80172-5
53
Costa A Jan E Sarnow P Schneider D . The Imd Pathway Is Involved in Antiviral Immune Responses in Drosophila. PLoS One (2009) 4(10):e7436. doi: 10.1371/journal.pone.0007436
54
Hillyer JF . Mosquito Immunity. In: Invertebrate Immunity (2010). p. 218–38.
55
Hillyer JF . Insect Immunology and Hematopoiesis. Dev Comp Immunol (2016) 58:102–18. doi: 10.1016/j.dci.2015.12.006
56
Ciota AT Matacchiero AC Kilpatrick AM Kramer LD . The Effect of Temperature on Life History Traits of Culex Mosquitoes. J Med Entomol (2014) 51(1):55–62. doi: 10.1603/ME13003
57
Moore FC Obradovich N Lehner F Baylis P . Rapidly Declining Remarkability of Temperature Anomalies may Obscure Public Perception of Climate Change. Proc Natl Acad Sci USA (2019) 116(11):4905–10. doi: 10.1073/pnas.1816541116
58
Mordecai EA Caldwell JM Grossman MK Lippi CA Johnson LR Neira M et al . Thermal Biology of Mosquito-Borne Disease. Ecol Lett (2019) 22(10):1690–708. doi: 10.1111/ele.13335
59
Boukal DS Bideault A Carreira BM Sentis A . Species Interactions Under Climate Change: Connecting Kinetic Effects of Temperature on Individuals to Community Dynamics. Curr Opin Insect Sci (2019) 35:88–95. doi: 10.1016/j.cois.2019.06.014
60
Harvey JA Heinen R Gols R Thakur MP . Climate Change-Mediated Temperature Extremes and Insects: From Outbreaks to Breakdowns. Global Change Biol (2020) 26:6685–701. doi: 10.1111/gcb.15377
61
Arnold PA Levin SC Stevanovic AL Johnson KN . Drosophila Melanogaster Infected With Wolbachia Strain Wmelcs Prefer Cooler Temperatures. Ecol Entomol (2018) 44:287–90. doi: 10.1101/352872
62
Chrostek E Martins N Marialva MS Teixeira L . Wolbachia -Conferred Antiviral Protection Is Determined by Developmental Temperature. mBio (2021) 12:e0292320. doi: 10.1128/MBIO.02923-20
63
Hague MTJ Shropshire JD Caldwell CN Statz JP Stanek KA Conner WR et al . Temperature Effects on Cellular Host-Microbe Interactions Explain Continent-Wide Endosymbiont Prevalence. Curr Biol (2021). doi: 10.1016/J.CUB.2021.11.065
64
Hague MTJ Caldwell CN Cooper BS . Pervasive Effects of Wolbachia on Host Temperature Preference. mBio (2020) 11(5):1–15. doi: 10.1128/MBIO.01768-20
65
Ware-Gilmore F Sgrò CM Xi Z Dutra HLC Jones MJ Shea K et al . Microbes Increase Thermal Sensitivity in the Mosquito Aedes Aegypti, With the Potential to Change Disease Distributions. PLoS Negl Trop Dis (2021) 15(7):e0009548. doi: 10.1371/JOURNAL.PNTD.0009548
66
Zha X Zhang W Zhou C Zhang L Xiang Z Xia Q . Detection and Characterization of Wolbachia Infection in Silkworm. Genet Mol Bio (2014) 37:573–80. doi: 10.1590/S1415-47572014000400014
67
Caporaso JG Lauber CL Walters WA Berg-Lyons D Lozupone CA Turnbaugh PJ et al . Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences Per Sample. Proc Natl Acad Sci USA (2011) 108(SUPPL. 1):4516–22. doi: 10.1073/pnas.1000080107
68
Caporaso JG Lauber CL Walters WA Berg-Lyons D Huntley J Fierer N et al . Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J (2012) 6(8):1621–4. doi: 10.1038/ismej.2012.8
69
Apprill A McNally S Parsons R Weber L . Minor Revision to V4 Region SSU rRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquat Microb Ecol (2015) 75(2):129–37. doi: 10.3354/ame01753
70
Parada AE Needham DM Fuhrman JA . Every Base Matters: Assessing Small Subunit rRNA Primers for Marine Microbiomes With Mock Communities, Time Series and Global Field Samples. Environ Microbiol (2016) 18(5):1403–14. doi: 10.1111/1462-2920.13023
71
Walters W Hyde ER Berg-Lyons D Ackermann G Humphrey G Parada A et al . Improved Bacterial 16s rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems (2016) 1(1):e00009–15. doi: 10.1128/msystems.00009-15
72
Knight R Vrbanac A Taylor BC Aksenov A Callewaert C Debelius J et al . Best Practices for Analysing Microbiomes. Nat Rev Microbiol (2018) 16:410–22. doi: 10.1038/s41579-018-0029-9
73
Thompson LR Sanders JG McDonald D Amir A Ladau J Locey KJ et al . A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity. Nature (2017) 551(7681):457–63. doi: 10.1038/nature24621
74
The Earth Microbiome Project- Protocols and Standards (2020). Available at: http://www.earthmicrobiome.org/protocols-and-standards/16s/.
75
Quast C Pruesse E Yilmaz P Gerken J Schweer T Yarza P et al . The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res (2012) 41(D1):D590–6. doi: 10.1093/nar/gks1219
76
Yilmaz P Parfrey LW Yarza P Gerken J Pruesse E Quast C et al . The SILVA and ‘All-Species Living Tree Project (LTP)’ Taxonomic Frameworks. Nucleic Acids Res (2014) 42(Database issue):D643–8. doi: 10.1093/nar/gkt1209
77
Smartt CT Shin D Anderson SL . The Effect of West Nile Virus Infection on the Midgut Gene Expression of Culex Pipiens Quinquefasciatus Say (Diptera: Culicidae). Insects (2016) 7(4). doi: 10.3390/insects7040076
78
Farkas JZ Gourley SA Liu R Yakubu AA . Modelling Wolbachia Infection in a Sex-Structured Mosquito Population Carrying West Nile Virus. J Math Biol (2017) 75(3):621–47. doi: 10.1007/s00285-017-1096-7
79
Woodhams DC Bletz MC Becker CG Bender HA Buitrago-Rosas D Diebboll H et al . Host-Associated Microbiomes Are Predicted by Immune System Complexity and Climate. Genome Biol (2020) 21(1):1–20. doi: 10.1186/s13059-019-1908-8
80
Wang YH Chang MM Wang XL Zheng AH Zou Z . The Immune Strategies of Mosquito Aedes Aegypti Against Microbial Infection. Dev Comp Immunol (2018) 83:12–21. doi: 10.1016/j.dci.2017.12.001
81
Jupatanakul N Sim S Dimopoulos G . The Insect Microbiome Modulates Vector Competence for Arboviruses. Viruses (2014) 6(11):4294–313. doi: 10.3390/v6114294
Summary
Keywords
climate change, culex quinquefasciatus, disease ecology, vector-borne disease, Wolbachia
Citation
Tokash-Peters AG, Jabon JD, Fung ME, Peters JA, Lopez SG and Woodhams DC (2022) Trans-Generational Symbiont Transmission Reduced at High Temperatures in a West Nile Virus Vector Mosquito Culex quinquefasciatus. Front. Trop. Dis 3:762132. doi: 10.3389/fitd.2022.762132
Received
21 August 2021
Accepted
28 February 2022
Published
14 April 2022
Volume
3 - 2022
Edited by
Kounbobr Roch Dabiré, Research Institute for Health Science-Direction Régionale de l’Ouest (IRSS-DRO), Burkina Faso
Reviewed by
Adrian Diaz, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina; Olivier Gnankiné, University of Ouagadougou, Burkina Faso; Etiennne Bilgo, Research Institute for Health Sciences (IRSS), Burkina Faso
Updates
Copyright
© 2022 Tokash-Peters, Jabon, Fung, Peters, Lopez and Woodhams.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amanda G. Tokash-Peters, amanda.tokash-peters@centenaryuniversity.edu
†ORCID: Amanda G. Tokash-Peters, orcid.org/0000-0002-9274-6218; Megan E. Fung, orcid.org/0000-0002-9053-0915; Jessica A. Peters, orcid.org/0000-0003-0526-2280; Douglas C. Woodhams, orcid.org/0000-0003-4559-1046
This article was submitted to Vector Biology, a section of the journal Frontiers in Tropical Diseases
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.