- 1Department of Internal Medicine, Kampala International University, Kampala, Uganda
- 2Department of Medicine, Kisumu County Referral Hospital, Kisumu, Kenya
- 3Department of Microbiology and Immunology, Kampala International University, Kampala, Uganda
- 4Department of Research and Training, Jaramogi Oginga Odinga Teaching and Referral Hospital, Kisumu, Kenya
- 5Department of Medical Services, Public Health and Sanitation, County Government of Kisumu, Kisumu, Kenya
Introduction: There is paucity of data on the prevalence, bacterial profile, antimicrobial resistance (AMR) pattern, and predictors of blood culture positive sepsis among adults admitted at Jinja Regional Referral Hospital (JRRH). The aim of the present study was to determine the prevalence, bacterial profile, AMR pattern, and predictors of blood culture positive sepsis among adults admitted at JRRH.
Methods: This was a cross-sectional study between June and August 2023 where patients were recruited using the quick Sepsis Related Organ Failure Assessment (qSOFA) criteria. Data (Sociodemographic and clinical), and blood was collected from eligible patients. Blood was incubated, sub-cultured, and their antimicrobial susceptibility was determined using the disc diffusion method. Data was analyzed on the statistical package for the social sciences (SPSS version 26) and summarized using descriptive statistics and logistic regression.
Results: Most patients were male (132/261, 50.6%), aged 18-36 (155/261, 71.6%), single (146/261, 55.9%), Christian (224/261, 85.8%), had secondary level of education (129/261, 49.4%), were students (93/261, 35.6%), and consumed alcohol (35/261, 13.4%). The prevalence of blood culture-positive sepsis was 53 out of 261 (20.32%), the most isolated pathogens were Staphylococcus aureus (20.8%, 11/53), Streptococcus spp. (18.9%, 10/53), and Klebsiella pneumoniae (13.2%, 7/53). S. aureus exhibited 100% resistance to gentamicin, tetracycline, and minocycline. Streptococci spp was resistant to tetracycline, while K. pneumoniae was resistant to gentamicin. Blood culture positive sepsis was associated with the duration of symptoms for>5 days (aOR=2.010, CI=1.602-6.705, p=0.026), central nervous system (CNS) symptoms (aOR=3.058, CI=1.365-6.849, p=0.007), and low peripheral oxygen saturation (aOR=3.837, CI=1.733-8.496, p=0.001).
Conclusion: The findings suggest that blood culture positive sepsis occurred most frequently among male patients, with most patients aged 18–36 years, though further analysis is needed to establish strong demographic associations. The findings highlight notable antimicrobial resistance in Staphylococcus aureus, Streptococcus spp., and Klebsiella pneumoniae, which should prompt further investigation into antimicrobial stewardship programs. Furthermore, prolonged symptom manifestation, CNS involvement, and hypoxemia seem to be key predictors of blood culture positive sepsis in the study area. Public health interventions in the study area should focus on improving early detection and management of sepsis, with a particular emphasis on promoting awareness in high-risk groups.
Background
Sepsis is a life-threatening condition resulting from the body’s dysregulated response to infection (1–4). It is typically managed with source control, antimicrobial therapy, and supportive care for organ function (5, 6). A 2017 study by Rudd et al. reported that there were 11.0 million sepsis related fatalities, and 48.9 million sepsis cases globally with the highest burden recorded in sub-Saharan Africa, Oceania, and South Asia (7). Sepsis is a major cause of morbidity and mortality in sub-Saharan Africa, with limited data available on its bacterial etiology and resistance profiles (8, 9). Studies on the burden of sepsis in Uganda have reported values ranging from 11% to 14.7% (10, 11). Moreover, Krepiakevich and colleagues investigated the economic burden of sepsis in Uganda and reported that the mean out-of-pocket costs of managing sepsis was $124.50 at private facilities and $ 44.60 for post-discharge care, forcing many families to sell assets, particularly livestock, to cover hospital expenses (12). Moreover, pathogens which are responsible for sepsis in Uganda are increasingly developing resistance to commonly used antimicrobials (10, 13–15). A study at Mulago National Referral Hospital identified Staphylococcus aureus, Escherichia coli, and Klebsiella pneumoniae as predominant pathogens in neonatal sepsis (13). Furthermore, most of the clinical isolates showed resistance to ampicillin, gentamicin, and ceftriaxone (13). Similar observations have been made in other parts of Uganda (10, 14–18).
Jinja Regional Referral Hospital (JRRH) was established in the 1930’s as a health unit for World War II combatants and prisoners of war from Kimaka (MoH, Uganda) (19). It then went on to become a district hospital and was upgraded to a regional referral for Eastern Uganda in 1995 (MoH, Uganda) (19). The hospital currently serves 4.5 million people across a vast catchment area comprising one city and eleven districts (MoH, Uganda) (19). Despite the growing burden of sepsis in Uganda, there is limited information on the prevalence, bacterial etiology, and antimicrobial resistance patterns at Jinja Regional Referral Hospital. This study addresses these gaps and aims to provide valuable insights for better clinical management and public health interventions.
Materials and methods
Study setting
JRRH is a regional referral hospital In Eastern Uganda and is located 110km from Kampala. The hospital receives patients from several districts within Busoga, Central, Mbale, and some parts of Kenya. It is a 500-bed hospital with a bed occupancy of 42% (210/500) adults per daily. The hospital has a functional intensive care unit, an accidents and emergency department, and an internal medicine department. Patients with sepsis present at the accidents and emergency department and are initially evaluated and admitted to the medical or intensive care unit. Blood culture and antimicrobial sensitivity testing is carried out by a qualified microbiologist at the hospital laboratory and sepsis is managed by physicians and senior house officers.
Inclusion and exclusion criteria
All patients aged 18 years and above with at least 2 of the Quick Sepsis Related Organ Failure Assessment (qSOFA) criteria i.e. respiratory rate ≥22, altered mental status, systolic blood pressure ≤100 and at the intensive care unit or medical, accident and emergency departments at JRRH during the study period were recruited. Patients who had used antibiotics in the previous 2 weeks, trauma and surgical patients, those who didn’t consent, and those who could not understand English or Lusoga were excluded. Although the use of translators was considered, it was not feasible due to resource constraints.
Sample size
The Leslie Kish Formula was used to calculate sample size (20)
Where, N= Estimated sample size required for the study
Z-alpha = 1.96, standard normal value corresponding to 95% confidence level
p= Estimated prevalence of Sepsis among adult patients = 21.7% (21)
d = Precision (0.05)
Substituting,
The sample size required was 261.
Sampling technique
Consecutive sampling was conducted on all patients with suspected sepsis until the sample size of 261 participants was achieved. Patients who scored 2 or 3 based on the qSOFA criteria (Respiratory rate ≥22, Altered mental status, Systolic blood pressure ≤100) were approached and the aims of the study was communicated to them. Informed consent was then administered for patients willing to participate.
Data collection
A study questionnaire was designed to collect sociodemographic and clinical characteristics from patients or their caregivers. Patient codes were used to maintain confidentiality and the validity and reliability of the questionnaire was tested through a pilot study at the Kampala International University Teaching and Referral Hospital (KIU-TH). Questionnaire validity was evaluated using content validation using two physicians who analyzed the domains and items in the questionnaire and assigned scores. The threshold for the content validity index was set at 0.80 (22). Questionnaire reliability was evaluated on 10 patients at KIU-TH based on a Cronbach’s alpha of <0.70.
Sample collection
Ten (10) ml of blood was collected aseptically into two separate blood culture bottles for aerobic and anaerobic blood culturing. Four milliliters of blood were collected into purple top ethylene diamine tetra acetic acid (EDTA) vacutainers for HIV and complete blood count (CBC) testing. Random blood sugar was also conducted at bedside.
Sample analysis
Laboratory analysis was carried out at the microbiology laboratory of JRRH. Blood culture bottle details were entered in the Laboratory information system, and the culture bottles were onboarded onto the blood culture machine (BACTEC FX40). The blood culture bottles were incubated at 37°C for up to 7 days, with the BACTEC FX40 machine monitoring for bacterial growth. Once flagged as positive, subcultures were performed on blood agar, chocolate agar, and MacConkey agar. The cultures were incubated at 37°C and examined for bacterial growth after 24–48 hours. The Kirby-Bauer disc diffusion method was used to determine the susceptibility of the microorganisms to standard antibiotic discs (23, 24). Interpretation of the zones of inhibition was done based on the Clinical Laboratory Standard Institute (CLSI) criteria (25).
Data analysis
Data on sociodemographic variables were summarized using descriptive statistics on Statistical Package for the Social Sciences (SPSS Inc., Chicago, USA, version 26.0 for Windows). Cross tabulation was used to report the sensitivities of the different microorganisms to the tested antibiotics. Bivariable and multivariable analysis was conducted using binary logistic regression reporting both odds ratios and P values. Variables with p values ≤ 0.2 after bivariable analysis were reanalyzed multivariably using backward stepwise binary logistic regression. p≤ 0.05 was considered significant.
Results
Recruitment of study participants
Two hundred and eighty (280) adult patients who met the qSOFA criteria were managed at Jinja Regional Referral Hospital (JRRH) during the study period. Of these, 16 had taken antibiotics prior to presentation and were therefore excluded. Of the remaining 264, 3 did not consent to participate in the study. Therefore, 261 patients were recruited into the study but only 53 had blood culture positive sepsis.
Sociodemographic characteristics of the study participants
Table 1 describes the sociodemographic characteristics of study participants. Most of the study participants were male (132/261, 50.6%), had a mean age of 36.2 (SD=17.8), and were from the rural area (187/261, 71.6%). The mean duration of symptoms prior to presentation was 6.1(SD=3.5) days, thirty-two of the study participants were HIV positive with 31 on antiretroviral treatment. All participants had elevated temperature with leukocytosis but normal platelet count.
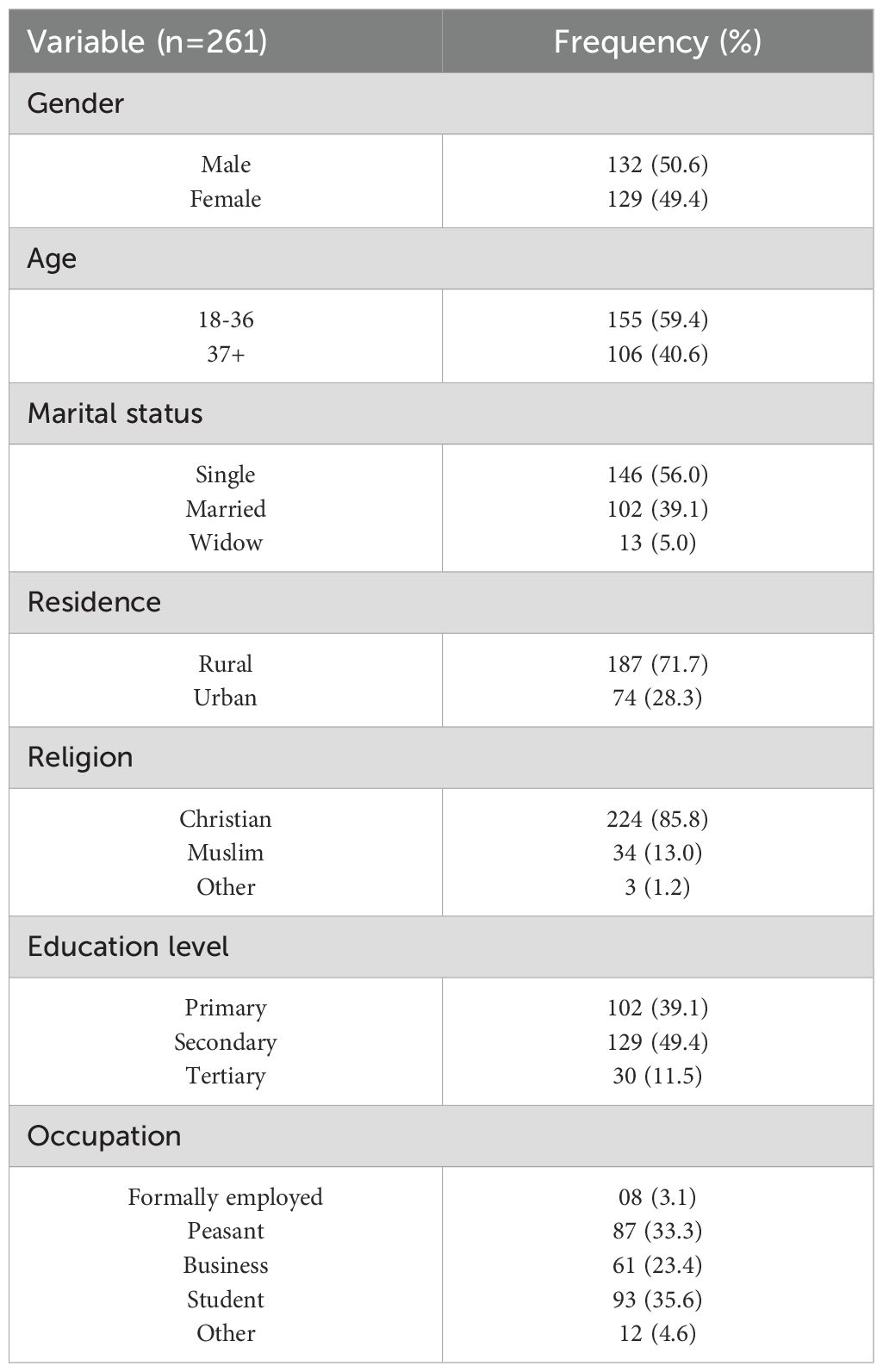
Table 1. Sociodemographic characteristics of study participants at the Jinja Regional Referral Hospital who were recruited into the study during the study period.
Baseline clinical characteristics of the study participants
Table 2 is a summary of the baseline clinical characteristics of the study participants. The mean duration of symptoms prior to presentation was 6.1 days (SD=3.5), with 126 participants (48.3%) experiencing symptoms for more than five days. The most common symptoms were respiratory issues, reported by 187 participants (71.7%), followed by gastrointestinal symptoms in 120 individuals (46.0%), and central nervous system symptoms in 118 participants (45.2%). All participants exhibited leukocytosis, with a white blood cell count exceeding 12 x 109/L. Furthermore, all patients presented with elevated body temperature, and the majority had normal platelet counts.
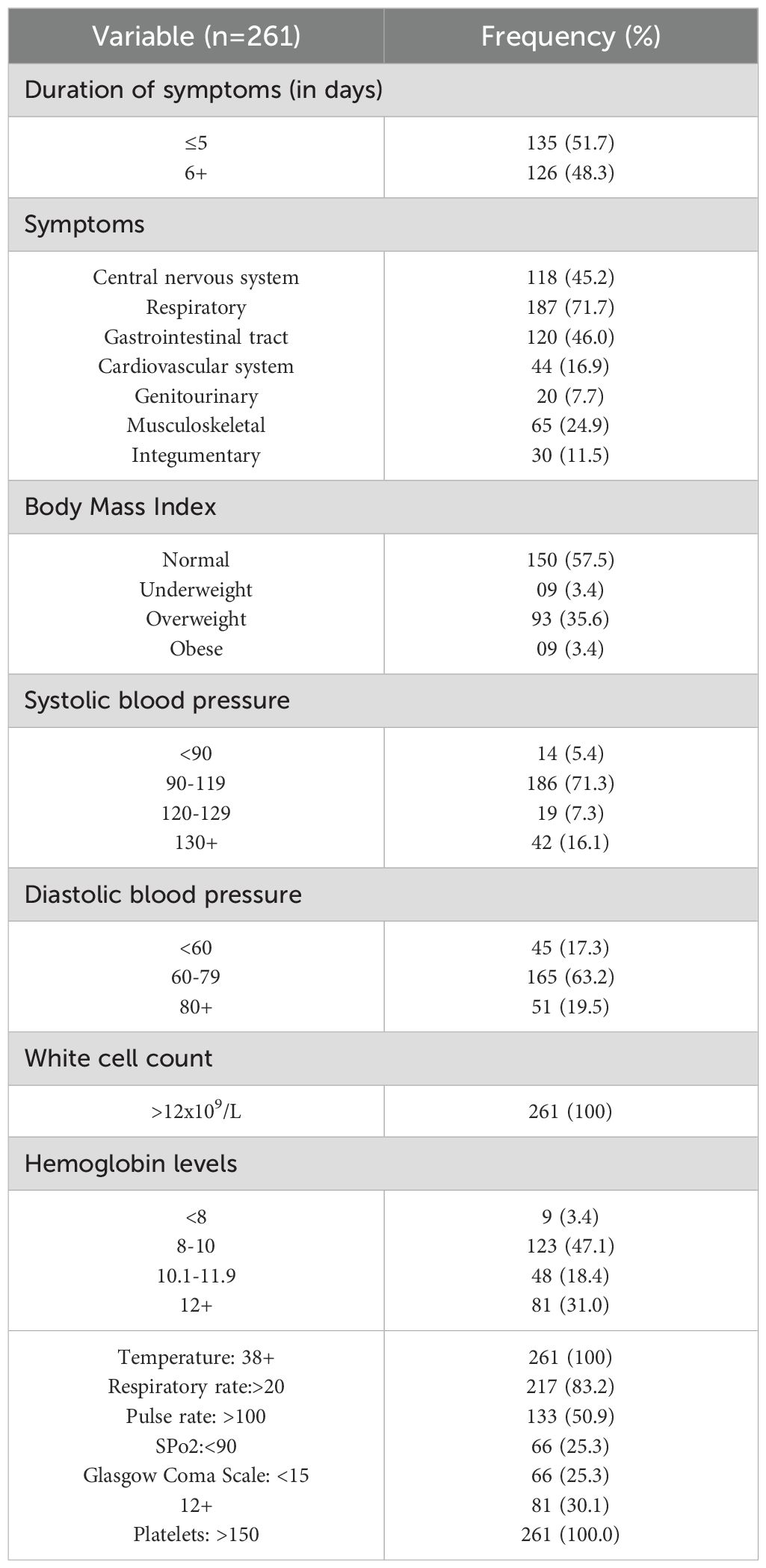
Table 2. Baseline clinical characteristics of study participants who were recruited into the study during the study period.
Table 3 is a summary of the history of chronic illnesses and medication use among the study participants. Of the 261 patients, 84 (32.2%) reported having a chronic illness. Thirty-two participants (12.3%) were HIV positive, of whom 31 were on antiretroviral treatment. Other chronic conditions included diabetes (11.9%), malignancy (3.4%), sickle cell disease (2.7%), and tuberculosis (2.7%). In terms of medication use, 77 participants (29.5%) were receiving treatment for chronic diseases.
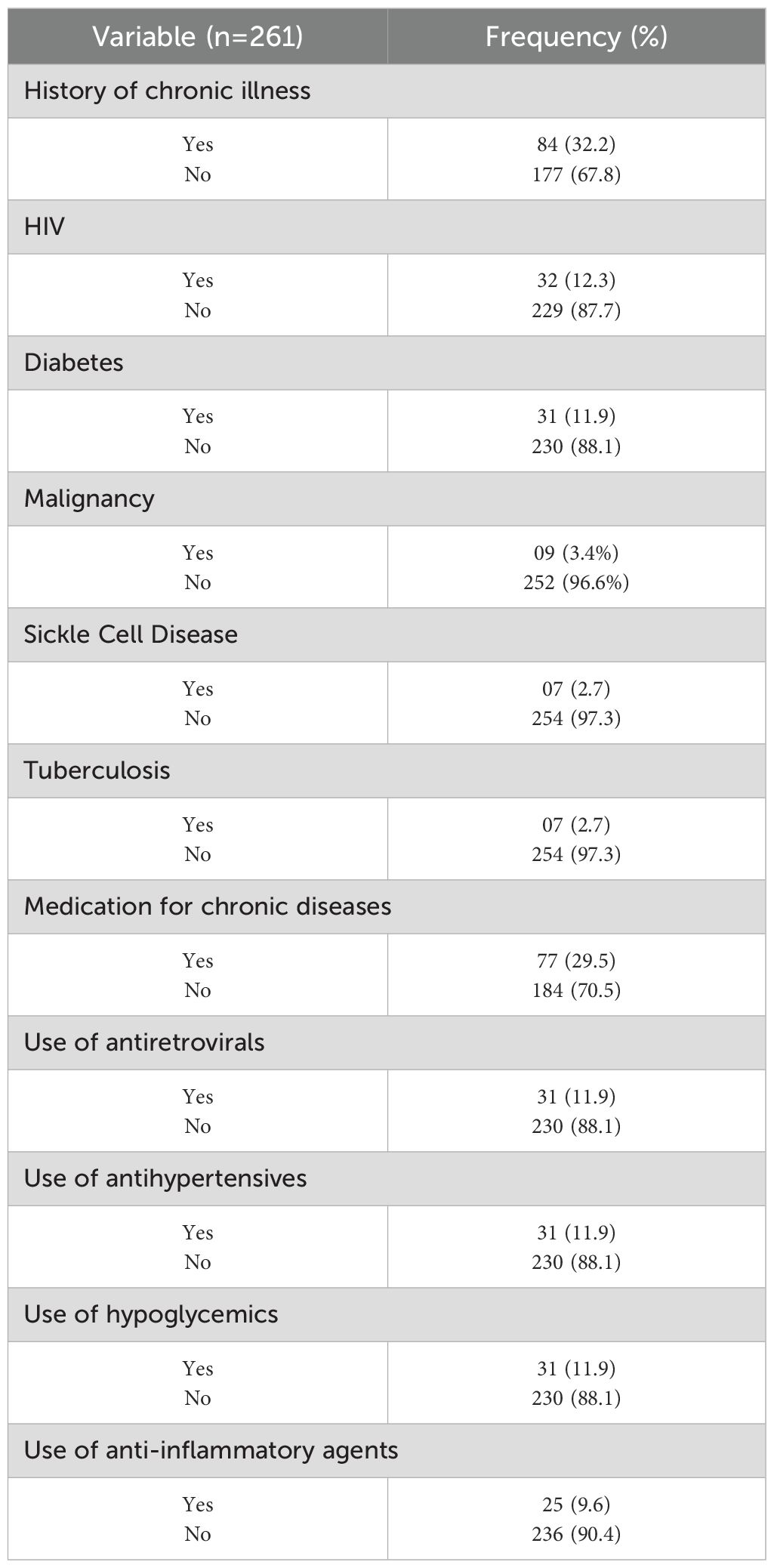
Table 3. Chronic illness and medication history of study participants who were recruited into the study during the period of study.
Profile of bacteria which were isolated from blood samples collected from study participants
Table 4 shows the profile of bacteria isolated from the blood samples collected from study participants at the Jinja Regional Referral Hospital. Bacterial growth was observed in 53/261 (20.31%) of the study participants with Staphylococcus aureus was the most common microorganism isolated from patient blood samples (11/53, 20.8%), followed by Streptococcus spp (10/53, 18.9%), and Klebsiella pneumoniae (7/53, 13.2%).

Table 4. Profile of the bacteria which were isolated from the blood samples of study participants at the Jinja Regional Referral Hospital during the study period.
Resistance patterns of bacteria isolated from the blood samples of study participants
Table 5 shows the resistance patterns of bacteria isolated from study participants at the Jinja Regional Referral Hospital during the study period. Staphylococcus aureus was 100% sensitive to Doxycycline, Penicillin, Azithromycin, Clindamycin and Cefoxitin but completely resistant to Gentamicin, Tetracycline and Minocycline. Streptococcus Spp was 100% sensitive to Levofloxacin, Cefepime, Clindamycin and Erythromycin but completely resistant to Tetracycline. Klebsiella pneumoniae was 100% sensitive to Ciprofloxacin, Imipenem, Cefalexin and Ceftazidime-Tazobactam but completely resistant to Gentamicin.
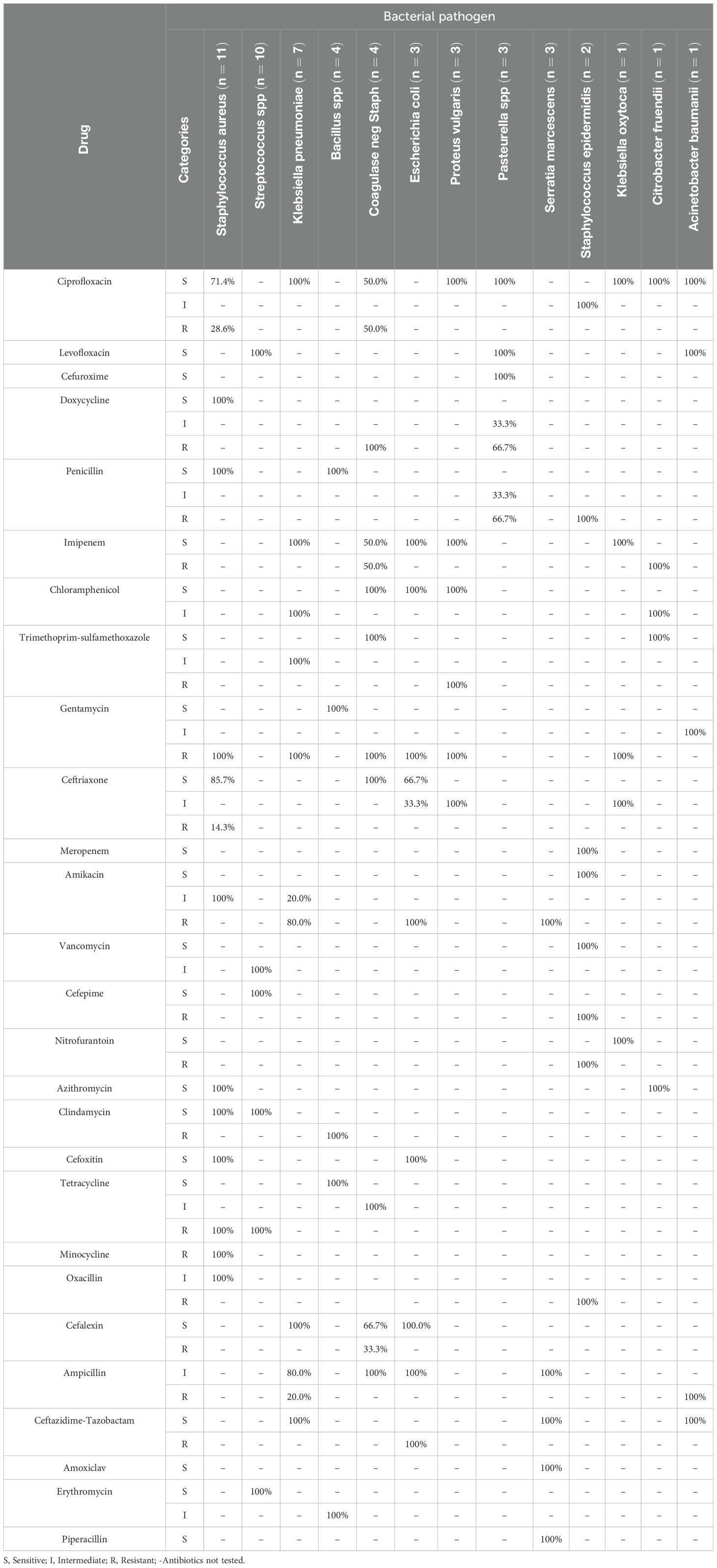
Table 5. Resistance patterns of bacteria isolated from study participants at the Jinja Regional Referral Hospital during the study period.
Bivariate and multivariate analysis of the factors associated with blood culture positive sepsis among study participants
Table 6 shows the results of the bivariate and multivariate analysis of the factors associated with blood culture positive sepsis among study participants. According to bivariate analysis, age (p=0.189), gender (p=0.198), marital status (p=0.156), education level (p=0.179, 0.140), occupation (p=0.147, 174), alcohol use (p=0.169), use of anti-inflammatory drugs (p=0.064), duration of symptoms (p=0.024), presence of central nervous system symptoms (p=0.006), body mass index category (p=0.011), peripheral oxygen saturation (p<0.001) and level of consciousness based on Glasgow coma scale (p<0.001) had p values less than 0.2. According to multivariate analysis, the duration of symptoms (p=0.026), the presence of central nervous system symptoms (p=0.007), and having low peripheral oxygen saturation (p=0.001) were significantly associated with blood culture positive sepsis. Moreover, patients who had the symptoms for more than 5 days were 2.010 times more likely to have blood culture positive sepsis compared to patients who had the symptoms for 5 days or less (aOR=2.010, CI=1.602-6.705, p=0.026). Patients who displayed CNS symptoms were 3.058 times more likely to have blood culture positive sepsis than patients who had no CNS symptoms (aOR=3.058, CI=1.365-6.849, p=0.007). Furthermore, patients who had low oxygen saturation were 3.837 times more likely to have blood culture positive sepsis than patients whose oxygen saturation was normal (aOR=3.837, CI=1.733-8.496, p=0.001).
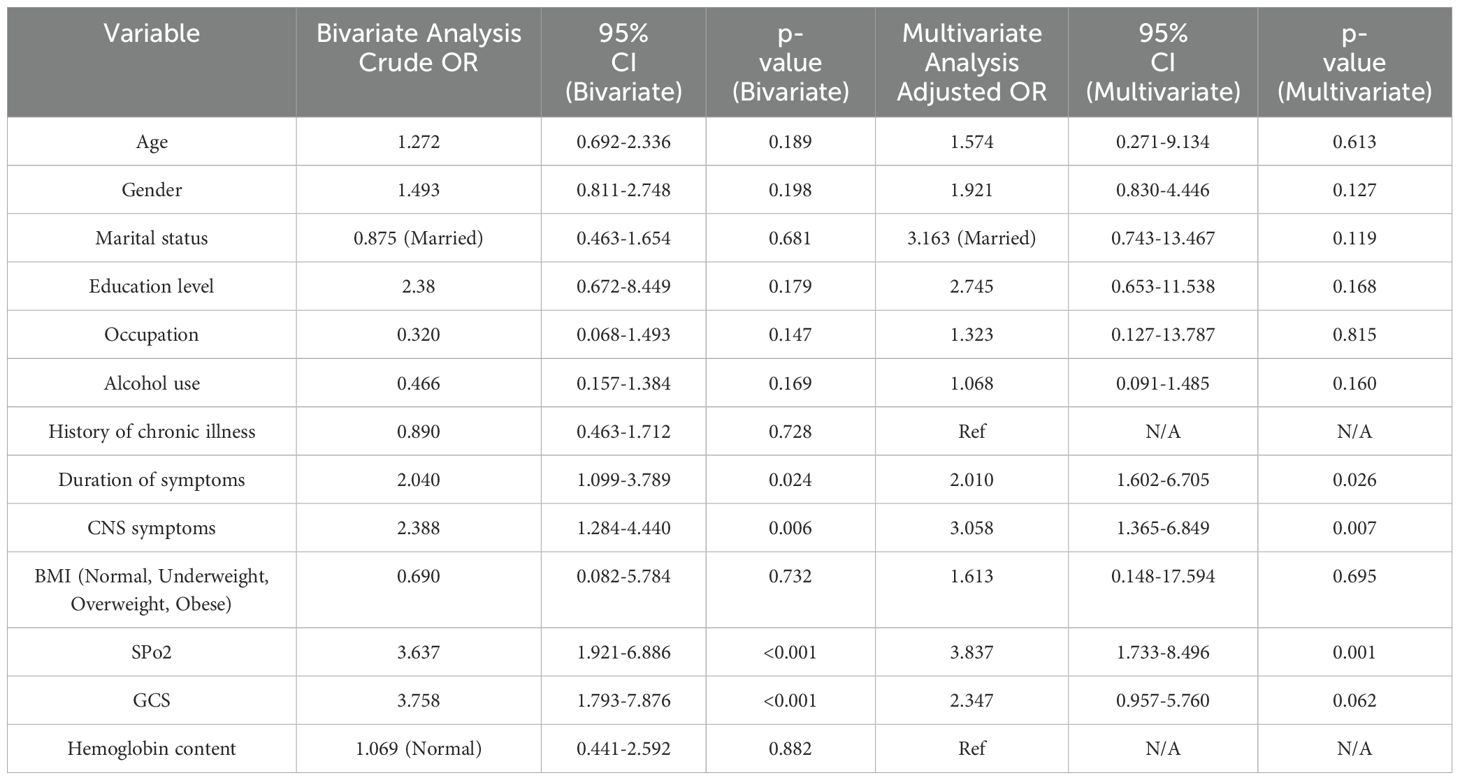
Table 6. Output of bivariate and multivariate analysis of the factors associated with blood culture positive sepsis among study participants.
Discussion
This study sought to evaluate the bacterial profile, resistance patterns, and predictors of blood culture positive samples collected from patients who presented to the Jinja Regional Referral Hospital, a major hospital in Eastern Uganda. The study observed that most participants were from the rural areas which was expected, as most patients in the region are typically referred from rural regions, despite the hospital’s urban location. Moreover, the time taken by patients to develop symptoms and when they presented to the hospital suggests a concerning trend of poor health-seeking behavior, as patients tended to present late. Moreover, 32 participants were HIV positive, with 31 receiving antiretroviral treatment, suggesting effective HIV care services. All participants exhibited elevated temperatures and leukocytosis, consistent with the study’s aim to enroll patients suspected of having sepsis based on the qSOFA criteria.
Despite the suspicion of sepsis in all enrolled patients, only 53 out of 261 (20.3%) had culture-positive sepsis. This value was higher than the 14.7% prevalence rate reported by Nyesiga et al. at the Kirrudu National Referral Hospital in Uganda (10) and the 11.0% reported by a community-based study on sepsis by Kayom and colleagues (11). However, the value was lower than the 35% reported by Kiwanuka et al. among neonates at Mbarara Regional Hospital (17) and the 21.8% reported among neonates at Kidera Health Center in Eastern Uganda by John and co-workers (16).
The prevalence of Staphylococcus aureus (20.8%) in the study setting was consistent with the 20% observed at Kidera Health Center (17). The prevalence of Streptococcus spp. (18.9%) was much higher than the 2.6% reported by John and colleagues (16) and the 1.3% reported by Kiwanuka and co-workers (17). Moreover, the prevalence of Klebsiella pneumoniae (13.2%) was much lower than what was reported by John et al. (2.6%) and Kiwanuka et al. (2.5%) (16, 17). The resistance patterns observed in the present study do not make for good reading. S. aureus was 100% resistant to gentamicin and tetracyline while Klebsiella pneumoniae was 100% resistance to gentamicin. This is in contrast to the study by Tumuhamye et al. who observed 24.1%, and 44.8% resistance of S. aureus to gentamicin and tetracyline, and 40% resistance of K. pneumoniae to gentamicin (13).
S. aureus is an ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp) group pathogen which are important bacteria responsible for multidrug resistance (26). Resistance of S. aureus to aminoglycosides has been linked to the synthesis of transferases that modify the aminoglycoside molecule or the lack of enzymes to transport aminoglycosides into the bacterial cell (26–28). Ida and colleagues reported that resistance to gentamicin was specifically due to the two-domain acetyltransferase/phosphotransferase (AAC(6′)-Ie/APH(2”)-Ia), encoded by the aacA-aphD gene (29). Resistance to tetracyclines in S. aureus is mainly due to the Tet(M) protein but tetracycline resistance in Staphylococcus spp. conditioned by Tet(O), Tet(W) and Tet(44) proteins conditioning ribosomal protection has also been described (26, 30). Klebsiella pneumoniae was 100% resistance to gentamicin. This is in contrast to the study by Tumuhamye et al. who observed 40% resistance of K. pneumoniae to gentamicin (13). Resistance of K. pneumoniae to gentamicin has been reported to be due to the expression of 16 S rNA methylase by the armA gene, the modification of cell permeability through changes in the A crab-TolC and KpnEF efflux pump system or the loss of the putative porin KpnO (31–34). All these lead to therapeutic failure.
This study observed that the duration of symptoms, presence of central nervous system symptoms, and low peripheral oxygen saturation (SpO2) were independently associated with blood culture-positive sepsis. This is an important finding especially in the context of a resource-limited setting. From a clinical standpoint, central nervous system symptoms and low oxygen saturation suggest a severe infection likely to enter the bloodstream, as central nervous system infections often originate from other systems (35). It could be argued that low oxygen saturation may indicate reduced tissue perfusion due to septic shock or severe respiratory infections, which compromise the body’s ability to fight off infections. Su and colleagues conducted a prospective cohort study at the emergency department of the National Taiwan University Hospital and reported that the predictors of bacteremia were fever ≥ 38.3°C, tachycardia ≥ 120/minute, lymphopenia < 0.5x103/L:., aspartate transaminase > 40 IU/L, C-reactive protein >10 mg/dL, and pro Calcitonin > 0.5 ng/mL and a presumptive diagnosis of respiratory tract infection (36). Another study by Chase and others identified respiratory failure, vasopressor use, neutrophilia, bandemia, abnormal temperature, suspected line or urinary infection and endocarditis to be associated with all-case bacteremia among patients at the emergency department at the Beth Israel Deaconess Medical Center in Boston, Massachusetts (37). No specific co-infections were reported in the study population. However, it is important to consider that bacterial co-infections could influence the clinical outcome of sepsis and complicate treatment strategies. Future studies should investigate the role of co-infections in sepsis cases to determine their impact on sepsis management and patient outcomes.
Previous authors have reported that HIV status is significantly associated with blood-culture positive sepsis (38, 39). However, this study presents contrary findings. It may be argued that because most of the HIV-positive participants in this study were on antiretroviral therapy (31 out of 32), these participants did not have advanced disease that is routinely associated with sepsis.
Limitations
The study adopted a cross-sectional design which measured data over a single point in time. Therefore, it was not possible to determine causal relationships between variables. Moreover, while a control group was not included, future studies could benefit from incorporating healthy controls or comparing sepsis prevalence and resistance patterns between different populations to better understand the broader implications of these findings.
Conclusion
The findings suggest that blood culture positive sepsis occurred most frequently among male patients, with most patients aged 18–36 years, though further analysis is needed to establish strong demographic associations. The findings highlight notable antimicrobial resistance in Staphylococcus aureus, Streptococcus spp., and Klebsiella pneumoniae, which should prompt further investigation into antimicrobial stewardship programs. The findings further suggest that prolonged symptom manifestation, CNS involvement, and hypoxemia are key predictors of blood culture positive sepsis in the study area. Public health interventions should focus on improving early detection and management of sepsis, with a particular emphasis on promoting awareness in high-risk groups.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Makerere University School of Public Health REC (SPHREC) (REC No. SPH-2023-435). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
NK: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. AP: Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. CA: Investigation, Methodology, Project administration, Resources, Software, Visualization, Writing – original draft, Writing – review & editing. AM: Data curation, Investigation, Methodology, Resources, Software, Validation, Writing – original draft, Writing – review & editing. JM: Conceptualization, Investigation, Methodology, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. KA: Investigation, Methodology, Project administration, Resources, Software, Visualization, Writing – original draft, Writing – review & editing. AA: Project administration, Resources, Writing – original draft, Writing – review & editing. IN: Project administration, Resources, Software, Writing – original draft, Writing – review & editing. SV: Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. MO: Data curation, Methodology, Project administration, Resources, Software, Validation, Writing – original draft, Writing – review & editing, Visualization.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to acknowledge the research participants and hospital administrators at Jinja Regional Referral Hospital and Kampala International University Teaching Hospital.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Delaloye J, Calandra T. Invasive candidiasis as a cause of sepsis in the critically Ill patient. Virulence. (2014) 5(1):161–9. doi: 10.4161/viru.26187
2. Gu X, Zhou F, Wang Y, Fan G, Cao B. Respiratory viral sepsis: epidemiology, pathophysiology, diagnosis and treatment. Eur Respiratory Rev. (2020) 29(157):200038. doi: 10.1183/16000617.0038-2020
3. Minasyan H. Sepsis: mechanisms of bacterial injury to the patient. Scandinavian J Trauma Resuscitation Emergency Med. (2019) 27:19. doi: 10.1186/s13049-019-0596-4
4. Rello J, Valenzuela-Sánchez F, Ruiz-Rodriguez M, Moyano S. Sepsis: A review of advances in management. Adv Ther. (2017) 34:2393–411. doi: 10.1007/S12325-017-0622-8
5. Plata-Menchaca EP, Ferrer R, Rodríguez JCR, Morais R, Póvoa P. Antibiotic treatment in patients with sepsis: A narrative review. Hosp Pract. (2022) 50(3):203–13. doi: 10.1080/21548331.2020.1791541
6. Coccolini F, Sartelli M, Kirkpatrick AW. What do we mean by source control and what are we trying to accomplish with an open abdomen in severe complicated intra-abdominal sepsis? J Trauma Acute Care Surg. (2023) 96(5):e39-40. doi: 10.1097/TA. 0000000000004253
7. Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality 1990–2017: analysis for the global burden of disease study. Lancet. (2020) 395:200–11. doi: 10.1016/s0140-6736(19)32989-7
8. Kiya GT, Mekonnen Z, Melaku T, Tegene E, Gudina EK, Cools P, et al. Prevalence and mortality rate of sepsis among adults admitted to hospitals in sub-saharan africa: A systematic review and meta-analysis. J Hosp Infection. (2024) 144:1–13.
9. Kwizera A, Urayeneza O, Mujyarugamba P, Baelani I, Meier J, Mer M, et al. Epidemiology and outcome of sepsis in adults and children in a rural, sub-sahara african setting. Crit Care Explor. (2021) 3:e05925.
10. Nyesiga S, Nakibuuka J, Kajumbula H, Ssenyonga R, Byakika-Kibwika P. Burden and bacteriological profile of sepsis among adult medical emergencies presenting to a national referral hospital in kampala, Uganda. MedRxiv. (2022). doi: 10.1101/2022.04.26.22274311
11. Kayom VO, Mugalu J, Kakuru A, Kiguli S, Karamagi C. Burden and factors associated with clinical neonatal sepsis in urban Uganda: A community cohort study. BMC Pediatr. (2018) 18:1–8. doi: 10.1186/S12887-018-1323-4
12. Krepiakevich A, Khowaja AR, Kabajaasi O, Nemetchek B, Ansermino JM, Kissoon N, et al. Out of pocket costs and time/productivity losses for pediatric sepsis in Uganda: A mixed-methods study. BMC Health Serv Res. (2021) 21:1–9. doi: 10.1186/ S12913-021-07272-9
13. Tumuhamye J, Sommerfelt H, Bwanga F, Ndeezi G, Mukunya D, Napyo A, et al. Neonatal sepsis at mulago national referral hospital in Uganda: etiology, antimicrobial resistance, associated factors and case fatality risk. PloS One. (2020) 15:e02370855. doi: 10.1371/JOURNAL.PONE.0237085
14. George M, Iramiot JS, Muhindo R, Olupot-Olupot P, Nanteza A. Bacterial aetiology and antibiotic susceptibility profile of post-operative sepsis among surgical patients in a tertiary hospital in rural eastern Uganda. Microbiol Res J Int. (2018) 24:41690. doi: 10.9734/MRJI/2018/41690
15. Lubwama M, Holte SE, Zhang Y, Mubiru KR, Katende G, Orem J, et al. Etiology, risk factors, and outcomes of bacteremia in patients with hematologic Malignancies and febrile neutropenia in Uganda. Open Forum Infect Dis. (2024) 11(12):1–10. doi: 10.1093/OFID/OFAE682
16. John B, David M, Mathias L, Elizabeth N. Risk factors and practices contributing to newborn sepsis in a rural district of eastern Uganda, august 2013: A cross sectional study. BMC Res Notes. (2015) 8:339. doi: 10.1186/S13104-015-1308-4.
17. Kiwanuka J, Bazira J, Mwanga J, Tumusiime D, Nyesigire E, Lwanga N, et al. The microbial spectrum of neonatal sepsis in Uganda: recovery of culturable bacteria in mother-infant pairs. PloS One. (2013) 8:e727755. doi: 10.1371/JOURNAL.PONE.0072775
18. Zamarano H, Musinguzi B, Kabajulizi I, Manirakiza G, Guti W, Muhwezi I, et al. Bacteriological profile, antibiotic susceptibility and factors associated with neonatal septicaemia at kilembe mines hospital, kasese district western Uganda. BMC Microbiol. (2021) 21:1–11. doi: 10.1186/S12866-021-02367-Z
19. Jinja Regional Referral Hospital, Ministry of Health, Government of Uganda. Available online at: https://jinjahospital.go.ug/ (Accessed December 18, 2024).
20. Kish L. Sampling organizations and groups of unequal sizes. Am Sociological Rev. (1965) 30:564–72. doi: 10.2307/2091346
21. Herman AM, Massenga G, Chilonga KS, Philemon RN, Katundu D. Surgical site infection: the rate and antimicrobial sensitivity pattern in electively operated surgical and gynecological patients at kilimanjaro christian medical centre, northern Tanzania. J Surg Surg Res. (2017) 3:001–5. doi: 10.17352/2455-2968.000034
22. Almanasreh E, Moles R, Chen TF. Evaluation of methods used for estimating content validity. Res Soc Administrative Pharm. (2019) 15:214–5. doi: 10.1016/J.SAPHARM.2018.03.066
23. Bauer AW, Perry DM, Kirby WMM. Single-disk antibiotic-sensitivity testing of staphylococci: an analysis of technique and results. A.M.A. Arch Internal Med. (1959) 104:208–165. doi: 10.1001/ARCHINTE.1959.00270080034004
24. Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. (1966) 45:493–96. doi: 10.1093/AJCP/45.4_TS.493
25. Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, et al. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. (2018) 56(4):1–9. doi: 10.1128/JCM.01934-17
26. Mlynarczyk-Bonikowska B, Kowalewski C, Krolak-Ulinska A, Marusza W. Molecular mechanisms of drug resistance in staphylococcus aureus. Int J Mol Sci. (2022) 23:8088. doi: 10.3390/IJMS23158088
27. Emaneini M, Taherikalani M, Eslampour M-A, Sedaghat H, Aligholi M, Jabalameli F, et al. Phenotypic and genotypic evaluation of aminoglycoside resistance in clinical isolates of staphylococci in tehran, Iran. Microbial Drug Resistance. (2009) 15:129–325.
28. Melter O, Radojevič B. Small colony variants of staphylococcus aureus. Folia Microbiologica. (2010) 55:548–58.
29. Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A, Inoue M. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant staphylococcus aureus in Japan. J Clin Microbiol. (2001) 39:3115–215.
30. Zhu Y, Wang C, Schwarz S, Liu W, Yang Q, Luan T, et al. Identification of a novel tetracycline resistance gene, tet (63), located on a multiresistance plasmid from staphylococcus aureus. J Antimicrobial Chemotherapy. (2021) 76:576–81.
31. Srinivasan VB, Venkataramaiah M, Mondal A, Vaidyanathan V, Govil T, Rajamohan G. Functional characterization of a novel outer membrane porin kpnO, regulated by phoBR two-component system in klebsiella pneumoniae NTUH-K2044. PloS One. (2012) 7:e415055.
32. Poulikakos P, Falagas ME. Aminoglycoside therapy in infectious diseases. Expert Opin Pharmacotherapy. (2013) 14:1585–975.
33. Doi Y, Wachino J-i, Arakawa Y. Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect Dis Clinics North America. (2016) 30:5235.
34. Li Y, Kumar S, Zhang L, Wu H, Wu H. Characteristics of antibiotic resistance mechanisms and genes of klebsiella pneumoniae. Open Med (Poland). (2023) 18(1):1–8. doi: 10.1515/med-2023-0707
35. Fathi M, Markazi-Moghaddam N, Ramezankhani A. A systematic review on risk factors associated with sepsis in patients admitted to intensive care units. Aust Crit Care. (2019) 32:155–645. doi: 10.1016/J.AUCC.2018.02.005
36. Su CP, Chen THH, Chen SY, Ghiang WC, Wu GHM, Sun HY, et al. Predictive model for bacteremia in adult patients with blood cultures performed at the emergency department: A preliminary report. J Microbiology Immunol Infection. (2011) 44:449–55. doi: 10.1016/J.JMII.2011.04.006
37. Chase M, Klasco RS, Joyce NR, Donnino MW, Wolfe RE, Shapiro NI. Predictors of bacteremia in emergency department patients with suspected infection. Am J Emergency Med. (2012) 30:1691–975. doi: 10.1016/J.AJEM.2012.01.018
38. Muchemwa L, Shabir L, Andrews B, Bwalya M. High prevalence of mycobacterium tuberculosis bacteraemia among a cohort of HIV-infected patients with severe sepsis in lusaka, Zambia. Int J STD AIDS. (2016) 28(6):584–93. doi: 10.1177/0956462416640963
Keywords: antimicrobial resistance, blood culture, sepsis, antimicrobial susceptibility, Uganda
Citation: Kara N, Peris A, Abonga C, Muyinda A, Muhumuza J, Akaba K, Abdi A, Nur I, Vidya S and Okumu M (2025) Prevalence, bacterial profile, antimicrobial resistance pattern, and predictors of blood culture positive sepsis among adults admitted at Jinja Regional Referral Hospital in Uganda. Front. Trop. Dis. 6:1552693. doi: 10.3389/fitd.2025.1552693
Received: 28 December 2024; Accepted: 14 April 2025;
Published: 08 May 2025.
Edited by:
Sylvia Opanga, University of Nairobi, KenyaReviewed by:
Ketema Tafess Tulu, Adama Science and Technology University, EthiopiaGayathri Govindaraju, Rutgers, The State University of New Jersey, United States
Copyright © 2025 Kara, Peris, Abonga, Muyinda, Muhumuza, Akaba, Abdi, Nur, Vidya and Okumu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naqeeb Kara, bmFxcnVsekBnbWFpbC5jb20=
†ORCID: Naqeeb Kara, orcid.org/0009-0000-4421-3794
Alina Peris, orcid.org/0000-0001-8356-8168
Charles Abonga, orcid.org/0000-0002-1001-5586
Asad Muyinda, orcid.org/0000-0003-4376-6527
Joshua Muhumuza, orcid.org/0000-0002-5121-264X
Kingsley Akaba, orcid.org/0000-0002-4573-1187
Awil Abdi, orcid.org/0009-0001-1983-3470
Ibrahim Nur, orcid.org/0009-0004-6097-0102
Sankarapandian Vidya, orcid.org/0000-0002-2783-1766
Mitchel Okumu, orcid.org/0000-0002-9316-990X
 Naqeeb Kara
Naqeeb Kara Alina Peris1†
Alina Peris1† Joshua Muhumuza
Joshua Muhumuza Awil Abdi
Awil Abdi Mitchel Okumu
Mitchel Okumu