- 1Department of Internal Medicine, School of Medicine, St. Paul‘s Hospital Millennium Medical College, Addis Ababa, Ethiopia
- 2Department of Medicine, Khairpur Medical College, Khairpur mir‘s, Pakistan
- 3Department of Medicine, D. G. Khan Medical College, Dera Ghazi Khan, Pakistan
- 4Department of Medicine, Riphah International University, Islamabad, Pakistan
- 5School of Medicine, St. Paul‘s Hospital Millennium Medical College, Addis Ababa, Ethiopia
- 6College of Health Science, School of Medicine, Addis Ababa University, Addis Ababa, Ethiopia
- 7Department Internal Medicine, College of Health Science, School of Medicine, Addis Ababa University, Addis Ababa, Ethiopia
The gastrointestinal (GI) microbiome, crucial for host health through its roles in digestion, immunity, and pathogen protection, is significantly disrupted by tropical infections. This disruption, termed dysbiosis, manifests as a loss of beneficial microbes, an increase in harmful bacteria, and altered microbial composition. This review synthesizes recent evidence (2019–2025) on how these infections impact the gut microbiome, influence host recovery, and contribute to long-term health outcomes. A structured literature search was conducted across PubMed, Scopus, and Web of Science, focusing on human GI microbiome dynamics, dysbiosis patterns, and recovery mechanisms in the context of tropical infectious diseases. Findings indicate that common tropical infections, such as cholera, giardiasis, and ascariasis, consistently lead to dysbiosis, characterized by decreased microbial diversity, an increase in opportunistic pathogens like Proteobacteria, and impaired gut barrier function. While natural host processes contribute to recovery, this is significantly influenced by host immunological status, infection severity, and environmental factors. Therapeutic interventions, including probiotics and fecal microbiota transplantation (FMT), show promise in aiding microbiome restoration. Understanding these intricate interactions is crucial for developing effective strategies to manage and treat the associated long-term consequences, including post-infectious GI disorders and malnutrition, particularly in vulnerable populations.
1 Introduction
Gastrointestinal (GI) microbiome serves as the vital community of different microorganisms found inside the human digestive tract, which helps maintain several core bodily functions, including digestion and nutrient breakdown and immune system development, while simultaneously preventing harmful organisms and maintaining intestinal wall strength. A healthy gut functions best when its microbial diversity maintains beneficial commensal microorganisms in high numbers. Disruption of this balance is called dysbiosis which leads to multiple health issues that include metabolic conditions and inflammatory disorders as well as infectious and neurological problems (1, 2).
Tropical infections represent diseases that exist solely in tropical and subtropical climate zones. These diseases cover bacteria, viruses, protozoa, helminths and other pathogens which can spread through water contamination along with poor sanitation and insufficient hygiene practices. Such infections tend to affect primarily people in countries with limited access to healthcare and poor infrastructure (3). The prevalence of parasitic infections including giardiasis, amebiasis, ascariasis and trichuriasis continues to be high in these regions, leading to both severe morbidity and mortality numbers (4).
Gut microbiome functions as a crucial component in controlling how hosts react to pathogenic infection through their immune system. The microbiome controls both infections vulnerability and treatment success and disease progression while experiencing significant transformation from infections. This two-way relationship between infections and the gut microbiome helps explain disease causation as well as intervention development specifically for gastrointestinal diseases found frequently in tropical areas (5, 6).
Gut microbial ecosystem’s reaction to tropical infections remains an area of active and evolving investigation. These infections result in direct mucosal damage while simultaneously disrupting microbial environments and modifying local immune reactions. A combination of pathogen-driven inflammation and immunological activation, and medication use that includes antibiotics and antiparasitic drugs result in major changes to microbial diversity and composition (5). Scientists have yet to uncover the particular microbiological signatures that correspond to different tropical infections, combined with the responsiveness of host systems to establish gut homeostasis. The conventional exposure patterns within tropical endemic zones lead to progressive deterioration of the microbiome. Long-lasting dysbiosis results in persistent inflammation that impairs nutrient digestion and can lead to chronic post-infectious diseases. Children face particular concern because their developing microbiomes are more sensitive to disruptions, which may impact their growth alongside immune maturation and neurodevelopmental health (7).
The purpose of this narrative review is to bring together literature concerning the impacts of tropical infections on the gastrointestinal microbiome, the perturbation and re-establishment of homeostasis, if any, by the host, and the potential long-term implications of these disturbances on health. It analyzes the patterns of dysbiosis for the most important groups of tropical infections, assess the other factors affecting the recovery and the wider clinical and public health impacts. This review will help in guiding further research and appropriate management strategies with regard to tropical diseases, especially in economically disadvantaged tropical countries, by bringing together different findings into one holistic framework.
2 Methods
A structured literature search of three prominent research databases - PubMed, Scopus, and Web of Science - allowed for the development of an evidence-based synthesis of gastrointestinal microbiota changes due to tropical infections. The chosen databases provided comprehensive access to biomedical research along with microbiological and global health studies. Two individual authors conducted the peer-reviewed journal article search from April 14 to April 25, 2025 while focusing on articles published between January 1, 2019 and April 2025. The authors chose this six-year period to capture recent research documenting microbiome sequencing advancements and host-microbe understanding progress.
The search used combination strategies together with free-text keywords which included gut microbiome, gastrointestinal microbiota, intestinal dysbiosis, microbial diversity, in conjunction with tropical infections, diarrheal diseases, protozoan infections, helminth infections, and neglected tropical diseases. The research included recovery and restoration terms together with long-term effects alongside specific disease names (cholera, giardiasis, ascariasis) along with other relevant terminology. Boolean operators together with syntax from each database helped narrow down search results.
Only English-language articles were included. Editorials and commentaries and preprints as well as non-peer-reviewed materials were excluded to guarantee high evidence standards. Two independent reviewers examined titles and abstract reviews before moving toward complete evaluation of articles that satisfied the criteria. Manual searches were conducted on reference lists from important studies and reviews in order to identify relevant articles that had not been discovered by electronic database queries.
Inclusion of studies followed these specific criteria:
● Research that is published in peer-reviewed journals appeared between 2019 and 2025.
● Studies which examined human gastrointestinal microbiome dynamics within the framework of tropical infectious diseases.
● Studies that included data describing microbiome composition alongside diversity levels and functional parameters as well as dysbiosis patterns and restorative mechanisms.
● Studies that investigated host responses throughout infection phases including acute infection periods and the post-infectious state.
● Research that focused on human populations who live primarily in tropical or resource-limited areas.
Exclusion criteria included:
● Articles published prior to 2019.
● The review excluded all materials without peer-review including preprints and dissertations along with conference abstracts.
● Studies that only examined in vitro or animal models which lacked human-relevant findings.
● The research did not investigate gastrointestinal microbiome or tropical infections or study unrelated aspects.
● Articles not available in English.
Although formal quality appraisal tools (e.g., Cochrane RoB, AMSTAR, CASP) were not applied, two independent reviewers informally assessed methodological clarity, relevance, and data reporting during full-text screening to ensure the inclusion of scientifically credible studies.
The authors used this specific selection approach to base their analysis on contemporary research which is peer-reviewed and relevant to the field.
3 The healthy gastrointestinal microbiome: a primer
A human body needs a balanced gut microbiome that demonstrates resilience for proper health status (8). A thorough understanding of normal gut bacterial communities helps to determine how tropical infections affect human wellness because these diseases often disrupt this delicate microbial network.
3.1 Composition and diversity
The gut microbiome consists of trillions of microorganisms, where bacteria make up the greatest volume fraction (9). Out of all gut microbiota bacterial phyla, the most common ones include Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria, and Firmicutes along with Bacteroidetes make up around 90% of the population (8, 9). The different phyla include many genera that perform specialized functions essential to human well-being. Some of these genera of bacteria, which stabilize our gut ecosystem, consist of Bacteroides together with Prevotella, Corynebacterium and Bifidobacterium, Lactobacillus and Fecalibacterium (8).
An expanded diversity level correlates with better resistance and stability conditions within the microbial system. A low microbial diversity, however, leads to multiple health conditions, such as inflammatory bowel disease (IBD) and metabolic disorders. The human microbiome undergoes a persistent evolutionary process that affects the host system. The baseline diversity of the microbiome reacts significantly to multiple factors, including age, dietary choices, geographic location, drug usage, lifestyle habits, hormonal changes, genetic background, and ongoing medical conditions (10–12).
3.2 Key functions of the gut microbiome
The gut microbiome carries out numerous vital physiological functions that are basic for human health maintenance.
3.2.1 Immune system development and modulation
The gut microbiota is essential for immune system development and regulation, training it to identify beneficial microbes while preserving its ability to detect pathogens. Butyrate and other SCFAs, for instance, facilitate the maturation of regulatory T cells that work to sustain immune tolerance while suppressing excessive inflammatory responses. It has been suggested that changes in cytokine biosynthesis as well as other immunological effectors are modulated through the influence of the gut microbiota (8, 10, 11, 13).
3.2.2 Barrier function against pathogens
Normal microbiome function protects the body from damage through its competition with pathogens for essential nutrients and wall-attachment surfaces within the digestive system. The production of antimicrobial peptides by helpful bacteria also protects the body from damaging microorganisms. Additionally, the gut microbiome strengthens the intestinal epithelium through excretion of mucus alongside tight junction forming proteins that fill cellular spaces, thus creating a barrier that pathogenic agents must overcome to cross through (8, 14).
3.2.3 Production of SCFAs and their roles
The gut bacteria transform soluble fibers into short chain fatty acids (SCFAs), which include butyrate and propionate, and acetate. Various metabolic products of gut bacteria, including SCFA, act as cellular energy while simultaneously controlling inflammation and defending the gut lining and modifying entire body metabolic processes. For example, it has been demonstrated that butyrate promotes anti-inflammatory reactions while strengthening gut barriers and influencing immune functions, making it especially significant for gut wellness (8, 12, 13). In addition, gut microbes participate in fundamental metabolic processes, such as producing B vitamins and Vitamin K, which are vital for many bodily operations (8, 12, 13).
3.3 Homeostasis and stability
Microbial stability is defined by two key elements: resistance to changes and the capability of protection against pathogens. Furthermore, achievement of homeostasis depends on three complementary factors: microbial competition, antimicrobial metabolism and evolutionary control from the immune system. Maintaining a resilient microbiome is achieved by following an organized approach, which includes consumption of prebiotics and probiotics, routine exercise, and restriction of antibiotic use to emergency medical situations (9, 11, 15, 16).
4 Dysbiosis of the gastrointestinal microbiome in tropical infections
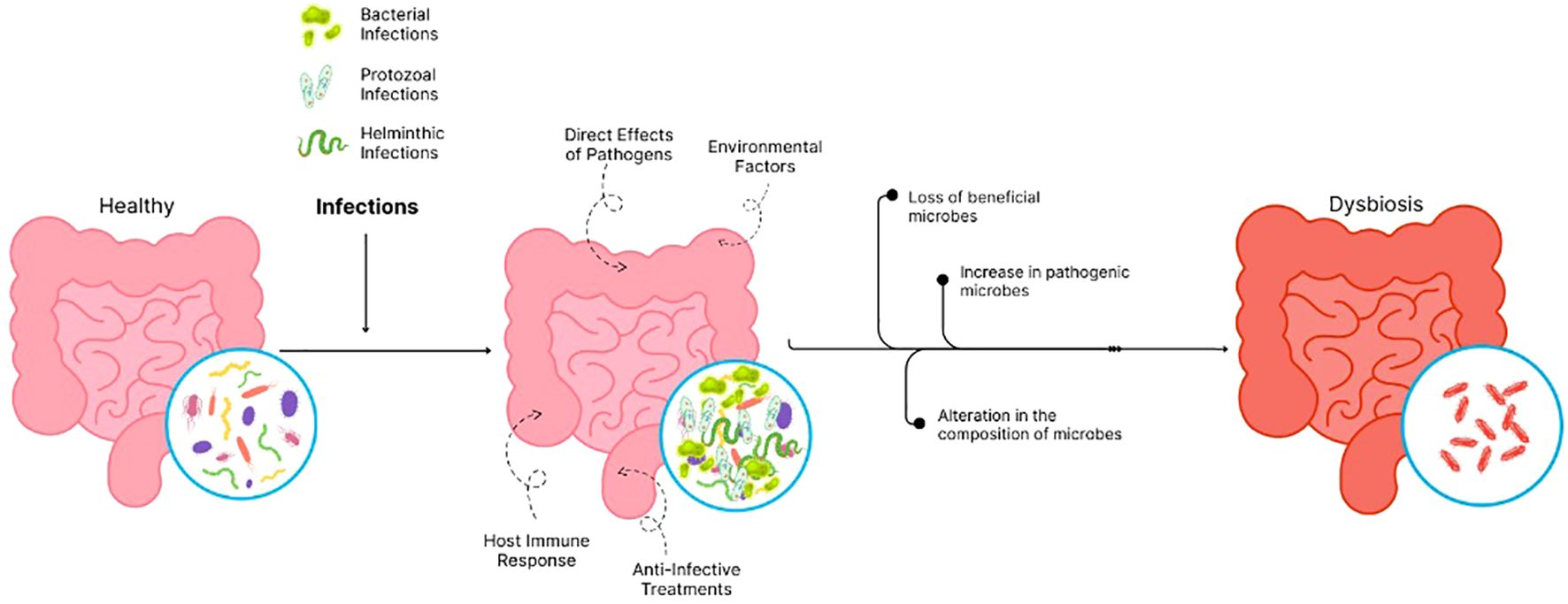
Infections due to bacteria, protozoa, and helminths create profound effects on the gut microbiome while leading to dysbiosis, which means an imbalance in the gut microbial composition. The dysbiotic state features three main components, which include the loss of beneficial microbes, an increase in pathogenic bacteria, and an alteration in the composition of microbes. These modifications lead to destructive impacts throughout the digestive system, the immune system, and on the health of infected people (17).
4.1 Mechanisms of microbiome disruption
4.1.1 Direct effects of pathogens
Both the environment in the gut and the makeup of the microbiome can shift when an infection occurs. Bacterial, viral, or parasitic agents may be responsible for these infections. When pathogens reach the intestines, they can impact the microbiome in various ways. They may deplete certain microbial compounds, produce bioactive substances, and trigger an inflammatory response, all of which contribute to modifications within the microbial ecosystem (12, 18).
4.1.2 Host immune responses
Inflammation and antimicrobial peptide release as an immune response to infection also influence microbial populations. Long-term inflammation typically fosters a decline in beneficial microorganisms while favoring pathogenic microorganisms to grow. For example, the trophozoites of E. histolyticaproliferate in intestinal lumen and phagocytose normal gut flora like Lactobacillus ruminus (19, 20).
4.1.3 Effects of anti-infective treatments
The control of an infection depends heavily on the utilization of anti-infective agents, including antibiotics. Antibiotics create detrimental side effects because they destroy all existing microbes without distinction, so they produce both lower microbial diversity and more resistant microbial strains (17).
4.1.4 Environmental factors
Insufficient hygiene and unhygienic surroundings are believed to contribute to the development of environmental enteropathy, which mostly affects children and leads to growth faltering (21). A study from Nicaragua found that the presence of animal feces on household floors influenced gut bacteria, especially in children, causing a change in the population of helpful and harmful bacteria. Clostridium perfringens, S. infantarius, and Campylobacter jejuni have been identified as potentially harmful microbes. At the same time, beneficial organisms such as Anaerostipes s that produce anti-inflammatory butyrate have also been found to be influenced by animal fecal contamination in the household environment (22). Additionally, diarrhea in children can work together with stunting to alter the intestinal microflora. As diarrhea upsets the gut’s microbial balance, stunting related to recurrent diarrhea further slows the restoration of gut bacteria, which leads to less resistance to new infection and a repeated cycle as shown in Figure 1 (23).
4.2 Specific examples of dysbiosis in key tropical infections
4.2.1 Bacterial diarrheal diseases
Conditions such as cholera, shigellosis, and enterotoxigenic E. coli infections usually entail marked changes to the gut microbiome. Further, these infections reduce the abundance of favorable microbes such as Lactobacillus and Bifidobacterium while increasing pathogenic members like Enterobacteriaceae. Dysbiosis is characterized by decreased microbial diversity, which further impairs the gut’s protective response against infections. Often, metabolic pathways that help produce butyrate are lost, thus impairing gut health (23–26).
4.2.2 Protozoan infections
Protozoan infections modify the host-microbiota interactions, immune system, and bacterial constituents of the gut ecosystem. Giardia intestinalis, Entamoeba histolytica, and Blastocystis are examples of protozoa that metabolically engage with bacteria to shape the microbiota composition. The bacterial composition change brought about by Toxoplasma gondii also alters the gut microbiota. The interplay between intestinal parasites and the gut microbiome shapes immune responses and the disease course (27).
4.2.3 Helminth infections
Helminth parasites, especially the soil-transmitted ones, Ascaris lumbricoides and Strongyloidesstercoralis, Ancylostoma duodenale, Necator americanus, Trichuris trichiura, andEnterobius vermicularis tend to cause chronic intestinal infections and alter the gut microbe composition. For example, Trichuris-infected individuals showed a marked shift from Bacteroidetes to Firmicutes and Clostridia (28, 29). Helminth infections have been linked to greater abundance of Lactobacillaceae species, which evoke host regulatory responses that could mediate the retention of helminths, hinting at a synergistic association of these bacteria with parasites. Furthermore, the helminths change the profile of intestinal nutrients and niches by disrupting glucose transport across the epithelium and changing mucus viscosity, which could be advantageous for the populating and glucose metabolizing bacteria like Mucispirillum and Clostridia. All these factors illustrate the intricate microbial changes brought about by helminths and their deeper consequences on the ecology of the gut microbiome (30).
4.2.4 Neglected tropical diseases with gastrointestinal involvement
Findings on neglected tropical diseases indicate that strongyloidiasis can affect the gut microbiome, thus explaining ongoing chronic symptoms, yet the evidence remains limited and sparse. Results from studies on strongyloidiasis patients infected with Strongyloidesstercoralis demonstrate that chronic infection causes microbial changes in which the Ruminococcus torques group bacteria become dominant, which destructively break down mucin and disrupt the protective barriers in the gut. It was also found that chronic infection leads to modified metabolic processes while removing beneficial SCFAs, which include acetate, from the microbiome. The observed modifications in microbial populations could help explain the gastrointestinal symptoms that commonly occur in chronic cases (31–35).
4.3 Common patterns of dysbiosis across tropical infections
The development of dysbiosis in different tropical infections exhibits several shared characteristics:
4.3.1 Decreased diversity
Having sparse microbial diversity represents the most enduring and advancing element of dysbiosis that occurs with tropical infections. Microbiota equilibrium suffers when diversity is absent, which makes the microbiota more exposed to infection and inflammation (36).
4.3.2 Elevated proteobacteria
The dysbiotic condition frequently reveals an increase of Proteobacteria, which links to both inflammation and infection. Many tropical infections yield this gut microbial change because their pathogenic agents, including Salmonella and Escherichia coli, thrive best when the gut is inflamed (37–39).
4.3.3 Altered barrier function and SCFA production
During dysbiosis in tropical infection environments, intestinal integrity weakens, and permeability rises, through which harmful microbes can enter the bloodstream. In addition, the production of SCFAs remains typically low in the gut, which generates an intestinal setting that does not effectively restrict pathogenic microbes (31, 32).
These identified patterns enable healthcare professionals to develop targeted therapeutic interventions that improve gut microbiome stability as well as patient clinical results for patients with tropical infections a comparison is shown in Table 1.
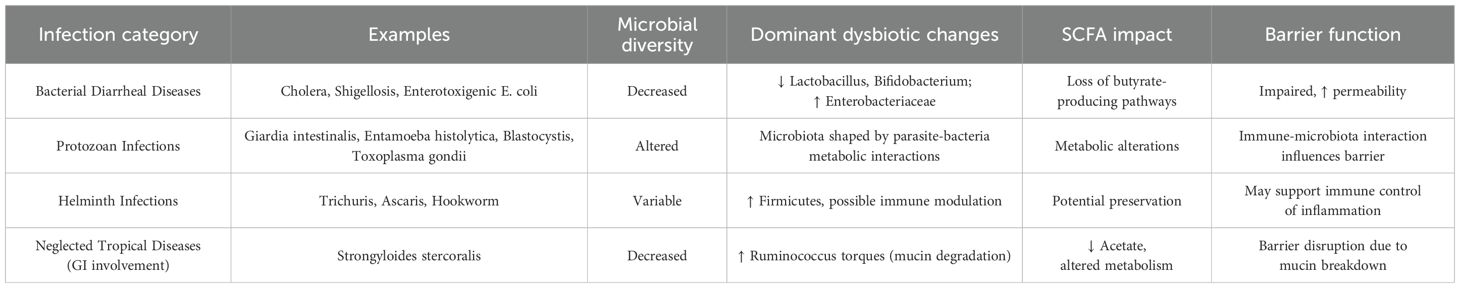
Table 1. Comparison of common dysbiotic features across different categories of tropical infections.
5 Host recovery mechanisms and microbiome restoration
5.1 Natural resolution of dysbiosis
After the gut flora is disrupted, restoration occurs in an orderly fashion. The early bacteria that colonize the GI tract are the ones that can replicate at a higher rate, tolerate an inflammatory environment, and have metabolic versatility like the Enterobacteriaceae (40). The pioneer species modify the environment and allow the emergence of intermediate colonizers. After the inflammation completely resolves, late colonizers appear, and the microbiome resembles the pre-infection environment (41).
The gut microbiome is a dynamic ecosystem with natural coping mechanisms to restore itself (42). There are bacteria in the gut microbiome that have functional redundancy. This allows the continuation of nutrient absorption and production of short-chain fatty acids, i.e., gut stability despite the disappearance of some bacteria species (43). The other mechanism through which the gut microbiome restores itself is through the survival of a reservoir of beneficial microbes. The adaptive mechanisms these bacteria use to survive disruptive events include biofilm production, spore formation, and colonization of a specific niche in the intestine, including the mucosa, crypts, or even host cells (44). These microbes persist in lower numbers in specific areas of the GI tract and proliferate after the infection is resolved. Since the GI tract is not uniformly affected during infection, bacteria from less affected areas could also migrate to re-establish the gut microbiome (45).
The host immune system exerts selective pressure, aids in infection resolution, and promotes commensal bacteria growth. The immune system not only initiates the necessary inflammatory response to clear the offending pathogen but also plays an active role in the recolonization process by interacting with commensal bacteria (46). Innate immune cells exert selective pressure by producing pattern recognition receptors, secretory IgA, and antimicrobial peptides like defensins and cathelicidines. This prevents the overgrowth of opportunistic pathogens and aids commensal bacteria growth (47, 48). Early epithelial cell repair is also essential to reverse dysbiosis. Epithelial cells prevent bacterial translocation through their tight cell junctions and the production of mucus. They also aid in the detection of pathogen-associated molecular patterns and release chemokines and cytokines that alter immune response and microbial proliferation (49).
5.2 Factors influencing microbiome recovery
Interaction and succession between different species of the gut microbiome is complex, and its recovery is affected by several factors.
5.2.1 Host-related factors
These are the factors that affect microbiome recovery include age, genetics, the presence of comorbidities, and immune status. Elderly patients and infants may have incomplete recovery because the first group has a declining microbiome diversity, and the latter has an immature microbiome (50). Host genetics determines the composition of the gut microbiome, immune response to pathogens, and recovery. Animal studies have shown that host genetics is responsible for the variation in the composition of the gut microbiome between individuals. Genes like NOD2 and FUT2 are conserved across different species and are heritable (51). The presence of comorbidities like inflammatory bowel disease also affects the ability of the GI tract to reverse dysbiosis. As discussed in the previous section, an intact immune system is also necessary for fast and complete recovery (52).
5.2.2 Infection-related factors
In addition to host factors, infection-related factors like the type of pathogen, severity of infection, site of infection, and duration of infection also affect the recovery process (52, 53). Some bacteria cause toxin-mediated or direct epithelial damage, while some parasites dwell in the gut lumen for a long time, competing with commensal bacteria for nutrients, which alters the microbiome, and some viruses, like the Norovirus, can directly kill commensal bacteria. Animal studies have also shown that helminth infections promote the development of Treg cells, this alters the balance between these cells and Th1 cells, which results in reduced immune response to and increased proliferation of invading bacteria. Some viruses, like the Norovirus, can directly kill commensal bacteria (53). Bacteria like vibrio-cholera which induce a massive secretion of water accelerates the removal of beneficial bacteria (24).
5.2.3 Lifestyle and environmental factors
Recovery is also affected by dietary habits, antibiotic use, probiotic supplementation, etc. A diet rich in vegetables, fruits, and fermented foods allows the re-establishment of the gut flora, while processed and sugary foods could hinder it (54). Antibiotic use can result in loss of beneficial bacteria and reduced short fatty acid synthesis, which alters mucus production and intestinal permeability (55). Environmental exposure and re-exposure alter immune response to pathogens, and the metabolism and activity of existing flora. It also accelerates microbiome recovery and diversification. Exposure to plant microbiomes in green spaces, to microbiomes in agricultural soil and livestock in farms, or contact with pets allows the exchange and colonization of the human gut with new microbiomes (56).
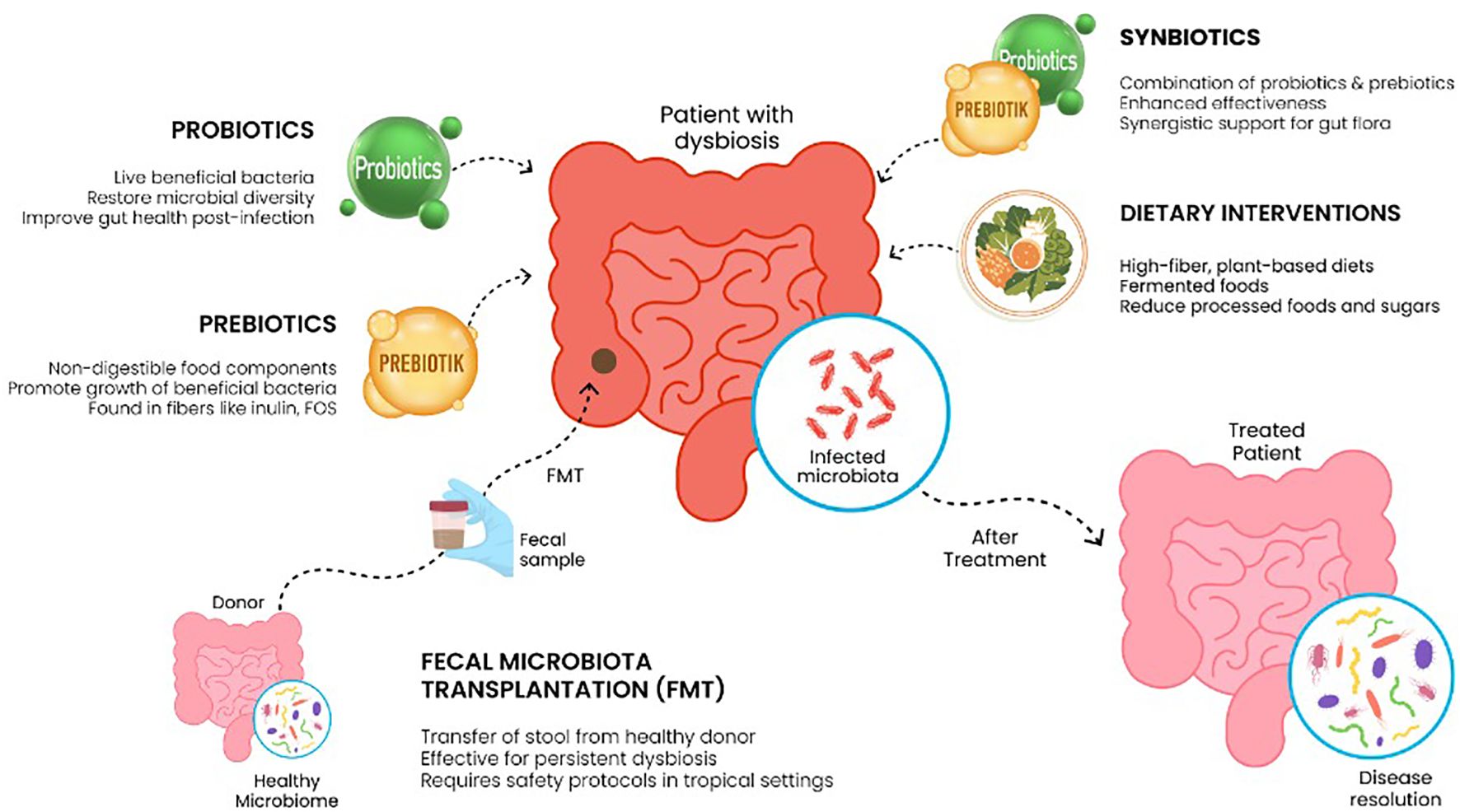
5.3 Potential therapeutic interventions for microbiome restoration
Some of the methods that can be applied for restoring balance to the gut microbiome include probiotics, prebiotics, or synbiotic administration, and in advanced cases, fecal microbiota transplants. Probiotics can be defined as immunomodulatory and anti-neoplastic microorganisms with the capability of restoring gut microbiota. The probiotics that are well studied with a confirmed benefit for microbiome recovery are currently in use include Lactobacilli, Bifidobacteria, Bacillus s, etc. Probiotics like Lactobacilli produce inducible polyphosphate, which enhances the integrity of the intestinal epithelium and reduces its permeability. They also produce glyceraldehyde-3-phosphate, which they use to adhere to the intestinal epithelia and prevent GI colonization by pathogenic bacteria. They also produce peptides that directly target the cell membranes of pathogenic bacteria (57, 58).
Prebiotics to note are fructo- and galacto-oligosaccharides as well as inulin. They are considered indigestible and promote better gut flora. They can be used to synthesize short-chain fatty acids, which have anti-inflammatory effects and are essential for epithelial cohesion. They also selectively regulate the growth of commensal saprophytic bacteria (59). The co-administration of a prebiotic and a probiotic, also called synbiotics, increases the chance of probiotic survival and colonization efficiency and amplifies all the positive effects these two agents exert (59, 60).
Fecal microbiota transplantation (FMT) is indicated in severe cases and usually for patients with inflammatory bowel disease, recurrent Clostridium Difficile infection, and antibiotic-associated dysbiosis. It results in direct reconstitution of the gut flora, production of fermentation products that are necessary for gut epithelia integrity, normal colonocyte function, and immune modulation. It also reduces the risk of colonization by pathogenic bacteria. In tropical settings, the higher prevalence of immunocompromised and malnourished patients and the increased risk of transmitting antibiotic-resistant bacteria make it less safe. Also, since there is a higher burden of parasitic, viral, and bacterial GI infections in this area, stool screening should be expanded. In addition to this, the cultural acceptance of the procedure, the shortage of trained health professionals who can do the procedure, and the cost of stool screening and processing make FMT less feasible in resource-strained regions (61).
Several dietary compositions can be used to enhance the restoration of the gut microbiome. They include the Mediterranean, low FODMAP, and vegan diets. The Mediterranean diet is low in processed foods and is mainly composed of fruits, vegetables, legumes, and olive oil. Since it is rich in fibers, which serve as prebiotic and anti-inflammatory polyphenols, this dietary pattern is suitable to promote microbiome recovery and diversity. Vegan diets contain a high level of anti-inflammatory phytochemicals. They also supply a good amount of fiber to the gut flora. The low FODMAP diet avoids the short-chain carbohydrates like oligosaccharides that are fermented to gas in the large intestine and cause bloating. Shown in Figure 2 (62).
6 Long-term clinical consequences of microbiome dysbiosis following tropical infections
6.1 Post-infectious gastrointestinal disorders
6.1.1 Post-infectious irritable bowel syndrome
The mechanisms of post-infectious IBS development include altered gut mobility, visceral hypersensitivity, low-grade inflammation, and microbiota dysbiosis. Enteric pathogens like Giardia Lamblia disrupt epithelial integrity, bacteria like Salmonella typhimurium induce IL-18 and IFN-γ release and alter gut microbiota (63). Pathogens like Campylobacter Jejuni and Shigella Flexneri induce NF-κB release, inflammation, and CD68 cell reduction. Studies have also revealed mast cell accumulation and altered serotonin signaling after GI infection (64). The persistent low-grade GI inflammation associated with altered GI signaling and visceral hypersensitivity could result in IBS. In a study done on patients after shigella infection, female sex, older age, having abdominal cramps, and more than 4 days of diarrhea were associated with a higher risk of developing post-infectious IBS (65).
6.1.2 Post-infectious functional dyspepsia
Post-infectious dyspepsia occurs in 12.7% of patients after acute gastroenteritis (66). Persistent low-grade inflammation, the altered gut microbiome, altered gut-brain axis, altered gut motility, and visceral hypersensitivity all contribute to post-infectious functional dyspepsia. Patients with IBS are at higher risk of developing post-infectious dyspepsia compared to those without. Altered gastric sensory motor function and immune system alteration are thought to be responsible for this overla (67) Prolonged abdominal pain and vomiting during gastroenteritis are predictive of post-infectious functional dyspepsia (68).
6.1.3 Chronic abdominal pain and altered bowel habits
Microbiome dysbiosis associated with chronic low-grade inflammation, altered gut motility and permeability, mast cell accumulation and activation in the mucosa contributes to the development of chronic abdominal pain after a gastrointestinal infection (68, 69). post-infectious bowel habit changes result from dysbiosis induced overgrowth of some bacteria. This results in excessive fermentation and production of gas, reduced short-chain fatty acid production, and disrupted bile acid metabolism. Altered gut permeability and motility also play a role (70).
6.2 Malnutrition and growth stunting
Persistent gut microbiome alteration results in impaired availability, production, and absorption of nutrients. The reduced synthesis of short-chain fatty acids, which are the main sources of energy for colonocytes, results in impaired nutrient absorption by these cells. Dysbiotic bacteria consumes nutrients and also releases peptides that interfere with the absorption of nutrients, which reduces the amount of available nutrients for absorption. Alteration of gut pH and impaired bile acid metabolism result in decreased absorption of fats and minerals. Microbiome dysbiosis could also result in a decrease in the vitamin-producing bacterial population (70, 71). The chronic inflammation associated with microbiome dysbiosis interferes with the signaling of growth factors like insulin derived growth factor and human growth hormone. These factors result in chronic malabsorption, malnutrition, and impaired growth during childhood. It could also lead to obesity later in life (72).
6.3 Increased susceptibility to secondary infections
Short-chain fatty acids like propionates and butyrates reduce the production of inflammatory cytokines by inhibiting histone deacetylase and activating G-protein coupled receptors that enhance the production of anti-inflammatory cytokines like IL-10 and TGF-β. This makes them essential for gut mucosal immunity and integrity. The microbiome also produces antimicrobial peptides that prevent the overgrowth of opportunistic bacteria (73). Commensal bacteria-derived p40 protein activates epidermal growth factor signaling and its downstream components, resulting in the proliferation of intestinal epithelial cells and the production of tight junctions. Short-chain fatty acids upregulate the expression of claudin and actin-binding proteins. They also induce the production of mucus secretion, which contains antibacterial substances like cathelicidines, ubiquitin, defensins, etc., by goblet cells, which helps maintain the integrity of the gut barrier. Lack of these beneficial effects alters the response and integrity of the gut epithelium to offending microbes (73, 74).
6.4 Potential extraintestinal manifestations
The gut microbiome is crucial in maintaining the balance between proinflammatory and anti-inflammatory actions of the intestinal epithelia. Microbiome dysbiosis would lead to the loss of anti-inflammatory effects of beneficial gut flora and its metabolites. Increased gut permeability to bacterial products like lipopolysaccharides cause pattern recognition receptor activation and subsequent inflammation. Some bacterial products can mimic self-antigens and trigger autoimmune responses (75). Short-chain fatty acids play a role in glucose metabolism by increasing the release of glucagon-like peptide 1 (GLP-1). Lack of this effect, combined with chronic inflammation, results in the dysfunction of the insulin signaling pathway. This increases the likelihood of insulin resistance and type II diabetes. Altered gut permeability and persistent immune stimulation also contribute to the development of autoimmune diseases like inflammatory bowel disease and systemic lupus erythematosus (76).
The gut microbiome also plays an important role in the regulation of the gut-brain axis. Metabolites like short-chain fatty acids induce the production of neurotrophic factors and downregulate the production of neuroinflammatory mediators and play a crucial role in brain recovery after traumatic brain injury and stroke (77). Short fatty acids also play a role in cognition by limiting the amount of kynurenic acid that reaches the hippocampus by reducing its production from tryptophan in the gut. The gut microbiome also plays a role in the production of neurotransmitters like dopamine, glutamate, gamma-aminobutyric acid, norepinephrine, and serotonin, and it also regulates tryptophan metabolism (78, 79). Microbiome dysbiosis has been associated with neurodevelopmental disorders like autistic spectrum disorder, neurodegenerative diseases like Alzheimer’s disease, mood disorders, and eating disorders (80).
6.5 Impact on vaccine efficacy
Oral vaccination effectiveness is impacted by microbiome dysbiosis in a number of ways. Oral vaccination effectiveness is dependent on well-functioning antigen-presenting cells. A change in the gut microbiome may decrease these cells, change how co-stimulatory signals are expressed, and affect the phenotype of differentiating antigen-presenting cells. Additionally, vaccine antigens may be broken down by opportunistic microbes. Furthermore, the establishment of secondary germinal centers and the immunological response to vaccine antigens could be hampered by altered gut permeability and a deficiency of microbial metabolites (81, 82).
7 Challenges and future directions
7.1 Challenges in studying the microbiome in tropical settings
Understanding the gut microbiome in tropical environments offers significant advantages and disadvantages that will dictate how planned study designs are conducted as well as how data produced from this research is interpreted. Studying microbiomes in tropical settings has its own challenges. One major factor, compromising sample integrity and data reliability, would be logistics, specifically sample collections, transportation, and processing with limited resources. Polyparasitism and other co-infections are also prevalent in these regions, making data interpretation more complicated. This makes difficult to pinpoint the microbiome response to a specific environmental component or a single infection (83).
Environmental conditions including sanitation or specific diet practices of the local area may vary the character of the microbiome. Such pronounced inherent variability complicates efforts to establish a clear healthy microbial baseline applicable across these diverse populations. A significant portion of current knowledge stems from cross-sectional studies, inherently limiting the capacity to determine causal relationships. Research efforts are frequently hampered by insufficient sample sizes, often draw participants solely from hospital settings, and exhibit a lack of uniform methods for sampling, sequencing, data analysis, and defining outcomes, thereby obstructing meaningful comparisons across different studies. A critical gap exists in the form of longitudinal research capable of tracking microbial community shifts throughout infection and subsequent recovery. Additionally, the influence of socio-environmental factors is commonly neglected in research designs, and information gathered from rural community settings remains notably scarce (83).
7.2 Future research priorities
Overcoming these obstacles demands a clear, strategic orientation for upcoming research initiatives. Executing large-scale longitudinal cohort studies, designed with rigorous controls and encompassing varied tropical locales, stands out as a chief priority. Studies of this nature are indispensable for thoroughly mapping the dynamics of the microbiome prior to, during, and following infectious episodes, linking observed microbial changes to patterns of clinical progression, recovery pathways, and the eventual onset of long-term health consequences. Simultaneously, dedicated research efforts must aim to pinpoint distinct microbial indicators, whether specific taxa, functional capabilities, or metabolic products, that could function as dependable biomarkers for diagnosing conditions, predicting prognoses, or guiding therapeutic choices for prevalent tropical diseases; insights like the potentially protective microbial profiles noted in malaria studies offer a foundation (84). To progress beyond merely describing microbial communities, it’s imperative to explore the functional impacts of dysbiosis through integrated multi-omics strategies. Effectively combining metagenomic data with meta-transcriptomic, metabolomic, and host immunological analyses is vital for untangling the complex mechanistic connections involving the microbiome, host immune functions, and the ways diseases develop, exemplified by investigating how gut-derived metabolites influence systemic immune activity (85).
Another area that will need to be balanced as an example through trial phase methods and evaluation is the initiation of microbiome focused intervention. There would be value in designing clinical trials to assess the effectiveness of probiotics, prebiotics, synbiotics or postbiotics to reduce rates of pathogen infections, lessen disease burden, or enable recovery to help further clinical and community life - as well as a possible way to phase or strengthen other treatment practices, for example in cholera and oral rehydration therapy (86), or in cases of amoebiasis and the effect of antibiotics while emphasizing the importance of stability of the microbial communities (87).
One major research paradigm continues to be how microbiota resident of the gut will influence vaccine efficacy through enhancing the immune response in future work in contexts in which vaccination has previously not been successful in leading to an immune response. Future work and follow-ups, ultimately, depend on and continue to be rooted in context, cultural sensitivity and ethics. This requires collaborative efforts drawing together diverse specialists and community representatives, ensuring research plans align with regional realities like dietary norms and traditional health practices. Such an approach enhances the practical relevance and potential longevity of both research insights and health interventions. Advancing the field will also depend on developing standardized methodologies and encouraging shared access to microbiome data, particularly covering less-represented tropical areas, which is necessary for conducting more powerful meta-analyses and enabling broader scientific comparisons (82).
8 Discussion
This review integrates evidence of GI microbiome being an important part of the pathogen-host interactions in tropical infections. Dysbiosis is heavily involved in the pathogenesis of infections like amoebiasis, dengue, malaria, and cholera, rather than being a consequence of disease (83). This influence on the disease outcomes is supported by clinical as well as experimental data. For instance, research shows that initial GI microbial composition has an effect on malaria susceptibility. A study in Mali showed children with particular gut microbiota profile got less episodes of febrile malaria implying that these microbial patterns offer protection (84). Research using experimental models confirmed that cerebral malaria disrupts intestinal structure and alters microbial communities which suggest involvement of the gut-brain-immune axis (88). Waide and Schmidt (87) demonstrated that metabolites produced by the gut can affect both systemic immune reactions and the elimination of parasites. The gut microbiome’s integrity serves as an evident factor that impacts cholera disease progression. Vibrio cholerae successfully colonizes environments with dysbiotic microbiota which is commonly found in areas with endemic disease and populations experiencing undernutrition and previous infections. Animal studies using microbiota transplant models demonstrate that rebalancing gut microorganisms reduces cholera toxin-induced disease and stops pathogen adhesion mainly through changes in bile acid metabolism and nutrient competition (86).
Patients with severe dengue exhibit both enhanced gut permeability and microbial elements translocating into their bloodstream. Suwannakarn et al. A study by Suwannakarn et al. (89) found that severe dengue patients had increased endotoxin and microbial DNA levels in their blood and these levels related to stronger inflammatory reactions and negative health outcomes. The GI barrier and microbiome play significant roles in the immunopathogenesis of dengue. The severity of amoebiasis infections can increase when patients experience dysbiosis resulting from antibiotic treatment or an inadequate diet. Research using animals demonstrates that Entamoeba histolytica infection worsens colitis in hosts with depleted microbiota because of reduced neutrophil recruitment and defective mucosal immunity (87). Soil-transmitted helminth infections in India led to reduced Lactobacillus levels alongside modified short-chain fatty acid profiles which potentially impaired immune function and nutrient absorption (90). Helminths play a dual role in which they both influence the gut microbiota while being influenced by it. The current literature has both strengths and limitations which need consideration. Next-generation sequencing platforms including 16S rRNA gene sequencing and whole-genome metagenomics have become essential for modern research by enabling precise taxonomic and functional microbiome analysis particularly in tropical environments. The adoption of advanced sequencing platforms has enabled researchers to discover microbial patterns linked to infection severity as well as recovery and treatment results (87).
Nonetheless, the field faces several limitations. Cross-sectional study designs dominate the field and create obstacles for drawing causal conclusions. Research studies commonly use limited sample sizes that focus only on hospital-based cohorts and lack consistent protocols for sampling and sequencing as well as unified definitions for outcomes. The scientific literature lacks longitudinal studies which follow the microbiome from initial infection to full recovery to evaluate both resilience and ongoing dysbiosis. Establishing a standard healthy microbiome for tropical populations proves difficult because individual differences in microbiome composition arise from dietary habits, sanitation conditions, co-infections, and exposure to antimicrobials. Research designs frequently overlook socio-environmental factors that shape microbiome composition and data from rural communities is particularly limited. Microbiome dysbiosis manifests in both tropical infections and non-tropical diseases such as influenza and COVID-19. Alterations in microbiome occur beyond tropical diseases and scientists have found similar changes in non-tropical infections including influenza, COVID-19 and Clostridioides difficile infection. Microbiome dysbiosis shows decreased protective Firmicutes alongside increased proinflammatory Enterobacteriaceae taxa. The pathophysiology of COVID-19 includes gut barrier disruption and systemic endotoxemia which resembles the condition seen in serious dengue cases (87, 89).
Tropical infections typically create complex interactions of stressors alongside malnutrition and chronic parasitic loads which lead to environmental enteropathy that results in more intricate or permanent dysbiosis. The influence of cultural and dietary factors such as fiber consumption and herbal medicine use on microbiome recovery trajectories complicates direct comparisons between tropical and non-tropical populations. The study of microbiome involvement in tropical diseases presents extensive practical applications for healthcare professionals and public health policies. Medical professionals can use microbiome profiles as indicators to predict disease outcomes and monitor treatment effectiveness. The discovery of protective microbiota related to malaria will lead to specific probiotic development approaches. Disease control plans can definitely benefit from public health strategies primarily focusing on microbiome restoration and preservation. Similarly, in areas where cholera is quite prevalent, several gut-stabilizing methods can serve as additional support to oral rehydration therapy. Nutritional and probiotic interventions guided by microbiome data show great potential for malnourished children suffering from frequent diarrhea and delayed recovery periods. The application of post-antibiotic microbiota recovery methods can lead to improved results in amoebiasis management while deworming programs that consider microbiome preservation minimize adverse effects during helminth treatment. Better gut health can increase vaccine effectiveness which becomes especially important in areas where vaccination rates are low (88).
Upcoming studies must focus on longitudinal research that follows infection progression through recovery stages and evaluates lasting results across various tropical environments. Critical assessment through interventional trials of probiotics, synbiotics, and postbiotics effect on infection outcomes remains an urgent research priority. Microbiome datasets that are standardized and openly accessible from tropical regions which are currently underrepresented would enable researchers to conduct comparative studies across different research projects and perform meta-analyses. Integrated multi-omics approaches that combine microbiome data with metabolome and host immune information are crucial for discovering mechanistic connections. Future research efforts need to address specific contexts and demonstrate cultural sensitivity. More collaboration is required among clinicians, microbiologists, local communities and healthcare workers for successful research. Research approaches need to represent the dietary habits and traditional medical practices of specific regions (89, 90).
9 Conclusion
Gastrointestinal microbiome dislocations are constantly linked with tropical infections like cholera, malaria, amoebiasis, and dengue. Consequently, these may impact host strength, susceptibility to disease, and clinical recovery. Plenty of evidence is available highlighting clinical benefits of maintaining or restoring a balanced microbiota. As shown in Table 2.
The gut microbiome is not merely an epiphenomenon of tropical disease but an active mediator of host-pathogen dynamics. Its effect ranges from susceptibility to recovery and possibly to long-term consequences. Recognizing this major role allows new avenues for diagnostics, prevention, and treatment. We need more rigorous research on how to use the GI microbiome to help treat tropical infections. This research should bring together different kinds of experts, focus on the needs of each region, and fit into global health plans. If we can understand and use the microbiome better, it could be a game-changer for helping people who are most affected by these diseases.
Author contributions
BA: Writing – original draft, Writing – review & editing, Conceptualization, Project administration. MU: Project administration, Writing – original draft, Writing – review & editing. MS: Project administration, Writing – original draft, Writing – review & editing, Visualization. AA: Methodology, Writing – original draft, Writing – review & editing, Investigation. KB: Investigation, Methodology, Writing – original draft, Writing – review & editing. TS: Investigation, Visualization, Writing – original draft, Writing – review & editing. YZ: Writing – original draft, Writing – review & editing, Investigation, Methodology. EG: Investigation, Writing – original draft, Writing – review & editing. ZA: Investigation, Writing – original draft, Writing – review & editing. AS: Writing – original draft, Writing – review & editing, Methodology, Supervision.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ruan W, Engevik MA, Spinler JK, and Versalovic J. Healthy human gastrointestinal microbiome: composition and function after a decade of exploration. Digestive Dis Sci. (2020) 65:695–705. doi: 10.1007/s10620-020-06118-4
2. Trakman GL, Fehily S, Basnayake C, Hamilton AL, Russell E, Wilson-O'Brien A, et al. Diet and gut microbiome in gastrointestinal disease. J Gastroenterol Hepatol. (2022) 37(2):237–45. doi: 10.1111/jgh.15728
3. Cook GC. Tropical gastroenterological problems. Manson's Trop Diseases. (2020) 22:107. doi: 10.1016/B978-1-4160-4470-3.50014-8
4. Assemie MA, Shitu Getahun D, Hune Y, Petrucka P, Abebe AM, Telayneh AT, et al. Prevalence of intestinal parasitic infection and its associated factors among primary school students in Ethiopia: A systematic review and meta-analysis. PloS Neglected Trop Diseases. (2021) 15(4):e0009379. doi: 10.1371/journal.pntd.0009379
5. Wiertsema SP, van Bergenhenegouwen J, Garssen J, and Knippels LM. The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients. (2021) 13(3):886. doi: 10.3390/nu13030886
6. Kogut MH, Lee A, and Santin E. Microbiome and pathogen interaction with the immune system. Poultry Sci. (2020) 99(4):1906–13. doi: 10.1016/j.psj.2019.12.011
7. Iacob S and Iacob DG. Infectious threats, the intestinal barrier, and its trojan horse: dysbiosis. Front Microbiol. (2019) 10:1676. doi: 10.3389/fmicb.2019.01676
8. Bidell MR, Hobbs ALV, and Lodise TP. Gut microbiome health and dysbiosis: A clinical primer. Pharmacother: J Hum Pharmacol Drug Ther. (2022) 42(11):849–57. doi: 10.1002/phar.2731
9. Madhogaria B, Bhowmik P, and Kundu A. Correlation between human gut microbiome and diseases. Infect Med. (2022) 1(3). doi: 10.1016/j.imj.2022.08.004
10. Ogunrinola GA, Oyewale JO, Oshamika OO, and Olasehinde GI. The human microbiome and its impacts on health. Int J Microbiol. (2020) 2020:8045646. doi: 10.1155/2020/8045646
11. Ruan W, Engevik MA, Spinler JK, and Versalovic J. Healthy human gastrointestinalmicrobiome: composition and function after a decade of exploration. Dig Dis Sci. (2020) 65(3):695–705. doi: 10.1007/s10620-020-06118-4
12. Gupta A, Singh V, and Mani I. Dysbiosis of human microbiome and infectious diseases. Prog Mol Biol Transl Sci. (2022) 192(1):33–51. doi: 10.1016/bs.pmbts.2022.06.016
13. Chen L, Sun M, Wu W, Yang W, Huang X, Xiao Y, et al. Microbiota metabolite butyrate differentially regulates Th1 and Th17 cells' differentiation and function in induction of colitis. Inflammation Bowel Dis. (2019) 25(9):1450–61. doi: 10.1093/ibd/izz046
14. Ghosh S, Whitley CS, Haribabu B, and Jala VR. Regulation of intestinal barrier function by microbial metabolites. Cell Mol Gastroenterol Hepatol. (2021) 11(5):1463–82. doi: 10.1016/j.jcmgh.2021.02.007
15. Lee JY, Tsolis RM, and Bäumler AJ. The microbiome and gut homeostasis. Science. (2022) 377(6601):eabp9960. doi: 10.1126/science.abp9960
16. Yang H, Sun Y, Cai R, Chen Y, and Gu B. The impact of dietary fiber and probiotics in infectious diseases. Microb Pathog. (2020) 140:103931. doi: 10.1016/j.micpath.2019.103931
17. Ramirez J, Guarner F, Bustos Fernandez L, Maruy A, Sdepanian VL, and Cohen H. Antibiotics as major disruptors of gut microbiota. Front Cell Infect Microbiol. (2020) 10:572912. doi: 10.3389/fcimb.2020.572912
18. Brown EM, Sadarangani M, and Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. (2013) 14(7):660–7. doi: 10.1038/ni.2611
19. Campbell C, Kandalgaonkar MR, Golonka RM, Yeoh BS, Vijay-Kumar M, and Saha P. Crosstalk between gut microbiota and host immunity: impact on inflammation and immunotherapy. Biomedicines. (2023) 11(2):294. doi: 10.3390/biomedicines11020294
20. Brosschot TP and Reynolds LA. The Impact of a Helminth-modified microbiome on host immunity. Mucosal Immunol. (2018) 11(4):1039–46. doi: 10.1038/s41385-018-0008-5
21. Regassa R, Tamiru D, Duguma M, and Belachew T. Environmental enteropathy and its association with water sanitation and hygiene in slum areas of Jimma Town Ethiopia. PloS One. (2023) 18(6):e0286866. doi: 10.1371/journal.pone.0286866
22. Mills M, Lee S, Piperata BA, Garabed R, Choi B, and Lee J. Household environment and animal fecal contamination are critical modifiers of the gut microbiome and resistome in young children from rural Nicaragua. Microbiome. (2023) 11(1):207. doi: 10.1186/s40168-023-01636-5
23. Chung The H and Le SH. Dynamic of the human gut microbiome under infectious diarrhea. Curr Opin Microbiol. (2022) 66:79–85. doi: 10.1016/j.mib.2022.01.006
24. Macbeth JC, Liu R, Alavi S, and Hsiao A. A dysbiotic gut microbiome suppresses antibody mediated-protection against Vibrio cholerae. iScience. (2021) 24(12):103443. doi: 10.1016/j.isci.2021.103443
25. Das O, Masid A, Chakraborty M, Gope A, Dutta S, Bhaumik M, et al. Butyrate driven raft disruption trots off enteric pathogen invasion: possible mechanism of colonization resistance. Gut Pathog. (2023) 15:19. doi: 10.1186/s13099-023-00545-0
26. Cho JY, Liu R, Macbeth JC, and Hsiao A. The interface of vibrio cholerae and the gut microbiome. Gut Microbes. (2021) 13(1):1937015. doi: 10.1080/19490976.2021.1937015
27. Saad G and Shams El-Din H. An overview on intestinal parasites and gut microbiome: a bidirectional relationship. J Egyptian Soc Parasitol. (2024) 54(1):1–10. doi: 10.21608/jesp.2024.351281
28. Lawson MAE, Roberts IS, and Grencis RK. The interplay between Trichuris and the microbiota. Parasitology. (2021) 148(14):1–8. doi: 10.1017/S0031182021000834
29. Kupritz J, Angelova A, Nutman TB, and Gazzinelli-Guimaraes PH. Helminth-induced human gastrointestinal dysbiosis: a systematic review and meta-analysis reveals insights into altered taxon diversity and microbial gradient collapse. mBio. (2021) 12(6):e0289021. doi: 10.1128/mBio.02890-21
30. Walusimbi B, Lawson MAE, Nassuuna J, Kateete DP, Webb EL, Grencis RK, et al. The effects of helminth infections on the human gut microbiome: a systematic review and meta-analysis. Front Microbiomes. (2023) 2:1174034. doi: 10.3389/frmbi.2023.1174034
31. Hai NT, Hongsrichan N, Intuyod K, Pinlaor P, Yingklang M, Chaidee A, et al. Strongyloides stercoralis infection induces gut dysbiosis in chronic kidney disease patients. PloS Negl Trop Dis. (2022) 16(9):e0010302. doi: 10.1371/journal.pntd.0010302
32. Nguyen HT, Hongsrichan N, Intuyod K, Pinlaor P, Yingklang M, Chaidee A, et al. Investigation of gut microbiota and short-chain fatty acids in Strongyloides stercoralis-infected patients in a rural community. Biosci Microbiota Food Health. (2022) 41(3):121–9. doi: 10.12938/bmfh.2021-054
33. Jenkins TP, Formenti F, Castro C, Piubelli C, Perandin F, Buonfrate D, et al. A comprehensive analysis of the faecal microbiome and metabolome of Strongyloides stercoralis infected volunteers from a non-endemic area. Sci Rep. (2018) 8(1):15651. doi: 10.1038/s41598-018-33937-3. Erratum in: Sci Rep. 2019 Jun 7;9(1):8571. doi: 10.1038/s41598-019-43508-9.
34. Afrin T, Murase K, Kounosu A, Hunt VL, Bligh M, Maeda Y, et al. Sequential changes in the host gut microbiota during infection with the intestinal parasitic nematode strongyloides venezuelensis. Front Cell Infect Microbiol. (2019) 9:217. doi: 10.3389/fcimb.2019.00217
35. Pane S, Sacco A, Iorio A, Romani L, and Putignani L. Strongyloides stercoralis infestation in a child: how a nematode can affect gut microbiota. Int J Mol Sci. (2021) 22(4):2131. doi: 10.3390/ijms22042131
36. Kedia S, Rampal R, Paul J, and Ahuja V. Gut microbiome diversity in acute infective and chronic inflammatory gastrointestinal diseases in North India. J Gastroenterol. (2016) 51(7):660–71. doi: 10.1007/s00535-016-1193-1
37. Mizutani T, Aboagye SY, Ishizaka A, Afum T, Mensah GI, Asante-Poku A, et al. Gut microbiota signature of pathogen-dependent dysbiosis in viral gastroenteritis. Sci Rep. (2021) 11(1):13945. doi: 10.1038/s41598-021-93345-y
38. Baldelli V, Scaldaferri F, Putignani L, and Del Chierico F. The role of enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms. (2021) 9(4):697. doi: 10.3390/microorganisms9040697
39. Shin NR, Whon TW, and Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. (2015) 33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011
40. Ng KM, Aranda-Díaz A, Tropini C, Frankel MR, Van Treuren W, O'Loughlin CT, et al. Recovery of the gut microbiota after antibiotics depends on host diet, community context, and environmental reservoirs. Cell Host Microbe. (2019) 26(5):650–665.e4. doi: 10.1016/j.chom.2019.10.011
41. Wetherington MT, Nagy K, Dér L, Ábrahám Á, Noorlag J, Galajda P, et al. Ecological succession and the competition-colonization trade-off in microbial communities. BMC Biol. (2022) 20(1):262. doi: 10.1186/s12915-022-01462-5
42. Fassarella M, Blaak EE, Penders J, Nauta A, Smidt H, and Zoetendal EG. Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut. (2021) 70(3):595–605. doi: 10.1136/gutjnl-2020-321747
43. Tian L, Wang X-W, Wu A-K, Fan Y, Friedman J, Dahlin A, et al. Deciphering functional redundancy in the human microbiome. Nat Commun. (2020) 11(1):6217. doi: 10.1038/s41467-020-19940-1
44. Johansson MEV, Sjövall H, and Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. (2013) 10(6):352–61. doi: 10.1038/nrgastro.2013.35
45. Dethlefsen L and Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. (2011) 108 Suppl 1(Suppl 1):4554–61. doi: 10.1073/pnas.1000087107
46. Chen Y, Cui W, Li X, and Yang H. Interaction between commensal bacteria, immune response and the intestinal barrier in inflammatory bowel disease. Front Immunol. (2021) 12:761981. doi: 10.3389/fimmu.2021.761981
47. Rumpret M, von Richthofen HJ, Peperzak V, and Meyaard L. Inhibitory pattern recognition receptors. J Exp Med. (2022) 219(1):e20211463. doi: 10.1084/jem.20211463
48. Zong X, Fu J, Xu B, Wang Y, and Jin M. Interplay between gut microbiota and antimicrobial peptides. Anim Nutr. (2020) 6(4):389–96. doi: 10.1016/j.aninu.2020.09.002
49. Larsen SB, Cowley CJ, and Fuchs E. Epithelial cells: liaisons of immunity. Curr Opin Immunol. (2020) 62:45–53. doi: 10.1016/j.coi.2019.11.004
50. Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. (2012) 488(7410):178–84. doi: 10.1038/nature11319
51. Bubier JA, Chesler EJ, and Weinstock GM. Host genetic control of gut microbiome composition. Mamm Genome. (2021) 32(4):263–81. doi: 10.1007/s00335-021-09884-2
52. Dogra SK, Doré J, and Damak S. Gut microbiota resilience: definition, link to health and strategies for intervention. Front Microbiol. (2020) 11:572921. doi: 10.3389/fmicb.2020.572921
53. Su C, Su L, Li Y, Long SR, Chang J, Zhang W, et al. Helminth-induced alterations of the gut microbiota exacerbate bacterial colitis. Mucosal Immunol. (2017) 11(1):144–57. doi: 10.1038/mi.2017.20
54. Cusumano G, Flores GA, Venanzoni R, and Angelini P. The impact of antibiotic therapy on intestinal microbiota: dysbiosis, antibiotic resistance, and restoration strategies. Antibiotics. (2025) 14(4):371. doi: 10.3390/antibiotics14040371
55. Zhang P. Influence of foods and nutrition on the gut microbiome and implications for intestinal health. Int J Mol Sci. (2022) 23(17):9588. doi: 10.3390/ijms23179588
56. Sarkar A, McInroy CJA, Harty S, Raulo A, Ibata NGO, Valles-Colomer M, et al. Microbial transmission in the social microbiome and host health and disease. Cell. (2024) 187(1):17–43. doi: 10.1016/j.cell.2023.12.014
57. Gagliardi A, Totino V, Cacciotti F, Iebba V, Neroni B, Bonfiglio G, et al. Rebuilding the gut microbiota ecosystem. Int J Environ Res Public Health. (2018) 15(8):1679. doi: 10.3390/ijerph15081679
58. Raheem A, Liang L, Zhang G, and Cui S. Modulatory effects of probiotics during pathogenic infections with emphasis on immune regulation. Front Immunol. (2021). doi: 10.3389/fimmu.2021.616713
59. You S, Ma Y, Yan B, Pei W, Wu Q, Ding C, et al. The promotion mechanism of prebiotics for probiotics: A review. Front Nutr. (2022) 9:1000517. doi: 10.3389/fnut.2022.1000517
60. Vyas U and Ranganathan N. Probiotics, prebiotics, and synbiotics: gut and beyond. Gastroenterol Res Pract. (2012) 2012, 1–16. doi: 10.1155/2012/872716
61. Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS, et al. Recovery of the gut microbiome following fecal microbiota transplantation. mBio. (2014) 5(3):e00893-14. doi: 10.1128/mBio.00893-14
62. Rinninella E, Tohumcu E, Raoul P, Fiorani M, Cintoni M, Mele MC, et al. The role of diet in shaping human gut microbiota. Best Pract Res Clin Gastroenterol. (2023) 62-63:101828. doi: 10.1016/j.bpg.2023.101828
63. Ghoshal UC and Ranjan P. Post-infectious irritable bowel syndrome: The past, the present and the future. J Gastroenterol Hepatol. (2011) 26:94–101. doi: 10.1111/j.1440-1746.2011.06643.x
64. Beatty JK. Post-infectious irritable bowel syndrome: Mechanistic insights into chronic disturbances following enteric infection. World J Gastroenterol. (2014) 20(14):3976. doi: 10.3748/wjg.v20.i14.3976
65. Soheilipour M, Chahichi A, Mohajer H, Ghomashi N, Roohafza H, and Adibi P. Risk factors of developing postinfectious irritable bowel syndrome in shigellosis patients, 5 years after hospitalization during the outbreak. Open Forum Infect Dis. (2024) 11(3):ofae032. doi: 10.1093/ofid/ofae032
66. Porcari S, Ingrosso MR, Maida M, Eusebi LH, Black C, Gasbarrini A, et al. Prevalence of irritable bowel syndrome and functional dyspepsia after acute gastroenteritis: systematic review and meta-analysis. Gut. (2024) 73(9):1431–40. doi: 10.1136/gutjnl-2023-331835. Erratum in: Gut. 2025 Jan 17;74(2):e12. doi: 10.1136/gutjnl-2023-331835corr1.
67. Suzuki H and Hibi T. Overlap syndrome of functional dyspepsia and irritable bowel syndrome - are both diseases mutually exclusive? J Neurogastroenterol Motil. (2011) 17(4):360–5. doi: 10.5056/jnm.2011.17.4.360
68. Mearin F, Pérez-Oliveras M, Perelló A, Vinyet J, Ibañez A, Coderch J, et al. Dyspepsia and irritable bowel syndrome after a Salmonella gastroenteritis outbreak: one-year follow-up cohort study. Gastroenterology. (2005) 129(1):98–104. doi: 10.1053/j.gastro.2005.04.012
69. Shimbori C, De Palma G, Baerg L, Lu J, Verdu EF, Reed DE, et al. Gut bacteria interact directly with colonic mast cells in a humanized mouse model of IBS. Gut Microbes. (2022) 14(1):2105095. doi: 10.1080/19490976.2022.2105095
70. Wang Y, Gao X, Zhang X, Xiao Y, Huang J, Yu D, et al. Gut microbiota dysbiosis is associated with altered bile acid metabolism in infantile cholestasis. mSystems. (2019) 4(6):e00463-19. doi: 10.1128/mSystems.00463-19
71. DeGruttola AK, Low D, Mizoguchi A, and Mizoguchi E. Current understanding of dysbiosis in disease in human and animal models. Inflammatory Bowel Dis. (2016) 22(5):1137–50. doi: 10.1097/mib.0000000000000750
72. Fakharian F, Thirugnanam S, Welsh DA, Kim W-K, Rappaport J, Bittinger K, et al. The role of gut dysbiosis in the loss of intestinal immune cell functions and viral pathogenesis. Microorganisms. (2023) 11(7):1849. doi: 10.3390/microorganisms11071849
73. Kaur H, Ali SA, and Yan F. Interactions between the gut microbiota-derived functional factors and intestinal epithelial cells – implication in the microbiota-host mutualism. Front Immunol. (2022) 13:1006081. doi: 10.3389/fimmu.2022.1006081
74. Antoni L, Nuding S, Weller D, Gersemann M, Ott G, Wehkamp J, et al. Human colonic mucus is a reservoir for antimicrobial peptides. J Crohn’s Colitis. (2013) 7(12):e652–64. doi: 10.1016/j.crohns.2013.05.006
75. Shaheen WA, Quraishi MN, and Iqbal TH. Gut microbiome and autoimmune disorders. Clin Exp Immunol. (2022) 209(2):161–74. doi: 10.1093/cei/uxac057
76. Yoo J, Groer M, Dutra S, Sarkar A, and McSkimming D. Gut Microbiota and Immune System Interactions. Microorganisms. (2020) 8(10):1587. doi: 10.3390/microorganisms8101587
77. Yan S, Ji Q, Ding J, Liu Z, Wei W, Li H, et al. Protective effects of butyrate on cerebral ischaemic injury in animal models: a systematic review and meta-analysis. Front Neurosci. (2024) 18:1304906. doi: 10.3389/fnins.2024.1304906
78. Gao K, Mu CL, Farzi A, and Zhu WY. Tryptophan metabolism: a link between the gut microbiota and brain. Adv Nutr. (2020) 11(3):709–23. doi: 10.1093/advances/nmz127
79. Ahmed H, Leyrolle Q, Koistinen V, Kärkkäinen O, Layé S, Delzenne N, et al. Microbiota-derived metabolites as drivers of gut-brain communication. Gut Microbes. (2022) 14(1):2102878. doi: 10.1080/19490976.2022.2102878
80. Clapp M, Aurora N, Herrera L, Bhatia M, Wilen E, and Wakefield S. Gut microbiota’s effect on mental health: The gut-brain axis. Clin Pract. (2017) 7(4):987. doi: 10.4081/cp.2017.987
81. Wu H-J and Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. (2012) 3(1):4–14. doi: 10.4161/gmic.19320
82. Zimmermann P and Curtis N. The influence of the intestinal microbiome on vaccine responses. Vaccine. (2018) 36(30):4433–9. doi: 10.1016/j.vaccine.2018.04.066
83. Singh R, Zogg H, Wei L, Bartlett A, Ghoshal UC, Rahaman M, et al. Gut microbial dysbiosis in the pathogenesis of gastrointestinal dysmotility and metabolic disorders. J Neurogastroenterol Motil. (2021) 27(1):19–34. doi: 10.5056/jnm20149
84. Yooseph S, Kirkness EF, Tran TM, Harkins DM, Jones MB, Torralba MG, et al. Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genomics. (2015) 16:631. doi: 10.1186/s12864-015-1839-8
85. Waide EH and Schmidt NW. The gut microbiome and malaria: a new frontier in host-pathogen interactions. Curr Trop Med Rep. (2021) 8(1):22–30. doi: 10.1007/s40475-021-00229-2
86. Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA Jr, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. (2014) 515(7527):423–6. doi: 10.1038/nature13738
87. Burgess SL, Gilchrist CA, Lynn TC, and Petri WA Jr. Parasitic protozoa and interactions with the host intestinal microbiota. Infect Immun. (2017) 85(8):e00101-17. doi: 10.1128/IAI.00101-17
88. Taniguchi T, Miyauchi E, Nakamura S, Hirai M, Suzue K, Imai T, et al. Plasmodium berghei ANKA causes intestinal malaria associated with dysbiosis. Sci Rep. (2015) 5:15699. doi: 10.1038/srep15699
89. Suwannakarn K, Punyadee N, Vongpunsawad S, Guntapong R, Theamboonlers A, and Poovorawan Y. Microbial translocation in patients with dengue hemorrhagic fever. J Infect Dis. (2020) 221(3):478–86. doi: 10.1093/infdis/jiz489
Keywords: gastrointestinal microbiome, dysbiosis, tropical infections, microbiota, microbial diversity
Citation: Ayalew BD, Umar M, Saeed M, Ali A, Berhane KA, Sharew TM, Zewdie YA, Getachew EY, Alemayehu ZG and Shewaye AB (2025) Tropical infections and the gut microbiome: dysbiosis, recovery, and clinical implications. Front. Trop. Dis. 6:1620822. doi: 10.3389/fitd.2025.1620822
Received: 30 April 2025; Accepted: 14 July 2025;
Published: 07 August 2025.
Edited by:
Vivek Podder, Mount Sinai Medical Center, United StatesReviewed by:
Diptaraj Sangramsing Chaudhari, Wayne State University, United StatesIana Malasevskaia, Private Clinic, Yemen
Azza Fahmy, Theodor Bilharz Research Institute, Egypt
Copyright © 2025 Ayalew, Umar, Saeed, Ali, Berhane, Sharew, Zewdie, Getachew, Alemayehu and Shewaye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abate Bane Shewaye, YWJhdGUuYmFuZUBhYXUuZWR1LmV0
 Biruk Demisse Ayalew1
Biruk Demisse Ayalew1 Muhammad Umar
Muhammad Umar Ahtisham Ali
Ahtisham Ali Kaleb Assefa Berhane
Kaleb Assefa Berhane Yonatan Abbawa Zewdie
Yonatan Abbawa Zewdie Eskeatnaf Yosef Getachew
Eskeatnaf Yosef Getachew Abate Bane Shewaye
Abate Bane Shewaye