Abstract
Introduction:
Human African Trypanosomiasis (HAT) is a Neglected Tropical Disease targeted for elimination by 2030. Control efforts have reduced prevalence, but diagnostic challenges for field surveillance persist. Rapid Diagnostic Test kits (RDTs), used as frontline diagnostic tools for field surveillance, are often based on Variant Surface Glycoproteins (VSGs), which undergo constant mutations and exhibit considerable geographical diversity. Some VSGs are absent in Cameroon and Nigerian trypanosome strains/isolates.
Method:
This study evaluated the reliability of HAT RDTs using human and animal blood samples from Nigeria. Seropositive samples from ELISA and CATT were tested with Abbott Bioline HAT 2.0 RDT.
Result:
All ELISA and CATT seropositive human samples tested negative on the RDT, whereas two seropositive animal samples were positive on the RDT. The animal samples were positive on the test line 1, which is the ISG 65. This implies that the RDT kit was unable to detect seropositive samples from Nigeria, and therefore raises concerns about the reliability/suitability of this RDT for HAT field surveillance in Nigeria.
Conclusion:
This study emphasizes the importance of incorporating diverse Trypanosoma strains into RDT development and ensuring validation across all endemic areas for effective field surveillance and disease control.
Introduction
Human African Trypanosomiasis (HAT), also known as sleeping sickness, is caused by two subspecies of Trypanosoma brucei: Trypanosoma brucei gambiense (Tbg), which causes gHAT in West and Central Africa, and Trypanosoma brucei rhodesiense (Tbr), which causes rHAT in East and Southern Africa. The parasite is transmitted by tsetse flies of the Glossina species (1). gHAT is characterized by a long incubation period (up to several years), chronic/intermittent fever and low parasitemia in the early stage; and severe neurological symptoms in the advanced stages. Some infected people remain asymptomatic (2). These features complicate early diagnosis and treatment (3). HAT primarily affects vulnerable populations in remote and underserved areas with limited access to healthcare infrastructure (4). Despite successful control efforts leading to reduced prevalence, disease detection is still challenging due to inadequate diagnostic tools, especially in rural regions where advanced diagnostic techniques are inaccessible. This is particularly important as the WHO and health authorities work toward the Neglected Tropical Disease (NTD) roadmap, which targets the elimination of these NTDs by 2030.
Although, HAT is considered endemic in Nigeria, the World Health Organization (WHO) database shows no reported case since 2012, except for a case diagnosed in the United Kingdom in a Nigerian traveler in 2016 (3). This lack of reported cases does not indicate the absence of disease, as it is unlikely that only one patient was infected in a country of over 200 million people by the vector that transmits the disease. Moreover, in the same period, several researchers have published evidence of HAT infections from different parts of the country, but were not captured in the WHO database/reports (2, 5). This raises concerns regarding local surveillance, diagnostic capacity and disease reporting. Odebunmi et al., 2024 highlighted the possibility of cryptic or silent infections, with the prevalence of HAT in Nigeria at 3.6%, emphasizing the limitations in diagnostic tools availability and efficacy, as well as irregular field surveillance, are likely contributors to an underestimation of the true disease burden in the country (6).
Field surveillance of HAT is hindered by limited availability of field diagnostic tools, and low sensitivity/specificity of available serologic assays. These challenges can result in false negatives and a false sense of low or no disease prevalence, further truncating control efforts. Rapid Diagnostic Tests (RDTs) have been integrated into surveillance programs to enhance disease detection (7). Recent multicountry evaluations have raised additional concerns regarding the reliability of current diagnostic tools in low-prevalence and geographically diverse settings. For instance, studies by Tablado Alonso et al. (9) and N’Djetchi et al. (8) have demonstrated inconsistencies in the performance of rapid diagnostic tests (RDTs), particularly in regions where local parasite strains differ from those used in assay development (8, 9). These findings underscore the critical need for context-specific validation of serological assays before widespread deployment. Its sensitivity and specificity, which are crucial for accurate diagnosis, vary across regions due to differences in parasite strains and geographic locations (8, 9). While these RDT kits offer practical solutions for field use, concerns persist about their reliability, especially in some low-prevalence settings with atypical isolates and Variant Surface Glycoprotein (VSG) patterns.
The Card Agglutination Test for Trypanosomiasis (CATT) is a widely used tool for field screening due to its simplicity and quick turnaround (10). It detects antibodies against the variant surface glycoprotein (VSG) Litat 1.3. However, this antigen is absent in some T. b. gambiense strains, including those found in countries like Cameroon and Nigeria (11, 12). This limits the test’s effective use in disease monitoring due to concerns for false negatives in these areas.
The Abbott Bioline HAT 2.0 RDT was developed as an improvement over earlier versions and detects antibodies using two recombinant antigens: invariant surface glycoprotein (ISG) 65 and VSG LiTat 1.5 (7). While it offers advantages such as affordability and field applicability, sensitivity and specificity remain an issue, particularly in malaria-endemic regions like Nigeria (7–9).
In alignment with the 2030 NTD roadmap, the WHO has prioritized the elimination of HAT transmission (zero cases), with emphasis on early case detection, improved diagnostic accuracy, and integration of tools suited for field surveillance (13). This includes expanding the use of RDTs and strengthening their reliability through localized validation. Therefore, understanding how RDTs perform in field conditions in Nigeria is not only relevant for national policy but is also essential for achieving global elimination targets.
Method
Ethical approval
Ethical approvals for the study were obtained from Enugu State Ministry of Health Research Ethics Committee (Ref No: MH/MSD/REC21/782), and the University of Nigeria, Nsukka Institutional Animal Care and Use Committee (IACUC). Informed consent was obtained from all human participants before sample collection.
Sample collection, storage and transport conditions
Blood samples were collected in EDTA-coated sample bottles from hospital diagnostic laboratories (for human samples) and abattoirs (for animal samples) from two states in Southern Nigeria (Delta and Enugu States). After collection, samples were transported in ice packs (2–8 °C) to the laboratory. Upon arrival, all samples were processed within few hours. Serum was extracted by centrifugation at 3000 rpm for 10 minutes and aliquoted into sterile, labeled vials. Serum samples not analyzed immediately were stored at 4 °C to preserve sample integrity.
Inclusion and exclusion criteria
Only samples that tested positive on both ELISA (1:200 serum dilution) and CATT (1:64) dilution were selected for subsequent evaluation with the RDT kit. The use of higher dilution levels was to minimize false-positive results and enhance specificity (14). Samples that failed to meet these criteria were excluded from RDT testing.
Test reproducibility and quality control
To ensure the reliability of results, each serologic test; ELISA, CATT, and RDT, was performed strictly according to standard protocol or manufacturer’s instructions. ELISA and CATT tests were performed in duplicates at each dilution rate, with appropriate controls. Standardized positive and negative controls supplied with the kits were included in every batch of ELISA and CATT assays to monitor test performance. Additionally, all test results were independently reviewed by trained laboratory personnel to minimize observer bias and ensure accurate interpretation.
Sample screening
The serum samples were first screened for HAT with ELISA using plates coated with native antigens. The samples that tested positive on ELISA at 1:40 and 1:200 serum dilution were then screened with CATT kit (obtained from the Institute of Tropical Medicine, Belgium) at 1:4, 1:8, 1:16, 1:32, 1:64 serum dilutions. Due to low availability of RDT kits, 11 human samples and 5 animal samples that tested positive on both ELISA (1:200 dilution) and CATT (1:64 dilution) following the flowchart (Figure 1) were further tested with the RDT kit (Abbott Bioline HAT 2.0), which was obtained from the Federal Ministry of Health.
Figure 1

Flowchart showing the sample analysis methodology. Image showing the flowchart of the methological approach for the serologic assays.
Results
182 human sera and 91 animal sera were screened on ELISA at 1:40 and 1:200. 60 human sera and 26 animal sera were ELISA-positive and were subsequently screened on CATT at 1:4 and 1:8 serum dilution. A strong agglutination was observed with CATT at these serum dilutions for 27 human and 8 animal ELISA-positive samples (Figure 2). These samples were also consistently positive at 1:16, 1:32 and 1:64 serum dilution of CATT test. 11 out of the 27 human sera and 5 out of the 8 animal sera positive on both ELISA (1:200) and CATT (1:64) were selected and screened using the RDT kits. However, none of the human samples was positive on the RDT, but 2 of the animal samples (both from pigs) were positive on the RDT kit at Test Line 1 (Figure 3). Test line 1 represents the Invariant Surface Glycoprotein antigen (ISG 65) component of the RDT kit. A summary of the samples tested and the results from all serologic tests done is shown in Table 1.
Figure 2
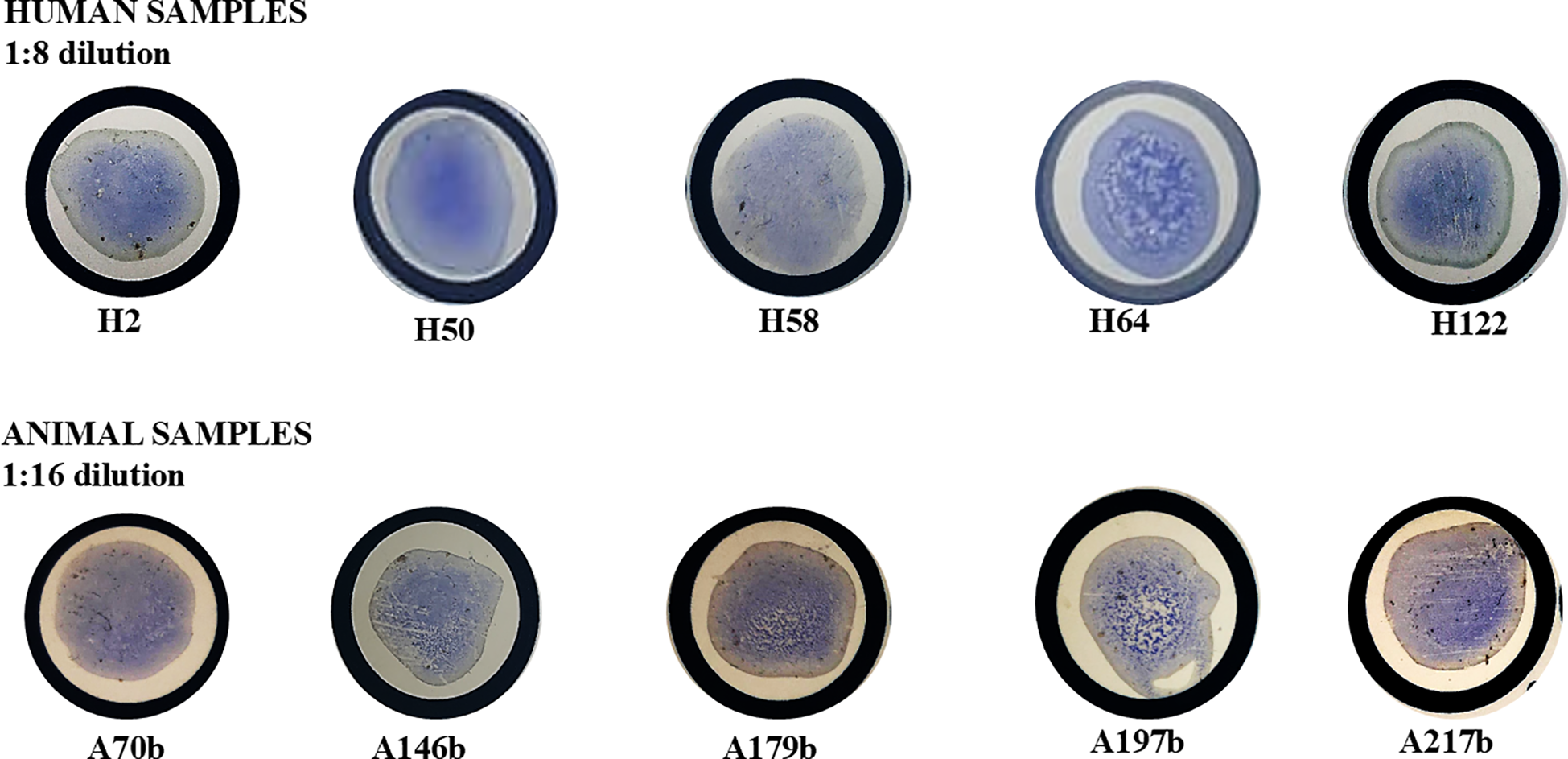
Representative positive results of CATT for ELISA-positive human and animal samples showing agglutination of the CATT antigen in the test serum. Samples showed strong agglutination (positive) up to 1:64 serum dilution. Alt text: Images of CATT assays of ELISA-positive serum samples showing agglutination (positive results) in all human and animal samples.
Figure 3
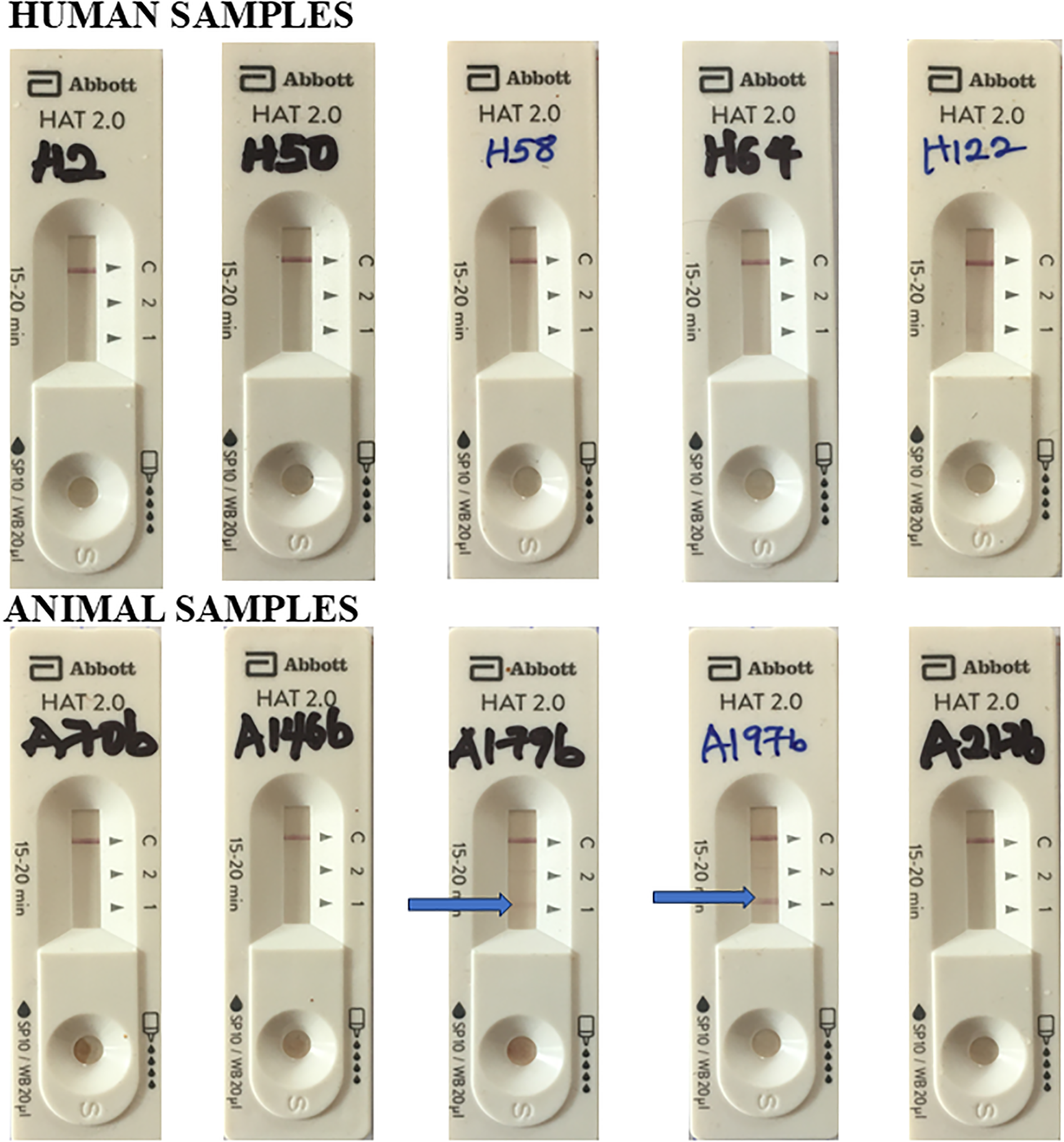
Representative results of Abbott HAT 2.0 RDT Kit for human and animal samples that were positive on ELISA and CATT assays. Arrow indicates positive result at test line 1. Test Line 1 = ISG 65; Test Line 2 = VSG LiTat 1.5. Alt text: Images of HAT RDT kit assays of seropositive human and animal samples showing positive result on test line 1 for only 2 of the animal samples and no positive human sample.
Table 1
| Sample ID | ELISA (1:200) | CATT (up to 1:64) | Abbott Bioline HAT 2.0 RDT KIT | Location in Nigeria | Specie | Age | Sex |
|---|---|---|---|---|---|---|---|
| H2 | + | + | – | Delta State | Human | 38 | M |
| H50 | + | + | – | Delta State | Human | 25 | M |
| H58 | + | + | – | Delta State | Human | 45 | F |
| H64 | + | + | – | Delta State | Human | 21 | F |
| H80 | + | + | – | Delta State | Human | 68 | F |
| H91 | + | + | – | Delta State | Human | 60 | F |
| H122 | + | + | – | Delta State | Human | 38 | M |
| H230b | + | + | – | Enugu State | Human | 15-30 | F |
| H398b | + | + | – | Enugu State | Human | 31-45 | M |
| H410b | + | + | – | Enugu State | Human | 16-30 | F |
| H525b | + | + | – | Enugu State | Human | 45-60 | M |
| A70b | + | + | – | Enugu State | Goat | >2yrs | M |
| A146b | + | + | – | Enugu State | Pig | >2yrs | M |
| A179b | + | + | + | Enugu State | Pig | <1yr | M |
| A197b | + | + | + | Enugu State | Pig | <1yr | F |
| A217b | + | + | – | Enugu State | Goat | >2yrs | F |
Summary of test results for human and animal samples using different serologic assays.
All human and animal samples tested negative on Abbott Bioline HAT 2.0 RDT KIT except for two animal samples, A179b and A197b.
+ = Positive; – = Negative.
Discussion
The WHO HAT epidemiological database shows that only one case was reported from Nigeria in 2016 in the last decade. This case was diagnosed in 2016 from a Nigerian visiting the United Kingdom (3), hence raising questions regarding HAT diagnosis and reporting in Nigeria. There is very little or no HAT surveillance or epidemiological data from Nigeria due to a complex combination of several factors, including the lack of appropriate diagnostic tools, inefficient disease reporting protocols, and little/no surveillance activities by health authorities. This study observed the inadequacies in the performance of the Abbott Bioline HAT 2.0 Rapid Diagnostic Test (RDT) in field conditions in Nigeria, using samples previously identified as seropositive by ELISA and CATT.
Interpretation of findings
The observed failure of the RDT kit to detect any of the ELISA/CATT seropositive human samples whereas it was able to detect two seropositive animal samples is both surprising and unexpected. This raises important concerns regarding the sensitivity and specificity of the RDT in this geographic location, suggesting potential limitations in its sensitivity and diagnostic reliability for Human African Trypanosomiasis in Nigeria. It has been demonstrated that Trypanosoma brucei gambiense VSGs and immune-reactivity tests are clearly correlated with geographical origin, and Nigerian samples have exhibited variable responses to immunologic/serologic tests as the targeted Variable Antigen Type (VAT) for these serologic tests are known to be absent in isolates from certain geographical locations such as Nigeria and Cameroon (11, 12). It is becoming apparent that variations in local strains, as indicated by the genetic diversity of Trypanosoma species across different geographical regions, may compromise the effectiveness of serologic tests generally, including RDTs (11). These serologic tools, which were developed and validated elsewhere, may not reliably detect Nigeria’s local parasite strains, thereby increasing the risk of false negatives (7–9). This RDT kit has previously been shown to perform poorly with unreliable results in passive surveillance studies in Nigeria (15). However, it is still being used for case detection and surveillance by the health authorities. This could contribute largely to the failure to detect cases in Nigeria, leading to the misleading prevalence reports/data recorded by WHO for policy implementation (6). Hence, there is a need to take into consideration the geographic diversity of Trypanosoma strains in the development of diagnostic tools.
Specificity and sensitivity
The RDT kits are specifically intended for use in the diagnosis of human Trypanosoma brucei gambiense infections, and are being used by the Federal Ministry of Health for HAT surveillance in Nigeria, though with variable results in different regions (16). The usual concern for HAT RDTs in many endemic areas is the occurrence of false positive reactions (low specificity), which are believed to be associated with cross-reacting antibodies in malaria co-endemic areas (7, 17). In high-prevalence settings, a test with low specificity will produce more false positives. Hence, increasing specificity ensures that positive results are more likely to be true cases. This improves the positive predictive value (PPV) which is the likelihood that a person who tests positive actually has the disease. It is widely believed that RDTs using recombinant antigens have higher specificity than those with native antigens (9, 18). Hence, the use of recombinant antigens in the Abbot Bioline HAT 2.0 RDT kit was supposed to improve specificity from previous RDTs. However, the findings in this report show that this may compromise sensitivity in some cases, especially in low-prevalence settings where a higher sensitivity is more desirable to ensure that all positive cases are detected. This is particularly important when disease elimination is targeted, as is the case for HAT. Nigeria has low HAT prevalence, and the use of a low-sensitive surveillance tool like this RDT kit portends great dangers for false negatives, thereby increasing the risk of missed infections, continued disease transmission, and the possibility of disease/epidemic re-emergence. This is in addition to the well-established absence of some VATs used in HAT serologic tests like RDTs in isolates from certain geographical locations such as Nigeria and Cameroon (11, 12).
While indigenous researchers have reported HAT case detection in Nigeria using several serologic, molecular, and biological means (2, 19, 20), these reports are hardly validated by the health authorities for appropriate disease reporting. This is because WHO largely relies on a particular serologic test (trypanolysis) for the validation of detected cases before official acceptance for reporting, despite the identified drawbacks of these serologic tests with regards to VSG and VAT geographical variability (10, 14). It is interesting to note that even the WHO-recorded case of 2016 was also negative in a trypanolysis test (3). Although these serologic tests have been acclaimed to be specific for the detection of human infection by T. brucei gambiense, Ilboudo et al., 2022 had demonstrated using pigs that the VATs used in the trypanolysis test is not specific for Tbg infection (14). Similarly, Matovu et al. (18) reported that serological tests designed for gambiense HAT can detect antibodies in cattle, further supporting the possibility of cross-reactivity or non-specific binding in animal hosts (18). The positive test results in animal samples obtained in this study raises important questions about the specificity of these diagnostic tools. One may argue that it is possible these positive animal samples were infected with Tbg. But the question remains as to why the RDT kit is not detecting the human samples that were also positive on ELISA and CATT like the animal samples. On the other hand, the animal samples were positive in the invariant surface glycoprotein (ISG) test line of the RDT (test line 1), and it is possible that this antigen is not specific to human or Tbg infection. It is also possible that the RDT was detecting ISG from other animal trypanosomes in the animal samples. However, these RDTs were designed to specifically detect Tbg infection in human samples. This particularly highlights the need to investigate the specificity of antigens used in these serologic tests for human Trypanosoma infections (14).
Implications
The use of this RDT for HAT surveillance may significantly underestimate the burden of HAT in Nigeria because failure to detect seropositive human samples and the detection of animal samples in this study location indicates significant shortcomings in the sensitivity and specificity of the RDT kit. Hence, there is a need to validate diagnostic tools in various geographical regions before field deployment. Although this study used a small number of samples with the RDT kit, the observations are consistent with the results previously reported that indicated the unreliability of the RDT kit in passive HAT surveillance in Nigeria (15).
Conclusion
The findings of this study highlight critical limitations in the performance of the Abbott Bioline HAT 2.0 RDT in field conditions in Nigeria. To address these gaps, we recommend that locally tailored validation studies be conducted before the deployment of any HAT RDTs. Such studies should assess diagnostic performance in field settings, considering the diverse epidemiological profiles within endemic regions. In addition, combining serological tests like ELISA and CATT with molecular techniques such as PCR could enhance diagnostic accuracy, particularly in areas with suspected silent or low-level transmission. Furthermore, HAT surveillance programs must be strengthened through structured training, diagnostic quality assurance, and the integration of context-specific diagnostic algorithms, to guide evidence-based policies and support the 2030 HAT elimination targets.
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by University of Nigeria Health Research Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by University of Nigeria Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CC: Resources, Writing – original draft, Data curation, Validation, Writing – review & editing, Conceptualization, Investigation, Supervision, Methodology. OA: Validation, Formal Analysis, Data curation, Writing – review & editing, Methodology, Investigation, Writing – original draft. JC: Formal Analysis, Methodology, Writing – original draft, Investigation. AE: Investigation, Writing – original draft, Formal Analysis, Methodology. CI: Writing – original draft, Investigation, Methodology, Formal Analysis. UA: Data curation, Formal Analysis, Writing – original draft, Investigation. CU: Investigation, Writing – original draft, Data curation. MJ: Investigation, Writing – original draft, Data curation. GJ: Investigation, Writing – original draft, Data curation. AO: Supervision, Conceptualization, Writing – original draft, Project administration. SA: Conceptualization, Project administration, Supervision, Writing – review & editing, Resources.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This document has been produced with the financial assistance of the Bill and Melinda Gates Foundation Investment ID INV-007175 (formerly OPP1191735) through the African Postdoctoral Training Initiative (APTI) programme. APTI is implemented by the African Academy of Sciences with support from the Bill and Melinda Gates Foundation and in Partnership with the US National Institutes of Health, where the grantees receive mentorship during their first and second years of the fellowship. The contents of this document are the sole responsibility of the author(s) and can under no circumstances be regarded as reflecting the position of the Bill and Melinda Gates Foundation, the African Academy of Sciences, and the US National Institutes of Health.
Acknowledgments
We acknowledge the support of the African Academy of Sciences for the Vaccine Research Centre, University of Nigeria.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
NTD, Neglected Tropical Disease; CATT, Card Agglutination Test for Trypanosomiasis; ELISA, Enzyme Linked Immunosorbent Assay; HAT, Human African Trypanosomiasis; RDT, Rapid Diagnostic Test; VSG, Variant Surface Glycoprotein; ISG, Invariant Surface Glycoprotein; VAT, Variable Antigen Type.
References
1
Papagni R Novara R Minardi ML Frallonardo L Panico GG Pallara E et al . Human African Trypanosomiasis (sleeping sickness): Current knowledge and future challenges. Front Trop Dis. (2023) 4. doi: 10.3389/fitd.2023.1087003
2
Ngutor KS Idris LA Oluseyi Oluyinka O . Silent Human Trypanosoma brucei gambiense Infections around the Old Gboko Sleeping Sickness Focus in Nigeria. J Parasitol Res. (2016) 2016:2656121. Available online at: https://pmc.ncbi.nlm.nih.gov/articles/PMC4752997/.
3
Luintel A Lowe P Cooper A MacLeod A Büscher P Brooks T et al . Case of Nigeria-acquired human African trypanosomiasis in the United Kingdom, 2016. Emerg Infect Dis. (2016) 23:1225–7. doi: 10.3201/eid2307.170695
4
Ozioko K Okoye C Obiezue R Idika I Awudu R Ezewudo B et al . Accelerating towards human African trypanosomiasis elimination: Issues and opportunities. J Vector Borne Dis. (2020) 57:105. doi: 10.4103/0972-9062.310860
5
Nmorsi OPG Isaac C Igbinosa IB Umukoro DO Aitaikuru DP . (2010). Human African trypanosomiasis in endemic focus of Abraka, Nigeria. Asian Pac J Trop Med. 3(6):448–50. doi: 10.1016/s1995-7645(10)60107-1
6
Odebunmi EO Ibeachu C Chukwudi CU . Prevalence of human and animal African trypanosomiasis in Nigeria: A scoping review. medRxiv. (2024) 2024. doi: 10.1101/2024.04.21.24306055v1
7
Camara O Kaboré JW Soumah A Leno M Bangoura MS N’Diaye D et al . Conducting active screening for human African trypanosomiasis with rapid diagnostic tests: The Guinean experience (2016–2021. PLoS Negl Trop Dis. (2024) 18:11985. doi: 10.1371/journal.pntd.0011985
8
N’Djetchi MK Camara O Koffi M Camara M Kaba D Kaboré J et al . Specificity of serological screening tests and reference laboratory tests to diagnose gambiense human African trypanosomiasis: a prospective clinical performance study. Infect Dis Poverty. (2024) 13. doi: 10.1186/s40249-024-01220-5
9
Tablado Alonso S Biéler S Luz R Verlé P Büscher P Hasker E . Retrospective clinical performance evaluation of the Abbott Bioline HAT 2.0, a rapid diagnostic test for human African trypanosomiasis based on recombinant antigens. Trop Med Int Health. (2025) 30:135–42. doi: 10.1111/tmi.14077
10
Chappuis F Loutan L Simarro P Lejon V Büscher P . Options for field diagnosis of Human African trypanosomiasis. Clin Microbiol Rev. (2005) 18:133–46. doi: 10.1128/CMR.18.1.133-146.2005
11
Dukes P Gibson W Gashumba J Hudson K Bromidge T Kaukus A et al . Absence of the LiTat 1.3 (CATT antigen) gene in Trypanosoma brucei gambiense stocks from Cameroon. Acta Trop. (1992) 51:123–34. doi: 10.1016/0001-706X(92)90054-2
12
Meirvenne N Magnus E Büscher P . Evaluation of variant specific trypanolysis tests for serodiagnosis of human infections with Trypanosoma brucei gambiense. Acta Trop. (1995) 60:189–99. doi: 10.1016/0001-706x(95)00127-z
13
Franco JR Priotto G Paone M Cecchi G Ebeja AK Simarro PP et al . The elimination of human African trypanosomiasis: Monitoring progress towards the 2021–2030 WHO road map targets. PLoS Negl Trop Dis. (2024) 18. doi: 10.1371/journal.pntd.0012111
14
Ilboudo K Hounyeme RE Kabore J Boulangé A Gimonneau G Salou E et al . Experimental evidence that immune trypanolysis using the LiTat 1.3 and LiTat 1.5 variant antigen types is not specific to Trypanosoma brucei gambiense in pigs. Parasite. (2022) 29. doi: 10.1051/parasite/2022063
15
Zongo K Emmanuel RT . Advancing diagnosis and treatment for human African trypanosomiasis in Nigeria: challenges and future directions. Front Trop Dis. (2024) 5:1–4. doi: 10.3389/fitd.2024.1503421
16
Lumbala C Bessell PR Lutumba P Baloji S Biéler S Ndung’u JM . Performance of the SD BIOLINE® HAT rapid test in various diagnostic algorithms for gambiense human African trypanosomiasis in the Democratic Republic of the Congo. PLoS One. (2017) 12. doi: 10.1371/journal.pone.0180555
17
Jamonneau V Camara O Ilboudo H Peylhard M Koffi M Sakande H et al . Accuracy of individual rapid tests for serodiagnosis of gambiense sleeping sickness in West Africa. PLoS Negl Trop Dis. (2015) 9:3480. doi: 10.1371/journal.pntd.0003480
18
Matovu E Kitibwa A Picado A Biéler S Bessell PR Ndung’u JM . Serological tests for gambiense human African trypanosomiasis detect antibodies in cattle. Parasit Vectors. (2017) 10. doi: 10.1186/s13071-017-2487-8
19
Eneh CI Uwaezuoke SN Okafor HU Edelu B Ogbuka S . Human African trypanosomiasis (sleeping sickness) in a Nigerian male adolescent and the treatment challenges: A case report. J Epidemiol Res. (2016) 2. doi: 10.5430/jer.v2n2p31
20
Onah DN Ebenebe OO . Isolation of a human serum-resistant Trypanosoma brucei from a naturally infected pig in the Nsukka area of Enugu State. Niger Vet J. (2003) 24:37–43. doi: 10.4314/nvj.v24i1.3435
Summary
Keywords
Human African Trypanosomiasis, CATT, HAT RDT, field surveillance, variant surface glycoprotein, NTD
Citation
Chukwudi CU, Adenekan OS, Chinwendu JC, Enwonwu AO, Iyi CC, Anyaorah UH, Ugwu CV, John MO, Joseph GN, Ogugua AJ and Anika SM (2025) Field observations on the use of rapid diagnostic tests for Human African Trypanosomiasis in Nigeria. Front. Trop. Dis. 6:1638558. doi: 10.3389/fitd.2025.1638558
Received
30 May 2025
Accepted
13 August 2025
Published
19 September 2025
Volume
6 - 2025
Edited by
Illich Mombo, Centre International de Recherches Médicales de Franceville, Gabon
Reviewed by
Leticia Gomes De Pontes, Departamento de Bioquímica e Imunologia da Universidade de Minas Gerais, Brazil
Larson Boundenga, Centre International de Recherches Médicales de Franceville, Gabon
Updates
Copyright
© 2025 Chukwudi, Adenekan, Chinwendu, Enwonwu, Iyi, Anyaorah, Ugwu, John, Joseph, Ogugua and Anika.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chinwe U. Chukwudi, chinwe.chukwudi@unn.edu.ng; cuchukwudi@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.