- 1Jiangsu Co-innovation Center for the Prevention and Control of Important Animal Infectious Disease and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou, China
- 2Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou, China
- 3Jiangsu Research Center of Engineering and Technology for Prevention and Control of Poultry Disease, Yangzhou, China
The contamination of Salmonella Enteritidis in eggs and chicken meat via vertical transmission has become a worldwide public health concern. Biofilm formation by S. Enteritidis further enhances its antibacterial resistance. However, whether genes related to biofilm formation affect the level of vertical transmission is still unclear. Here, S. Enteritidis mutants ΔcsgD, ΔcsgA, ΔbcsA, and ΔadrA were constructed from wild type strain C50041 (WT), and their biofilm-forming ability was determined by Crystal violet staining assay. Then the median lethal dose (LD50) assay was performed to determine the effects of the selected genes on virulence. The bacterial load in eggs produced by infected laying hens via the intraperitoneal pathway or crop gavage was determined for evaluation of the vertical transmission. Crystal violet staining assay revealed that S. Enteritidis mutants ΔcsgD, ΔcsgA, and ΔbcsA, but not ΔadrA, impaired biofilm formation compared with WT strain. Furthermore, the LD50 in SPF chickens showed that both the ΔcsgD and ΔbcsA mutants were less virulent compared with WT strain. Among the intraperitoneally infected laying hens, the WT strain-infected group had the highest percentage of bacteria-positive eggs (24.7%), followed by the ΔadrA group (16%), ΔcsgA group (9.9%), ΔbcsA group (4.5%), and ΔcsgD group (2.1%). Similarly, among the crop gavage chickens, the WT strain group also had the highest infection percentage in eggs (10.4%), followed by the ΔcsgA group (8.5%), ΔadrA group (7.5%), ΔbcsA group (1.9%), and ΔcsgD group (1.0%). Our results indicate that the genes csgD and bcsA help vertical transmission of S. Enteritidis in chickens.
Introduction
Salmonella Enteritidis is one of the most prevalent serotypes of Salmonella isolated from poultry and is the most commonly reported cause of human salmonellosis (1, 2). Salmonella can cause many acute and chronic infections in poultry, and mortality from these infections in poultry causes low yield and economic losses (3, 4). Poultry and poultry products are regarded as the main source of S. Enteritidis (5). Notably, chickens infected with S. Enteritidis frequently exhibit no clinical symptoms (6). Therefore, chicken meat and eggs from seemingly healthy animals can be contaminated by S. Enteritidis, which is subsequently transmitted to humans along the food cycle, resulting in a continuing public health problem (7, 8). Furthermore, vertical transmission is an essential way in which S. Enteritidis spreads during poultry production period (9). Previous work demonstrated that internal contamination of eggs by S. Enteritidis could be caused by penetration through the eggshell or by direct contamination of the egg contents before oviposition, resulting in the vertical transmission of S. Enteritidis from breeding chickens to commercial chickens (10). The ovaries and oviducts of the laying hens are the major S. Enteritidis colonization sites in which vertical transmission to eggs occurs (11).
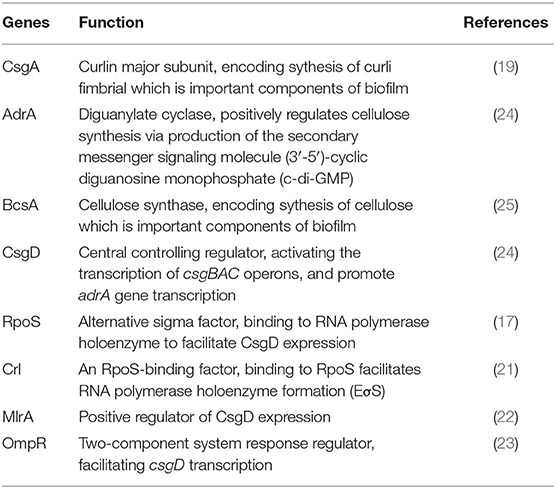
Biofilm formation is one of the most important mechanisms utilized by Salmonella to survive in host cells (12). It also contributes to bacterial resistance to adverse environments and helps the bacteria to evade host immune responses (13, 14). The in vivo pathogenicity of S. Enteritidis isolated from avian sources is partially related to the biofilm formation (15). Effective biofilm formation by S. Enteritidis also prolongs the survival time of these bacteria and enhances their resistance to host defenses (15). The biofilm-forming ability could facilitate the spread of S. Enteritidis. And the consumption of poultry-derived foods is an important route of human infection by S. Enteritidis (16). However, it remains unclear whether the ability of S. Enteritidis to form effective biofilms is related to vertical transmission among laying hens. Curli fimbria (17) and cellulose (18) are important components for biofilm formation. The gene csgD is a central controlling regulator that can activate the transcription of csgBAC operons and encode the synthesis of curli fimbriae (19). This gene also promotes adrA gene transcription, whose product interacts with bcsABZC-bcsEFG operons to synthesize cellulose (20). Crl, an RpoS-binding factor, binding to alternative sigma factor RpoS, facilitates RNA polymerase holoenzyme formation (EσS), further to enhance CsgD expression (21). Other regulators such as MlrA or OmpR could promote CsgD expression or transcription (22, 23). Overall, the four genes csgD, csgA, adrA, and bcsA are all directly related to biofilm components and finally are selected for investigation (Table 1).
In this study, deletion mutants of csgD, csgA, adrA, and bcsA were constructed by using the λred homologous recombination method. These mutants were used to explore whether the level of S. Enteritidis vertical transmission in chickens is influenced by biofilm-formation-related genes.
Materials and Methods
Animals and Ethics Statement
Eighty one-day-old specific pathogen-free (SPF) chickens were purchased from Merial Beijing Experimental Animal Technology Co., Ltd. (Beijing, China). Ninety-six 6-month-old laying hens were purchased from Jiangsu Lihua Co., Ltd. (Changzhou, China). All in vivo bird experiments were performed in the negative-pressure isolators of the authorized animal biosafety level 2 (ABSL-2) facilities at Yangzhou University. All bird experiments were approved by the Jiangsu Administrative Committee for Laboratory Animals and were conducted in compliance with the guidelines of laboratory animal welfare and ethics of the Jiangsu Administrative Committee for Laboratory Animals (Permission number: SYXKSU-2016-0020).
Bacterial Strains, Plasmids, and Growth Conditions
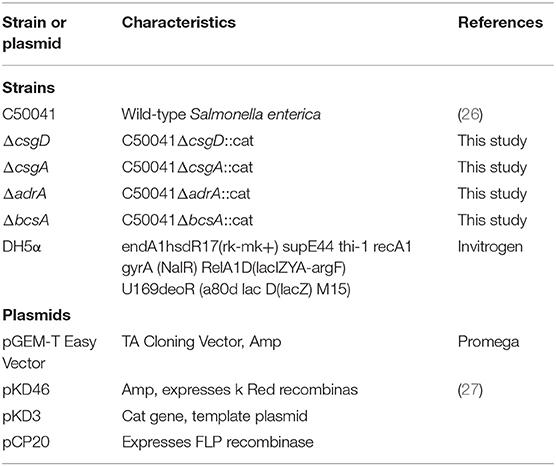
The strains and plasmids used in this study are listed in Table 2. Bacterial strains were routinely grown at 37°C in Luria Bertani (LB) broth or tryptic soy broth (TSB) with aeration. For strain selection, antibiotics were added at the following concentrations: 30 μg/mL chloramphenicol and 60 μg/mL ampicillin (28). Growth assays were performed in LB broth at 37°C with shaking at 220 rpm. Samples from each bacterial culture were spectrophotometrically monitored hourly for 8 h.
Construction of S. Enteritidis csgD, csgA, adrA, and bcsA Deletion Mutants
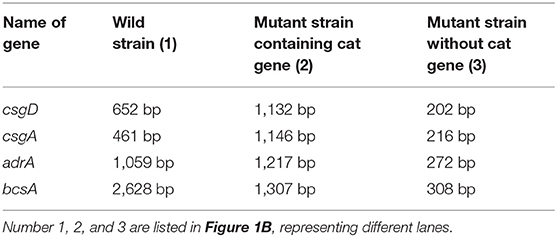
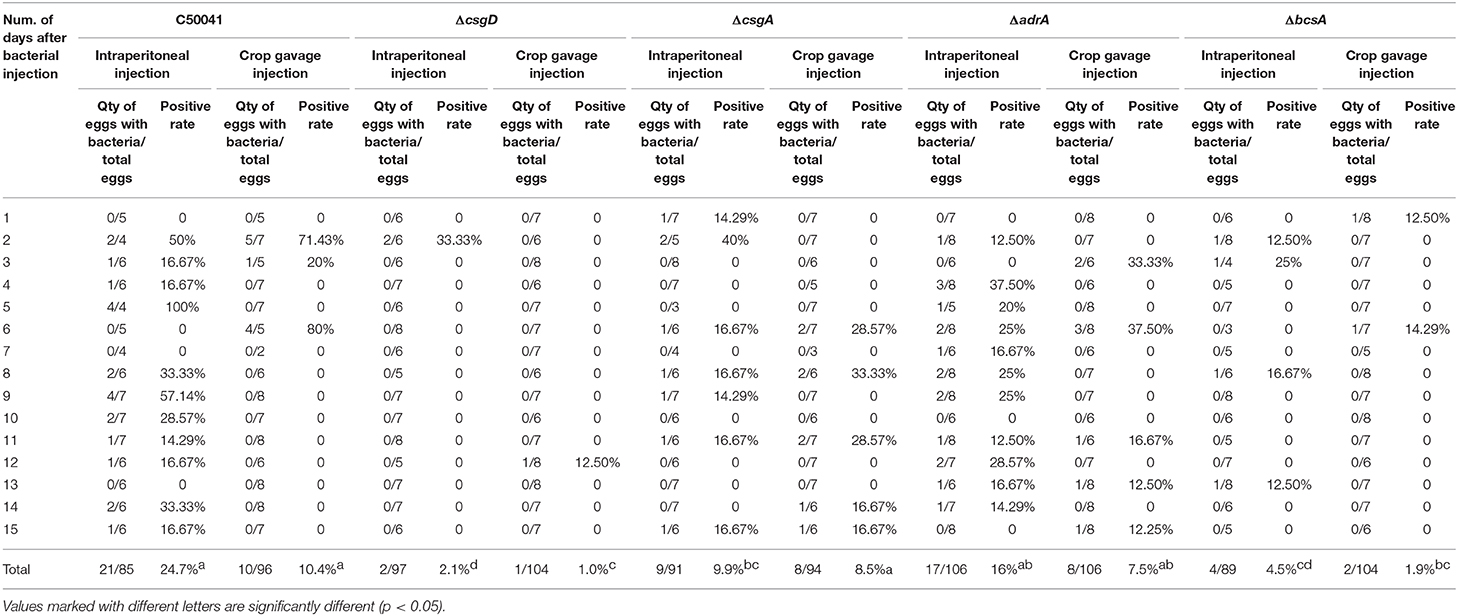
Deletions of csgD, csgA, adrA, or bcsA from the chromosome of S. Enteritidis C50041 were performed by using gene replacement methods based on the λRed recombinase system (29); the primers used in these protocols are listed in Table 3. The S. Enteritidis mutant strains C50041ΔcsgD, C50041ΔcsgA, C50041ΔadrA, and C50041ΔbcsA were constructed as shown in Figure 1A. The four genes csgD, csgA, adrA, and bcsA were amplified by PCR. The pKD3-encoded chloramphenicol resistance cassette was amplified using the primers csgD-D1/csgD-D2, csgA-D1/csgA-D2, adrA-D1/adrA-D2, and bcsA-D1/bcsA-D2 (Table 3). C50041 harboring plasmid pKD46 was electroporated with the csgD cat, csgA cat, adrA cat, or bcsA cat amplicon with 0.2% (wt/vol) L-arabinose at 30°C. Chloramphenicol-resistant transformants were selected at 37°C and were confirmed to have lost pKD46 on the basis of sensitivity to ampicillin conferred by the auxiliary plasmid pCP20. The successful creation of the mutants C50041ΔcsgD, C50041ΔcsgA, C50041ΔadrA, and C50041ΔbcsA was subsequently confirmed via PCR using primers in Table 3, respectively, followed by sequencing of the PCR products for verification.
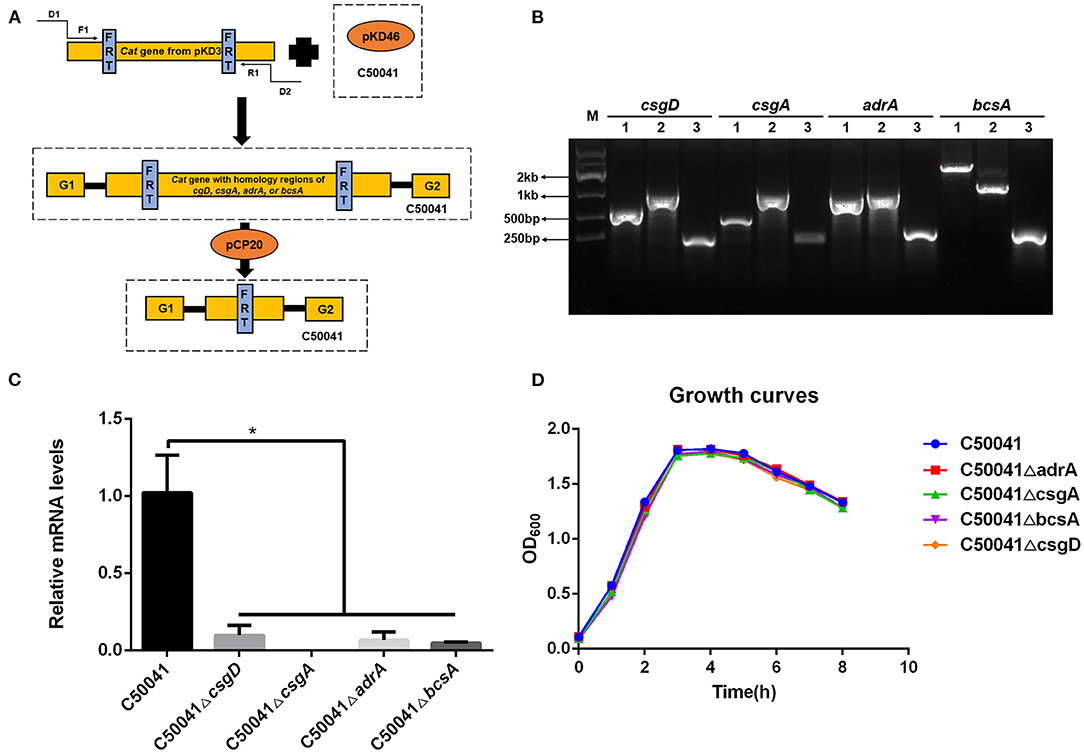
Figure 1. Construction of csgD, csgA, adrA, and bcsA deletion mutants. (A) Schematic diagram of the gene disruption strategy. D1 and D2 refer to the homology extensions or regions of csgD, csgA, adrA, or bcsA. F1 and R1 refer to the chloramphenicol resistance cassette priming sites that contain FRT sites. G1 and G2 refer to the upstream or downstream gene of the target genes. The specific steps used for constructing the mutants are described in section Construction of S. Enteritidis csgD, csgA, adrA, and bcsA Deletion Mutants. (B) PCR identification of the S. enteritidis mutant strains. (1) Wildtype strain; (2) Mutant strain containing the cat gene; (3) Mutant strain without the cat gene; M, marker. (C) Transcriptional levels of csgD, csgA, adrA, and bcsA genes. Means and standard deviations from three independent experiments are shown; *p < 0.05. (D) Growth curves of S. enteritidis WT and mutant strains (C50041ΔcsgD, C50041ΔcsgA, C50041ΔbcsA, and C50041ΔadrA).
Quantitative Real-Time PCR (qRT-PCR) Analysis
Bacteria were grown in 1/10 TSB medium at 28°C for 24 h in 60 mm dishes (Corning). The supernatant was discarded and the bacteria accumulated in the biofilms under the dishes were scraped. The total RNA was extracted using a Bacterial RNA Kit (Omega). The cDNA was synthesized using a PrimeScript RT reagent Kit with gDNA Eraser (Takara) and quantified via TB Green Premix Ex Taq (Takara). The gene transcript levels were tested in triplicate for real-time PCR in a Linegene 9600 Plus machine (Bioer). Primer pairs of Qgyrb-F/R, QcsgD-F/R, QcsgA-F/R, QadrA-F/R, and QbcsA-F/R (Table 3) were used for the mRNA detection of gyrB, csgD, csgA, adrA, and bcsA, respectively. The target genes' mRNA levels were normalized to the gyrB mRNA levels (2−ΔΔCt) (30).
Detection of Biofilm Formation
Biofilm formation by S. Enteritidis was detected by using the crystal violet staining quantitative method (28). For biofilm formation, a single bacterial colony was inoculated into 3 mL of TSB culture medium and subjected to shaking cultivation at 220 rpm at 37°C overnight, then this culture fluid was diluted 1/100 with 1/10 TSB diluent and then transferred to a 96-well U-shaped cell culture plate (100 μL/well) for static cultivation at 28°C for 24 h. For detection, the biofilm was stained with crystal violet, a 3:1 alcohol-acetone solution was used to dissolve the crystals, and the optical density at 550 nm (OD550 value) was measured on a microplate reader (Tecan, Switzerland).
Challenge of SPF Chickens With Wild Type Strain or Mutant S. Enteritidis Strains
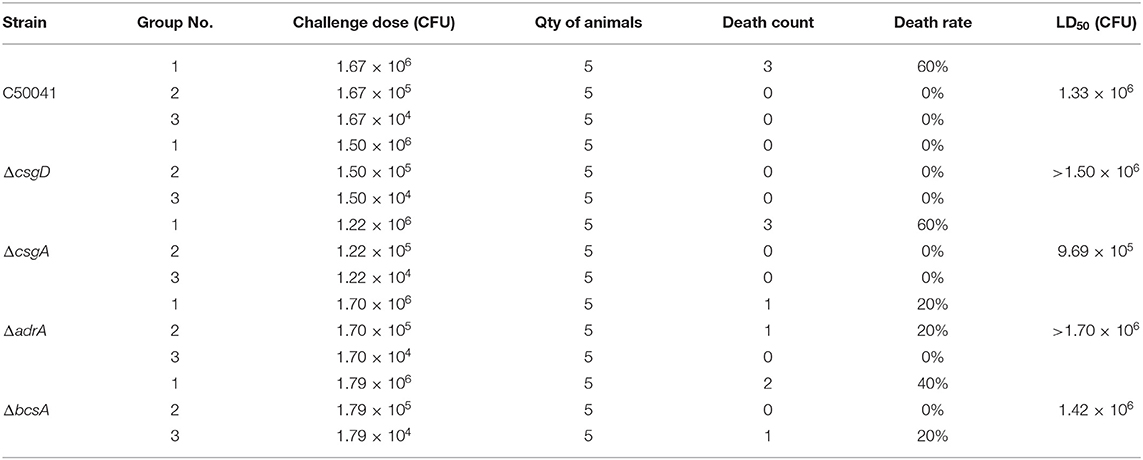
WT strain and its mutants were inoculated on a solid LB plate and statically cultivated at 37°C for 16–18 h. A few rich bacterial colonies were processed by the streak plate method and statically cultivated at 37°C for 3 h. Phosphate-buffered saline (PBS) containing 15% glycerol was used to wash the bacterial lawn. Eighty one-day old chickens were randomly divided into 16 groups (n = 5/group), with three groups per bacterial strain and one negative control group. Chickens from the challenged groups were intraperitoneally injected with 0.1 mL of bacterial fluid containing differing amounts of colony-forming units (CFU, Table 5). Chickens from the negative control group were intraperitoneally injected with 0.1 mL of PBS containing 15% glycerol. After 2 weeks of continual observation, the median lethal dose (LD50) was calculated by applying a modified Karber's method (31).
Experiments to Test S. Enteritidis Vertical Transmission Among Laying Hens
The Grouping of Laying Hens
Laying hens (96 birds, 6-month-old) in a peak period of egg production were randomly divided into six groups, each of which included a crop gavage subgroup (n = 8/subgroup, 109 CFU/chicken) and an intraperitoneal injection subgroup (n = 8/subgroup, 2 × 107 CFU/chicken). The negative control group was injected with PBS containing 15% glycerol. To determine whether the hens were previously exposed to Salmonella, blood samples were collected before challenge, and the resulting sera were tested with a diagnostic antigen (Diagnostic antigen for salmonella, China Veterinary Drug Supervision Institute, China) for Salmonella.
Detection of Eggs Contaminated by Wild Type Strain or Mutant S. Enteritidis Strains
The egg production quantity was recorded for 9 days before the injection. Two weeks after bacterial injection, the egg production was quantified for 9 days. For evaluating the level of vertical transmission, the S. Enteritidis load was detected in eggs produced every day (2 weeks). After sterilizing eggshells with 75% ethyl alcohol, the contents of the eggs were collected and sealed in sterile bags, and then incubated at 37°C for 48 h. The egg contents were subsequently inoculated onto MacConkey agar plates for 24 h at 37°C. The resulting suspected bacterial colonies were verified by using diagnostic serum (Diagnostic sera for salmonella, Sanshui Inspection Equipment Co. LTD, China) for Salmonella. This detection was performed continuously over 2 weeks.
Statistics
The results were analyzed with GraphPad Prism 8 software (San Diego, CA, USA) and are expressed as the means ± s.d. An unpaired Student's two-sided t-test was employed to determine the differences between the two groups. Differences with a p < 0.05 were considered to be significant. A chi-square test was applied to analyze the percentages of eggs in which bacteria were positively detected.
Results
Identification of Generated S. Enteritidis Mutants
The successful generation of S. Enteritidis mutant strains C50041ΔcsgD, C50041ΔcsgA, C50041ΔadrA, and C50041ΔbcsA was confirmed by using PCR (Figure 1B). After each target gene had been replaced by the chloramphenicol resistance gene, the size of the altered chromosome was estimated to be about 1.2 kb. After the resistance gene had been knocked out, the size of the gene was observed to be about 200–400 bp, which is consistent with the expected value (Table 4). The transcriptional levels of csgD, csgA, adrA, and bcsA genes were determined by relative qRT-PCR. The results showed that the four genes' transcriptional levels were significantly reduced in the mutants compared with the WT strain (Figure 1C). The growth rates of the S. Enteritidis mutant strains were similar to that of the wild type (WT) strain (Figure 1D).
Biofilm Formation of the S. Enteritidis Mutants
To examine the effects of the deleted genes on wild type strain biofilm formation, crystal violet staining tests were conducted. The WT strain C50041 and its mutant strains C50041ΔadrA and C50041ΔbcsA had similar amounts of circular staining in the plate well walls, whereas C50041ΔcsgD and C50041ΔcsgA had almost no circular staining (Figure 2A).
Figure 2. Measurement of S. enteritidis biofilms. (A) Crystal violet staining of bacteria grown on 96-well-plates. Overnight cultures were diluted in 1/10 TSB, and 100 μL of each bacterial suspension was added to a 96-well-plate, then incubated at 28°C for 24 h. Results were observed after staining with 0.4% crystal violet. (B) Quantification of the crystal violet staining by optical density (OD550) measurement. Means and standard deviations from three independent experiments are shown; **p < 0.01. Number 1, 2, and 3 means repetition of different strains in the same detection.
Quantifying the crystal violet staining results revealed that the OD550 values of C50041ΔcsgD, C50041ΔcsgA, and C50041ΔbcsA, but not that of C50041ΔadrA, were significantly lower than the OD550 value of the wild type strain (all p < 0.01), suggesting that biofilm formation was blocked after single mutations of csgD, csgA, or bcsA (Figure 2B).
Bacterial Virulence in SPF Chickens
Mortality Rate/LD50
As shown in Table 5, the group challenged with a dose of 1.67 × 106 CFU of the wild type strain had 60% mortality. In contrast, none of the SPF chickens died after challenge with C50041ΔcsgD at any of the tested infection doses. Furthermore, the LD50 value of the wild type strain was 1.33 × 106 CFU, whereas that of C50041ΔcsgD was >1.50 × 106 CFU, suggesting that the ability of S. Enteritidis to form a biofilm increased its bacterial virulence. Notably, the LD50 value of C50041ΔbcsA (1.42 × 106 CFU) and C50041ΔadrA (>1.70 × 106), but not that of C50041ΔcsgA (9.69 × 105 CFU), was also higher than the LD50 value of the wild type strain.
Detection of S. Enteritidis Vertical Transmission
Egg Production From the Infected Laying Hens
Before the laying hens were injected with bacteria, none of their serum samples agglutinated with diagnostic Salmonella antigens, suggesting these hens did not have a Salmonella infection. Two weeks after bacterial injection, the egg production was quantified for 9 days. As shown in Figure 3, the egg production capacity of the group infected with C50041 intraperitoneally (6.8 ± 1.0) was significantly lower than that of the non-infected group (p < 0.05). In contrast, there was no significant difference between the egg production of the non-infected group and the groups infected with any of the other mutants (p > 0.05).
Figure 3. Quantity comparison of egg production by the laying hens before and after injection of wild type strain and deletion mutants. Laying hens (96 birds, 6-month-old) in a peak period of egg production were randomly divided into six groups, each of which included a crop gavage subgroup (n = 8/subgroup, 109 CFU/chicken) and an intraperitoneal injection subgroup (n = 8/subgroup, 2 × 107 CFU/chicken). The negative control group was injected with PBS containing 15% glycerol. The egg production quantity was recorded for 9 days before and after the bacterial injection. Finally, the average of egg production was compared (*p < 0.05).
Bacterial Load in Eggs Produced by the Infected Laying Hens
After bacterial challenge, the bacterial load count in eggs produced by the infected laying hens was assessed daily. As shown in Table 6, among the intraperitoneally infected animals, the wild type-infected group had the highest infection percentage in eggs (24.7%), followed by the C50041ΔadrA group (16%), C50041ΔcsgA group (9.9%), C50041ΔbcsA group (4.5%), and C50041ΔcsgD group (2.1%). Similarly, among the crop gavage chickens, the wild type group also had the highest infection percentage in eggs (10.4%), followed by the C50041ΔcsgA group (8.5%), C50041ΔadrA group (7.5%), C50041ΔbcsA group (1.9%), and C50041ΔcsgD group (1.0%). Interestingly, the percentages of produced eggs were higher in the intraperitoneal injection subgroup than in the crop gavage subgroup. Together, these data indicate that the genes csgD and bcsA are closely related to the level of S. Enteritidis vertical transmission from infected laying hens to eggs.
Discussion
Bacteria within biofilms can enhance the resistance to adverse environments and prolonged survival. Cellulose and curli fimbria are both very important for biofilm formation by Salmonella bacteria (32). A review by Simm et al. suggests that most chronic infections are associated with the biofilm formation of microorganisms (19). In a S. Typhimurium model, the immune response to curli is site specific, and oral administration of curli ameliorates the damaged intestinal epithelial barrier and reduces the severity of colitis (33). Mauricio et al. found that preventing cellulose synthesis increased S. Typhimurium virulence, whereas stimulation of cellulose synthesis inside macrophages decreased the virulence (34). So far, whether biofilm-associated genes regulate the virulence in S. Enteritidis is unclear. Therefore, in this study, the S. Enteritidis genes bcsA and csgA, which encode cellulose and curli fimbria, respectively (19), as well as the genes csgD and adrA, which regulate the biofilm-related genes (20), were selected for investigation. We report here that mutants of S. Enteritidis with deletion in csgD, csgA, and bcsA, but not of adrA, display defects in the level of biofilm formation by S. Enteritidis. Our results also suggest that genes related to biofilm formation (csgD, csgA, bcsA, and adrA) can alter the virulence of S. Enteritidis differently. The mortality rates of chickens infected with these strains indicate that deletions of csgD or bcsA attenuated the virulence of WT strain whereas a deletion of csgA yielded the opposite result. Although the deletion of adrA had limited effects on the biofilm formation of S. Enteritidis, the LD50 value of the C50041ΔadrA mutant was much higher than that of the WT strain. These data reveal that biofilm formation is related to bacterial virulence, and in S. Enteritidis, synthesis of curli and cellulose could enhance its virulence.
S. Enteritidis can be transmitted vertically through laying hens (35) and may cause persistent infection. Although previous studies have shown that the biofilms may be related to persistent Salmonella infections (36), the relationship between biofilms and the vertical transmission of S. Enteritidis is still unclear. Here, we deleted four genes known to be related to biofilm formation and studied their roles in the vertical transmission of S. Enteritidis among laying hens. The results of our vertical transmission assay indicate that the genes csgD and bcsA significantly enhance the level of S. Enteritidis vertical transmission, whereas the genes csgA and adrA have limited effects. Within groups infected with the same S. Enteritidis strain, the percentages of produced eggs were higher in the intraperitoneal injection subgroup than in the crop gavage subgroup, which is consistent with the trends reported previously (37). Considering the biofilm makes the bacteria stuck somewhere, we speculated that biofilm could help the Salmonella better and longer survival in the reproductive tract or on the egg or associated environment. In adverse, deletion of cellulose encoded by bcsA prevented biofilm formation, further decreased the adaption of S. Enteritidis in produced eggs, resulting in a decrease in bacterial penetration through the eggshell or by direct contamination of the egg contents before oviposition.
As the central regulator of biofilm formation, csgD regulates the expression of CsgA and AdrA, AdrA further controls the BcsA expression (17). Therefore, deletion of csgD could affect the biofilm formation. However, bcsA, in addition to being controlled by adrA, also can be regulated by other regulators, including the second messenger c-di-GMP and sigma factor RpoS (19). Therefore, deletion of adrA had limited effects on biofilm formation. Interestingly, cellulose, encoded by bcsA, might be more important for the vertical transmission of S. Enteritidis compared with csgA-encoded curli fimbria, another biofilm component. These data are in line with our above virulence results and imply a different biological function between these two components. Overall, of the four genes studied here, csgD and bcsA had the strongest impacts on S. Enteritidis vertical transmission, the potential mechanisms will be studied in future.
In conclusion, we studied the impacts of four S. Enteritidis biofilm-associated genes on the vertical transmission in chickens by constructing gene-deletion mutants. Our results indicate that biofilm-associated genes csgD and bcsA may play important roles in the vertical transmission of S. Enteritidis. These findings lay the foundation for a better understanding how to control the vertical transmission of S. Enteritidis.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Jiangsu Administrative Committee for Laboratory Animals.
Author Contributions
SC, HS, and DP conceived research. HS and RZ performed research. SC, ZF, HS, and DP analyzed data. SC, ZF, HS, and TQ wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31572530), the Key R&D Project of Jiangsu Province (BE2018315), Six Talent Peaks Project in Jiangsu Province (NY-131), the Jiangsu Provincial Natural Science Fund for Excellent Young Scholars (BK20200105), the High-Level Talent Support Plan of Yangzhou University, and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Olaimat AN, Holley RA. Effects of changes in pH and temperature on the inhibition of Salmonella and listeria monocytogenes by allyl isothiocyanate. Food Control. (2013) 34:414–9. doi: 10.1016/j.foodcont.2013.05.014
2. Li QC, Wang X, Yin KQ, Hu YC, Xu HY, Xie XL, et al. Genetic analysis and CRISPR typing of Salmonella enterica serovar Enteritidis from different sources revealed potential transmission from poultry and pig to human. Int J Food Microbiol. (2018) 266:119–25. doi: 10.1016/j.ijfoodmicro.2017.11.025
3. Zhang-Barber L, Turner AK, Barrow PA. Vaccination for control of Salmonella in poultry. Vaccine. (1999) 17:2538–45. doi: 10.1016/S0264-410X(99)00060-2
4. Hurley D, McCusker MP, Fanning S, Martins M. Salmonella-host interactions - modulation of the host innate immune system. Front Immunol. (2014) 5:481. doi: 10.3389/fimmu.2014.00481
5. Chousalkar K, Gole VC. Salmonellosis acquired from poultry. Curr Opin Infect Dis. (2016) 29:514–9. doi: 10.1097/QCO.0000000000000296
6. Guard-Petter J. The chicken, the egg and Salmonella enteritidis. Environ Microbiol. (2001) 3:421–30. doi: 10.1046/j.1462-2920.2001.00213.x
7. Maciel BM, Dias JC, Romano CC, Sriranganathan N, Brendel M, Rezende RP. Detection of Salmonella enteritidis in asymptomatic carrier animals: comparison of quantitative real-time PCR and bacteriological culture methods. Genet Mol Res. (2011) 10:2578. doi: 10.4238/2011.October.24.1
8. Michael O, John B. Pass or fail: economic incentives to reduce Salmonella contamination in ground beef sold to the national school lunch program. Am J Agric Econ. (2018) 100:414–33. doi: 10.1093/ajae/aax088
9. Chousalkar K, Gast R, Martelli F, Pande V. Review of egg-related Salmonellosis and reduction strategies in United States, Australia, United Kingdom and New Zealand. Crit Rev Microbiol. (2018) 44:290–303. doi: 10.1080/1040841X.2017.1368998
10. Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Gast R, Humphrey TJ, et al. Mechanisms of egg contamination by Salmonella enteritidis. FEMS Microbiol Rev. (2009) 33:718–38. doi: 10.1111/j.1574-6976.2008.00161.x
11. Withanage GS, Sasai K, Fukata T, Miyamoto T, Lillehoj HS, Baba E. Increased lymphocyte subpopulations and macrophages in the ovaries and oviducts of laying hens infected with Salmonella enterica serovar enteritidis. Avian Pathol. (2003) 32:583–90. doi: 10.1080/03079450310001610631
12. Steenackers H, Hermans K, Vanderleyden J, De Keersmaecker SCJ. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res Int. (2012) 45:502–31. doi: 10.1016/j.foodres.2011.01.038
13. Jensen PØ, Givskov M, Bjarnsholt T, Moser C. The immune system vs. pseudomonas aeruginosa biofilms. Pathog Dis. (2010) 59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x
14. Bridier A, Briandet R, Thomas V, Dubois-Brissonnet F. Resistance of bacterial biofilms to disinfectants: a review. Biofouling. (2011) 27:1017–32. doi: 10.1080/08927014.2011.626899
15. Borges KA, Furian TQ, de Souza SN, Menezes R, de Lima DA, Fortes FBB, et al. Biofilm formation by Salmonella Enteritidis and Salmonella typhimurium isolated from avian sources is partially related with their in vivo pathogenicity. Microb Pathogenesis. (2018) 118:238–241. doi: 10.1016/j.micpath.2018.03.039
16. Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. (2012) 272:541–61. doi: 10.1111/joim.12004
17. Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium escherichia coli with respect to operon structure regulation. J. Bacteriol. (1998) 180:722–31. doi: 10.1128/JB.180.3.722-731.1998
18. Lamas A, Miranda JM, Vázquez B, Cepeda A, Franco CM. Biofilm formation, phenotypic production of cellulose and gene expression in Salmonella enterica decrease under anaerobic conditions. Int J Food Microbiol. (2016) 238:63–7. doi: 10.1016/j.ijfoodmicro.2016.08.043
19. Simm R, Ahmad I, Rhen M, Guyon SL, Römling U. Regulation of biofilm formation in Salmonella typhimurium. Future Microbiol. (2014) 9:1261–82. doi: 10.2217/fmb.14.88
20. Liu Z, Niu H, Wu S, Huang R. CsgD regulatory network in a bacterial trait-altering biofilm formation. Emerg Microbes Infect. (2014) 3:1–5. doi: 10.1038/emi.2014.1
21. Robbe-Saule V, Jaumouille V, Prevost MC, Guadagnini S, Talhouarne CH, Mathout H, et al. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar typhimurium J Bacteriol. (2006) 188:3983–94. doi: 10.1128/JB.00033-06
22. Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R. III. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium Mol Microbiol. (2001) 41:349–63. doi: 10.1046/j.1365-2958.2001.02529.x
23. Gerstel U, Kolb A, Romling U. Regulatory components at the csgD promoter–additional roles for OmpR and integration host factor and role of the 5' untranslated region. FEMS Microbiol Lett. (2006) 261:109–17. doi: 10.1111/j.1574-6968.2006.00332.x
24. Zakikhany K, Harrington CR, Nimtz M, Hinton JC, Romling U. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol Microbiol. (2010) 77:771–86. doi: 10.1111/j.1365-2958.2010.07247.x
25. Omadjela O, Narahari A, Strumillo J, Melida H, Mazur OV, Bulone V, et al. BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc. Natl. Acad. Sci. USA. (2013) 110:17856–61. doi: 10.1073/pnas.1314063110
26. Li Q, Xu Y, Jiao X. Identification of Salmonella pullorum genomic sequences using suppression subtractive hybridization. J Microbiol Biotechnol. (2009) 19:898–903. doi: 10.4014/jmb.0812.694
27. He S, Yuan Z, Zhu G. Construction and characterization of type I fimbriae fimH deletion mutant from avian pathogenic Escherichia coli. Wei Sheng Wu Xue Bao. (2008) 48:252–6
28. El Hag M, Zheng F, Su Y, Xiao W, Liu X. Contribution of the csgA and bcsA genes to Salmonella enterica serovar pullorum biofilm formation and virulence. Avian Pathol. (2017) 46:1–31. doi: 10.1080/03079457.2017.1324198
29. Bugarel M, Tudor A, Loneragan GH, Nightingale KK. Molecular detection assay of five Salmonella serotypes of public interest: Typhimurium, Enteritidis, Newport, Heidelberg, and Hadar. J Microbiol Methods. (2016) 134:14. doi: 10.1016/j.mimet.2016.12.011
30. Liu H, Xiao Y, Nie H, Huang Q, Chen W. Influence of (p)ppGpp on biofilm regulation in Pseudomonas putida KT2440. Microbiol Res. (2017) 204:1–8. doi: 10.1016/j.micres.2017.07.003
31. Gilles HJ. [Calculation of the index of acute toxicity by the method of linear regression. comparison with the method of “Karber and Behrens”]. Eur J Toxicol. (1974) 7:77–84.
32. Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. (2001) 39:1452–63. doi: 10.1046/j.1365-2958.2001.02337.x
33. Tursi SA, Tukel C. Curli-containing enteric biofilms inside and out: matrix composition, immune recognition, disease implications. Microbiol Mol Biol Rev. (2018) 82:e00028–18. doi: 10.1128/MMBR.00028-18
34. Pontes MH, Lee EJ, Choi J, Groisman EA. Salmonella promotes virulence by repressing cellulose production. Proc Natl Acad Sci USA. (2015) 112:5183–8. doi: 10.1073/pnas.1500989112
35. Anastasiadou M, Michailidis G. Cytokine activation during embryonic development and in hen ovary and vagina during reproductive age and Salmonella infection. Res Vet Sci. (2016) 109:86–93. doi: 10.1016/j.rvsc.2016.09.016
36. Vestby LK, Moretro T, Langsrud S, Heir E, Nesse LL. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet Res. (2009) 5:20. doi: 10.1186/1746-6148-5-20
37. Okamura M, Tachizaki H, Kubo T, Kikuchi S, Suzuki AK, Takehara K, et al. Comparative evaluation of a bivalent killed Salmonella vaccine to prevent egg contamination with Salmonella enterica serovars enteritidis, typhimurium, and gallinarum biovar pullorum, using 4 different challenge models. Vaccine. (2007) 25:0–4844. doi: 10.1016/j.vaccine.2007.03.004
Keywords: eggs, poultry, vertical transmission, biofilm, Salmonella Enteritidis
Citation: Chen S, Feng Z, Sun H, Zhang R, Qin T and Peng D (2021) Biofilm-Formation-Related Genes csgD and bcsA Promote the Vertical Transmission of Salmonella Enteritidis in Chicken. Front. Vet. Sci. 7:625049. doi: 10.3389/fvets.2020.625049
Received: 02 November 2020; Accepted: 11 December 2020;
Published: 14 January 2021.
Edited by:
Min Yue, Zhejiang University, ChinaReviewed by:
Ali Raza Jahejo, Shanxi Agricultural University, ChinaAlaa Sewid, The University of Tennessee, Knoxville, United States
Feng Li, Tianjin University, China
Copyright © 2021 Chen, Feng, Sun, Zhang, Qin and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daxin Peng, cGVuZ2R4QHl6dS5lZHUuY24=
†These authors have contributed equally to this work
 Sujuan Chen
Sujuan Chen Zheng Feng1†
Zheng Feng1† Ruonan Zhang
Ruonan Zhang Daxin Peng
Daxin Peng