- 1Joint International Research Laboratory of Agriculture and Agri-Product Safety of Ministry of Education of China, Yangzhou University, Yangzhou, China
- 2College of Animal Science and Technology, Yangzhou University, Yangzhou, China
- 3Jiangsu Key Laboratory of Zoonosis, Yangzhou University, Yangzhou, China
Extrachromosomal circular DNA (eccDNA), existing as double-stranded circular DNA, is derived and free from chromosomes. It is common in eukaryotes but has a strong heterogeneity in count, length, and origin. It has been demonstrated that eccDNA could function in telomere and rDNA maintenance, aging, drug resistance, tumorigenesis, and phenotypic variations of plants and animals. Here we review the current knowledge about eccDNA in category, biogenesis, recognition, and functions. We also provide perspectives on the potential implications of eccDNA in life science.
Introduction
It was previously thought that genetic variation and V(D)J recombination were the main reasons for genome heterogeneity of different tissues from the same individual or different cells from the same tissue. However, recent studies have shown that extrachromosomal circular DNA (eccDNA) is an additional source of genomic heterogeneity. EccDNAs are a group of double-stranded circular DNA molecules that are derived and free from eukaryotic genome DNA. They could function in genome evolution and environmental adaptation, which depend on eccDNA sequence features. The high copy number and the significant transcriptional activity of eccDNA lead to the overexpression of the inhabiting genes (1). Additionally, eccDNAs could serve as mobile enhancers to trans-regulate chromosomal genes (2). Small eccDNAs are more widespread, but less is known about their function in cell biology. They are too small to contain protein-coding genes. MicroDNA can be released from normal and tumor tissues to plasma and serum, implying their roles in cell communication (3, 4). Transcription factor sponge is another speculated function of microDNA, where the accumulation of specific microDNA could titrate components of the replication or transcription machinery and lead to an inability to replicate or transcribe genomic DNA (5). Thus, establishing the associations of microDNA with economic phenotype or disease is an important direction for future exploration.
History Notes of EccDNA
EccDNA was first detected and measured in pig sperm using electron microscopy technology, with a length of ~0.5–16.8 μm (1 μm ≈ 3,100 bp) (6, 7). The length of eccDNA in the Hela cell nucleus was estimated to be 0.2–19.8 μm, compared with 4.81 ± 0.24 μm (mean ± SD) of mitochondrial DNA, and the eccDNA count was 20% of the mitochondrial DNA count (8). When analyzing a set of tumor karyotypes, scientists found many small double chromatin bodies (double-minutes, DMs), sometimes in large numbers, in addition to the apparently structurally intact chromosome (9).
Small polydisperse circular DNA (spcDNA) was an obsolete concept to commonly characterize small eccDNAs with a length of ~0.05–2.00 μm (10). From the 1980s to the 1990s, the repetitive sequences of spcDNAs were widely observed, including short interspersed nuclear element (SINE), long interspersed nuclear element (LINE), tandem repeats, transposons, rDNA, and telomere DNA (11–18). Subsequently, spcDNAs with rDNA and telomere DNA were exclusively termed as extrachromosomal rDNA circles (ERCs) and extrachromosomal telomeric circles (t-circles), respectively. However, their lengths are larger than that of spcDNA, meaning that some ERCs and t-circles are not covered by spcDNA (19–21). With the help of high-throughput sequencing, microDNAs were identified. They have non-repetitive sequences with a length of about 200–400 bp and derive from 5′ UTRs, exons, and CpG islands (22). It is abundant, with several 100 to several 1,000 counts per cell (22, 23). Recently, the concept of extrachromosomal DNA (ecDNA) was developed to exclusively specify larger eccDNA in tumors, typically covering intact oncogenes, and 30% of ecDNAs exist as DMs (24). Further studies revealed that the deletion of large genomic fragments could be circularized into episomes and subsequently polymerized into DMs. Therefore, episomes are the precursors of DMs (25–30). In this review, we divide general eccDNA into two categories: narrow sense eccDNA with length < 100 kb and ecDNA covering DMs and episomes (1, 31, 32). Notably, eccDNA refers to general eccDNA in the following parts unless stated otherwise. Accordingly, we make a clear classification for eccDNAs in Figure 1.
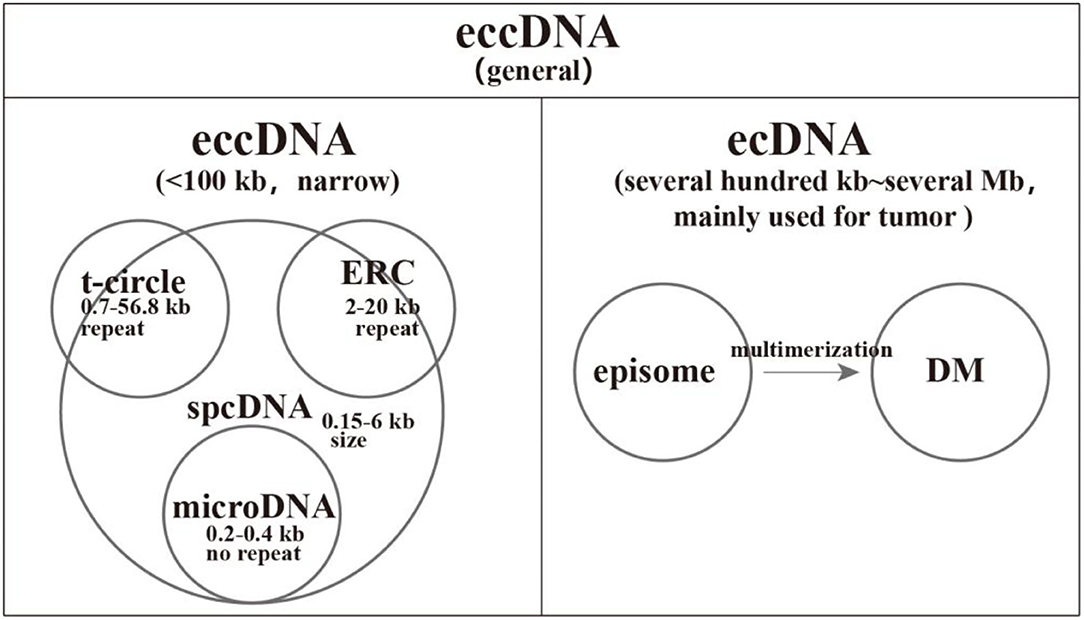
Figure 1. A comprehensive classification of extrachromosomal circular DNAs (eccDNAs). We divide the general eccDNAs into two categories: narrow sense eccDNAs with length < 100 kb and ecDNAs common in tumors with size ranging from several 100 kb to several megabases. Small polydisperse circular DNA (spcDNA) was an obsolete concept to commonly characterize small eccDNAs with repetitive sequences. The length of extrachromosomal rDNA circles (ERCs) and t-circles is larger than that of spcDNA, which means that some ERCs and t-circles were not covered by spcDNA. The deletion of large genomic fragments could be circularized into episomes and subsequently polymerized into double-minutes.
Biogenesis of EccDNA
Given the heterogeneity of eccDNAs in terms of sequence features, various molecular mechanisms may contribute to eccDNA biogenesis. Interestingly, all these mechanisms seem to be associated with DNA repair (33). We generalize these mechanisms into four categories: homologous recombination (HR), non-homologous end joining (NHEJ), DNA replication, and transcription (Figure 2). However, these potential models underlying the formation of different kinds of eccDNAs require further tests and verification.
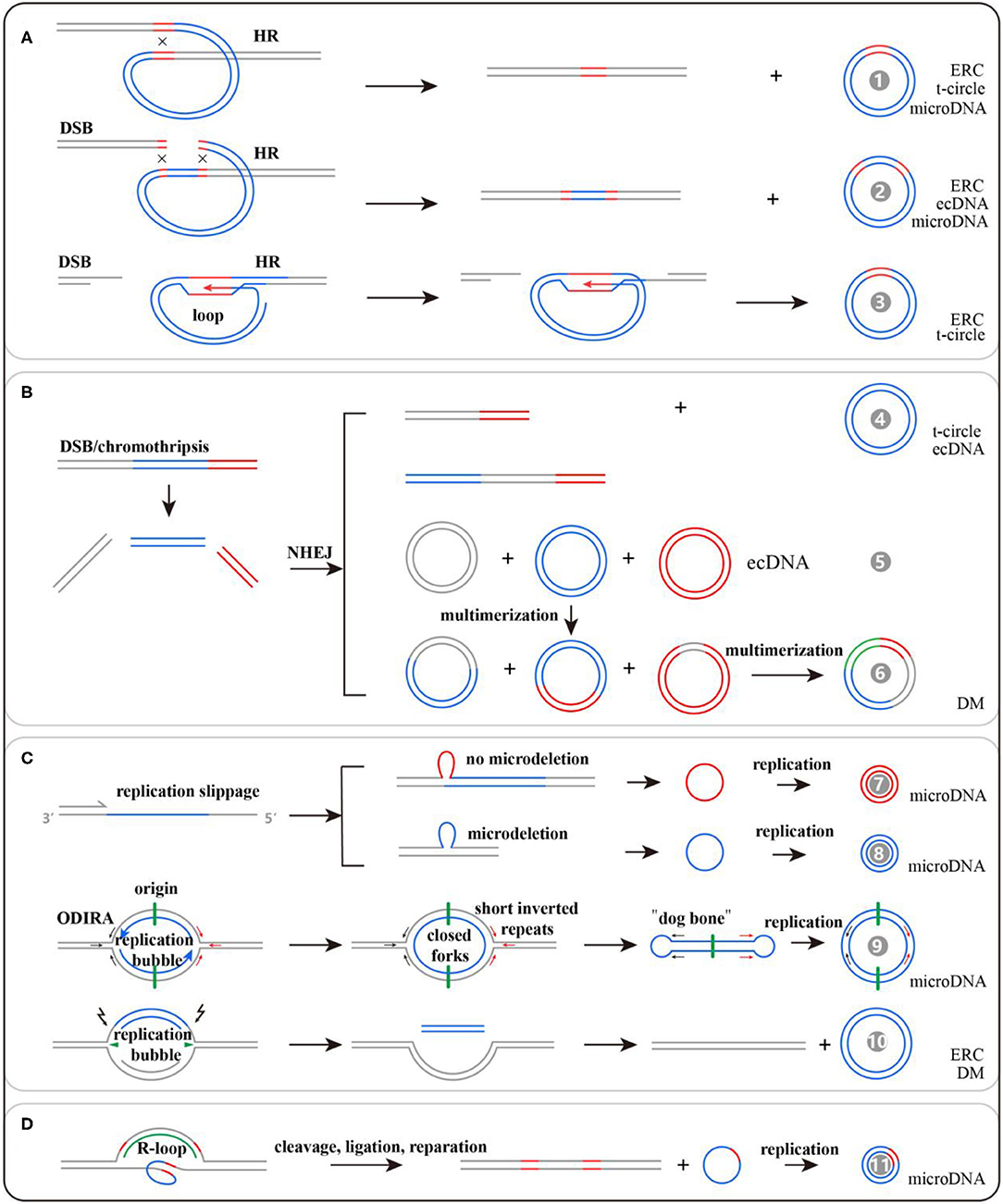
Figure 2. Potential models for extrachromosomal circular DNA (eccDNA) generation: (A) formed by HR, (B) formed by NHEJ, (C) formed by DNA replication, and (D) formed by transcription. There are 11 kinds of potential mechanisms for eccDNA formation, and their detailed information can be found in the references listed in Table 1.
DNA double-strand breaks could induce rDNA and tDNA to generate ERCs and t-circles, respectively, via loop structures mediated by HR (34). Among the sequencing reads mapped to ~100,000 eccDNAs in human muscle, only 8.1% are from genomic rDNA, 3.5% from LINE, 3.1% from SINE, 1.2% from satellite sequences, and 0.8% from tDNA. Most of the remaining reads are mapped to non-repetitive regions that could produce microDNA (23). The generation of microDNA is strongly associated with DNA mismatch repair. To comprehensively probe microDNA biogenesis, researchers knocked out the key proteins of NHEJ, HR, and mismatch repair (MMR) in a chicken DT40 cell line. They found that knocking out the MMR key protein, MSH3, could cause an 81% decrease of microDNA amount (35). Notably, the microdeletions by MMR were rare, occurring in one of ~400–4,000 alleles from the brain, and would be missed if genomic sequencing was not done at high depth. Nevertheless, the presence of microDNA from >100,000 sites in mouse, human, and chicken cells makes it unlikely that all of them are created by an excision event that leaves behind >100,000 somatically mosaic deletions on the chromosomes (22). Besides this, transcription in GC-rich regions and UTRs could generate triple-stranded DNA/RNA hybrids that function in DNA damage and repair processes and lead to microDNA formation (36). Origin-dependent inverted-repeat amplification may be involved in microDNA production as well. The nascent DNA strand could be circularized via short reverted repeats at both ends of the replication bubble (37). Inhibiting the expressions of BRCA1 and PRKDC, the key proteins for HR and NHEJ, respectively, lead to the reduction of ecDNA count in colon cancer cells, suggesting that HR and NHEJ activated by double-strand breaks and macrodeletions may be involved in ecDNA production (38, 39). The summarized mechanisms are listed in Table 1, and detailed information can be obtained from the corresponding references.
Toolbox For EccDNA Identification
Large eccDNAs could be observed with a light microscope in karyotype analysis, but as for the small eccDNAs, electron microscopy is necessary, and their lengths can be estimated all at once (Figure 3) (8, 53). This estimation could be achieved by 2D electrophoresis as well, but its detection power ranges from 0.7 to 56.8 kb (19). Southern blotting enables 2D electrophoresis to reveal the sequence features of eccDNA (60). Interestingly, software, such as ECdetect, has been developed for moderate-through counting of eccDNAs in DAPI-stained cells (24).
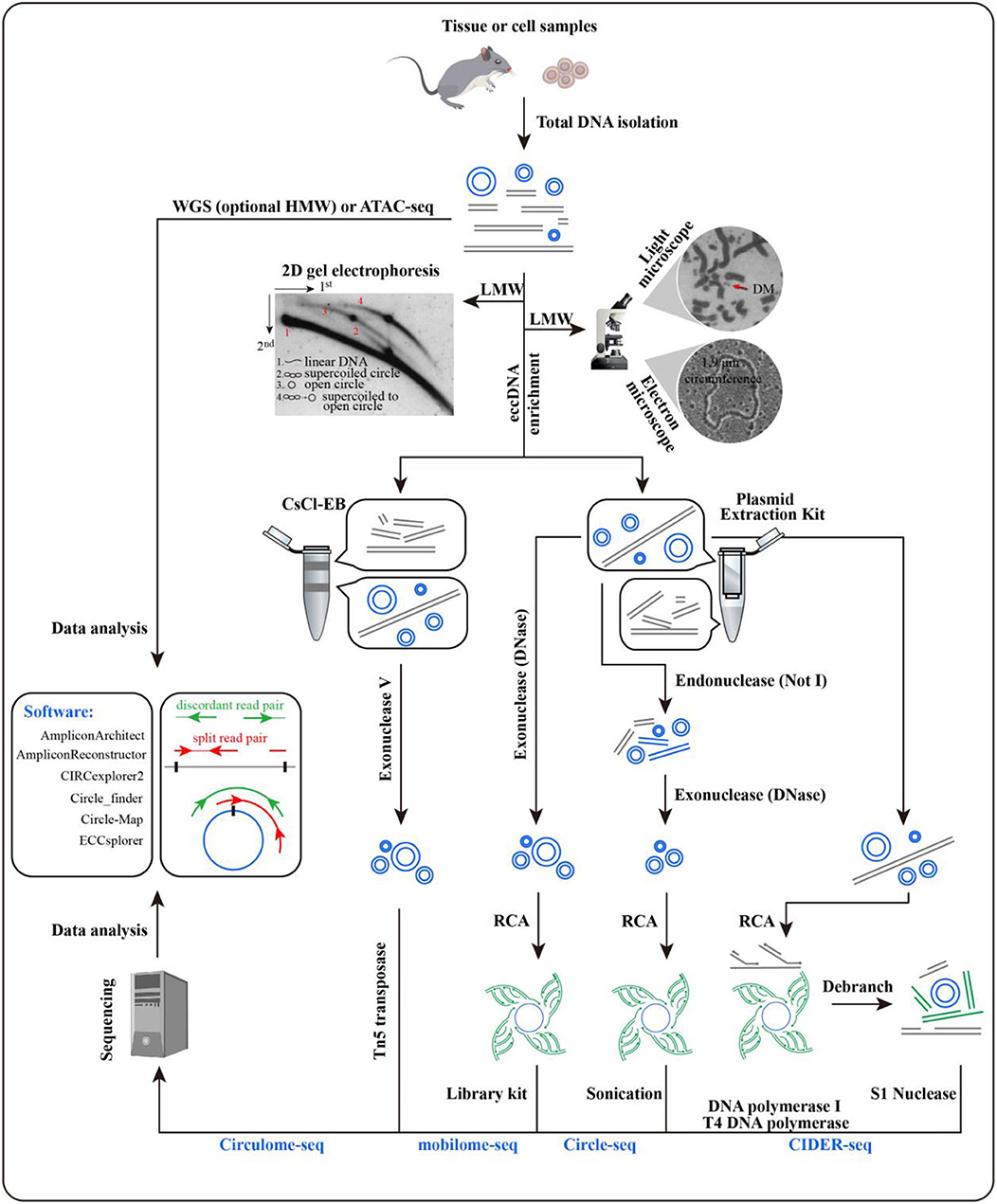
Figure 3. Methods for extrachromosomal circular DNA (eccDNA) identification. Microscopy and electrophoresis are used for eccDNA detection in total DNA after the enrichment of low-molecular-weight DNA [pictures were from Radloff et al. (8), Hahn (53), and Cohen et al. (54)]. Before rolling circle amplification or Tn5 treatment, eccDNAs with target size are enriched by CsCl-EB or a plasmid extraction kit. Split and discordant read pairs are crucial for eccDNA detection, which makes WGS and ATAC-seq data available as well. Various software packages have been developed to call eccDNA with sequencing data, including AmpliconArchitect, AmpliconReconstructor, CIRCexplorer2, Circle_finder, Circle-Map, and ECCsplorer (32, 55–59). Comparisons have been made for some of these packages, and detailed information can be found in Prada-Luengo et al. (57).
Initiated with a large amount of cells, the CsCl-EB method is time consuming and labor intensive for eccDNA enrichment, where most nicked circles are missed. Therefore, this approach is being replaced by plasmid extraction kits (60–62). The enriched eccDNAs can be subjected to high-throughput sequencing to determine their locations and junction sites by Circulome-seq, mobilome-seq, Circle-seq, or CIDER-seq (Figure 3). Circulome-seq adopts Tn5 transposition-based fragmentation and a tagging system, which simplify the sequencing library construction. This method could detect eccDNAs in length ranging from several hundreds of base pairs to several hundreds of kilobase pairs (62). Mobilome-seq is highly suitable for the detection of retrotransposon eccDNAs. It removes linear DNA with exonuclease DNase and then performs rolling circle amplification (RCA) (63). Circle-seq combines endonuclease NotI with exonuclease DNase to fully eliminate linear DNA, potentially leading to an unexpected damage on eccDNAs; its detection power is ~1–38 kb (64). As for CIDER-seq, RCA is performed straightforwardly for the eccDNAs enriched by plasmid extraction kits without the removal of linear DNA. Given the undesired amplification of linear DNA, long-read single-molecule real-time sequencing is used to guarantee the amount of valid reads (i.e., split and discordant reads), which are essential for eccDNA calling (65). CIDER-seq has a similar power to Circulome-seq, but the former has a more accurate detection for <10 kb eccDNAs. Notably, it is advisable to introduce plasmids as a control or internal reference during eccDNA library construction.
Despite the above-mentioned methods, whole-genome sequencing (WGS) and assay for transposase-accessible chromatin using sequencing (ATAC-seq) can be used for eccDNA calling, given the presence of split and discordant reads in their sequencing data (24, 66, 67). It has been demonstrated that 100% of ecDNAs and 30% of eccDNAs (narrow sense) called from WGS were supported by Circle-seq (32). Prior enrichment of high-molecular-weight DNA is helpful in ecDNA identification (1). To date, various software packages have been developed to call eccDNA with sequencing data (Figure 3).
Advances of EccDNA Function
EccDNA are able to self-replicate (unknown for microDNA) and evenly segregate to daughter and germ cells during mitosis and meiosis, respectively, because of lacking centromeres. Some eccDNAs can be reintegrated into genomic homogeneously staining regions (HSRs). The strong accessibility of ecDNA leads to highly frequent interactions between regulatory elements. These features equip cells with high heterogeneity and environmental adaptability (1, 31, 68).
Telomere and rDNA Copy Number Maintenance
Telomeric arrays can be maintained through various mechanisms, such as telomerase activity or recombination. T-circles function in recombination-dependent maintenance pathways by serving as templates for the rolling circle synthesis of telomere DNA. This may be the same case for animal and plant t-circles (20, 69). It is estimated that 15% of human immortalized cell lines may maintain telomere length through alternative lengthening of telomeres (70). A total of ~100–1,000 copies of eukaryotic rDNA are tandemly organized in the genome to satisfy the need for ribosome synthesis. The production of ERC reduces the copy number of rDNA in the Drosophila genome that could recover in germ cells. Studies have found that ERC could reintegrate into the genome to maintain rDNA copy number by self-replication (40).
Aging
Asymmetric inheritance and self-replication lead to ERC accumulation in yeast mother cells. The number of ERCs per mother cell after 15 generations has been estimated at between 500 and 1,000. Mutations in SGS1 could result in ERC accumulation and age-associated phenotypes in addition to a shortened lifespan. Conversely, loss of the replication fork blocking protein Fob1 decreases the formation of ERCs and extends the lifespan of mother cells by 30–40% (71). Interestingly, genes present on low-copy plasmids confer fitness effects rarely and of much lower magnitude than those on multi-copy plasmids. Therefore, young yeast populations contain about 1,800 circular DNA species, and it is only with substantial accumulation of any given circular DNA that major phenotypic effects are likely to manifest (64, 71, 72). These facts demonstrate that ERC accumulation functions in yeast aging. It was proposed that yeast senescence may be the result of health sacrifice to environmental adaption by accumulating specific eccDNAs, which could titrate components of the replication or transcription machinery and lead to an inability to replicate or transcribe genomic DNA and, thus, growth arrest and eventual death (5). According to this assumption, CUP1 eccDNA enrichment in a CuSO4 environment may contribute to yeast aging, but further research is still necessary (34).
Tumorigenesis and Drug Resistance
DMs, typically bearing intact oncogenes, are tumor specific and have been detected in 182/200 kinds of tumors, and ~0.26–44% of cancer patients and 7–100% of cancer cells have DMs (50, 53, 73–79). ecDNA exclusively refers to larger eccDNA in tumors where 30% of ecDNAs existed as DMs (24). The survival rate of cancer patients with ecDNAs is significantly lower than those without ecDNAs, making ecDNA a potential prognostic marker (80, 81). As a carrier of oncogene amplification, ecDNAs are subjected to non-Mendelian inheritance, which enables tumors to achieve very high intratumoral genetic heterogeneity and evolve rapidly in response to changing conditions (24, 66, 82)—for example, EGRF, MET, or MYC ecDNAs can make tumor cells proliferate rapidly and further develop into tumor invasion and migration (51, 66). There is a substantial quantity of cell-free microDNAs in the plasma and serum from both healthy individuals and cancer patients (3, 4). Surprisingly, tumor-derived human microDNAs are detected in mouse circulation in a mouse xenograft model of human ovarian cancer, and tumor excision alters the length of these small molecules (3). Thus, circular DNA in the circulation is a previously unexplored pool of nucleic acids that could complement miRNA and linear DNA for diagnosis and intercellular communication (3).
It had been accepted that the high copy number of ecDNA leads to oncogene overexpression. However, the significant transcriptional activity of ecDNA does matter as well (1). The deficiency of chromatin high-order structure and suppressing histone modification make ecDNAs more accessible than their genome parallels, which instigates strong promoter–enhancer interactions. Furthermore, ecDNA enhancers can shake off the insulator shackles and lead to novel interactions with oncogenes, which causes additional expression (83, 84). More recently, researchers have reported the chromatin connectivity networks of ecDNA in cancer, revealing that ecDNA can function as mobile super-enhancers, which drive genome-wide transcriptional amplification, including that of oncogenes. These findings support an expanded role for ecDNA in trans-regulating chromosomal genes in promoting tumor growth (2).
EGFR VIII, an oncogenic variant, could accelerate glioblastoma growth, but it also makes cells more sensitive to the EGFR tyrosine kinase inhibitor (TKI) (85). After TKI treatment, the proportion of TKI-sensitive tumor cells with a high expression of EGFR VIII was significantly decreased, whereas cells with low EGFR VIII expression were increased (85). Studies have demonstrated that tumor TKI resistance is caused by the elimination of DMs containing EGFR VIII, which could reintegrate into the genome HSRs (86). However, after drug withdrawal, the reemergence of clonal EGFR mutations on ecDNA follows quickly (86). Through this mechanism, cancer cells can escape targeted oncogene therapy. Therefore, pulsatile intermittent treatment with much higher doses of TKI could potentially lead to better target inhibition and even possibly less toxicity relative to continuous dosing (24). Notably, the self-replication of EGFR DMs could also generate EGFR mutations, which would provide additional heterogeneity (86).
Phenotypic Effects on Animals and Plants
To our current knowledge, eccDNAs have been associated with animal phenotypes, including cattle color sideness (Cs) and pigeon muscle development. Cs is a dominantly inherited trait characterized by a white band along cattle spines. The dominance of the Cs allele is expected to reflect a gain of function resulting from the dysregulated expression of the translocated KIT gene. A 492-kb fragment containing the KIT gene on chromosome 6 produces a circular intermediate (now referred to as eccDNA) and integrates into chromosome 29 to form the Cs29 allele. Then, a 575-kb fragment containing the partial Cs29 allele is circularized and translocated to chromosome 6 as Cs6 allele (87). Regenberg et al. found that the number of eccDNA in king pigeon muscle is nine-fold higher than that of homing pigeons. Interestingly, eccDNAs bearing the AGRIN gene were identified (88). This gene encodes a membrane protein that is involved in the development of neuromuscular junctions, and its variations could lead to abnormal muscle development (88).
As for plants, Amaranthus palmeri can develop herbicide resistance to glyphosate by amplification of the EPSPS gene as eccDNA, the molecular target of glyphosate. These circular molecules can be transmitted to germ cells and drive rapid glyphosate resistance through genome plasticity and adaptive evolution (89, 90). Moreover, retrotransposons, such as EVD and Tos17, can produce eccDNAs and insert into the genome to improve the response to environmental stress through promoting DNA methylation and gene silencing at the transcriptional level (63).
The above-mentioned facts suggest that eccDNAs may be promising molecular markers in life science. However, the inability to provide biopsies of some tissues limits the use of large eccDNAs as biomarkers. MicroDNAs released to the circulation represent a previously unexplored pool of nucleic acids; although they are too small to contain protein-coding genes, they are sufficiently long to code for regulatory elements. Possible acting mechanisms of microDNA have been assumed based on indirect evidence, including cell communication, transcription factor sponges, and mobile enhancers to trans-regulate chromosomal genes (Figure 4). Thus, establishing the associations of microDNA with a particular phenotype or disease is an important direction for future exploration.
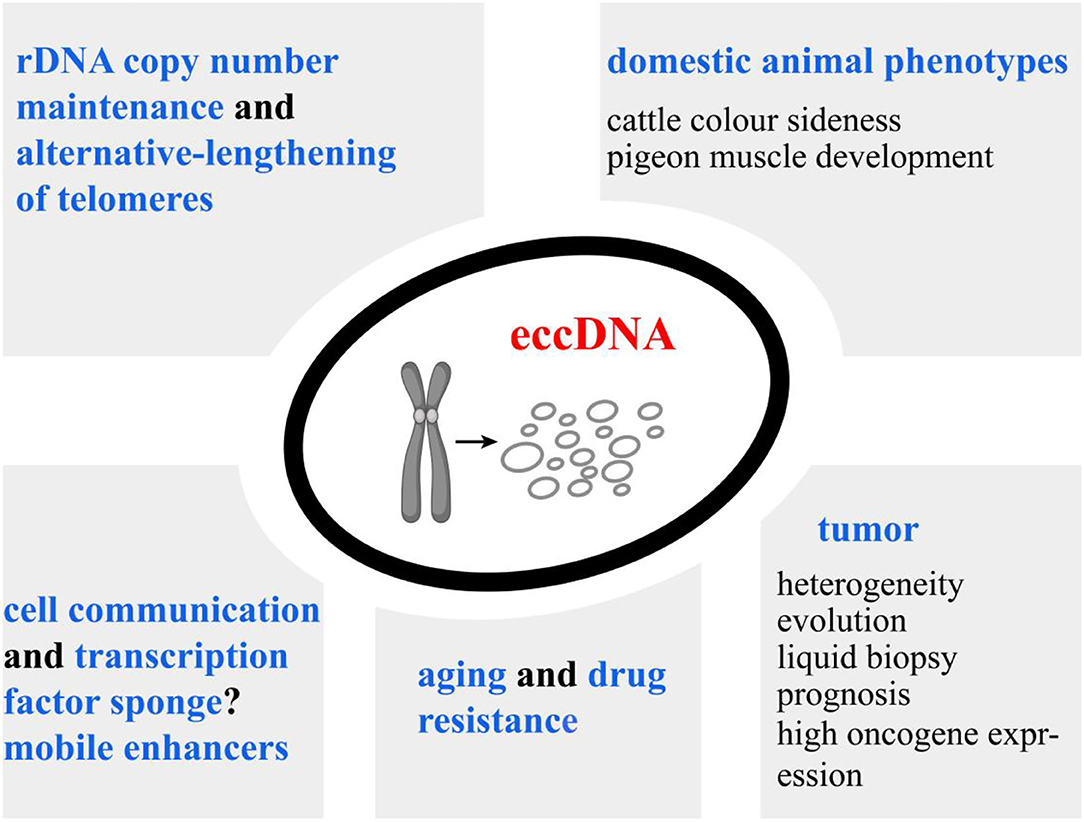
Figure 4. Overview of our current understanding of extrachromosomal circular DNA (eccDNA) functions. High copy number and significant transcriptional activity of eccDNAs lead to the overexpression of the inhabiting genes. Additionally, eccDNAs could serve as mobile enhancers to trans-regulate chromosomal genes. eccDNAs have been associated with cancer prognosis, drug resistance of plants, and phenotypic variations of animals, implying their potential implications in life science. MicroDNA can be released from normal tissues. Establishing the associations of microDNA copy numbers with economic traits is an important direction for future exploration.
Conclusions and Perspectives
The occurrence of eccDNA is a ubiquitous, normal phenomenon in eukaryotic systems, including those of plants, yeasts, and animals. There are various types of eccDNA according to sequence feature and size. Several models have been proposed to explain the formation and proliferation of eccDNAs, but the underlying mechanisms and direct evidence for each model are still required. Regardless of the above-mentioned issues, the CRISPR-hapC system has been developed for genome haplotyping based on the generation of eccDNA in cells (91). This system can map haplotypes from a few 100 bases to over 200 Mb and will be important for genome research and haplotype-specific gene therapy.
ecDNA could drive oncogene amplification and has become a hotspot of research in tumor pathogenesis and evolution. Additionally, the survival rate of cancer patients with ecDNAs is significantly lower than those without ecDNAs, making ecDNA a potential prognostic marker (80, 81). However, an interesting question arises: Are tumor suppressor genes present in ecDNA? If they exist, what are their functions? In plants and animals, large eccDNAs bearing intact genes have been identified, and they play important roles in environmental stress response and phenotypic variations, respectively (87, 89, 90). These facts make eccDNA a particularly promising molecular marker for breeding. However, given the unavailability of some tissue biopsies, it may be difficult to use large eccDNAs as biomarkers for early diagnosis and breeding.
MicroDNA represents the majority of eccDNAs. It has been detected as abundant cell-free DNA in plasma and serum released both by normal and tumor tissues (3, 4). Thus, establishing the associations between microDNAs and economic phenotype or diseases is an important direction for future exploration. Fortunately, an eccDNA database (eccDNAdb, http://www.eccdnadb.net/) has been set up. It has recorded a total of 1,700,000 eccDNAs for humans, mice, and chickens, which will provide supporting data for association analyses. If there are positive results, then the mechanisms of their regulatory roles can be validated, including serving as sponges of transcription factors, carriers of regulatory RNAs in intercellular communication, or mobile enhancers to globally amplify chromosomal transcription. These mechanisms may also provide novel insights into the phenotypic effects of genome copy number variations (41, 45, 92, 93). All of these questions require a further in-depth exploration.
Author Contributions
XC proposed the topic, retrieved literatures, provided outline, tables, and figures, and revised the manuscript. SW, LG, and WZ wrote the manuscript. JH and WS reviewed the final manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by the Key Research and Development Plan (modern agriculture) in Jiangsu Province (BE2018354), Jiangsu Agricultural Science and Technology Innovation Fund [XC(18)2003], major projects of Natural Science Research of colleges and universities in Jiangsu Province (17KJA230001), National Natural Science Foundation of China (31872333), major new varieties of agricultural projects in Jiangsu Province (PZCZ201739), National Natural Science Foundation of Cooperation on International Agricultural Research Organization (32061143036), and Open Project of Joint International Research Laboratory of Agriculture and Agri-Product Safety and the Ministry of Education of China (JILAR-KF202103).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thanks to all the people for their time and effort on the improvement of this paper.
Abbreviations
ALT, alternative lengthening of telomeres; ATAC-seq, transposase-accessible chromatin using sequencing; BRCA1, BRCA1 DNA repair-associated; CsCl-EB, cesium chloride ethidium bromide; CUP1, copper resistance-associated metallothionein; DAPI, 4′,6-diamidino-2-phenylindole; DM, double-minute; EccDNA, Extrachromosomal circular DNA; ecDNA, extrachromosomal DNA; EGRF, epidermal growth factor receptor; EPSPS, 3-phosphoshikimate 1-carboxyvinyltransferase 2; ERC, extrachromosomal rDNA circle; HMW, high molecular weight; HR, homologous recombination; HSR, homogeneously staining region; KIT, KIT proto-oncogene, receptor tyrosine kinase; LINE, long interspersed nuclear element; LMW, low molecular weight; MET, MET proto-oncogene, receptor tyrosine kinase; MMR, mismatch repair MSH3, MutS Homolog 3; MYC, MYC proto-oncogene, bHLH transcription factor; NHEJ, non-homologous end joining; ODIRA, origin-dependent inverted-repeat amplification; PRKDC, protein kinase, DNA-activated, catalytic subunit; RCA, rolling circle amplification; rDNA, ribosomal DNA; SGS1, ATP-dependent DNA helicase SGS1; SINE, short interspersed nuclear element; SMRT, long-read single-molecule real-time sequencing; spcDNA, small polydisperse circular DNA; t-circle, extrachromosomal telomeric circle; tDNA, telomere DNA; TKI, EGFR tyrosine kinase inhibitor; WGS, whole genome sequencing.
References
1. Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, et al. Circular ecdna promotes accessible chromatin and high oncogene expression. Nature. (2019) 575:699–703. doi: 10.1038/s41586-019-1763-5
2. Zhu Y, Gujar AD, Wong C-H, Tjong H, Ngan CY, Gong L, et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell. (2021) 39:694–707. doi: 10.1016/j.ccell.2021.03.006
3. Kumar P, Dillon LW, Shibata Y, Jazaeri AA, Jones DR, Dutta A. Normal and cancerous tissues release extrachromosomal circular DNA (eccDNA) into the circulation. Mol Cancer Res. (2017) 15:1197–205. doi: 10.1158/1541-7786.MCR-17-0095
4. Sin ST, Jiang P, Deng J, Ji L, Cheng SH, Dutta A, et al. Identification and characterization of extrachromosomal circular DNA in maternal plasma. Proc Natl Acad Sci USA. (2020) 117:1658–65. doi: 10.1073/pnas.1914949117
5. Sinclair DA, Guarente L. Extrachromosomal rdna circles—a cause of aging in yeast. Cell. (1997) 91:1033–42. doi: 10.1016/S0092-8674(00)80493-6
6. Hotta Y, Bassel A. Molecular size and circularity of DNA in cells of mammals and higher plants. Proc Natl Acad Sci USA. (1965) 53:356–62. doi: 10.1073/pnas.53.2.356
7. Yamagishi H, Kunisada T, Tsuda T. Small circular DNA complexes in eucaryotic cells. Plasmid. (1982) 8:299–306. doi: 10.1016/0147-619X(82)90067-1
8. Radloff R, Bauer W, Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in hela cells. Proc Natl Acad Sci USA. (1967) 57:1514–21. doi: 10.1073/pnas.57.5.1514
9. Cox D, Yuncken C, Spriggs A. Minute chromatin bodies in malignant tumours of childhood. Lancet. (1965) 286:55–8. doi: 10.1016/S0140-6736(65)90131-5
10. Smith CA, Vinograd J. Small polydisperse circular DNA of hela cells. J Mol Biol. (1972) 69:163–78. doi: 10.1016/0022-2836(72)90222-7
11. Bertelsen AH, Humayun MZ, Karfopoulos SG, Rush MG. Molecular characterization of small polydisperse circular DNA from an african green monkey cell line. Biochemistry. (1982) 21:2076–85. doi: 10.1021/bi00538a015
12. Krolewski JJ, Bertelsen AH, Humayun MZ, Rush MG. Members of the alu family of interspersed, repetitive DNA sequences are in the small circular DNA population of monkey cells grown in culture. J Mol Biol. (1982) 154:399–415. doi: 10.1016/S0022-2836(82)80003-X
13. Schindler CW, Rush MG. The kpni family of long interspersed nucleotide sequences is present on discrete sizes of circular DNA in monkey (bsc-1) cells. J Mol Biol. (1985) 181:161–73. doi: 10.1016/0022-2836(85)90082-8
14. Kunisada T, Yamagishi H. Sequence organization of repetitive sequences enriched in small polydisperse circular dnas from hela cells. J Mol Biol. (1987) 198:557–65. doi: 10.1016/0022-2836(87)90199-9
15. Pont G, Degroote F, Picard G. Some extrachromosomal circular dnas from drosophila embryos are homologous to tandemly repeated genes. J Mol Biol. (1987) 195:447–51. doi: 10.1016/0022-2836(87)90665-6
16. Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia a resulting from de novo insertion of l 1 sequences represents a novel mechanism for mutation in man. Nature. (1988) 332:164–6. doi: 10.1038/332164a0
17. Misra R, Matera AG, Schmid CW, Rush MG. Recombination mediates production of an extrachromosomal circular DNA containing a transposon-like human element, the-1. Nucleic Acids Res. (1989) 17:8327–41. doi: 10.1093/nar/17.20.8327
18. Regev A, Cohen S, Cohen E, Bar-Am I, Lavi S. Telomeric repeats on small polydisperse circular DNA (spcDNA) and genomic instability. Oncogene. (1998) 17:3455–61. doi: 10.1038/sj.onc.1202250
19. Cesare AJ, Griffith JD. Telomeric DNA in alt cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. (2004) 24:9948–57. doi: 10.1128/MCB.24.22.9948-9957.2004
20. Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol. (2009) 16:1010–5. doi: 10.1038/nsmb.1660
21. Cohen S, Agmon N, Sobol O, Segal D. Extrachromosomal circles of satellite repeats and 5s ribosomal DNA in human cells. Mob DNA. (2010) 1:11. doi: 10.1186/1759-8753-1-11
22. Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, et al. Extrachromosomal microdnas and chromosomal microdeletions in normal tissues. Science. (2012) 336:82–6. doi: 10.1126/science.1213307
23. Møller HD, Mohiyuddin M, Prada-Luengo I, Sailani MR, Halling JF, Plomgaard P, et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun. (2018) 9:1069. doi: 10.1038/s41467-018-03369-8
24. Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. (2017) 543:122–5. doi: 10.1038/nature21356
25. Carroll S, DeRose M, Gaudray P, Moore C, Needham-Vandevanter D, Von Hoff D, et al. Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol Cell Biol. (1988) 8:1525–33. doi: 10.1128/MCB.8.4.1525
26. Ruiz J, Choi K, Von Hoff D, Roninson I, Wahl G. Autonomously replicating episomes contain mdr1 genes in a multidrug-resistant human cell line. Mol Cell Biol. (1989) 9:109–15. doi: 10.1128/MCB.9.1.109
27. Schoenlein PV, Barrett JT, Kulharya A, Dohn MR, Sanchez A, Hou D-Y, et al. Radiation therapy depletes extrachromosomally amplified drug resistance genes and oncogenes from tumor cells via micronuclear capture of episomes and double minute chromosomes. Int J Radiation Oncol Biol Phys. (2003) 55:1051–65. doi: 10.1016/S0360-3016(02)04473-5
28. Storlazzi CT, Fioretos T, Surace C, Lonoce A, Mastrorilli A, Strömbeck B, et al. Myc-containing double minutes in hematologic malignancies: evidence in favor of the episome model and exclusion of myc as the target gene. Hum Mol Genet. (2006) 15:933–42. doi: 10.1093/hmg/ddl010
29. Shimizu N. Molecular mechanisms of the origin of micronuclei from extrachromosomal elements. Mutagenesis. (2011) 26:119–23. doi: 10.1093/mutage/geq053
30. Ozols RF. Molecular and Clinical Advances in Anticancer Drug Resistance, Vol. 57. New York, NY: Springer Science & Business Media (2012).
31. Verhaak RG, Bafna V, Mischel PS. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer. (2019) 19:283–8. doi: 10.1038/s41568-019-0128-6
32. Koche RP, Rodriguez-Fos E, Helmsauer K, Burkert M, MacArthur IC, Maag J, et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat Genet. (2020) 52:29–34. doi: 10.1038/s41588-019-0547-z
33. Chiu RW, Dutta A, Henssen AG, Lo YD, Mischel P, Regenberg B. What is extrachromosomal circular DNA and what does it do? Clin Chem. (2020) 66:754–9. doi: 10.1093/clinchem/hvaa096
34. Hull RM, King M, Pizza G, Krueger F, Vergara X, Houseley J. Transcription-induced formation of extrachromosomal DNA during yeast ageing. PLoS Biol. (2019) 17:e3000471. doi: 10.1371/journal.pbio.3000471
35. Dillon LW, Kumar P, Shibata Y, Wang Y-H, Willcox S, Griffith JD, et al. Production of extrachromosomal microdnas is linked to mismatch repair pathways and transcriptional activity. Cell Rep. (2015) 11:1749–59. doi: 10.1016/j.celrep.2015.05.020
36. Crossley MP, Bocek M, Cimprich KA. R-loops as cellular regulators and genomic threats. Mol Cell. (2019) 73:398–411. doi: 10.1016/j.molcel.2019.01.024
37. Brewer BJ, Payen C, Di Rienzi SC, Higgins MM, Ong G, Dunham MJ, et al. Origin-dependent inverted-repeat amplification: tests of a model for inverted DNA amplification. PLoS Genet. (2015) 11:e1005699. doi: 10.1371/journal.pgen.1005699
38. Meng X, Qi X, Guo H, Cai M, Li C, Zhu J, et al. Novel role for non-homologous end joining in the formation of double minutes in methotrexate-resistant colon cancer cells. J Med Genet. (2015) 52:135–44. doi: 10.1136/jmedgenet-2014-102703
39. Cai M, Zhang H, Hou L, Gao W, Song Y, Cui X, et al. Inhibiting homologous recombination decreases extrachromosomal amplification but has no effect on intrachromosomal amplification in methotrexate-resistant colon cancer cells. Int J Cancer. (2019) 144:1037–48. doi: 10.1002/ijc.31781
40. Nelson JO, Watase GJ, Warsinger-Pepe N, Yamashita YM. Mechanisms of rdna copy number maintenance. Trends Genet. (2019) 35:734–42. doi: 10.1016/j.tig.2019.07.006
41. Yerlici VT, Lu MW, Hoge CR, Miller RV, Neme R, Khurana JS, et al. Programmed genome rearrangements in oxytricha produce transcriptionally active extrachromosomal circular DNA. Nucl Acids Res. (2019) 47:9741–60. doi: 10.1093/nar/gkz725
42. Tomaska L, McEachern MJ, Nosek J. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. (2004) 567:142–6. doi: 10.1016/j.febslet.2004.04.058
43. Park PU, Defossez, P-A, Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in saccharomyces cerevisiae. Mol Cell Biol. (1999) 19:3848–56. doi: 10.1128/MCB.19.5.3848
44. Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the gap1 locus. Proc Natl Acad Sci USA. (2010) 107:18551–6. doi: 10.1073/pnas.1014023107
45. Paulsen T, Kumar P, Koseoglu MM, Dutta A. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. (2018) 34:270–8. doi: 10.1016/j.tig.2017.12.010
46. Gu X, Yu J, Chai P, Ge S, Fan X. Novel insights into extrachromosomal DNa: redefining the onco-drivers of tumor progression. J Exp Clin Cancer Res. (2020) 39:215. doi: 10.1186/s13046-020-01726-4
47. Yan Y, Guo G, Huang J, Gao M, Zhu Q, Zeng S, et al. Current understanding of extrachromosomal circular DNA in cancer pathogenesis and therapeutic resistance. J Hematol Oncol. (2020) 13:124. doi: 10.1186/s13045-020-00960-9
48. Liao Z, Jiang W, Ye L, Li T, Yu X, Liu L. Classification of extrachromosomal circular DNA with a focus on the role of extrachromosomal DNA (ecDNA) in tumor heterogeneity and progression. Biochim Biophys Acta Rev Cancer. (2020) 1874:188392. doi: 10.1016/j.bbcan.2020.188392
49. Mansisidor A, Molinar Jr T, Srivastava P, Dartis DD, Delgado AP, Blitzblau HG, et al. Genomic copy-number loss is rescued by self-limiting production of DNA circles. Mol Cell. (2018) 72:583–93. doi: 10.1016/j.molcel.2018.08.036
50. Vogt N, Lefèvre SH, Apiou F, Dutrillaux AM, Cör A, Leuraud P, et al. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci USA. (2004) 101:11368–73. doi: 10.1073/pnas.0402979101
51. Wei J, Wu C, Meng H, Li M, Niu W, Zhan Y, et al. The biogenesis and roles of extrachromosomal oncogene involved in carcinogenesis and evolution. Am J Cancer Res. (2020) 10:3532–50.
52. Ain Q, Schmeer C, Wengerodt D, Witte OW, Kretz A. Extrachromosomal circular DNA: current knowledge and implications for cns aging and neurodegeneration. Int J Mol Sci. (2020) 21:2477–508. doi: 10.3390/ijms21072477
53. Hahn PJ. Molecular biology of double-minute chromosomes. Bioessays. (1993) 15:477–84. doi: 10.1002/bies.950150707
54. Cohen S, Regev A, Lavi S. Small polydispersed circular DNA (spcdna) in human cells: association with genomic instability. Oncogene. (1997) 14:977–85. doi: 10.1038/sj.onc.1200917
55. Zhang X-O, Dong R, Zhang Y, Zhang J-L, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular rnas. Genome Res. (2016) 26:1277–87. doi: 10.1101/gr.202895.115
56. Deshpande V, Luebeck J, Nguyen N-PD, Bakhtiari M, Turner KM, Schwab R, et al. Exploring the landscape of focal amplifications in cancer using ampliconarchitect. Nat Commun. (2019) 10:392. doi: 10.1038/s41467-018-08200-y
57. Prada-Luengo I, Krogh A, Maretty L, Regenberg B. Sensitive detection of circular dnas at single-nucleotide resolution using guided realignment of partially aligned reads. BMC Bioinformatics. (2019) 20:663. doi: 10.1186/s12859-019-3160-3
58. Luebeck J, Coruh C, Dehkordi SR, Lange JT, Turner KM, Deshpande V, et al. Ampliconreconstructor integrates ngs and optical mapping to resolve the complex structures of focal amplifications. Nat Commun. (2020) 11:4374. doi: 10.1038/s41467-020-18099-z
59. Mann L, Seibt KM, Weber B, Schmidt T, Heitkam T. Eccsplorer: detection of extrachromosomal circular DNA (eccDNA) from ngs data. Biopolis Dresden PhD Symposium 2020: CRTD. Dresden: Zenodo (2020).
60. Cohen S, Segal D. Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res. (2009) 124:327–38. doi: 10.1159/000218136
61. Hollis M, Hindley J. Human sau3a repeated DNA is enriched in small polydisperse circular DNA from normal lymphocytes. Gene. (1986) 46:153–60. doi: 10.1016/0378-1119(86)90399-9
62. Shoura MJ, Gabdank I, Hansen L, Merker J, Gotlib J, Levene SD, et al. Intricate and cell type-specific populations of endogenous circular DNA (eccDNA) in caenorhabditis elegans and homo sapiens. G3 Genes Genomes Genet. (2017) 7:3295–303. doi: 10.1534/g3.117.300141
63. Lanciano S, Carpentier M-C, Llauro C, Jobet E, Robakowska-Hyzorek D, Lasserre E, et al. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genet. (2017) 13:e1006630. doi: 10.1371/journal.pgen.1006630
64. Møller HD, Parsons L, Jørgensen TS, Botstein D, Regenberg B. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci USA. (2015) 112:3114–22. doi: 10.1073/pnas.1508825112
65. Mehta D, Cornet L, Hirsch-Hoffmann M, Zaidi SS-e-A, Vanderschuren H. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using cider-seq. Nat Protoc. (2020) 15:1673–89. doi: 10.1038/s41596-020-0301-0
66. Decarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet. (2018) 50:708–17. doi: 10.1038/s41588-018-0105-0
67. Kumar P, Kiran S, Saha S, Su Z, Paulsen T, Chatrath A, et al. Atac-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci Adv. (2020) 6:eaba2489. doi: 10.1126/sciadv.aba2489
68. Qiu H, Shao Z-Y, Wen X, Zhang L-Z. New insights of extrachromosomal DNA in tumorigenesis and therapeutic resistance of cancer. Am J Cancer Res. (2020) 10:4056–65.
69. Mazzucco G, Huda A, Galli M, Piccini D, Giannattasio M, Pessina F, et al. Telomere damage induces internal loops that generate telomeric circles. Nat Commun. (2020) 11:5297. doi: 10.1038/s41467-020-19139-4
70. Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. (2003) 194:155–62. doi: 10.1016/S0304-3835(02)00702-4
71. Hull RM, Houseley J. The adaptive potential of circular DNA accumulation in ageing cells. Curr Genet. (2020) 66:889–94. doi: 10.1007/s00294-020-01069-9
72. Payen C, Sunshine AB, Ong GT, Pogachar JL, Zhao W, Dunham MJ. High-throughput identification of adaptive mutations in experimentally evolved yeast populations. PLoS Genet. (2016) 12:e1006339. doi: 10.1371/journal.pgen.1006339
73. Benner SE, Wahl GM, Von Hoff DD. Double minute chromosomes and homogeneously staining regions in tumors taken directly from patients versus in human tumor cell lines. Anticancer Drugs. (1991) 2:11–25. doi: 10.1097/00001813-199102000-00002
74. Nielsen JL, Walsh JT, Degen DR, Drabek SM, McGill JR, Von Hoff DD. Evidence of gene amplification in the form of double minute chromosomes is frequently observed in lung cancer. Cancer Genet Cytogenet. (1993) 65:120–4. doi: 10.1016/0165-4608(93)90219-C
75. Fletcher JA, Gebhardt MC, Kozakewich HP. Cytogenetic aberrations in osteosarcomas: nonrandom deletions, rings, and double-minute chromosomes. Cancer Genet Cytogenet. (1994) 77:81–8. doi: 10.1016/0165-4608(94)90154-6
76. Gebhart E. Double minutes, cytogenetic equivalents of gene amplification, in human neoplasia—a review. Clin Transl Oncol. (2005) 7:477–85. doi: 10.1007/BF02717000
77. Reader J, Zhao X, Butler M, Rapoport A, Ning Y. Rel-positive double minute chromosomes in follicular lymphoma. Leukemia. (2006) 20:1624–6. doi: 10.1038/sj.leu.2404289
78. Villa O, Salido M, Pérez-Vila ME, Ferrer A, Arenillas L, Pedro C, et al. Blast cells with nuclear extrusions in the form of micronuclei are associated with myc amplification in acute myeloid leukemia. Cancer Genet Cytogenet. (2008) 185:32–6. doi: 10.1016/j.cancergencyto.2008.04.014
79. Gibaud A, Vogt N, Hadj-Hamou NS, Meyniel JP, Hupe P, Debatisse M, et al. Extrachromosomal amplification mechanisms in a glioma with amplified sequences from multiple chromosome loci. Hum Mol Genet. (2010) 19:1276–85. doi: 10.1093/hmg/ddq004
80. Tandon I, Pal R, Pal JK, Sharma NK. Extrachromosomal circular dnas: an extra piece of evidence to depict tumor heterogeneity. Future Sci OA. (2019) 5:FSO390. doi: 10.2144/fsoa-2019-0024
81. Kim H, Nguyen NP, Turner K, Wu S, Gujar AD, Luebeck J, et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet. (2020) 52:891–7. doi: 10.1038/s41588-020-0678-2
82. Bailey C, Shoura MJ, Mischel PS, Swanton C. Extrachromosomal DNA–relieving heredity constraints, accelerating tumour evolution. Ann Oncol. (2020) 31:884–93. doi: 10.1016/j.annonc.2020.03.303
83. Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell. (2019) 179:1330–41. doi: 10.1016/j.cell.2019.10.039
84. Ott CJ. Circles with a point: new insights into oncogenic extrachromosomal DNA. Cancer Cell. (2020) 37:145–6. doi: 10.1016/j.ccell.2020.01.008
85. Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant egfr DNA. Science. (2014) 343:72–6. doi: 10.1126/science.1241328
86. Nikolaev S, Santoni F, Garieri M, Makrythanasis P, Falconnet E, Guipponi M, et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma. Nat Commun. (2014) 5:5690. doi: 10.1038/ncomms6690
87. Durkin K, Coppieters W, Drögemüller C, Ahariz N, Cambisano N, Druet T, et al. Serial translocation by means of circular intermediates underlies colour sidedness in cattle. Nature. (2012) 482:81–84. doi: 10.1038/nature10757
88. Møller HD, Ramos-Madrigal J, Prada-Luengo I, Gilbert MTP, Regenberg B. Near-random distribution of chromosome-derived circular DNA in the condensed genome of pigeons and the larger, more repeat-rich human genome. Genome Biol Evol. (2020) 12:3762–77. doi: 10.1093/gbe/evz281
89. Koo D-H, Molin WT, Saski CA, Jiang J, Putta K, Jugulam M, et al. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proc Natl Acad Sci USA. (2018) 115:3332–7. doi: 10.1073/pnas.1719354115
90. Molin WT, Yaguchi A, Blenner M, Saski CA. The eccDNA replicon: a heritable, extranuclear vehicle that enables gene amplification and glyphosate resistance in Amaranthus palmeri. Plant Cell. (2020) 32:2132–40. doi: 10.1105/tpc.20.00099
91. Yu J, Xiang X, Huang J, Liang X, Pan X, Dong Z, et al. Haplotyping by crispr-mediated DNA circularization (crispr-hapc) broadens allele-specific gene editing. Nucleic Acids Res. (2020) 48:e25. doi: 10.1093/nar/gkz1233
92. Gaines TA, Patterson EL, Neve P. Molecular mechanisms of adaptive evolution revealed by global selection for glyphosate resistance. New Phytol. (2019) 223:1770–5. doi: 10.1111/nph.15858
Keywords: eccDNA, microDNA, tumor, livestock, molecular marker
Citation: Cao X, Wang S, Ge L, Zhang W, Huang J and Sun W (2021) Extrachromosomal Circular DNA: Category, Biogenesis, Recognition, and Functions. Front. Vet. Sci. 8:693641. doi: 10.3389/fvets.2021.693641
Received: 11 April 2021; Accepted: 30 July 2021;
Published: 09 September 2021.
Edited by:
Eveline M. Ibeagha-Awemu, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Vratislav Peska, Academy of Sciences of the Czech Republic, CzechiaSonia Garcia, Instituto Botánico de Barcelona, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Rakesh Kumar, Shri Mata Vaishno Devi University, India
Copyright © 2021 Cao, Wang, Ge, Zhang, Huang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Sun, ZGt4bXN1bndlaUAxNjMuY29t
†These authors have contributed equally to this work
 Xiukai Cao
Xiukai Cao Shan Wang2†
Shan Wang2† Jinlin Huang
Jinlin Huang Wei Sun
Wei Sun