- 1Key Laboratory of New Animal Drug Project, Gansu Province, Key Laboratory of Veterinary Pharmaceutical Development, Ministry of Agriculture and Rural Affairs, Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS, Lanzhou, China
- 2Institute of Microbiology, Faculty of Veterinary Science, University of Agriculture Faisalabad, Faisalabad, Pakistan
- 3Department of Medicine, Cholistan University of Veterinary and Animal Sciences, Bahawalpur, Pakistan
- 4Institute of Biochemistry and Biotechnology, University of Veterinary and Animal Sciences, Lahore, Pakistan
- 5Department of Veterinary Medicine, University of Veterinary and Animal Sciences, Lahore, Pakistan
- 6Institute of Molecular Biology and Biotechnology, The University of Lahore, Lahore, Pakistan
- 7Department of Clinical Sciences, KBCMA, College of Veterinary and Animal Sciences, Narowal, University of Veterinary and Animal Sciences, Lahore, Pakistan
- 8Directorate General Farms and Feed Resources, Livestock and Dairy Development Department, Quetta, Pakistan
- 9Laboratory of Biochemistry and Immunology, College of Veterinary Medicine, Chungbuk National University, Cheongju, South Korea
- 10Department of Clinical Medicine and Surgery, University of Agriculture, Faisalabad, Pakistan
- 11College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
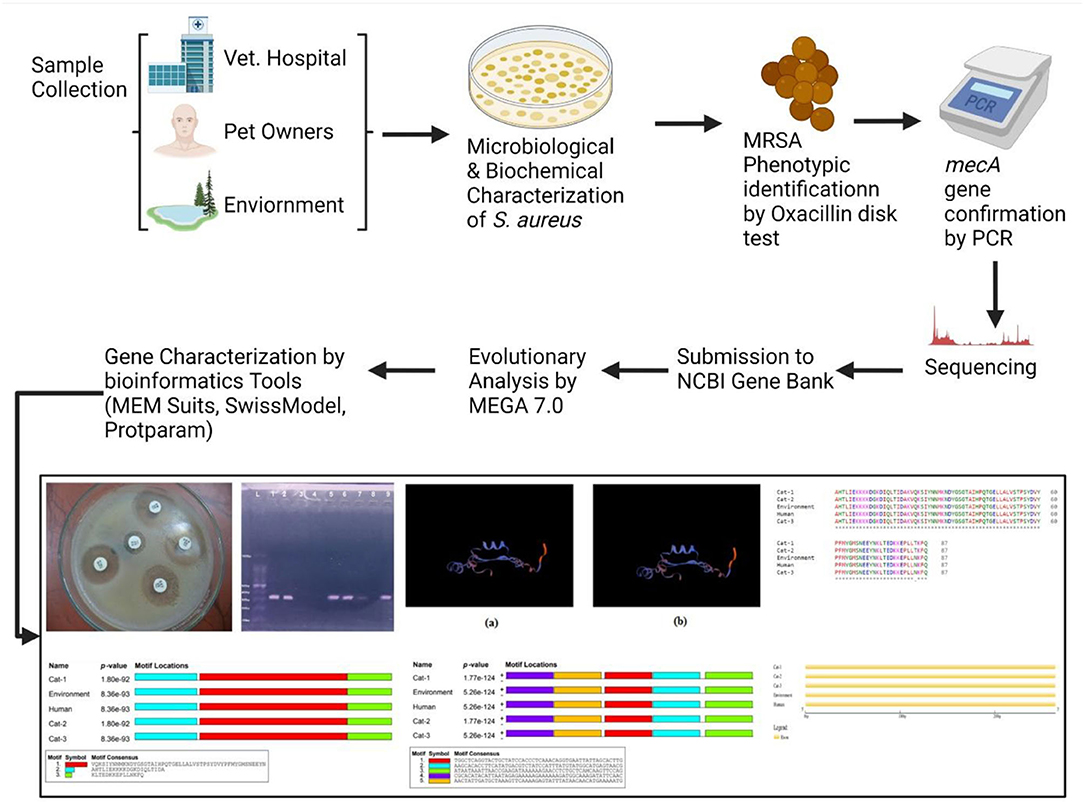
Staphylococcus aureus (S. aureus) has become a leading animal and public health pathogen that keeps on transferring from one host to other, giving rise to newer strains by genetic shifts. The current study was designed to investigate the epidemiology and genetic relatedness of mecA gene in S. aureus isolated from pets, immediate individuals in contact with pets, and veterinary clinic environments. A total of n = 300 samples were collected from different veterinary hospitals in Pakistan using convenience sampling. The collected samples were subjected to microbiological and biochemical examination for the isolation of S. aureus. Methicillin resistance was investigated by both phenotypically using oxacillin disk diffusion assay and by genotypically targeting mecA gene by PCR. PCR amplicons were subjected for sequencing by Sanger method of sequencing, which were subsequently submitted to NCBI GenBank under the accession numbers MT874770, MT874771, and MT874772. Sequence evolutionary analysis and mecA gene characterization was done using various bioinformatics tools. Overall, 33.66% mecA genes harboring S. aureus strains were isolated from all sources (33.33% from pets, 46.0% from surrounding, and 28.0% from immediate contact individuals). The bioinformatics analysis noted that one SNP was identified at position c.253C>A (Transvertion). The phylogenetic tree (two clades) of S. aureus mecA revealed a possibility of inter-transmission of disease between the environment and pets. Frequency of adenine and thymine nucleotide in motifs were found to be the same (0.334). Cytosine and guanine frequency were also the same (0.166). Threonine was replaced by asparagine (p.T84D) in each sample of cat, environment, and human. On the other hand, protein structures ofcat-1 and cat-2 proteins were found identical while cat-3, environmental, and human proteins shared identical structures. The study thus concludes rising circulation of methicillin-resistant S. aureus (MRSA) strains in animal-human-environment interfaces, forecasting the development of novel strains withmodified range of resistance.
Introduction
Over the lastdecade, with growing trends in pet or companion adoption, pedigree animals are imported for various purposes (shows, sports, pets, and breeding), and public awareness of animal welfare have raised concerns among veterinary researchers of Pakistan for treatment of animal diseases and/or common human and veterinary pathogens in Pakistan. Staphylococcus aureus (S. aureus) is a key pathogen that causes illnesses ranging from mild skin infections to life-threatening diseases in humans and animals, such as sepsis, pneumonia, endocarditis, deep rooted abscesses, and toxic shock syndrome. Staphylococcus aureus is an opportunistic pathogen that is capable of colonizing the skin, mucosal surface, gastrointestinal tract (GIT), urogenital tract, and respiratory tract (1). Within a couple of years, S. aureus has emerged to be a well-known pathogen due to its ability to develop resistance to commonly used antimicrobials and infect a significantly wider range of hosts (2–4). After horizontal gene transfer (HGT) and recombination, it becomes the storehouse of antibiotic resistance genes (ARG) and other virulence factor-encoding genes. The genomic divergence of S. aureus has led to the development of highly resistant strains that may cause serious problems with antibiotic treatment (5). Methicillin-resistant S. aureus (MRSA) that harbors the mec A gene has emerged to be a serious problem worldwide and is becoming more common in humans and animals. These strains are now recognized worldwide as livestock-associated (LA-MRSA), healthcare-associated (HA-MRSA), and community-associated (CA-MRSA) infections (6).
The transmission of S. aureus occurs either through direct human-to-human and/or human-to-animals contact with asymptomatic carriers (7). Until 2003, most of the identified MRSA clones belong to multi-locus sequence clones associated with human transmission and infection. The outbreak of the CC398 clone, considered as LA-MRSA clone, was found in farm animals and farm workers, indicating that certain MRSA strains are not strictly restricted to host species (8). A study conducted in the Netherlands found a high prevalence of MRSA in slaughtering pigs despite of reduction in antibiotic usage among pigs. Among the detected MRSA, 39% belonged to the ST398 complex (9). MRSA ST398 can cause infections in humans, suggesting that close contact with animals is an important risk factor for transfer of MRSA clones between human and animals (10).
Methicillin-resistant S. aureus harboring mecA gene is present within Staphylococcal Chromosomal Cassette mec (SCCmec) (11), and exhibits a decreased affinity to β-lactam and penicillin classes of antibiotics (12). SCCmec is a mobile element involved in the development of resistance to major antibiotics (e.g., β-lactams, macrolides, aminoglycosides, etc.). The phylogenetic patterns of the resistance genes encoded by staphylococcus SCCmec significantly differ from the phylogenetic patterns of the main genes, indicating that these genes are frequently switched between staphylococcal species. Due to the large population and the short replication time of bacteria, resistance develops within 2–4 years after the introduction of new antibiotics (13). Majority of isolated S. aureus strains have been identified using disk diffusion test. Further, mec A gene confirmation can done through PCR after preliminary identification by disk diffusion test (14).
Many reports of MRSA infections from animals to humans have been documented, but in Pakistan, there is lack of knowledge on MRSA transmission at the pet-human and environment cadre. The present study was planned to study the evolutionary phylogenetic analysis of mecA gene in S. aureus to analyze the distribution of S. aureus-causing infections within the same geographical environment within a certain period. We also discuss the phylogenetic placement of sequenced strains from Pakistan and worldwide by investigating the evolutionary histories of methicillin resistance in a phylogenetic framework.
Materials and Methods
Ethical Concern
A consent form was filled before taking samples by nasal swabs from nostrils of humans and animals in accordance with the standard ethical guidelines. The study was approved by departmental and faculty research committee and completion of study was notified via no.: CE/1701/2019.
Collection of Samples
A total of 300 samples were collected including cats (n = 150), cat owners (n = 100), and environment (n = 50) from public and private veterinary hospitals located in Faisalabad, Punjab, Pakistan. During sampling, sterilized swabs were inserted into the nostril at a depth of approximately 1 cm before being rotated five times, while environmental samples were taken from the surface of instruments, tables, and animal keepings. Samples were transported in Amie's medium to the laboratory of Institute of Microbiology, University of Agriculture Faisalabad and maintained in a cold chain (4°C) for further investigation.
Phenotypic Identification of MRSA
Each sample (10 μl) was disseminated on blood agar and incubated at 37°C for 24 h. These were then subsequently placed on Mannitol Salt Agar (MSA) for identification of S. aureus which was confirmed through culture characteristics, biochemical tests, and microscopic examination as recommended in the Bergey's Handbook of Determinative Bacteriology (15). Phenotypic identification of MRSA was carried out by oxacillin disk diffusion test in accordance with the recommendations of Clinical Laboratory Standard Institute (16).
Molecular Detection and Sequencing of MecA Gene
The DNA of S. aureus isolates were extracted through WizPrep™ gDNA Mini Kit (Cell/Tissue; Wizbiosolutions Inc. South Korea) following the manufacturer's guidelines. For PCR amplification, a total of 20 μl reaction mixture containing 10 μl of master mix (GeneDireX, Inc. USA), 3 μl extracted DNA (50 ng/μl) that was used as template, 3 μl of DNA free water, and 2 μl (20 pmoL) of each forward and reverse primer of mecA gene (Table 1) was prepared. The thermocycler conditions were as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of final denaturation 95°C for 30 s, annealing at 58°C for 30 s, initial extension at 72°C for 30 s, and a final extension at 72°C for 10 min. The presence of amplicons was determined by gel electrophoresis of 10 μl of products in 1.5% agarose gel containing 10 μl ethidium bromide for staining of DNA at 200 amperes and 120 volts for 30 min. The stained gel was visualized under UV light. The amplicons were subjected for sequencing to a 1st BASE DNA sequencing company (Singapore) that uses Applied Biosystems™ BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, USA) using Sanger method of sequencing.
Sequence Analysis
The mecA gene sequences from each source (cat, cat owner, and environment) was subjected to evolutionary analysis by MEGA 7.0 software. For this, the sequence was first analyzed through BLAST to get highly similar nucleotide sequences (n = 36) from NCBI nucleotide GenBank, and alignment was done through Clastal W software. The evolutionary history was accomplished by employing the Minimum Evolution (ME) methodology and replicating tree percentage, in which the associated taxa was clustered together in the bootstrap test (1,000 replicates). The distance of evolution was calculated in relation to the number of base substitutions at each site using the 2-parameter Kimura method. The initial phylogenetic tree was developed with mecA gene sequences from all sources by using neighbor-joining with a bootstrap value of 1,500, minimum evolution algorithm at 1.91 optimal tree branch length, and 0.1 evolutionary distance. The final phylogeny tree was constructed using a MEGA 7.0 software to investigate the evolution of mecA homologs.
Single nucleotide polymorphisms were identified in sequencing chromatograms using chromas software. Multiple Expectation maximizations for Motif Elicitation (MEME) Suit was used for the construction of nucleic acid and protein motifs. Gene structure display server software was used to construct the gene structure. Protein structures (3D) were predicted by Swiss model. Protparam was used to predict the proteins physical and chemical parameters.
Submission of Sequence to NCBI GenBank
The mecA gene sequences analyzed by Sanger method of sequencing from all sources were subjected to submission under NCBI GenBank. After complete submission and evaluation, sequences were accepted for publication in their repository. Accession numbers were issued as identifiers for each submitted sequence, namely, MT874770 (Environment), MT874771 (pet-owner), and MT874772 (Cat).
Analysis of Data
The data were analyzed using SPSS statistical computer software version 26, and sequence analyses was done using various online and offline bioinformatics tools.
Results
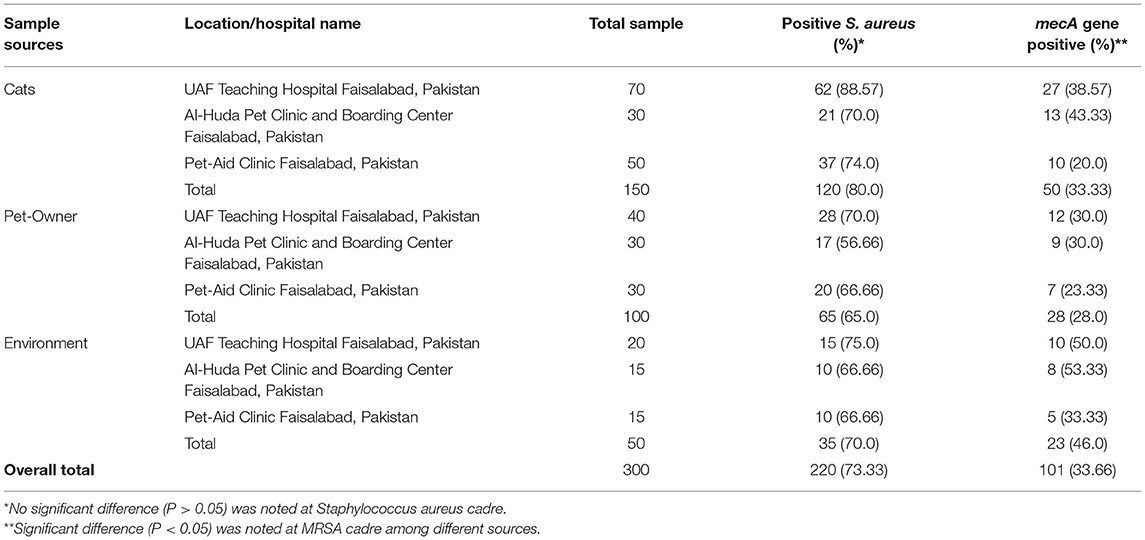
The present study found 101 (33.66%) mecA gene positive (MRSA) isolates from 300 pets (cats), individuals having direct contact with pet, and pet environment. The phenotypic identification of S. aureus and MRSA was done by MSA culture and oxacillin disk diffusion test, respectively (Figure 1). The mecA gene was identified by amplification through PCR, and mecA positive isolates from different sources were presented (Figure 2). Thirty-three-point-thirty-three percent (50/150) of mecA gene harboring S. aureus isolates were found in cats, 46.0% (23/50) from environment, which were twice to the isolates from pet owners [28.0% (28/100)]. In this study, there were no significant differences between S. aureus in animal, animal, and human interactions (P > 0.05), but a significant difference was noted in mecA gene prevalence among members (P < 0.05; Table 2).
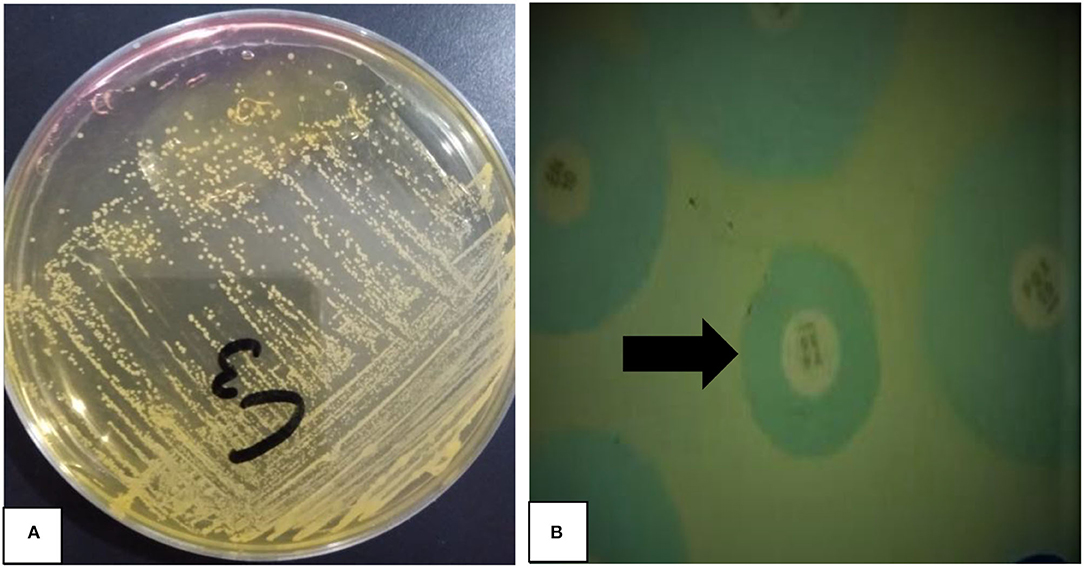
Figure 1. Phenotypic identification of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA). (A) A Yellow color growth of S. aureus (pinpoint colonies) on mannitol salt agar, NB: C3 mentioned on plate is sample number tested (B) methicillin resistant S. aureus (MRSA) identified by oxacillin disc (black arrow) diffusion assay.
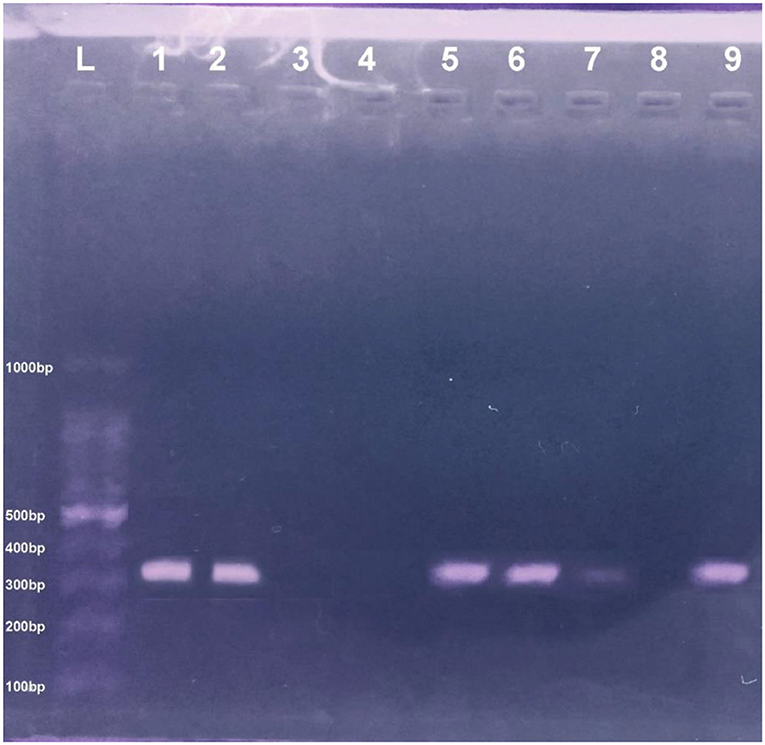
Figure 2. PCR pic of mecA gene at 310 bp. Lane L showing the ladder DNA, where Lane 8 and 9 are negative and positive control, respectively, and Lane 1–2 and 5–7 show mecA positive samples while Lane 3–4 express negative mecA isolates.
In present study, bacteriological and biochemical analysis of samples from cats, cat owners, and environments depicted the harboring of the mecA positive S. aureus. The conventional PCR results show the significant bands of mecA gene isolated from cats, cat owners, and environments (310 bp), as shown in Figure 2. The retrieved nucleotide sequences of targeted mecA genes were submitted to the NCBI GenBank with accession numbers MT874770 (Environment origin), MT874771 (pet-owner origin), and MT874772 (Cat origin).
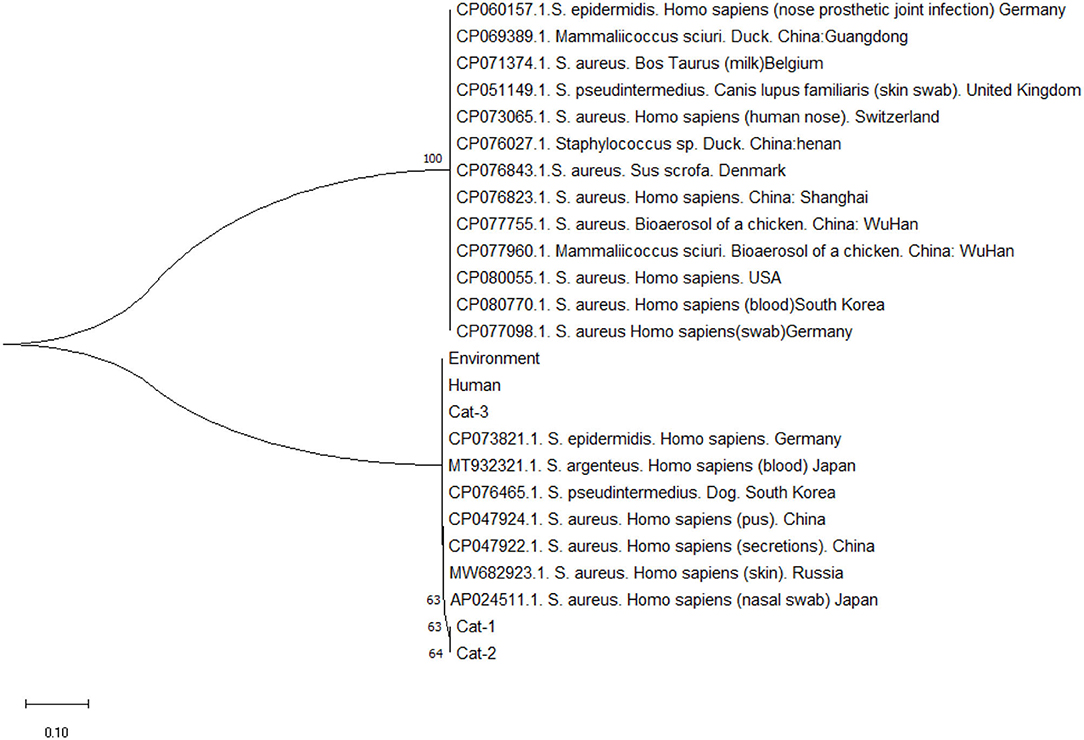
Nucleic acid sequence alignment are given in Figure 3, and protein sequences alignment are given in Figure 4. One SNP was identified at position c.253C>A (Transvertion; Table 3). Cat-1 and Cat-2 sequences were 100% identical, while Cat-3, environmental and human sequences, were 100% identical. The phylogenetic tree (two clades) of S. aureus mecA gene was observed. Staphylococcus aureus mecA genes of cat, environment, and human samples were compared with NCBI database sequences. Local isolates of S. aureus mecA (Cat-1 and Cat-2) gene sequences were closely related to the S. aureus mecA gene isolated from Homo sapiens (nasal swab) from Japan. Environmental and human mecA gene sequences were closely related to Staphylococcus epidermidis. All samples (Cat-1, Cat-2, Cat-3, Environmental, and Human) cluster together with S. aureus from H. sapiens (pus, secretions, skin and nasal swab), Staphylococcus argenteus from H. sapiens, Staphylococcus pseudintermedius (dog), and Staphylococcus epidermidis (H. sapiens; Figure 5).

Figure 3. Alignment of Staphylococcus aureus mecA gene (Cat-1, Cat-2, Cat-3, Environmental, and Human sequence).
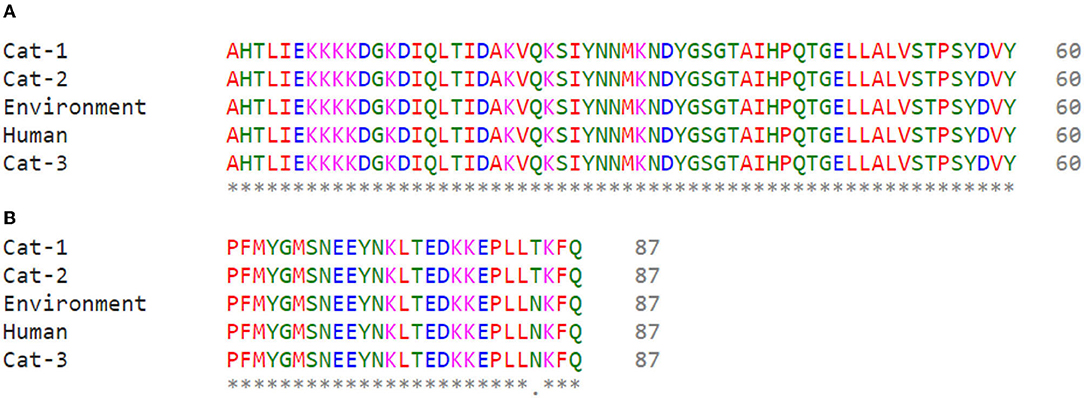
Figure 4. Alignment of Staphylococcus aureus mecA protein sequences (A) Cat-1 and Cat-2 (B) Environment, Human, and Cat-3.
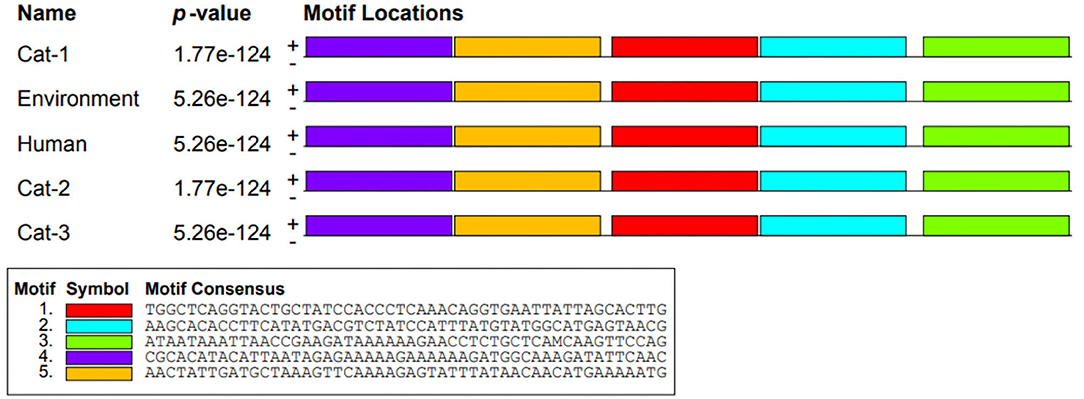
Nucleic acid motifs (1,320 bp sequence) of Cat-1 and Cat-2 sequences P-value were found to be the same (1.77e-124), while Cat-3, environmental, and human sequences share same P-value (5.26e-124; Figure 6). The P-values of protein motifs (435 bp sequence) of Cat-1 and Cat-2 sequences were also found to be the same (1.80e-92). Similarly, Cat-3, environmental, and human sequences share same P-value (8.36e-93; Figure 7). Frequency of adenine and thymine nucleotide in motifs (0.334) and that of cytosine and guanine were the same (0.166). The coding region in yellow color was involved in the nucleotide structure (Figure 8). Threonine was replaced by asparagine (p.T84D) in Cat-3, environmental, and human samples (Table 4).
The protein structure of Cat-1 and Cat-2 proteins is found to be identical while Cat-3, environmental, and human proteins share identical structures (Figure 9). Physical and chemical properties of proteins Cat-1 and Cat-2 were found to be identical, while Cat-3, environmental, and human proteins have same physical and chemical properties (Table 5).
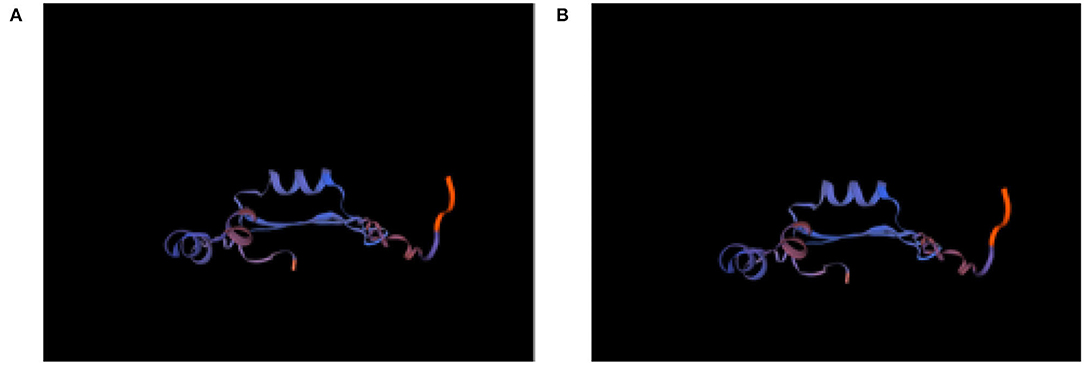
Figure 9. Protein structure of Staphylococcus aureus mecA gene (A) Cat-1 and Cat-2, and (B) Environment, Human, and Cat-3.
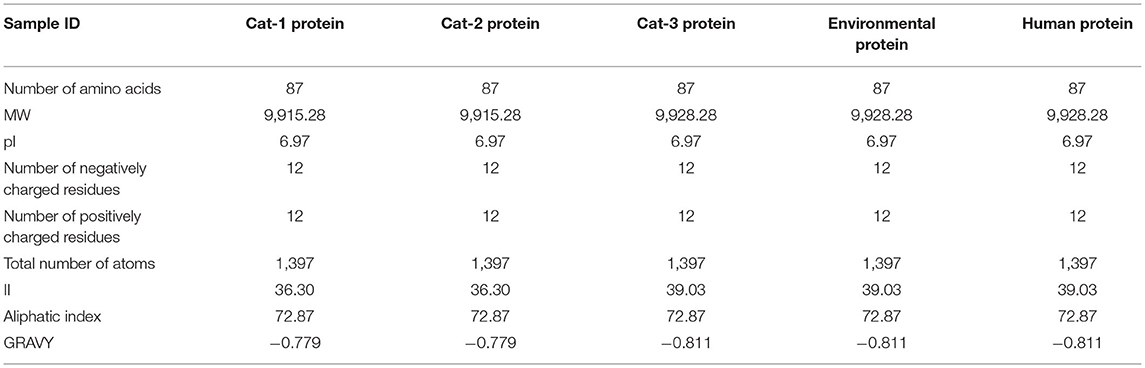
Table 5. Staphylococcus aureus mecA protein physical and chemical properties of proteins (Cat-1. Cat-2, Cat-3, Environmental, and Human).
Discussion
The human–animal bond has changed over the time. For example, the job of pets has shifted from working animals, e.g., catching mice and protecting houses, to animals that play roles as social pets, providing companionship. Pets may be important for their owners because they provide physical and mental health support but can also transmit number of zoonotic diseases (18). Staphylococcus aureus is one of the zoonotic pathogens that colonizes in the nasal cavity of pets and humans. Staphylococcus aureus is an infectious pathogen that acquires the ability to horizontally transfer genes between humans and animals (19). Staphylococcus aureus is recognized as major pathogen that causes major health problems in a community and is characterized by high rate of treatment failure worldwide (20). The One Health program is a global initiative plan for enhancing collaborations in every aspect of health care that involves humans, animals, and the environment. However, the role of dogs and cats in One Health communication is often underestimated. Therefore, the present study was conducted to investigate epidemiology and genetic relatedness of S. aureus by targeting the mecA gene at pet, pet owner, and environment interface. Nowadays, pets have become the major source of transmission of this pathogen to other animals and humans (21). In the last few years, much consideration has been paid on pets due to them serving as reservoir of antibiotic resistant bacteria and potential source of transfer of resistant genes from pets to humans (22). A study was conducted in Spain which reported co-carriage of ST398 strain of S. aureus in dogs and dog owners (23). Meanwhile, another study reported indistinguishable MRSA strains in humans and pets, suggesting the interspecies transmission of MRSA and S. aureus strains (24). Habibullah et al. (25) reported S. aureus that the prevalence in dogs (42.62%) and cats (37.50%) is twice higher than the current study. The prevalence of reported S. aureus contamination and/or infection in pet care workers is about six times less compared to current study findings (10% vs. 65.0%) (26). The high prevalence may be associated with unhygienic management practices in the study area, geographical differences, and unprotected interactions with pets (27). Staphylococcus aureus nasal colonization is associated with a number of factors, such as population type, geographical location, environmental factors, genetic factors, hormonal status, cell-wall composition, nasal secretions, antibiotic use, other infections, and immune level, which have been reported to be correlated with colonization (28). MRSA, increasingly being identified in both healthy and ill dogs and cats. It has been reported that mecA gene is the major evidence for the detection of MRSA isolates in Sudan (29). Consistent with current research, the results from two previous studies reported 46% and 49% MRSA in companion animals, respectively (30, 31). The relatively high prevalence in current studies and the high infection rate in hospitals and clinics are justified. High population density at clinics can cause the spread of pathogenic microorganisms into the environment.
Potential risks of carrying MRSA may include veterinary hospital staff, exposure to suppressant drugs, repeated utilization of the same antibiotics, use of non-specific antibiotics, and exposure to sources of infection carrying MRSA. MRSA can persist due to its antibiotic resistance ability, evolutionary adoption of biofilm covering, and its ability to bypass the immune system of hosts by utilizing specific molecular structures. Moreover, typical CA-MRSA, HA-MRSA, and LA-MRSA infecting non-specific hosts with MRSA strains is an additional indication for prolonged and persistent spread of MRSA strains (17). The high incidence of antibiotic resistance may be associated with the acquisition of resistance determinants (e.g., integrin's, plasmids, transposons, etc.) by vertical or horizontal gene transfer and, to a certain extent, with the misuse of antibiotics (5).
To curb the global spread of penicillin-resistant S. aureus, methicillin β-lactam antibiotics and subsequently oxacillin were manufactured. However, shortly after the use of methicillin, a MRSA emerged, making it one of the most life-threatening antibiotic-resistant pathogen (32). Methicillin resistance results from two distinct mechanisms. One is production of β-lactamases, which leads to a decrease in the antibiotic activity of β-lactams, and production of penicillin-binding protein 2a (PBP2a). PBP2a is an enzyme that is actively involved in the synthesis of peptidoglycan and promotes bacterial cell wall resistance. However, it does not have access to its active site, which binds to β-lactams, thereby interfering with its action and disrupting the normal process of bacterial cell wall synthesis (33). It is encoded by the mec gene, while the β-lactamase is encoded by the blaZ gene. The origin of the mecA gene is unknown. However, in some studies, the homologs of the mecA gene are found in S. sciuri, S. lentus, and S. vitulinus species. Since then, it has been hypothesized that these resistance determinants are derived from certain coagulase-negative staphylococci (CoNs). The species vitulinus suggests that this group may be the evolutionary ancestor of mecA. The mec genes are contained in the staphylococcal chromosome cassette mec (SCCmec), which is a mobile genetic component of staphylococci (34).
Another study conducted by Neamah et al. (1) found that mecA sequences from two clinically isolated cattle and human strains belong to the same group. Furthermore, another study conducted by Rolo et al. (35) studied the distribution pattern of the mecA gene, and showed that the earliest Staphylococcus species, including S. sciuri, S. vitulinus, and S. fleurettii, had an inclusive role in the stepwise aggregation of SCCmec elements and subsequently transferred to S. aureus. John et al. (36) conducted research to study 4,562 protein families and 1,764 unique genes of S. aureus and found 47 protein families that belonged to genes encoded by SCCmec, which include mecA gene, regulation genes (mecR and mecI), recombinase gene (ccrABC), transposons, insertion elements, heavy metals, and drug-resistant genes. Of the 152 S. aureus isolates, 73% were drug resistant and positive for mecA gene, while the remaining isolates were categorized as sensitive ones. Unsurprisingly, mecA was noted to be absent in most of the exposed strains, but, surprisingly, in scientist's dataset, 10% of sensitive S. aureus strains were composed of SCCmec elements that do not contain the mecA gene. The findings suggest that the presence of SCCmec element is not limited to only resistant strains of S. aureus.
The genomic composition of the core SCCmec gene, mecA-mecR-mecI was found in resistant strains of S. aureus and some species of Staphylococcaceae, such as S. pseudointermedius, S. epidermidis, S. sciuri, S. argenteus, S. schleiferi, S. haemolyticus, and M. caseolyticus, with mec-box genes found at 51 kb genes. The phylogenetic analysis based on the 16S rRNA gene sequence is widely used to study the evolutionary relationship of microorganisms (37). Interestingly, our study highlights the importance of those isolates as potential zoonotic agents. Hence, there is need to control and prevent this transmission by adopting clean living habits of pets and humans. Furthermore, the results of current study recommend more studies on whole genome sequence analysis along with multi locus sequence type (ST) approach in the future analyses to rule out the ST of each strain and the probability of raising a novel ST from the strains under study.
Conclusion
The study witnessed rise in MRSA from pets, pet owners, and the environment as threat for public and animal health. Phylogenetic tree of submitted sequences revealed significant relatedness of MRSAs of animals, humans, and environment to each other. Replacement of amino acids and variation in structural proteins of MRSAs of different sources are indicative of genetic shifts. In such scenarios, novel strains of MRSAs are expected to emerge with modified resistance to antibiotics if stern precautions are not adopted.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human and animal participants were reviewed and approved by the faculty and advanced studies board before the start of work, while approval of the completed research notified CE/1701/M.Phil., 2019 dated 11/10/2019. Post study ethical permission certificate was also obtained vide FVS/379/26.02.2020. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
MS did research and wrote the original manuscript draft. AA, MI, and WP contributed in project design and check the validity of the research. MA and HS executed the bioinformatics analysis. AG and MSH helped in laboratory work. ZB and MK did review and editing of manuscript. KA helped in clinical data collection. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (25-LZIHPS-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
S. aureus, Staphylococcus aureus; MRSA, Methicillin-resistant Staphylococcus aureus; HGT, horizontal gene transfer; ARG, antibiotic resistance genes; VFG, virulence factors-encoding genes; LA-MRSA, livestock-associated MRSA; HA-MRSA, healthcare-associated MRSA; CA-MRSA, community-associated MRSA; SCCmec, Staphylococcal Chromosomal Cassette mec; MIC, minimum inhibitory concentration; NCBI, National Center for Biotechnology Information; ST, sequence type.
References
1. Neamah AJ, Ayyez HN, Klaif SF, Khudhair YI, Hussain MH. Molecular and phylogenetic study of Staphylococcus aureus isolated from human and cattle of Al-Qadisiyah Governorate, Iraq. Vet World. (2019) 12:1378. doi: 10.14202/vetworld.2019.1378-1382
2. Javed MU, Ijaz M, Fatima Z, Anjum AA, Aqib AI, Ali MM, et al. Frequency and antimicrobial susceptibility of methicillin and vancomycin-resistant Staphylococcus aureus from bovine milk. Pak Vet J. (2021) 41:463–8. doi: 10.29261/pakvetj/2021.060
3. Altaf M, Ijaz M, Iqbal MK, Rehman A, Avais M, Ghaffar A, et al. Molecular characterization of methicillin resistant Staphylococcus aureus (MRSA) and associated risk factors with the occurrence of goat mastitis. Pak Vet J. (2020) 40:1–6. doi: 10.29261/pakvetj/2019.079
4. Sarwar I, Ashar A, Mahfooz A, Aqib AI, Saleem MI, Butt AA, et al. Evaluation of antibacterial potential of raw turmeric, nano-turmeric, and NSAIDs against multiple drug resistant Staphylococcus aureus and E. coli isolated from animal wounds. Pak Vet J. (2021) 41:209–14. Available online at: http://pvj.com.pk/pdf-files/41_2/209-214.pdf
5. Naorem RS, Urban P, Goswami G, Fekete C. Characterization of methicillin-resistant Staphylococcus aureus through genomics approach. 3 Biotech. (2020) 10:1–19. doi: 10.1007/s13205-020-02387-y
6. Lee AS, de Lencastre H, Garau J, Kluytmans J, Malhotra-Kumar S, Peschel A, et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers. (2018) 4:1–23. doi: 10.1038/nrdp.2018.33
7. Matuszewska M, Murray GG, Harrison EM, Holmes MA, Weinert LA. The evolutionary genomics of host specificity in Staphylococcus aureus. Trends Microbiol. (2020) 28:465–77. doi: 10.1016/j.tim.2019.12.007
8. Ballhausen B, Kriegeskorte A, van Alen S, Jung P, Köck R, Peters G, et al. The pathogenicity and host adaptation of livestock-associated MRSA CC398. Vet Microbiol. (2017) 200:39–45. doi: 10.1016/j.vetmic.2016.05.006
9. Dierikx CM, Hengeveld PD, Veldman KT, de Haan A, van der Voorde S, Dop PY, et al. Ten years later: still a high prevalence of MRSA in slaughter pigs despite a significant reduction in antimicrobial usage in pigs the Netherlands. J Antimicrob Chemother. (2016) 71:2414–8. doi: 10.1093/jac/dkw190
10. Haag AF, Fitzgerald JR, Penadés JR. Staphylococcus aureus in animals. Microbiol Spectr. (2019) 7:1–10. doi: 10.1128/microbiolspec.GPP3-0060-2019
11. Singh G, Broor S, Agarwal P. Staphylococcus cassette chromosome mec types among methicillin-resistant Staphylococcus aureus isolates from Haryana, India. Indian J Health Sci Care. (2017) 4:47–56. doi: 10.5958/2394-2800.2017.00010.4
12. Mistry H, Sharma P, Mahato S, Saravanan R, Kumar PA, Bhandari V. Correction: prevalence and characterization of oxacillin susceptible meca-positive clinical isolates of Staphylococcus aureus causing bovine mastitis in India. PLoS ONE. (2020) 15:e0232348. doi: 10.1371/journal.pone.0232348
13. Harkins CP, Pichon B, Doumith M, Parkhill J, Westh H, Tomasz A, et al. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. (2017) 18:1–11. doi: 10.1186/s13059-017-1252-9
14. Rahman M, Amin K, Rahman S, Khair A, Rahman M, Hossain A, et al. Investigation of methicillin-resistant Staphylococcus aureus among clinical isolates from humans and animals by culture methods and multiplex PCR. BMC Vet Res. (2018) 14:1–6. doi: 10.1186/s12917-018-1611-0
15. Bergey DH. Bergey's Manual of Determinative Bacteriology. Philadelphia, PA: Lippincott Williams & Wilkins (1994). ISBN: 0683006037.
16. Schwarz S, Silley P, Simjee S, Woodford N, van Duijkeren E, Johnson AP, et al. Assessing the antimicrobial susceptibility of bacteria obtained from animals. J Antimicrob Chemother. (2010) 65:601–4. doi: 10.1093/jac/dkq037
17. Shoaib M, Rahman SU, Aqib AI, Ashfaq K, Naveed A, Kulyar MF-e-A, et al. Diversified epidemiological pattern and antibiogram of mecA gene in Staphylococcus aureus isolates of pets, pet owners and environment. Pak Vet J. (2020) 40:331–6. doi: 10.29261/pakvetj/2020.039
18. Overgaauw PA, Vinke CM, van Hagen MA, Lipman LJ. A one health perspective on the human–companion animal relationship with emphasis on zoonotic aspects. Int J Environ Res Public Health. (2020) 17:3789. doi: 10.3390/ijerph17113789
19. Crespo-Piazuelo D, Lawlor PG. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir Vet J. (2021) 74:1–12. doi: 10.1186/s13620-021-00200-7
20. Islam MA, Parveen S, Rahman M, Huq M, Nabi A, Khan ZUM, et al. Occurrence and characterization of methicillin resistant Staphylococcus aureus in processed raw foods and ready-to-eat foods in an urban setting of a developing country. Front Microbiol. (2019) 10:503. doi: 10.3389/fmicb.2019.00503
21. Bierowiec K, Płoneczka-Janeczko K, Rypuła K. Is the colonisation of Staphylococcus aureus in pets associated with their close contact with owners? PLoS ONE. (2016) 11:e0156052. doi: 10.1371/journal.pone.0156052
22. Ye X, Wang X, Fan Y, Peng Y, Li L, Li S, et al. Genotypic and phenotypic markers of livestock-associated methicillin-resistant Staphylococcus aureus CC9 in humans. Appl Environ Microbiol. (2016) 82:3892–9. doi: 10.1128/AEM.00091-16
23. Gomez-Sanz E, Torres C, Ceballos S, Lozano C, Zarazaga M. Clonal dynamics of nasal Staphylococcus aureus and Staphylococcus pseudintermedius in dog-owning household members. Detection of MSSA ST398. PLoS ONE. (2013) 8:e69337. doi: 10.1371/journal.pone.0069337
24. Faires MC, Tater KC, Weese JS. An investigation of methicillin-resistant Staphylococcus aureus colonization in people and pets in the same household with an infected person or infected pet. J Am Vet Med Assoc. (2009) 235:540–3. doi: 10.2460/javma.235.5.540
25. Habibullah A, Rahman A, Haydar M, Nazir K, Rahman M. Prevalence and molecular detection of methicillin-resistant Staphylococcus aureus from dogs and cats in Dhaka City. Bangl J Vet Med. (2017) 15:51–7. doi: 10.3329/bjvm.v15i1.34055
26. Tarazi YH, Almajali AM, Ababneh MMK, Ahmed HS, Jaran AS. Molecular study on methicillin-resistant Staphylococcus aureus strains isolated from dogs and associated personnel in Jordan. Asian Pac J Trop Biomed. (2015) 5:902–8. doi: 10.1016/j.apjtb.2015.06.015
27. El-Deeb W, Fayez M, Elmoslemany A, Kandeel M, Zidan K. Methicillin resistant Staphylococcus aureus among goat farms in Eastern province, Saudi Arabia: Prevalence and risk factors. Prev Vet Med. (2018) 156:84–90. doi: 10.1016/j.prevetmed.2018.05.005
28. Sakr A, Brégeon F, Mège J-L, Rolain J-M, Blin O. Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front Microbiol. (2018) 9:2419. doi: 10.3389/fmicb.2018.02419
29. Elhassan MM, Ozbak HA, Hemeg HA, Elmekki MA, Ahmed LM. Absence of the mecA gene in methicillin resistant Staphylococcus aureus isolated from different clinical specimens in Shendi City, Sudan. Biomed Res Int. (2015) 2015. doi: 10.1155/2015/895860
30. Hogan PG, Mork RL, Boyle MG, Muenks CE, Morelli JJ, Thompson RM, et al. Interplay of personal, pet, and environmental colonization in households affected by community-associated methicillin-resistant Staphylococcus aureus. J Infect. (2019) 78:200–7. doi: 10.1016/j.jinf.2018.11.006
31. Ng W, Faheem A, McGeer A, Simor AE, Gelosia A, Willey BM, et al. Community-and healthcare-associated methicillin-resistant Staphylococcus aureus strains: an investigation into household transmission, risk factors, and environmental contamination. Infect Control Hosp Epidemiol. (2017) 38:61–7. doi: 10.1017/ice.2016.245
32. Zarazaga M, Gómez P, Ceballos S, Torres C. Molecular epidemiology of Staphylococcus aureus lineages in the animal–human interface. In: Fetsch A, editor. Staphylococcus aureus. Cambridge, MA: Academic Press (2018), p. 189–214. doi: 10.1016/B978-0-12-809671-0.00010-3
33. Fishovitz J, Hermoso JA, Chang M, Mobashery S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life. (2014) 66:572–7. doi: 10.1002/iub.1289
34. Silva V, Capelo JL, Igrejas G, Poeta P. Molecular epidemiology of Staphylococcus aureus lineages in wild animals in Europe: a review. Antibiotics. (2020) 9:122. doi: 10.3390/antibiotics9030122
35. Rolo J, Worning P, Nielsen JB, Bowden R, Bouchami O, Damborg P, et al. Evolutionary origin of the staphylococcal cassette chromosome mec (SCCmec). Antimicrob Agents Chemother. (2017) 61:302–16. doi: 10.1128/AAC.02302-16
36. John J, George S, Nori SRC, Nelson-Sathi S. Phylogenomic analysis reveals the evolutionary route of resistant genes in Staphylococcus aureus. Genome Biol Evol. (2019) 11:2917–26. doi: 10.1093/gbe/evz213
Keywords: S. aureus, mecA gene, pets, MRSA, phylogenetic analysis, pet owners
Citation: Shoaib M, Aqib AI, Ali MM, Ijaz M, Sattar H, Ghaffar A, Sajid Hasni M, Bhutta ZA, Ashfaq K, Kulyar MF-e-A and Pu W (2022) Tracking Infection and Genetic Divergence of Methicillin-Resistant Staphylococcus aureus at Pets, Pet Owners, and Environment Interface. Front. Vet. Sci. 9:900480. doi: 10.3389/fvets.2022.900480
Received: 20 March 2022; Accepted: 12 April 2022;
Published: 02 June 2022.
Edited by:
Fazul Nabi, Lasbela University of Agriculture, Water and Marine Sciences, PakistanReviewed by:
Ahsan Naveed, University of Kentucky, United StatesImmane Lamraoui, Ecole Nationale Supérieure de Biotechnologie, Algeria
Copyright © 2022 Shoaib, Aqib, Ali, Ijaz, Sattar, Ghaffar, Sajid Hasni, Bhutta, Ashfaq, Kulyar and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanxia Pu, cHV3YW54aWFAY2Fhcy5jbg==; Amjad Islam Aqib, YW1qYWRpc2xhbWFxaWJAY3V2YXMuZWR1LnBr
 Muhammad Shoaib
Muhammad Shoaib Amjad Islam Aqib
Amjad Islam Aqib Muhammad Muddassir Ali
Muhammad Muddassir Ali Muhammad Ijaz
Muhammad Ijaz Huma Sattar6
Huma Sattar6 Zeeshan Ahmad Bhutta
Zeeshan Ahmad Bhutta Muhammad Fakhar-e-Alam Kulyar
Muhammad Fakhar-e-Alam Kulyar Wanxia Pu
Wanxia Pu