- 1Department of Animal Health, Experimental Zooprophylactic Institute of Southern Italy, Portici, Italy
- 2Osservatorio Faunistico Venatorio—Campania Region, Naples, Italy
- 3Koret School of Veterinary Medicine, Hebrew University of Jerusalem, Rehovot, Israel
- 4Department of Veterinary Medicine and Animal Productions, University of Naples Federico II, Naples, Italy
Introduction: Following the increase of wild boar (Sus scrofa) populations in Europe, a potential risk of emerging infections by vector-borne pathogens may occur. Despite this, the circulation of piroplasmid species in these ungulates is still a neglected topic, particularly in the Mediterranean basin. Therefore, this study aimed to investigate the presence of Babesia/Theileria spp. in wild boars from southern Italy to assess the epidemiological role of these ungulates in the circulation of piroplasmids.
Methods: By using a citizen science approach among hunters and veterinarians, wild boar spleen samples were collected in the Campania region (southern Italy) between 2016 and 2022. A combined semi-nested PCR/sequencing analysis targeting the V4 hyper-variable region of 18S rRNA was run to detect Babesia/Theileria spp. DNA.
Results: Out of 243 boars, 15 (i.e., 6.2, 95% CI: 3.4–9.9) tested positive to Babesia/Theileria spp., Babesia vulpes (n = 13, 5.3, 95% CI: 3.1–8.9) the most prevalent, followed by Babesia capreoli (n = 2, 0.8, 95% CI: 0.2–2.9). Three different B. vulpes sequence types were identified (i.e., ST1, ST2, ST3), with the most representative as ST1 (60%), and a single B. capreoli sequence type. No statistically significant difference (p > 0.05) were found between the presence of the pathogens and boar age, sex, province and sample collection year.
Discussion: Data demonstrate for the first time the occurrence of B. vulpes and B. capreoli in wild boars, which may play a role in the biological cycle of piroplasmids. We emphasize the importance of monitoring these ungulates to prevent potential foci of infection. The engagement of hunters in epidemiological scientifically based surveys can constitute a technically sound control strategy of piroplasmids in a One Health perspective.
1. Introduction
Piroplasmids of the genus Babesia and Theileria (Aconoidasida, Piroplasmida) are global emerging tick-borne apicomplexan protozoa infecting multiple wild species, as well as domestic animals and humans (1, 2). Among more than 100 different species identified so far, some of these intracellular parasites display a high host specificity in wild mammals (3). For instance, the role of some wildlife species has been ascertained in the maintenance of certain Babesia spp., such as red foxes (Vulpes vulpes) for Babesia vulpes, red deer (Cervus elaphus) for the zoonotic Babesia divergens and roe deer (Capreolus capreolus) for Babesia capreoli and the zoonotic Babesia venatorum (3–6). Despite this, piroplasmid surveillance in wild boar (Sus scrofa) populations is a neglected topic due to their apparent absence in this ungulate in Europe (7). The only two Babesia spp. infecting boars, also common in pigs, Babesia trautmanni and Babesia perroncitoi, have been detected mostly in the 1990s via morphology without any molecular confirmation (7). The unique cases of molecular detection of piroplasmids in boars are to date reported as unspecified Theileria spp. in Italy (n = 3 out of 117) (8) and Portugal (n = 3 out of 65) (9), Babesia bigemina in Italy (n = 2 out of 257) (10) and a single finding of B. divergens out of 550 in the Czech Republic (7). This negligible occurrence of piroplasmids in boars is likely due to a low prevalence and parasitaemia and low number of tested animals (i.e., <100 in several epidemiological surveys) (11–13), despite the use of highly sensitive qPCR/conventional PCR protocols (7, 14). However, the role of boars in the epidemiology of piroplasmids in Europe cannot be ruled out considering that their high density (15), territorial expansion (16) and spatial overlap with other wildlife populations may increase the chance of tick infestation and piroplasmid transmission (8). Indeed, a considerable risk for new foci of emerging Babesia and Theileria infections is now evident in Europe, especially in the south and in the Mediterranean basin where great diversity of piroplasmid species (17) and high biodiversity of ixodid ticks occur (18). Some areas in these regions are also associated with a great vocation for outdoor recreational activities exposing to the risk of piroplasmid infection, as demonstrated by the high seroprevalence in hunting dogs from rural areas of southern Italy (19), where wildlife, ticks and related pathogens overlap (20). Therefore, this study aimed to investigate the occurrence of Babesia/Theileria spp. in wild boars from southern Italy and to assess the epidemiological role of these ungulates in the circulation of piroplasmids.
2. Materials and methods
2.1. Study area and sampling
The study was run in the Campania region, southern Italy, characterized by a typical Mediterranean temperate climate and progressively continental features of mainland and mountainous landscapes. Under the frame of a surveillance plan of wildlife by the Italian Ministry of Health (authorization no. IZSME RC 05/16), spleens of wild boars were collected from October 2016 to December 2022. Field activities were carried out in collaboration with “trained persons” (i.e., regular boar hunters educated specifically on hunting hygiene, health and food safety through specific theorical and practical courses, according to Reg. EU 853/2004) (21). Hunters culled boars and collected spleens and information (age, sex, geographic origin) under supervision of veterinarians affiliated with the University of Naples Federico II and regional health systems. In order to minimize the risk of cross-contamination, whole spleens were collected and stored at ±4°C in separate plastic biohazard bags and delivered to the necropsy room of the Department of Animal Health, Experimental Zooprophylactic Institute of southern Italy (Portici, Italy). Each spleen was flamed on the surface before sampling an aliquot from the inner portion for DNA extraction. Classes related to boar age (i.e., piglet <1 years old, juvenile 1–2 years old, adult >2 years old) were estimated by the examination of the teeth (i.e., primary and permanent teeth eruption times and root hole diameter of incisors), according to Massei and Toso (22).
2.2. Sample size calculation
A minimum sample size of 243 wild boars was estimated using the opensource software OpenEpi (23), inserting the following data: a population size of 84,000 boars (data supplied by the regional emergency plan of wild boars in Campania region); expected prevalence of Babesia/Theileria spp. infection in the population of 5% ±3 (i.e., 2%–8%), according to Zanet et al. (10); confidence limits of 5% and desired absolute precision of 3%.
2.3. DNA extraction, PCR protocol, and sequencing
One gram of spleen was individually homogenized by tissue lysis (Qiagen) in sterile PBS buffer with two 4.8 mm glass beads (Diatech Lab Line, Salerno, Italy). Each DNA extraction session included a negative extraction control (represented by an equal volume of RNase/DNase free water instead of DNA extraction elute). From 200 μL of homogenized sample, extraction of nucleic acid was obtained using a commercial kit (QIAampDNA Blood & Tissue; Qiagen, Hilden, Germany), according to the manufacturer’s instructions. A semi-nested PCR protocol targeting the V4 hyper-variable region of the 18S ribosomal RNA gene was used for the direct detection of Babesia/Theileria spp. DNA (10). In the first round, primers RLB-F2 (5’-GACACAGGGAGGTAGTGACAAG-3′) and RLB-R2 (5’-CTAAGAATTTCACCTCTGACAGT-3′) were used in a final reaction volume of 25 μL, using Promega PCR Master Mix (Promega Corporation, WI, United States), 20pM of each primer, and ≈100 ng of DNA template measured with the Biofhotometer plus (Eppendorf, Hamburg, Germany), according to the manufacturer’s instructions. The thermocycling conditions included initial denaturation for 5 min at 95°C, followed by 25 cycles of denaturation for 30s at 95°C, 45 s annealing at 50°C and 90s extension at 72°C and a final extension of 10 min at 72°C. Amplicons (1 μL) of the first PCR round were used as template in the second round with the same primer RLB-R2 plus RLB-FINT (5’-GACAAGAAATAACAATACRGGGC-3′). The reaction mix and cycling conditions were identical in first and second rounds, except for the total number of cycles (i.e., 40) and annealing temperature (55°C) in the second round. In all PCR runs, positive (i.e., Babesia canis DNA of fox spleen from Italy) and negative (reaction mix plus sterile water) controls were used. All PCR products were examined on 2% agarose gels stained with GelRed (VWR International PBI, Milan, Italy) and visualized on a GelLogic 100 gel documentation system (Kodak, New York, United States). Amplicons were purified by the QIAquick PCR Purification kit (Qiagen, Hilden, Germany) and sequenced in both directions using the same primers of the second round by the BigDye Terminator v.3.1 chemistry in a 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, United States). Consensus sequences were obtained by the Geneious software version 9.0 (Biomatters Ltd., Auckland, New Zealand) (24) and compared with those available in the GenBank database by the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.4. Statistical analysis
An exact binomial 95% confidence interval (95% CI) was established for the proportions of infection found herein. The Chi-squared or Fisherˈs exact test were used, depending on the population size, to assess any statistical differences of infection by animal age, sex, province of origin and sample collection year, while odds ratio was used for the infection risk by sex. A value of p < 0.05 was considered statistically significant. Statistical analyses were performed by using the online software Epitools—Epidemiological Calculators (25). The distribution of Babesia-positive wild boars according to provincial borders of the study area was determined using aerial imagery from Bing aerial maps software (Microsoft, Redmond, Washington, United States).
3. Results
A total number of 243 wild boar spleen samples from southern Italy between 2016 and 2022 were analyzed. Fifteen animals (i.e., 6.2, 95% CI: 3.4–9.9) tested positive to Babesia spp. DNA, 13 (i.e., 5.3, 95% CI: 3.1–8.9) and two (i.e., 0.8, 95% CI: 0.2–2.9) with B. vulpes and B. capreoli, respectively, using the combined semi-nested PCR/sequencing approach. The geographic distribution of Babesia-positive wild boars according to provincial borders of the study area is illustrated in Figure 1. Detailed data on prevalence, confidence intervals and statistical analyses are listed in Table 1. No statistically significant differences (i.e., p > 0.05) were found according to the boar’s age, sex, province and collection year. Three different 18S rRNA partial sequences of B. vulpes were identified (sequence types ST1, ST2, ST3), with the most representative type being ST1 (60%) and a single sequence of B. capreoli. Compared to ST1, there were single nucleotide polymorphisms in ST2 (a T instead of C in position 220) and ST3 (a C instead of G in position 71). All sequences had 99–100% nucleotide identity with those available in GenBank. Sequences obtained in this study were deposited in GenBank under the following accession numbers: OQ520218 for B. vulpes ST1, OQ520219 for ST2, OQ520220 for ST3 and OQ520222 for B. capreoli.
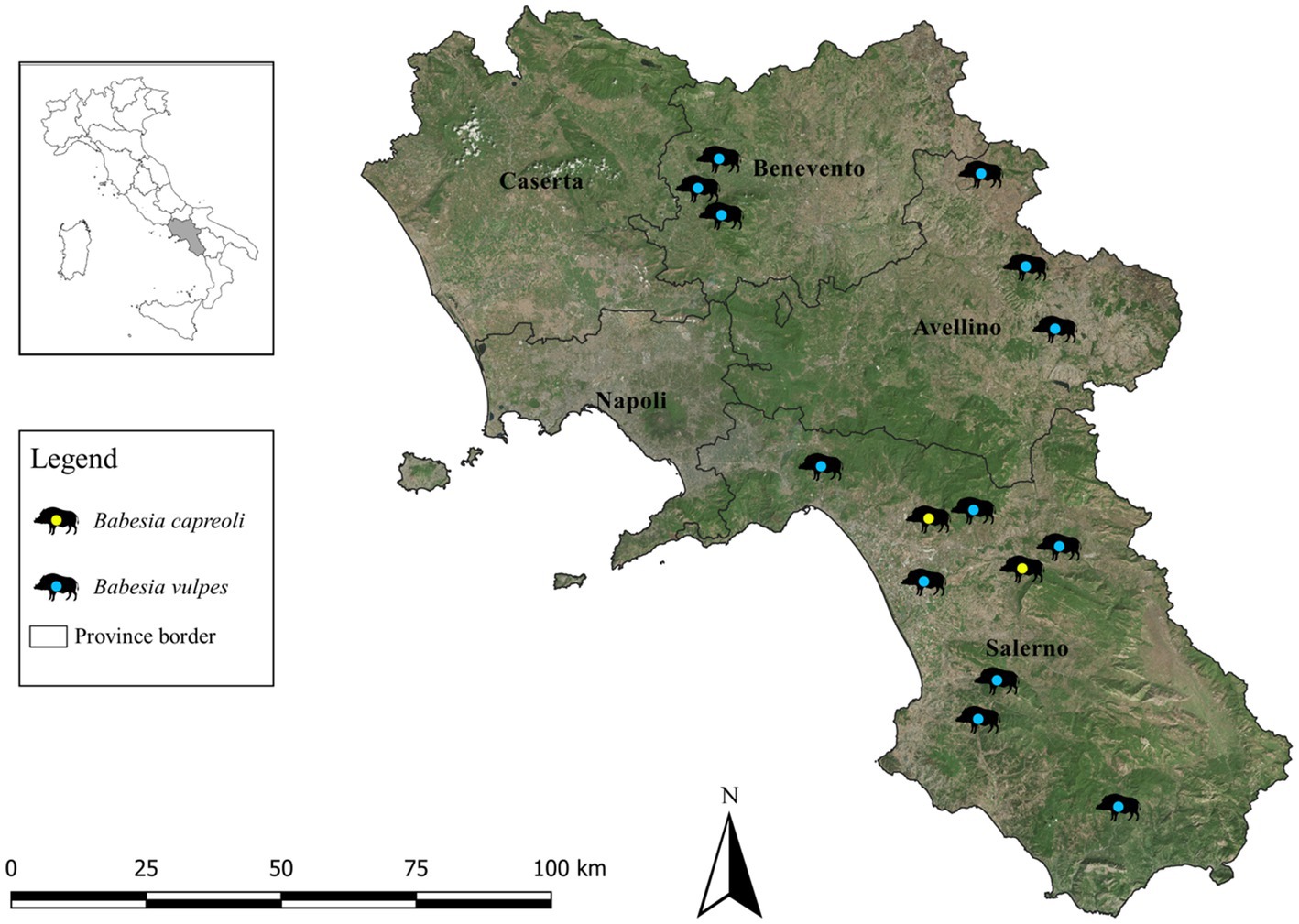
Figure 1. Distribution of wild boars (n = 15) positive to Babesia vulpes and Babesia capreoli by provinces (Avellino, Benevento, Caserta, Naples, Salerno) of the Campania region, southern Italy, 2016–2022.
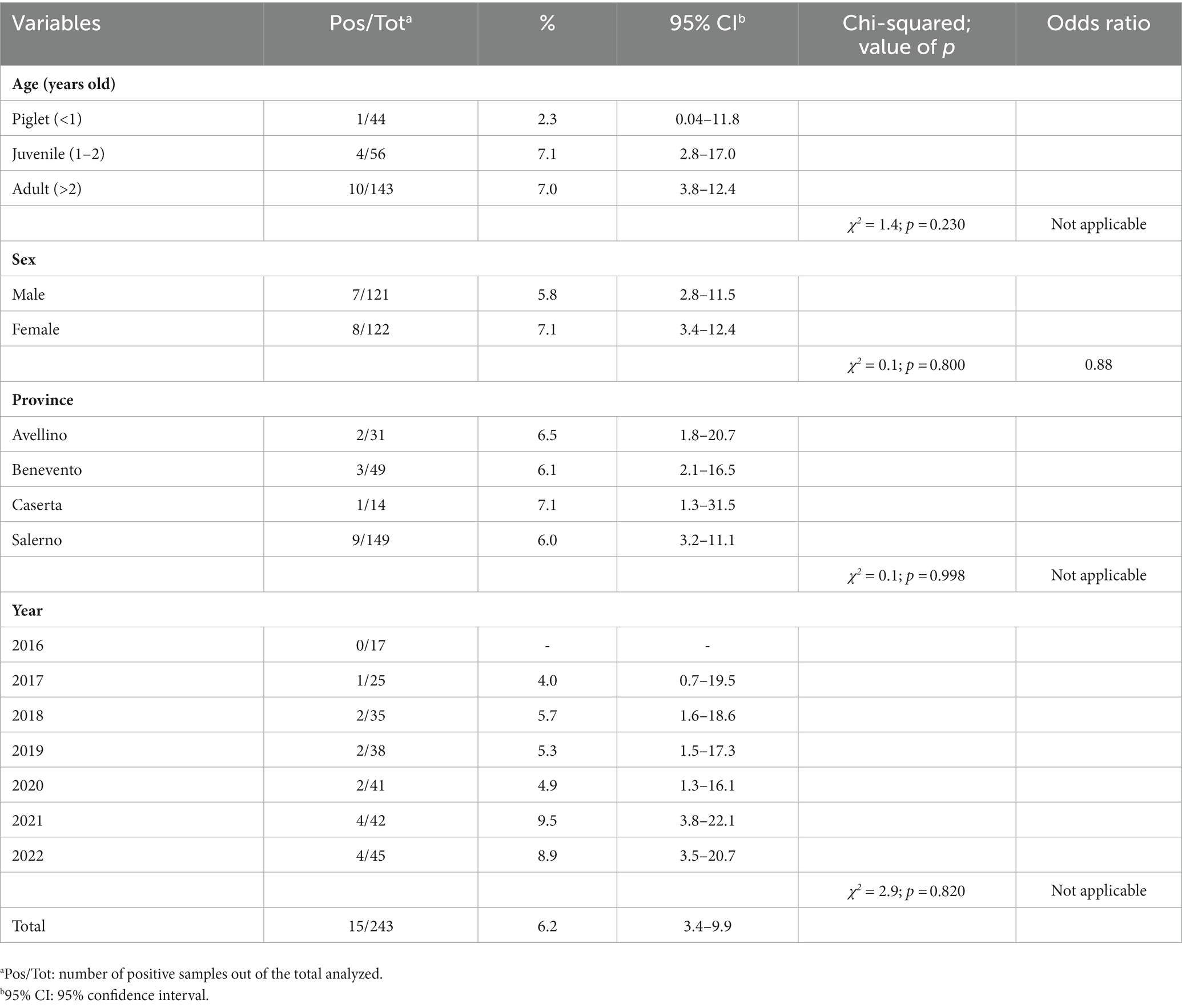
Table 1. Wild boar spleen samples (n = 243) tested for Babesia spp. DNA in southern Italy, 2016–2022.
4. Discussion
This study helps to fill the gap on Babesia spp. presence in wild boars, as well as suggesting cooperation of health stakeholders and trained persons (citizen science approach) as an effective tool for monitoring wildlife and related pathogens (26, 27).
To date, the only piroplasmid DNA in boars of Europe have been reported in a publication of B. bigemina in Italy (10), unspecified Theileria spp. in Italy and Portugal (8, 9) and B. divergens in the Czech Republic (7).
However, the moderate infection prevalence of Babesia spp. herein found in southern Italy (6.2%), and northern regions of the country (from 2.6% to 4.7%) (8, 10), suggests an involvement of boars in the sylvatic life cycle of the parasite. The absence of statistically significant difference in prevalence by boar’s age and sex in this study confirms that these variables do not influence the infection frequency, similar to findings of Zanet et al. (10). Again, the absence of significant differences in Babesia prevalence by province and collection year of samples suggests a stable circulation of infection in the study area.
Regarding B. vulpes, although the fox is the main reservoir in Europe (4, 28), the similar infection prevalence of this piroplasmid species in boars within this survey (13/243, 5.3%) and in foxes from the same study area (8/187, 4.3%) (29) indicates a potential involvement of this ungulate in pathogen maintenance. Despite roe deer being the most common host observed previously to be infected with B. capreoli (30, 31), its low prevalence (0.8%) in boars from this study should not exclude a role of these latter hosts in maintaining the pathogen considering the scant presence of other ungulate species in southern Italy, including roe deer (32). The potential pathogenic implications of B. capreoli infection in boars should be assessed in the future given that, although commonly asymptomatic in wildlife (6), cases of fatal babesiosis by this protozoan have been outlined in other wild ungulates, such as reindeer Rangifer tarandus (33) and Alpine chamois Rupicapra rupicapra (34, 35). Lastly, although not observed among boars in this study, the presence of suspected vectors of B. vulpes (i.e., Ixodes hexagonus and Ixodes canisuga) (36, 37) and B. capreoli (i.e., Ixodes ricinus) (38, 39) cannot be ruled out, considering that these tick species are commonly found on foxes (40) and hunting dogs (41) which live in sympatry with these ungulates. Indeed, due to the extensive time spent within sylvatic areas, hunting dogs show a higher prevalence of tick-borne pathogens compared to companion dogs (42). An example includes B. vulpes (43), capable to cause severe (4, 44) or fatal disease in dogs (45).
Despite the 18S rRNA gene is widely employed as a target for the molecular detection of piroplasmids (7, 14, 46, 47), the use of other genetic markers is recommended for species differentiation given the very high similarity of Babesia spp. sequences, such as B. capreoli and B. divergens which differ in just three positions (6, 30). Indeed, future studies on a larger sample size, including other wild ungulate species, and multiple genetic targets are needed to investigate the occurrence of piroplasmids in southern Italy.
The spread of wild boar populations may enhance the chance of transmission for emerging tickborne pathogens, including piroplasmids. More research is required to clarify the role of these ungulates in the maintenance of B. vulpes and B. capreoli in other epidemiological scenarios.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/nuccore; OQ520218, OQ520219, OQ520220, and OQ520222.
Ethics statement
The animal study was approved by the project “Ricerca corrente” (grant number: IZSME RC 05/16) by the Italian Ministry of Health. Written informed consent was not required for this study in accordance with national legislation and institutional requirements.
Author contributions
GS and VV conceptualized and designed the study. GS wrote the first draft of the manuscript. ND’A, CA, and HS wrote sections of the manuscript. AG, MGR, and FA performed molecular analyses. SR, SS, MO, and CDM were involved in sampling and database curation. GF and MGL managed project administration and resources. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the project “Ricerca corrente” (grant number: IZSME RC 05/16) funded by the Italian Ministry of Health.
Acknowledgments
The authors are grateful to all veterinarians and trained volunteers involved in this study for the kind collaboration in the sampling activities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Schnittger, L, Rodriguez, AE, Florin-Christensen, M, and Morrison, DA. Babesia: a world emerging. Infect Genet Evol. (2012) 12:1788–809. doi: 10.1016/j.meegid.2012.07.004
2. Kumar, A, O'Bryan, J, and Krause, PJ. The global emergence of human babesiosis. Pathogens. (2021) 10:1447. doi: 10.3390/pathogens10111447
3. Gray, A, Capewell, P, Zadoks, R, Taggart, MA, French, AS, Katzer, F, et al. Wild deer in the United Kingdom are a potential reservoir for the livestock parasite Babesia divergens. Curr Res Parasitol Vector Borne Dis. (2021) 1:100019. doi: 10.1016/j.crpvbd.2021.100019
4. Baneth, G, Cardoso, L, Brilhante-Simões, P, and Schnittger, L. Establishment of Babesia vulpes n. sp. (Apicomplexa: Babesiidae), a piroplasmid species pathogenic for domestic dogs. Parasit Vectors. (2019) 12:129. doi: 10.1186/s13071-019-3385-z
5. Cafiso, A, Bazzocchi, C, Cavagna, M, Di Lorenzo, E, Serra, V, Rossi, R, et al. Molecular survey of Babesia spp. and Anaplasma phagocytophilum in roe deer from a wildlife rescue center in Italy. Animals. (2021) 11:3335. doi: 10.3390/ani11113335
6. Fanelli, A. A historical review of Babesia spp. associated with deer in Europe: Babesia divergens/Babesia divergens-like, Babesia capreoli, Babesia venatorum, Babesia cf. odocoilei. Vet Parasitol. (2021) 294:109433. doi: 10.1016/j.vetpar.2021.109433
7. Hrazdilová, K, Lesiczka, PM, Bardoň, J, Vyroubalová, Š, Šimek, B, Zurek, L, et al. Wild boar as a potential reservoir of zoonotic tick-borne pathogens. Ticks Tick Borne Dis. (2021) 12:101558. doi: 10.1016/j.ttbdis.2020.101558
8. Tampieri, MP, Galuppi, R, Bonoli, C, Cancrini, G, Moretti, A, and Pietrobelli, M. Wild ungulates as Babesia hosts in northern and Central Italy. Vector Borne Zoonotic Dis. (2008) 8:667–74. doi: 10.1089/vbz.2008.0001
9. Pereira, A, Parreira, R, Nunes, M, Casadinho, A, Vieira, ML, Campino, L, et al. Molecular detection of tick-borne bacteria and protozoa in cervids and wild boars from Portugal. Parasit Vectors. (2016) 9:251. doi: 10.1186/s13071-016-1535-0
10. Zanet, S, Trisciuoglio, A, Bottero, E, de Mera, IG, Gortazar, C, Carpignano, MG, et al. Piroplasmosis in wildlife: Babesia and Theileria affecting free-ranging ungulates and carnivores in the Italian Alps. Parasit Vectors. (2014) 7:70. doi: 10.1186/1756-3305-7-70
11. Silaghi, C, Pfister, K, and Overzier, E. Molecular investigation for bacterial and protozoan tick-borne pathogens in wild boars (Sus scrofa) from southern Germany. Vector Borne Zoonotic Dis. (2014) 14:371–3. doi: 10.1089/vbz.2013.1495
12. Hornok, S, Sugár, L, Fernández de Mera, IG, de la Fuente, J, Horváth, G, Kovács, T, et al. Tick- and fly-borne bacteria in ungulates: the prevalence of Anaplasma phagocytophilum, haemoplasmas and rickettsiae in water buffalo and deer species in Central Europe, Hungary. BMC Vet Res. (2018) 14:98. doi: 10.1186/s12917-018-1403-6
13. Kazimírová, M, Hamšíková, Z, Špitalská, E, Minichová, L, Mahríková, L, Caban, R, et al. Diverse tick-borne microorganisms identified in free-living ungulates in Slovakia. Parasit Vectors. (2018) 11:495. doi: 10.1186/s13071-018-3068-1
14. Hrazdilová, K, Myśliwy, I, Hildebrand, J, Buńkowska-Gawlik, K, Janaczyk, B, Perec-Matysiak, A, et al. Paralogs vs. genotypes? Variability of Babesia canis assessed by 18S rDNA and two mitochondrial markers. Vet Parasitol. (2019) 266:103–10. doi: 10.1016/j.vetpar.2018.12.017
15. Pittiglio, C, Khomenko, S, and Beltran-Alcrudo, D. Wild boar mapping using population density statistics: from polygons to high resolution raster maps. PLoS One. (2018) 13:e0193295. doi: 10.1371/journal.pone.0193295
16. Fulgione, D, and Buglione, M. The boar war: five hot factors unleashing boar expansion and related emergency. Land. (2022) 11:887. doi: 10.3390/land11060887
17. Bajer, A, Beck, A, Beck, R, Behnke, JM, Dwużnik-Szarek, D, Eichenberger, RM, et al. Babesiosis in southeastern, central and northeastern Europe: an emerging and re-emerging tick-borne disease of humans and animals. Microorganisms. (2022) 10:945. doi: 10.3390/microorganisms10050945
18. Dantas-Torres, F, and Otranto, D. Species diversity and abundance of ticks in three habitats in southern Italy. Ticks Tick Borne Dis. (2013) 4:251–5. doi: 10.1016/j.ttbdis.2012.11.004
19. Veneziano, V, Piantedosi, D, Ferrari, N, Neola, B, Santoro, M, Pacifico, L, et al. Distribution and risk factors associated with Babesia spp. infection in hunting dogs from southern Italy. Ticks Tick Borne Dis. (2018) 9:1459–63. doi: 10.1016/j.ttbdis.2018.07.005
20. Sgroi, G, Iatta, R, Lia, RP, Napoli, E, Buono, F, Bezerra-Santos, MA, et al. Tick exposure and risk of tick-borne pathogens infection in hunters and hunting dogs: a citizen science approach. Transbound Emerg Dis. (2022) 69:e386–93. doi: 10.1111/tbed.14314
21. Union, European. (2004). Regulation (EC) no 853/2004 of the European Parliament and of the council laying down specific hygiene rules for food of animal origin of 29 April 2004. Available at: https://eur-lex.europa.eu/legal-content/en/ALL/?uri=CELEX%3A32004R0853 (Accessed 31 March 2023).
22. Massei, G, and Toso, S. Biologia e gestione del cinghiale. Bologna: Istituto Nazionale per la Fauna Selvatica (1993). 75 p.
23. Dean, AG, Sullivan, KM, and Soe, MM. (2003). OpenEpi: open source epidemiologic statistics for public health. http://www.openepi.com (Accessed 17 May 2023).
24. Kearse, M, Moir, R, Wilson, A, Stones-Havas, S, Cheung, M, Sturrock, S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. (2012) 28:1647–9. doi: 10.1093/bioinformatics/bts199
25. Sergeant, ESG. Epitools: Epidemiological Calculators (2018). Available at: https://epitools.ausvet.com.au (Accessed 20 January 2023).
26. Lawson, B, Petrovan, SO, and Cunningham, AA. Citizen science and wildlife disease surveillance. EcoHealth. (2015) 12:693–702. doi: 10.1007/s10393-015-1054-z
27. Hamer, SA, Curtis-Robles, R, and Hamer, GL. Contributions of citizen scientists to arthropod vector data in the age of digital epidemiology. Curr Opin Insect Sci. (2018) 28:98–104. doi: 10.1016/j.cois.2018.05.005
28. Baneth, G, Florin-Christensen, M, Cardoso, L, and Schnittger, L. Reclassification of Theileria annae as Babesia vulpes sp. nov. Parasit Vectors. (2015) 8:207. doi: 10.1186/s13071-015-0830-5
29. Sgroi, G, Iatta, R, Veneziano, V, Bezerra-Santos, MA, Lesiczka, P, Hrazdilová, K, et al. Molecular survey on tick-borne pathogens and Leishmania infantum in red foxes (Vulpes vulpes) from southern Italy. Ticks Tick Borne Dis. (2021) 12:101669. doi: 10.1016/j.ttbdis.2021.101669
30. Malandrin, L, Jouglin, M, Sun, Y, Brisseau, N, and Chauvin, A. Redescription of Babesia capreoli (Enigk and Friedhoff, 1962) from roe deer (Capreolus capreolus): isolation, cultivation, host specificity, molecular characterisation and differentiation from Babesia divergens. Int J Parasitol. (2010) 40:277–84. doi: 10.1016/j.ijpara.2009.08.008
31. Yabsley, MJ, and Shock, BC. Natural history of zoonotic Babesia: role of wildlife reservoirs. Int J Parasitol Parasites Wildl. (2012) 2:18–31. doi: 10.1016/j.ijppaw.2012.11.003
32. Freschi, P, Fascetti, S, Riga, F, Rizzardini, G, Musto, M, and Cosentino, C. Feeding preferences of the Italian roe deer (Capreolus capreolus italicus Festa, 1925) in a coastal Mediterranean environment. Animals. (2021) 11:308. doi: 10.3390/ani11020308
33. Bos, JH, Klip, FC, Sprong, H, Broens, EM, and Kik, MJL. Clinical outbreak of babesiosis caused by Babesia capreoli in captive reindeer (Rangifer tarandus tarandus) in the Netherlands. Ticks Tick Borne Dis. (2017) 8:799–801. doi: 10.1016/j.ttbdis.2017.06.006
34. Hoby, S, Robert, N, Mathis, A, Schmid, N, Meli, ML, Hofmann-Lehmann, R, et al. Babesiosis in free-living chamois (Rupicapra rupicapra) from Switzerland. Vet Parasitol. (2007) 148:341–5. doi: 10.1016/j.vetpar.2007.06.035
35. Hoby, S, Mathis, A, Doherr, MG, Robert, N, and Ryser-Degiorgis, MP. Babesia capreoli infections in alpine chamois (Rupicapra rupicapra), roe deer (Capreolus capreolus) and red deer (Cervus elaphus) from Switzerland. J Wildl Dis. (2009) 45:748–53. doi: 10.7589/0090-3558-45.3.748
36. Camacho, AT, Pallas, E, Gestal, JJ, Guitián, FJ, Olmeda, A, Telford, SR, et al. Ixodes hexagonus is the main candidate as vector of Theileria annae in Northwest Spain. Vet Parasitol. (2003) 112:157–63. doi: 10.1016/S0304-4017(02)00417-X
37. Obsomer, V, Wirtgen, M, Linden, A, Claerebout, E, Heyman, P, Heylen, D, et al. Spatial disaggregation of tick occurrence and ecology at a local scale as a preliminary step for spatial surveillance of tick-borne diseases: general framework and health implications in Belgium. Parasit Vectors. (2013) 6:190. doi: 10.1186/1756-3305-6-190
38. Estrada-Peña, A, Mihalca, AD, and Petney, TN. Ticks of Europe and North Africa: A guide to species identification. Cham: Springer Nature (2017). 368 p.
39. Bajer, A, and Dwużnik-Szarek, D. The specificity of Babesia-tick vector interactions: recent advances and pitfalls in molecular and field studies. Parasit Vectors. (2021) 14:507. doi: 10.1186/s13071-021-05019-3
40. Lorusso, V, Lia, RP, Dantas-Torres, F, Mallia, E, Ravagnan, S, Capelli, G, et al. Ixodid ticks of road-killed wildlife species in southern Italy: new tick-host associations and locality records. Exp Appl Acarol. (2011) 55:293–300. doi: 10.1007/s10493-011-9470-4
41. Maurelli, MP, Pepe, P, Colombo, L, Armstrong, R, Battisti, E, Morgoglione, ME, et al. A national survey of Ixodidae ticks on privately owned dogs in Italy. Parasit Vectors. (2018) 11:420. doi: 10.1186/s13071-018-2994-2
42. Pacifico, L, Braff, J, Buono, F, Beall, M, Neola, B, Buch, J, et al. Hepatozoon canis in hunting dogs from southern Italy: distribution and risk factors. Parasitol Res. (2020) 119:3023–31. doi: 10.1007/s00436-020-06820-2
43. Miró, G, Checa, R, Paparini, A, Ortega, N, González-Fraga, JL, Gofton, A, et al. Theileria annae (syn. Babesia microti-like) infection in dogs in NW Spain detected using direct and indirect diagnostic techniques: clinical report of 75 cases. Parasit Vectors. (2015) 8:217. doi: 10.1186/s13071-015-0825-2
44. Checa, R, López-Beceiro, AM, Montoya, A, Barrera, JP, Ortega, N, Gálvez, R, et al. Babesia microti-like piroplasm (syn. Babesia vulpes) infection in red foxes (Vulpes vulpes) in NW Spain (Galicia) and its relationship with Ixodes hexagonus. Vet Parasitol. (2018) 252:22–8. doi: 10.1016/j.vetpar.2018.01.011
45. Unterköfler, MS, Pantchev, N, Bergfeld, C, Wülfing, K, Globokar, M, Reinecke, A, et al. Case report of a fatal Babesia vulpes infection in a splenectomised dog. Parasitologia. (2023) 3:59–68. doi: 10.3390/parasitologia3010008
46. Santoro, M, Auriemma, C, Lucibelli, MG, Borriello, G, D'Alessio, N, Sgroi, G, et al. Molecular detection of Babesia spp. (Apicomplexa: Piroplasma) in free-ranging canids and mustelids from southern Italy. Front Vet Sci. (2019) 6:269. doi: 10.3389/fvets.2019.00269
Keywords: Babesia capreoli , Babesia vulpes , Italy, public health, wild boar
Citation: Sgroi G, D’Alessio N, Auriemma C, Salant H, Gallo A, Riccardi MG, Alfano F, Rea S, Scarcelli S, Ottaviano M, De Martinis C, Fusco G, Lucibelli MG and Veneziano V (2023) First molecular detection of Babesia vulpes and Babesia capreoli in wild boars from southern Italy. Front. Vet. Sci. 10:1201476. doi: 10.3389/fvets.2023.1201476
Edited by:
Georgiana Deak, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Dean Konjević, University of Zagreb, CroatiaLuciana-Catalina Rus, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, Romania
Copyright © 2023 Sgroi, D’Alessio, Auriemma, Salant, Gallo, Riccardi, Alfano, Rea, Scarcelli, Ottaviano, De Martinis, Fusco, Lucibelli and Veneziano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Gabriella Lucibelli, bWFyaWFnYWJyaWVsbGEubHVjaWJlbGxpQGl6c21wb3J0aWNpLml0
 Giovanni Sgroi
Giovanni Sgroi Nicola D’Alessio
Nicola D’Alessio Clementina Auriemma1
Clementina Auriemma1 Harold Salant
Harold Salant Marita Georgia Riccardi
Marita Georgia Riccardi Stefano Scarcelli
Stefano Scarcelli Claudio De Martinis
Claudio De Martinis Giovanna Fusco
Giovanna Fusco Maria Gabriella Lucibelli
Maria Gabriella Lucibelli