- 1Department of Large Animal Clinical Sciences, College of Veterinary Medicine, Michigan State University, East Lansing, MI, United States
- 2Departamento de Ciencias Veterinarias y Salud Pública, Universidad Católica de Temuco, Temuco, Chile
- 3Agriculture and Agribusiness Institute, Michigan State University Extension, Michigan State University, East Lansing, MI, United States
- 4Veterinary Diagnostic Laboratory, College of Veterinary Medicine, Michigan State University, East Lansing, MI, United States
Salmonella enterica serovar Dublin (S. Dublin) is a bacterium host-adapted to cattle with increasing prevalence in dairy facilities. It can severely affect cattle health, producing high morbidity and mortality in young calves and reducing the performance of mature animals. Salmonella Dublin is difficult to control and eradicate from herds, as it can be shed from clinically normal animals. In addition, S. Dublin is a zoonotic bacterium that can be lethal for humans and pose a risk for human and animal health due to its multi-drug resistant characteristics. This review provides an overview of S. Dublin as a pathogen in dairy facilities, the risk factors associated with infection, and current strategies for preventing and controlling this disease. Furthermore, current gaps in knowledge are also discussed.
1 Introduction
Salmonella enterica subspecies enterica serovar Dublin (S. Dublin) is a Gram-negative bacterium commonly affecting dairy cattle. Salmonella Dublin is host-adapted to cattle, where it can cause severe disease and compromise the welfare of young and mature bovine, and the economic return of the producer (1–4). Moreover, S. Dublin is a zoonosis that can cause severe disease leading to hospitalizations and mortality of consumers, farm personnel, calf handlers, veterinarians, and their families (5, 6). In France, major outbreaks had occurred as a result of the consumption of raw bovine cheese (7). Some countries like Denmark initiated a surveillance and control program since 2002, and as a result, the prevalence of S. Dublin in cattle was reduced from 25 to 7% from 2002 to 2015 (8). In countries without a control program, however, the prevalence of infections is high. For example, reports from Great Britain and Canada document apparent prevalence of infected herds ≥25% (9, 10). Also, S. Dublin has been the most frequently identified serotype among bovine Salmonella isolates from clinical samples submitted to veterinary diagnostic laboratories in the US and UK (11–14), and the second most common serovar in registered salmonellosis outbreaks in Germany, accounting for 30%−40% of cases (15). In the European Union, Dublin was also the second most common Salmonella serovar in cattle following Typhimurium (16).
In the US, S. Dublin has become one of the most important multi-drug resistant (MDR) bacteria in cattle (5, 17). The MDR has complicated the treatment of clinically sick animals and has become a threat to human medicine (18). In addition, S. Dublin may be difficult to control and eradicate from positive herds, as infection may persist in latent carriers and intermittently be shed to the environment (2). For the reasons provided, this review will focus primarily on risk factors for infection, prevention, and control of S. Dublin in dairy herds.
2 Salmonella Dublin in animal and human health
2.1 Prevalence in dairy farms
Salmonella Dublin is present worldwide but estimates of the proportion of S. Dublin infected herds vary greatly by country (Table 1). Some European countries have established a S. Dublin control and eradication that includes routine testing of all farms (19, 25, 26). Although no country is free from salmonellosis, 9 European countries report only sporadic cases. For example, S. Dublin has not been detected in Finland since early reports in the 1980s (26). Some countries, namely Finland, Norway, and Sweden, have additional restrictions for cattle trade in place (27). Conversely, more limited information regarding the prevalence of S. Dublin is available in countries without control programs. However, S. Dublin has been identified as one of the most common isolates of Salmonella spp. in dairy farms in the US, Germany, and the UK (11–15). In 2014, the USDA's National Animal Health Monitoring System (NAHMS) conducted a cross-sectional study including 234 farms nationwide. Salmonella Dublin was present in 0.7%, 6.7%, and 1.8% of the operations, milk samples, and milk filters, respectively (23). Additionally, the University of Minnesota Veterinary Diagnostic Laboratory (VDL) determined that S. Dublin was the most prevalent serotype isolated from bovine samples between 2005 and 2014, representing 31.8% of all isolates examined from 880 dairy farms from the upper Midwest (11). Likewise, S. Dublin was the most prevalent serotype in bovine samples in the University of Wisconsin VDL, accounting for 23% of all isolates from 2006 to 2015 (12). Similarly, S. Dublin has been the most common Salmonella serovar isolated from bovine samples at the Michigan State University VDL between 2018 and 2022, representing 10%−20% of all bovine Salmonella isolations (Table 2). In Germany and Italy, however, Salmonella Typhimurium was the most frequently isolated serovar in cattle samples collected as part of official outbreak investigations, followed by serovar Dublin accounting for 30%−40% of samples (15, 28). Nevertheless, there is some inherent bias in using veterinary diagnostic submissions to infer the prevalence of an organism on the broader population; therefore, these results need to be evaluated carefully.
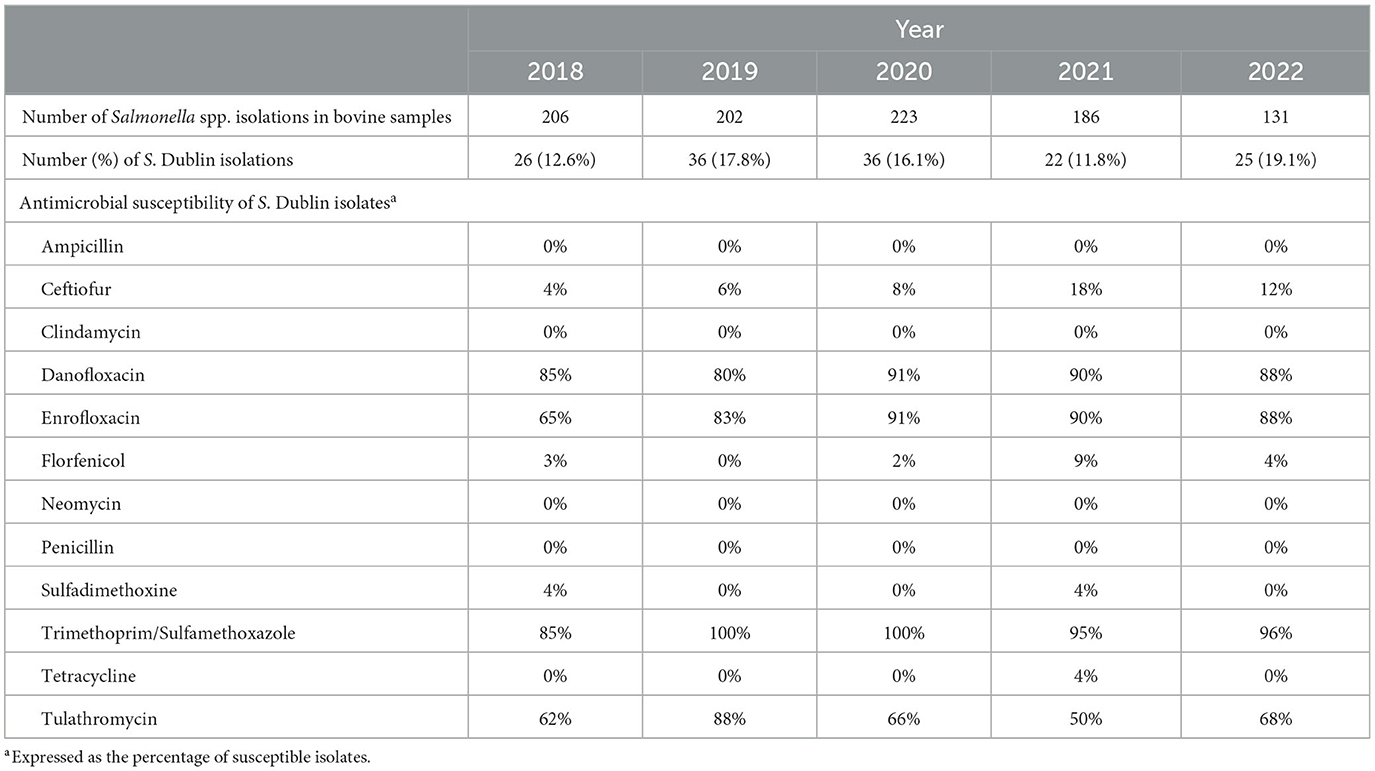
Table 2. Prevalence and antimicrobial susceptibility patterns of Salmonella Dublin in bovine isolates at the Michigan State University Veterinary Diagnostic Laboratory from 2018 to 2022.
For determining the prevalence of S. Dublin, different approaches may be taken, depending if the focus is the herd or within-herd prevalence. The use of fecal culture or serology has different strengths and limitations when determining prevalence. This will be discussed in the diagnosis section. However, sampling must be performed serially to categorize an animal as a carrier, transiently infected, or negative (29). Similarly, herd sampling must occur over time, as different factors may affect the shedding of the bacteria in the infected animals (29). For example, the prevalence of S. Dublin in Danish herds varied between 8 to 76% when samples were measured every 3 months for 1 year from 2000 to 2002 (29). Studies where S. Dublin has not been isolated should be taken cautiously due to intermittent shedding or animal selection. For example, Cummings et al. (30) did not isolate S. Dublin during the years 2004–2005 in the northeastern US because the samples in the study were collected from animals with clinical digestive disease and not respiratory; therefore, cases might have been missed. In addition, studies including samples from clinically ill animals may overestimate regional or within-herd prevalence. Nonetheless, it is still a valuable approach to differentiate positive and negative herds. Serology has been widely used to determine prevalence in herds (31–33). However, the age of the animal and the type of sample used may affect the correct classification of herds and animals. For example, if only milk is submitted for testing, non-lactating animals may be missed, and the within-herd prevalence may be underestimated.
2.2 Human health hazard
Salmonella Dublin is a zoonotic bacterium that can cause rare but severe illness in humans, and it is characterized by acute gastroenteritis and bacteremia (5). The case fatality for S. Dublin has been reported as the highest compared to other Salmonella enterica serotypes and has been described as six times greater than S. Typhimurium (34). The consumption of raw milk or raw dairy products has been associated with outbreaks of human salmonellosis caused by serovar Dublin (7, 35–37). However, farm personnel, veterinarians, and any person in direct contact with cattle are at risk of infection by accidentally ingesting animal feces or fluids (38). Moreover, S. Dublin has been isolated from the hide of dairy cull cows in processing meat plants, where it can constitute a risk for carcass contamination and a human health hazard (39).
In the US, the Foodborne Disease Active Surveillance Network determined an increase in the incidence of human S. Dublin by 7.6 times from 1968 to 2013 (5). The same study determined an increase in hospitalization from 68 to 78%, and an increase in mortality from 2.7 to 4.2%, when comparing the periods 1996–2004 with 2005–2013 (5). In a case-control study performed in Denmark, human salmonellosis due to S. Dublin was associated with an adjusted mortality of 12.4%, which was 12 times greater than the mortality in the control group (40). In the UK, S. Dublin was associated with 2% of human salmonellosis between 1949 and 1951 (41). Furthermore, Salmonella Dublin can cause long-lasting disease: in 17.7% of the cases, the condition lasted over 14 days with a 3.9% of mortality, which was higher than any other Salmonella serotype (41). As discussed in Section 2.4., S. Dublin has been characterized as an MDR bacterium to common antibiotics used for the treatment of bacterial infections in humans and animals. Therefore, S. Dublin is a pathogen that can affect human health severely and compromise the medical treatment. For that reason, it is fundamental to prevent and reduce the risk of infection from cattle to farm workers, animal care takers, and from animal derived food to humans.
2.3 Pathogenesis and clinical signs in cattle
Salmonella Dublin is a rod-shaped, facultative anaerobic, Gram-negative bacterium host adapted to cattle. It can produce enteritis and septicemia in bovine and severe disease in humans (5). The routes of infection for S. Dublin are shown in Figure 1. Briefly, the most common route of infection for S. Dublin is oral (42). Susceptible animals may uptake this bacterium by ingesting feces or body fluids (milk, saliva, and nasal secretion) from infected animals or contaminated environments (2, 42). In addition, vertical transmission with abortion in the last trimester of gestation or the birth of a congenitally infected calf has been reported (4, 43–45). One study determined the presence of S. enterica in the newborn calf. The results suggested that S. enterica was isolated from 50% of the enrolled calves and from samples that included lymph nodes and intestinal tissue (45).
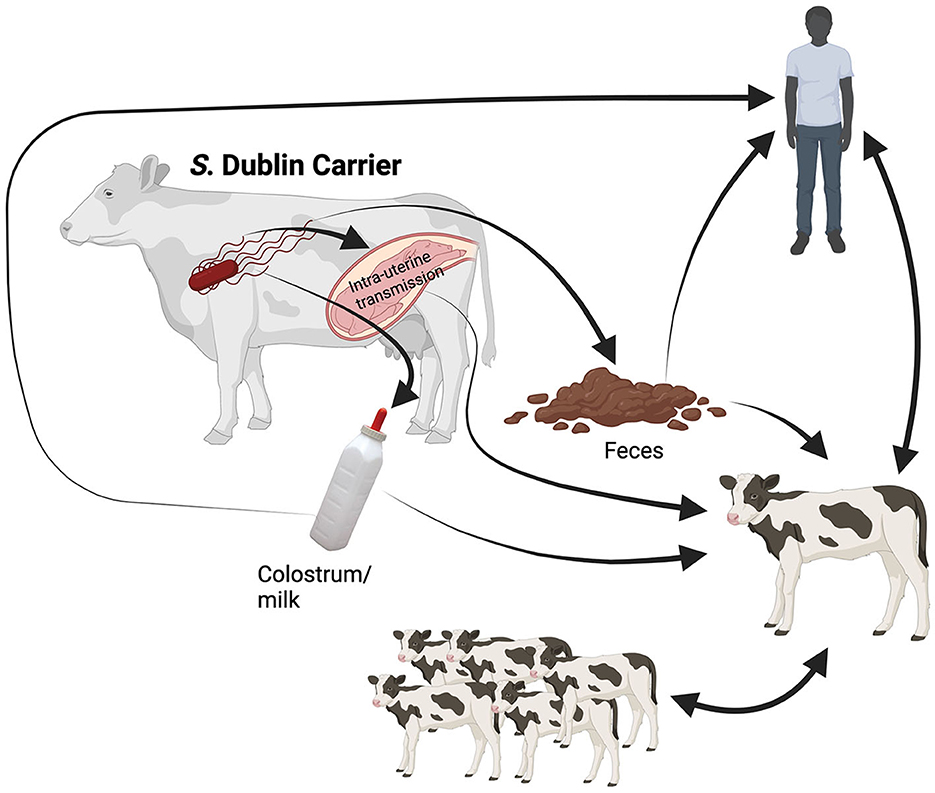
Figure 1. Illustration of the transmission routes for Salmonella Dublin. Symptomatic infected animals and latent carriers shed the bacterium to the environment under stress conditions, primarily in the peripartum. Once S. Dublin is shed in feces and secretions (saliva, colostrum, and milk) can survive in the environment. The newborn calf may uptake the bacterium via fecal-oral route at calving or by consumption of raw colostrum or milk from infected cows. Moreover, the infected calf will shed the bacterium to the environment, where susceptible calves will ingest S. Dublin through direct contact or fomite (contaminated surfaces or objects). In addition, the intrauterine infection of the fetus in the last trimester of gestation may occur, resulting in abortion or the birth of an infected calf. Finally, the zoonotic route will occur mainly in caretakers working with symptomatic animals and latent carriers at calving. The human will uptake S. Dublin from feces and secretions during calving assistance, cleaning equipment or facilities, manipulating raw colostrum and milk, or close contact with sick animals. Created with BioRender.com.
Additionally, experimental evidence suggests less common routes of infection through the upper respiratory tract (42) and mammary gland in cows under 60 days of lactation (46), although this study injected the bacteria directly into the udder in higher numbers which might not reflect natural exposure situations. In experimental conditions, it has been described that the minimum dose for infection that results in clinical disease and shedding of S. Dublin in animals younger than 6 months of life is 106 colony-forming unit (CFU) (2). However, this dose might depend on factors such as the strain used for infection (2, 47). In addition, experimental infection of mature cows with the administration of 1010 and 1011 CFU oral or intravenously resulted in diverse clinical manifestations, ranging from no signs to severe disease and abortion, which might be dependent on the strain used in the studies (44, 48). The incubation period has been determined as 12–72 h (49). The infectious dose might have importance in individual susceptibility to the pathogen and in the subclinical presentation of this disease.
Once S. Dublin enters the animal, it colonizes the digestive tract invading and multiplying in the enterocytes. From the distal ileum, it is translocated by efferent lymphatics from the mesenteric lymph node (50). After the translocation, Salmonella can rapidly disseminate and produce systemic disease. Furthermore, Salmonella can survive as facultative intracellular bacteria in numerous organs (spleen, liver, and lungs) and lymph nodes, allowing them to elude the adaptive immune response (38). The adaptation of S. Dublin to cattle has been linked to the selection of variants that can evade the innate response in the host and reduce the inflammatory response in the intestine's mucosa, which facilitates the dissemination (51). This process has been achieved by mutation within the host, resulting in the acquisition of genetic elements that encode specific virulence factors or the loss of specific genes to survive the particular conditions in the host environment (51, 52). In contrast, the infection of other animal species is accidental (52).
Several virulence factors have been associated with Salmonella, such as Salmonella Pathogenicity Islands (SPI), toxins, flagella, fimbriae, and virulence plasmids (53). Salmonella Dublin encodes the Type III Secretion System from SPI-1 and SPI-2, which allows it to invade the intestine and spread to systemic sites, respectively (54). Additionally, S. Dublin encodes the Type VI Secretion System from SPI-6 and SPI-19, which allows the injection of effector proteins into cells, increases the virulence, and in experimental settings, contributes to interbacterial competition (55). In addition, S. Dublin has the pSDV plasmid with a spv operon that encodes a toxin associated with the host's cellular apoptosis (53). Furthermore, the plasmid contributes to increased virulence, the systemic presentation of the disease, and encoding of antimicrobial-resistant genes (47, 53). Moreover, S. Dublin expresses flagella encoded by the gene fliC (56). The flagella allow motility and, through chemotaxis, enables the bacterium to respond to changes in the host environment (56). Salmonella Dublin also has fimbriae that aid in the adhesion to the host cells and virulence plasmids (53). Finally, S. Dublin virulence factors had been associated with an enhanced intracellular proliferation in intestinal and extraintestinal tissues, leading to severe diarrhea and mortality in experimental conditions (57). However, virulence factors will influence the infection depending on the host's immunity, and immunity is crucial in the outcome of clinical salmonellosis, especially in young calves with an immature immune system (58).
Compared to other Salmonella serovars, the clinical manifestation of S. Dublin is more severe due to a more invasive capacity in cattle (59). However, the clinical signs related to S. Dublin infection in cattle will depend on the animal's age and the endemicity of the pathogen on the farm, as endemic farms will have persistent infected animals (29). The clinical manifestation of S. Dublin infection is most common in calves 2–12 weeks old (42, 60); although it can affect cattle of all ages. A peracute presentation may occur in calves with sudden death in 1–2 days due to endotoxic shock. This presentation is more common in naïve herds (2, 3). Even though S. Dublin is an intestinal bacterium, the most distinctive clinical signs in acute infections of calves are pneumonia, respiratory distress, and hyperthermia (3, 17). In addition, calves show signs of obtundation, lack of appetite, diarrhea, arthritis, and meningoencephalitis. Chronic infection is more common in calves 6–8 weeks old that survived an acute infection and is characterized by growth retardation, loose stool, and lameness due to arthritis (2). The morbidity in outbreaks of S. Dublin has been reported between 10.5 and 34.8%, mortality between 2.3 and 18.2%, and case fatality of 26.4% for dairy calves (3, 61). As a point of comparison, respiratory disease is a common cause of morbidity and mortality in pre-weaned calves. It has been reported that respiratory disease mortality ranges from 2.8 to 14% and a case fatality of 6.0% (62–64). Considering the information in published studies, S. Dublin exceeds the mortality and case fatality of the common calf respiratory disease.
In unexposed adult animals, S. Dublin may lead to fever, bloody diarrhea, and in extreme cases to death. However, signs might be unnoticed in herds with a history of S. Dublin; the typical presentation is a slight fever and mild diarrhea, which can be accompanied by a sudden reduction in milk production (2, 38). As previously mentioned, abortion in the last trimester of gestation is another clinical manifestation in mature cows due to bacteremia (4).
Nonetheless, S. Dublin infection may generate persistent infections without clinical signs of disease, except for a reduction in the milk yield (2). These animals have been called “latent carriers.” These carriers host the bacteria in lymphoid tissue and periodically shed the pathogen in feces or fluids when the immune system is compromised or challenged, as in calving or transportation (29, 43, 65). Therefore, latent carriers are a source of infection for the herd, and identifying and culling these animals is essential for controlling the spread of the disease within and between herds (66, 67). Factors associated with an animal becoming a latent carrier are described in the Section 3.
2.4 Antimicrobial resistance
The prevalence of MDR S. Dublin is associated with geographical location. For example, although S. Dublin is considered one of the most common MDR serotypes in the US (17), MDR is not a common phenomenon in the European S. Dublin isolates (68). However, S. Dublin MDR can reduce the success of treatments, delay recovery, and increase mortality and costs in humans and cattle (18). As mentioned in the previous section, the infection of dairy calves with S. Dublin may produce a severe disease that may require antibiotic treatment. However, if antimicrobial treatment is administered without testing for drug susceptibility, the animal will not only fail to recover, but this practice will contribute to enhancing pathogen resistance (6). Hence, this will have negative consequences for human health due to the zoonotic nature of this agent (6). Furthermore, there are limited antimicrobial options to treat animals with clinical signs effectively, and some of those drugs are not labeled for treating S. Dublin infections, as will be discussed later in this section.
In the US, a study performed between 1996 and 2013 found that S. Dublin had a 43% higher prevalence of MDR compared to other Salmonella isolates (5). The National Antimicrobial Resistance Monitoring System (NARMS) reported that among S. Dublin isolates, 84% were resistant to five or more classes of antimicrobial drugs, and 57% were resistant to seven or more antimicrobial classes (5). Furthermore, an increase from 29 to 79% was observed in the proportion of isolates resistant to one or more antimicrobial classes when comparing 1996–2004 with 2005–2013 (5). Studies including samples from cattle, retail meat, and humans have reported S. Dublin antimicrobial resistance (AMR) to ampicillin, chloramphenicol, neomycin, tetracycline, streptomycin, sulfonamide, amoxicillin/clavulanic acid, and ceftriaxone (5, 6, 13, 17, 69). A similar situation has been described in other countries; in China, bovine isolates had AMR between 92 and 98% for tetracycline, sulfamethoxazole, and ampicillin (70). In the same study, human isolates were resistant to ceftiofur, chloramphenicol, tetracycline, and sulfamethoxazole in 40%, 50%, 50%, and 70% (70). Furthermore, in Canada, a study determined a high resistance to streptomycin, β-lactams, gentamicin, chloramphenicol, sulfisoxazole, and tetracycline from bovine samples (71). In Germany and the UK, S. Dublin has not been associated with significant AMR in bovine samples collected from cattle (68, 72) or beef products (73). This might be explained by a difference in the expression of plasmids and genes associated with AMR and MDR between regions, as specific AMR plasmids have been reported in the US and China but not in Europe (68).
In the US, it was reported that S. Dublin is susceptible to gentamicin, amikacin, cefoxitin, cephalothin, enrofloxacin, meropenem, and azithromycin (6, 17). Even though this pathogen is susceptible to enrofloxacin, this drug is only allowed to treat specific bovine respiratory disease pathogens in non-lactating cows and dairy replacements younger than 20 months. Hence, enrofloxacin is not labeled as a treatment for S. Dublin infections, and the extra-label use of this drug is prohibited for food animals in the US. Although most producers and veterinarians would treat respiratory disease without a pathogen isolation diagnosis, current US regulations imply that enrofloxacin cannot be used when S. Dublin is suspected or confirmed. This complicates the proper treatment of sick calves and potentially might increase the use of drugs to which S. Dublin has reduced susceptibility. The antimicrobial susceptibility pattern of S. Dublin isolated has largely remained unchanged in recent years (Table 2), with S. Dublin being generally susceptible to only four antimicrobials. Among those four, only Trimethoprim/Sulfamethoxazole has been labeled for treating Salmonella infections.
Different mechanisms have been described for the observed antimicrobial resistance, such as the production of enzymes that degrade or produce structural changes in antimicrobial agents, membrane impermeability, activation of antimicrobial efflux pumps, modification of cellular target for antibiotics, and biofilm formation (74, 75). Additionally, S. Dublin MDR has been linked to plasmid-borne resistant genes (6, 70, 75–77). Through whole genome sequencing, Srednik et al. (6) determined that the most common genes associated with AMR in the US were sulf2, tetA, aph(3″)-Ib, floR, and blaCMY−2. In China, similar genes were associated with AMR, and in a lower proportion aph(3′)-Ia and blaTEM−1B (70). In Canada, the AMR was associated with strAB, blaCMY−2/blaTEM−1, aadB, floR/cmlA, sul2, and a class A tetA (71). The mentioned genes conferred resistance to sulfonamide, tetracycline, aminoglycosides, chloramphenicol, and beta-lactams. Furthermore, in the mentioned studies, the MDR plasmid, IncA/C2, was present in 81.8% and 98.6% of all bovine isolates (6, 70). Hence, IncA/C2 might represent a critical MDR plasmid and a distinctive feature from bovine isolates with AMR (68, 70).
2.5 Economic impact
To date, a formal assessment of the financial impact of S. Dublin in the US has not been conducted. However, the economic importance of this pathogen in dairy farms is associated with the treatment of sick animals, reduction in milk yield, abortion, culled animals, mortality, cost of disinfection of facilities, and increased working hours during outbreaks (78, 79). Additionally, latent carriers contribute to the endemicity of the pathogen in the herd, spreading the pathogen and infecting susceptible animals, increasing the risk of outbreaks and associated costs. In Denmark, Nielsen et al. (78) utilized a simulation model to estimate the economic effects of S. Dublin for 10 years in a herd. The model included 12 scenarios based on three herd sizes and four management levels. The results indicated that the more significant economic losses occurred in the first year after the introduction of S. Dublin. The losses increased as the herd size increased, and the management practices decreased. From this analysis, milk yield losses in active and latent carriers were the most impactful. Even though these scenarios were adapted to dairy herds in Denmark, and some information might differ from other dairy systems, it provides an estimation of the economic implication of this pathogen. For example, in a 200 head herd with poor management practices, losses were estimated at 326 EUR per stall in the first year and 188 EUR per stall as an annual average of 10 years (78). Furthermore, this simulation reinforces the impact of basic management practices on the herd's economy. Those practices will be discussed in the prevention and control section.
3 Risk factors for infection
The identification of risk factors for infection with S. Dublin in animals and herds has come from epidemiological studies primarily conducted in Europe. These studies have reported factors associated with the host, farm practices/environment, and the agent. A summary list of risk factors can be found in Table 3.
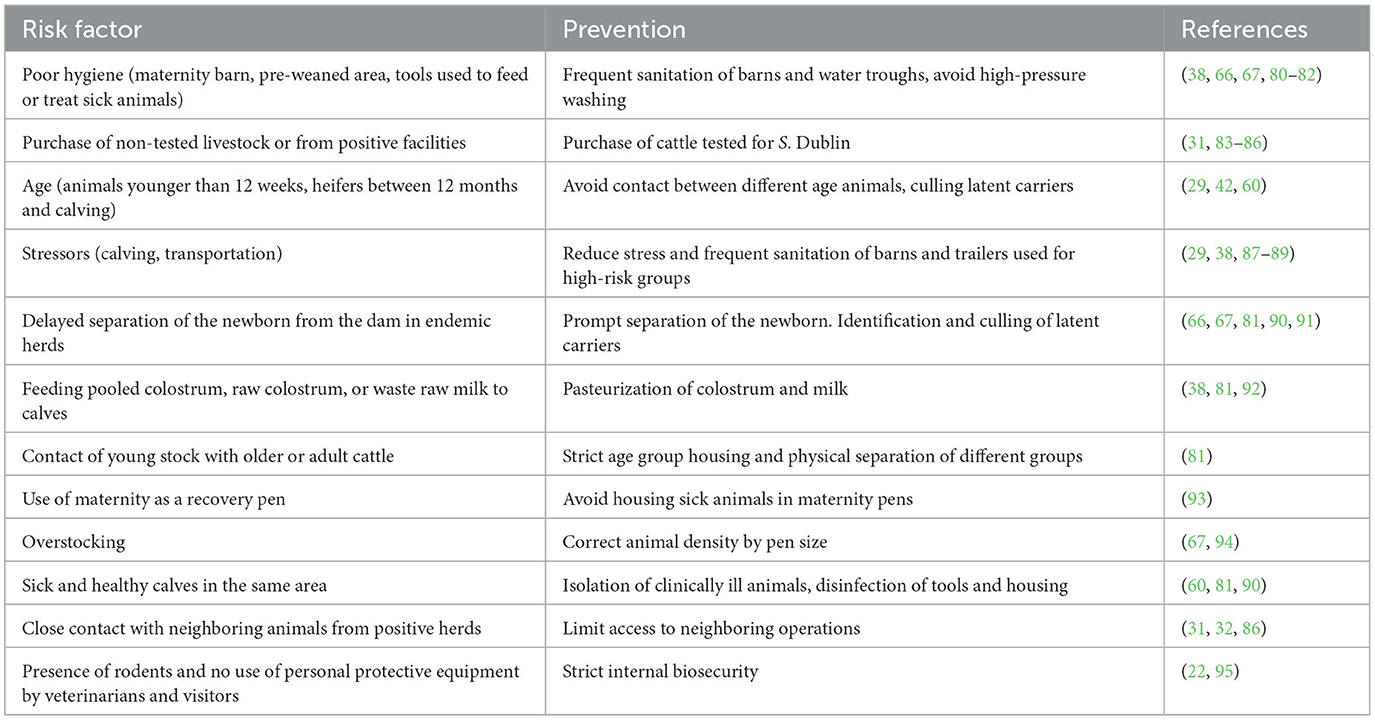
Table 3. Summary table for risk factors associated with infection and shedding of Salmonella Dublin and preventative strategies for dairy herds.
3.1 Host factors
The animal's immune status has a fundamental role in the control of new infections. Calves are born with an immature immune system that makes them highly susceptible to diseases early in life (96). In endemic herds, therefore, newborn calves exposed to contaminated environments or infected cows are at high risk of uptake S. Dublin by oral ingestion of feces, colostrum, or other body fluids and developing clinical disease (38). A similar situation is described for pre-weaned calves, as components of the adaptive immune system start to mature progressively from the second week of life (96); therefore, contact with S. Dublin in the environment or feed may lead to infection (38).
Moreover, animals experiencing concomitant diseases are at greater risk of infection or shedding of Salmonella spp. (97). In mature cows, liver fluke has been associated with an increase of over 14 times in the likelihood of S. Dublin infection compared to healthy cows (83). Likewise, concurrent infection with Bovine Viral Diarrhea Virus, an immunosuppressant virus, has been associated with a more severe clinical presentation of S. Dublin in calves (98).
3.2 Latent carrier factors
The importance of latent carriers in the epidemiology of S. Dublin is critical as they are defined as animals with a persistent infection without clinical signs but with an intermittent shedding of the pathogen to the environment through feces or secretions (2). However, latent carriers may be clinically ill or reactivate the shedding when the immune system is compromised (2). There is an association between age and the productive stage of the cattle with the risk of new infections and latent carrier status. For example, calves younger than 12 weeks of age are highly susceptible to infection of S. Dublin (42, 60). Repeated infection in young calves leads to a high proportion of resistant adult cattle and an increase in abortions in the last third of gestation (80). Moreover, it was reported that heifers infected between 1 year of life and calving were 11 times more likely to become a latent carrier than cows infected in mid or late lactation (29). A similar situation was described for cows infected in the peripartum, as they were 4 times more likely to become latent carriers (29).
Additionally, stress is an important factor associated with the infection of susceptible animals and the shedding of S. Dublin in latent carriers (29). Calving and transportation have been identified as periods of increased stress and shedding of S. Dublin (29, 87–89). Furthermore, any management practice that may increase the stress of the animals and the direct contact with infected animals that are actively shedding S. Dublin may result in infection of susceptible animals or outbreaks of disease (38).
3.3 Management practices and environmental factors
Different management strategies increase the prevalence of the herd and the susceptibility to infection for calves and cows. One of the most important risk factors for introducing and spreading infection into a susceptible herd is the purchase of livestock from facilities positive to S. Dublin or the purchase of non-tested animals (31, 83–85). The purchase from test-positive facilities has been associated with an increase between 2.5 and 4.2 times in the likelihood of infection of the new herd compared to animals purchased from negative facilities (31, 83). Furthermore, comparing the purchase of 1–10 animals with the purchase of more than 20 animals from test-positive herds, the likelihood of herd infection increased from 3.8 to 7.4 (86). The basic reproduction ratio (R0) for S. Dublin has been estimated at 1.1–2.7, meaning that 1–2 susceptible animals will be infected by introducing an infectious cow (84, 99). However, the screening and diagnosis of infected animals and latent carriers before purchase may be complex, as discussed in the diagnosis section.
In addition, poor hygiene has been identified as a highly influential risk factor, especially in maternity and calving pens (80). As mentioned previously, the shedding of S. Dublin increases around calving, and there is a greater probability of contact between the pathogen, newborn calves, and susceptible cows. Poor hygiene has been defined as visible or buildup of manure in the environment, the legs and udder of the cows, wet bedding, and infrequent manure removal (80). In fact, farms that added bedding material to calving areas once or twice per week had lower odds of being S. Dublin positive compared with farms that added bedding less than once weekly (10). A study comparing different levels of hygiene through a simulation model, determined that in a herd of 200 cows the probability of S. Dublin spread was 77 vs. 92% when comparing excellent with poor hygiene (80). Management practices associated with increased risk of Salmonella spp. infection of newborn calves are pooling and feeding raw colostrum in endemic herds (38, 92), delayed separation of the newborn calf from the dam (81), overstocking of the maternity pen (67, 94), increased number of cows calving outside the maternity area, and the use of the maternity as recovery pen (93). All these managements increase the exposure of newborn calves to feces and body fluids that might be contaminated with S. Dublin. In calf facilities, overstocking, poor nutrition, different age groups in a pen, and poor hygiene might lead to a fast spread of diseases. Additionally, clinically ill calves that are not isolated represent a hazard for pen mates or closely housed calves, as they are active shedders (38, 60, 81, 90). In endemic herds, feeding waste raw milk or the provision of contaminated water or feed has been associated with a greater probability of infection (38, 81).
There is no agreement in published studies if herd size is a risk factor for S. Dublin infection and spread. Several studies have determined that larger herds have a greater herd susceptibility (31, 83, 85, 86). However, other studies have not found herd size influential in infection, shedding, or spread of Salmonella (93, 100). Despite herd size, the lack of personal protective equipment may significantly increase the risk of introduction and infection of herds when veterinarians and visitors enter a facility (22). In addition, Tablante and Lane (95) suggested that wild mice could act as a reservoir and potentially transmit the pathogen to cattle or allow the persistence of the pathogen in the environment.
Concerning the environment, the prevalence of S. Dublin in nearby dairy facilities was associated with a greater risk of introduction into the herd (31, 32, 86). For example, a dairy farm with two or more positive neighboring facilities in a radius of 2 km is 1.7 times more likely to be infected than farms with no positive neighbors (86). This information may be critical in systems where cows have access to pasture and the possibility of close contact with neighboring herds exists. In systems where cows are housed in barns with no pasture access and higher biosecurity, this factor might be less influential in introducing S. Dublin. However, local herd prevalence was associated with 1.8 times greater risk of S. Dublin introduction (31). In addition, surface water contaminated with S. Dublin can lead to a fast spread of infection in animals with access to ponds, lakes, or rivers (83). Lactating and dry cows were 2.1 times more likely to shed Salmonella when having access to surface water than cows with no access (100). Finally, in the US, a seasonal association has been described; cows have a greater likelihood to test positive in summer, spring, and fall when compared to winter months (100). However, the authors speculate that this difference may be due to the intensity of the sampling in comparison to other studies, and the states included in the study had relatively similar weather conditions.
3.4 Agent factors
The main virulence factors associated to S. Dublin in cattle were presented in Section 2.3. In addition, S. Dublin is able to cross the placenta and infect the fetus. Therefore, maintaining latent carriers in the herd may contribute to the prenatal infection of newborn calves and keep the infection in the herd (4, 43–45). Hanson et al. (45) performed a study and found that 50% of the newborn calves enrolled were positive to S. enterica immediately after birth and before consuming colostrum. However, neither the dams nor the calves were tested to determine the serotype, and the sample size was small.
In addition, S. Dublin can survive in the environment, increasing the risk of infection of susceptible animals (101). Dairy cows in intense grazing herds had 13.2 times greater odds of infection with S. Dublin than cows from less intense grazing systems (83). This increase was explained by a greater fecal contamination of the pasture and a reduction in the time between manure spread and grazing. Likewise, a greater risk for shedding of S. Dublin has been identified in cows that consumed roughage from fields where manure was applied in solid or liquid form but not plowed in the same season (100).
4 Diagnosis
There are two main approaches for the diagnosis of S. Dublin: the detection of bacteria and the detection of antibodies. Both methods have advantages and limitations, and they can be performed in individual animals or the herd. This review will provide a basic approach to diagnosis, as techniques have been described in depth elsewhere (2).
4.1 Detection of bacteria
Bacteriological culture has been useful for isolating and identifying S. Dublin to trace infections and active shedders (2, 102). Bacteriological culture can be performed utilizing a variety of samples, including feces and fluids from live animals, organs from necropsies, aborted fetuses, or environmental samples. This method aims to isolate live bacteria (2). Thus, the procedure involves a pre-enrichment and a selective enrichment to allow bacterial growth, followed by plating and confirmation (2). This method has been described as more relevant in acute infections and clinically ill animals, as the correct isolation will depend on the number of bacteria in the sample (2, 102, 103). For that reason, the sensitivity of this assay has been described as low (104), and it has a limitation that latent carriers might be undetected due to the intermittent fecal shedding of S. Dublin. Bacteriological culture using samples from manure pits, drinking water, milk filters, and feces of clinically ill animals was associated with a sensitivity of 45%, 5%, 7%, and 38% for detecting S. Dublin, respectively (105). In post-mortem examination of clinically ill animals, the collection of tissues from the lungs, spleen, liver, intestine loops, gallbladder, intestinal content, and lymph nodes increases the probability of bacteria isolation (3, 106). A potentially more sensitive and faster method for the detection of genetic material of Salmonella is the polymerase chain reaction test (PCR) or real-time PCR (107). Persson et al. (108) described an S. Dublin-specific real-time PCR. The procedure for this method requires a pre-enrichment of the sample from lysates or extracted DNA (107). To increase sensitivity, a DNA extraction is recommended (107). However, the specificity of the assay in comparison to the numerous other Salmonella serotypes is yet to be determined. Currently, the most commonly utilized PCR assays do not allow serotype determination, and bacterial culture is recommended after performing a PCR (2). There is evidence of a multiplex PCR to discriminate Salmonella Enteritidis, Salmonella Pollorum, and S. Dublin (109). This assay is based on detecting three genes (tcpS, lygD, and flhB), where tcpS exists only in the serovars mentioned (109).
4.2 Detection of antibodies
The detection of immunoglobulins against S. Dublin is performed through an Enzyme-linked immunosorbent assay (ELISA). This method has a lower cost than bacteriological culture, and it can be used as a monitoring strategy in the herd to identify latent carriers during programs of control and eradication (67, 110). Salmonella Dublin is part of the D-serogroup of Salmonella and has the antigenic factors O1, O9, and O12; therefore, cross-reaction between serovars sharing O antigens may occur (111). The ELISA is based on detecting immunoglobulins directed to the LPS O- antigen from serum, milk, and bulk tank milk (BTM) samples (112, 113). The kit is commercially available in several countries for monitoring and surveillance of Salmonella infections in cattle herds (Applied Biosystems™, Massachusetts, USA). The results provided in this ELISA are semi-quantitative for antibody concentration as they are expressed in ODC% (optical density coefficient). The interpretation of the result is based on an estimated cut-off point to determine positive animals depending on the sample. The ODC% cut-off for serum, milk from an individual, or BTM is 35 ODC%. A positive correlation exists between the ODC% and antibody concentration in a sample. In BTM, the greater the ODC%, the higher the spread of infection in the herd (32). To identify latent carriers and the intermittent shedding of S. Dublin, sequential samples should be obtained from individual animals by using milk or serum samples. For example, studies from countries with eradication plans recommend that cows be sampled quarterly (31, 32). The limitations of this assay include that the sensitivity and specificity are age-dependent, as it performs better as a diagnostic test in animals older than 100 days (104). Additionally, milk samples have the limitation that only lactating cows can be tested (2, 105). Furthermore, when BTM samples are used, there is the probability of diluting immunoglobulins in cows' milk with low or no titers (110). Even though the aim of using BTM samples is to estimate herd prevalence, it is still unclear what is the limit of detection of the assay in BTM samples (i.e., the lowest proportion of positive animals needed for the sample to be correctly classified as positive). However, simulation models suggest that the apparent herd prevalence would need to be between 0.3 and 0.55 for a bulk tank milk sample to be declared positive with a 35 ODC% (110).
4.3 Postmortem examination
There are no pathognomonic lesions in internal organs for infections with S. Dublin. However, while considering the age of the animal and the clinical signs, necropsy may be helpful to guide diagnosis or for sample collection. In calves with clinical presentation, the gross pathologic findings in the lungs include pulmonary congestion, suppurative pneumonia, and chronic bronchopneumonia, depending on the severity of the clinical case (17, 106). The intestinal lesions may include diffuse catarrhal hemorrhagic enteritis, ileitis, and mesenteric lymphadenitis (3, 106). The intestinal content is watery, malodorous, and may contain mucous, blood, or fibrin clots (3, 106). Moreover, the liver is enlarged with rounded edges and hemorrhagic areas on the capsular surface, and gelatinous edema of the gallbladder (3). Jaundice and splenomegaly are also common postmorten findings in cases of S. Dublin (114). In some cases, swollen joints may be a finding (17).
5 Treatment
The treatment of S. enterica infection is directed to provide supportive therapy by correcting the electrolyte imbalance, dehydration, and management of inflammation (106, 115). However, salmonellosis due to serovar Dublin may be difficult to treat and fatal. Electrolyte therapy can be provided orally or intravenously, depending on the calf's degree of dehydration and suckle reflex. Calves with strong suckle reflex and dehydration below 8% can be offered oral electrolytes, while calves with over 8% dehydration should be administered intravenous fluid (116). Calves experiencing systemic infection should be administered non-steroidal anti-inflammatory drugs to manage inflammation. Antimicrobial use to treat the clinical presentation of S. Dublin is controversial, as discussed in the section on antimicrobial resistance. Ideally, the appropriate selection of antimicrobials should be based on susceptibility testing (106). However, this might not always be an option in the face of an outbreak. Moreover, the nutrition of the sick calf is important for recovery as it is the source of calories. Milk can be offered in smaller portions throughout the day to encourage milk consumption (117). Additionally, the calf's environment should be clean with dry bedding, have access to water, and be protected from adverse weather conditions.
6 Management strategies for prevention and control
The strategies for prevention and control of S. Dublin should focus on reducing disease transmission by changing management practices and restrictions in the purchase of animals, detecting and managing infectious animals, and continued surveillance (81). This section will discuss the most critical management for preventing and controlling S. Dublin on dairy farms. Table 3 lists strategies to reduce the risk of infection and shedding of S. Dublin.
6.1 Sanitation
Research has demonstrated that practices associated with the cleaning and disinfection of the environment are key elements in the prevention and control of S. Dublin (66, 67, 81). The first step in decontaminating the environment and equipment is removing organic material (i.e., food, manure, bedding), as this can inactivate disinfectants. Secondly, it is necessary to rinse the surfaces with water and apply a detergent in all the areas to clean. A thorough rinse with water should follow this step. High-pressure washing should be avoided, especially in indoor housing, as it may spread contaminants and aerosols to the environment (81). The final step is the application of the disinfectant in the concentration and contact time described on the label, as any variation in the use of a disinfectant may affect its effectiveness. Salmonella is susceptible to most disinfectants if steps one and two have been appropriately performed (118).
Besides cleaning and disinfection, improvement in management practices, such as cleaning water troughs with chlorinated disinfectant twice a week, replacing bedding weekly, and not recycling the water used for flushing pens, have been associated with a decrease in Salmonella incidence in a herd experiencing an outbreak (66). Furthermore, S. Dublin in endemic herds from Denmark has been controlled by daily removal of manure, the cleaning and disinfection of the calving area at least twice a month, and new bedding added weekly (67). Although these practices may be unpractical in some herds, the sanitation of calving areas should be performed according to their use. Moreover, trailers used for the transport of animals should be cleaned and disinfected consistently (38). In the case of calves, it has been recommended that after disinfection, pens should be kept empty for a minimum of 2 days before new animals are housed in them (81). Furthermore, cleaning and disinfection should be a priority for all equipment used to manage sick calves or to feed calves, including tools used to harvest, store, and provide colostrum and milk, such as esophageal tubes, nipples, bottles, and buckets (38, 66). In addition, buckets used to feed water and starter in pre-weaned calves should be cleaned daily and positioned to reduce the risk of fecal contamination (106).
Even though feeding waste milk can be cost-effective, the provision of raw milk may increase calf morbidity and mortality due to the ingestion of pathogens (82). Therefore, another hygiene strategy to prevent and control Salmonella infection in pre-weaned calves is the pasteurization of colostrum and milk as it has been shown to reduce microbial populations including S. enterica species (92). However, adequate maintenance of the equipment and regular sampling of colostrum and milk is highly recommended as a tool to monitor the correct functioning of the pasteurizer (38, 82).
6.2 Stocking density and isolation of sick animals
Maintaining an adequate stocking density in mature cow pens (i.e., close-up, maternity, fresh cow), heifer pens, and pre-weaned calves housed in groups is a practice that can reduce the contact between animals, the contamination of the environment, and new infections. As mentioned in the latent carrier factors section, it is necessary to consider the existence of groups of animals that are more susceptible to new infections or to shed S. Dublin. For instance, in those groups, it is critical not to overcrowd. In addition, the calving area should not be used to house sick animals due to the risk of environmental contamination and infection of newborn calves (67). Moreover, young calves should not have access to or contact with older animals; therefore, strict age group housing in conjunction with adequate stocking density has been recommended to prevent and control S. Dublin (81). Finally, animals exhibiting clinical signs of S. Dublin infection should be isolated, and strict cleaning procedures should be in place (38).
6.3 Newborn management
The management of the newborn and the calving area is critical to prevent S. Dublin infections. Some considerations must be taken as latent carriers can reactivate the shedding of bacteria around calving (29, 89). Newborn calves should be separated from the dam as soon as possible after birth to avoid oral infection due to consumption of colostrum or feces from the dam or other adult cows. The correct management of colostrum is fundamental to preventing infectious diseases in young calves (119). The newborn calf should receive the first colostrum feeding within 4 h after birth (119). Ideally, calves in endemic herds should be provided with pasteurized colostrum. Moreover, having fewer personnel in charge of calving and colostrum handling was associated with preventing S. Dublin infections (67). However, this might be challenging and not be suitable for all farms. Thus, it is fundamental to provide training to the personnel to keep farm practices according to the established protocols.
6.4 Vaccination
Commercial and autologous vaccines have been used to control S. Dublin in herds. However, autologous vaccines have not been evaluated in published studies for their efficacy in preventing and reducing the clinical signs or the shedding of S. Dublin in dairy animals. A commercially available modified-live vaccine available in various countries (EnterVene-D, Boehringer Ingelheim) is recommended for animals older than 2 weeks with a booster after 12–16 days. The benefits of an attenuated-live S. Dublin vaccine are associated with a robust response at mucosal level due to its action on lymphoid tissue in the gut, and a robust cell-mediated immune response due to intracellular proliferation (120, 121).
The age for the first dose can be too late as calves may get infected with S. Dublin at birth or in the first hours of life. Moreover, limited research addresses the dam vaccination as an approach for producing antibodies that can be delivered to the newborn calf through colostrum (122). The evidence suggests that specific antibodies for S. Dublin are in a higher concentration in the colostrum of cows vaccinated 30 days before dry-off than in non-vaccinated cows (122). However, it remains unknown if those antibodies have a protective effect on the newborn calf. In addition, research evaluating intranasal and oral vaccination of 4-day-old calves suggests that those are safe routes (123, 124). Using these extra-label routes of administration reduced the disease severity as calves administered the vaccine had a reduced mortality rate compared to unvaccinated calves (124). However, the incidence of pneumonia, abnormal fecal scores, and the fecal shedding of S. Dublin were not reduced (123, 124). Furthermore, no differences were observed in the average daily gain or antibody concentration at 10 weeks and 10 months of life compared to control calves (124).
Additionally, few studies assessed the cross-protection between S. enterica with modified-live vaccines. Mohler et al. (106) found that calves younger than 2 weeks of life orally vaccinated with modified-live S. Typhimurium had less severe clinical signs, improved appetite, and reduced fecal shedding when challenged with S. Dublin compared to control calves. However, calves in that study were challenged with a dose of S. Dublin to induce disease and minimize mortality, and respiratory clinical signs were not assessed. Similar results were found using an attenuated-live S. Typhimurium vaccine on diarrhea and shedding of Salmonella Newport and Salmonella Cerro (120). Moreover, there is a study assessing the vaccination of the dry cow with an S. Newport bacterin to provide cross-protection in an S. Typhimurium challenge in calves fed colostrum from vaccinated dams. Despite higher serological titers, no difference in mortality, clinical signs, hematology, and fecal cultures were observed in calves fed colostrum from vaccinated cows and the control group (125). Based on this research, the cross-protection between Salmonella spp. and potential protection against S. Dublin in dairy herds is still in development.
6.5 Farm biosecurity
Although the S. Dublin status of the herd is not frequently known in countries without a control program, several European studies have found that avoiding the purchase of cattle from test-positive herds or herds with unknown infection status should be a consistent practice in dairy facilities to prevent the introduction of S. Dublin in the herd (31, 67, 83, 86). Similarly, pasture-based operations should prevent close contact with cattle from neighboring farms, as the introduction of S. Dublin can occur through direct contact (22, 32, 83, 86). The vertical transmission of S. Dublin can perpetuate the disease even in closed herds (43, 45, 83). Therefore, culling of latent carriers should be considered for control or eradication of S. Dublin (66, 67). However, this strategy might not be suitable for all herds as seroprevalence within the herd may range from 3 to 70% (105, 126, 127). Additionally, persistently infected herds may have a high percentage of seroprevalence for S. Dublin without clinical signs or frequent outbreaks (127). Therefore, producers might find it economically counterproductive to cull valuable animals.
For surveillance, the collection of serial samples from the BTM is an easier, non-invasive, and less expensive method to determine the status of a herd concerning the presence or absence of S. Dublin antibodies (33, 91). In addition, for control of S. Dublin in a herd, it is helpful to collect serial samples from individual animals to diagnose the pathogen in an outbreak or identify latent carriers, this can be done with individual ELISA o bacteriological culture (90, 91). Additionally, the proper use of personal protective equipment for visitors and veterinarians, the control of rodents, along with the prevention of cross-contamination by not using the same equipment for different tasks are encouraged (22, 38, 95). As part of the eradication plan in Denmark, herds positive for S. Dublin had to maintain a high level of internal and external biosecurity for an average of 3 years to become test negative and prevent the recurrence of infection (31). Therefore, consistency in management practices is critical throughout the time to control S. Dublin.
6.6 Farm personnel
Training should be provided to personnel working with animals regarding the risk of zoonotic diseases and their prevention. Particular attention should be put on personnel handling animals during an outbreak of S. Dublin or personnel handling animals in periods of stress, when latent carriers may reactivate the shedding of S. Dublin. While working with animals or cleaning equipment, personal protective equipment (coveralls, washable boots, gloves, masks, and goggles) should be used. In addition, personnel should remove personal protective equipment before leaving the farm, and boots should be cleaned and disinfected. During an outbreak of salmonellosis or in eradication programs, it is recommended to have personnel working exclusively with clinically ill animals or isolated carriers to avoid potential spread and cross-contamination to other animals or areas. Finally, it is crucial to consider the mental and moral cost of an outbreak of S. Dublin on animal handlers, as it is a disease that might result in high morbidity and mortality, extended working hours, and depletion of morale due to low treatment success.
6.7 Gene therapy and gene editing
Recent research has evaluated the possibility to use gene therapy or gene editing to inhibit virulence gene expression in Salmonella spp. (54, 128). The use of certain lactic acid bacteria (LAB) combinations and their degradation products has been associated with the downregulation of virulence genes in different pathogens associated with neonatal calf diarrhea, including S. Dublin (54). Specifically for S. Dublin, a combination of 61 LAB strains was able to downregulate the expression of virulence factor fliC, which was assessed with RT-qPCR (54). Additionally, current research explores using CRISPR/Cas9 to delete the plasmid-based SpvB gene by using a modified pCas9 plasmid in pathogenic strains of S. Gallinarum (128). The results are promising, as the manipulated strain did not induce clinical disease or gross pathological lesions in broiler chickens 36 days after the challenge (128). Even though there are no current reports on S. Dublin, this procedure might be applied to produce nonvirulent strains that could be used in vaccines.
6.8 Next-generation sequencing
Whole genome sequencing (WGS) has become a tool to investigate the epidemiology of diseases, and S. Dublin has not been the exception. This methodology makes it possible to compare the whole genome of pathogens, which is a tremendous contribution to understanding disease dynamics, and it is a tool that can be used for the surveillance and control of diseases (129). WGS has been used to characterize the proximity in S. Dublin clades and differentiate the AMR and MDR genes between different regions and continents (68, 130). In addition, it has been used to determine the clonal relationship of S. Dublin strains in cattle and food animal products, with the potential to track zoonotic outbreaks (7, 73, 131). Furthermore, it has been used to study the proximity between cattle and human strains of S. Dublin, their virulence, and AMR genes (70). WGS has been and will be critical to understanding the adaptation of S. Dublin in different areas and the challenges associated with specific gene expression (68).
7 Future work
There is significant research related to S. Dublin in dairy cattle and operations. However, some areas remain less explored. Estimates of regional and national prevalence in many dairy production areas worldwide have yet to be determined. This type of research is associated with high costs, extended periods of sampling, and regional variation that might make the comparison difficult. Even though there are published studies, these have included specific regions, and in some cases, samples have not originated only from dairy farms. In addition, some studies have focused on determining several serovars of S. enterica; therefore, the study designs have not been specifically developed for S. Dublin. Moreover, studies determining the prevalence of S. Dublin have been performed mainly with samples submitted to VDLs, which might bias prevalence estimation (11, 12). Without knowing the local situation concerning S. Dublin, it is difficult to understand the real impact of this pathogen on cattle health and establish appropriate prevention strategies. Whole genome sequencing is becoming a tool of increasing importance in epidemiological studies, and it is an adequate method to investigate outbreaks, virulence factor, and their similarities between different regions and continents.
Currently, there are no published studies assessing the economic impact of S. Dublin in other production systems beyond Denmark. Worldwide, few studies have addressed the financial losses related to S. Dublin using simulation models (78, 79). However, the results of these studies might not be extrapolated to productive systems different from those described in the models. The simulation models might be an approach to estimating the economics of S. Dublin in a herd.
Another area with growing but limited information is gene editing or gene downregulation to prevent the severity of S. Dublin infections. Also, there is limited information is the use of vaccines to prevent S. Dublin in calves. As mentioned in the vaccination section, few studies have evaluated the effectiveness of the passive immunity provided to newborn calves by vaccinating the dam prepartum. Moreover, no studies evaluated the effect of vaccinating pregnant latent carriers on the disease transmission and severity in the offspring. Finally, few studies have addressed the effectiveness of extra-labeled routes of administration of vaccines against S. Dublin administered to young calves in field trials. Currently, no studies have addressed an integrated approach to prevent and reduce the devastating consequences of S. Dublin in neonatal and pre-weaned calves.
8 Conclusions
S. Dublin severely affects cattle and human health. Recent reports indicate that its prevalence has increased in several countries in the last several years, making it an emergent pathogen. Information is available on pathogenicity, antimicrobial resistance, risk factors, and preventive management practices. However, more research is still needed on the economic impact of outbreaks or endemic disease on herds and the effectiveness of strategies that could be implemented in dairy facilities to prevent and control S. Dublin.
Author contributions
AV-M: Conceptualization, Formal analysis, Methodology, Writing—original draft. RC-V: Methodology, Writing—review & editing. FC-N: Supervision, Writing—review & editing. RM: Resources, Writing—review & editing. AA: Conceptualization, Funding acquisition, Methodology, Supervision, Visualization, Writing—review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work has been supported by competitive grant 2022-68008-36354 of the US Department of Agriculture National Institute for Food and Agriculture (Kansas City, MO) and a grant from the Michigan Alliance for Animal Agriculture (East Lansing, MI), both awarded to AA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Nielsen TD, Green LE, Kudahl AB, Ostergaard S, Nielsen LR. Evaluation of milk yield losses associated with Salmonella antibodies in bulk tank milk in bovine dairy herds. J Dairy Sci. (2012) 95:4873–85. doi: 10.3168/jds.2011-4332
2. Nielsen LR. Review of pathogenesis and diagnostic methods of immediate relevance for epidemiology and control of Salmonella Dublin in cattle. Vet Microbiol. (2013) 162:1–9. doi: 10.1016/j.vetmic.2012.08.003
3. Guizelini CC, Tutija JF, Morais DR, Bacha FB, Ramos CAN, Leal CRB, et al. Outbreak investigation of septicemic salmonellosis in calves. J Infect Dev Ctries. (2020) 14:104–8. doi: 10.3855/jidc.12087
4. Hezil D, Zaidi S, Bensehir H, Zineddine R, Benamrouche N, Ghalmi F, et al. Salmonella Dublin associated with abortion in dairy cattle in Algiers and comparison of different diagnostic methods. Afr J Clin Exp Microbiol. (2021) 22:211–22. doi: 10.4314/ajcem.v22i2.14
5. Harvey RR, Friedman CR, Crim SM, Judd M, Barrett KA, Tolar B, et al. Epidemiology of Salmonella enterica serotype Dublin infections among humans, United States, 1968-2013. Emerg Infect Dis. (2017) 23:1493–501. doi: 10.3201/eid2309.170136
6. Srednik ME, Lantz K, Hicks JA, Morningstar-Shaw BR, Mackie TA, Schlater LK, et al. Antimicrobial resistance and genomic characterization of Salmonella Dublin isolates in cattle from the United States. PLoS ONE. (2021) 16:e0249617. doi: 10.1371/journal.pone.0249617
7. Ung A, Baidjoe AY, Van Cauteren D, Fawal N, Fabre L, Guerrisi C, et al. Disentangling a complex nationwide Salmonella Dublin outbreak associated with raw-milk cheese consumption, France, 2015 to 2016. Euro Surveill. (2019) 24:1700703. doi: 10.2807/1560-7917.ES.2019.24.3.1700703
8. de Knegt LV, Kudirkiene E, Rattenborg E, Sorensen G, Denwood MJ, Olsen JE, et al. Combining Salmonella Dublin genome information and contact-tracing to substantiate a new approach for improved detection of infectious transmission routes in cattle populations. Prev Vet Med. (2020) 181:104531. doi: 10.1016/j.prevetmed.2018.09.005
9. Henderson K, Mason C, Brulisauer F, Williams P. Determining the prevalence of antibodies to Salmonella Dublin in dairy herds in Great Britain by quarterly bulk tank testing. Prev Vet Med. (2022) 208:105776. doi: 10.1016/j.prevetmed.2022.105776
10. Perry KV, Kelton DF, Dufour S, Miltenburg C, Sedo SGU, Renaud DL, et al. Risk factors for Salmonella Dublin on dairy farms in Ontario, Canada. J Dairy Sci. (2023). doi: 10.3168/jds.2023-23517
11. Hong S, Rovira A, Davies P, Ahlstrom C, Muellner P, Rendahl A, et al. Serotypes and antimicrobial resistance in Salmonella enterica recovered from clinical samples from cattle and swine in Minnesota, 2006 to 2015. PLoS ONE. (2016) 11:e0168016. doi: 10.1371/journal.pone.0168016
12. Valenzuela JR, Sethi AK, Aulik NA, Poulsen KP. Antimicrobial resistance patterns of bovine Salmonella enterica isolates submitted to the Wisconsin Veterinary Diagnostic Laboratory: 2006-2015. J Dairy Sci. (2017) 100:1319–30. doi: 10.3168/jds.2016-11419
13. Adhikari B, Besser TE, Gay JM, Fox LK, Davis MA, Cobbold RN, et al. Introduction of new multidrug-resistant Salmonella enterica strains into commercial dairy herds. J Dairy Sci. (2009) 92:4218–28. doi: 10.3168/jds.2008-1493
14. APHA Animal and Plant Health Agency. Salmonella in Animals and Feed in Great Britain 2021. Weybridge: APHA, Animal and Plant Health Agency (2022).
15. Garcia-Soto S, Tomaso H, Linde J, Methner U. Epidemiological Analysis of Salmonella enterica subsp. enterica serovar Dublin in German cattle herds using whole-genome sequencing. Microbiol Spectr. (2021) 9:e0033221. doi: 10.1128/Spectrum.00332-21
16. European Food Safety, EFSA. Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. (2016) 14:e04634. doi: 10.2903/j.efsa.2016.4634
17. McDonough PL, Fogelman D, Shin SJ, Brunner MA, Lein DH. Salmonella enterica serotype Dublin infection: an emerging infectious disease for the northeastern United States. J Clin Microbiol. (1999) 37:2418–27. doi: 10.1128/JCM.37.8.2418-2427.1999
18. Davidson KE, Byrne BA, Pires AFA, Magdesian KG, Pereira RV. Antimicrobial resistance trends in fecal Salmonella isolates from northern California dairy cattle admitted to a veterinary teaching hospital, 2002-2016. PLoS ONE. (2018) 13:e0199928. doi: 10.1371/journal.pone.0199928
19. Nielsen LR, Houe H, Nielsen SS. Narrative review comparing principles and instruments used in three active surveillance and control programmes for non-EU-regulated diseases in the Danish cattle population. Front Vet Sci. (2021) 8:685857. doi: 10.3389/fvets.2021.685857
20. BLV Bundesamt für Verbraucherschutz und Lebensmittelsicherheit. BVL-Report·17, 3. Berichte zur Lebensmittelsicherheit: Zoonosen-Monitoring 2021. Berlin: BVL (2022).
21. Agren EC, Sternberg Lewerin S, Wahlstrom H, Emanuelson U, Frossling J. Low prevalence of Salmonella in Swedish dairy herds highlight differences between serotypes. Prev Vet Med. (2016) 125:38–45. doi: 10.1016/j.prevetmed.2015.12.015
22. van Schaik G, Schukken YH, Nielen M, Dijkhuizen AA, Barkema HW, Benedictus G, et al. Probability of and risk factors for introduction of infectious diseases into Dutch SPF dairy farms: a cohort study. Prev Vet Med. (2002) 54:279–89. doi: 10.1016/S0167-5877(02)00004-1
23. Sonnier JL, Karns JS, Lombard JE, Kopral CA, Haley BJ, Kim SW, et al. Prevalence of Salmonella enterica, Listeria monocytogenes, pathogenic Escherichia coli in bulk tank milk and milk filters from US dairy operations in the National Animal Health Monitoring System Dairy 2014 study. J Dairy Sci. (2018) 101:1943–56. doi: 10.3168/jds.2017-13546
24. Cummings KJ, Virkler PD, Wagner B, Lussier EA, Thompson BS. Herd-level prevalence of Salmonella Dublin among New York dairy farms based on antibody testing of bulk tank milk. Zoonoses Public Health. (2018) 65:1003–7. doi: 10.1111/zph.12523
25. Santman-Berends MH, Mars MF, Weber L, van Duijn HWF, Waldeck MM, Biesheuvel K, et al. Control and eradication programs for six cattle diseases in the Netherlands. Front Vet Sci. (2021) 8:670419. doi: 10.3389/fvets.2021.817576
26. Autio T, Tuunainen E, Nauholz H, Pirkkalainen H, London L, Pelkonen S, et al. Overview of control programs for cattle diseases in Finland. Front Vet Sci. (2021) 8:688936. doi: 10.3389/fvets.2021.688936
27. Hodnik JJ, Acinger-Rogic Z, Alishani M, Autio T, Balseiro A, Berezowski J, et al. Overview of cattle diseases listed under category C, D or E in the animal health law for which control programmes are in place within Europe. Front Vet Sci. (2021) 8:688078. doi: 10.3389/fvets.2021.674515
28. Tamba M, Pallante I, Petrini S, Feliziani F, Iscaro C, Arrigoni N, et al. Overview of control programs for twenty-four infectious cattle diseases in Italy. Front Vet Sci. (2021) 8:665607. doi: 10.3389/fvets.2021.665607
29. Nielsen LR, Schukken YH, Grohn YT, Ersboll AK. Salmonella Dublin infection in dairy cattle: risk factors for becoming a carrier. Prev Vet Med. (2004) 65:47–62. doi: 10.1016/j.prevetmed.2004.06.010
30. Cummings KJ, Warnick LD, Alexander KA, Cripps CJ, Grohn YT, McDonough PL, et al. The incidence of salmonellosis among dairy herds in the northeastern United States. J Dairy Sci. (2009) 92:3766–74. doi: 10.3168/jds.2009-2093
31. Nielsen LR, Dohoo I. Survival analysis of factors affecting incidence risk of Salmonella Dublin in Danish dairy herds during a 7-year surveillance period. Prev Vet Med. (2012) 107:60-9. doi: 10.1016/j.prevetmed.2012.06.002
32. Wedderkopp A, Stroger U, Lind P. Salmonella Dublin in Danish dairy herds: frequency of change to positive serological status in bulk tank milk ELISA in relation to serostatus of neighbouring farms. Acta Vet Scand. (2001) 42:295–301. doi: 10.1186/1751-0147-42-295
33. Um MM, Castonguay MH, Arsenault J, Bergeron L, Cote G, Fecteau G, et al. Estimation of the accuracy of an ELISA test applied to bulk tank milk for predicting herd-level status for Salmonella Dublin in dairy herds using Bayesian latent class models. Prev Vet Med. (2022) 206:105699. doi: 10.1016/j.prevetmed.2022.105699
34. Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D'Angelo M, Hurd S, et al. Salmonellosis outcomes differ substantially by serotype. J Infect Dis. (2008) 198:109–14. doi: 10.1086/588823
35. Taylor DN, Bied JM, Munro JS, Feldman RA. Salmonella Dublin infections in the United States, 1979-1980. J Infect Dis. (1982) 146:322–7. doi: 10.1093/infdis/146.3.322
36. Fang FC, Fierer J. Human infection with Salmonella Dublin. Medicine. (1991) 70:198–207. doi: 10.1097/00005792-199105000-00004
37. Maguire H, Cowden J, Jacob M, Rowe B, Roberts D, Bruce J, et al. An outbreak of Salmonella Dublin infection in England and Wales associated with a soft unpasteurized cows' milk cheese. Epidemiol Infect. (1992) 109:389–96. doi: 10.1017/S0950268800050378
38. Holschbach CL, Peek SF. Salmonella in dairy cattle. Vet Clin North Am Food Anim Pract. (2018) 34:133–54. doi: 10.1016/j.cvfa.2017.10.005
39. Brichta-Harhay DM, Arthur TM, Bosilevac JM, Kalchayanand N, Shackelford SD, Wheeler TL, et al. Diversity of multidrug-resistant Salmonella enterica strains associated with cattle at harvest in the United States. Appl Environ Microbiol. (2011) 77:1783–96. doi: 10.1128/AEM.01885-10
40. Helms M, Vastrup P, Gerner-Smidt P, Molbak K. Short and long term mortality associated with foodborne bacterial gastrointestinal infections: registry based study. BMJ. (2003) 326:357. doi: 10.1136/bmj.326.7385.357
41. Taylor J, Lovell R. Discussion on the epidemiology and treatment of Salmonella infections in man and animals with special reference to Salmonella Dublin. P Roy Soc Med. (1954) 46:445–7. doi: 10.1177/003591575304600610
42. Nazer AH, Osborne AD. Experimental Salmonella Dublin infection in calves. Br Vet J. (1977) 133:388–98. doi: 10.1016/S0007-1935(17)34040-X
43. Richardson A. The transmission of Salmonella Dublin to calves from adult carrier cows. Vet Rec. (1973) 92:112–5. doi: 10.1136/vr.92.5.112
44. Hall GA, Jones PW. A study of the pathogenesis of experimental Salmonella Dublin abortion in cattle. J Comp Pathol. (1977) 87:53–65. doi: 10.1016/0021-9975(77)90079-2
45. Hanson DL, Loneragan GH, Brown TR, Nisbet DJ, Hume ME, Edrington TS, et al. Evidence supporting vertical transmission of Salmonella in dairy cattle. Epidemiol Infect. (2016) 144:962–7. doi: 10.1017/S0950268815002241
46. Spier SJ, Smith BP, Cullor JS, Olander HJ, Roden LD, Dilling GW, et al. Persistent experimental Salmonella Dublin intramammary infection in dairy cows. J Vet Intern Med. (1991) 5:341–50. doi: 10.1111/j.1939-1676.1991.tb03148.x
47. Wallis TS, Paulin SM, Plested JS, Watson PR, Jones PW. The Salmonella Dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect Immun. (1995) 63:2755–61. doi: 10.1128/iai.63.7.2755-2761.1995
48. Hall GA, Jones PW. Experimental oral infections of pregnant heifers with Salmonella Dublin. Br Vet J. (1979) 135:75–82. doi: 10.1016/S0007-1935(17)32991-3
49. Cox JM, Pavic A. Salmonella | Introduction, Encyclopedia of Food Microbiology. Amsterdam: Elsevier (2014), p. 322–31. doi: 10.1016/B978-0-12-384730-0.00294-9
50. Pullinger GD, Paulin SM, Charleston B, Watson PR, Bowen AJ, Dziva F, et al. Systemic translocation of Salmonella enterica serovar Dublin in cattle occurs predominantly via efferent lymphatics in a cell-free niche and requires type III secretion system 1 (T3SS-1) but not T3SS-2. Infect Immun. (2007) 75:5191–9. doi: 10.1128/IAI.00784-07
51. Tanner JR, Kingsley RA. Evolution of Salmonella within hosts. Trends Microbiol. (2018) 26:986–98. doi: 10.1016/j.tim.2018.06.001
52. Evangelopoulou G, Kritas S, Govaris A, Burriel AR. Animal salmonelloses: a brief review of “host adaptation and host specificity” of Salmonella spp. Vet World. (2013) 6:703–8. doi: 10.14202/vetworld.2013.703-708
53. Cheng RA, Eade CR, Wiedmann M. Embracing Diversity: differences in virulence mechanisms, disease severity, and host adaptations contribute to the success of nontyphoidal Salmonella as a foodborne pathogen. Front Microbiol. (2019) 10:1368. doi: 10.3389/fmicb.2019.01368
54. Liu G, Kragh ML, Aabo S, Jensen AN, Olsen JE. Inhibition of virulence gene expression in Salmonella Dublin, Escherichia coli F5 and Clostridium perfringens associated with neonatal calf diarrhea by factors produced by lactic acid bacteria during fermentation of cow milk. Front Microbiol. (2022) 13:828013. doi: 10.3389/fmicb.2022.828013
55. Amaya FA, Blondel CJ, Barros-Infante MF, Rivera D, Moreno-Switt AI, Santiviago CA, et al. Identification of Type VI Secretion systems effector proteins that contribute to interbacterial competition in Salmonella Dublin. Front Microbiol. (2022) 13:811932. doi: 10.3389/fmicb.2022.811932
56. Olsen JE, Hoegh-Andersen KH, Casadesus J, Rosenkranzt J, Chadfield MS, Thomsen LE, et al. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium BMC Microbiol. (2013) 13:67. doi: 10.1186/1471-2180-13-67
57. Libby SJ, Adams LG, Ficht TA, Allen C, Whitford HA, Buchmeier NA, et al. The Spv genes on the Salmonella Dublin virulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect Immun. (1997) 65:1786–92. doi: 10.1128/iai.65.5.1786-1792.1997
58. Mohammed M, Le Hello S, Leekitcharoenphon P, Hendriksen R. The invasome of Salmonella Dublin as revealed by whole genome sequencing. BMC Infect Dis. (2017) 17:544. doi: 10.1186/s12879-017-2628-x
59. Salaheen S, Kim SW, Haley BJ, Van Kessel JAS. Differences between the global transcriptomes of Salmonella enterica serovars Dublin and Cerro infecting bovine epithelial cells. BMC Genomics. (2022) 23:498. doi: 10.1186/s12864-022-08725-z
60. Segall T, Lindberg AA. Experimental oral Salmonella Dublin infection in calves. A bacteriological and pathological study. Zentralblatt Veterinarmed B. (1991) 38:169–85. doi: 10.1111/j.1439-0450.1991.tb00859.x
61. Richardson A, Watson WA. A contribution to the epidemiology of Salmonella Dublin infection in cattle. Br Vet J. (1971) 127:173–83. doi: 10.1016/S0007-1935(17)37634-0
62. Dubrovsky SA, Van Eenennaam AL, Karle BM, Rossitto PV, Lehenbauer TW, Aly SS, et al. Bovine respiratory disease (BRD) cause-specific and overall mortality in preweaned calves on California dairies: the BRD 10K study. J Dairy Sci. (2019) 102:7320–8. doi: 10.3168/jds.2018-15463
63. Zhao W, Choi CY, Li G, Li H, Shi Z. Pre-weaned dairy calf management practices, morbidity and mortality of bovine respiratory disease and diarrhea in China. Livest Sci. (2021) 251:104608. doi: 10.1016/j.livsci.2021.104608
64. Urie NJ, Lombard JE, Shivley CB, Adams AE, Kopral CA, Santin M, et al. Preweaned heifer management on US dairy operations: part III. Factors associated with Cryptosporidium. and Giardia in preweaned dairy heifer calves. J Dairy Sci. (2018) 101:9199–213. doi: 10.3168/jds.2017-14060
65. Wray C, Wadsworth QC, Richards DW, Morgan JH. A 3-year study of Salmonella Dublin infection in a closed dairy-herd. Vet Rec. (1989) 124:532–5. doi: 10.1136/vr.124.20.532
66. Kent E, Okafor C, Caldwell M, Walker T, Whitlock B, Lear A, et al. Control of Salmonella Dublin in a bovine dairy herd. J Vet Intern Med. (2021) 35:2075–80. doi: 10.1111/jvim.16191
67. Nielsen TD, Vesterbaek IL, Kudahl AB, Borup KJ, Nielsen LR. Effect of management on prevention of Salmonella Dublin exposure of calves during a one-year control programme in 84 Danish dairy herds. Prev Vet Med. (2012) 105:101–9. doi: 10.1016/j.prevetmed.2012.01.012
68. Fenske GJ, Thachil A, McDonough PL, Glaser A, Scaria J. Geography shapes the population genomics of Salmonella enterica Dublin. Genome Biol Evol. (2019) 11:2220–31. doi: 10.1093/gbe/evz158
69. Berge AC, Thornburg E, Adaska JM, Moeller RB, Blanchard PC. Antimicrobial resistance in Salmonella enterica subspecies enterica serovar Dublin from dairy source calves in the central San Joaquin Valley, California (1998-2002). J Vet Diagn Invest. (2008) 20:497–500. doi: 10.1177/104063870802000414
70. Paudyal N, Pan H, Elbediwi M, Zhou X, Peng X, Li X, et al. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. (2019) 19:226. doi: 10.1186/s12866-019-1598-0
71. Mangat CS, Bekal S, Irwin RJ, Mulvey MR. A novel hybrid plasmid carrying multiple antimicrobial resistance and virulence genes in Salmonella enterica Serovar Dublin. Antimicrob Agents Chemother. (2017) 61:e02601-16. doi: 10.1128/AAC.02601-16
72. Klose C, Scuda N, Ziegler T, Eisenberger D, Hanczaruk M, Riehm JM, et al. Whole-genome investigation of Salmonella Dublin considering mountain pastures as reservoirs in Southern Bavaria, Germany. Microorganisms. (2022) 10:885. doi: 10.3390/microorganisms10050885
73. Linde J, Szabo I, Tausch SH, Deneke C, Methner U. Clonal relation between Salmonella enterica subspecies enterica serovar Dublin strains of bovine and food origin in Germany. Front Vet Sci. (2023) 10:1081611. doi: 10.3389/fvets.2023.1081611
74. Foley SL, Lynne AM. Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J Anim Sci. (2008) 86:E173–87. doi: 10.2527/jas.2007-0447
75. Alexander KA, Warnick LD, Wiedmann M. Antimicrobial resistant Salmonella in dairy cattle in the United States. Vet Res Commun. (2009) 33:191–209. doi: 10.1007/s11259-008-9170-7
76. Ferris KE, Andrews RE Jr, Thoen CO, Blackburn BO. Plasmid profile analysis, phage typing, and antibiotic sensitivity of Salmonella Dublin from clinical isolates in the United States. Vet Microbiol. (1992) 32:51–62. doi: 10.1016/0378-1135(92)90006-F
77. Marrero-Ortiz R, Han J, Lynne AM, David DE, Stemper ME, Farmer D, et al. Genetic characterization of antimicrobial resistance in Salmonella enterica serovars isolated from dairy cattle in Wisconsin. Food Res Int. (2012) 45:962–7. doi: 10.1016/j.foodres.2011.04.013
78. Nielsen TD, Kudahl AB, Ostergaard S, Nielsen LR. Gross margin losses due to Salmonella Dublin infection in Danish dairy cattle herds estimated by simulation modelling. Prev Vet Med. (2013) 111:51–62. doi: 10.1016/j.prevetmed.2013.03.011
79. Agren EC, Johansson J, Frossling J, Wahlstrom H, Emanuelson U, Sternberg-Lewerin S, et al. Factors affecting costs for on-farm control of Salmonella in Swedish dairy herds. Acta Vet Scand. (2015) 57:28. doi: 10.1186/s13028-015-0118-y
80. Nielsen LR, Kudahl AB, Ostergaard S. Age-structured dynamic, stochastic and mechanistic simulation model of Salmonella Dublin infection within dairy herds. Prev Vet Med. (2012) 105:59–74. doi: 10.1016/j.prevetmed.2012.02.005
81. Nielsen LR, Nielsen SS. A structured approach to control of Salmonella Dublin in 10 Danish dairy herds based on risk scoring and test-and-manage procedures. Food Res Int. (2012) 45:1158–65. doi: 10.1016/j.foodres.2011.02.027
82. Stabel JR, Hurd S, Calvente L, Rosenbusch RF. Destruction of Mycobacterium paratuberculosis. Salmonella spp, and Mycoplasma spp in raw milk by a commercial on-farm high-temperature, short-time pasteurizer. J Dairy Sci. (2004) 87:2177–83. doi: 10.3168/jds.S0022-0302(04)70038-7
83. Vaessen MA, Veling J, Frankena K, Graat EA, Klunder T. Risk factors for Salmonella Dublin infection on dairy farms. Vet Quart. (1998) 20:97–9. doi: 10.1080/01652176.1998.9694848
84. Van Schaik G, Klinkenberg D, Veling J, Stegeman A. Transmission of Salmonella in dairy herds quantified in the endemic situation. Vet Res. (2007) 38:861–9. doi: 10.1051/vetres:2007036
85. Nielsen LR, Dohoo I. Time-to-event analysis of predictors for recovery from Salmonella Dublin infection in Danish dairy herds between 2002 and 2012. Prev Vet Med. (2013) 110:370–8. doi: 10.1016/j.prevetmed.2013.02.014
86. Nielsen LR, Warnick LD, Greiner M. Risk factors for changing test classification in the Danish surveillance program for Salmonella in dairy herds. J Dairy Sci. (2007) 90:2815–25. doi: 10.3168/jds.2006-314
87. Gronstol H, Osborne AD, Pethiyagoda S. Experimental Salmonella infection in calves. 1. The effect of stress factors on the carrier state. J Hyg. (1974) 72:155–62. doi: 10.1017/S0022172400023342
88. Counter DE, Gibson EA. Salmonella Dublin infection in self contained dairy herds in East Anglia: excretion at calving. Vet Rec. (1980) 107:191–3. doi: 10.1136/vr.107.9.191
89. Wray C, Todd N, McLaren IM, Beedell YE. The epidemiology of Salmonella in calves: the role of markets and vehicles. Epidemiol Infect. (1991) 107:521–5. doi: 10.1017/S0950268800049219
90. Nielsen LR. Salmonella Dublin faecal excretion probabilities in cattle with different temporal antibody profiles in 14 endemically infected dairy herds. Epidemiol Infect. (2013) 141:1937–44. doi: 10.1017/S0950268812002579
91. Stockmarr A, Bodker R, Nielsen LR. Dynamic changes in antibody levels as an early warning of Salmonella Dublin in bovine dairy herds. J Dairy Sci. (2013) 96:7558–64. doi: 10.3168/jds.2012-6478
92. Edrington TS, Garcia Buitrago JA, Hagevoort GR, Loneragan GH, Bricta-Harhay DM, Callaway TR, et al. Effect of waste milk pasteurization on fecal shedding of Salmonella in preweaned calves. J Dairy Sci. (2018) 101:9266–74. doi: 10.3168/jds.2018-14668
93. Fossler CP, Wells SJ, Kaneene JB, Ruegg PL, Warnick LD, Bender JB, et al. Herd-level factors associated with isolation of Salmonella in a multi-state study of conventional and organic dairy farms II. Salmonella shedding in calves. Prev Vet Med. (2005) 70:279–91. doi: 10.1016/j.prevetmed.2005.04.002
94. Losinger WC, Wells SJ, Garber LP, Hurd HS, Thomas LA. Management factors related to Salmonella shedding by dairy heifers. J Dairy Sci. (1995) 78:2464–72. doi: 10.3168/jds.S0022-0302(95)76874-6
95. Tablante NL, Lane VM. Wild mice as potential reservoirs of Salmonella Dublin in a closed dairy-herd. Can Vet J. (1989) 30:590–2.
96. Chase CC, Hurley DJ, Reber AJ. Neonatal immune development in the calf and its impact on vaccine response. Vet Clin North Am Food Anim Pract. (2008) 24:87–104. doi: 10.1016/j.cvfa.2007.11.001
97. Fossler CP, Wells SJ, Kaneene JB, Ruegg PL, Warnick LD, Eberly LE, et al. Cattle and environmental sample-level factors associated with the presence of Salmonella in a multi-state study of conventional and organic dairy farms. Prev Vet Med. (2005) 67:39–53. doi: 10.1016/j.prevetmed.2004.10.005
98. Wray C, Roeder PL. Effect of bovine virus diarrhoea-mucosal disease virus infection on Salmonella infection in calves. Res Vet Sci. (1987) 42:213–8. doi: 10.1016/S0034-5288(18)30688-X
99. Nielsen LR, van den Borne B, van Schaik G. Salmonella Dublin infection in young dairy calves: transmission parameters estimated from field data and an SIR-model. Prev Vet Med. (2007) 79:46–58. doi: 10.1016/j.prevetmed.2006.11.006
100. Fossler CP, Wells SJ, Kaneene JB, Ruegg PL, Warnick LD, Bender JB, et al. Herd-level factors associated with isolation of Salmonella in a multi-state study of conventional and organic dairy farms I. Salmonella shedding in cows. Prev Vet Med. (2005) 70:257–77. doi: 10.1016/j.prevetmed.2005.04.003
101. Kirchner MJ, Liebana E, McLaren I, Clifton-Hadley FA, Wales AD, Davies RH, et al. Comparison of the environmental survival characteristics of Salmonella Dublin and Salmonella Typhimurium. Vet Microbiol. (2012) 159:509–14. doi: 10.1016/j.vetmic.2012.04.009
102. Richardson A, Fawcett AR. Salmonella Dublin infection in claves: the value of rectal swabs in diagnosis and epidemiological studies. Br Vet J. (1973) 129:151–6. doi: 10.1016/S0007-1935(17)36539-9
103. Baggesen DL, Nielsen LR, Sorensen G, Bodker R, Ersboll AK. Growth inhibitory factors in bovine faeces impairs detection of Salmonella Dublin by conventional culture procedure. J Appl Microbiol. (2007) 103:650–6. doi: 10.1111/j.1365-2672.2007.03292.x
104. Nielsen LR, Toft N, Ersboll AK. Evaluation of an indirect serum ELISA and a bacteriological faecal culture test for diagnosis of Salmonella serotype Dublin in cattle using latent class models. J Appl Microbiol. (2004) 96:311–9. doi: 10.1046/j.1365-2672.2004.02151.x
105. Veling J, Barkema HW, van der Schans J, van Zijderveld F, Verhoeff J. Herd-level diagnosis for Salmonella enterica subsp. enterica serovar Dublin infection in bovine dairy herds. Prev Vet Med. (2002) 53:31–42. doi: 10.1016/S0167-5877(01)00276-8
106. Mohler VL, Izzo MM, House JK. Salmonella in calves. Vet Clin North Am Food Anim Pract. (2009) 25:37–54. doi: 10.1016/j.cvfa.2008.10.009
107. Goodman LB, McDonough PL, Anderson RR, Franklin-Guild RJ, Ryan JR, Perkins GA, et al. Detection of Salmonella spp. in veterinary samples by combining selective enrichment and real-time PCR. J Vet Diagn Invest. (2017) 29:844–51. doi: 10.1177/1040638717728315
108. Persson S, Jacobsen T, Olsen JE, Olsen KE, Hansen F. A new real-time PCR method for the identification of Salmonella Dublin. J Appl Microbiol. (2012) 113:615–21. doi: 10.1111/j.1365-2672.2012.05378.x
109. Xiong D, Song L, Tao J, Zheng H, Zhou Z, Geng S, et al. An efficient multiplex PCR-based assay as a novel tool for accurate inter-serovar discrimination of Salmonella Enteritidis, S. Pullorum/Gallinarum and S Dublin. Front Microbiol. (2017) 8:420. doi: 10.3389/fmicb.2017.00420
110. Nielsen LR, Ersboll AK. Factors associated with variation in bulk-tank-milk Salmonella Dublin ELISA ODC% in dairy herds. Prev Vet Med. (2005) 68:165–79. doi: 10.1016/j.prevetmed.2004.12.006
111. Konrad H, Smith BP, Dilling GW, House JK. Production of Salmonella serogroup D (O9)-specific enzyme-linked immunosorbent assay antigen. Am J Vet Res. (1994) 55:1647–51.
112. Smith BP, Oliver DG, Singh P, Dilling G, Martin PA, Ram BP, et al. Detection of Salmonella Dublin mammary gland infection in carrier cows, using an enzyme-linked immunosorbent assay for antibody in milk or serum. Am J Vet Res. (1989) 50:1352–60.
113. Spier SJ, Smith BP, Tyler JW, Cullor JS, Dilling GW, Dapfaff L, et al. Use of ELISA for detection of immunoglobulins G and M that recognize Salmonella Dublin lipopolysaccharide for prediction of carrier status in cattle. Am J Vet Res. (1990) 51:1900–4.
114. Henderson K, Mason C. Diagnosis and control of Salmonella Dublin in dairy herds. In Pract. 39:158–68. doi: 10.1136/inp.j1160
115. Constable PD. Antimicrobial use in the treatment of calf diarrhea. J Vet Intern Med. (2004) 18:8–17. doi: 10.1111/j.1939-1676.2004.tb00129.x
116. Constable PD, Trefz FM, Sen I, Berchtold J, Nouri M, Smith G, et al. Intravenous and oral fluid therapy in neonatal calves with diarrhea or sepsis and in adult cattle. Front Vet Sci. (2020) 7:603358. doi: 10.3389/fvets.2020.603358
117. McGuirk SM. Disease management of dairy calves and heifers. Vet Clin North Am Food Anim Pract. (2008) 24:139–53. doi: 10.1016/j.cvfa.2007.10.003
118. Moretro T, Vestby LK, Nesse LL, Storheim SE, Kotlarz K, Langsrud S, et al. Evaluation of efficacy of disinfectants against Salmonella from the feed industry. J Appl Microbiol. (2009) 106:1005–12. doi: 10.1111/j.1365-2672.2008.04067.x
119. Godden SM, Lombard JE, Woolums AR. Colostrum management for dairy calves. Vet Clin North Am Food Anim Pract. (2019) 35:535–56. doi: 10.1016/j.cvfa.2019.07.005
120. Dueger EL, House JK, Heithoff DM, Mahan MJ. Salmonella DNA adenine methylase mutants elicit early and late onset protective immune responses in calves. Vaccine. (2003) 21:3249–58. doi: 10.1016/S0264-410X(03)00252-4
121. Jones BD. Salmonellosis: host imunne responses and bacterial virulence determinants. Annu Rev Immunol. (1996) 14:533–61. doi: 10.1146/annurev.immunol.14.1.533
122. Smith GW, Smith F, Zuidhof S, Foster DM. Short communication: characterization of the serologic response induced by vaccination of late-gestation cows with a Salmonella Dublin vaccine. J Dairy Sci. (2015) 98:2529–32. doi: 10.3168/jds.2014-8972
123. Habing GG, Neuder LM, Raphael W, Piper-Youngs H, Kaneene JB. Efficacy of oral administration of a modified-live Salmonella Dublin vaccine in calves. J Am Vet Med Assoc. (2011) 238:1184–90. doi: 10.2460/javma.238.9.1184
124. Cummings KJ, Rodriguez-Rivera LD, Capel MB, Rankin SC, Nydam DV. Short communication: oral and intranasal administration of a modified-live Salmonella Dublin vaccine in dairy calves: clinical efficacy and serologic response. J Dairy Sci. (2019) 102:3474–9. doi: 10.3168/jds.2018-14892
125. Foster D, Jacob M, Stowe D, Smith G. Exploratory cohort study to determine if dry cow vaccination with a Salmonella Newport bacterin can protect dairy calves against oral Salmonella challenge. J Vet Intern Med. (2019) 33:1796–806. doi: 10.1111/jvim.15529
126. House JK, Smith BP, Dilling GW, Roden LD. Enzyme-linked immunosorbent assay for serologic detection of Salmonella Dublin carriers on a large dairy. Am J Vet Res. (1993) 54:1391–9.
127. Nielsen LR. Within-herd prevalence of Salmonella Dublin in endemically infected dairy herds. Epidemiol Infect. (2013) 141:2074–82. doi: 10.1017/S0950268812003007
128. Basit A, Tahir H, Haider Z, Tariq H, Ullah A, Rehman SU, et al. CRISPR/Cas9-based deletion of SpvB gene from Salmonella Gallinarum leads to loss of virulence in chicken. Front Bioeng Biotechnol. (2022) 10:885227. doi: 10.3389/fbioe.2022.885227
129. Kao RR, Haydon DT, Lycett SJ, Murcia PR. Supersize me: how whole-genome sequencing and big data are transforming epidemiology. Trends Microbiol. (2014) 22:282–91. doi: 10.1016/j.tim.2014.02.011
130. Campioni F, Vilela FP, Cao G, Kastanis G, Dos Prazeres Rodrigues D, Costa RG, et al. Whole genome sequencing analyses revealed that Salmonella enterica serovar Dublin strains from Brazil belonged to two predominant clades. Sci Rep. (2022) 12:10555. doi: 10.1038/s41598-022-14492-4
Keywords: latent carrier, risk factors, prevention, calf health, zoonosis
Citation: Velasquez-Munoz A, Castro-Vargas R, Cullens-Nobis FM, Mani R and Abuelo A (2024) Review: Salmonella Dublin in dairy cattle. Front. Vet. Sci. 10:1331767. doi: 10.3389/fvets.2023.1331767
Received: 01 November 2023; Accepted: 19 December 2023;
Published: 09 January 2024.
Edited by:
David C. B. Taras, Boehringer Ingelheim, GermanyReviewed by:
Soraya Chaturongakul, Mahidol University, ThailandAruna Pal, West Bengal University of Animal and Fishery Sciences, India
Ulrich Methner, Friedrich Loeffler Institut, Germany
Copyright © 2024 Velasquez-Munoz, Castro-Vargas, Cullens-Nobis, Mani and Abuelo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angel Abuelo, YWJ1ZWxvQG1zdS5lZHU=
 Ana Velasquez-Munoz
Ana Velasquez-Munoz Rafael Castro-Vargas
Rafael Castro-Vargas Faith M. Cullens-Nobis3
Faith M. Cullens-Nobis3 Angel Abuelo
Angel Abuelo