- 1Key Laboratory of Animal Medicine at Southwest Minzu University of Sichuan Province, Chengdu, China
- 2Qinghai-Tibetan Plateau Animal Genetic Resource Reservation and Utilization, Key Laboratory of Sichuan Province, Chengdu, China
Background: Infectious ecthyma is a severe and highly contagious disease caused by ORF virus (ORFV). The virus is responsible for significant economic losses in the goat industry and threatens humans. Regarding complement component 1, q subcomponent binding protein (C1QBP), we previously showed that C1QBP can interact with ORFV129. However, the role of C1QBP in regulating the apoptosis of goat fetal turbinate cells is unknown.
Methods: After pcDNA3.1-C1QBP and siRNA were transfected into goat fetal turbinate cells (GFTCs), the expression of C1QBP was detected by western blot and cell cycle and apoptosis using flow cytometry. The expression of cycle-related genes cyclin-dependent protein kinase 2 (CDK2) and cyclin-dependent kinase inhibitors 1A (p21) and apoptosis-related genes cysteinyl aspartate specific proteinase 3 (Caspase 3), cysteinyl aspartate specific proteinase 7 (Caspase 7), p53, poly (ADP-ribose) polymerase 1 (PARP1), B-Cell lymphoma-2 Like-11 (BCL2L11), B-cell lymphoma 2 associated X protein (Bax), and B-cell lymphoma 2 (Bcl-2) were tested by real-time quantitative PCR (RT-qPCR). The effect of MTT on the proliferation of GFTCs was detected. The localization of C1QBP in GFTCs was detected by inverted fluorescence microscope. Finally, transcriptome sequencing was performed and validated by siRNA treatment knockdown of C1QBP, and screening two genes tripartite motif containing 5 (TRIM5) and tumor necrosis factor superfamilymember10 (TNFSF10) with significant changes in expression levels and relevance to cell apoptosis, and to verify their roles in C1QBP-induced cell apoptosis.
Results: Knockdown of C1QBP significantly increased cell viability; cells remained in the G0/G1 phase and reduced apoptosis. Knockdown of C1QBP reduced the mRNA expression of CDK2, p21, Caspase3, Caspase7, Bax, PARP1, BCL2L11, and p53, up regulated the mRNA expression of the Bcl-2. Except for Bcl-2, The opposite effect was observed when C1QBP was overexpressed in GFTCs and the mRNA levels of Bcl-2 had no significant effect. Immunostaining revealed intracellular localization of C1QBP, primarily in the cytoplasm of the GFTCs. Furthermore, gene expression profiling analysis in C1QBP depleted cells compared to the control revealed that a total of 236 differential expression genes (DEGs), including 119 up regulated DEGs and 117 down regulated DEGs, and the expression of TRIM5 and TNFSF10 genes were significantly upregulated. Pathway analysis were predicted to be enriched in Herpes simplex virus 1 infection, Pathways in cancer, PI3K-Akt signaling pathway, Apoptosis, Apoptosis-multiple species and p53 signaling pathway. C1QBP reduced the expression of TRIM5 and TNFSF10 genes. Knockdown of TRIM5 promoted apoptosis in GFTCs, but silencing of TNFSF10 had no change in apoptosis rate. In particular, the apoptosis rate was significantly increased in the TRIM5-siRNA2 or TNFSF10-siRNA2 and C1QBP-siRNA2 group compared to the only C1QBP-siRNA2 group.
Conclusion: These findings provide a deeper understanding of the role of C1QBP in apoptosis and could pave the way for further study investigating the role and mechanism of C1QBP protein in mediating the regulation of cell apoptosis by ORFV.
Highlights
• Knockdown of C1QBP by siRNA treatment increased cell viability, cells remained in G0/G1 phase, and reduced cell apoptosis. Whereas, the opposite effect was observed when C1QBP was overexpressed.
• The intracellular localization of C1QBP occurred mainly in the cytoplasm of GFTCs.
• Gene expression profiling and pathway analysis in C1QBP depleted cells compared to the control revealed that a total of 236 differential expression genes (DEGs), including 119 upregulated DEGs and 117 downregulated DEGs, and were predicted to enrich in Herpes simplex virus 1 infection, Pathways in cancer, PI3K-Akt signaling pathway, Apoptosis, Apoptosis-multiple species and p53 signaling pathway.
• The role of C1QBP in cell apoptosis could be regulated by the expression of TRIM5 or TNFSF10.
1 Introduction
Infectious ecthyma (Orf) is a zoonotic infectious disease caused by goat's aphthous virus (ORFV) infection. This virus mainly infects sheep and goats, especially lambs. Humans can also be infected through contact with virus-laden blood and tissues (1). The incidence rate of ORFV infection in newborn and young lambs can reach up to 100%, with a mortality rate of 93%. Although the mortality rate is relatively lower in adult goat infected with ORFV, it can still lead to weight loss, slow growth, and secondary infections. The vaccines currently available on the market cannot provide full immune protection, and there is no specific treatment (2). In recent years, with the increase in goat farming and cross-border circulation in China, ORFV causes considerable economic losses in the agriculture industry and threatens human health. ORFV is a double-stranded DNA virus in the genus Parapoxvirus (3). The ORFV genome comprises a central genomic region (ORFVs 001–008 and ORFVs 112–134) and a terminal genomic region (ORFVs 009–111). Ankyrin-repeat (ANK) protein, as a virulence factor encoded by ORFV, plays a significant role in mediating viral infection. Firstly, Ankyrin-repeat (ANK) protein can activate the ubiquitin-dependent programmed degradation of cellular proteins by interacting with cellular ubiquitin ligases, thereby regulating the cellular response to viral infection (4). Secondly, Ankyrin-repeat (ANK) protein can regulate the activity of the NF-κB signaling pathway by interacting with NF-κB pathway proteins, and thereby trigger viral immune evasion (5). Finally, the ankyrin-repeat (ANK) protein of poxviruses can regulate the survival and replication of the virus within cells by binding to host proteins (6). The terminal regions of the ORFV genomes are responsible for encoding five ankyrin-repeat (ANK) proteins, namely ORFV008, ORFV123, ORFV126, ORFV128, and ORFV129 (7). However, there is currently limited research on the functions of these ORFV ankyrin proteins. ORFV129 has six ANKs at its N-terminal end and one F-box-like domain at the C-terminal (8). Our previous study showed that ORFV129 can interact with proteins such as complement component 1, q subcomponent binding protein (C1QBP), and overexpression of C1QBP played a cellular antiviral role during ORFV infection, while C1QBP knockdown can facilitate the replication of the ORFV viral genome and the production of progeny viruses (9). This suggests that goat C1QBP has a regulatory role in the replication of ORFV. However, the specific mechanism by which C1QBP regulates ORFV replication remains unclear.
Complement 1q binding protein (C1QBP, also known as p32 or globular C1q binding protein or HABP1) was initially thought to be a splicing factor of intracellular pre-mRNA, interacting with the intracellular splicing factor ASF/SF2 (splicing factor-2) and regulating the function of SF2 (10, 11). Subsequently, C1QBP was found to be a counterpart of the yeast p30 gene, and it was shown that C1QBP and p30 proteins were localized in the mitochondrial matrix and were necessary for the maintenance of mitochondrial oxidative phosphorylation (12). In 2004, C1QBP was found to interact with platelets to regulate and repair vascular injury and inflammation (13, 14). C1QBP is one of the receptors of C1q, which combines with the spherical head of C1q to form the complement system in the body (15). The activation of the complement system regulates the defense function of the host immune system and produces a series of biological responses (16). Over time, C1QBP was found to be involved in the regulation of cellular immunity, metabolism, proliferation, apoptosis, and other biological processes (17–19). For example, C1QBP regulates the immune response by binding to the globular head domain of C1q. C1QBP improves vascular permeability by up regulating the expression of bradykinin receptor 1 (20). The purified porcine C1QBP protein increased mRNA levels of LPS/PRRSV (Porcine Reproductive and Respiratory Syndrome Virus)-induced inflammatory factors such as IL-1β, IL-6, IL-8, TNF-α, and RANTES. Also, it enhanced PRRSV-induced NF-κB activation and p65 phosphorylation, inducing inflammatory responses (21). C1QBP regulates the secretion of cytokines IL-6, IL-1β, and IFN-γ via interaction with ORFV129 (9). C1QBP expression of Larimichthys crocea was increased upon infection with Pseudomonas polyglossia, which caused the immune response to visceral leukoplakia (22). Meanwhile, C1QBP also plays a significant role in regulating cell apoptosis. Such as, C1QBP was reported to induce mitochondrial dysfunction, growth retardation, and apoptosis when stably overexpressed in normal fibroblasts (23). The high levels of expression of RPAIN in preeclampsia may affect the biological function such as proliferation, apoptosis, and invasion of placental trophoblast cells through C1QBP, which participates in the occurrence and development of preeclampsia (24). C1QBP promoted ROS production, apoptosis, and the expression of Bax and active Caspase 3 (25).
ORFV infection can induce cell apoptosis (26), and the ORFV129 protein is an ankyrin-repeat (ANK) protein that plays a significant role during viral infection. Based on our previous findings, there is an interaction between ORFV129 and C1QBP. Given that C1QBP is a gene related to cell apoptosis, we speculate that C1QBP may be involved in the phenomenon of cell apoptosis caused by ORFV infection. However, the specific mechanism of C1QBP in regulating cell cycle and apoptosis is unclear. This has also seriously affected the discussion on the role of C1QBP in regulating cell apoptosis caused by ORFV infection. The goat host protein C1QBP as the material in the present study. We examined the role of C1QBP in the cell cycle, apoptosis, and proliferation by RNA silencing and over expression. In addition, RNA-seq was performed to identify differential transcripts affected by C1QBP loss of function and to identify potential genes and signaling pathways that could mediate C1QBP-associated apoptosis.
2 Materials and methods
2.1 Animals and cell culture
A 2-day-old male Jianzhou big-eared goat was purchased from Dagda Animal Husbandry Co., Ltd. (Jianyang City, Sichuan Province, China). Animal care and experiments were performed according to the regulations of the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in June 2004), and all experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee, Southwest Minzu University (Chengdu, Sichuan, China).
The primary culture of goat fetal turbinate cells (GFTCs) was carried out according to Dan et al. (9) and was preserved in our laboratory. Briefly, a 2-day-old goat was fed freely in the house with milk and performed well during experiments. The turbinate bone tissues were collected after slaughter and immediately frozen in liquid nitrogen until it was transported back to the laboratory. In a laminar flow cabinet, the turbinate bone tissues were washed several times with phosphate-buffered saline. The smaller pieces of tissue blocks were maintained in Dulbecco's minimal essential medium (Hyclone, Beijing, China) containing 15% fetal bovine serum and supplemented with 10 kU/L penicillin and streptomycin (Boster, Wuhan, China) in 6-well plates for cell culture in a 5% CO2 incubator (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C, and the medium changed after 48 h. The primary GFTCs reached ~80% confluence and were passaged at a 1:2 ratio using 0.25% trypsin.
2.2 Plasmid constructs and chemical synthesis of siRNA
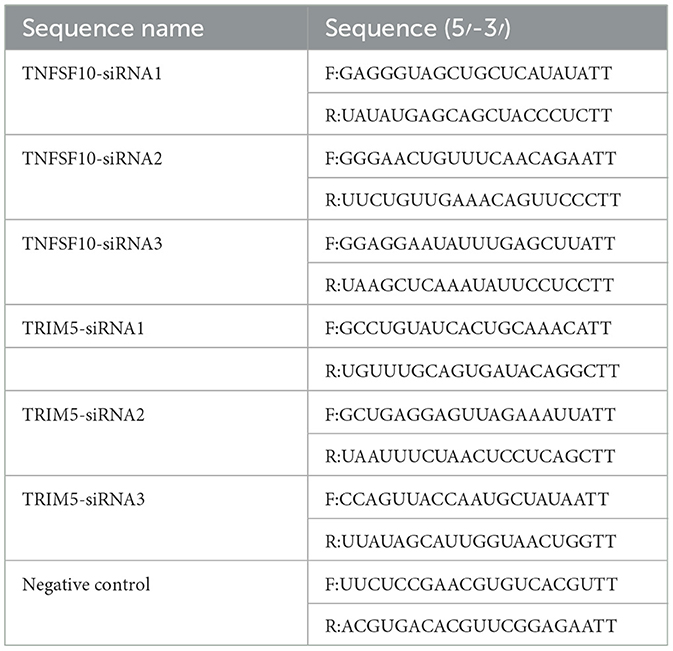
These expression vectors pcDNA3.1-C1QBP and C1QBP-siRNA2, which have been shown to silence C1QBP expression in GFTCs, were constructed and conserved in our laboratory (9, 27). The siRNAs were designed and synthesized using the goat gene TRIM5 (JX649920.1) and predicted gene TNFSF10 (XM_005675308.3) sequence as a target by Shanghai GenePharma Co., Ltd. Three specific siRNA pairs were designed as follows Table 1.
2.3 Goat fetal turbinate cells transfection
GFTCs from passage six were used for subsequent experiments, and the experimental treatment was performed when the GFTCs reached 70–80% confluence. The transient transfection cells were treated with Lipofectamine3000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA). Cells were collected after 48 h of transfection. Total RNA was extracted using trizol reagent (Takara, Dalian, China), and 1 μg of total RNA was reverse transcribed using a Revert Aid First Strand cDNA Synthesis Kit (Thermo, USA) according to the manufacturer's protocol.
2.4 Real-time quantitative PCR (RT-qPCR)
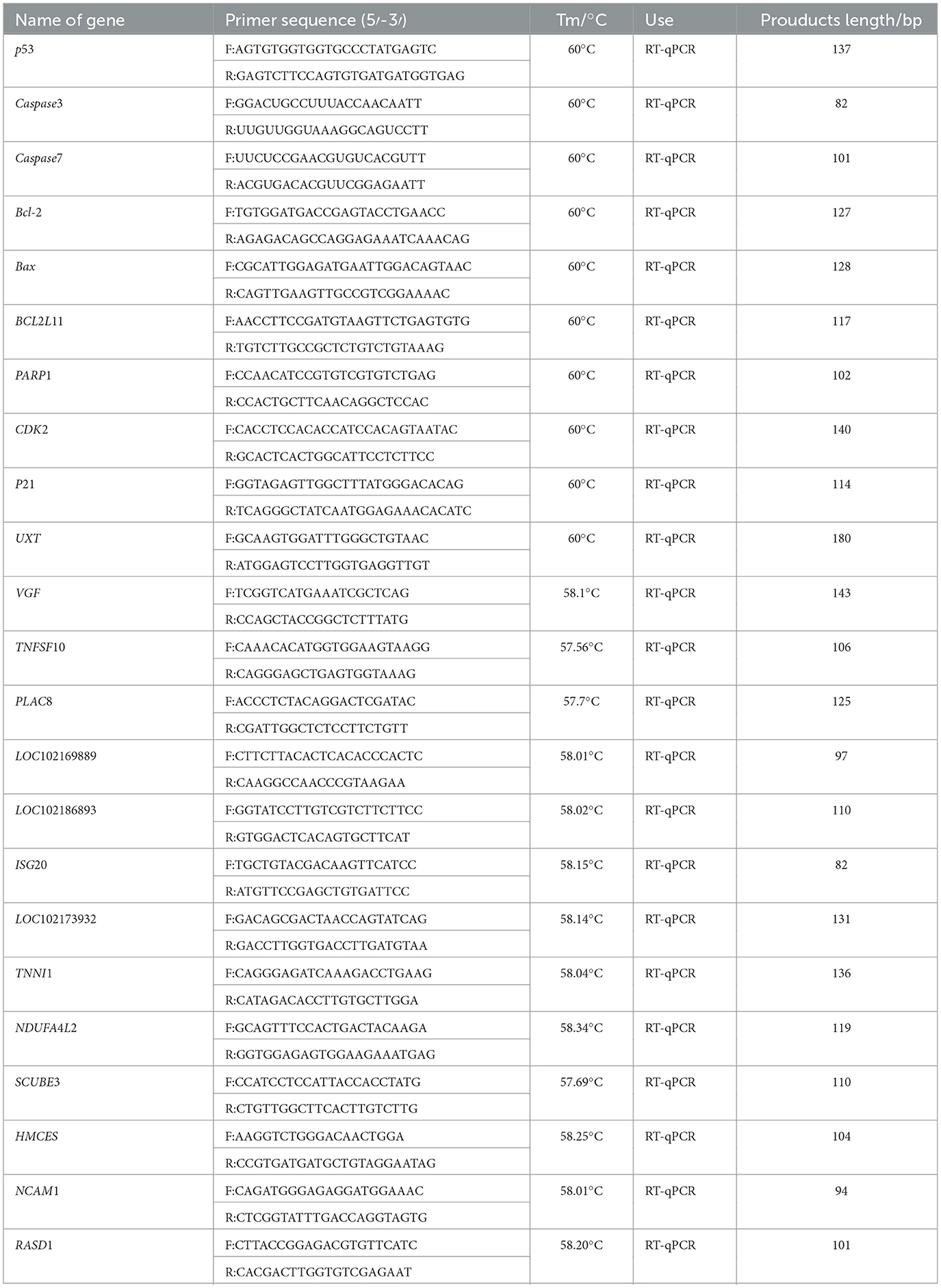
RT-qPCR assays for related gene expression were performed using the SYBR Green kit. The ubiquitously expressed transcript gene was used as an internal reference gene, and the relative expression was calculated using the 2−ΔΔCt method. The total volume of RT-qPCR was 20 μL: 10 μL SYBR® Premix Ex Taq TM (Takara, Dalian, China), 1 μL template cDNA, 1 μL upstream primer, 1 μL downstream primer, and 7 μL ddH2O. The PCR conditions included an initial step at 95°C for 3 min and 95°C for 10 s, followed by 40 cycles of amplification and quantification (60°C for 30 s, 72°C for 30 s). The primer information for RT-qPCR information is listed in Table 2.
2.5 Western blot
SDS-PAGE was performed using a 10-well gel, and sample volumes were 20 μg per well. The electrophoresis conditions were 80 V, 50 min; 120 V, 30 min. After SDS-PAGE electrophoresis, the total protein of the cells was transferred to polyvinylidene fluoride (PVDF) membranes using a semi-dry transfer printer and the transfer printer conditions were 12 V, 20 min, using 5%BSA blocked for 2 h. Mouse monoclonal Flag antibody (Boster, Wuhan, China, M30971-2) and rabbit beta Actin antibody (zenbio, Chengdu, China, T200068-8F10) were then added and incubated overnight at 4°C. Goat anti-mouse IgG (Boster, Wuhan, China, BA1050) and goat anti-rabbit IgG (Boster, Wuhan, China) were labeled with HRP and incubated for 2 h on a room-temperature shaker, followed by imaging using ECL reagent (4Ablotech, Suzhou, China).
2.6 Immunofluorescence localization analysis
GFTCs were transfected with pcDNA3.1-C1QBP and empty vectors pcDNA3.1(+) as control. After a 48 h incubation, the cells were fixed with formaldehyde for 15 min, blocked with 5% BSA for 1 h at 37°C. Mouse monoclonal Flag antibody (Boster, Wuhan, China, M30971-2) were then added and incubated overnight at 4°C. Goat anti-mouse IgG (Boster, Wuhan, China, BA1101) were labeled with FITC and incubated for 2 h in the dark on a room-temperature shaker, and with DAPI (Boster, Wuhan, China) in the dark for 15 min. Then, cells were observed under an inverted fluorescence microscope (Olympus, Japan).
2.7 MTT for goat fetal turbinate cells proliferation analysis
The MTT assay was used to evaluate GFTC proliferation. The sixth-generation GFTCs were inoculated in 96-well plates (LABSELECT, Anhui, China) and transfected after 24, 48, 72, and 96 h of treatment with 10 μL of MTT solution (Boster, Wuhan, China) for 4 h at 37°C and the addition of 100 μL of dimethyl sulfoxide (Boster, Wuhan, China) per well. The plates were left for 10 min and incubated for long periods. Finally, the OD of each well was measured using a microplate reader at a wavelength of 490 nm. Five biological replicates were performed, and the experiment was repeated three times.
2.8 Goat fetal turbinate cells cycle and cycle-related gene analysis
The sixth-generation GFTCs inoculated into twelve-well plates (LABSELECT, Anhui, China) were transfected using the same method as for the MTT assay. After 48 h of transfection, samples were collected according to the steps of the cell cycle detection kit (Multi Sciences, Hangzhou, China). Cell cycle analysis was then performed using flow cytometry. In addition, RT-qPCR was used to analyze the effects of the cycle-related genes CDK2 and p21. The primer information for RT-qPCR is listed in Table 2. Three biological replicates were performed, and the experiment was repeated three times.
2.9 Goat fetal turbinate cells apoptosis and apoptosis-related gene analysis
Sixth-generation GFTCs inoculated in 12-well plates were transfected using the same method as the cell cycle assay. After 48 h of transfection, samples were collected according to the instructions of the PI Apoptosis Kit (MultiSciences, Hangzhou, China). The effects of apoptosis-related genes p53, Caspase7, Caspase3, Bcl-2, Bax, BCL2L11, and PARP1 were analyzed by RT-qPCR. Information about RT-qPCR primers information were listed in Table 2. Three biological replicates were performed, and the experiment was repeated three times.
2.10 RNA sequencing (RNA-seq)
The total RNA of each sample (Negative Control and C1QBP knockdown) was extracted with trizol reagent. RNA purity, quantification, and integrity were assessed, and RNA quantification, library preparation, clustering, and sequencing were performed at Oebiotech. The libraries were sequenced on a Lumina Novaseq 6000 platform, generating 150 bp paired-end reads. Approximate raw data was generated for each sample. Raw reads in Fast Quality (Fastq) format were first processed using Fastp, and the low-quality reads were removed to obtain clean reads. Approximately clean readings for each sample were then retained for subsequent analysis. The clean reads were mapped to the reference genome using HISAT2. The FPKM of each gene was calculated, and the read counts of each gene were obtained using HT Seq counting. PCA analyses were performed using R (version 3.2.0) to assess the biological duplication of samples. Differential expression analysis was performed using DESeq2.A q value < 0.05 and fold-change >1.5 or < 0.5 were set as the threshold for significant DEGs. The radar map of the top 30 genes was constructed to show the expression of upregulated or downregulated DEGs using R packetage Gradar. Based on the hypergeometric distribution, GO, KEGG Pathway, Reactome, and Wiki pathways enrichment analyses of DEGs were performed to determine the significantly enriched term using R (version 3.2.0). Then, differentially expressed genes based on transcriptome data were selected to check the accuracy of the experimental transcriptome data by RT-qPCR. we used correlation screens between DEGs to analyze genes that may play a key role in the regulation of apoptosis. Finally, the role of apoptosis-related genes TRIM5 and TNFSF10 in C1QBP promoting apoptosis were studied using flow cytometry.
2.11 Statistical analysis
The expression level of C1QBP and cycle-related or apoptosis-related gene data from RT-qPCR in goat tissues were processed with the 2−ΔΔCt method. The ubiquitously expressed transcript was used as an internal control primer. One-way analysis of variance was performed using SPSS 26.0 software, followed by Duncan's multiple comparison test; p < 0.05 and p < 0.01 represent significant and highly significant differences, respectively. Spearman correlation analysis is used to analyze the correlation between indicators. All experiments were repeated three times.
3 Results
3.1 Knockdown of C1QBP inhibits apoptosis in goat turbinate bone cells
MTT assays were used to evaluate the proliferation and the results showed a significant increase in cell viability (OD 490 nm) in the knockdown C1QBP group compared to the negative control group (p < 0.05), and the promotion increased with time (Figure 1A). Flow cytometry was used to demonstrate the function of C1QBP in goat turbinate bone cell cycle. Flow cytometry revealed that 76.9 ± 0.021% of GFTCs represented progressed through G0/G1 when C1QBP knockdown, which was much higher than when the cells were transfected with negative control group. The G2/M+S phase of cells treated with C1QBP siRNA was lower than that of the control group (Figure 1B). Thus, a G1 phase arrest was observed in cells when C1QBP knockdown. In our study, the mRNA expression levels of cell cycle-related genes were examined, and the results showed that knockdown of C1QBP significantly reduced the mRNA levels of CDK2 and p21 (p < 0.01; Figure 1C).
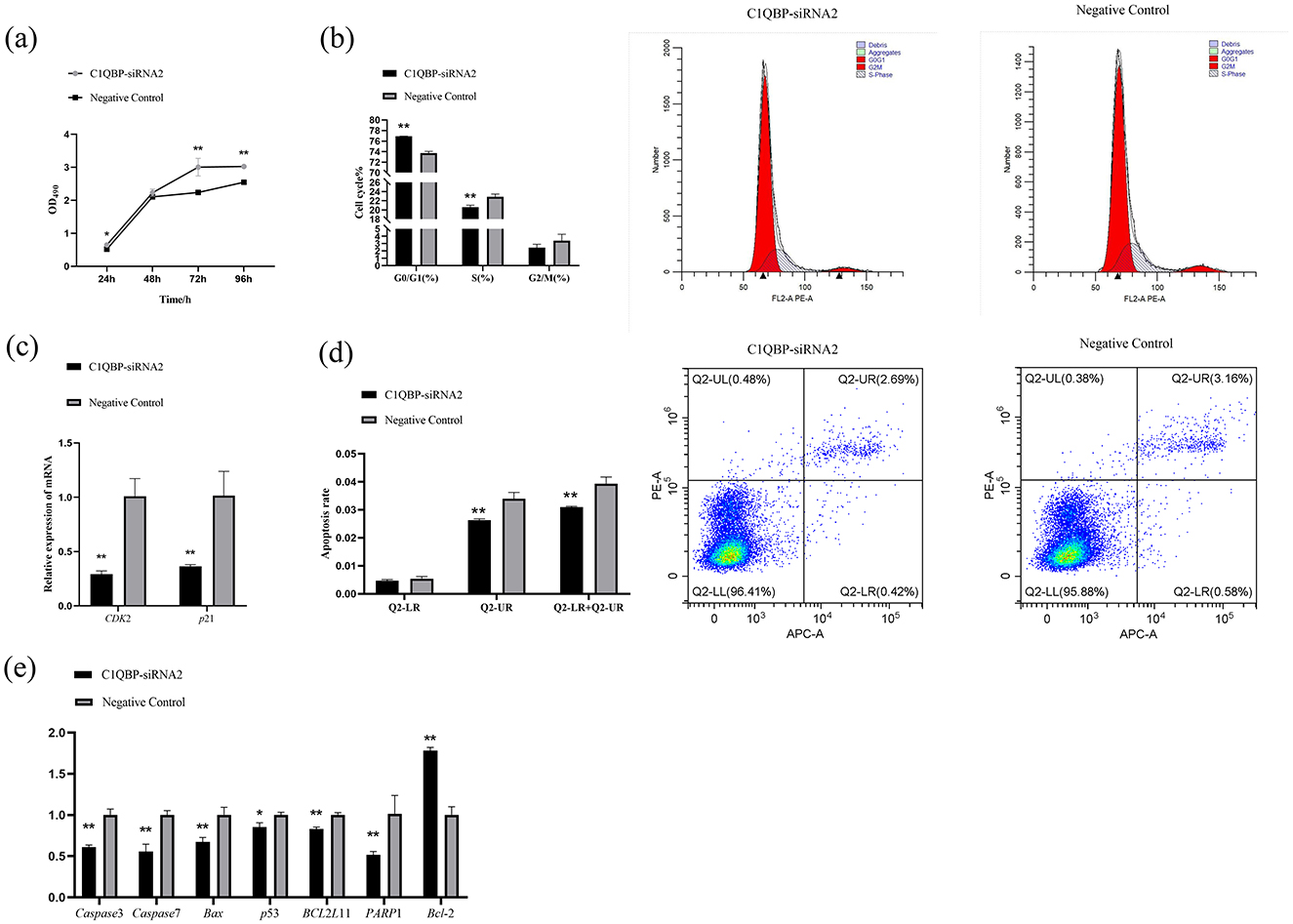
Figure 1. Effect of C1QBP reduction on the proliferation, cycle, and apoptosis of GFTCs. (A) Knockdown of C1QBP promotes the cell proliferation of GFTCs; OD 490 nm characterizes cell viability. (B) Effect of C1QBP deficiency on the cycle in GFTCs. (C) Effect of C1QBP deficiency on the expression of cycle-related genes in GFTCs. (D) Effect of C1QBP deficiency on the apoptosis in GFTCs. (E) Effect of C1QBP deficiency on expression of apoptosis-related genes in GFTCs. *represents 0.01 < p < 0.05, **represents p < 0.01.
We also examined the responses of apoptosis and apoptosis-related genes to the transfection with C1QBP-siRNA2, as compared to the negative control group. As a result, the knockdown of C1QBP inhibited apoptosis in goat turbinate bone cells (p < 0.01; Figure 1D). The mRNA expression levels of the apoptosis-related genes were examined. The results showed that disruption of the expression of the C1QBP gene significantly downregulated the mRNA expression of the genes Caspase3, Caspase7, Bax, PARP1, and BCL2L11 (p < 0.01), down regulated the mRNA expression of the p53 gene (p < 0.05), and significantly up regulated the mRNA expression of the Bcl-2 gene (p < 0.01; Figure 1E).
3.2 Over expression of C1QBP promotes apoptosis in goat turbinate bone cells
Based on the results that silencing of C1QBP increases cell proliferation, to verify whether over expression of C1QBP could inhibit cell viability. Using β-actin as an internal reference gene, western blotting results showed that the tag protein Flag (30.5 ku) expression was detected in the cells transfected with the recombinant plasmid pcDNA3.1-C1QBP. In contrast, the cells transfected with empty pcDNA3.1 (+) were detected (Figure 2A). Immunostaining revealed intracellular localization of C1QBP primarily in the cytoplasm of the GFTCs with red light (Figure 2B).
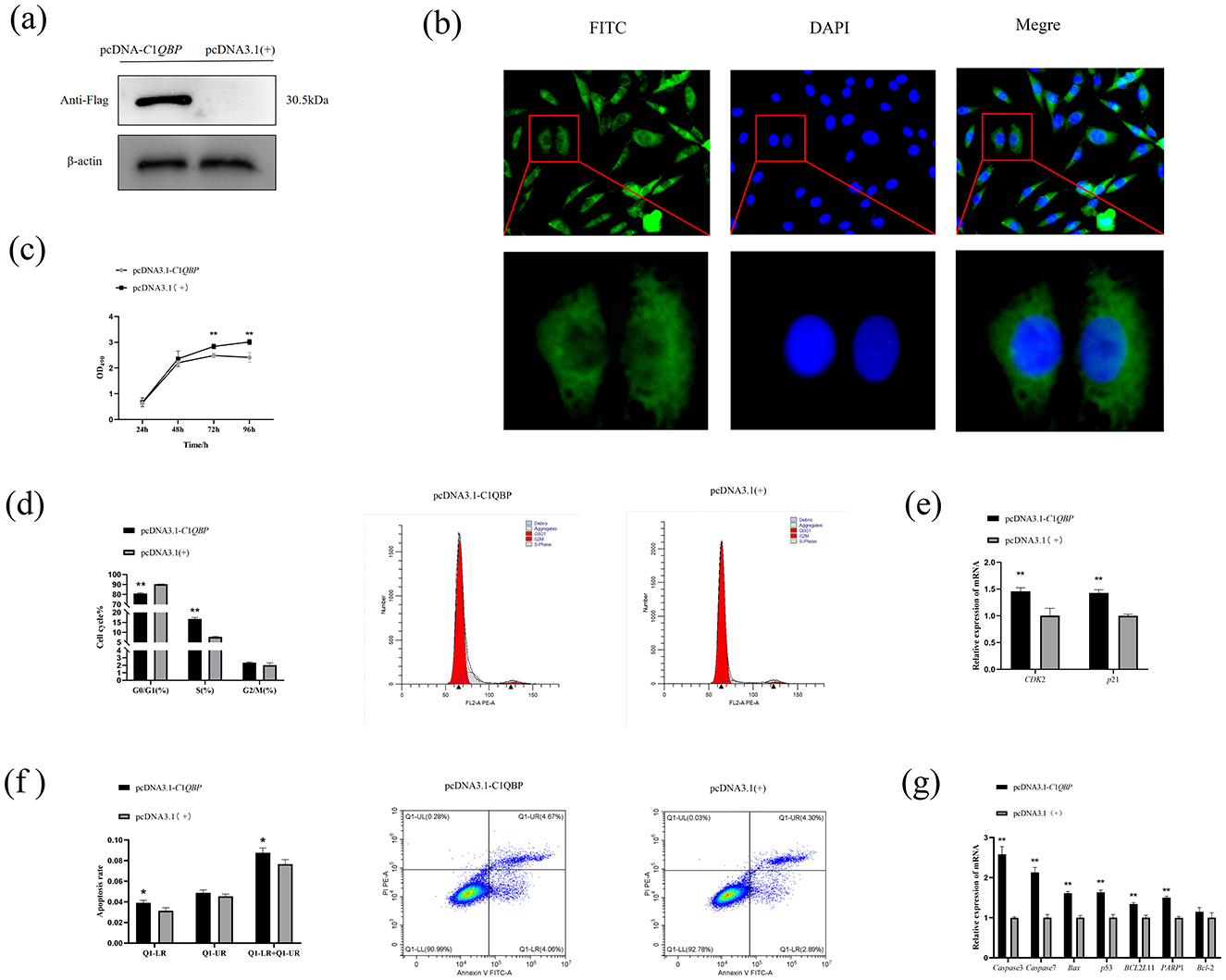
Figure 2. Effect of over expression of C1QBP on the proliferation, cycle, and apoptosis of GFTCs and Protein expression and subcellular localization of C1QBP. (A) Expression of C1QBP in GFTCs. (B) Localization of C1QBP in GFTCs. (C) Over expression of C1QBP inhibits cell proliferation in GFTCs. (D) Effect of C1QBP over expression on the cycle in GFTCs. (E) Effect of C1QBP over expression on the expression of cycle-related genes in GFTCs. (F) Effect of C1QBP over expression on the apoptosis in GFTCs. (G) Effect of C1QBP over expression on expression of apoptosis-related genes in GFTCs. *represents 0.01 < p < 0.05, **represents p < 0.01.
MTT assay was also performed with cells over expressing pcDNA3.1-C1QBP or pcDNA3.1(+). The results showed that over expression of C1QBP resulted in a significant decrease in cell growth compared to the negative control (Figure 2C). Flow cytometry showed that goat turbinate bone cells were in the G2/M+S phase when C1QBP was over expressed, significantly higher than the control group. The G0/G1phase of cells treated with C1QBP overexpression was lower than that of the pcDNA3.1(+) group (Figure 2D). The mRNA expression levels of CDK2 and p21 genes were increased after over expression of C1QBP (p < 0.01; Figure 2E). C1QBP-overexpressing cells were more susceptible to apoptosis than those expressing pcDNA3.1(+) (Figure 2F). In addition, C1QBP over expression could significantly up regulate the mRNA expression of Caspase3, Caspase7, p53, PARP1, BCL2L11, and Bax gene (p < 0.01), but had no significant effect on Bcl-2 (Figure 2G).
3.3 Silencing of C1QBP changes cellular gene expression profile in goat turbinate bone cells
To investigate the mechanism of action of C1QBP in increasing apoptosis, we sequenced and analyzed the transcriptome of total RNA from C1QBP knockdown and negative controls in goat turbinate bone cells. Principal component analysis showed a significant separation between the interference C1QBP and negative control groups (Supplementary Figure S1). A total of 236 differentially expressed genes (DEGs) were screened out on the volcano map (Figure 3A; Supplementary Table S1) (p < 0.05, multiplex change >1.5), of which 119 were up regulated, and 117 were down regulated. In the GO database, DEGs were divided into three directories (Supplementary Figure S3; Supplementary Table S2): molecular functions (MF, 237), cellular components (CC, 175), and biological processes (BP, 666). Go enrichment analysis showed that DEGs were enriched by the response to the virus, activation of the innate immune response, negative regulation of the viral genome, and defense response to the virus.
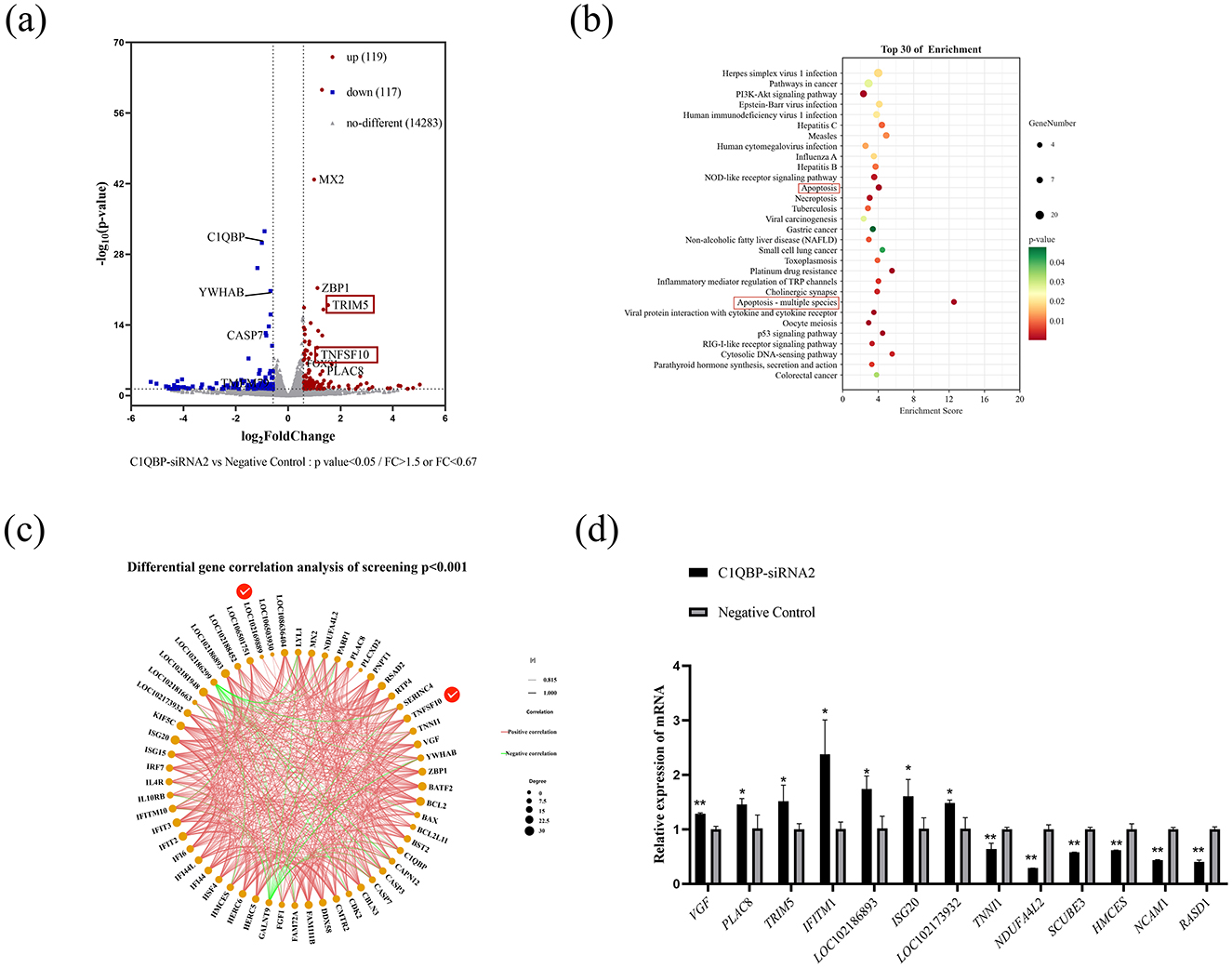
Figure 3. DEGs analysis after C1QBP knockdown. (A) Differential representation of volcano diagrams. (B) The top 30 enriched pathways of DEGs. (C) Visual network diagram for correlation analysis of differentially expressed gene expression (D). RT-qPCR verification of some differential genes. *represents 0.01 < p < 0.05, **represents p < 0.01.
The figure shows the first 30 significantly enriched KEGG signaling pathways, such as pathways in Herpes simplex virus 1 infection, Pathways in cancer, PI3K-Akt signaling pathway, Apoptosis, Apoptosis-multiple species and p53 signaling pathway (Figure 3B; Supplementary Table S3). Furthermore, we used correlation screens between DEGs to analyze genes that may play critical roles in regulating apoptosis (Figure 3C; Supplementary Table S4). The results showed that the essential genes may be YWHAB, TNFSF10, LOC102169889 (TRIM5), VGF, and PARP1 etc. In addition, we randomly selected eight up regulated genes VGF, TNFSF10, PLAC8, TRIM5, LOC106501751 (IFITM1), LOC102186893, ISG20, LOC102173932 (p < 0.05, FC > 2, FPKM > 3, FPKM < 50) and seven down regulated genes TNNI1, RPS25, NDUFA4L2, SCUBE3, HMCES, NCAM1, RASD1 (p < 0.05, FC < 0.67, FPKM > 2) of RNA-seq results for reliability analysis. The results showed that RT-qPCR trends generally agreed well with the RNA-seq expression profiles of randomly selected genes (Figure 3D).
3.4 Verification of signaling pathways
RNA-seq analysis showed that the expression of TRIM5 and TNFSF10 was enriched in the analysis of DEGs. To examine whether C1QBP regulate the expression of TNFSF10 and TRIM5, the pcDNA3.1-C1QBP or pcDNA3.1(+) was transfected in GFTCs. After 48 h, the cells were collected was analyzed by RT-qPCR. Compared with pcDNA3.1(+) cells, TNFSF10 and TRIM5 mRNA was reduced in pcDNA3.1-C1QBP cells (Figure 4A). And the opposite result was obtained after interfering with the expression of C1QBP (Figure 4B). This finding suggests that the TRIM5 or TNFSF10 gene may mediate the promoting effect of C1QBP on apoptosis. To investigate this, we knocked down the expression of C1QBP followed by treatment with TRIM5 or TNFSF10 to observe whether knockdown of TRIM5 or TNFSF10 could promote apoptosis through the low expression of TRIM5 or TNFSF10. Specific siRNAs (TRIM5-siRNA1, TRIM5-siRNA2, TRIM5-siRNA3, and negative control) knocked down the expression of TRIM5, with TRIM5-siRNA2 reduced the relative expression of TRIM5 mRNA by ~70.49% (p < 0.01; Figure 4C), which is significantly more than TRIM5-siRNA1 (no interference efficiency) and TRIM5-siRNA3 (There is no interference efficiency). Thus, TRIM5-siRNA2 was used to silence TRIM5 expression in GFTCs. We examined the cell apoptosis responses to transfection with TRIM5-siRNA2 or negative control, TRIM5 knockdown cells were more prone to apoptosis than those expressing negative control. Meanwhile, the results showed that the apoptosis rate was significantly increased in the TRIM5-siRNA2 and C1QBP-siRNA2 group compared to the only C1QBP-siRNA2 group (Figure 4D). These findings suggest that the expression of TRIM5 regulates C1QBP and thus influences the apoptosis of GFTCs. Like TRIM5, TNFSF10-siRNA2 reduced the relative expression of TNFSF10 mRNA by ~74.348% (p < 0.01; Figure 5A), which was used to silence TNFSF10 expression in GFTCs. Meanwhile, we demonstrated no change in apoptosis rate in the TNFSF10-siRNA2 group compared to the negative control. Interestingly, the apoptosis rate was significantly increased in the TNFSF10-siRNA2 plus C1QBP-siRNA2 group compared to the only C1QBP-siRNA2 group (Figure 5B). These results suggest that the expression of TNFSF10 does not affect the apoptosis rate of GFTCs, but the expression of TNFSF10 may can regulate C1QBP, thereby affecting the apoptosis of GFTCs.
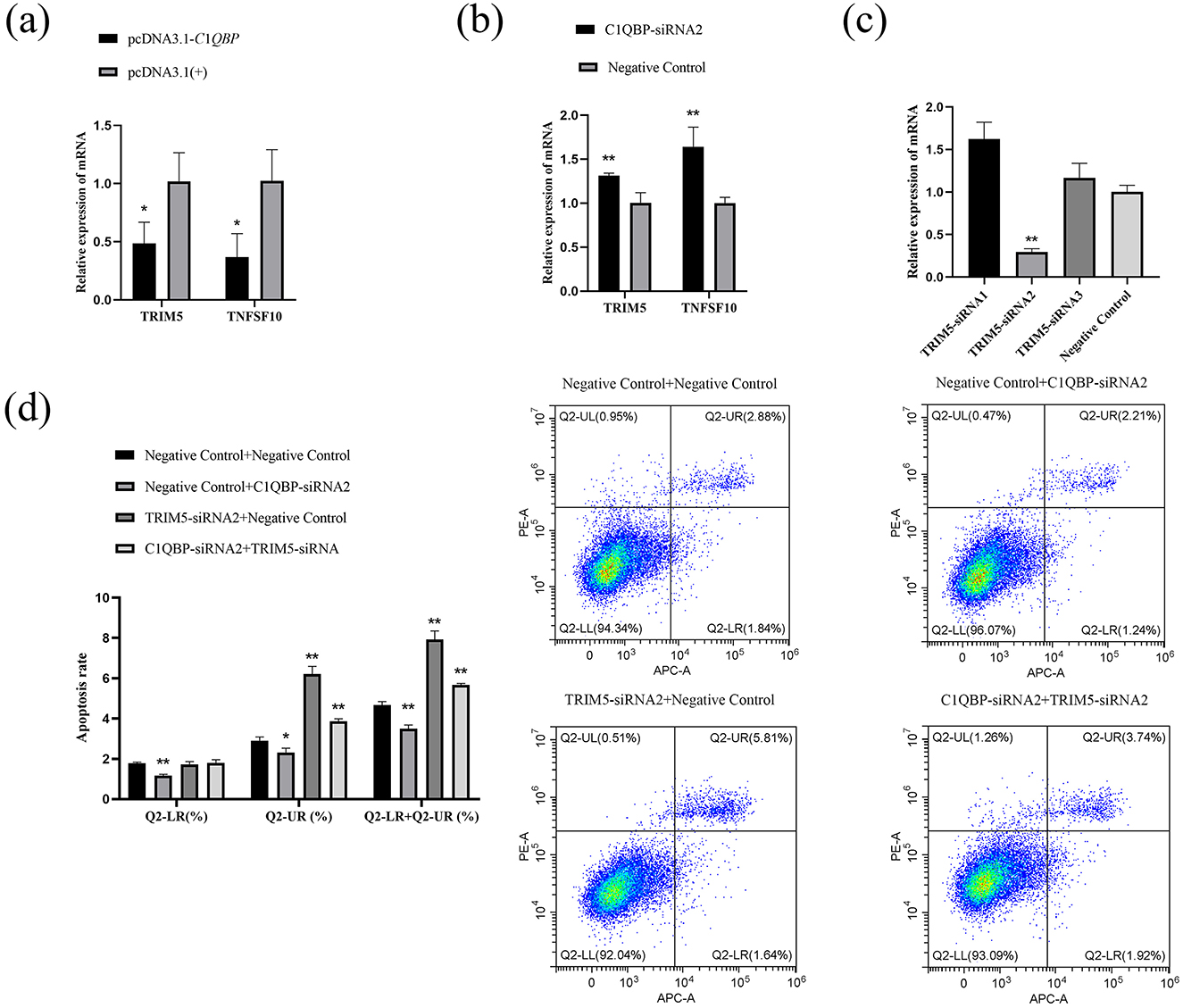
Figure 4. C1QBP deficiency inhibits apoptosis by regulating TRIM5 expression in GFTCs. (A) Effect of over expression of C1QBP on TNFSF10 and TRIM5. (B) Effect of knockdown of C1QBP on TNFSF10 and TRIM5. (C) Screening of interfering fragments for TRIM5. (D) Effect of TRIM5 and C1QBP on apoptosis of GFTCs. *represents 0.01 < p < 0.05, **represents p < 0.01.
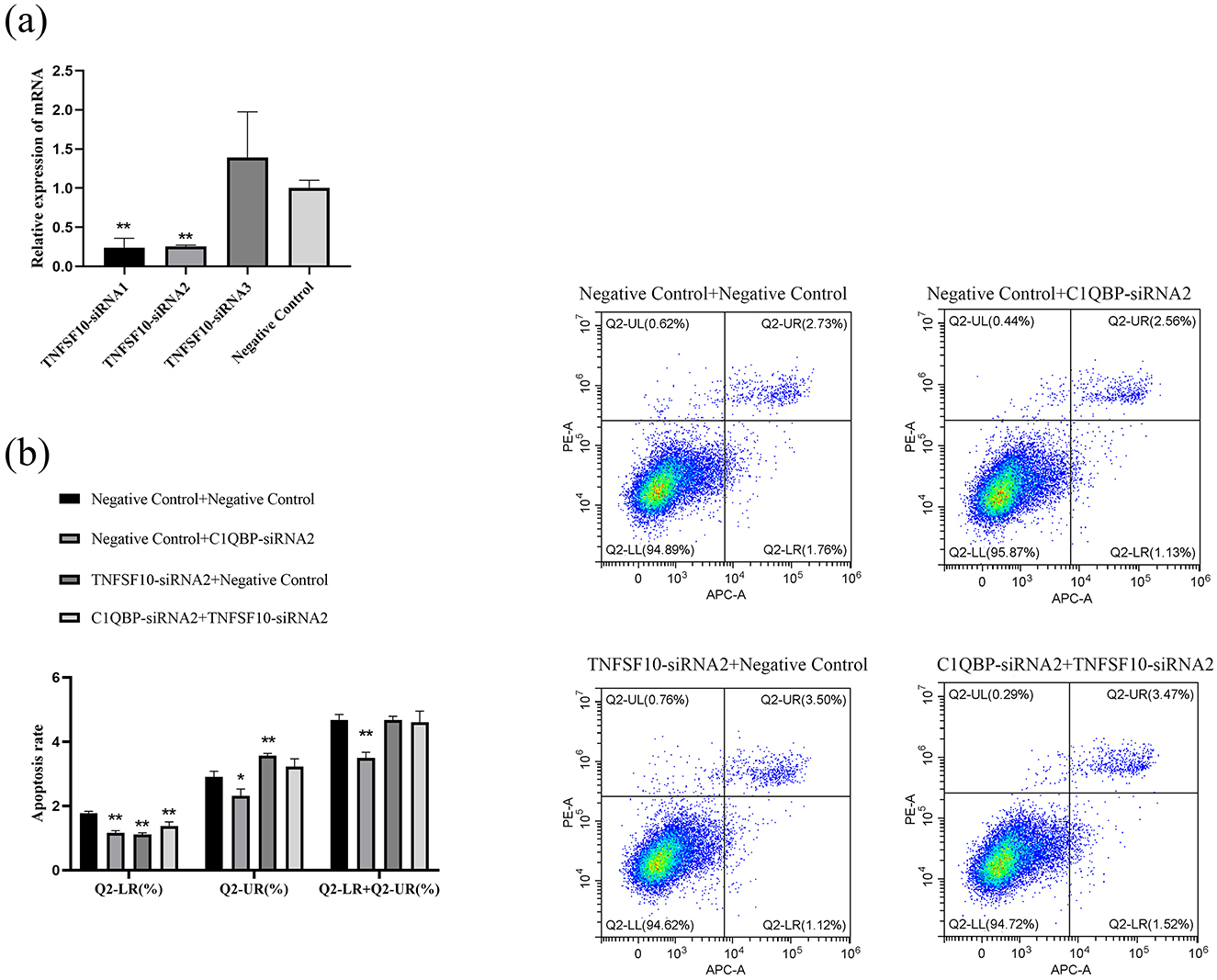
Figure 5. C1QBP deficiency inhibits apoptosis by regulating TNFSF10 expression in GFTCs. (A) Screening of interfering fragments for TNFSF10. (B) Effect of TNFSF10 and C1QBP on apoptosis of goat turbinate bone cells. *represents 0.01 < p < 0.05, **represents p < 0.01.
4 Discussion
Due to ORFV's unique immune mechanism, current vaccines cannot provide complete immune protection to goats. In addition, the existing treatment options for Orf are primarily palliative, as they provide symptomatic treatment and are not able to fundamentally cure the disease. The difficult-to-cure properties of Orf and the recurring outbreaks have become the bottleneck constraining the development of the goat industry. Uncovering the molecular mechanisms of ORFV infection will contribute to developing drugs against Orf, cultivating vaccine candidates, and then managing the current situation in which Orf is only treated symptomatically. Our previous study found that ORFV129 binding to C1QBP and C1QBP can affect ORFV replication. However, the specific mechanism by which C1QBP regulates ORFV replication remains unclear and there currently needs to be a report on the sequence of the goat C1QBP gene, its protein, and its role in regulating GFTCs. Here, our results revealed that C1QBP negatively regulates cell proliferation while positively regulating apoptosis in GFTCs. What happened can be controlled by both TNFSF10 and TRIM5 expression.
Currently, studies on C1QBP are focused on humans and mice (28–30), and there is no study on the goat C1QBP gene. Based on the cloned goat C1QBP gene sequence, we investigate the role of C1QBP on cell proliferation utilizing gene over expression and RNA silencing. Our results showed that silencing of C1QBP can lead to cell proliferation in GFTCs. Our results are also consistent with previous reports in which down regulation of C1QBP was observed to increase cell proliferation in renal carcinoma cell 786-0 (31). The increased C1QBP expression significantly reduced PK-15 cell proliferation (32). On the other hand, as demonstrated in our present study, over expression of the C1QBP protein triggered the opposite implication by inhibiting cell viability in GFTCs. Likewise, over expression of C1QBP has been reported to decrease cell proliferation in SkBr3 cells (33). A study reported that C1QBP silencing induces G1 to S phase arrest in prostate cancer cells (34). In our study, silencing of C1QBP induces the G1 to S phase progression in GFTCs, meaning that cells are progressing to a proliferative stage. At the same time, it was C1QBP over expression led to the opposite result in GFTCs. This finding suggests that C1QBP influences the cell cycle of GFTCs. Cell cycle progression was regulated by the expression of cyclin-dependent kinase (CDK) and cyclin-dependent kinase inhibitors (CK) (35). It has been found that over expression of C1QBP can regulate the transcription of many critical cyclin genes, including cyclin A, photosphere-like kinase 1 (PLK1), cyclin B1, cyclin B2, cyclin-dependent kinase 1 (CDK1), and CDC20 (36). Sensor of cellular mitosis, cyclins, along with their dependent kinases, will collectively regulate the cell cycle progression when DNA damage occurs. This regulation results a G1 phase arrest was observed in cells and facilitates the transition of cells from G1 to S phase, which is conducive to DNA replication (37). In our study, silencing of C1QBP results in the cells to be arrested at the GO/G1 phase, positively affecting cell proliferative. This finding suggests that the change in C1QBP expression might regulate the expression of cellular CDK and cell cycle progression.
Apoptosis is a typical procedural death pathway that can ensure the normal growth and development of the body by eliminating damaged or redundant cells. In our present study, over expression of C1QBP can promote the apoptosis of GFTCs. It has been demonstrated that C1QBP silencing can reduce apoptosis, and this confirmed the function of the C1QBP gene in the opposite direction, which is consistent with Ma, Itahana, and others (38, 39). Apoptosis involves internal (mitochondria) and external (death receptor pathways). Caspase family genes ultimately regulated apoptosis progression, although the signaling pathways were different, and many proteins were involved in regulating apoptosis, for example, Bcl-2, Caspase, and p53 (25, 40, 41). In our study, silencing of C1QBP can significantly alleviate the expression of gene Caspase3, Caspase7, Bax, PARP1, BCL2L11, and p53 genes and increase the expression of Bcl-2. This phenomenon was consistent with the change in apoptosis and thus inhibited apoptosis. In the previous study, the Bcl-2/Bax ratio was considered as a molecular switch that can regulate apoptosis. When the Bcl-2/Bax ratio decreases, Bax is more likely to form homodimers, thereby inducing apoptosis. On the other hand, Bcl-2 inhibits apoptosis when the Bcl-2/Bax ratio increases (42). In our study, the Bcl-2/Bax ratio increased relatively when silencing C1QBP, evidence that silencing C1QBP can inhibit apoptosis. Meanwhile, there is no apparent change in Bcl-2 expression after over expression of C1QBP. However, the mRNA expression of Bax relatively increased, and the Bcl-2/Bax ratio decreased, confirming that over expression of C1QBP can promote apoptosis.
The present study analyzed the potential mechanism of C1QBP to regulate apoptosis using RNA-seq. Among these 236 differentially expressed genes, we used correlation screens between DEGs to analyze genes that may play critical roles in regulating apoptosis. The results showed that the essential genes may be YWHAB, TNFSF10, TRIM5, VGF, and PARP1 etc. Apoptosis is a process characterized by chromatin condensation, nuclear shrinkage and apoptosis body production. In the process of apoptosis, numerous signal transduction systems play a pivotal role in delivering apoptotic signals and triggering cell death. During this series of complex reactions, various proteins are involved in regulating the apoptotic signaling pathway, jointly maintaining the balance between cell survival and death. For example, Bcl-2, Caspases, p53, BCL2L11, PARP1, and Caspase-3 serves as the ultimate executor, and its activation can promote cell death (40). The Bcl-2 family (Bcl-2, BCL2L11) regulates cell apoptosis by inhibiting the release of cytochrome C from mitochondria (43). Research demonstrated that C1QBP knockdown reduced the recruitment of the anti-apoptotic proteins, including Bcl-2 and Bcl-XL, and repressed Caspase3 activation and poly (ADP-ribose) polymerase cleavage, which consequently accelerated the T cell apoptotic process (44). C1QBP promotes the catabolism of hypoxanthine and elevates the apoptosis of renal cell carcinoma cells by modulating xanthine dehydrogenase (XDH)-mediated oxygen species (ROS) generation (25). This indicated that there was a connection between the function of Bcl-2, Caspase3, Bax and the functionality of C1QBP. Knockdown of C1QBP can inhibit the proliferation of human embryonic stem cells and promote their apoptosis (45). MiR-128-3p promoted chicken granulosa cell apoptosis through 14-3-3β/FoxO pathway via repressing YWHAB (46). YWHAB inhibits PCV2-induced endoplasmic reticulum stress (ERS), autophagy, reactive oxygen species (ROS) production and apoptosis (47). Aan-miR-181b-3p can induce sensitive cell apoptosis by regulating the expression of TNFSF10, thereby inhibiting Anguillid herpesvirus 1 replication (48). TRIM5 can induce apoptosis in human hepatocellular carcinoma cells by inhibiting the expression level of Bcl-2 and activating the expression of Caspases3 (49). IFIT2 can enhance the apoptotic effect of curcumin-induced leukemia cells (50). This study we demonstrated C1QBP promotes apoptosis in goat turbinate bone cells and showed the effect of C1QBP on the expression of some apoptosis-related genes. Therefore, based on the result of RNA-seq analysis, we further speculate that C1QBP may generally interact with more apoptosis-related proteins, thereby regulating the cell apoptosis.
Go enrichment analysis showed that DEGs were enriched by the response to the virus, activation of the innate immune response, negative regulation of the viral genome, and defense response to the virus, indicating that C1QBP is closely associated with virus infection. And in the previous study, we have already proven that C1QBP has a direct impact on the proliferation of ORFV (9). However, there are still gaps in its role in regulating ORFV infection, and its exploration is necessary for future research. The juncture between cell death and proliferation is a general target for viral infections because viruses need to hinder apoptosis to exploit cells for their own replication (51). ORFV infection can induce cell apoptosis (26) and we have established that C1QBP can promoted apoptosis in goat turbinate bone cells in our study. Therefore, we speculate that C1QBP may play an important role in the cell apoptosis induced by ORFV infection and will conduct experiment to verify this in the further. KEGG enrichment analysis of DEGs showed significant enrichment of Herpes simplex virus 1 infection, Pathways in cancer, PI3K-Akt signaling pathway, Apoptosis, Apoptosis-multiple species and p53 signaling pathway, suggesting that C1QBP is involved in apoptosis. Previous studies have found that stable knockdown of the C1QBP gene has also been previously shown to inhibit apoptosis in hepatocellular carcinoma cells (52). It was also demonstrated that C1QBP induces apoptosis in cervical cancer cells via the p53-dependent mitochondrial damage pathway (53). However, the specific mechanism of C1QBP in regulating apoptosis is still unclear, and there are no studies on the role of goat C1QBP in regulating apoptosis. Based on the results of RNA-seq analysis, we selected several proteins that may be involved in the regulation of apoptosis and whose functions are unclear. TRIM5 and TNFSF10 are two of them to investigate the role of C1QBP in regulating apoptosis. The TRIM5 protein family has more than 70 members in the human body and is an evolutionarily conservative family involved in many physiological and pathological processes, such as apoptosis and antiviral infections (54, 55). However, to date, the functions of most members of the TRIM5 family are unknown. Our study showed that TRIM5 interference promoted the apoptosis of goat turbinate bone cells and the apoptosis rate was significantly increased in the TRIM5-siRNA2 and C1QBP-siRNA2 groups compared to the only C1QBP-siRNA2 group. These results suggest that the expression of TRIM5 regulates C1QBP and thus influences the apoptosis of GFTCs. TNFSF10 is a tumor necrosis factor ligand super family member that can activate apoptosis in many tumor cell lines (56). EFLDO was identified to sensitize HepG2 cells to TNF super family member 10 (TNFSF10)-induced apoptosis (57). This study showed that compared with the C1QBP-siRNA2 group, the apoptosis rate of the TNFSF10-siRNA2 group plus C1QBP-siRNA2 group was significantly increased, but there was no significant change in the TNFSF10-siRNA2 group compared with the Negative Control group. These results suggest that the expression of TNFSF10 does not affect the apoptosis rate of GFTCs, but the expression of TNFSF10 may can regulate C1QBP, thereby affecting the apoptosis of GFTCs. In summary, we have confirmed that the expression of TRIM5 or TNFSF10 regulates C1QBP and thus influences the apoptosis of GFTCs. And, whether there is a direct interaction between TRIM5 and C1QBP or TNFSF10 and C1QBP remains unclear. We will conduct experiment such as yeast two-hybrid technology and co-immunoprecipitation (Co-IP) assays to verify this in the further.
5 Conclusions
Our current work demonstrates that C1QBP plays a significant role in maintaining metabolic activities and that its depletion leads to increased cell viability, cells remaining in the G0/G1 phase, and reduced apoptosis. Conversely, the opposite effect was observed when C1QBP was over expressed in GFTCs. Furthermore, we demonstrated that the expression of TRIM5 or TNFSF10 could regulate the role of C1QBP in apoptosis. This work provides an understanding of the role of C1QBP in apoptosis and could pave the way for further study investigating the role and mechanism of C1QBP protein in mediating the regulation of cell apoptosis by ORFV.
Data availability statement
Raw data have been deposited to National Center for Biotechnology Information (NCBI) under the BioProject number PRJNA1251641.
Ethics statement
All animal procedures used in this study were approved by the Institutional Animal Care and Use Committee, South-West Minzu University (Permit Number: S2020-013, revised in June 2004).
Author contributions
QL: Data curation, Investigation, Methodology, Software, Validation, Writing – original draft, Visualization, Writing – review & editing. ZT: Formal analysis, Methodology, Software, Writing – review & editing. HX: Funding acquisition, Resources, Supervision, Writing – review & editing. YD: Methodology, Supervision, Validation, Writing – review & editing. YR: Resources, Supervision, Writing – review & editing. HZ: Supervision, Writing – review & editing. FY: Investigation, Writing – review & editing. JZ: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Sichuan Science and Technology Program (Chengdu, China, 2021YFYZ0003, 2024NSFSC0029), Sichuan Provincial Science and Technology Program (Chengdu, China, 2023YFN0043), The Innovation Team Development Funds for Sichuan Mutton Goat and Sheep (SCCXTD-2024-14), and National Natural Science Foundation of China (Beijing, China, 32302702).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1524734/full#supplementary-material
References
1. Thompson HJ, Harview CL, Swick B, Powers JG. Orf virus in humans: case series and clinical review. Cutis. (2022) 110:e0567. doi: 10.12788/cutis.0567
2. Bergqvist C, Kurban M, Abbas O. Orf virus infection. Rev Med Virol. (2017) 27:e1932. doi: 10.1002/rmv.1932
3. Zhou Y, Guan J, Gao F, Li Z, Lan Y, Lu H, et al. Orf virus ORF120 protein positively regulates the NF-κB pathway by interacting with G3BP1. J Virol. (2021) 95:e0015321. doi: 10.1128/JVI.00153-21
4. Mercer AA, Fleming SB, Ueda N. F-Box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes. (2005) 31:127–33. doi: 10.1007/s11262-005-1784-z
5. An X, Jia H, Feng Y, Chen G, He X, Jing Z, et al. Regulating effect of ORF128 on NF-κB signaling pathway of HEK293T cells and its mechanism. J Cell Mol Immunol. (2019) 35:243–9. doi: 10.13423/j.cnki.cjcmi.008759
6. Herbert MH, Squire CJ, Mercer AA. Poxviral ankyrin proteins. Viruses. (2015) 7:709–38. doi: 10.3390/v7020709
7. Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. Poxvirus ankyrin repeat proteins are a unique class of f-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc Nat Acad Sci. (2008) 105:10955–60. doi: 10.1073/pnas.0802042105
8. Bai G, Jia HJ, He XB, Wang YY, He YP, Wu R, et al. Construction and expression of the recombinant plasmid of virus ORF129 gene in BHK-21 cells. J Gansu Agricult Univ. (2013) 48:8–13. doi: 10.13432/j.cnki.jgsau.2013.05.002
9. Dan Y, Yang L, Zhang H, Ren Y, He H, Yang F, et al. The orf virus 129 protein can inhibit immune responses by interacting with host complement C1q binding protein in goat turbinate bone cells. Vet Microbiol. (2023) 283:109782. doi: 10.1016/j.vetmic.2023.109782
10. Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. (1984) 36:993–1005. doi: 10.1016/0092-8674(84)90049-7
11. Peerschke EI, Reid KB, Ghebrehiwet B. Platelet activation by c1q results in the induction of alpha IIb/beta 3 integrins (GPIIb-IIIa) and the expression of p-selectin and procoagulant activity. J Exp Med. (1993) 178:579–87. doi: 10.1084/jem.178.2.579
12. Muta T, Kang D, Kitajima S, Fujiwara T, Hamasaki N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J Biol Chem. (1997) 272:24363–70. doi: 10.1074/jbc.272.39.24363
13. Peerschke EI, Minta JO, Zhou SZ, Bini A, Gotlieb A, Colman RW, et al. Expression of gC1q-R/p33 and its major ligands in human atherosclerotic lesions. Mol Immunol. (2004) 41:759–66. doi: 10.1016/j.molimm.2004.04.020
14. Peerschke EI, Ghebrehiwet B. Human blood platelet gC1qR/p33. Immunol Rev. (2001) 180:56–64. doi: 10.1034/j.1600-065X.2001.1800105.x
15. Pugsley MK, Abramova M, Cole T, Yang X, Ammons WS. Inhibitors of the complement system currently in development for cardiovascular disease. Cardiovasc Toxicol. (2003) 3:43–70. doi: 10.1385/CT:3:1:43
16. Harboe M, Thorgersen EB, Mollnes TE. Advances in assay of complement function and activation. Adv Drug Deliv Rev. (2011) 63:976–87. doi: 10.1016/j.addr.2011.05.010
17. Zhong JC, Zhong M, Yi XKZ, Wang JK, Chai ZX, Wang H, et al. Gene and protein expression patterns of C1QBP in maiwa yak, tibetan cattle and holstein cow. J Southwest Univ National. (2021) 47:221–8. doi: 10.11920/xnmdzk.2021.03.001
18. Wang Y, Su J, Yuan B, Fu D, Niu Y, Yue D. The role of C1QBP in CSF-1-dependent PKCζ activation and macrophage migration. Exp Cell Res. (2018) 362:11–6. doi: 10.1016/j.yexcr.2017.09.038
19. Xiao MM. Study of C1QBP-Interacting Proteins in Mitochondria. Tianjin: Tianjin Medical University (2017).
20. Ghebrehiwet B, Geisbrecht BV, Xu X, Savitt AG, Peerschke EIB. The C1q receptors: focus on gC1qR/p33 (C1qBP, p32, HABP-1)1. Semin Immunol. (2019) 45:101338. doi: 10.1016/j.smim.2019.101338
21. Li Y, Wei Y, Hao WJ, Zhao WK, Zhou YR, Wang D, et al. Porcine reproductive and respiratory syndrome virus infection promotes C1QBP secretion to enhance inflammatory responses. Vet Microbiol. (2020) 2:108563. doi: 10.1016/j.vetmic.2019.108563
22. Peng J, Zhang S, Han F, Wang Z. C1QBP is a critical component in the immune response of large yellow croaker (Larimichthys crocea) against visceral white spot disease caused by pseudomonas plecoglossicida. Fish Shellfish Immunol. (2024) 146:109372. doi: 10.1016/j.fsi.2024.109372
23. Saha P, Chowdhury AR, Dutta S, Chatterjee S, Ghosh I, Datta K. Autophagic vacuolation induced by excess ROS generation in HABP1/p32/gc1qr overexpressing fibroblasts and its reversal by polymeric hyaluronan. Public Lib Sci One. (2013) 8:e78131. doi: 10.1371/journal.pone.0078131
24. Song XJ. Effect and Mechanism of Differentially Expressed LncRNA RPAIN in Preeclampsia on the Biological Behavior of Placental Trophoblast Cells Through Complement Protein c1q. Nanjing: Nanjing Medical University (2018).
25. Wang YT, Liu S, Tian SP, Du RX, Lin TY, Xiao XS, et al. C1QBP regulates apoptosis of renal cell carcinoma via modulating xanthine dehydrogenase (XDH) mediated ROS generation. Int J Med Sci. (2022) 19:842–57. doi: 10.7150/ijms.71703
26. Long QQ, Wei M, Wang YT, Wen M, Pang F. The battle between orf virus and host: immune response and viral immune evasion mechanisms. Acta aeterinaria et zootechnica sinica. (2023) 54:1845–53. doi: 10.11843/j.issn.0366-6964.2023.05.008
27. Dan YX, Xiang H, Zhang HR, Yang L, Ren YP, Xu SW, et al. Screening of plasma virus 129 protein-interaction host protein and cloning analysis of C1QBP gene. China Anim Husb Vet Med. (2023) 50:1–14. doi: 10.16431/j.cnki.1671-7236.2023.01.001
28. Hong X, Yu Z, Chen Z, Jiang H, Niu Y, Huang Z. High molecular weight fibroblast growth factor 2 induces apoptosis by interacting with complement component 1 Q subcomponent-binding protein in vitro. J Cell Biochem. (2018) 119:8807–17. doi: 10.1002/jcb.27131
29. Wang WJ, Wu W, Xue XC, Sheng L, Wang QP. C1QBP regulation of mitochondrial function mediates the role of glioblastoma in the study of temozolomide tolerance. J Clin Neurosurg. (2023) 20:340–343+347. doi: 10.3969/j.issn.1672-7770.2023.03.021
30. Chowdhury AR, Ghosh I, Datta K. Excessive reactive oxygen species induces apoptosis in fibroblasts: role of mitochondrially accumulated hyaluronic acid binding protein 1 (HABP1/p32/gC1qR). Exp Cell Res. (2008) 314:651–67. doi: 10.1016/j.yexcr.2007.10.033
31. Chen YQ. The Role of C1QBP in the Development of Kidney Cancer. Tianjin: Tianjin Medical University (2017).
32. Ma X, Lv C, Wang Q, Li C, Wang P, Luo C, et al. C1QBP inhibits proliferation of porcine circovirus type 2 by restricting nuclear import of the capsid protein. Arch Virol. (2021) 166:767–78. doi: 10.1007/s00705-020-04950-7
33. Kandov E, Kaur A, Kishore U, Ji P, Williams J, Peerschke EIB, et al. C1q and C1q receptors (gC1qR and cC1qR) as potential novel targets for therapy against breast cancer. Immunology. (2018) 19:59–76.
34. Amamoto R, Yagi M, Song Y, Oda Y, Tsuneyoshi M, Naito S, et al. Mitochondrial p32/C1QBP is highly expressed in prostate cancer and is associated with shorter prostate-specific antigen relapse time after radical prostatectomy. Cancer Sci. (2011) 102:639–47. doi: 10.1111/j.1349-7006.2010.01828.x
35. Pan JF, Shang FZ, Ma R, Rong YJ, Zhang YJ. Research progress on the regulatory mechanism of cyclin and cyclin-dependent kinases and related kinase inhibitors in cell cycle progression. Chin J Biotechnol. (2023) 39:1525–47. doi: 10.13345/j.cjb.220478
36. Engeland K. Cell cycle arrest through indirect transcriptional repression by p53:i have a dream cell death and differentiation. Cell Death Differ. (2018) 25:114–32. doi: 10.1038/cdd.2017.172
37. Chen S, Luo T, Yu Q, Dong W, Zhang H, Zou H. Isoorientin plays an important role in alleviating cadmium-induced DNA damage and G0/G1 cell cycle arrest. Ecotoxicol Environ Saf. (2020) 15:109851. doi: 10.1016/j.ecoenv.2019.109851
38. Ma J, Ren C, Yang H, Zhao J, Wang F, Wan Y. The expression pattern of p32 in sheep muscle and its role in differentiation, cell proliferation, and apoptosis of myoblasts. Int J Mol Sci. (2019) 20:5161. doi: 10.3390/ijms20205161
39. Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. (2008) 13:542–53. doi: 10.1016/j.ccr.2008.04.002
40. Li M, Gao P, Zhang J. Crosstalk between autophagy and apoptosis: potential and emerging therapeutic targets for cardiac diseases. Int J Mol Sci. (2016) 17:332. doi: 10.3390/ijms17030332
41. Miyamae Y, Mochizuki S, Shimoda M, Ohara K, Abe H, Yamashita S, et al. ADAM28 is expressed by epithelial cells in human normal tissues and protects from C1q-induced cell death. Feder Eur Biochem Soc J. (2016) 283:1574–94. doi: 10.1111/febs.13693
42. Lalier L, Cartron PF, Juin P, Nedelkina S, Manon S, Bechinger B, et al. Bax activation and mitochondrial insertion during apoptosis. Apoptosis. (2007) 12:887–96. doi: 10.1007/s10495-007-0749-1
43. Marquez RT, Xu L. Bcl-2: beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res. (2012) 2:214–21.
44. Tian H, Wang G, Wang Q, Zhang B, Jiang G, Li H, et al. Complement C1q binding protein regulates T cells' mitochondrial fitness to affect their survival, proliferation, and anti-tumor immune function. Cancer Sci. (2022) 113:875–90. doi: 10.1111/cas.15261
45. Li RM, Li M, Chen L. Knockdown of placental-specific protein 8 (PLAC8) inhibits proliferation and promotes apoptosis of human embryonic stem cells. J Cell Mol Immunol. (2020) 36:1089–94. doi: 10.13423/j.cnki.cjcmi.009113
46. Ning Z, Deng X, Li L, Feng J, Du X, Amevor FK, et al. MiR-128-3p regulates chicken granulosa cell function via 14-3-3β/FoxO and PPAR-γ/LPL signaling pathways. Int J Biol Macromol. (2023) 30:124654. doi: 10.1016/j.ijbiomac.2023.124654
47. Guo K, Zhang X, Hou Y, Liu J, Feng Q, Wang K, et al. novel PCV2 ORF5-interacting host factor YWHAB inhibits virus replication and alleviates PCV2-induced cellular response. Vet Microbiol. (2020) 251:108893. doi: 10.1016/j.vetmic.2020.108893
48. Guo R. Regulation of Apoptosis by Host miRNA Targeting TRAIL During Anguillid herpesvirus Infection. Fuzhou: Fujian Agriculture and Forestry University (2023).
49. Yu Q, Zhang WM, Sun YY, Gong YY, Li DS, Wang XL. The effect of TRIM5α on the proliferation and apoptosis of hepatocellular carcinoma cells. J Pract Oncol. (2016) 30:103–8. doi: 10.11904/j.issn.1002-3070.2016.02.002
50. Zhang Y, Kong Y, Liu S, Zeng L, Wan L, Zhang Z. Curcumin induces apoptosis in human leukemic cell lines through an IFIT2-dependent pathway. Cancer Biol Ther. (2017) 18:43–50. doi: 10.1080/15384047.2016.1276129
51. Watthanasurorot A, Jiravanichpaisal P, Söderhäll K, Söderhäll I. A calreticulin/gC1qR complex prevents cells from dying: a conserved mechanism from arthropods to humans. J Mol Cell Biol. (2013) 5:120–31. doi: 10.1093/jmcb/mjt005
52. Hou G, Lu Z, Wang Z, Yang X. The mitochondrial protein C1QBP promotes hepatocellular carcinoma progression by enhancing cell survival, migration and invasion. J Cancer. (2022) 13:2477–89. doi: 10.7150/jca.69379
53. Zhao W. Study on the Mechanism of Apoptosis in Cervical Cancer Cells Induced by gC1qR Gene. Nanjing: Nanjing Medical University (2018).
54. Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. (2017) 42:297–311. doi: 10.1016/j.tibs.2017.01.002
55. Gu TT. TRIM25 Ubiquitination Modulates the IFN-β Production Through RIG-I Signaling Pathway in Duck. Yangzhou: Yangzhou University (2021).
56. Hu ZX. Association Study of TNFSF10 With the Prognosis and Clinical Features of Cutaneous Melanoma. Shantou: Shantou University (2022).
Keywords: C1QBP, apoptosis, RNA-seq, TRIM5, TNFSF10
Citation: Li Q, Tang Z, Xiang H, Dan Y, Ren Y, Zhang H, Yang F and Zhu J (2025) C1QBP promotes apoptosis of goat fetal turbinate cells via inhibiting the expression of TRIM5 or TNFSF10. Front. Vet. Sci. 12:1524734. doi: 10.3389/fvets.2025.1524734
Received: 08 November 2024; Accepted: 14 February 2025;
Published: 16 May 2025.
Edited by:
Penghao Wang, Murdoch University, AustraliaReviewed by:
Joanna Zarzynska, Warsaw University of Life Sciences, PolandXiaolong Zhou, Zhejiang Agriculture and Forestry University, China
Yong Ruan, Guizhou University, China
Copyright © 2025 Li, Tang, Xiang, Dan, Ren, Zhang, Yang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Xiang, eGlhbmdodWEyMDA4NDExQDE2My5jb20=
†ORCID: Zhengding Tang orcid.org/0009-0008-9594-5918
 Qiuyun Li
Qiuyun Li Zhengding Tang1†
Zhengding Tang1† Hua Xiang
Hua Xiang Yupeng Ren
Yupeng Ren Falong Yang
Falong Yang Jiangjiang Zhu
Jiangjiang Zhu