- 1Istituto Zooprofilattico Sperimentale del Mezzogiorno, Portici, Italy
- 2Dipartimento di Medicina Veterinaria, Università degli Studi di Bari, Valenzano, Italy
- 3University of Veterinary Medicine, Budapest, Hungary
CDV has been detected in a wide range of domestic and wild animal also in Italy and it is highly prone to cross-species transmission, therefore representing a significant health risk. In this study the presence of CDV and other coinfecting selected viruses, in wild carnivorans of the family Mustelidae and Canidae and rodents of the family Hystricidae, collected in Southern Italy (Campania region), in 2022–2024, was investigated. Over a period of 3 years (2022–2024), tissue samples from 136 wild animals including stone martens, porcupines, otters, wolves, martens, badgers and foxes were examined. CDV RNA was detected in 14 (10.3%) animals encompassing badgers (n = 6), foxes (n = 5), wolves (n = 2), and marten (n = 1). The complete genome of a CDV strain was reconstructed from a spleen sample of a badger. On sequence and phylogenetic analyses, the novel CDV strain belonged to the Arctic clade, which has already been reported from badger and dog in Italy. Our study contributes to extend the knowledge on the epidemiology of CDV in wildlife and confirm the need for a continuous surveillance in wild animals to monitor the circulation in wildlife of viruses pathogenic for domestic carnivores and endangered wild species.
Introduction
Canine distemper (CD) is a highly contagious multi-systemic disease caused by canine distemper virus (CDV). The clinical disease can be sub-clinical to fatal, spreading through the bloodstream to multiple organs, with clinical signs that include respiratory distress, anxiety, conjunctivitis, anorexia, diarrhea, lymphopaenia, encephalitis, coughing, rhinorrhoea, ocular discharge and fever (1, 2).
CDV has been detected in a wide range of domestic and wild animal hosts worldwide including canids, felids, mustelids, procyonids, urisds and hyaenids (3–5). It is highly prone to cross-species transmission between domestic and wildlife reservoir hosts, therefore representing a significant conservational and animal health risk around the globe (6, 7). CDV transmission primarily occurs via various body fluids, such as respiratory droplets, ocular and nasal discharge, saliva, urine and feces (8). In Italy, CDV has been reported in both domestic and free-ranging canids, as well as in mustelids. The virus poses a significant threat to wildlife conservation and domestic animal health, making its study crucial for disease management and control. CDV is an enveloped virus with a non-segmented, negative sense, single-stranded RNA genome and belongs to the family Paramyxoviridae, genus Morbillivirus (3). His genome encodes six structural proteins: nucleocapsid (N), matrix (M), fusion (F), hemagglutinin (H), polymerase (L), and phosphoprotein (P) (9). Based on H-gene sequences, CDV strains have been classified into 19 genetic lineages: America-1, America-2, North America-3, South America/North America-4, America-5 (formerly regarded as a sub-genotype of America-2), Asia-1, Asia- 2, Asia-3, Asia-4, Asia-5, Europe Wildlife, Arctic, Africa-1, Africa-2, Europe-1/South America-1, South America-2, South America-3, Rockborn-like, and Asia-6 (5, 10). Recently, Lanszki et al. (11) proposed a distinct CDV classification in 16 genetic lineages based on full-genome sequences. In Italy, the presence of CDV has been reported within the Canidae family, including domestic dogs (12) and free-ranging canids, like foxes and wolves (13, 14), and also within the family Mustelidae (15–17).
In recent decades, three CDV genetic lineages have been reported to circulate in Italy: the Europe/South America-1, EuropeWildlife, and Arctic-like lineages (10). The first to be detected in both domestic dogs and wild carnivores was the Europe/South America-1 lineage, while the Europe Wildlife have been reported in wildlife and only sporadically in domestic dogs (13, 18). Conversely, Arctic-like lineage has been reported mainly in Italian dogs and, less frequently, in wildlife (14, 16, 19). These data indicate the circulation of the three different CDV strains across the domestic/wildlife interface in Italy.
This study aimed at investigating the presence of CDV and other selected viruses (rabies virus, canine coronavirus, canine adenoviruses, canine herpesvirus type 1, rotavirus, protoparvovirus carnivoran 1) from internal organs of wild carnivorans of the family Mustelidae and Canidae and rodents of the family Hystricidae, collected in Southern Italy (Campania region) in 2022–2024CDV was consistently detected in this study and genetic characterization of the identified CDV strains was performed.
Methods
Sample collection
A total of 1,088 tissue samples (lungs, livers, hearts, spleens, kidneys, intestines, brains and muscles) were collected from 136 wild animals including 4 stone martens (Martes foina), 12 porcupines (Erethizon dorsatum), 6 otters (Lutra lutra), 11 wolves (Canis lupus italicus), 3 martens (Martes martes), 34 badgers (Meles meles), and 66 foxes (Vulpes vulpes).
Samples were collected over a period of 3 years (2022–2024) at the Istituto Zooprofilattico Sperimentale del Mezzogiorno (IZSM), a ministerial institution that operates within the National Health Service. All the tested animals were found dead in the Campania region during passive surveillance and samples were collected at necropsy and were tested in this study in the framework within the diagnostic activity of IZSM. For each animal, information on gender, age, place, and date of collection were recorded. Subjects were classified as juveniles (6–12 months of age), subadults (between 12 and 24 months) and adults (>24 months).
Nucleic acids extraction
The collected samples were processed just after their arrive in laboratory or, stored for a short time at −20°C, or for long time at −80°C, before NAs extraction. After NAs extraction the samples were immediately analyzed. The samples were subjected to NAs extraction by means of an automatic extractor (QIAsymphony, Qiagen, Hilden, Germany), using the “Virus/Pathogen” kit (Qiagen) according to the manufacturer’s protocol. The DNA and RNA quality were monitored using an exogenous Internal Positive Control (IPC) added to each sample to supervise the presence of potential PCR and RT-PCR inhibitors (VetMAX™ Xeno™ Internal Positive Controls and Assays for PCR and RT-PCR, Thermo Fisher Scientific). A sample with nuclease-free water instead of homogenate was used as a negative process control (NPC).
Molecular screening for CDV
Identification of CDV was performed using a reverse transcription (RT) protocol followed by a quantitative real-time PCR (qPCR) assay (20).
Molecular screening for other pathogens
All the samples were tested for rabies virus for monitoring (21). In addition, the extracts were also analyzed for the following viruses, to evaluate the presence of co-infections: canine coronavirus (CCoV) (22–24), canine adenoviruses (CAdVs) type 1 and 2 (25), canine herpesvirus type 1 (CaHV-1) (26), rotavirus A (RVA) (27), protoparvovirus carnivoran 1 (28, 29). For the latter, a qPCR able to discriminate between field variants and the vaccine virus was performed (30).
Full-length genome sequencing of CDV
The complete genome sequencing of CDV strains identified in this study was obtained through an amplicon-based sequencing method performed according to the protocol described by Lanszki et al. (11). PCR products were pooled in equimolar ratios, quantified by Qubit dsDNA HS assay (Thermo Fisher Scientific, Waltham, MA) and used for library preparation and adapter-ligation by Ligation kit SQK-LSK110 (Oxford Nanopore Technologies, ONT, Oxford UK) following manufacturer’s guidelines. Libraries were purified by means of Agencourt AMPure XP magnetic beads (Beckman Coulter™) and sequenced employing flowcell flongle FLO-FLG001 version R10.4.1 adapted in a MinION Mk1C (ONT, Oxford UK) platform for 24 h. FastQ MinION files underwent quality control, trimming and reference assembly by Minimap2 plugin implemented in the software package Geneious Prime v. 2021.2.2 (Biomatters Ltd., Auckland, New Zealand).
Sequence and phylogenetic analyses
Sequence analyses were performed by web-based tool BLAST1 applying default values to find homologous hits in the Genbank database. Sequence alignment was performed by MAFFT (31). The correct substitution model parameters for the phylogenetic analysis were obtained using “Find the best protein DNA/Protein Models” implemented in MEGA X version 10.0.5 software (32). Phylogenetic analysis was performed using the maximum likelihood (ML) method implemented in MEGAX version 10.0.5 software.
GenBank sequence submission
The obtained sequence of CDV strain ITA/2022/badger/33340 were deposited in the Genbank database under accession number PQ584613.
Results
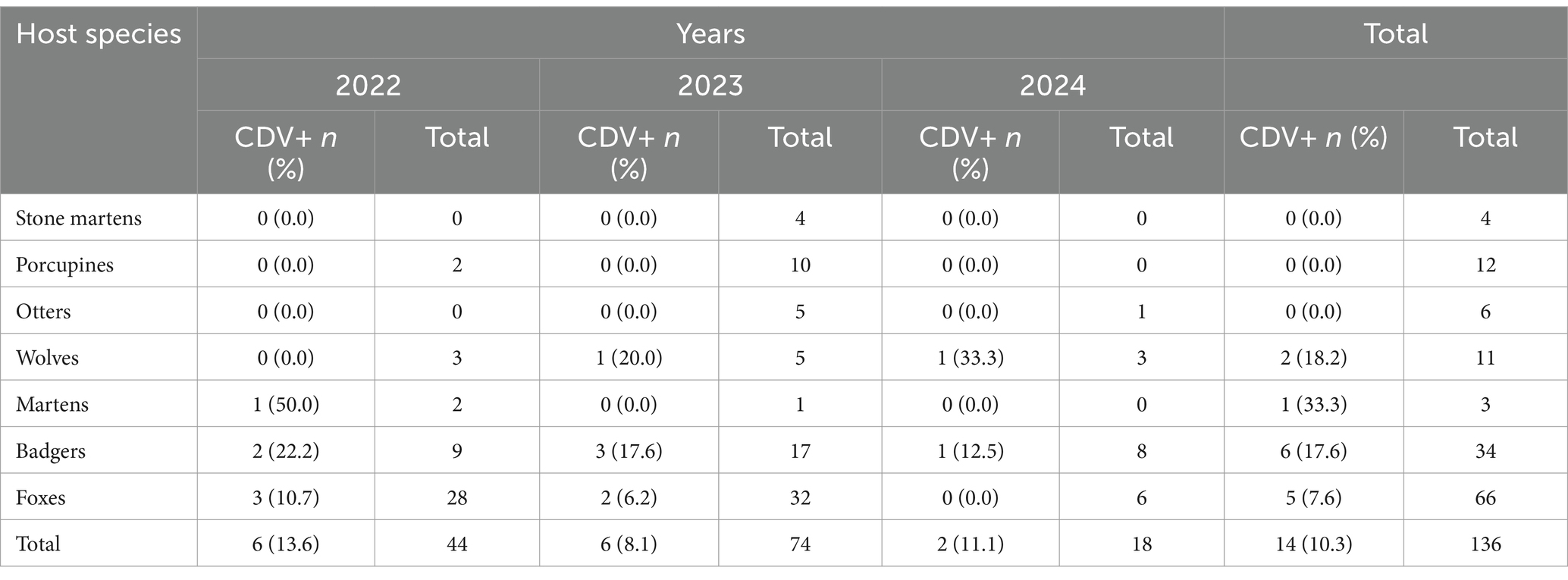
Overall, out of 136 wild animals collected in Campania region in a period spanning from 2022 to 2024, 14 (10.3%) tested positive for CDV RNA by RT-qPCR (Table 1) with a cycle threshold (Ct) ranging from 17.8 to 42.5. CDV infections were observed in badgers (6/34, 17.6%), foxes (5/66, 7.6%), wolves (2/11, 18.2%) and martens (1/3, 33.3) while CDV was not retrieved from stone martens, porcupines and otters (Table 2). Six out 44 (13.6%) CDV positive cases (3 foxes, 2 badgers and 1 marten) were identified in 2022, 6/74 (8.1%) (3 badgers, 2 foxes and 1 wolf) in 2023, 2/18 (11.1%) (1 wolf and 1 badger) in 2024. CDV was unevenly identified in the lung, intestine, brain, spleen, liver, heart, muscle and kidney of the positive animals. The 14 CDV positive animals did not present co-infections with other analyzed pathogens. The 6 CDV-positive badgers were 3 males and 3 females and comprised 3 adults, 2 subadults and 1 young. The 5 CDV-positive foxes were 3 males and 2 females and included 3 adults, 1 subadult and 1 young. The 2 wolves were 1 male and 1 female and encompassed 1 adult and 1 subadult. The marten was an adult male. Out of 14 CDV-positive animals, the most frequent clinical observations at necroscopy were congested lungs and possibly also trachea in 4 (28.5%) animals followed by enteritis in 3 (21.4%) animals. The cause of death of the 14 CDV-positive animals were either trauma or infection (6/14, 42.8% for both), or gunshot and bite wounds (1/14, 7.1% for both). All the samples were tested for rabies virus and found to be negative.
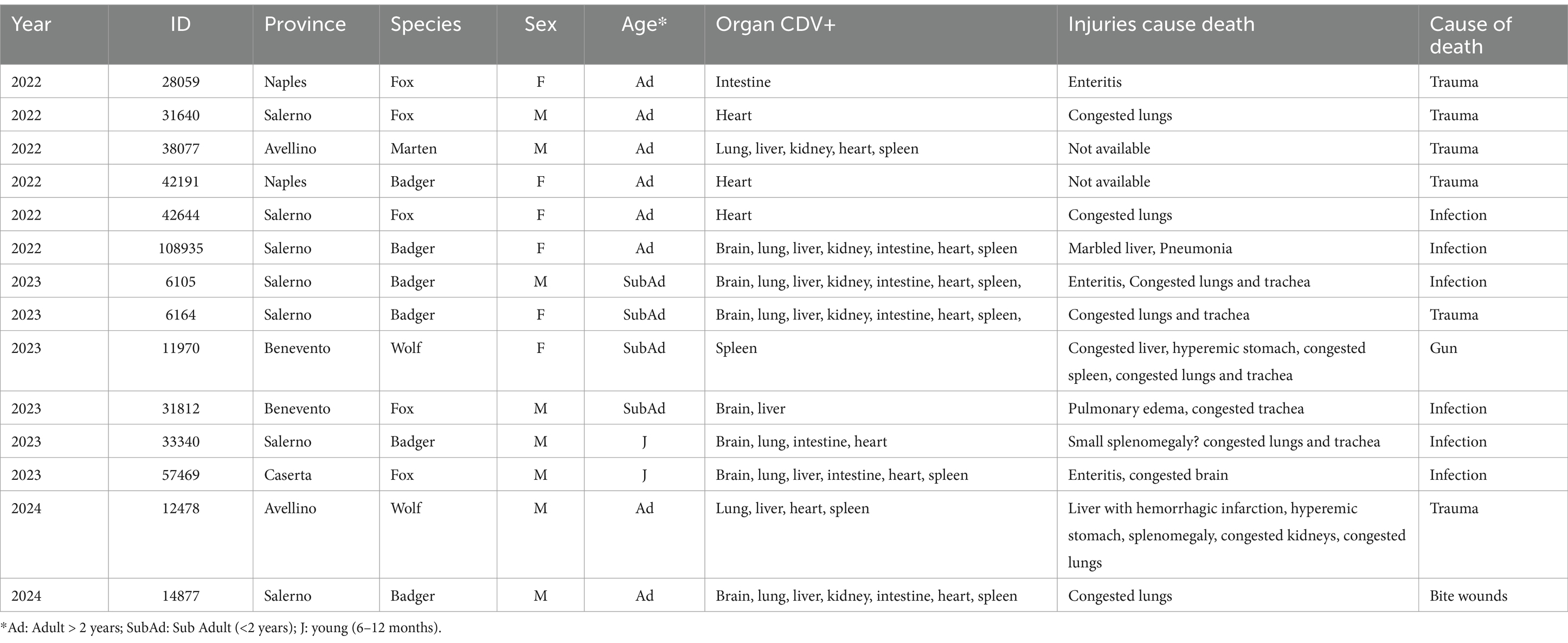
Table 1. Information on animals infected by CDV: species, sex, age, organ CDV+, injuries, cause of death.
CDV strain ITA/2022/badger/33340, retrieved from spleen of an adult male badger in 2022 (Ct = was subjected to full genome amplification and ONT sequencing protocol).
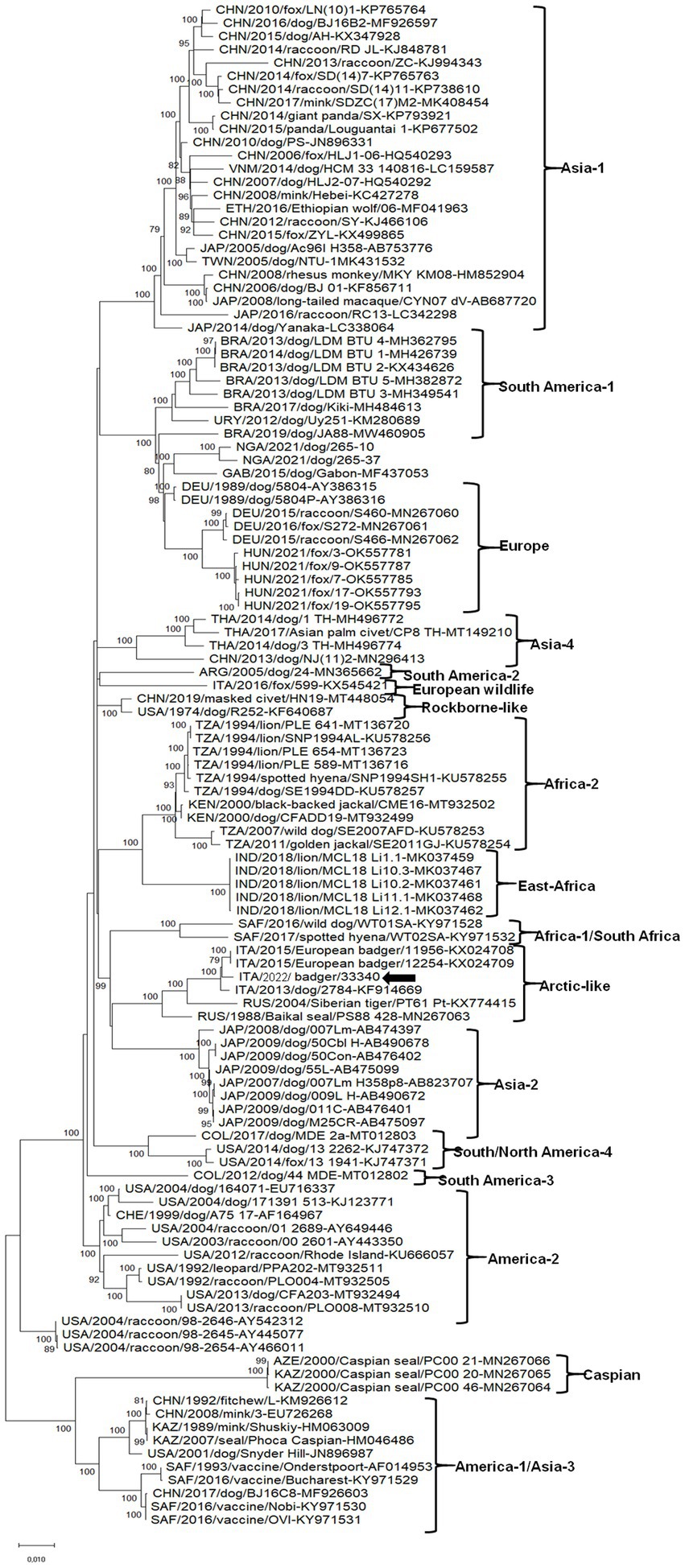
On full genome level, the CDV strain retrieved in this study displayed the highest nt identity (99.4%) to the isolate CDV11956/2015 (KX024708). The genomic sequence of the CDV strain ITA/2022/badger/33340 was aligned with cognate CDV sequences available in GenBank. Upon ML phylogenetic tree the CDV strain ITA/2022/badger/33340 segregated with other CDV strains included in the Arctic-like cluster (Figure 1).
Discussion
The results of this study demonstrated the circulation of CDV in the wildlife in Southern Italy with infection rate equal to nearly 10%. No co-infections with other pathogens (i.e., CCoV, CAdVs type 1 and 2, CaHV-1, RVA and protoparvovirus carnivoran 1) were detected in the CDV-positive animals. In a previous report CDV infection rate reached 16% in tissue samples of foxes and badgers collected in Northern Italy (33). In this study, also, no other pathogens were detected in the animals that tested positive for distemper. In some cases, we can hypothesize, that the absence of other pathogens could also be related to the poor conditions of the carcasses.
About the rabies virus our territory has been declared free since 2013, checks are currently carried out only for constant monitoring, due to the presence of rabies in Eastern Europe. The complete genome sequence of the CDV strain, that we obtained from an adult male badger, phylogenetically characterized as an Arctic-like lineage, clustered along with other CDV strains retrieved from badger and dog from Italy.
The arctic lineages and the EuropeanWildlife have been mostly detected in Central and Southern Italy (14–16) while the Europe/South America-1 lineage is peculiarly spread in Northern Italy (13, 17, 34).
The Arctic CDV strains were first reported in animals of the Arctic ecosystem but this lineage appears common in European territories in both domestic and wildlife carnivores. In Italy, CDV strains belonging to the Arctic lieage were first reported in dogs in the early 21th century (35) and subsequently in the United States (36) and other European countries (2, 12, 37), raising questions on the origin of these unusual strains.
A large distemper epidemic, sustained by an Arctic-lineage (38), occurred in Italy during 2013 and involved primarily the Abruzzi region and neighboring regions, affecting foxes (Vulpes vulpes), badgers (Meles meles), beech martens (Martes foina) and European polecats (Mustela putorius) (14, 34).
Compared to other morbilliviruses, the CDV shows the highest genetic variability (39). The main factors fostering CDV spread, cross-species infection (40, 41) and increasing virulence (17, 42–45) are genetic variability, broad spectrum of hosts and the uncontrolled animal movements of stray and domestic dogs (2, 46–48) and other wild animals (49).
As previously observed in CDV infected animals, traumatic lesions appear a frequent cause of death in wild animals, probably due to vehicular collisions (33). In this study an equal number of CDV-positive animals dying from trauma and infections were observed.
A possible correlation between neurological disease due to either traumatic or infective etiology, and a delay in the animal’s response to external stimuli could be hypothesized (49).
Although the presence of specific clinical signs or anatomopathological lesions in live or dead animals could be related to CDV infection, confirmatory laboratory tests are needed to roule out other diseases. Molecular approaches could help monitoring CDV spread in susceptible wild animal hosts, particularly endangered species, thus supporting global animal welfare.
To date, the presence of CDV in Southern Italy has been described in both domestic dogs (2, 35, 46, 50) and wildlife (51). Our study contributes to extend the knowledge on the epidemiology of CDV in wildlife.
Continuous surveillance of CDV occurrence in wildlife will help depicting a better picture the ecology of CDV in domestic and wildlife animals and safeguarding endangered species.
Data availability statement
The obtained sequence of CDV strain ITA/2022/badger/33340 were deposited in the Genbank database under accession number PQ584613.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because Ethical review and approval were waived for this study, as samples were part of the diagnostic activity of Istituto Zooprofilattico Sperimentale del Mezzogiorno, statal public institution that operate within the national health service.
Author contributions
FA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MGL: Investigation, Methodology, Writing – original draft. ND’A: Investigation, Methodology, Writing – original draft. CA: Investigation, Methodology, Writing – original draft. SR: Investigation, Methodology, Writing – original draft. GS: Investigation, Methodology, Writing – original draft. MSL: Investigation, Methodology, Writing – original draft. FP: Investigation, Methodology, Writing – original draft. GD: Investigation, Methodology, Writing – original draft. EC: Writing – review & editing. ND: Writing – review & editing. GL: Conceptualization, Data curation, Formal analysis, Writing – review & editing. VM: Writing – review & editing. GF: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by the Italian Ministry of Health: Ricerca Corrente 2023, IZS ME 08/23 RC, recipient Flora Alfano.
Acknowledgments
Vito Martella was funded by National Laboratory for Infectious Animal Diseases, Antimicrobial Resistance, Veterinary Public Health and Food Chain Safety, RRF-2.3.1-21-2022-00001.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Ke, G-M, Ho, C-H, Chiang, M-J, Sanno-Duanda, B, Chung, C-S, Lin, M-Y, et al. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet Res. (2015) 11:164. doi: 10.1186/s12917-015-0491-9
2. Mira, F, Purpari, G, Di Bella, S, Vicari, D, Schirò, G, Di Marco, P, et al. Update on canine distemper virus (CDV) strains of Arctic-like lineage detected in dogs in Italy. Vet Ital. (2018) 54:225–36. doi: 10.12834/VetIt.1455.7862.2
3. Loots, AK, Mitchell, E, Dalton, DL, Kotzé, A, and Venter, EH. Advances in canine distemper virus pathogenesis research: a wildlife perspective. J Gen Virol. (2017) 98:311–21. doi: 10.1099/jgv.0.000666
4. Martinez-Gutierrez, M, and Ruiz-Saenz, J. Diversity of susceptible hosts in canine distemper virus infection: a systematic review and data synthesis. BMC Vet Res. (2016) 12:1–11. doi: 10.1186/s12917-016-0702-z
5. Wang, R, Wang, X, Zhai, J, Zhang, P, Irwin, DM, Shen, X, et al. A new canine distemper virus lineage identified from red pandas in China. Transbound Emerg Dis. (2022) 69:e944–52. doi: 10.1111/tbed.14370
6. Ludlow, M, Rennick, LJ, Nambulli, S, de Swart, RL, and Paul, DW. Using the ferret model to study morbillivirus entry, spread, transmission and cross-species infection. Curr Opin Virol. (2014) 4:15–23. doi: 10.1016/j.coviro.2013.11.001
7. McCarthy, AJ, Shaw, M-A, and Goodman, SJ. Pathogen evolution and disease emergence in carnivores. Proc R Soc B Biol Sci. (2007) 274:3165–74. doi: 10.1098/rspb.2007.0884
8. Greene, CE. Infectious diseases of the dog and cat Elsevier (2012). doi: 10.1111/j.1748-5827.2008.00601.x
9. Martella, V, Elia, G, and Buonavoglia, C. Canine distemper virus. Vet Clin N Am Small Anim Pract. (2008) 38:787–97. doi: 10.1016/j.cvsm.2008.02.007
10. Guercio, A, Mira, F, Di Bella, S, Gucciardi, F, Lastra, A, Purpari, G, et al. Biomolecular analysis of canine distemper virus strains in two domestic ferrets (Mustela putorius furo). Vet Sci. (2023) 10:375. doi: 10.3390/vetsci10060375
11. Lanszki, Z, Tóth, GE, Schütz, É, Zeghbib, S, Rusvai, M, Jakab, F, et al. Complete genomic sequencing of canine distemper virus with nanopore technology during an epizootic event. Sci Rep. (2022) 12:4116. doi: 10.1038/s41598-022-08183-3
12. Martella, V, Elia, G, Lucente, MS, Decaro, N, Lorusso, E, Banyai, K, et al. Genotyping canine distemper virus (CDV) by a hemi-nested multiplex PCR provides a rapid approach for investigation of CDV outbreaks. Vet Microbiol. (2007) 122:32–42. doi: 10.1016/j.vetmic.2007.01.005
13. Bianco, A, Zecchin, B, Fusaro, A, Schivo, A, Ormelli, S, Bregoli, M, et al. Two waves of canine distemper virus showing different spatio-temporal dynamics in alpine wildlife (2006–2018). Infect Genet Evol. (2020) 84:104359. doi: 10.1016/j.meegid.2020.104359
14. Di Sabatino, D, Lorusso, A, Di Francesco, CE, Gentile, L, Di Pirro, V, Bellacicco, AL, et al. Arctic lineage-canine distemper virus as a cause of death in Apennine wolves (Canis lupus) in Italy. PLoS One. (2014) 9:e82356. doi: 10.1371/journal.pone.0082356
15. Balboni, A, Savini, F, Scagliarini, A, Berti, E, Naldi, M, Urbani, L, et al. Natural distemper infection in stone martens (Martes foina): from infection to neutralizing antibodies. Res Vet Sci. (2021) 138:196–200. doi: 10.1016/j.rvsc.2021.06.015
16. Di Sabatino, D, Di Francesco, G, Zaccaria, G, Malatesta, D, Brugnola, L, Marcacci, M, et al. Lethal distemper in badgers (Meles meles) following epidemic in dogs and wolves. Infect Genet Evol. (2016) 46:130–7. doi: 10.1016/j.meegid.2016.10.020
17. Monne, I, Fusaro, A, Valastro, V, Citterio, C, Pozza, MD, Obber, F, et al. A distinct CDV genotype causing a major epidemic in alpine wildlife. Vet Microbiol. (2011) 150:63–9. doi: 10.1016/j.vetmic.2011.01.009
18. Balboni, A, De Lorenzo, DG, Scagliarini, A, Prosperi, S, and Battilani, M. Occurrence of different canine distemper virus lineages in Italian dogs. Vet Ital. (2014) 50:227–31. doi: 10.12834/VetIt.52.2173.2
19. Di Francesco, CE, Smoglica, C, and Angelucci, S. Infectious diseases and wildlife conservation medicine: the case of the canine distemper in European Wolf population. Animals (Basel). (2020) 10:2426. doi: 10.3390/ani10122426
20. Elia, G, Decaro, N, Martella, V, Cirone, F, Lucente, MS, Lorusso, E, et al. Detection of canine distemper virus in dogs by real-time RT-PCR. J Virol Methods. (2006) 136:171–6. doi: 10.1016/j.jviromet.2006.05.004
21. WOAH Terrestrial Manual. Chapter 3.1.19. – rabies (infection with rabies virus and other lyssaviruses) In:. Real-time reverse-transcription polymerase chain reaction (real time RT-PCR) (2023)
22. Decaro, N, Pretelli, A, Campolo, M, Elia, G, Martella, V, Tempesta, M, et al. Quantitation of canine coronavirus RNA in the feaces of dogs by TaqMan RT-PCR. J Virol Methods. (2004) 119:145–50. doi: 10.1016/j.jviromet.2004.03.012
23. Decaro, N, Martella, V, Ricci, D, Elia, G, Desario, C, Campolo, M, et al. Genotype-specific fluorogenic RT-PCR assays for detection and quantitation of canine coronavirus type I and type II RNA in fecal sample of dogs. J Virol Methods. (2005) 130:72–8. doi: 10.1016/j.jviromet.2005.06.005
24. Decaro, N, Cordonnier, N, Demeter, Z, Egberink, H, Elia, G, Grellet, A, et al. European surveillance for pantropic canine coronavirus. J Clin Microbiol. (2013) 51:83–8. doi: 10.1128/JCM.02466-12
25. Dowgier, G, Mari, V, Losurdo, M, Larocca, V, Colaianni, ML, Cirone, F, et al. A duplex real-time PCR assay based on TaqMan technology for simultaneous detection and differentiation of canine adenovirus types 1 and 2. J Virol Methods. (2016) 234:1–6. doi: 10.1016/j.jviromet.2016.03.011
26. Decaro, N, Amorisco, F, Desario, C, Lorusso, E, Camero, M, Bellacicco, AL, et al. Development and validation of a real-time PCR assay for specific and sensitive detection of canid herpesvirus 1. J Virol Methods. (2010) 169:176–80. doi: 10.1016/j.jviromet.2010.07.021
27. Gutiérrez-Aguirre, I, Steyer, A, Boben, J, Gruden, K, Poljsak-Prijatelj, M, and Ravnikar, M. Sensitive detection of multiple rotavirus genotypes with a single reverse transcription-real-time quantitative PCR assay. J Clin Microbiol. (2008) 46:2547–54. doi: 10.1128/JCM.02428-07
28. Decaro, N, Elia, G, Martella, V, Desario, C, Campolo, M, Di Trani, L, et al. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet Microbiol. (2005) 105:19–28. doi: 10.1016/j.vetmic.2004.09.018
29. Decaro, N, Desario, C, Lucente, MS, Amorisco, F, Campolo, M, Elia, G, et al. Specific identification of feline panleukopenia virus and its rapid differentation from canine parvoviruses using minor grove probes. J Virol Methods. (2008) 147:67–71. doi: 10.1016/j.jviromet.2007.08.006
30. Decaro, N, Elia, G, Desario, C, Roperto, S, Martella, V, Campolo, M, et al. A minor groove binder probe real-time PCR assay for discrimination between type 2-based vaccines and field strains of canine parvovirus. J Virol Methods. (2006) 136:65–70. doi: 10.1016/j.jviromet.2006.03.030
31. Katoh, K, Misawa, K, Kuma, KI, and Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. (2002) 30:3059–66. doi: 10.1093/nar/gkf436
32. Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
33. Trogu, T, Castelli, A, Canziani, S, Tolini, C, Carrera, M, Sozzi, E, et al. Detection and molecular characterization of canine distemper virus in wildlife from northern Italy. Pathogens. (2022) 11:1557. doi: 10.3390/pathogens11121557
34. Lorusso, A, and Savini, G. Old diseases for new nightmares: distemper strikes back in Italy. Vet Ital. (2014) 50:151–4. doi: 10.12834/VetIt.66.191.2
35. Martella, V, Cirone, F, Elia, G, Lorusso, E, Decaro, N, Campolo, M, et al. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet Microbiol. (2006) 116:301–9. doi: 10.1016/j.vetmic.2006.04.019
36. Pardo, ID, Johnson, GC, and Kleiboeker, SB. Phylogenetic characterization of canine distemper viruses detected in naturally infected dogs in North America. J Clin Microbiol. (2005) 43:5009–17. doi: 10.1128/JCM.43.10.5009-5017.2005
37. Demeter, Z, Lakatos, B, Palade, EA, Kozma, T, Forgách, P, and Rusvai, M. Genetic diversity of Hungarian canine distemper virus strains. Vet Microbiol. (2007) 122:258–69. doi: 10.1016/j.vetmic.2007.02.001
38. Marcacci, M, Ancora, M, Mangone, I, Teodori, L, Di Sabatino, D, De Massis, F, et al. Whole genome sequence analysis of the arctic-lineage strain responsible for distemper in Italian wolves and dogs through a fast and robust next generation sequencing protocol. J Virol Methods. (2014) 202:64–8. doi: 10.1016/j.jviromet.2014.02.027
39. Pomeroy, LW, Bjornstad, ON, and Holmes, EC. The evolutionary and epidemiological dynamics of the paramyxoviridae. J Mol Evol. (2008) 66:98–106. doi: 10.1007/s00239-007-9040-x
40. Beineke, A, Baumgartner, W, and Wohlsein, P. Cross-species transmission of canine distemper virus-an update. One Health. (2015) 1:49–59. doi: 10.1016/j.onehlt.2015.09.002
41. Weckworth, JK, Davis, BW, Dubovi, E, Fountain-Jones, N, Packer, C, Cleaveland, S, et al. Cross-species transmission and evolutionary dynamics of canine distemper virus during a spillover in African lions of Serengeti National Park. Mol Ecol. (2020) 29:4308–21. doi: 10.1111/mec.15449
42. Di Blasio, A, Irico, L, Caruso, C, Miceli, I, Robetto, S, Peletto, S, et al. Canine distemper virus as an emerging multihost pathogen in wild carnivores in Northwest Italy. J Wildl Dis. (2019) 55:844–56. doi: 10.7589/2018-09-226
43. Nikolin, VM, Wibbelt, G, Michler, FUF, Wolf, P, and East, ML. Susceptibility of carnivore hosts to strains of canine distemper virus from distinct genetic lineages. Vet Microbiol. (2012) 156:45–53. doi: 10.1016/j.vetmic.2011.10.009
44. Origgi, FC, Plattet, P, Sattler, U, Robert, N, Casaubon, J, Mavrot, F, et al. Emergence of canine distemper virus strains with modified molecular signature and enhanced neuronal tropism leading to high mortality in wild carnivores. Vet Pathol. (2012) 49:913–29. doi: 10.1177/0300985812436743
45. Sekulin, K, Hafner-Marx, A, Kolodziejek, J, Janik, D, Schmidt, P, and Nowotny, N. Emergence of canine distemper in Bavarian wildlife associated with a specific amino acid exchange in the haemagglutinin protein. Vet J. (2011) 187:399–401. doi: 10.1016/j.tvjl.2009.12.029
46. Alfano, F, Lanave, G, Lucibelli, MG, Miletti, G, D'Alessio, N, Gallo, A, et al. Canine distemper virus in Autochtonous and imported dogs, southern Italy (2014-2021). Animals (Basel). (2022) 12:2852. doi: 10.3390/ani12202852
47. Jo, WK, Peters, M, Kydyrmanov, A, van de Bildt, MWG, Kuiken, T, Osterhaus, A, et al. The canine morbillivirus strain associated with an epizootic in Caspian seals provides new insights into the evolutionary history of this virus. Viruses. (2019) 11:e894. doi: 10.3390/v11100894
48. Nikolin, VM, Olarte-Castillo, XA, Osterrieder, N, Hofer, H, Dubovi, E, Mazzoni, CJ, et al. Canine distemper virus in the Serengeti ecosystem: molecular adaptation to different carnivore species. Mol Ecol. (2017) 26:2111–30. doi: 10.1111/mec.13902
49. Alfano, F, Dowgier, G, Valentino, MP, Galiero, G, Tinelli, A, Nicola, D, et al. Identification of pantropic canine coronavirus in a Wolf (Canis lupus italicus) in Italy. J Wildl Dis. (2019) 55:504–8. doi: 10.7589/2018-07-182
50. Alfano, F, Fusco, G, Mari, V, Occhiogrosso, L, Miletti, G, Brunetti, R, et al. Circulation of pantropic canine coronavirus in autochthonous and imported dogs, Italy. Transbound Emerg Dis. (2020) 67:1991–9. doi: 10.1111/tbed.13542
Keywords: canine distemper virus, wildlife, badger, passive surveillance, Italy, phylogenetic analysis
Citation: Alfano F, Lucibelli MG, D’Alessio N, Auriemma C, Rea S, Sgroi G, Lucente MS, Pellegrini F, Diakoudi G, De Carlo E, Decaro N, Lanave G, Martella V and Fusco G (2025) Detection of canine distemper virus in wildlife in Italy (2022–2024). Front. Vet. Sci. 12:1527550. doi: 10.3389/fvets.2025.1527550
Edited by:
Jesús Hernández, National Council of Science and Technology (CONACYT), MexicoReviewed by:
Elena Maria Bozzetta, Istituto Zooprofilattico Soerimenata Piemonte Liguria Valle d’Aosta, ItalyJelena Prpić, Croatian Veterinary Institute, Croatia
Copyright © 2025 Alfano, Lucibelli, D’Alessio, Auriemma, Rea, Sgroi, Lucente, Pellegrini, Diakoudi, De Carlo, Decaro, Lanave, Martella and Fusco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Flora Alfano, ZmxvcmEuYWxmYW5vQGl6c21wb3J0aWNpLml0
 Flora Alfano
Flora Alfano Maria Gabriella Lucibelli
Maria Gabriella Lucibelli Nicola D’Alessio
Nicola D’Alessio Clementina Auriemma1
Clementina Auriemma1 Giovanni Sgroi
Giovanni Sgroi Maria Stella Lucente
Maria Stella Lucente Francesco Pellegrini
Francesco Pellegrini Georgia Diakoudi
Georgia Diakoudi Esterina De Carlo
Esterina De Carlo Nicola Decaro
Nicola Decaro Gianvito Lanave
Gianvito Lanave Vito Martella
Vito Martella Giovanna Fusco
Giovanna Fusco