- 1Department of Veterinary Medical Science, University of Parma, Parma, Italy
- 2Clinica Veterinaria Modena Sud, Modena, Italy
Introduction: Mesenchymal stromal cells (MSCs) hold great clinical potential in veterinary regenerative medicine. However, a notable gap exists in the literature regarding the isolation and characterization of these cells in reptiles. The objective of this study was to evaluate the feasibility of isolating adipose tissue-derived mesenchymal stem cells (MSCs) from pond slider (Trachemys scripta) tissue samples collected during routine neutering procedures.
Methods: Adipose tissue samples were obtained from five animals and processed using an enzymatic procedure. The resulting cell suspension was subsequently cultured at 28°C in a controlled atmosphere with 5% CO2. The cell growth rates were evaluated through direct counting of cells up to passage 7. The colony-forming unit (CFU) capacity of MSCs was evaluated in low-density cell cultures, and the ability of the cells to differentiate into adipogenic, chondrogenic and osteogenic lineages was assessed. The cell phenotype was characterized at the molecular level using reverse transcription-polymerase chain reaction (RT–PCR) and amplicon sequencing, with a focus on markers commonly used for gene expression profiling of mammalian MSCs.
Results: The cells demonstrated the capacity to differentiate into adipogenic, chondrogenic, and osteogenic lineages. RT–PCR revealed the expression of CD105, CD73, CD44, and CD90, whereas CD34 and HLA-DRA were not expressed. Sequence homology analysis demonstrated that the amplicons matched the sequences reported in the Trachemys scripta whole-genome shotgun sequence. This study represents the first investigation aimed at the isolation, in vitro expansion, and characterization of reptile adipose tissue-derived MSCs.
Discussion: The results demonstrate the feasibility of isolating MSC-like cells from chelonian adipose tissue and underscore their potential for application in regenerative medicine for both companion reptiles and endangered wild species.
1 Introduction
Mesenchymal stromal cells (MSCs) are multipotent cells that can be isolated and expanded from a range of tissues, including bone marrow, adipose tissue, the umbilical cord, and the placenta (1). Initially investigated as pro-regenerative therapeutic agents based on their capacity to differentiate into multiple cell types, including osteoblasts, chondrocytes, and adipocytes, they have since been evaluated primarily for their immunomodulatory potential (2, 3). MSCs have the capacity to modulate immune responses, reduce inflammation, and promote tissue healing through paracrine signaling, rendering them an appealing option for the treatment of a multitude of diseases (4). In veterinary medicine, MSCs have been applied in the treatment of musculoskeletal diseases, as well as in organ and systemic pathologies where acute and chronic inflammation processes play pathogenic roles. A non-exhaustive list of veterinary pathologies investigated with MSCs includes orthopedic and musculoskeletal diseases (osteoarthritis, tendon and ligament injuries, desmopathies), neurological diseases (spinal cord injuries, degenerative myelopathy) and immune-mediated and inflammatory diseases (canine and feline inflammatory bowel disease, feline chronic gingivostomatitis, atopic dermatitis, chronic kidney disease) (5). Additionally, MSCs have been utilized in the management of ophthalmic and cardiac diseases, as well as chronic non-healing wounds (5). One of the most noteworthy characteristics of MSCs is their capacity to serve dual roles in both regenerative and immunomodulatory therapies. They are capable of secreting a diverse array of bioactive soluble molecules, including immunomodulatory cytokines and growth factors, and producing extracellular vesicles (EVs), which facilitate the modulation of the local immune environment, promote healing, and prevent excessive scarring (3). MSCs are isolated and expanded from a variety of tissues, including bone marrow, adipose tissue, peripheral venous blood, endometrium, skin, dental pulp, and perinatal tissues such as the umbilical cord, amniotic membranes, and fluid. Adipose tissue and bone marrow represent the most extensively investigated sources of cells for clinical application, offering an accessible route for the isolation of autologous MSCs from adult patients (1, 6).
Although the literature has documented the use of blood-derived products such as thrombocyte–leukocyte-rich plasma (TLRP) as adjunctive regenerative therapies for various lesions in reptiles (7–9), there is a significant gap in research concerning the isolation and characterization of MSCs within this taxonomic class. This study represents the first attempt to isolate, expand, and characterize MSCs derived from reptilian adipose tissue. The feasibility of isolating turtle MSCs with characteristics similar to those described for mammalian species was explored starting from adipose tissue samples collected during routine Trachemys scripta neutering. When possible, the cells were characterized using the methods described for isolation from species of veterinary interest. The objective of this study was to evaluate the possibility of applying MSC-based regenerative medicine protocols to companion reptiles and endangered wild species.
2 Materials and methods
2.1 Ethics statement, species, and animal selection criteria
Ethical approval for the study was given by the University of Parma ethical committee (PROT. No. 15/CESA/2022). Five clinically healthy adult female pond sliders (Trachemys scripta) aged 20–30 years were presented to the Department of Veterinary Science, University of Parma, Italy, for routine spaying. T. scripta is not a reptile species of relevant clinical interest in the veterinary clinic but was used in the present study as a model reptile. In Italy, T. scripta is considered an alien invasive species because it competes with native species for food resources and habitats. To manage the Trachemys scripta population, a national control plan (National Plan for the Management of the American Marsh Tortoise (Trachemys scripta) 2020 [Decree of the Minister for Ecological Transition No. 370 of September 28, 2022 (10)] has been approved that requires the sterilization or culling of individuals in the territory (11, 12). The study involved the enrolment of subjects at the University of Parma Teaching Veterinary Hospital (OVUD) by the local ENPA, an Italian national association for the protection of animals that have been rescued from various situations (abandonment, found in parks, and public places). During the sterilization process, peri-ovarian adipose tissue samples were recovered. The tissue would have otherwise been discarded as a consequence of the sterilization procedure. No patients underwent surgery with the specific purpose of harvesting adipose tissue. The animals were weighed with an analytical balance (VEVOR® Digital Precision Scale 5,000 g × 0.01 g, Taicang Vevor Industry Co., Ltd. 9448 Richmond Pl, Rancho Cucamonga, CA, 91730, USA) from 1 to −23 kg [2.20 to −50.70 lb; median, 1.68 g (3.70 lb)] and measured from 22 to −28 cm (8.66 to −11.02 inches) [median 25.9 cm (10.19 inches)] along the curved carapace length (CCL). All the subjects underwent blood sampling from the jugular vein (13) of ~0.5 mL (1% live weight) (14). The sample was collected in tubes containing lithium heparin as an anticoagulant and used to perform a microhaematocrit for the evaluation of packed cell volume (PCV%), white blood cell count (WBC) using the Natt and Herrick stain method (15) and biochemical examination of the plasma prior to surgery to quantify analytes such as blood urea nitrogen (BUN), K+, total protein (TP), bile acids (BAs) and uric acid (UA). Exclusion criteria: For each animal, a dorsoventral X-ray projection was performed. Animals with eggs, a PCV < 20% or >35%, a WBC >19 × 103/μL, and a TP < 1.5 or >8 g/dL were excluded from the study (15). Ultimately, no animals were excluded.
2.2 Surgical procedure
The animals were hospitalized 1 week before surgery, housed individually in plastic containers, and acclimatized with light and heat from 50-W metal halide lamps (Reptifood S.r.l., Roma, Italia). Food was withheld 48 h before surgery. Room temperature was maintained between 25 and 28°C, and body temperature was monitored using a cloacal digital thermometer and maintained at 28°C. Each animal was identified with a unique number, and candidates were selected for surgery through simple randomization using randomizer software (16). A combination of ketamine (10 mg/kg) (Acme S.r.l., Milano, Italy), dexmedetomidine (50 μg/kg) (Vétoquinol Italia S.r.l., Bertinoro, FC, Italy), and midazolam (1 mg/kg) (Hameln Pharmaceuticals, Gloucester, UK) was administered intramuscularly in the left forearm. For multimodal analgesia, meloxicam (0.2 mg/kg) was given 1 h before anesthesia (17, 18). A venous catheter was placed in the jugular vein for access, and the subjects were monitored with Doppler, ECG, and capnography to assess anesthesia depth. The eyelid reflex, mandibular tone, neck extension, and limb retraction were checked every 5 min to evaluate analgesia and recovery. Loss of limb retraction indicated adequate anesthesia, whereas mandibular tone loss allowed intubation. In the event of failure to lose mandibular tone, propofol was used as an induction agent at 5 mg/kg (18, 19). During surgery, the subjects were maintained on halogenated gases (1.5% isoflurane, Zoetis Italia S.r.l., Rome, Italy) and manually ventilated at 4 breaths per min. At the end of the procedure, dexmedetomidine was antagonized with atipamezole (0.5 mg/kg) intramuscularly (20). The subjects were positioned in left lateral recumbency and secured with foam support and tape, with their hind limbs extended and taped together to expose both flank fossae. A surgical scrub was performed on the prefemoral fossa and adjacent areas. A 2 cm cranio-caudal incision was made in the center of the prefemoral fossa using a no. 11 blade (Mealli S.r.l., Firenze, Italy), followed by subcutaneous dissection. Approximately 0.5–1 g of adipose tissue was aseptically collected from each animal before it reached the aponeurosis of the abdominal muscles for coelomic access (Figure 1). The right ovary and oviduct were identified using a 30° oblique telescope with an operative sheath (Endoscopia Italia S.r.l., Verona, Italy) and then exteriorized with sterile swabs. Vessels of the ovarian pedicle, mesovarium, and mesosalpinx were cauterized and excised with an electrocoagulation device (EnSeal®, Alcyon Italia, Rome, Italy). The oviduct was ligated ~3 cm from the cloacal opening and removed. The muscle layers were closed with simple continuous sutures, and the skin layers were closed with interrupted sutures, both of which used a 2/0 absorbable monofilament (Braun Avitum Italy S.p.A. Mirandola, Italy). The procedure was repeated on the opposite side. Finally, 0.5 mg/kg atipamezole (Fatro, Ozzano dell'Emilia, Italy) was administered as a reversal agent (21), along with a single 10 mg/kg dose of tramadol IM and 0.2 mg/kg meloxicam SC once daily for 5 days. No complications were observed during the surgical procedure or during recovery in any of the animals.
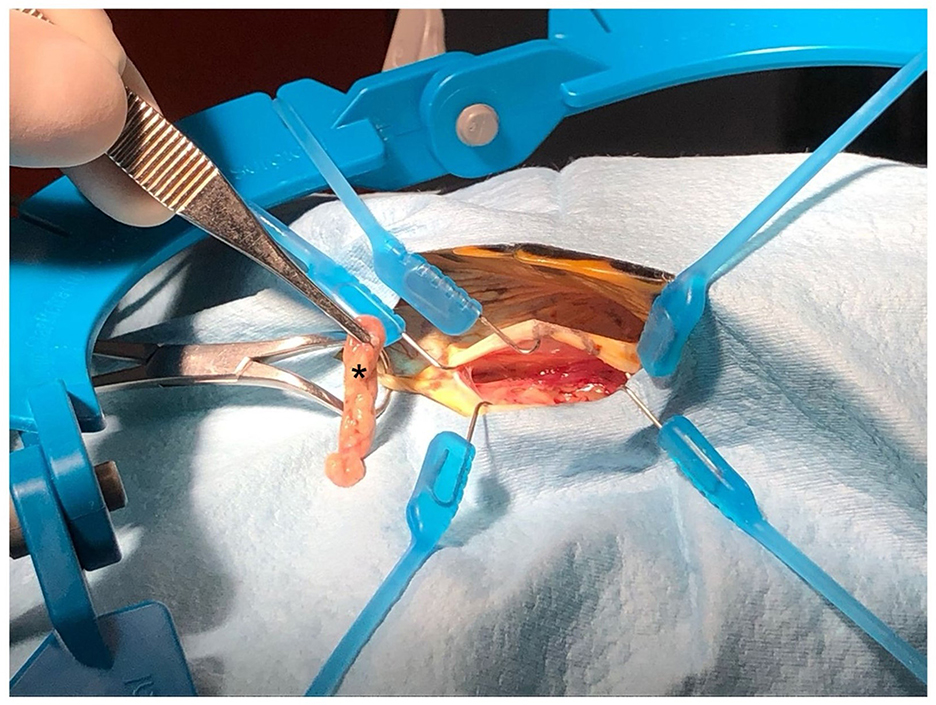
Figure 1. Collection of adipose tissue samples (fat pads of the prefemoral fossa) from Trachemys scripta during neutering surgery. Note the brownish colouration of the tissue (asterisk).
2.3 Histological evaluation of the adipose tissue sample
To confirm the actual nature of the obtained adipose tissue, a small fragment of the sample measuring ~0.5 cm3 was fixed with 10% neutral buffered formalin solution, dehydrated in a graded series of ethanol to absolute alcohol, cleared with xylene and embedded in paraffin. Five-micrometer-thick sections were stained with haematoxylin and eosin (22).
2.4 Isolation and culture of Trachemys scripta AD-MSCs (Ts-AD-MSCs)
The media, supplements, and other cell culture reagents used for cell culture were obtained from Gibco (Thermo Fisher Scientific Inc., Waltham, MA, USA) unless otherwise specified. The plastic labware used was from VWR (Avantor, Radnor, OR, USA).
2.5 Enzymatic digestion and setup of bidimensional and three-dimensional cultures
To isolate MSCs from the adipose tissue of Trachemys scripta, an enzymatic digestion protocol adapted from established mammalian methods was employed (23). The adipose tissue samples (0.56 ± 0.22 g) were briefly washed in 70% ethanol to remove bacterial contaminants and then transferred into phosphate-buffered saline (PBS), pH 7.4, to completely remove any residual ethanol. Before digestion, the tissue samples were mechanically minced into ~1 mm3 fragments using a no. 19 sterile surgical blade. The fragments were then placed in 15 mL Falcon tubes containing 5 mL of prewarmed DMEM enriched with 0.1% type I collagenase (w/v), and the tubes were placed at 37°C in an orbital shaking incubator set at 80 RPM for 1 h. After digestion, 5 mL of DMEM supplemented with 10% fetal bovine serum (FBS), penicillin (100 μg/mL), streptomycin (100 μg/mL), and amphotericin B (2.5 μg/mL) (maintenance DMEM, mDMEM) was added to the tubes to stop collagenase digestion, after which the tubes were centrifuged at 400 × g for 10 min. The supernatant was removed, and the cell pellet was resuspended in 1 ml of mDMEM. The cell suspension was then divided into two 0.5 mL aliquots. Each aliquot was seeded in a 3.5 cm diameter Petri dish containing 2 mL of mDMEM. The cell cultures were then placed in a dedicated incubator with a controlled atmosphere of 5% CO2 at 28°C. At the time of adipose tissue processing, small fragments were seeded into a three-dimensional gel to test the ability of the cells to migrate outwards from the tissue. Prior to collagenase digestion, the fragments were collected and seeded in a 3D gel composed of 10% calcium gluconate, 40% serum-free DMEM, and 50% plasma. Tissue fragments were cultured in a 3D environment exclusively to assess their viability. All characterization analyses of the obtained MSC-like populations were performed on cells obtained by digestion using the collagenase-based protocol.
2.6 Cell culture and amplification
The first medium change occurred 48 h after the seeding of the cell suspension. The maintenance DMEM was then replaced every 48 h to facilitate cell expansion. Once the cells reached ~80–90% confluence, they were detached with 0.3 mL of 0.05% trypsin-EDTA. Cell counting was performed manually at each passage using a Burker chamber. After counting, 8,000 cells/cm2 were seeded into two 25 cm2 cell culture flasks, which were maintained at 28°C in an atmosphere with 5% CO2. This process was repeated whenever the cells reached 90% confluence until passage 8.
2.7 Cryopreservation
After trypsinization, the cells were collected by centrifuging the cell suspension at 190 × g for 10 min. The supernatant was discarded, and the resulting pellet was resuspended in 1 mL of a freezing mixture composed of high-glucose DMEM supplemented with 50% FBS, penicillin/streptomycin (100 U/mL, 100 μg/mL), amphotericin B (2.5 μg/mL), and 10% dimethyl sulfoxide (DMSO). The cell suspension was then transferred into cryovials and placed in a freezing container (Mr Frosty, Fisher Scientific Italia, Segrate, Italy) at −80°C. The cells were then transferred to liquid nitrogen for long-term storage.
2.8 White blood cell isolation
White blood cells were used as a positive control to assess the expression of negative MSC markers (24). Two milliliters of venous blood was collected in sodium citrate from the caudal or cervical vein and processed within 1 h, following a protocol proposed by Di Ianni et al. in 2015 (9). Briefly, whole blood was carefully layered over a prewarmed Histopaque 1.077 solution at 28°C, avoiding mixing of the two fluids. The blood-to-histopaque volume ratio was maintained at 1:1. Each sample was then centrifuged at 150 × g for 20 min without using brakes. After centrifugation, the WBC cellular fraction was visually identified as a white ring at the interface with the Histopaque solution and was carefully collected to avoid contamination with the underlying layers. The cells were washed twice with 5 mL of sterile PBS and then centrifuged at 250 × g for 10 min. The resulting pellet was frozen at −80°C until it was used for gene expression analysis.
2.9 In vitro expansion potential
The growth curves of adipose tissue samples from five different individuals were analyzed to assess the in vitro expansion potential of the isolated cells. The evaluation of cellular growth parameters was conducted from passage 1 (P1) to passage 7 (P7). In our experimental setup, the seeding of the cell population freshly isolated from the tissue was considered P0. P1 indicates the first passage of the cells, meaning the first time they were detached, split, and reseeded into new flasks.
For each passage, 8,000 cells per square centimeter (8,000 cells/cm2) were seeded into two 25 cm2 flasks. Once the cells reached a semiconfluent state (80–90%), they were trypsinized and counted. The cells were then reseeded at a density of 8,000 cells/cm2, and this process continued through passage 7 (P7). The cell-doubling number (CDn) and cell-doubling time (DT) were calculated as suggested by Vidal et al. (25) and Roth (26). The two parameters were evaluated as follows: cell doubling number (CDn): ln (Nf/Ni)/ln2, and doubling time (DT): CT/CDn.
2.10 Colony-forming unit (CFU) assay
The CFU assay was performed on five different cell preparations at P2 and P5. For the CFU assay, 6-well plates were used; 100 and 200 cells/well were seeded in triplicate in mDMEM. The plates were incubated at 28°C with 5% CO2 for 14 days, and the DMEM was replaced every 72 h. After 14 days, the DMEM was removed, and the cells were fixed in 4% v/v formaldehyde for 1 h. The wells were subsequently stained using May–Grunwald Giemsa staining (22) to enable CFU counting. A CFU was defined as a cluster of at least 50 cells, and the counts were performed following the protocol described by Secunda et al. (27). Briefly, the wells were washed with distilled water, and 2 mL of May–Grunwald Giemsa stain (0.2 mL of Giemsa azure–eosin–methylene blue solution per mL of distilled water) was added for 12 min. After that, the wells were washed with distilled water, and colony counting was performed.
2.11 Trachemys scripta AD-MSC differentiation
Due to the lack of studies evaluating the differentiation capacity of adipose-derived cells in reptiles, protocols routinely used in mammals have been employed (27, 28). Differentiation assays were conducted to evaluate the potential for adipogenic, osteogenic, and chondrogenic differentiation. Each assay was performed in 6-well plates, with an initial seeding density of 6,000 cells/cm2. For each differentiation assay (n = 3; cell passage P3), a corresponding control was prepared, with a glass coverslip placed at the bottom of each well to facilitate cell staining and subsequent microscopic observation. To this aim glass coverslips were rinsed with distilled water and then immersed in a 0.1 N HCl solution for 2 h. Afterward, the coverslips were washed several times with distilled water and subsequently immersed in a 70% ethanol solution for 10 min. Finally, the coverslips were rinsed with sterile water and sterilized in an oven at 180°C for 2 h. Adipogenic differentiation treatment started once the cells reached ~70–80% confluence. The differentiation medium consisted of high-glucose DMEM supplemented with 10% FBS, antibiotics, and antifungal agents as described above and a mixture of 1 μM dexamethasone, 0.2 mM indomethacin, 0.5 μM IBMX (3-isobutyl-1-methylxanthine), and 1.7 μM insulin (10 μg/mL). This differentiation medium was maintained for 3 days, after which it was replaced with maintenance medium containing high-glucose DMEM supplemented with 10% FBS, antibiotics, antifungal agents, and 1.7 μM insulin (10 μg/mL) on the 4th day. On the 5th day, the medium was replaced with differentiation medium. This cycle continued until Day 24, when the cells were fixed with formalin and stained with oil red O (29). For chondrogenic differentiation, the StemPro® Chondrogenesis Differentiation Kit (GIBCO) was used, with medium changes occurring every 3 days according to the manufacturer's guidelines. After the differentiation period (21 days), the cells were fixed in formalin and stained with Alcian blue (29). Osteogenic differentiation was induced when the cells reached 80–90% confluence using a medium consisting of high-glucose DMEM supplemented with 10% FBS, antibiotics, antifungal agents, 100 nM dexamethasone, 10 mM glycerophosphate, and 0.250 mM ascorbic acid. The medium was replaced every 48 h until Day 31. Following treatment, the cells were fixed with 95% methanol and stained with Alizarin red (29).
2.12 Gene expression analysis
To evaluate the phenotype of the cell populations, the expression levels of a panel of genes were evaluated by reverse transcription–PCR (RT–PCR), following the pattern of characterization suggested for mammalian cells (30). For total RNA extraction, the Macherey–Nagel NucleoSpin RNA Kit was used (31). RNA was extracted from P3 cell cultures (0.8–1 × 106 cells) as well as from leukocytes isolated using a previously described procedure following the manufacturer's protocol. The total RNA obtained was quantified using the GeneQuant-pro RNA/DNA Calculator (Cambridge Scientific, Watertown, MA, USA), and 2 μg of RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™, Cheshire, UK) following the manufacturer's guidelines. The primers used for RT–PCR were designed using the NCBI database with a dedicated software tool (Primer-BLAST, https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on March 18, 2024). The sequences were derived from the NCBI Reference Sequence database, based on the genome of Trachemys scripta, which was fully sequenced and published in 2020 (32). The primers, shown in Table 1, were synthesized by Macrogen Europe, Milan, Italy.
For gene expression analysis, nine genes were evaluated. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene to validate the experimental results. CD44, CD73, CD90, and CD105 were used as positive markers for MSCs, whereas CD31, CD34, CD45, and HLA-DRA were used as negative markers. As a positive control for the expression of negative markers, cDNA from leukocytes was used (33). For RT–PCR master mix preparation, 1 unit of DreamTaq™ DNA Polymerase (Thermo Fisher Scientific, Baltics UABA V.A. Vilnius, Lithuania) was used, and 2 μg of cDNA, 1.125 μL per primer (from a 25 μM stock) and 1 μL of dNTPs (from a 10 mM stock) were added, resulting in a final reaction volume of 50 μL. Before sequencing, the presence of the expected amplicons was verified by 1.5% agarose gel electrophoresis. Amplicons were visualized using a UV transilluminator.
2.13 Sequencing
After confirming the expression of the individual genes by gel electrophoresis, an aliquot of each amplicon was sequenced using the Sanger sequencing service at Eurofins Genomics, Milan, Italy. The sequencing results were validated by comparing the obtained sequences with the specific sequences published on the NCBI website. The specific reference number for each gene is reported in Table 1. Sequence comparison was performed using the NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on September 16, 2024). A manual evaluation of the chromatographic traces from the sequencing was also conducted to confirm the results.
2.14 Statistical analysis
Statistical analysis was performed using the software Jamovi (version 2.5.6). Data from the cell proliferation assays were assessed for normality (Shapiro–Wilk test) and homogeneity of variance (Levene's test). Because the data were normally distributed, statistical analysis was conducted using one-way ANOVA, followed by Tukey's post-hoc test when resulted statistically significant (p ≤ 0.05). Data from the CFU counts were analyzed using a t-test. Given that these datasets were not normally distributed, a non-parametric Mann–Whitney U-test was applied for analysis.
3 Results
3.1 Histological evaluation of the samples and cell adhesion, migration, and CFU assays
To confirm the actual nature of the adipose tissue samples, five-micrometer-thick sections were subjected to haematoxylin and eosin (H&E) staining. The analysis revealed numerous clusters of mature adipocytes associated with fibrous tissue, blood vessels, lymphatics, and perivascular chromatophores (Figure 2). After confirming the adipose tissue nature of the collected samples, the behavior of the cell population obtained following enzymatic digestion was evaluated.
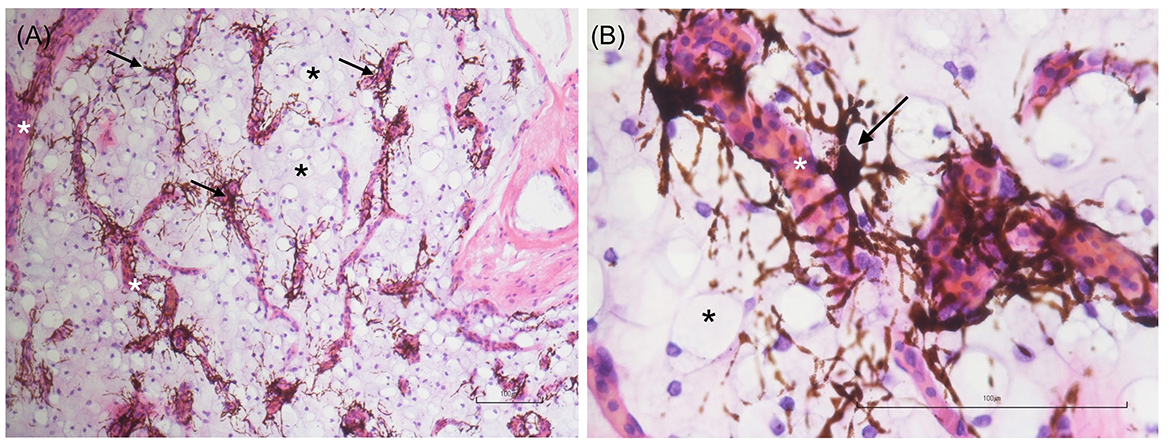
Figure 2. Low- (A) and high-magnification (B) images of adipose tissue sections stained with hematoxylin and eosin (H&E). Numerous mature adipocytes (black asterisks) and chromatophores (black arrows) arranged in a perivascular pattern are evident. The white asterisk indicates a blood vessel. Scale bar: 100 μm.
Spindle-shaped cells adhering to the culture plate surface were observed in all examined samples (Figure 3A) ~5–7 days after seeding the cellular pellets derived from the enzymatic digestion of adipose tissue. This morphology remained consistent throughout all the cell passages (P1 to P7). Over time, the adherent cells tended to form CFUs, eventually leading to full plate colonization (Figure 3B). In 3D cultures derived from the direct fragmentation and seeding of undigested adipose tissue, the first cellular entities began migrating from the fragments ~5–6 days postseeding (Figure 3C), fully colonizing the surrounding area in the following days. As shown in the image, the cells continued expanding until they encountered cellular populations from other fragments (Figure 3D). For the CFU formation assay, colony assessments were conducted at two different passages (P2 and P5). The average values obtained from seeding 100 and 200 cells across three different passages are shown in Figure 4. No statistically significant differences were observed in CFU counts between the two passages.
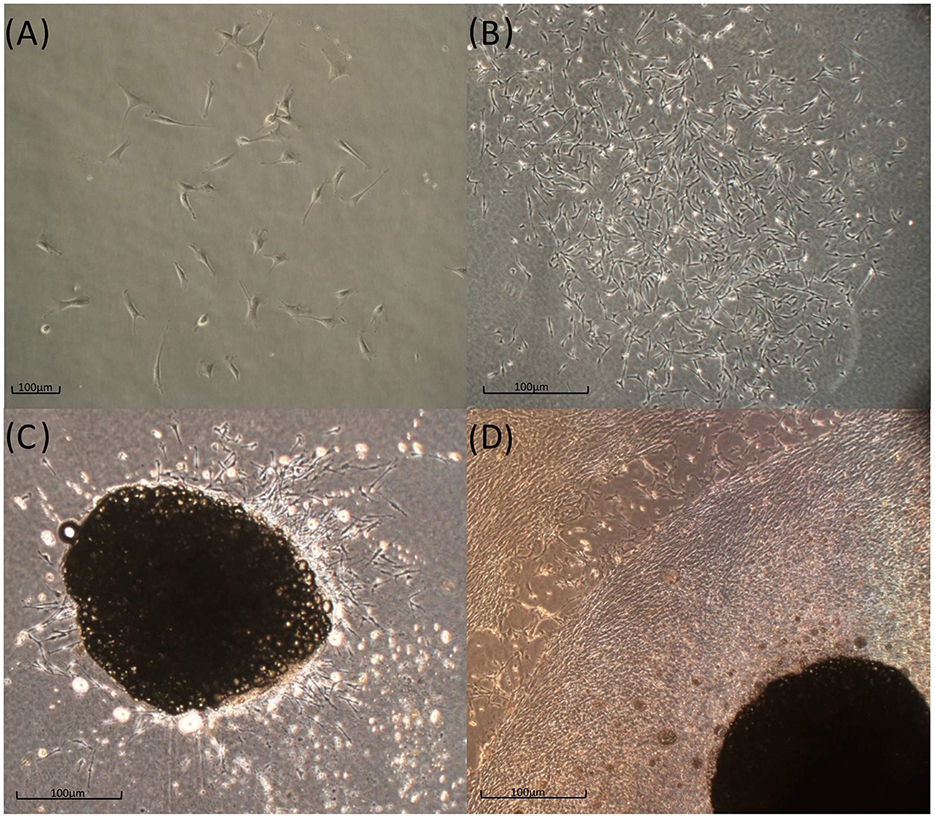
Figure 3. In vitro T. scripta MSC phenotype. Cells that were expanded in the 2D environment had a fibroblast-like appearance and tended to form small CFUs 5–7 days postseeding (A), which gradually expanded in the following week (B). Tissue fragments cultured in a 3D fibrin-derived environment show an outflow of fibroblast-like cells (C, Day 7) that gradually tend to occupy all the available space (D, 3 weeks).
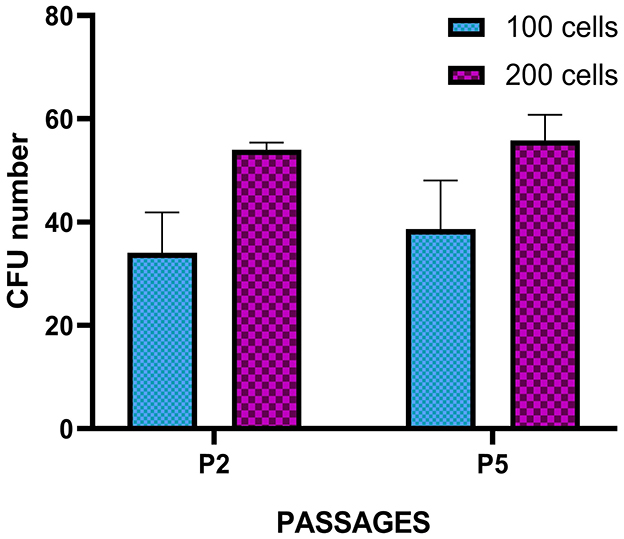
Figure 4. Colony-forming unit (CFU) assay of T. scripta MSCs. Colony formation was observed in MSCs seeded at 100 and 200 cells/well at P2 and P5. The results represent the mean ± standard deviation of three independent experiments. No significant difference was observed between P2 and P5 or between the different cell densities evaluated.
3.2 Growth curves
Following the protocol described in Section 2.9, cell growth curves were evaluated by analyzing the growth curves of the five samples across successive passages, up to P7. For each passage, key parameters were evaluated, including the cell doubling number (CDn: ln (Nf/Ni)/ln2), the number of cell doublings per 24-h period (CDn/24 h), the cumulative number of cell doublings across all passages (total CDn: CDnP1 + CDnP2… + CDnPx), and the population doubling time (DT: CT/CDn). The average CDn/24 h, total CDn and DT values from passages P1–P7 were compared, and the results are reported in Figures 5A–C. Statistical analysis revealed no significant differences in cell growth rates between passages. Additionally, the consistent cell growth observed for total CDn (Figure 5B) suggested a continuous proliferation rate from P1–P7, with no plateau phase reached. Similarly, no statistically significant differences were observed in the doubling times (DTs, Figure 5C), further confirming the ability of these cell populations to sustain steady replication through the final passage tested. Furthermore, no statistically significant differences were identified in the parameters evaluated across the different animals considered.
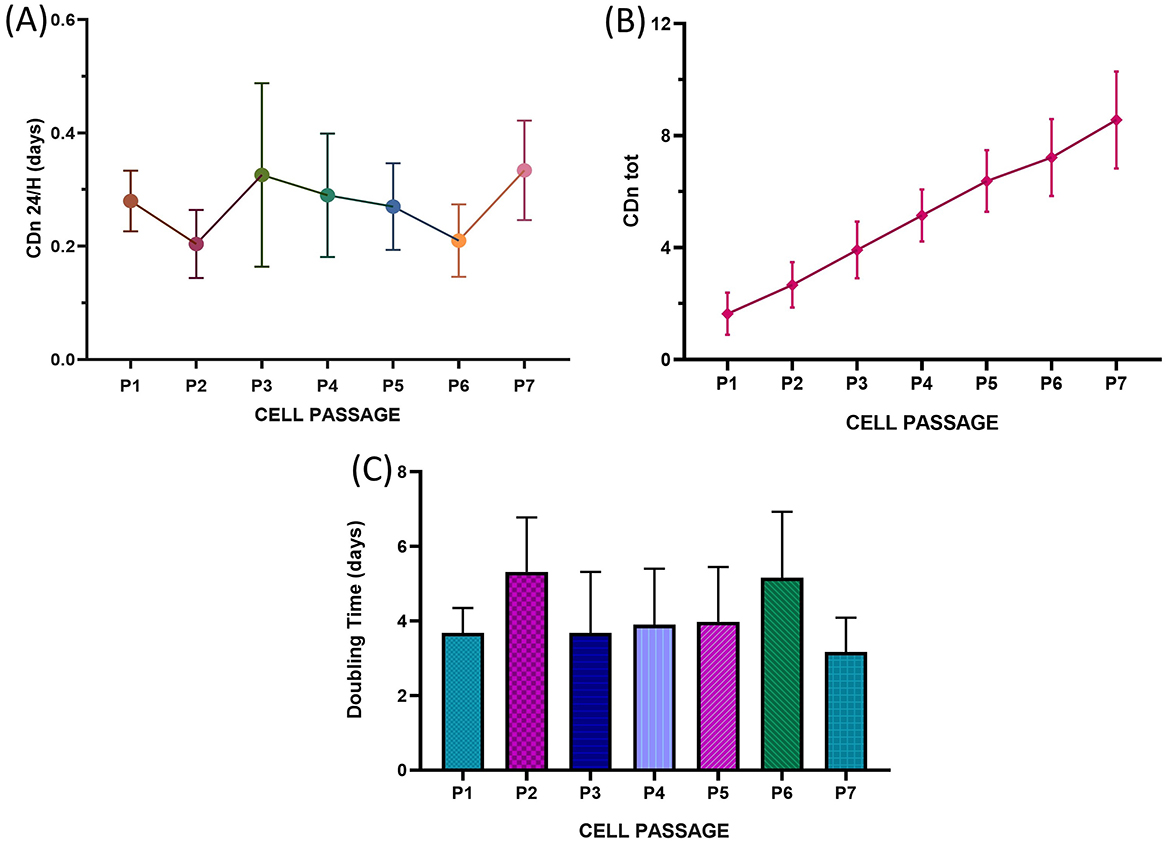
Figure 5. Long-term growth curves of T. scripta MSC-like cells. The fraction of cells that underwent cell replication at 24 h (CDn 24 h) (A), total cell doubling number (CDn tot) (B) and mean doubling time (days, C) are reported from P1 to P7. No statistically significant differences were observed between the different cell passages.
3.3 Differentiation capacity of TS-ADMSCs
According to the differentiation protocols and staining procedures utilized, Ts-AD-MSCs are able to differentiate toward the adipogenic, osteogenic, and chondrogenic lineages. Figure 6A shows the accumulation of intracellular lipids, which is indicative of successful adipogenic differentiation. Figures 6E, E′ display the presence of cartilaginous matrix deposits positive for Alcian blue staining, indicating the chondrogenic differentiation of the MSCs. Figures 6C, C′ reveal bone matrix-like deposits, indicating the osteogenic differentiation of MSCs. A negative, unstimulated control was evaluated for each differentiation protocol (Figures 6B, D, F).
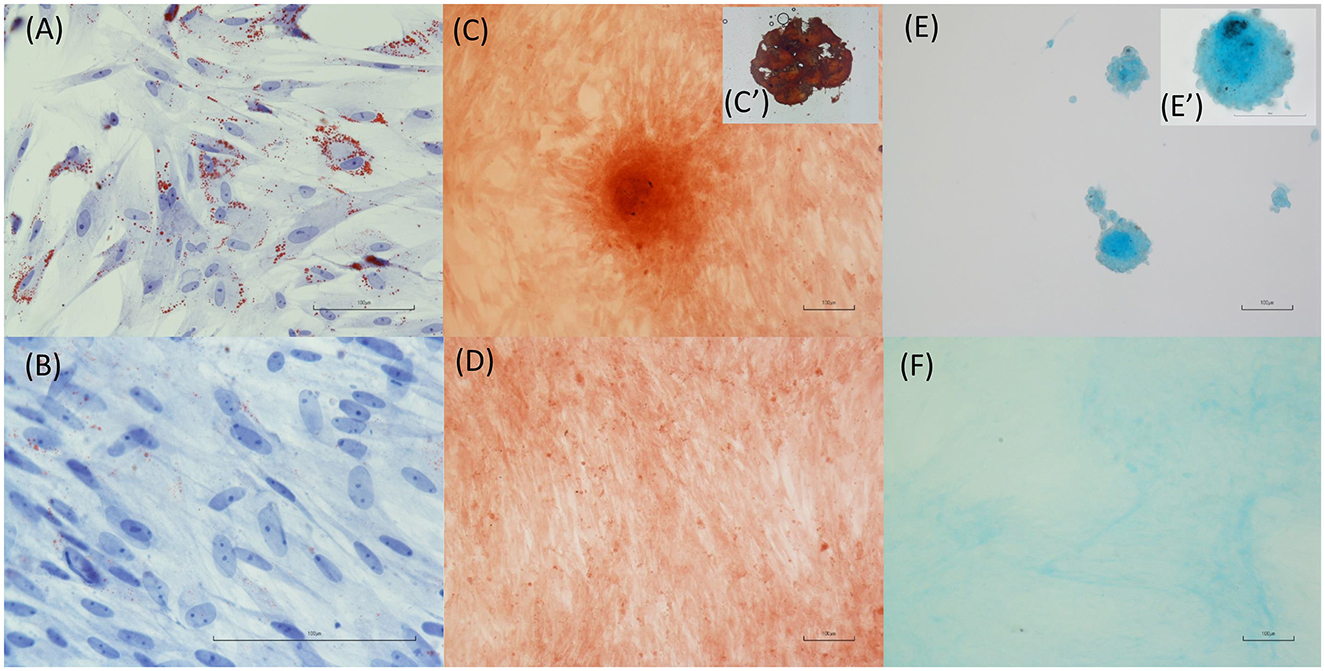
Figure 6. Differentiation capacity of T. scripta MSCs. Intracellular lipid accumulation (A, oil red O staining) is indicative of successful adipogenic differentiation. Osteogenic differentiation is supported by the staining of bone matrix-like deposits (C, C′, Alizarin red staining) in cells exposed to osteogenic medium. Finally, cartilage matrix formation (E, E′) is evidenced by Alcian blue staining of cell cultures treated with chondrogenic differentiation medium. Unstimulated cell cultures were used as a control (B, D, F).
3.4 Gene expression analysis and sequencing
The identification of MSC populations in mammals typically relies on the analysis of gene expression for a panel of markers defined by the International Society for Cellular Therapy (ISCT) (30, 34). Given the unavailability of species-specific antibodies for Trachemys scripta, gene expression was analyzed using reverse transcription polymerase chain reaction (RT–PCR). Specific primers for the relevant markers were designed in accordance with the methodology outlined in the Materials and Methods. Polymerase chain reaction (PCR) was conducted on four distinct samples between passages P4 and P7, with three leukocyte samples serving as positive controls for markers anticipated to be negative in mesenchymal stem cells (MSCs). The resulting amplicons were subjected to sequencing to verify their alignment with known gene sequences in databases. For marker genes whose expression was not expected in MSCs (e.g., CD45, CD34, and HLA-DRA), expression was evaluated in white blood cell populations to validate the primers. The cells derived from adipose tissue and expanded in culture presented positive expression of CD90, CD105, CD44, and CD73, whereas CD34 and HLA-DRA expression was absent (Figure 7A). The presence of CD34 and HLA-DRA in white blood cells served to validate the accuracy of the primers (Figure 7B). The correspondence between the obtained sequences and the original sequences in the NCBI database was 100% for CD44, CD34, and CD73, 99% for CD105, CD90, and GAPDH, confirming that the obtained amplicons matched the expected sequences. The correspondence level of HLA-DR was 95% suggesting the possibility of natural polymorphisms or genetic diversity within the species. Amplicon sequences are reported in Supplementary material. CD45 expression was undetectable in both MSCs and leukocytes. CD31 was weakly expressed in MSCs, but sequencing did not confirm a match with the database sequences (data not shown). In this case, a possible discrepancy between the database sequence used for primer picking and the primers used is a potential explanation.
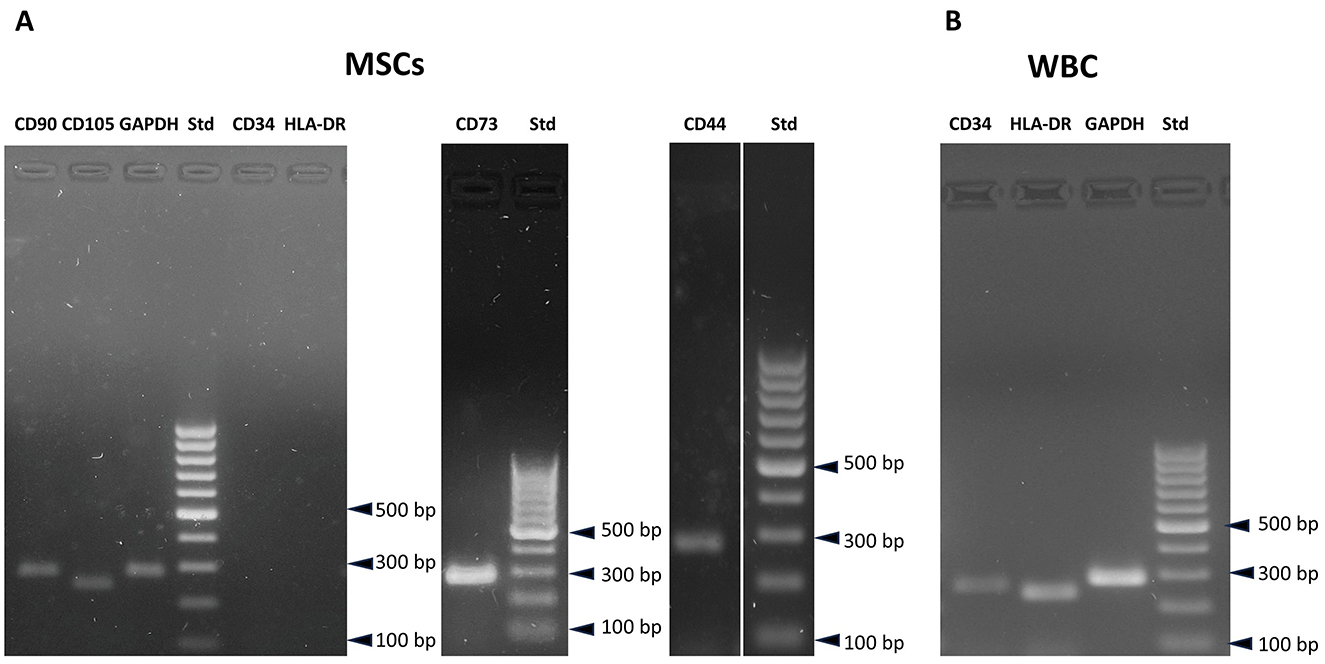
Figure 7. Gene expression analysis of T. scripta MSCs. MSCs are positive for CD90, CD73, CD105, and CD44 expression, while CD34 and HLA-DRA expression is not observed (A). CD34 and HLA-DRA expression is positive in white blood cells (B).
4 Discussion
In this study, MSC-like cells were isolated and expanded from the adipose tissue of red-eared sliders (Trachemys scripta) obtained during routine ovariosalpingectomy. The aim of this study was to evaluate the potential use of this species as a model for developing laboratory protocols to collect, isolate, expand, and characterize MSCs for future applications in other reptile species. We propose a method for adipose tissue collection that, while necessarily requiring a surgical procedure, can be considered minimally invasive for the animal. In these turtles, adipose deposits accumulate subcutaneously in the prefemoral region, which serves as the preferred access site for laparoscopic celiotomy procedures. This anatomical feature allows for efficient tissue sampling with minimal trauma (35). There is currently no detailed description in the literature of the structure of subcutaneous adipose deposits in the genus Trachemys. To confirm the nature of the collected adipose tissue, sample fragments were processed for histological analysis. Haematoxylin and eosin (H&E) staining revealed numerous clusters of mature adipocytes associated with fibrous tissue, blood vessels, lymphatics, and chromatophores (see Figure 2). The variable presence of chromatophores in the samples might be responsible for the brownish color typical of the adipose deposits collected. To evaluate the basic biological characteristics of the isolated cells, their cellular morphology, growth rate, ability to generate CFUs, trilineage differentiation capacity, and expression of a panel of gene markers suggested by the ISCT were analyzed (30). The protocol for in vitro isolation and expansion used in this study has been validated in mammals (36, 37) but not in reptiles. For example, reptiles are ectothermic animals (38); therefore, incubation parameters typically used for mammalian cells (37°C) are not applicable. To the best of our knowledge, to date, no studies have been published on the isolation, in vitro expansion, and identification of MSCs in this class of animals. However, studies in the literature have focused on toxicology involving fibroblast cultures from loggerhead turtles (Caretta caretta) (39, 40). Webb et al. (39) successfully isolated and characterized fibroblasts from the skin of loggerhead turtles. In their study, they analyzed various incubation temperatures for cultured cells (25, 30, 35°C) and demonstrated that 30°C was optimal for cell proliferation. Other studies suggest incubation temperatures between 26 and 30°C for marine turtle cell cultures (41). These data support the hypothesis that the ideal incubation temperature for cells, which allows for proliferation while minimizing stress and ensuring maximum survival during various passages, varies among not only different classes of animals but also different species. In the absence of validated protocols for obtaining MSCs from Trachemys scripta adipose tissue, we chose 28°C as the incubation temperature, which is the average value of the optimal thermal range for this species (41). It is also not surprising that this incubation temperature (28°C) has been used to expand cell cultures from other classes of ectothermic animals (amphibians and fish) (24, 42). Recently, a study by Otsuka-Yamaguchi et al. (24) demonstrated the feasibility of obtaining and characterizing MSC-like cells from the bone marrow of Xenopus laevis, an amphibian used as an experimental model in various fields of medicine owing to its ease of breeding, prolific nature, and excellent tissue regenerative capabilities (43). In the study by Otsuka-Yamaguchi et al., bone marrow was directly extracted from the bones of euthanized animals by mechanical fragmentation. In chelinators, bone marrow is contained within the spongy bone not only of the long bones and vertebrae but also of the carapace, bridge, and plastron (14, 44). Extracting bone marrow from these species therefore requires drilling into these structures to reach the bone cavities and access various cellular components. This procedure is invasive and painful, and it would also be challenging to obtain a sufficient sample size to isolate a significant number of MSCs for clinical application. Therefore, for ethical reasons, this method was excluded from this study. Instead, we evaluated the possibility of harvesting adequate quantities of adipose tissue during neutering surgeries. This approach was found to be feasible and not overly traumatic for the animals. We used two methods for cell isolation: collagenase digestion and mechanical fragmentation of adipose tissue followed by 3D culture. Both methods have been validated and described in mammals (28, 45, 46). Given the average size of a Trachemys scripta, it was not possible to obtain large amounts of adipose tissue from a single subject ( ≤ 1 g per subject). For the culture medium, DMEM was chosen because it is widely used for culturing MSCs in mammals, zebrafish (47), and loggerhead turtles (Caretta caretta) (39). Webb and colleagues evaluated different culture media (DMEM, MEM, and RPMI 1640) during the isolation and in vitro expansion of Caretta caretta fibroblasts and reported no statistically significant differences in cell growth among the various media (39). For this study, we also used DMEM supplemented with 20% fetal bovine serum (FBS), following the protocol used by Otsuka-Yamaguchi et al. (24) in Xenopus laevis. In terms of cell growth in culture, we observed a significant difference in the growth rate of the cells compared with those of mammals, fish, and amphibians. The average doubling time (DT) of the Ts-AD-MSCs at P3 was 127.2 h (5.3 days), which was greater than the DTs for bone marrow-derived MSCs from Xenopus laevis (25.6 h; 1.06 days) (24), MSCs isolated from the heart (50.67 h; 2.11 days) and liver (46.6 h; 1.94 days) of zebrafish (Danio rerio) at P4 (47), and adipose tissue MSCs from dogs (~36 h; 1.5 days at P3) (48–51). This difference could be related to the slower chelonian cellular metabolism (52) compared with those of other animal species, or it may be due to suboptimal culture conditions. However, no statistically significant differences were observed in the replicative potential of the cells from P1–P7, with both CDn and DT remaining constant across passages. In the Xenopus laevis study, a decrease in cellular replicative activity was noted from P5 onwards (24), whereas in zebrafish and dogs, this decrease was observed from P4 (47, 53). Several variables can influence the replicative capacity of stem cells in culture, such as the donor's age and health status (54, 55). The animals used in this study were provided by ENPA Parma, an animal protection organization, and were rescued from various situations (abandonment, found in parks and public places) and sterilized according to the Italian National Plan for the Management of the American Marsh Tortoise (Trachemys scripta). It was therefore not possible to determine the actual ages of the subjects, nor was it possible to estimate their ages accurately through morphometric investigations (56, 57). All the subjects included in the study were considered clinically healthy following a thorough clinical examination and routine hematological tests (58). However, reptiles with subclinical conditions may sometimes be asymptomatic or not show abnormalities in laboratory tests. Additionally, numerous variables (such as sex, breed, season, hormonal status, and captivity vs. wild state) can influence the normal reference values for each species (58). Therefore, it was not possible to definitively exclude underlying pathological conditions potentially affecting cell replication or other biological features. Cells were successfully differentiated into adipocytes, chondrocytes, and osteoblasts using differentiation medium validated for use with mammalian MSCs. This result is particularly interesting, as it demonstrates that Ts-AD-MSCs are responsive to the same differentiation factors. Similarly, in Xenopus laevis and Danio rerio, the differentiation of cells derived from the bone marrow (24), myocardium, and liver (47) has been shown to be positively influenced by chondrogenic, osteogenic, and adipogenic differentiation using human differentiation kits. With respect to the phenotypic characterization of the cells, it was not possible to use the validated method for mammalian MSCs, namely, flow cytometry (59), owing to the lack of validated antibodies for identifying Trachemys scripta CD markers. Consequently, the gene expression levels of various CD markers were assessed by RT–PCR, similar to the approach described by Otsuka-Yamaguchi et al. (24) for isolating MSCs from Xenopus laevis bone marrow. The Trachemys scripta genome was recently sequenced (32). To the best of our knowledge, no studies have measured the expression levels of the genes of interest for MSC characterization in the cells or tissues of the American pond turtle. Therefore, the primers used in this study were designed de novo based on gene sequences available on the NCBI website. The primer pairs used successfully amplified the genes of interest, with the exception of CD45 and CD31. Both the CD45- and CD31-encoding genes were investigated not only in MSCs but also in leukocytes isolated from blood, and the results were also negative. It is currently unclear whether the lack of expression of CD45 and CD31 is due to their actual absence in T. scripta leukocytes or analytical error. It is known from the literature that these genes are expressed in mammalian leukocytes, but there are no published studies regarding their expression in reptile leukocytes. Some recent studies have described CD45 expression in bird leukocytes (60, 61). As birds are phylogenetically close to reptiles (62, 63), this expression may be conserved. However, further studies are needed to verify its expression in this animal class. The cellular morphology, differentiation capacity into various mesenchymal cell types, and expression patterns of markers corresponding to mammalian features suggest that cells isolated from the adipose tissue of the flank of Trachemys scripta are likely to correspond to the mesenchymal stromal cell phenotype. Although some uncertainties remain concerning the results for CD45 and CD31, gene expression was substantially consistent with the ISCT expectations: the cells were positive for CD90, CD73, CD105, and CD44, whereas CD34 and HLA DRA expression was not observed.
We believe that our data suggest a substantial correspondence between the obtained cell populations and mammalian MSCs and support the hypothesis that applying regenerative medicine protocols in chelonian species will be possible in the future. Sea turtles belonging to various genera (e.g., Caretta caretta, Chelonia mydas, and Dermochelys coriacea) sustain significant traumatic injuries due to collisions with boat hulls or propellers. These injuries are frequently extensive and necessitate prolonged hospitalization, and some injured turtles ultimately cannot be released back into their natural environment (64). A similar fate affects thousands of terrestrial turtles worldwide each year, many of which are listed as “critically endangered” on the IUCN Red List (https://www.iucnredlist.org). Such injuries have been reported in wild turtles (65, 66) and in turtles kept as pets (67), including those injured by lawnmowers or other garden tools (67). The utilization of MSCs in such patients may prove advantageous, as evidenced by findings in other domestic animal species and humans.
The scientific literature on the use of regenerative medicine approaches for traumatic injuries in reptiles remains limited. Thrombocyte-leukocyte-rich plasma (TLRP) has been suggested as a treatment for traumatic injuries in various reptilian species, including chelonians, snakes, and lizards (7–9), with promising results. However, no data is available regarding MSC therapies. In contrast, an extensive body of research supports the application of MSCs in both soft and hard tissue injuries in mammals. In veterinary clinics, the use of biologics is gaining increasing interest for the treatment of both acute and chronic cutaneous lesions. MSCs have been shown to secrete bioactive factors that promote the proliferation of resident cells, facilitate extracellular matrix deposition and remodeling, enhance angiogenesis, and modulate the inflammatory response at the injury site, thereby accelerating wound healing (68, 69). Alongside soft tissue injuries, shell and bone fractures pose a significant threat to the survival of turtles and tortoises. In most chelonians, the carapace and plastron are structurally composed of both bone and dermal elements of the skin (70). Mesenchymal stem cells (MSCs) have been extensively studied for their differentiation potential toward the osteogenic lineage. However, their role in bone regeneration is likely mediated not only by direct differentiation but also through the secretion of immunoregulatory cytokines, the regulation of angiogenesis, and the modulation of the local microenvironment via trophic factor release (71, 72). Despite the absence of established clinical protocols for the use of MSCs in treating bone lesions in veterinary medicine, preliminary applications of cell-based therapies suggest promising indications of their safety and efficacy (73, 74). It can be hypothesized that, in the future, MSC-based therapies may also represent a viable treatment strategy for other vertebrates, including reptiles, complementing existing therapeutic approaches.
We are aware that the development of biological assays will be necessary to confirm that the population of MSC-like cells we have expanded maintains the key biological and therapeutic features of mammalian MSCs. Using biological assays to determine the potency of MSCs is crucial for evaluating their potential clinical application. Different assays have been proposed to assess the key functionalities of MSCs used in veterinary medicine, mainly aimed at demonstrating immunomodulation and tissue regeneration ability (6, 34). Future studies will be necessary to optimize standardized protocols, including cell sources, culture methods, passage numbers, and clinical handling protocols.
5 Conclusion
The results of the present study suggest that it is feasible to isolate and expand in vitro populations of MSC-like cells harvested from turtles starting from small samples of adipose tissue. Despite the absence of scientific literature demonstrating a functional correspondence between these cells and those isolated from mammals, which have been extensively studied for their immunomodulatory and proregenerative properties, the present work paves the way for further studies aimed at the application of regenerative medicine protocols in Chelonians.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal studies were approved by University of Parma Ethical Committee (PROT. No. 15/CESA/2022). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
VA: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. AV: Conceptualization, Investigation, Methodology, Visualization, Writing – original draft. VC: Investigation, Writing – review & editing. MG: Investigation, Writing – review & editing. PB: Investigation, Writing – review & editing. RR: Validation, Writing – review & editing. GB: Validation, Writing – review & editing. GN: Validation, Writing – review & editing. IP: Validation, Writing – review & editing. SG: Conceptualization, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. FD: Conceptualization, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We would like to express our gratitude to Professor Rosanna Di Lecce for her valuable scientific contribution to the study, and in particular for her careful analysis of the biological features of Trachemys scripta adipose tissue.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1546091/full#supplementary-material
References
1. Voga M, Adamic N, Vengust M, Majdic G. Stem cells in veterinary medicine—current state and treatment options. Front Vet Sci. (2020) 7:278. doi: 10.3389/fvets.2020.00278
2. Caplan AI. Mesenchymal stem cells: time to change the name! Stem Cells Transl Med. (2017) 6:1445–51. doi: 10.1002/sctm.17-0051
3. Wang Y, Fang J, Liu B, Shao C, Shi Y. Reciprocal regulation of mesenchymal stem cells and immune responses. Cell Stem Cell. (2022) 29:1515–30. doi: 10.1016/j.stem.2022.10.001
4. Rodríguez-Fuentes DE, Fernández-Garza LE, Samia-Meza JA, Barrera-Barrera SA, Caplan AI, Barrera-Saldaña HA. Mesenchymal stem cells current clinical applications: a systematic review. Arch Med Res. (2021) 52:93–101. doi: 10.1016/j.arcmed.2020.08.006
5. Williams ZJ, Pezzanite LM, Chow L, Rockow M, Dow SW. Evaluation of stem-cell therapies in companion animal disease models: a concise review (2015-2023). Stem Cells. (2024) 42:677–705. doi: 10.1093/stmcls/sxae034
6. Ivanovska A, Wang M, Arshaghi TE, Shaw G, Alves J, Byrne A, et al. Manufacturing mesenchymal stromal cells for the treatment of osteoarthritis in canine patients: challenges and recommendations. Front Vet Sci. (2022) 9:897150. doi: 10.3389/fvets.2022.897150
7. Treatment of a Degloving Lesion in a Green Bush Rat Snake (Gonyosoma prasinum) With Surgical Approach and Single Application of Thrombocyte-Leukocyte-Rich Plasma (TLRP)—Veterinaria|Official Magazine of SCIVAC. Available online at: https://veterinaria.scivac.org/2020/year-34-n-2-april-2020/treatment-of-a-degloving-lesion-in-a-green-bush-rat-snake-gonyosoma-prasinum-with-surgical-approach-and-single-application-of-thrombocyte-leukocyte-rich-plasma-tlrp.html (accessed September 20, 2024).
8. Bardi E, Vetere A, Aquaro V, Lubian E, Lauzi S, Ravasio G, et al. Use of thrombocyte–leukocyte-rich plasma in the treatment of chronic oral cavity disorders in reptiles: two case reports. J Exotic Pet Med. (2019) 29:32–9. doi: 10.1053/j.jepm.2018.06.008
9. Di Ianni F, Merli E, Burtini F, Conti V, Pelizzone I, Di Lecce R, et al. Preparation and application of an innovative thrombocyte/leukocyte-enriched plasma to promote tissue repair in chelonians. PLoS One (2015) 10:e0122595. doi: 10.1371/journal.pone.0122595
10. Decree of the Minister for Ecological Transition No. 370 of September 28, 2022. Available online at: https://www.mase.gov.it/sites/default/files/archivio/allegati/biodiversita/Piano_di_gestione_testuggine_palustre_americana.pdf (accessed November 13, 2024).
11. Cadi A, Joly P. Impact of the introduction of the red-eared slider (Trachemys scripta elegans) on survival rates of the European pond turtle (Emys orbicularis). Biodivers Conserv. (2004) 13:2511–8. doi: 10.1023/B:BIOC.0000048451.07820.9c
12. Ficetola GF, Scali S. Invasive amphibians and reptiles in Italy. VIII Congresso Nazionale. Societas Herpetologica Italica, Chieti: Ianieri Edizioni (2010). p. 335–40.
13. Doneley—Monks—Johnson—Carmel Reptile Medicine and Surgery in Clinical Practice Wiley. Available online at: https://www.libreriauniverso.it/ (accessed November 13, 2024).
14. Sykes JM, Klaphake E. Reptile hematology. Vet Clin North Am Exotic Anim Pract. (2008) 11:481–500. doi: 10.1016/j.cvex.2008.03.005
15. Campbell TW, Ellis CK. Avian and Exotic Animal Hematology and Cytology. 2nd ed. Hoboken, NJ: John Wiley & Sons. (2013). p. 562.
16. Research Randomizer. Available online at: https://www.randomizer.org/ (accessed November 14, 2024).
17. Scarabelli S, Di Girolamo N. Chelonian sedation and anesthesia. Vet Clin North Am Exot Anim Pract. (2022) 25:49–72. doi: 10.1016/j.cvex.2021.08.009
18. Greer LL, Jenne KJ, Diggs HE. Medetomidine-ketamine anesthesia in red-eared slider turtles (Trachemys scripta elegans). Contemp Top Lab Anim Sci. (2001) 40:9–11.
19. Masi M, Vetere A, Casalini J, Corsi F, Di Ianni F, Nardini G. Comparison of subcutaneous versus intramuscular dexmedetomidine-midazolam-ketamine-morphine (DMKM) mixture as chemical restraint for endoscopic sex determination in aldabra giant tortoises (Aldabrachelys gigantea). Animals. (2023) 13:3626. doi: 10.3390/ani13233626
20. Eshar D, Rooney TA, Gardhouse S, Beaufrère H. Evaluation of the effects of a dexmedetomidine-midazolam-ketamine combination administered intramuscularly to captive red-footed tortoises (Chelonoidis carbonaria). Am J Vet Res. (2021) 82:858–64. doi: 10.2460/ajvr.82.11.858
21. Ting AKY, Tay VSY, Chng HT, Xie S. A Critical review on the pharmacodynamics and pharmacokinetics of non-steroidal anti-inflammatory drugs and opioid drugs used in reptiles. Vet Anim Sci. (2022) 17:100267. doi: 10.1016/j.vas.2022.100267
22. Bancroft JD, Gamble M, editors. Theory and Practice of Histological Techniques. 6th ed. Philadelphia, PA: Churchill Livingstone/Elsevier. (2008). p. 742.
23. Suelzu CM, Conti V, Khalidy Y, Montagna S, Strusi G, Di Lecce R, et al. Xenobiotic-free medium guarantees expansion of adipose tissue-derived canine mesenchymal stem cells both in 3D fibrin-based matrices and in 2D plastic surface cultures. Cells. (2020) 9:2578. doi: 10.3390/cells9122578
24. Otsuka-Yamaguchi R, Kitada M, Kuroda Y, Kushida Y, Wakao S, Dezawa M. Isolation and characterization of bone marrow-derived mesenchymal stem cells in Xenopus laevis. Stem Cell Res. (2021) 53:102341. doi: 10.1016/j.scr.2021.102341
25. Vidal MA, Kilroy GE, Johnson JR, Lopez MJ, Moore RM, Gimble JM. Cell growth characteristics and differentiation frequency of adherent equine bone marrow–derived mesenchymal stromal cells: adipogenic and osteogenic capacity. Vet Surg. (2006) 35:601–10. doi: 10.1111/j.1532-950X.2006.00197.x
26. Roth V. Doubling Time—Online Computing With 2 Points (2024). Available online at: https://www.doubling-time.com/compute.php (accessed August 5, 2024).
27. Secunda R, Vennila R, Mohanashankar AM, Rajasundari M, Jeswanth S, Surendran R. Isolation, expansion and characterisation of mesenchymal stem cells from human bone marrow, adipose tissue, umbilical cord blood and matrix: a comparative study. Cytotechnology. (2015) 67:793–807. doi: 10.1007/s10616-014-9718-z
28. Neupane M, Chang C-C, Kiupel M, Yuzbasiyan-Gurkan V. Isolation and characterization of canine adipose-derived mesenchymal stem cells. Tissue Eng Part A. (2008) 14:1007–15. doi: 10.1089/ten.tea.2007.0207
29. Berni P, Andreoli V, Conti V, Ramoni R, Basini G, Scattini G, et al. Evaluation of a novel mechanical device for the production of microfragmented adipose tissue for veterinary regenerative medicine: a proof-of-concept. Int J Mol Sci. (2024) 25:11854. doi: 10.3390/ijms252111854
30. Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. (2006) 8:315–7. doi: 10.1080/14653240600855905
31. Grezella C, Fernandez-Rebollo E, Franzen J, Ventura Ferreira MS, Beier F, Wagner W. Effects of senolytic drugs on human mesenchymal stromal cells. Stem Cell Res Ther. (2018) 9:108. doi: 10.1186/s13287-018-0857-6
32. Brian Simison W, Parham JF, Papenfuss TJ, Lam AW, Henderson JB. An annotated chromosome-level reference genome of the red-eared slider turtle (Trachemys scripta elegans). Genome Biol Evol. (2020) 12:456–62. doi: 10.1093/gbe/evaa063
33. Sun Y, Liang M, Qu J, Jin C, Zhang Q, Li J, et al. Early diagnosis of novel SFTS bunyavirus infection by quantitative real-time RT-PCR assay. J Clin Virol. (2012) 53:48–53. doi: 10.1016/j.jcv.2011.09.031
34. Guest DJ, Dudhia J, Smith RKW, Roberts SJ, Conzemius M, Innes JF, et al. Position statement: minimal criteria for reporting veterinary and animal medicine research for mesenchymal stromal/stem cells in orthopedic applications. Front Vet Sci. (2022) 9:817041. doi: 10.3389/fvets.2022.817041
35. Coelioscopic and Endoscope-Assisted Sterilization of Chelonians—PubMed. Available online at: https://pubmed.ncbi.nlm.nih.gov/26336003/ (accessed September 27, 2024).
36. Andreoli V, Berni P, Conti V, Ramoni R, Basini G, Grolli S. Mesenchymal stromal cells derived from canine adipose tissue: evaluation of the effect of different shipping vehicles used for clinical administration. Int J Mol Sci. (2024) 25:63426. doi: 10.3390/ijms25063426
37. Mocchi M, Grolli S, Dotti S, Di Silvestre D, Villa R, Berni P, et al. Equine mesenchymal stem/stromal cells freeze-dried secretome (lyosecretome) for the treatment of musculoskeletal diseases: production process validation and batch release test for clinical use. Pharmaceuticals. (2021) 14:553. doi: 10.3390/ph14060553
38. Angilletta MJ, Niewiarowski PH, Navas CA. The evolution of thermal physiology in ectotherms. J Therm Biol. (2002) 27:249–68. doi: 10.1016/S0306-4565(01)00094-8
39. Webb SJ, Zychowski GV, Bauman SW, Higgins BM, Raudsepp T, Gollahon LS, et al. Establishment, characterization, and toxicological application of loggerhead sea turtle (Caretta caretta) primary skin fibroblast cell cultures. Environ Sci Technol. (2014) 48:14728–37. doi: 10.1021/es504182e
40. Fukuda T, Eitsuka T, Donai K, Kurita M, Saito T, Okamoto H, et al. Expression of human mutant cyclin dependent kinase 4, Cyclin D and telomerase extends the life span but does not immortalize fibroblasts derived from loggerhead sea turtle (Caretta caretta). Sci Rep. (2018) 8:9229. doi: 10.1038/s41598-018-27271-x
41. Finlayson KA, Madden Hof CA, van de Merwe JP. Development and application of species-specific cell-based bioassays to assess toxicity in green sea turtles. Sci Total Environ. (2020) 747:142095. doi: 10.1016/j.scitotenv.2020.142095
42. Lund TC, Patrinostro X, Kramer AC, Stadem P, Higgins LA, Markowski TW, et al. sdf1 expression reveals a source of perivascular-derived mesenchymal stem cells in zebrafish. Stem Cells. (2014) 32:2767–79. doi: 10.1002/stem.1758
43. Carotenuto R, Pallotta MM, Tussellino M, Fogliano C. Xenopus laevis (Daudin, 1802) as a model organism for bioscience: a historic review and perspective. Biology. (2023) 12:890. doi: 10.3390/biology12060890
44. Jacobson ER, ed. Infectious Diseases and Pathology of Reptiles: Color Atlas and Text. Boca Raton, FL: CRC Press (2014). p. 736.
45. Araña M, Mazo M, Aranda P, Pelacho B, Prosper F. Adipose tissue-derived mesenchymal stem cells: isolation, expansion, and characterization. Methods Mol Biol. (2013) 1036:47–61. doi: 10.1007/978-1-62703-511-8_4
46. Ghorbani A, Jalali SA, Varedi M. Isolation of adipose tissue mesenchymal stem cells without tissue destruction: a non-enzymatic method. Tissue Cell. (2014) 46:54–8. doi: 10.1016/j.tice.2013.11.002
47. Fathi E, Farahzadi R, Sheikhzadeh N. Immunophenotypic characterization, multi-lineage differentiation and aging of zebrafish heart and liver tissue-derived mesenchymal stem cells as a novel approach in stem cell-based therapy. Tissue Cell. (2019) 57:15–21. doi: 10.1016/j.tice.2019.01.006
48. Yaneselli KM, Kuhl CP, Terraciano PB, de Oliveira FS, Pizzato SB, Pazza K, et al. Comparison of the characteristics of canine adipose tissue-derived mesenchymal stem cells extracted from different sites and at different passage numbers. J Vet Sci. (2018) 19:13–20. doi: 10.4142/jvs.2018.19.1.13
49. Duan W, Lopez MJ. Canine adult adipose tissue-derived multipotent stromal cell isolation and characterization. In: Bunnell BA, Gimble JM, editors. Adipose-Derived Stem Cells: Methods and Protocols. New York, NY: Springer (2018). p. 189–202.
50. Arnhold S, Elashry MI, Klymiuk MC, Geburek F. Investigation of stemness and multipotency of equine adipose-derived mesenchymal stem cells (ASCs) from different fat sources in comparison with lipoma. Stem Cell Res Ther. (2019) 10:309. doi: 10.1186/s13287-019-1429-0
51. Kim I, Bang SI, Lee SK, Park SY, Kim M, Ha H. Clinical implication of allogenic implantation of adipogenic differentiated adipose-derived stem cells. Stem Cells Transl Med. (2014) 3:1312–21. doi: 10.5966/sctm.2014-0109
52. Wallace BP, Jones TT. What makes marine turtles go: a review of metabolic rates and their consequences. J Exp Mar Bio Ecol. (2008) 356:8–24. doi: 10.1016/j.jembe.2007.12.023
53. Marcoccia R, Nesci S, Merlo B, Ballotta G, Algieri C, Pagliarani A, et al. Biological characteristics and metabolic profile of canine mesenchymal stem cells isolated from adipose tissue and umbilical cord matrix. PLoS ONE. (2021) 16:e0247567. doi: 10.1371/journal.pone.0247567
54. Maslov AY, Barone TA, Plunkett RJ, Pruitt SC. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. J Neurosci. (2004) 24:1726–33. doi: 10.1523/JNEUROSCI.4608-03.2004
55. Alt EU, Senst C, Murthy SN, Slakey DP, Dupin CL, Chaffin AE, et al. Aging alters tissue resident mesenchymal stem cell properties. Stem Cell Res. (2012) 8:215–25. doi: 10.1016/j.scr.2011.11.002
56. Wilson DS, Tracy CR, Tracy CR. Estimating age of turtles from growth rings: a critical evaluation of the technique. Herpetologica. (2003) 59:178–94. doi: 10.1655/0018-0831(2003)059[0178:EAOTFG]2.0.CO;2
57. Armstrong DP, Brooks RJ. Estimating ages of turtles from growth data. Chelon Conserv Biol. (2014) 13:9–15. doi: 10.2744/CCB-1055.1
58. Stacy NI, Alleman AR, Sayler KA. Diagnostic hematology of reptiles. Clin Lab Med. (2011) 31:87–108. doi: 10.1016/j.cll.2010.10.006
59. Sober SA, Darmani H, Alhattab D, Awidi A. Flow cytometric characterization of cell surface markers to differentiate between fibroblasts and mesenchymal stem cells of different origin. Arch Med Sci. (2021) 19:1487–96. doi: 10.5114/aoms/131088
60. Vahsen T, Zapata L, Guabiraba R, Melloul E, Cordonnier N, Botterel F, et al. Cellular and molecular insights on the regulation of innate immune responses to experimental aspergillosis in chicken and turkey poults. Med Mycol. (2021) 59:465–75. doi: 10.1093/mmy/myaa069
61. De Boever S, Croubels S, Demeyere K, Lambrecht B, De Backer P, Meyer E. Flow cytometric differentiation of avian leukocytes and analysis of their intracellular cytokine expression. Avian Pathol. (2010) 39:41–6. doi: 10.1080/03079450903473574
62. Modesto SP, Anderson JS. The phylogenetic definition of reptilia. Syst Biol. (2004) 53:815–21. doi: 10.1080/10635150490503026
63. Modesto SP, Scott DM, MacDougall MJ, Sues H-D, Evans DC, Reisz RR. The oldest parareptile and the early diversification of reptiles. Proc R Soc B Biol Sci. (2015) 282:20141912. doi: 10.1098/rspb.2014.1912
64. Orós J, Torrent A, Calabuig P, Déniz S. Diseases and causes of mortality among sea turtles stranded in the Canary Islands, Spain (1998-2001). Dis Aquat Org. (2005) 63:13–24. doi: 10.3354/dao063013
65. Tucker-Retter EK, Lewbart GA. Corticosterone, lactate, and injury severity as short-term and long-term prognostic indicators in eastern box turtles (Terrapene carolina carolina) with traumatic injuries admitted to a wildlife rehabilitation clinic. hpms (2022) 32:281–90. doi: 10.5818/JHMS-D-22-00022
66. Biaggini M, Corti C. Facing habitat reduction in your own shell: patterns of non-lethal injuries in the endangered tortoise Testudo hermanni in Italy. Herpetol Conserv Biol. (2018) 13:539–50.
68. Buote NJ. Updates in wound management and dressings. Vet Clin North Am Small Anim Pract. (2022) 52:289–315. doi: 10.1016/j.cvsm.2021.12.001
69. Harman RM, Rajesh A, Van De Walle GR. Use of biologics and stem cells for wound healing in the horse. Vet Clin North Am Equine Pract. (2023) 39:525–39. doi: 10.1016/j.cveq.2023.06.003
70. Boyer TH, Innis CJ, editors. Chelonian Taxonomy, Anatomy, and Physiology. Reptile Medicine and Surgery. St. Louis, MO: Saunders Elsevier (2006). p. 31–35.
71. Chen Q, Su Y, Yang Z, Lin Q, Ke Y, Xing D, et al. Bibliometric mapping of mesenchymal stem cell therapy for bone regeneration from 2013 to 2023. Front Med. (2025) 11:1484097. doi: 10.3389/fmed.2024.1484097
72. Bowles-Welch AC, Jimenez AC, Stevens HY, Frey Rubio DA, Kippner LE, Yeago C, et al. Mesenchymal stromal cells for bone trauma, defects, and disease: Considerations for manufacturing, clinical translation, and effective treatments. Bone Rep. (2023) 18:101656. doi: 10.1016/j.bonr.2023.101656
73. Dobson LK, Zeitouni S, McNeill EP, Bearden RN, Gregory CA, Saunders WB. Canine mesenchymal stromal cell-mediated bone regeneration is enhanced in the presence of sub-therapeutic concentrations of BMP-2 in a murine calvarial defect model. Front Bioeng Biotechnol. (2021) 9:764703. doi: 10.3389/fbioe.2021.764703
Keywords: mesenchymal stromal cells, regenerative medicine, veterinary regenerative medicine, chelonia, reptile medicine, turtles, tortoise
Citation: Andreoli V, Vetere A, Conti V, Gavezzoli M, Berni P, Ramoni R, Basini G, Nardini G, Pelizzone I, Grolli S and Di Ianni F (2025) Mesenchymal stromal cell isolation from pond slider (Trachemys scripta) adipose tissue obtained during routine neutering: a model for turtle species. Front. Vet. Sci. 12:1546091. doi: 10.3389/fvets.2025.1546091
Received: 16 December 2024; Accepted: 25 February 2025;
Published: 19 March 2025.
Edited by:
Louis Charles Penning, Utrecht University, NetherlandsReviewed by:
Valerie Johnson, Michigan State University, United StatesLuca Melotti, University of Padua, Italy
Copyright © 2025 Andreoli, Vetere, Conti, Gavezzoli, Berni, Ramoni, Basini, Nardini, Pelizzone, Grolli and Di Ianni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefano Grolli, c3RlZmFuby5ncm9sbGlAdW5pcHIuaXQ=
†These authors share first authorship
‡These authors have contributed equally to this work and share senior authorship
 Valentina Andreoli
Valentina Andreoli Alessandro Vetere1†
Alessandro Vetere1† Virna Conti
Virna Conti Priscilla Berni
Priscilla Berni Roberto Ramoni
Roberto Ramoni Giuseppina Basini
Giuseppina Basini Stefano Grolli
Stefano Grolli Francesco Di Ianni
Francesco Di Ianni