- School of Public Health, Dali University, Dali, China
Background: Bartonellosis is a zoonotic infectious disease caused by Bartonella spp. Small mammals are the most important hosts of Bartonella and play an important role in its long-term maintenance and spread. The multi-organ studies help understand the Bartonella prevalence of hosts more systematically and comprehensively. This study aimed to investigate the prevalence of Bartonella in small mammals and explore the genetic diversity of the infected strains and the influencing factors from Mile City and Lianghe County, Yunnan Province.
Methods: Small mammals were captured in Mile City and Lianghe County of Yunnan Province from July to August 2019. Spleen and kidney tissues were collected and the gltA gene was amplified to detect and analyze the prevalence of Bartonella in two regions and two organs.
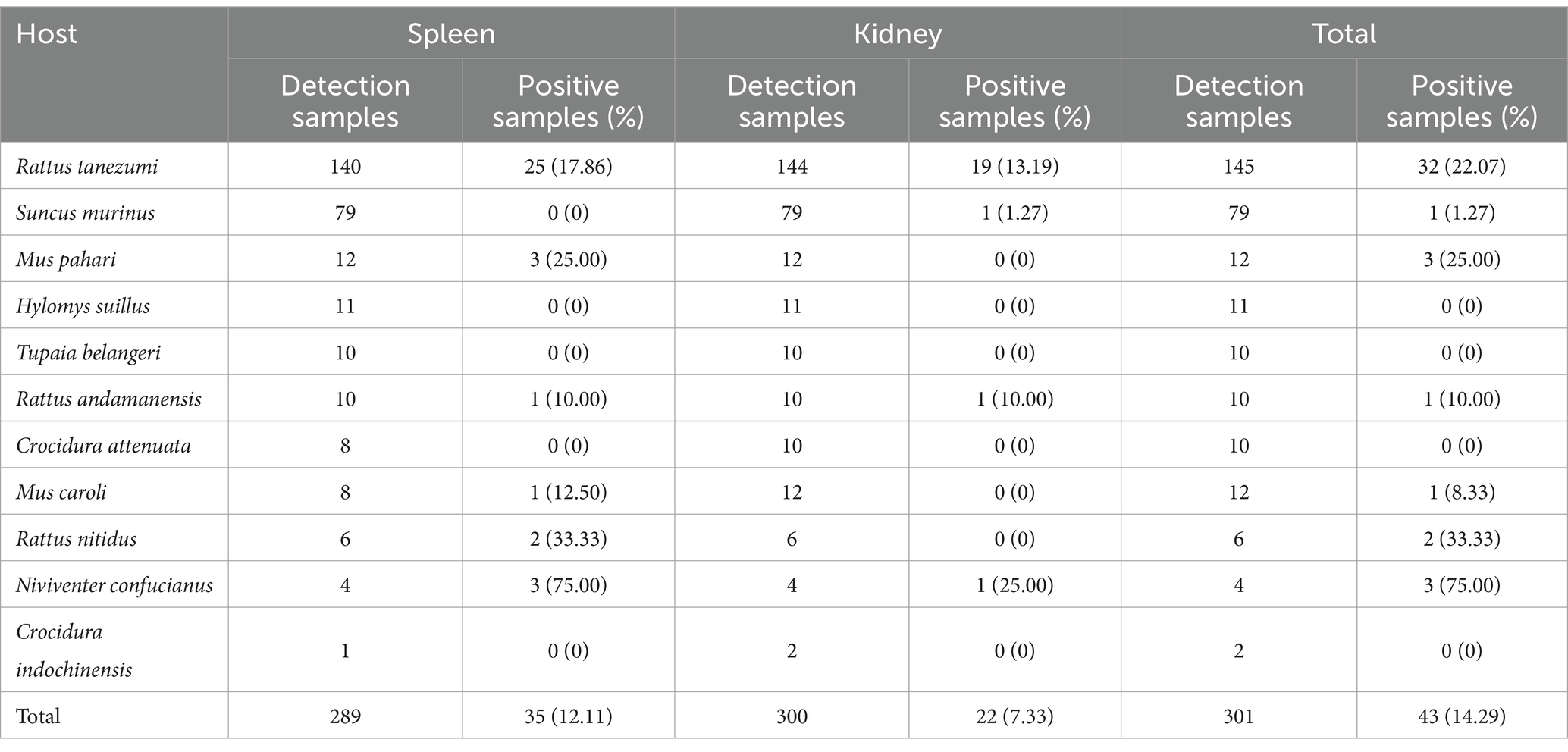
Results: The prevalence of Bartonella in small mammals was 14.29% (43/301). Lianghe County’s risk of infection was 3.79-fold (95%CI: 1.39–13.35) compared to that of Mile City. The risk of infection in Rattus tanezumi was increased by 90% compared to Suncus murinus (95%CI: 0.01–0.63). The small mammals with tail lengths > 132 mm infected by Bartonella were 6.34 folds than that with tail lengths ≤ 132 mm (95%CI: 1.87–23.39). The spleen had a higher infection rate of 12.11% (35/289) than the kidney at 7.33% (22/300) (χ2 = 4.966, p = 0.026). There were no statistically significant differences in the prevalence of Bartonella among small mammals with different habitats, sex, age, flea infestation status, body weight, body length, hindfoot length, and ear height. Five Bartonella species were isolated in seven species of small mammals. Bartonella tribocorum is the dominant species in both regions, and it has a genetic relationship with the zoonotic pathogen Bartonella elizabethae.
Conclusion: This study showed the prevalence of Bartonella in small mammals from Mile City and Lianghe County of Yunnan Province was high, and there were more types of Bartonella infection species. The spleen was more conducive to the growth and reproduction of Bartonella. The results of the study will help to prevent and control Bartonella infection and transmission to humans from small mammals in the two regions and provide a reference basis for further research on Bartonella infection in Yunnan or other similar regions.
1 Introduction
Bartonella spp. is a group of nutritionally demanding, facultative intracellular parasitic gram-negative aerobic bacteria belonging to the class Proteobacteria, subclass Alpha proteobacteria, order Rhizobiales, family Bartonellaceae, genera Bartonella. Bartonella is transmitted by blood-sucking arthropods, since its first isolation, new strains and species have been discovered, and more than 40 species and subspecies of Bartonella have been identified, 17 of which are associated with human infection (1, 2). With the continuous discovery of Bartonella species, the host animals suitable for storing Bartonella have increased exponentially, and due to the high heritability and diversity of Bartonella in rodents, rodents are the most important hosts of Bartonella and play an important role in its long-term maintenance and spread (1, 3). Previous studies have shown that the prevalence of Bartonella in foreign rodents ranges from 4.9% to 85%, and the common infection strains are Bartonella grahamii and Bartonella phoceensis (4–6). Rodents commonly associated with Bartonella infection in China are Rattus and Apodemus, with prevalence as low as 6.4% and as high as 57.7%, covering a wide range of areas, and the predominant strain of infection in many regions being Bartonella grahamii, which is a danger to human health, there is a need to investigate the prevalence of Bartonella in small mammals from different regions of the country (7–10). In addition, researchers performed multi-tissue Bartonella detection in rodents from northern and eastern China to obtain more accurate infection rates and more strain sequences for evaluation (9, 11).
Yunnan Province is located in the southwestern border mountainous area of China, the land area is large, the climate types are complex and diverse, and the special geographical environment and climate conditions provide an excellent growth environment for a variety of pathogenic microorganisms, increasing the risk of natural focal disease (8, 12). According to previous research findings, studies on Bartonella species prevalence in small mammals conducted in Heqing County and Gongshan County of Yunnan Province have primarily focused on assessing the genetic diversity and tissue tropism of Bartonella, while lacking cross-regional comparative analyses of regional differences (13). The study conducted in Yulong, Jianchuan, and Lianghe counties of Yunnan Province focused on comparing multifactorial differences across regions but did not involve multi-tissue testing (8). In light of the current research status on Bartonella infection in small mammals in Yunnan Province, it is imperative to integrate multi-tissue detection with cross-regional investigation of infection-influencing factors, building upon the key focus areas identified in previous studies. This comprehensive approach will contribute to a more accurate representation of Bartonella infection prevalence and its determinant factors among small mammals across different regions of Yunnan Province. The present study was based on two types of tissues for pathogen detection, aiming to investigate the infection status of Bartonella in small mammals from Mile City in eastern Yunnan Province and Lianghe County in western, to compare and analyze the epidemiological distribution of Bartonella in different regions, and to explore the influencing factors of the occurrence of Bartonella infections, to provide scientific basis for effective prevention and control of Bartonella in the region.
2 Materials and methods
2.1 Ethics statement
All methods were performed in accordance with the Medical Ethics Committee of Dali University. This Experimental Protocol has been reviewed and approved by the Medical Ethics Committee of Dali University (No. MECDU-201901-3) and conforms to the principles of medical experiment ethics and the relevant provisions of the national medical experiment ethics and welfare. Endangered or protected animal species were not included in this research.
2.2 Small mammal collection
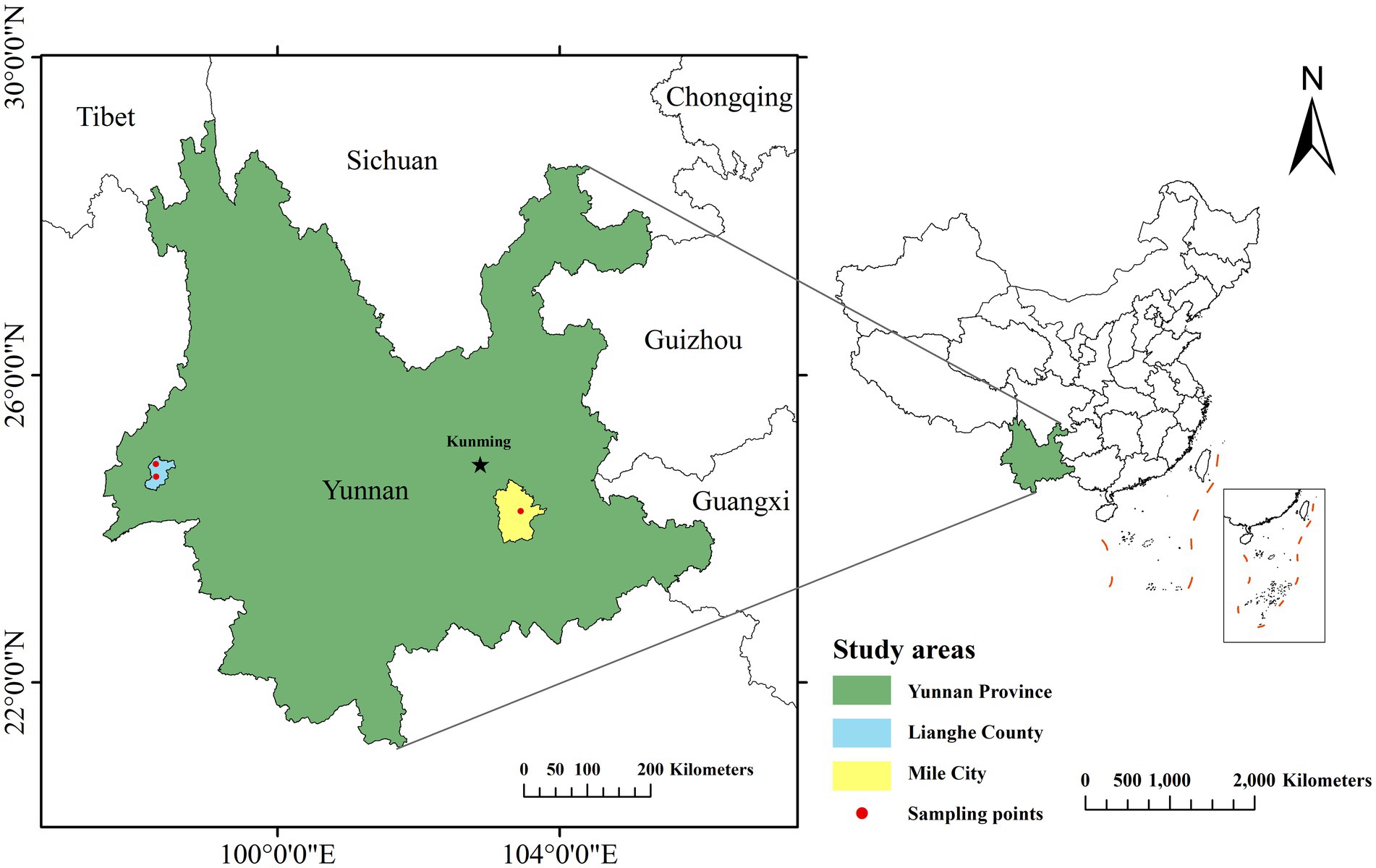
Small mammals in Mile City and Lianghe County of Yunnan Province were captured using the night clamping method from July to August 2019, employing dead traps (15 × 8 cm) baited with peanuts. The areas for the trapping setting were selected and suggested by experts from the Center for Disease Control and Prevention of Mile and Lianghe in different landscapes (Residential areas, Cultivation, Shrubland, and Woodland), they provided the setting where small mammals captured were the most based on annual small mammals monitoring data. The sampling area map was constructed by ArcGIS 10.8, the sampling points are specified down to the township level, as shown in Figure 1. Within the selected area, place dead traps at approximately 20-meter intervals based on the specific activity traces of small mammals, to ensure coverage of the target species’ activity range and minimize repeated captures. Each sampling point should have no fewer than 200 traps deployed daily. All dead traps were placed the whole night starting from the evening of the day to the next early morning. As small mammals captured were already deceased upon collection, no anesthesia or euthanasia was performed in this study. Then, the captured small mammals were recorded with the geographical landscape elements where they were located on notes and transported back to the laboratory in individual bags (“one mammal per bag”). Species of small mammals were identified by morphological methods, and the mammals were sequentially numbered while recording their characteristics. Repeated freeze–thaw cycles can rupture cellular structures, releasing more nucleases and leading to DNA degradation. Increased frequency of freeze–thaw cycles progressively compromises DNA quality, as repeated temperature fluctuations negatively impact the integrity of biomolecules in tissues. To minimize degradation, it is recommended to divide fresh tissue into small aliquots before freezing for optimal preservation of biological samples (14). In addition, ultra-low temperature storage can inhibit ice crystal growth through vitrification, reduce molecular motion, maintain tissue integrity, and prolong storage duration. Therefore, in this study, spleen and kidney tissues collected under aseptic conditions were divided into multiple tubes and preserved through low-temperature transportation to −80°C freezers for storage.
2.3 DNA extraction and PCR
Approximately 10 mg of spleen or kidney sample was taken, and DNA was extracted according to the manufacturer’s instruction of TIANamp Genomic DNA Kit (DP304; TIANGEN; Beijing; China). The concentration and purity of each DNA sample were determined by an ultraviolet–visible spectrophotometer. Qualified DNA samples have concentrations greater than 50 ng/μl (50 μg/mL), and the A260/A280 was between 1.8 to 2.1. DNA samples that met the criteria were stored at minus 80°C until subsequent molecular experiments, while those that did not meet the criteria were subjected to a secondary re-extraction.
The citrate synthase (gltA) gene sequence was amplified by polymerase chain reaction (PCR) with reference to the previous study (15). The primer sequences are BhCS.781p:5′-GGGGACCAGCTCATGGTGG-3′ and BhCS.1137n:5′-AATGCAAAAAGAACAGTAAACA-3′. The reaction mixture (25 μL) contains the following components: 1 μL (10 μmol/L) of each primer, DreamTaq Green PCR Master Mix (2X) 12.5 μL, 3 μL of DNA template, 7.5 μL double-distilled H2O. GltA amplification was performed under the following conditions: one cycle for 3 min at 94°C; 30 cycles for 30s at 94°C, 30s at 53°C, and 1 min at 72°C; and a final extension for 5 min at 72°C. Next, PCR products with 379 bp were identified by 1.5% agarose gel containing 4SGelred (Sangon Biotech (Shanghai) Co., Ltd.) and visualized under the Gel imaging system (G: BOX F3, Syngene, American). The samples were subsequently subjected to Sanger sequencing (dideoxy chain-termination method) performed by Sangon Biotech (Shanghai, China) Co., Ltd. The final sequencing was successfully determined to be positive, at the same time, the detection of Bartonella in one tissue of the small mammal was judged to be an infection.
2.4 Phylogenetic analysis
The successfully detected sequences were edited and trimmed by the SeqMan program in DNASTAR Lasergene (7.1 version). Reference sequences encoding gltA of Bartonella were retrieved from GenBank by using the Basic Local Alignment Search Tool at the National Center for Biotechnology Information website.1 Sample sequences were aligned with reference sequences using Sequence distance in MegAlign of DNASTAR Lasergene. Phylogenetic analysis was performed with Clustal W protocol (default parameters) by MEGA software (11.0 version). The phylogenetic tree was created by using the Neighbor-joining method, and bootstrap values were calculated with 1,000 replicates. The outgroup used was Brucella melitensis (gltA gene accession number: NZ_ACEM01000037). For Bartonella species, the following gltA gene sequences were included: Bartonella tribocorum (OR117609), Bartonella queenslandensis (MH748120), Bartonella phoceensis (AY515126), Bartonella mastomydis (OQ305211), and Bartonella clarridgeiae (MH019300). The phylogenetic tree was modified using iTOL v62 to add comments on host and strain isolation.
2.5 Statistical analysis
Statistical analysis was performed using R software (4.4.0 version). The precise probability approach was used to compute the overall 95% confidence intervals for each sample rate. A total of 301 small mammals were captured. Depending on their constituent ratio, small mammals were classified as dominant (>10%) or other (≤10%). Among them, 288 small mammals had their spleen and kidney collected simultaneously. McNemar’s test was used to compare the difference in infection rate among different organs of the same small mammal. The Chi-squared test was used to compare the difference in Bartonella infection rate among 301 small mammals in different regions, habitats, species, sex, ages, flea-carrying status, and appearance characteristics. The appearance characteristics of small mammals showed skewness and were grouped by median. In the analysis of infection influencing factors, variables with statistical significance (p < 0.05) were initially screened using univariate analysis (Chi-square test). These selected variables were then incorporated into a multivariate logistic regression model. Subsequently, the stepwise bidirectional regression method was employed for variable selection and model optimization. Multi-factor Logistic regression analysis was performed to identify the influencing factors of Bartonella infection in small mammals. The test level was α = 0.05.
3 Results
3.1 Species of small mammals and Bartonella detection
In total, 301 small mammals were trapped and identified as belonging to 3 orders, 4 families, 7 genera, and 11 species, with a wide range of species, as shown in Table 1. Overall, Rattus tanezumi was the dominant species accounting for 48.17% (145/301), with a 22.07% (32/145) infection rate in Bartonella, followed by Suncus murinus with 26.25% (79/301) account and a 1.27% (1/79) infection rate. In Mile City, the dominant species was Rattus tanezumi 44.12% (30/68) with an infection rate of 3.33% (1/30), followed by the Mus caroli 17.65% (12/68) with an infection rate of 8.33% (1/12). The dominant species in Lianghe County, consistent with the overall situation, were Rattus tanezumi 49.36% (115/233) with an infection rate of 26.96% (31/115) and the Suncus murinus 31.76% (74/233) with an infection rate of 1.35% (1/74). The prevalence of Bartonella in small mammals is shown in Table 1.
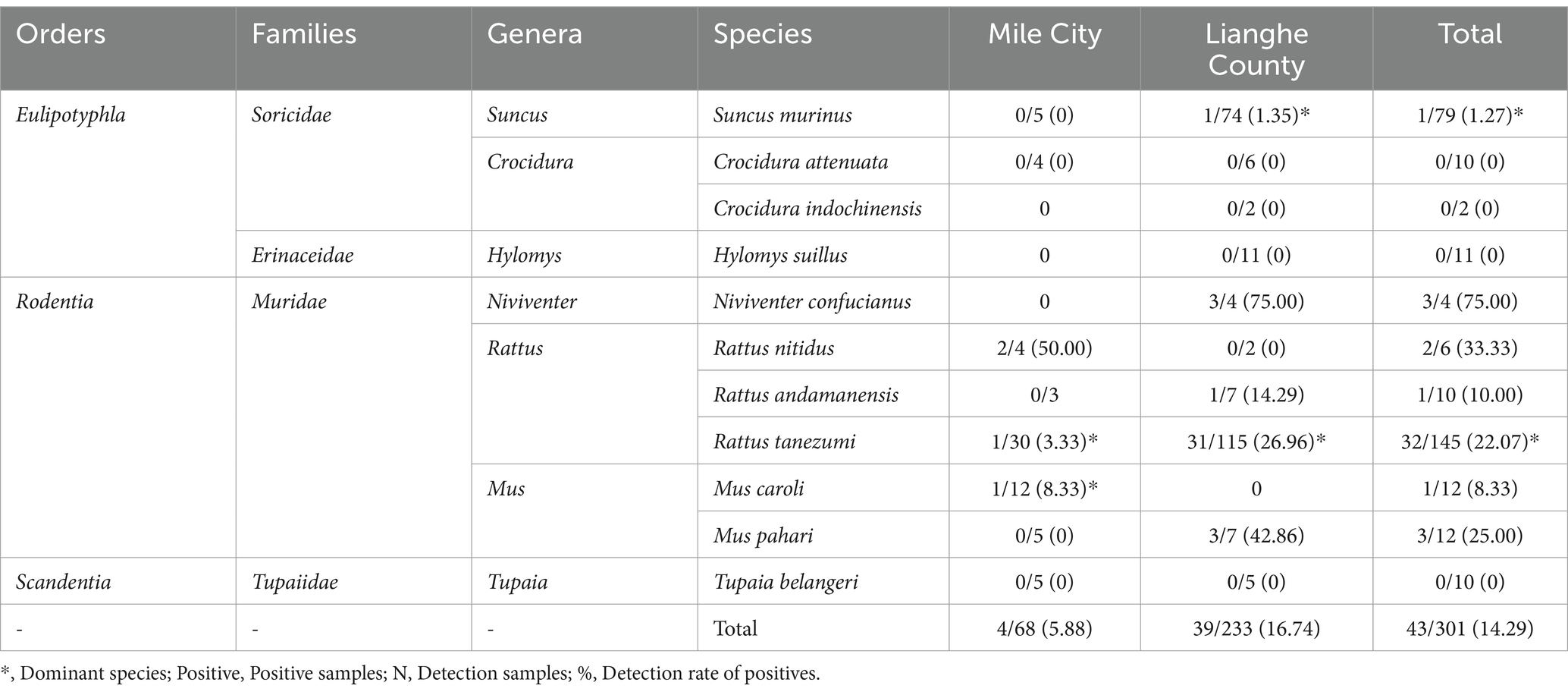
Table 1. Prevalence of Bartonella in small mammals captured from Mile City and Lianghe County, Yunnan province [Positive/N(%)].
3.2 Distribution of Bartonella in different tissues of small mammal
A total of 43 small mammals were judged to be infected with Bartonella, with an infection rate of 14.29% (43/301). There were 7 species of small mammals infected with Bartonella: Rattus tanezumi, Suncus murinus, Mus pahari, Rattus andamanensis, Mus caroli, Rattus nitidus, and Niviventer confucianus, as shown in Table 2.
In this study, 289 spleen samples and 300 kidney samples were tested. The infection rate of the spleen was 12.11% (35/289) and the kidney was 7.33% (22/300), which was higher in the spleen than in the kidney (χ2 = 4.966, p = 0.026), as shown in Table 2.
3.3 Analysis of infection difference and influencing factors in different characteristics of small mammals
Among 301 small mammals, the infection rate in Lianghe County was 16.74% (39/233) higher than that in Mile City at 5.88% (4/68) (χ2 = 5.066, p = 0.024). The infection rate of small mammals was 13.56% (8/59) in residential areas, 9.09% (12/132) in cultivation, 23.08% (18/78) in Shrubland and 15.63% (5/32) in woodland, with statistically significant rates of Bartonella infestation in the different habitats (χ2 = 7.905, p = 0.048), and the habitats of scrubland have the highest infection rate. The prevalence was 22.07% (32/145) in the dominant species of Rattus tanezumi, 1.27% (1/79) in the Suncus murinus, and 12.99% (10/77) in the other non-dominant species of small mammals, with statistically significant differences (χ2 = 18.216, p < 0.001), and Rattus tanezumi had the highest prevalence in this study. In terms of the physical characteristics of the small mammals, the prevalence with body weight > 68 g was 21.71% (33/152) higher than that of those with body weight ≤ 68 g, which was 6.71% (10/149) (χ2 = 13.824, p < 0.001). The infection rate of small mammals with body length > 141 mm was 18.37% (27/147) higher than that of body length ≤ 141 mm 10.39% (16/154) (χ2 = 3.909, p = 0.048); the tail length > 132 mm was 23.81% (35/147) higher than that of tail length ≤ 132 mm infection rate 5.19% (8/154) (χ2 = 21.283, p < 0.001); hindfoot length > 26 mm infection rate was 23.29% (34/146) higher than hindfoot length ≤ 26 mm 5.81% (9/155) (χ2 = 18.763, p < 0.001) and the infection rate of ear height > 17 mm was 22.14% (31/140) higher than ≤17 mm 7.45% (12/161) (χ2 = 13.196, p < 0.001). In addition, there was no statistical difference in Bartonella infection rate among different sex, ages, and flea-carrying status in small mammals, as shown in Table 3.
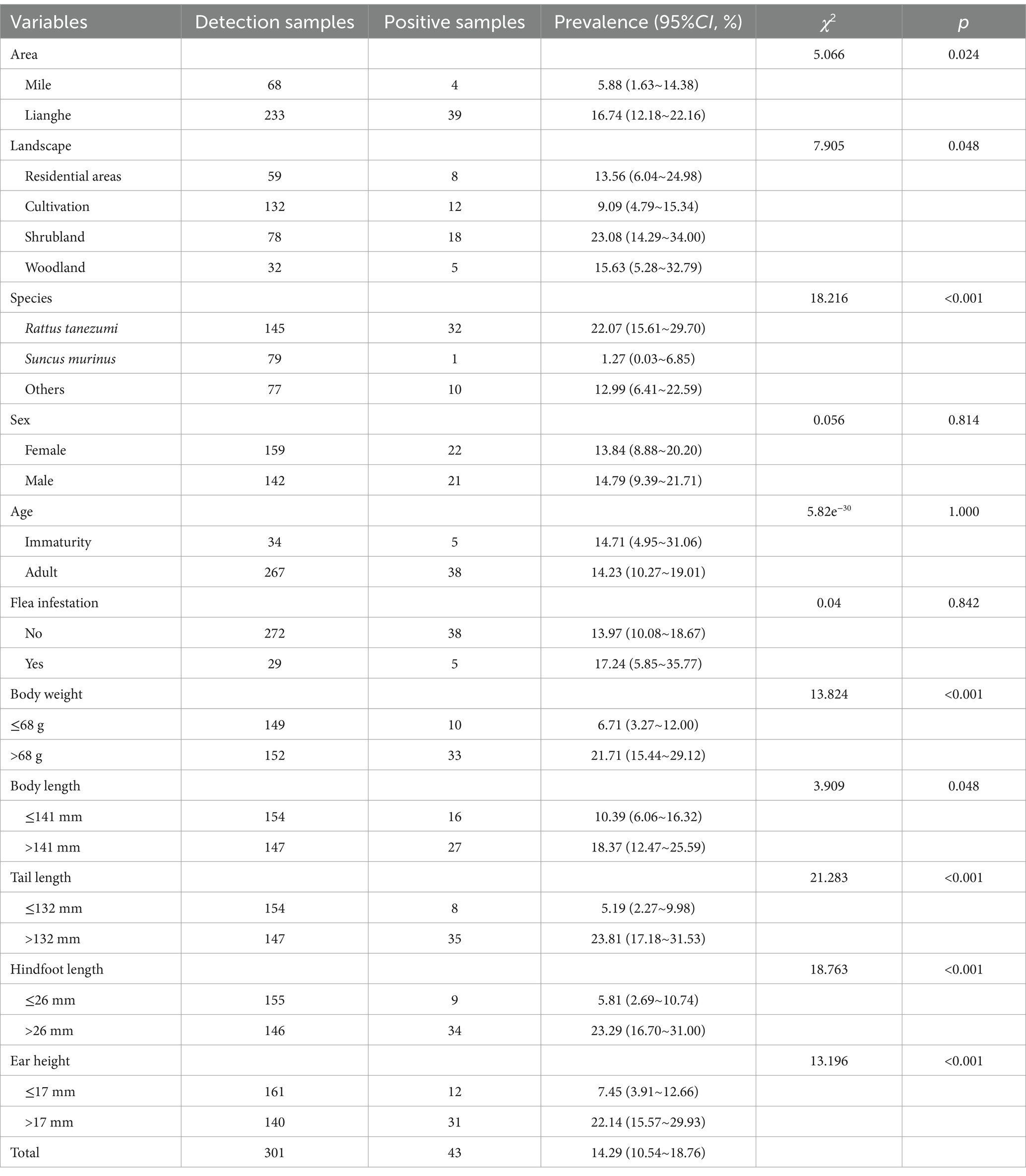
Table 3. Univariate analysis of Bartonella infection rates in small mammals collected from Mile City and Lianghe County, Yunnan province.
The results of the single factor Chi-square test for different characteristics of small mammals showed that there were statistical differences in the prevalence of Bartonella in different areas, habitats, species, body weight, body length, tail length, hind foot length, and ear height. The stepwise regression method was used to incorporate these variables into the model, and finally, three variables (areas, species, and tail length) entered the model. The risk of Bartonella infection in Lianghe County was 3.79 folds higher than that in Mile City (95%CI: 1.39–13.35). Rattus tanezumi had a 90% (95%CI:0.01–0.63) increased risk of Bartonella infection compared to Suncus murinus. The risk of Bartonella infection in small mammals with tail length > 132 mm was 6.34 folds that of ≤132 mm (95%CI: 1.87–23.39). The analysis results are shown in Table 4.
3.4 Identifications of Bartonella species and distribution in small mammals
Fifty-seven strain sequences were obtained from 43 small mammals infected with Bartonella, of which 5 strains were isolated from Rattus tanezumi, Rattus nitidus, and Mus caroli in Mile City, 52 strains were isolated from Rattus tanezumi, Suncus murinus, Niviventer confucianus, Rattus andamanensis and Mus pahari in Lianghe County. Five species were identified through gltA gene analysis, including 32 strains of Bartonella tribocorum, the homology ranged from 91.5% to 99.3%; 11 strains of Bartonella queenslandensis, exhibited a similarity of 92.5% to 98%; 8 strains are closely related to Bartonella phoceensis (98.3%~99.8%); 4 strains of Bartonella mastomydis with identities between 95.5%~99.3%; 2 strains of Bartonella clarridgeiae were 93.3% and 95.8% homologous, respectively. The bacterial strains of small mammals in different regions are shown in Table 5.
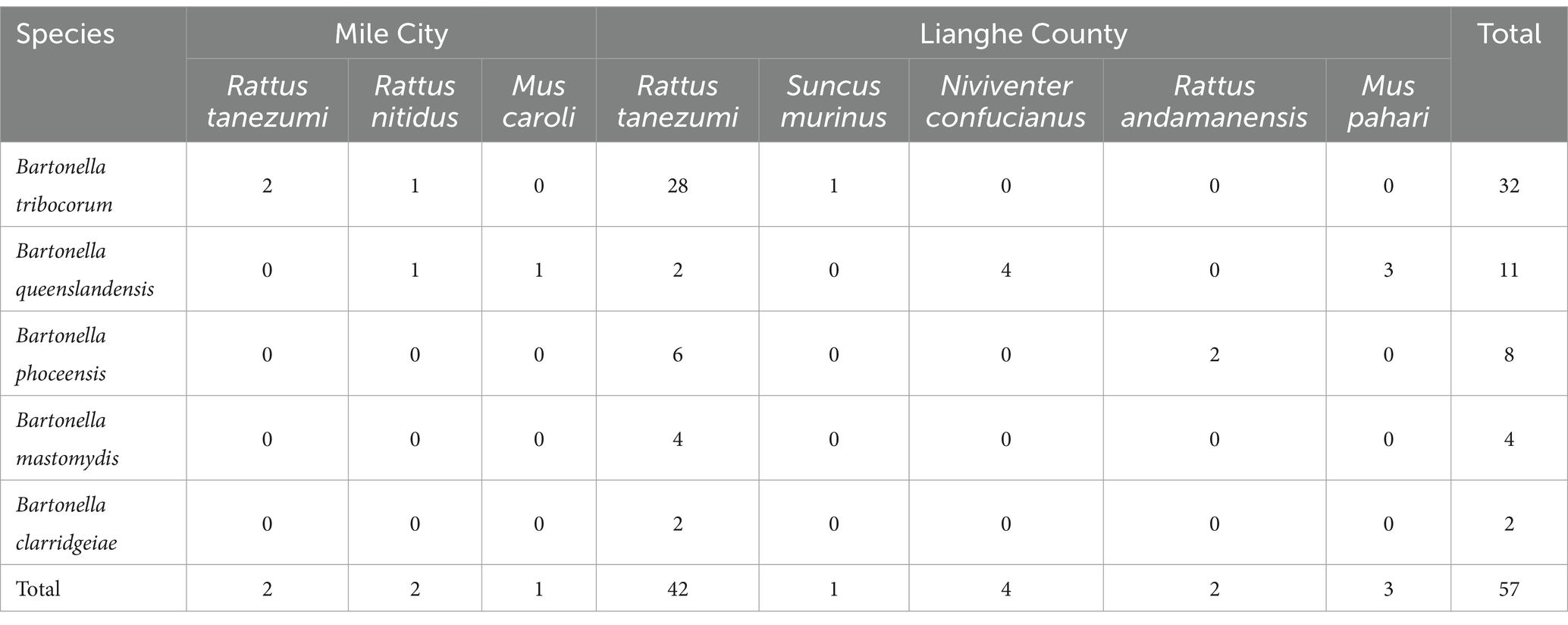
Table 5. Distribution of Bartonella species among small mammals in Mile City and Lianghe County, Yunnan Province.
According to the differences in tissues, four Bartonella species were isolated from kidney tissues, including 3 strains of Bartonella mastomydis, 2 strains of Bartonella phoceensis, 1 strain of Bartonella queenslandensis, and 16 strains of Bartonella tribocorum. Five Bartonella species were isolated from the spleen: 2 strains of Bartonella clarridgeiae, 1 strain of Bartonella mastomydis, 6 strains of Bartonella phoceensis, 10 strains of Bartonella queenslandensis and 16 strains of Bartonella tribocorum. Among them, Bartonella strains were isolated from both tissues of 14 small mammals, and the Bartonella species isolated from different tissues of 13 small mammals were the same, but different species of strains were isolated from different tissues of Rattus tanezumi No. P016. The strain isolated from the kidneys of Rattus tanezumi P016 was Bartonella mastomydis, while that from the spleen was Bartonella tribocorum.
3.5 Phylogenetic tree construction
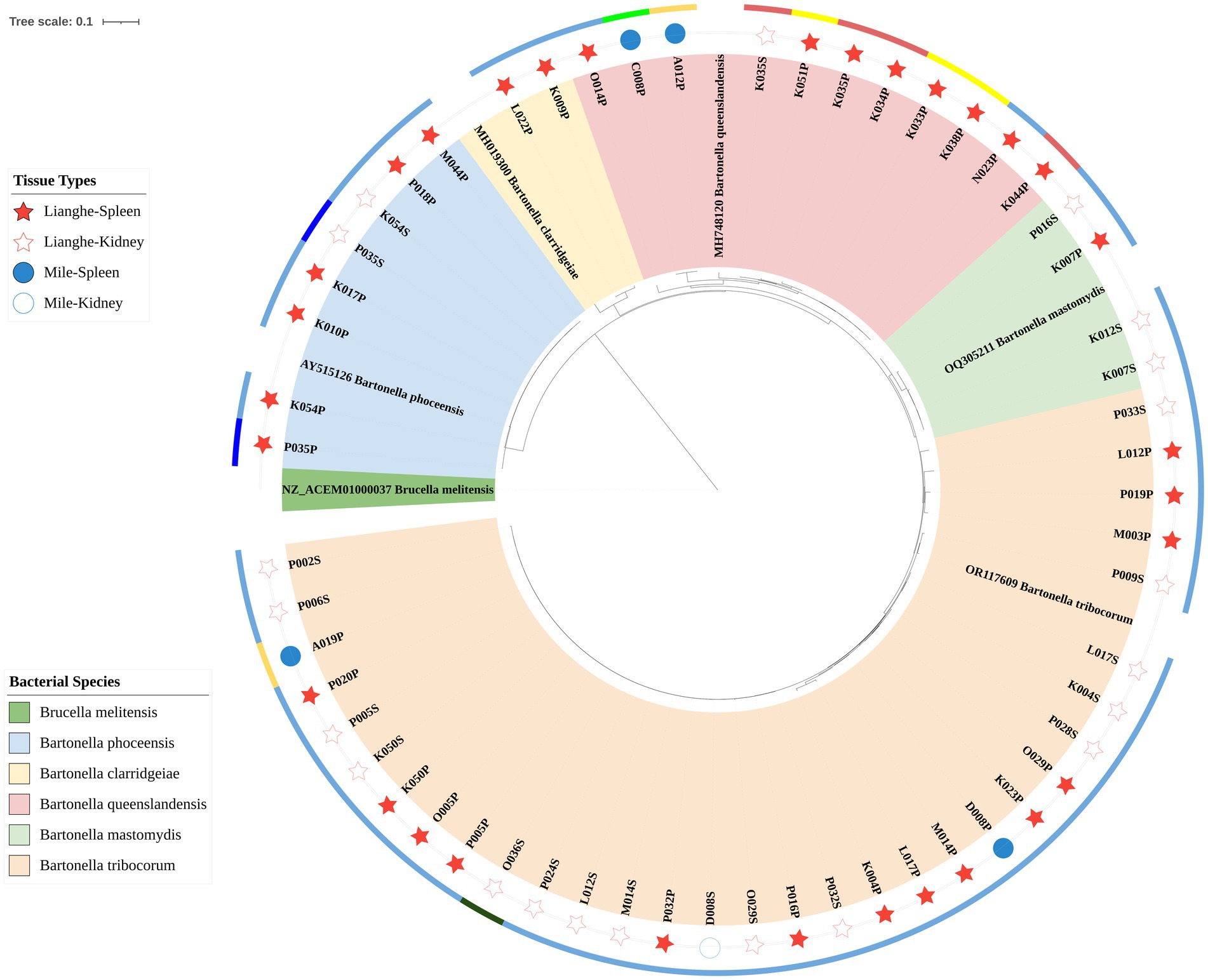
Fifty-seven samples were successfully sequenced, 5 reference strains based on gltA were selected in GenBank after BLAST to construct a phylogenetic tree, and Brucella, which was closely related to Bartonella, was selected as an outgroup. The phylogenetic tree of gltA showed that 57 strains were divided into five distinct clades, including 32 strains in the Bartonella tribocorum branch; 11 strains in the Bartonella queenslandensis branch; 8 strains in the Bartonella phoceensis branch; 4 strains in the Bartonella mastomydis branch; and 2 strains in the Bartonella clarridgeiae branch.
The phylogenetic tree constructed based on the gltA gene is shown in Figure 2, where the red pentagram represents the Lianghe sample, the blue circle represents the Mile sample, the solid is the spleen sample, and the hollow is the kidney sample. The different colors of the outer ring represent the different small mammals: dark blue for the Rattus andamanensis, fluorescent green for the Mus caroli, dark green for the Suncus murinus, light blue for the Rattus tanezumi, light red for the Niviventer confucianus, orange for the Rattus nitidus, and yellow for the Mus pahari. Different branching colors represent different species of Bartonella.
4 Discussion
Bartonella is an emerging zoonotic pathogen that can be transmitted by blood-sucking arthropods. Since Bartonella was first isolated, it has been studied in various countries around the world, and rodents are important reservoir hosts for Bartonella, with the prevalence of Bartonella varying by region (2, 16). In recent years, the prevalence of Bartonella has been studied in many provinces in China, and the results are different (8, 10, 17–20). Therefore, it is important to investigate the prevalence of Bartonella with multiple organs among rodents in different areas.
A total of 301 small mammals were captured in this study and identified as belonging to 11 species, indicating the rich composition of small mammal species in Mile City and Lianghe County, Yunnan Province. The overall dominant rodent species in the two regions are the Rattus tanezumi and the Suncus murinus, and Lianghe County is consistent with the overall situation, but the dominant rodent species in Mile City are the Rattus tanezumi and the Mus caroli, which may be affected by the geographic and climatic differences between Mile City and Lianghe County. Mile City,3 located in the karst landform area of the Eastern Yunnan Plateau (103°04′–103°49′E, 23°50′–24°39′N), sits at an elevation of approximately 1,400 m. Its subtropical plateau monsoon climate features distinct dry and wet seasons, with annual precipitation ranging from 900 to 1,000 mm. Lianghe County,4 situated in the southern extension of the Hengduan Mountains’ river valleys in southwestern Yunnan (98°06′–98°31′E, 24°31′–24°58′N), spans elevations from 500 to 2,600 meters, creating vertical climate zones, characterized by a south subtropical monsoon climate, the county receives 1,300–1,600 mm of annual rainfall, maintains year-round humidity above 75%, and exhibits more pronounced contrasts between its extreme dry and wet seasons. Resulting in the differences in the distribution of rodent species.
The spleen tissue is usually used for the detection of Bartonella, and in this study, both spleen and kidney tissues were used for the detection, and a positive test was sufficient for one of the two. There was a statistically significant difference in the infection rate of Bartonella between the two tissues, with a higher rate of infection in the spleen than in the kidney, and only four strains of species were isolated from kidney tissues, whereas five were isolated from spleen tissues, which suggests that the spleen is more conducive for the growth and reproduction of Bartonella. This is inconsistent with the study results of Yu et al. (11) and Rao et al. (20), which showed no significant difference in the infection rate of various tissues. For the detection of infection in Bartonella, priority can be given to spleen tissue, and when conditions permit, multi-tissue testing is more conducive to accurately describing the true infection situation in rodents and enabling the acquisition of as many strain sequences as possible for analytical traceability.
In this study, the total infection rate of Bartonella in small mammals was 14.29%, which was similar to the rate of 14.9% (169/1,137) in southeastern China (21), higher than the rates of 7.9%–8.38% in Shandong Province (19, 22), 6.4% in Guangdong Province (10), and slightly lower than the rate of rodent Bartonella infection of 16.67% in Beitun area of Xinjiang (9), and significantly lower than 38.61% in the Qaidam Basin in western China (20) and 57.7% in Heilongjiang Province, China (7). The regional differences in Bartonella infection rates among small mammal species are the result of the combined effects of ecological characteristics at sampling sites, the selection of target organs for detection, and the methodological sensitivity employed. Current studies indicate that high-latitude regions (e.g., Heilongjiang and Beitun, Xinjiang) typically exhibit elevated Bartonella infection rates (7, 9, 20), potentially associated with arthropod vector activity and bacterial load in host organisms. Although Yunnan is situated at a lower latitude (24°N), it maintains a moderate infection level of 14.29% in this study area, likely attributable to its high rodent population density and exceptional species diversity (13), suggesting a compensatory effect of host community complexity on pathogen transmission. Regarding detection strategies, this study employed traditional PCR targeting the gltA gene in spleen and kidney tissues. Research from the Qaidam Basin demonstrated comparable infection rates between spleen/brain tissues (20), while studies in Beitun, Xinjiang revealed similar culture-positive rates in liver/spleen tissues (9). Collectively, although the spleen remains the preferred target organ for Bartonella detection (23), with significantly higher bacterial loads than other tissues (13), the low-load characteristics of kidney tissue may dilute overall detection rates. Furthermore, methodological differences substantially impact final detection outcomes: investigations in Heqing and Gongshan counties, Yunnan, utilizing ssrA-qPCR technology, achieved markedly higher sensitivity (31.5%) compared to conventional PCR (9.0%–1.4%) (13); Southeastern China studies enhanced detection reliability (14.9%) through dual-gene (ssrA/gltA) amplification (21); whereas research in Heilongjiang and the Qaidam Basin, combining bacterial culture with multi-gene sequencing (gltA, ftsZ, rpoB, etc.), proved more effective in capturing high-load or viable strains (7, 20).
Through comparative analysis of the geographical distribution of Bartonella in different regions, the research has confirmed that Bartonella infection is associated with the geographical distribution of host animals and is influenced by geographical environmental factors (7). The karst topography of Mile City, characterized by its caves, fissures, and hilly terrain, provides concealed habitats for rodents. In Lianghe County, the densely vegetated mountainous environment at the tropical margin, combined with persistently high humidity and an annual average temperature of around 20°C, sustains stable rodent populations. The dominant rodent species composition differs between these two regions, as does the Bartonella infection status in small mammals. Mile City captured 8 rodent species with 3 species testing positive for Bartonella, while Lianghe County captured 10 rodent species with 5 infected species. In addition, the prevalence of Bartonella among small mammals in Lianghe County in this study was 16.74% (39/233), although higher than 5.88% (4/68) in Mile City, and 3.79 folds higher than the infection risk in Mile City (95%CI: 1.39–13.35), it was significantly lower than 56.27% (229/407) shown by Luo et al. (8), which was found by comparison that there were as many as 18 species of small mammals infected with Bartonella in Lianghe County in their study, suggesting that increased rodent species richness may increase the risk of Bartonella transmitted by rodents.
Rodents are considered to be infected reservoir hosts for Bartonella, and differences in dominant rodent species in different regions may result in different local primary hosts for Bartonella. The prevalence of Bartonella in Inner Mongolia, China, was 47.27% (52/110), with Eolagurus luteus having the highest prevalence of 85.71% (18/21), followed by the Spermophilus dauricus at 47.62% (10/21), and the dominant species, Meriones unguiculatus, with a prevalence of 35.82% (24/67) (18). In this study, the highest infection rate was 75% (3/4) in Niviventer confucianus, followed by 33.33% (2/6) in Rattus nitidus; the dominant species of Rattus tanezumi was 22.07% (32/145), and the rate of Suncus murinus was 1.27% (1/79) and the risk of Suncus murinus infection was only 10% of that of the Rattus tanezumi (95% CI: 0.01–0.63), considering that the captured numbers of Niviventer confucianus and Rattus nitidus were small, they could not truly reflect the prevalence of Bartonella in the two species, and further studies were needed to verify it. The tail length of small mammals is also an influential factor in the prevalence of Bartonella, the longer the tail length, the greater the contact area provided to the pathogen, increasing the risk of infection. Tail length is one of the important external features for morphological identification of rodent species, and in particular, the tail length of adult rodents correlates with the species, confirming the influence of rodent species on the prevalence of Bartonella, which is consistent with the results of the Rattus tanezumi has a higher infection rate than that of the Suncus murinus and other rodent species.
Based on gltA gene sequencing, 57 strains were obtained from 43 murine animals, which were classified into five Bartonella species: Bartonella tribocorum, Bartonella queenslandensis, Bartonella phoceensis, Bartonella mastomydis, and Bartonella clarridgeiae. Among them, Bartonella clarridgeiae detected from Rattus tanezumi in Lianghe County is considered to be a zoonotic strain that may cause endocarditis (24). In addition, Bartonella tribocorum is genetically related to Bartonella elizabethae, which was isolated from endocarditis patients and identified as a zoonotic agent (25). It is suggested that residents of Lianghe County are at risk of infection with zoonotic bacterial strains of Bartonella, which warrants heightened vigilance. Bartonella tribocorum can be detected in Rattus nitidus and Suncus murinus, especially in Rattus tanezumi, so the main host of Bartonella tribocorum in this study was Rattus tanezumi. Bartonella queenslandensis strain has strong adaptability to different rodents and can be detected in Rattus nitidus, Mus caroli, Rattus tanezumi, Niviventer confucianus, and Mus pahari. Bartonella phoceensis was detected in Rattus tanezumi and Rattus andamanensis, in addition, Bartonella mastomydis and Bartonella clarridgeiae were detected only in Rattus tanezumi. Overall, five strains were isolated from Rattus tanezumi, suggesting that Rattus tanezumi are an important reservoir host for infection with Bartonella strains. The detection of multiple Bartonella in small mammals indicates that Bartonella has a high species diversity in small mammals. The same Bartonella can be detected in a variety of small mammals, indicating that Bartonella has a high adaptability in small mammals.
Co-infection (or mixed infection) refers to the simultaneous presence of at least two genetically distinct pathogens within the same host, this phenomenon not only reflects the coexistence of multiple infectious agents in hosts but also involves complex interactions between pathogens (26). In such infections, horizontal gene transfer and competitive inter-genotypic interactions among pathogens serve as critical evolutionary drivers for their survival strategies (27). The impact of mixed infections on host fitness is dual: while they may directly lead to atypical disease manifestations or increased host mortality, they can also indirectly regulate the transmission dynamics of pathogens by altering host susceptibility, infection probability, or transmission rates (26). Taking Bartonella as an example, co-infection systems may exhibit synergistic enhancement due to immune suppression, while ecological exclusion phenomena may arise from resource competition or cross-immunity (28). Previous studies have indicated potentially high co-infection rates in rodents (3). Our study revealed the concurrent detection of Bartonella mastomydis and Bartonella tribocorum in the same host species, the Rattus tanezumi, confirming the existence of Bartonella co-infections in small mammals. This finding aligns with a similar observation from the Qaidam Basin study, where two Bartonella species were isolated from different tissues of a Meriones meridianus (20). Further investigations into multi-tissue infections are warranted to better clarify the co-infection patterns of Bartonella in small mammals.
The limitations of this study are that the bacteria content in some tissue samples was small, the gel electrophoresis imaging showed positive initial results but failed to sequence, and the deviation of sequencing results reduced the number of representative strains. Additionally, the relatively limited sample size (a total of 301 rodents in Mile City and Lianghe County) may affect the accuracy of Bartonella infection rate assessment. Compared with similar studies in Yunnan Province, such as research in Jianchuan County, Yulong County, and Lianghe County based on 1,605 single-organ samples revealing an overall infection rate of 47.85% (8) and a study in Heqing County and Gongshan County with 333 samples achieving a 31.5% infection rate through multi-organ detection (13), the 14.29% overall infection rate observed here suggests insufficient sample size may weaken statistical power, potentially underestimating true infection levels or misjudging regional heterogeneity. Furthermore, the conventional PCR targeting the gltA gene used in this study exhibits technical limitations in sensitivity, struggling to identify low pathogen-load samples and non-gltA-dominant species, which may contribute to lower observed infection rates compared to high-prevalence regions. Future studies should expand the sampling scope, enhance sample representativeness, and implement multi-organ detection where feasible. To improve detection efficacy, a two-step strategy is recommended: initial high-efficiency screening using Bartonella genus-specific qPCR, followed by confirmatory testing of multiple conserved genes (e.g., ssrA, rpoB, ftsZ) via conventional or multiplex real-time PCR. Additionally, integrating molecular detection of multiple tissues and establishing a multi-locus sequence typing (MLST)-based analytical system will help systematically elucidate Bartonella epidemiology and genetic diversity in rodent populations. More sensitive and accurate sequencing methods should also be adopted to obtain stable results, aiding in characterizing antibacterial prevalence in murine animals.
The prevalence of Bartonella was higher among small mammals in Mile City and Lianghe County, Yunnan Province. Five species of Bartonella were isolated from the kidney or spleen tissues of 7 small mammals (Rattus tanezumi, Suncus murinus, Niviventer confucianus, Rattus andamanensis, Mus pahari, Rattus nitidus, and Mus caroli), including Bartonella tribocorum, Bartonella queenslandensis, Bartonella phoceensis, Bartonella mastomydis and Bartonella clarridgeiae, where Bartonella tribocorum is the dominant species in both regions. It is genetically related to Bartonella elizabethae, which was identified as a zoonotic pathogen early, and Bartonella clarridgeiae is also a zoonotic strain. The infection rate and isolated genotype of Bartonella in the spleen were higher than those in the kidney, which was more conducive to the growth and reproduction of Bartonella. The prevalence of Bartonella is affected by region, rodent species, and tail length. Therefore, it is very important to monitor the prevalence of Bartonella in rodents in different regions, and it is necessary to obtain a whole genome sequence as far as possible to identify Bartonella species more accurately, to better take targeted measures to prevent the infection of Bartonella transmitted by rodents to humans.
Data availability statement
The original contributions presented in the study are publicly available. These data can be found in GenBank at the National Center for Biotechnology Information (NCBI), accession numbers PQ279835-PQ279891.
Ethics statement
The animal study was approved by the Medical Ethics Committee of Dali University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RF: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. J-XY: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – review & editing. PH: Data curation, Investigation, Methodology, Writing – review & editing. YC: Data curation, Investigation, Methodology, Writing – review & editing. YL: Data curation, Investigation, Validation, Writing – review & editing. P-GL: Data curation, Investigation, Validation, Writing – review & editing. S-LG: Data curation, Investigation, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the National Natural Science Foundation of China (no. 81860565), the project “Talent Support Program in Yunnan” (no. YNWR-MY-2019-008), and the Science and Technology Innovation Team of Natural Focal Diseases Epidemiology in the University of Yunnan Province [Yunnan Provincial Department of Education issued (2020) no.102].
Acknowledgments
The authors would like to thank the Lianghe Center for Disease Control and Prevention, and the Mile Center for Disease Control and Prevention for assistance in the progress of the project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Breitschwerdt, EB. Bartonellosis, one health and all creatures great and small. Vet Dermatol. (2017) 28:96. doi: 10.1111/vde.12413
2. Gutiérrez, R, Krasnov, B, Morick, D, Gottlieb, Y, Khokhlova, IS, and Harrus, S. Bartonella infection in rodents and their flea ectoparasites: an overview. Vector Borne Zoonotic Dis. (2015) 15:27–39. doi: 10.1089/vbz.2014.1606
3. Gutiérrez, R, Cohen, C, Flatau, R, Marcos-Hadad, E, Garrido, M, Halle, S, et al. Untangling the knots: co-infection and diversity of Bartonella from wild gerbils and their associated fleas. Mol Ecol. (2018) 27:4787–807. doi: 10.1111/mec.14906
4. Buhler, KJ, Fernando, C, Hill, JE, Galloway, T, Carriere, S, Fenton, H, et al. Combining deep sequencing and conventional molecular approaches reveals broad diversity and distribution of fleas and Bartonella in rodents and shrews from Arctic and subarctic ecosystems. Parasit Vectors. (2022) 15:366. doi: 10.1186/s13071-022-05446-w
5. Demoncheaux, J-P, Medkour, H, Louni, M, Laugier, L, Pasqualini, C, Fenollar, F, et al. Detection of potential zoonotic Bartonella species in African Giant rats (Cricetomys gambianus) and fleas from an urban area in Senegal. Microorganisms. (2022) 10:489. doi: 10.3390/microorganisms10030489
6. Mohd-Azami, SNI, Loong, SK, Khoo, JJ, Husin, NA, Lim, FS, Mahfodz, NH, et al. Molecular surveillance for vector-borne Bacteria in rodents and tree shrews of peninsular Malaysia oil palm plantations. Trop Med Infect Dis. (2023) 8:74. doi: 10.3390/tropicalmed8020074
7. Li, D-M, Hou, Y, Song, X-P, Fu, Y-Q, Li, G-C, Li, M, et al. High prevalence and genetic heterogeneity of rodent-borne Bartonella species on Heixiazi Island, China. Appl Environ Microbiol. (2015) 81:7981–92. doi: 10.1128/AEM.02041-15
8. Luo, Y-Y, Yu, D, Zhang, H-Z, Liu, Z-X, Hong, R-D, Hong, M, et al. Molecular detection of Bartonella species in wild small mammals in western Yunnan Province, China. Front Vet Sci. (2023) 10:1301316. doi: 10.3389/fvets.2023.1301316
9. Xu, A-L, Chen, Y-F, Mu, L, Liu, P-B, Wang, J, Li, R-X, et al. Bartonella prevalence and genome sequences in rodents in some regions of Xinjiang, China. Appl Environ Microbiol. (2023) 89:e0196422. doi: 10.1128/aem.01964-22
10. Yao, X-Y, Liu, H, Sun, J, Zhang, Y-Q, Lv, Z-H, Zhang, X-L, et al. Epidemiology and genetic diversity of Bartonella in rodents in urban areas of Guangzhou, southern China. Front Microbiol. (2022) 13:942587. doi: 10.3389/fmicb.2022.942587
11. Yu, J, Xie, B, Bi, G-Y, Zuo, H-H, Du, X-Y, Bi, L-F, et al. Prevalence and diversity of small rodent-associated Bartonella species in Shangdang Basin, China. PLoS Negl Trop Dis. (2022) 16:e0010446. doi: 10.1371/journal.pntd.0010446
12. Wang, N, Yin, J-X, Zhang, Y, Wu, L, Li, W-H, Luo, Y-Y, et al. Genetic evolution analysis and host characteristics of hantavirus in Yunnan Province, China. Int J Environ Res Public Health. (2022) 19:3433. doi: 10.3390/ijerph192013433
13. Han, P-Y, Xu, F-H, Tian, J-W, Zhao, J-Y, Yang, Z, Kong, W, et al. Molecular prevalence, genetic diversity, and tissue tropism of Bartonella species in small mammals from Yunnan Province, China. Animals. (2024) 14:1320. doi: 10.3390/ani14091320
14. Ji, X, Wang, M, Li, L, Chen, F, Zhang, Y, Li, Q, et al. The impact of repeated freeze-thaw cycles on the quality of biomolecules in four different tissues. Biopreserv Biobanking. (2017) 15:475–83. doi: 10.1089/bio.2017.0064
15. Norman, AF, Regnery, R, Jameson, P, Greene, C, and Krause, DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. (1995) 33:1797–803. doi: 10.1128/jcm.33.7.1797-1803.1995
16. Malania, L, Bai, Y, Osikowicz, LM, Tsertsvadze, N, Katsitadze, G, Imnadze, P, et al. Prevalence and diversity of Bartonella species in rodents from Georgia (Caucasus). Am J Trop Med Hygiene. (2016) 95:466–71. doi: 10.4269/ajtmh.16-0041
17. Jian, R, Ren, Q, Xue, J, Xie, G-C, Wang, J, Chen, G-Q, et al. Genetic diversity of Bartonella infection in residential and field rodents in Hebei, China. Front Microbiol. (2022) 13:1039665. doi: 10.3389/fmicb.2022.1039665
18. Li, J, Zhang, C, Lu, M, Wang, Y, Wang, W, Liu, F, et al. The diverse genetic genotypes of Bartonella species circulating in rodents from Inner Mongolia, northern China. PLoS Negl Trop Dis. (2023) 17:e0011462. doi: 10.1371/journal.pntd.0011462
19. Qin, X-R, Liu, J-W, Yu, H, and Yu, X-J. Bartonella species detected in rodents from eastern China. Vector Borne Zoonotic Dis. (2019) 19:810–4. doi: 10.1089/vbz.2018.2410
20. Rao, H, Li, S, Lu, L, Wang, R, Song, X, Sun, K, et al. Genetic diversity of Bartonella species in small mammals in the Qaidam Basin, western China. Sci Rep. (2021) 11:1735. doi: 10.1038/s41598-021-81508-w
21. Liu, H, Han, T, Liu, W, Xu, G, Zheng, K, and Xiao, F. Epidemiological characteristics and genetic diversity of Bartonella species in rodents from southeastern China. Zoonoses Public Health. (2022) 69:224–34. doi: 10.1111/zph.12912
22. Zhang, L, Peng, Q, Gu, X-L, Su, W-Q, Cao, X-Q, Zhou, C-M, et al. Host specificity and genetic diversity of Bartonella in rodents and shrews from eastern China. Transbound Emerg Dis. (2022) 69:3906–16. doi: 10.1111/tbed.14761
23. Gutiérrez, R, Vayssier-Taussat, M, Buffet, J-P, and Harrus, S. Guidelines for the isolation, molecular detection, and characterization of Bartonella species. Vector Borne Zoonotic Dis. (2017) 17:42–50. doi: 10.1089/vbz.2016.1956
24. Eremeeva, ME, Gerns, HL, Lydy, SL, Goo, JS, Ryan, ET, Mathew, SS, et al. Bacteremia, fever, and splenomegaly caused by a newly recognized bartonella species. N Engl J Med. (2007) 356:2381–7. doi: 10.1056/NEJMoa065987
25. Heller, R, Riegel, P, Hansmann, Y, Delacour, G, Bermond, D, Dehio, C, et al. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. (1998) 48:1333–9.
26. Hoarau, AOG, Mavingui, P, and Lebarbenchon, C. Coinfections in wildlife: focus on a neglected aspect of infectious disease epidemiology. PLoS Pathog. (2020) 16:e1008790. doi: 10.1371/journal.ppat.1008790
27. Abbot, P, Aviles, AE, Eller, L, and Durden, LA. Mixed infections, cryptic diversity, and vector-borne pathogens: evidence from Polygenis fleas and Bartonella species. Appl Environ Microbiol. (2007) 73:6045–52. doi: 10.1128/AEM.00228-07
Keywords: Bartonella, small mammals, prevalence, phylogenetic analysis, Yunnan Province
Citation: Fu R, Yin J-X, He P, Chen Y, Luo Y, Liu P-G and Guo S-L (2025) Comparative study on Bartonella infection in spleen and kidney of small mammals from Mile City and Lianghe County, Yunnan Province. Front. Vet. Sci. 12:1554633. doi: 10.3389/fvets.2025.1554633
Edited by:
Kevin Bown, Liverpool John Moores University, United KingdomReviewed by:
Marcela Suárez-Esquivel, National University of Costa Rica, Costa RicaQuan Liu, First Affiliated Hospital of Jilin University, China
Copyright © 2025 Fu, Yin, He, Chen, Luo, Liu and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia-Xiang Yin, Y2hpbmF5anhAaG90bWFpbC5jb20=
 Rong Fu
Rong Fu Jia-Xiang Yin
Jia-Xiang Yin Ping He
Ping He