- National Institutes for Food and Drug Control (NIFDC), Beijing, China
Lungworm disease caused by Dictyocaulus filaria is an infectious condition affecting sheep and goats worldwide in recent years. It causes significant economic losses and is considered a potential public health threat. To date, there have been few studies on Dictyocaulus filaria. Its pathogenic mechanism is still unclear, and there are no effective vaccines or drugs available for the prevention and control of the disease. Therefore, it is essential to develop a rapid and reliable molecular diagnostic method to facilitate the study of this novel parasite. In this study, we developed a TaqMan-minor groove binder (MGB) probe-based quantitative real-time polymerase chain reaction (qPCR) for the rapid detection of Dictyocaulus filaria for the first time. Specific primers and a TaqMan-MGB probe were designed targeting the internal transcribed spacer 2 (ITS2) region of Dictyocaulus filaria. The assay showed a strong specificity for detecting Dictyocaulus filaria, and had no cross-reactivity with other control pathogenic parasites. It also exhibited high sensitivity, with both the lower limit of quantification (LLOQ) and the limit of detection (LOD) determined to be 1.5 copies per reaction. The assay exhibited excellent repeatability and reproducibility, with an intra-assay coefficient of variation (CV) of 0.11–0.58% and an inter-assay CV of 0.33–2.19%. Finally, the developed TaqMan-MGB probe-based qPCR was used to detect 200 clinical samples. The results indicated that the positive detection rate of Dictyocaulus filaria was 3.5% (7/200) for Dictyocaulus filaria, and this finding showed good consistency with conventional PCR. The developed TaqMan-MGB probe-based qPCR assay offers several advantages, including high sensitivity, strong specificity, high throughput, speed, convenience, and cost-effectiveness. It is of great significance for safeguarding animal quality and protecting human health.
Introduction
Lungworm disease caused by Dictyocaulus filaria (Rudolphi, 1809) is an infectious condition affecting sheep and goats worldwide. It can pose serious threats to animal quality, human health, and overall public health security (1–3). Dictyocaulus filaria resides in the trachea and bronchioles, where the host's response to infection leads to bronchitis, eosinophilic pneumonia, and respiratory bronchiole obstruction. This can cause clinical signs such as dyspnea, coughing, fever, loss of appetite, poor growth and development, and even death (4–7).
Accurate diagnosis of lungworm disease using reliable detection techniques is essential for formulating targeted control measures, which are crucial for the effective management of Dictyocaulus filaria.
At present, the diagnosis of Dictyocaulus filaria depends on microscopic examination methods, such as the Baermann technique, which is used to identify first-stage larvae in feces (8, 9). However, microscopic examination may easily miss cases with less apparent parasite morphological characteristics, potentially resulting in environmental pollution and endangering the safety of inspectors.
The enzyme-linked immunosorbent assay (ELISA) is suitable for large-scale sample testing, but it involves multiple steps and requires a long experimental duration. It is usually used to detect antibodies in serum. However, in the early stages of infection, the body may not have produced sufficient relevant antibodies or antibody levels may not have reached the detection limit, increasing the likelihood of false negative results. In addition, cross-reactions between Dictyocaulus filaria and certain other parasites can easily lead to false positive results, thereby affecting the specificity of the detection method (10).
Therefore, reliable detection techniques are needed for the timely and accurate diagnosis of lungworm disease to prevent its widespread transmission. Due to the challenges associated with serologic testing, molecular testing has become the preferred diagnostic test for detecting certain pathogenic agent infections. In parasitological diagnosis, molecular detection offers high specificity because it can detect the specific nucleic acid of parasites. This approach also effectively addresses the “bottleneck” problem associated with traditional microscopic examination, which often misses early infection cases.
With advancements in molecular biology, polymerase chain reaction (PCR) has been increasingly used for the auxiliary diagnosis of lungworm disease and the detection and identification of Dictyocaulus lungworms (11–14). However, conventional PCR has some limitations, such as complex procedures, a high risk of contamination, and challenges in providing accurate and quantitative detection.
Random amplified polymorphic DNA (RAPD)-PCR and PCR/ restriction fragment length polymorphism (RFLP) methods compare banding patterns to assess the degree of genetic differences between different lungworms (Dictyocaulidae). However, these methods require PCR amplification, enzyme digestion identification, and agarose gel electrophoresis to analyze the results (15, 16).
Loop-mediated isothermal amplification (LAMP) offers high amplification efficiency and a short reaction time. However, due to its strong sensitivity, it is particularly prone to aerosol formation, which can lead to false positives and impact the reliability of test results. Therefore, there is an urgent need to develop a rapid, specific, sensitive, and high-throughput detection method for lungworm disease, particularly for early-stage infections of Dictyocaulus filarial, as well as for related drugs and biological products.
In recent years, TaqMan probe-based quantitative real-time polymerase chain reaction (qPCR) has been widely used for gene expression analysis, mutation and polymorphism research, and qualitative and quantitative detection of pathogens due to its advantages of high sensitivity, rapid speed, and strong specificity. Compared to conventional TaqMan probe-based qPCR, the minor groove binder (MGB) probe has two key features: First, the 3′ end of the probe is labeled with a non-fluorescent quencher (NFQ). This reduces the fluorescence background and greatly improves the fluorescence spectral resolution. Second, an MGB is attached to the 3′ end of the probe. The MGB can stabilize the hybridization between the probe and the template, increase the annealing temperature of the probe, shorten the length of the probe, and reduce the distance between the fluorophore and the quenching group, thereby enhancing the quenching effect. On the other hand, the specificity of the probe is also improved, resulting in more accurate results and higher resolution (17).
To date, there have been no reports of using TaqMan-MGB probe-based qPCR to detect Dictyocaulus filaria. In this study, a specific and sensitive TaqMan-MGB probe-based qPCR method was developed for the first time, which was successfully applied to the rapid detection of Dictyocaulus filaria in clinical samples. This provides an effective tool for the molecular diagnosis of lungworm disease.
Materials and methods
Design and synthesis of primers and a probe
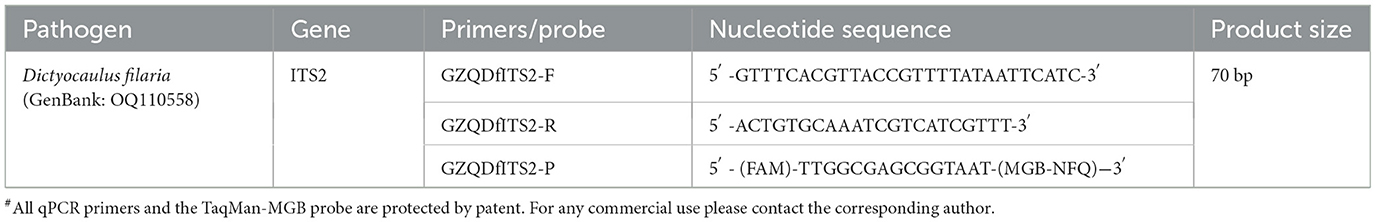
According to the whole genome sequences of Dictyocaulus filaria deposited in GenBank, multiple sequence alignment was performed with the DNASTAR software (version 7.1) to identify the highly conserved internal transcribed spacer 2 (ITS2) region of Dictyocaulus filaria as the target gene. Primers and a probe for the ITS2 gene sequences were designed with Primer Express™ Version 3.0 (Thermo Fisher Scientific, Waltham, MA, USA). The specificity of all primers and the probe was checked using the Basic Local Alignment Search Tool (BLAST) provided by the National Center for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). No significant similarities were observed. Specific primers and a probe for TaqMan-MGB probe-based qPCR were designed, with the 5′ end labeled with a FAM fluorescence reporter and the 3′ end labeled with an MGB NFQ. The primers and probe were synthesized using Applied Biosystems (ABI, Massachusetts, USA), and their sequences are shown in Table 1. To evaluate the specificity of the primer and probe sets, DNA from Dictyocaulus filaria was confirmed by PCR-specific primers serving as templates for amplification using the developed TaqMan-MGB probe-based qPCR. Primers and a probe demonstrating excellent specificity and high sensitivity were identified through experimental screening.
Parasites and clinical samples
Clinical samples collected between 2019 and 2023 were preserved at −80°C in our laboratory. These samples were mainly obtained from Beijing and Jilin in China. DNA preserved at −20°C was used as the PCR template. All positive samples were identified by conventional PCR in our laboratory and confirmed by DNA sequencing conducted by Takara Biomedical Technology (Beijing) Co., Ltd.
DNA extraction
According to the manufacturer's instructions, total DNA was extracted from blood, serum, and anal swab samples using the DNeasy Blood and Tissue Kit (Qiagen, Germany). Parasite DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Germany).
Construction of plasmid standards
The target fragments of Dictyocaulus filaria were amplified by PCR using the DNA extracted in the previous step. The primers used for amplification were the same as those used in the TaqMan-MGB probe-based qPCR method. The PCR fragments were then cloned into the pMD18-T vector (Takara Biomedical Technology (Beijing) Co., Ltd.) via TA cloning and confirmed by DNA sequencing. Plasmid concentrations were then converted to copy number using the following formula: {plasmid copy number/μL = [6.02 × 1023 × plasmid concentration (ng/μL) × 10−9] / [plasmid length × 660]}. TaqMan-MGB probe-based qPCR was performed for Dictyocaulus filaria using 10-fold diluted plasmids to generate standard curves, based on which, the E value (amplification efficiency), R2 (correlation coefficient), and the standard equation were calculated.
Optimization of the TaqMan-MGB probe-based qPCR assay
The primer and probe concentrations, along with the annealing temperatures of the TaqMan-MGB probe-based qPCR assay, were optimized to establish the optimal reaction system and conditions for detecting Dictyocaulus filaria. Briefly, TaqMan-MGB probe-based qPCR was performed in a 20 μL reaction volume, consisting of 10 μL of TaqMan™ Qiankun Platinum Multiple Premix (A55163, Applied Biosystem, USA), 1 μL of primer/probe mix (0.9 μM of the forward primer GZQDfITS2-F, 0.9 μM of the reverse primer GZQDfITS2-R, and 0.25 μM of the probe GZQDfITS2-P for Dictyocaulus filaria), 1 μL of template DNA, and 8 μL of nuclease-free water. Amplification reactions were performed using the QuantStudio™ 6 Real-time PCR System (Applied Biosystems, Massachusetts, USA). The amplification procedure was as follows: 50°C for 2 min (1 cycle), followed by 95°C for 10 min (1 cycle), then 40 cycles of 95°C for 15 s and 60°C for 1 min. Fluorescence signals for each sample were collected at the end of each step at 60°C. Quantification cycle (Cq) values were generated using the QuantStudio Design & Analysis Software with automated threshold settings. Positive and negative controls were included in each assay.
Specificity of the TaqMan-MGB probe-based qPCR assay
To assess potential cross-reactivity, DNA from other parasites, including Strongyloides stercoralis, Syphacia obvelata, Syphacia muris, and Aspiculuris tetraptera, was tested to evaluate the specificity of the TaqMan-MGB probe-based qPCR assay using the abovementioned procedure and parameters. Dictyocaulus filaria was used as the positive control, while nuclease-free water was used as the negative control.
The efficiency (E), lower limit of quantification (LLOQ), and limit of detection (LOD) of the TaqMan-MGB probe-based qPCR assay
To evaluate the linearity, dynamic range, amplification efficiency (E), lower limit of quantification (LLOQ), and limit of detection (LOD) of the TaqMan-MGB probe-based qPCR assay (18–21), serial dilutions of Dictyocaulus filaria DNA standards were used as templates. A final standard curve was generated based on the Cq value and the logarithm of the standard copy number. The slope of the linear regression of a plot of Cq (y-axis) vs. the log of the target concentration (x-axis) was determined by the amplification efficiency (E), which was calculated as %E = 100 × (−1 + 10 (−1/slope)).
Repeatability and reproducibility of the TaqMan-MGB probe-based qPCR assay
To assess the repeatability and reproducibility of the TaqMan-MGB probe-based qPCR assay, five 10-fold gradient dilutions (1.5 × 107-1.5 × 103 copies/μL) of the DNA standard were tested in triplicate on the same day to evaluate intra-assay repeatability. For inter-assay reproducibility, each dilution was tested in triplicate across three different days within 1 week. The coefficient of variation (CV) for intra- and inter-assay precision was calculated using the following formula: %CV = 100 × [standard deviation (SD) value / mean value].
Clinical performance of the TaqMan-MGB probe-based qPCR assay
DNA from clinical specimens was tested using the TaqMan-MGB probe-based qPCR assay. When detecting clinical specimens, positive controls (Dictyocaulus filaria DNA) and nuclease-free water were included in each run to monitor for false negative and false positive results, respectively.
Statistical analysis
Statistical analyses for the standard curves were carried out using SPSS Statistics (version: 21.0). Correlation coefficients (R2) and amplification efficiencies (E) of the standard curves were calculated, and the TaqMan-MGB probe-based qPCR amplification curves were visualized using the QuantStudio Design & Analysis Software (version: 1.4.1).
Results
Specificity of the TaqMan-MGB probe-based qPCR assay
The results showed that only Dictyocaulus filaria produced positive fluorescent signals and specific amplification curves, while Strongyloides stercoralis, Syphacia obvelata, Syphacia muris, and Aspiculuris tetraptera did not demonstrate any positive fluorescent signals or amplification curves, indicating the high specificity of the TaqMan-MGB probe-based qPCR assay (Figure 1).
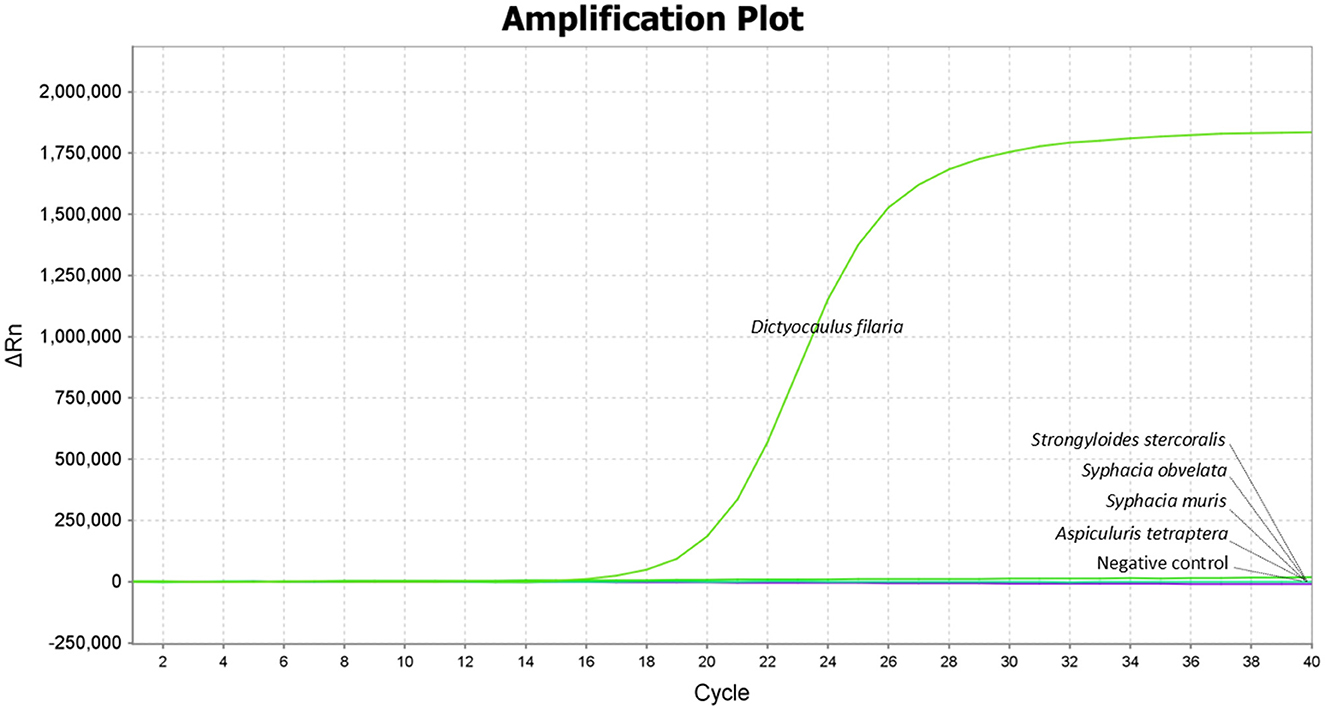
Figure 1. Specificity of the TaqMan-MGB probe-based qPCR assay for detecting Dictyocaulus filaria. Amplification curves show the samples positive for Dictyocaulus filaria as detected by the TaqMan-MGB probe-based qPCR assay. The negative samples include Strongyloides stercoralis, Syphacia obvelata, Syphacia muris, and Aspiculuris tetraptera, and nuclease-free water was used as the negative control.
Efficiency (E) of the TaqMan-MGB probe-based qPCR assay
To construct a standard curve with the logarithm of the DNA copy number and the measured Cq values, serial 10-fold gradient dilutions of the DNA standards at concentrations ranging from 1.5 × 1010 to 1.5 × 100 copies/μL were tested. Three replicates were tested for each dilution in a single reaction. The optimal curves were selected as the standard curves (Figure 2).
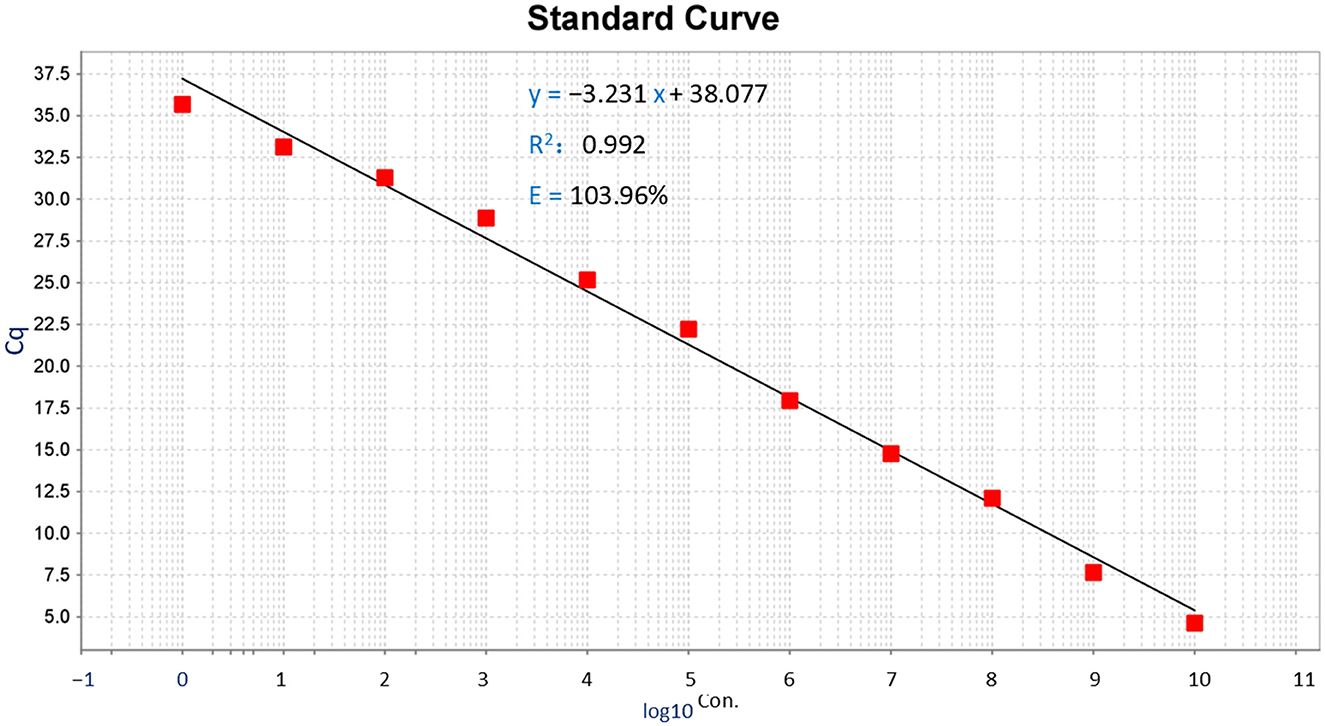
Figure 2. Standard curve of TaqMan-MGB probe-based qPCR for detecting Dictyocaulus filaria. Standard curves of Dictyocaulus filaria for plasmid standards at concentrations ranging from 1.5 × 101° to 1.5 × 10° copies per reaction.
Establishment of the Dictyocaulus filaria standard curve: The abscissa represents the logarithm of the copy number, and the ordinate represents the Cq value. The correlation coefficient was 0.992, the slope of the equation was −3.231, the intercept was 38.077, and the amplification efficiency (E) of Dictyocaulus filaria was 103.926%. The standard formula for Dictyocaulus filaria was y = −3.231x + 38.077 (Figure 2, the y-axis represents the Cq value, and the x-axis represents the logarithm of the concentration). The correlation coefficient of the standard curves for Dictyocaulus filaria was 0.992, suggesting that the correlation between each quantified concentration group of the standards was reliable. The amplification efficiency of Dictyocaulus filaria was 103.926%, which is considered satisfactory for TaqMan-MGB probe-based qPCR.
Lower limit of quantification (LLOQ), and limit of detection (LOD) of the TaqMan-MGB probe-based qPCR assay
The 10-fold gradient dilutions of the DNA standard were tested using the developed TaqMan-MGB probe-based qPCR. The lower limit of quantification (LLOQ) of the method was 1.5 × 100 copies/μL for detecting Dictyocaulus filaria (Figure 3), indicating that the TaqMan-MGB probe-based qPCR was highly sensitivity.
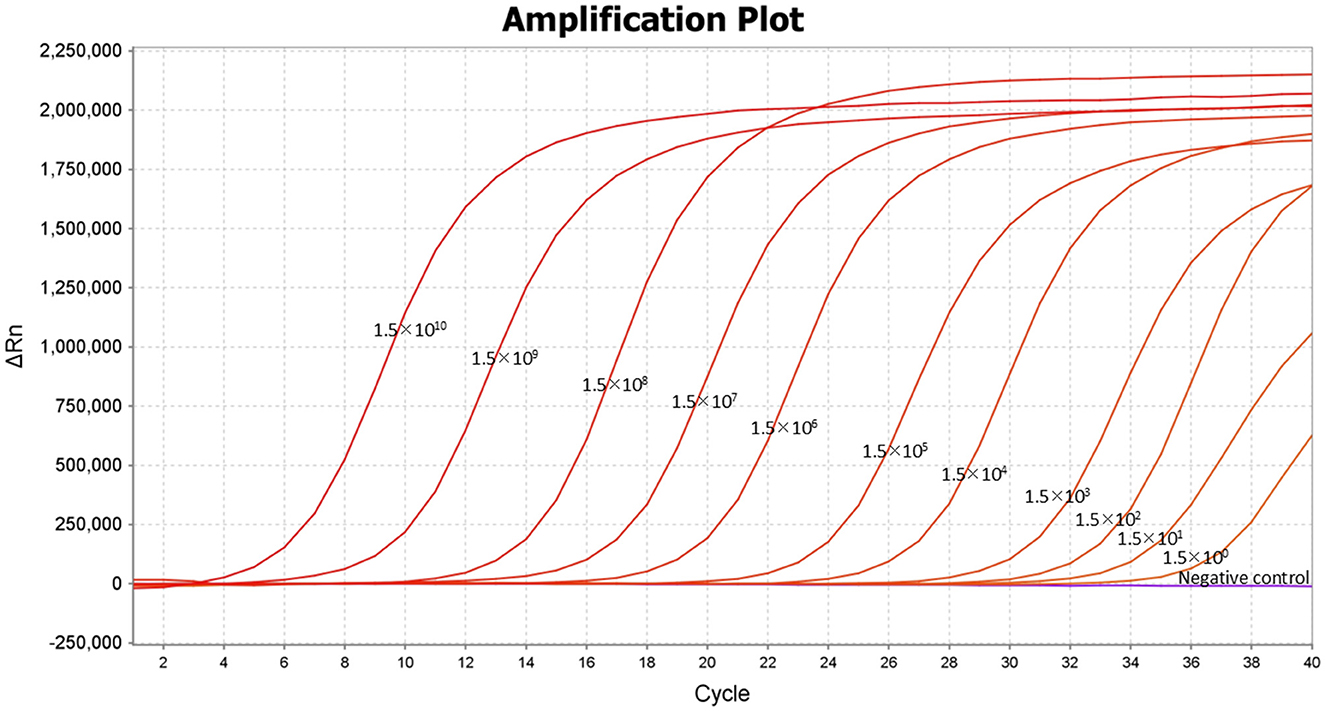
Figure 3. Lower limit of quantification (LLOQ) of the TaqMan-MGB probe-based qPCR assay for detecting Dictyocaulus filaria. Amplification curves of Dictyocaulus filaria obtained using serial 10-fold dilutions of the DNA standards, ranging from 1.5 × 1010 copies/μL to 1.5 × 100 copies/μL.
The two-fold gradient dilution of the DNA standard was tested using the developed TaqMan-MGB probe-based qPCR. The limit of detection (LOD) for this method was defined as the lowest concentration of the target analyte, which can be detected with a 95% detection rate. Follow-up experiments showed that the detection rate of the samples at 1.5 copies/μL for detecting Dictyocaulus filaria was 100% across 20 replicates (Table 2). Thus, the reliable LOD of the TaqMan-MGB probe-based qPCR assay was 1.5 copies per reaction for detecting Dictyocaulus filaria. In this experiment, the cutoff line for positivity was automatically determined by the QuantStudio™ 6 Real-time PCR Instrument. We set the cutoff line for positivity at 38, meaning that the samples with a Cq value less than or equal to 35 (≤35) were regarded as positive, those with Cq values greater than 35 but less than or equal to 38 (35 < and ≤ 38) were considered invalid, and those with Cq values greater than 38 (>38) were considered negative. The criteria were established based on two considerations. First, the LOD of the method was 1.5 copies/μL, with a corresponding Cq value of ~35. Second, although some samples at 1.5 copies/μL were detectable, their Cq values were ~35.71. All subsequent experiments were conducted in accordance with these criteria. Briefly, if there was a specific S-shaped curve and the Cq value was ≤ 35, the result was determined to be positive for Dictyocaulus filaria nucleic acid. If there was no Cq value and no specific fluorescence amplification curve, the result was interpreted as negative for Dictyocaulus filaria nucleic acid. If the Cq value was between 35 and 38 (35 < Cq value < 38) and there was a specific fluorescence amplification curve, the result was interpreted as suspicious for Dictyocaulus filaria nucleic acid, and the suspicious samples were resampled for DNA extraction and retesting. If the Cq value was < 38, the result was interpreted as positive; otherwise, the result was considered negative.
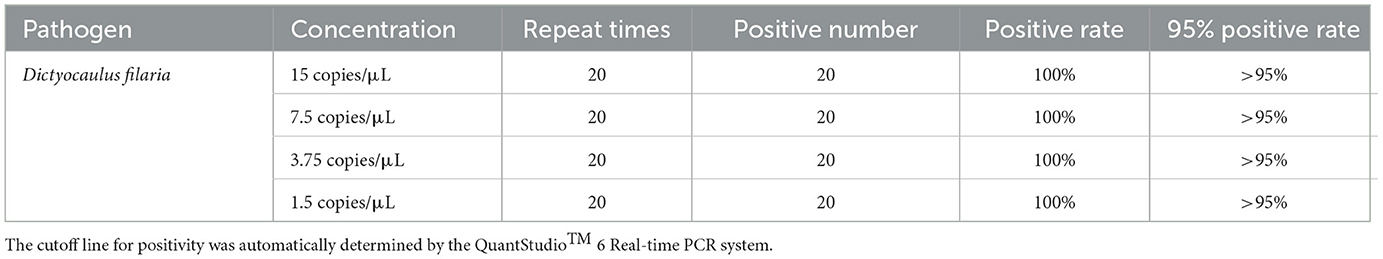
Table 2. Limit of detection (LOD) of the TaqMan-MGB probe-based qPCR assay for detecting Dictyocaulus filaria.
Repeatability and reproducibility of the TaqMan-MGB probe-based qPCR assay
As shown in Table 3, the TaqMan-MGB probe-based qPCR assay exhibited intra-assay CVs ranging from 0.11% to 0.58% for Dictyocaulus filaria. The TaqMan-MGB probe-based qPCR assay showed inter-assay CVs ranging from 0.33% to 2.19% for Dictyocaulus filaria. The results showed that both intra- and inter-assay CVs were below 3%, indicating good repeatability (intra-assay) and reproducibility (inter-assay) of the TaqMan-MGB probe-based qPCR assay.
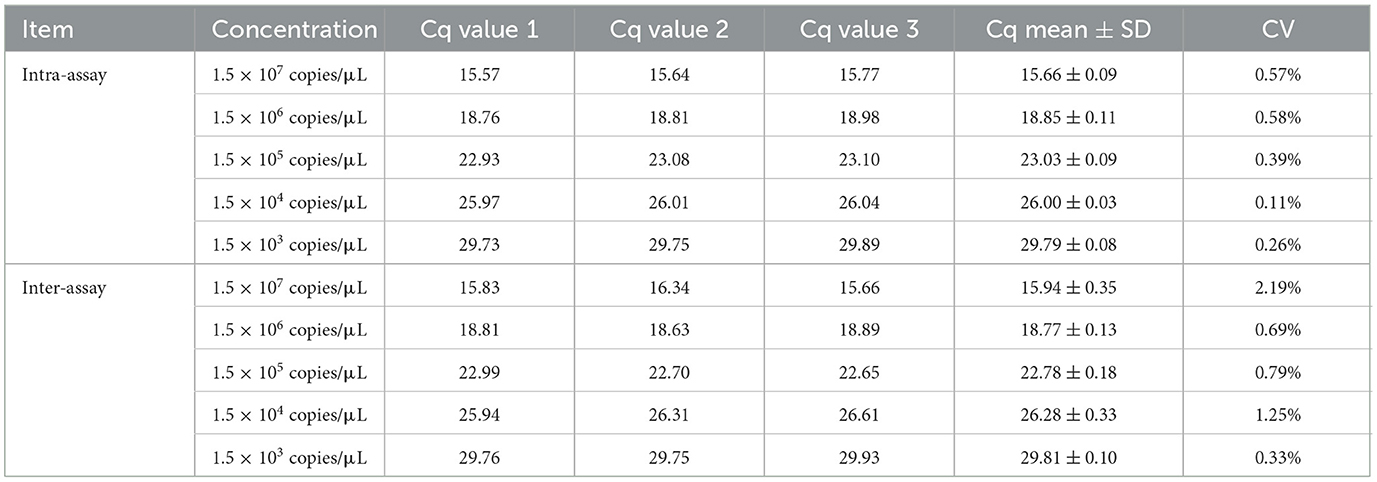
Table 3. Repeatability and reproducibility of the TaqMan-MGB probe-based qPCR assay for detecting Dictyocaulus filaria.
Clinical performance of the TaqMan-MGB probe-based qPCR assay
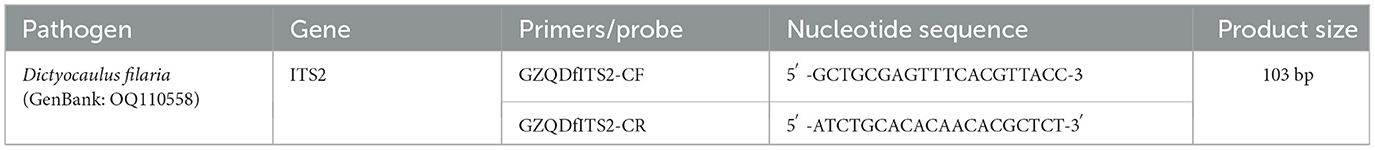
The application of the TaqMan-MGB probe-based qPCR assay to clinical samples detected Dictyocaulus filaria in seven blood samples, evidenced by positive fluorescent signals, specific amplification curves, and cycle threshold values ≤ 35 (Figure 4). The TaqMan-MGB probe-based qPCR assay showed that 3.5% (7/200) of the samples were positive for Dictyocaulus filariai. Notably, all seven samples also tested positive for Dictyocaulus filaria when confirmed using conventional PCR and sequencing methods (Figure 5). A brief, the total volume of the conventional PCR reaction was 50 μL, consisting of 5 μL of 10 × Buffer (Mg2+ Plus), 4 μL of dNTP mixture, 0.5 μL of the forward primer GZQDfITS2-CF (10 μM), 0.5 μL of the reverse primer GZQDfITS2-CR (10 μM), 0.25 μL of EX Taq (5 U/μL) [Takara Biomedical Technology (Beijing) Co., Ltd.], 2 μL of template DNA, and 37.75 μL of nuclease-free water. Amplification was carried out on the Veriti 96-Well Thermal Cycler equipment (Applied Biosystems) using the following program: 94°C for 5 min, 1 cycle; 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min, 35 cycles; and 72°C for 10 min, 1 cycle. The PCR amplification products were detected using 3% agarose gel electrophoresis. The positive PCR amplification products were confirmed using the sequencing method. The conventional PCR primers were synthesized by Takara Biomedical Technology (Beijing) Co., Ltd. and are shown in Table 4.
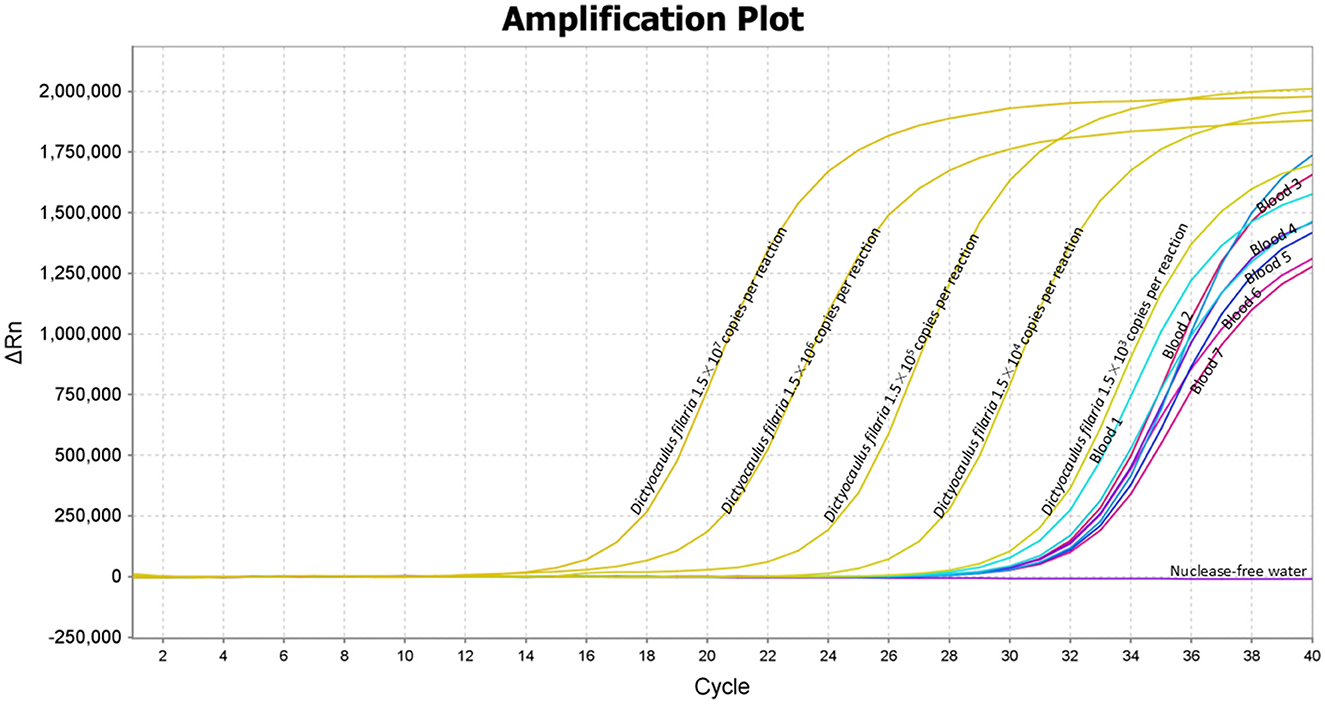
Figure 4. Application of the TaqMan-MGB probe-based qPCR assay for detecting Dictyocaulus filaria in the clinical samples. The clinical samples revealed positive results for Dictyocaulus filaria in the seven blood samples, indicated by positive fluorescent signals, specific amplification curves, and quantification cycle (Cq) values ≤ 35. Amplification curves of Dictyocaulus filaria for plasmid standards at concentrations ranging from 1.5 × 107 to 1.5 × 103 copies per reaction.
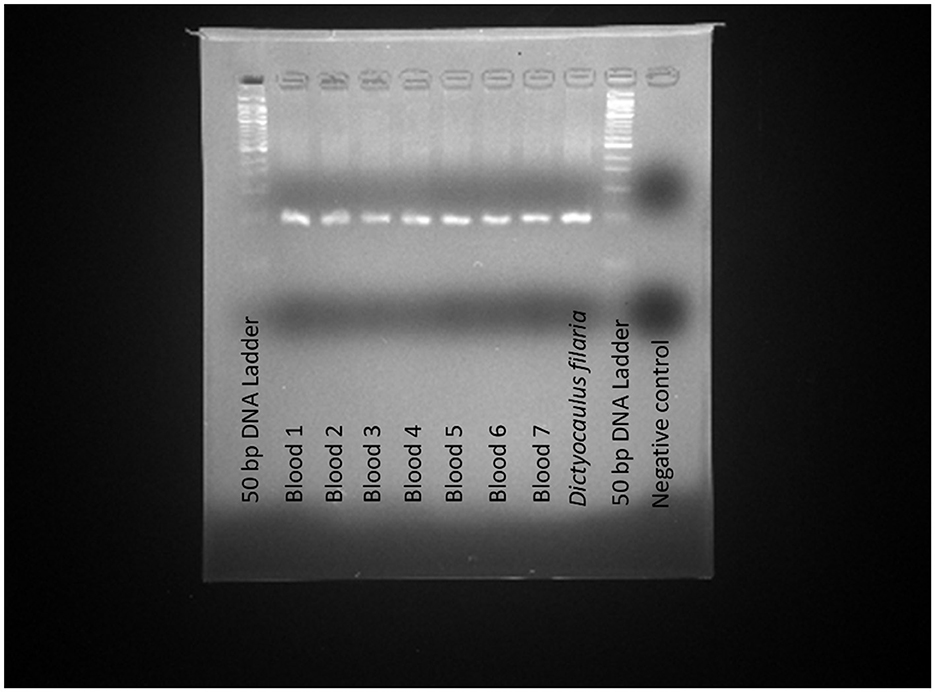
Figure 5. Application of conventional PCR for the detection of Dictyocaulus filaria in the clinical samples. The clinical samples revealed positive results for Dictyocaulus filaria in the seven blood samples, indicated by the presence of a positive 103 bp fragment on agarose gel electrophoresis. Positive control: Dictyocaulus filaria; Negative control: nuclease-free water.
Discussion
Dictyocaulus filaria infection remains a major financial concern due to its impact on both production losses and animal health and welfare (22). There is no effective vaccine available for the prevention and control of this disease (23–25). The control of Dictyocaulus filaria infestation in sheep relies on pasture management, the development of host immunity against the parasite, and the prudent use of a suitable anthelmintic (26–28). Indeed, in the context of increasing levels of resistance to anthelmintics (29) and growing concerns regarding the environmental impact of anthelmintic drug residues (30, 31), it is important to implement a safe strategy for parasitism management at the farm level that includes the accurate use of detection tools.
In view of this situation, an effective way to prevent the disease is to detect and eliminate positive individuals step by step. In this study, a novel TaqMan-MGB probe-based qPCR for detecting Dictyocaulus filaria was developed for the first time.
Specific primers and a probe targeting the conserved region of ITS2 were selected as detection molecular markers. The ITS2 region is moderately conserved, and its conservation is basically manifested as relative consistency within parasite species and distinct differences between different parasite species. This characteristic makes ITS2 suitable for the molecular identification of Dictyocaulus filaria. The method successfully detected Dictyocaulus filaria without any cross-reaction with other parasites, indicating the high specificity and reliability of this method for Dictyocaulus filaria detection.
For Dictyocaulus filaria DNA, the minimum detection limit of the developed TaqMan-MGB probe-based qPCR method was 1.5 copies per reaction. Accurate detection of Dictyocaulus filaria at lower concentrations enables early diagnosis and prevention of lungworm disease. TaqMan-MGB probe-based qPCR provides improved monitoring of Dictyocaulus filaria and is expected to enhance biosafety in sheep farm management.
However, the improvement in sensitivity also increases the risk of false positive results, which demands stricter contamination prevention during sampling and analysis. Good laboratory practices are necessary to obtain reliable results. An excellent and efficient detection method should accurately reflect epidemiological data—such as disease prevalence, transmission rate, and epidemic severity—thereby facilitating effective monitoring and prevention of the disease.
The developed TaqMan-MGB probe-based qPCR assay offers improved detection capabilities in a shorter time and at a lower labor cost. However, due to technical difficulties encountered during development, establishing this method remains more challenging compared to conventional PCR To the best of our knowledge, this is the first TaqMan-MGB probe-based qPCR detection method developed specifically to detect Dictyocaulus filaria, a parasite commonly found in sheep. This method can ensure good specificity and sensitivity and will undoubtedly save labor and material costs. The established detection method can detect Dictyocaulus filaria in clinical samples, making the detection process more convenient.
Conclusion
In this study, we designed specific primes and a probe based on the conserved region of the Dictyocaulus filaria sequence, constructed plasmid standards and standard curves, and developed a novel TaqMan-MGB probe-based qPCR detection method characterized by simple operation, high sensitivity, strong specificity, good stability, and a wide dynamic range. This assay can detect Dictyocaulus filaria qualitatively, quantitatively, rapidly, and accurately in different clinical samples. This is of great significance for safeguarding animal quality and human health. This assay has significant practical value in the quality control of animal-derived biological products intended for human use, as well as in the inspection and quarantine of animals for import and export. It deserves broad application in these fields.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
This study was conducted with the approval of the Animal Ethics Committee of National Institutes for Food and Drug Control [Approval ID: 2019 (A) 141]. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
Z-qG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. JX: Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key Research and Development Program of China (Project No. 2022YFF0710504).
Acknowledgments
We thank the editor and reviewers for their helpful comments regarding this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The experimental technique employed in this study has been the subject of a pending patent application (Patent No. 202411348950.3).
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Borji H, Azizzadeh M, Ebrahimi M, Asadpour M. Study on small ruminant lungworms and associated risk factors in northeastern Iran. Asian Pac J Trop Med. (2012) 5:853–56. doi: 10.1016/S1995-7645(12)60159-X
2. de Macedo LO, Lima TARF, Verocai GG, Alves LC, de Carvalho GA, Ramos RAN. Lungworms in ruminants from Brazil: a retrospective epidemiological study over four decades. Vet Parasitol Reg Stud Reports. (2021) 26:100645. doi: 10.1016/j.vprsr.2021.100645
3. Mohtasebi S, Sazmand A, Zafari S, Verocai GG, Otranto D. Lungworms of non-ruminant terrestrial mammals and humans in Iran. Pathogens. (2023) 12:759. doi: 10.3390/pathogens12060759
4. Ayalew L, Fréchette JL, Malo R, Beauregard C. Seasonal fluctuation and inhibited development of populations of Dictyocaulus filaria in ewes and lambs. Can J Comp Med. (1974) 38:448–56.
5. Panuska C. Lungworms of ruminants. Vet Clin North Am Food Anim Pract. (2006) 22:583–93. doi: 10.1016/j.cvfa.2006.06.002
6. Mangiola S, Young ND, Sternberg PW, Strube C, Korhonen PK, Mitreva M, et al. Analysis of the transcriptome of adult Dictyocaulus filaria and comparison with Dictyocaulus viviparus, with a focus on molecules involved in host-parasite interactions. Int J Parasitol. (2014) 44:251–61. doi: 10.1016/j.ijpara.2013.12.003
7. Gomez-Puerta LA, Angulo-Tisoc JM, Pacheco JI. The vicuna (Vicugna vicugna) as a natural host of Dictyocaulus filaria in Peru. Parasitol Int. (2024) 101:102897. doi: 10.1016/j.parint.2024.102897
8. Fesseha H, Mathewos M. Prevalence and risk factors of Bovine and Ovine lungworm infection at Durame district, southern Ethiopia. J Parasitol Res. (2021) 2021:6637718. doi: 10.1155/2021/6637718
9. Firouzivand Y. A field outbreak of Dictyocaulus filaria in sheep in the northwest of Iran. J Altern Vet Med. (2023) 6:1097–102.
10. Parfitt JW, Sinclair IJ. Cross resistance to Dictyocaulus viviparus produced by Dictyocaulus filaria infections in calves. Res Vet Sci. (1967) 8:6–13. doi: 10.1016/S0034-5288(18)34645-9
11. Höglund J, Wilhelmsson E, Christensson D, Mörner T, Waller P, Mattsson JG. ITS2 sequences of Dictyocaulus species from cattle, roe deer and moose in Sweden: molecular evidence for a new species. Int J Parasitol. (1999) 29:607–11. doi: 10.1016/S0020-7519(98)00229-X
12. Divina BP, Wilhelmsson E, Mörner T, Mattsson JG, Höglund J. Molecular identification and prevalence of Dictyocaulus spp. (Trichostrongyloidea: Dictyocaulidae) in Swedish semi-domestic and free-living cervids. J Wildl Dis. (2002) 38:769–75. doi: 10.7589/0090-3558-38.4.769
13. Pyziel AM, Laskowski Z, Höglund J. Development of a multiplex PCR for identification of Dictyocaulus lungworms in domestic and wild ruminants. Parasitol Res. (2015) 114:3923–26. doi: 10.1007/s00436-015-4657-y
14. Danks HA, Sobotyk C, Saleh MN, Kulpa M, Luksovsky JL, Jones LC, et al. Opening a can of lungworms: molecular characterization of Dictyocaulus (Nematoda: Dictyocaulidae) infecting North American bison (Bison bison). Int J Parasitol Parasites Wildl. (2022) 18:128–34. doi: 10.1016/j.ijppaw.2022.04.011
15. Epe C, Bienioschek S, Rehbein S, Schnieder T. Comparative RAPD-PCR analysis of lungworms (Dictyocaulidae) from fallow deer, cattle, sheep, and horses. Zentralbl Veterinarmed B. (1995) 42:187–91. doi: 10.1111/j.1439-0450.1995.tb00699.x
16. Schnieder T, Epe C, von Samson-Himmelstjerna G. Species differentiation of lungworms (Dictyocaulidae) by polymerase chain reaction/restriction-fragment-length polymorphism of second internal transcribed spacers of ribosomal DNA. Parasitol Res. (1996) 82:392–4. doi: 10.1007/s004360050134
17. Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, et al. 3'-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. (2000) 28:655–61. doi: 10.1093/nar/28.2.655
18. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. (2009) 55:611–22. doi: 10.1373/clinchem.2008.112797
19. Forootan A, Sjöback R, Björkman J, Sjögreen B, Linz L, Kubista M, et al. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomol Detect Quantif. (2017) 12:1–6. doi: 10.1016/j.bdq.2017.04.001
20. Ruijter JM, Barnewall RJ, Marsh IB, Szentirmay AN, Quinn JC, van Houdt R, et al. Efficiency correction is required for accurate quantitative PCR analysis and reporting. Clin Chem. (2021) 67:829–42. doi: 10.1093/clinchem/hvab052
21. Bustin SA, Ruijter JM, van den Hoff MJB, Kubista M, Pfaffl MW, Shipley GL, et al. MIQE 2.0: revision of the minimum information for publication of quantitative real-time PCR experiments guidelines. Clin Chem (2025) 24:hvaf043. doi: 10.1093/clinchem/hvaf043
22. Zafari S, Mohtasebi S, Sazmand A, Bahari A, Sargison ND, Verocai GG, et al. The prevalence and control of lungworms of pastoral ruminants in Iran. Pathogens. (2022) 11:1392. doi: 10.3390/pathogens11121392
23. Al-Saadi AA, Al-Samarrae SA, Altaif KI. Effect of anthelmintic treatment on the development of resistance in sheep vaccinated or experimentally infected with Dictyocaulus filaria. Res Vet Sci. (1984) 36:144–46. doi: 10.1016/S0034-5288(18)31969-6
24. Bain R. Irradiated vaccines for helminth control in livestock. Int J Parasitol. (1999) 29:185–91. doi: 10.1016/S0020-7519(98)00187-8
25. Claerebout E, Geldhof P. Helminth vaccines in ruminants: From development to application. Vet Clin North Am Food Anim Pract. (2020) 36:159–71. doi: 10.1016/j.cvfa.2019.10.001
26. Hidalgo-Argüello MR, Díez-Baños N, Rojo-Vázquez FA. Efficacy of moxidectin 1% injectable and 0.2% oral drench against natural infection by Dictyocaulus filaria in sheep. Vet Parasitol. (2002) 107:95–101. doi: 10.1016/S0304-4017(02)00116-4
27. Little PR, Hodge A, Maeder SJ, Wirtherle NC, Nicholas DR, Cox GG, et al. Efficacy of a combined oral formulation of derquantel-abamectin against the adult and larval stages of nematodes in sheep, including anthelmintic-resistant strains. Vet Parasitol. (2011) 181:180–93. doi: 10.1016/j.vetpar.2011.05.008
28. Hamel D, Visser M, Mayr S, Rauh R, Wang H, Fankhauser R, et al. Eprinomectin pour-on: Prevention of gastrointestinal and pulmonary nematode infections in sheep. Vet Parasitol. (2018) 264:42–6. doi: 10.1016/j.vetpar.2018.11.002
29. Abdolahzadeh S, Tavassoli M, Esmaeilnejad B, Jalilzadeh-Amin G. Evaluation of drug resistance to albendazole and levamisole against lung worms in goat flocks based on fecal larvae count reduction test. Vet Res Forum. (2024) 15:181–86. doi: 10.30466/vrf.2023.2010062.3991
30. Beynon SA. Potential environmental consequences of administration of anthelmintics to sheep. Vet Parasitol. (2012) 189:113–24. doi: 10.1016/j.vetpar.2012.03.040
Keywords: lungworm disease, Dictyocaulus filaria, TaqMan-MGB-probe, LLOQ, LOD
Citation: Gao Z-q and Xing J (2025) Development and application of a TaqMan-MGB probe-based quantitative real-time polymerase chain reaction assay for the rapid detection of Dictyocaulus filaria. Front. Vet. Sci. 12:1559088. doi: 10.3389/fvets.2025.1559088
Received: 14 January 2025; Accepted: 03 June 2025;
Published: 11 August 2025.
Edited by:
Mohammed Rohaim, Cairo University, EgyptReviewed by:
Alexandre Bendas, Federal Rural University of Rio de Janeiro, BrazilMayron Antonio Candia-Puma, Catholic University of Santa María, Peru
Tiago Feitosa Mota, Gonçalo Moniz Institute (IGM), Brazil
Fayez Salib, Faculty of Veterinary Medicine Cairo University, Egypt
Copyright © 2025 Gao and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng-qin Gao, Z2FvemhlbmdxaW5AMTI2LmNvbQ==; Z2FvemhlbmdxaW5AbmlmZGMub3JnLmNu; Jin Xing, eGp2ZXRAbmlmZGMub3JnLmNu
 Zheng-qin Gao
Zheng-qin Gao Jin Xing*
Jin Xing*