- Laboratory of Molecular Ecology, Nature Research Centre, Vilnius, Lithuania
Background: The genus Sarcocystis comprises a diverse group of apicomplexan parasites that infect reptiles, birds, and mammals. They are characterized by the formation of sarcocysts in the muscles of the intermediate host and the development of sporocysts in the intestines of the definitive host. Raptors usually act as definitive hosts for numerous Sarcocystis species; however there is a lack of studies on Sarcocystis in the muscles of raptorial birds. Therefore, we aimed to assess infection rates and identify Sarcocystis species in the muscles of raptors in Lithuania.
Methods: Muscle samples from 90 raptors (Accipitriformes, Falconiformes, and Strigiformes) were collected throughout Lithuania and analyzed for Sarcocystis spp. Sarcocysts isolated from fresh methylene blue-stained muscle samples were identified using the internal transcribed spacer region 1 sequence genetic marker.
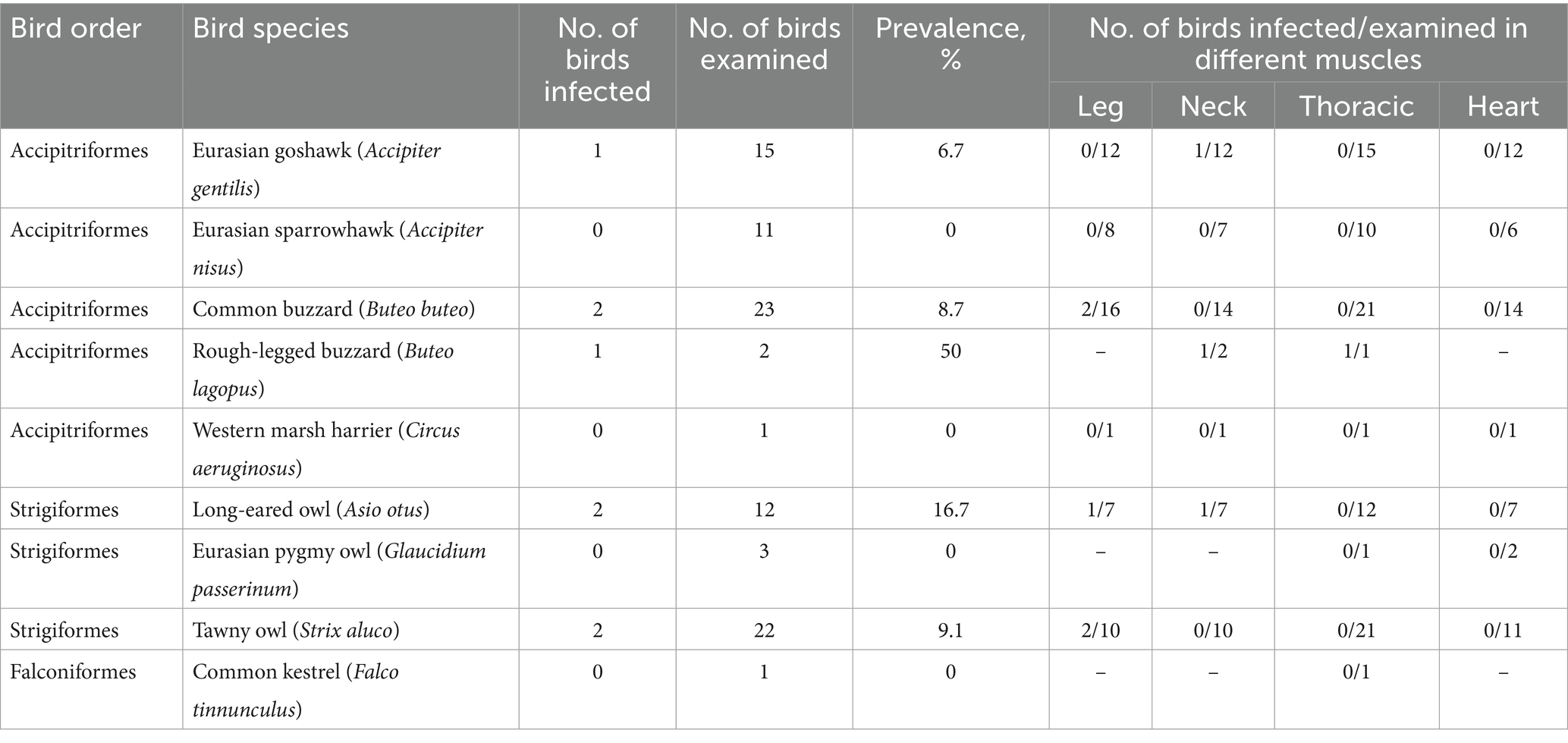
Results: Under the light microscope, sarcocysts were detected in 8.9% (8/90) of the raptors examined. Sarcocysts were found in the leg muscles of common buzzards (Buteo buteo), tawny owls (Strix aluco), and a long-eared owl (Asio otus); neck muscles of a Eurasian goshawk (Accipiter gentilis), rough-legged buzzard (Buteo lagopus), and long-eared owl; and thoracic muscles of a rough-legged buzzard. We observed no sarcocysts in the cardiac muscles. Representatives of one Sarcocystis species, S. halieti were molecularly identified in seven birds.
Conclusion: This is the first study to report five new intermediate hosts for S. halieti. Further investigations are needed to assess the possible pathogenicity of S. halieti in extra-intestinal organs of raptors.
1 Introduction
Over the last 500 years, 140 species of birds have become extinct, more than any other vertebrate group (1). Over half of raptors are seeing population reductions worldwide, and 18% of raptors are in danger of going extinct (2). The most common causes of morbidity in raptors are infectious and parasitic diseases, traumatic injuries, toxicosis, and metabolic or nutritional diseases (3). Raptors feature a large diversity of protozoan, helminth, and associated bacterial infections of the digestive system. For instance, the study conducted in 2000 has shown that even 89.2% of all raptors were infected with helminths such as Acanthocephala, Cestoda, Nematoda, and Trematoda (4). In addition, in the same study, coccidians were found in 31.4% of raptors studied. In general, there is still a lack of knowledge about parasite infections in the muscles of raptors (5, 6).
Sarcocystis spp. are apicomplexan intracellular parasites that have a mandatory two-host life cycle. These parasites are known for their characteristics of forming sarcocysts in the muscles of various animals, including poikilothermic ones, birds, and mammals. Two routes of Sarcocystis spp. infection are possible. The primary transmission route is fecal-oral, involving the ingestion of water or food contaminated with sporocysts/oocysts from the feces of the definitive host (DH). The second mode of transmission is ingestion of muscles or other tissues of intermediate hosts (IH) that harbor mature sarcocysts (7).
Globally, various bird species serve as hosts for more than 48 Sarcocystis species (8). Among these, 32 are named species described as utilizing birds as IHs. In contrast, at least 16 Sarcocystis species have been molecularly confirmed to use birds as DHs (6, 9, 10). Sarcocystis spp. are mainly asymptomatic for DHs, while these parasites can cause histopathological changes for tissues of IHs (11). Three species, Sarcocystis halieti, Sarcocystis falcatula, and Sarcocystis wobeseri, have been found in extra-intestinal tissues of birds belonging to orders Accipitriformes and Strigiformes (6, 10–15). Sarcocystis falcatula is highly pathogenic and can cause lung injuries and encephalitis, which can be fatal even to birds (8, 10, 11, 16). However, the prevalence of S. falcatula is restricted to North and South America, where their host species belonging to the genus Didelphis are distributed (7, 17, 18). Sarcocystis halieti is considered to be pathogenic due to a report of severe multifocal granulomatous encephalitis in the little owl (Athene noctua) (10). This Sarcocystis species is multi-host specific and has been identified in Old and New World birds (6, 10, 15, 19). Sarcocysts of S. wobeseri have been identified in muscles of Anseriformes, Charadriiformes, and Accipitriformes (14, 20, 21); however, the pathogenicity of this Sarcocystis species has not yet been fully investigated. Based on molecular analysis, DHs of S. halieti and S. wobeseri belong to the order Accipitriformes (19, 22–25).
In Lithuania, the muscles of birds belonging to Anseriformes, Charadriiformes, and Passeriformes have been the most extensively studied, while raptors have not yet been investigated for muscular sarcocystosis. Thus, our study aimed to evaluate infection rates and identify Sarcocystis species found in the muscles of raptors from Lithuania.
2 Materials and methods
2.1 Sample collection
Muscle samples of 90 raptorial birds belonging to orders Accipitriformes, Falconiformes, and Strigiformes were collected in different regions of Lithuania (54–55°N, 21–24°W) between 2014 and 2024 and examined for the presence of sarcocysts of Sarcocystis spp. (Table 1). Samples of birds were retrieved from Kaunas Tadas Ivanauskas Zoology Museum, which is the national authority that runs wildlife research programs and is responsible for monitoring dead animals found. Birds used for our study were killed along roadsides as well as after collisions with architectural structures and high-voltage wires. Muscle samples were kept frozen (−20°C) until microscopic examination of Sarcocystis parasites. The conducted study was approved by the Animal Welfare Committee of the Nature Research Center (no. GGT-9). It should be noted that only some muscle types were available for each bird examined. A total of 243 muscle samples, 54 leg muscle samples, 53 neck tissue samples, 83 thoracic muscles samples and 53 heart samples were tested for sarcocysts (Table 1).
2.2 Microscopical identification of Sarcocystis spp
Sarcocystis spp. infection was confirmed by microscopy of stained muscle samples (leg, neck, thoracic, and heart). Twenty-eight rice-sized pieces of each muscle (1 g ± 0.3) were cut from each bird, stained with 0.2% methylene blue for 35 min, soaked in 1.5% acetic acid for 25 min, squeezed into a glass compressor, and examined under light microscopy (LM) at a magnification of × 200. Infection intensity was quantified by counting sarcocysts within 28 tissue sections obtained from the muscle. The sarcocysts detected were removed with a preparation needle, each placed separately in a 1.5 mL microcentrifuge tube containing 1 mL of 96% ethanol and stored at 4°C. The ethanol was replaced once a week for 1 month or until the blue color faded.
The bird muscle samples in which Sarcocystis spp. infection was detected by methylene blue-staining were re-examined in fresh squashed preparations using a 0.9% NaCl solution. Observed sarcocysts were morphologically characterized at × 200 and × 400 magnifications. The discovered sarcocysts were free from myofibrils as much as possible, and a part of the bradyzoites was released by pressing the cyst firmly. The excreted sarcocysts were placed in individual microcentrifuge tubes filled with 96% ethanol and stored at −20°C until DNA isolation.
2.3 Molecular analysis of detected sarcocysts
The genomic DNA from each isolated sarcocyst was purified using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific Baltics, Vilnius, Lithuania) according to the manufacturer’s instructions. Obtained DNA samples were kept frozen at −20°C for further molecular analysis. Sarcocystis species identification was performed using the internal transcribed spacer 1 (ITS1) region, which was previously shown to be variable enough and could be used for the discrimination of avian Sarcocystis species (26).
Firstly, DNA fragments were amplified using nested PCR. Each reaction was carried out using DreamTaq PCR Master Mix (Thermo Fisher Scientific Baltics, Vilnius, Lithuania). In the first step, a forward NITSpauk1 (5′-TGTCCGGAATGGGAAGTTTT-3′) and reverse NITSpauk2 (5′-ACACCATCCDAAATTCTCAG-3′) primers were used. In the second step, the primer pair NITSpauk3 (5′-GGAAGGATCATTCACACGTT-3′)/NITSpauk4 (5′- ATCACTGCAAGTTCCAACCA-3′) was used (11). After a second round of nested PCR, the resulting 261 bp-long products were purified and sequenced as previously specified (11). Based on the short ITS1 fragments, S. halieti was detected in all cases of successful amplification and sequencing.
For the conclusive Sarcocystis species identification we used longer S. halieti specific primers, forward GsShalF1 (5′-GATAATTGACTTTACGCGCCATTAC-3′) and reverse GsShalR2 (5′-CCATCCCTTTTTCTAAAGGAGGTC-3′) (27) amplifying about a 645 bp-long ITS1 fragment. The generation of such fragments was carried out in two alternative ways, by using nested or direct PCR. In case of the nested PCR, SU1F/5.8SR2 (5′- GATTGAGTGTTCCGGTGAATTATT-3′/5’-AAGGTGCCATTTGCGTTCAGAA-3′) external primers (26) and DreamTaq PCR Master Mix were used following previously described procedures (27). Direct PCR was performed using PlatinumTM SuperFi II Green PCR Master Mix (Thermo Fisher Scientific Baltics, Vilnius, Lithuania). PCR cycling conditions were as follows: initial denaturation for 30 s at 98°C, 35 cycles of 10 s at 98°C, 10 s at 60°C, 30 s at 72°C, and final extension for 5 min at 72°C. Visualization and purification of PCR products and Sanger sequencing were performed as described in Prakas et al. (11).
2.4 Phylogenetic analysis
The obtained ITS1 sequences (GenBank accession numbers: PQ900168–PQ900180) were compared with those of various Sarcocystis spp. using nBLAST sequence similarity search algorithm (28). Phylogenetic analyses were carried out with the help of MEGA v. 11.0.13 software (29). Multiple sequence alignments were generated using the MUSCLE algorithm. Phylogenetic trees were constructed using the Maximum Likelihood (ML) method. The nucleotide substitution model that best suited the analyzed alignments was selected using the MEGA “Find Best DNA/Protein Models (ML).” The robustness of phylogeny was tested using the bootstrap method with 1,000 bootstrap replications.
3 Results
3.1 Infection rate and parasite load of sarcocysts in raptors
The infection rate of Sarcocystis spp. was established by examining methylene blue-stained muscle samples. Sarcocysts were detected in 8.9% (8/90) of muscles of Lithuanian raptors examined (Table 1). Sarcocysts were found in the leg muscles of two common buzzards (Buteo buteo) (12.5%, 2/16) and tawny owls (Strix aluco) (20%, 2/10), and one long-eared owl (Asio otus) (14.3%, 1/7). In addition, Sarcocystis spp. were found in the neck muscles of a single Eurasian goshawk (Accipiter gentilis) (8.3%, 1/12), rough-legged buzzard (Buteo lagopus) (1/2), and long-eared owl (14.3%, 1/7). Furthermore, Sarcocystis spp. have been observed in the thoracic muscles of one rough-legged buzzard (Table 1). No sarcocysts were observed in the hearts of birds examined. Overall, of the 243 muscle samples stained with methylene blue, sarcocysts were found in nine of them (3.7%).
Higher, but statistically not significant (χ2 = 0.26, p = 0.612), infection rate was identified in birds of the order Strigiformes (4/37; 10.8%) compared to that determined in birds of the order Accipitriformes (4/52; 7.7%). Most frequently, sarcocysts were detected in leg muscles (5/54; 9.3%), followed by neck (3/54; 5.6) and thoracic (1/83; 1.2) muscles.
The parasite load ranged from one to 15 cysts/g of muscles stained with methylene blue; the mean and median parasite load were two and four cysts/g of muscle, respectively.
3.2 Morphology of detected sarcocysts
Sarcocyts were observed in fresh muscle samples from only two birds. In methylene blue-stained samples, Sarcocystis spp. sarcocysts were found to be 4,952 × 34 μm (range: 2673–7,380 × 25–64 μm; n = 10) in size and looked like tiny threads (Figure 1A). The sarcocyst wall found in unstained squashed muscles appeared to be smooth and 1 μm thick (Figure 1B). Septa split sarcocysts into sections that were filled with 6.7 × 1.9 μm (4.9–8.7 × 1.2–2.5 μm; n = 7) banana-shaped bradyzoites (Figure 1C).
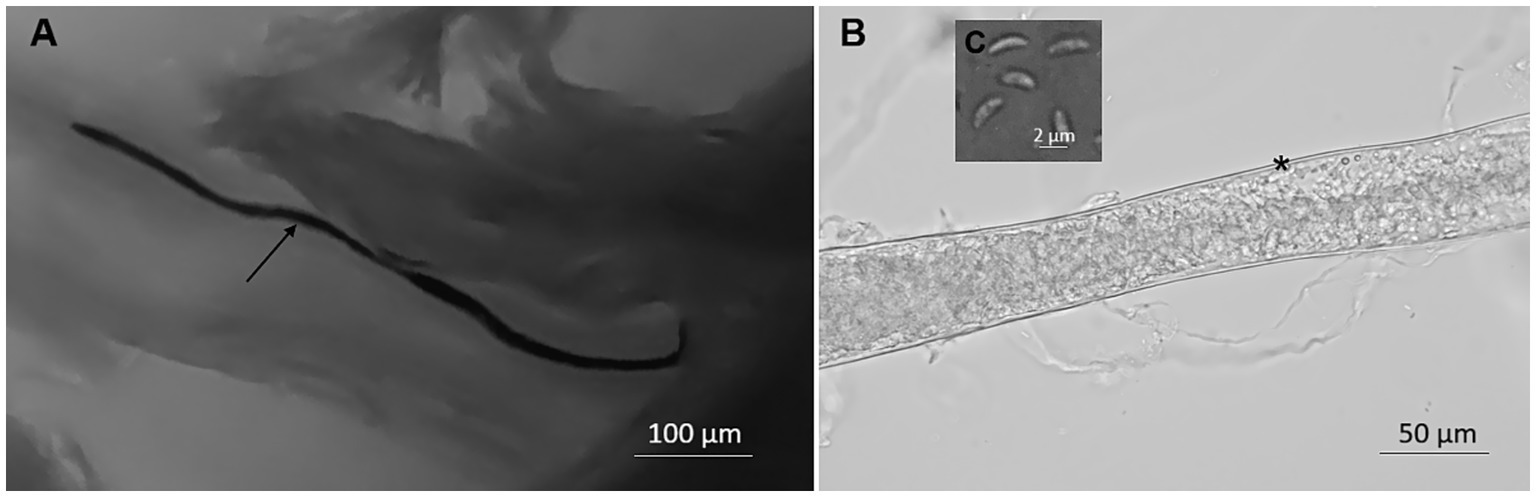
Figure 1. Morphology of Sarcocystis halieti in tissue sample taken from leg muscle of raptors. (A) Sarcocyst from common buzzard (Buteo buteo) stained with methylene blue; the arrow shows the cyst. (B) Fragment of the smooth cyst wall in fresh preparation from tawny owl (Strix aluco); *marked the cyst wall. (C) Banana-shaped bradyzoites in fresh sample from tawny owl.
3.3 Molecular identification of Sarcocystis halieti
In fresh samples, sarcocysts of Sarcocystis spp. were only observed and isolated from neck muscles of the rough-legged buzzard and leg muscles of the tawny owl. In eight more Sarcocystis spp. positive samples, sarcocysts were found only in methylene blue-stained preparations. One sarcocyst from each Sarcocystis spp. positive sample was subjected to molecular analysis. Using the Sarcocystis spp.-specific NITSpauk3/NITSpauk4 primer pair, we obtained three different 221 bp-long ITS1 genotypes differing in 1–2 SNPs (single nucleotide polymorphisms). These ITS1 sequences showed 95.9–100% identity to those of S. halieti, 94.6–95.0% similarity to that of Sarcocystis sp. isolate Skua-2016-CH (MW160469), 94.1–94.6% similarity to those of Sarcocystis sp. isolate 38P (PQ133336) and Sarcocystis sp. ex Accipiter cooperii (KY348755), similarity to that of 93.6–94.1% Sarcocystis sp. ex Corvus corax (MZ707151) and 92.9–93.3%% similarity to that of Sarcocystis corvusi (JN256119).
Using S. halieti-specific primers (GsShalF1/GsShalR2), we were able to amplify five samples when conventional PCR and PlatinumTM SuperFi polymerase were applied, whereas three samples were positive when nested PCR and DreamTaq polymerase were used (Table 2). The five 596 bp-long ITS1 sequences generated represented four distinct genetic variants that differed in one to three SNPs. The comparison of these 596 bp-long ITS1 sequences demonstrated 96.8–100% identity to those of S. halieti, 96.1–96.5% similarity to that of Sarcocystis sp. isolate Skua-2016-CH, 94.6–95.0% similarity to that of Sarcocystis sp. ex Corvus corax, 92.8–93.2% similarity to that of Sarcocystis sp. isolate 38P, 92.7–93.0% similarity to that of Sarcocystis sp. ex Accipiter cooperii, 91.7–92.3% similarity to those of S. columbae, and 90.9–91.2% similarity to that of S. corvusi. Thus, based on DNA sequence analysis S. halieti was identified in eight muscle samples of raptors from Lithuania (Table 2).
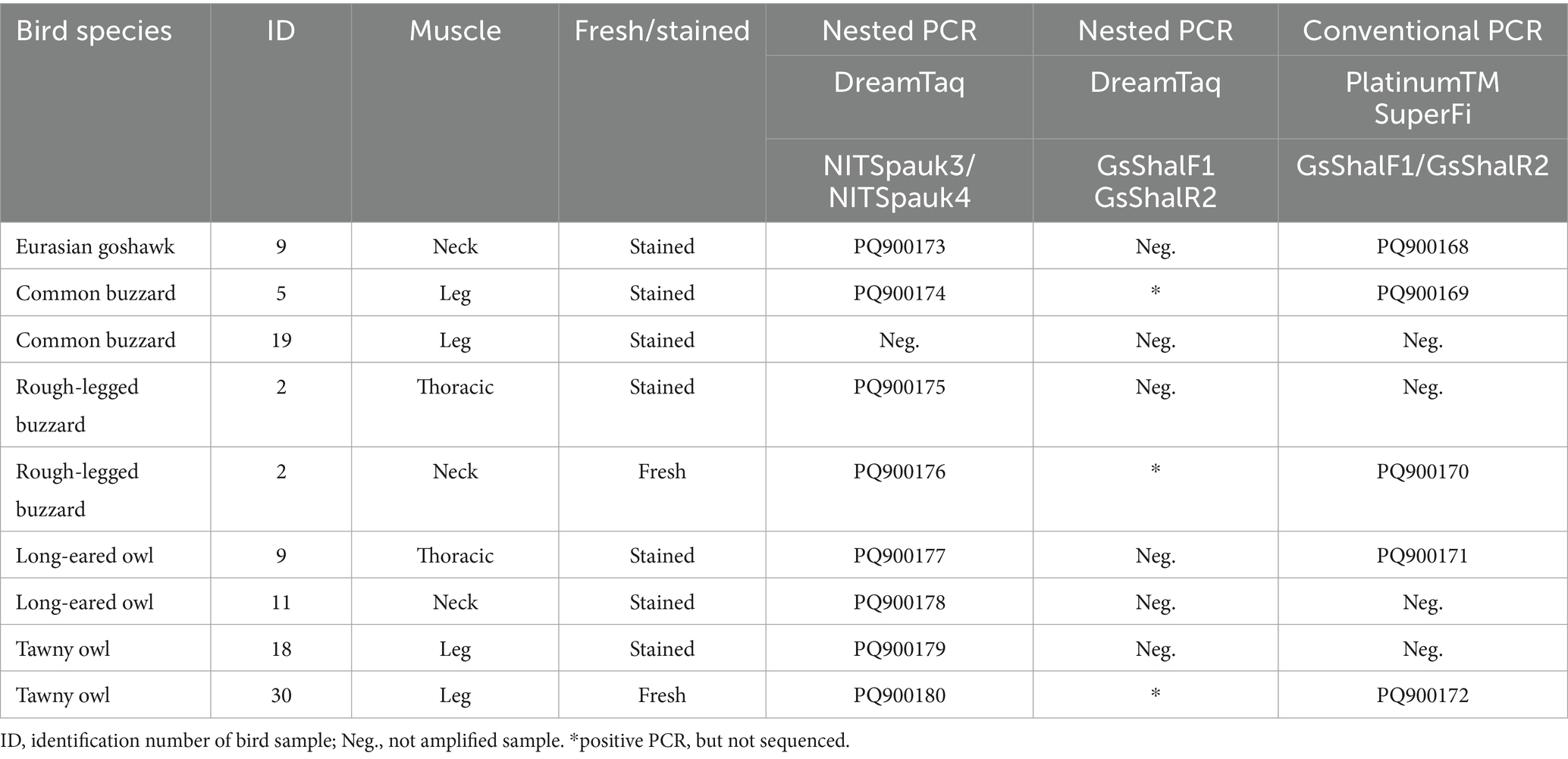
Table 2. Identification of Sarcocystis halieti in muscles of raptors using different muscle examination techniques, PCR methods and polymerases.
3.4 Phylogenetic results
For the phylogenetic analyses, all possible different genetic variants of S. halieti taken from GenBank were included. In addition to our S. halieti sequences, eight and 18 different ITS1 genotypes were analyzed, when sequences were obtained using NITSpauk3/NITSpauk4 and GsShalF1/GsShalR2 primers, respectively. Furthermore, sequences of other Sarcocystis spp. showing the highest similarity to our sequences were used to reveal phylogenetic relationships. The ITS1 sequences generated in the present study with fairly high bootstrap support values (84 and 94) were grouped with other sequences of S. halieti (Figure 2). However, using short 221 bp-long ITS1 fragments, a single sequence of S. halieti OP419624 formed a separate branch in the phylogram (Figure 2A). Based on longer ~596 bp-long ITS1 sequences, S. halieti genetic variants clustered into several clades (Figure 2B). Sequences obtained in the present work were grouped with those of S. halieti isolated from various IHs (birds classified as insectivorous, piscivores, omnivores, and raptors) and DHs of the order Accipitriformes collected in the Czeck Republic, Lithuania, and Spain. Both phylogenetic analyses showed that S. halieti was most related to Sarcocystis sp. from Chilean skua (Stercorarius chilensis) (MW160469).
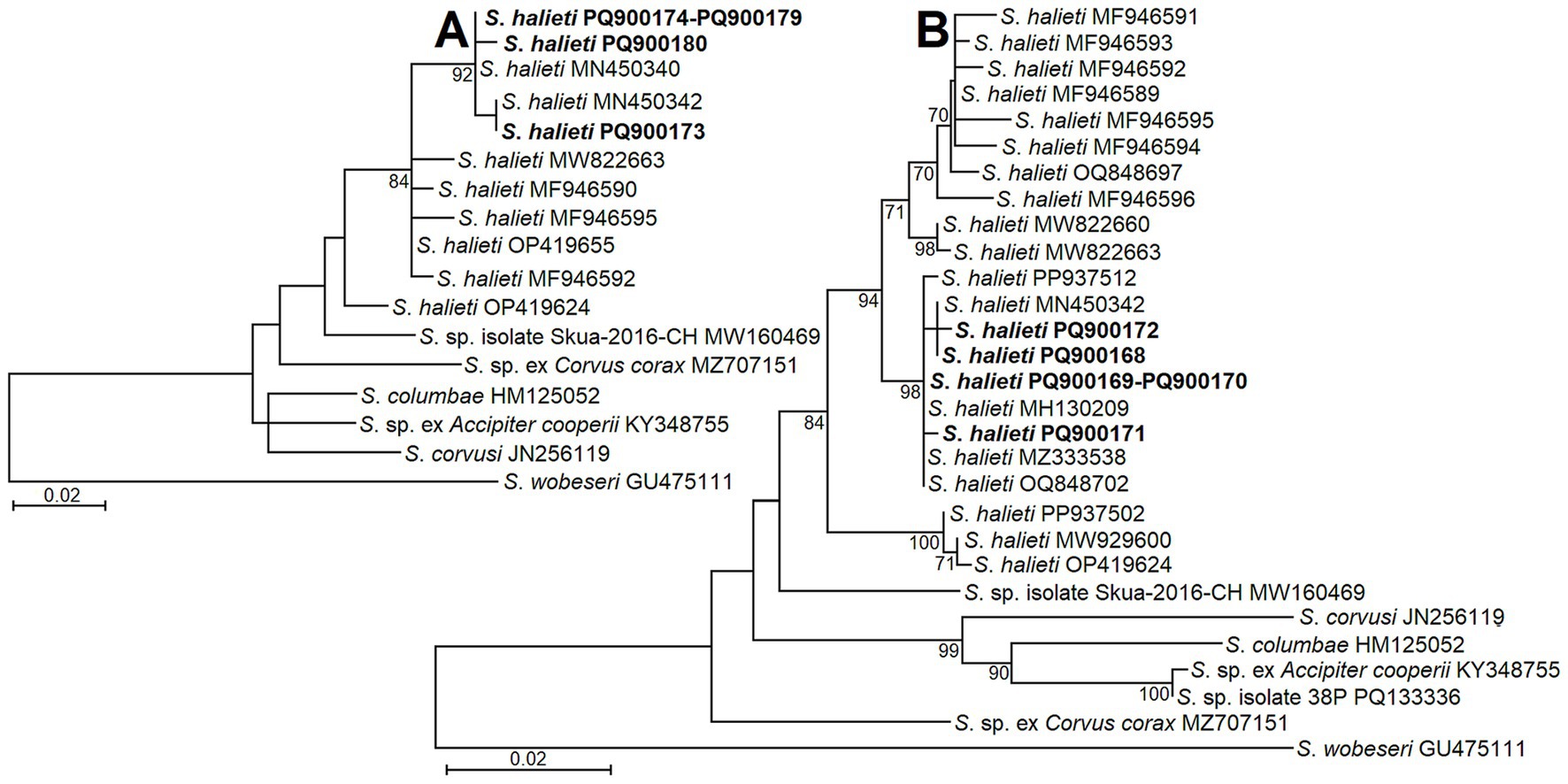
Figure 2. The phylogenetic placement of S. halieti isolates obtained from raptors in Lithuania. Phylogenetic trees were obtained analyzing ITS1 sequences obtained with the help of NITSpauk3/NITSpauk4 (A) and GsShalF1/GsShalR2 (B) primer pairs. Trees were constructed using ML method, scaled according to the branch length and rooted on S. wobeseri. In the left analysis (A) the alignment consisted of 17 sequences and 225 nucleotide positions; Tamura 3 parameter + G nucleotide substitution model was set. In the right analysis (B) the alignment contained of 29 sequences and 624 nucleotide positions; HKY nucleotide substitution model was set. Sequences generated in present study are indicated in boldface.
4 Discussion
In the current study, we provide the first data on Sarcocystis in the muscles of raptors from Lithuania. Based on methylene blue-staining, sarcocysts of Sarcocystis spp. were observed in 8.9% of 90 muscle samples of raptorial birds from Lithuania (Table 1). Sarcocysts were found in the leg, neck, and thoracic muscles of the birds studied, but not in the heart. Using the same method of microscopic analysis, birds of three families, i.e., Anatidae, Corvidae, and Laridae, were the most extensively studied for the presence of sarcocysts in Lithuania (20, 21, 30–36). Comparing the estimated infection rates, sarcocysts were much less frequently found in the muscles of raptors than in those of corvids (62/181, 34.3%), anatids (100/342, 29.2%), and larids (21/140, 15.0%) (20, 21, 36, 37).
It should also be mentioned that there is also a lack of comprehensive research on Sarcocystis in extra-intestinal tissues worldwide (6). Previously, Sarcocystis spp. were detected in the skeletal muscles, heart, and brain of raptors (5, 6, 10, 12, 13, 15, 38). In general, the observed infection rates of Sarcocystis spp. in muscles highly depend on the method applied (6), methylene blue-staining, histological examination, muscle digestion, or immunofluorescence antibody testing (5, 39–41). However, in most cases the infection rates observed in the muscles of raptors were in the range of 5 and 15%, despite studies having been carried out in different geographical regions, Australia, Brazil, Germany, Kazakhstan, and Spain (5, 6, 39, 41, 42).
By microscopical examination, thin and smooth sarcocysts, similar to those found in our study (Figure 1), were also detected in muscles of common kestrel (Falco tinnunculus) from Kazakhstan (42), in common buzzard from Germany (5), and in several species of raptors from the United States (38). However, several avian Sarcocystis spp. form sarcocysts with smooth and thin cyst walls. Furthermore, two Sarcocystis species (S. halieti and S. wobeseri) having thin sarcocysts walls have been confirmed in raptors (6, 14). Therefore, conclusive identification of sarcocysts found in raptors’ muscles requires molecular analysis methods.
In the present study, by using nested PCR and NITSpauk3/ NITSpauk4 primers, S. halieti was confirmed in muscles of seven birds belonging to five different species of orders Accipitriformes and Strigiformes (Table 2). Based on a comparison of ITS1 sequences, the obtained intraspecific and interspecific genetic variability values did not overlap, showing that the species was correctly identified. The detection of S. halieti in the samples analyzed was also confirmed by phylogenetic analysis, as investigated parasite isolates significantly clustered with other isolates of S. halieti (Figure 2). Thus, relatively small 221 bp-long and 596 bp-long fragments of ITS1 can be used for the identification of S. halieti. Sarcocystis spp. from birds have evolved more recently compared to species found in mammals and reptiles (7). Therefore, conservative markers, such as ribosomal 18S rRNA, the mitochondrial cytochrome c oxidase subunit 1 gene (cox1), and the RNA polymerase B gene of the apicoplast genome (rpoB), are not variable enough to discriminate all Sarcocystis species with birds as IH (21, 24, 26, 43). By contrast, numerous studies have confirmed that highly variable ITS1 is currently the most appropriate marker for distinguishing avian Sarcocystis species (6, 15, 21, 23, 36, 41, 43). In the current work, the GsShalF1/GsShalR2 primer pair has been confirmed in several studies to be specific for the amplification of S. halieti (25, 27). Other primers (internal NITSpauk1/NITSpauk2 and external NITSpauk3/NITSpauk4) we have applied in the present study were theoretically designed for the detection of Sarcocystis spp. employing birds and predatory mammals as their IHs (11). Due to short products amplified and due to Sarcocystis spp. conservative primers NITSpauk1, targeting 18S rRNA, the above-described primers can be used for the initial screening of muscles of birds and predatory mammals for the presence of Sarcocystis spp.
Based on ITS1 or 28S rRNA S. halieti was previously detected in muscles and brains of birds belonging to even six orders: Accipitriformes, Charadriiformes, Passeriformes, Procellariiformes, Strigiformes, and Suliformes (6, 10, 11, 15, 20, 21, 23, 36, 41, 43). Sarcocysts of S. halieti were found in birds from Europe, South America, and Asia (6, 11, 15, 20, 41; also see PQ270246 and PQ276104 GenBank records). In this study, we provide new host records of S. halieti in common buzzard, Eurasian goshawk, rough-legged buzzard (Accipitridae), long-eared owl and tawny owl (Strigidae). During this study, the material obtained from Kaunas Tadas Ivanauskas Zoology Museum was frozen, which prevented us from performing histopathological examinations. Prior to this study, S. halieti was confirmed in Lithuania in muscles of birds of Corvidae, Laridae, and Phalacrocoracidae families (20, 21, 36, 43). Furthermore, relatively high detection rates of S. halieti were established in intestinal scrapings of common buzzards, Eurasian goshawks and Eurasian sparrowhawks (Accipiter nisus) collected in Lithuania (24, 25). Therefore, we can expect to find this Sarcocystis species in understudied IH. According to the current knowledge, S. halieti is widely adapted to parasitise New World and Old World birds that live in terrestrial and aquatic environments and are prey or predators (6, 10, 11, 15, 20, 21, 23, 36, 41, 43). It is worrisome that S. halieti is thought to cause granulomatous encephalitis in little owl (10), but most of the studies that have been done so far have only looked at how to diagnose this parasite in muscle tissues (20, 21). Furthermore, neurological sarcocystosis was reported in several raptors, i.e., American goshawk (Astur atricapillus), bald eagle (Haliaeetus leucocephalus), and golden eagle (Aquila chrysaetos) from the United States (reviewed by 7). Thus, future extensive histopathological examinations of various birds are desirable to enclose pathogenicity and threat of S. halieti and potentially other Sarcocystis species parasitizing raptors.
To detect sarcocysts, we used a modified microscopic-compressor method initially designed to examine muscle samples of Trichinella spp. (44, 45). The method based on the analysis of methylene blue-stained muscle samples squeezed between glass compressors is superior to the analysis of fresh muscle samples in the case of low parasite loads of Sarcocystis spp. (46). Furthermore, in the case of our study, we obtained intact sarcocysts, which is not possible in the case of tissue digestion, a highly effective method for detecting low levels of Sarcocystis infection in muscle samples analyzed (47). Extraction of high-quality DNA from stained and/or fixed samples remains challenging (48). The fixation of tissues leads to fragmentation of the nucleic acids and has a negative impact on the quantity of recovered DNA. A lot of studies have been performed to optimize the extraction process from formalin-fixed paraffin-embedded (FFPE) samples (49). Several studies have successfully identified Sarcocystis species using molecular techniques from FFPE samples (12, 50, 51). Some of the studies have shown that PCR primers targeting relatively short DNA fragments (200–500 bp) facilitate the amplification of Sarcocystis species DNA from FFPE samples (11, 51). Initially, for the screening of Sarcocystis spp. in methylene blue samples, we selected to use a nested PCR approach. This PCR method is an attractive choice because it increases sensitivity and specificity and can amplify target sequences presented in low abundance (52, 53). Research has demonstrated the value of nested PCR in amplifying DNA fragments from FFPE samples (11, 54). However, nested PCR has the disadvantage of increasing the number of false positive results, mainly due to cross-contamination caused by the transfer of the products of the first amplification step to a second tube (55). Therefore, we subsequently tried direct PCR with S. halieti-specific primers. Based on direct PCR, 1/7 and 3/7 methylene blue samples were successfully amplified using standard and high-fidelity polymerases, respectively (Table 2). In each case, the Sanger sequencing confirmed S. halieti in PCR-positive samples. To the best of our knowledge, this study is the first successful attempt to identify Sarcocystis species by PCR-based methods from methylene blue-stained muscle samples. The applied method is attractive since Sarcocystis species are diagnosed from microscopically observed sarcocysts rather than from bradyzoite suspension obtained after digestion or from DNA extracted from host tissue (47, 56). We suggest this method when the muscle samples have been frozen for too long and when parasite load is low. However, optimisation of DNA extraction and PCR procedures from sarcocysts stained with methylene blue is still needed.
5 Conclusion
The present study is the first attempt to detect Sarcocystis parasites in the muscles of raptors from Lithuania. Based on methylene blue-staining, sarcocysts were detected in 8.9% (8/90) of the raptors tested, which is consistent with the low prevalence of Sarcocystis spp. in raptors reported worldwide. Using DNA sequence comparisons, the potentially pathogenic S. halieti was identified in five new intermediate host species. Due to its pathogenic potential, further extensive histopathological examinations of these hosts are recommended. In addition, for the first time, we managed to identify Sarcocystis species from methylene blue-stained muscle samples. We recommend using the suggested method when the available tissue samples are not fresh, or parasite load is low.
Data availability statement
The original data are available in a publicly accessible repository of the GenBank under the accession numbers: PQ900168–PQ900180.
Ethics statement
The animal study was approved by Animal Welfare Committee of the Nature Research Center. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
PP: Conceptualization, Formal analysis, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review & editing. TŠ: Investigation, Formal analysis, Writing – original draft, Writing – review & editing. EJ-N: Investigation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. DB: Conceptualization, Supervision, Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research was funded by the Research Council of Lithuania (grant no. S-MIP-23-4).
Acknowledgments
The authors are grateful to Kaunas T. Ivanauskas Zoology Museum for providing bird samples used in the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. McClure, CJW, Buij, R, Thorstrom, R, Vargas, FH, and Virani, MZ. The world’s most imperiled raptors present substantial conservation challenges. J Raptor Res. (2023) 57:79. doi: 10.3356/JRR-22-79
2. McClure, CJW, Westrip, JRS, Johnson, JA, Schulwitz, SE, Virani, MZ, Davies, R, et al. State of the world’s raptors: distributions, threats, and conservation recommendations. Biol Conserv. (2018) 227:390–402. doi: 10.1016/j.biocon.2018.08.012
3. Oliveira, AR, Souza, TD, Mol, JPS, Flecher, MC, Hiura, E, and Santos, RL. Pathological and molecular characterization of systemic isosporosis (atoxoplasmosis) in captive green-winged saltator (Saltator similis). Vet Parasitol. (2018) 255:98–101. doi: 10.1016/j.vetpar.2018.04.007
4. Krone, O. Endoparasites In: O Krone, editor. Raptor research and management techniques. 2nd ed. United States: Wilson Ornithological Society (2007)
5. Krone, O, Rudolph, M, and Jakob, W. Protozoa in the breast muscle of raptors in Germany. Acta Protozool. (2000) 39:35–42.
6. Prakas, P, Bea, A, Juozaitytė-Ngugu, E, Olano, I, Villanúa, D, Švažas, S, et al. Molecular identification of Sarcocystis halieti in the muscles of two species of birds of prey from Spain. Parasit Vectors. (2021) 14:414. doi: 10.1186/s13071-021-04921-0
7. Dubey, JP, Calero-Bernal, R, Rosenthal, BM, Speer, CA, and Fayer, R. Sarcocystosis of animals and humans. 2nd ed. Boca Raton, FL: CRC Press (2016).
8. Prakas, P, Calero-Bernal, R, and Dubey, JP. Sarcocystis infection in domestic and wild avian hosts: inseparable flight partners. Vet Parasitol. (2025) 335:110413. doi: 10.1016/j.vetpar.2025.110413
9. Maier-Sam, K, Kaiponen, T, Schmitz, A, Schulze, C, Bock, S, Hlinak, A, et al. Encephalitis associated with Sarcocystis halieti infection in a free-ranging little owl (Athene noctua). J Wildl Dis. (2021) 57:712–4. doi: 10.7589/JWD-D-20-00184
10. Prakas, P, Estruch, J, Velarde, R, Ilgūnas, M, Šneideris, D, Nicolás-Francisco, O, et al. First report of Sarcocystis halieti (Apicomplexa) in bearded vulture (Gypaetus barbatus). Vet Res Commun. (2024) 48:541–6. doi: 10.1007/s11259-023-10191-1
11. Fayer, R, Esposito, DH, and Dubey, JP. Human infections with Sarcocystis species. Clin Microbiol Rev. (2015) 28:295–311. doi: 10.1128/CMR.00113-14
12. Wünschmann, A, Rejmanek, D, Cruz-Martínez, L, and Barr, BC. Sarcocystis falcatula-associated encephalitis in a free-ranging great horned owl (Bubo virginianus). J Vet Diagn Invest. (2009) 21:283–7. doi: 10.1177/104063870902100223
13. Wünschmann, A, Rejmanek, D, Conrad, PA, Hall, N, Cruz-Martinez, L, Vaughn, SB, et al. Natural fatal Sarcocystis falcatula infections in free-ranging eagles in North America. J Vet Diagn Invest. (2010) 22:282–9. doi: 10.1177/104063871002200222
14. Shadbolt, T, Pocknell, A, Sainsbury, AW, Egerton-Read, S, and Blake, DP. Molecular identification of Sarcocystis wobeseri-like parasites in a new intermediate host species, the white-tailed sea eagle (Haliaeetus albicilla). Parasitol Res. (2021) 120:1845–50. doi: 10.1007/s00436-021-07103-0
15. Llano, HAB, Zavatieri Polato, H, Borges Keid, L, de Souza, F, Oliveira, TM, Zwarg, T, et al. Molecular screening for Sarcocystidae in muscles of wild birds from Brazil suggests a plethora of intermediate hosts for Sarcocystis falcatula. Int J Parasitol Parasites Wildl. (2022) 17:230–8. doi: 10.1016/j.ijppaw.2022.03.002
16. Konradt, G, Bianchi, MV, Leite-Filho, RV, da Silva, BZ, Soares, RM, Pavarini, SP, et al. Necrotizing meningoencephalitis caused by Sarcocystis falcatula in bare-faced ibis (Phimosus infuscatus). Parasitol Res. (2017) 116:809–12. doi: 10.1007/s00436-016-5341-6
17. Dubey, JP, Lindsay, DS, Rezende, PC, and Costa, AJ. Characterization of an unidentified Sarcocystis falcatula-like parasite from the south American opossum, Didelphis albiventris from Brazil. J Eukaryot Microbiol. (2000) 47:538–44. doi: 10.1111/j.1550-7408.2000.tb00087.x
18. Gondim, LFP, Soares, RM, Tavares, AS, Borges-Silva, W, de Jesus, RF, Llano, HAB, et al. Sarcocystis falcatula-like derived from opossum in northeastern Brazil: in vitro propagation in avian cells, molecular characterization and bioassay in birds. Int J Parasitol Parasites Wildl. (2019) 10:132–7. doi: 10.1016/j.ijppaw.2019.08.008
19. Rogers, KH, Arranz-Solís, D, Saeij, JPJ, Lewis, S, and Mete, A. Sarcocystis calchasi and other Sarcocystidae detected in predatory birds in California, USA. Int J Parasitol Parasites Wildl. (2021) 17:91–9. doi: 10.1016/j.ijppaw.2021.12.008
20. Kutkienė, L, Prakas, P, Sruoga, A, and Butkauskas, D. The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis). Parasitol Res. (2010) 107:879–88. doi: 10.1007/s00436-010-1945-4
21. Juozaitytė-Ngugu, E, and Prakas, P. The richness of Sarcocystis species in the common gull (Larus canus) and black-headed gull (Larus ridibundus) from Lithuania. Parasitologia (Bratisl). (2023) 3:172–80. doi: 10.3390/parasitologia3020018
22. Gjerde, B, Vikøren, T, and Hamnes, IS. Molecular identification of Sarcocystis halieti n. sp., Sarcocystis lari and Sarcocystis truncata in the intestine of a white-tailed sea eagle (Haliaeetus albicilla) in Norway. Int J Parasitol Parasites Wildl. (2018) 7:1–11. doi: 10.1016/j.ijppaw.2017.12.001
23. Máca, O, and González-Solís, D. Role of three bird species in the life cycle of two Sarcocystis spp. (Apicomplexa, Sarcocystidae) in the Czech Republic. Int J Parasitol Parasites Wildl. (2022) 17:133–7. doi: 10.1016/j.ijppaw.2022.01.002
24. Šukytė, T, Butkauskas, D, Juozaitytė-Ngugu, E, Švažas, S, and Prakas, P. Molecular confirmation of Accipiter birds of prey as definitive hosts of numerous Sarcocystis species, including Sarcocystis sp., closely related to pathogenic S. calchasi. Pathogens. (2023) 12:752. doi: 10.3390/pathogens12060752
25. Šukytė, T, Juozaitytė-Ngugu, E, Švažas, S, Butkauskas, D, and Prakas, P. The genetic identification of numerous apicomplexan Sarcocystis species in intestines of common buzzard (Buteo buteo). Animals. (2024) 14:2391. doi: 10.3390/ani14162391
26. Gjerde, B. Molecular characterisation of Sarcocystis rileyi from a common eider (Somateria mollissima) in Norway. Parasitol Res. (2014) 113:3501–9. doi: 10.1007/s00436-014-4062-y
27. Juozaitytė-Ngugu, E, Švažas, S, Šneideris, D, Rudaitytė-Lukošienė, E, Butkauskas, D, and Prakas, P. The role of birds of the family Corvidae in transmitting Sarcocystis protozoan parasites. Animals. (2021) 11:3258. doi: 10.3390/ani11113258
28. Altschul, SF, Gish, W, Miller, W, Myers, EW, and Lipman, DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
29. Tamura, K, Stecher, G, and Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
30. Kutkienė, L, Prakas, P, Sruoga, A, and Butkauskas, D. Description of Sarcocystis anasi sp. nov. and Sarcocystis albifronsi sp. nov. in birds of the order Anseriformes. Parasitol Res. (2012) 110:1043–6. doi: 10.1007/s00436-011-2588-9
31. Kutkienė, L, Prakas, P, Sruoga, A, and Butkauskas, D. Sarcocystis in the birds family Corvidae with description of Sarcocystis cornixi sp. nov. from the hooded crow (Corvus cornix). Parasitol Res. (2009) 104:329–36. doi: 10.1007/s00436-008-1196-9
32. Prakas, P, Kutkienė, L, Butkauskas, D, Sruoga, A, and Žalakevičius, M. Molecular and morphological investigations of Sarcocystis corvusi sp. nov. from the jackdaw (Corvus monedula). Parasitol Res. (2013) 112:1163–7. doi: 10.1007/s00436-012-3247-5
33. Prakas, P, Kutkienė, L, Butkauskas, D, Sruoga, A, and Žalakevičius, M. Description of Sarcocystis lari sp. n. (Apicomplexa: Sarcocystidae) from the great black-backed gull, Larus marinus (Charadriiformes: Laridae), on the basis of cyst morphology and molecular data. Folia Parasitol Praha. (2014) 61:11–7. doi: 10.14411/fp.2014.002
34. Prakas, P, Butkauskas, D, and Juozaitytė-Ngugu, E. Molecular identification of four Sarcocystis species in the herring gull, Larus argentatus, from Lithuania. Parasit Vectors. (2020) 13:869. doi: 10.1186/s13071-019-3869-x
35. Prakas, P, Butkauskas, D, and Juozaitytė-Ngugu, E. Molecular and morphological description of Sarcocystis kutkienae sp. nov. from the common raven (Corvus corax). Parasitol Res. (2020) 119:4205–10. doi: 10.1007/s00436-020-06941-8
36. Juozaitytė-Ngugu, E, Butkauskas, D, Švažas, S, and Prakas, P. Investigations on Sarcocystis species in the leg muscles of the bird family Corvidae in Lithuania. Parasitol Res. (2022) 121:703–11. doi: 10.1007/s00436-021-07409-z
37. Kutkienė, L, and Sruoga, A. Sarcocystis spp. in birds of the order Anseriformes. Parasitol Res. (2004) 92:171–2. doi: 10.1007/s00436-003-1018-z
38. Von Dohlen, AR, Scott, D, Dubey, JP, and Lindsay, DS. Prevalence of Sarcocysts in the muscles of raptors from a rehabilitation center in North Carolina. J Parasitol. (2019) 105:11–6. doi: 10.1645/18-139
39. Munday, BL, Hartley, WJ, Harrigan, KE, Presidente, PJA, and Obendorf, DL. Sarcocystis and related organisms in Australian wildlife: II. Survey findings in birds, reptiles, amphibians and fish. J Wildl Dis. (1979) 15:57–73. doi: 10.7589/0090-3558-15.1.57
40. Lindsay, DS, and Blagburn, BL. Prevalence of encysted apicomplexans in muscles of raptors. Vet Parasitol. (1999) 80:341–4. doi: 10.1016/s0304-4017(98)00228-3
41. Sato, AP, Goulart, MA, Konell, AL, de Oliveira Koch, M, da Fonseca, FM, Morel, AP, et al. Serosurvey of Toxoplasma gondii, Neospora caninum and Sarcocystis neurona in raptors and risk factor analysis. Parasitol Int. (2021) 82:102312. doi: 10.1016/j.parint.2021.102312
42. Pak, SM, and Eshtokina, NV. Sarcosporidians of birds In: SM Pak, editor. Sarcosporidians of animals in Kazakhstan. Alma-Ata: Nauka (1984)
43. Prakas, P, Butkauskas, D, Švažas, S, and Stanevičius, V. Morphological and genetic characterisation of Sarcocystis halieti from the great cormorant (Phalacrocorax carbo). Parasitol Res. (2018) 117:3663–7. doi: 10.1007/s00436-018-6083-4
44. Angi, AH, Satrija, F, Lukman, DW, Sudarwanto, M, and Sudarnika, E. Microscopic examination of pig masseter muscle by pooled sample digestion method to identify the presence of Trichinella spp. larvae. Parasitol Vet. (2023) 28:1862. doi: 10.5380/avs.v28i4.91862
45. Sharma, R, Thompson, PC, Hoberg, EP, Scandrett, WB, Konecsni, K, Harms, NJ, et al. Hiding in plain sight: discovery and phylogeography of a cryptic species of Trichinella (Nematoda: Trichinellidae) in wolverine (Gulo gulo). Int J Parasitol. (2020) 50:277–87. doi: 10.1016/j.ijpara.2020.01.003
46. Ng, YH, Subramaniam, V, and Lau, YL. Modified use of methylene blue in the tissue compression technique to detect sarcocysts in meat-producing animals. Vet Parasitol. (2015) 214:200–3. doi: 10.1016/j.vetpar.2015.09.032
47. Prakas, P, Rehbein, S, Rudaitytė-Lukošienė, E, and Butkauskas, D. Molecular identification of Sarcocystis species in diaphragm muscle tissue of European mouflon (Ovis gmelini musimon) from Austria. Parasitol Res. (2021) 120:2695–702. doi: 10.1007/s00436-021-07212-w
48. Lin, J, Kennedy, SH, Svarovsky, T, Rogers, J, Kemnitz, JW, Xu, A, et al. High-quality genomic DNA extraction from formalin-fixed and paraffin-embedded samples deparaffinized using mineral oil. Anal Biochem. (2009) 395:265–7. doi: 10.1016/j.ab.2009.08.016
49. Ramesh, PS, Madegowda, V, Kumar, S, Narasimha, S, Parichay, SR, Manoli, NN, et al. DNA extraction from archived hematoxylin and eosin-stained tissue slides for downstream molecular analysis. World J Methodol. (2019) 9:32–43. doi: 10.5662/wjm.v9.i3.32
50. Gozalo, AS, Montali, RJ, St Claire, M, Barr, B, Rejmanek, D, and Ward, JM. Chronic polymyositis associated with disseminated Sarcocystosis in a captive-born rhesus macaque. Vet Pathol. (2007) 44:695–9. doi: 10.1354/vp.44-5-695
51. Hodo, CL, Whitley, DB, Hamer, SA, Corapi, WV, Snowden, K, Heatley, JJ, et al. Histopathologic and molecular characterization of Sarcocystis calchasi encephalitis in white-winged doves (Zenaida asiatica) and Eurasian collared doves (Streptopelia decaocto), east-Central Texas, USA, 2010-13. J Wildl Dis. (2016) 52:395–9. doi: 10.7589/2015-10-292
52. Yang, S, and Rothman, RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. (2004) 4:337–48. doi: 10.1016/S1473-3099(04)01044-8
53. Green, MR, and Sambrook, J. Nested polymerase chain reaction (PCR). Cold Spring Harb Protoc. (2019) 2019:pdb.prot095182. doi: 10.1101/pdb.prot095182
54. Dietrich, D, Uhl, B, Sailer, V, Holmes, EE, Jung, M, Meller, S, et al. Improved PCR performance using template DNA from formalin-fixed and paraffin-embedded tissues by overcoming PCR inhibition. PLoS One. (2013) 8:e77771. doi: 10.1371/journal.pone.0077771
55. Liu, L, Li, M, Liu, G, He, J, Liu, Y, Chen, X, et al. A novel, highly sensitive, one-tube nested quantitative real-time PCR for Brucella in human blood samples. Microbiol Spectr. (2023) 11:e0058223. doi: 10.1128/spectrum.00582-23
Keywords: Sarcocystis, Accipitriformes, Strigiformes, methylene blue staining, molecular identification, ITS1
Citation: Prakas P, Šukytė T, Juozaitytė-Ngugu E and Butkauskas D (2025) Detection of Sarcocystis halieti in muscles of raptors from Lithuania. Front. Vet. Sci. 12:1568013. doi: 10.3389/fvets.2025.1568013
Edited by:
Vikrant Sudan, Guru Angad Dev Veterinary and Animal Sciences University, IndiaReviewed by:
Deepak Sumbria, Guru Angad Dev Veterinary and Animal Sciences University, IndiaCalin Mircea Gherman, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, Romania
Horwald Bedoya, Remington University Corporation, Colombia
Copyright © 2025 Prakas, Šukytė, Juozaitytė-Ngugu and Butkauskas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petras Prakas, cHJha2FzcGV0cmFzQGdtYWlsLmNvbQ==
 Petras Prakas
Petras Prakas Tautvilė Šukytė
Tautvilė Šukytė Evelina Juozaitytė-Ngugu
Evelina Juozaitytė-Ngugu Dalius Butkauskas
Dalius Butkauskas