- 1Department of Public and Ecosystem Health, College of Veterinary Medicine, Cornell University, Ithaca, NY, United States
- 2Department of Veterinary Biomedical Sciences, Shreiber School of Veterinary Medicine, Rowan University, Mullica Hill, NJ, United States
- 3Central Diagnostic Laboratory, College of Veterinary Medicine, Animal Resources and Biosecurity, Makerere University, Kampala, Uganda
Introduction: African swine fever (ASF) is a contagious and hemorrhagic viral disease of pigs that may present as a per-acute, sub-acute or chronic disease. Prior to this study, the clinical and pathologic presentation of ASF in pigs slaughtered in Uganda had not been characterized, and studies varied in their findings regarding differential diagnoses. The objectives of this study were to: (1) describe the clinical and pathologic presentation of ASF in pigs sampled from abattoirs in the Kampala metropolitan area over the course of one year, and (2) determine the prevalence of swine influenza A viruses (S-IAV), porcine reproductive and respiratory syndrome virus (PRRSV), classical swine fever virus (CSFV), and Salmonella spp. in these pigs.
Methods: Clinical and pathological data and samples were collected from pig abattoirs located in the Kampala metropolitan area from May 2021 through June 2022. Confirmatory diagnostic testing for African swine fever virus (ASFV) was performed using the real-time PCR (qPCR) assay. Diagnostic testing for ASFV differential diagnoses were performed using serologic and molecular techniques.
Results: Severe fever was found in 3.3% (26/794) of all pigs that were ASFV positive by any of the sample types tested. Of 196 blood positive pigs, 26% (51) had widespread splenic hemorrhages compared to 15.2% (67/442) of the pigs positive based on testing of lymph nodes, 15.5% (72/464) of pigs positive based on tonsil samples, and 15.8% (61/385) of pigs with positive spleen samples. The median gross pathologic lesion score for all pigs that tested positive for any sample type was six out of 33 [interquartile range (IQR): 4, 9]. Overall, 89.3% of the pig samples (1,188/1,330) were seropositive for S-IAV, and 0.8% (11/1,329) were seropositive for PRRSV. As for Salmonella spp., 4.4% (40/903) were qPCR positive, and all samples tested for CSFV nucleic acid were negative.
Conclusion: ASF in pigs slaughtered in central Uganda presents with clinical signs and lesions that vary; they present as healthy pigs or pigs with subacute or acute disease. However, surveillance programs in Uganda will require confirmatory laboratory diagnosis due to the occurrence of pathogens that cause similar clinical signs and lesions.
1 Introduction
African swine fever (ASF) is a highly contagious and hemorrhagic disease of pigs caused by the African swine fever virus (ASFV) belonging to the family Asfarviridae (1, 2). African swine fever was first described in Kenya in 1921 (3) and is known to be endemic in Uganda (4). The clinical presentation and pathological lesions of ASF may vary based on the virulence of the infecting virus, the route and dose of infection, and the host characteristics (5). Clinically, ASF may present as per-acute, acute, sub-acute, or chronic disease (6). Highly virulent strains of ASFV cause per-acute and acute forms of the disease, acute and subacute disease is caused by moderately virulent strains, and the chronic form of ASF is associated with moderate to low virulence isolates (5).
The per-acute form of ASFV infection is characterized by a rapid clinical course, coupled with high fever, anorexia, lethargy, and occasionally sudden death with no clinical signs, and no gross lesions observed during post-mortem examination (6, 7). The acute form presents with clinical signs and pathologic lesions such as, loss of appetite, high fever, pulmonary edema, extensive necrosis and hemorrhage of lymphoid tissue, and hemorrhages in the skin (6). Hemorrhagic splenomegaly is the most characteristic pathologic lesion (6). The subacute form of ASF resembles the acute form although with less severe clinical signs and more intense hemorrhage and edema (5). In animals with subacute disease, hydropericardium, and ascites are observed post-mortem and multifocal edema of the wall of the gallbladder or the perirenal fat is also observed (6). A review of the key ASF gross and microscopic pathologic features showed that chronic ASF is characterized by multifocal skin necrosis, arthritis, growth retardation, loss of weight, respiratory distress, and abortion (6). This clinical form has been associated with naturally occurring ASFV isolates (8, 9) and low virulence isolates that are believed to have evolved from ASFV isolates employed in early vaccine trials carried out in the Iberian Peninsula in the 1960s (6, 10).
The clinical signs and lesions of ASF are similar to those observed with other pig diseases such as classical swine fever (CSF), septicemic salmonellosis, swine influenza, porcine reproductive and respiratory syndrome (PRRS) (5, 11). Clinically, both ASF and CSF present with high fever, inappetence, incoordination, erythema (reddening of the skin), and disseminated hemorrhages predominantly in the lymph nodes (5, 11, 12). Just like ASF, septicemic salmonellosis may present with signs of febrile disease, e.g., fever and reduced feed intake, lymph node and spleen enlargement, and skin cyanosis (5, 11). The clinical signs and pathologic lesions observed in both ASF and swine influenza include fever, depression, reduced feed intake, coughing, nasal and ocular discharges, conjunctivitis, difficulty in breathing, lesions in the respiratory system such as pulmonary edema, airway exudates, lack of lung collapse (7, 11–14). Clinically, both ASF and PRRS may present with signs of respiratory disease (11, 15) and skin cyanosis, swollen or marbled lymph nodes, petechial hemorrages in the kidneys are observed in both diseases (5). In light of these similarities, clinical diagnosis of ASF based on clinical signs and lesions alone is difficult and unreliable. Thus, confirmatory laboratory testing is required for a definitive diagnosis of ASFV (7). Prior to this study, the clinical and pathologic presentation of ASF in pigs slaughtered in the Kampala metropolitan area of Uganda had not been characterized. Additionally, in Uganda, little was known about the occurrence of pathogens such as swine influenza A viruses (S-IAV), porcine reproductive and respiratory syndrome virus (PRRSV), classical swine fever virus (CSFV), and Salmonella spp., all of which cause clinical disease resembling ASF. The objectives of this study were to: (1) describe the clinical & pathologic presentation of ASF in pigs sampled from abattoirs in the Kampala metropolitan area over the course of one year, and (2) determine the prevalence of S-IAV, PRRSV, CSFV, and Salmonella spp., in pigs slaughtered in central Uganda.
2 Results
2.1 Characteristics of the pigs
Data and samples were collected from 1,334 sampled pigs that originated from 44 out of 146 (30.1%) districts of Uganda. Most of the ASFV positive pigs were sampled at the Wambizi (33%) and Lusanja (31.9%) abattoirs, 54.4% of the 794 positive pigs were exotic breed type, and most of the pigs (43.8%) came from smallholder farmers raising 1–3 pigs. Table 1 summarizes the characteristics of the ASF positive pigs.
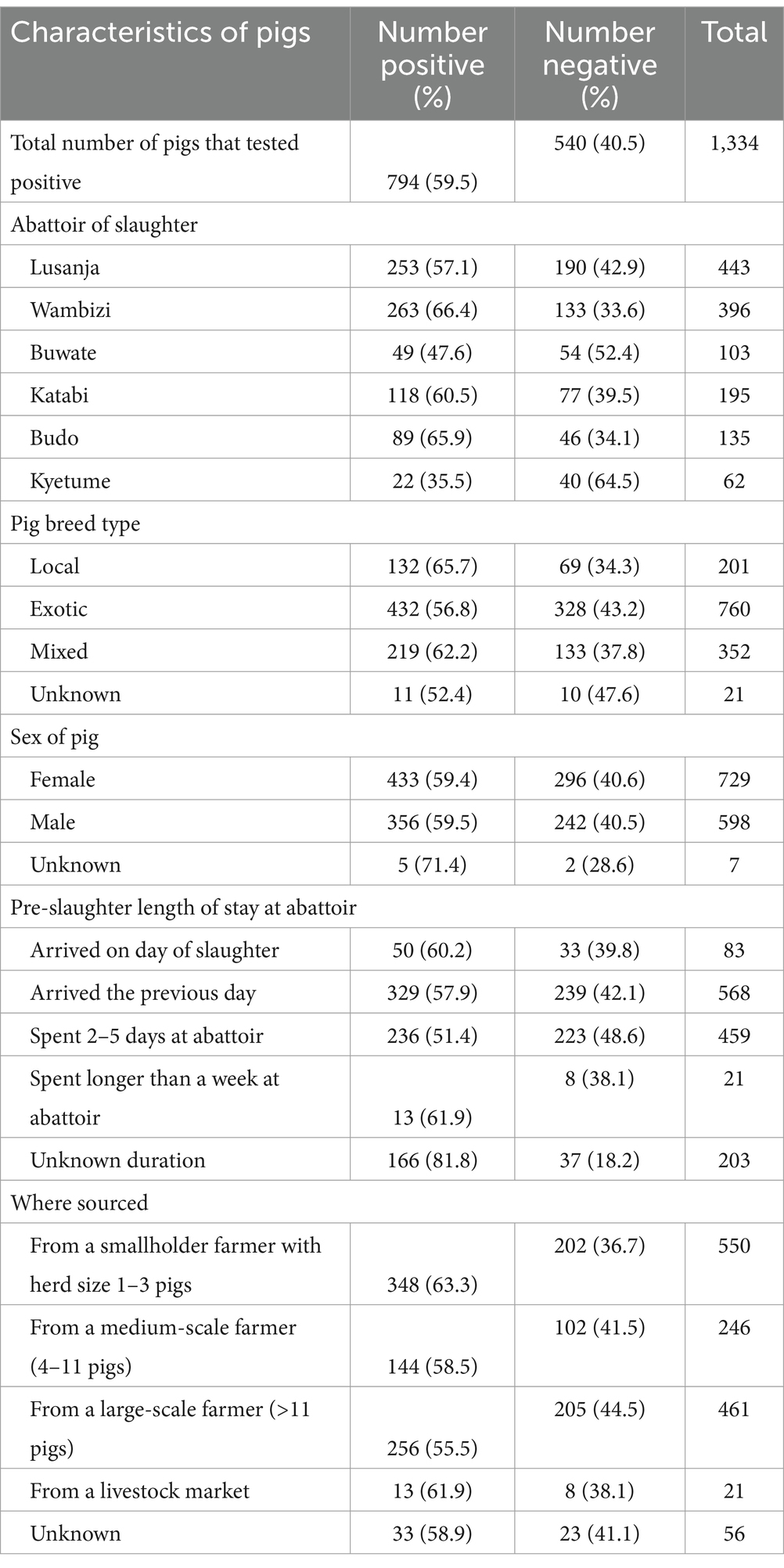
Table 1. Summary of the characteristics of ASFV qPCR positive pigs sampled from May 2021 through June 2022 from six abattoirs located in the Kampala metropolitan area of Uganda.
2.2 Clinical signs observed antemortem
Of the 1,334 pigs sampled and tested for ASFV using real-time PCR (qPCR), 59.5% (794) tested positive for ASFV for any sample type (blood, tonsil, lymph node, or spleen) collected. Figures 1–3 show the distribution of clinical signs observed in pigs that were ASFV qPCR positive by any of the sample types tested following antemortem examination. The majority of ASFV positive pigs were lively with no signs of depression (>90% regardless of sample type), had well-coordinated movements (>90% regardless of sample type), and had a normal or over-conditioned body score (>80% regardless of sample type).

Figure 1. Distribution of clinical signs (depression, gait, diarrhea, and vomiting) in pigs that were ASFV qPCR positive by any of the sample types tested.
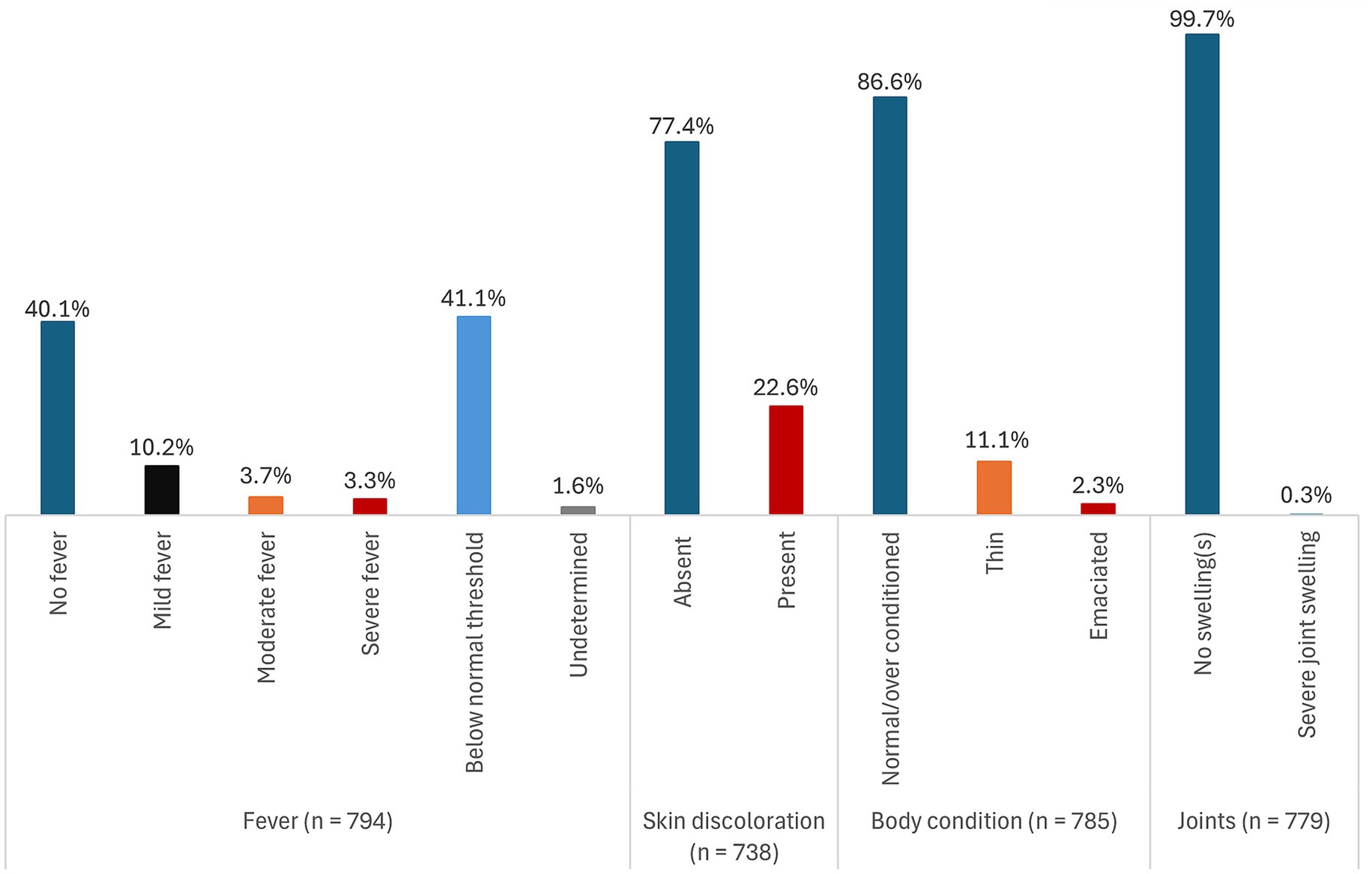
Figure 2. Distribution of clinical signs (fever, skin discoloration, body condition, and joint characteristics) in pigs that were ASFV qPCR positive by any of the sample types tested.

Figure 3. Distribution of clinical signs (breathing, cough, nasal discharges, eyes/conjunctiva, and skin necrosis) in pigs that were ASFV qPCR positive by any of the sample types tested.
Rarely found clinical signs regardless of sample type included evidence of non-bloody diarrhea (<2%), conjunctivitis (<1%), labored breathing (<2%), skin necrosis (<4%), and severe fever (<5%). No pigs had signs of vomiting and coughing. Overall, 41.1% (326/794) pigs had rectal temperatures below the normal threshold. However, there was no evidence that subnormal temperatures were associated with severe illness.
There were very little clinical signs associated with chronic disease detected. Severe joint swellings and lameness were observed in 0.2% (1/441) of lymph node positive pigs and in 0.4% (2/464) of tonsil positive pigs. Joint swellings were absent in blood and spleen positive pigs. The most common clinical sign was reddening of the skin. It was observed in 22.6% (167/738) of all pigs that were ASFV positive by any of the sample types tested, 31.5% (58/184) of blood positive pigs, 22.5% (95/423) of lymph node positive pigs, 25.3% (110/435) of tonsil positive pigs, and in 26.7% (98/367) of the spleen positive pigs. A detailed summary of the distribution of the clinical signs stratified by each of the sample types tested is presented in Supplementary file 1 (Supplementary Table S1).
2.3 Gross pathologic lesions
Pathologic lesions typical of acute ASF were found and are summarized in Supplementary file 1 (Supplementary Table S2). This included edema of the lungs with 64.1 to 73.2% of having lung edema based on the sample type used to detect ASFV. There were lesser amounts of hemorrhage in the lung with 34 to 36.1% showing some signs of hemorrhage depending on the sample type used to diagnose them. Hydropericardium was found less commonly. It was seen most often in pigs diagnosed using spleen samples (21.2%) and least often in pigs diagnosed using blood samples (16.6%). Marked splenomegaly was found in 15.5% (31/200) of blood positive pigs, 8.4% (38/451) of lymph node positive pigs, 9.3% (44/473) of tonsil positive pigs, and 10.7% (42/392) of spleen positive pigs. Of 196 blood positive pigs with data on spleen hemorrhages, 26% (51) had very dark red to almost black hemorrhages over the entire spleen compared to 15.2% (67/442) of the lymph node positive, 15.5% (72/464) of tonsil positive pigs, and 15.8% (61/385) of spleen positive pigs.
Across sample types, a large proportion of pigs had hemorrhagic lymphadenitis. Of the 197 blood positive pigs, 25.9% (51) had enlarged and diffusely hemorrhagic gastro-hepatic lymph nodes compared to 16.9% (75/443) of lymph node positive pigs, 16.9% (79/468) of tonsil positive pigs, and 19.5% (75/385) of spleen positive pigs. Hemorrhages found in other lymph node types are presented in Supplementary Table S2 in Supplementary file 1. Widespread petechial hemorrhages on the kidney surface were found in 22.9% (46/201) of blood positive pigs, 22.3% (100/448) of lymph node positive pigs, 20.5% (96/469) of tonsil positive pigs, and 25% (98/393) of spleen positive pigs.
Other hemorrhages were most commonly found on the intestinal serosa (6 to 7.5%) compared to low rates of detection of hemorrhages on the urinary bladder (≤1.7%), pericardium (≤1.1%), renal fat (≤1.8%) and gall bladder (≤1.1%). Necrosis of the tongue that is previously reported as lesions seen in chronic ASF was absent in all pigs evaluated. However, pathologic tonsil lesions previously described in chronic ASF were observed in a few pigs. Thirteen out of 196 (7%) blood positive pigs, 3.4% (15/444) of lymph node positive pigs, 3% (14/463) of tonsil positive pigs and 3.7% (14/384) of spleen positive pigs had reddened tonsil and/or tonsil with marked exudate.
Table 2 gives a summary of the median gross pathologic severity scores by sample type and overall. The median lesion score for all pigs that tested positive for any sample type was six out of 33 [interquartile range (IQR): 4, 9]. For specific sample types, the median lesion for blood was eight (IQR: 4, 12), six for both lymph nodes (IQR: 4, 9) and tonsil (IQR: 4, 10), and seven for spleen (IQR: 4, 10). All sample types had a score of four at the 25th percentile, and blood samples showed the greatest spread in its distribution of scores. The Kruskal–Wallis test showed a statistically significant difference in median scores (p = 0.025). Although no significant pairwise differences were found with the post-hoc test (p-value <0.05), differences between lymph nodes and blood (p = 0.059) and lymph nodes and spleen (p = 0.0752) were influential. Lymph nodes had a lower median and 75th percentile score than both blood and spleen.
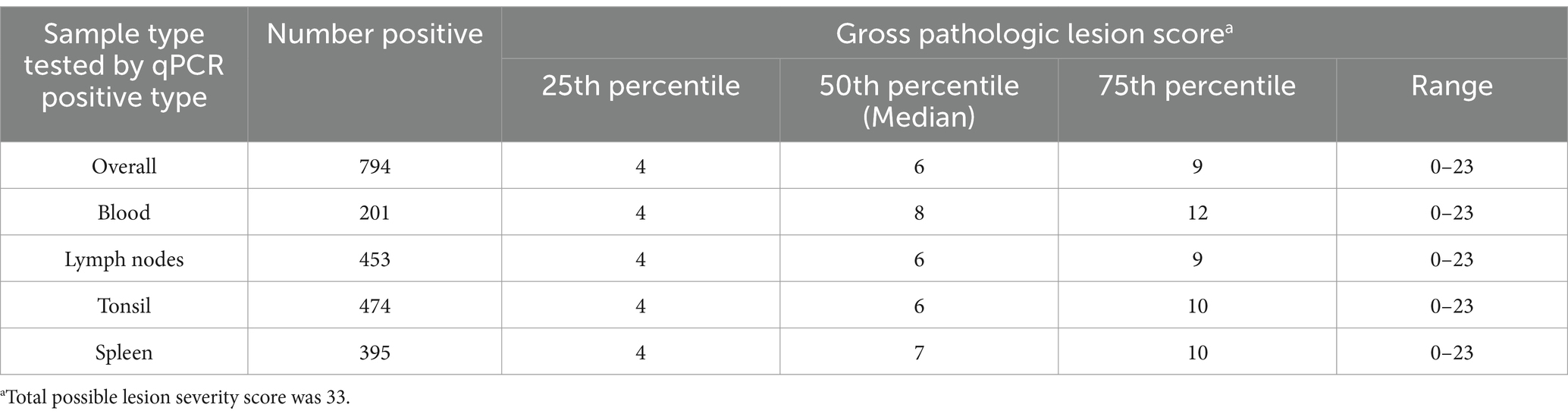
Table 2. Scores for gross pathologic lesions for all ASFV positive pigs sampled from Kampala metropolitan area abattoirs and for positive pigs by sample type tested, May 2021 through June 2022.
2.4 Differential diagnoses
Table 3 summarizes the proportions of all positive samples for ASFV differential diagnoses and the proportions of ASFV positive pigs that tested positive for ASFV differential diagnoses. Overall, 89.3% of the pig samples (1,188/1,330) were seropositive for S-IAV and 0.8% (11/1,329) were seropositive for PRRSV. In addition, 4.4% (40/903) were positive for Salmonella spp. using qPCR and all the 559 samples tested for CSFV nucleic acid using reverse transcriptase qPCR (rt-qPCR) were negative. A high proportion (89.9%) of ASFV positive pigs were exposed to S-IAV, 4.6% had Salmonella spp. nucleic acid detected, while just 0.9% were exposed to PRRSV. The median gross pathologic lesion score for the Salmonella spp. positive pigs was 3.5 (IQR: 0, 9). The 25th percentile, median and maximum (16) pathologic lesion scores were lower than for the ASFV positive pigs.
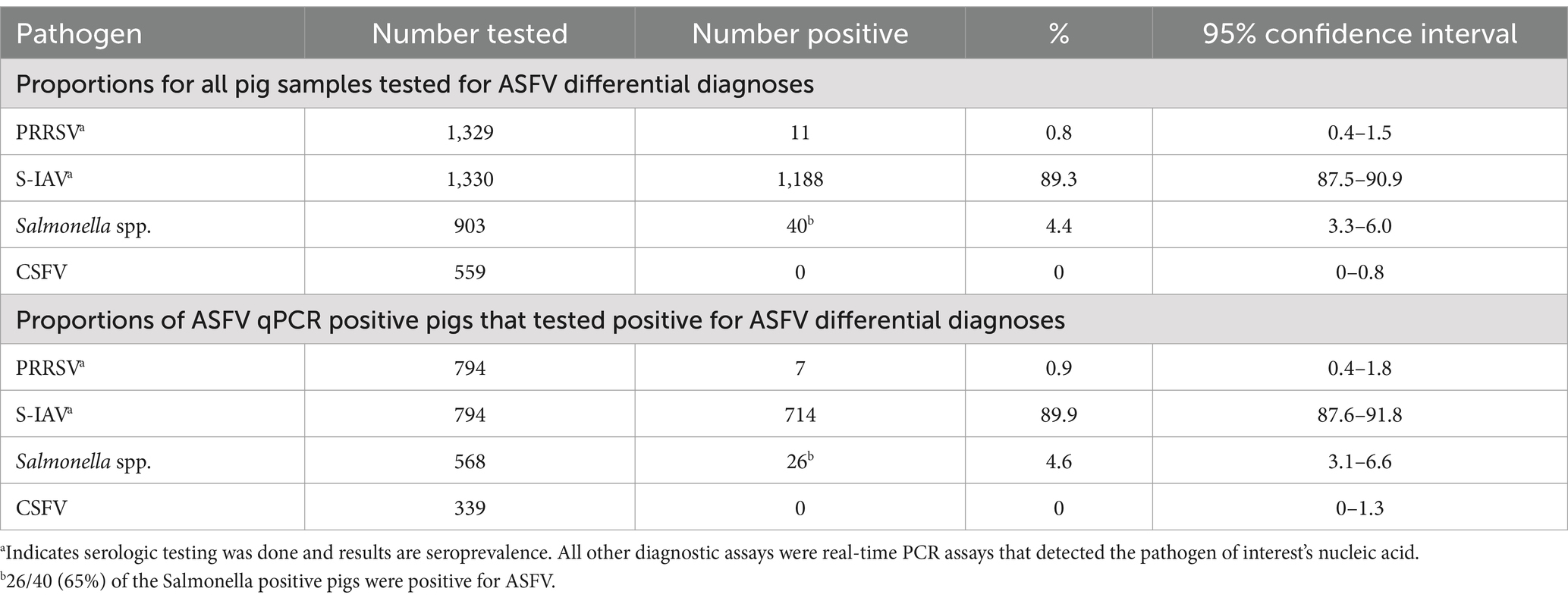
Table 3. Prevalence of ASFV differential diagnoses among pigs sampled from May 2021 through June 2022 from six abattoirs located in the Kampala metropolitan area of Uganda.
3 Discussion
This study was part of a larger project that evaluated African swine fever presentation and distribution in Uganda. To our knowledge this study is the first to describe the clinical signs and pathologic lesions of ASFV and the prevalence of its differential diagnoses in slaughtered pigs in Uganda. The findings described in this study are based on comprehensive data collected over a period of 13 months at six pig abattoirs that have a wide catchment area in the country. We found mild to moderate clinical signs in ASFV positive pigs and severe pathologic lesions in ASFV positive pigs slaughtered in the Kampala metropolitan area of Uganda. There were generally no signs of chronic ASFV lesions or signs. We also found pigs with no clinical signs. These findings show that the ASFV genotype and strains circulating in Uganda can cause subacute to acute disease in domestic pigs. As part of this project, we analyzed 31 samples by Whole Genome Sequencing, and they were all identified as genotype IX, indicating that ASFV genotype IX is currently circulating in Uganda (17). The clinical presentation reported in this present study could be associated with genotype IX. It is important to note that the historic ASFV genotype IX has been maintained as genotype 9 in a proposed new genotype classification (18). Pigs without clinical signs or with mild clinical signs may represent pigs that were exposed and sent for slaughter, potentially as part of outbreak sell-offs. Since these pigs were slaughtered and not followed over time, it is not clear if they were pre-clinical or if the disease was sub-clinical. Selling of pigs during outbreaks following outbreaks (panic sales) is a commonly reported management strategy used by farmers to reduce economic losses due to ASF (16, 19, 20). Pig abattoirs in Uganda could be used as surveillance sites for identifying ASF outbreaks and pathologic lesions seem more reliable for syndromic surveillance than clinical signs. However, it is important to note that some pigs that tested negative for ASFV had hemorrhages in the spleen, lymph nodes, kidneys, lungs as well as lung edema and tracheal exudates. This is indicative of the presence of ASFV differential diagnoses, hence confirmation will require laboratory diagnosis.
We found a very high level of exposure to S-IAV and a very low level of exposure to PRRSV among the slaughtered pigs. This high level of exposure to S-IAV and very low exposure to PRRSV may reflect immunologic responses to natural infections as the authors are not aware of any pig vaccination programs in Uganda against any of the pathogens tested. A previous Ugandan study conducted between 2010 and 2011 found a seroprevalence of 4.6% for S-IAV and a prevalence of 1.4% using reverse transcriptase polymerase chain reaction (21). The difference in seroprevalence found between the present study and that of Kirunda et al. (21) could be due to the difference in the type of ELISA test used or could be reflective of the temporal changes in the epidemiology of S-IAV given close to a 10-year difference between the study periods. The indirect S-IAV ELISA used in the present study had a sensitivity of 87 and 89% specificity (22). Another study conducted in Lira and Masaka districts of Uganda in 2013 found S-IAV seroprevalence of 8.5% in Lira and 2% in Masaka, and for PRRSV they found a 1.7% seroprevalence in Lira and 1.3% in Masaka (23). When compared to the Lira and Masaka study, our study sampled pigs originating from a wider geographic area over a longer period (13 months of sampling), hence giving us a more comprehensive picture of the seroprevalence of these pathogens in pigs at Kampala metropolitan area abattoirs. A recent cross-sectional study conducted in Lira, northern Uganda found a PRRSV seroprevalence of 7.5% (24), and a prevalence of 24.65% for PRRSV type 1 and 2.73% for PRRSV type 2 using molecular techniques (25).
In the present study we found a Salmonella spp. prevalence of 4.4% (40/903) among all pigs tested for Salmonella and 4.6% (26/568) among ASFV qPCR positive pigs tested for Salmonella spp. Furthermore, of the 40 Salmonella positive pigs, 65% (26/40) were ASFV qPCR positive. Our findings on Salmonella spp. prevalence are similar to those of a previous study (26) that found a 4% Salmonella spp. prevalence in swine fecal samples collected from Wambizi pig abattoir in Kampala. However, other previous studies in Uganda found higher prevalence of Salmonella spp. One study conducted on samples collected at Wambizi abattoir found a prevalence of 16.5% (33/100) in pig fecal matter and 10.5% (21/100) in muscle tissue (27). Another previous study conducted in 2011 and 2012 in northern and Eastern Uganda found an overall Salmonella spp. prevalence of 12% in suckling and weaned pigs (28). Although our findings show a relatively low occurrence of Salmonella spp. in slaughtered pigs, we were not able to culture and enrich samples before testing as was done as in these studies cited here and in other previous work (29). Salmonellosis needs to be considered an important differential diagnosis for ASF in Uganda that should be ruled out during ASF surveillance. Additionally, the reason 26/40 Salmonella positive pigs were ASFV qPCR positive is perhaps due to secondary Salmonella infections due to immunosuppression following ASFV infection. However, this needs to be further investigated.
In the present study, all the samples tested were negative for classical swine fever virus (CSFV), suggesting that CSFV is not circulating in Uganda. A previous study conducted in 2010–2011 that evaluated 239 pig samples found no CSFV positive pigs (30), which was similar to our results. This is an important finding in that efforts should be made to prevent the introduction of CSFV to Uganda through importation of pork products and illegal entry of pork products from endemic areas. An introduction of CSFV to Uganda would further cripple the pig industry that is struggling to contain ASFV. Since CSFV infection resembles ASFV infection, it is important to support any findings with diagnostics to ensure a definitive diagnosis is reached and a new disease is not missed during investigations.
This study had some limitations. It is possible that some misclassification bias was introduced during the qualitative analysis of free text entries in the dataset. However, such bias if any, is minimal because the qualitative analyses were performed by veterinarians and epidemiologists in the research team (JE and KH) who are knowledgeable of the clinical and pathologic presentation of ASF and in the analysis of qualitative data. It was necessary to include free-text entries into the abattoir data collection form to allow for the collection of clinical and pathologic information that would otherwise have been missed if it did not fit into pre-determined clinical signs and pathologic lesions severity categories. Also, the proportion of ASFV positive pigs with skin discolorations may be over-represented because some of the reddening observed could have resulted from insect/bug bites, and pre-slaughter pig restraint methods used at the abattoirs such as twisting of the ears and tails, dragging of pigs on the abattoir floors. It was difficult to observe skin discolorations in black/dark colored pigs.
4 Conclusion
ASF in pigs slaughtered in central Uganda presents with no clinical signs or pathologic lesions or with clinical signs and lesions typical of subacute to acute disease. It is not clear if the normal pigs are pre-clinical or have subclinical disease. Nonetheless, pig abattoirs in Uganda could be used as surveillance sites for identifying ASF outbreaks and pathologic lesions seem more reliable for syndromic surveillance than clinical signs. There is a high-level of exposure of pigs to S-IAV, a very low exposure to PRRSV, a relatively low number of pigs with detectable Salmonella spp., and no pigs with detectable CSFV among pigs slaughtered in and around Kampala. Due to the occurrence of other pathogens causing similar clinical signs and lesions, ASF surveillance programs in Uganda will require confirmatory laboratory diagnosis. There is no evidence that CSFV is currently circulating in Uganda and is not a differential diagnosis of concern at this time, but diagnostic testing to confirm suspect cases should be done to properly diagnose hemorrhagic disease cases in pigs and to identify an introduction should it occur.
5 Methods
5.1 Training of research assistants, and abattoir data collection form
Prior to data collection and sampling, a team of research assistants that comprised veterinarians and laboratory technologists were provided with a tailored manual (Supplementary file 2) on African swine fever, its clinical signs and lesions and the standard operating procedures for collecting blood and tissue samples for ASFV laboratory diagnosis. A veterinary pathologist familiar with ASF trained the research assistants on ASF and appropriate methods for sample collection, handling, and storage using the training manual as a guide. An abattoir data collection form (Supplementary file 3) that captured the clinical signs and lesions of the sampled pigs was developed and checked for validity and reliability by a team of veterinary pathologists in Uganda and the United States who are known to be experts in ASF. The abattoir data collection form was further modified to ease data capture (Supplementary file 4).
The abattoir data collection form had three sections: pig biodata, clinical scoring, and pathological scoring. The pig biodata section captured pig breed type (local, exotic, mixed), pig sex, district of origin, among other variables, while the clinical scoring section captured pre-slaughter clinical findings such as rectal temperature, clinical signs such as depression, abnormal gait, diarrhea, vomiting, body condition, cough, skin reddening (cyanosis). The clinical scoring scheme used was a modification of previously described ASF clinical scores (31–34). ASF body condition scoring was as described previously (35). The pathologic scoring section captured gross pathological lesions observed post-mortem and included lung, kidney, spleen, lymph node lesions and other lesion types as described previously (6, 32, 36, 37).
Following their training, the research assistants pretested the data collection form at three pig abattoirs under the guidance of the veterinary pathologist. Adjustments were made to the data collection form based on the feedback received from the abattoir pretesting. The data collection form was further modified and reformatted after the initial data collection to capture the number of days arriving pigs spent at the abattoir and to ease data entry.
5.2 Sampling and data collection
Clinical and pathological data and samples were collected from Lusanja, Buwate, Kyetume, Budo, Katabi, and Wambizi pig abattoirs (Figure 4) located in the Kampala metropolitan area from May 2021 through June 2022. A stratified, systematic sampling approach that weighted sample sizes for each abattoir by the average annual slaughter rates was used. For purposes of systematic sampling, adjustments were made to the sample sizes in that monthly sample size was rounded to an even number if the determined monthly sample size was an odd number and a minimum sampling size per visit was set at four. Table 4 gives a summary of the monthly sampling frequency and associated sample sizes for each abattoir. Two to four sampling days were randomly selected each month for each abattoir. On the day of sampling, a systematic sampling approach was used to select pigs at the site until the desired sample size for that day was achieved. Typically, for each slaughterhouse visit, an estimate of the number of pigs to be slaughtered was determined and a sampling interval was computed by dividing the day’s estimated slaughter total by the required sample size for that visit.
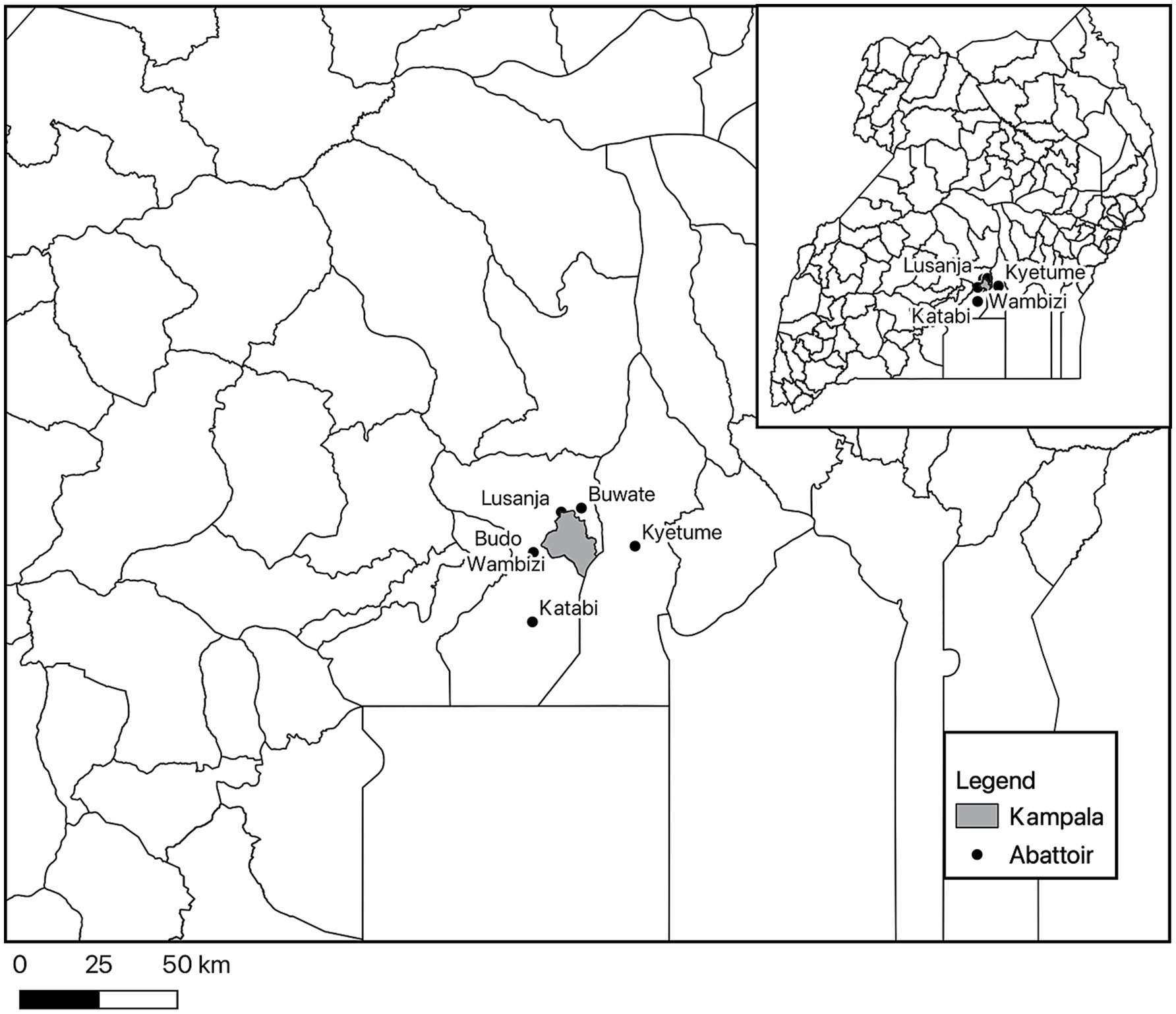
Figure 4. Map showing the location of the six abattoirs where pigs were sampled in the Kampala metropolitan area of Uganda.
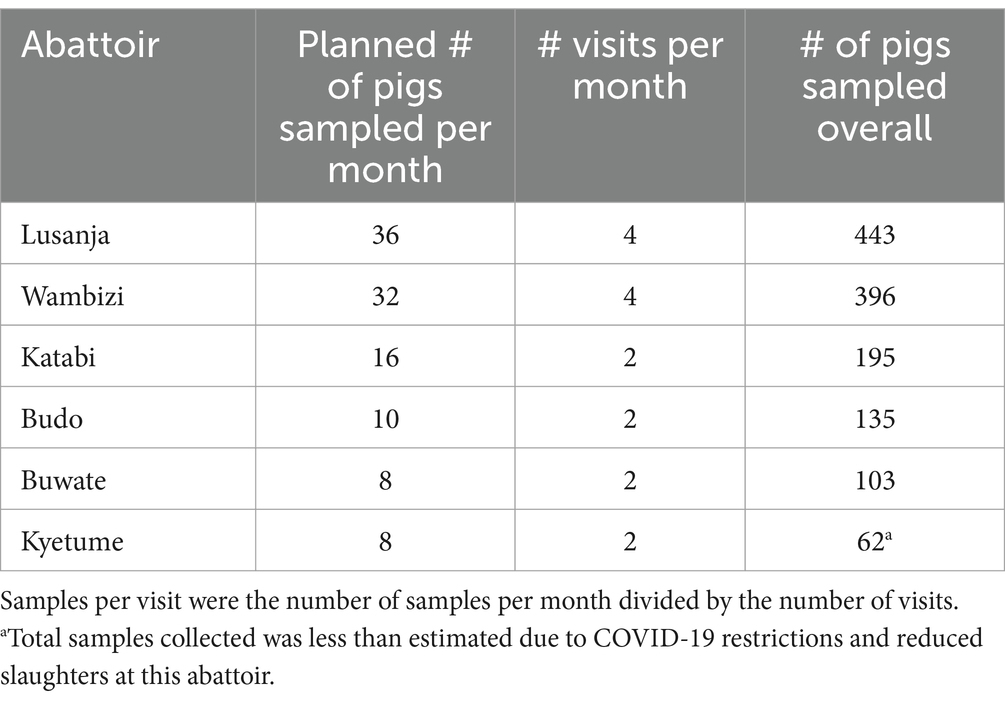
Table 4. Distribution of the number for pigs sampled each month at six abattoirs in the Kampala metropolitan area, May 2021 through June 2022.
At the study design stage, it was determined that 100 positive pigs were needed to reasonably characterize the clinical and pathologic presentation of ASF at the abattoirs. The prevalence of ASF was expected to be 11.5% (4) and the sample size needed to detect this prevalence at a 95% confidence level and 5% error rate was determined to be 157 pigs (openepi.com; Accessed July 2018). This would result in 18 positive pigs, and we sampled 1,200 pigs needed to ensure 100 positive pigs are sampled.
5.3 Confirmatory diagnostic testing for ASFV
Confirmatory diagnostic testing for ASFV was performed using the real-time PCR (qPCR) assay as we previously described (38). In brief, the samples tested included whole blood, pooled lymph nodes (submandibular, renal and gastro-hepatic), tonsil, and spleen. All samples were tested following standard operating procedures (SOPs) from the Foreign Animal Disease Diagnostic Laboratory (Plum Island Animal Disease Center, New York, United States), a World Organisation for Animal Health African swine fever virus reference laboratory. Only the clinical and pathologic lesions of qPCR positive pigs are described in this paper.
5.4 Diagnostic testing for ASFV differential diagnoses
5.4.1 Serologic testing
Serum samples were prepared from blood collected into a clotting tube (Becton, Dickinson and Company, Franklin Lakes, New Jersey, United States). The tube was transported on ice to the diagnostic laboratory and stored at 4°C overnight to incubate. They were then centrifuged at 1,000 × g (Eppendorf 5804, Hamburg, Germany) for 10 min and then the serum was aliquoted into cryovials and stored at −20°C until testing occurred. Serum samples were tested for antibodies against swine influenza A virus (S-IAV) and porcine reproductive and respiratory syndrome virus (PRRSV). The INgezim PRRS 2.0 indirect ELISA for the detection of antibodies against North American and European PRRSV variants (Ingenasa, Madrid, Spain) and the indirect INgezim swine influenza 2.0 ELISA (Ingenasa, Madrid, Spain), which detects antibodies against influenza A viruses in swine, were used, and manufacturer’s instructions included in the kit were followed.
5.4.2 Molecular testing
Pooled lymph nodes (submandibular, renal and gastro-hepatic) were tested for Salmonella spp. DNA, and tonsils for classical swine fever virus RNA. Tissue samples were stored at −20°C after collection until they were processed. Tissue processing occurred as follows. For each tissue type, one gram was washed in 1X phosphate buffered saline (PBS) solution (Thermo Fisher Scientific, Waltham, Massachusetts, United States) and homogenized using the Stomacher 80 Biomaster (Seward Ltd., West Sussex, United Kingdom). After the tissue was homogenized, 9 mL of 1X PBS was added and the mixture was centrifuged for 10 min at 1,000 × g. The supernatant was collected and stored at −20°C until extraction occurred. These procedures followed the US Department of Agriculture (USDA) Foreign Animal Disease Diagnostic Laboratory (FADDL) sample preparation standard operating procedures (SOPs) except that a stomacher was used rather than a tissue lyser for homogenization (39).
Lymph nodes underwent DNA extraction using the Qiagen DNeasy tissue and blood kit (Qiagen, Hilden, Germany) following the standard operating procedures developed by the USDA FADDL (39) and tonsils underwent RNA extraction using the Qiagen RNeasy mini kit (Qiagen, Hilden, Germany) also following the standard operating procedures developed by the USDA FADDL (40). Both DNA and RNA extraction aligned with the manufacturer’s instructions.
Real-time PCR (qPCR) assays were run on a Quantstudio 5 thermocycler (Thermo Fisher Scientific, Waltham, Massachusetts, United States). The Salmonella qPCR procedure has been previously described (29). The primers target highly conserved regions of the Salmonella-ttr locus (4). The samples were not enriched prior to testing. The forward primer of 5′-CTCACCAGGAGATTACAACATGG-3′, reverse primer of 5′-AGCTCAGACCAAAAGTGACCATC-3′, and a probe of 5′-FAM-CACCGACGGCGAGACCGACTTT-3′-BHQ1 (Eurofin Genomic, Munich, Germany) were used along with TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific, Waltham, Massachusetts, United States). The VetMax Xeno DNA internal positive control (IPC) (Thermo Fisher Scientific, Waltham, Massachusetts, United States) was added to each sample prior to extraction and detected during qPCR using the VetMax Zeno IPC LIZ assay (Thermo Fisher Scientific, Waltham, Massachusetts, United States). CSFV detection was completed following USDA FADDL SOPs (41). The same master mix and VetMax Xeno Liz assay as was used for the Salmonella qPCR were used, except the VetMax Xeno IPC was an RNA control. The forward primer was 5′-TGCCCAAGACACACCTTAACC-3′, reverse primer was 5′-GGCCTCTGCAGCGCCCTAT-3′, and the probe was 5′-FAM-TGATGGGAGTACGACCTG-3′-MGBEQ (Eurofin Genomic, Munich, Germany). Not all tonsils and lymph nodes were tested due to a work stoppage implemented by the funding entity for political reasons.
5.5 Data management and analyses
Clinical and pathologic data captured in the data collection forms were entered into Microsoft Excel version 16 (Microsoft, Redmond, Washington, United States) and the data entry validated. Gross pathologic lesion data captured as free text was qualitatively assessed and re-aligned with existing score categories in the dataset where possible or discarded if they could not fit. The clinical signs and pathologic lesions data were collated in Microsoft Excel Version 2,306 Build 16.0.16529.20100 (Microsoft, Redmond, Washington, United States).
Diagnostic results were first evaluated by assessing the controls. For serology, this was the negative and positive controls and for molecular diagnostics this included negative and positive extraction and amplification controls as well as the IPC. Any results with failed controls were excluded. Any samples with a negative IPC and negative results were excluded. For any duplicate samples, results were compared across the CSFV or Salmonella spp. results as well as across ASFV results, if the results for the two pathogens agreed across the duplicate samples, the first sample tested was included. If the results differed, including Ct values with a difference >3 cycle threshold (Ct) values, then those samples were excluded. Data was collated in Microsoft Excel v 16.74 (Microsoft, Redmond, Washington, United States) in preparation for analysis.
Stata 18.0 and Stata 16.1 IC (Stata Corp, College Station, Texas, United States) were used for statistical analyses of the clinical signs and pathologic lesions and the differential diagnoses data, respectively. The clinical signs and gross pathologic lesion scores data were summarized using frequencies and proportions by sample type tested (blood, lymph nodes, tonsil, and spleen) and for all pigs that were ASFV positive by any of the sample types tested. The gross pathologic lesions were scored as described in Supplementary file 2, and the total sum of scores per pig sampled was calculated. The total possible severity score for the gross pathologic lesions was 33. Normality of the pathologic lesion scores data was evaluated using a visual assessment of a histogram and the Shapiro–Wilk test, and both showed the data were not normally distributed. Percentiles of the pathologic lesion scores were calculated. The Kruskal–Wallis test followed by Dunn’s test of multiple comparisons was used to evaluate for differences in median scores among the different sample types tested by qPCR and the level of significance for this test was 0.05, but influential variables were described as those with a probability value of less than 0.1. Percent positivity for the serologic and molecular results were summarized and 95% confidence intervals calculated using the Agresti-Coull method (42). The denominators used in the calculation of the proportions varied based on the completeness of the data for each variable in the dataset. The map showing the location of the pig abattoirs visited was created in QGIS Firenze version 3.28.11 and the map shape file for Ugandan administrative districts were from the United Nations High Commissioner for Refugee Operations Data Portal (https://data.unhcr.org/en/documents/details/83043; Accessed March 30, 2023, published on November 17, 2020).
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by the College of Veterinary Medicine, Animal Resources, and Biosecurity, Makerere University Higher Degrees Research Committee (Reference Number: SBLS.EWM.2020), approved by the Uganda National Council for Science and Technology (Registration Number: NS266ES). Animal work was approved by Cornell University’s Institutional Animal Care and Use Committee (Protocol Number: 2019-0108), and the US Army Medical Research and Development Command’s Animal Care and Use Review Office (Protocol Number: CT-2020-38). The abattoir data collection form was reviewed and determined to be exempt by the Cornell University’s Institutional Review Board (Protocol Number: 2012010020) with a concurrence from the Human Research Protection Office of the US Army Medical Research and Development Command (Log Number: A-21116). All methods were performed in accordance with the relevant guidelines and regulations. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JE: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing, Software. AN: Data curation, Investigation, Writing – review & editing. CH: Data curation, Investigation, Resources, Writing – review & editing. KO: Data curation, Investigation, Writing – review & editing. DN: Conceptualization, Methodology, Project administration, Resources, Writing – review & editing, Funding acquisition. RO: Data curation, Investigation, Validation, Writing – review & editing. EK: Data curation, Investigation, Validation, Writing – review & editing. EW: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing. KH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was sponsored by the U.S. Department of the Defense, Defense Threat Reduction Agency, under Grant Number HDTRA1-20-1-0007. The content of the information does not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred.
Acknowledgments
The authors would like to thank all the veterinarians, slaughterhouse staff and laboratory technologists who assisted with data and sample collection at the pig abattoirs. The authors would also like to thank the US Department of Agriculture’s Foreign Animal Disease Diagnostic Laboratory’s guidance on RNA extraction techniques.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1568095/full#supplementary-material
Footnotes
References
1. Alonso, C, Borca, M, Dixon, L, Revilla, Y, Rodriguez, F, Escribano, JM, et al. ICTV virus taxonomy profile: Asfarviridae. J Gen Virol. (2018) 99:613–4. doi: 10.1099/jgv.0.001049
2. Kennedy, M, Delhon, G, McVey, DS, Vu, H, and Borca, M. Asfarviridae and Iridoviridae In. Veterinary microbiology. Hoboken, NJ: John Wiley & Sons (2022). 478–83.
3. Eustace, MR. On a form of swine fever occurring in British East Africa (Kenya Colony). J Comp Pathol Ther. (1921) 34:159–91. doi: 10.1016/S0368-1742(21)80031-4
4. Atuhaire, DK, Afayoa, M, Ochwo, S, Mwesigwa, S, Mwiine, FN, Okuni, JB, et al. Prevalence of African swine fever virus in apparently healthy domestic pigs in Uganda. BMC Vet Res. (2013) 9:263. doi: 10.1186/1746-6148-9-263
5. Sánchez-Vizcaíno, JM, Mur, L, Gomez-Villamandos, JC, and Carrasco, L. An update on the epidemiology and pathology of African swine fever. J Comp Pathol. (2015) 152:9–21. doi: 10.1016/j.jcpa.2014.09.003
6. Salguero, FJ. Comparative pathology and pathogenesis of African swine fever infection in swine. Front Vet Sci. (2020) 7:282. doi: 10.3389/fvets.2020.00282
7. Sánchez-Vizcaíno, JM, Laddomada, A, and Arias, ML. African swine fever virus In. Diseases of swine. Hoboken, NJ: John Wiley & Sons (2019). 443–52.
8. Boinas, FS, Hutchings, GH, Dixon, LK, and Wilkinson, PJ. Characterization of pathogenic and non-pathogenic African swine fever virus isolates from Ornithodoros erraticus inhabiting pig premises in Portugal. J Gen Virol. (2004) 85:2177–87. doi: 10.1099/vir.0.80058-0
9. Gallardo, C, Soler, A, Nurmoja, I, Cano-Gómez, C, Cvetkova, S, Frant, M, et al. Dynamics of African swine fever virus (ASFV) infection in domestic pigs infected with virulent, moderate virulent and attenuated genotype II ASFV European isolates. Transbound Emerg Dis. (2021) 68:2826–41. doi: 10.1111/tbed.14222
10. Sun, E, Huang, L, Zhang, X, Zhang, J, Shen, D, Zhang, Z, et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg Microbes Infect. (2021) 10:2183–93. doi: 10.1080/22221751.2021.1999779
11. Merck & Co., Inc.. African swine fever—generalized conditions. Merck veterinary manual. Available online at: https://www.merckvetmanual.com/generalized-conditions/african-swine-fever/african-swine-fever. (Accessed March 25, 2025)
12. Constable, PD, Hinchcliff, KW, Done, SH, and Gruenberg, W. Systemic and multi-organ diseases In. Veterinary medicine: a textbook of the diseases of cattle, horses, sheep, pigs and goats. 11th ed. St. Louis, MO: Elsevier (2017). 2002–214.
13. Merck & Co., Inc.. Influenza A virus in swine—respiratory system. Merck veterinary manual. Available online at: https://www.merckvetmanual.com/respiratory-system/respiratory-diseases-of-pigs/influenza-a-virus-in-swine. (Accessed April 25, 2025)
14. Constable, PD, Hinchcliff, KW, Done, SH, and Grünberg, W. Diseases of the respiratory system In. Veterinary medicine: a textbook of the diseases of cattle, horses, sheep, pigs and goats. 11th ed. St. Louis, MO: Elsevier (2016). 845–1090.
15. Merck & Co., Inc.. Porcine reproductive and respiratory syndrome—generalized conditions. Available online at: https://www.merckvetmanual.com/generalized-conditions/porcine-reproductive-and-respiratory-syndrome/porcine-reproductive-and-respiratory-syndrome. (Accessed April 25, 2025)
16. Dione, M, Ouma, E, Opio, F, Kawuma, B, and Pezo, D. Qualitative analysis of the risks and practices associated with the spread of African swine fever within the smallholder pig value chains in Uganda. Prev Vet Med. (2016) 135:102–12. doi: 10.1016/j.prevetmed.2016.11.001
17. Okwasiimire, R, Flint, JF, Kayaga, EB, Lakin, S, Pierce, J, Barrette, RW, et al. Whole genome sequencing shows that African swine fever virus genotype IX is still circulating in domestic pigs in all regions of Uganda. Pathogens. (2023) 12:912. doi: 10.3390/pathogens12070912
18. Spinard, E, Dinhobl, M, Tesler, N, Birtley, H, Signore, AV, Ambagala, A, et al. A re-evaluation of African swine fever genotypes based on p72 sequences reveals the existence of only six distinct p72 groups. Viruses. (2023) 15:2246. doi: 10.3390/v15112246
19. Dione, MM, Akol, J, Roesel, K, Kungu, J, Ouma, EA, Wieland, B, et al. Risk factors for African swine fever in smallholder pig production systems in Uganda. Transbound Emerg Dis. (2017) 64:872–82. doi: 10.1111/tbed.12452
20. Muhangi, D, Masembe, C, Berg, M, Ståhl, K, and Ocaido, M. Practices in the pig value chain in Uganda; implications to African swine fever transmission. Livestock Research for Rural Development (2014). Available at: http://www.lrrd.org/lrrd26/5/muha26094.html (Accessed May 21, 2025).
21. Kirunda, H, Erima, B, Tumushabe, A, Kiconco, J, Tugume, T, Mulei, S, et al. Prevalence of influenza a viruses in livestock and free-living waterfowl in Uganda. BMC Vet Res. (2014) 10:50. doi: 10.1186/1746-6148-10-50
22. Ingenasa. Ingezim Influenza Porcina R.11.FLU.K1 (Technical Sheet). Available online at: https://www.goldstandarddiagnostics.com/pub/media/productattachments/files/11FLUK1_Technical_sheet_Influenza_Porcina.pdf. (Accessed July 3, 2023)
23. Dione, M, Masembe, C, Akol, J, Amia, W, Kungu, J, Lee, HS, et al. The importance of on-farm biosecurity: sero-prevalence and risk factors of bacterial and viral pathogens in smallholder pig systems in Uganda. Acta Trop. (2018) 187:214–21. doi: 10.1016/j.actatropica.2018.06.025
24. Oba, P, Dione, MM, Wieland, B, Mwiine, FN, and Erume, J. Correlations between lung pneumonic lesions and serologic status for key respiratory pathogens in slaughtered pigs in northern Uganda. Porc Health Manag. (2021) 7:53. doi: 10.1186/s40813-021-00233-y
25. Oba, P, Dione, MM, Erume, J, Wieland, B, Mutisya, C, Ochieng, L, et al. Molecular characterization of porcine reproductive and respiratory syndrome virus (PRRSv) identified from slaughtered pigs in northern Uganda. BMC Vet Res. (2022) 18:176. doi: 10.1186/s12917-022-03272-x
26. Afema, JA, Byarugaba, DK, Shah, DH, Atukwase, E, Nambi, M, and Sischo, WM. Potential sources and transmission of Salmonella and antimicrobial resistance in Kampala, Uganda. PLoS One. (2016) 11:e0152130. doi: 10.1371/journal.pone.0152130
27. Tinega, GM, Magiri, E, Kinyua, J, Njahira, M, Erume, J, Ejobi, F, et al. Characterization of Salmonella isolates obtained from pigs slaughtered at Wambizzi Abattoir in Kampala, Uganda. J. Agric. Sci. Technol. (2016) 17:99–120. doi: 10.4314/jagst.v17i1
28. Ikwap, K, Erume, J, Owiny, DO, Nasinyama, GW, Melin, L, Bengtsson, B, et al. Salmonella species in piglets and weaners from Uganda: prevalence, antimicrobial resistance and herd-level risk factors. Prev Vet Med. (2014) 115:39–47. doi: 10.1016/j.prevetmed.2014.03.009
29. Malorny, B, Paccassoni, E, Fach, P, Bunge, C, Martin, A, and Helmuth, R. Diagnostic real-time PCR for detection of Salmonella in food. Appl Environ Microbiol. (2004) 70:7046–52. doi: 10.1128/AEM.70.12.7046-7052.2004
30. Muhangi, D, Masembe, C, Emanuelson, U, Boqvist, S, Mayega, L, Ademun, RO, et al. A longitudinal survey of African swine fever in Uganda reveals high apparent disease incidence rates in domestic pigs, but absence of detectable persistent virus infections in blood and serum. BMC Vet Res. (2015) 11:106. doi: 10.1186/s12917-015-0426-5
31. Mittelholzer, C, Moser, C, Tratschin, JD, and Hofmann, MA. Analysis of classical swine fever virus replication kinetics allows differentiation of highly virulent from avirulent strains. Vet Microbiol. (2000) 74:293–308. doi: 10.1016/s0378-1135(00)00195-4
32. Galindo-Cardiel, I, Ballester, M, Solanes, D, Nofrarías, M, López-Soria, S, Argilaguet, JM, et al. Standardization of pathological investigations in the framework of experimental ASFV infections. Virus Res. (2013) 173:180–90. doi: 10.1016/j.virusres.2012.12.018
33. Howey, EB, O’Donnell, V, de Carvalho Ferreira, HC, Borca, MV, and Arzt, J. Pathogenesis of highly virulent African swine fever virus in domestic pigs exposed via intraoropharyngeal, intranasopharyngeal, and intramuscular inoculation, and by direct contact with infected pigs. Virus Res. (2013) 178:328–39. doi: 10.1016/j.virusres.2013.09.024
34. McCleary, S, Strong, R, McCarthy, RR, Edwards, JC, Howes, EL, Stevens, LM, et al. Substitution of warthog NF-κB motifs into RELA of domestic pigs is not sufficient to confer resilience to African swine fever virus. Sci Rep. (2020) 10:8951. doi: 10.1038/s41598-020-65808-1
35. Olesen, AS, Lohse, L, Boklund, A, Halasa, T, Gallardo, C, Pejsak, Z, et al. Transmission of African swine fever virus from infected pigs by direct contact and aerosol routes. Vet Microbiol. (2017) 211:92–102. doi: 10.1016/j.vetmic.2017.10.004
36. Sánchez-Vizcaíno, JM, and Neira, AM. African swine fever virus. 10th ed. Chichester: Wiley-Blackwell (2012). 983 p.
37. Guinat, C, Reis, AL, Netherton, CL, Goatley, L, Pfeiffer, DU, and Dixon, L. Dynamics of African swine fever virus shedding and excretion in domestic pigs infected by intramuscular inoculation and contact transmission. Vet Res. (2014) 45:93. doi: 10.1186/s13567-014-0093-8
38. Okwasiimire, R, Kayaga, EB, Ekakoro, JE, Ndoboli, D, Schumann, K, Faburay, B, et al. Spatiotemporal description of African swine fever virus nucleic acid and antibodies detected in pigs sampled at abattoirs in the greater Kampala metropolitan area, Uganda from May 2021 through June 2022. Porcine Health Manag. (2023) 9:51. doi: 10.1186/s40813-023-00345-7
39. Schumann, K. Extraction of RNA from Swabs, Tissues, Tissue Culture Supernatant a Using Qiagen RNeasy Mini Kit (Standard Operating Procedure No. SOP-FADDL-0015 Revision A). Greenport, New York: Foreign Animal Disease Diagnostic Laboratory, National Veterinary Services Laboratory, US Department of Agriculture, Plum Island Animal Disease Center (2014).
40. Schumann, K. Classical Swine Fever and Foot and Mouth Disease rRT-PCR Using the TaqMan Fast Virus 1-Step Master Mix on the Applied Biosystems Quantstudio 5 Real-time PCR Platform (Standard Operating Procedure No. SOP-FADDL-0072 Revision 1). Greenport, New York: Foreign Animal Disease Diagnostic Laboratory, National Veterinary Services Laboratory, US Department of Agriculture, Plum Island Animal Disease Center (2020).
Keywords: African swine fever, clinical signs and lesions, differential diagnoses, pigs, Uganda
Citation: Ekakoro JE, Nassali A, Hauser C, Ochoa K, Ndoboli D, Okwasiimire R, Kayaga EB, Wampande EM and Havas KA (2025) A description of the clinical signs and lesions of African swine fever, and its differential diagnoses in pigs slaughtered at selected abattoirs in central Uganda. Front. Vet. Sci. 12:1568095. doi: 10.3389/fvets.2025.1568095
Edited by:
Mariangela Caroprese, University of Foggia, ItalyReviewed by:
Rachel Jean Derscheid, Iowa State University, United StatesKaichuang Shi, Guangxi Center for Animal Disease Control and Prevention, China
Copyright © 2025 Ekakoro, Nassali, Hauser, Ochoa, Ndoboli, Okwasiimire, Kayaga, Wampande and Havas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John E. Ekakoro, ZWtha29yb0Byb3dhbi5lZHU=
 John E. Ekakoro
John E. Ekakoro Aisha Nassali1
Aisha Nassali1 Edrine B. Kayaga
Edrine B. Kayaga Karyn A. Havas
Karyn A. Havas