- 1Department of Animal Sciences, University of Illinois Urbana-Champaign, Urbana, IL, United States
- 2Gastrointestinal Laboratory, Department of Small Animal Clinical Sciences, Texas A&M University, College Station, TX, United States
- 3Division of Nutritional Sciences, University of Illinois Urbana-Champaign, Urbana, IL, United States
- 4Department of Veterinary Clinical Medicine, University of Illinois Urbana-Champaign, Urbana, IL, United States
There is great interest in studying the canine gastrointestinal microbiome. In healthy dogs versus those with acute and chronic enteropathies, specific bacterial taxa have been identified that are consistently associated with shifts in the microbiome. A qPCR-based dysbiosis index (DI) that assesses microbiome shifts was developed based on a subset of these taxa. Because most dogs consume kibble diets, published data on core bacteria and the DI were largely derived from dogs consuming that diet form. Because dietary composition impacts the microbiome, it was unknown whether data from dogs consuming other diet types would adhere to reported core taxa abundance and DI guidelines. The study’s aim was to determine the fecal abundance of core bacteria and DI of dogs fed commercially available kibble vs. mildly-cooked human-grade (fresh) diets. Fecal samples collected from adult dogs across four experiments were used (4 kibble diets, n = 10–12/treatment; 4 fresh diets, n = 10–24/treatment). Moderate correlations were observed between total dietary fiber (TDF) and Fusobacterium (positive correlation), Lactobacillus (negative), and DI (negative). Dietary protein was correlated with fecal Ruminococcus gnavus (negative), while dietary fat was correlated with fecal Bacteroides and C. perfringens abundance (both positive). Dogs fed fresh diets exhibited higher (p < 0.01) abundances of Streptococcus, Escherichia coli, and Clostridium perfringens, while those fed kibble diets had higher (p < 0.05) abundances of Fusobacterium, Clostridium hiranonis, and Bacteroides. Dogs fed fresh diets had a greater (p < 0.0001) DI, but the majority of scores remained within the normal range. Dogs fed animal protein-based kibble diets had higher (p < 0.05) fecal Faecalibacterium and Fusobacterium, while dogs fed animal protein-based fresh diets had higher (p < 0.05) Streptococcus, E. coli, and C. perfringens. Bifidobacterium and Bacteroides were more abundant (p < 0.01) in dogs fed animal protein-based kibble and plant protein-based fresh diets. Dogs fed animal protein-based fresh diets had a greater (p < 0.0001) DI. Even though microbiota populations were statistically different among diets, all mean DI were <0, with only a few individual dogs consuming fresh diets having DI >0 (5 dogs >0; 1 dog >2). Overall, these data demonstrate the utility of the DI across different diet types in healthy dogs.
1 Introduction
Over the past decade, there has been significant interest in characterizing the gastrointestinal microbiome of dogs because of its role in digestion, energy extraction, pathogen defense, support for enterocytes, and immune stimulation (1–3). Most canine microbiome studies have focused on assessing the effects of various ingredients present in extruded kibble diets, which dominate the dog food market (4). The pet food industry frequently mirrors human dietary trends and has resulted in a great interest in the feeding of fresh diet formats. Unlike conventional extruded diets, fresh diets utilize methods like kettle cooking and steam cooking, with many cooking individual ingredients before blending. Conventional pet foods usually include animal and plant byproducts, but an emerging preference among pet owners is to choose byproduct-free diets (5). The macronutrient composition of these diets may differ based on these preferences or manufacturing method, with potential influences on the nutritional and health status of animals.
Dietary differences in manufacturing method, ingredient composition, and nutrient concentration may impact nutrient digestibility and consequent influences on the microbiome. Alpha and beta diversity results have been inconsistent across previous microbiome studies. For instance, dogs fed fresh diets have been shown to have lower species richness than those fed kibble diets (6). However, other studies found no differences in alpha diversity between dogs fed fresh diet and those fed kibble diets (7–9). Some studies have reported significant shifts in beta diversity when comparing fresh diets to kibble diets (8); however, other studies have not found significant separation between microbial communities (6, 7, 9). These findings underscore that diet type may affect gut microbiota composition, although there are major inconsistencies in the reported microbiome results across studies.
Eubiosis, or microbial ecosystem balance, is present in a healthy state, where the symbiotic relationship between the host and microorganisms maintains a state of equilibrium. Disturbances may lead to a state of dysbiosis, often characterized by reduced bacterial diversity, depletion of beneficial bacteria, and the proliferation of pathobionts, resulting in altered metabolic functionality (10, 11). Dysbiosis is implicated in various diseases such as inflammatory bowel disease (IBD), obesity, allergy, and diabetes in humans, rodent models, and companion animals (12). The collection of studies testing the gut microbiota populations of individuals with these conditions, however, have not described a consistent microbial shift. For instance, in dogs with chronic enteropathies, some have reported greater Gammaproteobacteria and reduced Erysipelotrichia, Clostridia, and Bacteroidia (13). Another study noted elevated Sutterella and Clostridium perfringens alongside reduced Blautia, Ruminococcaceae, and Turicibacter (14). Others reported decreases in bacterial groups within the phyla Firmicutes and Bacteroidetes, increases within Proteobacteria, and reduction in Lachnospiraceae and Clostridium coccoides subgroups (15). Despite these disparities across studies, they highlight that a reduction of Clostridium clusters XIVa and IV are associated with IBD in dogs (13–16). These results underscore the pivotal role of these bacterial clusters, which are known short-chain fatty acid (SCFA) producers, in maintaining gastrointestinal health (15).
A major challenge in interpreting results of microbiome data, which has recently been highlighted, is the lack of standardization and high inter-assay variability of next-generation sequencing (NGS) methods that are commonly used in microbiome studies (17, 18). This leads to lack of reproducibility and contributes to the variation reported across different studies when evaluating dietary or health effects. For example, even when the same samples are analyzed, the choices of sample preparation, sequencing protocols, and bioinformatics (i.e., databases, statistical tests) leads to significantly different reported results and interpretations between laboratories (17, 18). Next-generation sequencing data shows high assay variability (19), and due to the compositional data of results (i.e., data is reported as relative percentages), analytical-induced changes in some taxa per definition leads to changes in all other taxa due to method variation rather than because of biological variation (20). Furthermore, some important bacterial taxa (i.e., Clostridium hiranonis) remain undetected by NGS, while targeted quantitative PCR is able to detect these taxa with higher analytical sensitivity and reproducibility (21–23).
A dysbiosis index (DI) was developed by employing qPCR analysis of some core bacterial taxa, particularly in the context of healthy dogs vs. those with chronic enteropathies (24). The DI is used with clinical outcomes to aid in disease diagnosis and management, dividing samples into four groups. A DI <0 and with all targeted taxa within the reference interval are considered normal, a DI <0 but with any of the targeted taxa outside the reference interval are defined as having a minor shift in the microbiome, a DI between 0 and 2 is defined as a mild to moderate microbiome shift, and a DI >2 is considered to be significant dysbiosis. Development of the DI was largely based on samples from dogs consuming extruded kibble diets, as it is the most popular diet format among United States dog owners (25, 26). As stated above, the dietary composition (including ingredients and nutrients) and processing methods have an impact on nutrient digestibility and the fecal microbiome (6–9). Given the impact that diet has on the microbiome, it is important to evaluate whether the DI guidelines are appropriate for dogs fed non-kibble diets.
Furthermore, evaluating dietary induced changes in core bacterial taxa in dogs using the same reproducible method by quantitative PCR would allow better comparison across future studies (23, 27). Therefore, the primary aim of the current study was to compare the fecal core bacteria and DI of dogs fed commercially available fresh diets versus those fed extruded kibble diets. A secondary aim was to compare these core bacteria and DI of dogs fed fresh animal protein-based diets (FAP), fresh plant protein-based diets (FPB), and kibble animal protein-based diets (KAP).
2 Materials and methods
All procedures were approved by the University of Illinois Institutional Animal Care and Use Committee prior to experimentation and were performed in accordance with the U. S. Public Health Service Policy on Humane Care and Use of Laboratory Animals.
2.1 Experimental design
Fresh fecal samples from 37 adult spayed female Beagle dogs from previous studies (8, 9, 28) were used for the analyses. All dogs were adult spayed females in a healthy condition (between 3–7 years old) and were within a healthy body weight range at the time of sample collection. To be included in this experiment, dogs had not been administered any medications (antibiotics, antacids, anti-inflammatory medications, or corticosteroids) that could potentially impact their gut microbiota for a minimum of 4 weeks prior to and during the course of the study. Dogs were housed individually in pens (1.22 m wide × 1.85 m long) in a temperature-controlled room under a 12 h light: 12 h dark cycle at the University of Illinois Urbana-Champaign. As per the University of Illinois IACUC requirements, all dogs received a minimum of two socialization sessions per week with two trained personnel. Additionally, they were bathed every other week (every 14 days) and had continuous access to at least one enrichment toy in their run at all times. Dogs had free access to fresh water and were fed to maintain body weight throughout the studies. Dogs were fed once (28) or twice (8, 9) daily. Before collecting fresh fecal samples, dogs were acclimated to the diet for a minimum of 7 days.
2.2 Diets
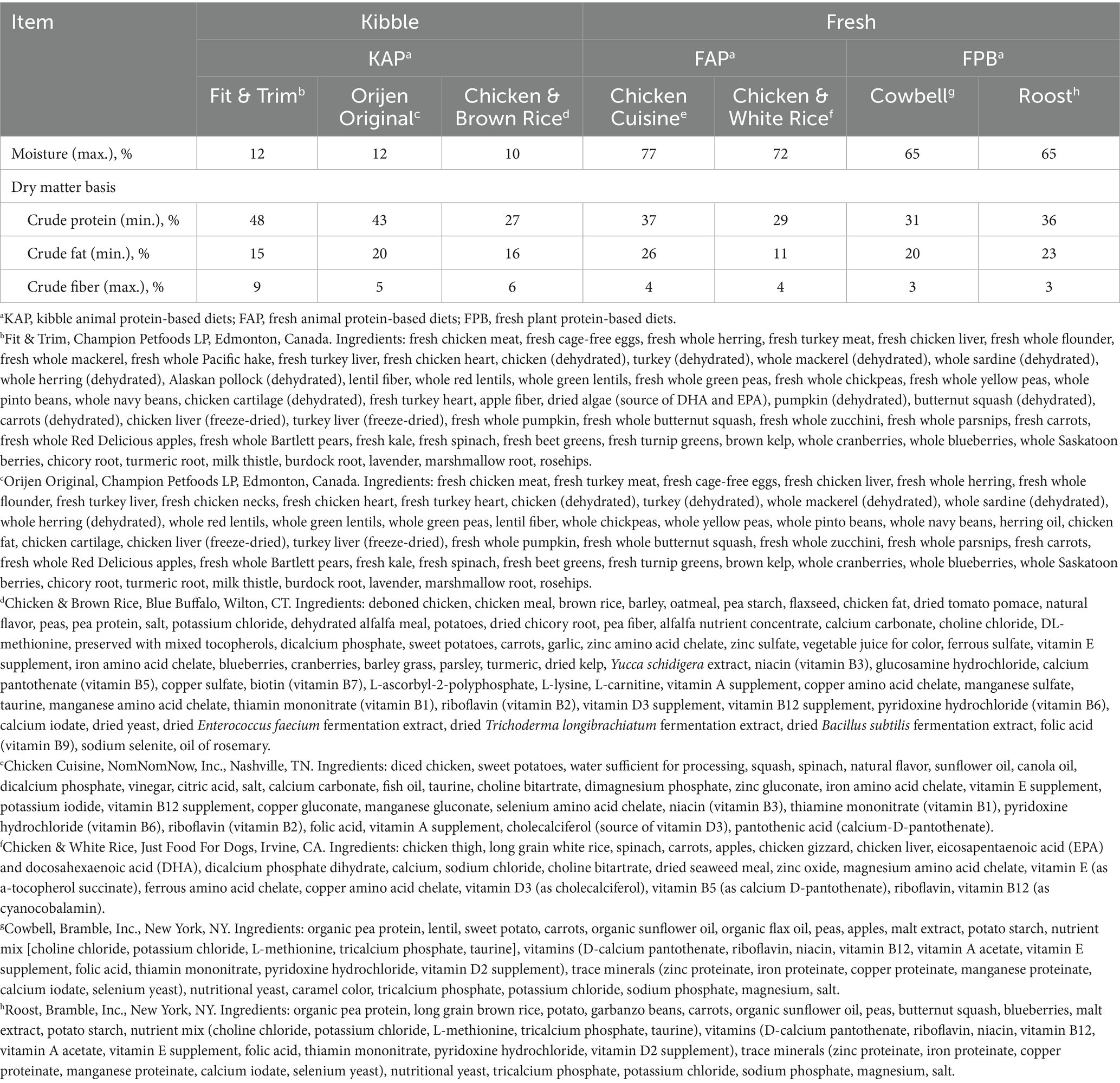
All diets tested were commercially available and formulated to meet Association of American Feed Control Officials (63) nutrient profiles for adult dogs at maintenance. Test diets included those listed below (Table 1).
2.2.1 KAP diets (n = 45 samples)
• Fit & Trim, Champion Petfoods LP, Edmonton, Canada.
• Orijen Original, Champion Petfoods LP, Edmonton, Canada.
• Chicken & Brown Rice, Blue Buffalo, Wilton, CT.
2.2.2 FAP diets (n = 34 samples)
• Chicken & White Rice, Just Food For Dogs, Irvine, CA.
• Chicken Cuisine, NomNomNow, Inc., Nashville, TN.
2.2.3 FPB diets (n = 22 samples)
• Cowbell, Bramble, Inc., New York, NY.
• Roost, Bramble, Inc., New York, NY.
2.3 Fecal sample collection
One fresh fecal sample from each dog was collected within 15 min of defecation and immediately transferred to sterile cryogenic vials (Nalgene, Rochester, NY). Cryovials were stored on dry ice until being transferred to a −80°C freezer. At a later date, samples were placed in dry ice and shipped to the Gastrointestinal Laboratory at Texas A&M University for analysis.
2.4 Quantitative PCR and dysbiosis index
DNA was extracted from an aliquot of 100–120 mg fecal sample using a bead-beating method with a MoBio Power soil DNA isolation kit. qPCR analysis of core bacterial taxa (23, 27) was performed with specific primers targeting Faecalibacterium, Fusobacterium, Blautia, universal bacteria, Turicibacter, Escherichia coli, Clostridium hiranonis, and Streptococcus as described in (24). In addition to the bacterial groups included in the DI calculation, Bacteroides, Bifidobacterium, Clostridium perfringens, Collinsella, Lactobacillus, Prevotella copri, and Ruminococcus gnavus were also quantified by qPCR as described before (23). Specific primers and PCR conditions are listed in Supplementary File 1. Briefly, the conditions for qPCR were as follows: initial denaturation at 98°C for 2 min, then 40 cycles with denaturation at 98°C for 3 s, and annealing for 3 s. Melt curve analysis was performed to validate the specific generation of the qPCR product using these conditions: 95°C for 1 min, 55°C for 1 min, and increasing incremental steps of 0.5°C for 80 cycles for 5 s each. Each reaction was run in duplicate. The qPCR data were expressed as the log amount of DNA (fg) for each particular bacterial group/10 ng of isolated total DNA as reported previously (14, 29). The degree of dysbiosis is represented as a single numerical value that measures the closeness of a taxa compared with the mean abundances derived from healthy and diseased populations and is calculated by an Euclidean distance model, as detailed previously (24). The reference intervals for each taxa and the dysbiosis index were based on the study conducted by AlShawaqfeh et al. (24).
2.5 Statistical analyses
The data underwent analysis through R (version 4.3.1, R Core Team). Given the lack of normality in the dataset, pairwise comparisons between the groups were conducted using Wilcoxon tests with Bonferroni correction for multiple testing from the ggpubr package (version 0.6.0). Correlation tests between nutrient concentrations, bacterial abundance, and the DI were performed using Spearman’s rank correlation from the stats package (version 4.3.3). Because the chemical composition of the NomNomNow diet was not analyzed, samples from that diet were excluded from the correlation analyses. Statistical significance was determined at a threshold of p < 0.05. Plot graphics were generated using ggplot2 package (version 3.5.1).
3 Results
Abundances of fecal Fusobacterium, C. hiranonis, Streptococcus, and E. coli (log DNA/g feces) and the DI were statistically impacted by diet type, although the majority of samples remained in the reported reference intervals (Figure 1). Fecal abundances of Streptococcus (p = 0.0023) and E. coli (p < 0.0001) were higher in dogs fed fresh diets than those fed kibble diets. In contrast, fecal abundances of Fusobacterium (p = 0.0036) and C. hiranonis (p = 0.0380) were higher in dogs fed kibble diets than those fed fresh diets. The DI was greater (p < 0.0001) in dogs fed fresh diets than those fed kibble diets. Despite the large differences in fecal microbiota, all mean DI were <0. A few individual dogs fed fresh diets had a DI >0 (5 dogs >0 and 1 dog >2).
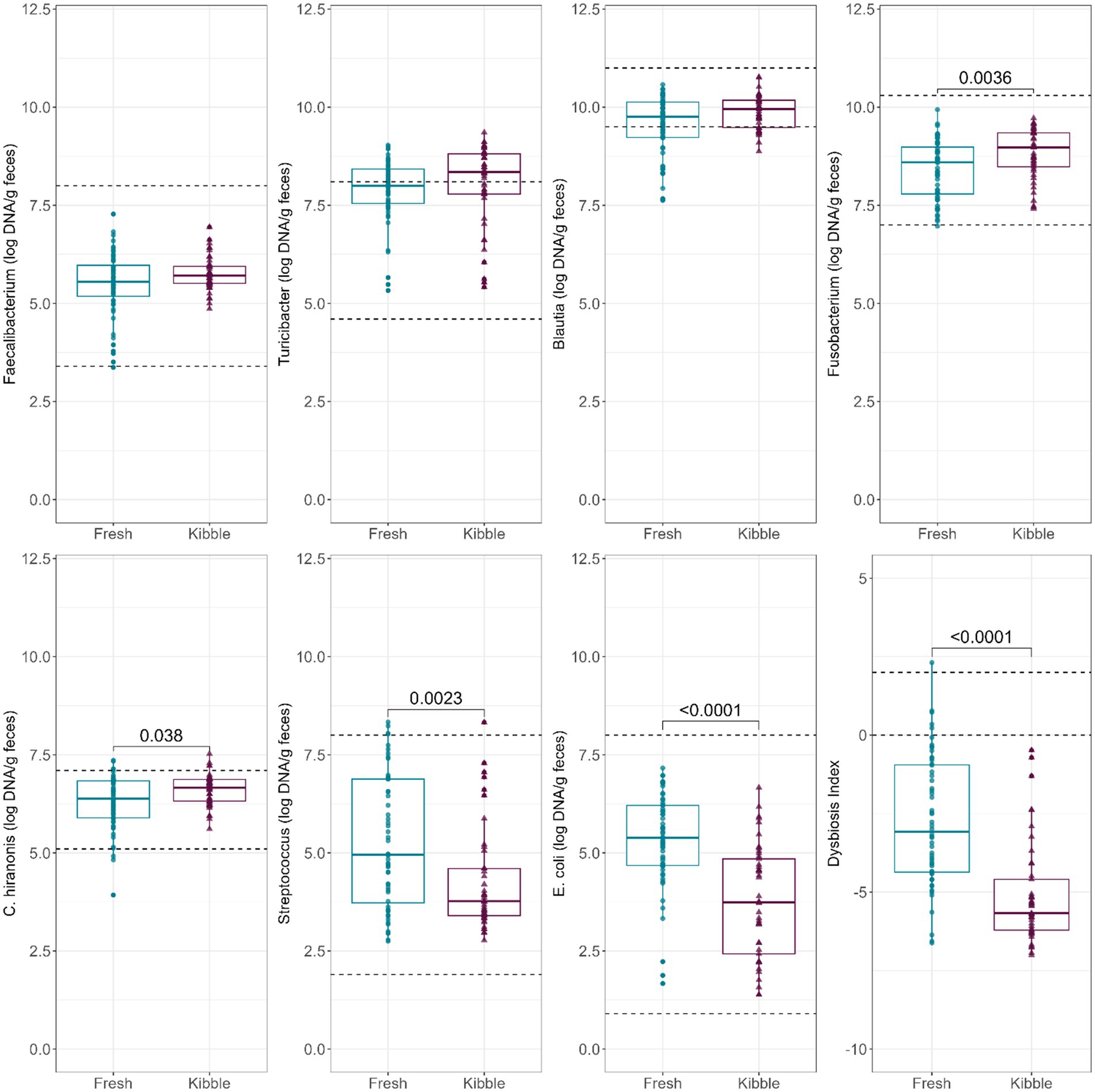
Figure 1. Abundances of fecal Faecalibacterium, Turicibacter, Blautia, Fusobacterium, C. hiranonis, Streptococcus, and E. coli (log DNA/g feces) and DI of dogs fed fresh or kibble diets. Horizontal lines are the normal reference ranges for each bacterial taxa. For DI, the horizontal lines indicate the thresholds for mild and severe dysbiosis.
Abundances of fecal Bacteroides and C. perfringens were also impacted by diet type (Figure 2). Fecal Bacteroides abundance was higher (p = 0.0180) in dogs fed kibble diets than those fed fresh diets. Fecal C. perfringens abundance was higher (p < 0.0001) in dogs fed fresh diets than those fed kibble diets.
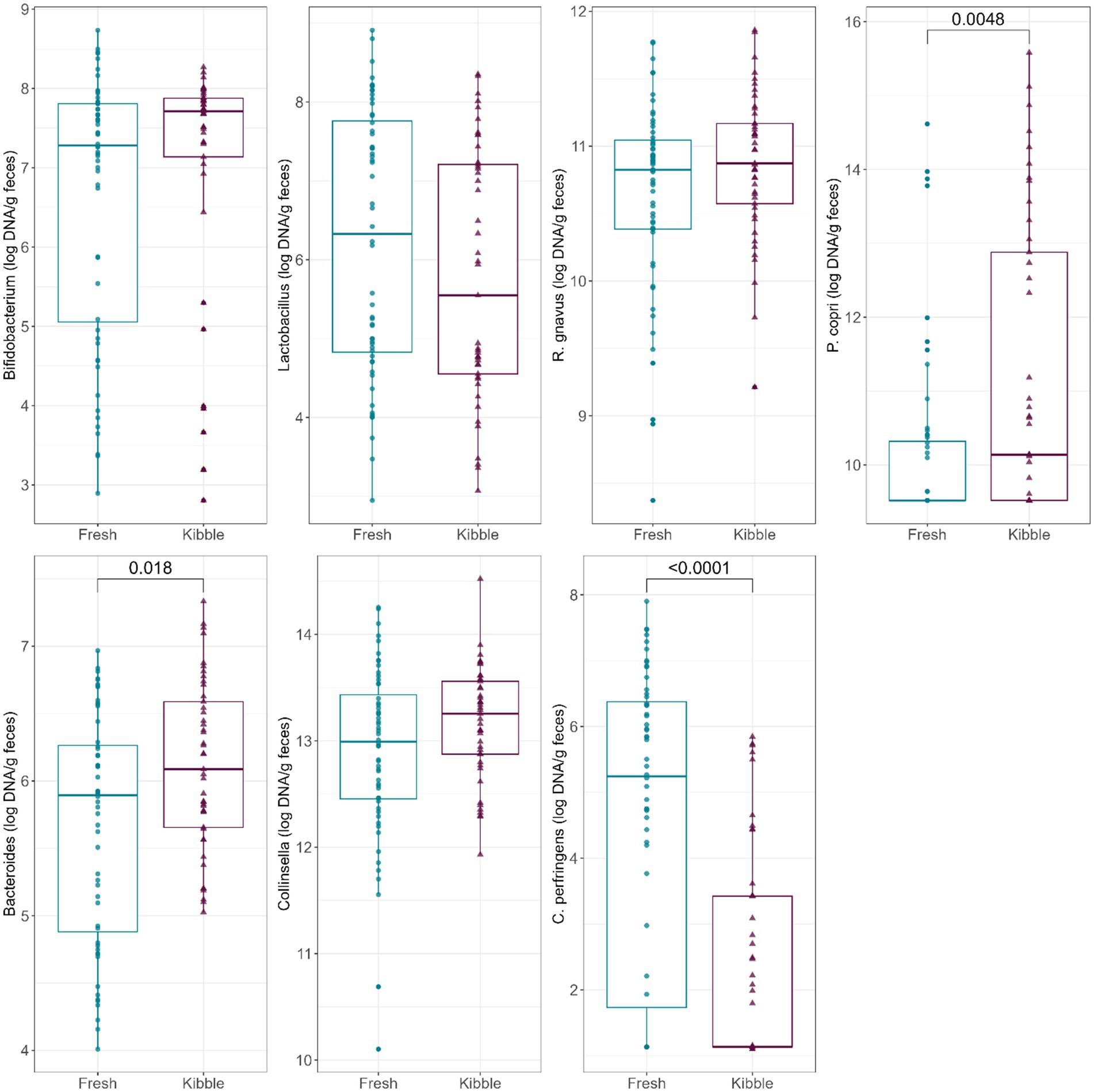
Figure 2. Abundances of fecal Bifidobacterium, Lactobacillus, R. gnavus, P. copri, Bacteroides, Collinsella, and C. perfringens (log DNA/g feces) of dogs fed fresh or kibble diets.
Abundances of fecal Faecalibacterium, Fusobacterium, C. hiranonis, Streptococcus, and E. coli (log DNA/g feces) and the DI were impacted by protein source and diet type (Figure 3). Fecal abundances of Faecalibacterium (p = 0.015) and Fusobacterium (p = 0.0010) were higher in dogs fed KAP diets than those fed FAP diets. Fecal abundance of C. hiranonis was higher in dogs fed FAP diets (p = 0.006) and KAP diets (p < 0.0001) than dogs fed FPB diets. Fecal abundance of Streptococcus was higher in dogs fed FAP diets (p = 0.012) and FPB diets (p = 0.009) than dogs fed KAP diets. Likewise, fecal abundance of E. coli was higher in dogs fed FAP diets (p < 0.0001) and FPB diets (p = 0.0002) than those fed than those fed KAP diets. The DI was greater (p < 0.0001) in dogs fed the FAP and FPB diets than those fed the KAP diets.
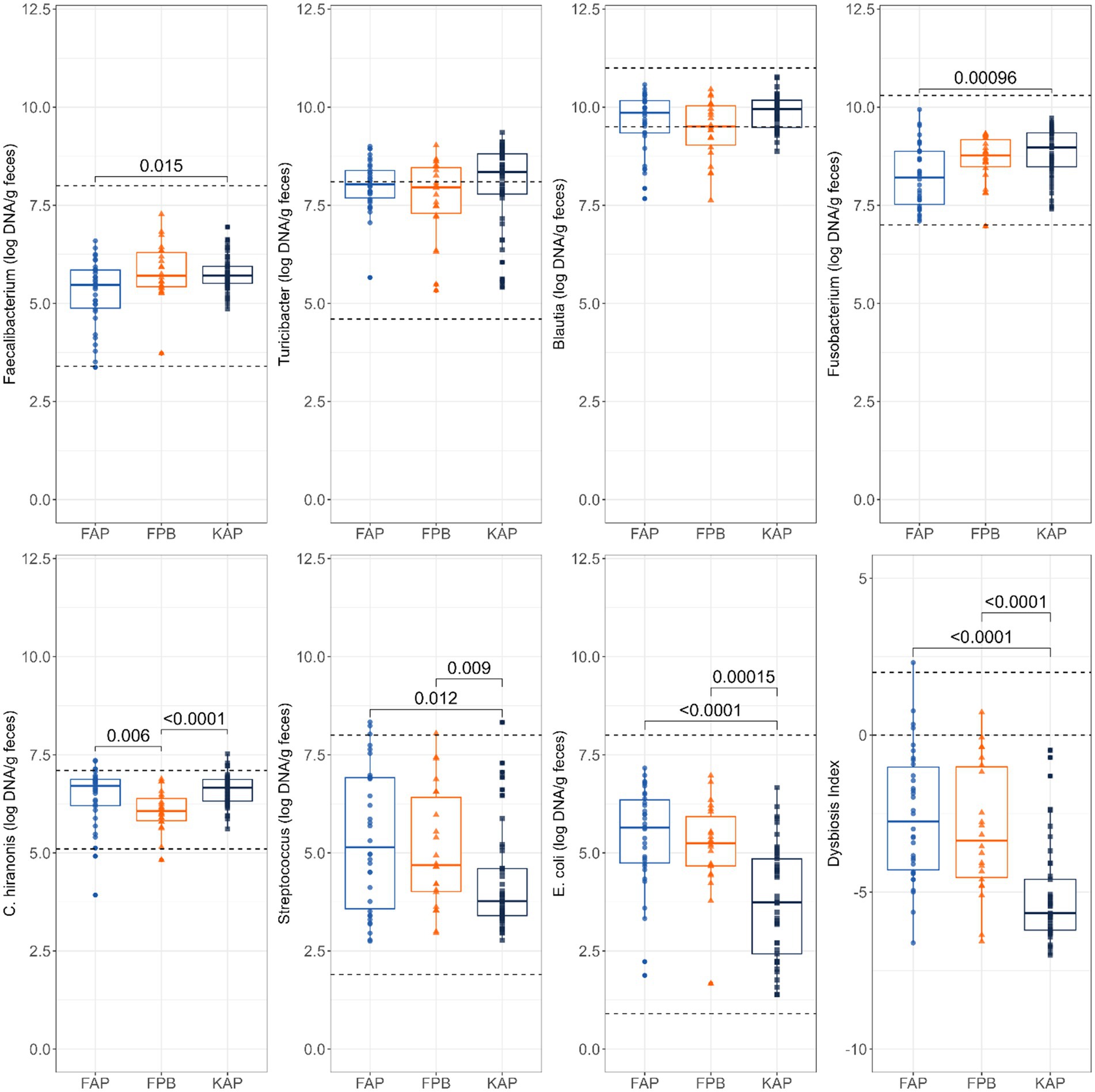
Figure 3. Abundances of fecal Faecalibacterium, Turicibacter, Blautia, Fusobacterium, C. hiranonis, Streptococcus, and E. coli (log DNA/g feces) and DI of dogs fed fresh animal protein-based (FAP), fresh plant protein-based (FPB), and kibble animal protein-based (KAP) diets. Horizontal lines are the normal reference ranges for each bacterial taxa. For DI, the horizontal lines indicate the thresholds for mild and severe dysbiosis.
Abundances of fecal Bacteroides, Bifidobacterium, and C. perfringens were also impacted by protein source and diet type (Figure 4). Fecal abundance of Bifidobacterium was lower in dogs fed FAP diets than those fed FPB diets (p = 0.003) and KAP diets (p = 0.008). Fecal abundance of Bacteroides had a similar pattern, being lower (p < 0.0001) in dogs fed FAP diets than those fed FPB and KAP diets. In contrast, fecal C. perfringens abundance was higher (p < 0.0001) in dogs fed FAP diets than those fed FPB and KAP diets.
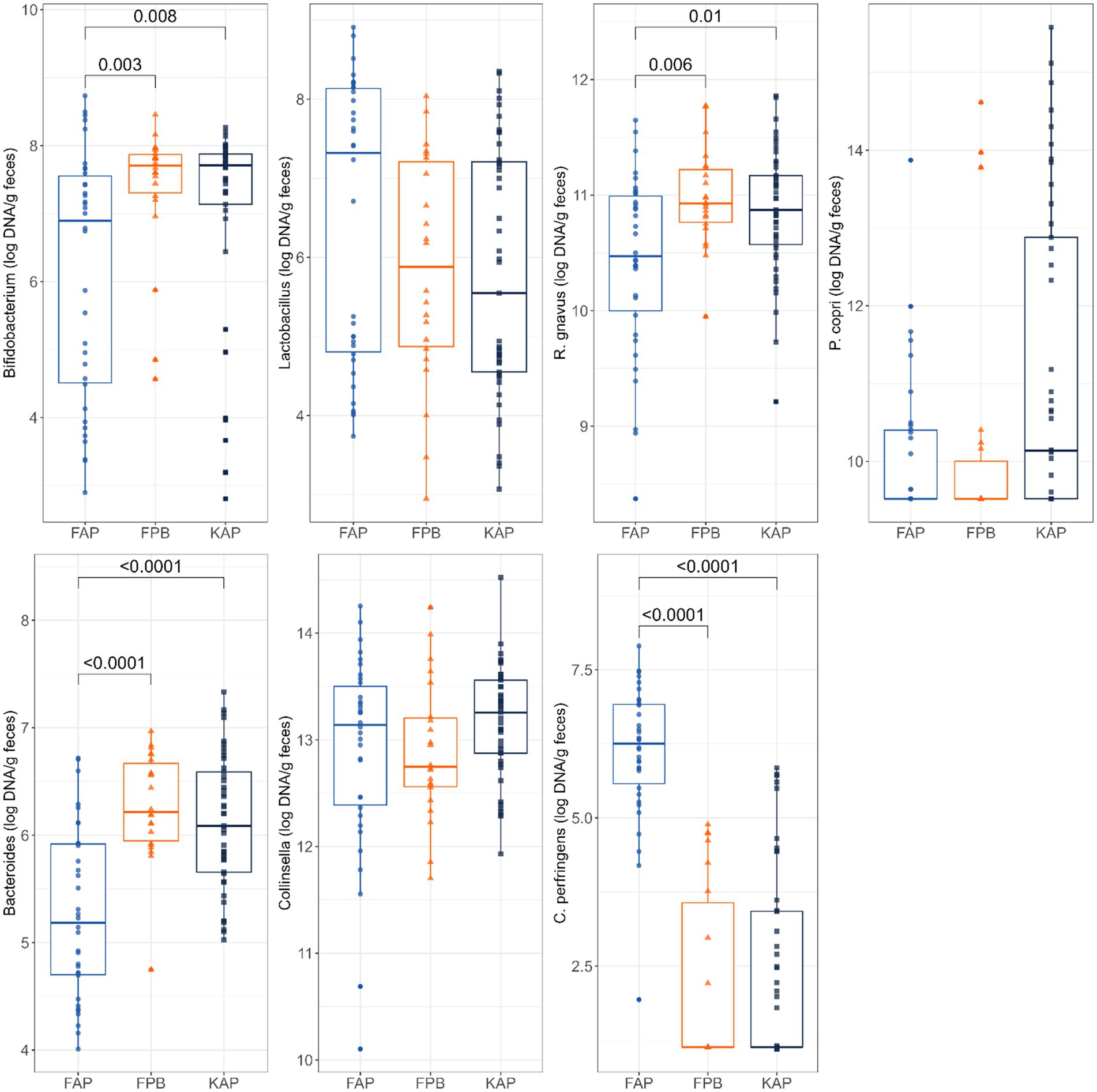
Figure 4. Abundances of fecal Bifidobacterium, Lactobacillus, R. gnavus, P. copri, Bacteroides, Collinsella, and C. perfringens (log DNA/g feces) of dogs fed fresh animal protein-based (FAP), fresh plant protein-based (FPB), and kibble animal protein-based (KAP) diets.
The Spearman correlation coefficients indicated weak positive correlations between dietary total dietary fiber (TDF) concentration and the abundances of fecal Blautia, C. hiranonis, and Bacteroides (R between 0.2 and 0.39), whereas fecal Fusobacterium had a moderate positive correlation (R between 0.4 and 0.59) (Figure 5). Additionally, dietary TDF concentration had low negative correlations with fecal Streptococcus, E. coli, and C. perfringens abundances, along with moderate negative correlations with fecal Lactobacillus and the DI (Figure 6). For dietary CP concentration, weak positive correlations were observed with Fusobacterium abundance (Figure 7). Dietary CP concentration had low negative correlations with fecal Faecalibacterium, Turicibacter, Streptococcus, and Collinsella abundances, as well as moderate negative correlations with fecal R. gnavus abundance (Figure 8). Dietary acid-hydrolyzed fat (AHF) concentration had weak positive correlations with fecal Faecalibacterium, Fusobacterium, and R. gnavus abundances, but a moderate positive correlation with fecal Bacteroides abundance (Figure 9). Finally, dietary AHF concentration had weak negative correlations with fecal C. hiranonis, P. copri, and Lactobacillus abundances, and a moderate correlation with C. perfringens abundance (Figure 10).
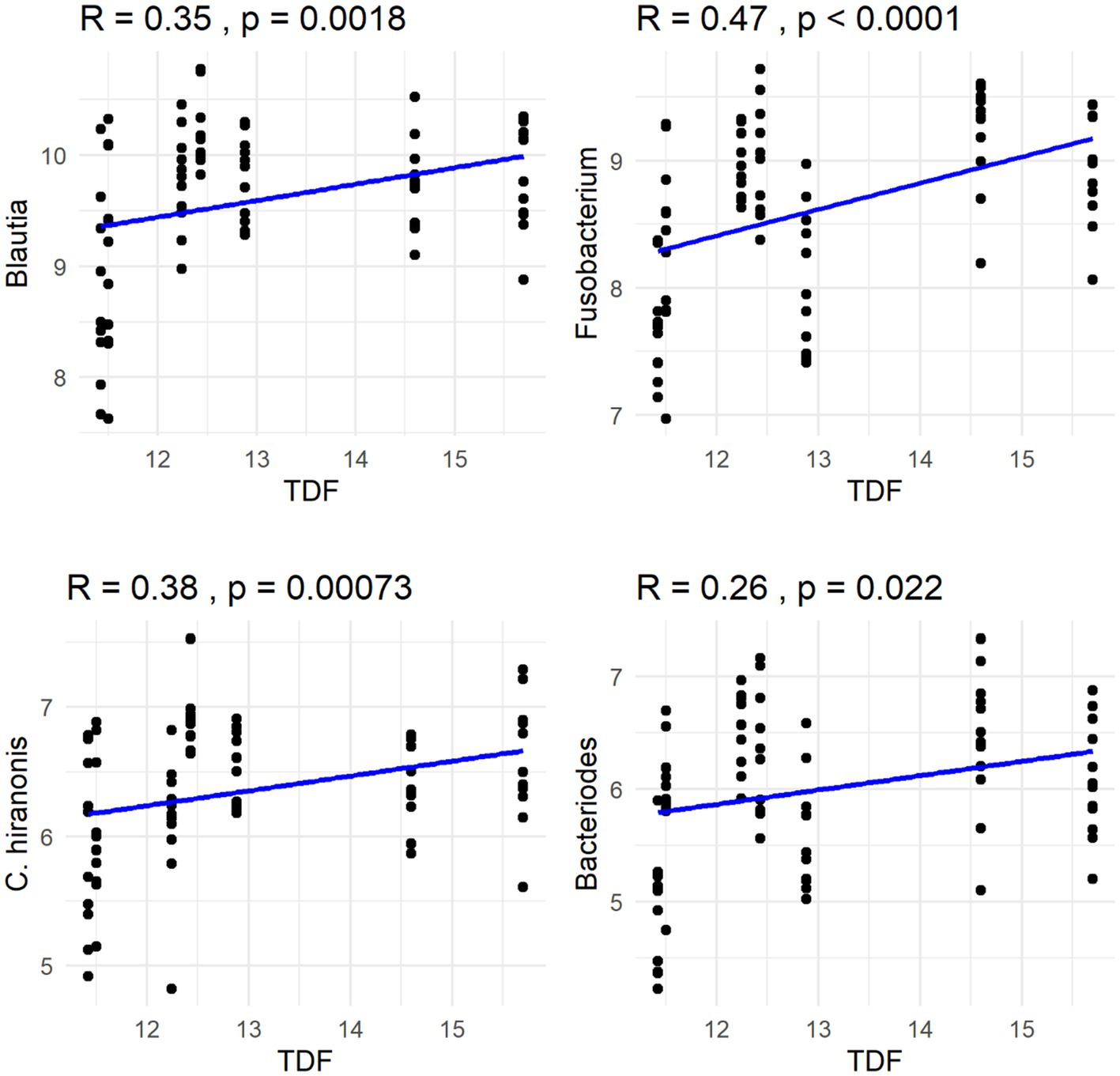
Figure 5. Dietary total dietary fiber (TDF) concentrations were positively correlated with abundances of fecal Blautia, Fusobacterium, C. hiranonis, and Bacteroides (log DNA/g feces).
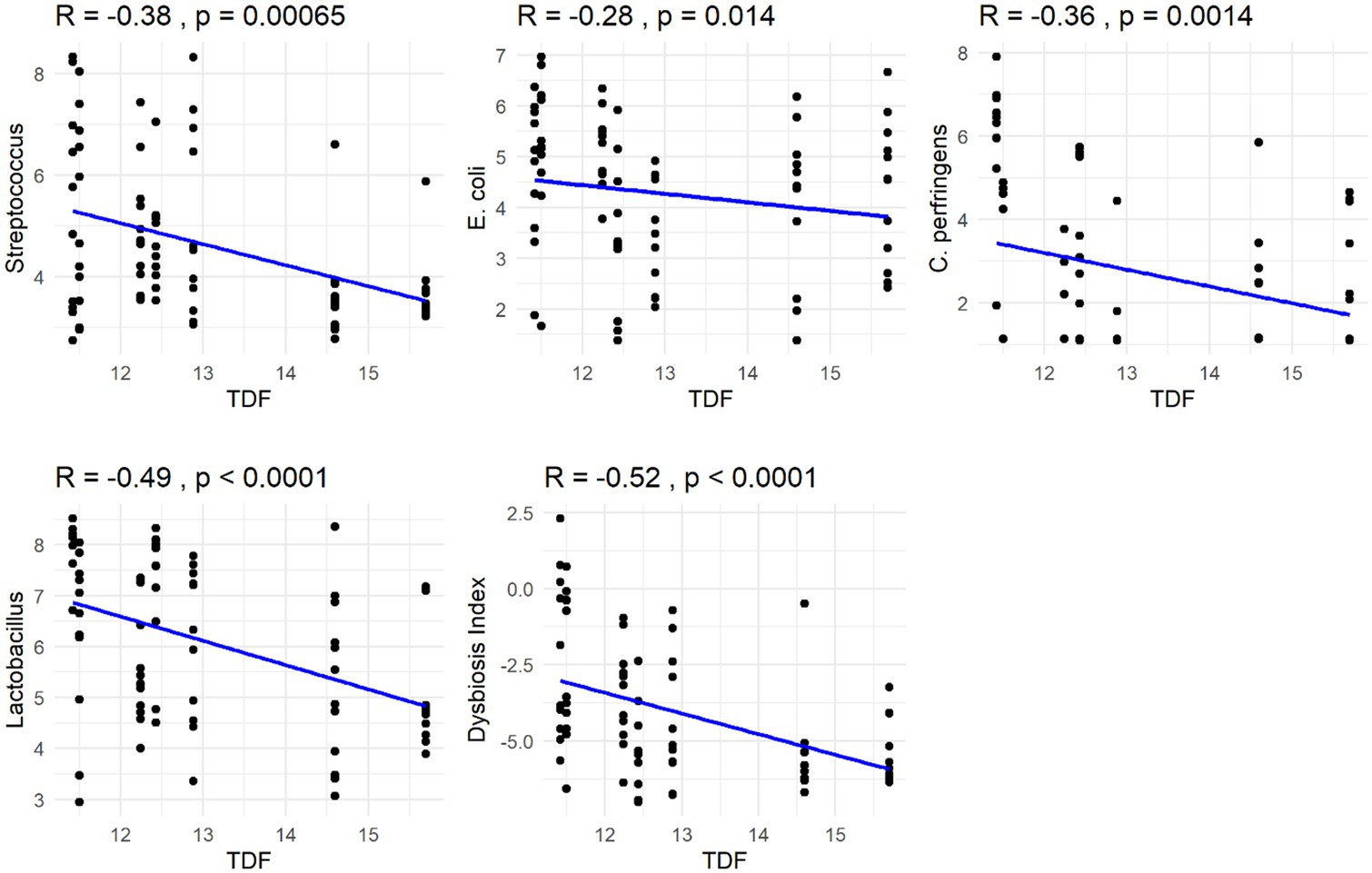
Figure 6. Dietary total dietary fiber (TDF) concentrations were negatively correlated with the abundances of fecal Streptococcus, E. coli, C. perfringens, and Lactobacillus (log DNA/g feces), and the dysbiosis index.
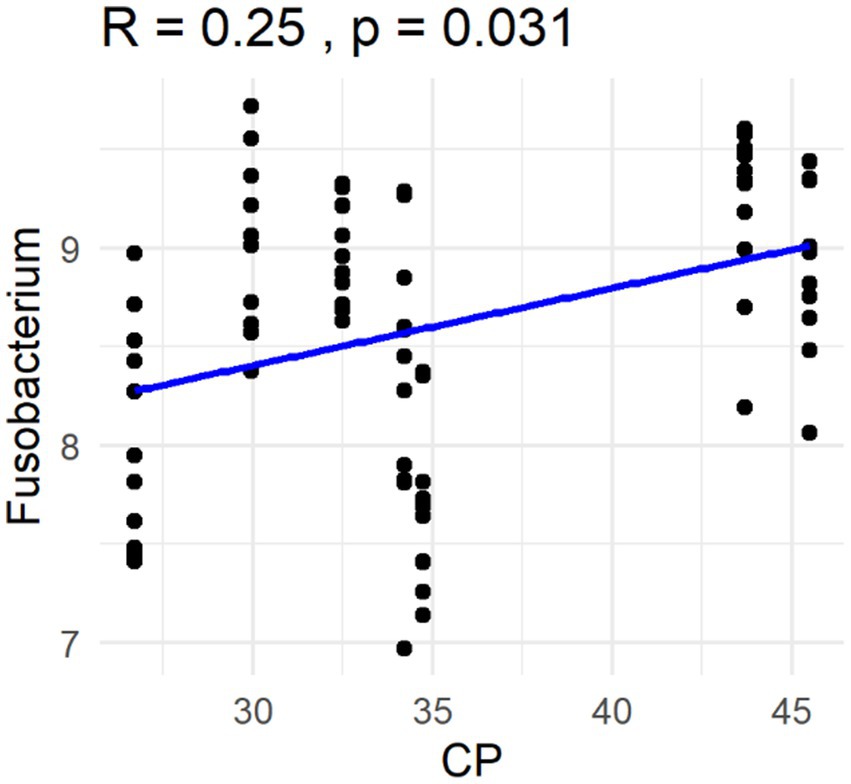
Figure 7. Dietary crude protein (CP) concentrations were positively correlated with the abundance of fecal Fusobacterium (log DNA/g feces) and the dysbiosis index.
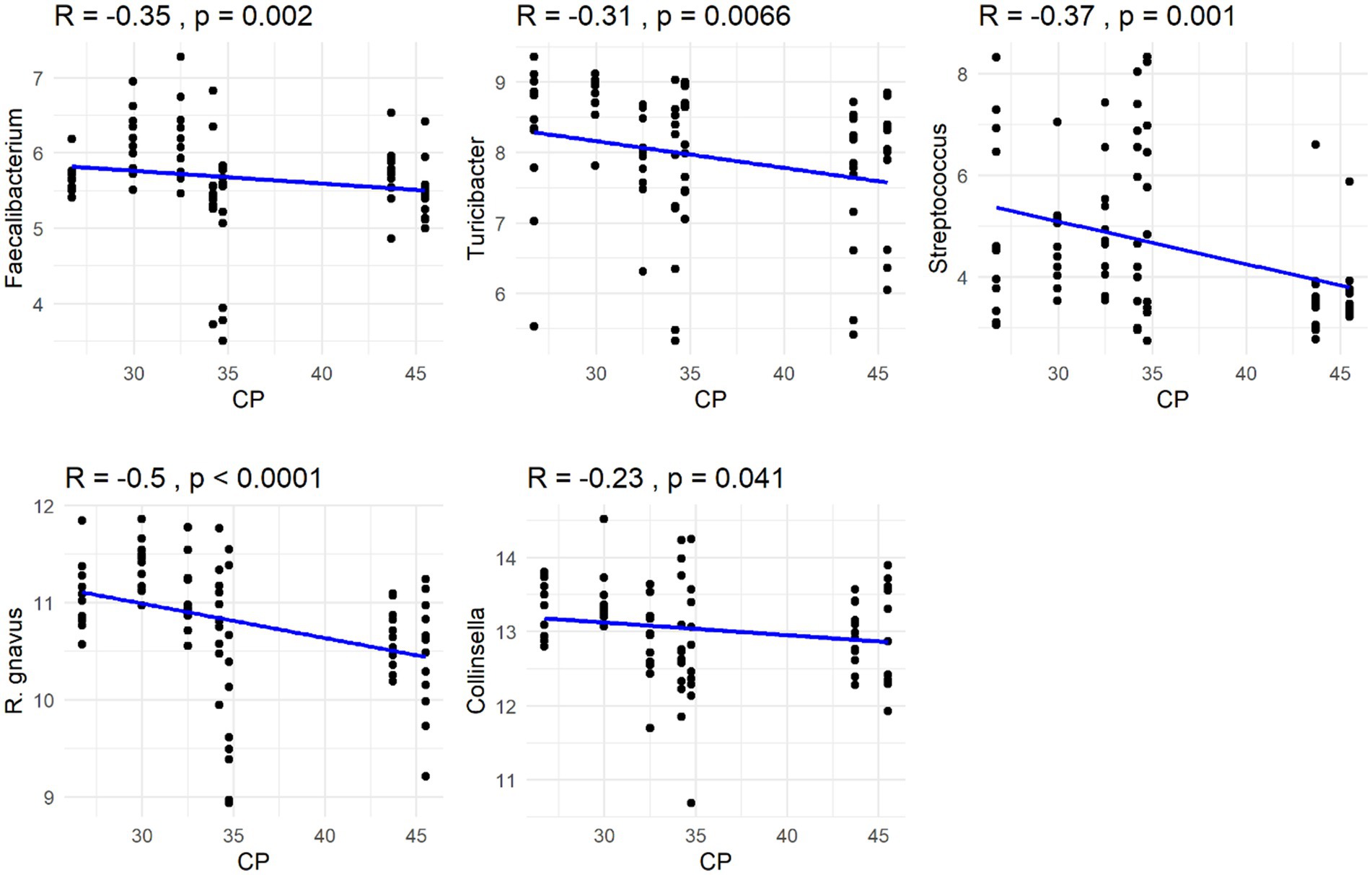
Figure 8. Dietary crude protein (CP) concentrations were negatively correlated with the abundances of fecal Faecalibacterium, Turicibacter, Streptococcus, R. gnavus, and Collinsella (log DNA/g feces).
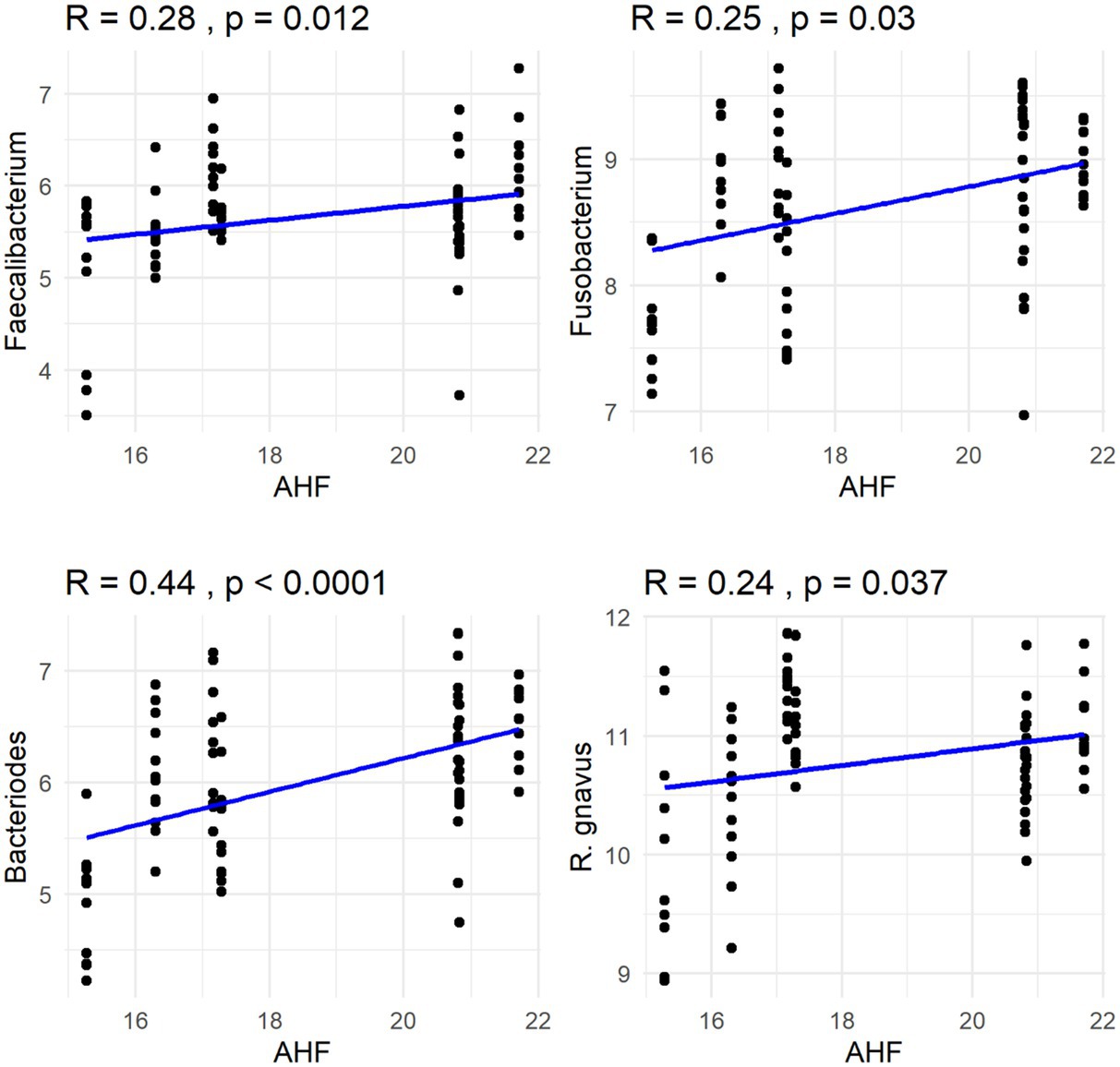
Figure 9. Dietary acid-hydrolyzed fat (AHF) concentrations were positively correlated with abundances of fecal Faecalibacterium, Fusobacterium, Bacteroides, and R. gnavus (log DNA/g feces).
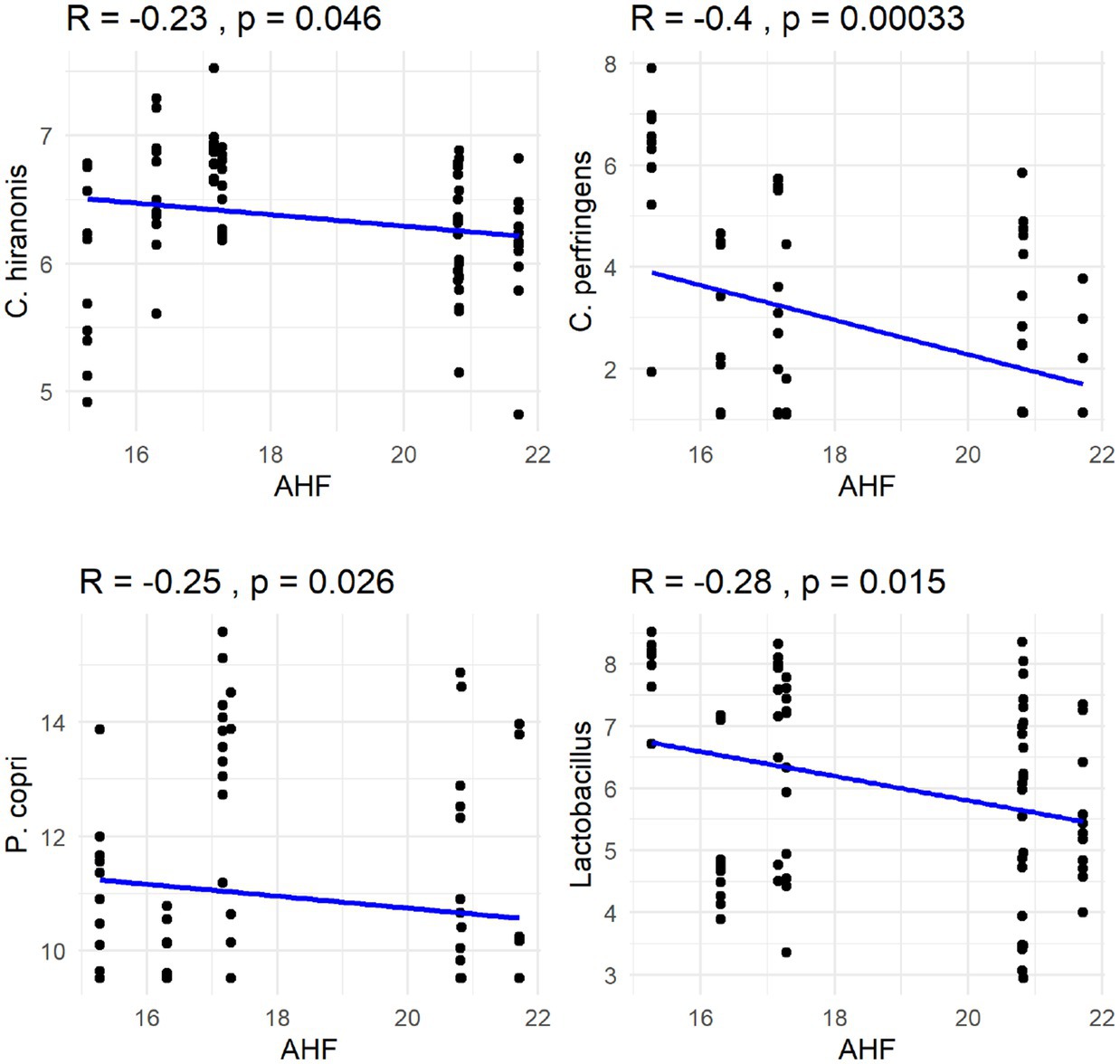
Figure 10. Dietary acid-hydrolyzed fat (AHF) concentrations were negatively correlated with abundances of fecal C. hiranonis, C. perfringens, P. copri, and Lactobacillus (log DNA/g feces).
4 Discussion
The DI was trained against the microbiota of dogs with chronic enteropathies, but has also been shown to increase in antibiotic-induced dysbiosis and to correlate well with overall shifts in the canine microbiome across different disease states (23, 30). The major bacterial taxa contributing to intestinal dysbiosis in dogs include E. coli (increased), Streptococcus (increased), and Faecalibacterium (decreased) (24, 31–34). Multiple studies have used the DI as a marker for assessing gastrointestinal health in dogs (24, 35–38). Nevertheless, the impact of diet on the canine microbiome questions the validity of the DI in dogs consuming varied diets. To our knowledge, this is the first study to use qPCR-based DI profiling to compare both animal- and plant-based fresh protein diets with kibble-based animal protein diets in healthy animals.
Recent studies have evaluated the impact that fresh diets have on the canine microbiome, but continued research is necessary to fully comprehend their influence on gastrointestinal and overall host health. One study analyzed fecal samples from 27 dogs fed a home-made high protein and high fat bones and raw food (BARF) and 19 dogs fed commercially available kibble foods using qPCR and the DI (39). Abundances of fecal E. coli, Streptococcus, and C. perfringens and the DI were greater, while abundance of fecal Faecalibacterium was lower, in dogs fed BARF than those fed kibble (39). The findings from that study and the current study demonstrate that raw or fresh diets, whether derived from animal- or plant-based proteins, lead to an elevation in fecal E. coli and Streptococcus abundances. Fresh diets based on animal proteins also resulted in greater fecal C. perfringens abundance but lower Faecalibacterium abundance. Faecalibacterium are SCFA producers, largely use dietary fibers as substrates (29, 40), and are considered to be beneficial to gastrointestinal health, while C. perfringens, E. coli and Streptococcus are considered to be potential pathogenic bacteria (41–43). All animals included in these studies were healthy, however, so the clinical relevance of these shifts is unknown.
High protein intake often elevates fecal protein catabolites (e.g., phenols, indoles, branched-chain fatty acids), reduces SCFA concentrations, and shifts gut microbiota towards potentially harmful genera like Clostridium, E. coli, and Streptococcus, while reducing beneficial bacteria such as Bifidobacterium and Faecalibacterium (44–46). Such changes may differ based on the source of protein and other nutrients that are also present (e.g., non-digestible oligosaccharides). Fecal Bifidobacterium abundance was lower in dogs consuming an animal protein kibble diet than those consuming a fresh plant-based protein diet tested. The opposite was true of fecal Fusobacterium abundance, which was lower in dogs consuming an animal protein kibble diet than those consuming a fresh plant-based protein diet (9). Similarly, fecal Bifidobacterium abundance was higher in animals consuming fresh animal protein diets than those consuming kibble animal protein diets, while fecal Bacteroides abundance was lower in animals consuming fresh animal protein diets than those consuming kibble animal protein diets (8). The results from the current and prior studies suggest that formulas based on animal proteins lead to a reduction in fecal Bifidobacterium, Bacteroides, and Fusobacterium abundances, which are all known SCFA producers (47–50).
The impact of dietary macronutrient content on the gastrointestinal health of animals is intriguing, yet further studies are needed to understand the optimal ratios and their effects. For instance, one study indicated that fecal microbial communities were not different between dogs consuming a plant-based kibble diet or animal protein kibble diet when macronutrient profiles were similar (51). Those results suggest that the significance of ingredients appears to be outweighed by the overall macronutrient content. In healthy adult dogs, consumption of four different types of kibble-based prescription diets – a weight-loss diet (33% protein, 10% fat, 31% fiber), a low-fat diet (24% protein, 8% fat, 10% fiber), a renal diet (15% protein, 20% fat, 8% fiber), and an allergenic diet (19% protein, 18% fat, 6% fiber, all on a dry matter basis)—was associated with changes in microbiome composition (52). Specifically, fecal Streptococcus abundances were lower in animals consuming the weight-loss (high fiber and protein) and low-fat diets than those consuming the anallergenic diet, while fecal Faecalibacterium abundances were greater in dogs consuming those diets. Additionally, fecal Fusobacterium abundance was greater in dogs consuming the weight-loss diet than those consuming the anallergenic diet (52).
In the present study, dietary TDF had a positive correlation with fecal Fusobacterium abundance and a negative correlation with fecal Streptococcus abundance, which was consistent with previous findings. In a study comparing a low-protein diet (28% CP, 16% fat, 9% TDF in DMB) and a high-protein diet (53% CP, 15% fat, 13% TDF in DMB), fecal C. hiranonis, Streptococcus, C. perfringens, and R. gnavus abundances were greater in dogs fed the high-protein diet, while fecal Fusobacterium, Turicibacter, and P. copri abundances were greater in dogs fed the low-protein diet (53). Similar to the previous study, fecal C. hiranonis abundance was positively correlated with dietary TDF concentration, while fecal Turicibacter abundance was negatively correlated with dietary CP concentration in the current study. Faecalibacterium is a SCFA producer and use a variety of fibers as subtrates (29, 40); C. hiranonis is capable of dehydroxylating primary bile acids into secondary bile acids, with high-fat diets inducing their proliferation in humans (54, 55). Similar dietary shifts are known to occur with dietary CP, with Fusobacterium, a taxa known to ferment amino acids, being positively correlated (56, 57). In the present study, fecal Bacteroides abundance was positively correlated with dietary TDF and AHF concentrations. Because Bacteroides possess a variety of polysaccharide and protein degradative capabilities (58–61), it is able to adapt to diets differing in nutrient content.
While dietary protein concentration is important, so is the source of protein (e.g., animal or plant). In this study, dogs consuming plant-based fresh and kibble diets had higher fecal Bifidobacterium, R. gnavus, and Bacteroides abundances and lower fecal C. perfringens abundance than those consuming a fresh animal-based diet. R. gnavus ferments non-digestible carbohydrates to produce acetate, formate, ethanol, propanol, and propionate (62). As noted earlier, Bacteroides and Bifidobacterium are also SCFA producers (47–50, 64).
This study had some strengths and limitations. One strength included the use of a dog population that was similar in regard to breed (all beagles), sex (all spayed females) and age (all middle aged). Another strength was the fact that all dogs were housed in an experimental facility on the University of Illinois campus, which allowed for strict control of dietary intake, environmental exposures, exercise allowance, light cycle, and other factors that may affect microbiota populations. One limitation was the sample sizes, which were variable (n = 10–24/diet) across diets and were modest in some cases. Another limitation was the fact that a single fecal sample was collected from dogs consuming a diet, which did not allow evaluation of microbial stability over time. Lastly, healthy dogs were tested in this study so the clinical relevance of fecal microbial differences were unknown. In addition, while diet categories (FAP, FPB, and KAP) were used to group the foods for analysis, we acknowledge that variability exists within each category, and the use of previously collected data meant that not all diets had complete compositional information available. This reflects the real-world diversity of commercially available diets and is an inherent limitation when evaluating broader dietary categories.
In conclusion, this study compared core bacterial taxa by qPCR and the dysbiosis index and identified several fecal bacterial taxa that differed among dogs fed fresh vs. kibble diets and those based on animal- or plant-based ingredients. In general, dogs fed fresh diets had elevated fecal abundances of potentially pathogenic bacteria (e.g., E. coli, Streptococcus, C. perfringens) and dysbiosis index, and lower fecal abundance of SCFA producers (e.g., Bacteroides, Fusobacterium). Dogs consuming fresh diets based on animal protein had greater fecal C. perfringens abundance and lower fecal Bacteroides abundance than those consuming animal kibble diets or plant-based fresh diets. Dietary macronutrient concentrations also affected fecal microbiota, as dietary TDF content was positively correlated with SCFA-producing bacteria (Blautia, Bacteroides, and Fusobacterium), and negatively correlated with potential pathogenic bacteria (Streptococcus, E. coli, and C. perfringens). Dietary protein content was negatively correlated with SCFA-producing bacteria (Faecalibacterium, Turicibacter, R. gnavus, and Collinsella). Dietary fat content was positively correlated with SCFA-producing bacteria (Faecalibacterium, Fusobacterium, R. gnavus, and Bacteroides), and negatively correlated with potentially pathogenic bacteria (C. perfringens). Despite the differences noted in bacterial taxa and dysbiosis across diets, it is important to highlight that average dysbiosis index values remained within the normal range, suggesting that a fresh diet alone does not induce dysbiosis in healthy animals. Further investigations are warranted to fully comprehend the distinct attributes of fresh diet formats and their implications for gastrointestinal health in dogs.
Data availability statement
All relevant data are contained within the article. The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by University of Illinois Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
PO: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Project administration. LR: Writing – review & editing, Investigation. EG: Writing – review & editing, Investigation. JS: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing. KS: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The funding for this project was provided by the USDA National Institute of Food and Agriculture.
Acknowledgments
The author acknowledges Champion Petfoods LP, Blue Buffalo, Just Food For Dogs, NomNomNow, Inc., and Bramble, Inc. for providing the diets used in this study.
Conflict of interest
JS is an employee of the Gastrointestinal Laboratory at Texas A&M University, which offers microbiome testing including the dysbiosis index on a fee-for-service basis.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1572875/full#supplementary-material
Abbreviations
BARF, Bones and raw food; DI, Dysbiosis index; FAP, Fresh animal protein-based diet; FPB, Fresh plant protein-based diet; KAP, Kibble animal protein-based diet; qPCR, Quantitative polymerase chain reaction; SCFA, Short-chain fatty acids.
References
1. Hsu, C, Marx, F, Guldenpfennig, R, Valizadegan, N, and de Godoy, MRC. The effects of hydrolyzed protein on macronutrient digestibility, fecal metabolites and microbiota, oxidative stress and inflammatory biomarkers, and skin and coat quality in adult dogs. J Anim Sci. (2024) 102:skae057. doi: 10.1093/JAS/SKAE057
2. Jha, AR, Shmalberg, J, Tanprasertsuk, J, Perry, LA, Massey, D, and Honaker, RW. Characterization of gut microbiomes of household pets in the United States using a direct-to-consumer approach. PLoS One. (2020) 15:e0227289. doi: 10.1371/JOURNAL.PONE.0227289
3. Liu, Y, Liu, B, Liu, C, Hu, Y, Liu, C, Li, X, et al. Differences in the gut microbiomes of dogs and wolves: roles of antibiotics and starch. BMC Vet Res. (2021) 17:1–8. doi: 10.1186/S12917-021-02815-Y
4. Phillips-Donaldson, D. (2024). US pet food sales: units and volume catching up to dollars. Available online at: https://www.petfoodindustry.com/pet-food-market/market-trends-and-reports/article/15707136/us-pet-food-sales-units-and-volume-catching-up-to-dollars (Accessed 19 May, 2025).
5. Carter, RA, Bauer, JE, Kersey, JH, and Buff, PR. Awareness and evaluation of natural pet food products in the United States. J Am Vet Med Assoc. (2014) 245:1241–8. doi: 10.2460/JAVMA.245.11.1241
6. Algya, KM, Cross, TWL, Leuck, KN, Kastner, ME, Baba, T, Lye, L, et al. Apparent total-tract macronutrient digestibility, serum chemistry, urinalysis, and fecal characteristics, metabolites and microbiota of adult dogs fed extruded, mildly cooked, and raw diets. J Anim Sci. (2018) 96:3670. doi: 10.1093/JAS/SKY235
7. Do, S, Phungviwatnikul, T, De Godoy, MRC, and Swanson, KS. Nutrient digestibility and fecal characteristics, microbiota, and metabolites in dogs fed human-grade foods. J Anim Sci. (2021) 99:skab028. doi: 10.1093/JAS/SKAB028
8. Geary, EL, Oba, PM, Applegate, CC, Clark, LV, Fields, CJ, and Swanson, KS. Effects of a mildly cooked human-grade dog diet on gene expression, skin and coat health measures, and fecal microbiota of healthy adult dogs. J Anim Sci. (2022) 100:skac265. doi: 10.1093/jas/skac265
9. Roberts, LJ, Oba, PM, and Swanson, KS. Apparent total tract macronutrient digestibility of mildly cooked human-grade vegan dog foods and their effects on the blood metabolites and fecal characteristics, microbiota, and metabolites of adult dogs. J Anim Sci. (2023) 101:skad093. doi: 10.1093/jas/skad093
10. Belchik, SE, Oba, PM, Lin, C-Y, and Swanson, KS. Effects of a veterinary gastrointestinal low-fat diet on fecal characteristics, metabolites, and microbiota concentrations of adult dogs treated with metronidazole. J Anim Sci. (2024) 102:skae297. doi: 10.1093/JAS/SKAE297
11. Lin, C-Y, Jha, AR, Oba, PM, Yotis, SM, Shmalberg, J, Honaker, RW, et al. Longitudinal fecal microbiome and metabolite data demonstrate rapid shifts and subsequent stabilization after an abrupt dietary change in healthy adult dogs. Anim Microbiome. (2022) 4:46. doi: 10.1186/s42523-022-00194-9
12. Degruttola, AK, Low, D, Mizoguchi, A, and Mizoguchi, E. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis. (2016) 22:1137–50. doi: 10.1097/MIB.0000000000000750
13. Minamoto, Y, Dhanani, N, Markel, ME, Steiner, JM, and Suchodolski, JS. Prevalence of Clostridium perfringens, Clostridium perfringens enterotoxin and dysbiosis in fecal samples of dogs with diarrhea. Vet Microbiol. (2014) 174:463–73. doi: 10.1016/J.VETMIC.2014.10.005
14. Suchodolski, JS, Dowd, SE, Wilke, V, Steiner, JM, and Jergens, AE. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS One. (2012) 7:e39333. doi: 10.1371/JOURNAL.PONE.0039333
15. Honneffer, JB, Minamoto, Y, and Suchodolski, JS. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J Gastroenterol. (2014) 20:16489–97. doi: 10.3748/wjg.v20.i44.16489
16. Mondo, E, Marliani, G, Accorsi, PA, Cocchi, M, and Di Leone, A. Role of gut microbiota in dog and cat’s health and diseases. Open Vet J. (2019) 9:253–8. doi: 10.4314/ovj.v9i3.10
17. Forry, SP, Servetas, SL, Kralj, JG, Soh, K, Hadjithomas, M, Cano, R, et al. Variability and bias in microbiome metagenomic sequencing: an interlaboratory study comparing experimental protocols. Sci Rep. (2024) 14:9785. doi: 10.1038/S41598-024-57981-4
18. Roume, H, Mondot, S, Saliou, A, Le Fresne-Languille, S, and Doré, J. Multicenter evaluation of gut microbiome profiling by next-generation sequencing reveals major biases in partial-length metabarcoding approach. Sci Rep. (2023) 13:22593. doi: 10.1038/S41598-023-46062-7
19. Hoisington, AJ, Stamper, CE, Ellis, JC, Lowry, CA, and Brenner, LA. Quantifying variation across 16S rRNA gene sequencing runs in human microbiome studies. Appl Microbiol Biotechnol. (2024) 108:367. doi: 10.1007/S00253-024-13198-Z
20. Gloor, GB, Macklaim, JM, Pawlowsky-Glahn, V, and Egozcue, JJ. Microbiome datasets are compositional: and this is not optional. Front Microbiol. (2017) 8:2224. doi: 10.3389/fmicb.2017.02224
21. Li, F, Liu, J, Maldonado-Gómez, MX, Frese, SA, Gänzle, MG, and Walter, J. Highly accurate and sensitive absolute quantification of bacterial strains in human fecal samples. Microbiome. (2024) 12:168. doi: 10.1186/S40168-024-01881-2
22. Sezgin, E, Terlemez, G, Bozkurt, B, Bengi, G, Akpinar, H, and Büyüktorun, İ. Quantitative real-time PCR analysis of bacterial biomarkers enable fast and accurate monitoring in inflammatory bowel disease. PeerJ. (2022) 10:e14217. doi: 10.7717/PEERJ.14217
23. Sung, CH, Pilla, R, Chen, CC, Ishii, PE, Toresson, L, Allenspach-Jorn, K, et al. Correlation between targeted qPCR assays and untargeted DNA shotgun metagenomic sequencing for assessing the fecal microbiota in dogs. Animals. (2023) 13:2597. doi: 10.3390/ani13162597
24. AlShawaqfeh, MK, Wajid, B, Minamoto, Y, Markel, M, Lidbury, JA, Steiner, JM, et al. A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiol Ecol. (2017) 93:136. doi: 10.1093/FEMSEC/FIX136
25. Dinallo, GK, Poplarski, JA, Van Deventer, GM, Eirmann, LA, and Wakshlag, JJ. A survey of feeding, activity, supplement use and energy consumption in North American agility dogs. J Nutr Sci. (2017) 6:e45. doi: 10.1017/JNS.2017.44
26. Laflamme, DP, Abood, SK, Fascetti, AJ, Fleeman, LM, Freeman, LM, Michel, KE, et al. Pet feeding practices of dog and cat owners in the United States and Australia. J Am Vet Med Assoc. (2008) 232:687–94. doi: 10.2460/JAVMA.232.5.687
27. Rojas, CA, Park, B, Scarsella, E, Jospin, G, Entrolezo, Z, Jarett, JK, et al. Species-level characterization of the core microbiome in healthy dogs using full-length 16S rRNA gene sequencing. Front Vet Sci. (2024) 11:1405470. doi: 10.3389/fvets.2024.1405470
28. Oba, PM, Kelly, J, Kostiuk, D, and Swanson, KS. Effects of weight loss and feeding specially formulated diets on the body composition, blood metabolite profiles, voluntary physical activity, and fecal metabolites and microbiota of obese dogs. J Anim Sci. (2023) 101:skad073. doi: 10.1093/jas/skad073
29. Panasevich, MR, Kerr, KR, Dilger, RN, Fahey, GC, Guérin-Deremaux, L, Lynch, GL, et al. Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. Br J Nutr. (2015) 113:125–33. doi: 10.1017/S0007114514003274
30. Li, Q, Larouche-Lebel, É, Loughran, KA, Huh, TP, Suchodolski, JS, and Oyama, MA. Gut dysbiosis and its associations with gut microbiota-derived metabolites in dogs with myxomatous mitral valve disease. mSystems. (2021) 6:e00111-21. doi: 10.1128/mSystems.00111-21
31. Manchester, AC, Hill, S, Sabatino, B, Armentano, R, Carroll, M, Kessler, B, et al. Association between granulomatous colitis in French Bulldogs and invasive Escherichia coli and response to fluoroquinolone antimicrobials. J Vet Intern Med. (2013) 27:56–61. doi: 10.1111/JVIM.12020
32. Minamoto, Y, Otoni, CC, Steelman, SM, Büyükleblebici, O, Steiner, JM, Jergens, AE, et al. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes. (2015) 6:33–47. doi: 10.1080/19490976.2014.997612
33. Suchodolski, JS, Markel, ME, Garcia-Mazcorro, JF, Unterer, S, Heilmann, RM, Dowd, SE, et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One. (2012) 7:e51907. doi: 10.1371/JOURNAL.PONE.0051907
34. Xenoulis, PG, Palculict, B, Allenspach, K, Steiner, JM, Van House, AM, and Suchodolski, JS. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol Ecol. (2008) 66:579–89. doi: 10.1111/J.1574-6941.2008.00556.X
35. Chaitman, J, Ziese, AL, Pilla, R, Minamoto, Y, Blake, AB, Guard, BC, et al. Fecal microbial and metabolic profiles in dogs with acute diarrhea receiving either fecal microbiota transplantation or oral metronidazole. Front Vet Sci. (2020) 7:527840. doi: 10.3389/fvets.2020.00192
36. Giaretta, PR, Rech, RR, Guard, BC, Blake, AB, Blick, AK, Steiner, JM, et al. Comparison of intestinal expression of the apical sodium-dependent bile acid transporter between dogs with and without chronic inflammatory enteropathy. J Vet Intern Med. (2018) 32:1918–26. doi: 10.1111/JVIM.15332
37. Minamoto, Y, Minamoto, T, Isaiah, A, Sattasathuchana, P, Buono, A, Rangachari, VR, et al. Fecal short-chain fatty acid concentrations and dysbiosis in dogs with chronic enteropathy. J Vet Intern Med. (2019) 33:1608–18. doi: 10.1111/JVIM.15520
38. Pilla, R, Gaschen, FP, Barr, JW, Olson, E, Honneffer, J, Guard, BC, et al. Effects of metronidazole on the fecal microbiome and metabolome in healthy dogs. J Vet Intern Med. (2020) 34:1853–66. doi: 10.1111/JVIM.15871
39. Schmidt, M, Unterer, S, Suchodolski, JS, Honneffer, JB, Guard, BC, Lidbury, JA, et al. The fecal microbiome and metabolome differs between dogs fed bones and raw food (BARF) diets and dogs fed commercial diets. PLoS One. (2018) 13:e0201279. doi: 10.1371/JOURNAL.PONE.0201279
40. Myint, H, Iwahashi, Y, Koike, S, and Kobayashi, Y. Effect of soybean husk supplementation on the fecal fermentation metabolites and microbiota of dogs. Anim Sci J. (2017) 88:1730–6. doi: 10.1111/ASJ.12817
41. Uzal, FA, Vidal, JE, McClane, BA, and Gurjar, AA. Clostridium Perfringens toxins involved in mammalian veterinary diseases. Open Toxinol J. (2013) 2:24. doi: 10.2174/1875414701003010024
42. Vázquez- Baeza, Y, Hyde, ER, Suchodolski, JS, and Knight, R. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat Microbiol. (2016) 1:16177. doi: 10.1038/nmicrobiol.2016.177
43. Weese, JS, Staempfli, HR, Prescott, JF, Kruth, SA, Greenwood, SJ, and Weese, HE. The roles of Clostridium difficile and enterotoxigenic Clostridium perfringens in diarrhea in dogs. J Vet Intern Med. (2001) 15:374–8. doi: 10.1111/J.1939-1676.2001.TB02332.X
44. Gebreselassie, EEE, and Jewell, DE. Long-term consumption of high protein disrupts dog gut microbiome and metabolites. FASEB J. (2019) 33:lb248. doi: 10.1096/FASEBJ.2019.33.1_SUPPLEMENT.LB248
45. Herstad, KMV, Gajardo, K, Bakke, AM, Moe, L, Ludvigsen, J, Rudi, K, et al. A diet change from dry food to beef induces reversible changes on the faecal microbiota in healthy, adult client-owned dogs. BMC Vet Res. (2017) 13:147. doi: 10.1186/S12917-017-1073-9
46. Sandri, M, Dal Monego, S, Conte, G, Sgorlon, S, and Stefanon, B. Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. BMC Vet Res. (2016) 13:65. doi: 10.1186/s12917-017-0981-z
47. Den Besten, G, Van Eunen, K, Groen, AK, Venema, K, Reijngoud, DJ, and Bakker, BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. (2013) 54:2325–40. doi: 10.1194/JLR.R036012
48. Flint, HJ, Bayer, EA, Rincon, MT, Lamed, R, and White, BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. (2008) 6:121–31. doi: 10.1038/nrmicro1817
49. Louis, P, and Flint, HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. (2017) 19:29–41. doi: 10.1111/1462-2920.13589
50. Palframan, RJ, Gibson, GR, and Rastall, RA. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr Issues Intest Microbiol. (2003) 4:71–5.
51. Bresciani, F, Minamoto, Y, Suchodolski, JS, Galiazzo, G, Vecchiato, CG, Pinna, C, et al. Effect of an extruded animal protein-free diet on fecal microbiota of dogs with food-responsive enteropathy. J Vet Intern Med. (2018) 32:1903–10. doi: 10.1111/JVIM.15227
52. Mori, A, Goto, A, Kibe, R, Oda, H, Kataoka, Y, and Sako, T. Comparison of the effects of four commercially available prescription diet regimens on the fecal microbiome in healthy dogs. J Vet Med Sci. (2019) 81:1783–90. doi: 10.1292/JVMS.19-0055
53. Li, Q, Lauber, CL, Czarnecki-Maulden, G, Pan, Y, and Hannah, SS. Effects of the dietary protein and carbohydrate ratio on gut microbiomes in dogs of different body conditions. mBio. (2017) 8:e01703-16. doi: 10.1128/mBio.01703-16
54. Kitahara, M, Takamine, F, Imamura, T, and Benno, Y. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7α-dehydroxylating activity. Int J Syst Evol Microbiol. (2001) 51:39–44. doi: 10.1099/00207713-51-1-39
55. O’Keefe, SJD, Li, JV, Lahti, L, Ou, J, Carbonero, F, Mohammed, K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. (2015) 6:6342. doi: 10.1038/ncomms7342
56. Loesche, WJ, and Gibbons, RJ. Amino acid fermentation by Fusobacterium nucleatum. Arch Oral Biol. (1968) 13:191-IN11. doi: 10.1016/0003-9969(68)90051-4
57. Rogers, AH, Chen, J, Zilm, PS, and Gully, NJ. The behaviour of Fusobacterium nucleatum chemostat-grown in glucose- and amino acid-based chemically defined media. Anaerobe. (1998) 4:111–6. doi: 10.1006/ANAE.1997.0140
58. Bartlett, A, and Kleiner, M. Dietary protein and the intestinal microbiota: an understudied relationship. iScience. (2022) 25:105313. doi: 10.1016/J.ISCI.2022.105313
59. Kleiner, M, Kouris, A, Violette, M, D’Angelo, G, Liu, Y, Korenek, A, et al. Ultra-sensitive isotope probing to quantify activity and substrate assimilation in microbiomes. Microbiome. (2023) 11:24. doi: 10.1186/s40168-022-01454-1
60. Reese, AT, Pereira, FC, Schintlmeister, A, Berry, D, Wagner, M, Hale, LP, et al. Microbial nitrogen limitation in the mammalian large intestine. Nat Microbiol. (2018) 3:1441–50. doi: 10.1038/s41564-018-0267-7
61. Starke, R, Oliphant, K, Jehmlich, N, Schäpe, SS, Sachsenberg, T, Kohlbacher, O, et al. Tracing incorporation of heavy water into proteins for species-specific metabolic activity in complex communities. J Proteome. (2020) 222:103791. doi: 10.1016/J.JPROT.2020.103791
62. Crost, EH, Coletto, E, Bell, A, and Juge, N. Ruminococcus gnavus: friend or foe for human health. FEMS Microbiol Rev. (2023) 47:fuad014. doi: 10.1093/FEMSRE/FUAD014
63. AAFCO. (2021). Official Publication. Association Of American Feed Control Officials, Oxford, IN.
Keywords: canine microbiome, canine nutrition, diet format, core microbiome, gastrointestinal microbiome
Citation: Oba PM, Roberts LJ, Geary EL, Suchodolski JS and Swanson KS (2025) Effects of diet type on the core fecal bacterial taxa and the dysbiosis index of healthy adult dogs. Front. Vet. Sci. 12:1572875. doi: 10.3389/fvets.2025.1572875
Edited by:
Susan Wernimont, Hill’s Pet Nutrition, Inc., United StatesReviewed by:
Michal Aureliusz Jank, Poznan University of Life Sciences, PolandPavlos G. Doulidis, University of Veterinary Medicine Vienna, Austria
Copyright © 2025 Oba, Roberts, Geary, Suchodolski and Swanson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kelly S. Swanson, a3Nzd2Fuc29AaWxsaW5vaXMuZWR1
 Patrícia M. Oba
Patrícia M. Oba Leah J. Roberts1
Leah J. Roberts1 Jan S. Suchodolski
Jan S. Suchodolski Kelly S. Swanson
Kelly S. Swanson