- 1College of Animal Science, Shanxi Agricultural University, Taigu, China
- 2Department of Obstetrics and Gynecology, Reproductive Medicine Center, Shanxi Bethune Hospital, Shanxi Academy of Medical Sciences, Third Hospital of Shanxi Medical University, Tongji Shanxi Hospital, Taiyuan, China
Introduction: Subclinical mastitis (SCM) is a common disease in dairy cows associated with dysbiosis of the gastrointestinal microbiota and systemic inflammatory response. Gongying San (GYS), a commonly used herbal formula for the treatment of mastitis, has anti-inflammatory, antibacterial and antioxidant effects, but the underlying mechanisms remain unknown. Therefore, we performed a multi-omics analysis to determine the effects of GYS on intestinal microbiota and metabolites in cows with SCM.
Methods: A total of 32 Holstein cows were divided into four groups of 8 cows each, including healthy control group, subclinical mastitis group, GYS treatment group (290 g/day) and ceftiofur treatment group (2.2 mg/kg bw).
Results: GYS significantly increased milk yield, lactose and milk protein, and decreased somatic cell count (SCC) in milk from cows with SCM. In the serum, GYS decreased the levels of lipopolysaccharide (LPS), interleukin-2 (IL-2), interleukin-4 (IL-4), interleukin-8 (IL-8), interleukin-10 (IL-10), tumor necrosis factor-α (TNF-α) and malondialdehyde (MDA) and increased the concentration of superoxide dismutase (SOD). In addition, there was an increase in UCG-010 and Blautia and a decrease in Bacteroides, Lachnospiraceae, and Agathobacter in feces after GYS treatment. Fecal untargeted metabolomics showed that GYS supplementation mainly downregulated inflammation-related metabolism, including arachidonic acid and choline metabolism.
Discussion: In the treatment of SCM, GYS showed multi-target therapeutic advantages of anti-inflammatory, antioxidant and immunomodulatory properties compared to antibiotics. Bacteroides, Lachnospiraceae and UCG-010 may be involved in the regulation of inflammation through 3-oxo-Δ4bile acids and phosphatidylcholine.
1 Introduction
Mastitis represents a significant challenge to the advancement of the global dairy industry and exerts a considerable influence on the economic viability of animal husbandry. In addition to impairing the quality and yield of milk, the disease also endangers the well-being and health of dairy cows (1). In particular, SCM has a high incidence in dairy cows due to its hidden nature and long incubation period, accounting for approximately 90–95% of mastitis cases (2). The total economic cost of dairy cattle diseases is estimated to be $65 billion per year globally, of which SCM alone contributes about $9 billion (3). SCM reduces milk production by 5.6–17.2% during lactation, with a loss of $38 per cow per lactation due to SCM, and up to $66.6 per cow on urban farms (4). However, the current therapeutic regimen faces serious challenges in terms of antibiotic dependence, the spread of resistance and the control of recurrence rates. Therefore, there is a need to find a more efficient treatment option that takes into account the pathogenesis of mastitis.
In examining the etiology of mastitis, researchers have come to recognize that the disease is not solely a consequence of a localized inflammatory response. Rather, it may also be influenced by alterations in the structure of the host’s microbial community. Some studies have demonstrated that the fecal microbiota of cows with mastitis can be transferred to germ-free mice, which then exhibit mastitis and serum inflammation [increased TNF-α, interleukin-17 (IL-17), LPS levels] (5). This evidence suggests that there may be a “gastroenterogenic mastitis” pathogenic pathway for the development of mastitis (6). The concept of the “intestinal-mammary axis” has also been proposed by some scientists (7). It can be surmised that intestinal microbiota and their metabolites may migrate to the mammary gland via the bloodstream, thereby modulating mastitis. Currently, antibiotics remain the primary choice of medication for clinical treatment (8). However, the indiscriminate use of antibiotics not only increases production costs, promotes the development of drug-resistant pathogens, and disrupts the intestinal microbiota, but also poses a significant threat to public health (9). With growing concerns about antibiotic dependency, exploring alternatives to antibiotics has become a key area in addressing mastitis. Therefore, we attempted to improve the intestinal microbiota structure through nutritional modulation in order to alleviate SCM in dairy cows.
Herbal medicine represents a promising avenue for the treatment of mastitis. The use of herbal combination therapy offers a practical approach to reducing inflammation and regulating intestinal microbiota (10). Taraxacum is a member of the Asteraceae family and contains a variety of phytochemicals that are beneficial to animal health. These include phenolic compounds, sesquiterpene lactones, polysaccharides, and flavonoids (11). It has been demonstrated that the administration of taraxacum to mid-lactation cows can enhance lactation performance and stimulate amino acid metabolism and rumen fermentation (12, 13). Taraxacum extract has been demonstrated to alleviate LPS-induced oxidative stress by scavenging cellular reactive oxygen species (ROS) and increasing antioxidant enzyme activities. Furthermore, it has been shown to activate the nuclear factor erythroid 2-related factor 2 (Nrf2) signaling pathway, which in turn promotes the expression of antioxidant genes (14). Additionally, the extract has been observed to significantly inhibit the expression of TNF-α and intercellular cell adhesion molecule-1 (ICAM-1), thereby alleviating inflammation (15). The anti-inflammatory effects of lonicerae japonicae flos have been shown to down-regulate the release of IL-1β, IL-6, and TNF-α (16). Forsythia suspensa is commonly used in the treatment of various inflammatory diseases, for example it significantly reduces the elevated TNF-α and IL-6 caused by colitis and improves colonic tissue damage (17). Gongying San is a classic herbal formula consisting of taraxacum, lonicerae japonicae flos, forsythia suspensa, retinervus luffae fructus, bulbus fritillariae thunbergii, hibisci mutabilis folium and tetrapanax papyrifer. In Gongying San, the combination of dandelion with other herbs can further enhance its efficacy through the ‘multi-component multi-target multi-pathway’ mode of action, which makes it more promising in the treatment of various inflammatory and infectious diseases. Network pharmacology and molecular-docking analyses revealed that quercetin, lignans and kaempferol were the main active components of Gongying San in the treatment of mastitis, and TNF, IL-6, IL1β, ICAM1, and CXCL8 were its key targets (18). However, there is a lack of standardized clinical validation of Gongying San. Accordingly, the objective of this study was to examine the impact of Gongying San, a Chinese herbal compound, on intestinal microbiota and their metabolites, lactation performance, and inflammatory response in cows with SCM.
2 Materials and methods
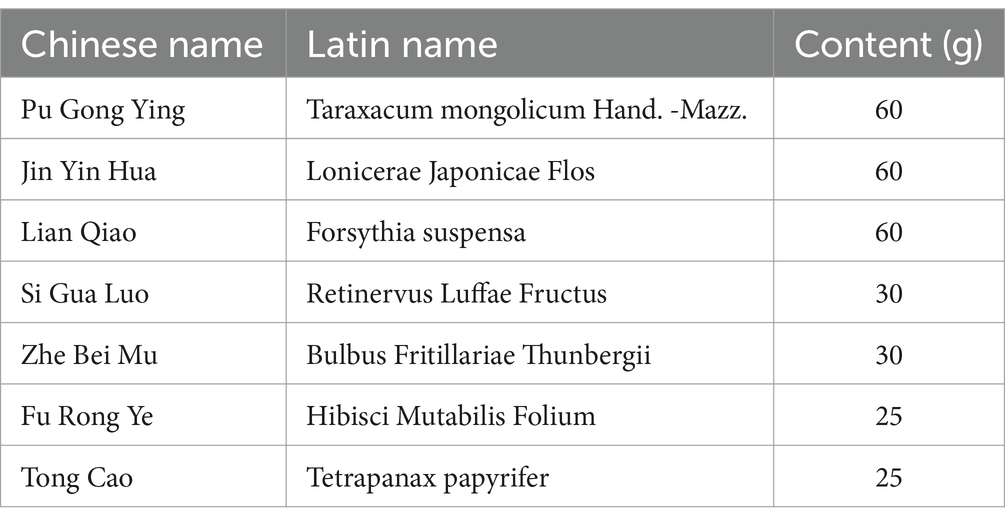
2.1 Animal, diets, and experiment design
The experimental design has been shown in Figure 1. This study used 32 Holstein dairy cows in mid-lactation (body weight 655 ± 18.7 kg, days to lactation 110 ± 12.4 d) reared at ZiLin Ranch of the Shanxi, China. These cows have not been treated with antibiotics or other drugs within the past year and have been raised in the same environment. The ingredients and chemical composition of basal diet are shown in Supplementary Table S1. SCM was diagnosed by the result of California mastitis test (CMT) was weakly positive (600,000 < SCC < 1,000,000 cells/mL) and no clinical symptoms in udders (19). The experimental design has been shown in Figure 1. Eight healthy cows were used as control group (CON; n = 8) (SCC < 100, 000cells/mL; no clinical symptoms in udders; CMT results were negative), Twenty-four cows with SCM were selected from the farm and randomly divided into the subclinical mastitis group (SCM; n = 8); Gongying San group (GYS; n = 8); ceftiofur hydrochloride group (CEF; n = 8). In GYS group, the Gongying San (New Century Pharmaceutical Co., Ltd., Hebei, China) was taken through the mouth 290 g/day per cow for 5 days; the detailed composition of GYS is listed in Table 1. In CEF group, antibiotic treatment consisted of daily intramuscular injections of ceftiofur hydrochloride (2.2 mg/kg bw) into the neck region for 5 days. Two weeks before the start of the experiment, the four groups of cows were kept in four different pens and fed the basal diet. The drug treatment started on the first day of the experiment and was carried out for five consecutive days. Milk, serum and fecal samples were collected on the sixth day. The drug treatment started on the first day of the experiment and was carried out for five consecutive days. Milk, serum and fecal samples were collected on the sixth day. This was followed by serum index tests, milk yield tests, milk composition tests, 16SrRNA sequencing and non-targeted metabolomics analyses, and finally correlation analyses.
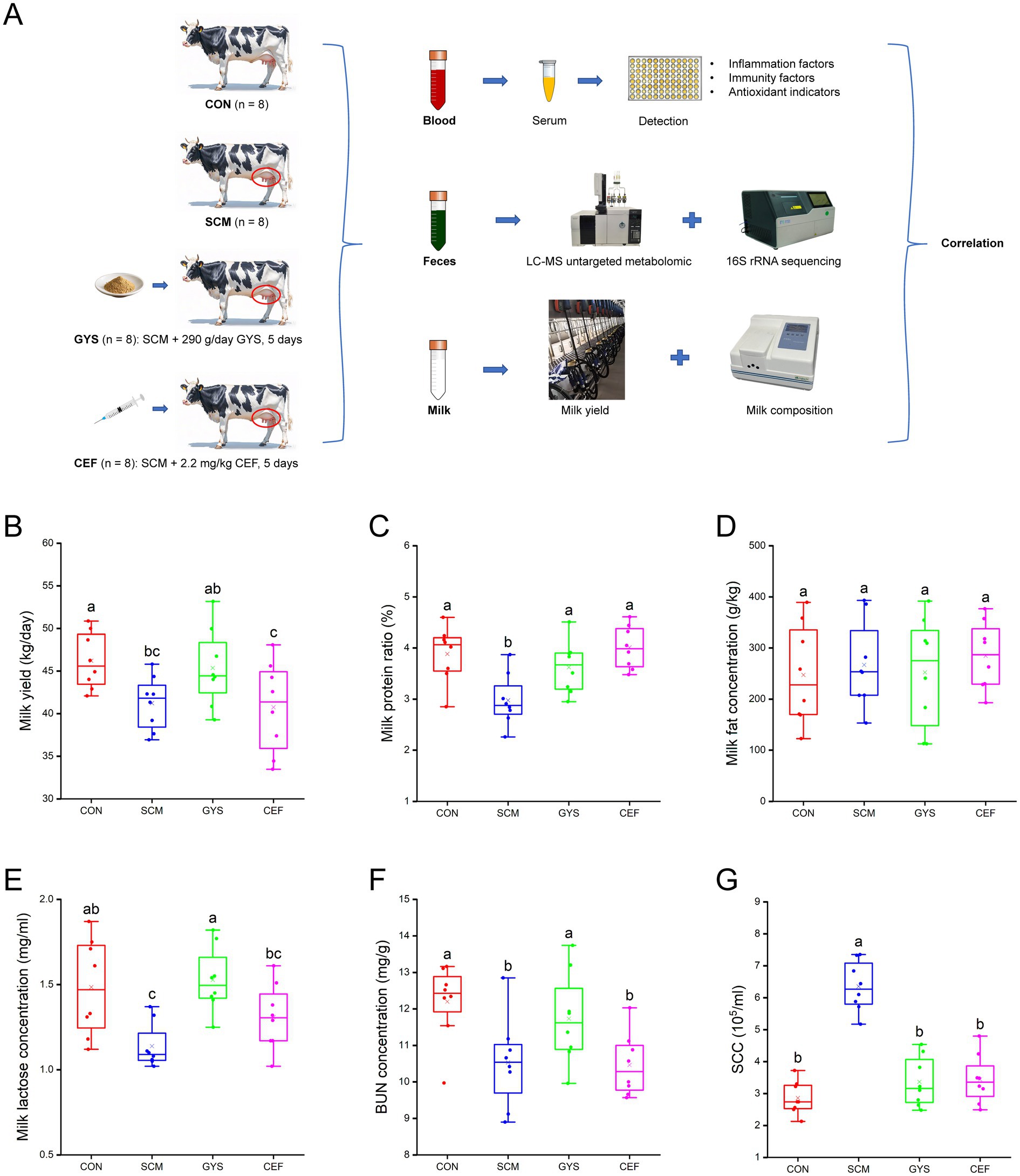
Figure 1. Experimental design and lactation performance of dairy cows. (A) Experimental design. (B) Milk yield. (C) Milk protein. (D) Milk fat. (E) Milk lactose. (F) BUN, blood urea nitrogen. (G) SCC, somatic cell counts. CON, control group; SCM, subclinical mastitis group; GYS, Gongying San group, the Gongying San addition level was 290 g/day per cow; CEF, antibiotic group, antibiotic treatment consisted of daily intramuscular injections of ceftiofur hydrochloride (2.2 mg/kg bw) into the neck region for 5 days. Differences in letters on the box-and-line plot indicate significant differences between groups (p < 0.05). a, b, c Different letters differed significantly (p < 0.05).
2.2 Milk sampling and analysis
Cows were milked three times a day at 07: 30, 13:30, and 19:30, utilizing an automated milking system. The data pertaining to the yield of milk were recorded on a daily basis by a dairy herd management software program. Milk samples from each cow were collected 7:00, 13:00, and 19:00 and mixed with a ratio of 4:3:3 on the sixth day of the experiment. The mixed milk samples from each cow were collected into a 15-ml sterile centrifuge tubes and stored at 4°C. The milk protein was determined by Kjeldahl method. The milk fat was determined by Soxhlet extractor method. The concentration of milk lactose was analyzed using a milk lactose test kit (Sangon Biotech Co., Ltd., Shanghai, China). The milk SCC was determined by Microscopic method. The concentration of milk urea nitrogen (MUN) was analyzed using a MUN test kit (Sangon Biotech Co., Ltd., Shanghai, China).
2.3 Serum collection and analysis
Blood samples were collected through the tail vein at 06: 30, on the sixth day of the experiment. One tube of blood sample was taken from each cow. Blood samples were allowed to stand for 30 min at room temperature and then centrifuged at 3,000 × g for 15 min at 4°C to separate the serum. The collected serum samples were stored at - 80°C for biochemical analysis. Blood urea nitrogen (BUN) was analyzed using a BUN test kit (Sangon Biotech Co., Ltd., Shanghai, China). The concentrations of total protein (TP) and albumin (ALB) were analyzed by a TP colorimetric test kit (BCA method) (Sangon Biotech Co., Ltd., Shanghai, China) and an ALB assay kit (bromocresol green colorimetry) (Sangon Biotech Co., Ltd., Shanghai, China), respectively. IL-2, IL-4, IL-6, IL-8, IL-10, TNF-α, prostaglandin E2 (PGE2), glutathione peroxidase (GSH-Px), SOD, MDA, and LPS concentrations were determined by the corresponding bovine enzyme-linked immunosorbent assay (ELISA) kits (Sangon Biotech Co., Ltd., Shanghai, China). The concentrations of IgA, IgG, and IgM were measured by bovine immunoglobulin ELISA kits (Sangon Biotech Co., Ltd., Shanghai, China).
2.4 Feces sample collection
Feces samples were collected from the rectum at 09:00, on the sixth day of the experiment. Fecal samples from each cow were stored in two sterile 5 mL tubes at - 80°C for microbiological and metabolite analysis.
2.5 DNA extraction, sequencing and data processing
Total community genomic DNA extraction was performed using a E. Z. N. A™ Mag-Bind Soil DNA Kit (#M5635-02, Omega, United States), following the manufacturer’s instructions. We measured the concentration of the DNA using a Qubit4.0 (Thermo, United States) to ensure that adequate amounts of high-quality genomic DNA had been extracted.
The objective of this study was to target the V3-V4 hypervariable region of the bacterial 16S rRNA gene. Polymerase chain reaction (PCR) was initiated immediately following the extraction of DNA. The 16S rRNA V3-V4 amplicon was amplified using 2 × Hieff® Robust PCR Master Mix (#10105ES03, Yeasen, China). Two universal bacterial 16S rRNA gene amplicon PCR primers (PAGE-purified) were utilized: the amplicon PCR forward primer (CCTACGGGNGGCWGCAG) and the amplicon PCR reverse primer (GACTACHVGGGTATCTAATCC). The reaction was configured as follows: microbial DNA (10 ng/μl) (2 μL); amplicon PCR forward primer (10 μM) (1 μL); amplicon PCR reverse primer (10 μM) (1 μL); 2 × Hieff® Robust PCR Master Mix (30 μL). The plate was then sealed and subjected to PCR in a thermal instrument (Applied Biosystems 9700, United States) using the following program: Initially, the program subjected the samples to a cycle of denaturing at 95°C for 3 min. This was followed by five cycles of denaturing at 95°C for 30 s, annealing at 45°C for 30 s, and elongation at 72°C for 30 s. The subsequent twenty cycles consisted of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 30 s. The program culminated with a final extension at 72°C for 5 min. The amplification products were subjected to electrophoresis in 2% (w/v) agarose gels in TBE buffer (Tris, boric acid, EDTA) containing ethidium bromide (EB) and were then visualized under UV light.
We used Hieff NGS™DNA Selection Beads (#10105ES03, Yeasen, China) to purify the free primers and primer dimer species in the amplicon product. Samples were delivered to Sangon BioTech for library construction using universal Illumina adaptor and index. Before sequencing, the DNA concentration of each PCR product was determined using a Qubit®4.0Green double-stranded DNA assay and it was quality controlled using a bioanalyzer (Agilent2100, United States). Depending on coverage needs, all libraries can be pooled for one run. The amplicons from each reaction mixture were pooled in equimolar ratios based on their concentration. Sequencing was performed using the Illumina MiSeq system (Illumina MiSeq, United States), according to the manufacturer’s instructions.
The primer junction sequences must be removed using Cutadapt (version 1.181), after which the PEAR software (version 0.9.82) will be used to merge pairs of reads into a single sequence based on the overlap relationship between PE reads. PRINSEQ (version 0.20.43) will then be used to filter the quality of each sample data for quality control purposes. The operational taxonomic units (OTU) clustering of non-repetitive sequences was conducted according to 97% similarity using Usearch (version 11.0.6674). The taxonomic analysis of OTU representative sequences was conducted in accordance with the Silva 16S rRNA database (version 138.25), utilizing the RDP classifier (version 2.126). The calculation of the alpha diversity index (Chao1 and Shannon) is performed using the Mothur software (version 1.43.07). The distance algorithms of the principal coordinates analysis (PCoA) were weighted normalized UniFrac and unweighted UniFrac, which were performed by using the vegan package in R (version 3.6.08). Linear discriminant analysis effect size (LEfSe) was performed using LEfSe software (version 1.1.09) to discover the differential microbiota between groups and the extent to which the differential microbiota influenced the differences between groups was expressed by Linear discriminant analysis (LDA). Significantly different microbiotas were defined as LDA > 3.5 and p < 0.05 (Student’s t-test), the FDR method was applied to adjust p-values.
2.6 Feces metabolomics analysis
After thawing the samples on ice, 20 mg (±1 mg) of feces from each cow was mixed with 400 μL of 70% methanol–water internal standard extract and vortexed for 3 min. The samples were ultrasonicated for 10 min in an ice-water bath, removed from the samples, vortexed for 1 min, and allowed to stand for 30 min in a refrigerator at −20°C. The supernatant was extracted by centrifugation at 12,000 r/min for 10 min at 4°C, and then centrifuged again at 12,000 r/min for 3 min at 4°C. 200 μL of supernatant was extracted for LC–MS analysis. Chromatographic separation was performed on a HSS T3 chromatographic column (2.1 mm × 100 mm, 1.8 μm, Waters). Mobile phase A contained 0.1% formic acid and water, and mobile phase B contained 0.1% formic acid and acetonitrile at a flow rate of 0.4 mL/min, with an instrumental column temperature of 40°C and a sample volume of 4 μL. Separation gradient: 95%: 5% at 0 min; 80%: 20% at 2 min; 40%: 60% at 5 min; 1%: 99% for 6–7.5 min; 95%: 5% for 7.6–10 min. Data were collected in positive and negative ion modes with a duration of 10 min for ESI + and ESI-, ion spray voltage of 5,000 and −4,000 V and temperature of 550 and 450°C.
Mass spectrum peaks were extracted, aligned, and retention time corrected from LC–MS raw data using the xcms package in R software. Peaks with > 50% missing in each set of samples were filtered, blanks were filled, and peak areas were corrected. The filtered peaks were used for metabolite identification by searching Human metabolome database (HMDB10). Principal component analysis (PCA) analysis was performed using the prcomp function in the R software, and Orthogonal projections to latent structures-discriminate analysis (OPLS-DA) analysis was performed using the MetaboAnalystR package OPLSR. Anal function in the R software. The criteria for determining differential metabolites were variable importance in projection (VIP) > 1 and p < 0.05 (Student’s t-test), the FDR method was applied to adjust p values. The metabolite content data were processed using unit variance scaling (UV), and all samples were analyzed by cluster analysis. Annotation of metabolic pathways using the KEGG database.11 Spearman’s correlation analysis was executed by employing the Scipy packages of Python (version 1.26.4), and the R heatmap package was utilized for the purpose of visualization.
2.7 Statistical analysis
A comprehensive analysis of lactation performance, serum markers, microorganisms, and metabolites were conducted using one-way analysis of variance (ANOVA) and Student’s t-test, with SPSS software (version 22.0) serving as the statistical analysis tool. Differences were statistically significant when p < 0.05.
3 Results
3.1 Effect of GYS treatment on milk yield and composition
As shown in Figures 1B,C,E,F, compared with the CON group, SCM group significantly decreased milk yield (p = 0.024), milk protein (p = 0.001), milk lactose (p = 0.002), BUN (p = 0.005) and significantly increased SCC (p < 0.001). However, there was a significant recovery of milk protein (p = 0.014), milk lactose (p < 0.001), BUN (p = 0.039) and SCC (p < 0.001) and a trend toward higher Milk yield (p = 0.058) after GYS treatment compared to the SCM group. Milk fat content did not differ significantly among the four groups (Figure 1D). Although CEF significantly reduced SCC (p < 0.001), it did not increase milk yield (Figures 1B,G).
3.2 Effect of GYS treatment on blood biochemical indexes
As shown in Table 2, TP, ALB and GLB did not change significantly in the four groups. In terms of inflammation, IL-2 (p = 0.013), IL-4 (p = 0.019), IL-6 (p = 0.028), IL-8 (p = 0.011), IL-10 (p = 0.016), and TNF-α (p = 0.018) levels were significantly higher in the SCM group compared to the CON group, with IL-2 (p = 0.023), IL-4 (p = 0.004), IL-8 (p = 0.041), IL-10 (p = 0.008), and TNF-α (p = 0.016) levels being significantly reduced after GYS treatment, while IL-4 (p = 0.045) and TNF-α (p = 0.009) levels were reduced by CEF treatment. In terms of immunity, SCM significantly increased IgA (p = 0.015) levels compared to the CON group, whereas there was a significant decrease in IgA (p = 0.009) levels after GYS treatment. In terms of antioxidants, SOD (p = 0.019) levels were significantly lower in the SCM group compared to the CON group, significantly higher in GYS and CEF groups (p < 0.050). MDA (p = 0.002) levels were significantly higher in the SCM group compared to the CON group, significantly lower in the GYS and CEF groups (p < 0.050).
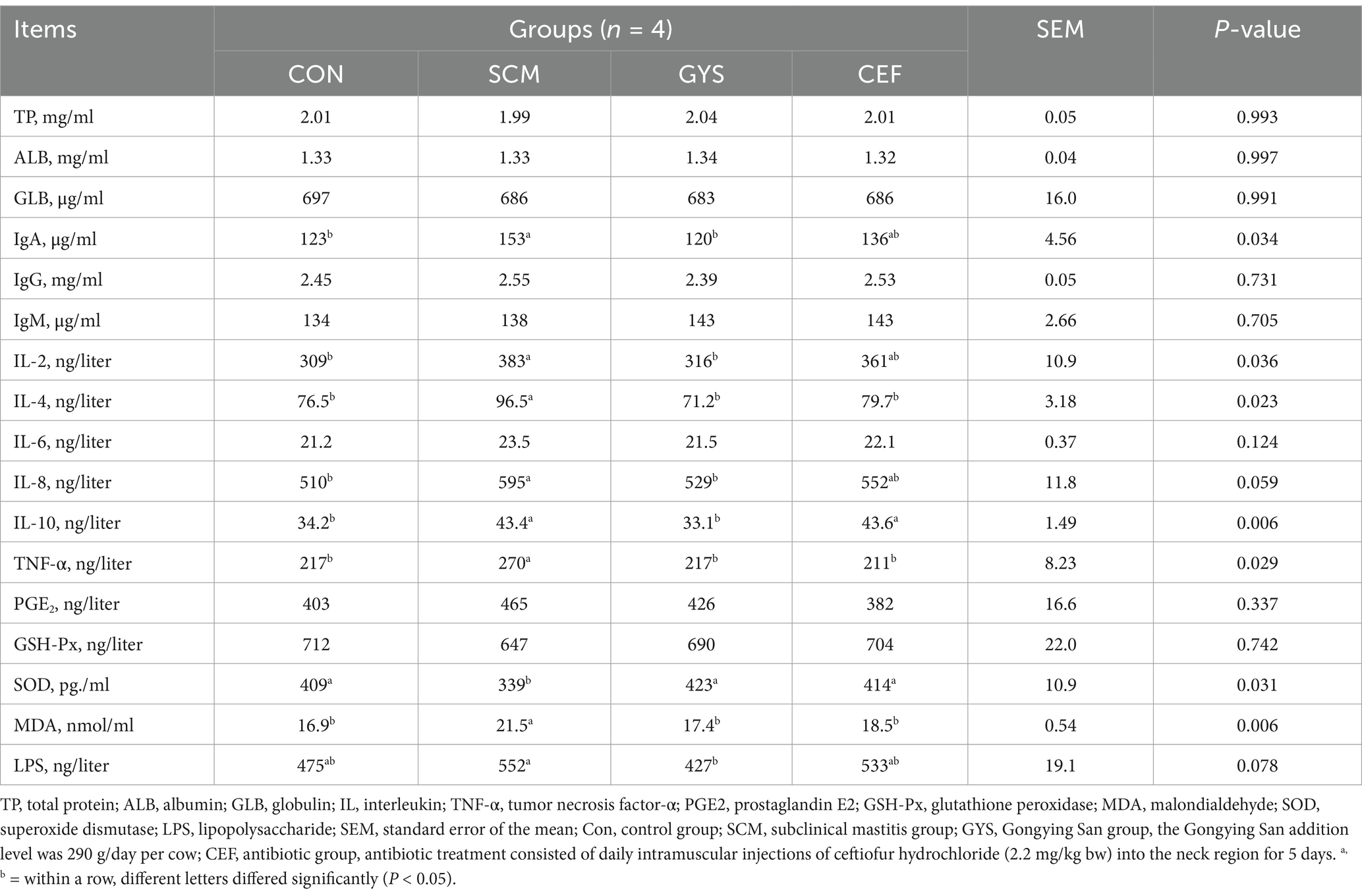
Table 2. Effect of GYS on the inflammatory cytokine, oxidative stress indexes and LPS in serum of dairy cows with SCM.
3.3 Differences in diversity, richness and composition of fecal microbiota
The effect of GYS on intestinal microbiota in dairy cows was determined by 16S rRNA high-throughput sequencing. As the number of reads sampled increases, the alpha diversity index gradually reaches a plateau, indicating that the amount of data for this sequencing is sufficient (Supplementary Figure S1). The results of the alpha diversity analysis indicated that there were no significant differences in the chao1 index and shannon index between the CON, SCM, and GYS groups. However, a significant decrease was observed in the CEF (p < 0.001) group (Figures 2A,B). In Figures 2C,D, Regarding beta diversity, PCOA of intestinal microbiota by unifrac and wunifrac algorithms. An analysis of similarity (Anosim) was used to test whether the differences between groups were significantly greater than the differences within groups, with R > 0 indicating that the groupings were meaningful. Significant differences in microbial composition were observed in the GYS group compared to the SCM group (p = 0.048), but similar to the CON group (p = 0.118). Also the microbial composition of the CEF group was significantly different from the other three groups (p < 0.001). At the phylum level, Firmicutes, Bacteroidota, Actinobacteriota, Proteobacteria, and Spirochaetota were the dominant groups in four groups (Figure 2E). At the genus level, the most prevalent intestinal microbiota include UCG-005, Rikenellaceae_RC9_intestinal_group, unclassified_Lachnospiraceae, Christensenellaceae_R_7_group, norank_Eubacterium_coprostanoligenes_group, Bifidobacterium, unclassified_Clostridia, and norank_Muribaculaceae (Figure 2F).
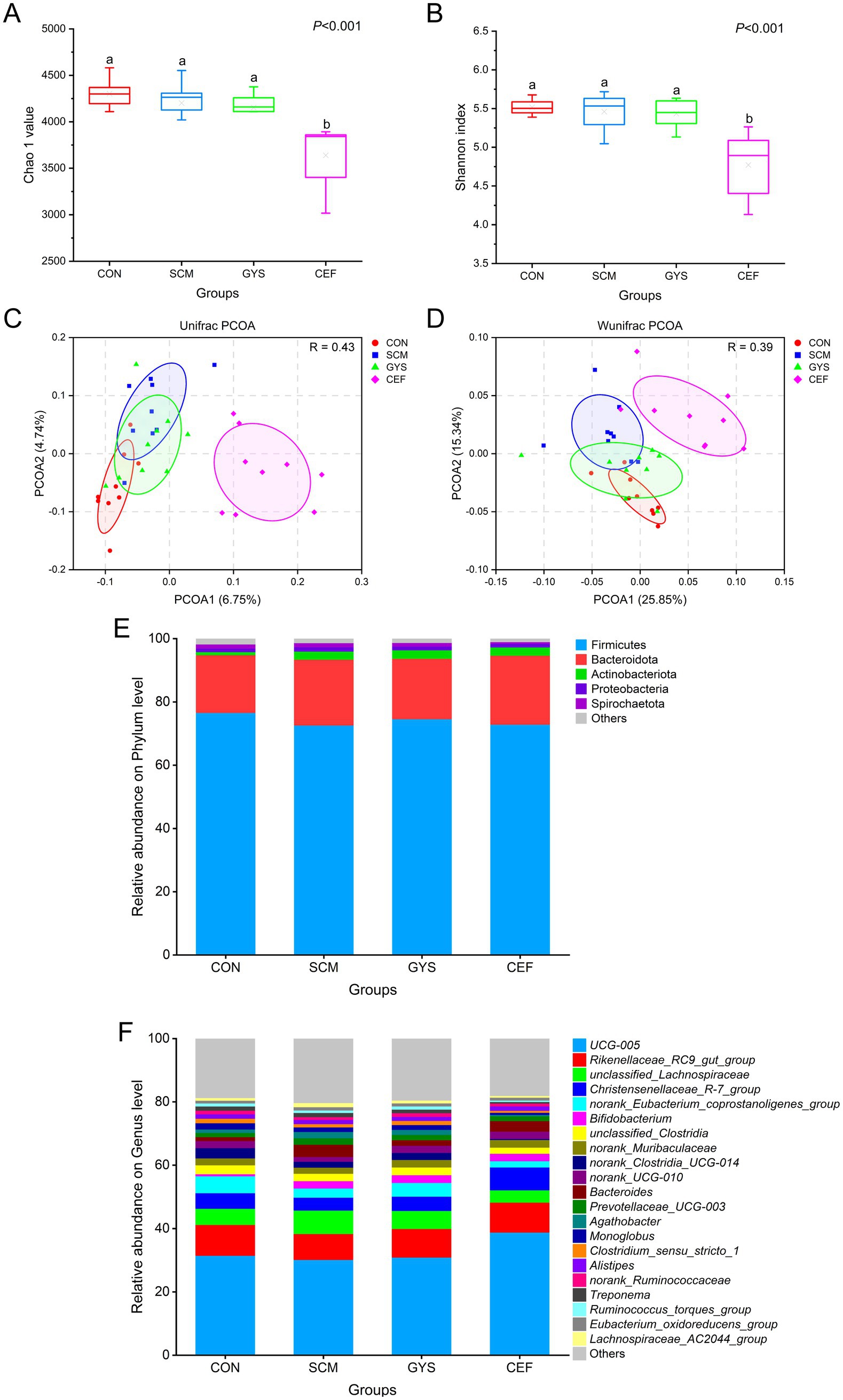
Figure 2. Changes in α-and β-diversity as well as feces bacterial community of dairy cows. Chao1 (A) and Shannon (B) index (α-diversity) reflect the diversity and richness of fecal microbiota. a,bDifferent letters differed significantly (p < 0.05). Unweighted unifrac distance (C) and weighted unifrac distance (D) (β-diversity), which showed the profile of fecal microbial communities among groups. Relative abundance of phylum (E) and genus (F) between four groups.
3.4 Effect of GYS on fecal microbiota content
To assess how GYS administration affected the dairy cow intestinal microbial composition, linear discriminant analysis effect size (LEfSe) analysis was utilized to analyse the bacterial microbiota. We found significant differences in the relative abundance of 10 genera in the comparison of the CON group with the SCM group, while 6 genera were found to be significantly different in the comparison of the SCM group with the GYS group, with 5 genera being common to the colony (Figures 3A,B). Compared with CON, the relative abundances of unclassified_Lachnospiraceae (p < 0.001), Agathobacter (p = 0.003), Bacteroides (p < 0.001) and Lachnospiraceae_AC2044_group (p = 0.007) in the SCM were increased and decreased in the GYS group (p < 0.01). The relative abundances of norank_UCG-010 (p = 0.026) were lower in the CFE than in the CON and increased in the GYS group (p = 0.019) (Figures 3C–G).
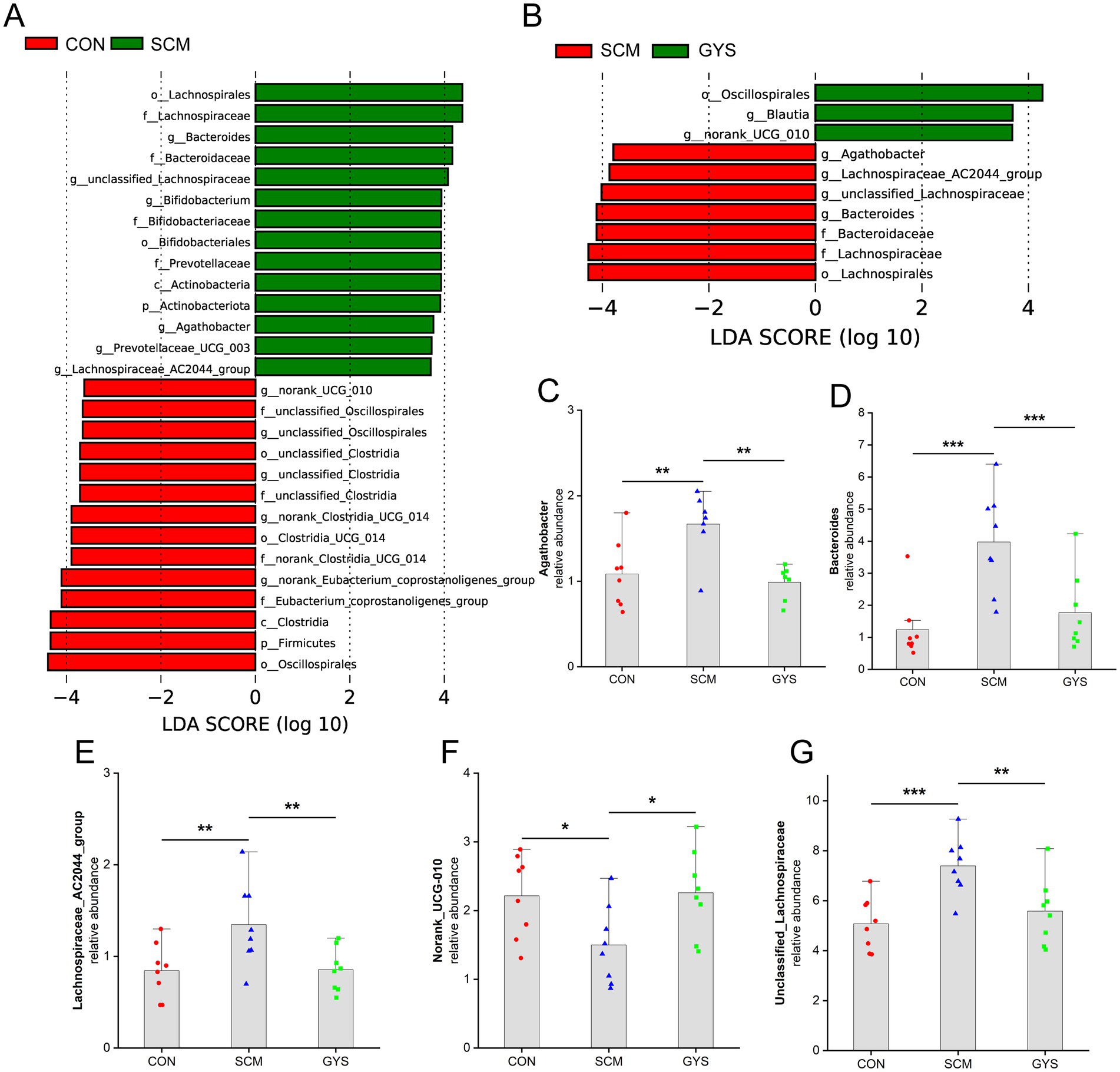
Figure 3. Composition of the feces bacterial community of dairy cows. (A,B) Bar chart of linear discriminant analysis (LDA) score of different bacterial taxa in the feces, respectively. LDA score > 3.5. (C–G) Bar charts showing the relative abundance of common differential microbiota in CON versus SCM and SCM versus GYS dairy cow fecal samples. *p < 0.05, **p < 0.01, ***p < 0.001.
3.5 Differences in fecal metabolites
Subsequently, we conducted an untargeted metabolomics analysis of fecal samples from the four groups. In order to elucidate the metabolite distinctions among the groups, an orthogonal partial least squares discriminant analysis (OPLS-DA) was conducted on fecal samples. As demonstrated in Figures 4A,B, the CON group can be distinguished from the SCM group, and the SCM group from the GYS group. This finding indicates that there have been substantial changes in the intestinal metabolites of dairy cows with subclinical mastitis treated with GYS. Following the filtration and optimization steps, a total of 4,467 metabolites were identified from 24 fecal samples. A total of 35 metabolites exhibited significant differences between the CON and SCM groups, including 34 metabolites that were up-regulated and 1 metabolite that was down-regulated in the SCM group (|log2FC| ≥ 2, VIP > 1 and p < 0.05) (Figure 4C). A total of 41 metabolites exhibited significant differences between the SCM and GYS groups, including 14 metabolites that were up-regulated and 27 metabolites that were down-regulated in the GYS group (Figure 4D). We identified 24 shared metabolites in the CON, SCM and GYS groups (Figure 4E), including Efavirenz, Benfuresate, Alangicine, Ethyl icosapentate, Asn-Ala-Leu-Ala-His, Lys-Ile-Glu, Diclofenac sodium, Docosahexaenoic acid methyl ester, Nomifensine maleate, 1-Oleoyl-2-myristoyl-sn-glycero-3-phosphocholine, 7alpha-Hydroxy-3-oxochol-4-en-24-oic acid, Oleoyltaurine, Lys-Pro-Lys, Fenaminosulf, Gentian violet, Cavipetin C, His-Arg-Lys-Glu, Arg-Gln-Arg, 5-O-beta-D-mycaminosyl-20-oxotylonolide, Phe-Asn-Leu, Malyngamide J, Ala-Thr-Ile-Lys, 3,6,9,12,15,18,21-Heptaoxatricosane-21,23-diol, Arg-Thr-Ala-Arg, and interestingly, all these metabolites were upregulated in the SCM group and significantly decreased after GYS treatment (Figure 4F).
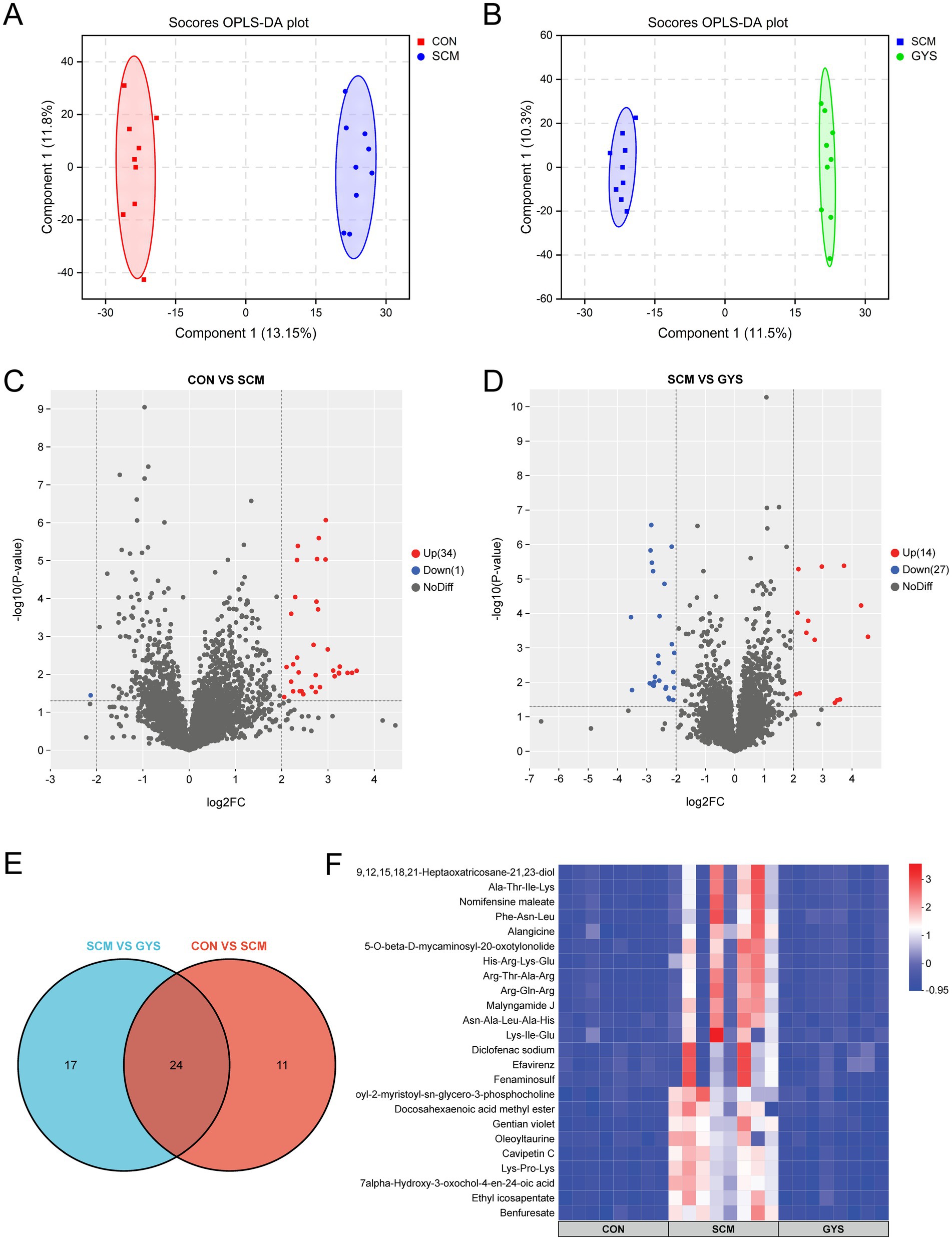
Figure 4. Fecal metabolome of CON, SCM and CFE cows. Scores plot of orthogonal partial least square discriminant analysis (OPLS-DA) for CON versus SCM (A) and SCM versus GYS (B) fecal metabolites. Volcano plot of CON versus SCM (C) and SCM versus GYS (D) differential metabolites. (E) Venn diagram outlining the shared differentially abundant metabolites in CON versus SCM and SCM versus GYS. (F) Heatmap showing differences in shared metabolites among the CON, SCM, and GYS groups.
Metabolic pathways were annotated based on different metabolites using KEGG database. Arachidonic acid metabolism, retrograde endocannabinoid signaling, choline metabolism in cancer, alpha-Linolenic acid metabolism, linoleic acid metabolism, and glycerophospholipid metabolism were the pathways that were significantly different between the CON and SCM groups (Figure 5A). In the metabolome of SCM and GYS groups arachidonic acid metabolism, retrograde endocannabinoid signaling, glycerophospholipid metabolism, alpha-Linolenic acid metabolism, choline metabolism in cancer, linoleic acid metabolism, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, etc. were significantly different pathways (Figure 5B). By comparison, the co-enriched pathways were found to be Arachidonic acid metabolism, Retrograde endocannabinoid signaling, Glycerophospholipid metabolism, alpha-Linolenic acid metabolism, Choline metabolism in cancer and Linoleic acid metabolism.
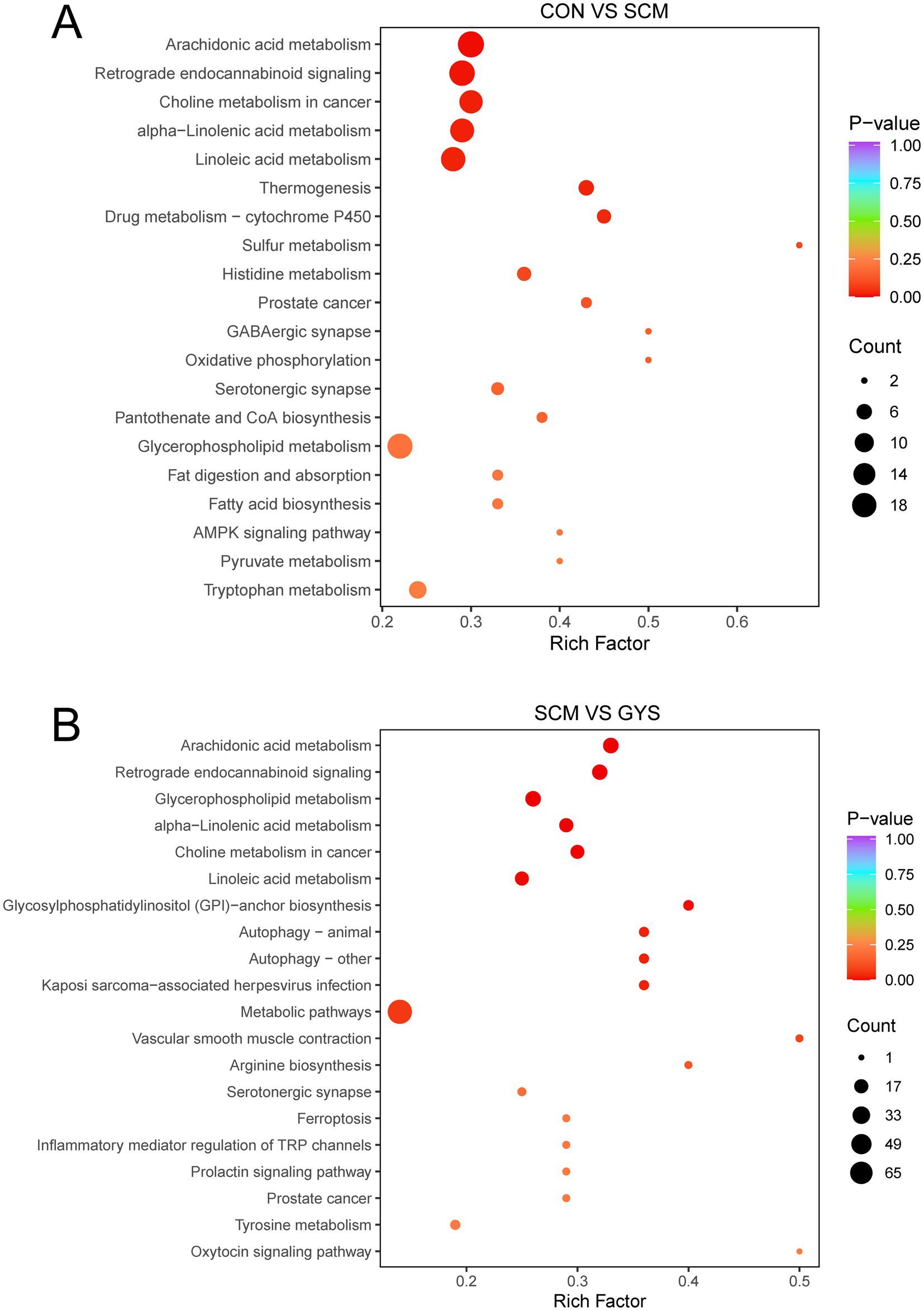
Figure 5. KEGG enrichment pathways analysis. Metabolic pathway analysis conducted with the differentially abundant fecal metabolites in the CON versus SCM (A) and SCM versus GYS (B) comparisons.
3.6 Significant correlations between different fecal microbiota and milk components, and between metabolites and fecal microbiota
First, we investigated the correlation between differential microbiota and lactation performance. Bacteroides was negatively correlated with milk yield (p = 0.003) and milk lactose (p = 0.001). Norank_UCG-010 (p = 0.030) was positively correlated with milk protein, by contrast, Unclassified_Lachnospiraceae (p = 0.009), Agathobacter (p = 0.020) and Lachnospiraceae_AC2044_group (p = 0.013) were negatively correlated with milk protein. Unclassified_Lachnospiraceae (p < 0.001), Agathobacter (p = 0.010), Bacteroides (p = 0.026) and Lachnospiraceae_AC2044_group (p < 0.001) were positively correlated with milk SCC, but norank_UCG-010 (p = 0.049) was negatively correlated with milk SCC (Figure 6A).
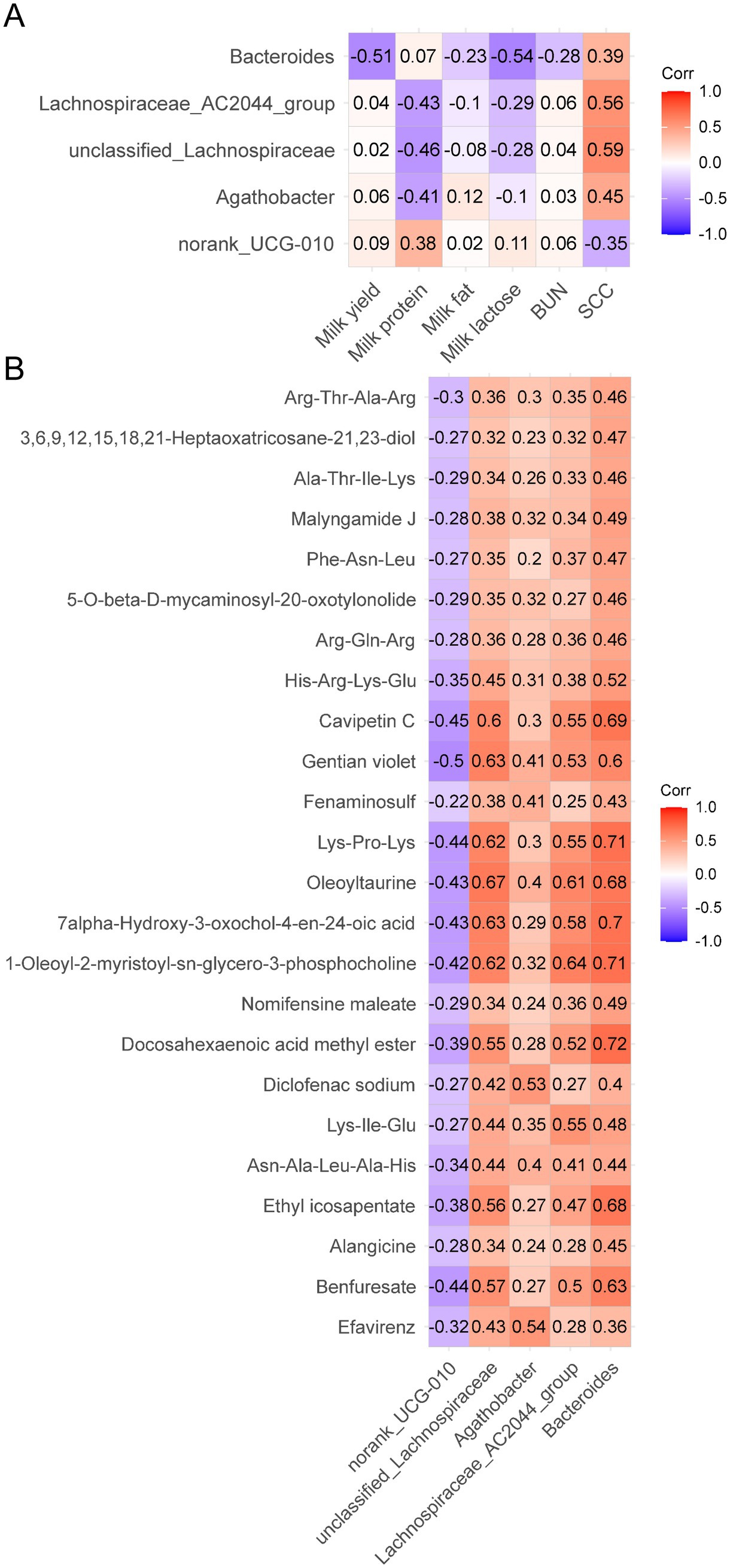
Figure 6. Analysis of the correlation between significant different fecal microbiota and milk compositions (A) and differential metabolites (B). Each lattice represents a Pearson correlation coefficient between a bacterium and a metabolite or milk compositions. Red represents a positive correlation, while blue represents a negative correlation.
For metabolites, Benfuresate (p = 0.031), 1-Oleoyl-2-myristoyl-sn-glycero-3-phosphocholine (p = 0.039), 7alpha-Hydroxy-3-oxochol-4-en-24-oic acid (p = 0.034), Oleoyltaurine (p = 0.036), Lys-Pro-Lys (p = 0.030), Gentian violet (p = 0.013) and Cavipetin C (p = 0.029) were negatively correlated with norank_UCG-010. Efavirenz (p = 0.007), Diclofenac sodium (p = 0.008), Fenaminosulf (p = 0.044) and Gentian violet (p = 0.048) were positively correlated with Agathobacter. Efavirenz (p = 0.036), Benfuresate (p = 0.003), Ethyl icosapentate (p = 0.005), Asn-Ala-Leu-Ala-His (p = 0.030), Lys-Ile-Glu (p = 0.032), Diclofenac sodium (p = 0.041), Docosahexaenoic acid methyl ester (p = 0.006), 1-Oleoyl-2-myristoyl-sn-glycero-3-phosphocholine (p = 0.001), 7alpha-Hydroxy-3-oxochol-4-en-24-oic acid (p < 0.001), Oleoyltaurine (p < 0.001), Lys-Pro-Lys (p = 0.001), Gentian violet (p < 0.001), Cavipetin C (p = 0.002) and His-Arg-Lys-Glu (p = 0.029) were positively correlated with unclassified_Lachnospiraceae. Benfuresate (p = 0.012), Ethyl icosapentate (p = 0.020), Asn-Ala-Leu-Ala-His (p = 0.044), Lys-Ile-Glu (p = 0.005), Docosahexaenoic acid methyl ester (p = 0.009), 1-Oleoyl-2-myristoyl-sn-glycero-3-phosphocholine (p < 0.001), 7alpha-Hydroxy-3-oxochol-4-en-24-oic acid (p = 0.003), Oleoyltaurine (p = 0.002), Lys-Pro-Lys (p = 0.006), Gentian violet (p = 0.007) and Cavipetin C (p = 0.005) were positively correlated with Lachnospiraceae_AC2044_group. All differential metabolites except Efavirenz and Diclofenac sodium were positively correlated with Bacteroides (p < 0.050) (Figure 6B).
4 Discussion
Although the udders and milk of cows with SCM do not show significant changes in appearance, the milk yield and composition can be severely compromised. Antibiotics are still the drug of choice for the treatment of mastitis, but the misuse of antibiotics in animal husbandry has led to a series of problems such as bacterial resistance, environmental pollution and harm to human health (20–22). Therefore, the appropriate addition of botanicals appears to be critical to the lactation performance of cows. Yue Wang et al. reported that supplementation with inulin improved lactation performance and milk quality in cows with SCM (23). Furthermore, the addition of 200 g/day of dandelion has been shown to enhance milk production and lactose levels in dairy cows (12). In herbal formulas, the interplay among components can exhibit agonistic, compatible, or antagonistic effects, resulting in the modulation of multiple signaling pathways in various organs, thereby contributing to disease alleviation (24). In this study, we observed a significant decrease in lactose levels in cows with SCM, which may be attributed to the inflammation-induced increase in blood-milk barrier permeability. GYS treatment led to a substantial reduction in serum LPS levels, suggesting that intestinal permeability was ameliorated, which leads to elevated lactose levels (25). Furthermore, the increase in milk production due to GYS can be attributed to the cure of SCM. Although CEF treatment group reduced milk SCC, the milk yield was still significantly lower than that of the CON group. Antibiotics destroy beneficial microbial communities in the cow’s gut, affecting fiber breakdown and nutrient absorption, leading to impaired energy metabolism, which in turn reduces the supply of raw materials needed for milk synthesis.
Mastitis in dairy cows is frequently accompanied by systemic low-grade inflammation, and increased inflammatory factors are important in compromising the integrity of the blood-milk barrier (26, 27). It has been demonstrated that TNF-α has the capacity to compromise the integrity of the intestinal barrier by inducing the shedding of intestinal epithelial cells (28). IL-6 activates the JAK–STAT signaling pathway, resulting in an increase in the permeability of the blood–brain barrier (29). LPS can enter the bloodstream from the intestinal and cause a range of inflammatory responses that disrupt barrier integrity by reducing the expression of tight junction proteins (30). The present study demonstrated that serum inflammatory factors were significantly elevated in cows with SCM. Furthermore, levels of IL-2, IL-4, IL-8, IL-10, and TNF-α were significantly reduced after GYS treatment, suggesting that GYS ameliorated the inflammatory response induced by SCM. The reduction in LPS suggests that the integrity of the intestinal and blood-milk barriers is also improved, but direct evidence is still needed for further proof. In addition, consistent with the findings of most studies, GYS has been shown to improve antioxidant activity by increasing SOD and decreasing MDA (12, 31–33). In the CEF group, we also found that the levels of IL-4 and TNF-α were reduced, which may be related to the alteration of intestinal microbiota by antibiotics (34). Surprisingly, TP, ALB, and GLB levels did not change during SCM. This result is consistent with the study by Yue et al. (23). Elevated immunoglobulin levels due to prolonged low-grade inflammation may be offset by lowering of other types of globulins, resulting in no change in GLB levels. Several studies have shown the inhibitory effect of IL-6 on albumin synthesis (35–37). In the present study, there was also no significant change in IL-6 levels, which may explain the lack of change in ALB. In conclusion, the addition of GYS was found to be more effective in alleviating the inflammatory response in SCM cows.
Alpha diversity analysis and PCOA revealed that the diversity of the microbiota in the CEF group was significantly lower and the structure of the microbiota was significantly altered compared to the other three groups. This is consistent with the results of most studies that antibiotics may have contributed to the imbalance of the intestinal microbiota (38, 39). A large body of evidence suggests that there are significant differences in the structure and function of the rumen and intestinal microbiota between healthy cows and cows with mastitis (19, 40). In this study, we found that Bacteroides was significantly elevated in the SCM group compared to the CON and GYS groups. It has been reported that Bacteroides can synthesize putrescine and spermidine in vitro and in vivo (41). Polyamines through c-Myc modulating intestinal epithelial barrier function (42). In addition, intestinal-derived enterotoxigenic Bacteroides fragilis caused mammary epithelial hyperplasia, suggesting that Bacteroides disrupt the mammary barrier (43). In addition, Lachnospiraceae, Prevotellaceae, and Bifidobacteriaceae, which were elevated in the SCM group, are fiber-degrading microbiota that produce short-chain fatty acids (SCFAs) by fermenting plant fibers and other carbohydrates. This may lead to an abnormal elevation of SCFAs, which in turn leads to a decrease in pH in the rumen, thus triggering subacute rumen acidosis (SARA) (44). It has been found that SARA is often accompanied by a systemic inflammatory response and mastitis (45). Although Lachnospiraceae and Agathobacter are generally considered to be beneficial microbiota, the present study found both to be in high abundance in the feces of cows with SCM (46, 47). Caijun et al. also found that Lachnospiraceae and Agathobacter were present in high levels in the intestinal tracts of cows with mastitis and mice with rumen microbial transplants (40, 48). Furthermore, these levels were found to be positively correlated with inflammatory factors. Norank_UCG-010 belongs to the group of rumenococci that are involved in the breakdown of starch and fiber in ruminants and contribute to the further digestion of feed in the intestinal (49). UCG-010 is negatively correlated with inflammatory factors (IL-6, IL-12, and IL-17) (50). Furthermore, an increase in Blautia abundance was observed in the GYS group. Blautia is defined as having potential probiotic properties, and it plays a role in reducing inflammation (51). GYS is characterized by its richness in flavonoids, including quercetin, luteolin, and kaempferol (18). These flavonoids can be metabolized by Blautia to form bioactive substances, which may account for the enrichment of Blautia (52). Therefore, the findings suggest that GYS can provide relief from SCM by influencing the inflammation-related microbiota restoring a healthy community structure. Among them, UCG-010 has potential application in the treatment of mastitis.
Several correlated metabolites (e. g., Efavirenz, Diclofenac sodium, Gentian violet) are synthetic drugs or chemicals not expected to be present in untreated dairy cows. Environmental contamination or dietary sources have not been confirmed, and their correlation with specific taxa may result from mis-annotation or database error. It is evident that alterations in the composition of intestinal microbes give rise to concomitant changes in metabolites. We saw that 1-Oleoyl-2-myristoyl-sn-glycero-3-phosphocholine was more abundant in the SCM group and less abundant in the GYS group. 1-Oleoyl-2-myristoyl-sn-glycero-3-phosphocholine belongs to the group called phosphatidylcholines, which have been shown to be converted by intestinal bacteria into a harmful substance called trimethylamine-N-oxide (TMAO) (53). This activates a pathway in the body called the NF-κB pathway, which triggers the production of cytokines (54). Furthermore, in the present study 7alpha-Hydroxy-3-oxochol-4-en-24-oic acid was found to be elevated in the SCM group compared to the CON and GYS groups. 7alpha-Hydroxy-3-oxochol-4-en-24-oic acid belongs to the group of 3-oxo-Δ4 bile acids (55). And the accumulation of 3-oxo-Δ4 bile acids replaces primary bile acid conjugates, resulting in a decrease in secondary bile acid synthesis (56). Deoxycholic acid-mediated activation of the G protein-coupled receptor reduces inflammation by inhibiting the NF-κB and NLRP3 pathways and improves the integrity of the blood-milk barrier, thereby reducing mastitis (57, 58). In the present study, phosphatidylcholine and 3-oxo-Δ4 bile acids were positively correlated with Lachnospiraceae and Bacteroides, negatively correlated with UCG-010, suggesting that they may be regulated by microbiota, which requires further study.
On the other hand, intestinal metabolic pathways were significantly altered after GYS treatment, and these changes were mainly related to lipid metabolism. We found that Arachidonic acid metabolism and Choline metabolism were downregulated in the GYS group. Arachidonic acid amplifies inflammatory signals to promote the production of leukocytes, pro-inflammatory cytokines and immune cells to fight and eliminate pathogens (59). Moreover, we found that phosphatidylcholine was elevated in the SCM group and that phospholipase could induce the release of arachidonic acid from phosphatidylcholine via TNF-α binding to its receptor (60). Endogenous cannabinoids are also a source of arachidonic acid (61). Although linoleic acid is an essential fatty acid, it can be elongated and desaturated to arachidonic acid, which in turn leads to inflammation (62). Phosphatidylcholine is synthesized from choline via the CDP-choline pathway (63), and elevated phosphatidylcholine implies increased cellular uptake of choline, which is often accompanied by an inflammatory response (64–66). Therefore, GYS may alleviate SCM by affecting Arachidonic acid metabolism and Choline metabolism, with Phosphatidylcholine being the key factor. Based on correlation analyses, it was hypothesized that UCG-010, Lachnospiraceae_AC2044_group and Bacteroides may influence secondary bile acid synthesis as well as choline metabolism via 3-oxo-Δ4 bile acids and phosphatidylcholine.
5 Conclusion
The present study demonstrated that GYS enhanced milk yield, and improved lactation performance in cows with SCM. GYS reduced serum inflammatory cytokines, LPS and MDA levels, elevated SOD levels, and reduced microbiota associated with inflammation and intestinal barriers in dairy cows with SCM, thus, GYS attenuated inflammation and oxidative stress in serum. Furthermore, we observed substantial alterations in arachidonic acid metabolism and choline metabolism, with the metabolite phosphatidylcholine being reduced by GYS. This suggests that GYS may alleviate inflammation through this pathway. In conclusion, the use of GYS could be considered as an effective alternative therapeutic strategy to antibiotics, with the potential to reduce the risk of systemic inflammation in cows with SCM.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI BioProject, accession PRJNA1268138.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of Shanxi Agricultural University (SXAU-EAW-2025C.IM.004006280). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
GZ: Conceptualization, Data curation, Visualization, Writing – original draft, Writing – review & editing. HL: Formal analysis, Project administration, Resources, Writing – original draft. LH: Investigation, Methodology, Writing – original draft. YC: Software, Writing – original draft. JL: Validation, Writing – review & editing. RS: Supervision, Writing – review & editing. XW: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Shanxi Livestock and Poultry Genetic Gene Bank Project of Biological Breeding Project of Shanxi Agricultural University (Cattle) (YZGC131), Shanxi Modern Agricultural Industrial Technology System Construction Project (2025CYJSTX13-04), and Establishment of Beef Cattle Breed Resource Bank and Big Data Platform for Molecular Breeding (202201140601026-1).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1589900/full#supplementary-material
Footnotes
1. ^https://cutadapt.readthedocs.io/en/stable/
2. ^https://cme.h-its.org/exelixis/web/software/pear/
3. ^https://prinseq.sourceforge.net/
4. ^https://drive5.com/uparse/
8. ^https://www.r-project.org/
References
1. Hogeveen, H, Huijps, K, and Lam, TJGM. Economic aspects of mastitis: new developments. N Z Vet J. (2011) 59:16–23. doi: 10.1080/00480169.2011.547165
2. Nesma Helmy, Y, Nagah Moustafa, H, Mohamady Ahmed, H, and Mena, FS. Influence of some hygienic measures on the prevalence of subclinical mastitis in a dairy farm. Int J Dairy Sci. (2019) 15:38–47. doi: 10.3923/ijds.2020.38.47
3. Philip, R, Herman, WB, Prince, PO, James, T, Alexandra, S, Beate, C, et al. Global losses due to dairy cattle diseases: a comorbidity-adjusted economic analysis. J Dairy Sci. (2024) 107:6945–70. doi: 10.3168/jds.2023-24626
4. Erick Ouma, M, Tenhagen, BA, Fikru, R, Kyule, MN, Yoseph, S, Tesfu, K, et al. Reduced Milk production in udder quarters with subclinical mastitis and associated economic losses in crossbred dairy cows in Ethiopia. Trop Anim Health Prod. (2005) 37:503–12. doi: 10.1007/s11250-005-7049-y
5. Castello, M, Zheng, S, Benhua, Z, Shi, H, Jie, Z, Yong, Z, et al. Cow-to-mouse fecal transplantations suggest intestinal microbiome as one cause of mastitis. Microbiome. (2018) 6:200. doi: 10.1186/s40168-018-0578-1
6. Xiaoyu, H, Zhaoqi, H, Caijun, Z, Yuhong, H, Min, Q, Kaihe, X, et al. Gut/rumen-mammary gland axis in mastitis: gut/rumen microbiota–mediated “gastroenterogenic mastitis”. J Adv Res. (2024) 55:159–71. doi: 10.1016/j.jare.2023.02.009
7. Juan, MR, Leónides, F, and Valérie, V. The gut–breast Axis: programming health for life. Nutrients. (2021) 13:606. doi: 10.3390/nu13020606
8. Tariq Hisham Beshara, H. Bioeconomic modeling of intervention against clinical mastitis caused by contagious pathogens. J Dairy Sci. (2012) 95:5740–9. doi: 10.3168/jds.2012-5470
9. Godfrey, SB, Norah, M, John, O, David, BK, and Muhammad, N. Antibiotics/antibacterial drug use, their marketing and promotion during the post-antibiotic golden age and their role in emergence of bacterial resistance. Health. (2014) 6:410–25. doi: 10.4236/health.2014.65059
10. Yupei, Z, Kairui, T, Yuxuan, D, Runsen, C, Liang, S, Huijun, X, et al. Effects of shenling baizhu powder herbal formula on intestinal microbiota in high-fat diet-induced NAFLD rats. Biomed Pharmacother. (2018) 102:1025–36. doi: 10.1016/j.biopha.2018.03.158
11. Schütz, K, Carle, R, and Schieber, A. Taraxacum—a review on its phytochemical and pharmacological profile. J Ethnopharmacol. (2006) 107:313–23. doi: 10.1016/j.jep.2006.07.021
12. Yan, L, Jie, M, Jiaqi, W, and Hongyun, L. Effects of dandelion (Taraxacum sp.,) supplements on lactation performance, antioxidative activity, and plasma metabolome in primiparous dairy cows. Anim Biosci. (2023) 36:229–37. doi: 10.5713/ab.22.0061
13. Yan, L, Mei, L, Jiaqi, W, Zhonghong, T, Bo, Y, Bing, W, et al. Dandelion (Taraxacum mongolicum hand.-Mazz.) supplementation-enhanced rumen fermentation through the interaction between ruminal microbiome and metabolome. Microorganisms. (2020) 9:83. doi: 10.3390/microorganisms9010083
14. Yawang, S, Yongjiang, W, Zili, W, Juncai, C, Yu, Y, and Guozhong, D. Dandelion extract alleviated lipopolysaccharide-induced oxidative stress through the Nrf2 pathway in bovine mammary epithelial cells. Toxins. (2020) 12:496. doi: 10.3390/toxins12080496
15. Ge, H, Junjie, W, Daojun, H, Tao, Z, Huiqin, D, Ming, X, et al. Effects of aqueous extracts of Taraxacum Officinale on expression of tumor necrosis factor-alpha and intracellular adhesion molecule 1 in LPS-stimulated RMMVECs. BMC Complement Altern Med. (2017) 17:38. doi: 10.1186/s12906-016-1520-3
16. Wenjiao, L, Zhang, L, Ping, H, Haiying, L, Peng, X, Weilong, Z, et al. Traditional uses, botany, phytochemistry, and pharmacology of Lonicerae japonicae flos and Lonicerae flos: a systematic comparative review. J Ethnopharmacol. (2024) 322:117278. doi: 10.1016/j.jep.2023.117278
17. Youn-Hwan, H, Dong-Gun, K, Wei, L, Hye Jin, Y, Nam-Hui, Y, and Jin, Y. Anti-inflammatory effects of Forsythia suspensa in dextran sulfate sodium-induced colitis. J Ethnopharmacol. (2017) 206:73–7. doi: 10.1016/j.jep.2017.05.011
18. Shuang, G, Liyun, T, Jiayi, M, Kaiming, W, Hua, Y, Jinjin, T, et al. Evaluation of the mechanism of gong Ying san activity on dairy cows mastitis by network pharmacology and metabolomics analysis. PLoS One. (2024) 19:e0299234. doi: 10.1371/journal.pone.0299234
19. Yue, W, Xuemei, N, Yiguang, Z, Linshu, J, Mengling, W, Hui, W, et al. Rumen microbiome structure and metabolites activity in dairy cows with clinical and subclinical mastitis. J Anim Sci Biotechnol. (2021) 12:36. doi: 10.1186/s40104-020-00543-1
20. Megan, C. Public health: The politics of antibiotics. Nature. (2014) 509:S16–7. doi: 10.1038/509S16a
21. Lisa, M, Camille, G, Jakob, W, Michael, K, Claudia, E, Mihaela, P, et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature. (2021) 599:120–4. doi: 10.1038/s41586-021-03986-2
22. The, L. Food industry must act to safeguard the future of antibiotics. Lancet. (2017) 390:2216. doi: 10.1016/S0140-6736(17)32900-8
23. Yue, W, Xuemei, N, Yiguang, Z, Linshu, J, Hui, W, Fan, Z, et al. Dietary supplementation of inulin ameliorates subclinical mastitis via regulation of rumen microbial community and metabolites in dairy cows. Microbiol Spectr. (2021) 9:e0010521. doi: 10.1128/Spectrum.00105-21
24. Ting-Yu, L, Cuihua, L, Wei-Fan, L, Ting-Shu, W, Jang-Jih, L, Young-Mao, C, et al. Role of gut microbiota in identification of novel TCM-derived active metabolites. Protein Cell. (2020) 12:394–410. doi: 10.1007/s13238-020-00784-w
25. Catherine, F, Ching Tat, L, Leon, RM, and Peter, H. Excretion of lactose in urine as a measure of increased permeability of the lactating breast during inflammation. Acta Obstet Gynecol Scand. (2006) 85:20–5. doi: 10.1080/00016340500324514
26. Wenjin, G, Bingrun, L, Yunhou, Y, Xingchi, K, Qian, G, Yanwei, L, et al. Licochalcone a protects the blood–Milk barrier integrity and relieves the inflammatory response in LPS-induced mastitis. Front Immunol. (2019) 10:287. doi: 10.3389/fimmu.2019.00287
27. Caijun, Z, Keyi, W, Lijuan, B, Luotong, C, Limin, F, Zhuoyu, L, et al. Kynurenic acid protects against mastitis in mice by ameliorating inflammatory responses and enhancing blood-milk barrier integrity. Mol Immunol. (2021) 137:134–44. doi: 10.1016/j.molimm.2021.06.022
28. Tieren, G, Chongyang, H, Jiaye, C, Bingxia, C, Yangyang, S, and Fachao, Z. An IRF1-dependent pathway of TNFα-induced shedding in intestinal epithelial cells. J Crohn's Colitis. (2021) 16:133–42. doi: 10.1093/ecco-jcc/jjab134
29. Jung, K, Hsiu-Chun, C, Natalie, KW, Christopher, JN, David, HR, Kaoru, S, et al. Tumor-induced disruption of the blood-brain barrier promotes host death. Dev Cell. (2021) 56:2712.
30. David, RS-P, Gaber, M, Alana, AA, Steven, MB, Nildris, CD, Ana, W, et al. Diet alters Entero-mammary signaling to regulate the breast microbiome and tumorigenesis. Cancer Res. (2021) 81:3890–904. doi: 10.1158/0008-5472.CAN-20-2983
31. Yan, Z, Musa, M, Ziqing, X, Yu, C, Juncai, C, and Yawang, S. Effects of dandelion extract on promoting production performance and reducing mammary oxidative stress in dairy cows fed high-concentrate diet. Int J Mol Sci. (2024) 25:6075. doi: 10.3390/ijms25116075
32. Dariusz, J, Bernadetta, L, Agata, R, Anna, S, and Beata, O. Comparative phytochemical, cytotoxicity, antioxidant and haemostatic studies of Taraxacum officinale root preparations. Food Chem Toxicol. (2019) 126:233–47. doi: 10.1016/j.fct.2019.02.017
33. Fen, L, Kang Lin, F, Jinlong, Y, Yuan, H, Hongliang, G, Sheng Peng, W, et al. Polysaccharides from dandelion (Taraxacum mongolicum) leaves: insights into innovative drying techniques on their structural characteristics and biological activities. Int J Biol Macromol. (2021) 167:995–1005. doi: 10.1016/j.ijbiomac.2020.11.054
34. Nitzan, O, Elias, M, Peretz, A, and Saliba, W. Role of antibiotics for treatment of inflammatory bowel disease. World J Gastroenterol. (2016) 22:1078–87. doi: 10.3748/wjg.v22.i3.1078
35. Lukas, Z, Melanie, K, Alexander, H, Sibylle, S, Sebastian, W, Mihail, T, et al. Sepsis induces interleukin 6, gp130/JAK2/STAT3, and muscle wasting. J Cachexia Sarcopenia Muscle. (2021) 13:713–27. doi: 10.1002/jcsm.12867
36. Sabrina Milan, M, Grazia Maria, V, Anna, C, Alessandra, B, Massimo de, C, Ilaria, T, et al. Pro-inflammatory cytokines: a possible relationship with dialytic adequacy and serum albumin in peritoneal dialysis patients. NDT Plus. (2015) 9:153–7. doi: 10.1093/ckj/sfv137
37. Liren, Z, Weiping, Y, Yuwu, Z, Xiaohua, C, Peng, W, Xiaohong, F, et al. Albumin infusion may improve the prognosis of critical COVID-19 patients with hypoalbuminemia in the intensive care unit: a retrospective cohort study. Infect Drug Resist. (2022) 15:6039–50. doi: 10.2147/IDR.S383818
38. Ethel, T, Ellen, RJ, Jose, AG, Hagevoort, GR, Keri, NN, Sara, DL, et al. Effects of two-dose ceftiofur treatment for metritis on the temporal dynamics of antimicrobial resistance among fecal Escherichia coli in Holstein-Friesian dairy cows. PLoS One. (2019) 14:e0220068. doi: 10.1371/journal.pone.0220068
39. Lindsey, RC, Ying, Y, Heather, L, Partha Pratim, R, Tong, Z, Amy, P, et al. Metagenomic analysis of antibiotic resistance genes in dairy cow feces following therapeutic Administration of Third Generation Cephalosporin. PLoS One. (2015) 10:e0133764. doi: 10.1371/journal.pone.0133764
40. Caijun, Z, Lijuan, B, Min, Q, Keyi, W, Yihong, Z, Limin, F, et al. Commensal cow Roseburia reduces gut-dysbiosis-induced mastitis through inhibiting bacterial translocation by producing butyrate in mice. Cell Rep. (2022) 41:111681. doi: 10.1016/j.celrep.2022.111681
41. Jutta, N, Dongowski, G, Ludger, H, and Michaël, B. The human gut Bacteria Bacteroides thetaiotaomicron and Fusobacterium varium produce putrescine and spermidine in cecum of pectin-fed Gnotobiotic rats. J Nutr. (2000) 130:1225–31. doi: 10.1093/jn/130.5.1225
42. Lan, L, Xin, G, Jaladanki, NR, Tongtong, Z, Lan, X, Tina, Y, et al. Polyamines regulate E-cadherin transcription through c-Myc modulating intestinal epithelial barrier function. Am J Phys Cell Phys. (2009) 296:C801–10. doi: 10.1152/ajpcell.00620.2008
43. Sheetal, P, Shaoguang, W, Sumit, S, Guannan, W, Nethaji, M, Arumugam, N, et al. A Procarcinogenic Colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and β-catenin axes. Cancer Discov. (2021) 11:1138–57. doi: 10.1158/2159-8290.CD-20-0537
44. Chenxu, Z, Gerd, B, Shaobin, W, Xinyue, Z, Zhibo, Z, Shiqi, Z, et al. Potential role of SLC5A8 expression in the etiology of subacute ruminal acidosis. Front Vet Sci. (2020) 7:394. doi: 10.3389/fvets.2020.00394
45. Caijun, Z, Xiaoyu, H, Min, Q, Lijuan, B, Keyi, W, Xiangyue, M, et al. Sialic acid exacerbates gut dysbiosis-associated mastitis through the microbiota-gut-mammary axis by fueling gut microbiota disruption. Microbiome. (2023) 11:78. doi: 10.1186/s40168-023-01528-8
46. Tom, Z, Solange, M, and Andrew, CT. Lachnospiraceae are emerging industrial biocatalysts and biotherapeutics. Front Bioeng Biotechnol. (2024) 11:1324396. doi: 10.3389/fbioe.2023.1324396
47. Xinhuang, L, Zhan, L, Ye, T, Huijia, X, Zhibo, C, Yan, L, et al. Gut commensal Agathobacter rectalis alleviates microglia-mediated neuroinflammation against pathogenesis of Alzheimer disease. iScience. (2024) 27:111116. doi: 10.1016/j.isci.2024.111116
48. Caijun, Z, Xiaoyu, H, Lijuan, B, Keyi, W, Yihong, Z, Kaihe, X, et al. Gut dysbiosis induces the development of mastitis through a reduction in host anti-inflammatory enzyme activity by endotoxemia. Microbiome. (2022) 10:205. doi: 10.1186/s40168-022-01402-z
49. Mengru, C, Wan, X, Shendong, Z, Nana, M, Yan, W, Jie, H, et al. A high-concentrate diet induces colonic inflammation and barrier damage in Hu sheep. J Dairy Sci. (2023) 106:9644–62. doi: 10.3168/jds.2023-23359
50. Xin, Y, Shoukun, J, Chunhui, D, Peizhi, T, Sisi, J, Hui, Y, et al. The succession of fecal bacterial community and its correlation with the changes of serum immune indicators in lambs from birth to 4 months. J Integr Agric. (2023) 22:537–50. doi: 10.1016/j.jia.2022.08.055
51. Yuxing, Z, Shiqiang, Y, Huiying, Z, Liuxue, L, Yuqin, L, Ming, L, et al. Integrated multi-omics analysis reveals the positive leverage of citrus flavonoids on hindgut microbiota and host homeostasis by modulating sphingolipid metabolism in mid-lactation dairy cows consuming a high-starch diet. Microbiome. (2023) 11:236. doi: 10.1186/s40168-023-01661-4
52. Xuemei, L, Bingyong, M, Jiayu, G, Jiaying, W, Shumao, C, Gang, W, et al. Blautia—a new functional genus with potential probiotic properties? Gut Microbes. (2021) 13:e1875796. doi: 10.1080/19490976.2021.1875796
53. Tang, WHW, Zeneng, W, Bruce, SL, Robert, AK, Earl, BB, Xiaoming, F, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. (2013) 368:1575–84. doi: 10.1056/NEJMoa1109400
54. Luis, ACJ, Yoshua, E-P, Alma Reyna, EM, and Paulina, H-R. Luis MA-G, Amedeo a, marı́a Magdalena a-Ga: contribution of trimethylamine N-oxide (TMAO) to chronic inflammatory and degenerative diseases. Biomedicines. (2023) 11:431. doi: 10.3390/biomedicines11020431
55. Tudor, M, Dong Wook, K, Billy, JM, Hyeonho, J, Zeno, S, Diren, B, et al. Plasma fetal bile acids 7α-hydroxy-3-oxochol-4-en-24-oic acid and 3-oxachola-4,6-dien-24-oic acid indicate severity of liver cirrhosis. Sci Rep. (2021) 11:8298. doi: 10.1038/s41598-021-87921-5
56. Jing, Z, Yi-Ling, Q, Li, W, Zhong-die, L, Xin-Bao, X, Yi, L, et al. Recurrent AKR1D1 c.580-13T>a variant. J Mol Diagn. (2023) 25:227–33. doi: 10.1016/j.jmoldx.2023.01.004
57. Caijun, Z, Keyi, W, Haoyang, H, Yihong, Z, Lijuan, B, Min, Q, et al. Gut microbiota-mediated secondary bile acid alleviates Staphylococcus aureus-induced mastitis through the TGR5-cAMP-PKA-NF-κB/NLRP3 pathways in mice. npj Biofilms Microbiomes. (2023) 9:8. doi: 10.1038/s41522-023-00374-8
58. Kazuaki, Y, Tadakazu, H, Katsuyoshi, S, Nobuhiko, K, Riko, I, Mina, TK, et al. TGR5 signalling inhibits the production of pro-inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn's disease. Immunology. (2013) 139:19–29. doi: 10.1111/imm.12045
59. Edward, AD, and Paul, CN. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. (2015) 15:511–23. doi: 10.1038/nri3859
60. Violette Said, H. Ebtisam Abdel Aziz H: synopsis of arachidonic acid metabolism: a review. J Adv Res. (2018) 11:23–32. doi: 10.1016/j.jare.2018.03.005
61. Kay, A, Michele, KM, and Benjamin, FC. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem Rev. (2008) 108:1687–707. doi: 10.1021/cr0782067
63. Zhaoyu, L, and Dennis, EV. Thematic review series: Glycerolipids. Phosphatidylcholine and choline homeostasis. J Lipid Res. (2008) 49:1187–94. doi: 10.1194/jlr.R700019-JLR200
64. Sanna, H, Johanna, MUS, Max, K, Heidi, L, Olli, M, Tapio, V, et al. Type 2 diabetes enhances arterial uptake of choline in atherosclerotic mice: an imaging study with positron emission tomography tracer 18F-fluoromethylcholine. Cardiovasc Diabetol. (2016) 15:26. doi: 10.1186/s12933-016-0340-6
65. Christian, MM, Matthias, TW, Patricia Stutzmann, M, Nicolas, S, Tobias von, L, Christine, L, et al. 18 F-choline images murine atherosclerotic plaques ex vivo. Arterioscler Thromb Vasc Biol. (2006) 26:584–9. doi: 10.1161/01.ATV.0000200106.34016.18
Keywords: dairy cows, subclinical mastitis, Gongying San, intestinal microbiota, metabonomics
Citation: Zhao G, Li H, Huang L, Cheng Y, Liu J, Song R and Wang X (2025) Integrated multi-omics analysis reveals that Gongying San ameliorates subclinical mastitis by modulating intestinal microbiota and metabolites in dairy cows. Front. Vet. Sci. 12:1589900. doi: 10.3389/fvets.2025.1589900
Edited by:
Deji Abiodun Ekunseitan, North Carolina Agricultural and Technical State University, United StatesReviewed by:
Abosede Tomilola Abolude, Alcorn State University, United StatesTunde Emmanuel Ogundare, North Carolina Agricultural and Technical State University, United States
Safiu Suberu, North Carolina Agricultural and Technical State University, United States
Copyright © 2025 Zhao, Li, Huang, Cheng, Liu, Song and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruigao Song, cmVnYW43MTFAMTYzLmNvbQ==; Xi Wang, d3hwaGlsaXBAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work
 Guoqing Zhao
Guoqing Zhao Hongxia Li2†
Hongxia Li2† Yu Cheng
Yu Cheng Ruigao Song
Ruigao Song Xi Wang
Xi Wang